Abstract
Tumor evolution drives tumor progression, therapeutic resistance, and metastasis. Therefore, new predictive medicine strategies that adapt with a tumor are needed to improve patient outcomes. The techniques used in weather prediction are mathematically proven to enable prediction of evolving systems, and thus provide a framework for a new predictive medicine paradigm for cancer.
When a hurricane develops, we recognize its complexity. Emergency management strategies are based on computational models of weather forecasts of the future storm locations, and continuously adapt to the inherent unpredictability of the system. Cancer is no less complex than a hurricane. From the time a premalignancy develops, the molecular and cellular pathways continually adapt to rewire the microenvironment, promote growth, survive through treatment, and metastasize. Despite this, precision medicine strategies take a static, gene-centric view, leveraging genetics and genomics profiling of pre-treatment tumors to match therapies to a tumor’s drivers. Although this approach has improved outcomes in some cancer subtypes, resistance is pervasive and limits durable treatment efficacy. In part, pervasive resistance arises because precision medicine strategies disregard the dynamics of complex adaptation of tumor cells and the microenvironment that are responsible for treatment response. Therefore, cancer requires new predictive medicine strategies that utilize patient datasets to adapt with the tumor.
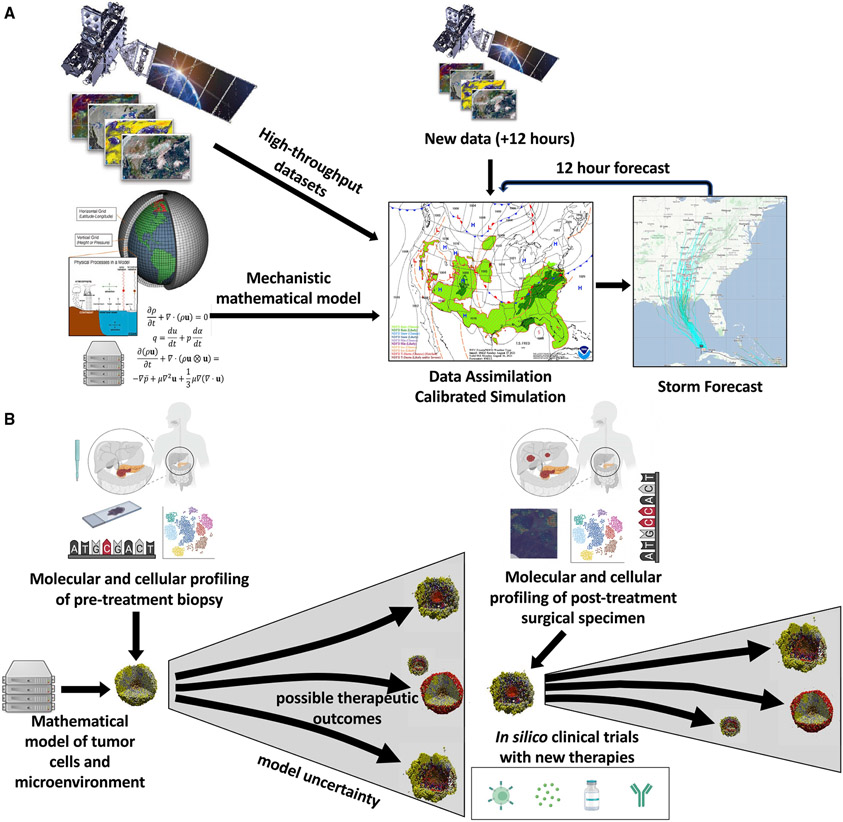
Beyond analogy, weather forecasting also provides a quantitative framework to implement predictive medicine. The techniques used to forecast the weather were designed to overcome the “butterfly effect,” in which small perturbations can limit later predictions in evolving complex systems, and are thus applicable to tumor biology.1 Accurate weather forecasts rely on regular updates made by integrating mechanistic knowledge with high-throughput data using data assimilation tools (Figure 1A). Data assimilation utilizes three components: mathematical models derived from the laws of the system, databases containing observations from continuous monitoring of the atmosphere, and computational methodologies for their integration. These components are mirrored in biomedical sciences by mechanistic inference from advanced molecular and cellular measurement technologies, clinical and molecular databases, and computational oncology algorithms. New technologies can now profile the spatial landscape of tumors across multiple molecular resolutions, and the widespread adoption of electronic medical records provide unprecedented characterization of tumors. For the first time, these datasets provide the necessary cancer observing systems that provide a foundation for the implementation of predictive medicine.
