Abstract
The ability of the Gram-negative outer membrane (OM) to act as a permeability barrier has long been appreciated, but recent studies have uncovered a more expansive and versatile role for the OM in cellular physiology and viability. Due to recent developments in microfluidics and microscopy, the structural, rheological, and mechanical properties of the OM are becoming apparent across multiple scales. In this review, we discuss recent experimental and computational studies that have revealed key molecular factors and interactions that give rise to the spatial organization, limited diffusivity, and stress-bearing capacity of the OM. These physical properties suggest broad connections between cellular structure and physiology, and we explore future prospects for further elucidation of the implications of OM construction on cellular fitness and survival.
Introduction
The architecture and makeup of the bacterial cell envelope have long been topics of intense interest, in part due to the importance of the envelope for viability, virulence, and mechanical integrity. Unlike Gram-positive bacteria, Gram-negative bacteria have two highly distinct membranes that delimit an aqueous cellular compartment called the periplasm1 (Figure 1A). The peptidoglycan (PG) cell wall, which determines cell shape2, resides in this extra-cytoplasmic compartment, as does a variety of proteins. Surrounding the cell wall is the outer membrane (OM), a hallmark of Gram-negative species. The OM is an atypical biological membrane: while it is a lipid bilayer, it is asymmetric, with phospholipids (PLs) in the inner leaflet and a glycolipid known as lipopolysaccharide (LPS) in the outer leaflet3. The OM also contains two major classes of proteins. Nearly all of the transmembrane proteins assume a β-barrel fold and are commonly referred to as outer membrane proteins (OMPs)1. The OM additionally contains lipoproteins, which are mostly soluble but have a lipid moiety at the amino terminus that enables their embedding in the OM4. Unlike typical biological membranes, which are impermeable to protons, the OM is freely permeable to small, water-soluble molecules like sugars and amino acids5. These molecules diffuse through the OM via channels formed by a major class of OMPs called porins5. Also unlike typical membranes, the OM is resistant to detergents and other hydrophobic toxins. The strong lateral interactions between LPS molecules, together with their saturated acyl chains, stabilize the OM, greatly hindering the passage of hydrophobic molecules5. These biochemical properties hint at the mechanical importance of the OM, yet many of its structural properties remain mysterious. Only recently have studies started to probe the magnitude, molecular determinants, and tuneability of OM physical properties.
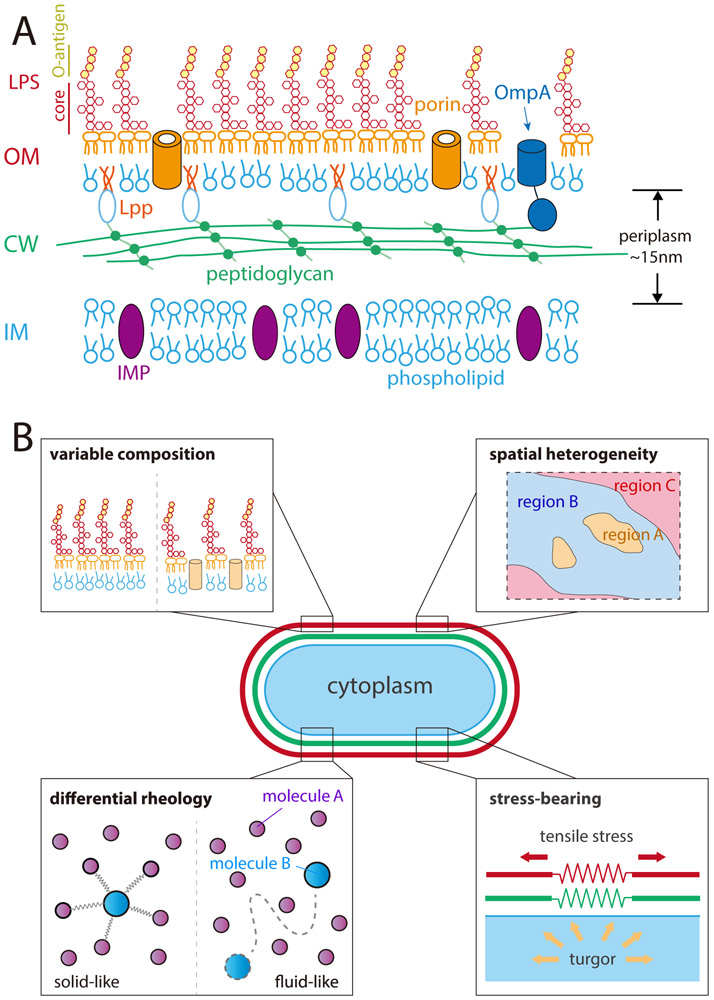
Figure 1: The Gram-negative outer membrane has a diverse makeup and a wide variety of physical and mechanical properties.
A) The cytoplasm of all bacterial cells is surrounded by an inner membrane (IM) composed of phospholipids and inner membrane proteins (IMPs), which is enclosed by the cell wall (CW), a macromolecular network of peptidoglycan (PG) that single-layered in E. coli29. In Gram-negative bacteria, the cell wall resides in the periplasm, a ~15-nm thick aqueous compartment enclosed by the inner and outer membranes. The outer membrane (OM) is asymmetric, composed of phospholipids in the inner leaflet and lipopolysaccharides (LPS), along with outer membrane proteins (OMPs) such as porins and lipoproteins such as Lpp, which links the OM and PG.
B) The OM (red) can exhibit variable composition, spatial heterogeneity, solid- or fluid-like rheology (diffusive dynamics), and the ability to bear mechanical stresses of similar magnitude to the cell wall (green). As a result, the chemical, physical, and mechanical properties of the OM can have broad impacts on bacterial physiology.
The OM is thought to be essential for viability in most Gram-negative bacteria1. Due to its adjacency with the extracellular milieu, the OM serves as the location of a substantial fraction of environmental sensors6-9 and as an anchor point for adhesive organelles10. The OM acts as a platform for interacting with host immune systems11-13, as well as with neighboring bacterial cells through surface contact14 and vesicles15. Moreover, the OM is an important permeability barrier providing Gram-negative bacteria resistance to large and hydrophobic antibiotics that are effective against Gram-positive bacteria with conserved targets5,16-18. Indeed, multi-drug resistant Gram-negative bacteria currently pose a serious threat to human health19,20, and the inability to overcome OM barrier function has, in part, hampered antibiotic discovery efforts21,22 aimed at avoiding a possible return to the time when infectious diseases were more feared than cancer23. The OM is also a barrier to large molecules, for example by sterically hindering antibodies and phages from binding to surface targets24-28. Although clearly useful, this barrier function does not explain why the OM is essential. In Escherichia coli for example, there are only a few enzymes present in the OM, and none of these enzymes performs an essential function. Indeed, the only essential proteins in the OM are ones necessary for building the OM1.
Besides the presence or absence of an OM, another major difference between Gram-negative and Gram-positive bacteria is the thickness of the PG cell wall, which is a monolayer in at least several Gram-negative model organisms29 and multi-layered (often tens of nanometers thick30,31) in Gram-positive organisms. The classical picture of the Gram-negative bacterial envelope has assigned responsibilities for stress-bearing to the cell wall and barrier functions to the OM. However, the two structures are physically connected: across the entire surface by proteins such as OmpA32,33, at the division site by the Tol/Pal complex34,35, and during cell wall synthesis by the LpoA/B proteins that bind to the wall synthesis enzymes PBP1A/B36,37. In certain enteric bacteria such as E. coli, the cell wall and the OM are also coupled covalently by Braun’s lipoprotein38. Moreover, in recent years, accumulating evidence has suggested that the essential function of the OM may be mechanical39-42, contributing physical strength to compensate for the thin cell wall. Indeed, the crosslinking of LPS molecules by divalent cations43 has been suggested as a mechanism of mechanical stabilization44. Moreover, although antibiotics targeting cell-wall biogenesis are effective against Gram-negative bacteria, they can often survive at least temporarily without a cell wall45,46, with certain mutations enabling stable, wall-less proliferation47. In this review, we will focus on the emerging physical and mechanical properties of the OM and the ways this unique membrane contributes strength to improve bacterial fitness.
Molecular components of the OM
The OM is made up of PLs, LPS, and proteins3,48-50, whose localization patterns and interactions determine the basic chemical and physical properties of this structure. The abundances of OMPs and lipoproteins51 as well as LPS offer insights into OM function. For decades, OM research has focused on bilayer asymmetry and the biochemical functions of the molecular components16,17,48,49,52-58. Now, advances in imaging and force spectroscopy are improving our understanding of the molecular organization and physical properties of the OM at higher spatial and temporal resolution (Figure 1B).
