Abstract
The challenge continues globally triggered by the absence of an approved antiviral drug against COVID-19 virus infection necessitating global concerted efforts of scientists. Nature still provides a renewable source for drugs used to solve many health problems. The aim of this work is to provide new candidates from natural origin to overcome COVID-19 pandemic. A virtual screening of the natural compounds database (47,645 compounds) using structure-based pharmacophore model and molecular docking simulations reported eight hits from natural origin against SARS-CoV-2 main proteinase (Mpro) enzyme. The successful candidates were of terpenoidal nature including taxusabietane, Isoadenolin A & C, Xerophilusin B, Excisanin H, Macrocalin B and ponicidin, phytoconstituents isolated from family Lamiaceae and sharing a common ent-kaurane nucleus, were found to be the most successful candidates. This study suggested that the diterpene nucleus has a clear positive contribution which can represent a new opportunity in the development of SARS-CoV-2 main protease inhibitors.
Keywords: COVID-19, Pharmacophore, Terpenoids, Ent-kaurane, Main protease inhibitors
Graphical abstract
1. Introduction
The current pandemic of coronavirus disease 2019 (COVID-19) pandemic has menaced human health with over 27.9 million infection cases and 905,000 death till September 10, 2020, with a continuous increase in number (Bai et al., 2020). The shortage of specific effective medications for COVID-19 necessitates serious and combined attempts of scientists through the world.
Several strains of COVID are causing infections in humans and animals, among the strains of human CoV causing severe symptoms, Middle East respiratory syndrome-related coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Su et al., 2016). The etiological agent of COVID-19 is SARS-CoV-2, which is genetically similar (78%) to SARS-CoV, which led to the SARS outbreak in 2003 (Sacco et al., 2020). Identification of drug targets is imperative to identify effective antivirals. SARS-CoV-2 initiates its infection via the interaction with angiotensin-converting enzyme 2 (ACE2) receptors and transmembrane protease, serine 2 (TMPRSS2) on the host cell membrane (Hoffmann et al., 2020; H. Zhang et al., 2020). SARS-COV-2 main protease (Mpro) enzyme is considered a promising drug target, as it is dissimilar to human proteases. The Mpro is very important for the proteolytic maturation of the virus. It is perceived as a possible target to inhibit the dissemination of infection by preventing the cleavage of viral polyprotein through blocking its active sites. The sequence and structure of Mpro are closely related to those from other beta coronaviruses. Mpro monomer consists of N-terminal domain-I, N-terminal domain-II, and C-terminal domain-III. The active site of the enzyme contains a catalytic dyad having Cys145 and His41 (Khan et al., 2020; Mirza and Froeyen, 2020; Ullrich and Nitsche, 2020).
The tough and changing nature of this virus unified the efforts of researchers from different disciplines across the world to deal with this pandemic through restudying the possible effects of currently available medications, examining passive immunity, and looking for vaccines against the virus. For viral infection control, it is possible to use a prophylactic technique and/or drug therapy. The major issue while designing a drug is that, it is very difficult to attack the virus without generating side effects in the host as the virus use the host it to survive and replicate, so most of the metabolic pathways are the same (Akram et al., 2018). The first tried drugs were, the antimalarials Chloroquine and hydroxychloroquine however, owing to its side effects, a shift has been made to the study of natural quinine to alleviate the symptoms. Herbal remedies have served humankind since antiquity as fruitful drug leads remedy for many diseases including viral infections, where more than 90 drugs belonging to different chemical classes were approved against HIV, hepatitis B, and CCV (Adhikari et al., 2020; De Clercq and Li, 2016; Khan et al., 2021). Hundreds of herbal products were screened for their possible effects against coronaviruses. After the first outbreak of SARS-CoV, glycyrrhizin (a saponin from Liquorice roots) demonstrated its ability to inhibit the replication of SARS-associated coronavirus (EC50 300–600 mg/L) providing a substitute to Ribavirin causing hemolysis and a significant drop in haemoglobin levels in SARS patients (Cinatl et al., 2003), while the inclusion of 2-acetamido-β-d-glucopyranosylamine into the glycoside chain of glycyrrhizin leads to a 10-fold increase in its activity (Hoever et al., 2005). Studies showed that pharmacological candidates either inhibit viral entry and replication or elicit an immune reaction to yield type I IFN against SARS-CoV (Báez-Santos et al., 2015; Kim et al., 2011). Terpenoids isolated from different plants were also reported to be able to block the S proteins of the virus that inhibits viral replication (Wen et al., 2007).
To discover the best drug candidate against SARS-CoV-2, bio-informatic tools play an important role relying on the evaluation of the available data and selecting the most promising drug candidates helped by the recent findings for the use of herbal medicines in handling the COVID-19 outbreak with the added benefit of reducing experimental costs and time. The drug repurposing strategy with various ways offers a successful tool to discover more drugs with antiviral activities against COVID-19. One of the modern virtual screening best ways to find new compounds that can bind strongly to the enzyme active site is to generate a pharmacophore model from various co-crystallized inhibitors, this helps to explore the essential features required for an inhibitor. Many valuable studies concerning drug repurposing strategies have been lately carried out against SARSCoV-2 (Cavasotto and Di Filippo, 2021; Fayed et al., 2021; Ibrahim et al., 2020, 2021; Meyer-Almes, 2020).
