Abstract
Background
Most cystic echinococcosis cases in Southern Brazil are caused by Echinococcus granulosus and Echinococcus ortleppi. Proteomic studies of helminths have increased our knowledge about the molecular survival strategies that are used by parasites. Here, we surveyed the protein content of the hydatid fluid compartment in E. granulosus and E. ortleppi pulmonary bovine cysts to better describe and compare their molecular arsenal at the host-parasite interface.
Methods
Hydatid fluid samples from three isolates of each species were analyzed using mass spectrometry-based proteomics (LC-MS/MS). In silico functional analyses of the identified proteins were performed to examine parasite survival strategies.
Results
The identified hydatid fluid protein profiles showed a predominance of parasite proteins compared to host proteins that infiltrate the cysts. We identified 280 parasitic proteins from E. granulosus and 251 from E. ortleppi, including 52 parasitic proteins that were common to all hydatid fluid samples. The in silico functional analysis revealed important molecular functions and processes that are active in pulmonary cystic echinococcosis, such as adhesion, extracellular structures organization, development regulation, signaling transduction, and enzyme activity.
Conclusions
The protein profiles described here provide evidence of important mechanisms related to basic cellular processes and functions that act at the host-parasite interface in cystic echinococcosis. The molecular tools used by E. granulosus and E. ortleppi for survival within the host are potential targets for new therapeutic approaches to treat cystic echinococcosis and other larval cestodiases.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-022-05232-8.
Keywords: Echinococcus granulosus, Echinococcus ortleppi, Secretome, Hydatid fluid, Parasite proteomics, Host-parasite interface
Background
Echinococcosis is caused by infection with flatworms from the genus Echinococcus. Depending on the species causing the infection, distinct morphological features can be observed because of differences in larval stage development [1]. Presently, the E. granulosus sensu lato (s.l.) complex is formed by five species: Echinococcus granulosus sensu stricto (s.s.; G1, G2 and G3), Echinococcus equinus (G4) Echinococcus ortleppi (G5), Echinococcus canadensis (G6-G8, G10), and Echinococcus felidis [2–5]. Echinococcus granulosus (s.s.) (G1, sheep strain) and E. ortleppi (G5, cattle strain) are etiological agents of cystic echinococcosis, which is characterized by the growth of the parasite’s larval stage (metacestode) as an unilocular, fluid-filled cyst (the hydatid cyst) in the viscera of suitable intermediate hosts (mainly cattle and sheep). Humans can be accidental hosts and develop cystic echinococcosis [3, 6]. In terms of epidemiology, E. granulosus (s.s.) is the most relevant species due to its worldwide occurrence, with high prevalence in domestic animals and humans [7]. Echinococcus ortleppi seems to be well adapted to cattle, although other intermediate hosts, including humans, can also be infected by this species [7, 8]. Echinococcus ortleppi ortleppi differs markedly in both larval and adult morphology from that of E. granulosus (s.s.), presenting a short development time in dogs [6, 9].
In E. granulosus and E. ortleppi life cycles [10], intermediate hosts become infected upon ingestion of parasite eggs. Egg hatching releases oncospheres, which develop into hydatid cysts in the host viscera (mainly liver and lungs). The hydatid cyst wall is formed by an external acellular, mucin-based laminated layer and an internal germinative layer. The germinative layer gives rise to brood capsules, where pre-adults (protoscoleces; PSCs) are produced by asexual reproduction. When PSCs are ingested by definitive hosts (canids, such as domestic dogs or wolves), they mature into adult worms within the small intestine, where they produce eggs that are released into the environment with host feces.
The hydatid cyst causes a chronic infection because it can survive and grow for decades in the host, in most cases remaining fertile, with full capacity to generate PSCs [4]. To achieve this, the parasites adopt a wide repertoire of molecular strategies to evade host defense mechanisms and acquire nutrients necessary for their development [11]. Such strategies allow parasite survival and development despite chronic exposure to a hostile environment created by the host response against infection. The liquid that fills the hydatid cyst, the hydatid fluid (HF), contains parasite excretory-secretory (ES) products and host proteins, making it a good component from which to analyze relevant molecules [12–14]. Although the HF is an inner component of the metacestode, it contains proteins that interact with the host. This can be evidenced by the humoral response to HF antigens detected in the host serum [11, 15]. Also, the germinative layer has secretory activity in its outer surface, since the presence of 14-3-3 and enolase in the laminated layer has already been observed [16, 17]. Recently, E. granulosus exosomes were detected in serum from patients with cystic echinococcosis [18], and the interaction of extracellular vesicles produced by Echinococcus with mammalian cells have been demonstrated in vitro [19]. Extracellular vesicles are carriers for different biomolecules and could act in the transfer of proteins through the hydatid cyst wall.
Despite its preference for ovine hosts, E. granulosus can also successfully infect, grow, and asexually reproduce in bovine hosts, although with less efficiency than E. ortleppi [4, 20]. For bovine hosts, the E. ortleppi cyst fertility rate is high (> 90%), while for E. granulosus, it normally does not exceed 30% [6, 21–23]. Moreover, E. ortleppi develops preferentially in bovine lungs, whereas E. granulosus cysts are located in the liver and lungs [8, 24–26]. Therefore, E. granulosus and E. ortleppi infections in bovines offer the opportunity to analyze two related species with different degrees of adaptation to a single host species.
Molecular characterization of the HF content is essential for a better understanding of Echinococcus spp infections. Proteomic studies of helminth ES products have been particularly valuable for identifying proteins involved in the host-parasite relationship [27–29]. Previous proteomic studies of Echinococcus ES products included analysis of different E. granulosus cyst components [12, 30], comparisons among hydatid cyst fluid of E. granulosus cysts from different hosts (sheep, cattle, and humans) [13], and comparison of HF from two different isolates of Echinococcus multilocularis, the etiological agent of alveolar echinococcosis [31]. Within the genus Echinococcus, proteomic studies involving interspecies comparisons have been performed only between E. granulosus and E. multilocularis [32]. These studies showed that analyses of the same species infecting different hosts and different genotypes/species/strains infecting a common host can provide valuable insight into molecular survival strategies adopted by parasites. The discovery of proteins shared by distinct species allows identification of conserved mechanisms involved in their interactions with the respective hosts. Furthermore, a species-specific set of proteins can provide molecular markers for parasite diagnosis.
In the present study, we generated MS protein profiles of HF samples from E. granulosus and E. ortleppi cattle pulmonary cysts. The identified proteins outlined a variety of molecular processes acting in cystic echinococcosis, helping to better understand different aspects of the infection, including parasite survival strategies and host defenses. The generated results will assist the selection of potential targets for new therapeutic approaches and of disease markers capable of differentiating between the two etiological agents.
Methods
Biologic material
Echinococcus ortleppi granulosus and E. ortleppi hydatid cysts were from lungs of cattle obtained at a commercial abattoir in the metropolitan region of Porto Alegre, RS (Brazil). Animal slaughtering was conducted according to Brazilian laws and under the supervision of the Serviço de Inspeção Federal (Brazilian Sanitary Authority) of the Brazilian Ministério da Agricultura, Pecuária e Abastecimento. Contaminated viscera, identified during mandatory meat inspection, were donated by the abattoir for use in this work.
Lungs were dissected, and HF was aspirated from the hydatid cysts. The HF recovered from individual cysts was centrifuged at 10,000 ×g for 15 min at 4 °C to sediment PSCs and debris [12]. Only HF samples from fertile cysts, i.e. with viable PSCs, were used in the study. The PSC DNAs were used for species identification by high-resolution melting (HRM), using a 444-bp fragment of the cytochrome c oxidase subunit I (cox1) gene, and the amplification was carried out with the primers 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ (forward) and 5′-TAAAGAAAG AACATAATGAAAATG-3′ (reverse), as previously described [33]. Thirty-four E. granulosus and 29 E. ortleppi HF samples were qualitatively evaluated using 12% SDS-PAGE gel. The intensity of the bovine albumin band, estimated by using IMAGEJ (https://imagej.nih.gov/ij/) to quantify band intensity, was correlated to the cyst volumes. Spearman's rank correlation test was used to estimate the correlation, as previously described [14] (Additional file 1: Figure S1). We selected three individual E. granulosus and three individual E. ortleppi HF samples (EG1–3 and EO1–3, respectively) with low quantity of albumin for the proteomic analysis.
Sample preparation and mass spectrometry analysis
Each HF sample protein concentration was determined using Qubit™ (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Proteins were digested in solution using trypsin and fractionated using strong cation exchange (SCX) [14]. To release peptides, 5 mM phosphate buffer (pH 3.0) was added to the SCX columns with a salt gradient, as follows: 75 mM KCl (fraction A), 125 mM KCl (fraction B), 200 mM KCl (fraction C), 300 mM KCl (fraction D), and 400 mM KCl (fraction E). Each fraction was lyophilized and stored at − 80 °C until liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
The five resulting SCX fractions from each one of the six biological samples were analyzed individually, totalizing 30 LC-MS/MS runs. The tryptic peptide mixture corresponding to each SCX fraction was automatically loaded onto a C18 Jupiter pre-column (Phenomenex; bead diameter 10 μm; 100 μm × 50 mm; Phenomenex, Torrance, CA, USA) by an Easy-nLCII nano HPLC system (Thermo Fisher Scientific, Inc., Waltham, MA, USA) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). After loading the samples in solvent A (0.1% formic acid), the peptides were subjected to chromatographic separation in reverse-phase using a C18 AQUA column (Phenomenex; beads diameter 5 μm; 75 μm × 100 mm). Both the pre-column and analytical column were packed in house. The peptides were eluted on a gradient of 5%–35% solvent B (0.1% formic acid in acetonitrile) for 60 min; 35%–85% B for 5 min; 85% B for 5 min; 85%–5% B for 2 min; and 5% B in 13 min, under a flow of 200 nl/min. Spray voltage was set at 1.8 kV and 200 °C, and the mass spectrometer was operated in the positive, data-dependent mode, in which one full MS scan was acquired in the m/z range of 300–1800 followed by MS/MS acquisition using collisional induced dissociation (CID) of the ten most intense ions from the MS scan using an isolation window width of 3 m/z. MS spectra were acquired in the Orbitrap analyzer at 30,000 resolution (at 400 m/z). Dynamic exclusion was defined by a list size of 500 and exclusion duration of 90 s at a repetition intervals of 30 s. For the survey (MS) scan, an automatic gain control (AGC) target value of 1,000,000 and maximum injection time of 100 ms were set whereas the target value for the fragment ion (MS/MS) spectra was set to 10,000 and maximum injection time of 100 ms. The lower threshold for targeting precursor ions in the MS scans was 200 counts per scan. The raw files (*.raw) from the MS and MS/MS spectra were converted to the extension *.mgf (mascot generic format) using the MSconvert software (available at http://proteowizard.sourceforge.net).
Database search and MS data analysis
For protein identification, the generated LC-MS/MS data were used to search local databases containing the known amino acid sequences from the E. granulosus genome assembly (PRJEB121), version WBPS11, available at WormBase ParaSite (http://parasite.wormbase.org), and the Bos taurus protein sequences obtained from UniProt/Swiss-Prot (Proteome ID: UP000009136).
Mascot Search Engine v. 2.3.02 (Matrix Science, London, UK) was used for peptide and protein identification. The search parameters consisted of carbamidomethylation as a fixed modification, oxidation of methionine as a variable modification, two trypsin missed cleavage, and a tolerance of 10 ppm for precursor and 1 Da for fragment ions. Ion type was set as monoisotopic, and 2 +, 3 +, and 4 + peptide charges were taken into account.
Peptide and protein identification was validated using Scaffold v. 4.8.7 (Proteome Software Inc., Portland, OR, USA). The peptide identification was accepted if it could be established with > 95% probability. Protein identification was accepted if it could be established at > 99% probability and contained two unique identified peptides. The false discovery rate (FDR) was 0.9% and 0.0% for proteins and peptides, respectively. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [34] partner repository with the dataset identifier PXD019314 and https://doi.org/10.6019/PXD019314.