The LPS that occupies the outer leaflet3 consists of three moieties: the hydrophobic lipid A, a conserved oligosaccharide core, and a variable polysaccharide called the O-antigen59. The negatively charged LPS molecules are neutralized and bridged (often referred to as “cross-linked”) by divalent cations such as Mg2+. At roughly 1 million molecules per cell53, LPS makes up a substantial fraction of the OM and hence has the potential to be a major contributor to maintaining structural integrity of the OM. LPS localization to the OM outer leaflet was first determined in Salmonella3 and is now thought to generally apply to Gram-negative bacteria. Some antibacterial agents specifically target LPS: LPS molecules can be stripped out of the OM by the divalent cation chelator EDTA52, and are bound stoichiometrically by the cationic cyclic lipopeptide polymyxin B55,60. By examining the ability of uncharged, hydrophilic molecules to penetrate the OM16, it was found that disruption of the negative charge of LPS substantially increased permeability, suggesting a central role for LPS in OM barrier function55,61.
OMPs are transmembrane β-barrels that also have important structural and functional implications for the OM. AFM revealed that OMPs cover ~70% of the cell surface62. Many studies have revealed diverse and critical roles played by OMPs, including signaling, nutrient import, virulence, and OM biogenesis34,37,63,64. In addition to these biological functions, the high abundance of OMPs, at roughly 500,000 per E. coli cell51, suggests a role in maintaining the physical properties of the OM. Indeed, AFM measurements revealed OMPs packed into islands rather than existing as single proteins that diffuse freely in the membrane, a sign of strong interactions that restrict mobility62,65,66 (Figure 2A,B). Coarse-grained molecular dynamics simulations suggested that specific interaction surfaces between OMPs are key to island formation67.
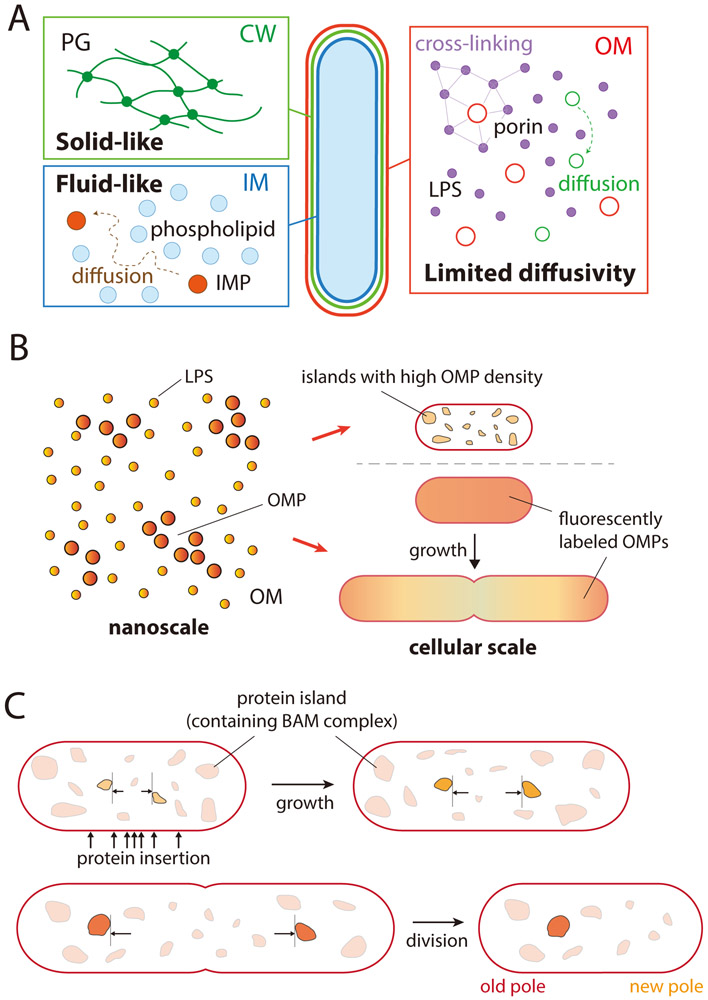
Figure 2: Outer membrane rheology and spatial organization are highly distinct from that of the PG or IM.
A) While the PG behaves as a relatively immobile solid due to its highly crosslinked nature, and the IM behaves as a liquid with rapid diffusion of lipids, the OM exhibits limited diffusivity. LPS molecules (purple) and OMPs (red) appear largely immobile, although there is some evidence of diffusive motion in the OM79. Green circles represent proteins with relatively high diffusivity.
B) At the nanoscale, LPS molecules and OMPs exist in small clusters. At the cellular scale, the density of fluorescently labeled OMPs along the cylindrical portion of the cell is diluted by growth, while the density at the poles remains high.
C) At the mesoscale, islands (represented by irregular shapes) including proteins such as the BAM complex expand and spread apart due to insertion of new materials along the cylindrical portion of the cell. After division, clusters at the old pole remain in one daughter cell, while clusters at the new pole have been trapped there by cytokinesis.
Lipoproteins, which are anchored in the membrane with a lipid moiety, are another important class of protein in the OM with over 1 million per E. coli cell51. As noted above, in certain enteric bacteria such as E. coli, the most abundant is Braun’s lipoprotein (Lpp), whose C-terminus is covalently bound to the PG layer, providing coupling between the OM and the cell wall38 (Figure 1A). Lipoproteins are involved in various pathways including capsular synthesis regulation7, colonization, immune system evasion, peptidoglycan synthesis36,37, OMP biogenesis, and LPS transport68,69. The extent to which lipoproteins affect OM organization is largely unknown, although evidence of their transfer between Myxococcus xanthus cells suggests that they are highly mobile14. Taken together, every component of the OM has the potential to impact its physical properties.
Molecular interactions between LPS and OM proteins
In the OM, proteins and LPS interact in multiple ways. Insertion of LPS into the outer leaflet requires the Lpt transport proteins63,69. Recent evidence increasingly supports a physical picture of the outer leaflet made up of LPS and porins as a partially ordered structure with strong lateral interactions at the molecular scale (Figure 2A). In X-ray reflectivity and grazing incidence X-ray diffraction measurements, LPS monolayers at water-air interfaces showed lateral ordering highly distinct from phospholipid membranes, and this organization could be perturbed by the ionic strength of the aqueous phase; Ca2+ ions increased the rigidity of the monolayer by cross-linking LPS molecules43. Conversely, treatment with EDTA, which chelates divalent cations, induces release of LPS from the OM52. Atomic force microscopy (AFM) measurements on living cells showed that OM porins distributed among LPS molecules formed a densely packed, net-like structure that diffused slowly62,66. Mutagenesis and structural studies (X-ray, neutron scattering) together revealed specific interactions between LPS and porins that stabilize the ordered network of LPS molecules and maintain permeability64. Molecular dynamics simulations have provided further support for strong interactions among LPS molecules and between LPS and proteins70-73, and have suggested that the LPS environment can affect the accessibility of certain small molecules to the passive transport porin OmpF74, as well as access to surface epitopes by antibodies28 and phage26,75,76.
Physical structure and properties of the OM
The dynamics of and interactions between molecules in the OM lead to a variety of structural characteristics and physical properties, including limiting the diffusion of molecules within the OM and producing spatial heterogeneity. Thus, molecular composition, structure, and dynamics can generate physical behaviors at the cellular scale. Recent studies integrating experimental and computational approaches are beginning to reveal the mechanisms relating these properties to molecular-scale organization and envelope mechanics, and to connect them to cellular fitness and survival.
The lipids and proteins within a bilayer such as the bacterial inner membrane typically diffuse quickly, with fluorescence recovery after photobleaching (FRAP) occurring within seconds77. However, the unusual asymmetric bilayer structure of the OM can give rise to distinct diffusive properties, altering the motion of constituent molecules and hence their spatial distribution. FRAP experiments and pulse-chase labeling with a general OM label, fluorescent succinimidyl ester (flSE), suggested that LPS and some proteins in the OM are largely immobile while other molecules can diffuse relatively quickly78,79, suggesting that the OM behaves more like a gel (a semi-solid with dilute crosslinks) than a fluid (Figure 2A). Proteins with limited mobility that are inserted locally must rely on the heterogeneity of OM growth and on cell division to cover the entire OM (Figure 2B). In addition to proteins, the limited motility of LPS molecules may impact the diffusivity of the OM as a whole given their high abundance and lateral interactions79.
