Abstract
We present a novel algorithm, implemented in the software ARGinfer, for probabilistic inference of the Ancestral Recombination Graph under the Coalescent with Recombination. Our Markov Chain Monte Carlo algorithm takes advantage of the Succinct Tree Sequence data structure that has allowed great advances in simulation and point estimation, but not yet probabilistic inference. Unlike previous methods, which employ the Sequentially Markov Coalescent approximation, ARGinfer uses the Coalescent with Recombination, allowing more accurate inference of key evolutionary parameters. We show using simulations that ARGinfer can accurately estimate many properties of the evolutionary history of the sample, including the topology and branch lengths of the genealogical tree at each sequence site, and the times and locations of mutation and recombination events. ARGinfer approximates posterior probability distributions for these and other quantities, providing interpretable assessments of uncertainty that we show to be well calibrated. ARGinfer is currently limited to tens of DNA sequences of several hundreds of kilobases, but has scope for further computational improvements to increase its applicability.
Author summary
One of the important challenges in population genetics is to reconstruct the historical mutation, recombination and shared ancestor events that underly a sample of DNA sequences drawn from a population. Aspects of this history can inform us about evolutionary processes, ages of mutations and times of common ancestors, and historical population sizes and migration rates. Performing such inferences is difficult, and progress has been slow over the past two decades. Recently, a new and more efficient way to store sequence data has led to improved simulations and also a fast way to reconstruct some aspects of the history. We augment the new data structure to infer many more details of the history, including the times of events. We also provide approximations of the full probability distributions for all the unknowns, not just plausible values. Because this task is highly challenging, we are limited to relatively small data sets, but we show that our inference algorithm represents an important step forward over those currently available in terms of the accuracy of its inferences.
This is a PLOS Computational Biology Methods paper.
Introduction
A core problem of population genetics is to infer the genealogical history of a sample of homologous DNA sequences, including the recombination, mutation and branching events that produced the currently-observed sample. The Coalescent with Recombination (CwR) [1] provides a simple yet powerful prior distribution for the genealogical history of a set of sequences. A sample path of the CwR can be represented in an Ancestral Recombination Graph (ARG) [2], which embeds the genealogical trees at each genome site into a single graph, incorporating information about the recombination events that cause the genealogy to differ between sites. Knowledge of the true ARG underlying a sample facilitates many evolutionary and demographic inferences [3], and hence inferring the ARG from a set of sequences has been a major challenge for over two decades [4–6]. Note that the term ARG was originally introduced for a stochastic process equivalent to the CwR, but we will here follow the current practice of using the term to refer to a realization of the CwR, a fixed graph that represents a possible genealogical history of the sample.
The CwR includes many discrete and continuous parameters with complex relationships among them. The sequence data can be poorly informative about some parameters, so that multiple topologically-different ARGs have similar likelihoods. For these reasons, only limited progress has been made in the ARG inference problem, resulting in little use of ARG-based inference in population genetics. Instead, inference is often based on summary statistics, leading to both information loss and lack of the quantification of uncertainty that model-based probabilistic inference offers.
Early efforts to tackle the ARG inference problem used importance sampling based on CwR simulations, or Markov Chain Monte Carlo (MCMC) with the CwR as prior [7–12]. Although they produced useful ideas for ARG inference, the algorithms scaled poorly both with sample size and sequence length. The Sequentially Markov Coalescent (SMC) model [13], which simplifies the CwR by assuming that the genealogical trees at each site form a Markov process along the genome, allowed computational advances. However, the SMC does not model “trapped” non-ancestral material (TNAM, genome segments that connect ancestral segments but are not ancestral to the observed sample). By adopting the more realistic CwR, we model the evolution of TNAM due to coalescence and recombination events, which provides information about their rates that is unavailable to SMC-based inference.
The current state-of-art ARG-inference algorithm ARGweaver [14] assumes the SMC model and also discretizes time. These assumptions, combined with an ingenious ‘re-threading’ algorithm, allow ARGweaver to be relatively efficient, at the cost of the SMC and time approximations. Arbores [15] is another MCMC algorithm that also uses the SMC. It takes a different approach that does not discretize time, and performs similarly to ARGweaver.
The Succinct Tree Sequence data structure [16, 17], or tree sequence for short, has recently revolutionised simulation of the CwR and some ARG-based inferences [18], due to enormous efficiency gains obtained by storing only a single copy of a subtree conserved across multiple sites. More recently, the tsinfer software [18] exploits the efficiency of the tree sequence to estimate the ARG for very large sample sizes (∼105 sequences). However, tsinfer only generates a single point estimate, with no underlying statistical model to assess uncertainty, and it does not infer branch lengths. Relate [19], another recent heuristic algorithm, can also generate point estimates for marginal trees, but not the full ARG, and also lacks measures of uncertainty.
ARGinfer is the first probabilistic ARG inference method that exploits the efficiencies of the tree sequence. However, the original tree sequence structure is not sufficiently rich for ARG inference, and we first develop the augmented tree sequence (ATS) data structure to remedy this deficiency. Our other key developments are algorithms to construct an initial ARG compatible with the observed data, to evaluate the likelihood, and to traverse ARG space within an MCMC algorithm. We show in a simulation study that ARGinfer can accurately infer, with well-calibrated probability intervals, ARG properties including the topology and branch lengths of each genealogical tree, the number of recombinations, both ancestral and non-ancestral, time since common ancestors and mutation age. We provide detailed comparisons with ARGweaver, showing that our algorithm provides gains in accuracy at some cost in computational effort.
