Abstract
Genetic determinants of speciation in closely related species are poorly understood. We sequenced and analyzed transcriptomes of swallowtail butterflies Heraclides cresphontes (northeastern species) and Heraclides rumiko (southwestern species), a pair of mostly allopatric sister species whose distribution ranges overlap narrowly in central Texas. We found that the two swallowtails confidently differ (FST > 0.5 for both species) in about 5% of genes, similarly to the divergence in another pair of swallowtail species Pterourus glaucus (southern species) and Pterourus canadensis (northern species). The same genes tend to diverge in both species pairs, suggesting similar speciation paths in Heraclides and Pterourus. The most significant differences for both species pairs were found in the circadian clock genes that were conserved within each species and diverged strongly between species (P-value < 0.01 and FST > 0.7). This divergence implied that adaptations to different climates and photoperiod at different latitudes or differences in mating behavior, including mating time and copulation duration, may be possible factors in ecological or behavioral-based speciation. Finally, we suggest several nuclear DNA regions that consistently and prominently differ between the sister swallowtail species as nuclear barcodes for swallowtail identification, with the best barcode being an exon from the protein TIMELESS.
Keywords: biodiversity, evolutionary genomics, cryptic species, ecological speciation, eclosion rhythm
Résumé :
Les déterminants génétiques de la spéciation chez des espèces proches sont peu connus. Les auteurs ont séquencé et analysé les transcriptomes de deux espèces de grands porte-queues l’Heraclides cresphontes (espèce du Nord-Ouest) et l’Hercalides rumiko (espèce du Sud-Est), une paire d’espèces sœurs largement allopatriques dont les aires de distribution se chevauchent légèrement dans le centre du Texas. Les auteurs ont trouvé que les deux espèces diffèrent de manière convaincante (FST > 0,5 pour les deux espèces) pour environ 5 % des gènes, une divergence semblable à celle rencontrée chez une autre paire de grands porte-queues, le Pterourus glaucus (espèce méridionale) et le Pterourus canadensis (espèce septentrionale). Les mêmes gènes tendaient à diverger chez les deux paires d’espèces, ce qui suggère des voies de spéciation semblables chez les deux genres. Les différences les plus importantes chez les deux paires d’espèces ont été trouvées au sein des gènes de l’horloge circadienne, lesquels étaient fortement conservés au sein de chaque espèce, mais présentaient une forte divergence entre les espèces (valeur P < 0,01 et FST > 0,7). Cette divergence implique que les adaptations aux différents climats et aux diverses photopériodes des différentes latitudes ou encore des différences dans les comportements reproducteurs (p. ex. le moment de l’accouplement, la durée de la copulation) pourraient constituer des facteurs dans la spéciation basée sur des différences écologiques ou comportementales. Finalement, les auteurs suggèrent que plusieurs régions de l’ADN nucléaire qui diffèrent de manière reproductible et marquée entre les espèces sœurs de grands porte-queues pourraient servir de codes à barres nucléaires pour l’identification des grands porte-queues, le meilleur code à barre étant un exon au sein du gène codant pour la protéine TIMELESS. [Traduit par la Rédaction]
Keywords: biodiversité, génomique évolutive, espèces cryptiques, spéciation adaptative, rythme d’éclosion
Introduction
Adaptation and speciation drive the generation of biodiversity. Much attention has been paid to deciphering the genetic basis for speciation in model organisms, and a number of genes involved in reproductive isolation between species have been identified (Maheshwari and Barbash 2011; Phadnis et al. 2015). However, with recent advances in sequencing techniques there has been a growing interest in a broader spectrum of organisms, in the roles of ecological factors and adaptation in speciation, and in the genetic bases for the early stages of speciation (Seehausen et al. 2014; Wolf et al. 2010). Incipient species pairs, representing very closely related sister species that may not yet reach complete reproductive isolation, are particularly suitable for such studies (Feder et al. 2012).
Becasue of their small genome size, diverse phenotypes, and abundance of many closely related species, Lepidoptera are well-suited for genetics and evolutionary studies. Studies in the genus Heliconius showed that gene introgression can lead to similarity in wing patterns of different species and is one possible evolutionary mechanism of mimicry (Dasmahapatra et al. 2012). Using the transcriptomes of tiger swallowtails, we found that only a small fraction of genes can distinguish Pterourus glaucus from P. canadensis (Cong et al. 2015a), which is in agreement with that observed in other animals (Harr 2006; Turner et al. 2005) and plants (Rieseberg and Blackman 2010).
Recently, we described a new swallowtail, Heraclides rumiko, which is a southwestern (from USA to Panama) sister species of the eastern US H. cresphontes (Shiraiwa et al. 2014). These two species are sympatric in central Texas and may hybridize with each other. Heraclides rumiko and H. cresphontes can be identified by male genitalia (although it is not known whether these differences reduce interspecies hybridization), the shape and size of yellow spots on the neck, the wing shape, and the details of wing patterns. They exhibit a nearly 3% divergence in the mitochondrial DNA barcode sequence that encodes the N-terminal half of mitochondrial cytochrome oxidase I (COI).
Here we sequence and analyze the transcriptomes from 10 specimens (5 for each species) of Heraclides (Table 1). Nuclear DNA divergence between H. rumiko and H. cresphontes is comparable to that between two species of Pterourus (P. glaucus and P. canadensis) and is in agreement with the divergence in their COI barcodes. We detect the most divergent nuclear genes between each pair of sister species and term these genes divergence hotspots. Interestingly, the functions of the divergence hotspots in both species pairs overlap significantly, suggesting common mechanisms behind the two independent speciation events. The circadian clock system stands out most strongly in the divergence hotspots of both Heraclides and Pterourus, with all four central components being significantly more divergent between species than within. Finally, we propose several long exons that can most confidently identify sister species of Pterourus and Heraclides as nuclear barcodes for swallowtail species identification.
Table 1.
Specimens used in this study.
| Voucher | Species name | Sex | Status | Locality (USA) | Date | Transcriptsa | Mappedb | Orthologsc |
|---|---|---|---|---|---|---|---|---|
| NVG-2565 | Heraclides rumiko | M | Holotype | Texas, Duval County, Benavides, | 30 May 2014 | 26 611 | 7948 | 9488 |
| NVG-2559 | H. rumiko | M | Paratype | CR306, 2.9 km west of SH339 | 26 May 2014 | 24 325 | 6950 | 8007 |
| NVG-2564 | H. rumiko | F | Paratype | 29 May 2014 | 23 451 | 7450 | 8706 | |
| NVG-3369 | H. rumiko | M | — | Texas, Hidalgo County, Penitas, | 23 May 2015 | 40 202 | 10 673 | 14 289 |
| NVG-3373 | H. rumiko | M | — | railroad tracks | 23 May 2015 | 41 066 | 11 112 | 14 810 |
| NVG-2740 | H. cresphontes | M | — | Texas, Denton County, Flower | 8 July 2014 | 26 503 | 8073 | 9617 |
| NVG-2741 | H. cresphontes | F | — | Mound, Grapevine Lake, | 15 July 2014 | 30 155 | 8904 | 10 800 |
| NVG-2760 | H. cresphontes | M | — | Murrell Park | 17 July 2014 | 28 211 | 8678 | 10 685 |
| NVG-5323 | H. cresphontes | F | — | Florida, Monroe County, Key West | 19 December 2015 | 67 447 | 11198 | 13 970 |
| NVG-5360 | H. cresphontes | M | — | Florida, Miami-Dade County, Florida City | 20 December 2015 | 51445 | 9288 | 10 368 |
Number of de novo assembled transcripts by Trinity in each specimen.