By contrast to the homogenizing tendency of rapid diffusion, low diffusivity has the potential to produce spatial heterogeneity at the cellular scale by limiting exploration of individual molecules. Indeed, pulse-chase labeling of the abundant OM protein LamB in E. coli cells revealed the insertion of large clusters along the cylindrical region, not at the poles, that were essentially immobile80. Local interactions of LPS and proteins, together with expansion of the OM during cell growth are predicted to result in a spatial pattern wherein newly inserted clusters near mid-cell push older clusters toward the poles80 (Figure 2C). Pulse-chase flSE labeling showed that localization of OM components at the poles is more stable than along the cylindrical region78. A separate study demonstrated that synthesis of clusters of OMPs that include the β-barrel assembly machine (BAM) is biased away from the poles, with clusters migrating to the poles due to growth81 (Figure 2C). Heterogeneity of the OM is accompanied by heterogeneous growth of the PG82,83, with bursts of similar sizes84. While E. coli has been the predominant model for studying OM synthesis and properties, evidence from many other species such as Agrobacterium tumefaciens85, Shigella flexneri86,87, and Salmonella Typhimurium17 indicates that the OM may be spatially heterogeneous across Gram-negative bacteria.
Given the asymmetric structure of the OM, its molecular organization may play a critical role in its biochemical functions. Interestingly, Lpp was found to occupy distinct subcellular compartments in which molecules were either surface-exposed or periplasmic and PG-bound88. This transition between bound and unbound states is reversible, and detachment from the PG has been proposed to be beneficial under certain stress conditions89,90. It remains unclear why free-form Lpp is spatially distinct from its PG-bound counterparts, and whether alteration of the balance between the two states alters OM organization and/or mechanics.
Despite continuing knowledge gaps, recent studies have highlighted the impact of OM physical organization and properties on cellular physiology and interactions with the environment. For instance, binding of phages to the OM and subsequent motion before internalization is impacted by the heterogeneous distribution of its receptors91. Moreover, the physical organization of molecules within the OM can contribute to its structural integrity and mechanical strength. Characterization of the rheological properties of an in vitro monolayer showed that LPS undergoes a viscous-to-elastic transition upon compression44. This finding suggests that the molecular interaction between LPS molecules provides an important basis for the viscosity, stiffness, and/or strength of the OM. Similarly, it has been speculated that protein interactions can contribute to OM mechanical integrity92 and biosynthesis93. In addition, Lpp defines the periplasmic width94, and AFM measurements showed that mutants with disrupted Lpp crosslinking to the PG or increased Lpp length have weaker cell envelopes95. In some species lacking Lpp, β-barrel proteins were found to take on the role of covalently linking the OM to the cell wall, potentially also providing mechanical coupling96,97. Taken together, OM integrity and mechanics are likely intrinsically tied to the assembly and movement of its molecular components.
Evidence for the OM as a mechanical structure
Historically, the cell wall has been assumed to be the sole agent responsible for the task of maintaining mechanical integrity. However, over the past few decades, a growing appreciation for the mechanical role of the OM has emerged, driven in large part by advances in experimental methodologies. Studies of perturbations that lead to cell lysis revealed that Gram-negative bacterial cells can survive temporarily when the cell wall is disrupted or degraded, suggesting a role of the OM in maintaining cell integrity. For example, when phages induce breakdown of the wall, cells round up rapidly but persist in a viable state for up to an hour before lysis98. Upon vancomycin treatment of E. coli mutants with a permeable OM, the cytoplasm bulges through holes in the cell wall, but nevertheless lysis does not occur for tens of minutes99. In a similar study of bulging, cells treated with both β-lactam antibiotics and EDTA to disrupt OM integrity lysed more rapidly than during β-lactam treatment alone, implicating the OM in mechanical integrity41. Under certain conditions, spheroplasts (cell wall-deficient cells) can even survive indefinitely without a wall45,47,100-102. Importantly, Mg2+ is often required for generation of spheroplasts, suggesting that in the absence of the cell wall the OM must be stabilized by LPS crosslinking in order for cells to survive100.
However, direct evidence for a mechanical role of the OM had been lacking until recently. Spurred by the advent of microfluidics, which allows for the tracking of changes to single-cell morphology during perturbations, a recent study subjected E. coli cells to hyperosmotic shock (Figure 3A). The shock caused cells to decrease in length, consistent with the removal of turgor-mediated stress on the cell envelope. Under the hypothesis that the cell wall bears most (if not all) stress, in the plasmolyzed state after hyperosmotic shock the cell wall would be stress-free. However, after cells were subsequently treated with detergent to remove the IM and OM, a further reduction in length of up to 40% occurred40. This second contraction indicated that the cell wall was still under stretching forces after hyperosmotic shock, and that removal of the membranes allowed the cell wall to relax to its true rest length. To test whether the OM was the element contributing to cell-wall stretching, cells were treated before and during hyperosmotic shock with EDTA to cause rapid removal of LPS from the OM52. Now, there was significantly larger contraction after hyperosmotic shock, while the rest length of the cell wall after detergent treatment remained the same as without EDTA treatment. These two contractions (post-shock and post-detergent) provided key input to a biophysical model predicting that the relative stiffness of the OM is comparable to that of the PG cell wall40. This conclusion is supported by molecular dynamics simulations of a model of the OM as an asymmetric bilayer with a mixture of E. coli PLs in the inner leaflet and LPS in the outer leaflet103.
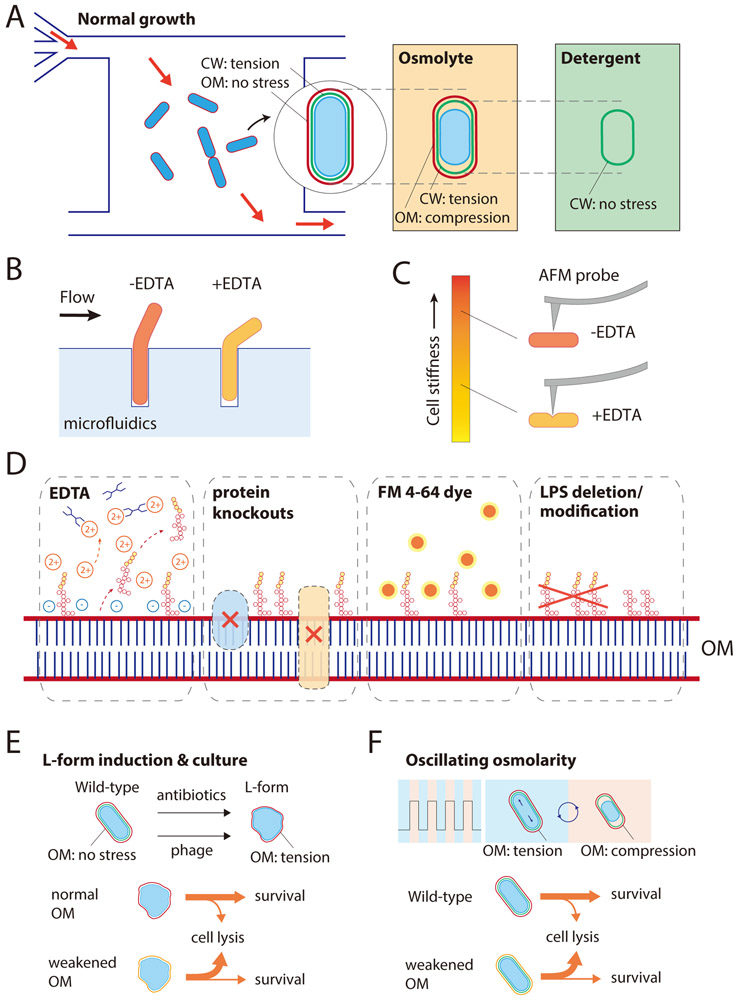
Figure 3: Probing the magnitude and impact of OM stiffness.
A) In a microfluidic flow cell, exponentially growing E. coli cells are under mechanical stress due to turgor pressure, which is born predominantly by the PG rather than the OM. Cells shrink suddenly when exposed to a hyperosmotic shock due to water efflux, decreasing the overall stress on the cell envelope. In this shrunken state, the cell wall remains partially extended as the OM experiences compression. Removal of the OM by detergent or EDTA treatment allows the cell wall to fully relax to its rest length.
B) Growth of E. coli cells initially embedded within a narrow channel leads to exposure of part of the cell body to fluid flow, whose hydrodynamic force can be tuned. EDTA-treated cells deflect more than untreated cells for a given flow strength40, highlighting the strength of the OM.
C) AFM measurements directly confirm the loss of cell stiffness by EDTA treatment. EDTA-treated cells indented more than untreated cells for the same amount of applied force; several outer membrane mutants also exhibited more indentation40.
D) In addition to EDTA treatment, OM stiffness can be compromised by deletion of various OM proteins, intercalation of the OM-specific dye FM4-64, or LPS modification such as deletion of the O-antigen.
