Abstract
Lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine] reduces woodchuck hepatitis virus (WHV) titers in the sera of chronically infected woodchucks by inhibiting viral DNA synthesis. However, after 6 to 12 months, WHV titers begin to increase toward pretreatment levels. Three WHV variants with mutations in the active site of the DNA polymerase gene are present at this time (W. S. Mason et al., Virology 245:18–32, 1998). We have asked if these mutant viruses were responsible for the lamivudine resistance and if their emergence caused an immediate rise in virus titers. Cell cultures studies implied that the mutants were resistant to lamivudine. Emergence of mutant WHV was not always associated, however, with an immediate rise in virus titers in the serum. One of the three types of mutant viruses became prominent in serum up to 7 months before titers in serum actually began to increase, at a time when wild-type virus was still predominant in the liver. The two other mutants did not show this behavior but were detected in serum and liver later, just at the time that virus titers began to rise. A factor linking all three mutants was that a similar duration of drug administration preceded the rise in titers, irrespective of which mutant ultimately prevailed. A simple explanation for these results is that the increase in virus titers following emergence of drug-resistant mutants can occur only as the preexisting wild-type virus is cleared from the hepatocyte population, allowing spread of the mutants. Thus, prolonged suppression of virus titers in the serum may sometimes be a measure of the stability of hepatocyte infection rather than of a successful therapeutic outcome.
Exposure to hepatitis B virus (HBV) can lead to chronic infection, which is a cause of cirrhosis and liver cancer. The treatment of chronic HBV infection to block this progression is still in its infancy. Alpha interferon is the only drug widely used to treat virus carriers, and the response rate is probably about 20% (10, 21). Lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine], a nucleoside analogue, has now been approved by the U.S. Food and Drug Administration for the treatment of hepatitis B. The administration of lamivudine induces a significant drop in the amount of virus in the serum of most HBV carriers and, unlike alpha interferon, causes no significant side effects in most patients. The emergence of drug-resistant HBV may, unfortunately, begin after 6 to 12 months of therapy (2–4, 9, 15, 19, 20, 25). As with human immunodeficiency virus (23), mutations related to lamivudine resistance are invariably mapped to the methionine residue in the YMDD motif of the HBV polymerase active site (amino acid 552), with a switch to YIDD or YVDD. The YVDD mutation has also been shown to confer lamivudine resistance when it is introduced into duck HBV (7). In addition, the change from YMDD to YVDD in HBV is associated with an upstream amino acid replacement of leucine (amino acid 528) with methionine (2, 19). This upstream change is localized to the B region, a stretch of amino acids found in the active site of many polymerases, including that of HBV and woodchuck hepatitis virus (WHV) (Fig. 1). Moreover, resistance of HBV to famciclovir maps to the B region (3).
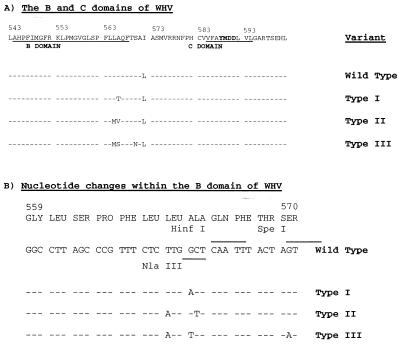
FIG. 1.
Mutations in the B region of the WHV polymerase arise during lamvidine therapy. (A) Amino acid changes. (B) Nucleotide changes and cleavage sites for the three restriction endonucleases used for genotyping of clones of PCR-amplified viral DNA. The sequence of WHV, as reported by earlier investigators (5), is presented in the top line of each panel. It should be noted that this polymerase sequence is one amino acid longer than that reported by Kodama et al. (14), as the result of an insertion at position 15 (see Fig. 4).
We recently reported on the development of drug resistance in WHV-infected woodchucks treated with lamivudine (17). The WHV genomes, present after virus titers had increased toward pretreatment levels, had a normal YMDD motif. However, mutations in the active site of the viral DNA polymerase were detected in the upstream B region (Fig. 1). All mutants were altered at amino acid 566, which corresponds to position 529 in HBV. One had an alanine-to-threonine mutation (type I variant), one had a mutation to valine (type II variant), and the other had a mutation to serine (type III variant). The nucleotide change that produces the alanine-to-threonine change in polymerase also introduces a stop codon into the overlapping S gene, potentially truncating 55 amino acids from the carboxy termini of the three viral envelope proteins. The mutation to valine at position 566 occurred together with a leucine-to-methionine change at position 565 (HBV position 528). The mutation to serine at position 566 occurred with a leucine-to-methionine change at position 565 and a serine-to-asparagine change at position 570.
The present study was undertaken to address two issues. First, were these mutant viruses responsible for the rise in virus titers seen in virtually all woodchucks treated for up to a year with lamivudine? Second, did the rise in virus titers coincide with the rapid spread of a new drug-resistant strain of WHV through the liver? Alternatively, was the spread of mutant virus retarded by the wild-type virus present in hepatocytes when therapy was initiated, with the rise in titers occurring only after a sufficient time for clearance of wild-type DNA? We present evidence that the mutant viruses had a drug-resistant polymerase and were probably responsible for the emergence of the drug-resistant phenotype in vivo. Our data also indicated that at least one of the three strains of lamivudine-resistant WHV could be present months before virus titers increased. These data support the hypothesis that wild-type virus infection of hepatocytes prevents emergence of drug resistance, as reflected by rising virus titers, until there is a substantial depletion of wild-type covalently closed circular DNA (cccDNA). A recent study on the appearance of drug-resistant strains of HBV, in which mutant virus was present in the serum of patients 4 months prior to an increase in virus titers, points to the same conclusion (4).
MATERIALS AND METHODS
Woodchucks.