Some histones (proteins that are highly conserved in eukaryotes) did not fulfill the criteria of at least two unique peptides when the identifications obtained using each database, E. granulosus, or B. taurus were compared (Additional File 2: Table S1). Because we were unable to definitively determine their organism of origin, histones H4 (EgrG_000323100 and E1BBP7), H2A (EgrG_002051500 and A0A0A0MP90), and H2B (E1BGW2) were removed from further analysis.
Normalized spectral abundance factor (NSAF), acquired using Scaffold, was used to quantify the differences in protein abundance between samples [35]. To determine statistical differences between E. granulosus and E. ortleppi shared protein NSAF values, we performed a Student’s t-test and P-value correction using the Benjamini and Hochberg FDR. A heat map analysis was performed using the Matrix2png web interface (https://matrix2png.msl.ubc.ca/) with NSAF values for all identified proteins.
Prediction of secretion pathways
The identified parasite proteins were searched for the presence of a secretion signal peptide using SignalP 4.1, PrediSi, and SecretomeP 2.0. The presence of an alternative signal for exportation was verified using SecretomeP 2.0. A protein was considered to contain a classical signal peptide when two of the three software programs detected a signal peptide sequence. Proteins that did not meet this criterion, but showed a neural network score (NN score) > 0.6 in SecretomeP, were considered to be alternatively secreted proteins. Those that did not meet any of the previous parameters comprised the group of proteins with an unidentified secretion pattern.
Functional annotation
Parasitic and bovine proteins were subjected to Gene Ontology (GO) enrichment analysis. The analysis was performed using the total protein repertoire from each species, using the Cytoscape plugin BiNGO [36]. The ontology files were retrieved from GO database, while Wellcome Trust Sanger Institute (UK) kindly provided the files associated with E. granulosus protein annotation. Functional enrichment analyses were performed using hypergeometric distribution and P-value correction with Benjamini and Hochberg FDR. Values of P ≤ 0.05 were considered statistically significant.
The software ESG (extended similarity group) and PFP (protein function prediction), both available at https://kiharalab.org/web/software.php, were used to functionally annotate proteins with an unknown function [37]. The GO terms predicted for a determined protein were considered valid results when they were identified in both ESG and PFP.
The platform REVIGO (http://revigo.irb.hr/) was used to remove redundant GO terms and summarize GO term lists [38]. The semantic similarity of the GO terms was calculated using SimRel (allowed similarity = 0.5).
Results
Protein profiles from E. granulosus and E. ortleppi hydatid fluid samples
A proteomic survey was performed to describe the HF protein components of E. granulosus and E. ortleppi. Because E. ortleppi develops predominantly in lungs [8, 25] and to minimize differences in the protein profiles due to hydatid cyst location or the host species, we only used samples from pulmonary bovine infections. The number of identified proteins in the three biological replicates, i.e. HF samples from individual fertile hydatid cysts (EG1–3, for E. granulosus; EO1–3, for E. ortleppi) are summarized in Fig. 1. To visualize the overall sample composition, a heat map analysis was performed using NSAF values of all identified proteins (Additional File 3: Figure S2). The number of proteins identified varied among individual samples from each species. We identified 207, 230, and 78 parasitic proteins in EG1, EG2, and EG3 HF samples, respectively, and overall, 280 E. granulosus unique proteins were identified (Additional File 4: Table S2A). In E. ortleppi HF samples, we identified 251 unique parasitic proteins, of which 194 were found in EO1, 224 in EO2, and 123 in EO3 (Additional File 4: Table S2B). Overall, 214 proteins were shared between E. granulosus and E. ortleppi, 66 proteins were found exclusively in E. granulosus, and 37 proteins were found exclusively in E. ortleppi, totaling 317 proteins. Exclusive proteins identified in E. granulosus and E. ortleppi are shown in Tables 1 and 2, respectively. Proteins in the shared group did not show differences in abundance between E. granulosus and E. ortleppi, indicating that the two species may employ similar molecular strategies at the host-parasite interface (Additional File 3: Figure S2 and Additional File 5: Table S3).
Fig. 1.
Parasitic proteins identified in HF samples from pulmonary cystic echinococcosis. Venn diagrams showing the number of proteins identified: a in E. granulosus biological replicates; b in E. ortleppi biological replicates; c in each species or shared between them. The overall numbers of proteins detected are indicated below the sample/species identification
Table 1.
Proteins exclusively identified in HF samples from E. granulosus hydatid cysts
| Accession number1 | Protein Name | NSAF2 | SD3 | GO terms associated |
|---|---|---|---|---|
| EgrG_002002600 | Alpha mannosidase 2 | 0.000213 | 0.000368 | Catalytic activity; carbohydrate metabolic process |
| EgrG_000888900 | Anosmin 1 | 0.000153 | 0.000266 | Regulation of peptidase activity |
| EgrG_001134100 | Aspartate aminotransferase mitochondrial | 0.001567 | 0.002201 | Catalytic activity; transferase activity |
| EgrG_000297300 | BC026374 protein S09 family | 0.000176 | 0.000304 | Hydrolase activity |
| EgrG_000741700 | Beta-galactosidase | 0.000275 | 0.000476 | Carbohydrate metabolic process; hydrolase activity |
| EgrG_000678900 | Bifunctional heparan sulfate | 0.000216 | 0.000374 | Hydrolase activity; transferase activity |
| EgrG_000887000 | Cadherin | 0.000264 | 0.000458 | Cell adhesion; calcium ion binding |
| EgrG_000722600 | Calcium binding protein | 0.000139 | 0.000241 | Calcium ion binding |
| EgrG_000904400 | Carbonic anhydrase | 0.000401 | 0.000694 | Carbonate dehydratase activity |
| EgrG_000477200 | Cathepsin L | 0.000249 | 0.000432 | Cysteine-type peptidase activity |
| EgrG_000989200 | Cathepsin L1; cathepsin L cysteine peptidase | 0.000777 | 0.001346 | Cysteine-type peptidase activity |
| EgrG_000644850 | Cell adhesion molecule | 0.000731 | 0.000634 | Protein binding |
| EgrG_000111700 | Complement C1q tumor necrosis factor | 0.000801 | 0.001387 | Protein binding |
| EgrG_000654600 | Cysteine protease | 0.001284 | 0.002224 | Cysteine-type peptidase activity |
| EgrG_000061600 | Discoidin domain containing receptor 2 | 0.000232 | 0.000201 | Integral component of membrane |
| EgrG_001069200 | Ectonucleotide pyrophosphatase:phosphodiesterase | 0.000200 | 0.000346 | Lipid metabolic process; catalytic activity |
| EgrG_001096100 | EF hand domain containing protein | 0.000200 | 0.000174 | Metal ion binding |
| EgrG_000524400 | EGF region | 0.000304 | 0.000527 | Cell communication |
| EgrG_000824100 | Estrogen regulated protein EP45; Serpin B9 | 0.000207 | 0.000358 | Extracellular space |
| EgrG_000227300 | Expressed conserved protein | 0.001550 | 0.002684 | – |
| EgrG_000656900 | Expressed conserved protein | 0.000446 | 0.000773 | Integral component of membrane |
| EgrG_000814100 | Expressed conserved protein | 0.000502 | 0.000870 | Integral component of membrane |
| EgrG_000956500 | Expressed conserved protein | 0.000755 | 0.001307 | – |
| EgrG_000647100 | Expressed protein | 0.001002 | 0.001736 | – |
| EgrG_000253000 | Glutaminyl peptide cyclotransferase | 0.000615 | 0.000554 | Transferase activity |
| EgrG_000768900 | Glycosyl transferase family 8 | 0.000220 | 0.000382 | Transferase activity |
| EgrG_000418900 | Glycosyltransferase 14 family member | 0.000319 | 0.000553 | Transferase activity |
| EgrG_000778400 | Glypican | 0.000231 | 0.000201 | Regulation of signal transduction |
| EgrG_000545700 | Hexosyltransferase | 0.001173 | 0.002031 | Transferase activity; protein glycosylation |
| EgrG_000655200 | Inositol monophosphatase | 0.000277 | 0.000479 | Phosphatidylinositol phosphorylation |
| EgrG_000357600 | Lipase | 0.001776 | 0.001790 | Hydrolase activity; lipid catabolic process |
| EgrG_001157000 | Lymphocyte antigen 75 | 0.000103 | 0.000178 | Integral component of membrane; carbohydrate binding |
| EgrG_000116900 | Lysosomal protein NCU G1 B | 0.002407 | 0.003044 | Integral component of membrane |
| EgrG_000144800 | N acyl phosphatidylethanolamine hydrolyzing | 0.000477 | 0.000826 | Hydrolase activity |
| EgrG_000115400 | N/A | 0.000609 | 0.001056 | Integral component of membrane |
| EgrG_000237600 | N/A | 0.000433 | 0.000750 | Integral component of membrane |
| EgrG_000334500 | N/A | 0.000566 | 0.000981 | – |
| EgrG_000759860 | N/A | 0.000430 | 0.000744 | Integral component of membrane |
| EgrG_000522900 | Neurexin 1 alpha | 0.000139 | 0.000120 | Integral component of membrane; multicellular organism development |
| EgrG_000119200 | Neuroendocrine protein 7b2 | 0.000410 | 0.000710 | Neuropeptide signaling pathway, regulation of proteolysis |
| EgrG_000926700 | Peptide methionine sulfoxide reductase | 0.000926 | 0.001604 | Oxidoreductase activity |
| EgrG_000591200 | Pfam-B_8122 and DUF4381 domain containing protein | 0.000199 | 0.000345 | Integral component of membrane |
| EgrG_000443300 | Procollagen lysine2 oxoglutarate 5 dioxygenase | 0.000119 | 0.000205 | Oxidoreductase activity |
| EgrG_000443800 | Prohormone 4 | 0.001334 | 0.001175 | Protein binding |
| EgrG_001022300 | Protein disulfide-isomerase | 0.000121 | 0.000210 | Isomerase activity |
| EgrG_000211300 | Protein Wnt | 0.000167 | 0.000290 | Signaling receptor binding |
| EgrG_000228100 | Protocadherin | 0.000141 | 0.000245 | Cell adhesion |
| EgrG_000861900 | Protocadherin 11; Protocadherin-11 X-linked | 0.000111 | 0.000192 | Cell adhesion |
| EgrG_000878500 | Protocadherin 9 | 0.000138 | 0.000239 | Cell adhesion |
| EgrG_000112900 | Protocadherin alpha 6 | 0.000196 | 0.000340 | Cell adhesion |
| EgrG_000075800 | Receptor protein-tyrosine kinase | 0.000082 | 0.000143 | Protein kinase activity |
| EgrG_000461400 | Receptor protein-tyrosine kinase | 0.000286 | 0.000301 | Protein kinase activity |
| EgrG_000655700 | Receptor type tyrosine protein phosphatase | 0.000131 | 0.000226 | Phosphatase activity |
| EgrG_000136400 | Semaphorin 5B | 0.000144 | 0.000250 | Semaphorin receptor binding; multicellular organism development |
| EgrG_000961100 | Slit 2 protein | 0.000056 | 0.000097 | Calcium ion binding; multicellular organism development |
| EgrG_001127800 | Speract scavenger receptor | 0.000382 | 0.000661 | Scavenger receptor activity; endocytosis |
| EgrG_000814400 | Subfamily M14A unassigned peptidase | 0.000222 | 0.000384 | Hydrolase activity |
| EgrG_000381100 | Tapeworm specific antigen B (AgB8/2) | 0.026035 | 0.045093 | – |
| EgrG_000381600 | Tapeworm specific antigen B (AgB8/3) | 0.017258 | 0.029891 | – |
| EgrG_000381400 | Tapeworm specific antigen B (AgB8/4) | 0.009642 | 0.016701 | – |
| EgrG_000381800 | Tapeworm specific antigen B (AgB8/5) | 0.002018 | 0.003496 | – |
| EgrG_000178100 | TGF beta family | 0.000784 | 0.000923 | Signal transduction |
| EgrG_000359800 | Thioredoxin fold | 0.000451 | 0.000782 | – |
| EgrG_000092800 | Transaldolase | 0.000264 | 0.000457 | Carbohydrate metabolic process; transferase activity |
| EgrG_001004900 | Transgelin | 0.000179 | 0.000311 | Protein binding |
| EgrG_000959800 | Voltage dependent calcium channel subunit | 0.000430 | 0.000572 | Calcium channel activity |
aAccording to E. granulosus genome annotation (PRJEB121, version WBPS11) available at WormBase ParaSite
NSAF Normalized spectral abundance factor, SD standard deviation
Table 2.