The current research work aims to propose hits that can bind effectively at the active sites of the main protease of SARS-CoV-2, for drug development and lead optimization. A previously tested pharmacophore (Refaey et al., 2021) is used in this study to test a large database of natural constituents (CMAUP)(Zeng et al., 2019) the found hits were filtered through drug-like filters, and then a molecular docking study was performed to confirm the results.
2. Experimental
2.1. Virtual screening
The used database was downloaded from (http://bidd.group/CMAUP/). Molecular Operating Environment (MOE, 2019.0102) software was used to prepare the database through washing and energy minimization. All compounds were subjected to pharmacophore search using a previously published pharmacophore model (Refaey et al., 2021). The hits were subjected to filtering on drug-likeness properties based on Lipinski's rule of five. The final list of compounds has those with a maximum of only one violation to that rule.
2.2. Molecular docking
Molecular Operating Environment (MOE, 2019.0102) software was used to carry out all the molecular modeling studies. All minimizations were performed by MOE until a root-mean-squared-deviation (RMSD) gradient of 0.05 kcal mol−1 Å−1 with the MMFF94x force field and the partial charges were automatically calculated.
The X-ray crystallographic structure of SARS-COV-2 main protease enzyme co-crystalized with its bound ligand X77 (PDB ID: 6W63) was downloaded from the protein data bank (https://www.rcsb.org/structure/6W63). For each co-crystallized enzyme, water molecules and ligands which are not involved in the binding were removed, the protein was prepared for the docking study using Protonate 3D protocol in MOE with default options. The co-crystalized ligand (X77) was used to define the binding site for docking. Triangle Matcher placement method and London dG scoring function were used for docking (Jin et al., 2020). The amino acid interactions were visualized by Discovery Studio Visualizer v17.2.0.16349.
2.3. Molecular dynamics
Molecular dynamics simulations were carried out using GROMACS 2020.3 (GROningen Machine for Chemical Simulations) (Abraham et al., 2015) molecular dynamics package on the docked complexes of the three top compounds; NPC 472282, NPC 184170 and NPC 107385, in order to confirm the docking results and further examine the behaviour of the ligands in the binding pocket. Pdb2gmx was used to generate the topology file for the protein, using Amber 94 Force field (Hornak et al., 2006). On the other hand, the topology and parameter files of the ligand using the same force field were generated using the Antechamber Python parser interface (ACPYPE) tools (Sousa da Silva and Vranken, 2012). The TIP3P model was used to solvate the complexes in an orthorhombic box and 72 Na+ and 68 Cl- ions were added to all three complexes to neutralize the system and achieve a physiological NaCl concentration of 0.150 mM. Energy minimization was carried out using steepest decent for 50000 steps. The minimized system was the subjected to two equilibration runs, a 100 ps NVT (constant number of particles, volume and temperature at 300 K) and a 100 ps NPT (constant number of particles, pressure at 1 bar and temperature at 300 K) were carried out. This was followed by a 100 ns production run where the Particle Mesh Ewald method (PME) (Cerutti et al., 2009) was used to calculate the electrostatic interactions and Linear constraint solver algorithm (LINCS) (Hess, 2007) was used to constrain bond length. The resultant trajectory was then examined using visual molecular dynamics 1.9.3 (VMD) (Humphrey et al., 1996) and UCSF Chimera 1.13.1 (Pettersen et al., 2004).. The root mean square deviation (RMSD) and the number of hydrogen bonds were calculated and XMgrace was then used to generate the 2D plots.
3. Results and discussion
3.1. Virtual screening
A database of natural compounds (Collective Molecular Activities of Useful Plants, CMAUP) was downloaded, which was developed by the Bioinformatics & Drug Design group (BIDD), Department of Pharmacy, National University of Singapore, Singapore (Zeng et al., 2019). The previously tested pharmacophore model for SARS-COV-2 main protease was used for virtual screening of the selected database (47,645 compounds). Sixteen molecules were hits for the pharmacophore (Fig. 1 ) and were selected (Table 1 ). To refine the resulted hits, a drug-likeness filter (Lipinski's rule of five) was applied to choose the compounds with suitable pharmacokinetic properties (Lipinski, 2004; Lipinski et al., 1997; Walters and Murcko, 2002). The results are summarized in Table 2 . Eight compounds were selected and the deviation from the pharmacophore model was expressed as root mean square deviation (RMSD) (Table 3 ) while superimposition on the pharmacophore model was shown in (Fig. 2 ).
Fig. 1.
Pharmacophoric features for SARS-COV-2 main protease enzyme (ligand-based pharmacophore).
Table 1.
Sixteen selected molecules through virtual screening of the pharmacophore.
| NPC157929 | NPC309388 |
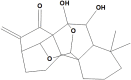 Xerophilusin B |
 Excisanin H |
| NPC17165 | NPC94141 |
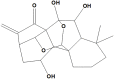 Macrocalin B |
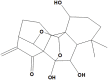 Ponicidin |
| NPC184170 | NPC107385 |
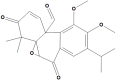 Taxusabietane B |
 Isoadenolin C |
| NPC470166 | NPC472282 |
 Isoadenolin A |
 Swielimonoid B |
| NPC476139 | NPC71905 |
 7-Methoxygambogellic Acid |
 Trichilin A |
| NPC470181 | NPC268760 |
 Xyloccensin E |
 |
| NPC136920 | NPC268760 |
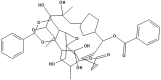 |
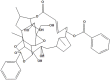 |
| NPC270946 | NPC134452 |
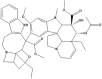 |
 |
Table 2.
