Abstract
Some proteins, like the lac repressor (LacI), mediate long-range loops that alter DNA topology and create torsional barriers. During transcription, RNA polymerase generates supercoiling that may facilitate passage through such barriers. We monitored E. coli RNA polymerase progress along templates in conditions that prevented, or favored, 400 bp LacI-mediated DNA looping. Tethered particle motion measurements revealed that RNA polymerase paused longer at unlooped LacI obstacles or those barring entry to a loop than those barring exit from the loop. Enhanced dissociation of a LacI roadblock by the positive supercoiling generated ahead of a transcribing RNA polymerase within a torsion-constrained DNA loop may be responsible for this reduction in pause time. In support of this idea, RNA polymerase transcribed 6-fold more slowly through looped DNA and paused at LacI obstacles for 66% less time on positively supercoiled compared to relaxed templates, especially under increased tension (torque). Positive supercoiling propagating ahead of polymerase facilitated elongation along topologically complex, protein-coated templates.
INTRODUCTION
DNA in the cell is complexed with proteins and adopts a highly compact structure including protein-mediated loops and supercoiling (1–11). Genome-bound proteins can be roadblocks that hinder elongation by RNA polymerase (RNAP) during transcription (12). The strength of such roadblocks may vary if the protein in question wraps DNA or secures a DNA loop. Several recent investigations have been conducted to understand how RNA polymerase navigates through nucleosomes (13–15). Nucleosomes contain histones which interact with DNA non-specifically and are substrates for combinatorial post-translational modifications that regulate chromatin remodelling and transcription of DNA. However, many transcription factors from organisms spanning all kingdoms shape genomes and influence transcription without such extensive chemical modification. Often, their activity is regulated by concentration, DNA supercoiling, and for the many which recognize specific sites on DNA, by the presence of multiple binding sites with different affinities. These transcription factors may bend, wrap, bridge, and loop DNA segments (5,16–24). The effects of these topologies have not been addressed in earlier studies on transcription roadblocks in vivo and are just beginning to be investigated in vitro (25).
In this study, the Escherichia coli lac repressor (LacI) was used as a transcriptional roadblock (26). The bivalent LacI tetramer binds specific sites (operators) with up to nanomolar affinity, depending on the sequence, and can mediate a DNA loop. The strength of the roadblock depends on the affinity for the binding site(s), on tethering, which increases the effective local concentration of the protein near a binding site(s) (27), and on the loop thermodynamic stability. Using a DNA template containing a weak binding site (O2) near a promoter and a high-affinity binding site (O1) further downstream, it was shown that LacI bound to O2 was a much stronger roadblock when securing a loop between the two operators (25). However, transcription of the loop segment by RNAPs that bypass the promoter-proximal LacI roadblock and the effect of the loop on the roadblocking capacity of promoter-distal roadblock have not been previously investigated. Thus, we used the tether particle motion (TPM) technique, to monitor the process of transcription through a LacI-mediated loop.
Surprisingly, we found that RNAP paused for long times within the loop region and that the loop weakened the promoter-distal roadblock. According to the twin-domain model (28), an elongating RNAP generates negative supercoiling behind and positive supercoiling ahead. In addition, a protein-mediated loop is a barrier against diffusion of supercoiling (29). Therefore, we hypothesized that while negative supercoiling behind RNAP inside the loop stabilizes the binding of LacI at the promoter-proximal operator (30,31), the positive supercoiling accumulated ahead of RNAP may destabilize LacI at the promoter-distal binding site, reducing its strength as a roadblock. Although it has been shown that transcription-generated positive supercoiling can destabilize nucleosomes (32), it is still not clear how supercoiling affects the binding affinity of other transcription factors or the topologies these factors induce. Thus, we used magnetic tweezers (MTs) to test the effect of supercoiling on LacI roadblocks. We found that RNAP paused more briefly in front of an O1-bound LacI when the DNA template was positively supercoiled than in the absence of supercoiling. This observation supports the idea that transcription-induced supercoiling within a LacI-mediated loop stabilizes LacI binding to the operator behind while destabilizing LacI binding to the operator ahead of the transcription complex.
In summary, our study reveals complex effects of protein-stabilized loops on the kinetics of RNA chain elongation. The LacI-mediated loops lengthen the pause time in front of the promoter-proximal binding site, shorten the pause time in front of the promoter-distal binding site, and increase the frequency and duration of pausing within the loop. We conclude that DNA loops can be potent transcription roadblocks that can temporarily sequester RNAP, until positive supercoiling build-up breaks the loop and enables RNAP escape. The significance of these findings is that in vivo, although RNAP elongation may be hindered by proteins, or protein-mediated long-range interactions, positive supercoiling generated ahead of the enzyme tends to clear the path ahead.
MATERIALS AND METHODS
Preparation of DNA constructs
DNA tethers for TPM experiments (Figure 1A) were amplified in PCR reactions with plasmid templates containing various combinations of LacI operators. Tethers with O1 and O2 in the proximal and distal position from the promoter, respectively, were amplified from template pWX_12_400 (33). A tether with O2 and O1 in the opposite order was amplified from pZV_21_400 (25). Constructs with a single O1 operator at the promoter-proximal, or distal, site were amplified from pRS_1N_400, or pDM_N1_400 (33). The reactions contained dNTPs (Fermentas-Thermo Fisher Scientific Inc., Pittsburgh, PA, USA), biotin-labeled, or unlabeled anti-sense primers (Eurofins Genomics, Louisville, KY or Integrated DNA Technologies, Coralville, IA, USA) and Taq DNA Polymerase (New England BioLabs, Ipswich, MA, USA).
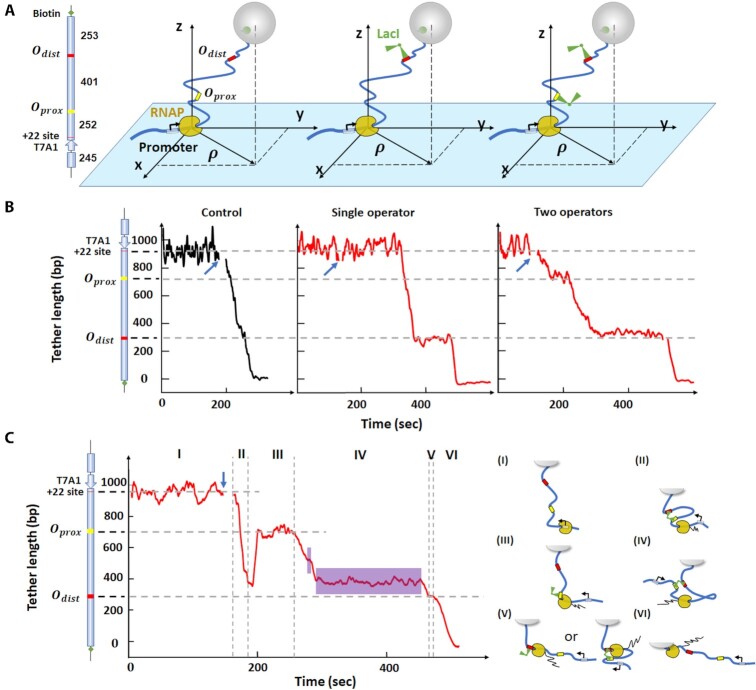
Figure 1.