Number of Pterourus glaucus proteins that the de novo assembled transcripts can map to.
Number of Heraclides reference transcripts that are covered (coverage ≥50%) by this specimen.
Materials and methods
RNA-seq library preparation and sequencing
Information about the 10 specimens of Heraclides is presented in Table 1, and they will be deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM). Upon capture or eclosion, a specimen was euthanized either by thorax pinching or injection with a 30% NH3 water solution. A piece of muscle cut out from the thorax of reared specimens (including H. rumiko holotype) or the whole specimen body, except wings and genitalia, were preserved in RNAlater solution. Total RNA was extracted from the specimen using QIAGEN RNeasy Plus Mini Kit. We further isolated mRNA using NEBNext Poly(A) mRNA Magnetic Isolation Module. RNA-seq library for each specimen was prepared with NEBNext Ultra Directional RNA Library Prep Kit for Illumina following manufacturer’s protocol. Six specimens (NVG-2559, NVG-2564, NVG2565, NVG-2740, NVG-2741, and NVG-2760) were pooled at an equal ratio and sequenced together on a 3/4 illumina Hiseq2500 lane for 50 bp at the single end in the first run and for 100 bp at both ends in another run. Four other specimens were prepared in another two batches (NVG-3369 and NVG-3373 in one batch, and NVG-5323 and NVG-5360 in another batch), and each library was sequenced on a 1/8 illumina Hiseq2500 lane for 150 bp at both ends. The sequence reads have been deposited to NCBI SRA database under accession numbers: SRR3138026–SRR3138039.
Assembling reference transcriptome for Heraclides
After removal of contamination from TruSeq adapters by Mirabait (version 3.4.0) (Chevreux et al. 1999) and trimming the low quality portion (trimming from the beginning and the end until the first base with quality score >20) at the beginning and the end of each read using an in-house python script, we applied Trinity (version r20140413p1) (Haas et al. 2013) to de novo assemble the transcriptome. The transcripts from all specimens were mapped to the protein set of P. glaucus by BLASTX (e-value: 0.00001) (version 2.2.31+) (Altschul et al. 1990). Transcripts that could not find a confident hit (e-value ≤ 0.00001) among P. glaucus proteins were discarded. We filtered the BLASTX hits requiring the aligned positions between the transcript and the hit to cover at least 50% of the residues in the hit or at least 50% of the nucleotides in the transcript, and the remaining hits were ranked primarily by e-value and secondarily by bit score. From the ranked list we identified the best hits that were aligned to non-overlapping regions in the transcript. Usually there was only one best hit, and in cases where multiple non-overlapping best hits were identified, the transcript was split to multiple segments corresponding to multiple best hits.
Each transcript (or a segment) was considered to map to the top hit from the P. glaucus protein set, and the Heraclides transcripts mapping to the same P. glaucus protein were aligned against each other using BLASTN (version 2.2.31+) (Altschul et al. 1990) to remove redundancy and to merge partial transcripts to a complete transcript. We wanted to represent alternatively spliced isoforms with just the longest isoform, and we removed other isoforms and redundant transcripts from different specimens using the following criteria: (i) if two transcripts were over 95% identical to each other and the aligned region covered at least 80% of one transcript, the shorter transcript was removed; (ii) if two transcripts were over 90% identical to each other and the aligned region covered at least 80% of one transcript and the two transcripts share at least one identical 40mer, the shorter transcript was removed. To merge the partial transcripts, we referred to the alignment between transcripts and the P. glaucus proteins. If two transcripts were aligned to different regions of the protein with at least 20 overlapping residues, the sequences were merged, and in the overlapping region, the transcript that was more similar to the P. glaucus protein were taken. The above procedure produced a representative transcriptome for Heraclides consisting of 17 968 transcripts.
Obtaining sequence alignments and estimation of divergence time
We aligned the reads from each specimen to the reference transcriptome using BWA (version 0.6.2-r126) (Li and Durbin 2009) and performed SNP calling with GATK (version 3.3–0) (DePristo et al. 2011). The sequences of each specimen were derived from the GATK results, and 13 050 reference transcripts covered by at least two H. rumiko and two H. cresphontes specimens for at least 20 amino acids were used in downstream analyses. The alignments of orthologous transcripts in Pterourus were prepared similarly, except that the coding sequences of the annotated P. glaucus proteins were used as reference instead. We calculated the pair-wise divergence (percent of different positions) for each orthologous group in both the DNA and protein sequences and obtained the average values for intra- and interspecific pairs. The distribution of the intra- and interspecific divergence for each orthologous group was plotted and the means over all groups were calculated with Scipy (version 1.1.0, http://www.scipy.org/). The significance level for the difference between intra- and interspecific divergence was estimated using Mann–Whitney U-test implemented in the Stats package of Scipy.
A subset of the orthologous groups in Heraclides that satisfy the following criteria were used to estimate the divergence time for H. cresphontes and H. rumiko using the Isolation-with-migration model (version 8.26.12) (Hey and Nielsen 2007): (i) there were more than 200 positions in the alignment after removing any positions with gaps; (ii) the alignment contained all eight specimens; (iii) there were no significant (P < 0.05) signs of recombination in the PHI permutation test performed with PhiPack (Bruen et al. 2006). The qualified alignments were randomly divided into 20 data sets. IMa2 (parameters: -s5437330 -b200000 -t10.0 -m0.40 -q24.8 -l540 -hfg -hn100 -ha0.99 -hb0.75 -r245 -z100 -p35 -u0.5 –hfg) was applied to each data set, and we took the average for the estimates from the 20 data sets as the final estimated divergence time.
Phylogenetic analysis
From the multiple sequence alignment of each orthologous group of Heraclides transcripts, the consensus sequence for this group was obtained. The consensus sequence for each Heraclides orthologous group was aligned to its closest sequence in the transcript set derived from the P. glaucus reference genome using BLASTN (version 2.2.31+), and thus a common alignment for transcripts from both genera was obtained. In each alignment, we represented one specimen with only one sequence, and in cases of heterozygous positions, one possible allele was randomly chosen. The alignments for individual orthologous groups consisting of sequences from all the specimens of Heraclides and Pterourus were concatenated and positions with any gaps were removed. The concatenated alignment was used to construct a maximum likelihood tree using RAxML (version 8.1.17, model: GTRGAMMA) (Stamatakis 2014). Bootstrap resampling of the concatenated alignment was performed to assign confidence levels for the branches in the maximum likelihood tree.
To fully use the information from heterozygous positions in each specimen, since in early speciation most of the difference between sister species are probably a result of genetic drift, we also inferred the evolutionary history of Heraclides using TreeMix (version 1.13) (Pickrell and Pritchard 2012), which is designed to work with closely related population (or species) and fully utilize the heterozygous positions.
Identification and in-depth analysis of the divergence hotspots
The Heraclides transcripts in each specimen were translated to protein sequences according to their mapping to the reference P. glaucus genome. The alignments of 13 050 orthologous proteins containing sequences from at least two specimens of each species and at least 20 aligned positions were used in this analysis. We used two criteria to identify the diverged proteins between H. rumiko and H. cresphontes that may be important for their speciation.
First, we estimated the fixation indices for both H. rumiko and H. cresphontes using the following formula: FST = (πbetween – πwithin) / πbetween, where πbetween is the average divergence between species and πwithin is the average divergence within species. We required the divergence hotspots to have fixation index above 0.5, i.e., the interspecific divergence is at least two times that of the intraspecific divergence in both species. Second, we detected all the positions that are conserved (sharing a common amino acid in over 80% of sequences) within but different between species, and required the divergence hotspots to be significantly enriched (p < 0.05) in such positions. The enrichment was quantified using a binomial test (p = rate of divergent positions in the alignment, m = the number of divergent positions in a protein, n = the total number of aligned positions in a protein).
