Abstract
Protein model refinement is the last step applied to improve the quality of a predicted protein model. Currently the most successful refinement methods rely on extensive conformational sampling and thus, take hours or days to refine even a single protein model. Here we propose a fast and effective model refinement method that applies GNN (graph neural networks) to predict refined inter-atom distance probability distribution from an initial model and then rebuilds 3D models from the predicted distance distribution. Tested on the CASP (Critical Assessment of Structure Prediction) refinement targets, our method has comparable accuracy as two leading human groups Feig and Baker, but runs substantially faster. Our method may refine one protein model within ~11 minutes on 1 CPU while Baker needs ~30 hours on 60 CPUs and Feig needs ~16 hours on 1 GPU. Finally, our study shows that GNN outperforms ResNet (convolutional residual neural networks) for model refinement when very limited conformational sampling is allowed.
Editor summary:
Deep graph neural networks can refine a predicted protein model efficiently with less computing resources. The accuracy is comparable to that of the leading physics-based methods that rely on time consuming conformation sampling.
Introduction
High-accuracy protein structure prediction can facilitate the understanding of biological processes at the molecular level. In the past few years, protein structure prediction has been greatly improved, mainly due to deep convolutional residual networks (ResNet)1–4 introduced by RaptorX and lately Transformer-like networks implemented in AlphaFold2. However, some predicted protein structural models still deviate much from their native structures, which limits their value in downstream applications. To further improve model quality, much effort has been devoted into developing model refinement methods5–7. The main goal is to refine an initial model towards its native structure and then, to generate new models of higher quality. This is very challenging since the space of worse models is much larger than that of better models. Many refined models submitted to Critical Assessment of Structure Prediction (CASP) have worse quality than their starting models5.
A typical model refinement method involves side-chain repacking, energy minimization and constrained structure sampling8–11. Since it is challenging to optimize energy function, large-scale conformational sampling is often resorted. Currently, the most successful refinement methods use large-scale conformational sampling either through molecular dynamics (MD) simulations6 or fragment assembly7,12. For example, the Feig group employs iterative MD simulation with flat-bottom harmonic restraints to sample conformations. The Baker group7 uses local error estimation to guide conformational sampling by fragment assembly, and iteratively refines the models by recombining secondary structure segments and replacing torsional angles. DeepAccNet13, developed by the Baker group recently, uses both 3D and 2D convolution networks to estimate residue-wise accuracy and inter-residue distance error, which are then converted into restraints to guide conformational sampling. GalaxyRefine212 developed by the Seok group employs multiple conformation search strategies. Although performing well on some proteins, these methods rely on extensive conformational sampling and thus, a lot of computing resources for even refining a single protein model 13,14. There are also some fast refinement methods such as ModRefiner, 3DRefine and ReFold that do not use extensive conformational sampling8–11, but their performances lag far behind Feig’s and Baker’s.
Here we propose a model refinement method GNNRefine that may quickly improve model quality with only limited conformational sampling. GNNRefine represents an initial protein model as a graph and then employs graph neural networks (GNN) to refine it. Compared to contact and distance matrix representation (used by ResNet) of a protein model, graph representation may capture multiple-residue correlation and the global information of a protein more easily. GNN has been used to predict protein model quality15,16, but not to refine protein models so far. GNNRefine predicts inter-atom distance probability distribution and converts it into distance potential, which is then fed into PyRosetta17 FastRelax18 to produce refined models. Our experimental results show that GNNRefine may improve model quality with very limited confirmation sampling, outperforms a majority number of existing methods and is slightly worse than Feig’s method that uses large-scale conformational sampling. Another advantage is that GNNRefine produces fewer degraded models than the others.
In practice it is important to be able to refine protein models quickly. This is because many biologists do not have human and computational resources for in-house protein structure modeling and thus, have to rely on protein structure prediction web servers. In order to respond to many users in a short time, a web server shall be able to refine protein models quickly without sacrificing accuracy. It is challenging for a structure prediction server to run Baker’s or Feig’s refinement programs without a large number of GPUs or CPUs.
Results
Overview of the method
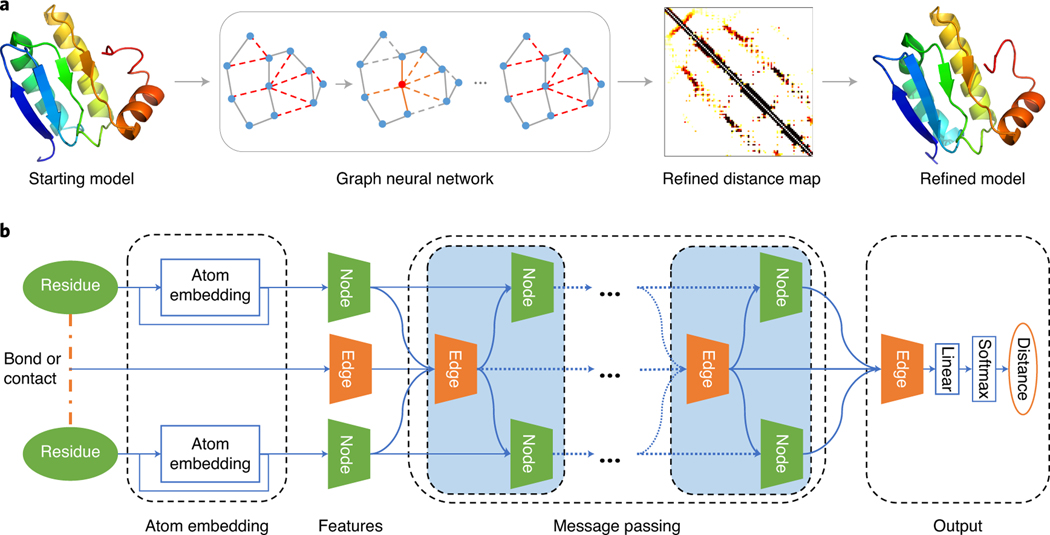
As shown in Fig. 1a, our method GNNRefine mainly includes three steps: 1) represent the initial model as a graph and extract atom, residue, and geometric features from the initial model, 2) predict refined distance distribution for each edge using GNN, and 3) convert the predicted distance probability into distance potential and feed it into PyRosetta17 FastRelax18 to produce refined models by side-chain packing and energy minimization. Meanwhile, the GNN-based distance prediction is the key to the refined model quality. As shown in Fig. 1b, GNNRefine mainly consists of three modules: an atom embedding layer, multiple message passing layers, and an output layer. The atom embedding layer learns atom-level structure information of one residue and its output is concatenated with other residue features to form the final feature of a residue. The protein graph is built on the residue feature (node) and bond or contact feature (edge) between residue pairs. The multiple message passing layers iteratively update the node and edge features to capture global structural information. Finally, a linear layer and a softmax function are used to yield distance probability distribution from the edge feature.