Proteins exclusively identified in HF samples from E. ortleppi hydatid cysts
| Accession numbera | Protein name | NSAF | SD | GO terms associated |
|---|---|---|---|---|
| EgrG_001104800 | Acidic leucine-rich nuclear phosphoprotein | 0.000557 | 0.000964 | Protein binding |
| EgrG_000528900 | Actin depolymerizing factor | 0.000600 | 0.001040 | Actin cytoskeleton |
| EgrG_000501600 | Alpha-1,4 glucan phosphorylase | 0.000193 | 0.000334 | Carbohydrate metabolic process; transferase activity |
| EgrG_000041200 | Annexin | 0.000823 | 0.001426 | Calcium ion binding |
| EgrG_000193700 | Annexin | 0.001538 | 0.001436 | Calcium ion binding |
| EgrG_000244000 | Annexin | 0.001183 | 0.002048 | Calcium ion binding |
| EgrG_000911200 | Calpain-A | 0.000366 | 0.000321 | Calcium-dependent cysteine-type endopeptidase activity |
| EgrG_000936600 | Cytoskeleton associated protein CAP Gly containing ankyrin repeats | 0.000202 | 0.000349 | Protein binding |
| EgrG_000564000 | Diagnostic antigen gp50 | 0.000620 | 0.001074 | – |
| EgrG_000566700 | Diagnostic antigen gp50 | 0.001858 | 0.001630 | – |
| EgrG_000940900 | Dynein light chain | 0.001240 | 0.002149 | Microtubule-based process |
| EgrG_000941100 | Dynein light chain | 0.005642 | 0.001419 | Microtubule-based process |
| EgrG_000946900 | Dynein light chain | 0.000674 | 0.001168 | Microtubule-based process |
| EgrG_000113800 | Elongation factor 1-gamma; eukaryotic translation elongation factor 1 | 0.000196 | 0.000339 | Translation elongation factor activity |
| EgrG_000865300 | Elongation factor 2 | 0.000097 | 0.000168 | Translation elongation factor activity |
| EgrG_000261600 | Fructose 1,6 bisphosphatase 1 | 0.000222 | 0.000384 | Carbohydrate metabolic process; phosphatase activity |
| EgrG_000476900 | GDP L fucose synthase | 0.000725 | 0.000636 | Nucleotide-sugar biosynthetic process |
| EgrG_000882300 | Gelsolin; Severin | 0.002067 | 0.001813 | Actin filament binding |
| EgrG_000485800 | H17g protein tegumental antigen | 0.000937 | 0.000270 | Actin binding; localization of cell |
| EgrG_002016600 | Histone | 0.001589 | 0.001433 | DNA binding |
| EgrG_000906000 | Histone H1 delta | 0.002786 | 0.003000 | DNA binding |
| EgrG_000799300 | Insulin growth factor binding; Kazal-type serine protease inhibitor domain-containing protein | 0.001389 | 0.002406 | Regulation of cell growth |
| EgrG_000634800 | L-lactate dehydrogenase | 0.000458 | 0.000793 | Carbohydrate metabolic process; oxidoreductase activity |
| EgrG_000142500 | Major vault protein | 0.000456 | 0.000472 | Protein binding |
| EgrG_000631600 | N/A | 0.000348 | 0.000603 | – |
| EgrG_000838600 | N/A | 0.006931 | 0.006472 | – |
| EgrG_000736050 | NAD(P)H-hydrate epimerase | 0.000214 | 0.000370 | Isomerase activity |
| EgrG_000763300 | Paramyosin | 0.000714 | 0.000715 | Myosin complex |
| EgrG_000334550 | Peptidylprolyl isomerase | 0.001265 | 0.002192 | Isomerase activity |
| EgrG_000943900 | Phosphoglucomutase | 0.000901 | 0.000783 | Carbohydrate metabolic process |
| EgrG_000122100 | Profilin | 0.002285 | 0.003957 | Actin binding |
| EgrG_001046200 | Subfamily S1A unassigned peptidase S01 family | 0.001233 | 0.001069 | Serine-type peptidase activity |
| EgrG_000607900 | Superoxide dismutase | 0.000372 | 0.000645 | Superoxide metabolic process |
| EgrG_001001800 | Tegumental antigen | 0.000490 | 0.000848 | Microtubule-based process |
| EgrG_000355700 | Tetraspanin | 0.000445 | 0.000771 | Integral component of membrane |
| EgrG_000471600 | Transitional endoplasmic reticulum ATPase | 0.000103 | 0.000179 | ATPase activity |
| EgrG_000416400 | Triosephosphate isomerase | 0.002899 | 0.000913 | Glycolytic process; isomerase activity |
aAccording to E. granulosus genome annotation (PRJEB121, version WBPS11) available at WormBase ParaSite
NSAF Normalized spectral abundance factor, SD standard deviation
A large group of proteins of unknown function (35 unique sequences) was identified (Additional File 4: Table S2). They were annotated as “expressed conserved protein,” “expressed protein,” or “N/A (non-annotated).” Some of these proteins of unknown function were identified in all six samples, and some are among the most abundant proteins considering each species separately.
The sequences of proteins of unknown function were subjected to automated function prediction using ESG and PFP software [37]. For ten of these proteins, GO terms predicted by ESG software were further predicted in PFP, and these results are listed in Additional File 6: Table S4. Some molecular function ontologies predicted were calcium channel regulator activity (EgrG_000236300 and EgrG_000296900), RNA binding (EgrG_000316400), DNA binding (EgrG_000471400), and acetylcholine receptor binding (EgrG_000956500). For biological process, chemical synaptic transmission (EgrG_000236300 and EgrG_000296900), regulation of neurotransmitter receptor activity (EgrG_000956500), synapse organization (EgrG_001058700), and protein transport (EgrG_001024500) were some of the ontologies predicted.
As expected, host proteins were also identified in E. granulosus and E. ortleppi HF samples. Fewer host proteins were identified compared to parasite proteins. Overall, 58 distinct B. taurus proteins were identified, with 40 (13 exclusive) of them being identified in E. granulosus HF samples and 45 (18 exclusive) in E. ortleppi samples, and 27 proteins were common to both samples (Additional File 7: Table S5). Variable numbers of bovine proteins were found in each biological sample, 12, 13, and 28 for EG1, EG2, and EG3, respectively, and 21, 11, and 37 for EO1, EO2, and EO3, respectively (Additional File 8: Figure S3).
Main proteins identified in hydatid fluid samples from E. granulosus and E. ortleppi
To highlight the most frequent parasitic proteins in HF, we selected those detected in at least two samples of each species, totaling 217 proteins, among which 13 and 15 were detected exclusively in E. granulosus and E. ortleppi samples, respectively (Additional File 9: Table S6). For each species, the proteins detected in the three biological samples were selected as HF common proteins (Additional File 10: Table S7). The E. granulosus and E. ortleppi HF common proteins comprised, respectively, 61 and 105 proteins, and 52 were shared by the two species (Table 3).
Table 3.
Identification and relative abundance of proteins present in HF samples from E. granulosus and E. ortleppi bovine pulmonary hydatid cysts
| Accession numbera | Protein name | Molecular Massb | EG | EO | ||
|---|---|---|---|---|---|---|
| NSAF | SD | NSAF | SD | |||
| EgrG_000144400 | Abnormal EMBroygenesis family member emb 9 | 168 kDa | 0.00382 | 0.00227 | 0.00254 | 0.00064 |
| EgrG_000061200 | Actin | 42 kDa | 0.01910 | 0.01909 | 0.01722 | 0.00630 |
| EgrG_000156400 | Aldo keto reductase family 1 member B4 | 42 kDa | 0.00199 | 0.00101 | 0.00277 | 0.00102 |
| EgrG_000704400 | Alpha-mannosidase | 118 kDa | 0.01298 | 0.00567 | 0.01019 | 0.00120 |
| EgrG_000530400 | Amine oxidase | 84 kDa | 0.00932 | 0.00324 | 0.00531 | 0.00204 |
| EgrG_001032200 | Aminotransferase class III; Ornithine aminotransferase | 46 kDa | 0.01093 | 0.00898 | 0.00551 | 0.00226 |
| EgrG_000184900 | Antigen 5 | 55 kDa | 0.04773 | 0.00409 | 0.05786 | 0.02534 |
| EgrG_000575900 | Basement membrane specific heparan sulfate | 860 kDa | 0.01233 | 0.00292 | 0.00869 | 0.00066 |
| EgrG_000701800 | Basement membrane specific heparan sulfate | 96 kDa | 0.00857 | 0.00187 | 0.00467 | 0.00036 |
| EgrG_000879900 | Beta D xylosidase 2 | 92 kDa | 0.00252 | 0.00108 | 0.00271 | 0.00146 |
| EgrG_000789900 | Beta mannosidase | 108 kDa | 0.00220 | 0.00084 | 0.00115 | 0.00032 |
| EgrG_000903100 | Calsyntenin 1 | 130 kDa | 0.00405 | 0.00445 | 0.00285 | 0.00157 |
| EgrG_000970500 | Cathepsin D lysosomal aspartyl protease | 47 kDa | 0.01626 | 0.00742 | 0.01783 | 0.00509 |
| EgrG_000144350 | Collagen alpha 1(IV) chain | 172 kDa | 0.00553 | 0.00266 | 0.00351 | 0.00066 |
| EgrG_000417600 | Collagen alpha 1(IV) chain | 182 kDa | 0.00280 | 0.00195 | 0.00154 | 0.00066 |
| EgrG_000203400 | Collagen alpha 1(V) chain | 172 kDa | 0.00630 | 0.00263 | 0.00512 | 0.00185 |
| EgrG_000144300 | Collagen alpha 1(V) chain | 177 kDa | 0.00453 | 0.00246 | 0.00309 | 0.00012 |
| EgrG_000729300 | Collagen alpha 1(XV) chain | 191 kDa | 0.00398 | 0.00232 | 0.00250 | 0.00054 |
| EgrG_000823800 | Collagen alpha 2(I) chain | 131 kDa | 0.00914 | 0.00326 | 0.00605 | 0.00239 |
| EgrG_001190600 | Collagen type I II III V XI alpha | 123 kDa | 0.00786 | 0.00374 | 0.00445 | 0.00175 |
| EgrG_000524200 | Collagen type XI alpha 2 | 163 kDa | 0.00445 | 0.00130 | 0.00191 | 0.00115 |
| EgrG_000766600 | Cysteine-rich secretory protein LCCL domain-containing; Peptidase inhibitor 16 | 29 kDa | 0.01633 | 0.01258 | 0.00751 | 0.00292 |
| EgrG_000255800 | EGF domain protein | 267 kDa | 0.00385 | 0.00187 | 0.00193 | 0.00014 |
| EgrG_000682900 | Epididymal secretory protein E1; Niemann Pick C2 protein | 20 kDa | 0.00778 | 0.00123 | 0.00369 | 0.00151 |
| EgrG_000824000 | Estrogen regulated protein EP45 | 45 kDa | 0.01048 | 0.00136 | 0.00722 | 0.00093 |
| EgrG_001061900 | Expressed conserved protein | 74 kDa | 0.00762 | 0.00602 | 0.01483 | 0.00569 |
| EgrG_000412500 | Expressed conserved protein | 14 kDa | 0.01324 | 0.00948 | 0.01267 | 0.00320 |
| EgrG_000523100 | Expressed conserved protein | 53 kDa | 0.00638 | 0.00255 | 0.00600 | 0.00175 |
| EgrG_000596300 | Expressed conserved protein | 25 kDa | 0.01830 | 0.01295 | 0.01610 | 0.00609 |
