Abstract
Long before pathogenic interactions with eukaryotic cells evolved, bacteria were competing with one another for limited resources. In this issue of Cell, Ting and colleagues identify previously unappreciated players in the interbacterial arms race that may be the evolutionary ancestors of eukaryotic cell-targeting ADP-ribosyltransferase toxins.
PREVIEW
Bacteria are locked in constant inter- and intraspecies competition to accrue limiting nutrients, space, and resources. Apart from determining the fate of individual cells, interbacterial competition also drives emergent properties of microbial ecosystems, including community structure and dynamics, pathogenicity, and evolutionary diversification. Contact dependent inhibition (CDI) systems, including the EXS/type 7 secretion system (T7SS) in Gram-positive bacteria and the type 6 secretion system (T6SS) in Gram-negative bacteria, are the primary weaponry in this never ending war. In CDI systems, complex molecular machines transverse the cell envelopes of both the donor and recipient cells to translocate toxic effectors directly into the target cell. The effectors are delivered indiscriminately and principally target conserved, essential macromolecules or co-factors (e.g. peptidoglycan, membrane lipids, DNA, and NAD), thereby conferring broad protection against a swath of environmental competitors. To protect themselves from intoxication, donor cells encode cognate immunity proteins that physically block the toxin active site until effector translocation. Interbacterial effectors are postulated to serve as evolutionary “stepping stones” from which eukaryotic cell-targeting effectors and toxins have evolved. Indeed, many prokaryotic and eukaryotic cell-targeting toxins—including phospholipases and NADases—share similar mechanisms despite disparate delivery systems (Sun et al., 2015; Whitney et al., 2015). A subset of T6S effectors, including the recently identified Pseudomonas aeruginosa phospholipase Tpe1, are even able to intoxicate both prokaryotic and eukaryotic cells (Jiang et al., 2016). In this issue of Cell, Ting et al. further advance this narrative and describe a novel interbacterial effector that has hallmarks typically associated with potent eukaryotic cell-targeting toxins.
Combining bioinformatics with functional characterization, the authors identify a widespread family of effector-immunity pairs encoding an ADP ribosyltransferase (ART) and an ADP-ribosylhydrolase (ARH), respectively. Although ADP-ribosylation is among the most prevalent (and earliest described) mechanisms of eukaryotic cell intoxication by bacterial pathogens (e.g. diphtheria toxin and cholera toxin), this modification was not thought to play a role in interbacterial competition (Simon et al., 2014). Using the predicted ART/ARH pair Tre1/Tri1 of Serratia proteamaculans as a model, the authors systematically validate the role of both the toxin and immunity protein in both intra- and interspecies antagonism in a T6S-dependent manner. Importantly, ADP ribosylation as a mechanism of intoxication is unlikely to be restricted to T6S competent bacteria: bioinformatic analysis identified ART/ARH pairs in Gram-positive species, including Listeria monocytogenes, in association with genes encoding components of the EXS machinery.
To identify the targets of ADP-ribosylation, and thus the proximal cause of intoxication, the authors subjected lysates of E. coli cells expressing functional homologs of Tre1 to mass spectrometry, identifying seven ribosylated targets, including the highly conserved, essential cell division protein FtsZ. A tubulin homolog, FtsZ polymerizes into treadmilling filaments that serve as a foundation for the cell wall synthesis machinery during cytokinesis (Bisson-Filho et al., 2017; Yang et al., 2017). Although FtsZ assembly is controlled by a host of endogenous regulatory proteins, the only exogenous factors previously known to target it are virally derived, including the 45-residue Kil peptide from bacteriophage λ (Haeusser et al., 2014). Significantly, FtsZ embodies many qualities of an attractive target: cell division is an essential process, and unlike most of the division machinery, FtsZ is highly conserved among bacteria.