RNAP can transcribe through a LacI-mediated loop after a pause. (A) The DNA tethers for TPM include DNA (thick, blue line), LacI binding sites (yellow and red) and LacI (green, V-shape). The black arrow at the promoter indicates the direction of transcription. RNAP is stalled at the + 22 site before resuming RNA synthesis in the presence of all NTPs. The cartoons on the right illustrate three scenarios: RNAP elongation without LacI (left), with LacI bound to single operator (middle), or with two LacI bound to two operators separately (right). Each cartoon corresponds to the trace shown below it in Panel B. (B) Representative transcription records. Left: control record without LacI. Center: transcription in the presence of 10 nM LacI on a template with the distal operator only. Right: transcription in the presence of 10 nM LacI and two LacI binding sites. At this concentration, each binding site is likely to be occupied by a different LacI tetramer, so no looping is likely. The blue arrows indicate the time at which NTPs were introduced. (C) Transcription in the presence of 0.2 nM LacI to promote looping. The vertical dash lines identify six intervals (I - VI) in the progress of RNAP along the DNA template and the cartoons on the right depict the likely conformation of the transcription elongation complex in each interval. The purple areas in region IV indicate random pauses between operators. The cartoon on the left of the y-axis in panels B and C shows the features of the DNA template used in TPM measurements. From top to bottom: a T7A1 promoter, a stall site at + 22, a promoter-proximal binding site (Oprox) and promoter-distal binding site (Odist). The horizontal dashed lines indicate the position of the LacI binding site(s) in the construct, and the expected location of pauses in the record.
DNA tethers used in the MTs measurements were built by ligating a 4098 bp-long main fragment containing the promoter-proximal O1 operator to a ∼150 bp-long multiple biotin-labeled DNA fragment at the promoter-distal end with T7 DNA ligase (New England Bio- Labs, Ipswich, MA, USA). The main fragment was amplified from pRS_1N_400 using an equimolar dNTP mix and a primer containing an ApaI restriction site, digested with ApaI, and purified on a PCR clean up column. A 302 bp-long biotin-labeled amplicon was also produced from pBluKSP and using dATP, dCTP, dGTP, dTTP (Fermentas-Thermo Fisher Scientific Inc., Pittsburgh, PA, USA) and biotin-11-dUTP (Invitrogen, Life Technologies, Grand Island, NY, USA) in a molar ratio of 1:1:1:0.7:0.3. Digestion of this fragment with ApaI generated ∼150 bp biotin-labeled ‘tail’ fragments for ligation. The biotinylated ‘tail’ fragment anchored the tether, through multiple attachment sites, to a streptavidin-coated bead, allowing torque to be applied to the DNA with the magnetic tweezer (Supplementary Figure S1). On all constructs, RNAP could be stalled 22 bp from the transcription start site (TSS) by withholding CTP. Supplementary Table S1 summarizes the plasmids, primers, and restriction enzymes used.
Preparation of RNAP
Doubly-HA tagged E. coli RNAP was used in both TPM and MTs experiments. This enzyme was purified as described previously (34).
Microchamber preparation
The bottom coverslip of the microchamber (Fisherbrand, Thermo Fisher Scientific, Waltham, MA, USA) supported a parafilm gasket produced with a laser cutter (Universal Laser Systems, VLS 860, Middletown, CT) with a central observation area connected through narrow inlet and outlet channels to inlet and outlet reservoirs, extending beyond the edges of the top coverslip, (Supplementary Figure S2). The coverslips with parafilm assembly were heated to seal its components together and form the microchamber. The narrow inlet and outlet reduced evaporation of buffer, while the triangular shape of the chamber, confined the reaction in a relatively small volume and provided a gradient of tether densities to optimize throughput.
The entire sample preparation was performed at room temperature and materials were kept on ice. Beads were first washed with phosphate buffered saline (PBS), and then washed and resuspended in either transcription buffer (TXB: 20 mM Tris-glutamate (pH 8), 10 mM magnesium glutamate, 50 mM potassium-glutamate, 0.2 mg/mL α-casein (Sigma-Aldrich, St. Louis, MO), 1 mM DTT), or incubation buffer (IB) (20 mM Tris-Cl (pH 8), 50 mM KCl, 1 mM DTT). The chambers were first incubated with reference beads resuspended in IB for 5 minutes to let some beads adhere to the surface and serve as references during data acquisition and analysis. The chambers were then incubated with 10 μg/ml purified Anti-HA 11 Epitope tag antibody (BioLegend, San Diego, CA, USA) in IB at room temperature for 1 hour. They were then passivated with IB supplemented with 6 mg/ml α-casein at 4°C overnight. 60 nM doubly-HA tagged E. coli RNA polymerase was then drawn into the chamber and incubated 30 min at room temperature to allow it bind to the anti-HA coated surface. RNAP immobilization is commonly used in single molecule measurements of transcription and does not affect RNA chain elongation. 10 nM DNA template, 50 μM GpA (initiating dinucleotide, TriLink Bio Technologies, San Diego, CA, USA), and 10 μM ATP/UTP/GTP in TXB at 37 °C were introduced into the chamber for 30 min to produce the stalled elongation complexes (SECs) which were reactivated upon addition of all 4 nucleotides. For TPM experiments, the end of the DNA far from the promoter was labeled with beads (0.32 μm diameter, streptavidin-coated polystyrene beads (Roche Life Science, Indianapolis, IN, USA)) by flowing in the microchamber 0.03 mg/mL beads resuspended in TXB and letting them incubate for 10 min. For MTs experiments, instead, a 0.02 mg/ml solution of 1.0 μm diameter, streptavidin-coated superparamagnetic beads (Dynabead MyOne Streptavidin T1, Invitrogen, Grand Island, NY)) in TXB was incubated for 5 min. The experimental construct for TPM/MTs is schematically illustrated in Figure 1A and Supplementary Figure S1, respectively. The extension of the DNA tether was monitored before, to control the original tether length, and after introducing 1 mM NTPs with/without LacI in TXB.