Similarly, we identified the divergence hotspot for Pterourus. The significance level for the overlap in divergence hotspots between Pterourus and Heraclides was evaluated by a binomial test (m = number of common divergence hotspots, N = number of divergence hotspots for a genus, p = probability for a gene that is shared by both genera to be a divergence hotspot of another genus).
We identified the enriched GO terms associated with these divergence hotspots using binomial tests (m = the number of divergence hotspots that were associated with this GO term, N = number of divergence hotspots, p = the probability for this GO term to be associated with any gene). GO terms with P-values lower than 0.01 were considered enriched. The significance level for the overlap in enriched (P-value < 0.01) GO terms between Pterourus and Heraclides was also evaluated by a binomial test (m = number of common enriched GO terms, N = number of enriched GO terms for a genus, p = probability for any GO term associated with the divergence hotspots in both genera to be enriched GO term for another genus). The four divergence hotspots related to circadian clock system were submitted to MESSA server (Cong and Grishin 2012) to perform secondary structure and disordered region prediction, domain identification, and 3D structure prediction.
Selection of nuclear barcodes
We selected nuclear DNA barcodes from nuclear exons that were longer than 100 bp and present in the transcriptomes of at least two specimens of each species: P. glaucus, P. canadensis, H. rumiko, and H. cresphontes. We calculated the percent of different positions between any pair of specimens from the same genus, and computed the difference between the minimal interspecific divergence and the maximal intraspecific divergence. This number is negative for most exons: the mean and standard deviation for Pterourus are −0.010 and 0.018, respectively; similarly the mean and standard deviation for Heraclides are −0.017 and 0.020, respectively. Only 121 exons have a minimal interspecific divergence higher than the maximal intraspecific divergence for both Heraclides and Pterourus and only 10 exons’ minimal interspecific divergence is 1.0% higher than the maximal intraspecific value (minimal interspecific – maximal intraspecific ≥ 1.0%) and they were selected as possible nuclear barcodes.
Results
Statistics of assembling transcripts
The specimens of Heraclides (5 H.rumiko and 5 H.cresphontes) used in this study are listed in Table 1. De novo transcriptome assembly resulted in a range of 20000–70000 transcripts per specimen. A majority of these transcripts (>90%) could be mapped to the P. glaucus reference protein set (Cong et al. 2015a) by BLASTX (version 2.2.31+) (Altschul et al. 1990), and the mapped transcripts covered 7000 – 12000 proteins annotated in the reference genome. We combined all the de novo assembled transcripts from the 10 specimens of Heraclides and removed redundancy to create a reference transcriptome consisting of 17 942 protein coding sequences. For alternatively spliced isoforms, the longest one was taken as the reference.
We mapped the reads from all the specimens of Heraclides to this reference and performed SNP calling using GATK (version 3.3–0) to obtain the sequences of each specimen. About 13 050 reference transcripts were covered by at least two H. rumiko and two H. cresphontes specimens for at least 20 amino acids. The Heraclides dataset used in the following analyses consisted of 5 288 104 residues in these 13 050 transcripts. They mapped to 10 272 (out of 15 685) proteins in the P. glaucus reference genome. Transcripts that were mapped to the same protein were usually different segments of the same protein, and they were combined into the same orthologous group.
This seemingly modest completeness likely results from that only a subset of proteins being expressed in adults. Additionally, some annotated genes in the P. glaucus reference genome could be pseudogenes. Out of the 15 685 annotated protein-coding genes in the P. glaucus genome, only 12 928 (82.4%) had annotated orthologs in two other swallowtail genomes (Nishikawa et al. 2015) or are present in the transcriptomes of Pterourus. In addition, we identified 9421 orthologous protein families from the specimens of Pterourus (3 P. glaucus and 2 P. canadensis), and these orthologous groups contained 3 417 956 residues that were covered by at least two P. glaucus and two P. canadensis specimens.
Divergence in two species of Heraclides is comparable to that in two species of Pterourus
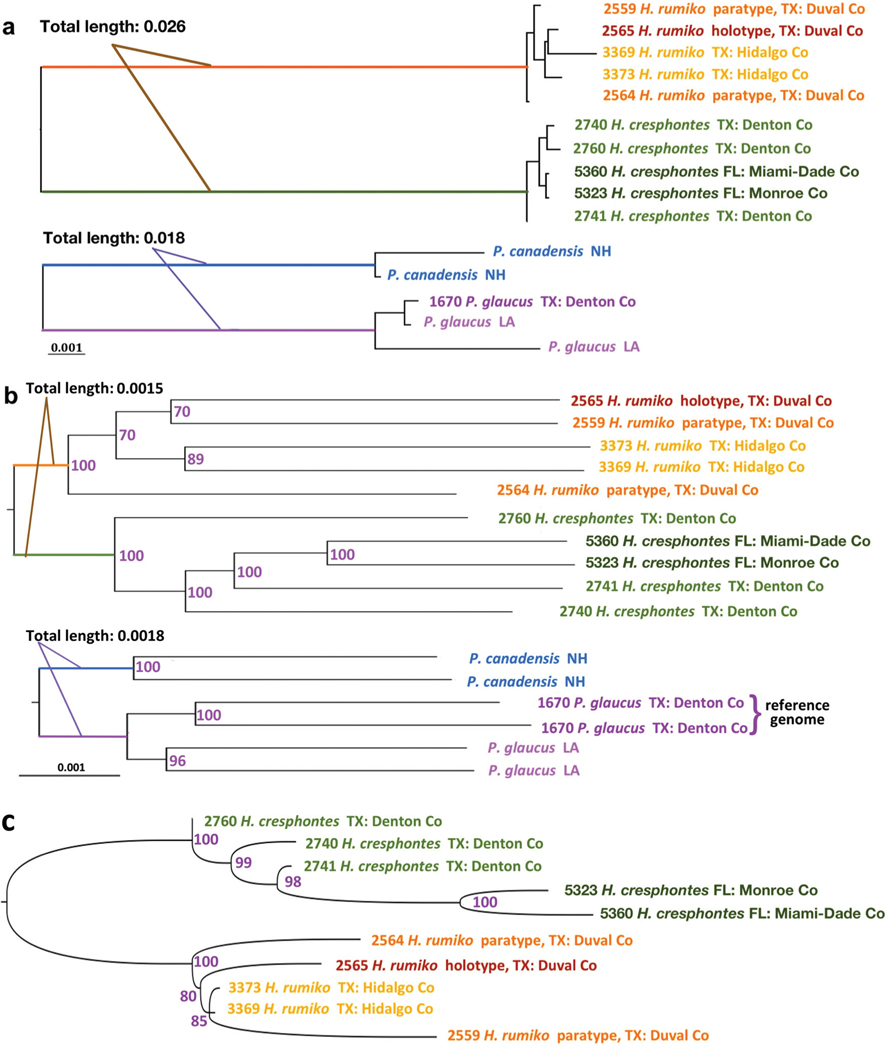
We previously showed that H. cresphontes and H. rumiko could be confidently distinguished by the COI barcodes: intraspecific divergence was less than 0.5%, while interspecific divergence was about 2.9% (Shiraiwa et al. 2014). Other mitochondrial genes revealed a similar separation between H. cresphontes and H. rumiko. In a tree based on the concatenated alignment of all mitochondrial protein-coding genes, the internal branch length between the two clades formed by both species is 0.026 (26 changes per 1000 positions). This divergence is comparable to that between P. glaucus and P. canadensis (18 changes per 1000 positions, shown in Fig. 1a).
