Abstract
An outbreak of the coronavirus disease 2019 (COVID-19) caused by an infection of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) occurred in Wuhan, China, in December 2019. This new virus belongs to the group of enveloped RNA beta-coronaviruses. Symptoms may differ in various infected persons, but major presentations include dry cough, nasal congestion, shortness of breath, fever, and general malaise. The disease appears to be more severe in patients above the age of 60 years and those with underlying conditions such as diabetes, cancer, cardiovascular diseases, chronic respiratory disease, and hypertension. There is still no approved vaccine against COVID-19, but more than a hundred are at different stages of development. It is known that the development of new drugs takes a relatively long time, so several known and already-approved drugs are being repurposed for the treatment of this disease. In this review, we explore the therapeutic and vaccine options that are available for COVID-19 6 months after its outbreak. Most noteworthy among the therapeutic options are dexamethasone, remdesivir, Avigan (favipiravir) and convalescent plasma.
Keywords: COVID-19, SARS-CoV-2, therapeutics, vaccines, drug repurposing
摘要
2019年12月に中国・武漢で、新型重症急性呼吸器症候群コロナウイルス2(severe acute respiratory syndrome coronavirus 2:SARS-CoV-2)の感染症に起因する新型コロナウイルス(coronavirus disease 2019:COVID-19)のアウトブレイクが発生した。この新型ウイルスは、エンベロープ型RNAベータコロナウイルス属に分類される。症状は感染者ごとに異なるが、主として空咳、鼻閉、息切れ、発熱、全身倦怠などを呈する。この疾患は、患者が60歳を過ぎている場合や、糖尿病、癌、心血管疾患、慢性呼吸器病、高血圧などの基礎疾患を有する場合に重症度が高くなるとみられている。新型コロナウイルスに対するワクチンは依然としていずれも未承認であるが、現在100種を超えるワクチンがさまざまな開発段階にある。新薬の開発は相対的に長時間を要することが知られていることから、すでに承認されている既知の複数の薬剤がこの疾患の治療目的で利用されている。本稿では、このアウトブレイクから6ヵ月を経た現時点でCOVID-19に利用できる治療法とワクチンの選択肢を検討する。治療選択肢の中で最も注目に値するものは、デキサメタゾン、レムデシビル、アビガン(ファビピラビル)、および回復期血漿から2つを併用する方法である。
초록
새로운 중증급성호흡기증후군 코로나바이러스2(SARS-CoV-2)의 감염으로 인한 코로나바이러스감염증-19(COVID-19) 발병이 2019년 12월 중국 우한에서 발생하였다. 이 새로운 바이러스는 외피보유 RNA 베타 코로나바이러스 군에 속한다. 감염자마다 증상이 상이할 수 있으나, 주요한 증상 발현에는 마른기침, 코막힘, 호흡곤란, 발열, 전신무력감이 포함된다. 이 질병은 60세 이상의 환자와 당뇨, 암, 심혈관 질환, 만성 호흡기 질환, 고혈압과 같은 기저 질환자들에서 더 중증으로 나타난다. 승인된 COVID-19 백신은 아직 없으나, 100개 이상의 백신이 서로 다른 개발 단계에 있다. 신약 개발은 상대적으로 오랜 시간이 걸리므로, 이 질병의 치료를 위해 이미 알려지거나 승인된 여러 약물을 재창출하고 있다. 이 리뷰논문에서는 COVID-19의 발병 이후 6개월 동안 이용 가능한 치료제와 백신의 선택지에 대해 탐구한다. 가장 주목할 만한 치료제 옵션은 덱사메타손, 렘데시비르, 아비간(favipiravir), 회복기혈장의 이중 병용요법이다.
抄録
由新型严重急性呼吸系统综合症冠状病毒2(SARS-CoV-2)感染引起的2019年冠状病毒病(COVID-19)于2019年12月在中国武汉爆发。这种新病毒属于包膜β属的RNA冠状病毒。不同感染者的症状可能有所不同,但其主要症状包括干咳、鼻塞、呼吸急促、发烧和全身不适。该疾病似乎在60岁以上的患者以及患有诸如糖尿病、癌症、心血管疾病、慢性呼吸系统疾病和高血压等潜在疾病的患者中更为严重。目前尚无任何针对COVID-19的疫苗被批准使用,但也有一百多种疫苗正处于不同的开发阶段。众所周知,新药的开发需要相对较长的时间,因此几种已知且已被批准使用的药物被转用于治疗该疾病。在本文中,我们探讨了COVID-19爆发6个月后可被使用的治疗方法和疫苗。在各种治疗方法中,最值得一提的是使用地塞米松、瑞德西韦、法匹拉韦(favipiravir)和恢复期血浆。
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel human pathogenic virus; it belongs to the Coronaviridae family, whose members are named after their crown-like appearance caused by the surface glycoproteins that decorate the virus. This novel virus, also referred to as 2019-nCoV, is responsible for the coronavirus disease 2019 (COVID-19).1 , 2 Coronaviruses including 229E, NL63, OC43, and HKU1 are common human pathogens that cause common cold-like symptoms. Other known pathogenic coronaviruses for humans include SARS-CoV (which causes the severe acute respiratory syndrome) and MERS-CoV (which causes the Middle Eastern respiratory syndrome). Coronaviruses are large (28–32 kb), single-stranded, positive-sense RNA viruses.3 Within the phylogenetic subgroups of the family Coronaviridae, SARS-CoV-2 belongs to the beta-coronavirus, along with SARS-CoV and MERS-CoV.4 Coronaviruses have similar proteins that are involved in replication at the 5′ end, and structural proteins encoded at the 3′ end of the genome.3 The RNA script allows expression of the replicase, which is expressed as two polyproteins, pp1a and pp1ab. These can include up to 16 nonstructural proteins.5 The nonstructural proteins are generated by processing of pp1a and pp1ab by two or three viral proteases encoded within the replicase. There are several accessory proteins that seem to be important for pathogenesis, but all are not functionally characterized.
Structural Proteins
A matured SARS-CoV-2 consists of four structural proteins, spike (S), envelope (E), nucleocapsid (N), and membrane (M), all of which constitute the complete structural viral particle6 ( Fig. 1). Each of these proteins plays a primary role in the structure of the viral particle.
Figure 1.
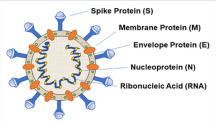
A schematic representation showing the structure of SARS-CoV-2.