Properties of the selected hits for drug-likeness filter.
| Ingredient ID | Molecular weight (g/mol) | Log P | HBD count | HBA count | Drug-Likeness |
|---|---|---|---|---|---|
| "NPC157929" | 346.42 | 1.65 | 2 | 5 | Yes; 0 violations |
| "NPC309388" | 348.44 | 1.40 | 3 | 5 | Yes; 0 violations |
| "NPC17165" | 362.42 | 0.83 | 3 | 6 | Yes; 0 violations |
| "NPC94141" | 362.42 | 0.55 | 3 | 6 | Yes; 0 violations |
| "NPC184170" | 384.43 | 3.32 | 0 | 6 | Yes; 0 violations |
| "NPC107385" | 394.46 | 0.11 | 3 | 7 | Yes; 0 violations |
| "NPC470166" | 406.47 | 0.91 | 3 | 7 | Yes; 0 violations |
| "NPC472282" | 586.68 | 2.65 | 1 | 10 | Yes; 1 violation: MW>500 |
| "NPC476139" | 658.79 | 5.75 | 2 | 9 | No; 2 violations: MW>500, Log P >5 |
| "NPC71905" | 674.74 | 1.20 | 3 | 13 | No; 2 violations: MW>500, HBA >10 |
| "NPC470181" | 702.71 | 1.24 | 0 | 15 | No; 2 violations: MW>500, HBA >10 |
| "NPC203670" | 800.90 | 2.82 | 5 | 13 | No; 2 violations: MW>500, HBA >10 |
| "NPC136920" | 814.92 | 3.10 | 5 | 13 | No; 2 violations: MW>500, HBA >10 |
| "NPC268760" | 814.92 | 3.08 | 5 | 13 | No; 2 violations: MW>500, HBA >10 |
| "NPC270946" | 820.98 | 3.51 | 2 | 13 | No; 2 violations: MW>500, HBA >10 |
| "NPC134452" | 1049.21 | 0.21 | 10 | 20 | No; 3 violations: MW>500, HBA>5, HBA >10 |
Table 3.
Root mean square deviation (rmsd) of the selected hits from the pharmacophoric features
| Ingredient ID | rmsd |
|---|---|
| "NPC157929" | 0.9854 |
| "NPC309388" | 0.8664 |
| "NPC17165" | 0.9855 |
| "NPC94141" | 0.8957 |
| "NPC184170" | 0.9673 |
| "NPC107385" | 0.9096 |
| "NPC470166" | 0.9775 |
| "NPC472282" | 1.0723 |
Fig. 2.
Superimposition of selected hits on pharmacophoric features.
3.2. Molecular docking
The catalytic dyad system of SARS-COV-2 main protease interactions is essential for inhibition activity. The main two amino acids that are involved in protease processing are Cys145 & His41 (Anand, 2002; Yang et al., 2003). Several other interactions were also noticed with other inhibitors that were reported recently (L. Zhang et al., 2020).
The X-ray crystallographic structure of SARS-COV-2 main protease enzyme co-crystallized with its bound ligand X77 (PDB ID: 6W63) was downloaded from the protein data bank.
Through the examination of the binding interactions of X77 to the active site of the enzyme, it shows H-bond interactions with the His41, Gly143, His163 & Glu166 amino acids & Pi-Sulfur interactions with Met49 & Cys145 (Fig. 3 ).
Fig. 3.
2D interactions of X77 within SARS-COV-2 main protease active site.
Docking setup was first validated by self-docking of the co-crystallized ligand (X77) in the vicinity of the binding site of the enzyme, the docking score (S) was -8.5061 kcal/mol and RMSD was 1.0543 Å indicating the validation of the docking process (Fig. 4 ).
Fig. 4.
3D representation of the superimposition of the co-crystallized (red) and the docking pose (green) of X77 in the SARS-COV-2 main protease active site.
The validated setup was then used in predicting the ligand-receptor interactions at the binding site for the compounds of interest. Docking results are summarized in Table (4) and Fig. 5 . All eight compounds were fit on the active site of SARS-COV-2 main protease with a docking score ranging between −5.7116 and −8.1579 kcal/mol. The amino acid interactions for all docked compounds were comparable to that of the co-crystallized ligand X77. However, not all compounds exhibited the essential interactions with the two key amino acids (Cys145 and His41). Compounds NPC157929 (Xerophilusin B), NPC309388 (Excisanin H), NPC184170 (Taxusabietane B), NPC107385 (Isoadenolin C), and NPC472282 showed interactions with the key amino acids as well as extra interactions similar to that of the co-crystallized ligand. The similarity in amino acid interactions between the docked compounds and the co-crystallized ligands are listed in Table (4) with red color for common amino acids while different amino acids were listed in black color.
Table 4.
