Abstract
Neurodegenerative disorders, i.e., Alzheimer's or Parkinson's disease, involve progressive degeneration of the central nervous system, resulting in memory loss and cognitive impairment. The intensification of neurodegenerative research in recent years put some molecules into clinical trials, but still there is an urgent need to develop effective therapeutic molecules to combat these diseases. Chromone is a well-identified privileged structure for the design of well-diversified therapeutic molecules of potential pharmacological interest, particularly in the field of neurodegeneration. In this short review, we focused on the recent advancements and developments of chromones for neurodegenerative therapeutics. Different small molecules were reviewed as multi-target-directed ligands (MTDLs) with potential inhibition of AChE, BuChE, MAO-A, MAO-B, Aβ plaque formation and aggregation. Recently developed MTDLs emphasized that the chromone scaffold has the potential to develop new molecules for the treatment of neurodegenerative diseases.
Chromones were proved as an important anti-neurodegenerative and neurogenesis agent. They act in numerous ways such as an efficient inhibitory activity against AChE, BuChE, MOA, Aβ aggregation, and neuroprotection activities.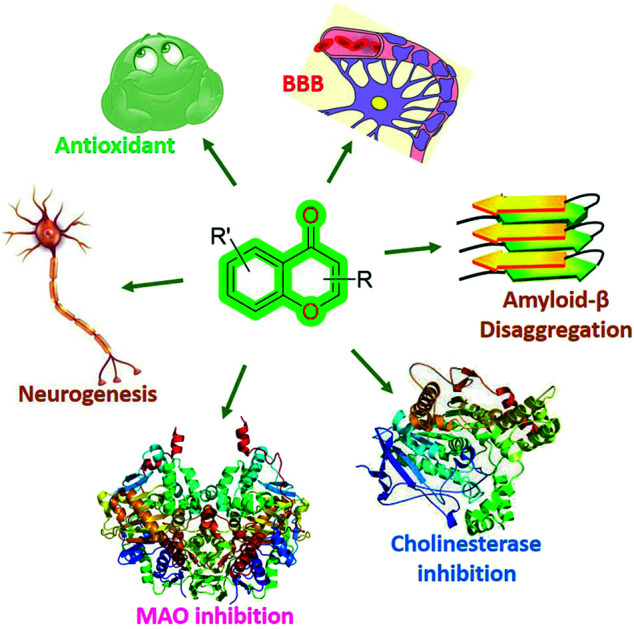
1. Introduction
Neurodegenerative disease is a broad term that can be defined as a group of disorders principally related to the neurons in the human brain.1 In general, these disorders could be understood as the progressive degeneration of the structure and function of the central nervous system or peripheral nervous system, also denoted as age-dependent disorders.2–4 Neurons are the building blocks of the nervous system which normally do not replicate or replace themselves;5 therefore, damage or death of neurons results in some memory and cognitive impairments in humans, while in some cases it affects a person's ability to move, speak and breathe.6 The most common neurodegenerative diseases are Alzheimer's disease (AD),7 Parkinson's disease (PD),8 and Huntington's disease along with other disorders such as prion disease, amyotrophic lateral sclerosis (ALS),9 motor neuron diseases (MNDs),10 frontotemporal dementia,11 spinal muscular atrophy (SMA), and spinocerebellar ataxia (SCA).12 In 2020, the World Health Organization (WHO)13 estimated that around 50 million people have dementia worldwide with an increase of nearly 10 million new cases every year. The WHO reported that AD may contribute to nearly 60–70% of cases of dementia.
Presently, the available treatments may help relieve some of the physical or mental symptoms associated but no effective treatment is available to cure neurodegenerative diseases or to slow the progression of these diseases,6,14,15 although several studies of new treatments are being investigated in various stages of clinical trials. However, effective treatments are desperately needed to support and improve the lives of people with dementia and their carers and families. As related to medicinal chemistry, several studies were reported on the development of new inhibitors of known biological targets related to neurodegenerative diseases, such as acetylcholinesterase (AChE),16,17 butyrylcholinesterase (BuChE),18 glycogen synthase kinase 3 beta (GSK3β),19 monoamine oxidase A and B (MAO-A and MAO-B),20 plasma membrane redox enzymes,21etc., and disaggregation of misfolded proteins,22,23i.e., τ-protein,24,25 amyloid-β (Aβ),26,27 TAR DNA binding protein 43 (TDP-43),28 α-synuclein,29,30etc., using small therapeutic molecules. Most of the small molecules were synthesized using privileged scaffolds of medicinal importance such as pyrimidine,31 pyrazine,32 acridine,33 triazolopyrimidine,34,35 triazene,36 coumarin,37 chromones, etc.
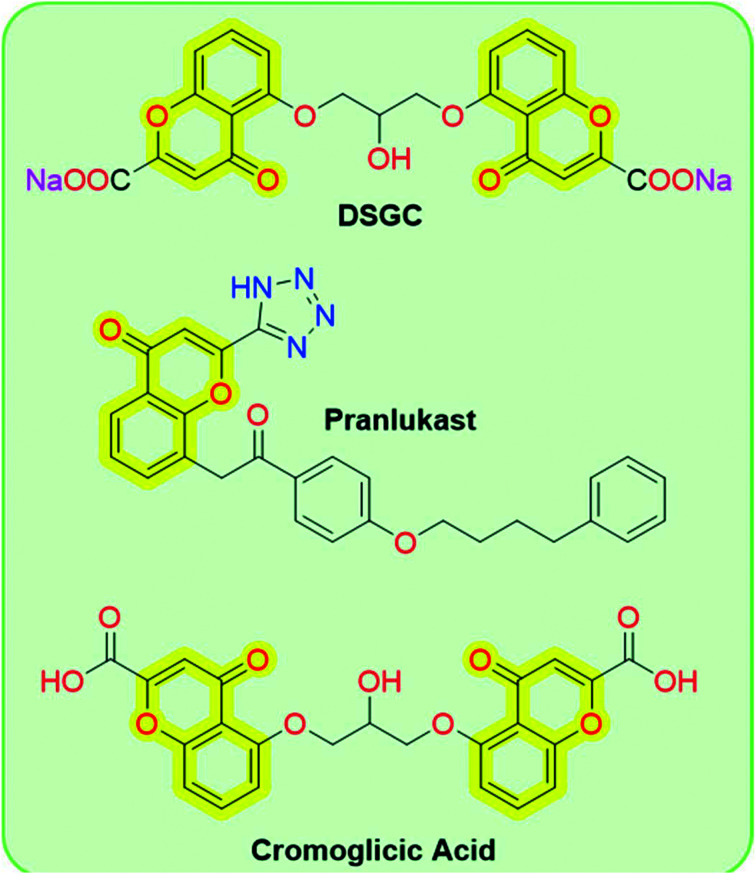
Chromones (4H-chromen-4-one, 4H-1-benzopyran-4-one, or benzo-γ-pyrone) are a large class of heterocyclic compounds that are ubiquitous in nature, especially in plants.38 The term chromone indicates its characteristic and is derived from the Greek word chroma, meaning “color”, because several chromones exhibit a variety of colors.39 The chromone moiety is the main scaffold of numerous flavonoids, such as flavones and isoflavones.40,41 From a chemistry perspective, the chromone scaffold represents itself as a privileged core domain that permits unlimited structural diversifications to develop chromone-based drug candidates. Due to their synthetic accessibility and structural diversity, these compounds particularly attracted the interest of medicinal chemists to develop new molecules of medicinal interest. Chromones exhibit a wide range of biological activities such as anti-allergic,42 anti-inflammatory,43,44 anti-diabetic,45 antitumor, antimicrobial, etc.46 Chromone-based FDA-approved cromoglicic acid is widely used for long-term management of bronchial asthma by acting as a mast cell stabilizer,47 while other chromone-based drugs such as disodium cromoglycate (DSGC) and pranlukast (Fig. 1) are used for the treatment of mild to moderate asthma and allergic rhinitis, respectively.41
Fig. 1. Chemical structures of chromone-based drugs.
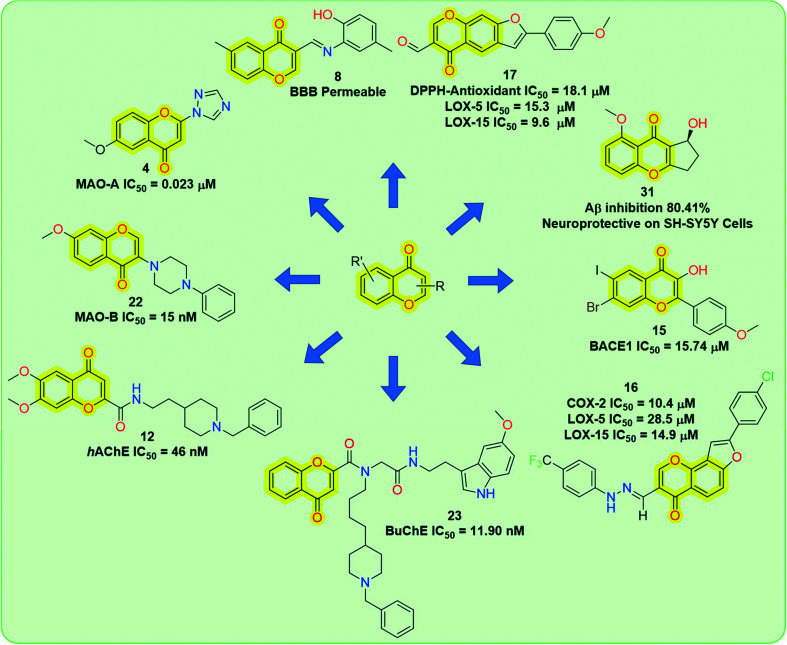
Among these biological activities, chromones are also important for neurodegenerative diseases (Fig. 2), i.e., inhibitory activities related to MOA, AChE, Aβ fibril formation, neuroprotection,48–50 neurogenesis properties,51,52etc.