The tethered particle motion technique and data analysis
The tethered particle motion (TPM) technique has been described in detail elsewhere (35). All TPM experiments were conducted at room temperature. The lamp of the microscope was turned on approx. 1–2 hours before recording the motion of the beads to thermally equilibrate the microscope. Five immobilized beads were recorded and at least one of them was used to calculate the drift, which was then subtracted from the tethered particle data (35,36). The absolute x, y positions of each bead were recorded at 50 Hz with a custom LabVIEW virtual instrument (National Instruments, Austin, TX, USA). The variance of the excursion of a tether was then calculated as 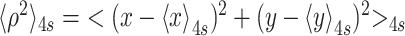 , in which
, in which 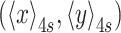 represents a four-second moving average of the coordinates that reveals the anchor point of the bead. Changes in the bead excursion reflected changes in the DNA tether length (37–39). The recorded x, y data were then analyzed for symmetry and initial tether length prior to the start of transcription. Beads that exhibited distributions of (x, y) positions with a ratio of the major to minor axes greater than 1.07, were discarded (35). The tether length was determined using a calibration curve relating tether length to <ρ2> values (Supplementary Figure S3). Tethers with anomalous length were excluded. Pauses in both TPM and MT traces were identified as previously described (25). Transcription rates were calculated as the slope between two points in smoothly processive regions. These measured rates are consistent with other single molecule and bulk measurements by Schafer et al. (40), as well as others (41,42) reported in the literature. They are 2-fold lower than those routinely measured in bulk at 1 mM NTPs 37 °C on a template devoid of strong pauses (40,43). This conforms to the general rule that the rates of chemical reactions decrease as the temperature is lowered.
represents a four-second moving average of the coordinates that reveals the anchor point of the bead. Changes in the bead excursion reflected changes in the DNA tether length (37–39). The recorded x, y data were then analyzed for symmetry and initial tether length prior to the start of transcription. Beads that exhibited distributions of (x, y) positions with a ratio of the major to minor axes greater than 1.07, were discarded (35). The tether length was determined using a calibration curve relating tether length to <ρ2> values (Supplementary Figure S3). Tethers with anomalous length were excluded. Pauses in both TPM and MT traces were identified as previously described (25). Transcription rates were calculated as the slope between two points in smoothly processive regions. These measured rates are consistent with other single molecule and bulk measurements by Schafer et al. (40), as well as others (41,42) reported in the literature. They are 2-fold lower than those routinely measured in bulk at 1 mM NTPs 37 °C on a template devoid of strong pauses (40,43). This conforms to the general rule that the rates of chemical reactions decrease as the temperature is lowered.
Magnetic Tweezers
Our magnetic tweezers parameters have been previously described (44,45). All MTs experiments were conducted at room temperature using a protocol similar to what already described (25). Twisting the DNA tether, anchored through RNAP (Supplementary Figure S1), produced plectonemes under low tension (<1 pN) after reaching the critical torque value. An extension-versus-twist curve displayed the extension (ΔZ) of DNA tether versus the number of magnet turns (Supplementary Figure S4, left) and was used to distinguish beads tethered by intact, single dsDNA tethers from those linked to nicked or multiple DNA tethers. Prior to adding NTPs to initiate transcription, tethers were twisted by −19 turns and the extension was recorded for about 1 minute. Then 1 mM NTPs with or without 10 nM LacI in TXB were introduced to resume transcription.
Estimation of RNAP progress within the loop
When a transcription elongation complex (TEC) is trapped inside a loop, torque will accumulate quickly, and a torque of +11 pN⋅nm ahead, or −11 pN⋅nm behind RNAP has been shown to stall its progress (46). Torque will quickly accumulate and stall TECs close to either of the LacI binding sites sealing the loop but will build up more slowly when the enzyme is in the middle of the loop. At this location, the torque in the flanking DNA can be expressed as:
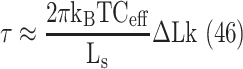 |
(1) |
in which Ls is the contour length of the flanking DNA. Ceff is the effective twist persistence length (twist modulus/kBT) determined as 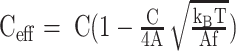 (47), where C = 100nm, A = 50nm, and the tension, f
(47), where C = 100nm, A = 50nm, and the tension, f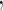 in this case is 0.45 pN. ΔLk is the linking number change in the DNA. In this study,
in this case is 0.45 pN. ΔLk is the linking number change in the DNA. In this study, 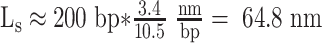 , Ceff ≈ 63 nm, kBT ≈ 4.1 pN⋅nm. Therefore, the maximum change in linking number that RNAP can induce before stalling (|τ| ≈ 11 pN⋅nm), is
, Ceff ≈ 63 nm, kBT ≈ 4.1 pN⋅nm. Therefore, the maximum change in linking number that RNAP can induce before stalling (|τ| ≈ 11 pN⋅nm), is 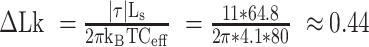 turns, which corresponds to
turns, which corresponds to 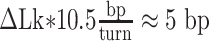 . Thus, 5 bp is the furthest that RNAP should be expected to transcribe before stalling when it operates inside a ∼400 bp loop. Such processivity should most likely occur when RNAP is halfway between the two operators.
. Thus, 5 bp is the furthest that RNAP should be expected to transcribe before stalling when it operates inside a ∼400 bp loop. Such processivity should most likely occur when RNAP is halfway between the two operators.
Estimation of torque at the O1 pause site
Mosconi et al. (48) have shown that under 0.45 pN or 0.8 pN of tension Ceff equals 63 nm or 72 nm, respectively. Since the initial twist of the DNA tether may be arbitrarily set and RNAP positively supercoils the downstream DNA, a DNA tether can be manipulated to have ΔLk = 5 just as a TEC reaches O1. The length of the DNA tether at that point is 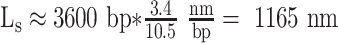 , and Equation (1) yields a torque of 7.0 or 8.0 pN⋅nm at tensions of 0.45 or 8 pN, respectively. Instead, under 0.25 pN tension, ΔLk = 5 occurs beyond the buckling transition, a phase in which the torque in the plectonemic DNA remains constant at approximately 5 pN⋅nm (48).
, and Equation (1) yields a torque of 7.0 or 8.0 pN⋅nm at tensions of 0.45 or 8 pN, respectively. Instead, under 0.25 pN tension, ΔLk = 5 occurs beyond the buckling transition, a phase in which the torque in the plectonemic DNA remains constant at approximately 5 pN⋅nm (48).
RESULTS
Monitoring elongation through LacI-mediated loops with tethered particle motion
First, TPM was used to study elongation through LacI-mediated loops. As RNAP transcribed the DNA template containing two binding sites for the LacI protein, three encounters were possible: RNAP might encounter an unencumbered binding site, a binding site bound by LacI in unlooped DNA (Figure 1A, B), or a LacI bridging two operators to secure a DNA loop (Figure 1C). In addition, during the progress of RNAP, LacI might randomly bind to/dissociate from either the promoter-proximal or -distal binding site, Oprox and Odist, respectively, producing/breaking intermittent loops. In data that satisfied TPM screening criteria (see material and methods), prior to addition of all four nucleotides the average excursion of the bead remained constant at a value consistent with the DNA tether length (Figure 1B). Addition of all NTPs without and with LacI, indicated by the blue arrow in Figures 1B and C, caused a short-lived disturbance, which was deleted from the record. Then, RNAP resumed elongation producing a progressive decrease in tether length that continued uninterrupted to the end of the template, unless LacI was present. Control experiments with 1 mM NTPs revealed no pausing by RNAP at either of the two LacI binding sites in the absence of LacI (Figure 1B, left). In contrast, in the presence of 10 nM LacI, RNAP clearly paused in front of Odist on a DNA template containing only this LacI binding site (Figure 1B, middle) or paused in front of both Oprox and Odist on a DNA template containing two LacI binding sites (Figure 1B, right).