| EgrG_000316400 | Expressed protein | 35 kDa | 0.00261 | 0.00088 | 0.00217 | 0.00114 |
| EgrG_000842900 | Fgfr protein | 80 kDa | 0.00302 | 0.00128 | 0.00273 | 0.00042 |
| EgrG_001060700 | Fibrillar collagen chain FAp1 alpha | 116 kDa | 0.00989 | 0.00164 | 0.00725 | 0.00357 |
| EgrG_000176400 | Fras1 related extracellular matrix protein | 263 kDa | 0.00187 | 0.00107 | 0.00147 | 0.00037 |
| EgrG_000905600 | Fructose-bisphosphate aldolase | 40 kDa | 0.01739 | 0.00955 | 0.01265 | 0.00119 |
| EgrG_000712600 | Gynecophoral canal protein | 97 kDa | 0.00883 | 0.00348 | 0.00577 | 0.00146 |
| EgrG_000824400 | Gynecophoral canal protein; Transforming growth factor-beta-induced protein ig-h3 | 73 kDa | 0.01180 | 0.00181 | 0.00923 | 0.00139 |
| EgrG_000422350 | Hemicentin 1 | 477 kDa | 0.00279 | 0.00100 | 0.00138 | 0.00065 |
| EgrG_001132400 | Laminin | 395 kDa | 0.00113 | 0.00081 | 0.00054 | 0.00029 |
| EgrG_000458400 | Laminin subunit gamma | 163 kDa | 0.00254 | 0.00085 | 0.00105 | 0.00044 |
| EgrG_000684200 | Lipid transport protein N terminal | 344 kDa | 0.00439 | 0.00189 | 0.00236 | 0.00186 |
| EgrG_000343000 | Neurogenic locus notch protein | 339 kDa | 0.00555 | 0.00140 | 0.00384 | 0.00030 |
| EgrG_001181950 | Papilin | 67 kDa | 0.00315 | 0.00094 | 0.00159 | 0.00057 |
| EgrG_000920600 | Peptidyl-prolyl cis–trans isomerase | 17 kDa | 0.01078 | 0.00123 | 0.01451 | 0.00122 |
| EgrG_000292700 | Phosphoenolpyruvate carboxykinase | 71 kDa | 0.01145 | 0.00131 | 0.01344 | 0.00424 |
| EgrG_001132700 | Poly(U) specific endoribonuclease | 29 kDa | 0.00748 | 0.00254 | 0.00659 | 0.00084 |
| EgrG_000849600 | Proteinase inhibitor I25 cystatin | 31 kDa | 0.03216 | 0.01174 | 0.02514 | 0.01025 |
| EgrG_001133400 | Protein-L-isoaspartate O-methyltransferase | 27 kDa | 0.00630 | 0.00374 | 0.00363 | 0.00209 |
| EgrG_000929500 | SPONdin extracellular matrix glycoprotein | 111 kDa | 0.00293 | 0.00178 | 0.00126 | 0.00016 |
| EgrG_000381200 | Tapeworm specific antigen B (AgB8/1) | 10 kDa | 0.09772 | 0.00805 | 0.19553 | 0.08363 |
| EgrG_000791700 | Thioredoxin peroxidase | 21 kDa | 0.01305 | 0.00927 | 0.01254 | 0.00268 |
| EgrG_001060600 | Type II collagen B | 154 kDa | 0.00352 | 0.00080 | 0.00206 | 0.00116 |
| EgrG_000317300 | Vesicular amine transporter | 49 kDa | 0.01186 | 0.00156 | 0.00688 | 0.00342 |
The listed proteins were identified in the three biological replicates from each species. Quantitative data are presented based on averaged NSAF values calculated for E. granulosus (EG) and E. ortleppi (EO)
NSAF Normalized spectral abundance factor, SD standard deviation
aAccording to E. granulosus genome annotation (PRJEB121, version WBPS11) available at WormBase ParaSite
bMolecular mass calculated from primary sequence
Top ten NSAF values in HF samples of each species are highlighted in bold
Within the HF common proteins, in the subgroup of proteins shared between the two species, we identified proteins associated with different biological processes, such as cathepsin D, laminin, thioredoxin peroxidase, poly(U) endoribonuclease, cystatin, fructose-bisphosphate aldolase, and antigens previously described as relevant in Echinococcus spp. biology, such as antigen B (AgB) and antigen 5 (Ag5). AgB and Ag5 are antigens with recognized significance in Echinococcus spp. biology by their abundance and immunogenicity.
AgB is an oligomeric lipoprotein, which can comprise up to five related subunits (AgB8/1 to 5). We detected subunit AgB8/1 in the shared subgroup of common proteins, while subunits AgB8/2 to 5 were detected in only one E. granulosus sample (Additional File 3: Figure S2 and Additional File 5: Table S3). These subunit levels in the other samples might be below the level of detection under our experimental conditions.
HF common proteins shared between E. granulosus and E. ortleppi are interesting study targets to understand molecular mechanisms at the host-parasite interface in cystic echinococcosis. Additionally, they are candidate targets for the development of new therapies for Echinococcus spp. infections.
Some B. taurus proteins were more frequently identified in our HF analysis. The host proteins found in at least two biological replicates in each species are listed in Table 4. The proteins actin, apolipoprotein A-1, heat shock cognate 71 kDa protein, hemoglobin subunit alpha, hemoglobin subunit beta, and serum albumin were identified in HF samples from both species.
Table 4.
Bovine proteins identified in at least two biological replicates from E. granulosus and E. ortleppi hydatid fluid
| Accession numbera | Protein name | Molecular massb | NSAF | SD | GO terms associated |
|---|---|---|---|---|---|
| E. granulosus | |||||
| 1433G_BOVIN | 14–3-3 protein gamma | 28 kDa | 0.0148 | 0.01632 | Regulation of biological quality; protein binding |
| ACTB_BOVIN | Actin, cytoplasmic 1 | 42 kDa | 0.06094 | 0.01676 | Protein binding; response to toxic substance |
| FETUA_BOVIN | Alpha-2-HS-glycoprotein | 38 kDa | 0.015 | 0.01725 | Endopeptidase regulator activity; defense response |
| APOA1_BOVIN | Apolipoprotein A-I | 30 kDa | 0.0141 | 0.01228 | Protein binding; regulation of protein transport |
| HSP7C_BOVIN | Heat shock cognate 71 kDa protein | 71 kDa | 0.0036 | 0.0032 | Nucleotide metabolic process; protein binding |
| HBA_BOVIN | Hemoglobin subunit alpha | 15 kDa | 0.19286 | 0.05119 | Detoxification; cellular response to chemical stimulus |
| HBB_BOVIN | Hemoglobin subunit beta | 16 kDa | 0.27648 | 0.05495 | Detoxification; cellular response to chemical stimulus |
| A0A140T897_BOVIN | Serum albumin | 69 kDa | 0.19232 | 0.10116 | Protein binding; cell killing |
| E. ortleppi | |||||
| ACTB_BOVIN | Actin, cytoplasmic 1 | 42 kDa | 0.09472 | 0.05639 | See above |
| ENOA_BOVIN | Alpha-enolase | 47 kDa | 0.0036 | 0.00314 | Glycolytic process; binding |
| APOA1_BOVIN | Apolipoprotein A-I | 30 kDa | 0.011 | 0.00995 | See above |
| CATA_BOVIN | Catalase | 60 kDa | 0.0048 | 0.00521 | Cellular response to toxic substance; detoxification |
| HSP7C_BOVIN | Heat shock cognate 71 kDa protein | 71 kDa | 0.00934 | 0.00643 | See above |
| HBA_BOVIN | Hemoglobin subunit alpha | 15 kDa | 0.20497 | 0.01557 | See above |
| HBB_BOVIN | Hemoglobin subunit beta | 16 kDa | 0.33547 | 0.06589 | See above |
| LDHA_BOVIN | L-lactate dehydrogenase A chain | 37 kDa | 0.0107 | 0.00933 | Carbohydrate metabolic process |
| F1MYX5_BOVIN | Lymphocyte cytosolic protein 1 | 70 kDa | 0.0028 | 0.00247 | Immune response; regulation of localization |
| PRDX1_BOVIN | Peroxiredoxin-1 | 22 kDa | 0.0125 | 0.01226 | Immune response; detoxification |
| A5D984_BOVIN | Pyruvate kinase | 58 kDa | 0.0034 | 0.00292 | Glycolytic process; binding |
| A0A140T897_BOVIN | Serum albumin | 69 kDa | 0.09619 | 0.02085 | See above |
| TBA1B_BOVIN | Tubulin alpha-1B chain | 50 kDa | 0.0049 | 0.00441 | Nucleotide binding |
| TBB5_BOVIN | Tubulin beta-5 chain | 50 kDa | 0.0051 | 0.00475 | Nucleotide binding |
| VIME_BOVIN | Vimentin | 54 kDa | 0.0128 | 0.01761 | Immune response; cellular response to chemical stimulus |
aAccording to Bos taurus reference proteome (ID: UP000009136) available at Uniprot/Swiss-Prot
bMolecular mass calculated from primary sequence
NSAF Normalized spectral abundance factor, SD standard deviation
Potential secretion pathways associated with parasitic proteins identified in E. granulosus and E. ortleppi hydatid fluid
All E. granulosus and E. ortleppi proteins identified in the corresponding HF samples were analyzed using bioinformatic tools to predict whether they would be secreted by a classical pathway (signal peptide) or by an alternative pathway, and the results are summarized in Fig. 2. In the E. granulosus protein repertoire (Fig. 2a), 54% (150/278) of the proteins were predicted to have a signal peptide, 11% (31/278) were predicted to be secreted by an alternative pathway, and 35% (97/278) were not predicted to be secreted. In the E. ortleppi repertoire (Fig. 2b), 45% (111/249) of the proteins were predicted to have a signal peptide, 13% (32/249) were predicted to be secreted by an alternative pathway, and 43% (106/249) were not predicted to be secreted.
Fig. 2.
In silico prediction of secretion pathways. Percentages of the total and absolute number of proteins in E. granulosus and E. ortleppi HF repertoires with probable classic or alternative signals for secretion are presented. Proteins with any identifiable signal for secretion were grouped under the term “Unidentified secretion pattern.” a E. granulosus HF proteins. b E. ortleppi HF proteins
Functional annotation of the protein repertoires from E. granulosus and E. ortleppi hydatid fluid
GO enrichment analyses were performed for all parasitic proteins identified in E. granulosus and E. ortleppi using the Cytoscape plugin BiNGO [36]. Functional classification with GO enrichment data is shown in Additional File 11: Table S8. Most proteins were functionally annotated for both E. granulosus (220/278 proteins) and E. ortleppi (203/249 proteins). GO enrichment (P ≤ 0.05) was found for 180 GO subcategories in E. granulosus (Additional File 11: Table S8A) and for 224 GO subcategories in E. ortleppi (Additional File 11: Table S8B), using the following three main GO categories: biological process, molecular function, and cellular component. Echinococcus granulosus and E. ortleppi showed the same profile regarding the most significant GO subcategories (P < 0.001).