Fig. 1.
Phylogenetic trees for Heraclides and Pterourus. (a) Maximal likelihood trees based on the concatenated alignments of mitochondrial genes. (b) The 50% majority-rule consensus tree of the maximum likelihood trees based on the bootstrap sampling of the concatenated alignment of orthologous transcripts from all specimens of Heraclides and Pterourus. The branch that connects the Heraclides and Pterourus clades is omitted from the figure. Specimen numbers, species names, and localities are labeled. The Pterourus glaucus specimen with a reference genome is represented by two sequences to reflect the heterozygous positions. Combined length of the internal branches that separate the two species from each genus is measured (approximately) and labeled in the figure. (c) Evolution history inferred by TreeMix based on random samples (bootstrap) of SNP markers in specimens of Heraclides. The middle point of the longest internal branch is inferred as the root of the tree.
We obtained a concatenated alignment of 5427 orthologous transcripts that were shared by all 10 specimens of Heraclides and 5 specimens of Pterourus. This alignment contained 2.8 million positions without gaps, and was used to build phylogenetic trees (Fig. 1). In both the maximum likelihood tree (Fig. 1b), based on concatenated alignment, and the TreeMix (version 1.13) tree, based on allele frequencies from randomly sampled SNPs, the specimens of Heraclides confidently (bootstrap: 100) partitioned into two clades: one clade for each species. The divergence between the two clades, measured by the total branch length of the two clades (1.5 expected substitutions per 1000 positions in coding regions in maximum likelihood tree), is comparable to that between P. glaucus and P. canadensis (1.8 expected substitutions per 1000 positions in coding regions in maximum likelihood tree). Finally, the estimated divergence time of the two species of Heraclides, using an isolation-with-migration model, is 0.8 million (95% confidence interval: 0.61–0.99) generations ago, which is again similar to the estimate for the two species of Pterourus at 0.77 million (95% confidence interval: 0.62–0.92) generations ago (supplemental data, Fig. S11).
Heraclides rumiko and H. cresphontes diverged in a small number of genes
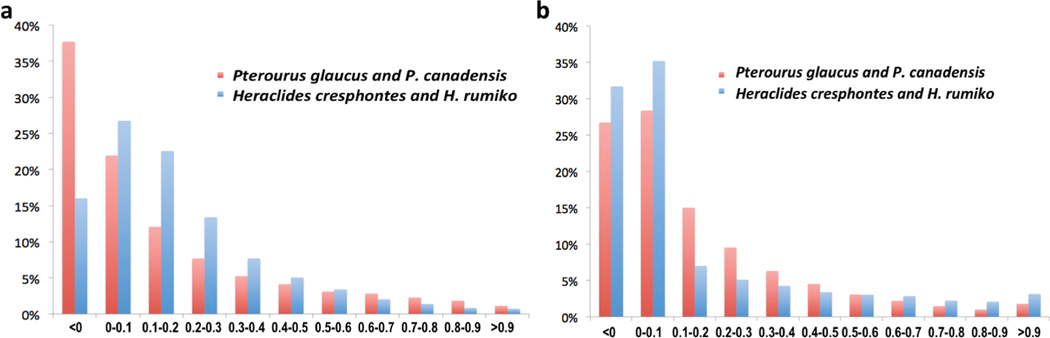
As in the case of P. glaucus and P. canadensis, H. rumiko and H. cresphontes can be confidently distinguished based on the whole-transcriptome data, but not by most individual genes. The average interspecific divergence for each gene is significantly higher than the intraspecific divergence in species of both Pterourus (p = 7.9e–79) and Heraclides (p = 2.3e–234). However, for both Pterourus and Heraclides, the intraspecific and interspecific distributions largely overlap (Fig. S21), and the overall interspecific divergence level is only 15% and 23%, respectively, higher than the intraspecific divergence level. Therefore, genetic variation within each species is high (above 0.8% in protein coding genes), and the divergence between species only slightly, albeit statistically significantly, exceeds intraspecific variation rate.
As quantified by fixation indices (FST), the majority of individual genes (Fig. 2a) and the proteins (Fig. 2b) they encode do not diverge between sister species in both genera. For over half (55% for Heraclides and 67% for Pterourus) of the proteins, the interspecific divergence is comparable (FST < 0.1) to the variability within species. However, a smaller fraction (13.3% for Pterourus and 9.5% for Heraclides) of proteins diverged noticeably between sister species (FST ≥ 0.5). Genomic regions encoding these proteins (divergence hotspots) may be candidates for genomic islands of speciation forming the reproductive barrier between species, resisting gene flow, and making each species distinct.
Fig. 2.
Fixation indices for sister species in genera Pterourus and Heraclides. (a) Distribution of FST for protein-coding genes in the species pairs of Pterourus (red) and Heraclides (blue) genera, respectively. (b) Distribution of FST for protein sequences in the species pairs of Pterourus (red) and Heraclides (blue) genera, respectively.
Divergence hotspots suggest common speciation paths in Heraclides and Pterourus
Out of 5 839 088 residues from 13 050 orthologous protein families encoded by the Heraclides transcripts, we identified 8729 positions that were conserved (showing the same amino acid in over 80% of the sequences) within both species of Heraclides but were divergent between them (the dominant amino acids are different). These divergent positions are not evenly distributed among proteins, and 883 Heraclides proteins (6.8%) are significantly enriched (P < 0.05) in such positions. These proteins are either important for speciation or are intrinsically fast evolving proteins that are also highly variable within a species. We further required the entire protein to be relatively conserved within both species (FST > = 0.5), which resulted in 432 proteins. These proteins show significantly (P < 0.001) lower intraspecific divergence than the rest. Their conservation within both species of Heraclides but elevated divergence between the species suggests their roles in speciation, and we named them divergence hotspots.
The Gene Ontology (GO) terms standing out (P < 0.01) as associated with these divergence hotspots are listed in Table 2. These GO terms suggest that the two species show differences in the circadian clock system, which might directly cause differences in timing of the mating behavior and contribute to prezygotic reproductive barrier. Other enriched GO terms suggest the two species diverged in proteins that are involved in transcription regulation and signal transduction. Divergence in these proteins may affect many downstream processes, and thus have a profound impact on the divergence and speciation. Using the same criteria, we identified 445 divergence hotspots for the two species of Pterourus.
Table 2.
Enriched GO terms associated with the divergence hotspots of specis of Heraclides.