Fig. 2. General structure of the chromone scaffold and its activities for different biological targets for neurodegenerative therapeutics.
In fact, an old dogma that neurons cannot generate or be replaced after birth has now been questioned during the past decades. Neurogenesis is the progression of the neurons and nervous system cells by neural stem cells (NSCs) which are found in all species of animals excluding placozoans and Porifera (sponges).53 Immunohistochemistry enables researchers to validate the generation of neurons by using bromodeoxyuridine (BrdU), an analog of thymidine that binds DNA in the S-phase in the brain of all adult mammals including humans.54 Neurogenesis continues in specific brain regions of adult mammals, known as neurogenic niches, which are extremely dynamic and are controlled by several physiological stimuli and pathological states.55 These two neurogenic niches are defined as the subventricular zone (SVZ) of the lateral ventricle and the subgranular zone (SGZ) of the dentate gyrus (DG) where NSCs proliferate, divide and differentiate into mature neurons. Recent studies indicated that adult neurogenesis could also occur in other areas of the brain along with the ventricular system, mostly in pathological conditions.56 A natural chromone baicalin 3, isolated from the root of Scutellaria baicalensis, found the ability to promote neuron differentiation of neural stem/progenitor cells (NSPCs) and stimulate neurogenesis, which also makes chromone a privileged scaffold for developing clinically relevant small molecules to manipulate NSPCs for brain repair.57–60
So far, the chromone scaffold has been reviewed by Gaspar et al.40 in detail for a medicinal chemistry perspective in 2014 and updated by Reis et al.41 in 2017; still, a focused literature review related to the recent exploration of chromones in the area of neurodegenerative disease is missing. Hence, the purpose of this short review is to analyze and summarize the recent work in the last five years related to chromones to look at their potential therapeutic application in medicinal chemistry to develop new anti-neurodegenerative agents. In addition, we have also summarized the synthetic strategies of the reported chromone derivatives.
2. Chromones with anti-neurodegenerative properties
Chromones with remarkable anti-neurodegenerative properties and that act in numerous ways such as having inhibitory activity against AChE, BuChE, MOA, Aβ aggregation, and having neuroprotection activities have recently been explored. By moving through different pieces of literature, several studies were found related to the development and isolation of natural and synthetic chromones for the treatment of neurodegenerative diseases. In this section, we have tried to put all studies of natural and synthetic chromones related to the neurodegenerative therapeutics reported in the last five years under the umbrella.
2.1. Natural chromones
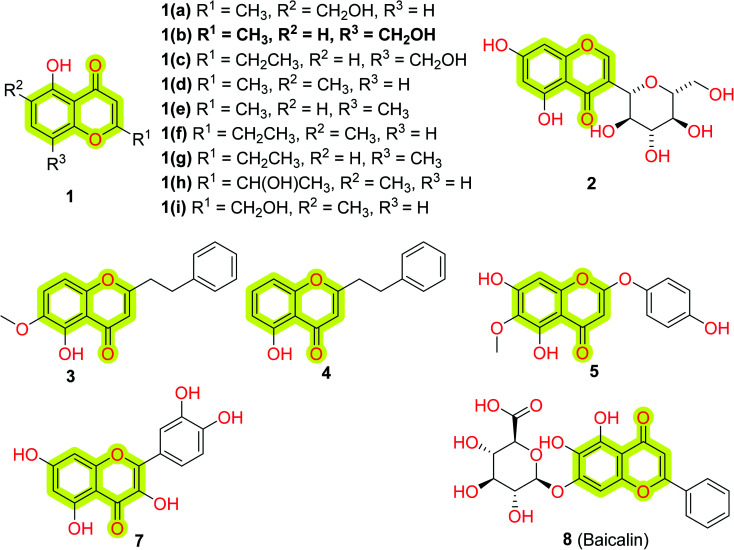
As we discussed, chromones originated from nature; therefore researchers working on natural products continuously try to identify and isolate new chromones from natural sources. For this, recently Kittisrisopit et al.48 identified and isolated nine new chromone analogs 1(a–i) from the soil actinomycete Microbispora sp. TBRC6027 as shown in Fig. 3.
Fig. 3. Chemical structure of natural chromone analogs 1 isolated from the soil actinomycete Microbispora sp. TBRC6027, macrolobin 2 from Macrolobium latifolium, 3, 4, 5, 7, and baicalin 8.
The authors established the chemical structures of the isolated chromones through NMR spectroscopy methods and examined their neuroprotective activity in vitro using P19-derived neurons. The reported results indicated that most of the compounds showed neuron viability against oxidative stress at the concentration of 1 ng mL−1 with a %viability of 44–53% without any significant neurotoxicity. The study suggested that the reported chromone analogs may be used as a model in designing potential neuroprotective agents. Another study of do Nascimento et al.61 also contributed towards the identification and isolation of a new unusual C-glycoside chromone from the crude methanolic extract of Macrolobium latifolium, named macrolobin 2, shown in Fig. 3. The authors used 1H, 13C, heteronuclear single quantum coherence (HSQC), and heteronuclear multiple-bond coherence (HMBC) NMR spectroscopy along with IR and mass spectrometry to establish the chemical structure of the isolated phytochemicals. The study indicated that macrolobin displayed significant inhibitory activity against AChE with an IC50 value of 0.8 μM along with some antimicrobial activity. The results suggested that further structural modification in 2 may turn it into a good inhibitor of AChE for the treatment of Alzheimer's disease. Recently, He et al.62 isolated two chromones, 3 and 4, from the resinous heartwood of Aquilaria sinensis (Thymelaeaceae) (Fig. 3) and evaluated them for their neuroprotective activities using models of BACE1 inhibition and PC12 cells with corticosterone- and 1-methyl-4-phenylpyridine ion (MPP+)-induced damage. Kim et al.63 analyzed the anti-oxidative and anti-inflammatory response of natural chromone capillarisin 5 (Fig. 3) by activating Nrf2/HO-1 signaling in neuroblastoma SHSY5Y cells and microglial BV2 cells. The authors reported that 5 leads to Nrf2 phosphorylation, upregulation of downstream molecules such as heme oxygenase-1 (HO-1) and NAD(P)H:quinone oxidoreductase 1, and subsequent activation of antioxidant response element (ARE)-mediated transcription. The study indicated that 5 possessed inflammatory responses in lipopolysaccharide-treated BV2 cells and protects SH-SY5Y cells from induced oxidative stress. The authors discussed that chromone 5 induces the activation of c-Jun N-terminal kinase in SH-SY5Y and BV2 cells which lead to the phosphorylation of Nrf2 and HO-1 upregulation.
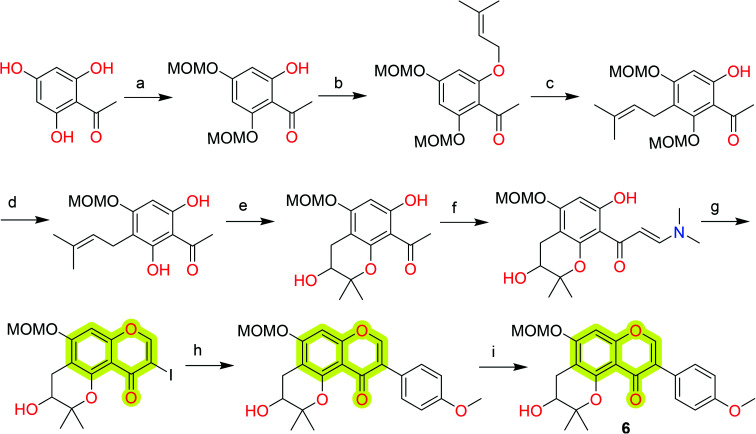
Hiep et al.64 isolated ten new isoflavones along with some known isoflavones from the extract of fruits of Cudrania tricuspidata and analyzed their neuroprotective effect against 6-hydroxydopamine-induced cell death in human neuroblastoma SH-SY5Y cells. The study came out with the most potent compound 6 with an EC50 value of 0.5 μM for human neuroblastoma SH-SY5Y cells. Later, Lu et al.65 reported the total synthesis of compound 6 using Claisen rearrangement and Suzuki coupling reaction as the key steps that are discussed in Scheme 1. They utilized a modified Mosher's method to elucidate the absolute configuration of naturally occurring chromone 6. A natural chromone, quercetin 7 (Fig. 3), also has been reviewed for its neuroprotective, MAO, and AChE inhibition properties.66,67
Scheme 1. Total synthesis of the natural chromone 6. Reagents and conditions: (a) MOMCl, DIPEA, DCM, 0 °C to room temperature (rt); (b) prenyl bromide, K2CO3, acetone, reflux; (c) N,N-diethylaniline, microwave; (d) 2N HCl, DCM, MeOH; (e) m-CPBA, M-K10, DCM; (f) DMF-DMA, reflux; (g) I2, MeOH, rt; (h) 4-methoxyphenylboronic acid, Pd(OAc)2, K2CO3, PEG-400; and (i) 3N HCl, DCM, MeOH, reflux.