The S protein mediates viral attachment to the host cell surface receptors and is responsible for the consequent fusion between the viral and host membranes to facilitate viral entry.6 , 7 The S protein has two subunits: S1, which contains the receptor-binding domains (RBDs) that facilitate virus–host binding, which then transitions to the S2 for virus–host fusion.1 , 8 Both SARS-CoV and SARS-CoV-2 recognize the human angiotensin-converting enzyme 2 (hACE2) as its host receptor binding to the S protein.9 A novelty of the SARS-CoV-2 S protein is the presence of a furin cleavage site at the S1/S2 boundary of the protein, which is then cleaved during biosynthesis. The functionality of this is still not fully understood. It should be emphasized that the indispensable function of the S glycoprotein makes it a key target for therapeutic antibodies, diagnostics, and tentative vaccines. The coronavirus N protein packages viral genomic RNA into a ribonucleoprotein complex and serves as an RNA chaperone.10 This protein localizes in the endoplasmic reticulum–Golgi region that is structurally bound to the nucleic acid material of the virus. The N protein is also involved in the host immunological response. It is also heavily phosphorylated, which may lead to structural changes enhancing the affinity for viral RNA.11 The M structural protein is the most abundant and plays a prominent role in determining the shape of the virus envelope.12 M is involved in facilitating interactions between all other structural proteins.13 The M protein stabilizes the N proteins and promotes the completion of viral assembly by stabilizing the N protein–RNA complex within the virion, promoting viral assembly.10 Finally, the E protein is the smallest structural protein and plays a role in the production and maturation of the virion.14 The majority of this protein is located within sites of intracellular trafficking to participate in coronavirus assembly and budding.15 The E protein is abundantly expressed inside infected cells, but minimal portions are incorporated within the envelope.14 Both the M and E proteins constitute the viral envelope of the coronavirus family.10 , 16
Key Nonstructural Proteins
There are 29 proteins known to be produced by SARS-CoV-2.17 Several of these proteins are critical nonstructural proteins that are valuable targets for antiviral drugs. Of these, the most druggable targets in this virus are several of its enzymes, some of which will be discussed below. The viral genome has 14 open reading frames, each of which encodes a variety of proteins. A viral replicase is used to translate most of the viral genomic RNA. From this, two polyproteins are synthesized (pp1a and pp1ab), which are further cleaved into nonstructural proteins.3 These two polyproteins are processed by two proteases, papain-like protease (PLpro) and 3 chymotrypsin-like protease (3CLpro), which are both essential for generating functional replication complexes. PLpro cleaves the N-terminal region of the polyprotein to generate three nonstructural proteins (1/2/3) and is thought to have deubiquitinating activity.18 , 19 3CLpro cleaves 11 different sites of the polyprotein to produce a mature protein that anchors replication and transcription complexes and releases mature nonstructural proteins. RNA-directed RNA polymerase (RdRp) is critical for host cell RNA replication in RNA viruses due to its functionality of catalyzing the template synthesis of polynucleotides. This protein was found to be critical for the infection cycle of all RNA viruses. Chien et al. demonstrated the requirement of RdRp activity for SARS-CoV pathogenesis by showing that without RdRp, there is a complete disruption of SARS-CoV both in vitro and in vivo, as indicated by stopping RNA replication and halting viral growth.20 These proteases have emerged as important drug targets because of their critical viral roles and low similarity with human proteases.21 , 22 2′-O-Methyltransferase (2′-O-MT) mediates mRNA cap 2′-O-ribose methylation of the 5′ cap of viral mRNAs, while 2′-O methylation is important for the host immune system to discern self-RNA from non-self-RNA. Viral helicase is essential for viral replication and therefore proliferation. Nonstructural uridylate-specific endoribonuclease (NendoU) is another nonstructural protein worth investigating as an antiviral target since its endoribonuclease is suspected to be similar in all coronaviruses.23 , 24 Angiotensin-converting enzyme 2 (ACE2) is an antigen receptor recognition enzyme that is located on the host cell surface. To gain entry into a target cell, the SARS-CoV S protein binds to the ACE2 receptor.25 hACE2 is present in a wide array of human tissues, including in the lung epithelia, kidney, testis, and small intestine.25 The S protein consists of three sections, an ectodomain, a single-pass transmembrane anchor, and a short intracellular tail.26 The ectodomain of the S protein consists of two subunits: S1 and S2. The S1 subunit contains an RBD residing on its C terminus that is involved in the ACE2-binding process.26 The SARS-CoV-2 S1 RBD has a substantially higher binding affinity to hACE2 in comparison with the SARS-CoV-1 RBD.27 Both SARS-CoV-1 and SARS-CoV-2 rely on proteolytic processing from cell surface transmembrane serine protease 2 (TMPRSS2) and lysosomal endopeptidase enzyme (cathepsin L) for the preactivation of the S protein. TMPRSS2 cleaves the S protein, allowing for transmission of the virus via the ACE2 receptor, while cathepsin L activates membrane fusion.9 , 28 , 29 There is strong evidence that SARS-CoV-2 has an additional novel preactivation mechanism through the proteasomal processing from proprotein convertase (PPC) furin.27 A study has revealed that furin-mediated preactivation of the SARS-CoV-2 S protein enhances its ability to enter target cells.27 This is significant as furin-mediated cleavage of the SARS-CoV-2 S protein allows SARS-CoV-2 to gain entry into cells with low expression of TMPRSS2 and/or cathepsin L.27
Drugs against SARS-CoV-2
There are many drugs that are either in development or in trial that target several of the SARS-CoV-2 structural and nonstructural candidates mentioned. Furthermore, there have been numerous reports of drug repurposing with the intent of finding already approved or nearly approved compounds that have efficient antiviral properties and are clinically safe. Repurposing is critical because it speeds up the amount of time for treatments to find their place in the clinical setting. Several compounds, such as remdesivir (RDV) and hydroxychloroquine (HQC), showed early promise, though opinions differ on HQC. Table 1 shows several candidates that either target SARS-CoV-2 directly or serve to reduce COVID-19 pathology by targeting human receptors.
Table 1.
Drugs That Have Been Either Repurposed or Synthesized to Show Antiviral Activity against SARS-CoV-2.
| Drug Name | 2D Structures | Target | Mechanism | Novel (NV) or Repurposed (RP) | References |
|---|---|---|---|---|---|
| Arbidol | 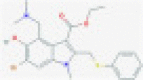 |
S glycoprotein and hACE2 | Blocks viral entry | RP | Vankadari73 |
| Aurine tricarboxylic acid | 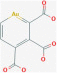 |
RdRp | Blocks viral replication | RP | Morse et al.74 |
| Benzopurpurin B |
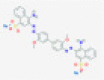 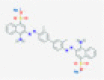
|
Endoribonuclease NSP15 | Causes viral RNA degradation | RP | Ortiz-Alcantara et al.5 |
| Baricitinib | 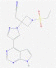 |
JAK kinase | Suppression of proinflammatory cytokines typically observed in people with COVID-19 | RP | Richardson et al.75 |
| Camostat mesylate | 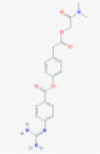 |
TMPRSS2 | Blocks nucleocapsid entry from phagosome to cytoplasm | RP | Hoffmann et al.9 |
| Chloroquine | 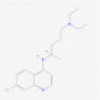 |
Endosome/ACE2 | Interferes with S protein processing by lysosomal enzymes as well as viral envelop assembly | RP | Vincent et al.37 |
| Colchicine | 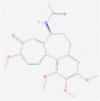 |
Host tubulin | Suppression of proinflammatory cytokines typically observed in people with COVID-19 | RP | Finkelstein et al.76 |
| Remdesivir | 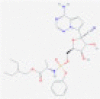 |
RdRp | Blocks viral replication | RP | Agostini et al.30 |
| Ribavirin | 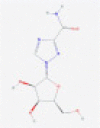 |
RdRp | Blocks viral replication | RP | Morse et al.74 |
| Favipiravir (Avigan) |  |
RdRp | Blocks viral replication | RP | Guo77 |
| Galidesivir | 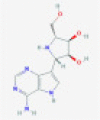 |
RdRp | Blocks viral replication | RP | Warren et al.78 |
| Gilenya (fingolimod) | 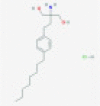 |
Host sphingosine 1-phosphate receptor | Anti-inflamatory | RP | Torjesen38 |
| Lopinavir | 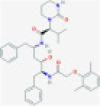 |
3CLpro and PLprop | Blocks viral replication by inhibiting polyprotein processing | RP | Sheahan et al.4 |
| Darunavir | 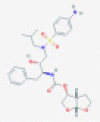 |
3CLpro and/or PLpro | Blocks viral replication by inhibiting polyprotein processing | RP | Liu et al.79 |
| Hirsutenone | 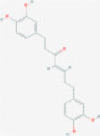 |
3CLpro and/or PLpro | Blocks viral replication by inhibiting polyprotein processing | RP | Kumar et al.,80 Zhou et al.81 |