Figure 1. Data-driven simulation and forecasting in weather and oncology.
(A) Data assimilation systems from weather provide a model for long-term prediction of complex systems to enable predictive medicine. In weather, imaging and sensors capture high-throughput data profiling the atmosphere, which then calibrates mechanistic mathematical models that are based on the physical laws of the atmosphere. These mathematical models are used to predict future weather conditions, with increasing uncertainty as they are extended forward. Every 6-12 h, new data are assimilated with the forecasted state at that time to recalibrate the simulation and improve subsequent forecasts. Weather maps and model images from noaa.gov and satellite image of from GOES-16 weather satellite.
(B) Implementation of predictive medicine in neo-adjuvant platform clinical trials. Molecular and cellular profiling data from pre-treatment biopsies can be used to calibrate the states of cell-based mathematical models of tumors. These forecasts are then updated based on additional high-throughput data obtained from post-treatment surgical specimens to enable in silico clinical trials modeling the impact of new treatment strategies selected at later time points to overcome mechanisms of therapeutic resistance. Platform studies enable model calibration with multiple therapeutic agents. Cell-based models created with PhysiCell,2 and figure created with Biorender.com.
Inferring the laws of tumor progression from high-throughput data
Whereas the physical laws of the atmosphere are known and codified in mathematical equations, many of the variables governing tumor biology and disease progression are still unknown. Single-cell technologies are poised to identify the molecular and cellular underpinnings of carcinogenesis, accelerating the discovery of the biological processes underlying disease progression and therapeutic response. New molecular and cellular profiling technologies are rapidly developing to characterize the molecular and cellular state of tumors, with current spatial molecular technologies expanding to enable multi-omics characterization. In spite of the promise of these technologies, the complexity of regulatory networks and high-dimensional nature of data from new profiling technologies challenge their direct human interpretation. Artificial intelligence (AI) methodologies, such as unsupervised learning methods for pattern detection, can reduce the effective dimensionality of high-throughput data to infer the underlying biological processes represented in a dataset.3 Although powerful, the data-driven nature of genomics analyses of single-cell multi-omics limits biological inference to the conditions under which a dataset is measured.
Tumors and the cells in their microenvironments are continuously evolving. Fully elucidating the mechanisms that underlie therapeutic response and resistance requires serial sampling of cancers as they evolve in response to therapy. Single-cell atlas projects for spatiotemporal profiling of cancer through consortia such as the Human Tumor Atlas Network are emerging.4 Longitudinal profiling of human tumors depends critically on a patient’s return to the clinic. Molecular and cellular profiling often require expensive, invasive procedures for re-sampling tissue, which may be infeasible and even unethical to obtain in all cases. Moreover, these may capture only one site of a tumor and thereby limit the ability to characterize its complete molecular and cellular heterogeneity. While imaging and blood-based biomarkers are more feasible for longitudinal sampling, they also are insufficient to capture the underlying cellular and molecular variables that govern tumor biology.
Human clinical trial platform studies are a novel approach to study changes in the tumor and its microenvironment with sequential interventions. When these studies are conducted prior to surgical resection of the tumor in neo-adjuvant or “window of opportunity” setting, they allow for sufficient tissue to apply multiple high-throughput assays for cellular and molecular characterization.5 Typically, treatment is continued after surgery, which allows for long-term follow up of patients throughout their treatment course to capture clinical outcomes for correlation with the profiling data. These studies can be conducted for all disease stages, providing the clinical foundation to combine state-of-the-art profiling with longitudinal patient data that are needed to develop predictive medicine paradigms.