| GO term | Q a | P b | Category | Definition | Associated divergence hotspots |
|---|---|---|---|---|---|
| Circadian rhythm | |||||
| GO:0008062 | 2.05E−02 | 2.45E−05 | BP | Eclosion rhythm | pgl483.12, pgl8568.1, pgl526.13, pgl856.9 |
| GO:0007622 | 2.25E−02 | 5.38E−05 | BP | Rhythmic behavior | pgl483.12, pgl8568.1, pgl526.13, pgl856.9 |
| GO:0048148 | 3.17E−02 | 1.14E−04 | BP | Behavioral response to cocaine | pgl483.12, pgl856.9, pgl526.13 |
| GO:0045187 | 3.89E−02 | 1.87E−04 | BP | Regulation of circadian sleep/wake cycle, sleep | pgl483.12, pgl8568.1, pgl526.13, pgl856.9 |
| GO:0060086 | 1.81E−01 | 1.74E−03 | BP | Circadian temperature homeostasis | pgl483.12, pgl8568.1 |
| GO:0032922 | 1.73E−01 | 2.69E−03 | BP | Circadian regulation of gene expression | pgl856.9, pgl526.13 |
| GO:0045475 | 1.96E−01 | 3.29E−03 | BP | Locomotor rhythm | pgl8568.1, pgl1965.12, pgl798.7, pgl856.9, pgl483.12, pgl526.13 |
| GO:0007623 | 2.22E−01 | 7.73E−03 | BP | Circadian rhythm | pgl483.12, pgl8568.1, pgl526.13, pgl856.9 |
| Gene regulation | |||||
| GO:0001047 | 1.84E−01 | 1.33E−03 | MF | Core promoter binding | pgl1629.18, pgl1613.5, pgl1785.2 |
| GO:2000620 | 2.07E−01 | 1.74E−03 | BP | Positive regulation of histone H4-K16 acetylation | pgl4056.3, pgl4056.2 |
| GO:0061085 | 1.61E−01 | 1.74E−03 | BP | Regulation of histone H3-K27 methylation | pgl352.17, pgl352.16 |
| GO:0043998 | 1.45E−01 | 1.74E−03 | MF | H2A histone acetyltransferase activity | pgl4056.3, pgl4056.2 |
| GO:2000678 | 1.32E−01 | 1.74E−03 | BP | Negative regulation of transcription regulatory region DNA binding | pgl483.12, pgl8568.1 |
| GO:0043968 | 2.13E−01 | 3.84E−03 | BP | Histone H2A acetylation | pgl4056.3, pgl4056.2 |
| GO:0043035 | 2.27E−01 | 5.98E−03 | BP | Chromatin insulator sequence binding | pgl4475.6, pgl1716.2, pgl352.14 |
| GO:0010485 | 2.23E−01 | 6.69E−03 | MF | H4 histone acetyltransferase activity | pgl4056.3, pgl4056.2 |
| GO:0042826 | 2.22E−01 | 7.44E−03 | MF | Histone deacetylase binding | pgl526.23, pgl5241.3, pgl352.20 |
| Signal transduction | |||||
| GO:0004871 | 1.65E−01 | 2.38E−03 | MF | Signal transducer activity | pgl856.9, pgl75.4, pgl4481.1, pgl483.12, pgl526.13 |
| GO:0016567 | 2.20E−01 | 5.02E−03 | BP | Protein ubiquitination | pgl1770.6, pgl6275.4, pgl306.7, pgl1282.9, pgl6275.2 |
| GO:0051282 | 2.15E−01 | 6.69E−03 | BP | Regulation of sequestering of calcium ion | pgl212.16, pgl16373.1 |
| GO:1901896 | 2.43E−01 | 6.69E−03 | BP | Positive regulation of calcium-transporting ATPase activity | pgl212.16, pgl16373.1 |
| GO:0006486 | 2.64E−01 | 9.49E−03 | BP | Protein glycosylation | pgl798.7, pgl1094.7, pgl903.31, pgl2713.9 |
| Others | |||||
| GO:0006505 | 2.00E−01 | 3.84E−03 | BP | GPI anchor metabolic process | pgl 1911.12, pgl4475.9 |
| GO:0016772 | 2.29E−01 | 5.48E−03 | MF | Transferase activity, transferring phosphorus-containing groups | pgl337.7, pgl1857.12, pgl2448.5, pgl1857.2 |
| GO:0006801 | 2.33E−01 | 6.69E−03 | BP | Superoxide metabolic process | pgl21.1, pgl2217.6 |
| GO:0015171 | 2.19E−01 | 4.47E−03 | MF | Amino acid transmembrane transporter activity | pgl44.17, pgl59.19, pgl1568.3, pgl1568.1, pgl7608.3 |
| GO:0017056 | 2.18E−01 | 4.71E−03 | MF | Structural constituent of nuclear pore | pgl2593.2, pgl3383.3, pgl21618.1 |
| GO:0003333 | 2.23E−01 | 5.62E−03 | BP | Amino acid transmembrane transport | pgl44.17, pgl59.19, pgl1568.3, pgl1568.1, pgl7608.3 |
Note:BP, biological process; MF, molecular function.
False discovery rate for GO terms at this significance level.
P-value for binomial test of GO term enrichment.
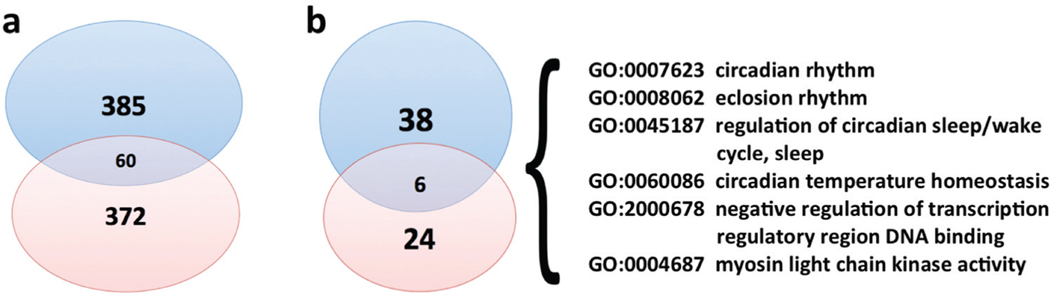
A binomial test shows that the divergence hotspots in Heraclides overlap very significantly (P = 9.7e–24, Fig. 3a, total number of genes in the reference genome: 15 685) with the ones for Pterourus. We identified the divergence hotspots from the Heraclides transcripts that could be mapped to the Pterourus protein set, which may artificially increase the overlap in divergence hotspots for both genera. Therefore, we performed another binomial test using only the orthologous groups shared by the two genera. This alternate non-reference test again reveals significant overlap between the divergence hotspots in both genera (P = 8.6e–20). The enriched (p < 0.01) GO terms associated with the Heraclides divergence hotspots also overlap significantly (p = 7.0e–9, total number of GO terms associated with proteins in the reference genome: 9337).
Fig. 3.
Significant overlap between the divergence hotspots for Heraclides and Pterourus. (a) Venn diagram of divergence hotspots for Pterourus (blue) and Heraclides (red), which overlap significantly (p = 8.6e–20). (b) Venn diagram for the enriched GO-terms associated with the divergence hotspots for Pterourus (blue) and Heraclides (red), which overlap significantly (p = 7.0e–9). The six GO-terms that are shared by both genera are listed.