The probability of looping in a DNA template containing two binding sites can be adjusted by varying the LacI concentration (27). For a 400-bp loop between the O2 and O1 binding sites, a maximum looping probability of ∼44% could be achieved with 0.2 nM LacI, while 10 nM LacI decreased the probability to ∼6% as the two binding sites became occupied by different LacI tetramers (Supplementary Figure S5). Indeed, with 10 nM LacI, RNAP paused at positions corresponding to the promoter-proximal and -distal LacI binding sites and no loops were observed (Figure 1B, right).
However, with 0.2 nM LacI, loops occurred during transcription (Figure 1C) and the pattern was more complex. The record can be divided into six intervals (I-IV). In interval (I) the DNA tether length is constant before introducing 1 mM NTPs and 0.2 nM LacI. Transcription begins shortly after introducing NTPs and in interval II, a loop forms, but it ruptures as RNAP approaches Oprox; since RNAP paused at this location, LacI likely was bound to Oprox. In interval (III), RNAP paused at Oprox for approximately 50 s. This particular pause was shorter than the average lifetime (derived from dissociation constants measured in vivo) of the LacI-O1 complex which is approximately 318 s (49), or 434 s (50), and 102 s (50) for O2. In interval (IV), RNAP proceeded to transcribe the segment between the LacI binding sites and paused at random locations (purple areas) before reaching the distal operator. Traces without loop formation (Figure 1B) did not reveal any random pauses. Instead, about half of pauses associated with transcription through looped segments occurred within the loop before reaching the distal operator, 53/(53 + 43) = 0.55 for O1proxO2dist and 46)/(46 + 41) = 0.53 for O2proxO1dist. Thus, pauses within the loop occur frequently and may be induced by the accumulation of supercoiling that takes place inside a loop, as depicted in Figure 1C, cartoon IV (see also ‘Materials and Methods/Estimation of RNAP pauses within the loop’). Elongation resumed after loop breakdown and RNAP transcribed to Odist. In interval (V), RNAP paused at Odist indicating that LacI was still associated with this operator. Then RNAP surpassed this obstacle and continued transcribing in interval (VI), finally reaching the end of the template. This record illustrates how transcription through a loop can be monitored using TPM.
RNAP pauses longer at entry-to than exit-from LacI-loops
Previous work showed that LacI bound to a promoter-proximal O2 operator blocked transcription more effectively when securing a loop (25), a conclusion drawn from an analysis of the distance traveled by RNAP before immobilization for static AFM imaging. In those experiments, stalled transcription elongation complexes, reactivated upon the addition of the missing ribonucleotide triphosphate, transcribed segments as long as the entire 906 bp template in 60 seconds, and were even observed inside the Lac-mediated loop. To measure the time required for transcription of the loop and determine whether looping affected the passage of RNAP through bound LacI, the pause times of RNAP at O1 or O2 binding sites in the Oprox position on unlooped templates (Figure 1B, right) were compared with those in looped templates (Supplementary Figure S6, interval III). On unlooped templates, pauses at proximal and distal O1(O2) operators occupied by LacI at 0.2 or 10 nM differed insignificantly (Supplementary Table S2). Their pause times were aggregated in Figure 2. By contrast, formation of a loop had profound effects on RNAP elongation. RNAP paused longer at promoter-proximal LacI-O1 or LacI-O2 obstacles that were part of a loop (Figure 2A, left). Pauses at LacI-O1prox obstacles were 77 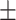 7 s (N = 215) without a loop, but 199
7 s (N = 215) without a loop, but 199 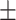 25 s (N = 92) with a loop. Pauses at LacI-O2prox obstacles were 39
25 s (N = 92) with a loop. Pauses at LacI-O2prox obstacles were 39 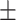 4 s (N = 133) without a loop, but 79
4 s (N = 133) without a loop, but 79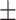 14 s (N = 58) with a loop. A likely reason for the increases is that the secondary, distal binding site increases the local concentration of LacI and the effective affinity for the proximal binding site (27). The steric hindrance caused by the loop itself may also contribute to the lengthening of the pause at the proximal binding site, although this effect is likely to be small (25). It is also informative that with respect to unlooped templates, looped LacI-O1prox obstacles obstructed transcription for a considerably longer time (199 versus 77 sec) than looped LacI-O2prox obstacles (79 versus 39 sec). Since LacI has higher affinity for the O1 than the O2 binding site, LacI is more likely to remain at O1 than at O2 after the loop break down, increasing the probability of blocking elongating RNAP at O1.
14 s (N = 58) with a loop. A likely reason for the increases is that the secondary, distal binding site increases the local concentration of LacI and the effective affinity for the proximal binding site (27). The steric hindrance caused by the loop itself may also contribute to the lengthening of the pause at the proximal binding site, although this effect is likely to be small (25). It is also informative that with respect to unlooped templates, looped LacI-O1prox obstacles obstructed transcription for a considerably longer time (199 versus 77 sec) than looped LacI-O2prox obstacles (79 versus 39 sec). Since LacI has higher affinity for the O1 than the O2 binding site, LacI is more likely to remain at O1 than at O2 after the loop break down, increasing the probability of blocking elongating RNAP at O1.
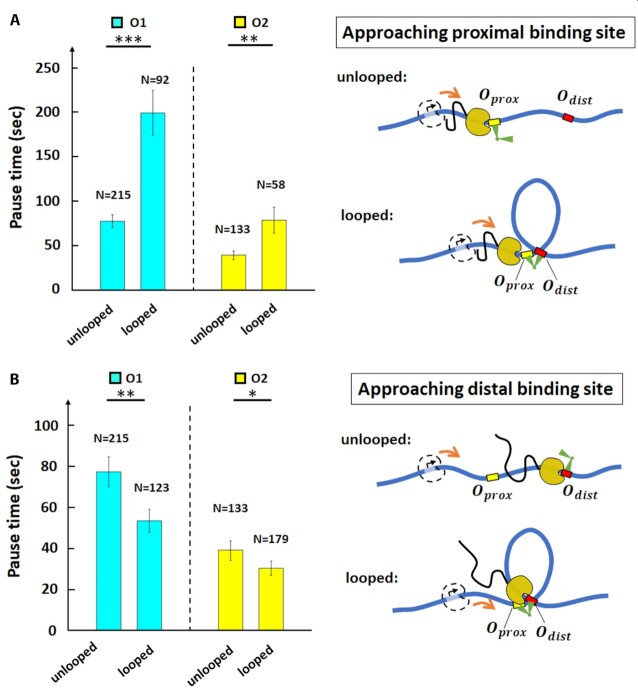
Figure 2.
The LacI-mediated loop enhances and attenuates RNAP pausing at the proximal and distal binding sites respectively. (A) Average pause durations were longer at the proximal operator consisting of either O1 (cyan) or O2 (yellow), in the looped with respect to the unlooped conformations depicted at right (***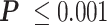 , **
, **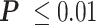 for two-sample t-tests). (B) Average pause durations were shorter at the distal operator consisting of either O1 (cyan) or O2 (yellow), in the looped with respect to the unlooped conformations depicted at right (**
for two-sample t-tests). (B) Average pause durations were shorter at the distal operator consisting of either O1 (cyan) or O2 (yellow), in the looped with respect to the unlooped conformations depicted at right (**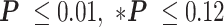 for two-sample t-tests). Standard errors and numbers of samples, N, are indicated. Data for pauses at proximal and distal operators in unlooped tethers were insignificantly different (see Supplementary Table S2) and aggregate values are shown.
for two-sample t-tests). Standard errors and numbers of samples, N, are indicated. Data for pauses at proximal and distal operators in unlooped tethers were insignificantly different (see Supplementary Table S2) and aggregate values are shown.