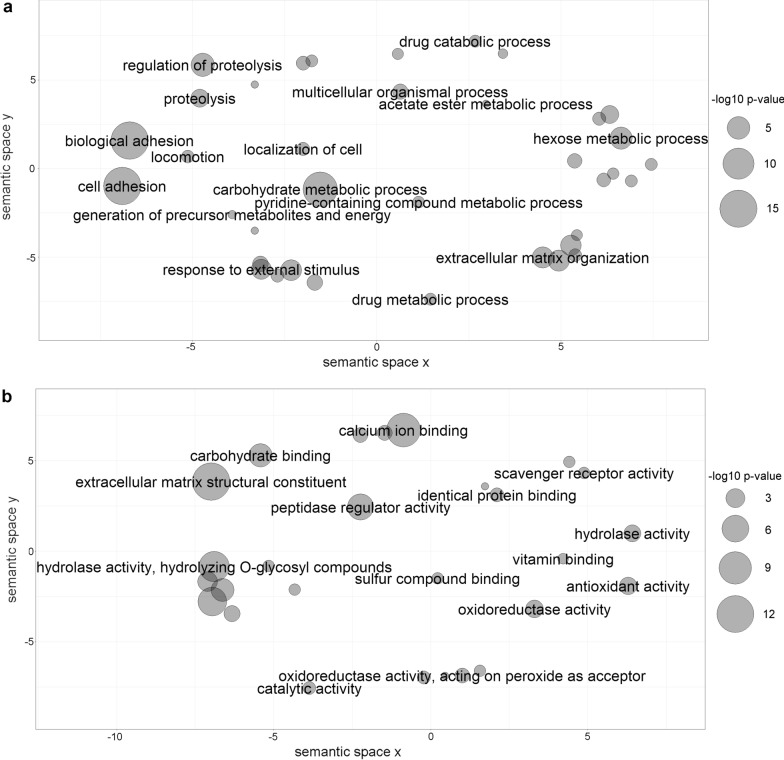
The enriched GO terms in the biological process and molecular function major categories for E. granulosus and E. ortleppi proteins were summarized using REVIGO [38]. The complete lists of summarized non-redundant terms are shown in Additional File 12: Table S9 and Additional File 13: Table S10. After the summary using REVIGO, 65 and 63 category clusters were generated for E. granulosus and E. ortleppi, respectively. For the biological process main category, the clusters “cell adhesion,” “carbohydrate metabolic process,” and “regulation of proteolysis” were among the most enriched clusters in both E. granulosus and E. ortleppi (Figs. 3a and 4a). In the molecular function main category, the clusters “extracellular matrix structural constituent,” “calcium binding,” and “hydrolase activity acting on glycosyl bonds” were among the most enriched clusters in both E. granulosus and E. ortleppi (Figs. 3b and 4b).
Fig. 3.
Summarized functional classification of proteins identified in E. granulosus HF. Scatterplot view of REVIGO category clusters of related GO terms obtained in functional enrichment analysis. a Biological process category clusters. b Molecular function category clusters. Sphere size is proportional to the P-value (larger spheres indicate more significant P-values, according to the scale)
Fig. 4.
Summarized functional classification of proteins identified in E. ortleppi HF. Scatterplot view of REVIGO category clusters of related GO terms obtained in functional enrichment analysis. a Biological process category clusters. b Molecular function category clusters. Sphere size is proportional to the P-value (larger spheres indicate more significant P-values, according to the scale)
Host proteins identified in E. granulosus and E. ortleppi HF samples were also subjected to GO enrichment analysis. An extensive list of GO terms were enriched (281 in E. granulosus and 378 in E. ortleppi), and they were summarized using the REVIGO platform (Additional File 14: Table S11 and Additional File 15: Table S12).
REVIGO category clusters generated for the bovine proteins in E. granulosus and E. ortleppi HF showed different host biological mechanisms. There were “carbohydrate metabolic process,” “defense response,” “cell killing,” “protein binding,” and “regulation of protein stability” among the shared category clusters. Some E. granulosus category clusters were “negative regulation of hydrolase activity,” “acute-phase response,” and “regulation of peptide transport.” Some E. ortleppi category clusters were “response to external stimulus,” “immune response,” and “regulation of cell death.”
Discussion
In our study, we performed a MS-based proteomic analysis of HF samples from three E. granulosus and three E. ortleppi hydatid cysts collected from B. taurus lungs. We identified 280 and 251 proteins in E. granulosus and E. ortleppi samples, respectively, and there were 317 different parasitic proteins overall.
Many proteins identified in our study do not have a signal to secretion, and because of that, they would be unexpected in HF. However, extracellular vesicles are described in the literature as carriers for a wide range of proteins, indicating that proteins without recognizable signal peptide can also be secreted to exert their function extracellularly. The composition of the extracellular vesicles is diverse, including several classes of proteins, like signaling proteins, membrane receptors, glycolytic enzymes, proteases, inhibitors, etc. A quick search in exocarta (http://exocarta.org) and vesiclepedia (http://www.microvesicles.org/) databases showed that several proteins from the HF repertoire of E. granulosus and E. ortleppi have been identified in extracellular vesicles from other organisms. Proteomic analyses of E. granulosus extracellular vesicles isolated from sheep [39] and human hosts [40] have shown several proteins in common with our results, supporting that this may be the mechanism of secretion for many proteins in Echinococcus metacestode.
The GO analysis showed the association of the protein profiles with a variety of ontology terms. The heterogeneity of functions assigned to the identified proteins, such as cell adhesion, extracellular matrix structural constituent, carbohydrate metabolic process, and calcium binding, indicates that many molecular mechanisms are active in E. granulosus and E. ortleppi larval infection. The heterogeneity in the function and number of proteins in our samples may result from differences in the cyst developmental stage or their physiological state.
Proteins associated with nutrient transport and metabolism were well represented in our analysis. These proteins may act in basic cellular functions, playing important roles in nutrient uptake and in structural constituent and energy production. Some of them were found in both E. granulosus and E. ortleppi HF protein repertoires, such as beta mannosidase, fructose-bisphosphate aldolase, phosphoenolpyruvate carboxykinase, aminotransferase class III, AgB, and lipid transport protein N terminal. The metacestode is very active and certainly requires a good supply of nutrients and energy to maintain the viability. Tapeworms have reduced synthesis capability, but an increased ability to absorb nutrients from host [41, 42]. Echinococcus do not synthesize fatty acids and cholesterol; instead, they scavenge them from the host. AgB is a lipoprotein acting in transport of host-derived fatty acids, triacylglycerols, and sterols to the parasite tissues [43, 44]. AgB is also the major antigen in HF, and it has important immunomodulatory properties [11]. Among all five AgB subunits, we detected only AgB8/1 in all six analyzed samples. AgB8/1 has been reported to be the most abundant subunit in the E. granulosus AgB oligomer [45], and the AgB subunit is consistently identified by MS-based HF analysis [12, 14, 32]. However, detection and abundance of AgB8/2–5 in HF are variable [12–14, 32]. A few works have analyzed the proportion of each subunit in the AgB pool in HF, and many questions remain unanswered, such as the dynamic of subunit production along the metacestode development or whether the production is modulated upon determined host responses. Additionally, AgB subunit representation in HF varies among different Echinococcus species and isolates in E. multilocularis, for example AgB8/3 is the most abundant subunit [32, 46].
Different carbohydrate-metabolizing enzymes were identified in E. granulosus and E. ortleppi HF. Such enzymes are repeatedly observed in the secretome of E. granulosus and other cestodes, including E. multilocularis [14, 31, 47]. In E. granulosus, they have been found in the HF of hydatids from cattle, sheep and human hosts [13, 14, 32, 48]. Previous studies indicated that some carbohydrate-metabolizing enzymes exerted other effects in addition to their primary biochemical roles [49]. In addition to their described function, the carbohydrate-metabolizing enzymes identified in this study might exert extracellular functions, protecting parasite tissues from host immune attack and aiding in metacestode development. Glycolytic enzymes were shown to exert many effects, such as binding to complement proteins and interference in their response, binding of host plasminogen with further increase in its activation and interaction with adhesins and the cytoskeleton to facilitate invasion [50–52]. In E. granulosus, fructose-bisphosphate aldolase was shown to interact with actin, and enolase was detected by immunolocalization in the laminated layer of hydatids from cattle [17]. These molecules do not have a signal peptide, but significant amounts appeared to be secreted through specific mechanisms such as extracellular vesicles [14, 17]. The glycolytic enzymes have been identified in extracellular vesicles of HF from sheep and human hydatids [39, 40].
Echinococcus ortleppi granulosus and E. ortleppi HF showed a diverse range of proteolytic enzymes. Our analysis identified enzymes such as aminopeptidases, carboxypeptidases, cysteine peptidases, metalloproteases, and an enteropeptidase. Proteolytic enzymes have pivotal roles at the host-parasite interface, especially related to nutrient acquisition, tissue migration, and protection against the host immune response [53–56]. Metalloproteases, a class of proteolytic enzymes frequently found in parasitic secretomes, function mainly in extracellular matrix degradation and tissue remodeling, and they also facilitate a diverse range of cellular processes, including regulation of stem cell proliferation in planarians [57]. Cathepsins are cysteine proteases that are widely described as molecular players in helminthic infections and suppress the host immune response at the host-parasite interface [56]. Three cathepsin L sequences were identified in E. granulosus. Calpain, a Ca2+-dependent cysteine protease, was identified in E. ortleppi HF. Calpains are associated with cell degeneration; studies have reported that under Ca2+ imbalances, calpains become activated and mediate apoptosis and necrosis [58–60]. Thus, a role for Calpain-A as a defense molecule inducing cell death at infiltrating and adjacent host cells in E. ortleppi infection is possible. Based on their importance in different processes of basic parasitic biology and their role at the host-parasite interface, some proteases have been proposed as therapeutic targets [61–64].
However, protease inhibitors such as cystatins, serpins, and proteins containing Kunitz and Kazal domains were also detected in E. granulosus and E. ortleppi HF. Proteases are part of defense mechanisms in mammals, and the presence of parasitic protease inhibitors suggests that modulation of host protease activities could be a mechanism of protection against elimination in Echinococcus spp. Proteases and inhibitors could also be associated with the same molecular processes in which the inhibitors regulate protease activity to avoid excessive tissue damage [65]. Thus, the parasite would produce inhibitors to modulate their own protease activity to minimize host tissue damage and avoid an increased immune response at the infection site. Important immunomodulatory roles have been described for protease inhibitors in other parasitic flatworms [65, 66]. In different invertebrates, Kunitz proteins have been described as acting in defense against microbial infection and with toxin activity mediated by ion channel blockade [67, 68].
A group of proteins related to the extracellular matrix and structure maintenance was identified in both E. granulosus and E. ortleppi HF. We highlight the presence of proteins associated with extracellular matrix structures and dynamics, such as collagen, laminin, hemicentin-1, SPONdin extracellular matrix glycoprotein, basement membrane specific heparan sulfate, and FRAS1-related extracellular matrix protein 1. These proteins may be related to maintenance of the hydatid cyst wall structural integrity, helping the metacestode to resist the host responses. The germinative layer inside the hydatid cyst plays a pivotal role in hydatid cyst development and survival, and its outward face is covered by a syncytial tegument that is also a physical barrier against the entrance of macromolecules into hydatid cysts [4, 69]. The laminated layer, an acellular, carbohydrate-rich sheath secreted by the germinative layer, shields the parasite from direct attack by host immune cells [70]. The extracellular matrix proteins and their regulators may be associated with a molecular network that both maintains the integrity of the cyst wall and allows tissue expansion that is necessary for hydatid growth.