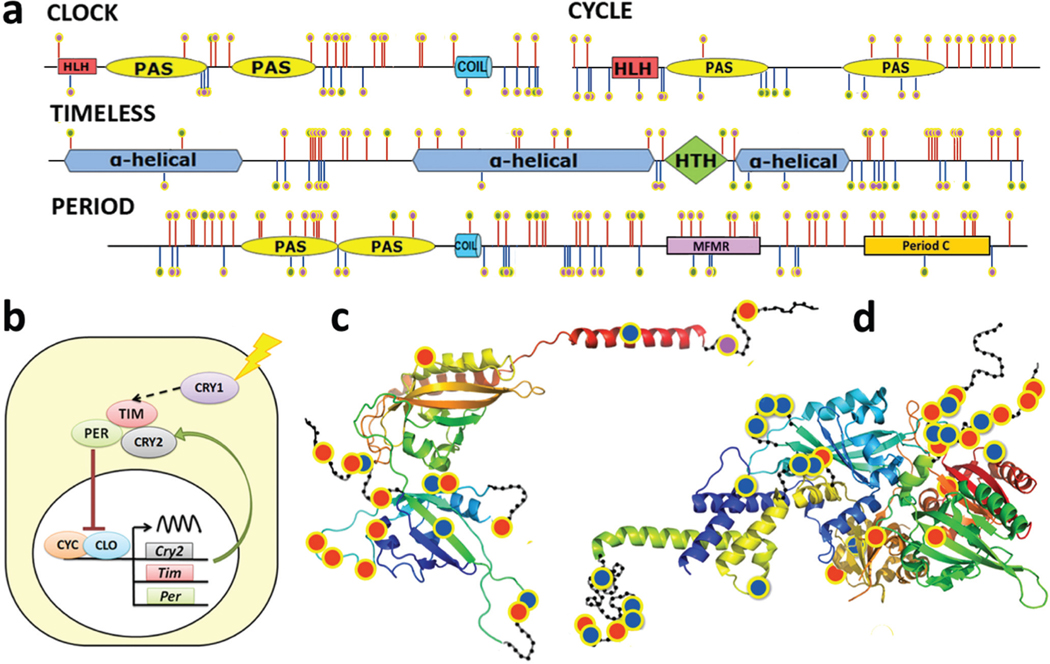
The largest group of enriched GO terms (Fig. 3b) associated with the divergence hotspots in both genera are related to the circadian clock system. All four central components of the circadian clock system, CLOCK, CYCLE, PEROID, and TIMELESS (Figs. 4a, 4b), are enriched in interspecific variation and belong to the divergence hotspots in both Heraclides and Pterourus. These four proteins are highly conserved within each species (on average 0.27% difference) but differ strongly between species (on average 2.5% difference). Moreover, the positions in these proteins that diverged between species of Heraclides and between species of Pterourus hardly overlap, suggesting that these positions are not intrinsically fast evolving. Instead, the divergence is probably due to adaptation to different environments. Two out of the four circadian clock proteins, CLOCK and PERIOD, are suggested to be under positive selection in Pterourus (Cong et al. 2015a).
Fig. 4.
Circadian clock proteins are divergent between species of Heraclides and Pterourus. (a) Domain diagram of CLOCK, CYCLE, PERIOD, and TIMELESS. Positions that are conserved within but differ between species of Heraclides are marked by pink dots on red stems; positions that are divergent within species of Heraclides are marked by green dots on red stems; positions that are conserved within but differ between species of Pterourus are marked by pink dots on blue stems; positions that are divergent within species of Pterourus are marked by green dots on blue stems. (b) Circadian clock system (CRY, cryptochrome proteins; CLO, CLOCK; CYC, CYCLE; PER, PERIOD; TIM, TIMELESS). (c and d) Map of interspecific mutations on the spatial structure templates (PDB ids: 4F3L and 3RTY) of the CLOCK/CYCLE complex and protein PERIOD. Positions that are conserved within but divergent between species of Heraclides, species of Pterourus, and species in both genera are marked by red, blue, and magenta dots, respectively. The approximate position of disordered loops is shown as black beads on threads.
Mapping divergent positions between the two species to their protein sequences and predicted structures shows that these mutations concentrate on one side of the protein PERIOD, forming clusters on the surface (Fig. 4c). A similar distribution of mutation sites is observed in the CLOCK–CYCLE complex (Fig. 4d). The surface clustering of mutations suggests that they may affect the interactions between the CLOCK–CYCLE complex and other regulators of the circadian clock system (Tataroglu and Emery 2015).
Nuclear barcodes to identify swallowtail species
The commonly used nuclear markers for insects include 18s rRNA, wingless, EF1a genes, and non-coding ITS1 and ITS2. However, these genes fail to distinguish incipient species such as P. glaucus versus P. canadensis and H. cresphontes versus H. rumiko, which can be clearly separated by COI barcodes. In a quest for nuclear barcodes, we searched for exons in protein coding genes that can clearly identify species in the two genera. Based on our analysis on Heraclides and Pterourus, we selected 10 nuclear barcodes (Table 3) using the following criteria: (i) a nuclear barcode should be a long exon of over 100 bp; (ii) the interspecific divergence for this barcode should be much higher than the intraspecific divergence in both genera; (iii) it should be from the divergence hotspots we identified above. Since the speciation mechanisms shared by these two swallowtail pairs may not be general for other species, we would not expect any single nuclear barcodes to be generally applicable. However, a combination of several nuclear barcodes we selected here might be useful in other butterflies. Only a single nuclear barcode appears to distinguish species of Heraclides and Pterourus even better (measured by the difference between interspecific divergence and intraspecific divergence) than the COI barcode. It is from a circadian clock protein, TIMELESS, reinforcing the divergence in the circadian clock system as an important player in both speciation events.
Table 3.
Nuclear gene markers for the identification of species of Pterourus and Heraclides.
|
Pterourus
|
Heraclides
|
||||||
|---|---|---|---|---|---|---|---|
| Protein ID | Exon No. | Flybase ID | Length (bp) | Max. intra.a (%) | Min. inter.b (%) | Max. intra.a (%) | Min. inter.b (%) |
| pgl 2000.8 | exon3 | CG8224-PB | 227 | 0.00 | 1.32 | 0.71 | 2.20 |
| pgl2046.5 | exon69 | CG33950-PAT | 195 | 0.00 | 1.03 | 0.00 | 1.03 |
| pgl2093.1 | exon2 | CG44162-PU | 3823 | 0.25 | 2.40 | 0.42 | 1.89 |
| pgl2093.4 | exon3 | CG3626-PA | 162 | 0.00 | 1.23 | 0.62 | 1.85 |
| pgl2093.4 | exon7 | CG3626-PA | 171 | 0.58 | 1.75 | 0.00 | 2.34 |
| pgl352.20 | exon15 | CG1868-PB | 192 | 0.52 | 1.56 | 0.00 | 1.56 |
| pgl352.36 | exon5 | CG8789-PA | 133 | 0.00 | 1.50 | 0.81 | 3.01 |
| pgl352.36 | exon7 | CG8789-PA | 365 | 0.27 | 2.19 | 1.21 | 3.03 |
| pgl8568.1 | exon13 | CG3234-PB | 165 | 1.21 | 3.64 | 1.21 | 2.42 |
| pgl8568.1 | exon4 | CG3234-PB | 246 | 0.41 | 2.85 | 0.41 | 2.94 |
| COI | — | — | 658 | 0.46 | 2.14 | 0.46 | 2.88 |
Maximal intraspecific divergence (percent of different positions).
Minimal interspecific divergence (percent of different positions).
Discussion
High divergence within butterfly species
The intraspecific divergence rate in the protein-coding region we observed is close to 1%, and this high divergence likely reflects a large effective population size. Given the mutation rate (μ) of Lepidoptera is about 3e–9 per generation (Keightley et al. 2015), intraspecific variation (θ) of 1% roughly corresponds to an effective population size (Ne) of about 1 million (θ = 4 μNe). Swallowtail butterflies are common, widely distributed, and are strong flyers. Therefore, their effective population sizes are likely to be large. In addition, the recent speciation in both sister species pairs and thus possible remaining gene flow between the incipient species (Mercader et al. 2009) may increase the intraspecific divergence. Also, several recently published butterfly genomes (Cong et al. 2015b, 2016) report similarly high (about 1%) polymorphism.
Divergence hotspots and proteins undergoing positive selection
A previous study found 21 positively selected proteins between P. glaucus and P. canadensis using McDonald–Kreitman Tests (Zhang et al. 2013), and about half of these proteins overlap with the divergence hotspots we identified. The lack of a larger overlap may be due to three reasons. First, previous results were based on 2225 genes, whereas our analysis, aided by the reference genome of P. glaucus, is performed on 9421 genes. Second, inference of positive selection using a small number of individuals could be influenced by the underlying statistical model and the sequences used in the test. For instance, the proteins identified using two methods, McDonald–Kreitman Test and Ka/Ks ratio, show very limited overlap (5%) (Zhang et al. 2013). Third, the drivers of speciation do not have to be positively selected (Wu and Ting 2004).