Leveraging clinical data and electronic medical records
Refining the mechanistic underpinnings of cancer evolution with emerging technologies has clinical potential to enhance treatment selection. Today, clinicians rely on case history and physical exams, standard laboratory and radiographic tests, and evaluation of tumor specimens that may have been removed months or years prior to a treatment decision point. Profiling tumors with emerging technologies also has the potential to prioritize a smaller set of mechanistic biomarkers that can provide a more dynamic evaluation to support predictive medicine. As an example, liquid biopsies using circulating tumor DNA can be used as blood-based biomarkers to guide treatment or monitor disease progression over time. Additional lifestyle and environmental perturbations further impact disease progression beyond the mechanistic underpinnings captured with high-throughput profiling technologies. These factors make clinical manifestations in disease symptoms, routine laboratory tests, and clinical data that are important for computational biology to model in automating strategies for durable patient care. Tools that can aid the clinician to synthesize symptoms, signs, and tests results into anticipated disease outcome are urgently needed and have the potential to ensure that patients are receiving treatment that is likely to provide benefit, while at the same time minimizing use of agents that are not likely to provide benefit but may be toxic and costly. Ideally, future decision making will also incorporate computational tools based upon the mechanistic underpinnings of a tumor informed from molecular, genomic, and cellular factors.
The transition to electronic medical records has provided the potential for large-scale databases of records containing clinical tests, disease states, adverse reactions, and associated clinical decisions. In the context of predictive medicine, the clinical data in these records provide the most practical means to generate observation systems with longitudinal monitoring of cancer. Patients routinely return for clinical follow up as part of their care, even when tumor tissue is not accessible for high-throughput profiling. When these data are available, incorporating the results of genetic and molecular assays has the potential to leverage established molecular drivers to alter treatment recommendations at distinct decision points. Even in the absence of genetic data, the predictive power of clinical expertise that relies on these data suggests that machine learning methodologies can still gain mechanistic insights from the data in clinical records. Notably, recent studies have demonstrated that clinical imaging data is predictive of genomic data and molecular biomarkers including microsatellite instability.6,7 In general, predicting high-throughput profiles based on low-dimensional clinical outcomes in electronic medical records provides an informative training data source for developing AI models that predict mechanistic biomarkers. Ultimately, applying these models to serial patient records could uncover the time-varying parameters that best fit a patient’s later outcomes and underlie the biology of their tumor’s evolution.
Just as computational tools are needed to interpret high-throughput molecular and cellular profiling into insights about the mechanistic underpinnings of disease, machine learning methods are also critical for analysis of clinical records. Many clinicians may be aware of which features from clinical data are pertinent to specific tumor type, yet features used in other cancer types or diseases may be relevant to a patient. Applying AI algorithms to large-scale databases can glean these pertinent features to alter clinical care and identify new clinical features that should be assessed in a disease subtype. These algorithms also have the potential to overcome clinician bias by automating treatment recommendations, although this requires robust training data and bias-aware algorithms to overcome bias in the machine learning algorithms themselves. For predictive medicine, real world clinical data can also monitor additional environmental and host factors such as socioeconomic factors, nutrition, physical activity, presence of co-morbid conditions, and pharmacogenomics that will impact disease progression and treatment efficacy that cannot be determined through molecular assays alone.
In spite of their promise for research and clinical decision making, most cancer research data are siloed by groups in academia, government, and the pharmaceutical industry. Cancer is composed of many different diseases with significantly different causes that influence their development and progression. Most individual cancers, each potentially deadly, are considered uncommon or rare when analyzed as a single entity. Furthermore, within each cancer type, environment, gender, geography, and race have been shown to impact disease development and progression, and treatment response, further limiting the power to assess data in small cohorts of individuals. Thus, to effectively uncover targetable cancer-specific pathways, pooling of data across cohorts and siloed groups is essential and requires data sharing. Still, many factors limit data sharing. A lack of accessible, user-friendly, and compatible databases is a major barrier. Most small groups are financially unable to invest in their development, and larger groups tend to develop their own databases that are not available to smaller groups and that are typically incompatible with publicly available databases. Government regulations also provide barriers in data sharing due to privacy guidelines. In the US, the Health Insurance Portability and Accountability Act (HIPAA) guidelines were established to protect patients from being discriminated against when applying for jobs or health insurance. These guidelines need to be updated to consider clinical use of large-scale databases, updating policies to account for the new protections provided from accessible health insurance through the Affordable Care Act while balancing other considerations for protection of patient privacy. The drug development and approval process at the US Food and Drug Administration (FDA) and other regulatory bodies also limits data sharing; companies must protect their data to maintain integrity during the drug development process in route to market. These issues vary by country and should be addressed to allow international data sharing. Finally, financial and academic credit is still a driving force in data protection across all sectors of the cancer research community. Providing database infrastructure with appropriate privacy protection and incentive structures for sharing are essential to usher in the data-driven age of clinical decision making for both precision and predictive medicine.