Fig. 1:
GNNRefine for protein model refinement. a. The flowchart includes feature extraction from the starting model, refined distance prediction using GNNs, and refined model building based on the refined distance prediction; b. the network with 10 message passing layers and 256 hidden neurons for both node and edge features. The atom embedding is concatenated with other residue features to form the node feature. The edge feature is derived for a pair of residues with Euclidean distance less than 10Å, including spatial distance, orientation and sequential separation of the two residues. The refined distance prediction is based on the final edge feature. PyMol 2.3.0 is used for structure visualization.
Evaluation metrics
We evaluate the quality of a protein model by Global Distance Test High Accuracy (GDT-HA), Global Distance Test Total Score (GDT-TS) and Local Distance Difference Test (lDDT)19, all ranging in [0, 100]. For all these three metrics, higher values indicate better quality and the value 100 means the predicted model is the same as the experimental structure. GDT-HA and GDT-TS measure the percentage of residues in the predicted model that deviate from the experimental structure by at most a small distance cutoff. GDT-HA is more sensitive than GDT-TS because its distance cutoffs are half of those used by GDT-TS. lDDT can also be interpreted similarly. To intuitively compare the performance of different methods, we mainly use the quality changes before and after refinement. A positive value (with positive sign) means the average quality of a refined model is better than its starting models, and a higher value indicates a larger improvement, while a negative value (with negative sign) has the opposite meaning. “Degradation” counts the number of refined models with worse quality than their initial models by a given threshold (0, −1 and −2). Meanwhile, 0 denotes that a refined model has worse GDT-HA than its starting model; −1 and −2 denote that a refined model is worse than its starting model by at least 1 and 2 GDT-HA units, respectively.
Performance on CASP13 refinement targets
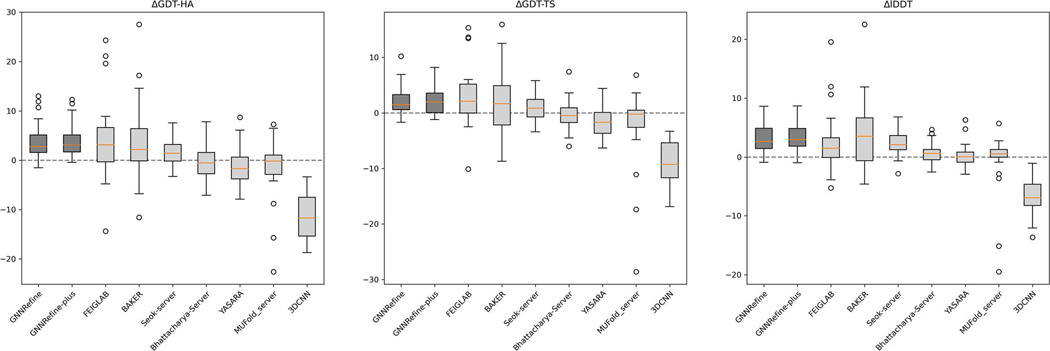
We compare our method with two leading human groups FEIGLAB and BAKER in the CASP13 refinement category5 and 5 server groups Seok-server, Bhattacharya-Server, YASARA, MUFold_server and 3DCNN, all of which are described in the CASP13 Abstract book20. Their refined models are available at the CASP official website. A human group has up to 3 weeks to refine one model while a server group has at most 3 days. A human group may use any extra information including all server models. For example, FEIGLAB selected refined models manually and BAKER chose their sampling strategy based upon the model quality provided by the CASP organizers. Note that our method was not blindly tested in CASP13. We show the average quality of the first submitted models in Table 1 and visualize the distribution of quality improvement in Extended Data Fig. 1. Even generating only 5 refined models for each initial model, GNNRefine obtains comparable performance as the two human groups and outperforms all the 5 servers. Seok-server is the only server that yielded improvement on the three metrics. Bhattacharya-Server and YASARA improved lDDT slightly, but degraded GDT-HA and GDT-TS. MUFold_server and 3DCNN degraded all the three metrics. Further, GNNRefine slightly worsens the quality of only 4 protein models, but all the others including the two human groups degraded many models. That is, it is very safe to use our method to refine models. Extended Data Fig. 1 shows that in terms of quality improvement the two human groups have a larger variance than ours because they used extensive conformational sampling, but we do not.
Table 1.
Performance on the CASP13 refinement targets. Our GNNRefine and GNNRefine-plus generate 5 and 50 refined models, respectively, from an initial model. Only the first-ranked refined models are evaluated. There are 28 targets in total. Seok-server and Bhattacharya-Server submitted refined models for all targets, YASARA submitted 27 models, MUFold_server submitted 26, and 3DCNN submitted 22.
| Type | Methods | GDT-HA | GDT-TS | lDDT | Degradation |
||
|---|---|---|---|---|---|---|---|
| 0 | −1 | −2 | |||||
| Starting | 52.27 | 71.51 | 61.74 | ||||
|
| |||||||
| Human | FEIGLAB | +4.04 | +2.97 | +2.48 | 8 | 6 | 4 |
| BAKER | +3.35 | +1.86 | +3.73 | 7 | 6 | 6 | |
|
| |||||||
| Server | GNNRefine | +3.83 | +2.31 | +3.19 | 3 | 1 | 0 |
| GNNRefine-plus | +3.90 | +2.31 | +3.33 | 4 | 0 | 0 | |
| Seok-server | +1.73 | +0.89 | +2.23 | 7 | 3 | 1 | |
| Bhattacharya-Server | −0.44 | −0.37 | +0.64 | 17 | 12 | 8 | |
| YASARA | −1.23 | −1.57 | +0.26 | 18 | 16 | 13 | |
| MUFold_server | −1.61 | −2.33 | −0.70 | 13 | 11 | 7 | |
| 3DCNN | −11.47 | −8.78 | −6.92 | 22 | 22 | 22 | |
The average performance of each group is calculated on its submitted models. GDT-HA: Global Distance Test High Accuracy, GDT-TS: Global Distance Test Total Score, lDDT: Local Distance Difference Test, Degradation: the number of refined models has quality worse than their initial models by a given threshold based on GDT-HA.