Docking results.
| Compound | S (kcal/mol) | Amino acids | Interacting groups | Type of interaction |
|---|---|---|---|---|
| X77 | −8.5061 | His41 | NH (Imidazole) | Conventional H-bond |
| Gly143 | N (Imidazole) | Conventional H-bond | ||
| Gly143 | O (C O) | Conventional H-bond | ||
| His163 | N (Pyridine) | Conventional H-bond | ||
| Glu166 | O (C O) | Conventional H-bond | ||
| Met49 | Phenyl | Pi-S Interaction | ||
| Cys145 | Pyridine | Pi-S Interaction | ||
| NPC157929 | −6.0030 | Glu166 | O (C O) | Conventional H-bond |
| Gln189 | O (Pyran) | Conventional H-bond | ||
| NPC309388 | −5.9784 | His41 | CH (Pyran) | Pi-Sigma |
| NPC17165 | −6.0431 | Glu166 | O (C O) | Conventional H-bond |
| Gln189 | O (Pyran) | Conventional H-bond | ||
| NPC94141 | −5.7116 | Met49 | O (C O) | Sulfur-X Interaction |
| NPC184170 | −6.8180 | Gln189 | O (C O) | Conventional H-bond |
| NPC107385 | −6.9672 | Glu166 | O (C O) | Conventional H-bond |
| NPC470166 | −6.7127 | Asn142 | OH | Conventional H-bond |
| NPC472282 | −8.1579 | Glu166 | O (C O) | Conventional H-bond |
| His41 | CH3 | Hydrophobic Interaction | ||
| Cys145 | CH3 | Hydrophobic Interaction |
Fig. 5.
3D interactions of the selected docked compounds in the SARS-COV-2 main protease active site a) NPC157929, b) NPC309388, c) NPC17165, d) NPC94141, e) NPC184170, f) NPC107385, g) NPC470166, h) NPC472282.
In a closer look at Table (4), we can assume that NPC472282 is the best candidate for further investigation based on having the best docking score among the tested compounds with a value of −8.1579 kcal/mol, the firm binding at the active site of the enzyme with the common interaction with six amino acid out of ten of the co-crystallized ligand and good interactions with Cys145 and His41 suggesting possible inhibition activity, The higher binding ability of is attributed to the hydrogen bond with Glu166 amino acid. NPC107385 & NPC184170 showed also a relatively high energy score of −6.9672 & −6.8180 kcal/mol, respectively with strong hydrogen bond interactions.
Taxusabietane B is an aromatic abietane-type diterpenoid, in the genus Clerodendrum (Xu et al., 2016). Although no studies were performed on Taxusabietane B, taxusabietane types revealed considerable inhibitory activity against lipoxygenase (LOX) (Khan et al., 2011). The aromatic abietanes displayed a wide spectrum of biological activities including antimicrobial, antileishmanial, antiplasmodial, antifungal, antitumor, cytotoxicity, antiviral, antiulcer, cardiovascular, antioxidant as well as anti-inflammatory activity (González, 2015). A representative example of aromatic abietane is ferruginol, isolated from the roots of Craniolaria annua (Martyniaceae) and showing trypanocidal activity (Herrera et al., 2008), potent antiviral effect against SARS-CoV (Wen et al., 2007), besides its ability to inhibit SARS-CoV protease, which is crucial in processing viral polyproteins and in the regulation of replicase complex. Ferruginol was nearly a fourfold more powerful suppressant than abietic acid, the parent abietane diterpenoid (Ryu et al., 2010). Other examples of aromatic abietane showed activity against Coxsackievirus B3 (IC50 7.0–22.2 μmol/mL). (Zhang et al., 2014), while carnosic acid showed inhibitory activity against the human respiratory syncytial virus (hRSV) where it successfully suppressed viral gene expression without inducing type-I interferon production or affecting cell viability (Shin et al., 2013). Other abietane types revealed powerful anti-hepatitis B activity with IC50 values smaller than that of the positive control (Yang et al., 2011).
Isoadenolin C (NPC107385) bind with a high dock score (−6.9 kcal/mol) where the catalytic dyad besides Met49 formed hydrophobic interactions while Asn142 and Glu166 formed H-bond. It is an 7,20:14,20-diepoxy-ent-kaur-15-one, isolated from Isodon adenolomus, (Lamiaceae), and showing certain antitumor effects (Zhao et al., 2011).
Isoadenolin A (NPC470166) showed a docking score −6.7 kcal/mol and binding with His41, Cys44, Met49 with hydrophobic bond while additional hydrogen bonds were formed with His41 and Asn142. It is also an ent-kauranoids isolated from the same plant (Zhao et al., 2011).
Xerophilusin B (NPC157929), B fits the receptor with a docking score −6.0 kcal/mol, Pi-alkyl interaction was formed with the catalytic dyad and Met49 besides a H-bond with Glu166. It is also 7,20:14,20-diepoxy-ent-kaurane diterpenoid isolated from Isodon xerophilus and reported for its antitneoplastic activity against human esophageal squamous cell carcinoma, through a strong antiproliferative as well as apoptotic activity by inhibiting telomerase activity in the K562 human leukemic cell line, besides an anti-inflammatory effect through lowering NF-κB triggering lipopolysaccharide-stimulated RAW 264.7 murine macrophages (Hou et al., 2000; Yao et al., 2015). Ent-kauranes are potent anti-inflammatory agents acting by suppression of NF-κB translocation and the subsequent decline of pro-inflammatory mediators (Aquila et al., 2009).