The authors present several lines of evidence supporting FtsZ as a physiologically relevant target of Tre1: 1) FtsZ is a target of Tre1 homologs from both S. proteamaculans and Pseudomonas putida, which share limited amino acid identity, 2) Tre1 intoxication leads to filamentous cells, a characteristic of malfunctioning division machinery, and 3) genetics indicating the modified arginine residue on FtsZ, Arg147 is likely essential for FtsZ function in the Enterobacteriaceae (Koppelman et al., 2003). Despite this, modification may be an artifact of overexpression, rather than a physiological target of Tre1 due to FtsZ high cellular abundance; furthermore, the morphological shift upon intoxication may reflect a stress response, downstream of a direct effect on the division machinery. To firmly establish FtsZ as a bona fide target of Tre1, the authors interrogate Z ring dynamics in vivo and FtsZ polymerization in vitro in the presence of physiological levels of effector. Compellingly, their findings are consistent with the modified FtsZ monomers failing to polymerize into nascent filaments and terminating polymerization of existing filaments—both activities that are likely to poison cell division. The authors suggest that ART factors might be leveraged to probe the mechanics of FtsZ assembly, dynamics, and polarity in vivo. At the same time, this idea might be premature. ART enzymes lack specificity—FtsZ is just one of seven modified several modified target proteins in Tre1 intoxicated cells – and overexpression of catalytically inactive Tre1 also leads to aberrant cell morphology, suggest an ADP-ribosylase independent mode of action.
During their analysis of the immunity protein Tri1, the authors make the unexpected observation that the enzyme removes ADP-ribosylation of the target protein FtsZ with the same efficiency as a control, non-target protein. Supporting atypical promiscuity of these ARH enzymes, a non-cognate ARH homology from L. monocyotgenes is capable of reversing this modification despite being unable to interact with S. proteamaculans ART effector. Based on the ability of the immunity factor to mediate nonspecific protection and the widespread nature of ART/ARH effector/immunity protein pairs, the authors postulate that this family of ARH enzymes may promote fitness even in the absence of a cognate effector. Indeed, bioinformatic analysis supports this hypothesis and suggests at least three separate instances of duplication and divergence of housekeeping ARH enzymes, which may function to detoxify toxic antagonistic ADP-ribosylation modifications.
Altogether, this study adds ART/ARH pairs to the arsenal of effectors in the interbacterial arms race. The key challenge remains reconstructing the evolutionary history of these molecules to establish whether they emerged as a result of selective pressure in natural competitor populations and how these interactions have shaped bacterial evolution in more recent timescales. One intriguing possibility is that evolution of promiscuous ARH enzymes in target bacteria catalyzed the expansion of redundant and synergistic effectors in antagonistic strains, curtailing the ability of the recipient cells to evolve resistance (LaCourse et al., 2018). Beyond interbacterial warfare, clarifying the relationship between prokaryotic and eukaryotic cell-targeting ART toxins also promises to shed light on the evolution of modern scourges of humanity—did formidable pathogens Vibrio cholerae and Corynebacterium diphtheriae evolve their namesake toxins from interbacterial effectors, or is ADP-ribosylation an example of convergent evolution? No doubt, Ting and colleagues have laid the groundwork for future studies to address these questions.