E) E. coli cells adopt a wall-less L-form or wall-deficient spheroplast state when exposed to phage or cell wall-targeting antibiotics, in which the OM bears stress to avoid envelope rupture.
F) Cells with compromised OM stiffness are more susceptible to death during osmotic-shock oscillations.
Several other complementary experimental approaches support the prediction of high stiffness of the OM. In a microfluidic device in which cells are placed in narrow channels to grow filamentously into a region subjected to tunable flow pressure, cells deflect under fixed hydrodynamic force104, and deflection was increased by treatment with EDTA (Figure 3B); fitting the data to a biophysical model of thin-shell bending enabled an estimate that Young’s modulus (Box 1) decreased ~3-fold40. AFM measurements of EDTA-treated cells, in which cells are subjected to local indentation in contrast to the longitudinal stretching induced by turgor pressure (Figure 3C), further supported the mechanical contribution of the OM40.
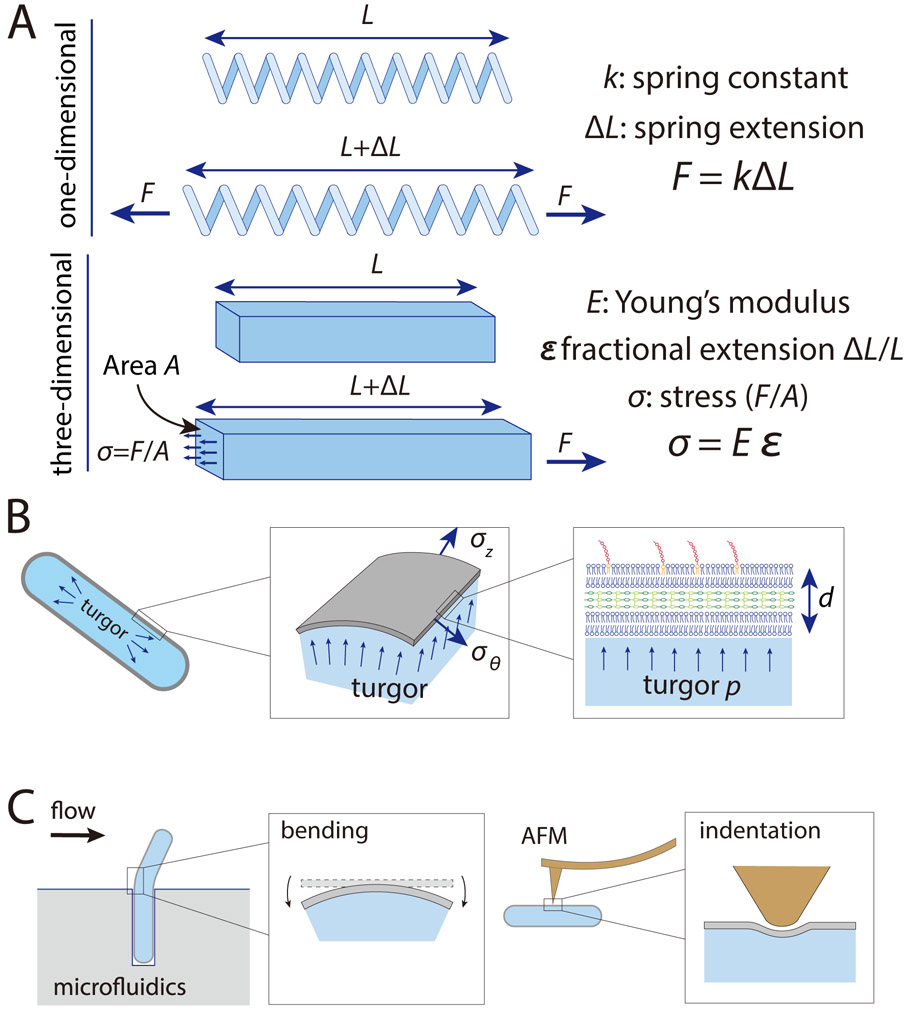
Box 1: Key concepts relevant to the mechanics of a bacterial cell.
The envelope of a bacterium is constantly subjected to mechanical perturbations such as turgor pressure. Understanding the magnitudes of these forces and the deformations that they cause provides insights into physical factors that dictate cellular function and survival. For solid objects, the simplest relationship between forces and deformation is linear elasticity, wherein the ratio between stress (force per unit area, σ) and strain (fractional change in size, ε) is a constant. This ratio characterizes the material’s stiffness and is known as Young’s modulus (E), such that σ=Eε. A one-dimensional analog of Young’s modulus is the spring constant k in the well-known Hooke’s law F=kx (Box Figure A).
To account for the three-dimensional nature of the stress-bearing cell envelope and the enclosed cytoplasm, a bacterial cell can be modeled as a thin shell under internal fluid pressure. For E. coli, the shell is rod-shaped, with length L several times larger than the cross-sectional radius r, and both are at least an order of magnitude larger than the thickness of the envelope d. In this case, along the cylindrical portion the stresses in the axial and circumferential directions are approximately and , respectively, where p is turgor pressure (Box Figure B). If we assume linear elasticity, the axial strain, which is the length extension ΔL relative to the rest length L0 when no forces are acting on the shell, is .
In addition to axial stress, rod-shaped objects can be subjected to other modes of deformation such as bending and indentation (Box Figure C). Bending of E. coli cells has been studied in microfluidic devices to reveal the coupling between the envelope strain and cell growth104. Indentation is typically studied via AFM using a nanoscale tip, through which Young’s modulus and other mechanical parameters of the cell surface can be inferred in combination with mechanical modeling39.
Although linear elasticity serves as a reasonable approximation for static loads and small strains, the true mechanical nature of cellular components can be more complicated and largely remains an open question. Generally, materials deviate from linear elasticity as the size of the deformation increases. Molecular dynamics simulations103 and AFM measurements39 have suggested that the cell wall stiffens under high tension, whereas the OM becomes softer103. Moreover, due to the fluid contents of the cytoplasm and specific aspects of membrane rheology, cells can exhibit viscoelasticity in which the strain is time-dependent148. As a result, much work remains to fully elucidate the mechanical properties of the bacterial cell envelope.
Box Figure: The mechanics of cylindrical thin shells.
A) For a one-dimensional spring with spring constant k, the force required to extend the spring by an amount ΔL is F = kΔL. For a three-dimensional material, Young’s modulus E is the analog of the spring constant, and is the ratio of the stress σ to the fractional extension ε.
B) The cell envelope of a rod-shaped bacterial cell with cross-sectional radius r can be modeled as a thin shell of thickness d (which is much smaller than r) under turgor pressure p. Along the cylindrical portion the stresses in the axial and circumferential directions are approximately and .
C) Bending in a microfluidic chamber (Figure 3B) and AFM indentation (Figure 3C) explore other modes of deformation such as bending and indentation, respectively.
In addition to EDTA treatment, a number of factors can tune OM stiffness (Figure 3D). Underscoring the importance of LPS properties, deleting the LPS O-antigen from an E. coli gut isolate dramatically reduced OM stiffness, and the OM of V. cholerae O139 was comparatively less stiff, consistent with a lack of LPS multivalency in this species40. Deletion of several abundant OM proteins, including OmpA, Pal, and Lpp, also reduced stiffness40. Interestingly, as mentioned above, all three of these proteins have roles in linking the PG and OM32-35,38,88. In the case of Lpp, which is the only protein covalently linked to the PG, its deletion, disruption of its crosslinking to PG, or modification of its length reduced cell stiffness as measured by AFM95. It remains to be seen whether the role of these proteins in OM mechanics is direct, or a consequence of modified OM composition, particularly PL and LPS levels, which can be highly responsive to the deletion of OMPs54. Consistent with this idea, the mutant allele lptD4213, which suppresses transport of LPS to the OM, increases PL content at the expense of LPS105 and decreases OM stiffness40. However, the inner membrane protein YhdP, which modulates anterograde PL flow and affects biosynthesis of the enterobacterial common antigen106, decreases OM stiffness without affecting LPS levels107.
The physiological importance of OM stiffness
The structural importance of the cell wall is made clear by the deleterious effects of chemical inhibition of its biogenesis. Treatment with β-lactam antibiotics, which inhibit penicillin binding proteins involved in PG synthesis, leads to swelling (increase in cell width)108 or bulging99,109, and eventually cell lysis at high concentrations. Nonetheless, cells can be propagated in a wall-less state, as so-called “L-forms”45, which survive via mechanical stabilization by the OM41 (Figure 3E). As with spheroplast generation, successful L-form propagation relies on the addition of Mg2+ to the medium100, likely due to the importance of LPS cross-linking in the absence of the wall, and proliferation has been postulated to rely on membrane blebbing and tubulation47, implicating membrane composition as an important factor in mechanical stability. Mutants with a mechanically weakened OM have dramatically lower spheroplast yields40,107. In fact, during the process of spheroplast generation, lysis of lptD4213 cells occurred as the cytoplasm started to escape from the cell wall40, the time at which the OM would be required to take up the stress. Similarly, after degradation of the cell wall during phage infection, disruption of the OM was necessary to enable virus release110,111. Thus, it is likely that the burden of mechanical integrity falls on the OM when the cell wall is compromised or removed.