| Rupintrivir | 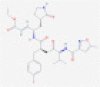 |
3CLpro and/or PLpro | Blocks viral replication by inhibiting polyprotein processing | RP | Anand et al.21 |
| Nitazoxanide | 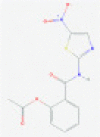 |
Unknown | Slows replication; unknown target | RP | Guo77 |
| NSC-306711 |  |
Endoribonuclease NSP15 | Viral genomic RNA degradation by host cellular innate immunity blocking | RP | Ortiz-Alcantara et al.5 |
| C-473872 | 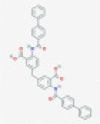 |
Endoribonuclease NSP15 | Viral genomic RNA degradation by host cellular innate immunity blocking | RP | Ortiz-Alcantara et al.5 |
| C-467929 | 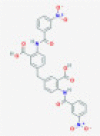 |
Endoribonuclease NSP15 | Viral genomic RNA degradation by host cellular innate immunity blocking | RP | Ortiz-Alcantara et al.5 |
Of the several compounds tested as potential COVID-19 treatments, few have come out as front-runners, most notably, RDV, HQC, and lopinavir (LPV)/ritonavir (RTV). RDV is a broad-spectrum antiviral prodrug that is metabolized into its active form, GS-441524. This compound has shown antiviral properties against several viruses, including Ebola and MERS-CoV.30 RDV does exhibit in vitro antiviral activity against SARS-CoV-2, warranting its use as a potential treatment for COVID-19.31 More recently, RDV was shown to be efficacious in shortening the recovery time of hospitalized COVID-19 patients in a study spanning several countries.32 RDV is a competitive inhibitor of RdRP, competing with adenosine triphosphate.33 The RDV prodrug undergoes several metabolic steps within the cell in the formation of the active RDV–triphosphate (GS-441524) compound.33 Recently, the claims of RDV’s efficacy as a mortality-reducing drug by a small U.S. trial32 were unfounded during a large trial by the World Health Organization (WHO)34 where RDV had little effect on mortality reductions. HCQ is an aminoquinoline that is commonly used as an antimalarial agent, but it has also been used against lupus and rheumatoid arthritis.35 It is an analog of chloroquine (CQ) in which one of the N-ethyl substituents of CQ is β-hydroxylated. The activity of HCQ against malaria is equivalent to that of CQ, and HCQ is preferred over CQ when high doses are required because of the lower level of ocular toxicity of HCQ.36 HCQ has had promising in vitro data against SARS-CoV-2, suggesting its use as a possible COVID-19 treatment. Mechanistically, HCQ can pass through the host cell’s membrane and aggregates within the host’s intracellular compartments, including lysosomes and other vesicles. This accumulation results in the increase of the vesicular pH, which consequently does not allow the virus to release the vesicle into the cytoplasm, thus resulting in a minimized viral load within the host cell.35 Also, HCQ might be involved in disruption of various enzymatic functions, including glycosylation of newly synthesized proteins.37 Several other mechanisms have been proposed for how HCQ interacts with SARS-CoV-2; however, HCQ is no longer considered a therapeutic option as it does not reduce mortality.38 LPV, commonly administered for treatment of HIV-1, is a retroviral protease inhibitor.39 LPV is often combined with RTV, which acts as an inhibitor against cytochrome P450 metabolism of LPV.39 LPV-RTV has also shown inhibitory activity against SARS-CoV-1 cysteine proteases, making it a drug of interest in combating SARS-CoV-2.40 In regard to the inhibition of the SARS-CoV-2 3CLpro, it has been suggested that both LPV and RTV inhibit 3CLpro activity.40
On June 16, 2020, a breakthrough repurposed drug was reported for critically ill hospitalized COVID-19 patients such as those on ventilators. The steroid, dexamethasone, was found to reduce deaths due to COVID-19 by one-third in a controlled clinical trial conducted in the UK.41 So far, it is not advisable to use dexamethasone for patients who are not critically ill since the steroid showed no effect.41
The most recent promising drug alternative emanated from a study by Bouhaddou et al.,42 who mapped the phosphorylation landscape of SARS-CoV-2 infection in Vero E6 cells and utilized the results to identify several drugs for treatment of the infection. Silmitasertib, gilteritinib, apilimod, dinaciclib, ARRY-797, and ralimetinib were among the compounds tested, and some of them were able to decimate 50% of coronavirus at a lower concentration than RDV.42 While some of these compounds are already at different stages of clinical trials for other applications, it remains to be seen in the near future if they will perform equally well in human patients as they did in cells.
Antibody Treatments
Antibodies have immense potential as treatments for COVID-19. This is because antibodies can neutralize the viral particle or target inflammatory factors, such as cytokines. Research groups at universities and companies have used antibodies from recovered COVID-19 patients to treat new patients. Many of these are at the clinical stage, while others are still in preclinical phases. The most popular of these was the use of convalescent plasma developed early in the course of the COVID-19 pandemic.43 Numerous case series and observational studies have since been published with variable results.44, 45, 46 Two published randomized controlled trials were halted early, one due to concern of a lack of benefit and the other due to low enrollment.47 , 48 To date, one randomized controlled trial has been completed but did not meet its composite endpoint of progression to severe disease or all-cause mortality at 28 days; the median time from symptom onset to administration was 8 days, and the median antibody titer was 1:40. The results of additional randomized controlled trials are forthcoming. Questions remain about the antibody titer that should be used when treating patients with COVID-19, and if timing of administration is an important consideration. In August 2020, the Food and Drug Administration (FDA) announced an Emergency Use Authorization (EUA) for convalescent plasma in patients with COVID-19. On September 23, 2020, the FDA issued an update on convalescent plasma therapy for COVID-19. The update included an analysis that supported the concept of an antibody dose–response effect; the FDA concluded convalescent plasma “may be effective.” Another strategy is through using lab-synthesized antibodies that can neutralize the virus. Most often, antibodies are synthesized through genetically modified mice that are able to express various antibodies. Our laboratory uses a ribosome display method to synthesize antibodies against various pathogens, including the Zika virus, Ebola, and Marburg virus,49 , 50 and most recently has focused on SARS-CoV-2. Antibody therapeutics are of critical importance in the fight against this pandemic, and thus, in the coming months, we expect rapid progress to be made through available synthesis methodologies.
Camelid-derived single-domain antibody fragments, also called VHHs or nanobodies (nAbs), offer several advantages over conventional antibodies as candidates for specific therapies. Despite being approximately one-tenth of the size of a conventional antibody, they retain specificity and affinity similar to conventional antibodies, while being far easier to clone, express, and manipulate. They are readily expressed in bacteria in large quantities and show high thermal stability and solubility, making them easily scalable and cost-effective. Their modularity means that they can be oligomerized to increase avidity or to increase serum half-life.51 Critical to their use as antivirals in humans, they can easily be humanized with existing protocols.52 Importantly, they have proven to be highly potent inhibitors of viral infections in vivo, particularly respiratory infections.53 , 54
nAbs may be an alternative source of treatment against COVID-19, and various avenues for antibody treatment ( Table 2) are currently being explored, with a surge in research findings. Unfortunately, the poor cross-neutralizing efficacy of SARS-CoV-derived antibodies against SARS-CoV-2 has required additional input to generate new antibodies and improve existing ones. Thus, the shift in attention toward producing SARS-CoV-2-specific antibodies that have demonstrated higher neutralizing potential is timely and imperative. Antibodies such as REGN-COV, BD-23, CB6-LALA, SARSVHH-72, S309, 47D11, 311 mAb-31B5, and 311 mAb-32D4 appear to be particularly promising for combating the COVID-19 pandemic in view of their potent in vitro neutralizing activities and/or in vivo protection efficacies in animal models55 (Table 2). Current structural and sequence comparison-based analyses have attempted to summarize the various possible mechanistic reasons why most SARS-CoV-2 and SARS-CoV-derived antibodies do not cross-react and/or cross-neutralize.55 Gavor et al.55 have offered some insights into what types of antibodies might cross-react and cross-neutralize SARS-CoV-2 and SARS-CoV, and these should be further addressed experimentally. Gavor et al.55 have also provided a perspective on the impact of Asp614Gly and other mutations on the neutralizing effect of current antibodies.
Table 2.
Different Antibodies, Their Intended Targets, and Stages of Clinical Development.