Results
We simulated data sets under the CwR using msprime [16], discretizing the continuous genome axis into L = 105 sites. We set the mutation rate at μ = 1 × 10−8/site/generation, and (haploid) population size N = 10000. At each site, the allelic state was recorded as ancestral or derived. We assigned three values to the per-site recombination rate r, such that R = μ/r = 1, 2, and 4. For each R, we generated 150 data sets, each with 10 sequences. We rejected and resimulated < 0.5% of data sets that had > 1 mutation at any site.
Convergence diagnostics
We applied ARGinfer to each of the 450 simulated data sets, with run length 2 × 106 iterations, of which 4 × 105 (20%) are discarded as burn-in, after which every 400th sample is retained, resulting in an output chain of length 4 000.
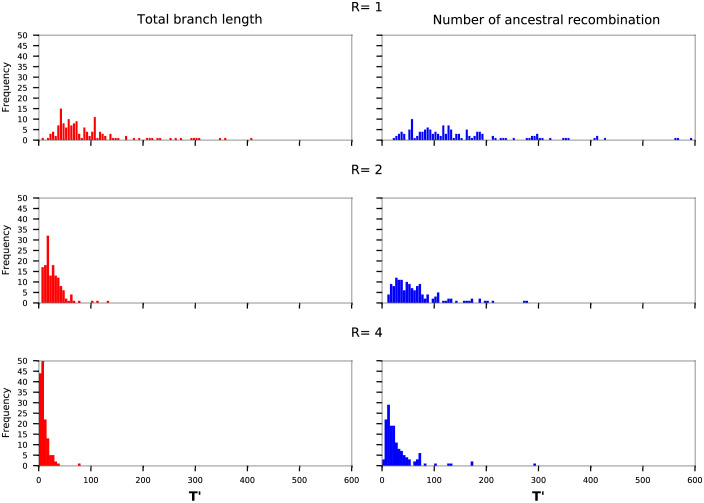
We assessed the convergence of ARGinfer using two heuristics applied to four ARG properties: total branch length, numbers of ancestral and of non-ancestral recombinations, and log(posterior density). First, we ran the MCMC algorithm 10 times on the same data and calculated the Gelman [20]. For all four properties, , indicating that each run reaches approximately the same posterior distribution. (S2 and S3 Figs). We also measured mixing by T′, the first lag for which the empirical autocorrelation was ≤ 0 (Fig 1). When R = 1, the median T′ is around 100, decreasing to roughly 50 and 25 when R = 2 and R = 4. We conclude that convergence is good for R = 2 or 4, and adequate for output chain lengths ≫103 when R = 1 (See S1 Fig for the other two ARG properties).
Fig 1. Plots of T′, the first time lag with autocorrelation ≤ 0, for the total branch length and number of ancestral recombinations for all 450 data sets.
Comparison with ARGweaver
We also applied ARGweaver to the same 450 simulated data sets, with run length 2 × 104 iterations, of which the first 10% are discarded as burn-in, and every 10th sample is retained, resulting in 1800 samples. All parameters were set to their default values, including the number of time points (default value 20). Both methods assumed the true values of μ, r, and N underlying the simulations.
Tables 1, 2 and 3 show that ARGinfer requires more computational time than ARGweaver for the parameter settings chosen, yet ARGweaver tends to generate larger Effective Sample Sizes (ESS) for the two ARG properties. We estimated the coverage of 50% equal-tailed posterior intervals as the fraction of the 150 data sets for which the true (simulation) ARG property value lies in the interval. The equal-tailed 95% interval for the coverage is (0.42, 0.58), and from Tables 2 and 3 we see that both methods appear well calibrated for all combinations of parameter and R shown. Lower root mean square error (RMSE) and higher ESS and Pearson coefficient are all indicators of better performance. For credible intervals, shorter length is better provided that coverage remains close to the target value of 0.5.
Table 1. Computation time for ARGinfer and ARGweaver.
| Method | Iterations | CPU time (hours) | ||
|---|---|---|---|---|
| R = 1 | R = 2 | R = 4 | ||
| ARGinfer | 2 × 106 | 19 | 6.5 | 3 |
| ARGweaver | 2 × 104 | 5 | 4 | 3 |
Table 2. The root mean square error (RMSE), coverage and average length of the posterior 50% equal-tailed intervals, and Effective Sample Size (ESS) for two ARG properties inferred by ARGinfer and ARGweaver.