Nonetheless, during steady-state growth, the E. coli OM is synthesized in a state such that the area of the OM does not change after plasmolysis and cell wall digestion40. Thus, OM area in a turgid state is the same as in the absence of turgor, indicating that the OM does not bear stress in this state. However, in environments such as the gastrointestinal tract and soil, cells are constantly subject to fluctuating osmolality108,112,113, in which the OM is likely to be relied upon for stress-bearing upon perturbation to avoid excessive compression or extension of the cell wall and IM (Figure 3F).
Modification of LPS, for example through addition of amino-arabinose or phosphoethanolamine114, can provide protection against AMPs and antibiotics115,116; cells likely have to balance the cost of these modifications on mechanical stability with the need to protect from molecules that can kill them. Osmotic oscillations led to increased lysis and greater amplitude of length change in situations with a compromised OM40, indicating that OM stiffness may be critical for survival in fluctuating environments.
In addition, bacterial membrane composition can be modulated by environmental cues such as culture conditions and temperature117-120, which will likely induce changes in physical properties121. For instance, the relative abundance of unsaturated or branched lipids in the IM can cause drastic changes in membrane viscosity122, and similar effects may occur in the OM123, given that the lipid A component of LPS can contain unsaturated and branched acyl chains under certain conditions124-127. Thus, it is likely that cells can tune the physical properties of the OM by adjusting its molecular composition, and thereby adapt to different growth stages, conditions, and environmental assaults.
Addressing knowledge gaps
From its structural and mechanical properties, it is clear that the OM can act as a versatile layer that provides both protection against chemicals and also resistance to turgor pressure and other mechanical loads. How general is a stiff OM across Gram-negative species, and across growth conditions? OM mechanical properties were similar across E. coli strains and in Pseudomonas aeruginosa as well as in minimal medium40, hinting at conservation across species and environments. Nonetheless, the decreased potential for crosslinking in V. cholerae was predictive of the comparatively low stiffness of its OM, suggesting that LPS may be the most informative signature of OM mechanics. Regardless, it will be important to understand the feedback mechanisms that regulate the fractions of LPS and OMPs in the OM, which may impact whether the OM behaves as a linear elastic material or one that shows strain stiffening like peptidoglycan39,103 (Box 1). It may be the case that cells can more easily tune the mechanical stiffness of the OM compared with that of the cell wall, since the OM harbors more components and is partially fluid. Certain environmental conditions, such as growth in a biofilm, may emphasize the importance of stiffness tunability. To pin down the structural role of the OM in other (particularly non-enteric) Gram-negative bacteria, key experiments such as determining the level of asymmetry, degree of PLs in the outer leaflet, and the chemical nature of the LPS will be highly informative.
How are OM mechanical properties related to other physical parameters, such as growth rate, cell shape, OM fluidity, and OM transport? For instance, in stationary phase, when cells are depleted of nutrients and as a result activate stress responses, the OM is more resistant to SDS than in log phase128 and it might be advantageous to construct a stiffer OM. Another possibility is a trade-off between the fluidity of the OM and its stiffness, based on the assumption that solid-like structures will be more rigid. One intriguing possibility is that perturbations known to disrupt physical properties such as fluidity also change OM mechanics; exploring such a potential coupling will be critical for determining the extent to which each property can be independently tuned. While cells with a less stiff OM are more sensitive to osmotic fluctuations, it remains to be determined what environmental mechanical forces have shaped the selective environment for OM properties. It is also possible that tradeoffs exists between the parallel functions of the OM as a load-bearing structure and a chemical barrier, in which modifications that enhance protection against AMPs or antibiotics114 may compromise OM stiffness.
Many aspects of OM physical properties remain unknown. From the compositional perspective, a large mystery is how PLs are transported from the IM to the OM. A recent study showed that the inner membrane protein YhdP modulates the rate of a high-flux diffusive anterograde PL flow pathway in E. coli107, but YhdP is non-essential so other mechanisms must also exist for PL transport. Although potentially an artifact of sample fixation for electron microscopy129, Bayer’s junctions are hypothesized connections between the IM and OM130 that, if they do exist, would allow for lipid transport via hemifusions. There may also be protein bridges for PLs like there are for LPS69,131-133. How are PLs linked to the physical properties of the OM? Although unproven, the natural hypothesis is that increasing PL concentration would make the OM less stiff based on the flexibility of PL vesicles, consistent with the finding that the FM4-64 OM-specific lipophilic stain disrupts OM stiffness40 (Figure 3D) as does lptD4213, which increases PL concentration by decreasing LPS concentration105. PLs may also be connected to the rheology/fluidity of the membrane, thereby affecting the localization of proteins within the OM. The cytoplasmic factors that dictate the pattern of OM protein insertion134 and localization have yet to be fully uncovered, but it is possible that the elongation machinery responsible for cell wall insertion also dictates OM insertion. Regardless of insertion, the relative lack of fluidity in the OM means that memory of the insertion pattern is maintained for long intervals.
From a mechanical perspective, a major knowledge gap is the magnitude of turgor pressure across organisms. There have not been any direct measurements of turgor in bacteria; in E. coli, the magnitude of turgor pressure was estimated indirectly from blebbing cells39. In the green algae Chara corallina, which are extremely large compared to a bacterium, turgor has been measured and controlled by piercing the cytoplasm with a needle135; there is some hope of a similar strategy being successful with bacteria using open AFM tips136. Without information about turgor, particularly regulation of turgor under conditions in which the cell wall is compromised, the distribution of stresses across the cell envelope is hard to discern. Moreover, it remains unclear whether turgor pressure is predominantly acting on the IM or the OM. One study provided evidence for the latter137, consistent with the idea that the IM would rupture at high turgor, although Gram-positive bacteria can maintain high turgor in the absence of an OM. Curiously, the cell wall can itself bear turgor after detergent treatment removes the membranes40; this ability may be due to the wall being clogged with cytoplasmic components, and hence behaving as a semipermeable membrane. The molecular mechanisms of OM construction itself (i.e., OMP and LPS insertion) are still not completely resolved, but will have to account for building this structure under mechanical pressure.
While the OM is specific to Gram-negative species, it remains possible (perhaps even likely) that elements beyond the PG provide mechanical/structural support in Gram-positive bacteria. One possibility is the teichoic acids, which are essential for shape determination in B. subtilis138 and may serve in a crosslinking capacity by binding divalent cations139. Consistent with this notion, sudden removal of Mg2+ from the extracellular medium caused B. subtilis cells to suddenly expand, suggesting that the cell envelope rapidly weakened140. Moreover, there are many other candidate structures that may contribute to cell mechanics. In response to cell-wall damage, E. coli cells produce a capsule that could be structurally important when the cell is mechanically compromised6,7. Similarly, gut commensals such as Bacteroides species produce capsular polysaccharide layers in response to different environmental cues. These layers can be extremely thick (>1 μm)141, and hence have potential for altering the physical properties of cells. Other Gram-negative bacteria, such as Caulobacter crescentus, have highly ordered S-layers surrounding the OM142-144. In fact, it seems likely that many structural layers contribute mechanically (it is even possible that the IM also plays a mechanical role). If so, then it is likely that they all have similar stiffnesses as the OM and cell wall because otherwise the stiffest layer would dominate mechanical responses.
It remains unresolved whether the OM is truly essential. No mutants of Gram-negative species have been identified that completely lack the OM, although the composition can be changed quite dramatically (e.g., removal of LPS) and still support growth. Removing the OM would likely also remove many periplasmic proteins, which could have serious consequences. Does perturbing the OM impact survival in different environments, for instance the transition between rich and poor nutrients? Ultimately, it may be that the role of the OM becomes most clear during survival of cell wall-deficient states.
Future prospects
Future studies will benefit from the recent expansion in experimental methods to probe cell mechanics, including direct methods such as AFM and indirect methods such as the degree of growth in a gel of known stiffness145,146, neutron reflectometry147, and microfluidic perturbations104. In addition, manipulation methods such as optical traps that can be used to pull tethers from the OM could provide information on OM viscoelasticity, particularly whether such tethers retract. Helping to interpret such experiments will be an enhanced understanding of cellular and single-molecule properties, such as the mechanics and folding of β-barrels.