ART/ARH enzymes modulate FtsZ assembly.
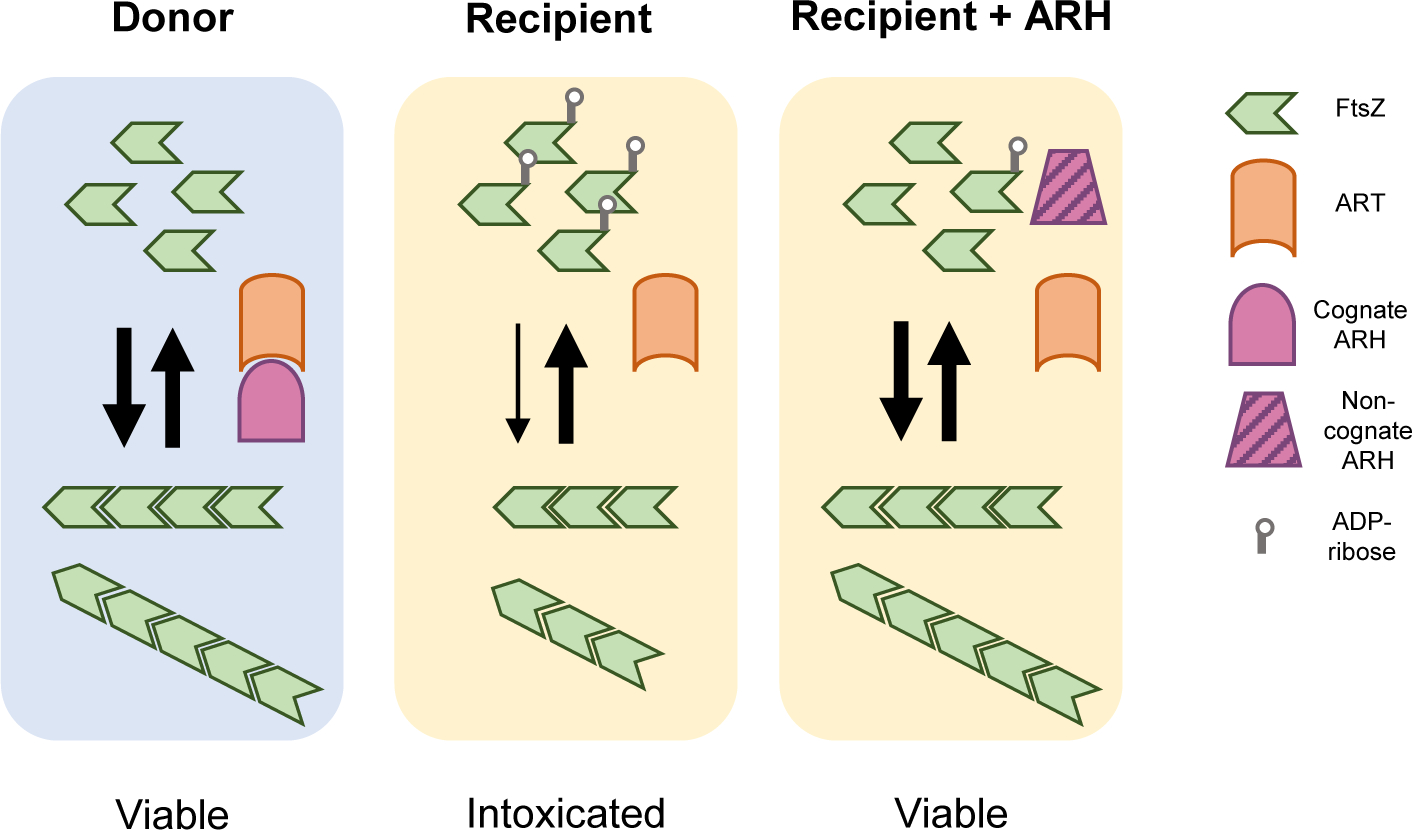
The cellular FtsZ pool exists in two states: unpolymerized monomers and treadmilling filaments. Although donor cells (blue background) encode a toxic ADP ribosyltransferase (ART) effector capable of introducing a toxic adduct on FtsZ, they also encode a cognate ADP hydrolase (ARH) immunity protein, which neutralizes effector activity enzymatically and physically. ART translocation into recipient cells (yellow background) results in ADP-ribosylation of FtsZ monomers, leading to impaired polymerization, cell division failure, and intoxication. However, some recipient cells encode a non-cognate, promiscuous ARH enzyme and are able to hydrolyze the toxic modification, restoring wild-type FtsZ assembly dynamics and broadly protecting against ART activity.
ACKNOWLEDGEMENTS
We acknowledge support from an NSF Graduate Research Fellowship DGE-1745038 to E.A.M. and NIH grant GM127331 to P.A.L. We thank Joseph Merriman and Corey Westfall for critical reading and comments on this commentary.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Hoashi M, Weaver A, Brown N, Pan J, Sawitzke JA, Thomason LC, Court DL, and Margolin W (2014). The Kil Peptide of Bacteriophage λ Blocks Escherichia coli Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly. PLoS Genet. 10, e1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Wang X, Wang B, Chen L, Zhao Z, Waterfield NR, Yang G, and Jin Q (2016). The Pseudomonas aeruginosa Type VI Secretion PGAP1-like Effector Induces Host Autophagy by Activating Endoplasmic Reticulum Stress. Cell Rep 16, 1502–1509. [DOI] [PubMed] [Google Scholar]
- Koppelman C-M, Aarsman MEG, Postmus J, Pas E, Muijsers AO, Scheffers D-J, Nanninga N, and Blaauwen, den T (2003). R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 51, 645–657. [DOI] [PubMed] [Google Scholar]
- LaCourse KD, Peterson SB, Kulasekara HD, Radey MC, Kim J, and Mougous JD (2018). Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat Microbiol 3, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon NC, Aktories K, and Barbieri JT (2014). Novel bacterial ADP-ribosylating toxins: structure and function. Nature Reviews Microbiology 2011 10:2 12, 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Siroy A, Lokareddy RK, Speer A, Doornbos KS, Cingolani G, and Niederweis M (2015). The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat. Struct. Mol. Biol. 22, 672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, Tran BQ, Robinson H, Goo YA, Goodlett DR, et al. (2015). An Interbacterial NAD(P)+ Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells. Cell 163, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, and Xiao J (2017). GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]