Interesting studies were found in the literature discussing the critical role of a natural chromone baicalin 8 (Fig. 3) to promote NSPCs toward neuronal differentiation68 and prevent depressive-like behaviors in a chronic mild stress (CMS) animal model.69 A study by Zhang et al.70 highlighted that 8 promotes hippocampal neurogenesis via SGK1- and FKBP5-mediated glucocorticoid receptor phosphorylation in a neuroendocrine mouse model of anxiety/depression. Gao et al.71 highlighted that 8 promotes neurogenesis through modulating APPL2/glucocorticoid receptor signaling cascade and reduced emotional and olfactory dysfunctions in chronic corticosterone-induced depression. Another study highlighted that 8 inhibits activation of the GSK3β/NF-κB/NLRP3 signal pathway and shows remarkable neuroprotective effects in a rat model of depression.72 Zhang et al.73 explored that 8 was able to promote neuronal differentiation and survival through the Akt/FOXG1 pathway and could reverse the reduction of p-Akt, FOXG1, and FGF2 caused by chronic unpredictable mild stress (CUMS)-induced depression; it was also supported by the study of Fang et al.74 Recent studies highlighted that 8 acts through several pathways such as the BDNF/ERK/CREB signaling pathway,75 Wnt/β-catenin pathway,76 and activation of TLR4/MYD88/caspase-3 pathway77 to improve cognitive dysfunctions induced by CUMS and promotes neurogenesis in an animal model of depression. Recently, Li et al.78 explored 8 to analyze its antiepileptic effects and found that it exhibited a significant antiepileptic effect by regulating astrocyte phenotype to maintain systemic homeostasis. The authors reported that 8 suppresses neuron autophagy and apoptosis in pentylenetetrazol-induced epileptic rats and PC12 cells. These results highlighted the importance of chromones in neurogenesis and it could be a prominent template to develop new molecules for neurogenesis by targeting different pathways.
2.2. Synthetic chromones
Synthetic chromones attracted medicinal chemists due to the widespread possibilities for the design and development of new therapeutic small molecules of structural versatility with suitable different biological properties. As evidenced from the literature, structurally versatile chromone derivatives were explored for their biological activities such as anti-neurodegenerative, anti-allergic, anti-inflammatory, anti-diabetic, antitumor, antimicrobial, etc. in the past decade. Accordingly, this review will report only chromones including the data related to anti-neurodegenerative activity and described for the first time in the last 5 years along with their synthetic route.
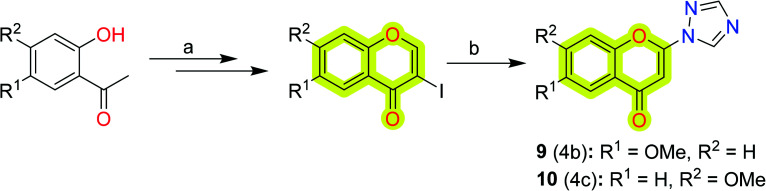
Towards the development of new potent chromone-based inhibitors of MAO-A and MAO-B for the treatment of neurodegenerative diseases, Takao et al.79 reported a series of 2-azolylchromone derivatives with their respective inhibitory activities, shown in Scheme 2. The study indicated that the derivatives showed potent inhibitory activities against MAO-A with an IC50 value of 0.023–0.32 μM with the most potent compound 9, while against MAO-B, the IC50 value was reported to be 0.019–0.73 μM with the most potent compound 10. The authors suggested that 6-methoxy substitution was good for MAO-A inhibition, and 7-methoxy substitution was feasible for inhibition of MAO-B. The study highlights the importance of 2-triazolylchromone in designing new MAO inhibitors.
Scheme 2. Synthetic protocol for the 2-azolylchromone derivatives. Reagents and conditions: (a) (i) DMF-DMA, 95 °C, (ii) pyridine, I2, CHCl3; (b) azole, K2CO3, DMF, 80 °C.
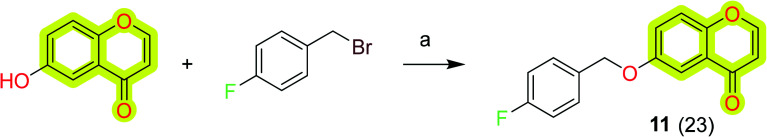
Wang et al.80 reported a series of structurally diverse benzyloxy substituted small molecules towards monoamine oxidase inhibition as anti-neurodegenerative agents. The study included the MAO-A and MAO-B inhibition data and neuroprotective effects investigated in 6-OHDA- and rotenone neurotoxin-treated PC12 cells. The results indicated that most of the compounds of the study exhibited potent inhibition activity against the MAO enzyme and an increase in the survival of PC12 cells treated with neurotoxins. The chromone-based compound 11 (Scheme 3) was reported with an IC50 value of 15.62 nM and 13.61 μM for MAO-B and MAO-A, respectively. Compound 11 shows 115% and 106% survival of 6-OHDA- and rotenone neurotoxin-treated PC12 cells. The results highlighted the importance of the chromone scaffold for the development of therapeutic molecules for Parkinson's disease.
Scheme 3. Synthesis of compound 11. Reagents and conditions: (a) K2CO3, acetonitrile, reflux, 8 h.
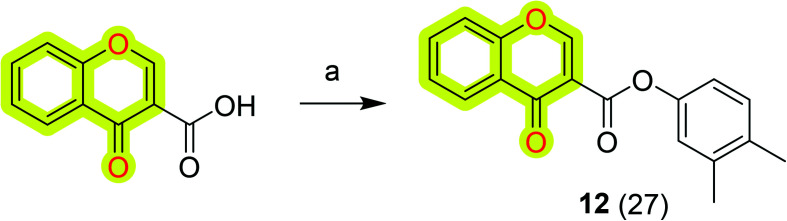
Further, Reis et al.81 synthesized and biologically evaluated chromone-based MAO-B inhibitors by following the lead optimization approach. The results highlighted that the compound N-(3′,4′-dimethylphenyl)-4-oxo-4H-chromene-3-carboxamide 12 (Scheme 4) was the most potent against MAO-B with an IC50 value of 0.67 nM that was acting as a non-competitive reversible inhibitor. The authors discussed that the amide spacer and the direct linkage of the carbonyl group to the γ-pyrone ring along with the presence of meta and para substituents in the exocyclic ring were highly important for its improved biological activities. Compound 12 also possessed a favorable toxicological profile and physicochemical properties that pointed toward BBB permeability; thus the authors suggested it as a valid candidate for subsequent animal studies.
Scheme 4. Synthetic route for compound 12. Reagents and conditions: (a) POCl3, DMF, N-methylaniline, rt, 1–5 h.
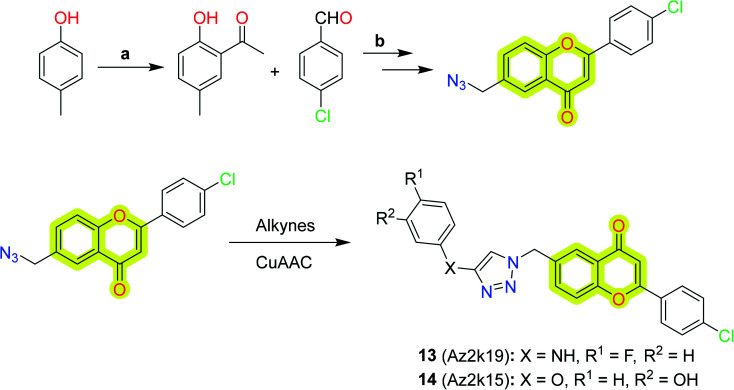
Jia et al.82 explored flavone-based MAO inhibitors with structural versatility by retaining the core chromone scaffold using click chemistry (CuAAC reaction) between 6-N3-2-phenyl chromones and alkynes, shown in Scheme 5. The study indicated that compound 13 was the best compound with inhibitory activity IC50 = 1.6 μm for MAO-A and 2.1 μm for MAO-B, while the compound with better selectivity (SI >14.0) towards MAO-B was 14. The study will be helpful in designing new MAO inhibitors from C6 substitution of flavone derivatives for the treatment of neurodegenerative diseases.