Woodchucks (Marmota monax) were housed in the laboratory animal facility of the Fox Chase Cancer Center. The experiments with the woodchucks were reviewed and approved by the Center’s Institutional Animal Care and Use Committee. Captive-born woodchucks 300, 302, and 315 were purchased from Northeastern Wildlife (South Plymouth, N.Y.), and woodchucks 4960 and 4961, a gift of B. Tennant, were from the breeding colony at Cornell University, Ithaca, N.Y. Chronic infection was achieved by neonatal inoculation with serum from WHV-infected woodchucks. Lamivudine treatment of woodchucks 300, 302, 315, and 4960 and placebo administration to woodchuck 4961 were initiated when the animals were 13 to 16 months of age, as described previously (17). The first three woodchucks received an initial dose of 40 mg per kg of body weight, which was escalated to 200 mg per kg after 3 months, while the last woodchuck was started on the dose of 200 mg per kg.
PCR amplification and direct sequencing of WHV DNA.
Serum samples collected from each woodchuck were stored at −80°C. To prepare virion DNA, 50 μl of woodchuck serum was layered on top of a 10 to 20% sucrose step gradient containing 0.15 M NaCl and 0.02 M Tris-HCl (pH 7.5), and virus was pelleted by centrifugation for 3 h at 50,000 rpm in a Beckman SW-60 rotor. The virus was resuspended in 200 μl of 0.1 M NaCl–0.01 M Tris-HCl (pH 7.5)–0.01 M EDTA–0.2% sodium dodecyl sulfate–1 mg of pronase per ml, followed by incubation for 1 h at 37°C. The mixture was then extracted two times with phenol-chloroform, and viral DNA was precipitated by the addition of 2 volumes of ethanol, with 10 μg of dextran used as the carrier. PCR amplification was then carried out in a reaction volume of 50 μl with KlenTaq polymerase (ClonTech Laboratories). The amplification conditions included 1 min of denaturation at 94°C and 30 cycles of 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C, followed by 4 min at 72°C. The forward primer (5′-AGATTGGTGGTGCACTTCTCTCAG-3′) spanned WHV nucleotides 385 to 408 (14). The reverse primer (5′-CCACGGAATTGTCAGTGCCCAACA-3′) spanned nucleotides 1474 to 1451. PCR products were purified with the QIAquick Kit (Qiagen, Inc., Hilden, Germany), as recommended by the manufacturer. The products were sequenced with 5′-GGATGTATCTGCGGCGTTT-3′ (nucleotides 510 to 528) as the sense primer and 5′-CCCAAATCAAGAAAAACAGAACA-3′ (nucleotides 953 to 931) as the antisense primer, respectively.
Proportions of wild-type and mutant viral DNA in serum and liver.
Reconstruction experiments revealed that direct sequencing of the PCR products could detect sequence variants that represented as little as 10 to 20% of the major species. However, where more than one minor variant is present, this approach cannot reveal whether minor sequences changes are associated with one or more than one variant. To obtain a truer representation of sequence variation, virion DNA isolated from woodchuck serum that was collected at different time points during lamivudine treatment was amplified by PCR with forward primer 5′-GACATACCACGTGGTTTAGTTCCG-3′ (nucleotides 8 to 31) and reverse primer 5′-AGGGAGATCCGAGTCGTCTGA-3′ (nucleotides 1664 to 1644). To estimate the proportions of the four different genetic variants of the B region of the viral polymerase (wild type and type I, type II, and type III variants) the PCR products were purified with the QIAquick Kit (Qiagen, Inc.) and were then cloned. Individual recombinant clones were subjected to PCR. The forward primer had the sequence 5′-TACCTATTAGTCCTGCTGCTGTGC-3′ (nucleotides 536 to 560), and the reverse primer had the sequence 5′-GCCCCACCATTTTGTTTTATTGAC-3′ (nucleotides 990 to 966). The mutations in variants types I, II, and III change the pattern of cleavage by HinfI, NlaIII, and SpeI, respectively (Fig. 1). HinfI cuts only type I, SpeI cuts the wild type and types I and II but not type III, and NlaIII cuts types II and III but not the wild type or type I. Therefore, digestion of the PCR product with these restriction endonucleases, followed by agarose gel electrophoresis, was used to determine the B-region genotype.
To determine the sequence of the B region of cccDNA extracted from woodchuck liver biopsy specimens, PCR amplification, cloning, and restriction digestions were carried out as described above. Prior to PCR amplification, the cccDNA was purified as reported previously (17) and was then subjected to alkaline extraction, as described by Yang et al. (29).
Cloning of WHV cccDNA.
Liver biopsy samples were obtained from woodchuck 302 before and during lamivudine therapy, and nucleic acids enriched for cccDNA were extracted as described previously (17). The cccDNA was then purified by agarose gel electrophoresis (detection was by subsequent blot hybridization), cleaved with EcoRI, and cloned into bacteriophage lambda gt11 (Promega). (Only a single linear cleavage product was detected after digestion of the cccDNA with EcoRI.) In some cases, the viral DNA was then subcloned, after EcoRI digestion, into pGEM-3Z, and the full sequence of the viral DNA was determined.
To compare the sequences of the DNA polymerase region of a large number of individual phage lambda gt11 cccDNA clones, PCR amplification was carried out, followed by sequencing of the PCR product.
Transfection of a human liver cell line, Huh7.
WHV expression vectors in which pregenomic RNA is transcribed from the cytomegalovirus immediate-early promoter (24) were used to test the effects of different mutations of the B region of the DNA polymerase on viral DNA synthesis in the presence and absence of lamivudine. For the wild type, we used an infectious clone of WHV (24), which was thought to be identical to the clone originally sequenced by Kodama et al. (14). In preliminary studies, a number of nucleotide differences between this clone and the published sequence were detected in the region under study. We therefore determined the complete sequence of this WHV clone. The predicted polymerase open reading frame (ORF) is shown in Fig. 4. Mutations were introduced into the B region of the wild type through site-directed mutagenesis by the PCR method of gene splicing by overlap extension (11, 12). Design of the oligonucleotides for introduction of mutations was based upon prior sequencing through this region of the cloned DNA, which was carried out as described above. All the constructs were sequenced. Ten micrograms of DNA was used to transfect Huh7 cells, which were growing in 60-mm tissue culture dishes, when the monolayers were about 70% confluent (26). Huh7 cells were cultured in F-12 minimal essential medium at 37°C with 5% CO2. To test for inhibition of viral DNA synthesis, lamivudine was added 24 h prior to transfection at a concentration of 100 μM. Culture medium with or without lamivudine was changed every day.
