ABSTRACT
Hemogenic endothelial (HE) cells in the dorsal aorta undergo an endothelial-to-hematopoietic transition (EHT) to form multipotent progenitors, lympho-myeloid biased progenitors (LMPs), pre-hematopoietic stem cells (pre-HSCs) and adult-repopulating HSCs. These briefly accumulate in intra-arterial hematopoietic clusters (IAHCs) before being released into the circulation. It is generally assumed that the number of IAHC cells correlates with the number of HSCs. Here, we show that changes in the number of IAHC cells, LMPs and HSCs can be uncoupled. Mutations impairing MyD88-dependent toll-like receptor (TLR) signaling decreased the number of IAHC cells and LMPs, but increased the number of HSCs in the aorta-gonad-mesonephros region of mouse embryos. TLR4-deficient embryos generated normal numbers of HE cells, but IAHC cell proliferation decreased. Loss of MyD88-dependent TLR signaling in innate immune myeloid cells had no effect on IAHC cell numbers. Instead, TLR4 deletion in endothelial cells (ECs) recapitulated the phenotype observed with germline deletion, demonstrating that MyD88-dependent TLR signaling in ECs and/or in IAHCs regulates the numbers of LMPs and HSCs.
KEY WORDS: Inflammation, AGM region, Toll-like receptor, Hematopoietic stem cells, Hemogenic endothelial cells
Summary: Toll-like receptor signaling in endothelial cells restricts the number of hematopoietic stem cells but increases the number of committed progenitors and intra-arterial hematopoietic cluster cells, and promotes their proliferation.
INTRODUCTION
Hematopoietic ontogeny occurs in multiple waves in which hematopoietic stem and progenitor cells (HSPCs) with different potentials differentiate from hemogenic endothelial (HE) cells via an endothelial-to-hematopoietic transition (EHT). HE cells in the extra-embryonic yolk sac undergo EHT to become committed erythro-myeloid progenitors (EMPs) and lymphoid progenitors, while HE cells in the caudal arteries (dorsal aorta in zebrafish; dorsal aorta, vitelline and umbilical in mouse) differentiate into multipotent, lympho-myeloid biased progenitors (LMPs), HSCs and HSC precursors called pre-HSCs (Auerbach et al., 1996; Hadland and Yoshimoto, 2018). Following EHT, the newly formed HSPCs in the major arteries briefly accumulate in intra-arterial hematopoietic clusters (IAHCs) consisting of cells that uniformly express endothelial markers, hematopoietic transcription factors (such as RUNX1) and high levels of KIT (Bollerot et al., 2005). At embryonic day (E) 10.5, when IAHC numbers peak, the IAHC cells contain several hundred LMPs, ∼25 pre-HSCs and ∼0.03 adult-repopulating HSCs (Kumaravelu et al., 2002; Li et al., 2014; Rybtsov et al., 2016). At E11.5, IAHCs contain ∼65 pre-HSCs and one adult repopulating HSC (Kumaravelu et al., 2002; Rybtsov et al., 2016). Hence, the IAHCs are heterogeneous, containing HSPCs with unique differentiation and engraftment potentials, and their composition changes during development.
Innate immune and inflammatory signaling pathways have been shown to regulate the number of HSPCs generated in the arteries of zebrafish and mouse embryos. Mutations in or morpholino knockdown of genes encoding type I and II interferons (Li et al., 2014; Sawamiphak et al., 2014), tumor necrosis factor (TNF) (Espín-Palazón et al., 2014), inflammasomes (Frame et al., 2020), interleukin 1β (IL1β) (Frame et al., 2020), granulocyte colony stimulating factor (G-CSF) (He et al., 2015), toll-like receptor 4 (TLR4) (He et al., 2015), nuclear factor κB (NF-kB) (Espín-Palazón et al., 2014; He et al., 2015) and RIG1-like receptors (Lefkopoulos et al., 2020) reduced the number of myb:GFP+ kdrl:mCherry+ HSPCs in transgenic reporter zebrafish embryos, and decreased the levels of runx1 and myb transcripts in the dorsal aorta (DA). Innate immune signaling is initiated by binding of ligands to pattern recognition receptors (PRRs). PRRs recognize pathogen-associated molecular patterns (PAMPs), which are small molecules common to a specific class of infectious agents, and host molecules referred to as danger-associated molecular patterns (DAMPs), which are endogenous proteins or small molecules that are released from the cell or the extracellular matrix in response to cell damage, stress or death. Although, in theory, PAMPs from infectious agents might be responsible for activating innate immune signaling in mammalian or pre-hatched zebrafish embryos, it is more likely that DAMPs are the PRR ligands in the relatively sterile environment of the mammalian embryo.
Activation of toll-like receptors (TLRs) drives the production of type I interferons, TNF, IL-1β and G-CSF, and therefore directly or indirectly stimulates multiple inflammatory pathways (Monlish et al., 2016). TLRs signal through two intracellular pathways that involve either the MyD88 (myeloid differentiation primary response protein) and TIRAP (toll-interleukin 1 receptor domain containing adaptor protein), or the TRIF [toll IL-1 receptor (TIR)-domain-containing adaptor inducing interferon beta] and TRAM (translocating chain-associated membrane protein) adaptor proteins. Signaling from TLR4 on the plasma membrane through MyD88/TIRAP results in the nuclear localization of the CREB, AP1 and NF-κB transcription factors, and expression of inflammatory cytokine genes such as TNF, IL1β and G-CSF. Signaling through endosomal TLR4, TRIF/TRAM and the interferon regulatory factors (IRFs) primarily activates the expression of interferon genes. Morpholino knockdown of the genes encoding TLR4 (tlr4bb) and MyD88 (myd88) in zebrafish embryos reduced the expression of runx1, the number of myb:GFP+ kdrl:mCherry+ HSPCs in the DA, and HSPCs in secondary sites of colonization, including the caudal hematopoietic tissue and thymus (He et al., 2015). However, markers that discriminate between committed progenitors (e.g. LMPs) and pre-HSCs/HSCs are not available for zebrafish embryos; therefore, determining how TLR4 signaling differentially affects them is difficult. Understanding how LMP and pre-HSC/HSC generation are regulated is important for selectively optimizing production of the desired cells from arterial HE cells ex vivo.
Here, we examine TLR signaling to determine in which cells it is required, through which intracellular adaptors it signals to regulate arterial HSPC ontogeny and the types of HSPCs it regulates. We show that signaling through the MyD88 arm of the TLR pathway increases the number of IAHC cells and LMPs, while restricting the number of multi-lineage adult-repopulating HSCs.