Scheme 5. Synthesis of flavone-based MAO inhibitors using click chemistry (CuAAC reaction). Reagents and conditions: (a) 1. CH3COCl, pyridine; 2. AlCl3, 130 °C; (b) 1. Ba(OH)2, EtOH; 2. I2, DMSO, 130 °C; 3. NBS, BPO, CCl4, reflux; 4. NaN3, DMF.
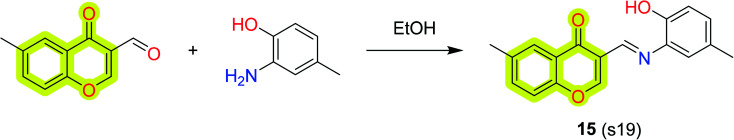
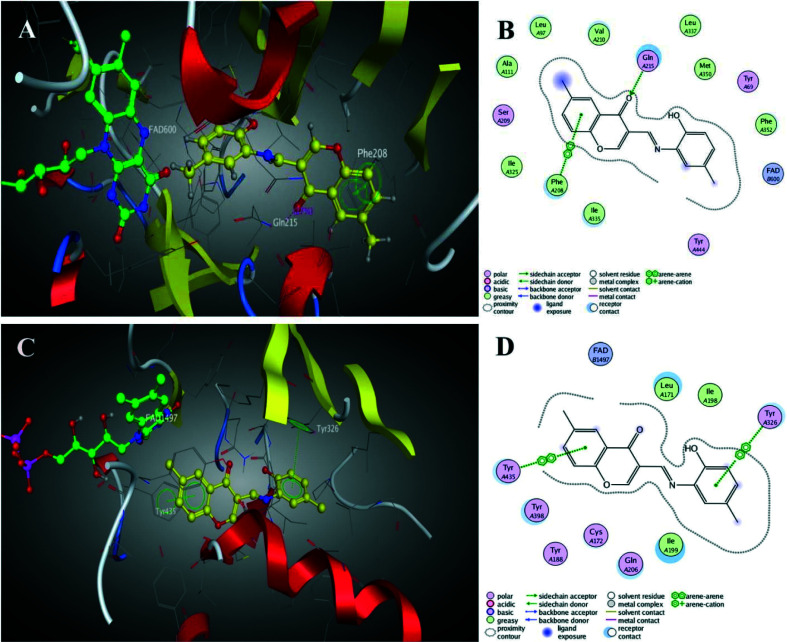
Li et al.83 reported the synthesis and pharmacological evaluation of novel chromone derivatives with multi-target activity as an anti-Alzheimer's agent, shown in Scheme 6. The authors reported that the best compound 15 exhibits good inhibitory potency for hMAO-A with IC50 values of 5.12 μM and 0.816 μM for hMAO-B. Moreover, the compound also inhibited 75% aggregation of amyloid-β at 20 μM with metal chelation, control of ROS generation, and antioxidant activity (ORAC = 3.62). The authors also reported molecular docking results using MOE 2008.10 and found that compound 15 established a π–π stacking interaction with Phe208, while a carbonyl group was involved with Gln215. The docking image shown in Fig. 4 indicated that the chromone moiety was sited within the substrate cavity of the enzyme, in close proximity to the flavin adenine dinucleotide (FAD) cofactor, while the phenyl group was involved in making a π–π stacking interaction with Tyr435. The other interactions were also discussed in the parent study which indicated that the compound has well interacted with the protein. The results indicated that the compound was able to reduce PC12 cell death induced by oxidative stress and penetrate the blood–brain barrier (BBB).
Scheme 6. Synthesis of the designed compound 15.
Fig. 4. (A) 3D docking model of 15 with MAO-A. (B) 2D schematic diagram of docking model of 15 with MAO-A. (C) 3D docking model of 15 with MAO-B. (D) 2D schematic diagram of docking model of 15 with MAO-B (adapted with permission from ref. 83. Copyright 2017 Elsevier).
Chromone-based hybrid analogs were also reported as an effective molecule against neurodegenerative disorders. Nesi et al.84 presented a series of chromone–rivastigmine-based new anti-Alzheimer's agents (Scheme 7) with both AChE and BuChE inhibitory activity and % inhibition of the self-mediated amyloid-β1–42 aggregation. The study indicated that 2-chromonecarboxylic acid hybrid 16 exhibited inhibitory activity with an IC50 for BuChE of 511 nM and able to inhibit 22% AChE at 500 nM concentration. Compound 16 was also able to inhibit self-mediated amyloid-β aggregation with 67% inhibition. The results indicated that further investigation and structural improvements may be helpful in elucidating the importance of their anti-neurodegenerative profile.
Scheme 7. Synthesis of chromone–rivastigmine hybrid compounds (a) 4-oxo-4H-1-benzopyran-2-carboxylic acid, BOP, TEA, rt, 12 h.
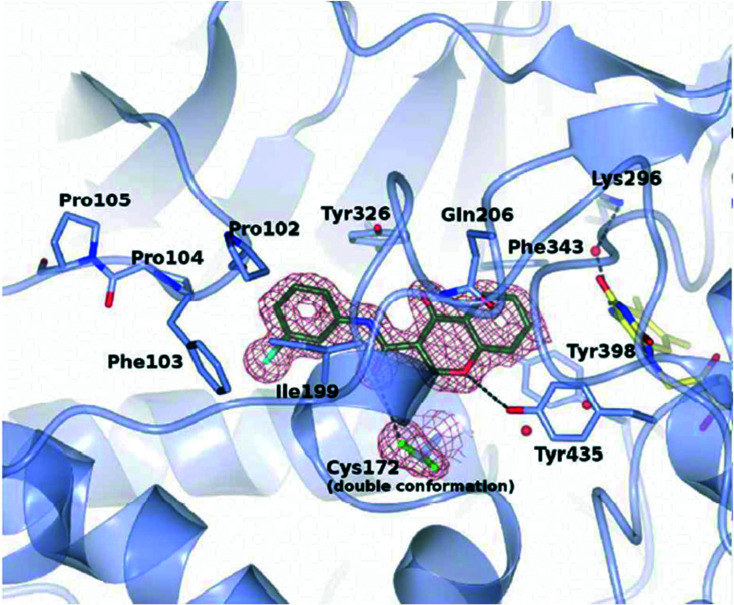
Reis et al.85 reported chromone derivatives as novel potent and reversible hMAO-B inhibitors and discussed their inhibition mechanism through crystallographic and biochemical analysis. The authors disclosed that the chromone moiety of the inhibitors is located in front of the FAD cofactor and all bind in the active site cavity of hMAO-B, as shown in Fig. 5.
Fig. 5. Crystal structure zoomed view of hMAO-B active site in complex with chromone inhibitor 17 (PDB code 6FW0) (adapted with permission from ref. 85. Copyright 2018 American Chemical Society).
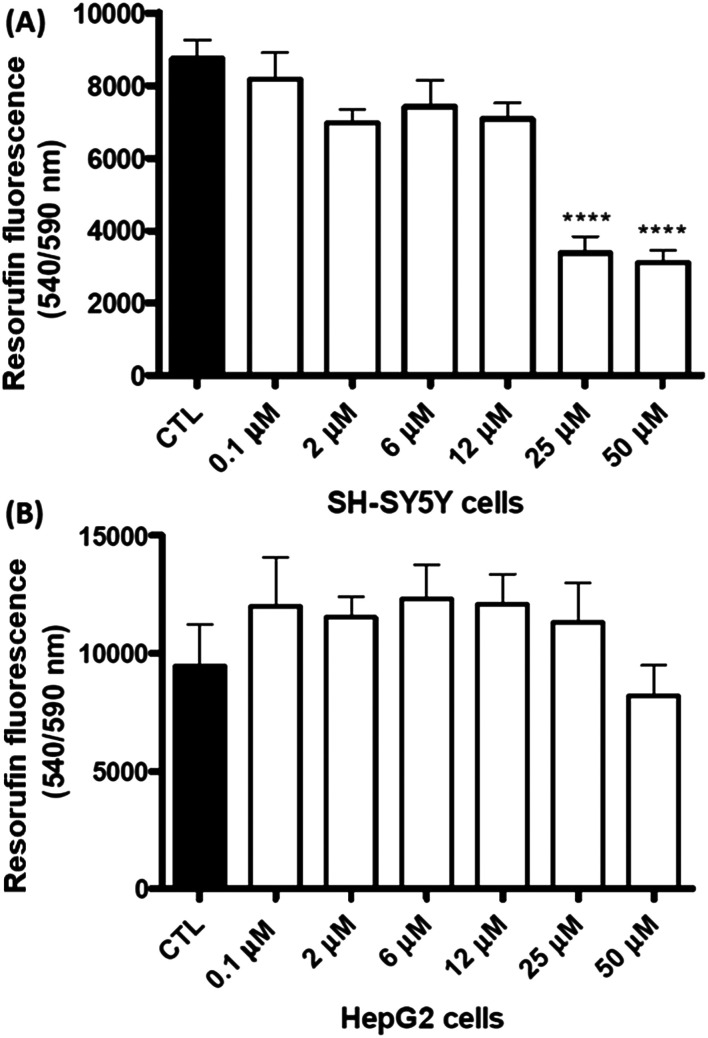
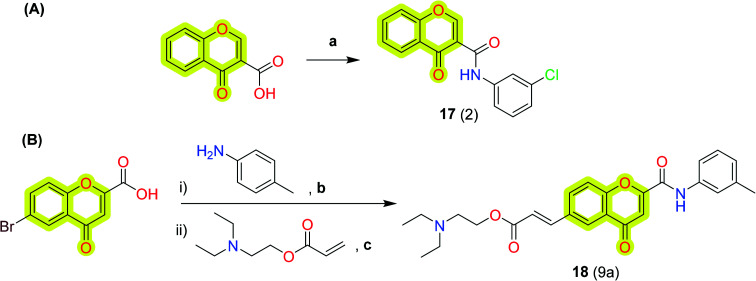
The results evidenced that the compounds are well fit with the hydrophobic site of the protein and formed two hydrogen bonds with Tyr435 and Cys172. The efficient inhibition of the compounds was also evidenced with a Ki value of 17 nM for the compound N-(3′-chlorophenyl)-4-oxo-4H-chromene-3-carboxamide 17. The authors stated that the reported compounds were 1000-fold more effective than l-deprenyl in reducing the cellular levels of reactive oxygen species (ROS). Later, the authors86 again reported a new study on chromone-based new derivatives with hAChE and MAO activities. The best compound 18 was reported with IC50 and Ki values of 0.21 μM and 0.19 μM for hAChE and dual inhibitory activity for hMAO-A and hMAO-B with IC50 and Ki values of 0.94, 0.057 μM and 3.81, 0.48 μM, respectively. The authors recorded the cytotoxicity profile of compound 18 in differentiated human neuroblastoma (SH-SY5Y) and human hepatocarcinoma (HepG2) cells after a 48 h incubation period as shown in Fig. 6 and found that compound 11 exhibited significant cytotoxic effects towards SH-SY5Y cells at 25 and 50 μM concentration, while it was safe on HepG2 cells, demonstrating low-risk drug-induced hepatotoxicity. The synthetic protocol of the reported inhibitors 17 and 18 is shown in Scheme 8.