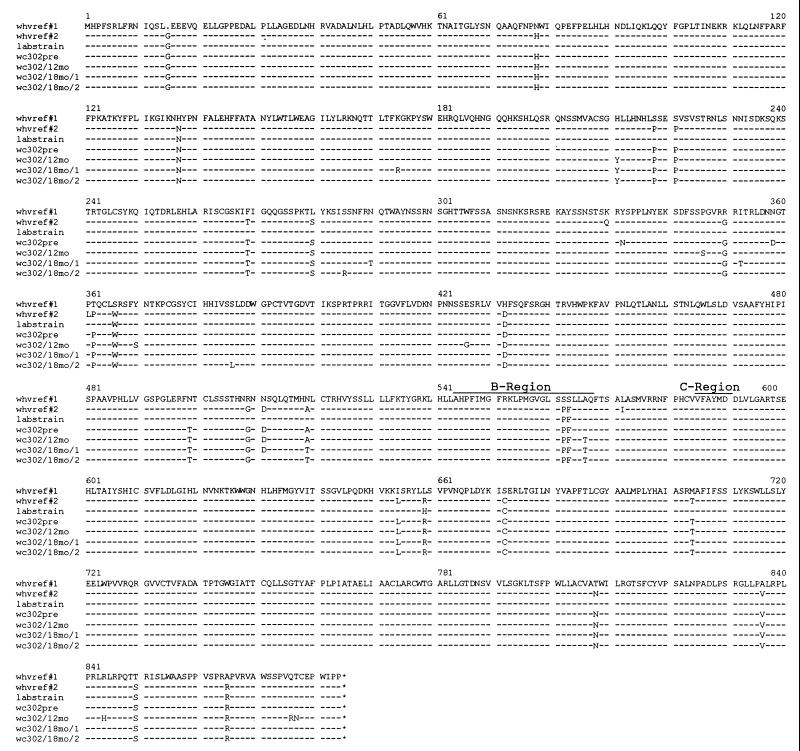
FIG. 4.
Sequences of the polymerase ORF for four clones of cccDNA isolated from woodchuck 302 (wc 302). Full-length viral DNA was cloned from preparations of cccDNA prepared from the liver before therapy and after 12 and 18 months of lamivudine treatment. The complete sequence of each was determined and was used to predict the amino acid sequence of the polymerase ORF. For comparison, two sequences from GenBank are shown (whvref#1, accession no. M11082 [14]; whvref#2, accession no. M19183 [5]), together with the sequence determined for the clone of WHV that is used in our laboratory. As shown, there were some discrepancies between the sequence reported by Kodama et al. (14) and the sequence of the laboratory strain of WHV DNA that was used in the present study, although it was believed that the strains were originally the same.
At 5 days posttransfection the cell monolayers were rinsed with phosphate-buffered saline and were stored at −80°C. For extraction of intermediates in viral DNA replication, the monolayers were thawed by the addition of 1 ml of lysis buffer containing 0.01 M Tris-HCl (pH 8.0), 0.001 M EDTA, 1% (vol/vol) Nonidet P-40, 0.05 M NaCl, and 8% (wt/vol) sucrose. The lysate was harvested and centrifuged at 11,000 × g for 2 min at 4°C, and the pellet was discarded. Ten microliters of 1 M magnesium acetate, 10 μl of 10 mg of DNase I (Boehringer) per ml, and 10 μl of 10 mg of microliters RNase A per ml were added to the supernatant; and the mixture was incubated for 30 min at 37°C. Following clarification by centrifugation at 11,000 × g for 4 min, polyethylene glycol, NaCl, and EDTA were added to the supernatant to final concentrations of 6.5%, 0.35 M, and 0.01 M, respectively. The mixture was then placed on ice for 1 h, after which the precipitate was collected by centrifugation at 11,000 × g. The pellet was then digested for 1 h at 37°C in 0.025 M Tris-HCl (pH 7.4)–0.01 M EDTA–0.1 M NaCl–0.5% (wt/vol) sodium dodecyl sulfate–0.5 mg of pronase per ml. The DNA was then extracted with a phenol-chloroform mixture (1:1) and was concentrated by ethanol precipitation.
One-quarter of the DNA extracted from each tissue culture dish was subjected to electrophoresis in a 1.5% agarose gel and was then transferred to a nitrocellulose membrane for hybridization (27) with a 32P-labeled probe representing the complete WHV genome. Following hybridization, bound radioactivity was measured with a Fuji Image Analyzer. A full-length, linear, cloned viral DNA served as an electrophoresis and hybridization standard.
Primary woodchuck hepatocyte cultures.
Primary hepatocytes prepared from WHV-negative woodchuck liver were seeded onto 60-mm tissue culture dishes coated with rat-tail collagen and were maintained in serum-free medium at 37°C (1). Prior to seeding, hepatocytes were partially purified by centrifugation through 90% Percoll (Sigma) for 15 min at 1,500 rpm (18). To directly evaluate the lamivudine-resistant properties of the WHV variants present in woodchuck serum, 50 μl of woodchuck serum was used to infect hepatocytes in the presence or absence of 500 μM lamivudine. Medium was changed daily, and the cells were harvested at 16 days postinfection. Intermediates in viral DNA replication were extracted and analyzed by electrophoresis in 1.5% agarose gels, transfer to nitrocellulose filters, and hybridization with a 32P-labeled WHV DNA probe (18).
RESULTS
Lamivudine resistance in woodchucks is due to the emergence of mutant strains of WHV.
Treatment of woodchucks with lamivudine leads to a transient suppression of virus titers. Within a year, however, titers begin to rise toward pretreatment levels (17). At this time, mutations are detected in the B region of the viral DNA polymerase (Fig. 1) (17), suggesting that resistance to therapy may be due to the emergence of drug-resistant strains of WHV.