Signaling pathway proteins were also identified, and many of them were shared between E. granulosus and E. ortleppi. Desert hedgehog protein (Dhh), noggin, notch, tyrosine protein kinase otk, and glypican-1 are examples of signaling proteins that play crucial roles in embryonic and morphological development in model organisms such as Caenorhabditis elegans, Drosophila melanogaster, and Mus musculus [71–74]. The germinative layer in fertile metacestodes comprises cells that actively participate in cyst development. These cells differentiate to generate brood capsules and PSC, secrete some HF components, and produce the required molecules to maintain cyst wall integrity [4, 75]. In this work, only viable fertile hydatid cysts were used, and thus, the germinative layer was probably very active and the signaling proteins we found could have a function in coordinating the events in this cell layer.
Some proteins identified here are linked to the major developmental pathways, Hedgehog, Notch, and Wnt, which are involved in many embryological development cascades, cell fate, cell polarity, and maintaining stemness of stem cells [73, 74, 76]. Because some cells in the germinative layer are stem cells responsible for generating other cell types and tissues in the metacestode, our findings suggest that such developmental pathways are active in the Echinococcus spp. hydatid cyst. Differential expression of signaling proteins among different E. granulosus and Hymenolepis microstoma developmental stages has been previously demonstrated [77, 78]. In E. multilocularis and H. microstoma, Wnt protein expression patterns during larval metamorphosis have been elucidated [79]. The roles played by the signaling transducing proteins might be necessary for proper metacestode development and growth.
Identification of extracellular matrix-related and signaling transduction proteins in the HF compartment indicates that they are secreted by germinative cells, brood capsules, or protoscoleces. Some of these proteins were identified in E. granulosus extracellular vesicles isolated from HF of sheep and human hydatid cysts [39, 40]. We hypothesized that production of extracellular vesicles containing these proteins could be a strategy to spread them to the entire cyst wall extension, as a form of coordinating processes at distinct positions in the germinative layer. Germinative layer secretion activity occurs in inward and outward directions in the hydatid cyst, so it is possible that these proteins could also act upon nearby host tissue.
Proteins discussed so far have also been identified by proteomic studies of E. granulosus total HF or extracellular vesicles in sheep or human infections [13, 32, 39, 40, 48]. Considering they are produced by E. granulosus in different hosts and by E. ortleppi too, these classes of proteins, i.e. carbohydrate-metabolizing enzymes, transporters, extracellular matrix-related proteins, signaling proteins, proteases, and inhibitors, seem to have pivotal roles in parasite biology.
Some proteins were identified for the first time, to our knowledge, in the HF from E. granulosus, such as speract scavenger receptor, hexosyltransferase, peptide-methionine sulfoxide reductase, ectonucleotide pyrophosphatase:phosphodiesterase, Cupin 2 barrel domain containing protein, armet protein, semaphorin 5B, EF hand domain containing protein, TGF beta family, and structural maintenance of chromosomes protein. These proteins are not characterized in Echinococcus sp., but they may represent molecular events associated to the lung location of the metacestode. Peptide-methionine sulfoxide reductase acts in an oxidation-reduction process that might protect the parasite tissues from oxidative damage. Scavenger receptors bind different molecules and facilitate endocytosis in mammals [80]. Semaphorins are involved in vesicular transport in C. elegans, which is an important mechanism for cell shape regulation during development [81]. TGF-β/Smad system is described playing a role in parasite tolerance and in liver fibrosis in E. multilocularis infection [82], so we reasoned whether the production of TGF beta family proteins could be a mechanism to modulate the fibrotic response in the host organ. These are possibilities that need to be verified in cystic echinococcosis.
We report for the first time a proteomic survey in E. ortleppi, the species best adapted to cattle as intermediate host. The exclusive repertoire of proteins identified in E. ortleppi HF shows three annexin sequences. There are some indications from studies in other helminths that annexins may act as defense molecules by inducing apoptosis in host immune cells [83–85]. Calpain-A (discussed before) is another protein exclusively found in E. ortleppi HF that mediates apoptosis [59, 60]. Higher levels of apoptotic proteins could be a characteristic of E. ortleppi to deal with host defenses, resulting in better development in bovine hosts. Further investigations will be necessary to determine the existence of differential patterns of apoptosis between E. granulosus and E. ortleppi.
The exclusive E. ortleppi repertoire has different proteins associated to cytoskeleton dynamics, for example: actin depolymerizing factor, cytoskeleton associated protein CAP, gelsolin, and profilin. These findings are interesting because the HF is an extracellular compartment; the roles of these proteins in the HF could be other than those related to actin and microtubule organization. This possibility needs to be investigated in the future, because currently there is no evidence of the function of these proteins in the HF.
Host proteins were also identified in the HF samples, but there were fewer than in the parasitic proteins. They were diverse among the biological replicates from E. granulosus and E. ortleppi, with only four bovine proteins identified from all the HF samples. Different classes of host proteins permeate into the hydatid cyst, and, as we highlight for the parasite protein profile, the cyst physiological state or developmental stage may be related to this heterogeneity. Host proteins can be part of the defense mechanisms that act to eliminate the parasite, as indicated by the enriched GO terms “defense response” and “immune response.” However, the parasite may also take up these proteins for its own use. The specific roles of host proteins in fertile HF are currently unknown, and further thorough studies are necessary to unveil them.
The balance of parasite-host protein content in HF has been associated with E. granulosus hydatid cyst fertility conditions, where fertile cysts have a predominant protein content from the parasite, while infertile hydatid cysts have a higher protein content from the host [14]. Samples collected in this study were from fertile hydatid cysts, so the low number of host proteins identified is consistent with other studies. Infertile hydatid cysts may have a weakened wall and are more susceptible to host protein entry. We identified a large set of parasitic proteins that are related to extracellular matrix and structure maintenance, which supports the idea that in fertile hydatid cysts, the wall is an important barrier to protect the parasite.
Conclusions
Our proteomic analysis highlighted proteins involved in molecular mechanisms, such as adhesion, extracellular structures organization, development regulation, signaling transduction, and enzyme activity, which are present at the host-parasite interface during E. granulosus and E. ortleppi infections in lungs from bovine hosts. The results provide valuable information on the E. granulosus and E. ortleppi molecular mechanisms during host chronic infection, helping to understand biological aspects of cystic echinococcosis caused by different parasite species. The data contribute to knowledge about E. ortleppi, a species that is still poorly characterized molecularly. The observed E. granulosus and E. ortleppi protein profiles can guide the choice of specific molecular processes to use in further studies on these two species. Some of the identified proteins and the pathways they belong to may be of clinical interest because they can be further explored to develop novel and more effective therapies against these and other Echinococcus species.
Supplementary Information
Additional file 1: Figure S1. E. granulosus and E. ortleppi HF protein comparison. (A) Correlation between cysts volume and intensity of bovine albumin band. Thirty-four E. granulosus and 29 E. ortleppi HF samples were qualitatively evaluated using 12% SDS-PAGE gel. The intensity of the bovine albumin band, estimated by using IMAGEJ (https://imagej.nih.gov/ij/) to quantify band intensity, was correlated to the cyst volumes. The six HF samples from cysts with similar sizes (4–6 cm diameter) used in the proteomic analysis are indicated by blue squares (E. granulosus) and red squares (E. ortleppi). (B) Analysis of HF proteins from the selected samples. 50 μg of HF proteins E. granulosus (EG1–3) and E. ortleppi (EO1–3) samples were evaluated by 12% SDS-PAGE gel. For each sample it was possible to identify stained proteins from 10 to 250 kDa. Markers are indicated on the left.
Additional file 2: Table S1. Histone peptides identified in the LC-MS analysis using E. granulosus and B. taurus database.
Additional file 3: Figure S2. Heat map of parasitic proteins identified in HF samples. All identified proteins are represented (blue: lower abundances; red: higher abundances), and their annotations are shown on the left.
Additional file 4: Table S2. Parasitic proteins identified by LC-MS in E. granulosus and E. ortleppi HF samples.
Additional file 5: Table S3. Comparative analysis of proteins identified by LC-MS in E. granulosus and E. ortleppi HF samples.
Additional file 6: Table S4. Predicted GO terms for proteins of unknown functions.
Additional file 7: Table S5. Bovine proteins identified by LC-MS in E. granulosus and E. ortleppi HF samples.
Additional file 8: Figure S3. Bovine proteins identified in hydatid fluid samples from pulmonary cystic echinococcosis. Venn diagrams showing the number of bovine proteins identified: a in E. granulosus HF samples; b in E. ortleppi HF samples; c in HF samples from each species or shared between them. The overall numbers of bovine proteins detected are indicated below the sample/species identification.
Additional file 9: Table S6. Comparative analysis of the proteins detected in at least two biological replicates in one of the species.
Additional file 10: Table S7. Common proteins from E. granulosus and E. ortleppi HF.
Additional file 11: Table S8. Functional classification and gene ontology (GO) enrichment analysis of proteins detected in hydatid fluid of E. granulosus and E. ortleppi.
Additional file 12: Table S9. Summarized GO categorization of proteins detected in E. granulosus hydatid fluid.
Additional file 13: Table S10. Summarized GO categorization of proteins detected in E. ortleppi hydatid fluid.
Additional file 14: Table S11. Gene ontology (GO) enrichment analysis of host proteins detected in hydatid fluid of E. granulosus.
Additional file 15: Table S12. Gene ontology (GO) enrichment analysis of host proteins detected in hydatid fluid of E. ortleppi.
Acknowledgements
Not applicable.
Abbreviations
- AgB
Antigen B
- Ag5
Antigen 5
- CID
Collisional induced dissociation
- Dhh
Desert Hedgehog Protein
- EG
Echinococcus granulosus
- ES
Excretory-secretory
- EO
Echinococcus ortleppi
- FDR
False discovery rate
- GO
Gene ontology
- HF
Hydatid fluid
- HPLC
High performance liquid chromatography
- HRM
High-resolution melting
- LC/MS–MS
Liquid chromatography tandem mass spectrometry
- NSAF
Normalized spectral abundance factor
- PSC
Protoscoleces
- SCX
Strong cation exchange
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Wnt
Wingless/Integrated
Authors' contributions
GBS, KMM, HBF and AZ conceived the study and designed the experiments. GBS and MEB collected and processed the biological material. ESK performed the LC-MS/MS. GBS, EDS, JCL and KMM analyzed the data. EDS and JCL prepared figures and tables. GBS, EDS and AZ wrote the original draft manuscript. EDS, KMM, JCL, ESK, HBF, SMTS and AZ reviewed and edited the final manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grant numbers 472316/2013-3 and 470716/2014-2, Fundação de Amparo à Pesquisa do Rio Grande do Sul (FAPERGS), grant number 001892-25.51/13-0, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), grant 2013/07467-1, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), grant PARASITOLOGIA-1278/2011 and grant Biologia Computacional-23038.010043/2013-02. GBS, EDS and JCL were funded by CAPES scholarships; MEB was funded by CNPq scholarship.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the ProteomeXchange Consortium repository, (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with the dataset identifier PXD019314 and https://doi.org/10.6019/PXD019314.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guilherme Brzoskowski dos Santos and Edileuza Danieli da Silva contributed equally to this work
Contributor Information
Guilherme Brzoskowski dos Santos, Email: guilhermebrzos@gmail.com.
Edileuza Danieli da Silva, Email: edidanieli@yahoo.com.br.
Eduardo Shigueo Kitano, Email: eduardosh.kitano@gmail.com.
Maria Eduarda Battistella, Email: mebattistella@gmail.com.
Karina Mariante Monteiro, Email: karina.monteiro@ufrgs.br.
Jeferson Camargo de Lima, Email: c.jothabiotec@gmail.com.
Henrique Bunselmeyer Ferreira, Email: henrique.bunselmeyer@ufrgs.br.
Solange Maria de Toledo Serrano, Email: solange.serrano@butantan.gov.br.
Arnaldo Zaha, Email: arnaldo.zaha@ufrgs.br.