RESULTS AND DISCUSSION
TLR4 signaling regulates RUNX1 levels and the number of IAHC cells
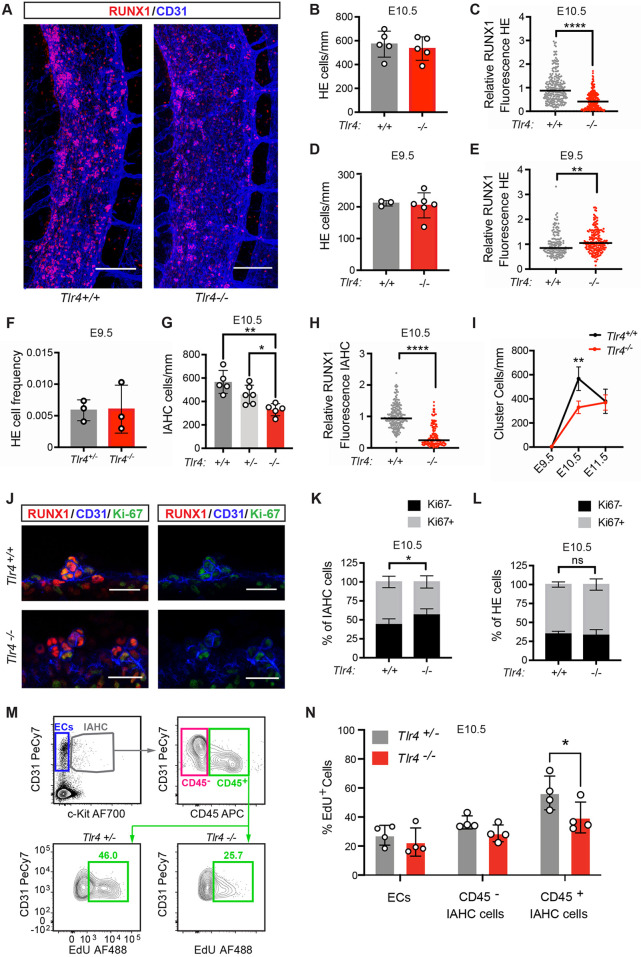
It was reported that loss of TLR4 reduced expression of the transcription factor runx1 in the DA of zebrafish embryos and the number of RUNX1+ cells in the DA of mouse embryos (He et al., 2015). RUNX1 is a specific marker of HE and IAHC cells, and is required for EHT (North et al., 1999). To examine whether the reduction in runx1 expression could be due to fewer RUNX1+ HE cells, to lower RUNX1 protein levels in HE cells or to a decrease in IAHC cells, we analyzed Tlr4−/− mouse embryos and their wild-type littermates by whole-mount confocal microscopy (Fig. 1A-I). IAHC cells were identified by their rounded appearance and expression of RUNX1 and CD31, and high levels of KIT, while HE cells were flatter, located within the endothelial layer, and expressed RUNX1, CD31 and very low or undetectable levels of KIT. We found that the relative fluorescence intensity of RUNX1 protein was lower in HE cells in the DA of E10.5 Tlr4−/− embryos compared with Tlr4+/+ littermates, but the numbers of RUNX1+ HE cells were equivalent (Fig. 1A-C). As the number of HE cells at a single snapshot in time is a function of the number that were originally specified minus the number that have undergone EHT, we enumerated HE cells at E9.5, prior to their major egress from the endothelial cell layer via EHT. The numbers of phenotypic RUNX1+ HE cells were equivalent in E9.5 Tlr4+/+ and Tlr4−/− embryos (Fig. 1D), and the levels of RUNX1 in Tlr4−/− HE cells were slightly increased (Fig. 1E). We compared the frequencies of functional HE cells by sorting limiting numbers of ECs (CD31hiCD144+ESAM+CD44+CD45−Ter119−Mac1−CD41−) into individual wells containing OP9 stromal cells (≤5 ECs/well) and counting the number of wells containing hematopoietic cells 10 days later. The frequencies of functional HE cells were equivalent in E9.5 Tlr4+/− and Tlr4−/− embryos (Fig. 1F, Fig. S1A). Therefore, loss of TLR4 decreases RUNX1 levels in E10.5 HE cells but has no effect on the number of HE cells.
Fig. 1.
TLR4 signaling increases the number and proliferation of IAHC cells but does not regulate the number of HE cells. (A) Representative images of confocal z-projections (interval=2 µM) of the dorsal aorta (DA) of E10.5 Tlr4+/+ (left) or Tlr4−/− (right) embryos immunostained for RUNX1 (red) and CD31 (blue). Scale bars: 100 µm. (B) Quantification of HE cells (CD31+RUNX1+KIT−) per mm length of the DA in immunostained E10.5 embryos. Data are mean±s.d.; each point represents one embryo; Student's t-test, unpaired, two-tailed; n=5 per genotype. The area counted centered on the intersection of the vitelline artery with the DA, and all cells within two or three somite pairs in both the rostral and caudal direction were counted. (C) Corrected total cell fluorescence (CTCF) was calculated to measure RUNX1 intensity in HE cells, identified as described in B. An integrated density value for each HE cell was measured using FIJI software and CTCF was calculated as [integrated density of cell−(area of selected cell×mean intensity of background)]. CTCF values were then normalized using the CTCF average for Tlr4+/+ HE cells (relative RUNX1 fluorescence). Horizontal lines indicate the mean, with individual data points plotted; Student's t-test, unpaired, two-tailed; n=248 for Tlr4+/+ and n=258 for Tlr4−/− with cells measured from three embryos per genotype. (D) Quantification of HE cells/mm at E9.5 in the DA of Tlr4+/+ and Tlr4−/− embryos. Cells were counted in the region spanning the caudal boundary of the heart to the intersection with the umbilical artery. Data are mean±s.d.; Student's t-test, unpaired, two-tailed; n=3 and 6 for Tlr4+/+ and Tlr4−/−, respectively. (E) CTCF values, indicating RUNX1 intensity, measured for HE cells in immunostained E9.5 Tlr4+/+ and Tlr4−/− embryos. Horizontal lines indicate the mean, with individual data points plotted; Student's t-test, two-tailed; n=141 for Tlr4+/+ and n=192 for Tlr4−/− with ∼40-50 cells measured from at least three embryos per genotype. (F) Frequency of HE cells within sorted ECs (CD31hiCD144+ESAM+CD44+CD45−Ter119−Mac1−CD41−) isolated from E9.5 Tlr4+/− and Tlr4−/− embryos, determined by plating limiting numbers of cells (≤5/well) on OP9 stromal cells. Tlr4+/− embryos generated from a cross of Tlr4+/− and Tlr4−/− mice were used in lieu of Tlr4+/+ embryos in order to generate a sufficient number of cells for the analysis. All embryos were 21-25 somite pairs. Plates were scored by eye for hematopoietic growth and positive wells were confirmed by flow cytometry (positive wells were CD45, CD41, Mac1, B220 and/or Ter119 positive). The frequency is the number of positive wells/total ECs plated. Data are mean±s.d.; Student's t-test, unpaired, two-tailed. Data are from three independent experiments with more than 500 cells sorted per genotype for each experiment. (G) Quantification of IAHC cells/mm (CD31+RUNX1+KIT+) in confocal images of E10.5 DAs from Tlr4+/+, Tlr4+/− and Tlr4−/− embryos. Data are mean±s.d.; one-way ANOVA and Tukey's multiple comparison test, n=5 or 6 embryos per genotype. (H) RUNX1 intensity determined by CTCF in IAHC cells in the DA measured as described in C. n=223 for Tlr4+/+ and n=157 for Tlr4−/− with approximately 50-60 cells measured from three or four embryos per genotype. Horizontal lines indicate the mean, with individual data points plotted. (I) Quantification of IAHC cells/mm (CD31+RUNX1+KIT+) in confocal images of E9.5, E10.5 and E11.5 DAs from Tlr4+/+ and Tlr4−/− embryos. Data are mean±s.d.; two-way ANOVA, Sidak correction for multiple comparisons, n=3 for E9.5 genotypes, n=5 for E10.5 genotypes, n=3 for E11.5 Tlr4+/+ and n=4 for E11.5 Tlr4−/−. (J) Confocal z-projections (2 µm intervals, four intervals total/image) of IAHC cells in E10.5 DAs of Tlr4+/+ (top row) and Tlr4−/− (bottom row) embryos immunostained for RUNX1 (red), CD31 (blue) and Ki67 (green). Scale bars: 25 µm. (K) Percentage of Ki67+ (gray) and Ki67− (black) IAHC cells, calculated as (Ki67+/− subset/total IAHC cells), observed in confocal images of E10.5 DAs of Tlr4+/+ and Tlr4−/− embryos. Data are mean±s.d.; two-way ANOVA, Sidak correction for multiple comparisons; n=6 or 7 per genotype. (L) Percentage of Ki67+ (gray) and Ki67− (black) HE cells, calculated as described in K. (M) Representative flow cytometry plots depicting gating for CD31+KIT− endothelial cells (ECs), CD31+KIT+CD45− IAHC cells and CD31+KIT+CD45+ IAHC cells. EdU labeling of CD45+ IAHC cells (bottom row) are shown for Tlr4+/− and Tlr4−/− embryos. Tlr4+/− embryos generated from a cross of Tlr4+/− and Tlr4−/− mice were used in lieu of Tlr4+/+ embryos in order to obtain a sufficient number of cells for the analysis. The number of IAHC cells in Tlr4+/− and Tlr4+/+ embryos was not significantly different, as shown in G. Cells in plots were previously gated on FSC versus SSC, live, lineage negative, CD41lo/− (see Fig. S1B). Images are representative of three independent experiments (four litters). Littermates were pooled in each experiment. (N) Percentage of EdU+CD31+KIT− endothelial cells (ECs), CD31+KIT+CD45− IAHC cells and CD31+KIT+CD45+ IAHC cells determined by flow cytometry. Data are mean±s.d.; two-way ANOVA, Sidak correction for multiple comparisons; n=4 pooled litters per genotype. Representative of three independent experiments. *P≤0.05, **P≤0.01, ****P≤0.0001, ns indicates not significant.