We next investigated RNAP approaching the promoter-distal operator, Odist. When RNAP is at the distal operator, the tether length will not be changed significantly by loop formation, that only affects the segment behind RNAP (Figure 1A and cartoon (V) on the right of Figure 1C). We used traces such as the ones in the rightmost panel of Figure 1B and in interval (V) of Figure 1C to compare pause durations at Odist (Figure 1C, interval V; Supplementary Figure S6, interval V), with or without looping. Under the 0.2 nM LacI concentration used, 41% of the LacI obstacles at Odist can be assumed to secure loops (Supplementary Figure S5). Surprisingly, and in contrast to Oprox, we observed that under these conditions, the average pause time of RNAP at LacI-O1dist and LacI-O2dist was shortened with respect to obstacles on unlooped templates to 54 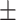 6 s (N = 123) and 30
6 s (N = 123) and 30 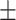 4 s (N = 179), respectively. Assuming 41% looped obstacles, RNAP inside the loop pauses at LacI for approximately
4 s (N = 179), respectively. Assuming 41% looped obstacles, RNAP inside the loop pauses at LacI for approximately 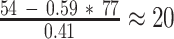 s at the distal O1 operator, and for
s at the distal O1 operator, and for 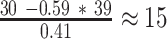 s at the distal O2 operator. These results suggest that transcription within the loop promotes the release of LacI from a distal operator site, despite the locally increased concentration of LacI and regardless of the operator affinity.
s at the distal O2 operator. These results suggest that transcription within the loop promotes the release of LacI from a distal operator site, despite the locally increased concentration of LacI and regardless of the operator affinity.
Transcription of looped segments is slower
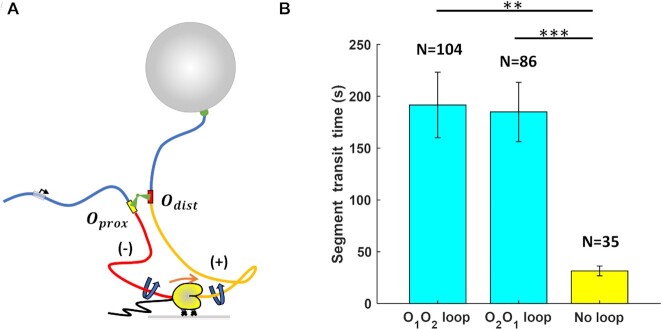
Inspection of interval IV in Figure 1C suggests that a protein-mediated loop can significantly delay transcription by RNAP. According to the twin-supercoiled-domain model (28), rotation of the DNA template unwinds DNA behind the transcribing RNAP, generating negative supercoiling (Figure 3A, red DNA), and winds DNA ahead, generating positive supercoiling (Figure 3A, yellow DNA). Furthermore, since a LacI-mediated loop constitutes a topological domain (51), transcription within the loop will generate torsional stress. Within a 400 bp-long loop, the torsional stress created by a transcribing RNAP can quickly accumulate to +11 pN⋅nm ahead or −11 pN⋅nm behind, stalling RNAP progress(46). We estimate that RNAP might translocate as few as five bp within the 400 bp loop before stalling (Materials and Methods). Stalled RNAP is prone to backtracking, and recovery from the backtracked state after a loop rupture further delays transcription. Thus, we measured the total time required to transcribe the loop segment in each trace (duration of interval IV in Figure 1C or intervals such as IV in Supplementary Figure S6), averaged it over all traces in the presence of LacI/looping and compared it with the average time required to transcribe between the two operators in the absence of LacI/looping (Figure 1B, left). The average transcription intervals for looped O1-O2 and O2-O1 segments were 192 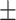 31 s (N = 104) and 185
31 s (N = 104) and 185 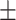 29 s (N = 86), respectively. Both these times were much longer than the average time without looping (32
29 s (N = 86), respectively. Both these times were much longer than the average time without looping (32  5 s, N = 35). We conclude that the LacI loop can significantly hinder RNAP progress by generating torsional stress.
5 s, N = 35). We conclude that the LacI loop can significantly hinder RNAP progress by generating torsional stress.
Figure 3.
RNAP transcribes a loop more slowly. (A) A cartoon depicting RNAP transcribing a loop. The right-angle black arrow indicates the promoter. The blue-colored DNA segments are torsionally relaxed. The red-colored DNA segment is unwound, while the yellow DNA segment is overwound by RNAP. Nascent RNA is the thin, black line emerging from RNAP. (B) RNAP requires almost tenfold more time to transcribe a looped segment (cyan) compared to the same unlooped segment (yellow). ***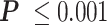 , **
, **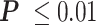 (two-sample t-tests). Standard errors and numbers of samples, N, are indicated.
(two-sample t-tests). Standard errors and numbers of samples, N, are indicated.
RNAP surpasses LacI obstacles faster on positively supercoiled templates
The strength of the LacI roadblock may be affected not only by loop formation, but also by torsional stress accumulating within the loop. In particular, the data in Figure 2 showed that the loop alleviates interference by the distal LacI obstacle. We hypothesized that the shorter pauses at the distal operator might result from destabilization of LacI-Odist complexes by transcription-generated positive supercoiling. In the TPM experiment, the DNA segments flanking the loop (blue in Figure 3A) quickly spin about the long DNA axis to release any torsional stress due transcription (Figure 1A). Within the loop, however, positive supercoiling ahead of RNAP builds torque and might cause LacI to dissociate from the DNA. To test this hypothesis, we used magnetic tweezers (MTs) to follow elongation of an RNAP transcribing toward a LacI-O1 obstacle on a construct where the DNA ahead could be positively supercoiled (Figure 4A, Supplementary Figure S1). In this experiment, the segment between RNAP and the tethered bead was rotationally immobilized by multiple biotin-streptavidin linkages to the bead. The promoter of this template was 252 base pairs (approximately 24 turns) upstream from the O1 binding site (Supplementary Figure S1). To create positive supercoiling just as RNAP arrived at the LacI obstacle, the DNA template was preloaded with negative plectonemes (Figure 4A) under forces ranging between approximately 0.25 and 0.8 pN. Transcription by RNAP is expected to change the DNA tether length as depicted in Figure 4A. We first verified that RNAP could transcribe a tether preloaded with −24 turns (gray and black trace in Figure 4B). After introducing NTPs (blue arrow in Figure 4B), the tether extension increased, elongating RNAP produced positive supercoiling that annihilated the pre-loaded negative supercoiling, until the DNA tether became torsionally relaxed. After that, RNAP continued introduce positive supercoiling until the bead was drawn to the surface of the flow-chamber, or RNAP stalled due to either steric hindrance by the plectonemes, or perhaps by large torsional stress. Once the ability of RNAP to transcribe a negatively supercoiled template by several turns was verified (Figure 4B, grey and black curves), transcription was recorded in the presence of LacI after pre-loading the template with −19 turns. RNAP was expected to reach the O1 binding site after having supercoiled the DNA template to ΔLk = +5, according to extension versus-twist-curves (Supplementary Figure S4) in which the tether length with + 5 turns can be clearly distinguished from that of the same torsionally-relaxed tether (ΔLk = 0). The blue and red traces in Figure 4B are the raw data and a 4 s moving average, respectively, of a measurement with 10 nM LacI. Although RNAP paused also at random positions along the trace, the expected pause at O1 was clearly distinguishable (Figure 4B, between vertical, black, dashed lines, see Supplementary Figure S4 for identification of the O1 position). For comparison, we acquired MT measurements of the RNAP pause in front of the LacI-O1 obstacle on torsionally relaxed (nicked) DNA under 0.45 pN tension (Figure 4C right, Supplementary Figure S1).