Performance on CASP14 refinement targets
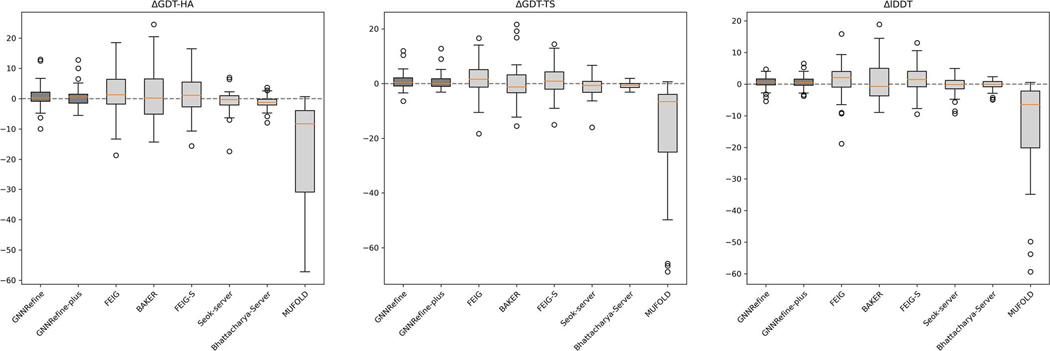
We test our GNNRefine on the 37 CASP14 refinement targets (not including the extended time refinement targets) and compare it with two human groups FEIG and BAKER and 4 server groups FEIG-S, Seok-server, Bhattacharya-Server and MUFold_server, all of which are described in the CASP14 Abstract book21. Note that we did not finish developing our method before CASP14, so it was not blindly tested in CASP14. FEIG-S is a server group, but not fully automated for some targets as mentioned in CASP14 Abstracts21. In CASP14 both the FEIG and BAKER groups used extra information in addition to the starting models to be refined. In contrast, our method only uses the starting models assigned by CASP14. For example, FEIG-S used some in-house template-based models as extra starting models for 6 targets. For 14 targets FEIG used the other models of the same server generating the starting model assigned by CASP14. BAKER used inter-residue distance predicted by trRosetta4 from MSAs (multiple sequence alignment) as an input feature of DeepAccNet13 but GNNRefine does not use MSAs. The CASP14 models are much harder to refine than CASP13 models. As shown in Table 2 and Extended Data Fig. 2, all the server groups except FEIG-S degraded the model quality in terms of GDT-HA, GDT-TS and lDDT, and all including the two human groups degraded the quality of more than 10 models. This is possibly because some starting models are already well refined, especially the 7 AlphaFold2 models. Excluding the AlphaFold2 models, the model quality improvement is comparable to CASP13, as shown in Supplementary Table 1.
Table 2.
Performance on all CASP14 refinement targets. GNNRefine and GNNRefine-plus generate 5 and 50 refined models, respectively, for each initial model. Only the first-ranked refined models are evaluated.
| Type | Methods | GDT-HA | GDT-TS | lDDT | Degradation |
||
|---|---|---|---|---|---|---|---|
| 0 | −1 | −2 | |||||
| Starting | 54.12 | 72.65 | 65.98 | ||||
|
| |||||||
| Human | FEIG | +2.01 | +1.49 | +1.13 | 14 | 12 | 9 |
| BAKER | +1.13 | −0.03 | +0.90 | 17 | 15 | 13 | |
|
| |||||||
| Server | GNNRefine | +0.84 | +0.82 | +0.50 | 17 | 9 | 7 |
| GNNRefine-plus | +0.80 | +0.77 | +0.67 | 14 | 10 | 6 | |
| FEIG-S | +1.59 | +1.05 | +1.16 | 15 | 14 | 11 | |
| Seok-server | −1.14 | −1.32 | −0.52 | 21 | 15 | 11 | |
| Bhattacharya-Server | −1.24 | −0.68 | −0.45 | 29 | 22 | 10 | |
| MUFOLD | −15.37 | −17.91 | −13.28 | 36 | 35 | 32 | |
GDT-HA: Global Distance Test High Accuracy, GDT-TS: Global Distance Test Total Score, lDDT: Local Distance Difference Test, Degradation: the number of refined models has quality worse than their initial models by a given threshold based on GDT-HA.
Overall, on the CASP14 targets our method performs worse than Feig’s methods (possibly because they used extra initial models), comparably to Baker’s method (which used MSAs) and better than the others. Our method degrades the least number of models. Note that even generating only 5 refined models for an initial model, our method has similar accuracy as generating 50 refined models for an initial model. When generating 250 refined models for an initial model, our method may further improve model quality, as shown in Supplementary Table 17.
As shown in Supplementary Table 2 and Fig. 1, on average all methods degrade the AlphaFold2 models, but our GNNRefine-plus degrades less than the others. On average GNNRefine-plus degrades the AlphaFold2 models by 2.69, 1.49 and 2.24 in terms of GDT-HA, GDT-TS and IDDT, respectively. FEIG degrades by 9.04, 7.06 and 8.39 in terms of GDT-HA, GDT-TS and IDDT, respectively. Baker degrades by 5.61, 4.69 and 5.26 in terms of GDT-HA, GDT-TS and IDDT, respectively. FEIG significantly degraded 4, 4, 6 AlphaFold2 models in terms of GDT-HA, GDT-TS and IDDT (<−5), respectively. Baker significantly degraded 5, 3, 4 AlphaFold2 models in terms of GDT-HA, GDT-TS and IDDT (<−5), respectively. GNNRefine-plus only significantly degrades 1, 0, 0 AlphaFold2 model in terms of GDT-HA, GDT-TS and IDDT (<−5), respectively. We further test our method on the first models submitted by AlphaFold2 for 88 CASP14 domains, as described in Supplementary Section 2. Supplementary Fig. 2 shows our method can only refine a small number of AlphaFold2 models with lDDT<88. One possible reason is that there are only a small percentage of protein models of high quality (i.e. lDDT>80) in our training set as shown in Supplementary Fig. 5.