Loss of TLR4 reduced the number of IAHC cells by 42% in E10.5 embryos, and IAHC cells had lower levels of RUNX1 (Fig. 1G,H). However, the numbers of IAHC cells in Tlr4+/+ and Tlr4−/− embryos were equivalent at E11.5, indicating that the TLR4 requirement for normal numbers of IAHC cells was transient (Fig. 1I). The decreased number of IAHC cells at E10.5 could be caused by inefficient EHT, decreased proliferation or increased apoptosis. We observed no decrease in specified HE cells at E9.5 or accumulation of HE cells at E10.5, suggesting that the decrease in IAHC cells at E10.5 is not due to inefficient EHT, although we cannot rule this out as inefficient EHT might cause HE cells to lose RUNX1 expression, to assume an alternative cell fate or to undergo apoptosis. Instead, our data suggest that TLR4 deficiency decreases the number of IAHC cells by reducing their proliferation, as reflected by a smaller fraction of IAHC cells that were Ki-67 positive (Fig. 1J,K). EdU labeling and flow cytometric analysis were also consistent with an effect of TLR4 loss on proliferation, specifically of CD45+ IAHC cells, which lack pre-HSCs but are enriched for LMPs at E10.5 (Rybtsov et al., 2011; Zhu et al., 2020) (Fig. 1M,N and Fig. S1B). TLR4 deficiency did not affect the proliferation of ECs, HE cells or CD45− IAHC cells, which include pre-HSCs at E10.5 (Fig. 1L-N). In summary, loss of TLR4 decreased RUNX1 protein levels in HE and IAHC cells, and reduced the proliferation of CD45+ IAHC cells (LMPs) at E10.5, but did not affect the number IAHC cells at E11.5 or the number of HE cells specified at E9.5.
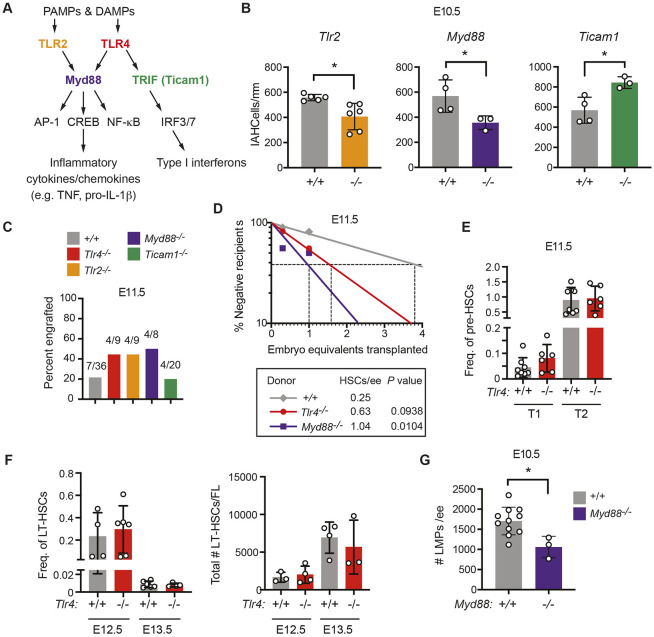
Both TLR4 and TLR2 are expressed in and regulate inflammatory signaling in ECs (Salvador et al., 2016); therefore, we determined whether TLR2 also regulates the number of IAHC cells. Loss of TLR2 reduced the number of IAHC cells in the DA to a similar extent to TLR4 deficiency (Fig. 2B). TLR2 signals exclusively through the MyD88 adaptor, while TLR4 signals through both the MyD88 and TRIF adaptors (Fig. 2A). To determine which signaling arm of TLR4 was important for regulating the number of IAHC cells, we enumerated IAHC cells in both Myd88−/− and TRIF-deficient (Ticam1−/−) embryos. Loss of MyD88 decreased, whereas loss of TRIF increased, the number of IAHC cells (Fig. 2B). Therefore, signaling via cell-surface TLR2 and TLR4 through the MyD88 adaptor increases the number of IAHC cells at E10.5, likely by promoting their proliferation, whereas endosomal TLR4 signaling through the TRIF adaptor constrains IAHC cell numbers.
Fig. 2.