Macrocalin B (NPC17165), showed a docking score −6.0 kcal/mol, and binding with His41 and Met49 through hydrophobic attachments and Glu166 with hydrogen bond. It is an ent-kaurane diterpenoid isolated from Isodon enanderianus that significantly inhibit the proliferation of tumor cells within the concentration range of 10−4–10−8 mol L−1 (Yang et al., 2009). In esophageal squamous cell carcinoma cells, acetyl-macrocalin B was found to induce cellular reactive oxygen species (ROS), initiate the p38 mitogen-activated protein kinase (MAPK) signaling pathway, and cause the caspase-9-dependent apoptosis cascade, as well as effectively suppresses xenograft growth without causing significant toxicity. (Wang et al., 2019).
Excisanin H (NPC309388) binds with a docking score −5.9 kcal/mol with His41 through Pi-sigma as well as hydrophobic bonds with Cys145 with VWV bond. It is a 14,20-epoxy-ent-kaurene diterpenoid, isolated from aerial parts of Rabdosia excise that displayed strong cytotoxic activity against P388 murine leukemia cells (Gui et al., 2004).
Ponicidin (NPC94141) binds with His41 with hydrophobic, Pi-donor H-bond, Met49 with sulfur X bond, and Glu166 with conventional H-bond as well as hydrophobic bonds with Cys145 with VWV bond. It is also an ent-kaurane diterpenoid isolated of many species of the genus Rabdosia and Isodon belonging to the Lamiaceae family and has been shown to exhibit potent antioxidant as well as anticancer effects due to its cytotoxicity, apoptotic cell death, cell cycle arrest, antiangiogenesis, and antiproliferative effects in cancer cells including leukemia, bladder, breast, gastric, nasopharyngeal epithelioid, and cervical carcinoma (HeLa). It was also proved to effectively inhibit the TNF-α-induced epithelial-mesenchymal transition, migration, and invasion of colorectal carcinoma in-vitro, and liver metastasis in-vivo, via AKT/GSK-3β/Snail signaling pathway (Islam et al., 2019; Zhang et al., 2019). Ponicidin also showed potent effect when administered synergistically in gene therapy with acyclovir/ganciclovir in Herpes simplex virus (HSV) by the selective stimulation of HSV-specific thymidine kinase (TK) activity and the suppression of the extracellular release of the degraded metabolite of the nucleoside analogs (Hayashi et al., 2000).
Most of the above-mentioned compounds are isolated from Isodon (Labiatae) plants, growing mainly in tropical regions in Africa and in subtropical regions in Asia. This genus has 150 species, but only thirty species showed promising effects, among which Isodon japonica (Burm.f) Hara var. galaucocalyx (maxin) Hara, commonly used as anticancer, antimastitis, and antiarthralgia agents in folk medicine, I. rubescens, is traditionally used in respiratory and gastrointestinal bacterial infections, inflammation, and cancer; I. ternifolia, I. lophanthoides, and I. megathyrsa are used as antimalarial and anti-inflammatory agents and also for the treatment of enteritis and jaundice, while I. amethystoides, were employed in the treatment of pneumonia (Sun et al., 2006). This genus is abundant in diterpenoids with diverse structural scaffolds (Liu et al., 2017).
Diterpenes are compounds that consist of twenty carbons originating from four isoprene units that are classified into 14 subclasses: phytane, labdane, clerodane, pimarane, cassane, abietane, lathyrane, taxane, beyarane, atisane, trachylobane, aconane, gibberellane, and kaurane. There are at least 2500 diterpenes distributed in a large number of plant families. Among these families, many genera are known for their diterpene content that can be associated with a great variety of biological activities. In our study, ent-kauranoid represented the predominant subclass. Ent-kaurane derivatives have been reported for their antiviral activity, where linearol showed significant anti-HIV effects in H9 lymphocyte cells with EC50 values in the range <0.1–3.11 μg/mL, while ent-kaur-16-en-19-oic acid showed a higher IC50 value (34.1 μg/mL) (Bruno et al., 2002; Saepou et al., 2010). Additionally, kaurane isolated from Sideritis lycia (Lamiaceae) were tested against human parainfluenza virus type 2 (HPIV-2), where the best EC50 was observed with isosidol (7.27 ± 1.59 mg/mL) (Kilic et al., 2020). A docking study performed to investigate the cytotoxic effect of en-kaurane revealed that, both hydrogen bond as well as interaction with the hydrophobic portion with the active sites of the protein is necessary for activity (Costa et al., 2020). Another study assessing their antioxidant and anti-tumor potential showed that compounds with less energy, had the largest number of hydrophilic regions (Scotti et al., 2014). Numerous Mpro inhibitors showed a better interaction in presence of hydrophobic substitutions at P2 position where cyclohexyl group can adapt in the same way. Additionally, P3 and P4 positions prefer hydrophobic substitutions (Sacco et al., 2020). This hypothesis supports the potency of the diterpenoid compounds in our study.