To directly test for lamivudine-resistant strains of WHV, we assayed for inhibition of viral DNA synthesis following infection of primary woodchuck hepatocyte cultures with serum-derived virus. For this experiment, we analyzed virus collected before lamivudine therapy and again when titers had increased after a period of suppression by lamivudine.
Lamivudine inhibited viral DNA replication (accumulation of viral DNA replication intermediates, detected as the more rapidly migrating species labeled SS [single stranded]) following infection with virus collected before therapy, as shown in Fig. 2A for four different woodchucks. Sera collected near the end of lamivudine therapy generally had a lower titer and sometimes a lower specific infectivity in primary hepatocyte cultures, and we were able to obtain results for only three of the four lamivudine-treated woodchucks. Sera from these three woodchucks contained lamivudine-resistant WHV (Fig. 2B). Thus, in these woodchucks, the failure of lamivudine to continuously suppress virus replication is apparently due to the eventual outgrowth of lamivudine-resistant strains of WHV.
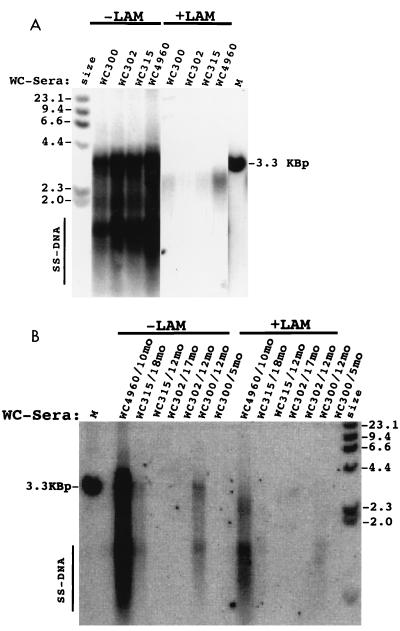
FIG. 2.
Lamivudine (LAM) resistance of WHV in primary hepatocyte cultures. Virus present in sera (50 μl) collected from four different woodchucks (WC) prior to lamivudine therapy (A) or after the indicated number of months of therapy (B) was tested for sensitivity to the drug. Virion titers per milliliter of serum (A), as determined by Southern blotting, were 8 × 109 (woodchuck 300), 8 × 109 (woodchuck 302), 8 × 109 (woodchuck 315), and 1.4 × 1010 (woodchuck 4960). Virion titers per milliliter of serum (B) were 1.3 × 109 (woodchuck 4960, 10 months), 2 × 109 (woodchuck 315, 18 months), <8 × 106 (woodchuck 315, 12 months), 2 × 109 (woodchuck 302, 17 months), 2 × 108 (woodchuck 302, 12 months), 5 × 108 (woodchuck 300, 12 months), and 3 × 107 (woodchuck 300, 5 months). Lamivudine (500 μM) was added 1 day before infection and was maintained in the culture medium until the cells were harvested at 16 days postinfection. DNA was extracted and assayed for WHV replication intermediates by Southern blot analysis. Markers included a 32P-labeled HindIII digest of phage lambda DNA and 75 pg of linear viral DNA (lane M). The viral DNA that migrated between 4.4 and 2.3 kbp may be derived, in part, from the inoculum. Therefore, the best indicator of virus DNA replication is the presence of species (single-stranded [SS] DNA) that migrated faster than the 2.0-kbp marker (16). By this criterion, replication of pretreatment virus was inhibited at least 40-fold. Replication of serum from woodchucks 300 (12 months), 315 (18 months), and 4960 (10 months) was inhibited approximately 2.5-, 2-, and 5-fold, respectively. cccDNA formation (data not shown) in the absence of lamivudine was detected only in cultures in which replicative intermediates were also detected and were found to be present in proportion to the amount of replicating DNA that accumulated in the cells. cccDNA formation was not inhibited by lamivudine in cultures infected with drug-resistant WHV and was inhibited between 3- and 30-fold in cultures infected with different stocks of nonresistant virus. In the latter instance, the cccDNA was presumably formed from infecting viral DNA, as no virus DNA replication was detected.
To obtain evidence that this might be the case for all of the woodchucks and to facilitate a more detailed analysis of the emergence of these lamivudine-resistant strains of WHV, we asked if the different mutations that have been observed in the B region could produce lamivudine resistance. If so, an analysis of virus genotypes could be used to characterize the emergence of drug-resistant strains of WHV during the course of therapy.
Site-directed mutagenesis was therefore used to introduce the type I, II, and III mutations (Fig. 1) into a laboratory strain of WHV. The wild-type DNA and the three different mutated viral DNAs were then used to transfect Huh7 cells (28). Expression of the WHV pregenome, which provides all the functions necessary for viral DNA synthesis (13), was under control of the cytomegalovirus immediate-early promoter, as WHV transcription is inefficient in many different liver cell lines (6). In the presence of lamivudine, accumulation of intermediates in the replication of wild-type WHV DNA was reduced to background levels (Fig. 3). In comparison, only a two- to threefold reduction was observed after introduction of the type I and II mutations into viral DNA, and a less than twofold reduction was observed with the type III mutations. Thus, all three classes of mutations produced resistance to lamivudine, implying that a direct assay for these genotypes could be used to assess the emergence of lamivudine-resistant WHV during the course of treatment. The results also suggest that the type I and II mutants may replicate less efficiently than the wild type; however, it is possible that other sequence differences between mutants and wild types of natural isolates may compensate for these differences.
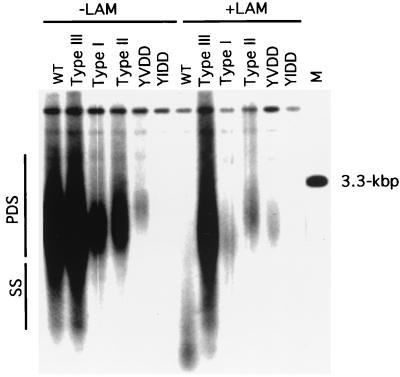
FIG. 3.