Fig. 6. Cytotoxicity profile of compound 18 measured by changes in cellular metabolic activity on (A) human neuroblastoma SH-SY5Y and (B) human hepatocarcinoma HepG2 cells (adapted with permission from ref. 86. Copyright 2018 Elsevier).
Scheme 8. Synthesis of the chromone-based hMAO-B inhibitors (A) 17 and (B) 18. Reagents and conditions: (a) aniline derivatives, POCl3, DMF, rt, 1–5 h; (b) POCl3, DCM, DMF; (c) NaHCO3, TBAB, Pd(OAc)2.
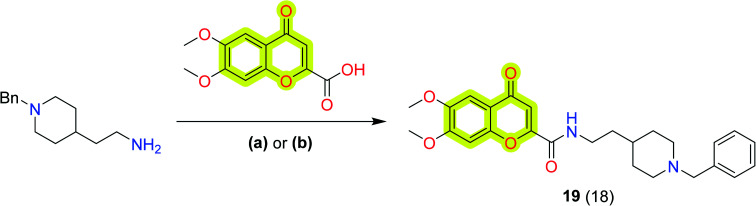
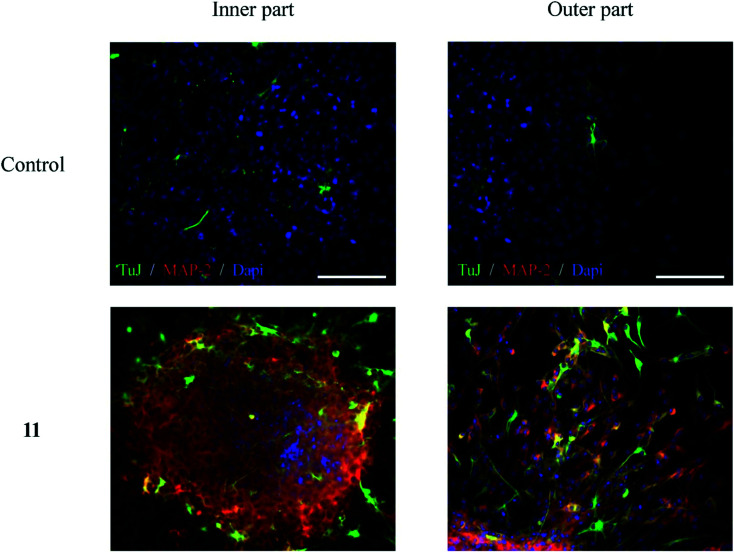
In another study, Valencia et al.87 synthesized a series of donepezil–chromone-based hybrid molecules for the treatment of Alzheimer's disease which exhibited nanomolar affinities for the sigma-1 receptor (s1R) and inhibition of key enzymes such as AChE, lipoxygenase-5 (LOX-5), and MAOs. The authors described compound 19 (Scheme 9) as the best compound of the study with IC50 values of 46 ± 4 nM, 74 ± 3 μM, 15 ± 1 μM, and 5 ± 0.3 μM for hAChE, LOX-5, hMAO-A, and hMAO-B, respectively. The affinity constant Ki towards hAChE, σ1, and σ2 receptors was reported as 39, 37 and 239 nM, respectively. The compound also reported neuroprotection activity against mitochondrial oxidative stress. Further, to evaluate compound 19 as a neurogenic agent, the authors isolated adult mice neural stem cells (NSCs) from the subgranular zone of the hippocampal dentate gyrus and cultured them as neurospheres (NSs) for 7 days in the presence and absence of the compound. As shown in Fig. 7, compound 19 promoted the differentiation of NSCs and their further maturation to a neuronal phenotype. To visualize early proliferation and neuronal maturation, β-III-tubulin (clone TuJ1; green) and microtubule-associated protein 2 (MAP-2: red) antibodies were used, respectively, inside the neurosphere (inner part) and in the distal area (outer part). The exploration of these hybrid compounds may turn them into a new clinical candidate for the treatment of Alzheimer's disease.
Scheme 9. Synthetic protocol for the preparation of donepezil–chromone-based hybrid molecule 19. Reagents and conditions: (a) (i) acid, CDI, DMF, mw, 120 °C, 10 min, (ii) amine, mw, 150 °C, 10 min; (b) BOP, Et3N, DMF, overnight, rt.
Fig. 7. In vitro neurogenic effect of hybrid 19 (10 μM) on mice hippocampal SGZ derived spheres (adapted with permission from ref. 87. Copyright 2018 Elsevier).
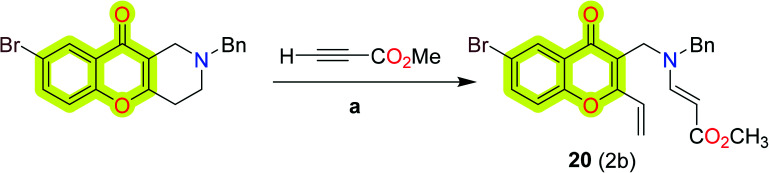
Makhaeva et al.88 reported a series of substituted chromeno[3,2-c]pyridines as selective BuChE inhibitors for potential Alzheimer's disease therapeutics. The enzyme inhibitory activity indicated that compound 20 was the most potent compound of the study with IC50 and Ki values of 2 and 2 μM for hBuChE, respectively. The synthetic protocol for compound 20 is shown in Scheme 10.
Scheme 10. Reactions of chromeno[3,2-c]pyridine with activated alkynes. Reagents and conditions: (a) (i) MeOH, rt.
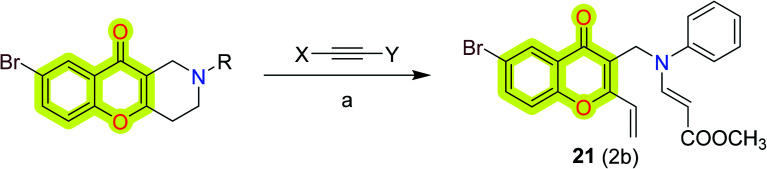
Makhaeva et al.89 synthesized and reported a series of substituted chromeno[3,2-c]pyridines for the inhibition of BuChE as anti-Alzheimer's agents. The study also contained the inhibitory assay of the synthesized compounds against AChE, BuChE, and carboxylesterase (CaE) by following the methods of enzyme kinetics and molecular docking. The results have shown that compound 21 (Scheme 11) was the most potent against BuChE with an IC50 value of 2.27 μM and Ki of 1.55 μM.
Scheme 11. Synthetic route for the compound 21. Reagents and conditions: (a) X = H, Y = CO2Me, MeOH, rt.