| Molecule/Description | Target | Neutralizing Mechanism | Stage | References |
|---|---|---|---|---|
| LY-CoV555 | SARS-2 S | Blocks viral attachment and entry into human cells | Phase 2 trials | NCT04427501 |
| REGN10987 and REGN10933 humanized and human mAb cocktail | REGN10933: RBM of SARS-2 REGN10987: RBD/S1BCD |
Block hACE2-RBD binding ADCC and ADCP | Clinical | Hansen et al.,82 Baum et al.83 |
| S309 human mAb | SARS and SARS-2 RBD/S1BCD | Targets a conserved glycan-containing epitope within S protein and shows Fc-dependent effector mechanisms | Clinical | Pinto et al.56 |
| 4A8, 1M-1D2, and 0304-3H3 human mAbs | 4A8: NTD of S1 1M-1D2: S1 domain 0304-3H3: S2 domain |
Likely restrain conformational change in S protein | Preclinical | Chi et al.84 |
| 47D11 human mAb | SARS-2 and SARS RBD/S1BCD | Binds to the conserved epitope of RBD without compromising spike–receptor interaction | Phase 1 trials expected | Wang et al.85 |
| CR3022 human mAb | SARS and SARS-2 RBD/S1BCD | Destabilizes and destroys the prefusion S trimer | Preclinical | Lan et al.,86 Tian et al.,87 Ter Meulen et al.88 |
| S230 human mAb | SARS-RBD/RBM | Blocks hACE2-RBD binding | Preclinical | Walls et al.89 |
| SARS-VHH-72 (HCAb) llama (camelid) mAb | SARS, SARS-2, and bat WIVI CoV RBD/S1BCD | Blocks hACE2-RBD binding Destabilizes the prefusion spike |
Preclinical | Wrapp et al.90 |
| ADI-55689 and ADI-56046 human mAbs | SARS, SARS-2, and bat WIV1 RBD/RBM/S1BCD | Block hACE2-RBD binding and induce S1 shedding | Preclinical | Wec et al.91 |
| P2C-1A3 and P2C-1C10 human mAbs | SARS-2 RBD | Block hACE2-RBD binding | Preclinical | Ju et al.92 |
| P2A-1A10 and P2A-1B3 human mAbs | SARS-2 RBD | Block hACE2-RBD binding | Preclinical | Ju et al.92 |
| P2C-1F11 and P2B-2F6 human mAbs | SARS-2 RBD/RBM | Block hACE2-RBD binding | Preclinical | Ju et al.92 |
| 311mab-31B5 and 311mab-32D4 human mAbs | SARS-2 RBD/RBM | Block hACE2-RBD binding | Preclinical | Chen et al.93 |
| B38 and H4 human mAbs | SARS-2 RBD/RBM/S1BCD, although at different sites | Block hACE2-RBD binding | Preclinical | Wu et al.59 |
| CA1 and CB6 human mAbs | SARS-2 RBD/RBM | Block hACE2-RBD binding | Preclinical | Shi et al.94 |
| BD-368-2, BD-218, and BD-23 human mAbs | SARS-2 RBD | Block hACE2-RBD binding | Preclinical | Cao et al.95 |
| EY6A mouse mAb | Both SARS and SARS-2 RBD/S1BCD | Might engage multiple mechanisms | Preclinical | Tian et al.,87 Zhou et al.96 |
| COV21 human Ab | SARS and SARS-2 | Blocks hACE2-RBD | Preclinical | Robbiani et al.,58 Lan et al.,86 Tian et al.,87 Walls et al.,89 Barnes et al.97 |
| C121, C135, C144, and C105 human mAbs | SARS-2 RBD | Block hACE2-RBD binding | Preclinical | Robbiani et al.,58 Lan et al.,86 Tian et al.,87 Walls et al.,89 Barnes et al.97 |
| COV2-2196, COV2-2130, COV2-2196, and COV2-2381 mAbs | SARS-2 RBD/RBM | Block hACE2-RBD binding | Preclinical | Zost et al.98 |
| 2-15, 2-7, 1-57, 1-20, and 2-4 human mAbs | SARS-2 RBD | Block hACE2-RBD binding | Preclinical | Liu et al.99 |
| H014 humanized mAb | SARS and SARS-2 RBD/S1BCD | Blocks hACE2-RBD binding | Preclinical | Lv et al.100 |
| CC12.1 and CC6.33 human mAbs | SARS-2 RBD/RBM and SARS RBD | Block hACE2-RBD binding | Preclinical | Hansen et al.,82 Rogers et al.,101 Lei et al.,102 Yuan et al.103 |
| 5C2 human scFv-Fc | SARS-2 S | Inhibits ACE2 from binding to S protein | Preclinical | Yuan et al.104 |
| n3088 and n3130 human nAbs | SARS-2 RBD | Target a cryptic epitope situated in RBD | Preclinical | Wu et al.105 |
| CV1/CV35 human mAb | SARS-2 RBD | Binds to an epitope distinct from the RBD | Preclinical | Seydoux et al.106 |
| CV30 human mAb | SARS-2 S | Inhibits the S-ACE2 interaction | Preclinical | Seydoux et al.106 |
| 31B5, 32D4, COVA1-18, and COVA2-15 human mAbs | SARS-2 RBD | Perturb the ACE2-RBD interaction | Preclinical | Chen et al.,93 Brouwer et al.107 |
| P2B-2F6 human mAb | SARS-2 RBD | Competes with ACE2 for binding to the RBD | Preclinical | Ju et al.92 |
| CB6 human mAb | SARS-2 RBD | Is overlapped with the binding epitopes of ACE2 | Preclinical | Ju et al.92 |
| H2 human mAb | SARS-2 RBD | Binds to the RBD but does not compete with ACE2 for RBD binding | Preclinical | Wu et al.59 |
| B5 human mAb | SARS-2 RBD | Binds to the RBD but displays partial competition with ACE2 | Preclinical | Wu et al.59 |
| B38 and H4 human mAb | SARS-2 RBD | Show complete competition with ACE2 for binding to RBD | Preclinical | Wu et al.59 |
| JS016 human mAb | SARS-2 RBD | Blocks SARS-CoV-2 RBD binding to ACE2 | Phase 1 clinical | Shi et al.94 |
| 414-1 and 553-15 human mAbs | RBD and S ectodomain of SARS-2 | Block hACE2-RBD binding | Preclinical | Wan et al.57 |
ADCC, antibody-dependent cell cytotoxicity; ADCP, antibody-dependent cellular phagocytosis.
Gavor et al.55 have considered a platform to easily identify and choose antibodies that might be tested in a cocktail against COVID-19 to overcome escape mutant strains. For example, promising cocktails might include REGN-COV, 414-1 + 553-15, COV2-2196 + COV2-2130, CR3022 + CR3014, or B38 + H4. The prospect of combining monoclonal antibodies (mAbs) 553-15 and S309 with other antibodies in a cocktail is particularly attractive because these mAbs demonstrate a potent synergistic neutralizing effect with many of the other antibodies.56 , 57 Moreover, mAb CR3022 might be combined with mAb COV21, C105, or B38 in a cocktail because CR3022 does not appear to compete with these three antibodies for binding to the SARS-CoV-2 S glycoprotein, and therefore might offer synergistic neutralizing effects.58 , 59 Similarly, the potent NTD-binding nAb 4A8 might also be considered in a cocktail with RBD-binding antibodies because 4A8 binding to the NTD leaves the RBD region of the S glycoprotein free for co-binding antibodies that might offer additive neutralizing effects. Of note, in addition to cocktail antibody therapies, a cocktail with other antiviral drugs such as RDV might be therapeutically explored against COVID-19. Moving forward, because antibody-dependent enhancement (ADE) of COVID-19 cannot be reliably predicted after vaccination or antibody treatment, careful analysis of safety will need to be conducted in humans.
A lot of antibodies have also been repurposed for use against COVID-19. Many of these antibodies do not have mechanisms of action relevant to SARS-CoV-2, but rather to COVID-19 pathology. Researchers have shown that the infection of SARS-CoV-2 activates CD4+ T lymphocytes, which consequently become pathogenic T-helper cells generating various cytokines.60 This then leads to high expression of interleukins (ILs) like IL-6 that accelerate inflammation. A report has shown that IL-6 levels in COVID-19 patients were significantly elevated,61 suggesting that antibodies targeting the IL-6 receptor may reduce COVID-19 pathology. Actemra is one of the repurposed antibodies against COVID-19. Actemra was first approved in Japan in 2005 as a treatment for rheumatoid arthritis and cytokine release syndrome.62 As severe cytokine release is part of COVID-19 pathology, Actemra may help reduce symptomatic expression of the disease. Kevzara is another antibody repurposed to treat COVID-19. It was first approved in the United States in 2017 for the treatment of rheumatoid arthritis.63 Several Kevzara clinical trials are ongoing or are expected to start in the near future (i.e., NCT04315298, NCT04321993, and NCT04324073).
Vaccines against SARS-CoV-2
A vaccine is of utmost importance to fully defeat the COVID-19 pandemic. Presently, there are more than 120 different vaccines being developed. There are different vaccine platforms that are currently being tested, some of which have not been tested in clinical settings before. Virus-like particle (VLP) vaccines consist of manipulated viral shells that mimic the viral structure but are not infectious because they lack the natural genetic material. They are used to prime the immune system by eliciting a strong immune response. Protein subunit-based vaccines consist of several viral proteins with an adjuvant that are expected to elicit an immune response, though several doses may be required. DNA- and RNA-based vaccines use modified nucleic acid scripts that encode a SARS-CoV-2 protein. Several of these vaccines encode and produce several copies of the SARS-CoV-2 S protein. There are also many vaccines that are viral vector based, including replicating viral vector and nonreplicating viral vector vaccine platforms. Replicating viral vector vaccines utilize weakened pathogens such as measles and horsepox, which can encode and express various structural proteins of the SARS-CoV-2 virus through viral replication. This methodology can provoke a strong immune response, but existing immunity to the viral vector can subdue the vaccine’s efficacy. Nonreplicating vector vaccines are like the latter but utilize different vectors, such as adenoviruses, which do not induce a great immune response. Although no licensed vaccines use this methodology, adenoviruses have been widely used in gene therapy. Vaccines will be necessary both for individual protection and for the safe development of population-level herd immunity.
Public–private partnership collaborative efforts, such as the Accelerating COVID-19 Therapeutic Interventions and Vaccines mechanism, are key to rapidly identifying safe and effective vaccine candidates as quickly and efficiently as possible. Table 3 shows several vaccines that are in preclinical or various phases of clinical trials. There are several more vaccines that are being developed that are in the preclinical phase. Several of these vaccines use methodologies that have never been used in a viral candidate, so there is uncertainty with regard to their utilization in the clinical sphere.