Biological and mathematical models can supplement serial sampling of human tumors
The heterogeneity between cancer patients and even within individual tumors limits the ability to evaluate the mechanistic and clinical impact that would have resulted from applying different therapeutic regimens, even in controlled designs of platform studies in clinical research studies. Pairing human profiling with parallel experiments in preclinical models can supplement some of these gaps, allowing for refined serial profiling under a breadth of therapeutic perturbations that are impossible to obtain directly from human studies. Preclinical platform studies in mouse and organoid models that are concurrent with human platform studies and use the same therapeutics and assays as human platform studies can allow for direct cross-species analysis, providing the opportunity for a deeper mechanistic evaluation of the molecular and cellular changes resulting from treatment. Because preclinical models are an imperfect representation of human disease, there is some skepticism in the clinical research community as to the value of these studies in preclinical biological models. For predictive medicine, these models are invaluable approaches for confirming and refining findings from data from clinical trial biospecimens that are limited in material and temporal analyses. Moreover, computational techniques for cross-species analysis can delineate the specific cell-dependent molecular pathways that are preserved between the systems and even across tumor types and subtypes from multi-omics single-cell datasets.8 Thus, databases integrating high-throughput datasets spanning preclinical models and human tumors are necessary to enable computational biology to infer the rules of tumor biology. These rules are key to transforming data into knowledge: we can know that a patient’s tumor cells divide in response to a specific growth factor, secrete a signal to recruit supporting stromal cells, or up-regulate a DNA damage repair pathway in response to chemotherapy.
Just as the knowledge of the physical rules of the atmosphere can be codified as equations to build computer models of weather, we can translate the rules of a patient’s tumor biology into mathematical rules to build in silico models that simulate tumor growth and progression. In contrast to the data-driven AI models or biostatistics, these models simulate and connect the dynamics of each of the underlying mechanistic variables. The evolving virtual tumor can drive changes in its microenvironment such as hypoxia or inflammation, while the microenvironmental changes drive further tumor evolution and affect response to treatments. These multiscale models afford us the opportunity to transform our biological knowledge at molecular, cellular, and tissue scales into virtual laboratories tailored to individual patients. If we silence a gene in a cancer cell or activate a gene in an immune cell, what changes should we expect in transcription, protein expression, and ultimately cell behavior, and how soon should they happen? How might these altered cell phenotypes interact with other cancer cells, and how will that change the overall patient state? The mathematical models can use those simulated states to predict patient outcomes to therapy over time.9 Moreover, simulation models can serve as in silico assays to augment human culture systems. After genomic sequencing and histopathologic assays guide initial selection of targeted drug candidates, simulation models can evaluate the therapeutic impact of 3D drug delivery limitations, competition between heterogeneous sub-clones, and potential interactions with immune cells. Thus, in silico assays have the potential to add mechanistic data that can be integrated with clinical decision making by defining the changes in direct cell-cell interactions following therapy.