MyD88-dependent TLR signaling increases the number of LMPs in the AGM region but limits the number of HSCs. (A) TLR signaling pathway highlighting cell surface receptors TLR2 and TLR4, their adaptors MyD88 and TRIF, and transcription factor effectors. (B) Quantification of CD31+RUNX1+KIT+ IAHC cells/mm in the DA of E10.5 embryos from confocal images. n=3-6 per genotype. Data are mean±s.d. Unpaired, two-tailed Student's t-test. Each dot represents one embryo. (C) Percentage of recipient mice (CD45.1+) exhibiting multi-lineage reconstitution following transplantation of one embryo equivalent (ee) of AGM regions from E11.5 embryos (CD45.2+). Multi-lineage reconstitution was assessed as >1% donor myeloid (Mac1+, Mac1+Gr1+), T-cells (CD3+) and B-cells (CD19+) in the peripheral blood 16 weeks post-transplantation (see Fig. S2A for details of flow cytometric analysis). (D) Quantification of HSCs in E11.5 wild-type (+/+), Tlr4−/− and MyD88−/− embryos by limiting dilution transplantation of 0.3 and 1.0 ee cells from the AGM region, umbilical and vitelline arteries (CD45.2+) into CD45.1+ adult recipients. Data are representative of at least eight recipients per group from seven transplants. The number of HSCs was calculated using ELDA software (Hu and Smyth, 2009). (E) Frequency of phenotypic type I (CD31+KIT+CD41loCD45−Ter119−) (T1) or type II (CD31+KIT+CD41loCD45+Ter119−) (T2) pre-HSCs in Tlr+/+ and Tlr4−/− E11.0 AGM regions measured by flow cytometry. Freq. represents the frequency of the population within live cells. All embryos were between 39 and 44 somite pairs. Data are mean±s.d.; one-way ANOVA and Tukey's multiple comparison test, n=8 for Tlr4+/+ and n=6 for Tlr4−/−. Data are representative of two independent experiments. Each dot represents one embryo. (F) Frequency and total number of phenotypic LT-HSCs at E12.5 and E13.5 within the fetal liver (FL) determined by flow cytometry. Freq. represents the frequency of the population within live cells. LT-HSCs were defined as CD150+CD48−KIT+Sca-1+Lin− (Lin includes B220, CD3, Gr-1, Nk1.1, Ter119). Total numbers of LT-HSCs are calculated from frequencies and the total numbers of cells in each FL. Data are mean±s.d.; one-way ANOVA and Tukey's multiple comparison test; E12.5, n=4 for Tlr4+/+ and n=6 for Tlr4−/−; E13.5, n=6 for Tlr4+/+ and n=4 for Tlr4−/−. Each dot represents one embryo. Representative of three independent experiments for both E12.5 and E13.5. (G) Frequency of LMPs in E10.5 +/+ and Myd88−/− embryos (n=11, n=3, respectively) determined by plating dissociated cells from the AGM region, umbilical and vitelline arteries in limiting dilution on OP9 and OP9-DLL1 stromal cells. +/+ embryos include offspring from intercrosses of mice heterozygous for mutations in other inflammatory mediators. The total number of progenitors that generated B cells when cultured on OP9 stromal cells and progenitors that generated T cells on OP9-DLL1 cells are plotted. Numbers of B and T cells were calculated using ELDA software (Hu and Smyth, 2009). Representative of at least three independent experiments. Data are mean±s.d. Unpaired, two-tailed Student's t-test. Each dot represents one embryo.*P≤0.05.
MyD88-dependent TLR signaling positively regulates the number of LMPs but restrains the number of adult repopulating HSCs
To determine whether loss of TLR signaling also decreased the number of adult-repopulating HSCs capable of multilineage engraftment, we transplanted one embryo equivalent of aorta-gonad-mesonephros (AGM) regions from E11.5 wild type, Tlr4−/−, Tlr2−/−, Myd88−/−, and Ticam1−/− embryos into irradiated adult recipient mice. One embryo equivalent (ee) of an E11.5 wild-type AGM region is a limiting dose of adult repopulating HSCs (Kumaravelu et al., 2002). We measured contribution to peripheral blood and bone marrow 16 weeks post-transplant (Fig. S2A). Despite the decrease in the number of IAHC cells, loss of TLR4, TLR2 and MyD88 increased the proportion of mice with multi-lineage engraftment (Fig. 2C), indicating that adult-repopulating HSCs were not depleted in the AGM regions of TLR4, TLR2 or MyD88-deficient mouse embryos and, if anything, appeared to be increased. Loss of Ticam1, on the other hand, did not affect the proportion of engrafted recipients (Fig. 2C).
To quantify the numbers of multi-lineage adult repopulating HSCs in embryos with mutations affecting MyD88-dependent TLR signaling, we transplanted a cohort of mice with limiting doses (0.3 ee and 1.0 ee) of dissociated cells from the AGM regions of E11.5 wild-type, Tlr4−/− and Myd88−/− embryos. There was a trend towards increased numbers of multi-lineage repopulating HSCs in the AGM regions of Tlr4−/− embryos and a significant 4.2-fold increase in Myd88−/− embryos (Fig. 2D), indicating that MyD88-dependent TLR signaling decreases the number of HSCs.
On the other hand, there were no differences in the frequencies of type I (CD31+KIT+CD41loCD45−Ter119−) or type II (CD31+KIT+CD41loCD45+Ter119−) pre-HSCs in the arteries (AGM region, vitelline and umbilical) of E11.5 Tlr4+/+ and Tlr4−/− embryos (Fig. 2E, Fig. S2B). We also observed no changes in the frequencies or absolute numbers of phenotypic long-term repopulating HSCs (LT-HSCs) in E12.5 or E13.5 fetal livers (Fig. 2F, Fig. S2C). Therefore, loss of TLR4 signaling did not decrease the number of phenotypic pre-HSCs or functional HSCs in the AGM region, or the number of phenotypic HSCs in the fetal liver, whereas loss of MyD88-dependent TLR signaling significantly increased the number of adult-repopulating HSCs in the AGM region.
LMPs with the potential to produce B and T cells in culture are >10-fold more abundant in AGM regions of E10.5 embryos compared with HSCs and pre-HSCs, and are enriched in the CD45+ population (Li et al., 2014; Rybtsov et al., 2016; Zhu et al., 2020), which we observed was less proliferative in Tlr4−/− embryos (Fig. 1M,N). To determine whether the number of LMPs was altered by mutations affecting MyD88-dependent signaling, we performed a limiting dilution analysis of cells from the AGM regions of wild-type and Myd88−/− embryos by plating them into wells containing OP9 stromal cells to enumerate progenitors with B cell potential, and on OP9 cells expressing the Notch ligand delta-like 1 (OP9-DLL1) to measure the number of T cell progenitors (Li et al., 2014; Schmitt and Zúñiga-Pflücker, 2002). Myd88−/− embryos contained significantly fewer progenitors capable of producing B or T cells compared with wild-type embryos (Fig. 2G). Therefore, MyD88-dependent TLR signaling positively regulates the number of LMPs while restraining the number of HSCs.
TLR4 signaling in endothelial cells and/or IAHCs, but not in primitive innate immune myeloid cells, regulates IAHC cell numbers
Professional innate immune myeloid cells are the primary producers of inflammatory cytokines and chemokines, and are proposed to be an important cellular source of the inflammatory molecules promoting the generation of HSPCs in the DA. Depletion of macrophages and granulocytes in zebrafish embryos using morpholinos targeting spi1a and spi1b, of macrophages alone with morpholinos against irf8, or of neutrophils using morpholinos to cebp1 decreased runx1 expression in the DA and the number of myb:GFP+ kdrl:mCherry+ HSPCs (Espín-Palazón et al., 2014; He et al., 2015; Li et al., 2014). Depletion of macrophages in explanted AGM regions from mouse embryos using clodronate also decreased HSPCs (Mariani et al., 2019). Vital imaging documented macrophages intimately contacting IAHC cells, and macrophages were occasionally observed to extravasate through ECs to reach the IAHCs (Mariani et al., 2019; Travnickova et al., 2015).