Specific examples.
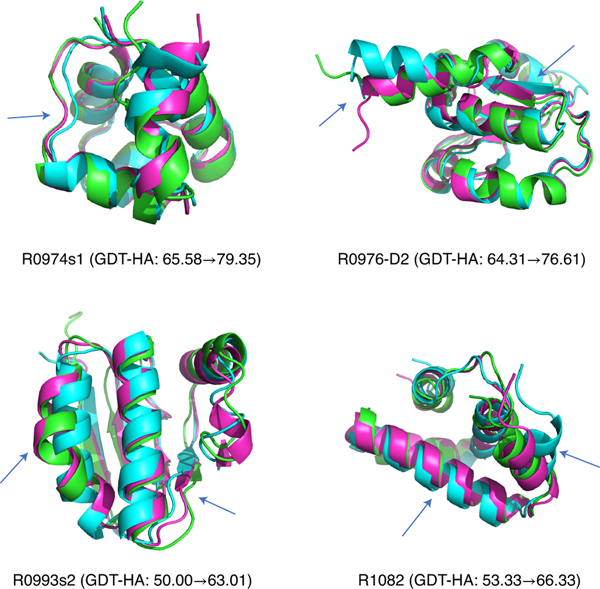
GNNRefine has successfully refined five CASP targets (3 CASP13 targets and 2 CASP14 targets) with ΔGDT-HA ≥10. Fig. 2 shows 4 of them with publicly available experimental structures and indicates that our method can refine models at different secondary structure regions.
Fig. 2:
Successful refinement examples by GNNRefine. These examples are for CASP13 targets R0974s1, R0976-D2 and R0993s2, and R1082 from CASP14. These starting models were provided by CASP13 and CASP14. Native structures, starting models, and refined models are shown in green, cyan, and magenta, respectively. The regions that are significantly refined are indicated with blue arrows. PyMol 2.3.0 is used for structure visualization.
Performance on the CAMEO and CASP13 FM data
We further evaluate our method on two large datasets. One has 208 starting models for the CAMEO targets and the other has 4193 models built by RaptorX-3DModeling (https://github.com/j3xugit/RaptorX-3DModeling) for the CASP13 FM (free modeling) targets. As shown in Supplementary Table 4, on the CAMEO targets, our methods on average improve model quality by ~2 GDT-HA units, much better than the others. Meanwhile, in terms of GDT-HA, GDT-TS and IDDT, ~78%, ~73% and ~92% of the refined models are better than their initial models, respectively. As shown in Supplementary Fig. 3 and Table 5, on the CAMEO targets there is a weak correlation (0.20) between the GNNQA-predicted quality of the starting model and the improvement by GNNRefine. Most of the CAMEO starting models have predicted quality (lDDT) from 47 to 80 and GNNRefine may refine most of them. GNNRefine has a slightly better chance to improve the quality of CAMEO targets when the predicted lDDT score is between 60 and 80. The performance of our method is not very sensitive to protein size. The correlation between protein sequence length and the improvement by GNNRefine is weak (0.0154, 0.0712 and 0.1020 in terms of GDT-HA, GDT-TS and IDDT, respectively). As shown in Supplementary Table 6, on the CASP13 FM models our method on average improves model quality by ~2 GDT-HA units.
Running time on CASP14 refinement targets
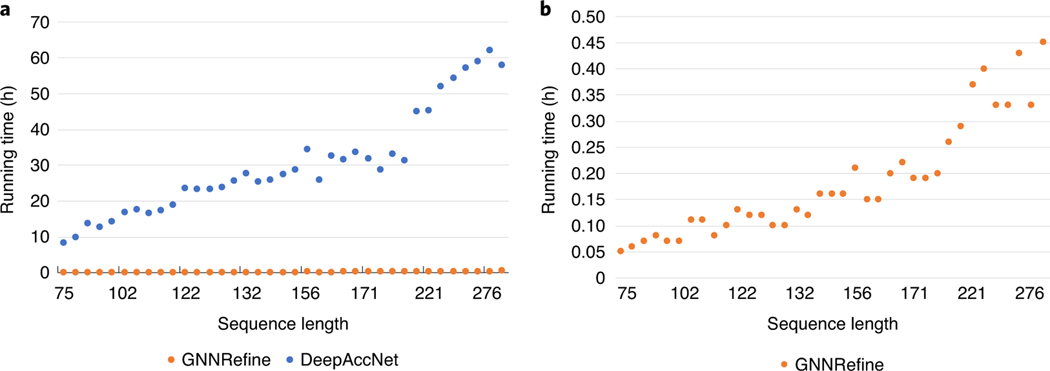
The Baker group has released the source code of their ResNet-based refinement protocol used in CASP14, i.e. DeepAccNet (https://github.com/hiranumn/DeepAccNet-TF), which allows us to accurately measure its running time. To ensure a fair comparison between our method and standard DeepAccNet, we ran them on the same Linux workstation (with 256 CPU cores and 2T memory) by their default configurations. As shown in Fig. 3, on average GNNRefine needs 0.18 hours (10.8 minutes) on 1 CPU to refine a single model while DeepAccNet needs 30.19 hours on 60 CPUs to do so, which implies that our method is about 10,000 times faster than DeepAccNet. The running time of both our method and DeepAccNet increases proportionally along with the protein sequence length. We are not able to install Feig’s program locally since it needs too many external packages. As reported in Feig’s CASP14 talk, it took 16 GPU hours for Feig’s method (running on RTX2080Ti) to refine a single protein model R1056 (of 169 residues)22. Our method is much more efficient because that it may predict inter-residue distance more accurately (see Supplementary Table 8) and thus, improve protein model quality without extensive conformational sampling while both Baker’s and Feig’s methods heavily rely on conformation sampling instead of better inter-residue distance constraints to generate protein models of higher quality. Further, we only consider those residue pairs with distance no more than 10Å in the starting model and thus, do not use too many distance restraints, which also helps to speed up.