Translational data assimilation to enable predictive oncology
Ultimately, the therapeutic outcomes in cancer span the molecular, cellular, tissue, and population scales. To address this complexity, the multiscale mathematical models of tumor dynamics described above are emerging alongside new data sources and databases.10 Similar to the example of weather prediction, the accuracy of these models can be enhanced and continuously refined through data-driven parameterization and data assimilation. In weather prediction, the key state variables (temperature, pressure, velocity, humidity) can be directly measured in the atmosphere at high resolution and integrated with known properties of air and water to improve predictive accuracy. For the first time, the analogous high-resolution measurements of cell and tissue states—along with cell and tissue dynamical parameters—are increasingly available to drive accurate biological predictions. Single-cell measurement technologies are measuring the state variables needed for mathematical models of biological systems, along with detailed preclinical and clinical characterizations of cell and tissue properties, allowing for direct integration of high-throughput data with mathematical models cancer growth dynamics, therapeutic response, and evolution of therapeutic resistance.11 The multi-scale nature of human tumors introduces greater complexity, requiring further incorporation of molecular, cellular, preclinical, and clinical data sources in mathematical models, while also learning from prior models (Figure 1B). As knowledge large-scale patient datasets, new data assimilation algorithms could also learn how the variables in mechanistic mathematical models relate to heterogeneous data sources, thus providing a clinically feasible path to enable predictive medicine. Based upon the success of weather prediction, we hypothesize that these integrative systems for tumor forecasting will one day have the potential to predict a patient’s response to therapy, when disease will recur, and the mechanistic underpinnings of that recurrence to adapt precision medicine strategies over time.
As we develop a new paradigm for predictive medicine in which treatment regimens adapt with patients, statistical method for complex and innovative designs (CIDs) becomes a cornerstone for modernizing medical product development.12 For example, Bayesian adaptive platform trial designs are making cancer drug development more efficient and “smarter” by simultaneously evaluating multiple treatment regimen candidates.13,14 However, CIDs come at the price of exacerbating and even introducing new challenges to clinical trial designs. With the intrinsic complexity in CIDs, patient selection, enrollment, and monitoring all need more sophisticated planning and execution. Biostatisticians and trialists are actively developing machine learning, deep learning, natural language processing, and other similar tools to enrich patients in the enrollment by prognostic and predictive biomarkers, personalize patient monitoring continuously through the trial, and improve compliance.15,16
Realizing the promise of computational decision-making tools that leverage serial profiling for predictive medicine to transform cancer diagnosis and treatment ultimately hinges on their adoption by the greater community. Understanding how to use these tools correctly and also understanding how they were developed and validated is critical to clinical adoption. Educational programs through institutional and community physician programs, foundation meetings, and web-based learning classes will be needed to demystify computational platforms for clinical decision making. The underlying computational methodologies must be developed in user-friendly software so that individuals with minimal computational backgrounds can easily learn to use them. Regulators must also develop guidelines and biostatistical standards for the developers of predictive medicine tools to ensure that the process for their evaluation for translation to clinical practice is transparent and will meet the standards for regulatory approval. Ultimately, these efforts to enable the use of mechanistic patient data for data assimilation in clinical studies will produce immediate results to improve survival outcomes for cancer patients by adapting treatments to the evolving trajectory of each patient’s tumor.
ACKNOWLEDGMENTS
Funding for the authors provided by Lustgarten Foundation (E.J.F. and E.M.J.), the Emerson Collective (E.J.F. and E.M.J.), the Johns Hopkins University Discovery Award (E.J.F.), Jayne Koskinas Ted Giovanis Foundation for Health and Policy (P.M.), Cancer MoonshotSM funds from the National Cancer Institute (P.M.), Leidos Biomedical Research Subcontract 21X126F (P.M.), and the NIH/NCI (U01CA232137 to P.M., U01CA212007 to E.J.F., U01CA253403 to E.J.F., and P01CA247886 to E.J.F. and E.M.J.).
Footnotes
DECLARATIONS OF INTERESTS
E.J.F. is on the Scientific Advisory Board of Viosera Therapeutics/Resistance Bio. E.M.J. is a paid consultant for Adaptive Biotech, CSTONE, Achilles, DragonFly, and Genocea; receives funding from the Lustgarten Foundation and Bristol Meyer Squibb; and is the chief medical advisor for Lustgarten and an SAB advisor to the Parker Institute for Cancer Immunotherapy (PICI) and for the C3 Cancer Institute. V.S. received research grants to the institution from Abbvie, Biocept, Pfizer, Novartis, and Puma Biotechnology and is a Member, Data Safety Monitoring Board, Immunomedics, Inc. C.W. has a professional affiliation with Regeneron Pharmaceuticals.