3.3. Molecular dynamics
The stability of the docked complexes of NPC472282, NPC184170, NPC107385 and X77 with the protein was monitored by means of the RMSD values of the complex over a period of 100 ns and comparing it to the unbound protein (Fig. 6 ). The RMSD trajectories of the analyzed structures were found to converge after around 10 ns for the complexes, while the protein converged to equilibrium after a longer period of time, 80 ns. However, the X77 complex showed slightly higher fluctuation values when compared to the other ligands, suggesting that the X77 complex shows less stability than the investigated compounds. The average RMSD was calculated for each structure and was found to be as follows; 0.2 nm, 0.107 nm, 0.056 nm, 0.045 nm and 0.26 nm for the protein, NPC472282, NPC184170, NPC107385 and X77 complexes respectively. The low values of the RMSD fluctuations of the complexes suggest that they are more stable than the protein.
Fig. 6.
RMSD plot of the protein (black), NPC472282 (red), NPC184170 (green) and NPC107385 (blue) and X77 (yellow) for 100 ns MD simulation.
The number of hydrogen bonds formed between the ligand and the protein were monitored over the period of the molecular dynamic simulation, 100 ns (Fig. 7 ). This shows that compounds NPC472282 was found to form an average of 4 hydrogen bonds across the 100ns simulation while NPC107385 forms an average of 3 hydrogen bonds across the 100 ns simulation. This extensive hydrogen bond network with the protein, suggesting that these compounds have the potential to act as inhibitory compounds.
Fig. 7.
The number of hydrogen bonds calculated over 100 ns molecular dynamics, where NPC472282 is shown in black, NPC184170 in red and NPC107385 in green.
4. Conclusion
This study provides a wide survey in the natural compounds database including about 47,645 compounds belonging to different classes of natural products screened using a structure-based pharmacophore model and drug-likeness filter to choose the compounds with suitable properties. Through this study, eight compounds belonging to ent-kaurane nucleus showed the best docking score and good amino acid interactions with a catalytic dyad of the main protease for SARS-COV-2. Ponicidin, Taxusabietane B and Isoadenolin C were found to be the most successful candidates. In addition, molecular dynamics studies were used to further verify the results obtained from the molecular docking studies. The molecular dynamics simulation suggested that the compounds with the highest docking scores, were found to form stable complexes with the protein, as well as an extensive hydrogen bond network. Those outcomes will provide a promising base for further research in the field of drug discovery against COVID-19. Those outcomes may provide a promising base for further research in drug discovery against COVID-19.
Credit Author Statement
All authors contributed equally in the concept and work design. Collection of data & clarification of phytochemical origins and taxonomy to which the screened compound belong were performed by R. O. Bakr & M. A. A. Fayed. Pharmacophore modeling, virtual screening & molecular docking were performed by Y. M. Nissan and M. K. El-Ashrey. R. H. Refaey performed the molecular dynamics study.
Declaration of competing interest
The authors declare no competing interests.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015;1–2:19–25. doi: 10.1016/J.SOFTX.2015.06.001. [DOI] [Google Scholar]
- Adhikari B., Marasini B.P., Rayamajhee B., Bhattarai B.R., Lamichhane G., Khadayat K., Adhikari A., Khanal S., Parajuli N. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID‐19: a review. Phyther. Res. ptr. 2020;6893 doi: 10.1002/ptr.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akram M., Tahir I.M., Shah S.M.A., Mahmood Z., Altaf A., Ahmad K., Munir N., Daniyal M., Nasir S., Mehboob H. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phyther. Res. 2018;32:811–822. doi: 10.1002/ptr.6024. [DOI] [PubMed] [Google Scholar]
- Anand K. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquila S., Weng Z.Y., Zeng Y.Q., Sun H.D., Ríos J.L. Inhibition of NF-κB activation and iNOS induction by ent-kaurane diterpenoids in LPS-stimulated RAW264.7 murine macrophages. J. Nat. Prod. 2009;72:1269–1272. doi: 10.1021/np9001465. [DOI] [PubMed] [Google Scholar]
- Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA, J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M., Rosselli S., Pibiri I., Kilgore N., Lee K.H. Anti-HIV agents derived from the ent-kaurane diterpenoid linearol. J. Nat. Prod. 2002;65:1594–1597. doi: 10.1021/np020029b. [DOI] [PubMed] [Google Scholar]
- Cavasotto C.N., Di Filippo J.I. In silico drug repurposing for COVID-19: targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol. Inform. 2021 doi: 10.1002/minf.202000115. [DOI] [PubMed] [Google Scholar]