The type I, II, and III mutations of the pol gene produce lamivudine (LAM) resistance. The type I, II, and III mutations (Fig. 1) were introduced into cloned WHV DNA as described in Materials and Methods. When present, lamivudine was added 24 h prior to transfection with 10 μg of DNA. Intracellular viral DNAs were harvested after 5 days and were subjected to Southern blot analysis. Unlike the infection experiment, the most likely source of nonreplicated viral DNA in this transfection experiment is small fragments of plasmid DNA that survived the DNase I treatment that was used during the isolation of viral particles. Therefore, partially double-stranded WHV DNA is a more reliable indicator of virus DNA replication in this experiment. PDS, partially double-stranded viral DNA; SS, single-stranded viral DNA; WT, wild type. Markers included a 32P-labeled HindIII digest of phage lambda DNA and 75 pg of linear viral DNA (lane M).
Therefore, before proceeding with a more detailed genotype analysis, we also carried out a preliminary study to determine if mutations outside the B region might contribute to the acquisition of lamivudine resistance. For this purpose, we focused on the type I mutant, which was the most commonly observed type of mutant in the previous study (17). cccDNA molecules present before therapy and after 12 and 18 months of lamivudine therapy were cloned from the liver of woodchuck 302. The genotypes of the B region of 47 of these clones are presented in Table 1. One clone with a wild-type B region and three clones with a B-region mutation (type I) were then fully sequenced. The predicted amino acid sequence of the polymerase ORF is shown in Fig. 4. A number of mutations that produced amino acid changes in addition to the alanine-to-threonine change at position 566 (type I) were observed; however, the only one that was found in all three of the clones with a type I mutation was the histidine-to-tyrosine change at position 211. This site is part of the tether region that separates the upstream protein that primes from the downstream active-site domains, and on this basis alone it seemed unlikely to affect substrate specificity. Moreover, analyses of additional type I clones did not reveal a complete correlation between this upstream mutation and the type I mutation in the B region (Table 1). Thus, amino acid changes outside the B region may contribute to the acquisition of lamivudine resistance and/or to the efficiency of replication of mutant virus in vivo, but these contributions do not appear to map to a unique site.
TABLE 1.
Genotypes of cccDNA clonesa
| Clone and time of sample collection | No. of clones with the following genotype/no. of clones tested:
|
||||
|---|---|---|---|---|---|
| Wild type | Variant I | Variant II | Variant III | Total | |
| Isolate from woodchuck 302 | |||||
| Pretherapy | 24/24 | 0/24 | 0/24 | 0/24 | 24/24 |
| 12 mo | 12/13 | 1/13 | 0/13 | 0/13 | 13/13 |
| 17 mo | 6/10 | 4/10 | 0/10 | 0/10 | 10/10 |
| Clones lacking histidine at position 211 | |||||
| Pretherapy | 1/2 | ||||
| 12 mo | 3/4 | 2/2 | |||
| 17 mo | 1/1 | 6/7 | |||
WHV cccDNA preparations from woodchuck 302 were cut with EcoRI and bacteriophage lambda gt11 clones of viral DNA were then obtained. Subsequent PCR amplification and sequencing through the B and C regions of the viral reverse transcriptase were carried out as described in Materials and Methods. Selected clones with a wild-type B region or a type I mutation in the B region were also amplified through the site encoding polymerase amino acid 211. The genotype at amino acid 211 was then estimated by digestion with Bsp1286I, as the H211Y mutation eliminates the restriction site at nucleotide 3057. These results are those for the clones that were sequenced (Fig. 4).
Emergence of lamivudine-resistant strains of WHV is not associated with an immediate rise in virus titers.
Having obtained evidence that the B-region mutations were able to produce lamivudine-resistant WHV, we determined the time course of appearance of mutant virus DNA in the liver and serum. The primary goal of this analysis was to determine the temporal relationship between the appearance of mutant virus in serum and liver and the increase in virus titers that occurs during prolonged lamivudine therapy.
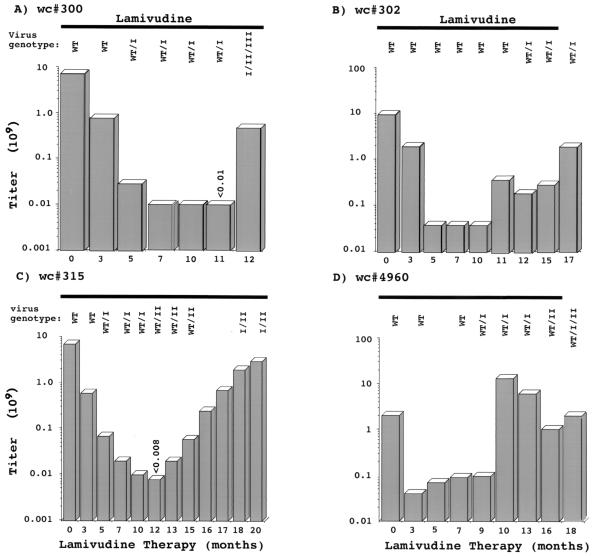
A region of the polymerase active site that includes regions B and C (Fig. 1) was amplified by PCR from virion DNA collected throughout the course of drug therapy and was then sequenced. Results for four woodchucks, woodchucks 300, 302, 315, and 4960 (also see Mason et al. [17]), are summarized in Fig. 5 and Table 2. With woodchuck 300 the type I variant was detected as early as 7 months before virus titers began to increase, while the type II and III variants were first prevalent at the time that the increase occurred. Similarly, for woodchuck 315, the type I mutant was detected 7 months before an increase in titers, and a shift to type II occurred at about the time that virus titers began to rise. For the two other woodchucks (woodchucks 302 and 4960), a type I mutant first became abundant in serum just about the time of the increase (woodchuck 4960) or shortly thereafter (woodchuck 302) (Fig. 5). However, cloning of the PCR products revealed that a type II variant was also abundant in the serum of woodchuck 4960 at the time that virus titers rose (Table 2), a result that was not apparent by direct sequencing of PCR-amplified viral DNA (Fig. 5). Likewise, cloning and sequencing of PCR products revealed the type I mutation in woodchuck 302 at 10 months, prior to the rise in virus titers (data not shown). (Unless a minor species represented at least 10 to 20% of viral DNA, it would not have contributed a detectable signal to the DNA sequencing ladders.) Thus, a type I mutant could persist for at least 7 months without producing an increase in titers, and in three of four woodchucks, this increase was associated with the nearly coincident emergence of type II and/or type III mutants. Moreover, the type II mutant appeared to be more efficient than the type I mutant at displacing the wild type (and type I mutant) from the liver, despite its much later appearance. One factor that may slow replacement of wild-type cccDNA is trans-complementation by mutant virus polymerase. Evidence for this was provided by the observation that both mutant and wild-type virus titers rose late in therapy (Table 2; Fig. 5).