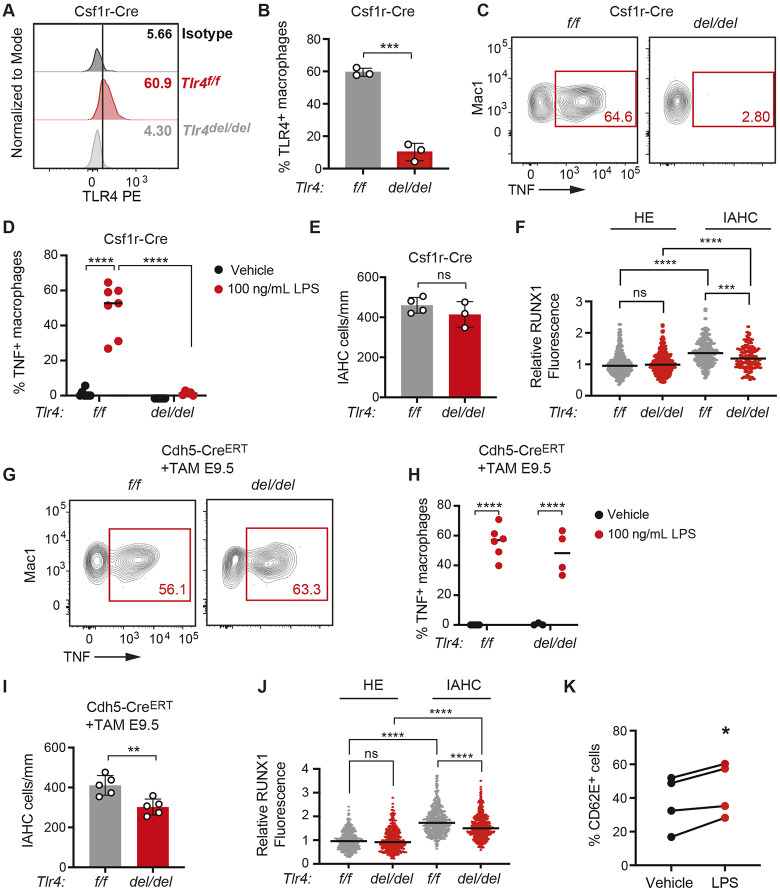
We determined whether TLR4 signaling in yolk sac-derived myeloid cells regulates the number of IAHC cells in the DA by deleting Tlr4 floxed alleles (Tlr4f/f) in EMPs using Csf1r-Cre (Gomez Perdiguero et al., 2015; Schulz et al., 2012). Csf1r-Cre should result in TLR4 loss in both yolk sac-derived macrophages and granulocytes, although as granulocytes are a minor population in the mouse embryo, macrophages are the more likely source of inflammatory cytokines (Mariani et al., 2019). We confirmed that cell surface TLR4 was depleted on macrophages following Csf1r-Cre deletion (Fig. 3A,B and Fig. S3A), and that macrophages had an impaired response to LPS stimulation ex vivo, as evidenced by a lower percentage of TNF+ cells (Fig. 3C,D and Fig. S3B). Deletion of TLR4 by Csf1r-Cre did not significantly impact the number of IAHC cells (Fig. 3E). RUNX1 levels were modestly decreased (12.1%) in IAHC cells (Fig. 3F), but whether this was due to Csf1r-Cre mediated deletion in EMPs, or in differentiating IAHC cells that expressed Csf1r-Cre could not be determined. The data indicate that the cytokines produced by primitive myeloid cells in response to TLR4 signaling do not regulate the number of IAHC cells. However, these results do not exclude the possibility of a role for primitive myeloid cells in regulating IAHC cell numbers, as other signaling pathways that regulate inflammatory cytokine production or other processes mediated by primitive myeloid cells may be responsible for their effect on hematopoietic ontogeny in the DA.
Fig. 3.
TLR4 signaling is required in endothelial and/or IAHC cells to promote IAHC cell numbers. (A) Representative histograms showing TLR4 surface expression on macrophages (CD45+Mac1+F4/80+Ter119−) in E10.5 Tlr4f/f (f/f) and Tlr4f/f;Csf1r-Cre embryos (del/del) compared with an isotype control. See Fig. S3A for gating strategy. (B) Percentage of TLR4+ macrophages (CD45+Mac-1+F4/80+) measured by flow cytometry in E10.5 Tlr4f/f (f/f) and Tlr4f/f;Csf1r-Cre embryos (del/del). Data are mean±s.d.; three independent experiments with pooled littermates; unpaired, two-tailed Student's t-test. (C) Representative flow plots of TNF+ macrophages following ex vivo stimulation of E10.5 Tlr4f/f and Tlr4f/f;Csf1r-Cre embryos with LPS. See Fig. S3B for gating strategy. (D) Percentage of TNF+ macrophages in E10.5 Tlr4f/f and Tlr4f/f;Csf1r-Cre embryos treated ex vivo with vehicle or LPS, determined by flow cytometry. Horizontal lines indicate the mean, with individual data points plotted; two-way ANOVA with Sidak's correction for multiple comparisons; n=5-7. Representative of three independent experiments. (E) Quantification of CD31+RUNX1+KIT+ IAHC cells in the DA of E10.5 Tlr4f/f and Tlr4f/f;Csf1r-Cre embryos. n=3. Data are mean±s.d. Unpaired, two-tailed Student's t-test. Each dot represents one embryo. Unpaired, two-tailed Student's t-test. (F) CTCF measuring RUNX1 intensity in HE and IAHC cells in E10.5 Tlr4f/f and Tlr4f/f;Csf1r-Cre embryos. Relative RUNX1 fluorescence was analyzed as described in Fig. 1C. HE cells, n=378 and 333; IAHC cells, n=209 and 119 for Tlr4f/f and Tlr4f/f;Csf1r-Cre, respectively. Horizontal lines indicate the mean, with individual data points plotted. Unpaired, two-tailed Student's t-test. (G) Representative flow plots of TNF+ macrophages in Tlr4f/f and Tlr4f/f;Cdh5-Cre embryos following ex vivo stimulation with LPS. Dams were injected with tamoxifen (TAM) at E9.5. (H) Percentage of TNF+ macrophages in vehicle and LPS-stimulated explanted E10.5 Tlr4f/f and Tlr4f/f;Cdh5-CreERT AGM regions. n=3-6. Representative of two independent experiments. Horizontal lines indicate the mean, with individual data points plotted. Two-way ANOVA with Sidak's correction for multiple comparisons. (I) Quantification of IAHC cells (CD31+RUNX1+KIT+) in E10.5 Tlr4f/f and Tlr4f/f;Cdh5-CreERT embryos. n=5. Data are mean±s.d. Each dot represents one embryo. Unpaired, two-tailed Student's t-test. (J) RUNX1 intensity in HE and IAHC cells in E10.5 Tlr4f/f and Tlr4f/f;Cdh5-CreERT embryos, determined as described in Fig. 1C. HE, n=389 and 739; IAHC, n=454 and 789 cells for Tlr4f/f and Tlr4f/f;Cdh5-CreERT, respectively. Horizontal lines indicate the mean, with individual data points plotted. Unpaired, two-tailed Student's t-test. (K) Percentage of CD62E+ ECs measured by flow cytometry following a 6 h stimulation of FACS-purified ECs with LPS (100 ng/ml). Data represent four independent experiments. Paired Student's t-test, two-tailed. *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001, ns indicates not significant.