| R | Method | RMSE | 50% credible interval | ESS | ||
|---|---|---|---|---|---|---|
| Coverage | Average length | |||||
| A. Total branch length per site (in generations) | 1 | ARGinfer | 6729 | 0.44 | 8300 | 360 |
| ARGweaver | 6620 | 0.41 | 7889 | 317 | ||
| 2 | ARGinfer | 6191 | 0.53 | 8714 | 1177 | |
| ARGweaver | 6430 | 0.52 | 8496 | 675 | ||
| 4 | ARGinfer | 6877 | 0.51 | 8827 | 2478 | |
| ARGweaver | 6931 | 0.51 | 8730 | 932 | ||
| B. Number of Ancestral recombination events | 1 | ARGinfer | 9.45 | 0.49 | 12.09 | 171 |
| ARGweaver | 9.53 | 0.54 | 11.73 | 565 | ||
| 2 | ARGinfer | 5.34 | 0.57 | 7.57 | 438 | |
| ARGweaver | 5.70 | 0.49 | 7.35 | 1086 | ||
| 4 | ARGinfer | 3.72 | 0.53 | 4.78 | 970 | |
| ARGweaver | 3.83 | 0.52 | 4.67 | 1398 | ||
Table 3. The root mean square error (RMSE), coverage and average length of the posterior 50% equal-tailed intervals, for two ARG properties inferred by ARGinfer and ARGweaver.
The ESS (reported in Table 2) is not available from ARGweaver for TMRCA and allele age, we report instead the Pearson correlation coefficient between posterior means and true values.
| R | Method | RMSE | 50% credible interval | Pearson coef. | ||
|---|---|---|---|---|---|---|
| Coverage | Average length | |||||
| A. TMRCA | 1 | ARGinfer | 7862 | 0.49 | 8949 | 0.54 |
| ARGweaver | 7968 | 0.49 | 9485 | 0.53 | ||
| 2 | ARGinfer | 6355 | 0.52 | 7769 | 0.61 | |
| ARGweaver | 6518 | 0.51 | 8113 | 0.60 | ||
| 4 | ARGinfer | 5271 | 0.51 | 6366 | 0.55 | |
| ARGweaver | 5517 | 0.45 | 6386 | 0.51 | ||
| B. Allele Age | 1 | ARGinfer | 4096 | 0.49 | 4225 | 0.85 |
| ARGweaver | 4697 | 0.49 | 4925 | 0.77 | ||
| 2 | ARGinfer | 3299 | 0.51 | 3517 | 0.90 | |
| ARGweaver | 3799 | 0.49 | 4077 | 0.85 | ||
| 4 | ARGinfer | 2528 | 0.50 | 2695 | 0.92 | |
| ARGweaver | 3109 | 0.46 | 3027 | 0.86 | ||
We ran the algorithms on Spartan, the University of Melbourne high performance computing system [21], with one Xeon(R) Gold 6154 CPU (1 core) and 15 GB RAM for each data set.
Total branch length
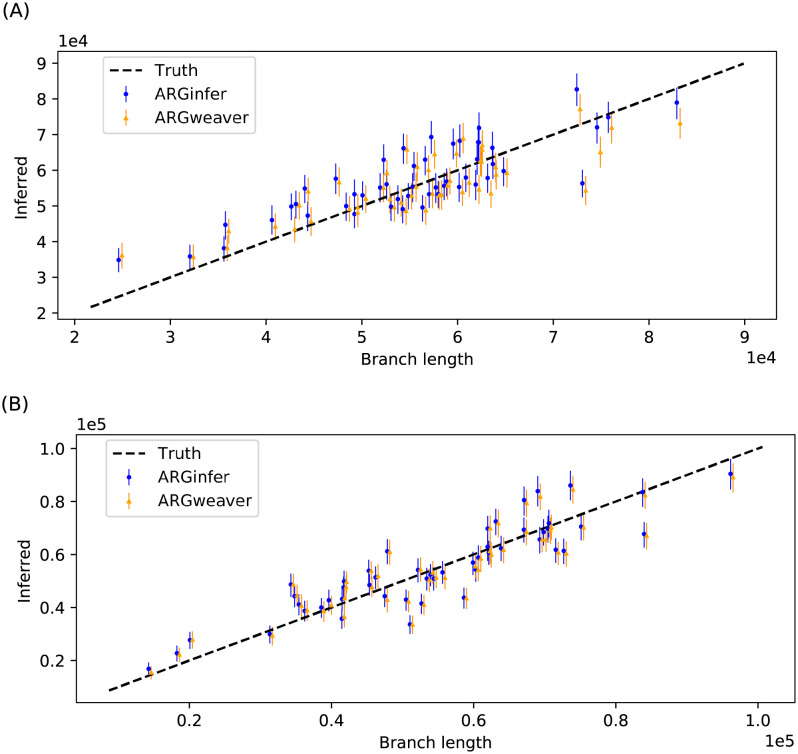
ARGinfer performs similarly to ARGweaver, with slightly better RMSE but longer credible intervals (Table 2A). Both methods show shrinkage towards the prior mean for both small and large values (Fig 2A).
Fig 2. Posterior mean and 50% equal-tailed intervals for the total branch length, measured in generations and averaged over the 105 sites, inferred by ARGinfer and ARGweaver in 50 randomly-chosen data sets.
(A) R = 1 and (B) R = 4.