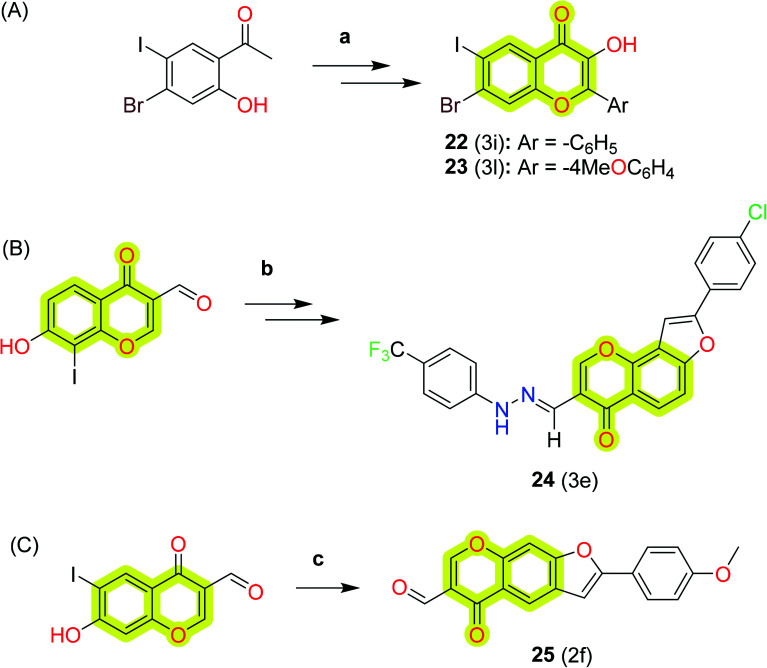
Mphahlele et al.90 synthesized a series of substituted 2-aryl-3-hydroxy-6-iodo-4H-chromen-4-ones and evaluated the inhibitory activities against AChE, BuChE, and β-secretase (BACE1) in vitro and reported that chromone-based chalcone derivative 22 exhibited a significant inhibitory effect against AChE and BuChE with an IC50 value of 10 and 6 μM, respectively, and compound 23 exhibited IC50 = 16 μM for BACE-1. The modes of binding of the reported inhibitors with the protein were evaluated using molecular docking studies and could be accessed from the original research article. Later, the authors also explored 4-oxo-4H-furo[2,3-h]chromene derivatives91 as potential multi-target inhibitors for cholinesterases, BACE1, and cyclooxygenase-2 and LOX-5 and LOX-15. The authors reported compound 24 as the most potent compound of the study with IC50 values of 5 μM, 10 μM, 14 μM, 10 μM, 15 μM, and 29 μM for AChE, BuChE, BACE1, COX-2, LOX-15, and LOX-5, respectively. Further, they reported 5-oxo-5H-furo[3,2-g]chromene-6-carbaldehyde derivatives92 as potential anti-Alzheimer's agents. The study suggested that the compound 2-(4-methoxyphenyl)-5-oxo-5H-furo[3,2-g]chromene-6-carbaldehyde 25 was the most potent against hAChE, hBuChE, LOX-15, and LOX-5 with in vitro IC50 values of 10 μM, 5 μM, 10 μM, and 15 μM, respectively. DPPH antioxidant activity was reported at 18 μM for compound 25. The results indicated that compound 25 was found to be cytotoxic against the breast cancer MCF-7 cell line. The synthetic protocol to obtain compounds 22, 23, 24, and 25 is shown in Scheme 12.
Scheme 12. The synthetic protocol to obtain compounds (A) 22 and 23, (B) 24, and (C) 25. Reagents and conditions: (a) (i) RC6H4CHO, 5% KOH (aq), EtOH, rt, 78 h, (ii) 3 M KOH (aq), H2O2, MeOH, 0 °C-rt, 3 h; (b) (i) alkyne, PdCl2(PPh3)2, CuI, PPh3, K2CO3, DMF (aq), 0 °C, 3 h, (ii) 4-CF3C6H4NHNH2, EtOH, pyridine, reflux, 6 h; (c) alkyne, PdCl2(PPh3)2, CuI, K2CO3, DMF (aq), 70 °C, 3 h.
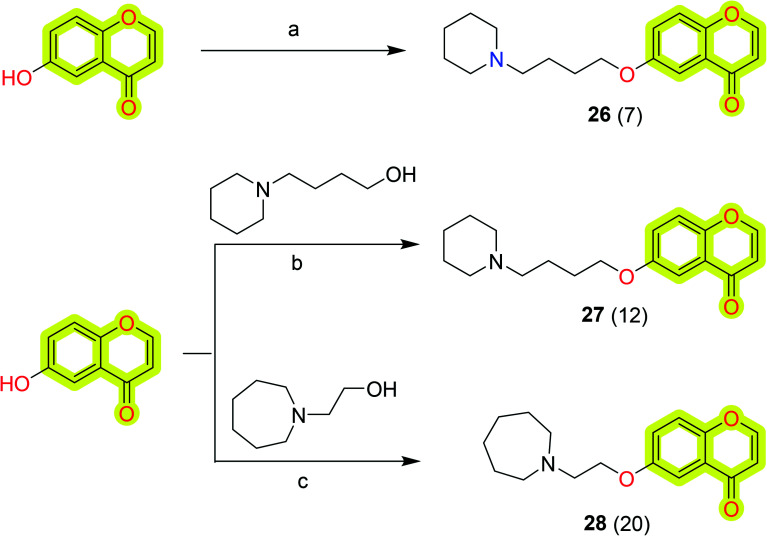
Further, Lemke et al.93 synthesized a library of substituted chromen-4-ones as multi-target-directed ligands for neurodegenerative therapy. The results highlighted the compound 6-(4-(piperidin-1-yl)butoxy)-4H-chromen-4-one 26 as the dual-target most potent inhibitor against AChE and MAO-B with IC50 values of 5.58 and 7.20 μM, respectively. The authors also developed the molecules 27 and 28 in the study that were later reported by Deuther-Conrad et al.94 as an extension of the study to analyze the affinity towards sigma (σ) receptors, potential agents for the treatment of several disorders, including Alzheimer's disease and neuropathic pain. The authors highlighted compound 6-((5-morpholinopentyl)oxy)-4H-chromen-4-one 27 as the most potent compound of the study with a Ki value of 19.6 nM to hσ1 receptor and selectivity index of 130 towards rat σ2 receptor. Another molecule of the study, 6-(3-(azepan-1-yl)propoxy)-4H-chromen-4-one 28, was reported with a Ki value of 27.2 nM for σ1 and selectivity (σ1/σ2) = 28, that combined the desired σ1 receptor affinity with a dual inhibitory capacity against both AChE and BuChE with an IC50 value of 10.6 and 25.0 μM, respectively. The study supported the future development of chromone-based molecules for neurodegenerative therapy. The synthetic procedure to achieve compounds 26, 27, and 28 is shown in Scheme 13.
Scheme 13. Synthesis of the compounds 26, 27, and 28. Reagents and conditions: (a) (i) (CH2)4Br2, CH3CN, K2CO3, 50 °C; (ii) piperidine, Et3N, 50 °C; (b) cyanomethylenetributylphosphorane (CMBP), toluene, 100 °C; (c) Ph3P, DCM, di-tertbutyl azodicarboxylate (DBAD), rt.
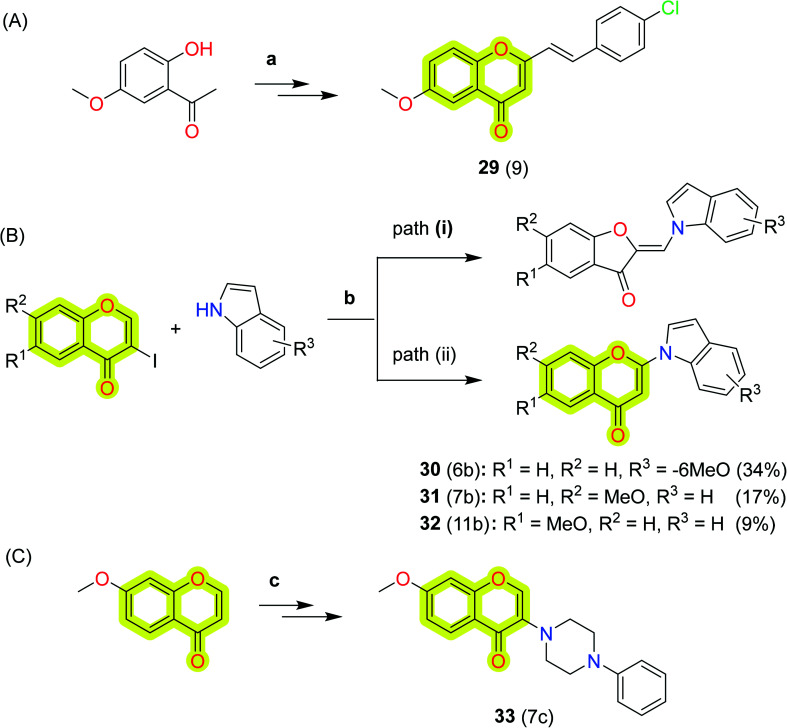
Takao et al.95 synthesized a series of 2-styrylchromone derivatives and reported their MAO-A and MAO-B inhibitory activities. The study highlighted compound 29 as the most potent against MAO-B with a minimal half-inhibitory activity of 17 nM and best selectivity over MAO-A of 1500. The mode of inhibition of compound 29 was reported as reversible, and quantitative structure–activity relationship (QSAR) analyses were also included with the study. These data suggested that the 2-styrylchromone structure might be a useful scaffold for the design and development of novel MAO-B inhibitors. The authors also explored 2-(indolyl)-4H-chromen-4-one derivatives96 as novel monoamine oxidase inhibitors for neurodegenerative therapeutics. The study indicated that compound 32 was active against both MAO-A and MAO-B with IC50 values of 0.32 μM and 0.63 μM, respectively, while compounds 30 and 31 were highly selective towards MAO-B with IC50 = 0.15 μM and selectivity index of >670. The authors reported molecular docking studies to explain the types of interaction of the reported inhibitors with the protein. Further, the authors explored 2- and 3-(N-cyclicamino)chromone derivatives97 as monoamine oxidase inhibitors and showed that the 3-(N-cyclicamino)chromone derivatives (with a few exceptions) significantly and selectively inhibit MAO-B. The authors marked compound 33, 7-methoxy-3-(4-phenyl-1-piperazinyl)-4H-1-benzopyran-4-one, as the most potent and selective inhibitor of MAO-B with an IC50 of 15 nM and an MAO-B selectivity index of more than 6700. To elucidate the mechanism of the inhibitory activity of compound 33, the authors used a molecular docking study which revealed that the mode of inhibition was competitive and reversible. The study revealed that the 3-(N-cyclicamino)-chromones are useful to lead compounds for the development of selective inhibitors of MAO-B. The synthetic protocol for the development of compounds 29, 30, 31, 32, and 33 is shown in Scheme 14.