Ecological speciation
If two populations of the same species have spent significant time in geographic isolation, mutations randomly accumulating in them could cause Dobzhansky–Muller hybrid incompatibilities, leading to reproductive isolation (Orr and Turelli 2001). Additionally, ecological factors could play a role. When two populations of the same ancestral species have been separated in different environments, they might undergo adaptive evolution (Dieckmann et al. 2004). It is possible that this environmental adaptation is the mechanism for the speciation of the sister species from Pterourus and Heraclides genera discussed here. Both speciation events are associated with the separation of populations by latitude with different temperature, photoperiod, and environment, which could have a profound impact on the populations. For instance, P. canadensis lives in a colder climate where winters cannot support a continuous life cycle. Therefore, it developed an obligate pupal diapause to overwinter, which is an adaptive behavior lacking in its sister species P. glaucus (Hagen et al. 1991).
Nuclear markers for species identification
The mitochondrial DNA COI barcode sequence is routinely used for insect identification and cryptic species discovery (Hebert et al. 2004). However, maternally inherited mitochondrial DNA can be transferred between species via cellular symbionts (Whitworth et al. 2007), hybridization, and backcrossing, and therefore they may have a history different from the whole organism (Bachtrog et al. 2006; Boratynski et al. 2011). Consequently, COI barcode studies need to be supplemented with work based on nuclear genes. However, it is not trivial to select proper nuclear barcodes to distinguish closely related species. The reproductive barrier is frequently not absolute for insects. While genes that contribute to Dobzhansky–Muller hybrid incompatibility may be less likely to flow between species, many other genes that do not contribute to the reproductive barrier, including the complete mitogenome, could be transferred. Proteins that are likely to play roles in speciation are better candidates for nuclear markers, such as the circadian clock proteins identified in this study.
Circadian clock and ecological speciation
The potential involvement of circadian clock proteins in speciation between sister species was unexpected. Therefore, we performed additional tests to rule out explanations not relevant to speciation. First, circadian clock proteins did not show a higher level of polymorphism within species comparative to other genes, indicating that they are not intrinsically more variable. Second, two out of the four circadian clock proteins, CLOCK and PERIOD, were under positive selection in Pterourus (Cong et al. 2015a), implying their divergence is likely a result of adaptive evolution. On the other hand, we see biological differences between the sister species that are likely affected by the divergence in the circadian clock system. Obligate pupal diapause controlled by these proteins in P. canadensis is a likely adaptation to colder northern climate this species lives in. Even in warmer years, P. canadensis would not have sufficient warm time within the year to develop more than a single brood. Conversely, southern species, i.e., P. glaucus, get a selective advantage by building up in numbers over longer warm periods of the year by going through several broods due to facultative pupal diapause induced by the photoperiod (Hagen et al. 1991). Biological differences between southwestern species H. rumiko and more northern and eastern H. cresphontes have not yet been studied. Heraclides rumiko inhabits more open and dryer areas with apparently more light, whereas H. cresphontes is more of a forest species, active in more shaded areas with less light. While both species possess facultative diapause, H. rumiko has more generations per year and may not diapause at all in the southern parts of its range from Mexico to Panama.
This divergence in circadian clock systems might result from adaptation to different latitudes. The proteins CLOCK, CYCLE, PERIOD, and TIMELESS directly interact with each other to regulate the circadian rhythm, and they are expected to evolve together in each species to maintain the functional interactions between proteins. Therefore, the components from one species may not be fully compatible with components from another species, contributing to Dobzhansky–Muller hybrid incompatibilities and post-zygotic isolation.
Meanwhile, this adaptive divergence might cause prezygotic reproductive isolation. Clock genes are known to directly play a role in mating behavior in Drosophila (Allada and Chung 2010). Similar molecular processes are expected to take place in Lepidoptera, as several studies on moths have suggested (Merlin et al. 2007; Quan et al. 2017; Wu et al. 2014). Circadian clock genes may regulate not only the timing of mating (Sakai and Ishida 2001), but also copulation duration (Beaver and Giebultowicz 2004) and frequency of vibratory signals (Medina et al. 2015). This intricate network of regulatory signals should be easy to break with differences in these genes, leading to mate rejection. Courtship rituals are highly elaborate in swallowtails (Lederhouse 1981; Scott 1986), and only a narrow range of behaviors (like flying in circles of a particular radius) would avoid mate rejection. This behavioral pressure is expected to evoke strong stabilizing selection on the genes involved in the circadian clock, resulting in low intraspecific variation. Interspecific differences will not be constrained and would increase further, driven by lower reproductive fitness of hybrids that would be selectively eliminated from the population in the contact zones due to their possibly unusual mating behavior.
Divergence in circadian clock genes may not be restricted to swallowtails and may be a more general trend for pairs of closely related species. Correlation between speciation and circadian proteins divergence has been recently reported for other species of Lepidoptera (Cong et al. 2016; Hänniger 2015; Tauber et al. 2003). It is possible that divergence in clock proteins may not be the driving event in speciation. However, this divergence is most likely an adaptive trait developed in the course of speciation. In addition, current study is based on transcriptomes of adult butterflies, and only about 60%–80% of total proteins are expressed. It is possible that some speciation genes fall outside this subset and future genomic studies will address this question. Regardless of causal relationship and the potential to miss important speciation drivers among non-coding regions in the genome and proteins that are not expressed in adults, circadian clock system recurrently comes up as a component strongly correlated with speciation in sister species and is expected to be an important player.
Supplementary Material
Acknowledgements
We acknowledge Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the permit #08-02Rev that makes research based on material collected in Texas State Parks possible. We thank Lisa N. Kinch for critical suggestions and proofreading of the manuscript and reviewers for helpful suggestions that improved this work. Qian Cong was a Howard Hughes Medical Institute International Student Research fellow when this project was carried out. This work was supported in part by the National Institutes of Health (GM094575 and GM127390 to N.V.G.) and the Welch Foundation (I-1505 to N.V.G.).
Footnotes
Supplementary data are available with the article through the journal Web site at http://nrcresearchpress.com/doi/suppl/10.1139/gen-2018-0084.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Qian Cong, Department of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390-8816, USA..
Nick V. Grishin, Department of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390-8816, USA; Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390-9050, USA.