Number of ancestral recombination events
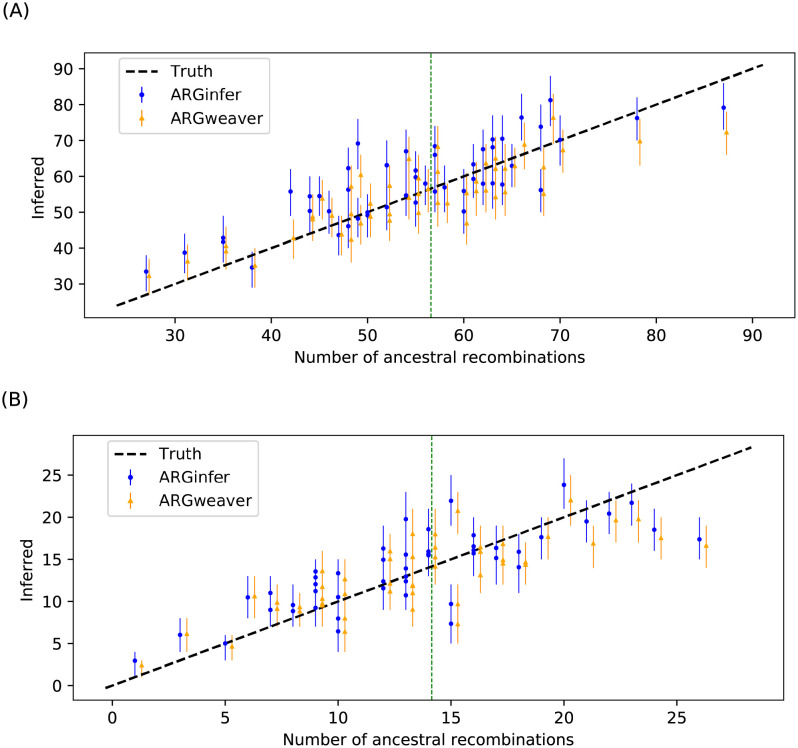
ARGinfer shows better RMSE than ARGweaver for all three R values, but its 50% credible intervals are again longer (Table 2B). Once again, both methods show shrinkage towards the prior mean (Fig 3), which in [14] was suggested to be due to the time discretization of ARGweaver but we suggest is inherent to Bayesian inference.
Fig 3. True versus inferred number of ancestral recombination events for the 50 data sets in Fig 2 for (A) R = 1 and (B) R = 4.
The vertical line segments are 50% credible intervals. The green vertical line indicates the prior mean.
Recombination rate
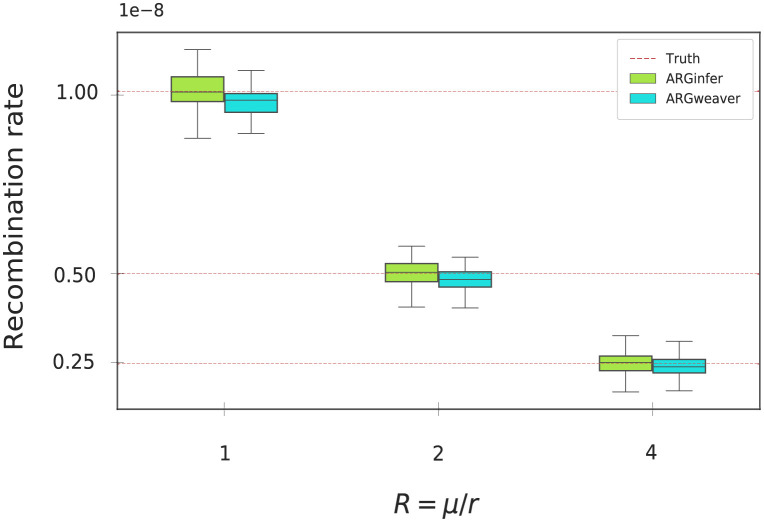
Estimating the recombination rate r as the ratio of the number of ancestral recombinations to the total branch length, ARGinfer is accurate for all three R values, whereas ARGweaver significantly underestimates (Fig 4 and S1 Table). When the number of time intervals in ARGweaver was increased from 20 to 40 to reduce the impact of time discretization, the level of bias was reduced but remained substantial (S1 Table).
Fig 4. Estimated recombination rates for 150 simulated data sets from ARGinfer and ARGweaver for R = 1, 2, and 4.
The red dashed lines (“Truth”) show the mean of the true number of ancestral recombination events divided by the true total branch length. See S1 Table for p-values from testing equality of true and inferred values.
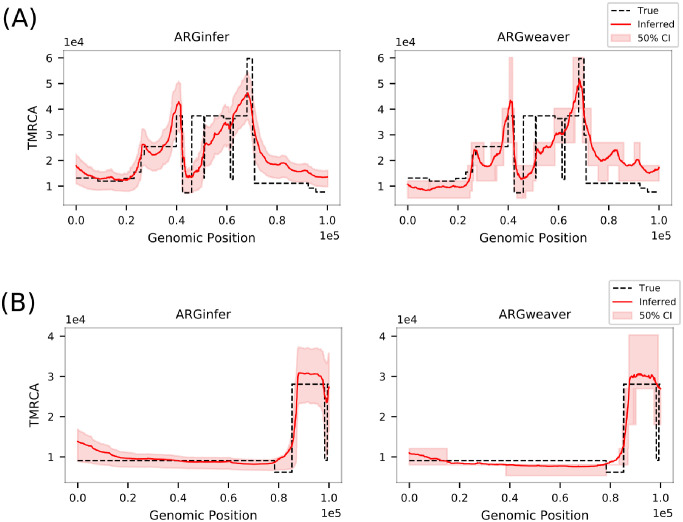
Time since the most recent common ancestor (TMRCA) at each genome site
ARGinfer has a higher correlation between the posterior mean and true TMRCA, lower RMSE, and shorter 50% credible intervals than ARGweaver for all R (Table 3A). One reason for its superior performance is that ARGinfer assumes the CwR with continuous time, whereas the impact of the time-discretized version of the SMC adopted by ARGweaver can be seen in the box-shaped 50% credible intervals in Fig 5, reflecting that the interval endpoints are limited to a relatively small number of pre-specified time points. For some genomic intervals, the 0.25 and 0.75 quantiles are identical, reflecting a high concentration of probability at a single time point.