Fig. 3:
The running time of GNNRefine and DeepAccNet on the CASP14 targets with respect to protein sequence length. a. comparison between GNNRefine and DeepAccNet. b. the running time of GNNRefine.
Comparison against standalone software and servers
Here we compare our method with some publicly available software and servers such as GalaxyRefine9, ModRefiner8, 3DRefine10, and ReFold23. We run GalaxyRefine locally by its default configuration. ModRefiner has a configurable parameter to control the strength of restraints extracted from the starting model, with 0 meaning no restraints at all while 100 indicating very tight restraints. We run ModRefiner locally with three different strength values: 0, 50 and 100. The refined models of 3DRefine were collected from the 3DRefine server and the refined models of ReFold were its CASP13 submissions. As a control, we also run PyRosetta FastRelax without using the distance restraints predicted by GNNRefine. As shown in Supplementary Table 7, on the CASP13 targets GNNRefine outperforms all the other methods by all metrics including running time.
Evaluation of distance prediction
To understand why GNNRefine works without extensive conformational sampling, we evaluate the distance predicted by GNNRefine in terms of top L contact precision and lDDT. For each residue pair, the predicted probabilities of distance bellow 8Å are summed up as predicted contact probability. We select top L contacts in the starting model by their respective Cβ-Cβ Euclidean distance ascendingly. To calculate the lDDT of the distance predicted by GNNRefine, for each residue pair we use the middle point of the bin with the highest predicted probability as its real-valued distance prediction. We only consider the Cβ-Cβ pairs with predicted distances less than 20Å. Supplementary Table 8 shows that the distance predicted by GNNRefine is better than the starting model in terms of both contact precision and lDDT.
Comparison against ResNet for model refinement
The convolutional residual neural network (ResNet) is widely used for protein contact and distance prediction. The Baker group developed a ResNet-based method DeepAccNet for model refinement. To test the performance of DeepAccNet when large-scale conformational sampling is not used, we feed the distance potential generated by standard DeepAccNet into PyRosetta FastRelax to build refined models, using exactly the same method as GNNRefine. We have also developed an in-house ResNet model (in Supplementary Section 6) to predict distance from initial models and test if the predicted distance can be used to refine models or not. To be fair, in this experiment we use only 1 instead of 5 deep GNNRefine models to do refinement. For each method, we generate 10 refined models from each starting model and select the lowest-energy model as the final refined model. Supplementary Table 9 shows that our GNN method greatly outperforms our in-house ResNet method, which in turn is better than DeepAccNet. That is, DeepAccNet is not able to refine models when extensive conformational sampling is not used, but our GNN method works. Our in-house ResNet differs from DeepAccNet in that our ResNet directly predicts distance distribution while DeepAccNet predicts the distribution of distance error. Supplementary Table 10 shows that our GNN method indeed can predict distance with better accuracy than ResNet.
One underlying reason that GNN outperforms ResNet for model refinement is that GNN is able to model the correlation of multiple residues more easily than ResNet. Most proteins have their radius of gyration proportional to the cube root of their length, so any two residues that are well separated along the primary sequence can be connected by a path in the protein graph shorter than the cube root of the protein length. As such, the correlation of multiple residues (spreading out in the distance matrix) can be modeled more effectively by (not so deep a) GNN, but not by a ResNet. That is, ResNet is good for inferring the initial inter-residue relationship and GNN is more suitable for refining it.
Ablation study
To assess the contribution of individual factors to GNNRefine, we evaluate the GNNRefine models trained by different data and features in Table 3. Supplementary Table 11 shows the quality of the distance predicted by these GNN models. In summary, the large training data, the inter-residue orientation and the DSSP-derived features are the three most important factors. The atom embedding does not help much, possibly because we did not make use of the chemical contexts of the atoms and the positions of these atoms in our training models are not very accurate. Supplementary Table 12 shows the performance of iterative refinements by 5 GNNRefine models on the CASP13 targets, which demonstrates that the GNNRefine models trained on different datasets are complementary to each other. We have also predicted the CαCα and NO distance and used them as restraints to build refined models, but did not observe significant improvement, as shown in Supplementary Table 13. Supplementary Table 14 shows the performance of GNNRefine with respect to the distance cutoff for graph edge definition, and that 10Å is the best distance cutoff. To verify if there are many incorrect or missing edges in the graph representation of an initial protein model, we evaluate the accuracy of the graph edges derived from the initial 3D models to be refined, as shown in Supplementary Table 15, ~80% of the graph edges derived from the initial models are correct when distance cutoff is set to 10Å. Supplementary Table 16 shows the impact of the number of message-passing layers and our method has the best performance with 10 message-passing layers. Supplementary Table 17 shows that when 250 refined models are generated for one initial model, more quality improvement may be achieved than when only 50 refined models are generated for one initial model.
Table 3.
GNNRefine’s performance with different features and training data on the CASP13 data. For the AtomEmb (with local frame), we use Cα, N, and C to define the reference frame of atom coordinates for each residue.
| Features | Training data | GDT-HA | GDT-TS | lDDT | Degradation |
||
|---|---|---|---|---|---|---|---|
| 0 | −1 | −2 | |||||
| All features | In-house | +3.15 | +1.96 | +2.88 | 1 | 0 | 0 |
| All features | DeepAccNet data | +3.19 | +1.75 | +2.74 | 3 | 1 | 1 |
| All features | CASP models only | +1.42 | +0.92 | +1.35 | 8 | 6 | 3 |
| no Orientation | In-house | +2.21 | +1.28 | +2.26 | 4 | 2 | 0 |
| no Dihedral&SS&RSA | In-house | +2.53 | +1.67 | +2.31 | 2 | 0 | 0 |
| no AtomEmb | In-house | +3.25 | +2.03 | +2.57 | 2 | 0 | 0 |
| AtomEmb (with local frame) | In-house | +3.05 | +1.82 | +2.50 | 3 | 1 | 1 |
GDT-HA: Global Distance Test High Accuracy, GDT-TS: Global Distance Test Total Score, lDDT: Local Distance Difference Test, Degradation: the number of refined models has quality worse than their initial models by a given threshold based on GDT-HA.