References
- 1.Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev. 2004;17:107–135. doi: 10.1128/CMR.17.1.107-135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakao M, Lavikainen A, Yanagida T, Ito A. Phylogenetic systematics of the genus Echinococcus (Cestoda: Taeniidae) Int J Parasitol. 2013;43:1017–1029. doi: 10.1016/j.ijpara.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Lymbery AJ. Phylogenetic pattern, evolutionary processes and species delimitation in the genus Echinococcus. Adv Parasitol. 2017;95:111–145. doi: 10.1016/bs.apar.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Thompson RCA. Biology and Systematics of Echinococcus. Adv Parasitol. 2017;95:65–109. doi: 10.1016/bs.apar.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Vuitton DA, McManus DP, Rogan MT, Romig T, Gottstein B, Naidich A, et al. International consensus on terminology to be used in the field of echinococcoses. Parasite. 2020;27:41. doi: 10.1051/parasite/2020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson RCA, Kumaratilake LM, Eckert J. Observations on Echinococcus granulosus of cattle origin in switzerland. Int J Parasitol. 1984;14:283–291. doi: 10.1016/0020-7519(84)90079-1. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez Rojas CA, Romig T, Lightowlers MW. Echinococcus granulosus sensu lato genotypes infecting humans - review of current knowledge. Int J Parasitol. 2014;44:9–18. doi: 10.1016/j.ijpara.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Balbinotti H, Santos GB, Badaraco J, Arend AC, Graichen DÂS, Haag KL, et al. Echinococcus ortleppi (G5) and Echinococcus granulosus sensu stricto (G1) loads in cattle from Southern Brazil. Vet Parasitol. 2012;188:255–260. doi: 10.1016/j.vetpar.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Thompson RCA, McManus DP. Towards a taxonomic revision of the genus Echinococcus. Trends Parasitol. 2002;18:452–457. doi: 10.1016/s1471-4922(02)02358-9. [DOI] [PubMed] [Google Scholar]
- 10.McManus DP, Zhang W, Li J, Bartley PB. Echinococcosis. Lancet. 2003;362:1295–1304. doi: 10.1016/S0140-6736(03)14573-4. [DOI] [PubMed] [Google Scholar]
- 11.Siracusano A, Delunardo F, Teggi A, Ortona E. Cystic echinococcosis: aspects of immune response, immunopathogenesis and immune evasion from the human host. Endocr Metab Immune Disord Targets. 2012;12:16–23. doi: 10.2174/187153012799279117. [DOI] [PubMed] [Google Scholar]
- 12.Monteiro KM, de Carvalho MO, Zaha A, Ferreira HB. Proteomic analysis of the Echinococcus granulosus metacestode during infection of its intermediate host. Proteomics. 2010;10:1985–1999. doi: 10.1002/pmic.200900506. [DOI] [PubMed] [Google Scholar]
- 13.Aziz A, Zhang W, Li J, Loukas A, McManus DP, Mulvenna J. Proteomic characterisation of Echinococcus granulosus hydatid cyst fluid from sheep, cattle and humans. J Proteomics. 2011;74:1560–1572. doi: 10.1016/j.jprot.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 14.dos Santos GB, Monteiro KM, da Silva ED, Battistella ME, Ferreira HB, Zaha A. Excretory/secretory products in the Echinococcus granulosus metacestode: is the intermediate host complacent with infection caused by the larval form of the parasite? Int J Parasitol. 2016;46:843–856. doi: 10.1016/j.ijpara.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Díaz A, Casaravilla C, Barrios AA, Ferreira AM. Parasite molecules and host responses in cystic echinococcosis. Parasite Immunol. 2016;38:193–205. doi: 10.1111/pim.12282. [DOI] [PubMed] [Google Scholar]
- 16.Teichmann A, Vargas DM, Monteiro KM, Meneghetti BV, Dutra CS, Paredes R, et al. Characterization of 14-3-3 isoforms expressed in the Echinococcus granulosus pathogenic larval stage. J Proteome Res. 2015;14:1700–1715. doi: 10.1021/pr5010136. [DOI] [PubMed] [Google Scholar]
- 17.Lorenzatto KR, Monteiro KM, Paredes R, Paludo GP, da Fonsêca MM, Galanti N, et al. Fructose-bisphosphate aldolase and enolase from Echinococcus granulosus: genes, expression patterns and protein interactions of two potential moonlighting proteins. Gene. 2012;506:76–84. doi: 10.1016/j.gene.2012.06.046. [DOI] [PubMed] [Google Scholar]
- 18.Fratini F, Tamarozzi F, Macchia G, Bertuccini L, Mariconti M, Birago C, et al. Proteomic analysis of plasma exosomes from Cystic Echinococcosis patients provides in vivo support for distinct immune response profiles in active vs inactive infection and suggests potential biomarkers. PLoS Negl Trop Dis. 2020;14:e0008586. doi: 10.1371/journal.pntd.0008586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolao MC, Rodriguez Rodrigues C, Cumino AC. Extracellular vesicles from Echinococcus granulosus larval stage: isolation, characterization and uptake by dendritic cells. PLoS Negl Trop Dis. 2019;13:e0007032. doi: 10.1371/journal.pntd.0007032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romig T, Deplazes P, Jenkins D, Giraudoux P, Massolo A, Craig PS, et al. Ecology and life cycle patterns of Echinococcus species. Adv Parasitol. 2017;95:213–314. doi: 10.1016/bs.apar.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Al Kitani FA, Al Riyami S, Al Yahyai S, Al Awahi AH, Al Aawali M, Hussain MH. Abattoir based surveillance of cystic echinococcosis (CE) in the Sultanate of Oman during 2010–2013. Vet Parasitol. 2015;211:208–215. doi: 10.1016/j.vetpar.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Tigre W, Deresa B, Haile A, Gabriël S, Victor B, Van PJ, et al. Molecular characterization of Echinococcus granulosus s.l. cysts from cattle, camels, goats and pigs in Ethiopia. Vet Parasitol. 2016;215:17–21. doi: 10.1016/j.vetpar.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Umhang G, Richomme C, Bastid V, Boucher JM, De Garam CP, Itié-Hafez S, et al. National survey and molecular diagnosis of Echinococcus granulosus sensu lato in livestock in France. Parasitology. 2012;2020:1–18. doi: 10.1017/S0031182020000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casulli A, Manfredi MT, La Rosa G, Di CAR, Genchi C, Pozio E. Echinococcus ortleppi and E. granulosus G1, G2 and G3 genotypes in Italian bovines. Vet Parasitol. 2008;155:168–172. doi: 10.1016/j.vetpar.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Urach Monteiro D, de Azevedo MI, Weiblen C, Correia Ribeiro T, Emmanouilidis J, Tonin AA, et al. Echinococcus granulosus sensu stricto, Echinococcus canadensis (G7), and Echinococcus ortleppi in fertile hydatid cysts isolated from cattle in Southern Brazil. Acta Trop. 2016;164:41–44. doi: 10.1016/j.actatropica.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 26.Corrêa F, Stoore C, Horlacher P, Jiménez M, Hidalgo C, Alvarez Rojas CA, et al. First description of Echinococcus ortleppi and cystic echinococcosis infection status in Chile. PLoS ONE. 2018;13:1–10. doi: 10.1371/journal.pone.0197620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X, Fu Z, Zhang M, Han Y, Han H, Han Q, et al. iTRAQ-based comparative proteomic analysis of excretory–secretory proteins of schistosomula and adult worms of Schistosoma japonicum. J Proteomics. 2016;138:30–39. doi: 10.1016/j.jprot.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Vendelova E, Camargo de Lima J, Lorenzatto KR, Monteiro KM, Mueller T, Veepaschit J, et al. Proteomic analysis of excretory-secretory products of Mesocestoides corti Metacestodes reveals potential suppressors of dendritic cell functions. PLoS Negl Trop Dis. 2016;10:1–27. doi: 10.1371/journal.pntd.0005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suttiprapa S, Sotillo J, Smout M, Suyapoh W, Chaiyadet S, Tripathi T, et al. Opisthorchisviverrini proteome and host-parasite interactions. Adv Parasitol. 2018;102:45–72. doi: 10.1016/bs.apar.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virginio VG, Monteiro KM, Drumond F, de Carvalho MO, Vargas DM, Zaha A, et al. Excretory/secretory products from in vitro-cultured Echinococcus granulosus protoscoleces. Mol Biochem Parasitol. 2012;183:15–22. doi: 10.1016/j.molbiopara.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Monteiro KM, Lorenzatto KR, de Lima JC, dos Santos GB, Förster S, Paludo GP, et al. Comparative proteomics of hydatid fluids from two Echinococcus multilocularis isolates. J Proteomics. 2017;162:40–51. doi: 10.1016/j.jprot.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Ahn C-S, Kim J-G, Han X, Kang I, Kong Y. Comparison of Echinococcus multilocularis and Echinococcus granulosus hydatid fluid proteome provides molecular strategies for specialized host-parasite interactions. Oncotarget. 2017;8:97009–97024. doi: 10.18632/oncotarget.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos GB, Espínola SM, Ferreira HB, Margis R, Zaha A. Rapid detection of Echinococcus species by a high-resolution melting (HRM) approach. Parasit Vectors. 2013;6:327. doi: 10.1186/1756-3305-6-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 36.Maere S, Heymans K, Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 37.Khan IK, Wei Q, Chapman S, Kihara D. The PFP and ESG protein function prediction methods in 2014: effect of database updates and ensemble approaches. Gigascience. 2015;4:43. doi: 10.1186/s13742-015-0083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siles-Lucas M, Sánchez-Ovejero C, González-Sánchez M, González E, Falcón-Pérez JM, Boufana B, et al. Isolation and characterization of exosomes derived from fertile sheep hydatid cysts. Vet Parasitol. 2017;236:22–33. doi: 10.1016/j.vetpar.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 40.Zhou X, Wang W, Cui F, Shi C, Ma Y, Yu Y, et al. Extracellular vesicles derived from Echinococcus granulosus hydatid cyst fluid from patients: isolation, characterization and evaluation of immunomodulatory functions on T cells. Int J Parasitol. 2019;49:1029–1037. doi: 10.1016/j.ijpara.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496:57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H, Zhang W, Zhang L, Zhang Z, Li J, Lu G, et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat Genet. 2013;45:1168–1175. doi: 10.1038/ng.2757. [DOI] [PubMed] [Google Scholar]
- 43.Obal G, Ramos AL, Silva V, Lima A, Batthyany C, Bessio MI, et al. Characterisation of the native lipid moiety of Echinococcus granulosus antigen B. PLoS Negl Trop Dis. 2012;6:e1642. doi: 10.1371/journal.pntd.0001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silva-Álvarez V, Franchini GR, Pórfido JL, Kennedy MW, Ferreira AM, Córsico B. Lipid-free antigen B subunits from Echinococcus granulosus: oligomerization, ligand binding, and membrane interaction properties. PLoS Negl Trop Dis. 2015;9:e0003552. doi: 10.1371/journal.pntd.0003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monteiro KM, Cardoso MB, Follmer C, da Silveira NP, Vargas DM, Kitajima EW, et al. Echinococcus granulosus antigen B structure: subunit composition and oligomeric states. PLoS Negl Trop Dis. 2012;6:e1551. doi: 10.1371/journal.pntd.0001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Folle AM, Kitano ES, Lima A, Gil M, Cucher M, Mourglia-Ettlin G, et al. Characterisation of antigen B protein species present in the hydatid cyst fluid of Echinococcus canadensis G7 genotype. PLoS Negl Trop Dis. 2017;11:e0005250. doi: 10.1371/journal.pntd.0005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y. Proteomic analysis of Taenia hydatigena cyst fluid reveals unique internal microenvironment. Acta Trop. 2017;176:224–227. doi: 10.1016/j.actatropica.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 48.Biosa G, Bonelli P, Pisanu S, Ghisaura S, Santucciu C, Peruzzu A, et al. Proteomic characterization of Echinococcus granulosus sensu stricto, Taenia hydatigena and Taenia multiceps metacestode cyst fluids. Acta Trop. 2022;226:106253. doi: 10.1016/j.actatropica.2021.106253. [DOI] [PubMed] [Google Scholar]