Fig 5. True (black dashed line) TMRCA, in units of 104 generations, and posterior mean (red line) inferred by ARGinfer (left) and ARGweaver (right) for randomly-selected, simulated data sets.
(A) R = 1, (B) R = 4, and red shading shows 50% credible intervals.
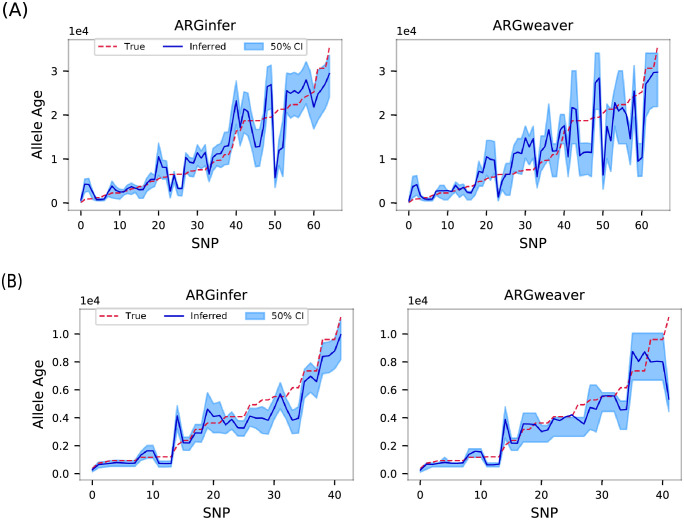
Allele age
We define the age of an allele as the mid-point of the tree branch on which the mutation occurred. In Table 3B, we observe that ARGinfer outperforms ARGweaver in terms of all four statistics for each value of R. The larger uncertainty for ARGweaver is due to the time discretization, because its time points are on a logarithmic scale so that more recent times are inferred more accurately, whereas Fig 6 shows that ARGweaver does not perform well for older mutations because older branch lengths are poorly measured.
Fig 6. True (red dashed line) allele ages, in units of 104 generations, and posterior mean (blue line) inferred by ARGinfer (left) and ARGweaver (right) for the simulated data sets used in Fig 5.
Along the x axis SNPs are ordered by increasing value of true allele age. (A) R = 1, (B) R = 4, and blue shading shows 50% credible intervals.
Discussion
ARGinfer represents important progress on a core problem in population genetics: inferring the evolutionary history underlying a sample of DNA sequences. It is the first practical method for inference under the CwR prior, rather than the SMC approximation. The CwR provides one of the simplest realistic models for the genealogy of a sample, under neutrality, constant population size, and random mating (no population structure). In our simulation study, ARGinfer estimates ARG parameters accurately and with well-calibrated credible intervals, improving on ARGweaver for inferences about the recombination rate, TMRCA, and allele age, while remaining approximately as accurate for other parameters.
The key innovation of ARGinfer is the ATS data structure, which extends the highly-successful tree sequence to incorporate mutation events and all details of the ancestral recombination graph (ARG). Other key developments include proposal steps that efficiently explore the ARG space, and algorithms for efficient likelihood evaluation and constructing an initial ARG compatible with the sequence data.
The main benefit of ARGinfer is well-calibrated probability distributions for features of the genealogical history of a set of DNA sequences. These include the topology and branch lengths of the genealogical tree at each site, allele age (time since mutation) at each polymorphic site, and the number of recombinations. By exploiting the efficiency properties of the ATS, ARGinfer is the first practical method to model trapped non-ancestral material (TNAM), which does not affect the marginal distributions at individual sites but is informative about multi-site joint distributions, including recombination rate and linkage disequilibrium (LD). Use of TNAM information helps ARGinfer improve recombination rate inferences, whereas ARGweaver ignores TNAM and underestimates the recombination rate.
The current implementation of ARGinfer can handle about 15 sequences of length up to 5 × 105 sites, compared with the original MCMC for CwR algorithm [12] that (at the time) handled 10 sequences with length ≈ 103 sites. This efficiency gain is largely due to the following reasons:
Recording mutations in the ATS allows likelihood evaluation without comparing sequences site by site.
Proposed ARGs that are incompatible with the dataset are rejected immediately, without costly likelihood evaluation.
Our simulation study shows that ARGinfer provides better inferences than the current state-of-art ARGweaver for evolutionary parameters, and in particular the recombination rate. This appears to be the result of directly modelling TNAM in ARGinfer: the non-ancestral recombinations ignored by ARGweaver are typically much more numerous than the ancestral recombinations, though more difficult to infer. ARGinfer is somewhat more computationally demanding than ARGweaver, but as the approach is novel we expect further advances, particularly in the proposal steps.