To select the best refined model, we have developed a model ranking tool by adapting it from our GNN model for refinement. Since our current refinement method does not use multiple sequence alignments (MSAs) as input, our GNN-based model ranking method (GNNQA) does not use MSAs either. As shown in Supplementary Table 18, GNNQA performs similarly as two recently developed quality assessment methods DeepAccNet13 and VoroCNN24 but outperforms a statistical potential RWplus25. Supplementary Table 19 shows the performance of GNNQA for GNNRefine-plus. Because we use GNNQA to rank the refined models, the final refined models are robust to the order of refinement, as shown in Supplementary Table 20.
Since GNNRefine uses only limited conformational sampling, a natural question to ask is how much GNNRefine may change the starting models and whether GNNRefine may improve significantly deviating regions in the starting models. As shown in Supplementary Table 21, on average our methods change structure as much as Feig’s but smaller than Baker’s. It should be noted that large structure change may lead to large quality degradation, especially when the initial models are of high quality. Supplementary Table 21 also lists the change of unreliable local regions (ULRs)26 proportion (i.e. the proportion of residues in ULRs) between the starting models and refined models. On the CASP13 targets, GNNRefine, GNNRefine-plus, FEIGLAB decrease the proportion by 0.71%, 1.14% and 2.19%, respectively, while BAKER increases the proportion by 1.28%. On the CASP14 targets, GNNRefine, GNNRefine-plus, FEIG decrease the average proportion by 0.19%, 0.72%, and 0.28%, respectively, while BAKER increases that by 5.87%. These results show that our methods can improve largely deviated regions although not too much.
Discussion
Our GNN-based method may estimate the distance probability distribution of existing edges more accurately, but cannot detect missing edges. So its performance may be impacted if there are many incorrect or missing edges in the graph representation of an initial protein model. Nevertheless, a refinement method usually focuses on protein models with reasonable quality, which are supposed to have a good percentage of correct graph edges.
Currently our method does not work well on the AlphaFold2 refinement models. To further deal with protein models of high quality, in addition to generating better training models, we are planning to improve our method by developing an end-to-end framework. We will revise our GNN method to take the MSA of a protein as input. The co-evolution information encoded in MSAs may help GNN to predict inter-residue interaction more accurately and thus, lead to better refinement. We will also study how useful the self-supervised learning of individual protein sequences and MSAs27,28 is. Currently the atom embedding did not help much. We will improve it by making use of the chemical contexts of atoms and generating training protein models with more accurate side chain atoms. Finally, we will use deep learning to directly predict 3D coordinates of (backbone and side-chain) atoms instead of inter-residue distance probability distribution. This will avoid using energy minimization methods to build 3D models and potentially improve model quality. We will also add more 3D protein models of higher quality into our training set so that deep learning may learn to refine a protein model of high quality.
Methods
Datasets
In-house training dataset.
It includes the CASP7-12 models and the models built by RaptorX for the ~29000 CATH domains. The CASP7-12 models are downloaded from http://predictioncenter.org/download_area/. There is only a small number (<600) of protein targets in CASP7-12. To increase the coverage, we select 28863 CATH domains (sequence identity <35%) released in March 201829, and build on average 13 template-based and template-free models for each domain using our in-house protein structure prediction software RaptorX. In total, there are 29455 proteins with 500255 models in this training set. About 5% of the proteins and their decoys are randomly selected to form the validation set and the remaining decoys are used to form the training set. We generate 3 different training and validation splits and accordingly train three different GNNRefine models.
DeepAccNet training dataset13.
It contains 7992 proteins (retrieved from the PISCES server30 and deposited to PDB by May 1, 2018) with 1104080 decoy models in total. Compared with our in-house training dataset, this dataset covers fewer protein targets (7992 v.s. 29455) but has many more decoy models for each target (~138 v.s. ~18). This set has a larger percentage of high-quality models. See Supplementary Fig. 5 for the model quality distribution of these two datasets. We generate two different training and validation splits on this dataset and then train two different GNN models.
Test data.
We use four test datasets to evaluate our method: the CASP13 refinement dataset, the CASP14 refinement dataset, the CAMEO dataset, and CASP13 FM dataset. The CASP13 refinement dataset includes 28 starting models in the CASP13 model refinement category5, excluding R0979 since it is an oligomeric target with three domains while our method is trained on individual domains. The CASP14 refinement dataset includes 37 starting models. The description of the CAMEO dataset and the CASP13 FM targets are available at the data availability statement. Note that all the training targets were released before May 1, 2018 and all the test targets were released after this date and thus, there is no overlap between our training and test datasets. The detailed information of our data is shown in the Supplementary Table 22.
Feature extraction and graph definition
From a protein model, we derive two types of features: residue feature and residue pair feature. The residue feature includes sequential and structural properties of a residue: 1) one-hot encoding of the residue (i.e., a binary vector of 21 entries indicating its amino acid type, including 20 standard amino acids plus 1 unknown or other amino acids); 2) the relative position of the residue in its sequence calculated as i/L (where i is the residue index and L is the sequence length); 3) dihedral angle (in radian), secondary structure (3-state), and relative solvent accessibility calculated by DSSP31; 4) one-hot encoding and relative coordinates of heavy atoms in the residue. The one-hot encoding is a four-dimensional vector representing four atom types (C, N, O and S) and the relative coordinate is a three-dimensional vector defined as: (x-xα, y-yα, z-zα), where S represents a sulphur atom, (x, y, z) is a heavy atom’s coordinate and (xα, yα, zα) is the Cα atom’s coordinate.
The residue pair feature is derived for a pair of residues with Euclidean distance less than 10Å, including: 1) spatial distances of three atom pairs (CαCα, CβCβ and NO) scaled by 0.1; 2) three types of inter-residue orientation (ω, θ dihedrals and φ angle) defined in trRosetta4; 3) the sequential separation of the two residues (i.e. the absolute difference between the two residue indices), which is discretized into 9 bins ([1, 2, 3, 4, 5, 6–10, 11–15, 16–20, >20]) and represented by one-hot encoding. All these features are summarized in Supplementary Table 23.
We represent an initial protein model as a graph, in which one node represents a residue and one edge represents a chemical bond or a contact between two residues. We say there is a contact between two residues if their Cβ Euclidean distance is no more than 10Å. It should be noted that this protein graph is equivariant (or symmetric) with respect to rotation and translation of atomic coordinates in the 3D space32.