Table 3.
Vaccine Candidates against SARS-CoV-2 and Clinical Phases.
| Vaccine Type | Vaccine | Developer | Clinical Stage | Number of Doses | Timing of Doses | Route of Administration |
Reported Results of Clinical Trials | References/Trial Registration Nos. |
|---|---|---|---|---|---|---|---|---|
| Inactivated vaccines | Inactivated | Institute of Medical Biology, Chinese Academy of Medical Sciences | Phase 1/2 | 2 | 0, 28 days | IM | Phase 1 data suggest the vaccine is safe and triggers an immune response, although a drop in neutralizing antibody titers from day 14 to day 28 is a potential cause for concern. | NCT04470609 |
| Inactivated | Wuhan Institute of Biological Products/Sinopharm | Phase 3 | 2 | 0, 14 or 0, 21 days | IM | A phase 2 trial showed that the geometric mean titres of nAbs were 121 and 247 at day 14 after two injections in participants receiving vaccine on days 0 and 14 and on days 0 and 21, respectively. Moreover, 7-day adverse reactions occurred in 6.0% and 19.0% of the participants receiving injections on days 0 and 14 vs on days 0 and 21. | Xia et al.108 | |
| Inactivated | Research Institute for Biological Safety Problems, Republic of Kazakhstan | Phase 1/2 | 2 | 0, 21 days | IM | The proportion of volunteers with increased levels of the immune response of specific neutralizing antibody titers in ELISA following the vaccination, compared with a placebo. | NCT04530357 | |
| Inactivated SARS-CoV-2 vaccine with aluminum hydroxide | Sinovac | Phase 3 | 2 | 0, 14 days | IM | A phase 2 trial showed that two doses of 6 μg/0.5 mL or 3 μg/0.5 mL of the vaccine were well tolerated and immunogenic in healthy adults, with the 3 μg dose eliciting 92.4% seroconversion under the day 0, 14 schedule and 97.4% under the day 0, 28 schedule. | Zhang et al.109 | |
| Inactivated | Beijing Institute of Biological Products/Sinopharm | Phase 3 | 2 | 0, 14 or 0, 21 days | IM | A phase 2 trial showed that the vaccine at a dose of 5 × 1010 viral particles per mL was safer than the vaccine at 1 × 1011 viral particles and elicited a comparable immune response. However, high preexisting Ad5 immunity reduced the nAb response and influenced the T-cell immune response. | ChiCTR2000031809 | |
| Whole-virion inactivated (BBV152A) | Bharat Biotech | Phase 2 | 2 | 0, 14 days | IM | N/A |
NCT04471519, CTRI/2020/07/026300 |
|
| RNA vaccines | mRNA | Curevac | Phase 2 | 2 | 0, 28 days | IM | N/A | NCT04449276, NCT04515147 |
| mRNA-1273 | Moderna/NIAID | Phase 3 | 2 | 0, 28 days | IM | A phase 1 study reported that the two-dose vaccine series was not seriously toxic, and it could elicit nAbs and Th1-biased CD4+ T-cell responses. | Jackson et al.64 | |
| mRNA | Arcturus/Duke-NUS | Phase 1/2 | N/A | N/A | IM | Phase 1/2 preclinical data have shown highly promising results with 100% seroconversion for neutralizing antibodies after a single administration using a very low 2 µg dose. Neutralizing antibodies continued to increase for 60 days after dosing. Preclinical results also demonstrated robust CD8+ T-cell induction and a Th1-biased T-helper cellular immune response. | NCT04480957 | |
| LNP-nCoVsaRNA | Imperial College London | Phase 1 | 2 | N/A | IM | N/A | SRCTN17072692 | |
| mRNA | People’s Liberation Army Academy of Military Sciences/Walvax Biotech | Phase 1 | 2 | 0, 14 or 0, 28 days | IM | N/A | ChiCTR2000034112 | |
| BNT162b1 (mRNA expressing a trimeric RBD) and BNT162b2 (mRNA expressing S protein) |
Pfizer/Fosun Pharma/BioNTech | Phase 3 | 2 | 0, 28 days | IM | A phase 1/2 study showed that the vaccine caused mild to moderate local and systematic symptoms in most vaccinators and geometric mean neutralizing titers after the 10 and 30 µg dose reached 1.8- to 2.8-fold that of the COVID-19 convalescent sera panel. Positive phase 3 study interim results showed >90% efficacy in preventing COVID-19 across study participants. |
Mulligan et al.110 | |
| DNA vaccines | DNA plasmid vaccine with electroporation | Inovio Pharmaceuticals/International Vaccine Institute | Phase 1/2 | 2 | 0, 28 days | ID | N/A |
NCT04447781, NCT04336410 |
| DNA plasmid vaccine + adjuvant | Osaka University/AnGes/Takara Bio | Phase 1/2 | 2 | 0, 14 days | IM | N/A |
NCT04463472, NCT04527081 |
|
| DNA plasmid vaccine | Cadila Healthcare Limited | Phase 1/2 | 3 | 0, 28, 56 days | ID | CTRI/2020/07/026352 | ||
| DNA Vaccine (GX-19) | Genexine Consortium | Phase 1/2 | 2 | 0, 28 days | IM | N/A | NCT04445389 | |
| Nonreplicating viral vector | Replication defective simian adenovirus (GRAd) encoding S | ReiThera/LEUKOCARE/Univercells | Phase 1 | 1 | N/A | IM | N/A | NCT04528641 |
| ChAdOx1 nCoV-19 | University of Oxford/AstraZeneca | Phase 3 | 1 | N/A | IM | A phase 1/2 trial reported that nAb responses were detected in 91% of participants after a single dose when measured in MNA80 and in 100% participants when measured in PRNT50. After a booster dose, all participants had neutralizing activity. Local and systemic reactions, including pain, fever, and muscle ache, could be reduced by paracetamol. | Folegatti et al.111 | |
| Adenovirus type 5 vector | CanSino Biological Inc./Beijing Institute of Biotechnology | Phase 3 | 1 | N/A | IM/mucosal | A phase 2 trial showed that the vaccine at a dose of 5 × 1010 viral particles per mL was safer than the vaccine at 1 × 10¹¹ viral particles and elicited a comparable immune response. However, high preexisting Ad5 immunity reduced the nAb response and influenced a T-cell immune response. | Zhu et al.67 | |
| Adeno based (rAd26-S + rAd5-S) (Sputnik V) | Gamaleya Research Institute | Phase 3 | 2 | 0, 21 days | IM | A phase 1/2 trial showed that administration of both rAd26-S and rAd5-S caused the production of nAbs in 100% of participants on day 42 for both the lyophilized and frozen vaccine formulations. Cellular immune responses were detected in all participants at day 28. Moreover, the preexisting immune response to the vectors rAd26 and rAd5 did not influence the titer of RBD-specific antibodies. | Logunov et al.68 | |
| Ad26COVS1 | Janssen Pharmaceutical Companies | Phase 3 | 2 | 0, 56 days | IM | Preclinical trials showed that a single immunization with an Ad26 vector encoding a prefusion stabilized S antigen triggered robust nAb responses and provided complete or near-complete protection in rhesus macaques. The immunogen contains the wild-type leader sequence, the full-length membrane-bound S, mutation of the furin cleavage site, and two proline stabilizing mutations. | Mercado et al.112 | |
| Ad5 adjuvanted oral vaccine platform |
Vaxart | Phase 1 | 2 | 0, 28 days Oral | Oral | N/A | NCT04563702 | |
| Replicating viral vector | Measles-vector based | Institute Pasteur/Themis/University of Pittsburgh CVR/Merck Sharp & Dohme | Phase 1 | 1 or 2 | 0, 28 days | IM | N/A | NCT04497298 |
| Intranasal flu-based RBD | Beijing Wantai Biological Pharmacy/Xiamen University | Phase 1 | 1 | N/A | IM | N/A | ChiCTR2000037782 | |
| Protein subunit | Full-length recombinant SARS-CoV-2 glycoprotein nanoparticle vaccine adjuvanted with Matrix M | Novavax | Phase 2/3 | 2 | 0, 21 days | IM | N/A | NCT04533399, 2020-004123-16 |
| Adjuvanted recombinant protein (RBD–dimer) | Anhui Zhifei Longcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences | Phase 2 | 2 or 3 | 0, 28 or 0, 28, 56 days | IM | N/A | NCT04550351, NCT04466085 | |
| RBD based | Kentucky Bioprocessing, Inc. | Phase 1/2 | 2 | 0, 21 days | IM | N/A | NCT04473690 | |
| S protein (baculovirus production) | Sanofi Pasteur/GSK | Phase 1/2 | 2 | 0, 21 days | IM | N/A | NCT04537208 | |
| Recombinant trimeric subunit S protein vaccine | Clover Biopharmaceuticals Inc./GSK/Dynavax | Phase 1 | 2 | 0, 21 days | IM | N/A | NCT04405908 | |
| Recombinant S protein with Advax adjuvant | Vaxine Pty. Ltd./Medytox | Phase 1 | 1 | N/A | IM | N/A | NCT04453852 | |
| Molecular clamp stabilized S protein with MF59 adjuvant | University of Queensland/CSL/Seqirus | Phase 1 | 2 | 0, 28 days | IM | N/A | ACTRN12620000674932p, ISRCTN51232965 |
|
| S-2P protein + CpG 1018 | Medigen Vaccine Biologics Corporation/NIAID/Dynavax | Phase 1 | 2 | 0, 28 days | IM | N/A | NCT04487210 | |
| RBD + adjuvant | Instituto Finlay de Vacunas, Cuba | Phase 1 | 2 | 0, 28 days | IM | N/A | IFV/COR/04 | |
| Peptide | FBRI SRC VB VECTOR, Rospotrebnadzor, Koltsovo | Phase 1 | 2 | 0, 21 days | IM | N/A | NCT04527575 | |
| RBD (baculovirus production expressed in Sf9 cells) | West China Hospital, Sichuan University | Phase 1 | 2 | 0, 28 days | N/A | ChiCTR2000037518 | ||
| SARS-CoV-2 HLA-DR peptides | University Hospital Tübingen | Phase 1 | 1 | N/A | SC | N/A | NCT04546841 | |
| S1-RBD-protein | COVAXX | Phase 1 | 2 | 0, 28 days | IM | N/A | NCT04545749 | |
| VLP | Plant-derived VLP adjuvanted with GSK or Dynavax adjuvants | Medicago Inc. | Phase 1 | 2 | 0, 21 days | IM | N/A | NCT04450004 |
ELISA, enzyme-linked immunosorbent assay; IM, intramuscular; N/A, not applicable/not available.