ARGinfer assumes that the ancestral allele is known at each site, which can often be accurately inferred from related species. It would be possible to also infer the ancestral allele at substantial computational cost. ARGinfer also assumes at most one mutation event in the history of each site, similar to the infinite-sites mutation model. This assumption means that ARGinfer cannot analyse data sets with more than two alleles at a site, Further, back-mutation (two mutation events, the second reversing the first) can adversely affect inferences. In the short term, these limitations can be addressed by removing sites with more than two alleles or that show signs of back-mutation. The resulting bias in estimates of the mutation rate will be low in human data of the scale considered here. A better, but computationally costly, solution is to use a mutation model that allows multiple hits per site, such as the Jukes-Cantor model [22] adopted by ARGweaver, which requires integration over all possible mutation events on each tree branch.
The current proposal steps in ARGinfer are relatively small rearrangements of the ARG, which helps to verify reversibility but can contribute to poor mixing for large sample sizes. This issue can be overcome, at least in principle, by employing non-reversible methods for sampling the posterior. An example is the zig-zag process, which has recently been introduced for the coalescent [23]. The zig-zag process resembles Probabilistic Path Hamiltonian Monte Carlo [24] in that continuous branch lengths and discrete tree topologies are embedded in a common continous space. Extensions of any of these methods to include recombination would present a significant challenge, but also potentially substantial gains.
In summary, the MCMC algorithm developed in this paper is the first probabilistic method that uses the tree sequence in the ARG inference problem based on the CwR. ARGinfer provides accurate estimates with well-calibrated measures of uncertainty for ARG parameters. Knowledge of these parameters can help understanding of biological processes such as gene-phenotype associations, disease processes, and genome structure [5, 25].
ARGinfer is useful for genome regions on the scale of 0.5 megabase, and can be used to infer both evolutionary parameters and properties of the genealogical history, such as the recombination rate and allele ages. It may also be useful to extend ARGinfer to multi-population models in order to estimate demographic parameters, including divergence times and effective population sizes. The fact that ARGinfer does not rely on a discretization of time facilitates ancient DNA studies with samples obtained at different times [4].
Methods
The coalescent with recombination
We begin by briefly reviewing the CwR and introducing some definitions. The CwR [1] is a stochastic process that reduces to the standard coalescent [26] at each site, but it includes recombination to allow joint modelling of all the coalescent trees in a genome interval. The Hudson algorithm [1] simulates the CwR backwards in time with two possible events: recombination and common ancestor (CA). A CA event merges two sequences into their common ancestor; if the sequences share ancestral material, we call it a coalescence event.
A recombination event breaks a sequence in two at a randomly-chosen point. Two newly-created sequences carry ancestral material to the left and to the right of the breakpoint. Both of the new sequences include sites that are not ancestral to any observed sequences (non-ancestral material, NAM). Sometimes a segment of NAM is trapped between two ancestral segments, called Trapped Non-ancestral Material (TNAM). Any sequence generated that consists entirely of NAM can be discarded with no loss of information. If a recombination breakpoint occurs either within ancestral material (ancestral recombination) or within a segment of TNAM, it alters correlations in the data [27] and can contribute to inferences.
To simulate the ARG of n DNA sequences, the Hudson algorithm starts from the current generation and simulates CA and recombination events backwards-in-time until a single sequence remains. If we continue to keep track of NAM lineages and segments which have reached their most common recent ancestor (MRCA), this is known as the “big ARG” [26, 28]. We keep track of a smaller “little ARG” (see [28]), which contains the same amount of information for inference but discards non-informative sequences.
Augmented Tree Sequence
Representing an ARG so that the embedded information is easily accessible and redundancies are avoided is challenging. Neighbouring coalescent trees within an ARG are often identical, or differ in just a subtree. In SMC-based methods, it is common to store the marginal trees separately, which results in much inefficient duplication. The tree sequence [16] represents marginal trees so that shared, neighboring subtrees are stored only once. This idea leads to a significant saving in storage and speed in processing and accessing ARG information [18]. Our goal is to employ this efficiency to improve the speed and accuracy of inferring the ARG under the CwR.
The tree sequence, however, does not contain all the information required for ARG inference. To be able to evaluate the CwR prior and explore the ARG space, more information is needed: recombination and non-coalescence CA events, recombination breakpoints and times, and the parent sequences of recombinations. We augment the tree sequence, retaining its efficient storage properties, to include this information, calling the new data structure the Augmented Tree Sequence (ATS). The ATS is a collection of linked branches that connect the ancestral sequences. Each branch consists of a sequence of segments that encode the genomic regions of the ancestral sequence on that branch, which are linked to extract and update the information efficiently.
Under the infinite sites model (ISM), there is exactly one mutation per segregating site [29]. The ATS assigns each mutation to a branch. If the mutation can be assigned to multiple branches consistent with the allele data, we assign it to the lowest of these branches (closest to the present time). This policy is useful for mixing and does not bias inferences of allele age, see the “Augmented tree sequence” section in S1 Text for more details.