GNNRefine architecture and training
Our GNN model contains an atom embedding layer, 10 message passing layers, and an output layer. The dimensions of the atom embedding, edge feature and node feature are all 256. As shown in Supplementary Fig. 6a, the atom embedding layer is used to extract the local structure information for each residue. Its input is the one-hot encoding of an amino acid and the relative coordinates of heavy atoms in the residue, and its output is the atom embedding with a fixed dimension. The atom embedding is concatenated with other residue features to form the input feature of one residue (i.e. node feature in the graph). Each message passing layer consists of a message block for edges and a reduce block for nodes. The message block for edges updates edge features and obtains edge attention values (Supplementary Fig. 6b) and the reduce block for nodes updates node features (Supplementary Fig. 6c).
For each edge, the inputs of its message block are the features of the two nodes connected by the edge and the edge feature itself. All these features go through an instance norm layer, a linear layer, and a LeakyReLU layer to generate an intermediate edge feature, which then goes through an LSTM cell to obtain the new edge feature. For each LSTM cell, its input is the intermediate edge feature, its hidden state is the output of its preceding LSTM cell, and its cell state is updated from its preceding LSTM cell (the cell state of the first LSTM cell is initialized to 0). The LSTM cell may help to capture the long-term dependency across layers, which enables us to build a deeper GNN33. The new edge feature also goes through a linear attention layer to obtain the attention value of the edge. For each node, the inputs of its reduce block are the reduced edge feature and the node feature. The reduced edge feature is a linear combination of all edge features weighted by their respective attention values. Similar to an edge block, the features go through an instance norm layer, a linear layer, and a ReLU layer to generate an intermediate node feature, and then the intermediate node feature together with the initial node feature and its preceding LSTM cell state pass through an LSTM cell to obtain the new node feature and new cell state.
The output layer uses a linear layer and a softmax function to estimate the distance probability distribution based on the edge feature. The distance probability distribution is a 37-dimensional vector with 36 bins representing the distances from 2 to 20 Å (0.5Å each) and one bin indicating the distance >20Å, as presented in trRosetta4. To evaluate the refined model quality, we train a GNN-based quality assessment model which uses the node feature to predict the global lDDT and residue-wise lDDT simultaneously.
To fit the deep model to a GPU with limited memory, when a protein has more than 400 residues, a sub-structure of 400 consecutive residues is randomly sampled. We implement GNNRefine with DGL34 for PyTorch35 and train it using the Adam optimizer with parameters: β1=0.9 and β2=0.999. We set the initial learning rate to 0.0001 and divide it by 2 every 5 epochs. One minibatch has 16 protein models. We use the cross-entropy loss to train GNNRefine at most 15 epochs and select the model with the minimum loss on the validation data as our final model. It takes ~3.5 days to train a model on our in-house training data and ~6.5 days on the DeepAccNet training dataset using one Tesla V100 or TITAN RTX GPU.
Building refined models
Building a refined full-atom model by FastRelax18 consists of the following steps: 1) use the initial model to initialize the pose in PyRosetta; 2) convert the predicted distance probability distribution into distance potential using the DFIRE36 reference state (similar to trRosetta4) and then add it onto the pose as spline restraints with relative weight 2; 3) conduct full-atom relaxation, side-chain packaging and gradient-based energy minimization with the built-in ref201537 scoring function. We have tried to add a backbone structure refinement step before running FastRelax, but it did not result in any backbone improvement. The GNN model training likely converges to a local minima. To deal with this, we train 5 different GNN models using five different training and validation data splits. Then we use the 5 GNN models to refine an initial model sequentially. That is, one GNN model is used to refine the protein model generated by the previous GNN model until all 5 GNN models are applied. In total we generate only 5 refined models from each initial model, which are then ranked by our GNN-based global model quality assessment (QA) method. See Supplementary Section 8 for evaluation of this QA method. We have tested another strategy (denoted as GNNRefine-plus) to see if we may improve refinement accuracy by generating 50 refined protein models in total. That is, for each GNN model we run FastRelax to generate 10 refined models and keep the lowest-energy protein model as the final refined model of this GNN model. Overall GNNRefine-plus generates 50 refined models but keeps only the 5 lowest-energy ones, which are then ranked by our GNN-based QA method. It turns out that GNNRefine-plus obtains slightly better performance than GNNRefine. We also find out that when 250 refined models are generated for one initial protein model, we may achieve even better refinement, as shown in Supplementary Table 17.
Extended Data
Extended Data Fig. 1. Quality improvement by different methods on the CASP13 refinement targets.
Boxplot of the distribution of ΔGDT-HA, ΔGDT-TS, and ΔlDDT on the CASP13 refinement targets. The five lines in each boxplot from top to bottom in turn mean: Maximum (Q3+1.5IQR), Third quartile (Q3, 75th percentile), Median (50th percentile), First quartile (Q1, 25th percentile), and Minimum (Q1−1.5IQR), where IQR is Q3–Q1. The precision is 2.
Extended Data Fig. 2. Quality improvement by different methods on the CASP14 refinement targets.
Box plot of the distribution of ΔGDT-HA, ΔGDT-TS, and ΔlDDT on the CASP14 refinement targets.The five lines in each boxplot from top to bottom in turn mean: Maximum (Q3+1.5IQR), Third quartile (Q3, 75th percentile), Median (50th percentile), First quartile (Q1, 25th percentile), and Minimum (Q1–1.5IQR), where IQR is Q3-Q1. The precision is 2.
Supplementary Material
Acknowledgements
The authors are grateful to Prof. David Baker’s team including Hahnbeom Park who provided us the DeepAccNet training data and helpful comments on our manuscript. The authors are also grateful to Dr. Lim Heo for explaining FEIG and FEIG-S to us. This work is supported by National Institutes of Health grant R01GM089753 to J.X. and National Science Foundation grant DBI1564955 to J.X. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Data Availability
Our in-house data is available at http://raptorx.uchicago.edu/download/. Click on this link and fill in your name, email address and organization name to obtain a data link, through which you may find a text file 0README.Data4GNNRefine.txt that specifies the names of the data files to be downloaded. The data is also available at Zenodo38. The DeepAccNet data is available at https://github.com/hiranumn/DeepAccNet. The CASP13 and CASP14 model s for refinement are available at https://predictioncenter.org/. The CAMEO models are available at https://www.cameo3d.org/modeling/. The CAMEO dataset includes 208 starting models for all the CAMEO hard targets released between May 1st 2018 and May 1st 2020. We keep only the targets with sequence length in [50, 500] and native structures containing at least 80% of sequence residues. Following CASPs we select the best -predicted models (in terms of GDT-HA) for each target as the starting models, and only keep the starting models with lDDT >50. For the CASP13 FM (free-modeling) dataset, there are 28 test targets corresponding to 32 official FM domains. For each target we build ~150 decoys as its starting models using our in -house template-free modeling software Raptor X-Contact. Source Data for Figures 2 and 3 and for Extended Data Figures 1 and 2 is available with this manuscript.