FIG. 5.
Time course of appearance of B-region mutants in lamivudine-treated woodchucks (wc). The presence of different variants was estimated by sequencing of PCR products, as described in Materials and Methods. For woodchuck 4960, only wild-type (WT) virus was detected at 8.5 months. In general, the results were confirmed by cloning and sequence analysis of selected PCR products (Table 1).
TABLE 2.
Genotyping of viral DNA in liver and seruma
| Woodchuck and time of sample collection | No. of the following cccDNA variants/total no. tested:
|
Virus titer | No. of the following variants from serum/total no. tested:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | I | II | III | Wild type | I | II | III | ||
| 300 | |||||||||
| Pretherapy | 26/26 | 0/26 | 0/26 | 0/26 | 7 × 109 | 24/24 | 0/24 | 0/24 | 0/24 |
| 5 Mo | 30/30 | 0/30 | 0/30 | 0/30 | 3 × 107 | 22/36 | 14/36 | 0/36 | 0/36 |
| 12 Mo | 11/29 | 12/29 | 4/29 | 2/29 | 5 × 108 | 0/34 | 12/34 | 9/34 | 13/34 |
| 302 | |||||||||
| Pretherapy | 28/28 | 0/28 | 0/28 | 0/28 | 1 × 1010 | 30/30 | 0/30 | 0/30 | 0/30 |
| 12 Mo | 20/20 | 0/20 | 0/20 | 0/20 | 2 × 108 | 21/30 | 9/30 | 0/30 | 0/30 |
| 17 Mo | 18/22 | 4/22 | 0/22 | 0/22 | 2 × 109 | 9/34 | 25/34 | 0/34 | 0/34 |
| 315 | |||||||||
| Pretherapy | 27/27 | 0/27 | 0/27 | 0/27 | 7 × 109 | 32/32 | 0/32 | 0/32 | 0/32 |
| 12 Mo | 9/32 | 12/32 | 11/32 | 0/32 | <8 × 106 | 4/8 | 0/8 | 4/8 | 0/8 |
| 18 Mo | 0/25 | 6/25 | 19/25 | 0/25 | 2 × 109 | 0/28 | 1/28 | 27/28 | 0/28 |
| 20 Mo | 1/29 | 14/29 | 14/29 | 0/29 | 3 × 109 | NDb | ND | ND | ND |
| 4960 | |||||||||
| Pretherapy | 30/30 | 0/30 | 0/30 | 0/30 | 2 × 109 | 31/31 | 0/31 | 0/31 | 0/31 |
| 3 Mo | 25/25 | 0/25 | 0/25 | 0/25 | 4 × 107 | 31/31 | 0/31 | 0/31 | 0/31 |
| 10 Mo | 6/18 | 8/18 | 3/18 | 1/18 | 1.3 × 1010 | 10/26 | 13/26 | 3/26 | 0/26 |
| 16 Mo | 4/26 | 17/26 | 5/26 | 0/26 | 1 × 109 | 3/34 | 13/34 | 17/34 | 1/34 |
| 18 Mo | 7/28 | 15/28 | 5/28 | 1/28 | 2 × 109 | ND | ND | ND | ND |
| 4961 | |||||||||
| Pretherapy | 28/28 | 0/28 | 0/28 | 0/28 | 3 × 109 | 30/30 | 0/30 | 0/30 | 0/30 |
| 3 Mo | 26/26 | 0/26 | 0/26 | 0/26 | 1 × 109 | 36/36 | 0/36 | 0/36 | 0/36 |
| 10 Mo | 16/16 | 0/16 | 0/16 | 0/16 | 2 × 109 | 35/35c | 0/35 | 0/35 | 0/35 |
Virus was partially purified from serum samples, DNA was isolated, and a region of the genome encoding the B and C domains of the reverse transcriptase was amplified by PCR. The PCR products were then cloned and genotyped by using restriction endonucleases, as described in Materials and Methods.
ND, not done.
Ten of 35 clones had the alanine-to-serine change at amino acid 566.
In addition to the mutations that arose during lamivudine treatment, we detected a spontaneous mutation of the B region in one woodchuck (woodchuck 4961) after 10 months of placebo treatment (Table 2). This mutation, which changed the alanine at position 566 to a serine, was previously observed in this animal but not in five other woodchucks receiving placebo for the same length of time (17). This is one of the three changes in the type III mutant. We therefore tested this virus stock for lamivudine resistance in primary hepatocyte cultures, following the procedure described in Fig. 2. This virus stock responded to lamivudine like the wild-type virus described in Fig. 2A; that is, there was no evidence that the mutation conferred lamivudine resistance to the infectious virus in this serum sample. Insertion of the mutation into wild-type viral DNA led to a slight increase in resistance to lamivudine in transfected Huh7 cultures, characterized as described above and shown in Fig. 3 (data not shown).
In an attempt to obtain additional information about the emergence of mutant virus in the liver, we determined the genotypes of cccDNAs isolated from biopsy specimens taken during the course of therapy (17). cccDNA was purified, and the active-site region of the polymerase was amplified by PCR. The genotypes of the PCR clones were then determined, with the results shown in Table 2. With two of the woodchucks (woodchucks 300 and 302), this analysis supported the earlier conclusion, derived from the results of cell culture studies, that the mutants were drug resistant in vivo and, therefore, replicated more efficiently than the wild type in the presence of lamivudine. For instance, after 5 months of treatment of woodchuck 300, all 30 clones obtained after PCR amplification of cccDNA were wild type, whereas 14 of 36 clones derived from amplification of virion DNA had a type I mutant genotype. Likewise, after 12 months, 11 of 29 cccDNA-derived clones were wild type, whereas 34 of 34 clones derived from virus had a B-region mutation.