Recent discoveries that highlight the physical properties of the OM and its mechanical importance complement the rich history of genetic and biochemical investigation into this versatile and complex structure, stimulating reflection on the evolution of cellular membranes in general. Physical stress clearly shapes bacterial physiology in profound ways, and mechanical factors were likely strong selective forces early on in the evolution of single-cell organisms. Thus, identifying the key determinants of OM physical properties and their association with other aspects of cellular physiology will provide a deeper understanding of microbial growth and fitness.
Acknowledgements
The authors thank the Huang, Rutherford, and Silhavy labs for helpful discussions. The authors acknowledge support from the Allen Discovery Center at Stanford on Systems Modeling of Infection (to K.C.H.), and National Institute of General Medical Sciences grants R35 GM118024 (to T.J.S.) and RM1 GM135102 (to K.C.H.). K.C.H. is a Chan Zuckerberg Biohub Investigator.
References
- 1.Silhavy TJ, Kahne D & Walker S The bacterial cell envelope. Cold Spring Harb Perspect Biol 2, a000414, doi: 10.1101/cshperspect.a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtje JV Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62, 181–203 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funahara Y & Nikaido H Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. Journal of bacteriology 141, 1463–1465 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konovalova A & Silhavy TJ Outer membrane lipoprotein biogenesis: Lol is not the end. Philos Trans R Soc Lond B Biol Sci 370, doi: 10.1098/rstb.2015.0030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SH et al. Detecting envelope stress by monitoring beta-barrel assembly. Cell 159, 1652–1664, doi: 10.1016/j.cell.2014.11.045 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Konovalova A, Perlman DH, Cowles CE & Silhavy TJ Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proceedings of the National Academy of Sciences of the United States of America 111, E4350–4358, doi: 10.1073/pnas.1417138111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belas R Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends in microbiology 22, 517–527 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Konovalova A, Mitchell AM & Silhavy TJ A lipoprotein/beta-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife 5, doi: 10.7554/eLife.15276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forero M, Yakovenko O, Sokurenko EV, Thomas WE & Vogel V Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS biology 4, e298, doi: 10.1371/journal.pbio.0040298 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaparakis-Liaskos M & Ferrero RL Immune modulation by bacterial outer membrane vesicles. Nature Reviews Immunology 15, 375–387 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kuehn MJ & Kesty NC Bacterial outer membrane vesicles and the host–pathogen interaction. Genes & development 19, 2645–2655 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Giordano NP, Cian MB & Dalebroux ZD Outermembrane lipid secretion and the innate immune response to Gram-negative bacteria. Infection and Immunity (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nudleman E, Wall D & Kaiser D Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309, 125–127, doi: 10.1126/science.1112440 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Kulp A & Kuehn MJ Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual review of microbiology 64, 163–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakae T & Nikaido H Outer membrane as a diffusion barrier in Salmonella typhimurium. Penetration of oligo- and polysaccharides into isolated outer membrane vesicles and cells with degraded peptidoglycan layer. The Journal of biological chemistry 250, 7359–7365 (1975). [PubMed] [Google Scholar]
- 17.Nikaido H Outer membrane of Salmonella typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochimica et biophysica acta 433, 118–132 (1976). [DOI] [PubMed] [Google Scholar]
- 18.Nikaido H & Vaara M Molecular basis of bacterial outer membrane permeability. Microbiological reviews 49, 1 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventola CL The antibiotic resistance crisis: part 1: causes and threats. Pharmacy and therapeutics 40, 277 (2015). [PMC free article] [PubMed] [Google Scholar]
- 20.Hofer U The cost of antimicrobial resistance. Nature Reviews Microbiology 17, 3–3 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Lewis K Platforms for antibiotic discovery. Nat Rev Drug Discov 12, 371–387, doi: 10.1038/nrd3975 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Payne DJ, Gwynn MN, Holmes DJ & Pompliano DL Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6, 29–40, doi: 10.1038/nrd2201 (2007). [DOI] [PubMed] [Google Scholar]
- 23.O'neill J Antimicrobial resistance. Tackling a Crisis for the Health and Wealth of Nations (2014). [Google Scholar]
- 24.Zgurskaya HI, Lopez CA & Gnanakaran S Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS infectious diseases 1, 512–522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delcour AH Outer membrane permeability and antibiotic resistance. Biochimica et biophysica acta 1794, 808–816, doi: 10.1016/j.bbapap.2008.11.005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bentley AT & Klebba PE Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. Journal of bacteriology 170, 1063–1068 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storek KM et al. Massive antibody discovery used to probe structure-function relationships of the essential outer membrane protein LptD. Elife 8, doi: 10.7554/eLife.46258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storek KM et al. Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal. Proceedings of the National Academy of Sciences of the United States of America 115, 3692–3697, doi: 10.1073/pnas.1800043115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan L, Chen S & Jensen GJ Molecular organization of Gram-negative peptidoglycan. Proceedings of the National Academy of Sciences 105, 18953–18957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matias VR & Beveridge TJ Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol 56, 240–251, doi: 10.1111/j.1365-2958.2005.04535.x (2005). [DOI] [PubMed] [Google Scholar]
- 31.Matias VR & Beveridge TJ Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. Journal of bacteriology 188, 1011–1021, doi: 10.1128/JB.188.3.1011-1021.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JS et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J 26, 219–228, doi: 10.1096/fj.11-188425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boags AT, Samsudin F & Khalid S Binding from Both Sides: TolR and Full-Length OmpA Bind and Maintain the Local Structure of the E. coli Cell Wall. Structure 27, 713–724 e712, doi: 10.1016/j.str.2019.01.001 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Gerding MA, Ogata Y, Pecora ND, Niki H & de Boer PA The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol 63, 1008–1025, doi: 10.1111/j.1365-2958.2006.05571.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray AN et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife 4, doi: 10.7554/eLife.07118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paradis-Bleau C et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143, 1110–1120, doi: 10.1016/j.cell.2010.11.037 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Typas A et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143, 1097–1109, doi: 10.1016/j.cell.2010.11.038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun V Covalent lipoprotein from the outer membrane of Escherichia coli. Biochimica et biophysica acta 415, 335–377, doi: 10.1016/0304-4157(75)90013-1 (1975). [DOI] [PubMed] [Google Scholar]
- 39.Deng Y, Sun M & Shaevitz JW Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Physical review letters 107, 158101 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Rojas ER et al. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621, doi: 10.1038/s41586-018-0344-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Z, Kahne D & Kishony R Distinct single-cell morphological dynamics under beta-lactam antibiotics. Molecular cell 48, 705–712, doi: 10.1016/j.molcel.2012.09.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young R Phage lysis: three steps, three choices, one outcome. J Microbiol 52, 243–258, doi: 10.1007/s12275-014-4087-z (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeworrek C et al. Effects of specific versus nonspecific ionic interactions on the structure and lateral organization of lipopolysaccharides. Biophysical journal 100, 2169–2177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrmann M, Schneck E, Gutsmann T, Brandenburg K & Tanaka M Bacterial lipopolysaccharides form physically cross-linked, two-dimensional gels in the presence of divalent cations. Soft matter 11, 6037–6044 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Klieneberger E The Natural Occurrence of Pleuro-Pneumonia-like Organisms in Apparent Symbiosis with Streptobacillus moniliformis and Other Bacteria. Journal of Pathology and Bacteriology 40, 93–105 (1935). [Google Scholar]
- 46.Leaver M, Dominguez-Cuevas P, Coxhead J, Daniel R & Errington J Life without a wall or division machine in Bacillus subtilis. Nature 457, 849–853 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Mercier R, Kawai Y & Errington J General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. Elife 3, doi: 10.7554/eLife.04629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ames GF, Spudich EN & Nikaido H Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. Journal of bacteriology 117, 406–416 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamio Y & Nikaido H Outer membrane of Salmonella typhimurium. Identification of proteins exposed on cell surface. Biochimica et biophysica acta 464, 589–601 (1977). [DOI] [PubMed] [Google Scholar]