References
- Allada R, and Chung BY 2010. Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol 72: 605–624. doi: 10.1146/annurev-physiol-021909-135815. PMID: 20148690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ 1990. Basic local alignment search tool. J. Mol. Biol 215(3): 403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Thornton K, Clark A, and Andolfatto P. 2006. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution, 60(2): 292–302. doi: 10.1554/05-337.1. PMID:16610321. [DOI] [PubMed] [Google Scholar]
- Beaver LM, and Giebultowicz JM 2004. Regulation of copulation duration by period and timeless in Drosophila melanogaster. Curr. Biol 14(16): 1492–1497. doi: 10.1016/j.cub.2004.08.022. PMID:15324667. [DOI] [PubMed] [Google Scholar]
- Boratynski Z, Alves PC, Berto S, Koskela E, Mappes T, and Melo-Ferreira J. 2011. Introgression of mitochondrial DNA among Myodes voles: consequences for energetics? BMC Evol. Biol 11: 355. doi: 10.1186/1471-2148-11-355. PMID:22151479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, and Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics, 172(4): 2665–2681. doi: 10.1534/genetics.105.048975. PMID:16489234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, and Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. In Computer Science and Biology: Proceedings of the German Conference on Bioinformatics, Vol. 99. pp. 45–56. [Google Scholar]
- Cong Q, and Grishin NV 2012. MESSA: MEta-Server for protein Sequence Analysis. BMC Biol. 10: 82. doi: 10.1186/1741-7007-10-82. PMID:23031578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Borek D, Otwinowski Z, and Grishin NV 2015a. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 10(6): 910–919. doi: 10.1016/j.celrep.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Borek D, Otwinowski Z, and Grishin NV 2015b. Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics, 16: 639. doi: 10.1186/s12864-015-1846-0. PMID:26311350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Warren AD, Borek D, Otwinowski Z, and Grishin NV 2016. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol. Evol 8(3): 915–931. doi: 10.1093/gbe/evw045. PMID:26951782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra KK, Walters JR, Briscoe AD, Davey JW, Whibley A, Nadeau NJ, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature, 487(7405): 94–98. doi: 10.1038/nature11041. PMID:22722851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43(5): 491–498. doi: 10.1038/ng.806. PMID:21478889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann U, Doebeli M, Metz JAJ, and Tautz D. 2004. Adaptive speciation. Cambridge Studies in Adaptive Dynamics, Cambridge University Press, Cambridge, UK. [Google Scholar]
- Feder JL, Egan SP, and Nosil P. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28(7): 342–350. doi: 10.1016/j.tig.2012.03.009. PMID:. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 8(8): 1494–1512. doi: 10.1038/nprot.2013.084. PMID:23845962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen RH, Lederhouse RC, Bossart JL, and Scriber JM 1991. Papilio canadensis and P. glaucus (Papilionidae) are distinct species. J. Lepidopt. Soc 45(4): 245–258. [Google Scholar]
- Hänniger S. 2015. Chasing sympatric speciation: The relative importance and genetic basis of prezygotic isolation barriers in diverging populations of Spodoptera frugiperda. Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam. [Google Scholar]
- Harr B. 2006. Genomic islands of differentiation between house mouse subspecies. Genome Res. 16(6): 730–737. doi: 10.1101/gr.5045006. PMID:16687734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Penton EH, Burns JM, Janzen DH, and Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A 101(41): 14812–14817. doi: 10.1073/pnas.0406166101. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, and Nielsen R. 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl. Acad. Sci. U.S.A 104(8): 2785–2790. doi: 10.1073/pnas.0611164104. PMID:17301231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Pinharanda A, Ness RW, Simpson F, Dasmahapatra KK, Mallet J, et al. 2015. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol. Biol. Evol 32(1): 239–243. doi: 10.1093/molbev/msu302. PMID: 25371432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederhouse RC 1981. Territorial defense and lek behavior of the Black Swallowtail Butterfly, Papilio polyxenes. Behav. Ecol. Sociobiol 10(2): 109–118. doi: 10.1007/BF00300170. [DOI] [Google Scholar]
- Li H, and Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25(14): 1754–1760. doi: 10.1093/bioinformatics/btp324. PMID: 19451168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, and Barbash DA 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet 45: 331–355. doi: 10.1146/annurev-genet-110410-132514. PMID:21910629. [DOI] [PubMed] [Google Scholar]
- Medina I, Casal J, and Fabre CC 2015. Do circadian genes and ambient temperature affect substrate-borne signalling during Drosophila courtship? Biol. Open, 4(11): 1549–1557. doi: 10.1242/bio.014332. PMID:26519517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader RJ, Aardema ML, and Scriber JM 2009. Hybridization leads to host-use divergence in a polyphagous butterfly sibling species pair. Oecologia, 158(4): 651–662. doi: 10.1007/s00442-008-1177-9. PMID:18949489. [DOI] [PubMed] [Google Scholar]
- Merlin C, Lucas P, Rochat D, Francois MC, Maibeche-Coisne M, and Jacquin-Joly E. 2007. An antennal circadian clock and circadian rhythms in peripheral pheromone reception in the moth Spodoptera littoralis. J. Biol. Rhythms, 22(6): 502–514. doi: 10.1177/0748730407307737. PMID:18057325. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, et al. 2015. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat. Genet 47(4): 405–409. doi: 10.1038/ng.3241. PMID:25751626. [DOI] [PubMed] [Google Scholar]
- Orr HA, and Turelli M. 2001. The evolution of postzygotic isolation: accumulating Dobzhansky–Muller incompatibilities. Evolution, 55(6): 1085–1094. doi: 10.1554/0014-3820(2001)0551085:TEOPIA2.0.CO;2. PMID:11475044. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Baker EP, Cooper JC, Frizzell KA, Hsieh E, de la Cruz AF, et al. 2015. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science, 350(6267): 1552–1555. doi: 10.1126/science.aac7504. PMID:26680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, and Pritchard JK 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11): e1002967. doi: 10.1371/journal.pgen.1002967. PMID:23166502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan WL, Liu W, Zhou RQ, Chen R, Ma WH, Lei CL, and Wang XP 2017. Difference in diel mating time contributes to assortative mating between host plant-associated populations of Chilo suppressalis. Sci. Rep 7: 45265. doi: 10.1038/srep45265. PMID:28338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, and Blackman BK 2010. Speciation genes in plants. Ann. Bot 106(3): 439–455. doi: 10.1093/aob/mcq126. PMID:20576737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, and Ishida N. 2001. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A 98(16): 9221–9225. doi: 10.1073/pnas.151443298. PMID:11470898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. 1986. The butterflies of North America. Stanford University Press, Stanford, California. [Google Scholar]
- Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, et al. 2014. Genomics and the origin of species. Nat. Rev. Genet 15(3): 176–192. doi: 10.1038/nrg3644. PMID:24535286. [DOI] [PubMed] [Google Scholar]
- Shiraiwa K, Cong Q, and Grishin NV 2014. A new Heraclides swallowtail (Lepidoptera, Papilionidae) from North America is recognized by the pattern on its neck. Zookeys, 468: 85–135. doi: 10.3897/zookeys.468.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9): 1312–1313. doi: 10.1093/bioinformatics/btu033. PMID: 24451623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroglu O, and Emery P. 2015. The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci 7: 51–57. doi: 10.1016/j.cois.2015.01.002. PMID:26120561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber E, Roe H, Costa R, Hennessy JM, and Kyriacou CP 2003. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr. Biol 13(2): 140–145. doi: 10.1016/S0960-9822(03)00004-6. PMID:12546788. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, and Nuzhdin SV 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3(9): e285. doi: 10.1371/journal.pbio.0030285. PMID:16076241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth TL, Dawson RD, Magalon H, and Baudry E. 2007. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proc. Biol. Sci 274(1619): 1731–1739. doi: 10.1098/rspb.2007.0062. PMID:17472911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Lindell J, and Backstrom N. 2010. Speciation genetics: current status and evolving approaches. Philos. Trans. R. Soc. B Biol. Sci 365(1547): 1717–1733. doi: 10.1098/rstb.2010.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, and Ting CT 2004. Genes and speciation. Nat. Rev. Genet 5(2): 114–122. doi: 10.1038/nrg1269. PMID:14735122. [DOI] [PubMed] [Google Scholar]
- Wu S, Refinetti R, Kok LT, Youngman RR, Reddy GV, and Xue FS 2014. Photoperiod and temperature effects on the adult eclosion and mating rhythms in Pseudopidorus fasciata (Lepidoptera: Zygaenidae). Environ. Entomol 43(6): 1650–1655. doi: 10.1603/EN14164. PMID:25479201. [DOI] [PubMed] [Google Scholar]
- Zhang W, Kunte K, and Kronforst MR 2013. Genome-wide characterization of adaptation and speciation in tiger swallowtail butterflies using de novo transcriptome assemblies. Genome Biol. Evol 5(6): 1233–1245. doi: 10.1093/gbe/evt090. PMID:23737327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.