Code Availability
The source code is available at Code Ocean 39.
References
- 1.Wang S, Sun S, Li Z, Zhang R. & Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLOS Computational Biology 13, e1005324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J. Distance-based protein folding powered by deep learning. PNAS 116, 16856–16865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senior AW et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Yang J. et al. Improved protein structure prediction using predicted interresidue orientations. PNAS 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Read RJ, Sammito MD, Kryshtafovych A. & Croll TI Evaluation of model refinement in CASP13. Proteins: Structure, Function, and Bioinformatics 87, 1249–1262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heo L, Arbour CF & Feig M. Driven to near-experimental accuracy by refinement via molecular dynamics simulations. Proteins: Structure, Function, and Bioinformatics 87, 1263–1275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park H. et al. High-accuracy refinement using Rosetta in CASP13. Proteins: Structure, Function, and Bioinformatics 87, 1276–1282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu D. & Zhang Y. Improving the Physical Realism and Structural Accuracy of Protein Models by a Two-Step Atomic-Level Energy Minimization. Biophysical Journal 101, 2525–2534 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo L, Park H. & Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41, W384–W388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya D, Nowotny J, Cao R. & Cheng J. 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res 44, W406–W409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharya D. refineD: improved protein structure refinement using machine learning based restrained relaxation. Bioinformatics 35, 3320–3328 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Lee GR, Won J, Heo L. & Seok C. GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res 47, W451–W455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiranuma N. et al. Improved protein structure refinement guided by deep learning based accuracy estimation. Nature Communications 12, 1340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirjalili V, Noyes K. & Feig M. Physics-based protein structure refinement through multiple molecular dynamics trajectories and structure averaging. Proteins: Structure, Function, and Bioinformatics 82, 196–207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanyal S, Anishchenko I, Dagar A, Baker D. & Talukdar P. ProteinGCN: Protein model quality assessment using Graph Convolutional Networks. bioRxiv 2020.04.06.028266 (2020) doi: 10.1101/2020.04.06.028266. [DOI] [Google Scholar]
- 16.Baldassarre F, Hurtado DM, Elofsson A. & Azizpour H. GraphQA: Protein Model Quality Assessment using Graph Convolutional Networks. Bioinformatics btaa 714 (2020) doi: 10.1093/bioinformatics/btaa714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhury S, Lyskov S. & Gray JJ PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway P, Tyka MD, DiMaio F, Konerding DE & Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci 23, 47–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mariani V, Biasini M, Barbato A. & Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 29, 2722–2728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Critical assessment of techniques for protein structure prediction Thirteenth round - Abstract book. https://predictioncenter.org/casp13/doc/CASP13_Abstracts.pdf (2018).
- 21.Critical assessment of techniques for protein structure prediction Fourteenth round - Abstract book. https://predictioncenter.org/casp14/doc/CASP14_Abstracts.pdf (2020).
- 22.Heo L, Arbour CF, Janson G. & Feig M. Improved Sampling Strategies for Protein Model Refinement Based on Molecular Dynamics Simulation. J. Chem. Theory Comput. 17, 1931–1943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shuid AN, Kempster R. & McGuffin LJ ReFOLD: a server for the refinement of 3D protein models guided by accurate quality estimates. Nucleic Acids Research 45, W422–W428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igashov I, Olechnovič liment, Kadukova M, Venclovas Č & Grudinin S. VoroCNN: Deep convolutional neural network built on 3D Voronoi tessellation of protein structures. Bioinformatics (2021) doi: 10.1093/bioinformatics/btab118. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J. & Zhang Y. A Novel Side-Chain Orientation Dependent Potential Derived from Random-Walk Reference State for Protein Fold Selection and Structure Prediction. PLOS ONE 5, e15386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Won J, Baek M, Monastyrskyy B, Kryshtafovych A. & Seok C. Assessment of protein model structure accuracy estimation in CASP13: Challenges in the era of deep learning. Proteins: Structure, Function, and Bioinformatics 87, 1351–1360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rives A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. PNAS 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao R. et al. MSA Transformer. bioRxiv 2021.02.12.430858 (2021). [Google Scholar]
- 29.Dawson NL et al. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res 45, D289–D295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G. & Dunbrack RL PISCES: a protein sequence culling server. Bioinformatics 19, 1589–1591 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. & Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 32.Thomas N. et al. Tensor field networks: Rotation- and translation-equivariant neural networks for 3D point clouds. arXiv:1802.08219 [cs] (2018). [Google Scholar]
- 33.Huang B. & Carley KM Residual or Gate? Towards Deeper Graph Neural Networks for Inductive Graph Representation Learning. arXiv:1904.08035 [cs, stat] (2019). [Google Scholar]
- 34.Wang M. et al. Deep Graph Library: A Graph-Centric, Highly-Performant Package for Graph Neural Networks. arXiv:1909.01315 [cs, stat] (2020). [Google Scholar]
- 35.Paszke A. et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. in Advances in Neural Information Processing Systems 32 (eds. Wallach H. et al.) 8026–8037 (Curran Associates, Inc., 2019). [Google Scholar]
- 36.Zhou H. & Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Science 11, 2714–2726 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park H. et al. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J. Chem. Theory Comput. 12, 6201–6212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J. data for protein model refinement and model quality assessment. (2021) doi: 10.5281/zenodo.4635356. [DOI] [Google Scholar]
- 39.Jing X. GNNRefine: Fast and effective protein model refinement by deep graph neural networks. (2021) doi: 10.24433/CO.8813669.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.