Scheme 14. Synthetic protocol for the development of compounds (A) 29; (B) 30, 31, and 32; (C) 33. Reagents and conditions: (a) (i) Na, AcOEt, rt, (ii) conc. HCl, MeOH, rt, (iii) 4-chlorobenzaldehyde, NaOMe, MeOH, reflux; (b) K2CO3, DMF, 80 °C; (c) (i) H2O2, PhCH2N+(CH3)3 OH−, Et2O, 0 °C, (ii) cyclic amine, CH3CN, and rt.
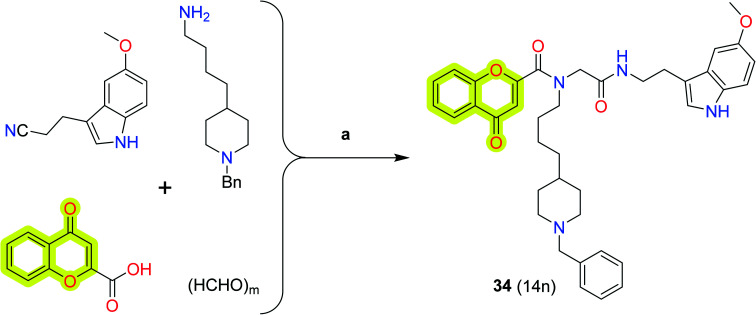
Pachón-Angona et al.98 synthesized new hybrid molecules from chromone–melatonin–donepezil for Alzheimer's disease therapeutics. The authors reported that their compounds exhibited multi-target activity. The study described that compound 34 (Scheme 15) was the most potent inhibitor for BuChE with an IC50 value of 12 nM along with moderate inhibition activity for hAChE, hMAO-A, and hMAO-B with IC50 values of 2 μM, 3 μM, and 21 μM, respectively, and strong antioxidant power (3.04 TE, ORAC test). The study is useful for the design and development of hybrid molecules for the treatment of neurodegenerative disorders.
Scheme 15. Synthesis of the donepezil–chromone–melatonin hybrid 34. Conditions: (a) MeOH/CH2Cl2 (3 : 1), rt, 24 h.
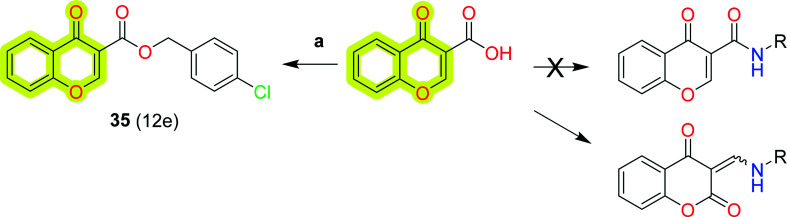
Mpitimpiti et al.99 reported the synthesis and MAO inhibition of chromone 3-carboxylic acid derivatives for neurodegenerative disease therapeutics. The authors reacted aromatic and aliphatic amines and alcohols with chromone 3-carboxylic acid in the presence of carbonyldiimidazole (CDI), which yielded chromane-2,4-dione and ester chromone derivatives as shown in Scheme 16. The study indicated that compound 35, 4-chlorobenzyl 4-oxo-4H-chromene-3-carboxylate, was a dual-acting inhibitor for both MAO-A and MAO-B with IC50 values of 19 μM and 10 μM, respectively. The study provides an insight to design MAO inhibitors for the treatment of neurodegenerative disease.
Scheme 16. Synthesis of chromone 3-carboxylic acid derivative 35. Reagents and conditions: (a) CDI, DMF, 60 °C, 2 h.
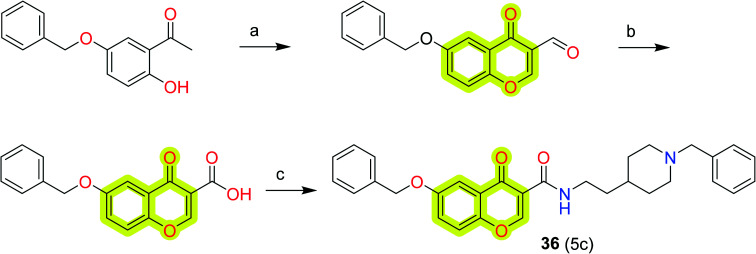
Wang et al.100 worked on a molecular hybridization approach to develop a new multi-target-directed ligand for the inhibition of cholinesterase and monoamine oxidase as anti-Alzheimer's agents. The study revealed compound 36 as the most potent inhibitor of BuChE, AChE, and MAO-B with IC50 values of 5.24, 0.37, and 0.272 μM, respectively. The authors discussed that compound 36 was a mixed-type inhibitor that binds concurrently to peripheral and active sites of AChE. It also acts as a competitive inhibitor of MAO-B which occupied the substrate and entrance cavities of the enzyme. The toxicity profile to rat pheochromocytoma (PC12) cells was reported within the limits that marked compound 36 as a multitarget drug lead molecule for the development of new potent anti-neurodegenerative agents. The synthetic route to achieve compound 36 is shown in Scheme 17.
Scheme 17. Synthesis of compound 36. Reagents and conditions: (a) POCl3, DMF, 0 °C, 2 h; (b) NaClO2, NH2HSO3, CH2Cl2, 0 °C, 3 h; (c) thionyl chloride, reflux; 2-(1-benzylpiperidin-4-yl)ethanamine.
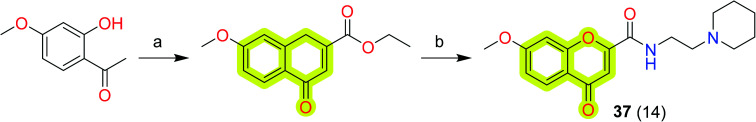
Suwanhom et al.101 synthesized chromone-2-carboxamido-alkylamines and reported them as potent acetylcholinesterase inhibitors for the treatment of neurodegenerative diseases. The authors marked compound 37 (Scheme 18) as the most potent compound with an IC50 value of 0.09 μM for AChE, which was higher than that of the clinical drug, tacrine. The results indicated that compound 37 was not cytotoxic against SH-SY5Y cells and was found to be neuroprotective. The authors concluded that compound 37 may be a promising lead candidate for the development of anti-Alzheimer's agents.
Scheme 18. Synthesis of compound 37. Reagents and conditions: (a) diethyl oxalate, NaH/THF, rt; (b) amine/CH2Cl2/reflux, then CH3COOH, 70 °C.
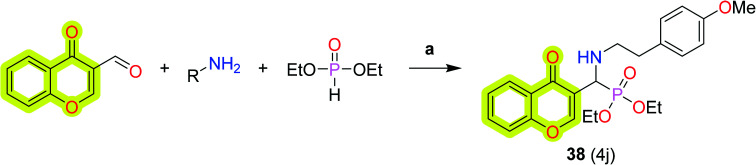
Further, a study by Shaikh et al.102 was related to the development of new chromone-derived aminophosphonates as a cholinesterase inhibitor using porcine pancreatic lipase (PPL) as a catalyst for the treatment of neurodegenerative diseases, as shown in Scheme 19. The authors found compound 38 as the most potent compound of the study with an IC50 value of 0.1 μM for AChE. The authors revealed that the aliphatic analogs of the study were efficiently inhibited AChE, while aromatic analogs were more potent for BuChE inhibition. The results defined that compound 38 was twofold more potent than tacrine, 35-fold more potent than galantamine, and 50-fold more potent than rivastigmine. The mode of binding with protein was explained using molecular docking studies which revealed that compound 38 binds to both the peripheral anionic site and the catalytic anionic site of AChE and BuChE. The antioxidant activities and DNA nicking activity were also performed which could be accessed from the original research.