We next assessed whether TLR4 signaling in endothelial or IAHC cells was necessary for generating normal numbers of IAHC cells by deleting Tlr4 with an endothelial cell-specific, tamoxifen-regulated Cre (Cdh5- CreERT) (Sørensen et al., 2009a). We timed the tamoxifen injections in pregnant dams to preferentially delete TLR4 in arterial ECs and IAHCs, but not in EMPs. EMPs differentiate from HE cells in the yolk sac vascular plexus between E8.5 and E9.5 (Frame et al., 2015), immediately prior to the major wave of EHT in arterial endothelium that produces IAHC cells (Tober et al., 2013; Yokomizo and Dzierzak, 2010). We previously performed a temporal analysis of Cdh5-CreERT activity following tamoxifen injection and found that deletion in EMPs could be mostly avoided by injecting tamoxifen at E9.0 or later in development (Tober et al., 2013). Therefore, to delete TLR4 in arterial ECs while sparing EMPs, we injected tamoxifen into pregnant dams at E9.5 and analyzed HE and IAHC cells 1 day later in Tlr4f/f and Tlr4f/f; Cdh5-CreERT embryos. We determined that the percentage of TNF+ macrophages was equivalent in E10.5 Tlr4f/f and Tlr4f/f; Cdh5-CreERT AGM regions stimulated ex vivo with LPS, confirming that deletion of Tlr4 using a carefully timed activation of Cdh5-CreERT has minimal impact on EMPs or their progeny (Fig. 3G,H). The number of IAHC cells at E10.5 was significantly decreased by 27% following deletion of Tlr4f/f alleles at E9.5 by Cdh5-CreERT. RUNX1 levels in IAHC cells were reduced by ∼14.0% , similar to the decrease caused by deletion with Csf1r-Cre (Fig. 3I,J). Deletion of Tlr4 in endothelial cells at E9.5 did not result in a phenotype as robust as that in Tlr4−/− embryos, suggesting that TLR4 function in either non-endothelial cells or endothelial cells prior to E9.5 contributes to the IAHC cell phenotype. Overall, however, the data indicate that TLR4 is required in arterial ECs and/or IAHC cells to generate normal numbers of IAHC cells.
To confirm that TLR4 signaling can be directly activated in embryonic ECs we purified CD31hiCD144+ESAM+CD44−CD45−Ter119−Mac1−CD41− ECs, stimulated them ex vivo with LPS and analyzed expression of the integrin CD62E (E-selectin, encoded by Sele), which is a direct NF-κB target gene (Collins et al., 1995), by flow cytometry. The percentage of CD62E+ ECs increased following stimulation with LPS, indicating that embryonic ECs respond to TLR4 signals (Fig. 3K and Fig. S3C). We were unable to reliably detect TLR4 by flow cytometry on the surface of ECs as its levels were low. However, as signaling through TLR4 on the cell surface requires MyD88, and the genetic data indicate that TLR signaling through MyD88 but not TRIF adaptors regulates IAHC numbers, we conclude that the relevant signaling is occurring through extracellular TLR4 signaling.
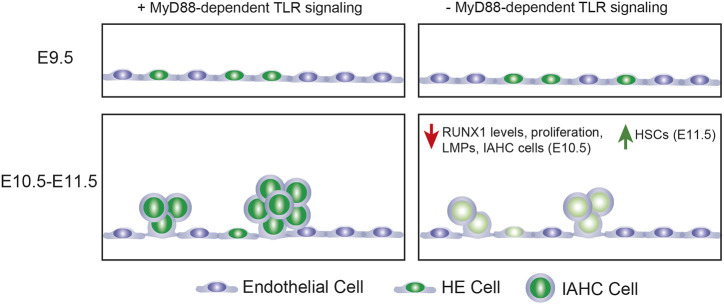
In summary, the number of IAHC cells and LMPs at E10.5 are positively regulated by MyD88-dependent TLR signaling (Fig. 4). The number of specified HE cells at E9.5, on the other hand, is not. The selective decrease in LMPs in the absence of MyD88-dependent signaling inversely correlates with an increase in adult-repopulating HSCs, indicating that MyD88-dependent TLR signaling oppositely regulates the numbers of stem cells and progenitors with different potentials (Fig. 4). MyD88-dependent TLR signaling is required in ECs or in the IAHC cells that differentiate from HE cells to regulate IAHC cell numbers. Recent work suggests multipotent progenitors and HSCs arise from distinct subsets of HE cells (Dignum et al., 2021). It is unclear whether loss of TLR signaling impacts the specification of these distinct subsets of HE cells or the formation of committed progenitors and HSCs from them. Alternatively (or additionally) MyD88-dependent TLR signaling may differentially affect the number of LMPs and HSCs by regulating their proliferation. TLR4 signaling in adult HSCs increases proliferation but reduces fitness (Takizawa et al., 2017), and the same may be true for embryonic HSCs.
Fig. 4.
Summary of results. Loss of Myd88-dependent TLR signaling does not affect the specification of HE cells at E9.5. At E10.5, loss of Myd88-dependent TLR signaling decreases RUNX1 levels in IAHC and HE cells, decreases the number of LMPs, and decreases the proliferation and number of IAHC cells. At E11.5, loss of Myd88-dependent TLR signaling increases HSCs.
Haploinsufficiency for RUNX1 decreases the number of IAHC cells and results in the precocious emergence of HSCs in the embryo (Cai et al., 2000). However, RUNX1 haploinsufficiency also decreases the number of HE cells (Zhu et al., 2020), whereas loss of MyD88-dependent TLR signaling does not. HE cells differentiate from a precursor population called pre-HE, and the efficiency at which pre-HE differentiates into HE is regulated by RUNX1 dose (Zhu et al., 2020). We believe the reason loss of MyD88-dependent TLR signaling does not reduce the number of HE cells is that RUNX1 levels are higher in mutant embryos at E9.5 when HE specification occurs. The subsequent decrease in RUNX1 levels in HE and IAHC cells at E10.5 may contribute to the lower number and proliferation of IAHC cells, and reflect a later requirement for RUNX1. RUNX1 promotes the G1- to S-phase transition in several contexts (Cai et al., 2011; Nimmo et al., 2005; Osorio et al., 2008), and loss of RUNX1 decreases proliferation, consistent with these observations.
Our data are consistent with those from zebrafish embryos that demonstrated lower levels of runx1 expression and decreased numbers of HSPCs upon loss of MyD88-dependent TLR signaling (He et al., 2015), but add an extra layer of complexity. They also provide a cautionary note for investigators studying HSC ontogeny – the level of RUNX1 expression, and the numbers of IAHC cells and progenitors do not always correlate with the number of HSCs.
MATERIALS AND METHODS
Mouse lines and embryo production
Tlr4+/− (B6(Cg)-Tlr4tm1.2Karp/J), Tlr4f/f (B6(Cg)-Tlr4tm1.1Karp/J), Csf1rCre (C57BL/6-Tg(Csf1r-cre)1Mnz/J), Ticam1+/− (C57BL/6J-Ticam1Lps2/J) and B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mice were purchased from Jackson Laboratory. Tg(Cdh5-cre/ERT2)1Rha mice (Sörensen et al., 2009b) were a gift from Ralf Adamson (Max Planck Institute for Molecular Biomedicine, Münster, Germany). Myd88+/− mice were a gift from Edward Behrens (Children's Hospital of Philadelphia, PA, USA).
Animal husbandry
The morning post-mating is considered embryonic day (E) 0.5. E9.5-E11.5 embryos were staged at the time of harvest by counting somites. Embryos that showed abnormal development were discarded. Mice were handled according to protocols approved by the University of Pennsylvania's Institutional Animal Care and Use Committee and housed in a specific-pathogen-free facility.
Cell sorting and flow cytometry
Antibodies used can be found in Table S1. All antibodies were used at a concentration of 1:200. DAPI, LIVE/DEAD Fixable Aqua (ThermoFisher L34957) or Fixable Viability Stain 780 (BD Biosciences 565388) were used to exclude dead cells. We used the Cytofix/Cytoperm Kit (BD Biosciences) for intracellular flow cytometry to detect TNF.