The Markov chain Monte Carlo method
We aim to sample from the posterior density
| (1) |
where D is a sample of n DNA sequences, G is the set of ARGs consistent with D, and Θ = (μ, r, N), where N is the effective population size, while r and μ are recombination and mutation rates, both per site per generation. The first term on the right-hand side of (1) is the likelihood. We use a discrete approximation of the ISM: the continuous genome interval is discretized into a finite number of sites, and if this results in >1 mutation at any site, the dataset is rejected and resimulated. The likelihood is calculated by
| (2) |
where T is the set of all distinct tree branches (branches spanning multiple sites are recorded only once), |mv| is the number of mutations on branch v, while lv is the branch length and gv is the length of genomic interval of branch v.
The second term on the right-hand side of (1) is the prior probability given by the CwR. For an ARG with E events at times t1, …, tE (in generations), we have
| (3) |
where
ki and are the total number of lineages and recombination links (a gap between sites where a recombination can occur) immediately before time ti, and Ic is an indicator function with value 1 for a CA, and 0 for a recombination.
We refer to the posterior distribution P(G|D; Θ) as the CwR+ and develop an MCMC algorithm to sample from it. The first step of the algorithm is to construct an initial ARG for D. Finding an ARG compatible with D can be challenging because most ARGs are incompatible with a given data set, even for a small number of sequences. Using some ideas from [30], we devised a heuristic algorithm to construct a compatible ARG from D (details are given in the “Initial ARG construction” section in S1 Text). The next step is to explore the state space of the ARG using a random walk in which steps from the current ARG (Gj) to a new ARG (Gj+1) are drawn from a proposal distribution Q(.). Gj+1 is accepted with probability
| (4) |
otherwise, Gj is kept. After a burn-in period, each ARG visited by the Markov chain can be regarded as a sample from the CwR+. The last term in Eq (4) is the Hastings term [31]. The numerator is the reverse transition probability, and the denominator is the forward transition probability.
We define Q(.) in terms of six proposal types (in short, proposals). Details of the proposals are discussed in the “Proposal types” section in the S1 Text. In brief, the six proposals are:
Subtree-Pruning-and-Regrafting (SPR), in which a branch is pruned from the ARG and reattached to the ARG at an older time. For this move to be reversible, we do not allow the pruned lineage to experience recombination. Thus, an SPR keeps the number of recombinations fixed.
Removing an existing recombination from the ARG, transferring the ancestry of one parent to the other parent.
Adding a new recombination to a lineage. This is the reverse of the second proposal.
Resampling the breakpoint of a recombination event.
Rearranging a subtree of the ARG in proportion to the prior probability, allowing changes on the number of recombinations with no limitation. This is a modification of the proposal introduced in [12] and we call it the “Kuhner move”.
Re-simulate the event times of the ARG, according to the CwR.
On average for our simulated data sets, the acceptance probability for ARGinfer is 0.3. The proposal types 1 to 6 are chosen with probabilities 1/14, 1/14, 1/14, 1/14, 5/14, and 5/14, respectively. We gave a higher chance to the time adjustment and the Kuhner move, because the former is the only proposal type that resamples the event times, and the latter introduces the biggest change to the ARG. We examined a range of values for these probabilities, and while the current values gave the best acceptance rate among those we considered, further improvement may be possible.
We calculated the ESS by
where ρ(h) is the autocorrelation at lag h, T the number of MCMC outputs, and T′ the time when the autocorrelation first becomes negative [32].
Further details on the algorithm are provided in S1 Text. It is implemented as a Python package ARGinfer, available at https://github.com/alimahmoudi29/ARGinfer.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
The vertical line segments are 50% credible intervals. The dotted green vertical lines indicate the prior means.
(PDF)
Red shading shows 50% credible intervals.
(PDF)
Along the x axis SNPs are ordered by increasing value of true allele age. Blue shading shows 50% credible intervals.
(PDF)
Data Availability
All code written in support of this publication is publicly available at https://github.com/alimahmoudi29/arginfer.
Funding Statement
A. M. was funded by the Melbourne Research Scholarship, the Xing Lei Scholarship, the Professor Maurice H. Belz Fund, and the Albert Shimmins Fund. J. Koskela was supported by the UK Engineering and Physical Sciences Research Council grant EP/R044732/1. J. Kelleher was supported by the Robertson Foundation. D.B., Y.C. and J. Kelleher were supported by the Australian Research Council grant DP210102168. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hudson RR. Properties of a neutral allele model with intragenic recombination. Theoretical Population Biology. 1983;23(2):183–201. doi: 10.1016/0040-5809(83)90013-8 [DOI] [PubMed] [Google Scholar]
- 2. Griffiths RC, Marjoram P. An ancestral recombination graph. Institute for Mathematics and its Applications. 1997;87:257. [Google Scholar]
- 3. Harris K. From a database of genomes to a forest of evolutionary trees. Nature Genetics. 2019;51(9):1306–1307. doi: 10.1038/s41588-019-0492-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubisz M. Inferring the population history of ancient hominins through use of the ancestral recombination graph [PhD dissertation]. Cornell University; 2019.