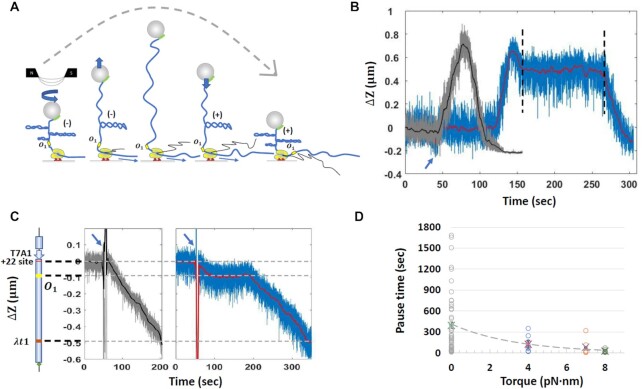
Figure 4.
Comparison of RNAP pause times at O1 with and without positive supercoiling. (A) A DNA tether was mechanically unwound forming plectonemes prior to the addition of NTPs. Subsequent transcription introduced positive supercoils that annihilated the mechanically induced, negative supercoils and lengthened the tether to a maximum. Further transcription and positive supercoiling eventually produced plectonemes that contracted the tether length. The dashed gray curve indicates the extension of the DNA tether during progressive conversion of negative to positive plectonemes due to transcription by RNAP. (B) Representative observations of extension versus time were recorded during transcription without (gray) and with (blue and red) LacI on a template pre-loaded with negative supercoiling. The blue arrow indicates the time at which all four NTPs or NTPs + LacI were introduced. The two vertical black dashed lines circumscribe a pause by RNAP at the LacI-O1 operator complex. (C) Representative observations of elongation were recorded in the absence of pre-loaded supercoiling without (left panel, gray and black) or with (right panel, blue and red) LacI. On the left is a schematic representation of the DNA template (D) Pause times by RNAP at the LacI obstacle varied as a function of torque on the DNA. Torque values were calculated using Equation (1), except that Ceff was estimated in 100 mM [Na+] instead of 50 mM [K+]. Circles represent measured pauses, while crosses represent averages.
Figure 4D shows how pause times changed with different torque on the DNA. The average RNAP pause at the LacI-O1 obstacle with no torque (0 pN⋅nm: gray circles, average: green cross) was 393 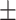 64 s (N = 49), much longer than the average pause time with positive torques ( ∼ 4 pN⋅nm: blue circles, average: red cross), ∼ 7 pN⋅nm (orange circles, average blue cross), and ∼ 8 pN⋅nm (green circles, average: purple cross), which pause RNAP for 125 ± 42 s (N = 8), 82 ± 50 s (N = 6) and 26 ± 8 s (N = 8), respectively. Thus, positive supercoiling significantly facilitated transcription through the LacI-O1 obstacle and likely disrupted the LacI-O2 obstacle as well. The decrease in pause duration as positive torque on the DNA increased suggests that positive supercoiling weakens LacI binding and transcriptional roadblocking. Positive supercoiling generated by RNAP translocation is also known to destabilize nucleosomes (32,52); it may, therefore, represent one means by which RNAP removes protein roadblocks along the DNA.
64 s (N = 49), much longer than the average pause time with positive torques ( ∼ 4 pN⋅nm: blue circles, average: red cross), ∼ 7 pN⋅nm (orange circles, average blue cross), and ∼ 8 pN⋅nm (green circles, average: purple cross), which pause RNAP for 125 ± 42 s (N = 8), 82 ± 50 s (N = 6) and 26 ± 8 s (N = 8), respectively. Thus, positive supercoiling significantly facilitated transcription through the LacI-O1 obstacle and likely disrupted the LacI-O2 obstacle as well. The decrease in pause duration as positive torque on the DNA increased suggests that positive supercoiling weakens LacI binding and transcriptional roadblocking. Positive supercoiling generated by RNAP translocation is also known to destabilize nucleosomes (32,52); it may, therefore, represent one means by which RNAP removes protein roadblocks along the DNA.
DISCUSSION
Here, we present evidence that the ability of DNA-bound LacI to act as barrier for the transcribing RNAP is strongly dependent on DNA topology. Our TPM measurements show that a DNA loop formed by LacI bridging two operator sequences alters the roadblocking effect of LacI-operator obstacles in opposite ways depending on their position relative to the promoter (proximal vs. distal). Approaching a loop from the outside, RNAP paused in front of LacI-proximal operator roadblock longer than it did when there was no loop. A loop effectively increases the LacI concentration in the vicinity of the operator (27), which increases the occupancy of the Oprox binding site and may also sterically hinder approaching RNAP (25). Once RNAP clears the proximal operator, the elongation rate decreases within the 400 bp looped segment, as compared to transcription of the same DNA in an unlooped configuration. This is likely due to torsion-induced stalling. Remarkably, once RNAP reaches the end of the loop, it clears the distal LacI roadblock faster than the same roadblock in the absence of a DNA loop.
Torsional disruption of LacI-DNA complexes might explain the shorter pauses at distal LacI securing a loop. Indeed, pauses at the distal LacI obstacle were shorter when magnetic tweezers were used to impose positive supercoiling on the DNA as RNAP arrived at the roadblock. This is strong evidence that positive supercoiling generated by transcription facilitates clearance of the LacI obstacle. In general, accumulated positive supercoiling ahead of RNAP may accelerate protein dissociation from DNA and shorten pauses at protein-mediated loops or other DNA structures.
Further control experiments using topoisomerase IB, or a nicking enzyme targeted to the loop region to artificially release the accumulating torsion, were not productive due to the inability to synchronize activities of those enzymes with RNAP elongation. Nonetheless, the results reported in this work strongly indicate that small loops of few hundred base pairs, such as the one considered here and those induced by many prokaryotic regulators, significantly slow transcription by RNAP, and that the positive torsional stress accumulated ahead of a transcription elongation complex may help to clear the path.