- 50.Narita S.-i. & Tokuda H Bacterial lipoproteins; biogenesis, sorting and quality control. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 1862, 1414–1423 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Li GW, Burkhardt D, Gross C & Weissman JS Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635, doi: 10.1016/j.cell.2014.02.033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leive L, Shovlin VK & Mergenhagen SE Physical, chemical, and immunological properties of lipopolysaccharide released from Escherichia coli by ethylenediaminetetraacetate. The Journal of biological chemistry 243, 6384–6391 (1968). [PubMed] [Google Scholar]
- 53.Smit J, Kamio Y & Nikaido H Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. Journal of bacteriology 124, 942–958 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamio Y & Nikaido H Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 (1976). [DOI] [PubMed] [Google Scholar]
- 55.Morrison DC & Jacobs DM Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13, 813–818 (1976). [DOI] [PubMed] [Google Scholar]
- 56.Nikaido H, Takeuchi Y, Ohnishi SI & Nakae T Outer membrane of Salmonella typhimurium. Electron spin resonance studies. Biochimica et biophysica acta 465, 152–164 (1977). [DOI] [PubMed] [Google Scholar]
- 57.Smit J & Nikaido H Outer membrane of gram-negative bacteria. XVIII. Electron microscopic studies on porin insertion sites and growth of cell surface of Salmonella typhimurium. Journal of bacteriology 135, 687–702 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyd A & Holland IB Regulation of the synthesis of surface protein in the cell cycle of E. coli B/r. Cell 18, 287–296 (1979). [DOI] [PubMed] [Google Scholar]
- 59.Raetz CR & Whitfield C Lipopolysaccharide endotoxins. Annu Rev Biochem 71, 635–700, doi: 10.1146/annurev.biochem.71.110601.135414 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hancock RE & Bell A in Perspectives in Antiinfective Therapy 42–53 (Springer, 1989). [Google Scholar]
- 61.Srimal S, Surolia N, Balasubramanian S & Surolia A Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharides and lipid A. Biochem J 315 ( Pt 2), 679–686 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jarosławski S, Duquesne K, Sturgis JN & Scheuring S High-resolution architecture of the outer membrane of the Gram-negative bacteria Roseobacter denitrificans. Molecular microbiology 74, 1211–1222 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Qiao S, Luo Q, Zhao Y, Zhang XC & Huang Y Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Arunmanee W et al. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proceedings of the National Academy of Sciences 113, E5034–E5043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casuso I et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nature nanotechnology 7, 525–529 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Yamashita H et al. Single-molecule imaging on living bacterial cell surface by high-speed AFM. Journal of molecular biology 422, 300–309 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Chavent M et al. How nanoscale protein interactions determine the mesoscale dynamic organisation of bacterial outer membrane proteins. Nat Commun 9, 2846, doi: 10.1038/s41467-018-05255-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konovalova A, Kahne DE & Silhavy TJ Outer Membrane Biogenesis. Annu Rev Microbiol 71, 539–556, doi: 10.1146/annurev-micro-090816-093754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N & Kahne D Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14, 337–345, doi: 10.1038/nrmicro.2016.25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel DS, Qi Y & Im W Modeling and simulation of bacterial outer membranes and interactions with membrane proteins. Curr Opin Struct Biol 43, 131–140, doi: 10.1016/j.sbi.2017.01.003 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Im W & Khalid S Molecular Simulations of Gram-Negative Bacterial Membranes Come of Age. Annual Review of Physical Chemistry 71, 171–188 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Gao Y, Lee J, Widmalm G & Im W Modeling and Simulation of Bacterial Outer Membranes with Lipopolysaccharides and Enterobacterial Common Antigens. The Journal of Physical Chemistry B (2020). [DOI] [PubMed] [Google Scholar]
- 73.Nascimento A Jr., Pontes FJ, Lins RD & Soares TA Hydration, ionic valence and cross-linking propensities of cations determine the stability of lipopolysaccharide (LPS) membranes. Chem Commun (Camb) 50, 231–233, doi: 10.1039/c3cc46918b (2014). [DOI] [PubMed] [Google Scholar]
- 74.Patel DS et al. Dynamics and Interactions of OmpF and LPS: Influence on Pore Accessibility and Ion Permeability. Biophys J 110, 930–938, doi: 10.1016/j.bpj.2016.01.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Domínguez-Medina CC et al. Outer membrane protein size and LPS O-antigen define protective antibody targeting to the Salmonella surface. Nature communications 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kortright KE, Chan BK & Turner PE High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proceedings of the National Academy of Sciences 117, 18670–18679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fleming TC et al. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes & development 24, 1160–1172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Pedro MA, Grunfelder CG & Schwarz H Restricted Mobility of Cell Surface Proteins in the Polar Regions of Escherichia coli. Journal of bacteriology 186, 2594–2602 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghosh AS & Young KD Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. Journal of bacteriology 187, 1913–1922, doi: 10.1128/JB.187.6.1913-1922.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ursell TS, Trepagnier EH, Huang KC & Theriot JA Analysis of surface protein expression reveals the growth pattern of the gram-negative outer membrane. PLoS computational biology 8, e1002680, doi: 10.1371/journal.pcbi.1002680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rassam P et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 523, 333–336, doi: 10.1038/nature14461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koch AL & Woldringh CL The metabolic inertness of the pole wall of a gram-negative rod. Journal of theoretical biology 171, 415–425 (1994). [Google Scholar]
- 83.De Pedro M, Quintela JC, Höltje J & Schwarz H Murein segregation in Escherichia coli. Journal of bacteriology 179, 2823–2834 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ursell TS et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proceedings of the National Academy of Sciences of the United States of America 111, E1025–1034, doi: 10.1073/pnas.1317174111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown PJ et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proceedings of the National Academy of Sciences of the United States of America 109, 1697–1701, doi: 10.1073/pnas.1114476109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Steinhauer J, Agha R, Pham T, Varga AW & Goldberg MB The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol Microbiol 32, 367–377 (1999). [DOI] [PubMed] [Google Scholar]
- 87.Charles M, Perez M, Kobil JH & Goldberg MB Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proceedings of the National Academy of Sciences of the United States of America 98, 9871–9876, doi: 10.1073/pnas.171310498 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cowles CE, Li Y, Semmelhack MF, Cristea IM & Silhavy TJ The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol 79, 1168–1181, doi: 10.1111/j.1365-2958.2011.07539.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winkle M et al. DpaA Detaches Braun's Lipoprotein from Peptidoglycan. mBio 12, doi: 10.1128/mBio.00836-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bahadur R, Chodisetti PK & Reddy M Cleavage of Braun's lipoprotein Lpp from the bacterial peptidoglycan by a paralog of l,d-transpeptidases, LdtF. Proceedings of the National Academy of Sciences of the United States of America 118, doi: 10.1073/pnas.2101989118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rothenberg E et al. Single-virus tracking reveals a spatial receptor-dependent search mechanism. Biophys J 100, 2875–2882, doi: 10.1016/j.bpj.2011.05.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lessen HJ, Fleming PJ, Fleming KG & Sodt AJ Building Blocks of the Outer Membrane: Calculating a General Elastic Energy Model for beta-Barrel Membrane Proteins. J Chem Theory Comput 14, 4487–4497, doi: 10.1021/acs.jctc.8b00377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kleanthous C, Rassam P & Baumann CG Protein–protein interactions and the spatiotemporal dynamics of bacterial outer membrane proteins. Current opinion in structural biology 35, 109–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asmar AT et al. Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS biology 15, e2004303, doi: 10.1371/journal.pbio.2004303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mathelie-Guinlet M, Asmar AT, Collet JF & Dufrene YF Lipoprotein Lpp regulates the mechanical properties of the E. coli cell envelope. Nat Commun 11, 1789, doi: 10.1038/s41467-020-15489-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Godessart P et al. β-Barrels covalently link peptidoglycan and the outer membrane in the α-proteobacterium Brucella abortus. Nature Microbiology, 1–7 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Sandoz KM et al. β-Barrel proteins tether the outer membrane in many Gram-negative bacteria. Nature Microbiology, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berry J, Rajaure M, Pang T & Young R The spanin complex is essential for lambda lysis. Journal of bacteriology 194, 5667–5674, doi: 10.1128/JB.01245-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang KC, Mukhopadhyay R, Wen B, Gitai Z & Wingreen NS Cell shape and cell-wall organization in Gram-negative bacteria. Proceedings of the National Academy of Sciences of the United States of America 105, 19282–19287, doi: 10.1073/pnas.0805309105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Billings G et al. De novo morphogenesis in L-forms via geometric control of cell growth. Mol Microbiol 93, 883–896, doi: 10.1111/mmi.12703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawai Y et al. Cell growth of wall-free L-form bacteria is limited by oxidative damage. Curr Biol 25, 1613–1618, doi: 10.1016/j.cub.2015.04.031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ranjit DK & Young KD The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. Journal of bacteriology 195, 2452–2462, doi: 10.1128/JB.00160-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hwang H, Paracini N, Parks JM, Lakey JH & Gumbart JC Distribution of mechanical stress in the Escherichia coli cell envelope. Biochim Biophys Acta Biomembr 1860, 2566–2575, doi: 10.1016/j.bbamem.2018.09.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Amir A, Babaeipour F, McIntosh DB, Nelson DR & Jun S Bending forces plastically deform growing bacterial cell walls. Proceedings of the National Academy of Sciences of the United States of America 111, 5778–5783, doi: 10.1073/pnas.1317497111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruiz N, Wu T, Kahne D & Silhavy TJ Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol 1, 385–395, doi: 10.1021/cb600128v (2006). [DOI] [PubMed] [Google Scholar]