- 5. Arenas M. The importance and application of the ancestral recombination graph. Frontiers in Genetics. 2013;4:206. doi: 10.3389/fgene.2013.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahmoudi A. Inference under the coalescent with recombination [PhD dissertation]. University of Melbourne; 2020.
- 7. Griffiths RC, Marjoram P. Ancestral inference from samples of DNA sequences with recombination. Journal of Computational Biology. 1996;3(4):479–502. doi: 10.1089/cmb.1996.3.479 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y, Rannala B. Bayesian inference of fine-scale recombination rates using population genomic data. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2008;363:3921–3930. doi: 10.1098/rstb.2008.0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nielsen R. Estimation of population parameters and recombination rates from single nucleotide polymorhisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fearnhead P, Donnelly P. Estimating recombination rates from population genetic data. Genetics. 2001;159(3):1299–1318. doi: 10.1093/genetics/159.3.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins PA. Importance sampling on the coalescent with recombination [PhD dissertation]. University of Oxford; 2008.
- 12. Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of recombination rates from population data. Genetics. 2000;156(3):1393–1401. doi: 10.1093/genetics/156.3.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McVean GA, Cardin NJ. Approximating the coalescent with recombination. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2005;360(1459):1387–1393. doi: 10.1098/rstb.2005.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rasmussen MD, Hubisz MJ, Gronau I, Siepel A. Genome-wide inference of ancestral recombination graphs. PLoS Genetics. 2014;10(5):e1004342. doi: 10.1371/journal.pgen.1004342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heine K, Beskos A, Jasra A, Balding D, De Iorio M. Bridging trees for posterior inference on ancestral recombination graphs. Proceedings of the Royal Society AL Mathematical, Physical and Engineering Sciences. 2018;474:20180568. doi: 10.1098/rspa.2018.0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kelleher J, Etheridge AM, McVean G. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Computational Biology. 2016;12(5):e1004842. doi: 10.1371/journal.pcbi.1004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelleher J, Thornton KR, Ashander J, Ralph PL. Efficient pedigree recording for fast population genetics simulation. PLoS Computational Biology. 2018;14(11):e1006581. doi: 10.1371/journal.pcbi.1006581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelleher J, Wong Y, Wohns AW, Fadil C, Albers PK, McVean G. Inferring whole-genome histories in large population datasets. Nature Genetics. 2019;51(9):1330–1338. doi: 10.1038/s41588-019-0483-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Speidel L, Forest M, Shi S, Myers SR. A method for genome-wide genealogy estimation for thousands of samples. Nature Genetics. 2019;51(9):1321–1329. doi: 10.1038/s41588-019-0484-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical Science. 1992;7(4):457–472. doi: 10.1214/ss/1177011136 [DOI] [Google Scholar]
- 21. Lafayette L, Sauter G, Vu L, Meade B. Spartan performance and flexibility: An HPC-cloud chimera. OpenStack Summit, Barcelona. 2016;27. [Google Scholar]
- 22. Jukes TH, Cantor CR, Munro H, et al. Evolution of protein molecules. Mammalian Protein Metabolism. 1969;3(21):132. [Google Scholar]
- 23. Koskela J. Zig-zag sampling for discrete structures and non-reversible phylogenetic MCMC. Journal of Computational and Graphical Statistics 2022, doi: 10.1080/10618600.2022.2032722 [DOI] [Google Scholar]
- 24.Dinh V, Bilge A, Zhang C, Matsen IV FA. Probabilistic path Hamiltonian Monte Carlo. In: Proceedings of the 34th International Conference on Machine Learning-Volume 70; 2017. p. 1009–1018.
- 25. Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayré SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415(6872):644–646. doi: 10.1038/415644a [DOI] [PubMed] [Google Scholar]
- 26. Nordborg M. Coalescent Theory. Chapter 5 of Handbook of Statistical Genomics, 4th ed. 2019; p. 145–175. [Google Scholar]
- 27. Wang Y, Zhou Y, Li L, Chen X, Liu Y, Ma ZM, et al. A new method for modeling coalescent processes with recombination. BMC Bioinformatics. 2014;15(1):273. doi: 10.1186/1471-2105-15-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hein J, Schierup M, Wiuf C. Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press, USA; 2004. [Google Scholar]
- 29. Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics. 1969;61(4):893. doi: 10.1093/genetics/61.4.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nguyen TTP, Le VS, Ho HB, Le QS. Building ancestral recombination graphs for whole genomes. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2017;14(2):478–483. doi: 10.1109/TCBB.2016.2542801 [DOI] [PubMed] [Google Scholar]
- 31. Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;. doi: 10.1093/biomet/57.1.97 [DOI] [Google Scholar]
- 32. Kass RE, Carlin BP, Gelman A, Neal RM. Markov chain Monte Carlo in practice: a roundtable discussion. The American Statistician. 1998;52(2):93–100. doi: 10.1080/00031305.1998.10480547 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
The vertical line segments are 50% credible intervals. The dotted green vertical lines indicate the prior means.
(PDF)
Red shading shows 50% credible intervals.
(PDF)
Along the x axis SNPs are ordered by increasing value of true allele age. Blue shading shows 50% credible intervals.
(PDF)
Data Availability Statement
All code written in support of this publication is publicly available at https://github.com/alimahmoudi29/arginfer.