- 49.Figuera L, Gómez-Arreaza A, Avilán L. Parasitism in optima forma: exploiting the host fibrinolytic system for invasion. Acta Trop. 2013;128:116–123. doi: 10.1016/j.actatropica.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 50.Jewett TJ, Sibley LD. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol Cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- 51.de la Torre-Escudero E, Manzano-Román R, Pérez-Sánchez R, Siles-Lucas M, Oleaga A. Cloning and characterization of a plasminogen-binding surface-associated enolase from Schistosoma bovis. Vet Parasitol. 2010;173:76–84. doi: 10.1016/j.vetpar.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 52.Sahoo S, Murugavel S, Devi IK, Vedamurthy GV, Gupta SC, Singh BP, et al. Glyceraldehyde-3-phosphate dehydrogenase of the parasitic nematode Haemonchus contortus binds to complement C3 and inhibits its activity. Parasite Immunol. 2013;35:457–467. doi: 10.1111/pim.12058. [DOI] [PubMed] [Google Scholar]
- 53.Halton DW. Nutritional adaptations to parasitism within the platyhelminthes. Int J Parasitol. 1997;27:693–704. doi: 10.1016/s0020-7519(97)00011-8. [DOI] [PubMed] [Google Scholar]
- 54.Verity CK, McManus DP, Brindley PJ. Vaccine efficacy of recombinant cathepsin D aspartic protease from Schistosoma japonicum. Parasite Immunol. 2001;23:153–162. doi: 10.1046/j.1365-3024.2001.00369.x. [DOI] [PubMed] [Google Scholar]
- 55.McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M. Proteases in parasitic diseases. Annu Rev Pathol Mech Dis. 2006;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- 56.Robinson MW, Dalton JP, Donnelly S. Helminth pathogen cathepsin proteases: it’s a family affair. Trends Biochem Sci. 2008;33:601–608. doi: 10.1016/j.tibs.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Dingwall CB, King RS. Muscle-derived matrix metalloproteinase regulates stem cell proliferation in planarians. Dev Dyn. 2016;245:963–970. doi: 10.1002/dvdy.24428. [DOI] [PubMed] [Google Scholar]
- 58.Syntichaki P, Xu K, Driscoll M, Tavernarakis N. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 59.Momeni HR. Role of calpain in apoptosis. Cell J. 2011;13:65–72. [PMC free article] [PubMed] [Google Scholar]
- 60.Joyce PI, Satija R, Chen M, Kuwabara PE. The atypical calpains: evolutionary analyses and roles in Caenorhabditis elegans cellular degeneration. PLoS Genet. 2012;8:e1002602. doi: 10.1371/journal.pgen.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alcala-Canto Y, Ibarra-Velarde F, Sumano-Lopez H, Gracia-Mora J, Alberti-Navarro A. Effect of a cysteine protease inhibitor on Fasciola hepatica (liver fluke) fecundity, egg viability, parasite burden, and size in experimentally infected sheep. Parasitol Res. 2007;100:461–465. doi: 10.1007/s00436-006-0308-7. [DOI] [PubMed] [Google Scholar]
- 62.Mulcahy G, Dalton JP. Cathepsin L proteinases as vaccines against infection with Fasciola hepatica (liver fluke) in ruminants. Res Vet Sci. 2001;70:83–86. doi: 10.1053/rvsc.2000.0425. [DOI] [PubMed] [Google Scholar]
- 63.Smooker PM, Jayaraj R, Pike RN, Spithill TW. Cathepsin B proteases of flukes: the key to facilitating parasite control? Trends Parasitol. 2010;26:506–514. doi: 10.1016/j.pt.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Sojka D, Hartmann D, Bartošová-Sojková P, Dvořák J. Parasite cathepsin D-like peptidases and their relevance as therapeutic targets. Trends Parasitol. 2016;32:708–723. doi: 10.1016/j.pt.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 65.Ranasinghe SL, McManus DP. Protease inhibitors of parasitic flukes: emerging roles in parasite survival and immune defence. Trends Parasitol. 2017;33:400–413. doi: 10.1016/j.pt.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 66.Yan Y, Liu S, Song G, Xu Y, Dissous C. Characterization of a novel vaccine candidate and serine proteinase inhibitor from Schistosoma japonicum (Sj serpin) Vet Parasitol. 2005;131:53–60. doi: 10.1016/j.vetpar.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 67.Ranasinghe S, McManus DP. Structure and function of invertebrate Kunitz serine protease inhibitors. Dev Comp Immunol. 2013;39:219–227. doi: 10.1016/j.dci.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 68.Fló M, Margenat M, Pellizza L, Graña M, Durán R, Báez A, et al. Functional diversity of secreted cestode Kunitz proteins: inhibition of serine peptidases and blockade of cation channels. PLOS Pathog. 2017;13:e1006169. doi: 10.1371/journal.ppat.1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lascano EF, Coltorti EA, Varela-Díaz VM. Fine structure of the germinal membrane of Echinococcus granulosus cysts. J Parasitol. 1975;61:853–860. [PubMed] [Google Scholar]
- 70.Díaz A, Casaravilla C, Allen JE, Sim RB, Ferreira AM. Understanding the laminated layer of larval Echinococcus II: immunology. Trends Parasitol. 2011;27:264–273. doi: 10.1016/j.pt.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 71.Artavanis-Tsakonas S, Matsuno K, Fortini M. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 72.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3:a004952–a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A, et al. The way Wnt works: components and mechanism. Growth Factors. 2013;31:1–31. doi: 10.3109/08977194.2012.752737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petrov K, Wierbowski BM, Salic A. Sending and receiving hedgehog signals. Annu Rev Cell Dev Biol. 2017;33:145–168. doi: 10.1146/annurev-cellbio-100616-060847. [DOI] [PubMed] [Google Scholar]
- 75.Hemer S, Konrad C, Spiliotis M, Koziol U, Schaack D, Förster S, et al. Host insulin stimulates Echinococcus multilocularis insulin signalling pathways and larval development. BMC Biol. 2014;12:5. doi: 10.1186/1741-7007-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chatterjee S, Sil PC. Targeting the crosstalks of Wnt pathway with Hedgehog and Notch for cancer therapy. Pharmacol Res. 2019;142:251–261. doi: 10.1016/j.phrs.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 77.Dezaki ES, Yaghoobi MM, Taheri E, Almani PG, Tohidi F, Gottstein B, et al. Differential expression of hox and notch genes in larval and adult stages of Echinococcus granulosus. Korean J Parasitol. 2016;54:653–658. doi: 10.3347/kjp.2016.54.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burchell P, Baillie A, Jarero F, James K, Chellappoo A, Zarowiecki M, et al. Genome-wide transcriptome profiling and spatial expression analyses identify signals and switches of development in tapeworms. EvoDevo. 2018;9:1–29. doi: 10.1186/s13227-018-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koziol U, Jarero F, Olson PD, Brehm K. Comparative analysis of Wnt expression identifies a highly conserved developmental transition in flatworms. BMC Biol. 2016;14:1–16. doi: 10.1186/s12915-016-0233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taban Q, Mumtaz PT, Masoodi KZ, Haq E, Ahmad SM. Scavenger receptors in host defense: from functional aspects to mode of action. Cell Commun Signal. 2022;20:2. doi: 10.1186/s12964-021-00812-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka H, Kanatome A, Takagi S. Involvement of the synaptotagmin/stonin2 system in vesicular transport regulated by semaphorins in Caenorhabditis elegans epidermal cells. Genes Cells. 2020;25:391–401. doi: 10.1111/gtc.12765. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, Zhang C, Wei X, Blagosklonov O, Lv G, Lu X, et al. TGF-β and TGF-β/Smad signaling in the interactions between Echinococcus multilocularis and its hosts. PLoS ONE. 2013;8:e55379. doi: 10.1371/journal.pone.0055379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan HL, Xue G, Mei Q, Ding FX, Wang YZ, Sun SH. Calcium-dependent proapoptotic effect of Taenia solium metacestodes annexin B1 on human eosinophils: A novel strategy to prevent host immune response. Int J Biochem Cell Biol. 2008;40:2151–2163. doi: 10.1016/j.biocel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 84.Carneiro-Santos P, Martins-Filho O, Alves-Oliveira LF, Silveira AMS, Coura-Filho P, Viana IRC, et al. Apoptosis: a mechanism of immunoregulation during human schistosomiasis mansoni. Parasite Immunol. 2000;22:267–277. doi: 10.1046/j.1365-3024.2000.00294.x. [DOI] [PubMed] [Google Scholar]
- 85.Nono JK, Pletinckx K, Lutz MB, Brehm K. Excretory/secretory-products of Echinococcus multilocularis larvae induce apoptosis and tolerogenic properties in dendritic cells in vitro. PLoS Negl Trop Dis. 2012;6:e1516. doi: 10.1371/journal.pntd.0001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. E. granulosus and E. ortleppi HF protein comparison. (A) Correlation between cysts volume and intensity of bovine albumin band. Thirty-four E. granulosus and 29 E. ortleppi HF samples were qualitatively evaluated using 12% SDS-PAGE gel. The intensity of the bovine albumin band, estimated by using IMAGEJ (https://imagej.nih.gov/ij/) to quantify band intensity, was correlated to the cyst volumes. The six HF samples from cysts with similar sizes (4–6 cm diameter) used in the proteomic analysis are indicated by blue squares (E. granulosus) and red squares (E. ortleppi). (B) Analysis of HF proteins from the selected samples. 50 μg of HF proteins E. granulosus (EG1–3) and E. ortleppi (EO1–3) samples were evaluated by 12% SDS-PAGE gel. For each sample it was possible to identify stained proteins from 10 to 250 kDa. Markers are indicated on the left.
Additional file 2: Table S1. Histone peptides identified in the LC-MS analysis using E. granulosus and B. taurus database.
Additional file 3: Figure S2. Heat map of parasitic proteins identified in HF samples. All identified proteins are represented (blue: lower abundances; red: higher abundances), and their annotations are shown on the left.
Additional file 4: Table S2. Parasitic proteins identified by LC-MS in E. granulosus and E. ortleppi HF samples.
Additional file 5: Table S3. Comparative analysis of proteins identified by LC-MS in E. granulosus and E. ortleppi HF samples.
Additional file 6: Table S4. Predicted GO terms for proteins of unknown functions.
Additional file 7: Table S5. Bovine proteins identified by LC-MS in E. granulosus and E. ortleppi HF samples.
Additional file 8: Figure S3. Bovine proteins identified in hydatid fluid samples from pulmonary cystic echinococcosis. Venn diagrams showing the number of bovine proteins identified: a in E. granulosus HF samples; b in E. ortleppi HF samples; c in HF samples from each species or shared between them. The overall numbers of bovine proteins detected are indicated below the sample/species identification.
Additional file 9: Table S6. Comparative analysis of the proteins detected in at least two biological replicates in one of the species.
Additional file 10: Table S7. Common proteins from E. granulosus and E. ortleppi HF.
Additional file 11: Table S8. Functional classification and gene ontology (GO) enrichment analysis of proteins detected in hydatid fluid of E. granulosus and E. ortleppi.
Additional file 12: Table S9. Summarized GO categorization of proteins detected in E. granulosus hydatid fluid.
Additional file 13: Table S10. Summarized GO categorization of proteins detected in E. ortleppi hydatid fluid.
Additional file 14: Table S11. Gene ontology (GO) enrichment analysis of host proteins detected in hydatid fluid of E. granulosus.
Additional file 15: Table S12. Gene ontology (GO) enrichment analysis of host proteins detected in hydatid fluid of E. ortleppi.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the ProteomeXchange Consortium repository, (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with the dataset identifier PXD019314 and https://doi.org/10.6019/PXD019314.