The vaccine mRNA-1273 was developed by the National Institute of Allergy and Infectious Diseases (NIAID) and the company Moderna. This vaccine uses messenger RNA to express SARS-CoV-2 proteins.64 This was the first vaccine to be tested in clinical trials in the United States. The first participant was administered this investigational vaccine on March 16, 2020. The ChadOx1 nCoV-19 vaccine candidate was developed at the University of Oxford Jenner Institute.65 This vaccine uses an adenovirus vector to induce a protective immune response. The ChadOx1 platform has been used to develop investigational vaccines against several different pathogens, including MERS-CoV. Recently, it was found that the vaccine was effective in tests on macaques and showed no viral replication within the lungs.66 Ad5-nCoV was the first SARS-CoV-2 vaccine tested in Chinese clinical trials. This vaccine candidate is also adenovirus vector based (type 5 vector) and expresses the SARS-CoV-2 S protein.67 It was developed by CanSino Biologics Inc. in Tianjin, China. The AAVCOVID vaccine candidate was developed in the laboratory of Luk Vandenberghe at Massachusetts General Hospital. This vaccine uses an adeno-associated virus (AAV) vector that expresses the SARS-CoV-2 S protein. AAV technology has been extensively used in the field of gene therapy, and this lab is a leader in the realm of AAV. This vaccine is expected to reach clinical trials by the end of 2020 (https://www.masseyeandear.org/news/press-releases/2020/05/mee-and-mgh-advancing-aavcovid-vaccine). In late June 2020, the clinical trial of an RNA-based vaccine, LNP-NCOVsaRNA, from Imperial College London started off in the United Kingdom (trial registration no.: SRCTN17072692). The self-replicating RNA vaccine relies on the encoded S protein from the envelope of SARS-CoV-2 and should induce immunity in recipients without causing COVID-19.
To date, just two coronavirus vaccines have been approved. Sputnik V—formerly known as Gam-COVID-Vac and developed by the Gamaleya Research Institute in Moscow—was approved by the Ministry of Health of the Russian Federation on August 1168 (Table 3). Experts have raised considerable concern about the vaccine’s safety and efficacy given it has not yet entered phase 3 clinical trials. A second vaccine in Russia, EpiVacCorona (ClinicalTrials.gov ID: NCT04527575), has also been granted regulatory approval, also without entering phase 3 clinical trials (Table 3).
Several antibodies have been identified to target different domains of SARS-CoV-2 and are effective in neutralizing SARS-CoV-2 (Table 2). These antibodies may have the potential to treat SARS-CoV-2-infected patients, and future work to define these antibody epitopes will further aid vaccine development. The experimental and clinical results of some vaccine candidates, such as BBIBPCorV and PiCoVacc, were reported, with most vaccines showing neutralizing capacity.69 For vaccine development, it is critical to generate protective T- and B-cell immune responses. The S protein has been shown to be the most potent antigen for SARS-CoV and MERS-CoV vaccines, and we hypothesize this may be similar for SARS-CoV-2 vaccines. However, the immunopathology induced by SARS-CoV or MERS-CoV vaccines was observed in animal models, which might be attributed to ADE, an aberrant Th2 response partially due to the N protein, as well as other unknown reasons.69
The mechanisms underlying this immunopathology deserve further investigation, which may provide instructive guidance for the future development of SARS-CoV-2 vaccines. Apart from immunopathology, other important questions remain to be addressed, such as how to protect the population vulnerable to lethal human CoVs, such as the elderly, and how best to provide protection against variant and heterologous CoV strains. Recently, hACE2 transgenic mice were developed that could be infected by SARS-CoV-2 and generated typical pathology that were similar to those of COVID-19 patients.70 , 71 Rhesus macaques infected by SARS-CoV-2 also exhibited humoral and cellular immune responses and were protected from rechallenge.72 In essence, it is equally important to identify the ideal animal model for evaluating potential SARS-CoV-2 vaccines.