- Cerutti D.S., Duke R.E., Darden T.A., Lybrand T.P. Staggered Mesh Ewald: an extension of the smooth particle-mesh Ewald method adding great versatility. J. Chem. Theor. Comput. 2009;5:2322–2338. doi: 10.1021/CT9001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R.A., da Silva J.N., Oliveira K.M.T., Dutra L.M., Costa E.V. Quantum chemical studies, vibrational analysis, molecular dynamics and docking calculations of some ent-kaurane diterpenes from Annona vepretorum: a theoretical approach to promising anti-tumor molecules. Struct. Chem. 2020;31:1223–1243. doi: 10.1007/s11224-020-01491-2. [DOI] [Google Scholar]
- De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016 doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayed M.A.A., El-Behairy M.F., Abdallah I.A., Abdel-Bar H.M., Elimam H., Mostafa A., Moatasim Y., Abouzid K.A.M., Elshaier Y.A.M.M. Structure- and ligand-based in silico studies towards the repurposing of marine bioactive compounds to target SARS-CoV-2. Arab. J. Chem. 2021 doi: 10.1016/j.arabjc.2021.103092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M.A. Aromatic abietane diterpenoids: their biological activity and synthesis. Nat. Prod. Rep. 2015;32:684–704. doi: 10.1039/c4np00110a. [DOI] [PubMed] [Google Scholar]
- Gui M.Y., Aoyagi Y., Jin Y.R., Li X.W., Hasuda T., Takeya K. Excisanin H, a novel cytotoxic 14,20-Epoxy-ent-Kaurene diterpenoid, and three new ent-Kaurene diterpenoids from Rabdosia excisa. J. Nat. Prod. 2004;67:373–376. doi: 10.1021/np030357r. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Hayashi T., Sun H.-D., Takeda Y. Cancer Gene Therapy; 2000. Potentiation of Ganciclovir Toxicity in the Herpes Simplex Virus Thymidine Kinase/ganciclovir Administration System by Ponicidin. [DOI] [PubMed] [Google Scholar]
- Herrera J.C., Troncone G., Henríquez D., Urdaneta N. Trypanocidal activity of abietane diterpenoids from the roots of Craniolaria annua. Zeitschrift fur Naturforsch. - Sect. C J. Biosci. 2008;63C:821–829. doi: 10.1515/znc-2008-11-1207. [DOI] [PubMed] [Google Scholar]
- Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theor. Comput. 2007;4:116–122. doi: 10.1021/CT700200B. [DOI] [PubMed] [Google Scholar]
- Hoever G., Baltina Lidia, Michaelis M., Kondratenko R., Baltina Lia, Tolstikov G.A., Doerr H.W., Cinatl J. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/PROT.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou A.J., Li M.L., Jiang B., Lin Z.W., Ji S.Y., Zhou Y.P., Sun H.D. New 7,20:14,20-diepoxy ent-kauranoids from Isodon xerophilus. J. Nat. Prod. 2000;63:599–601. doi: 10.1021/np9903705. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ibrahim M.A.A., Abdelrahman A.H.M., Allemailem K.S., Almatroudi A., Moustafa M.F., Hegazy M.E.F. Silico Evaluation of Prospective Anti-COVID-19 Drug Candidates as Potential SARS-CoV-2 Main Protease Inhibitors. 2021. Protein J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M.A.A., Abdelrahman A.H.M., Hussien T.A., Badr E.A.A., Mohamed T.A., El-Seedi H.R., Pare P.W., Efferth T., Hegazy M.E.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.T., Biswas S., Bagchi R., Khan M.R., Khalipha A.B.R., Rouf R., Uddin S.J., Shilpi J.A., Bardaweel S.K., Sabbah D.A., Mubarak M.S. Ponicidin as a promising anticancer agent: its biological and biopharmaceutical profile along with a molecular docking study. Biotechnol. Appl. Biochem. 2019;66:434–444. doi: 10.1002/bab.1740. [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M pro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1101/2020.02.26.964882. [DOI] [PubMed] [Google Scholar]
- Khan I., Nisar M., Shah M.R., Shah H., Gilani S.N., Gul F., Abdullah S.M., Ismail M., Khan N., Kaleem W.A., Qayum M., Khan H., Obaidullah Samiullah, Ullah M. Anti-inflammatory activities of taxusabietane A isolated from Taxus wallichiana Zucc. Fitoterapia. 2011;82:1003–1007. doi: 10.1016/j.fitote.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T., Khan M.A., Mashwani Z. ur R., Ullah N., Nadhman A. Therapeutic potential of medicinal plants against COVID-19: the role of antiviral medicinal metabolites. Biocatal. Agric. Biotechnol. 2021 doi: 10.1016/j.bcab.2020.101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic T., Topcu G., Goren A.C., Aydogmus Z., Karagoz A., Yildiz Y.K., Aslan I. Ent-kaurene diterpenoids from sideritis lycia with antiviral and cytotoxic activities. Record Nat. Prod. 2020;14:256–268. doi: 10.25135/rnp.163.19.08.1373. [DOI] [Google Scholar]
- Kim M.K., Yu M.S., Park H.R., Kim K.B., Lee C., Cho S.Y., Kang J., Yoon H., Kim D.E., Choo H., Jeong Y.J., Chong Y. 2,6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV) Eur. J. Med. Chem. 2011;46:5698–5704. doi: 10.1016/j.ejmech.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Liu M., Wang W.G., Sun H.D., Pu J.X. Diterpenoids from Isodon species: an update. Nat. Prod. Rep. 2017;34:1090–1140. doi: 10.1039/c7np00027h. [DOI] [PubMed] [Google Scholar]