For two of the four woodchucks (woodchucks 315 and 4960) whose results are presented in Table 2 and Fig. 5, a replication advantage of mutant virus was not evident simply by comparison of cccDNA and viral genotypes. (For woodchuck 315, the very low virus titers after 12 months were probably due to a transient immunological reaction to the infection rather than a direct simple consequence of the antiviral therapy [17].) For these two woodchucks, as for woodchucks 300 and 302, direct evidence that the mutants replicated more efficiently than the wild type was provided by the observation that mutant cccDNA supplanted the wild type as the prevalent species in the liver during the course of therapy. The presence of elevated titers of wild-type virus in the serum as total virus titers rise is presumably a consequence of trans-complementation of the wild-type pregenome by mutant WHV polymerase in a set of doubly infected hepatocytes.
Taken together, the results suggest that the emergence of the type I mutant and, by implication, of the type II and III mutants is impeded by preexisting infection of hepatocytes with wild-type virus. Moreover, this appears to be the case, despite the evidence from both cell culture and in vivo studies that the mutants replicate more efficiently than the wild type in the presence of lamivudine. Thus, a sustained suppression of virus titers in the serum may reflect the stability of wild-type cccDNA and of the hepatocytes that were infected by wild-type virus rather than a significant loss of infection from the liver.
Consequences of mutations in the YMDD motif of WHV.
One peculiar aspect of lamivudine-resistant WHV was the absence of mutations of the YMDD motif, which characterize lamivudine-resistant variants of HBV and human immunodeficiency virus that emerge during therapy. Mutations were therefore inserted into wild-type WHV to change the YMDD motif (Fig. 1) to either YIDD or YVDD. The mutant DNA was then transfected into Huh7 cells in the presence and absence of lamivudine, and the accumulation of intermediates in viral DNA synthesis was assayed (Fig. 3). The YVDD mutant of WHV was drug resistant, although it appeared to replicate poorly. Replication of the YIDD mutant was essentially undetectable in this experiment, but replication and drug resistance were demonstrated by more sensitive assays (data not shown). Thus, the data suggest that one reason that the YVDD and YIDD variants were not detected in vivo is that they may replicate inefficiently.
DISCUSSION
Our previous evaluation of lamivudine treatment of chronically infected woodchucks reveals three unexpected problems (17). First, the decline in virus titers in the serum is very slow, extending over several months. Second, the net suppression in virus titers is less than 1,000-fold in most woodchucks. Third, virus titers invariably rise again within 9 to 12 months.
The results presented above suggest that the overall decline in virus titers is limited, at least in some woodchucks, by the presence of drug-resistant WHV in the liver even early in therapy. This possibility was consistent with the detection of the type I mutant in some woodchucks as early as 5 months after the initiation of lamivudine treatment. Assuming that the type I mutant replicated in the presence of lamivudine with the same efficiency as wild-type WHV in the absence of lamivudine, mutant cccDNA would be 0.2 to 1% of the total cccDNA at this time. Indeed, analysis of the genotypes of cccDNA extracted from woodchuck 300 at this time revealed only wild-type cccDNA (30 of 30 clones; Table 2).
Unfortunately, none of our experiments shows why virus titers do not drop rapidly after initiation of lamivudine therapy, as they do in HBV-infected patients. A prolonged WHV half-life in blood is not an explanation. Rapid drops in WHV titer have been observed following even short-term therapy with other nucleoside analogs. For example, one group observed a 100-fold drop in WHV titers following initiation of therapy with BMS-200475, indicating that the half-life of WHV in blood is 1 day or even less (8). One possible explanation might be that type I mutant cccDNA is distributed evenly through the hepatocyte population. If this mutant cccDNA were a common variant in WHV carriers, representing 1% of the total at the start of therapy, and was spread evenly to hepatocytes containing an average of 20 cccDNA molecules (wild type plus mutant cccDNA molecules), there would be enough mutant cccDNA for 20% of the hepatocyte population. This could help to explain why virus titers dropped only about 10-fold in the first month even after treatment with very high doses of lamivudine (assuming efficient trans-complementation by the mutated polymerase).
This hypothesis for the slow decline in virus titers appears untenable, however, as it fails to explain why titers keep dropping as therapy is continued. If, instead, the type I variant is not initially spread among hepatocytes but is the predominant or only cccDNA in those hepatocytes in which it is initially present, a different picture emerges. The slow decline in virus titers reflects some peculiarity in uptake or phosphorylation of lamivudine in the woodchuck (22).
An even more complex issue is the eventual enrichment of type I cccDNA in the liver and the nearly complete replacement of wild-type cccDNA by the type II variant. If both are initially present in only a few hepatocytes, then the ultimate rise in virus titers and the enrichment of their respective cccDNAs requires spread to hepatocyte lineages originally infected with wild-type WHV. This requires that these hepatocytes either lose the wild-type virus or becomes susceptible to superinfection with the mutant virus. How this would occur is unknown. One possibility is that inhibition of viral DNA synthesis, plus elevated hepatocyte death and proliferation due to infection, leads to dilution of wild-type DNA in hepatocyte lineages, making them candidates for superinfection by the mutants. This type of mechanism would help explain why in almost every woodchuck there is an identical length of time between initiation of antiviral therapy and the eventual rise in titers whether it is due to the spread of type I or type II variants. That is, the eventual takeover by mutant virus is entirely dependent upon the loss of wild-type WHV, which, in turn, is governed by the rate of death and proliferation of hepatocytes. The preferential takeover by type II and III variants, when detectable, may reflect the fact that these two types have a higher replicative capacity than type I, plus the fact that the continued presence of some wild-type WHV is probably not required for their replication. (Because of the introduction by the pol mutation of the type I variant of a stop codon in the overlapping S gene [17], this virus is presumably dependent on coinfection with wild-type virus to form infectious virions; the effects of missense mutations in the S gene, which are associated with the type II and III mutations, on the formation of infectious virions are unknown.)