Summary
The spread of SARS-CoV-2 continues to cause problems to health systems and economies worldwide. There is currently no available vaccine against it that has passed the required clinical trials and received approval for use. However, only two drugs have emerged as effective treatments to combat it: the steroid drug dexamethasone, for critically ill patients on ventilators, and the antiviral drug RDV, for less critical cases, shortening the disease period. The international scientific community has intensified efforts on vaccines and therapeutic research at an unprecedented pace, and collaborations or formations of consortiums have allowed such speed in scientific advancement to take place. For antivirals against SARS-CoV-2, the development and clinical approval of novel compounds that specifically target SARS-CoV-2 will require an extended period of preclinical testing before they can enter clinical trials. The COVID-19 pandemic is a large-scale emergency and warrants the rapid use of already approved drugs that can be repurposed for its treatment. This strategy is what has facilitated the trials and uses of RDV, HQC, CQ, LPV, Avigan (favipiravir), and dexamethasone to treat COVID-19 in emergencies. It is expected that more effective drugs against SARS-CoV-2 will be found in the near future. Convalescent plasma may be used in the United States to treat hospitalized patients under an EUA or an Investigational New Drug (IND) application. “Adequate and well-controlled randomized trials remain necessary for a definitive demonstration of COVID-19 convalescent plasma efficacy and to determine the optimal product attributes and appropriate patient populations for its use,” according to updated guidance issued by the FDA on September 2. While the world is transfixed by the high-stakes race to develop a COVID-19 vaccine, an equally crucial competition is heating up to produce targeted antibodies that could provide an instant immunity boost against the virus. Clinical trials of these mAbs, which could both prevent and treat the disease, are already underway and could produce signs of efficacy in the next few months, perhaps ahead of vaccine trials. In conclusion, we have listed the possible therapies, many of which are being tested in clinical trials and some that still need more testing.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu A., Peng Y., Huang B., et al. Genome Composition and Divergence of the Novel Coronavirus (2019-NCoV) Originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheahan T.P., Sims A.C., Leist S.R., et al. Comparative Therapeutic Efficacy of Remdesivir and Combination Lopinavir, Ritonavir, and Interferon Beta against MERS-CoV. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortiz-Alcantara J., Bhardwaj K., Palaninathan S., et al. Small Molecule Inhibitors of the SARS-CoV Nsp15 Endoribonuclease. Virus Adapt. Treatment. 2010;2:125–133. [Google Scholar]
- 6.Naqvi A., Fatima K., Mohammad T., et al. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortorici M.A., Veesler D. Advances in Virus Research. Elsevier; Amsterdam: 2019. Structural Insights into Coronavirus Entry; pp. 93–116. Vol. 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.-J., Tortorici M.A., et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayanan K., Maeda A., Maeda J., et al. Characterization of the Coronavirus M Protein and Nucleocapsid Interaction in Infected Cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zúñiga S., Sola I., Moreno J.L., et al. Coronavirus Nucleocapsid Protein Is an RNA Chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan C.A.M., Rottier P.J.M. Advances in Virus Research. Elsevier; Amsterdam: 2005. Molecular Interactions in the Assembly of Coronaviruses; pp. 165–230. Vol. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman B.W., Kiss G., Kunding A.H., et al. A Structural Analysis of M Protein in Coronavirus Assembly and Morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatagopalan P., Daskalova S.M., Lopez L.A., et al. Coronavirus Envelope (E) Protein Remains at the Site of Assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieto-Torres J.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., et al. Severe Acute Respiratory Syndrome Coronavirus E Protein Transports Calcium Ions and Activates the NLRP3 Inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corse E., Machamer C.E. The Cytoplasmic Tails of Infectious Bronchitis Virus E and M Proteins Mediate Their Interaction. Virology. 2003;312:25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D., Lee J.-Y., Yang J.-S., et al. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barretto N., Jukneliene D., Ratia K., et al. The Papain-Like Protease of Severe Acute Respiratory Syndrome Coronavirus Has Deubiquitinating Activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S., Sharma P. P., Shankar U.; et al. Discovery of New Hydroxyethylamine Analogs against 3CLpro Protein Target of SARS-CoV-2: Molecular Docking, Molecular Dynamics Simulation and Structure-Activity Relationship Studies. J. Chem. Inf. Model.2020. DOI: 10.1021/acs.jcim.0c00326. [DOI] [PubMed]
- 20.Chien M., Anderson T. K., Jockusch S.; et al. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. bioRxiv2020. DOI: 10.1101/2020.03.18.997585. [DOI] [PMC free article] [PubMed]
- 21.Anand K., Ziebuhr J., Wadhwani P., et al. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Xu Q., Sun Z., et al. Current Targeted Therapeutics against COVID-19: Based on First-Line Experience in China. Pharmacol. Res. 2020:104854. doi: 10.1016/j.phrs.2020.104854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj K., Guarino L., Kao C.C. The Severe Acute Respiratory Syndrome Coronavirus Nsp15 Protein Is an Endoribonuclease That Prefers Manganese as a Cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta Y., Maciorowski D., Jones K.; et al. Bisindolylmaleimide IX: A Novel Anti-SARS-CoV2 Agent Targeting Viral Main Protease 3CLpro Demonstrated by Virtual Screening and In Vitro Assays. Res. Square2020. DOI: 10.21203/rs.3.rs-48709/v2. [DOI] [PMC free article] [PubMed]
- 25.Hamming I., Timens W., Bulthuis M., et al. Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M.-Y., Li L., Zhang Y., et al. Expression of the SARS-CoV-2 Cell Receptor Gene ACE2 in a Wide Variety of Human Tissues. Infect. Dis. Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang J., Ye G., Shi K., et al. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glowacka I., Bertram S., Muller M.A., et al. Evidence That TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch B.J., Bartelink W., Rottier P.J.M. Cathepsin L Functionally Cleaves the Severe Acute Respiratory Syndrome Coronavirus Class I Fusion Protein Upstream of Rather than Adjacent to the Fusion Peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agostini M.L., Andres E.L., Sims A.C., et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choy K.-T., Wong A.Y.-L., Kaewpreedee P., et al. Remdesivir, Lopinavir, Emetine, and Homoharringtonine Inhibit SARS-CoV-2 Replication In Vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beigel J.H., Tomashek K.M., Dodd L.E., et al. Remdesivir for the Treatment of Covid-19. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eastman R.T., Roth J.S., Brimacombe K.R., et al. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan H., Peto R., Karim Q. A.; et al. Repurposed Antiviral Drugs for COVID-19; Interim WHO SOLIDARITY Trial Results. medRxiv2020. DOI: 10.1101/2020.10.15.20209817. [DOI] [PMC free article] [PubMed]
- 35.Schrezenmeier E., Dörner T. Mechanisms of Action of Hydroxychloroquine and Chloroquine: Implications for Rheu-matology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 36.Yam J.C.S., Kwok A.K.H. Ocular Toxicity of Hydroxychloroquine. Hong Kong Med. J. 2006;12:294–304. [PubMed] [Google Scholar]
- 37.Vincent M.J., Bergeron E., Benjannet S., et al. Chloroquine Is a Potent Inhibitor of SARS Coronavirus Infection and Spread. Virology J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torjesen I. Covid-19: Hydroxychloroquine Does Not Benefit Hospitalised Patients, UK Trial Finds. BMJ. 2020;369:m2263. doi: 10.1136/bmj.m2263. [DOI] [PubMed] [Google Scholar]
- 39.Ratia K., Pegan S., Takayama J., et al. A Noncovalent Class of Papain-Like Protease/Deubiquitinase Inhibitors Blocks SARS Virus Replication. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nutho B., Mahalapbutr P., Hengphasatporn K., et al. Why Are Lopinavir and Ritonavir Effective against the Newly Emerged Coronavirus 2019? Atomistic Insights into the Inhibitory Mechanisms. Biochemistry. 2020;59:1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 41.Ledford H. Coronavirus Breakthrough: Dexamethasone Is First Drug Shown to Save Lives. Nature. 2020;582 doi: 10.1038/d41586-020-01824-5. 469–469. [DOI] [PubMed] [Google Scholar]
- 42.Bouhaddou M., Memon D., Meyer B., et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casadevall A., Pirofski L.A. The Convalescent Sera Option for Containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen C., Wang Z., Zhao F., et al. Treatment of 5 Critically Ill Patients with COVID-19 with Convalescent Plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B., Liu S., Tan T., et al. Treatment with Convalescent Plasma for Critically Ill Patients with Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Chest. 2020;158:e9–e13. doi: 10.1016/j.chest.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan K., Liu B., Li C., et al. Effectiveness of Convalescent Plasma Therapy in Severe COVID-19 Patients. Proc. Natl. Acad. Sci. U.S.A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gharbharan A., Jordans C. C. E, GeurtsvanKessel C.; et al. Convalescent Plasma for COVID-19. A Randomized Clinical Trial. medRxiv2020. DOI: 10.1101/2020.07.01.20139857.
- 48.Li L., Zhang W, Hu Y, et al. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients with Severe and Life-Threatening COVID-19: A Randomized Clinical Trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunamneni A., Clarke E.C., Ye C., et al. Generation and Selection of a Panel of Pan-Filovirus Single-Chain Antibodies Using Cell-Free Ribosome Display. Am. J. Trop. Med. Hyg. 2019;101:198–206. doi: 10.4269/ajtmh.18-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunamneni A., Ye C., Bradfute S.B., et al. Ribosome Display for the Rapid Generation of High-Affinity Zika-Neutralizing Single-Chain Antibodies. PloS One. 2018;13:e0205743. doi: 10.1371/journal.pone.0205743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bannas P., Hambach J., Koch-Nolte F. Nanobodies and Nanobody-Based Human Heavy Chain Antibodies as Antitumor Therapeutics. Front. Immunol. 2017;8:1603. doi: 10.3389/fimmu.2017.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincke C., Loris R., Saerens D., et al. General Strategy to Humanize a Camelid Single-Domain Antibody and Identification of a Universal Humanized Nanobody Scaffold. J. Biol. Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 53.Cardoso F.M., Ibañez L.I., Van den Hoecke S., et al. Single-Domain Antibodies Targeting Neuraminidase Protect against an H5N1 Influenza Virus Challenge. J. Virol. 2014;88:8278–8296. doi: 10.1128/JVI.03178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Detalle L., Stohr T., Palomo C., et al. Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 2016;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gavor E., Choong Y.K., Er S.Y., et al. Structural Basis of SARS-CoV-2 and SARS-CoV Antibody Interactions. Trends Immunol. 2020;41:1006–1022. doi: 10.1016/j.it.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinto D., Park Y.J., Beltramello M., et al. Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 57.Wan J., Xing S., Ding L., et al. Human-IgG Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 2020;32:107918. doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robbiani D.F., Gaebler C., Muecksch F., et al. Convergent Antibody Responses to SARS-CoV-2 in Convalescent Individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Y., Wang F., Shen C., et al. A Noncompeting Pair of Human Neutralizing Antibodies Block COVID-19 Virus Binding to Its Receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y., Fu B., Zheng X.; et al. Aberrant Pathogenic GM-CSF + T Cells and Inflammatory CD14+ CD16+ Monocytes in Severe Pulmonary Syndrome Patients of a New Coronavirus. Biorxiv2020. DOI: 10.1101/2020.02.12.945576v1.