- Meyer-Almes F.J. Repurposing approved drugs as potential inhibitors of 3CL-protease of SARS-CoV-2: virtual screening and structure based drug design. Comput. Biol. Chem. 2020 doi: 10.1016/j.compbiolchem.2020.107351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharm. Anal. 2020;10:320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., Ferrin T. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/JCC.20084. [DOI] [PubMed] [Google Scholar]
- Refaey R.H., El-Ashrey M.K., Nissan Y.M. Repurposing of renin inhibitors as SARS-COV-2 main protease inhibitors: a computational study. Virology. 2021;554:48–54. doi: 10.1016/j.virol.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Naguyen T.T.H., Park S.J., Chang J.S., Park K.H., Rho M.C., Lee W.S. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saepou S., Pohmakotr M., Reutrakul V., Yoosook C., Kasisit J., Napaswad C., Tuchinda P. Anti-HIV1 diterpenoids from leaves and twigs of Polyalthia sclerophylla. Planta Med. 2010;76:721–725. doi: 10.1055/s-0029-1240683. [DOI] [PubMed] [Google Scholar]
- Scotti L., Ishiki H., Mendonça F.J.B., Santos P.F., Tavares J.F., Silva M.S., Scotti M.T. Theoretical research into anticancer activity of diterpenes isolated from the paraiban flora. Nat. Prod. Commun. 2014;9:911–914. doi: 10.1177/1934578x1400900707. [DOI] [PubMed] [Google Scholar]
- Shin H.B., Choi M.S., Ryu B., Lee N.R., Kim H.I., Choi H.E., Chang J., Lee K.T., Jang D.S., Inn K.S. Antiviral activity of carnosic acid against respiratory syncytial virus. Virol. J. 2013;10:303. doi: 10.1186/1743-422X-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa da Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon parser interfacE. BMC Res. Notes. 2012;51(5):1–8. doi: 10.1186/1756-0500-5-367. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.D., Huang S.X., Han Q. Bin. Diterpenoids from Isodon species and their biological activities. Nat. Prod. Rep. 2006;23:673–698. doi: 10.1039/b604174d. [DOI] [PubMed] [Google Scholar]
- Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W.P., Murcko M.A. Prediction of “drug-likeness. Adv. Drug Deliv. Rev. 2002;54:255–271. doi: 10.1016/S0169-409X(02)00003-0. [DOI] [PubMed] [Google Scholar]
- Wang J.N., Che Y., Yuan Z.Y., Lu Z.L., Li Y., Zhang Z.R., Li N., Li R. Da, Wan J., Sun H.D., Sun N., Puno P.T., He J. Acetyl-macrocalin B suppresses tumor growth in esophageal squamous cell carcinoma and exhibits synergistic anti-cancer effects with the Chk1/2 inhibitor AZD7762. Toxicol. Appl. Pharmacol. 2019;365:71–83. doi: 10.1016/j.taap.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C., Hsiao P.W., Chien S.C., Shyur L.F., Yang N.S. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- Xu Mingfeng, Zhu Q., Wang H., Du Q., Xu Maojun. Separation of abietane-type diterpenoids from Clerodendrum kaichianum Hsu by high-speed counter-current chromatography using stepwise elution. J. Liq. Chromatogr. Relat. Technol. 2016;39:139–144. doi: 10.1080/10826076.2015.1137003. [DOI] [Google Scholar]
- Yang L. Bin, Li L., Huang S.X., Pu J.X., Zhao Y., Ma Y.B., Chen J.J., Leng C.H., Tao Z.M., Sun H.D. Anti-hepatitis B virus and cytotoxic diterpenoids from Isodon lophanthoides var. gerardianus. Chem. Pharm. Bull. 2011;59:1102–1105. doi: 10.1248/cpb.59.1102. [DOI] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sun H., Zhou Y., Ji S., Li M. Effects of three diterpenoids on tumour cell proliferation and telomerase activity. Nat. Prod. Res. 2009;23:1007–1012. doi: 10.1080/14786410802295149. [DOI] [PubMed] [Google Scholar]
- Yao R., Chen Z., Zhou C., Luo M., Shi X., Li J., Gao Y., Zhou F., Pu J., Sun H., He J. Xerophilusin b induces cell cycle arrest and apoptosis in esophageal squamous cell carcinoma cells and does not cause toxicity in nude mice. J. Nat. Prod. 2015;78:10–16. doi: 10.1021/np500429w. [DOI] [PubMed] [Google Scholar]
- Zeng X., Zhang P., Wang Y., Qin C., Chen S., He W., Tao L., Tan Y., Gao D., Wang B., Chen Z., Chen W., Jiang Y.Y., Chen Y.Z. CMAUP: a database of collective molecular activities of useful plants. Nucleic Acids Res. 2019;47:D1118–D1127. doi: 10.1093/nar/gky965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.J., Li Y.H., Jiang J.D., Yu S.S., Wang X.J., Zhuang P.Y., Zhang Y., Qu J., Ma S.G., Li Y., Liu Y.B., Yu D.Q. Diterpenes and sesquiterpenes with anti-Coxsackie virus B3 activity from the stems of Illicium jiadifengpi. Tetrahedron. 2014;70:4494–4499. doi: 10.1016/j.tet.2014.05.006. [DOI] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu J., Liu B., Chen F., Li J., Liu Y., Zhu J., Shen C. Ponicidin inhibits pro-inflammatory cytokine TNF-α-induced epithelial–mesenchymal transition and metastasis of colorectal cancer cells via suppressing the AKT/GSK-3β/Snail pathway. Inflammopharmacology. 2019;27:627–638. doi: 10.1007/s10787-018-0534-5. [DOI] [PubMed] [Google Scholar]
- Zhao W., Pu J.X., Du X., Su J., Li X.N., Yang J.H., Xue Y.B., Li Y., Xiao W.L., Sun H.D. Structure and cytotoxicity of diterpenoids from Isodon adenolomus. J. Nat. Prod. 2011;74:1213–1220. doi: 10.1021/np200140j. [DOI] [PubMed] [Google Scholar]