- 106.Mitchell AM, Srikumar T & Silhavy TJ Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. MBio 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grimm J et al. The inner membrane protein YhdP modulates the rate of anterograde phospholipid flow in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 117, 26907–26914, doi: 10.1073/pnas.2015556117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tropini C et al. Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations. Cell Rep 9, 1520–1527, doi: 10.1016/j.celrep.2014.10.027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goodell EW, Lopez R & Tomasz A Suppression of lytic effect of beta lactams on Escherichia coli and other bacteria. Proceedings of the National Academy of Sciences of the United States of America 73, 3293–3297, doi: 10.1073/pnas.73.9.3293 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajaure M, Berry J, Kongari R, Cahill J & Young R Membrane fusion during phage lysis. Proceedings of the National Academy of Sciences of the United States of America 112, 5497–5502, doi: 10.1073/pnas.1420588112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berry J, Summer EJ, Struck DK & Young R The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol Microbiol 70, 341–351, doi: 10.1111/j.1365-2958.2008.06408.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chowdhury N, Marschner P & Burns RG Soil microbial activity and community composition: impact of changes in matric and osmotic potential. Soil Biology and Biochemistry 43, 1229–1236 (2011). [Google Scholar]
- 113.Shiau YF, Feldman GM, Resnick MA & Coff PM Stool electrolyte and osmolality measurements in the evaluation of diarrheal disorders. Ann Intern Med 102, 773–775, doi: 10.7326/0003-4819-102-6-773 (1985). [DOI] [PubMed] [Google Scholar]
- 114.Cox AD et al. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. Journal of bacteriology 185, 3270–3277, doi: 10.1128/jb.185.11.3270-3277.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Simpson BW & Trent MS Pushing the envelope: LPS modifications and their consequences. Nature Reviews Microbiology 17, 403–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martynowycz MW et al. Salmonella Membrane Structural Remodeling Increases Resistance to Antimicrobial Peptide LL-37. ACS Infect Dis 5, 1214–1222, doi: 10.1021/acsinfecdis.9b00066 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alphen WV & Lugtenberg B Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. Journal of bacteriology 131, 623–630, doi: 10.1128/JB.131.2.623-630.1977 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lugtenberg B, Peters R, Bernheimer H & Berendsen W Influence of cultural conditions and mutations on the composition of the outer membrane proteins of Escherichia coli. Mol Gen Genet 147, 251–262, doi: 10.1007/BF00582876 (1976). [DOI] [PubMed] [Google Scholar]
- 119.Morein S, Andersson A, Rilfors L & Lindblom G Wild-type Escherichia coli cells regulate the membrane lipid composition in a "window" between gel and non-lamellar structures. The Journal of biological chemistry 271, 6801–6809, doi: 10.1074/jbc.271.12.6801 (1996). [DOI] [PubMed] [Google Scholar]
- 120.Sanders CR & Mittendorf KF Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry 50, 7858–7867, doi: 10.1021/bi2011527 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stokes JM et al. Cold Stress Makes Escherichia coli Susceptible to Glycopeptide Antibiotics by Altering Outer Membrane Integrity. Cell Chem Biol 23, 267–277, doi: 10.1016/j.chembiol.2015.12.011 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Budin I et al. Viscous control of cellular respiration by membrane lipid composition. Science 362, 1186–1189, doi: 10.1126/science.aat7925 (2018). [DOI] [PubMed] [Google Scholar]
- 123.Nichol CP, Davis JH, Weeks G & Bloom M Quantitative study of the fluidity of Escherichia coli membranes using deuterium magnetic resonance. Biochemistry 19, 451–457 (1980). [DOI] [PubMed] [Google Scholar]
- 124.Carty SM, Sreekumar KR & Raetz CR Effect of Cold Shock on Lipid A Biosynthesis inEscherichia coli: INDUCTION AT 12° C OF AN ACYLTRANSFERASE SPECIFIC FOR PALMITOLEOYL-ACYL CARRIER PROTEIN. Journal of Biological Chemistry 274, 9677–9685 (1999). [DOI] [PubMed] [Google Scholar]
- 125.Vorachek-Warren MK, Carty SM, Lin S, Cotter RJ & Raetz CR An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: Absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 C. Journal of Biological Chemistry 277, 14186–14193 (2002). [DOI] [PubMed] [Google Scholar]
- 126.Kumada H, Haishima Y, Umemoto T & Tanamoto K Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. Journal of bacteriology 177, 2098–2106 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reife RA et al. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra - and penta - acylated lipid A structures on E - selectin expression and TLR4 recognition. Cellular microbiology 8, 857–868 (2006). [DOI] [PubMed] [Google Scholar]
- 128.Mitchell AM, Wang W & Silhavy TJ Novel RpoS-Dependent Mechanisms Strengthen the Envelope Permeability Barrier during Stationary Phase. Journal of bacteriology 199, doi: 10.1128/JB.00708-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kellenberger E The 'Bayer bridges' confronted with results from improved electron microscopy methods. Mol Microbiol 4, 697–705, doi: 10.1111/j.1365-2958.1990.tb00640.x (1990). [DOI] [PubMed] [Google Scholar]
- 130.Bayer M Areas of adhesion between wall and membrane of Escherichia coli. Microbiology 53, 395–404 (1968). [DOI] [PubMed] [Google Scholar]
- 131.Coudray N et al. Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. Elife 9, e62518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ekiert DC et al. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285. e217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Isom GL et al. LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope. Cell 181, 653–664 e619, doi: 10.1016/j.cell.2020.03.030 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alvira S et al. Inter-membrane association of the Sec and BAM translocons for bacterial outer-membrane biogenesis. Elife 9, doi: 10.7554/eLife.60669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Proseus TE & Boyer JS Calcium pectate chemistry causes growth to be stored in Chara corallina: a test of the pectate cycle. Plant Cell Environ 31, 1147–1155, doi: 10.1111/j.1365-3040.2008.01829.x (2008). [DOI] [PubMed] [Google Scholar]
- 136.Chen P et al. Nanoscale probing the kinetics of oriented bacterial cell growth using atomic force microscopy. Small 10, 3018–3025, doi: 10.1002/smll.201303724 (2014). [DOI] [PubMed] [Google Scholar]
- 137.Cayley DS, Guttman HJ & Record MT Jr Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophysical journal 78, 1748–1764 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schirner K, Marles-Wright J, Lewis RJ & Errington J Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J 28, 830–842, doi: 10.1038/emboj.2009.25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kern T et al. Dynamics characterization of fully hydrated bacterial cell walls by solid-state NMR: evidence for cooperative binding of metal ions. J Am Chem Soc 132, 10911–10919, doi: 10.1021/ja104533w (2010). [DOI] [PubMed] [Google Scholar]
- 140.Rojas ER, Huang KC & Theriot JA Homeostatic Cell Growth Is Accomplished Mechanically through Membrane Tension Inhibition of Cell-Wall Synthesis. Cell Syst 5, 578–590 e576, doi: 10.1016/j.cels.2017.11.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martens EC, Roth R, Heuser JE & Gordon JI Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. The Journal of biological chemistry 284, 18445–18457, doi: 10.1074/jbc.M109.008094 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bharat TA et al. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nature microbiology 2, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Comerci CJ et al. Topologically-guided continuous protein crystallization controls bacterial surface layer self-assembly. Nature communications 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Herrmann J et al. Environmental calcium controls alternate physical states of the Caulobacter surface layer. Biophysical journal 112, 1841–1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Auer GK et al. Mechanical Genomics Identifies Diverse Modulators of Bacterial Cell Stiffness. Cell Syst 2, 402–411, doi: 10.1016/j.cels.2016.05.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tuson HH et al. Measuring the stiffness of bacterial cells from growth rates in hydrogels of tunable elasticity. Mol Microbiol 84, 874–891, doi: 10.1111/j.1365-2958.2012.08063.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hughes AV et al. Physical properties of bacterial outer membrane models: neutron reflectometry & molecular simulation. Biophysical journal 116, 1095–1104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vadillo-Rodriguez V, Beveridge TJ & Dutcher JR Surface viscoelasticity of individual gram-negative bacterial cells measured using atomic force microscopy. Journal of bacteriology 190, 4225–4232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]