Yolk sacs were removed from embryos, and vitelline vessels were retained with the embryonic tissue. The head, cardiac and pulmonary regions, liver, digestive tube, tail and limb buds were removed. The remaining tissue containing the aorta-gonad-mesonephros (AGM) region, parts of somite, umbilical and vitelline vessels was collected. Tissues were dissected in phosphate-buffered saline (PBS)/10% fetal bovine serum (FBS) and penicillin/streptomycin (Sigma), followed by dissociation in 0.125% collagenase (Sigma) for 1 h. Tissues were washed and filtered through a 40 μm filter and resuspended in antibody solution. Cells were sorted on a BD Influx equipped with a 100 μm nozzle, run at a pressure of 17 psi with flow rates of less than 4000 events/second, and collected in PBS/20% FBS/25 mM HEPES. Flow cytometry was performed on a LSR II (BD Biosciences) and data were analyzed using FlowJo software (v10.7, BD Biosciences).
Ex vivo LPS stimulation
TNF production by embryonic macrophages was analyzed by incubating dissected AGM regions from E10 embryos ex vivo with 100 ng/ml LPS for 4 h in RPMI medium in the presence of a Golgi inhibitor (BD Biosciences 555029). Collagenase (0.125%) was added for the last 30 min of incubation. After 4 h, dissociated cells were stained with extracellular antibodies, fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences), stained for intracellular TNF, and analyzed by flow cytometry.
T- and B-cell progenitor assays
OP9 and OP9-DLL1 stromal cells were maintained in αMEM containing 20% FBS and antibiotics, and plated into 96-well plates at a concentration of 5500 cells/well 1 day before dilutions of the dissociated embryonic tissues were added. Medium was supplemented with purified recombinant 5 ng/ml Flt3L and 10 ng/ml IL7 (OP9) or 5 ng/ml Flt3L and 1 ng/ml IL7 (OP9-DLL1) (Peprotech). Cells were harvested and analyzed by flow cytometry 12 days after culture. B cells were identified as CD19+ B220+, T cells as CD25+ Thy1.1+. The progenitor frequency was calculated using the method of maximum likelihood applied to Poisson distribution with L-Calc software (StemCell Technologies). OP9s were purchased from ATCC, and OP9-DLL1s were obtained from Juan Carlos Zún̋iga-Pflücker (Sunnybrook Research Institute, Toronto, Canada). Both OP9s and OP-Dll1 cells were tested for mycoplasma and found to be negative.
Limiting dose HE cell assay
OP9 stromal cells were cultured in αMEM with 20% FBS and antibiotics. One day before sorting, OP9s were plated at a concentration of 5000 cells/well in a 96-well plate. Tlr4+/− and Tlr4−/− embryos generated from a cross of Tlr4+/− and Tlr4−/− mice were used in this experiment in order to have a sufficient number of endothelial cells for the analysis. Endothelial cells were sorted on a BD Influx into individual wells with a maximum of five cells sorted into each well. Medium was supplemented with 10 ng/ml Flt3L, SCF, IL3 and IL7 (Peprotech). Cells were cultured for 10 days before being scored by eye for hematopoietic cells, and positive wells were harvested and analyzed by flow cytometry. Positive wells contained CD45+, CD41+, Mac1+, B220+ and/or Ter119+ cells.
EdU labeling
Yolk sacs were removed and intact embryos placed in αMEM containing 20% FBS and antibiotics. Embryos were incubated in a 24-well plate at 37°C for 2 h in EdU (5-ethynyl-2′-deoxyuridine, 100 µM). Immediately following incubation with EdU, embryos were dissociated using 0.125% collagenase (Sigma) for 30 min and stained for flow cytometry. EdU detection was performed using the Click-iT Plus EdU Flow Cytometry Assay kit (Thermo Fisher Scientific, C10632).
Transplant analyses
B6.SJL-PtprcaPepcb/BoyJ (CD45.1) mice were subjected to a split dose of 900 cGy, 3 h apart. Each recipient received 1.0 or 0.3 embryo equivalents (ee) of E11.5 AGM regions (CD45.2), vitelline and umbilical arteries with 2.5×105 carrier spleen cells by retro-orbital injection. We assessed donor (CD45.2) engraftment in peripheral blood at weeks 4, 8, 12 and 16, and in bone marrow at week 16 post-transplantation. HSC frequencies were determined by extreme limiting dilution analysis (ELDA) (Hu and Smyth, 2009).
Confocal microscopy
Embryos were prepared as previously described (Yokomizo et al., 2012). A Zeiss LSM 880 AxioObserver inverted confocal microscope with ZEN 2.3 SP1 software was used to acquire Z- and single optical projections. Images were processed using Fiji software (Schindelin et al., 2012). All primary antibodies were used at a concentration of 1:500, except for KIT, which was used at 1:250. All secondaries were used at a concentration of 1:1000. Corrected total cell fluorescence (CTCF) was calculated to measure RUNX1 intensity in HE and IAHC cells by measuring the integrated density value for each cell using FIJI software. CTCF was calculated as [integrated density of cell−(area of selected cell×mean intensity of background)]. Power analysis determined that detection of a 10% change in the intensity of RUNX1 levels at P≤0.01 required 220 cells per genotype.
Statistics
Significance for multiple comparisons was determined using ordinary one-way ANOVA with Tukey's test or two-way ANOVA with Sidak's correction for multiple comparisons. A two-tailed Student's t-test was used for pairwise comparisons. All statistics were performed using Prism software (v. 9.0.0). For all tests, *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
Supplementary Material
Acknowledgements
We thank Andrea Stout and Jasmine Zhao at the Cell and Developmental Biology Microscopy Core for confocal microscopy assistance. We thank Andrew Morschauser and Charles Pletcher at the Penn Cytometrics and Cell Sorting Resource Laboratory for their assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.A.S.; Formal analysis: L.F.B., M.D.M., Y.L.; Investigation: L.F.B., M.D.M., Y.L.; Writing - original draft: N.A.S.; Writing - review & editing: L.F.B., M.D.M., Y.L.; Project administration: N.A.S.; Funding acquisition: N.A.S.
Funding
This work was supported by the National Institutes of Health (R01HL091724, T32HL007439 to L.F.B and T32DK007780 to M.D.M.). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200025.