- 61.Chen X., Zhao B., Qu Y., et al. Detectable Serum SARS-CoV-2 Viral Load (RNAaemia) Is Closely Associated with Drastically Elevated Interleukin 6 (IL-6) Level in Critically Ill COVID-19 Patients. Clin. Infect. Dis. 2020;17:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oldfield V., Dhillon S., Plosker G.L. Tocilizumab: A Review of Its Use in the Management of Rheumatoid Arthritis. Drugs. 2009;69:609–632. doi: 10.2165/00003495-200969050-00007. [DOI] [PubMed] [Google Scholar]
- 63.Huizinga T.W.J., Fleischmann R.M., Jasson M., et al. Sarilumab, a Fully Human Monoclonal Antibody against IL-6Rα in Patients with Rheumatoid Arthritis and an Inadequate Response to Methotrexate: Efficacy and Safety Results from the Randomised SARIL-RA-MOBILITY Part A Trial. Ann. Rheum. Dis. 2014;73:1626–1634. doi: 10.1136/annrheumdis-2013-204405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson L.A., Anderson E.J., Rouphael N.G., et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Folegatti P.M., Ewer K.J., Aley P.K., et al. Safety and Immunogenicity of the ChAdOx1 nCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Doremalen N., Lambe T., Spencer A.; et al. ChAdOx1 NCoV-19 Vaccination Prevents SARS-CoV-2 Pneumonia in Rhesus Macaques. bioRxiv2020. DOI: 10.1101/2020.05.13.093195.
- 67.Zhu F.-C, Guan X.H., Li Y.H., et al. Immunogenicity and Safety of a Recombinant Adenovirus Type-5-Vectored COVID-19 Vaccine in Healthy Adults Aged 18 Years or Older: A Randomised, Double-Blind, Placebo-Controlled, Phase 2 Trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Logunov D.Y., Dolzhikova I.V., Zubkova O.V., et al. Safety and Immunogenicity of an rAd26 and rAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine in Two Formulations: Two Open, Non-Randomised Phase 1/2 Studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong Y., Dai T., Wei Y., et al. A Systematic Review of SARS-CoV-2 Vaccine Candidates. Signal Transduct. Target. Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun S.H., Chen Q., Gu H.J., et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang R.D., Liu M.Q., Chen Y., et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandrashekar A., Liu J., Martinot A.J., et al. SARS-CoV-2 Infection Protects against Rechallenge in Rhesus Macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vankadari N. Arbidol: A Potential Antiviral Drug for the Treatment of SARS-CoV-2 by Blocking Trimerization of the Spike Glycoprotein. Int. J. Antimicrob. Agents. 2020;56:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morse J.S., Lalonde T., Xu S., et al. Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-NCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richardson P., Griffin I., Tucker C., et al. Baricitinib as Potential Treatment for 2019-NCoV Acute Respiratory Disease. Lancet. 2020;395:e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finkelstein Y., Aks S.E., Hutson J.R., et al. Colchicine Poisoning: The Dark Side of an Ancient Drug. Clin. Toxicol. (Phila.) 2010;48:407–414. doi: 10.3109/15563650.2010.495348. [DOI] [PubMed] [Google Scholar]
- 77.Guo D. Old Weapon for New Enemy: Drug Repurposing for Treatment of Newly Emerging Viral Diseases. Virol. Sin. 2020;35:253–255. doi: 10.1007/s12250-020-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Warren T.K., Wells J., Panchal R.G., et al. Protection against Filovirus Diseases by a Novel Broad-Spectrum Nucleoside Analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu C., Zhou Q., Li Y., et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar V., Shin J.S., Shie J.-J., et al. Identification and Evaluation of Potent Middle East Respiratory Syndrome Coronavirus (MERS-CoV) 3CLPro Inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Y., Vedantham P., Lu K., et al. Protease Inhibitors Targeting Coronavirus and Filovirus Entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hansen J., Baum A., Pascal K.E., et al. Studies in Humanized Mice and Convalescent Humans Yield a SARS-CoV-2 Antibody Cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baum A., Fulton B.O., Wloga E., et al. Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Utational Escape Seen with Individual Antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chi X., Yan R., Zhang J., et al. A Neutralizing Human Antibody Binds to the N-Terminal Domain of the Spike Protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang C., Li W., Drabek D., et al. A Human Monoclonal Antibody Blocking SARS-CoV-2 Infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 87.Tian X., Li C., Huang A., et al. Potent Binding of 2019 Novel Coronavirus Spike Protein by a SARS Coronavirus-Specific Human Monoclonal Antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ter Meulen J., van den Brink E.N., Poon L.L., et al. Human Monoclonal Antibody Combination against SARS Coronavirus: Synergy and Coverage of Escape Mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Walls A.C., Xiong X., Park Y.J., et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wrapp D., De Vlieger D., Corbett K.S., et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wec A.Z., Wrapp D., Herbert A.S., et al. Broad Neutralization of SARS-Related Viruses by Human Monoclonal Antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ju B., Zhang Q., Ge J., et al. Human Neutralizing Antibodies Elicited by SARS-CoV-2 Infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 93.Chen X., Li R., Pan Z., et al. Human Monoclonal Antibodies Block the Binding of SARS-CoV-2 Spike Protein to Angiotensin Converting Enzyme 2 Receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi R., Shan C., Duan X., et al. A Human Neutralizing Antibody Targets the Receptor Binding Site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 95.Cao Y., Su B., Guo X., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou D., Duyvesteyn H., Chen C.P., et al. Structural Basis for the Neutralization of SARS-CoV-2 by an Antibody from a Convalescent Patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 97.Barnes C.O., West A.P., Jr., Huey-Tubman K.E., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zost S.J., Gilchuk P., Case J.B., et al. Potently Neutralizing and Protective Human Antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu L., Wang P., Nair M.S., et al. Potent Neutralizing Antibodies Directed to Multiple Epitopes on SARS-CoV-2 Spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 100.Lv Z., Deng Y.Q., Ye Q., et al. Structural Basis for Neutralization of SARSCoV-2 and SARS-CoV by a Potent Therapeutic Antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rogers T.F., Zhao F., Huang D., et al. Isolation of Potent SARS-CoV-2 Neutralizing Antibodies and Protection from Disease in a Small Animal Model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lei C., Qian K., Li T., et al. Neutralization of SARS-CoV-2 Spike Pseudotyped Virus by Recombinant ACE2-Ig. Nat. Commun. 2020;11:2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuan M., Liu H., Wu N.C., et al. Structural Basis of a Shared Antibody Response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuan A. Q., Zhao L., Bai L.; et al. Isolation of and Characterization of Neutralizing Antibodies to Covid-19 from a Large Human Naïve scFv Phage Display Library. BioRxiv2020. DOI: 10.1101/2020.05.19.104281.
- 105.Wu Y., Li C., Xia S., et al. Identification of Human Single-Domain Antibodies against SARSCoV-2. Cell Host Microbe. 2020;27:891–898.e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seydoux E., Homad L.J., MacCamy A.J., et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brouwer P.J.M., Caniels T.G., van der Straten K., et al. Potent Neutralizing Antibodies from COVID-19 Patients Define Multiple Targets of Vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xia S., Duan K., Zhang Y., et al. Effect of an Inactivated Vaccine against SARS-CoV-2 on Safety and Immunogenicity Outcomes: Interim Analysis of 2 Randomized Clinical Trials. JAMA. 2020;324:951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Y.-J., Zeng G., Pan H.-X.; et al. Immunogenicity and Safety of a SARS-CoV-2 Inactivated Vaccine in Healthy Adults Aged 18-59 Years: Report of the Randomized, Double-Blind, and Placebo-Controlled Phase 2 Clinical Trial. medRxiv2020. DOI: 10.1101/2020.07.31.20161216.
- 110.Mulligan M.J., Lyke K.E., Kitchin N., et al. Phase 1/2 Study of COVID-19 RNA Vaccine BNT162b1 in Adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 111.Folegatti P.M., Ewer K.J., Aley P.K., et al. Safety and Immunogenicity of the ChAdOx1 nCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mercado N.B., Zahn R., Wegmann F., et al. Single-Shot Ad26 Vaccine Protects against SARS-CoV-2 in Rhesus Macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