The energy contributed by RNAP-generated (+) supercoiling to the dissociation of operator-bound LacI can be calculated as follows. The dissociation rate of the LacI repressor from its binding site is given by the Arrhenius equation: k0 = A * exp (ΔG0 / kBT), which in the presence of transcription becomes: k1 = A * exp (ΔG1 / kBT), where A is a constant and ΔG0/1 is the LacI-operator binding energy without or with transcribing RNAP. The difference in binding energy due to elongating RNAP will then be: ΔG1 − ΔG0 = kBT * ln (k1/ k0). The LacI dissociation rate from the O1 operator is k0 = 0.0023 s–1 (50). In the absence of supercoiling or looping, the dissociation rate from O1 can be interpreted as the inverse of the average RNAP pause duration at this operator (Figure 2), and it equals k1 = 1/77 s–1 = 0.013 s–1. Thus, RNAP contributes ΔΔG01 = ΔG1 − ΔG0 = 1.7 kBT to dislodging the roadblock in conditions where (+) supercoiling cannot accumulate. Similarly, for the lower affinity O2 operator, k1 = 1/39 s–1 = 0.025 s–1 (see Figure 2), k0 = 0.0098 s–1 (50) and ΔΔG02 = 1.0 kBT. As expected, less energy is necessary to dislodge LacI from O2 than from O1.
When RNAP pauses at the O1 operator inside a loop, where (+) supercoiling can accumulate, k1 = 1/20 s–1 = 0.05 s–1 (see second paragraph after figure 2) and k0 = 0.0023 s–1 (50), so ΔΔG01,sc = 3.1kBT. Instead, for the O2 operator, k1 = 1/15 s–1 = 0.07 s–1 (see above) and k0 = 0.0098 s–1 (50) ΔΔG02,sc = 1.9kBT. Therefore, RNAP-produced supercoiling contributes (3.1 – 1.7) = 1.4 kBT to the dissociation of LacI at O1 and (1.9 − 1.0) = 0.9 kBT at O2.
Generally, destabilization of DNA roadblocks as positive supercoiling accumulates is likely to enhance RNAP progression along DNA with bound proteins. Simultaneously, negative supercoiling trailing the transcription complex may help dislodged proteins rebind and/or may stabilize proteins behind the complex. In vivo, transcription would generate supercoiling at rates of 3.9 − 5.5 turns/sec (39 – 55 bp/s) (53), a potent source of supercoiling for topoisomerases to manage. Looping transcription factors that can shift between sites ahead and behind transcription complexes would maintain at least one connection to the DNA as RNAP passes and avoid diffusing away from their binding sites. This adds a fine level of control to that exerted by the overall concentration of the transcription factors.
In vivo, the ability of protein-mediated loops to hinder RNAP elongation may be a critical factor in the regulation of transcription at the local level. In the eukaryotic organism Drosophila melanogaster, 4C-seq assays revealed that RNA polymerase II often paused near promoters involved in long-range interactions via several kilobase-pair-long loops with enhancers. The authors hypothesized that since promoter-proximal complexes can exert enhancer-blocking activity (54), the presence of paused polymerase could safeguard against premature transcriptional activation, and yet keep the system poised for activation (55). It is possible that RNAP pausing either in front of or within regulatory loops found along the template during elongation is part of a mechanism to (i) delay until additional factors disrupt the loop, or relieve supercoiling, as a signal to restart transcription, or (ii) avoid excessive transcription of a gene. Given the ubiquity of looping in any genome, stalled polymerases within loops may be targets for regulatory factors but the generation of positive supercoiling that accompanies transcription of the loop segment can disperse protein-DNA obstacles and facilitate exit from the loop.
DATA AVAILABILITY
Data is available on Dataverse at Emory at https://doi.org/10.15139/S3/UBWSVZ.
Supplementary Material
ACKNOWLEDGEMENTS
LacI was a generous gift from Kathleen Matthews, Rice University. We thank Karen Adelman, Harvard University, for generously providing RNAP for initial experiments.
Author contributions: W.X. Contributed measurements, to the design of experiments, and to writing of the paper. Y.Y. Contributed measurements, to the design of experiments, and to writing of the paper. I.A. Contributed RNA polymerase and to writing of the paper. D.D. Contributed to the design of experiments and to writing of the paper. L.F. Contributed to the design of experiments and to writing of the paper.
Notes
Present address: Yan Yan, Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, USA.
Contributor Information
Wenxuan Xu, Physics Department, Emory University, Atlanta, GA, USA.
Yan Yan, Physics Department, Emory University, Atlanta, GA, USA.
Irina Artsimovitch, Department of Microbiology, Ohio State University, Columbus, OH, USA.
David Dunlap, Physics Department, Emory University, Atlanta, GA, USA.
Laura Finzi, Physics Department, Emory University, Atlanta, GA, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Institutes of Health (NIH) grants [R01 GM084070 to LF and R01 GM067153 to IA].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ahmed S.M., Droge P.. Chromatin architectural factors as safeguards against excessive supercoiling during DNA replication. Int. J. Mol. Sci. 2020; 21:4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kantidze O.L., Razin S.V.. Weak interactions in higher-order chromatin organization. Nucleic Acids Res. 2020; 48:4614–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krogh T.J., Franke A., Moller-Jensen J., Kaleta C.. Elucidating the influence of chromosomal architecture on transcriptional regulation in prokaryotes - Observing Strong local effects of nucleoid structure on gene regulation. Front Microbiol. 2020; 11:2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnould C., Legube G.. The secret life of chromosome loops upon DNA double-strand break. J. Mol. Biol. 2020; 432:724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banigan E.J., Mirny L.A.. Loop extrusion: theory meets single-molecule experiments. Curr. Opin. Cell Biol. 2020; 64:124–138. [DOI] [PubMed] [Google Scholar]
- 6. Cacciatore A.S., Rowland B.D.. Loop formation by SMC complexes: turning heads, bending elbows, and fixed anchors. Curr. Opin. Genet. Dev. 2019; 55:11–18. [DOI] [PubMed] [Google Scholar]
- 7. Gramzow L., Ritz M.S., Theissen G.. On the origin of MADS-domain transcription factors. Trends Genet. 2010; 26:149–153. [DOI] [PubMed] [Google Scholar]
- 8. Hansen A.S., Cattoglio C., Darzacq X., Tjian R.. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus. 2018; 9:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lazniewski M., Dawson W.K., Rusek A.M., Plewczynski D. One protein to rule them all: the role of CCCTC-binding factor in shaping human genome in health and disease. Semin. Cell Dev. Biol. 2019; 90:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Umlauf D., Mourad R.. The 3D genome: from fundamental principles to disease and cancer. Semin. Cell Dev. Biol. 2019; 90:128–137. [DOI] [PubMed] [Google Scholar]
- 11. Hosokawa H., Rothenberg E.V.. How transcription factors drive choice of the t cell fate. Nat. Rev. Immunol. 2021; 21:162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavco P.A., Steege D. Elongation by escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem. 1990; 265:9960–9969. [PubMed] [Google Scholar]