References
- Auerbach, R., Huang, H. and Lu, L. (1996). Hematopoietic stem cells in the mouse embryonic yolk sac. Stem Cells 14, 269-280. 10.1002/stem.140269 [DOI] [PubMed] [Google Scholar]
- Bollerot, K., Pouget, C. and Jaffredo, T. (2005). The embryonic origins of hematopoietic stem cells: a tale of hemangioblast and hemogenic endothelium. APMIS 113, 790-803. 10.1111/j.1600-0463.2005.apm_317.x [DOI] [PubMed] [Google Scholar]
- Cai, Z., de Bruijn, M. F. T. R., Ma, X., Dortland, B., Luteijn, T., Downing, J. R. and Dzierzak, E. (2000). Haploinsufficiency of AML1/CBFA2 affects the embryonic generation of mouse hematopoietic stem cells. Immunity 13, 423-431. 10.1016/S1074-7613(00)00042-X [DOI] [PubMed] [Google Scholar]
- Cai, X., Gaudet, J. J., Mangan, J. K., Chen, M. J., De Obaldia, M. E., Oo, Z., Ernst, P. and Speck, N. A. (2011). Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS ONE 6, e28430. 10.1371/journal.pone.0028430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, T., Read, M. A., Neish, A. S., Whitley, M. Z., Thanos, D. and Maniatis, T. (1995). Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J. 9, 899-909. [PubMed] [Google Scholar]
- Dignum, T., Varnum-Finney, B., Srivatsan, S. R., Dozono, S., Waltner, O., Heck, A. M., Ishida, T., Nourigat-McKay, C., Jackson, D. L., Rafii, S.et al. (2021). Multipotent progenitors and hematopoietic stem cells arise independently from hemogenic endothelium in the mouse embryo. Cell Rep. 36, 109675. 10.1016/j.celrep.2021.109675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín-Palazón, R., Stachura, D. L., Campbell, C. A., Garcia-Moreno, D., Del Cid, N., Kim, A. D., Candel, S., Meseguer, J., Mulero, V. and Traver, D. (2014). Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 159, 1070-1085. 10.1016/j.cell.2014.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, J. M., Fegan, K. H., Conway, S. J., McGrath, K. E. and Palis, J. (2015). Definitive hematopoiesis in the Yolk Sac emerges from Wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells 34, 431-444. 10.1002/stem.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, J. M., Kubaczka, C., Long, T. L., Esain, V., Soto, R. A., Hachimi, M., Jing, R., Shwartz, A., Goessling, W., Daley, G. Q.et al. (2020). Metabolic regulation of inflammasome activity controls embryonic hematopoietic stem and progenitor cell production. Dev. Cell 55, 133-149.e36. 10.1016/j.devcel.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero, E., Klapproth, K., Schulz, C., Busch, K., Azzoni, E., Crozet, L., Garner, H., Trouillet, C., de Bruijn, M. F., Geissmann, F.et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547-551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland, B. and Yoshimoto, M. (2018). Many layers of embryonic hematopoiesis: new insights into B-cell ontogeny and the origin of hematopoietic stem cells. Exp. Hematol. 60, 1-9. 10.1016/j.exphem.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., Zhang, C., Wang, L., Zhang, P., Ma, D., Lv, J. and Liu, F. (2015). Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood 125, 1098-1106. 10.1182/blood-2014-09-601542 [DOI] [PubMed] [Google Scholar]
- Hu, Y. and Smyth, G. K. (2009). ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70-78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Kumaravelu, P., Hook, L., Morrison, A. M., Ure, J., Zhao, S., Zuyev, S., Ansell, J. and Medvinsky, A. (2002). Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 129, 4891-4899. 10.1242/dev.129.21.4891 [DOI] [PubMed] [Google Scholar]
- Lefkopoulos, S., Polyzou, A., Derecka, M., Bergo, V., Clapes, T., Cauchy, P., Jerez-Longres, C., Onishi-Seebacher, M., Yin, N., Martagon-Calderón, N. A.et al. (2020). Repetitive elements trigger RIG-I-like receptor signaling that regulates the emergence of hematopoietic stem and progenitor cells. Immunity 53, 934-951.e39. 10.1016/j.immuni.2020.10.007 [DOI] [PubMed] [Google Scholar]
- Li, Y., Esain, V., Teng, L., Xu, J., Kwan, W., Frost, I. M., Yzaguirre, A. D., Cai, X., Cortes, M., Maijenburg, M. W.et al. (2014). Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28, 2597-2612. 10.1101/gad.253302.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani, S. A., Li, Z., Rice, S., Krieg, C., Fragkogianni, S., Robinson, M., Vink, C. S., Pollard, J. W. and Dzierzak, E. (2019). Pro-inflammatory aorta-associated macrophages are involved in embryonic development of hematopoietic stem cells. Immunity 50, 1439-1452.e35. 10.1016/j.immuni.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlish, D. A., Bhatt, S. T. and Schuettpelz, L. G. (2016). The role of toll-like receptors in hematopoietic malignancies. Front. Immunol. 7, 390. 10.3389/fimmu.2016.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo, R., Antebi, A. and Woollard, A. (2005). mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development 132, 5043-5054. 10.1242/dev.02102 [DOI] [PubMed] [Google Scholar]
- North, T. E., Gu, T.-L., Stacy, T., Wang, Q., Howard, L., Binder, M., Marín-Padilla, M. and Speck, N. A. (1999). Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126, 2563-2575. [DOI] [PubMed] [Google Scholar]
- Osorio, K. M., Lee, S. E., McDermitt, D. J., Waghmare, S. K., Zhang, Y. V., Woo, H. N. and Tumbar, T. (2008). Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development 135, 1059-1068. 10.1242/dev.012799 [DOI] [PubMed] [Google Scholar]
- Rybtsov, S., Sobiesiak, M., Taoudi, S., Souilhol, C., Senserrich, J., Liakhovitskaia, A., Ivanovs, A., Frampton, J., Zhao, S. and Medvinsky, A. (2011). Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 208, 1305-1315. 10.1084/jem.20102419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtsov, S., Ivanovs, A., Zhao, S. and Medvinsky, A. (2016). Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development 143, 1284-1289. 10.1242/dev.131193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, B., Arranz, A., Francisco, S., Córdoba, L., Punzón, C., Llamas, M. A. and Fresno, M. (2016). Modulation of endothelial function by Toll like receptors. Pharmacol. Res. 108, 46-56. 10.1016/j.phrs.2016.03.038 [DOI] [PubMed] [Google Scholar]
- Sawamiphak, S., Kontarakis, Z. and Stainier, D. Y. R. (2014). Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev. Cell 31, 640-653. 10.1016/j.devcel.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, T. M. and Zúñiga-Pflücker, J. C. (2002). Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749-756. 10.1016/S1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- Schulz, C., Gomez Perdiguero, E., Chorro, L., Szabo-Rogers, H., Cagnard, N., Kierdorf, K., Prinz, M., Wu, B., Jacobsen, S. E., Pollard, J. W.et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86-90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Sørensen, A. L., Rumjantseva, V., Nayeb-Hashemi, S., Clausen, H., Hartwig, J. H., Wandall, H. H. and Hoffmeister, K. M. (2009a). Role of sialic acid for platelet life span: exposure of beta-galactose results in the rapid clearance of platelets from the circulation by asialoglycoprotein receptor-expressing liver macrophages and hepatocytes. Blood 114, 1645-1654. 10.1182/blood-2009-01-199414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörensen, I., Adams, R. H. and Gossler, A. (2009b). DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680-5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Takizawa, H., Fritsch, K., Kovtonyuk, L. V., Saito, Y., Yakkala, C., Jacobs, K., Ahuja, A. K., Lopes, M., Hausmann, A., Hardt, W.-D.et al. (2017). Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell 21, 225-240.e25. 10.1016/j.stem.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Tober, J., Yzaguirre, A. D., Piwarzyk, E. and Speck, N. A. (2013). Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765-3776. 10.1242/dev.094961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova, J., Tran Chau, V., Julien, E., Mateos-Langerak, J., Gonzalez, C., Lelièvre, E., Lutfalla, G., Tavian, M. and Kissa, K. (2015). Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 6, 6227. 10.1038/ncomms7227 [DOI] [PubMed] [Google Scholar]
- Yokomizo, T. and Dzierzak, E. (2010). Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development 137, 3651-3661. 10.1242/dev.051094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo, T., Yamada-Inagawa, T., Yzaguirre, A. D., Chen, M. J., Speck, N. A. and Dzierzak, E. (2012). Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat. Protoc. 7, 421-431. 10.1038/nprot.2011.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q., Gao, P., Tober, J., Bennett, L., Chen, C., Uzun, Y., Li, Y., Howell, E. D., Mumau, M., Yu, W.et al. (2020). Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 136, 845-856. 10.1182/blood.2020004801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.