As noted earlier, a recent study on the appearance of lamivudine-resistant strains of HBV, in which mutant virus was present in the serum of patients four months prior to an increase in virus titers, points to the same conclusions as our study (4): that wild-type virus that remains in the liver may temporarily inhibit outgrowth of the drug-resistant variant. Whether or not this conclusion will be supported by further studies of clinical specimens remains to be determined.
ACKNOWLEDGMENTS
We are grateful to G. Rall, C. Seeger, J. Summers (University of New Mexico), B. Tennant (Cornell University), and J. Taylor for helpful suggestions, to S. Berman for assistance in the preparation of the manuscript, and to A. Cywinski and the DNA Sequencing Facility of the Fox Chase Cancer Center for sequence determinations. Oligonucleotides were synthesized in the institutional DNA Synthesis Facility under the direction of T. Yeung.
This work was supported by Public Health Service grants AI-18641, 3P01-CA-4073711S1, and CA-06927 from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Aldrich C E, Coates L, Wu T T, Newbold J, Tennant B C, Summers J, Seeger C, Mason W S. In vitro infection of woodchuck hepatocytes with woodchuck hepatitis virus and ground squirrel hepatitis virus. Virology. 1989;172:247–252. doi: 10.1016/0042-6822(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 2.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K-A, Tyrrell D L J, Brown N, Condreay L D. Identification and characterization of mutations in HBV resistant to lamivudine. Hepatology. 1997;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomeusz A, Groenen L C, Locarnini S A. Clinical experience with famciclovir against hepatitis B virus. Intervirology. 1997;40:337–342. doi: 10.1159/000150566. [DOI] [PubMed] [Google Scholar]
- 4.Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type virus after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J I, Miller R H, Rosenblum B, Denniston K, Gerin J L, Purcell R H. Sequence comparison of woodchuck hepatitis virus replicative forms shows conservation of the genome. Virology. 1988;162:12–20. doi: 10.1016/0042-6822(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 6.Di Q, Summers J, Burch J B, Mason W S. Major differences between WHV and HBV in the regulation of transcription. Virology. 1997;229:25–35. doi: 10.1006/viro.1996.8422. [DOI] [PubMed] [Google Scholar]
- 7.Fischer K P, Tyrrell D L. Generation of duck hepatitis B virus polymerase mutants through site-directed mutagenesis which demonstrate resistance to lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine] in vitro. Antimicrob Agents Chemother. 1996;40:1957–1960. doi: 10.1128/aac.40.8.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovesi E V, Lamb L, Medina I, Taylor D, Seifer M, Innaimo S, Colonno R J, Standring D N, Clark J M. Efficacy of the carbocyclic 2′-carbocyclic 2′-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hepatitis B virus infection. Antimicrob Agents Chemother. 1998;42:3209–3217. doi: 10.1128/aac.42.12.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honkoop P, Niesters H G, de Man R A, Osterhaus A D, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B. Incidence and patterns. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 10.Hoofnagle J H, DiBisceglie A M. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 11.Horton R M, Cai Z L, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 12.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L A. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 13.Huang M J, Summers J. Infection initiated by the RNA pregenome of a DNA virus. J Virol. 1991;65:5435–5439. doi: 10.1128/jvi.65.10.5435-5439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodama K, Ogasawara N, Yoshikawa H, Murakami S. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J Virol. 1985;56:978–986. doi: 10.1128/jvi.56.3.978-986.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 16.Mason W S, Aldrich C, Summers J, Taylor J M. Asymmetric replication of duck hepatitis B virus DNA in liver cells: free minus-strand DNA. Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason W S, Cullen J, Moraleda G, Saputelli J, Aldrich C E, Miller D S, Tennant B, Frick L, Averett D, Condreay L D, Jilbert A R. Lamivudine therapy of WHV infected woodchucks. Virology. 1998;245:18–32. doi: 10.1006/viro.1998.9150. [DOI] [PubMed] [Google Scholar]
- 18.Moraleda G, Saputelli J, Aldrich C E, Averett D, Condreay L, Mason W S. Lack of effect of antiviral therapy in nondividing heptocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol. 1997;71:9392–9399. doi: 10.1128/jvi.71.12.9392-9399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 20.Nowak M A, Bonhoeffer S, Hill A M, Boehme R, Thomas H C, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perrillo R P, Schiff E R, Davis G L, Bodenheimer H C, Lindsay K, Payne J, Dienstag J L, O’Brien C, Tamburro C, Jacobson I M, Sampliner R, Feit D, Lefkowitch J, Kuhns M, Meschievitz C, Sanghvi B, Albrecht J, Gibas A. A randomized, controlled trial of interferon alfa-2b alone and after prednisone withdrawal for the treatment of chronic hepatitis B. N Engl J Med. 1990;323:295–301. doi: 10.1056/NEJM199008023230503. [DOI] [PubMed] [Google Scholar]
- 22.Rajagopalan P, Boudinot F D, Chu C K, Tennant B C, Baldwin B H, Schinazi R F. Pharmacokinetics of (−)-2′-3′-dideoxy-3′-thiacytidine in woodchucks. Antimicrob Agents Chemother. 1996;40:642–645. doi: 10.1128/aac.40.3.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuurman R, Nijhius M, van Leeumen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C A B. Rapid changes in human immunodeficiency virus type I RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine. J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 24.Seeger C, Baldwin B, Tennant B C. Expression of infectious woodchuck hepatitis virus in murine and avian fibroblasts. J Virol. 1989;63:4665–4669. doi: 10.1128/jvi.63.11.4665-4669.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L J. Mutation in HBV DNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 26.van der Eb A J, Graham F L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65:826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]
- 27.Wahl G M, Stern M, Stark G R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci USA. 1979;76:3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]