- 13. Petesch S.J., Lis J.T.. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012; 28:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kulaeva O.I., Hsieh F.-K., Chang H.-W., Luse D.S., Studitsky V.M.. Mechanism of transcription through a nucleosome by RNA polymerase iI. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2013; 1829:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanders T.J., Lammers M., Marshall C.J., Walker J.E., Lynch E.R., Santangelo T.J.. TFS and spt4/5 accelerate transcription through archaeal histone-based chromatin. Mol. Microbiol. 2019; 111:784–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dodd I.B., Egan J.B.. Action at a distance in cl repressor regulation of the bacteriophage 186 genetic switch. Mol. Microbiol. 2002; 45:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gramzow L., Theissen G.. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010; 11:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guerra R.F., Imperadori L., Mantovani R., Dunlap D.D., Finzi L.. DNA compaction by the nuclear factor-Y. Biophys. J. 2007; 93:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee B.K., Iyer V.R.. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 2012; 287:30906–30913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mack C.P., Thompson M.M., Lawrenz-Smith S., Owens G.K.. Smooth muscle alpha-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ. Res. 2000; 86:221–232. [DOI] [PubMed] [Google Scholar]
- 21. Mendes M.A., Guerra R.F., Berns M.C., Manzo C., Masiero S., Finzi L., Kater M.M., Colombo L.. MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in arabidopsis. Plant Cell. 2013; 25:2560–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miano J.M. Deck of CArGs. Circ. Res. 2008; 103:13–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miwa T., Boxer L.M., Kedes L.. CARG Boxes in the human cardiac alpha-actin gene are core binding sites for positive trans-acting regulatory factors. Proc. Natl. Acad. Sci. USA. 1987; 84:6702–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dorman C.J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004; 2:391–400. [DOI] [PubMed] [Google Scholar]
- 25. Voros Z., Yan Y., Kovari D.T., Finzi L., Dunlap D. Proteins mediating DNA loops effectively block transcription. Protein Sci. 2017; 26:1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Becker N.A., Peters J.P., Maher L.J. 3rd, Lionberger T.A.. Mechanism of promoter repression by lac repressor-DNA loops. Nucleic Acids Res. 2013; 41:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Priest D.G., Cui L., Kumar S., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of the DNA tethering effect in long-range DNA looping in vivo and in vitro using the lac and lambda repressors. Proc. Natl. Acad. Sci. USA. 2014; 111:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu L.F., Wang J.C.. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 1987; 84:7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan Y., Ding Y., Leng F., Dunlap D., Finzi L.. Protein-mediated loops in supercoiled DNA create large topological domains. Nucleic Acids Res. 2018; 46:4417–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Whitson P.A., Hsieh W.T., Wells R.D., Matthews K.S.. Supercoiling facilitates lac operator-repressor-pseudooperator interactions. J. Biol. Chem. 1987; 262:4943–4946. [PubMed] [Google Scholar]
- 31. Wang J.C., Barkley M.D., Bourgeois S.. Measurements of unwinding of lac operator by repressor. Nature. 1974; 251:247–249. [DOI] [PubMed] [Google Scholar]
- 32. Sheinin M.Y., Li M., Soltani M., Luger K., Wang M.D.. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat. Commun. 2013; 4:2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu W., Yan Y., Artsimovitch I., Dunlap D., Finzi L.. Replication Data for: Xu/Yan et al., “Positive supercoiling favors transcription”, NAR 2022. Emory Dataverse. 2022; University of North Carolina; 10.15139/S3/UBWSVZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Svetlov V., Artsimovitch I.. Purification of bacterial RNA polymerase: tools and protocols. Methods Mol. Biol. 2015; 1276:13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar S., Manzo C., Zurla C., Ucuncuoglu S., Finzi L., Dunlap D. Enhanced tethered-particle motion analysis reveals viscous effects. Biophys. J. 2014; 106:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han L., Lui B.H., Blumberg S., Beausang J.F., Nelson P.C., Phillips R.. Mathematics of DNA structure, function and interactions. 2009; Springer; 123–138. [Google Scholar]
- 37. Ucuncuoglu S., Schneider D.A., Weeks E.R., Dunlap D., Finzi L.. Multiplexed, tethered particle microscopy for studies of DNA-enzyme dynamics. Methods Enzymol. 2017; 582:415–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kovari D.T., Yan Y., Finzi L., Dunlap D. Tethered particle motion: an easy technique for probing DNA topology and interactions with transcription factors. Methods Mol. Biol. 2018; 1665:317–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Priest D.G., Kumar S., Yan Y., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of interactions between two DNA loops demonstrates loop domain insulation in e. coli cells. Proc. Natl. Acad. Sci. 2014; 111:E4449–E4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schafer D.A., Gelles J., Sheetz M.P., Landick R.. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991; 352:444–448. [DOI] [PubMed] [Google Scholar]
- 41. Wang M.D., Schnitzer M.J., Yin H., Landick R., Gelles J., Block S.M.. Force and velocity measured for single molecules of RNA polymerase. Science. 1998; 282:902–907. [DOI] [PubMed] [Google Scholar]
- 42. Davenport R.J., Wuite G.J., Landick R., Bustamante C.. Single-molecule study of transcriptional pausing and arrest by e. coli RNA polymerase. Science. 2000; 287:2497–2500. [DOI] [PubMed] [Google Scholar]
- 43. Svetlov V., Belogurov G.A., Shabrova E., Vassylyev D.G., Artsimovitch I.. Allosteric control of the RNA polymerase by the elongation factor rfaH. Nucleic Acids Res. 2007; 35:5694–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Finzi L., Dunlap D.D.. Single-molecule approaches to structure, kinetics and thermodynamics of transcriptional regulatory nucleoprotein complexes”. J. Biol. Chem. 2010; 285:18973–18978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kovari D.T., Dunlap D., Weeks E.R., Finzi L.. Model-free 3D localization with precision estimates for brightfield-imaged particles. Opt. Express. 2019; 27:29875–29895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma J., Bai L., Wang M.D.. Transcription under torsion. Science. 2013; 340:1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moroz J.D., Nelson P.. Torsional directed walks, entropic elasticity, and DNA twist stiffness. Proc. Natl. Acad. Sci. USA. 1997; 94:14418–14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mosconi F., Allemand J.F., Bensimon D., Croquette V.. Measurement of the torque on a single stretched and twisted DNA using magnetic tweezers. Phys. Rev. Lett. 2009; 102:078301. [DOI] [PubMed] [Google Scholar]
- 49. Hammar P., Wallden M., Fange D., Persson F., Baltekin O., Ullman G., Leroy P., Elf J.. Direct measurement of transcription factor dissociation excludes a simple operator occupancy model for gene regulation. Nat. Genet. 2014; 46:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hao N., Krishna S., Ahlgren-Berg A., Cutts E.E., Shearwin K.E., Dodd I.B.. Road rules for traffic on DNA-systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res. 2014; 42:8861–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leng F., Chen B., Dunlap D.D.. Dividing a supercoiled DNA molecule into two independent topological domains. Proc. Natl. Acad. Sci. 2011; 108:19973–19978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Teves S.S., Henikoff S.. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 2014; 21:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vogel U., Jensen K.F.. The RNA chain elongation rate in escherichia coli depends on the growth rate. J. Bacteriol. 1994; 176:2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chopra V.S., Cande J., Hong J.W., Levine M.. Stalled hox promoters as chromosomal boundaries. Genes Dev. 2009; 23:1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014; 512:96–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available on Dataverse at Emory at https://doi.org/10.15139/S3/UBWSVZ.