References
- 1.Kostelich EJ, Kuang Y, McDaniel JM, Moore NZ, Martirosyan NL, and Preul MC (2011). Accurate state estimation from uncertain data and models: an application of data assimilation to mathematical models of human brain tumors. Biol. Direct 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, and Macklin P (2018). PhysiCell: an open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput. Biol e1005991. 10.1371/journal.pcbi.1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein-O’Brien GL, Arora R, Culhane AC, Favorov AV, Garmire LX, Greene CS, Goff LA, Li Y, Ngom A, Ochs MF, et al. (2018). Enter the Matrix: Factorization Uncovers Knowledge from Omics. Trends Genet. 34, 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozenblatt-Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, Ashenberg O, Cerami E, Coffey RJ, Demir E, et al. ; Human Tumor Atlas Network (2020). The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell 181, 236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liudahl SM, Betts CB, Sivagnanam S, Morales-Oyarvide V, da Silva A, Yuan C, Hwang S, Grossblatt-Wait A, Leis KR, Larson W, et al. (2021). Leukocyte heterogeneity in pancreatic ductal adenocarcinoma: phenotypic and spatial features associated with clinical outcome. Cancer Discov. 11, 2014–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, et al. (2019). Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med 25, 1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmauch B, Romagnoni A, Pronier E, Saillard C, Maillé P, Calderaro J, Kamoun A, Sefta M, Toldo S, Zaslavskiy M, et al. (2020). A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun 11, 3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis-Marcisak EF, Fitzgerald AA, Kessler MD, Danilova L, Jaffee EM, Zaidi N, Weiner LM, and Fertig EJ (2021). Transfer learning between preclinical models and human tumors identifies a conserved NK cell activation signature in anti-CTLA-4 responsive tumors. Genome Med. 13, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady R, and Enderling H (2019). Mathematical Models of Cancer: When to Predict Novel Therapies, and When Not to. Bull. Math. Biol 81, 3722–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madhavan S, Beckman RA, McCoy MD, Pishvaian MJ, Brody JR, and Macklin M (2021). Envisioning the future of precision oncology trials. Nat. Cancer 2, 9–11. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KE, Howard GR, Morgan D, Brenner EA, Gardner AL, Durrett RE, Mo W, Al’Khafaji A, Sontag ED, Jarrett AM, et al. (2020). Integrating transcriptomics and bulk time course data into a mathematical framework to describe and predict therapeutic resistance in cancer. Phys. Biol 18, 016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FDA (2020). Interacting with the FDA on complex innovative trial designs for drugs and biological products cancer vaccines. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/interacting-fda-complex-innovative-trial-designs-drugs-and-biological-products.
- 13.Collins LM, Murphy SA, and Strecher V (2007). The multiphase optimization strategy (MOST) and the sequential multiple assignment randomized trial (SMART): new methods for more potent eHealth interventions. Am. J. Prev. Med 32 (5, Suppl), S112–S118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nanda R, Liu MC, Yau C, Shatsky R, Pusztai L, Wallace A, Chien AJ, Forero-Torres A, Ellis E, Han H, et al. (2020). Effect of pembrolizumab plus neoadjuvant chemotherapy on pathologic complete response in women with early-stage breast cancer: an analysis of the ongoing phase 2 adaptively randomized I-SPY2 trial. JAMA Oncol. 6, 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrer S, Shah P, Antony B, and Hu J (2019). Artificial intelligence for clinical trial design. Trends Pharmacol. Sci 40, 577–591. [DOI] [PubMed] [Google Scholar]
- 16.Mak KK, and Pichika MR (2019). Artificial intelligence in drug development: present status and future prospects. Drug Discov. Today 24, 773–780. [DOI] [PubMed] [Google Scholar]