Scheme 19. Synthesis of chromone-based α-aminophosphonate 38. Reagents and conditions: (a) PPL, solvent-free, 50 °C.
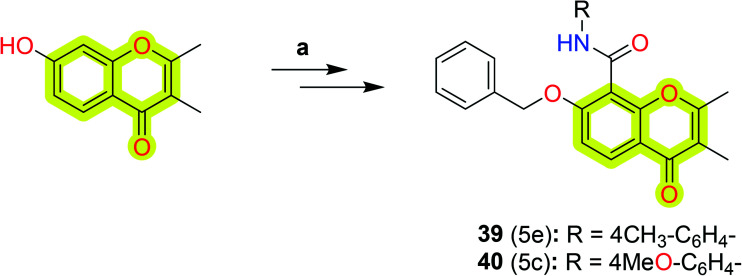
Rao et al.103 synthesized a series of bifunctional 7-benzyloxy-2,3-dimethyl-4-oxo-4H-chromene-8-carboxamide derivatives (Scheme 20) as potent and selective hMAO-B inhibitors using Jones oxidation and HBTU/HOBt as selective amide coupling agents. The authors marked compound 39 as the most potent against hMAO-B with an IC50 value of 0.1 μM (while 4 μM for hMAO-A) and compound 40 as the most selective towards hMAO-B with an IC50 value of 0.1 μM and selectivity index of 323.97. The study indicated that compound 39 showed reversibility of MAO-B inhibition in a dialysis method with a relative recovery percentage of up to 70%.
Scheme 20. Synthetic route of 7-benzyloxy-2,3-dimethyl-4-oxo-4H-chromene-8-carboxamides (39 and 40). Reagents and conditions: (a) (i) AcOH, HMTA, 90 °C, 10 h; (ii) PhCH2Br, K2CO3, 0–80 °C, 5 h; (iii) Jones reagent, 0 °C, 4 h; (iv) R-NH2, HBTU–HOBT, rt, 20 h.
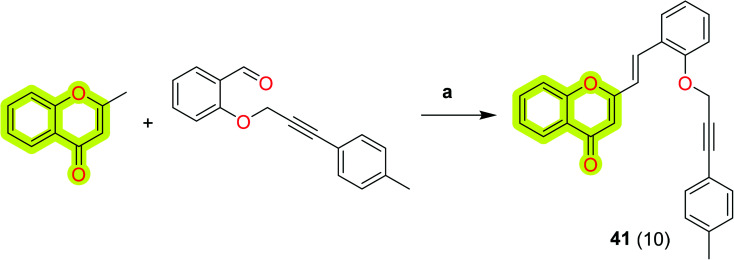
Malafaia et al.104 reported the design, synthesis, and biological evaluation of a family of 2-styrylchromones as AChE and amyloid-β aggregation dual inhibitors. The authors reported that compound 41 (Scheme 21) was observed to be a well-balanced dual-target inhibitor with an IC50 value of 3 μM for eeAChE and inhibitory percentage of 66% for amyloid-β aggregation. The molecular docking results of the study indicated that most of the compounds were bound to the AChE via H-bonds with the residues of the catalytic triad and π-stacking interactions. The promising activities of the developed compounds will be useful in designing new multi-target molecules for Alzheimer's disease therapeutics. The SAR profile of the compound towards eeAChE is shown in Fig. 8.
Scheme 21. Synthesis of (E)-2-styrylchromone 41. Reagents and conditions: (a) Na, EtOH, rt, 5 h.
Fig. 8. SAR profile towards eeAChE of (E)-2-styrylchromones (adapted with permission from ref. 104. Copyright 2021, licensed under CC BY 4.0).
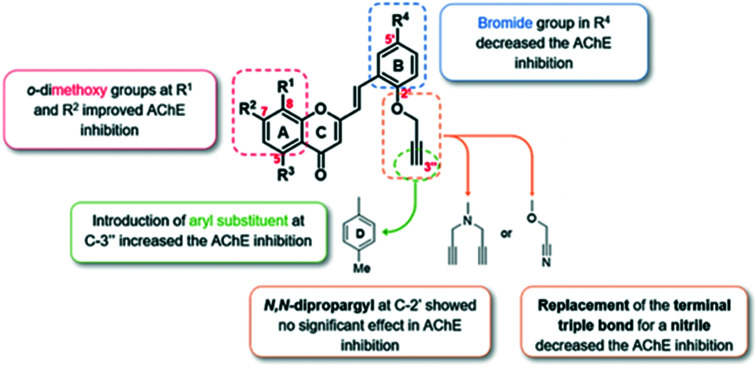
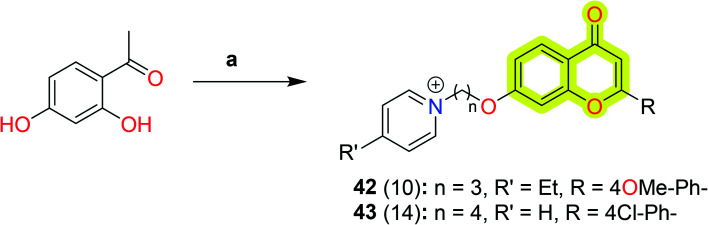
Abdpour et al.105 explored a family of 7-hydroxy-chromone derivatives bearing a pyridine moiety as multi-target-directed ligands against Alzheimer's disease. The authors reported that most of the compounds were found to be competitive AChE inhibitors with an IC50 value of 10–0.7 μM and potent inhibitors of BuChE with an IC50 value of 2–0.006 μM. The results indicated that compounds 42 and 43 showed the best multi-target efficacy with IC50 values of 3 μM and 0.7 μM for eeAChE, respectively, and 0.006 μM and 2 μM for equine serum BuChE, respectively. Both compounds 42 and 43 also showed remarkable amyloid-β inhibition against both self-induced amyloid-β aggregation and AChE-induced amyloid-β inhibition with a percent inhibition of 32% and 37% and 28% and 22%, respectively. The study indicated that both compounds showed acceptable neuroprotective activity on H2O2- and amyloid-β-induced neurotoxicity in PC12 cells, more than that of standard drugs, which suggested that the compounds may be a pioneer in designing chromone-based multi-target ligands for the treatment of Alzheimer's disease. The synthetic protocol to obtain compounds 42 and 43 is shown in Scheme 22.
Scheme 22. Synthetic routes to compounds 42 and 43. Reagents and conditions: (a) (i) KOH (30%), MeOH, appropriate benzaldehyde, stir, 72 h, then DMSO, I2, reflux, 45 min; (ii) Br(CH)nBr (n = 3, 4), anhydrous K2CO3, acetone, reflux, 4 h; (iii) pyridine, 110 °C, 24 h.
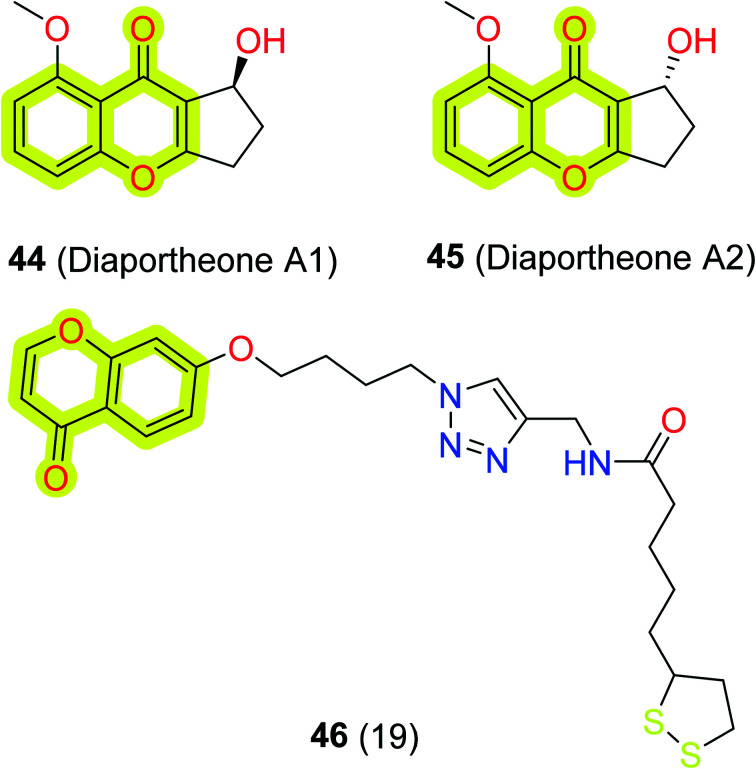
To identify chromones as neuroprotective and amyloid-β disaggregating agents, Tan et al.106 investigated the efficacy of synthetic chromones, diaportheones 44 and 45, shown in Fig. 8. The authors used thioflavin T (ThT) assay to evaluate the impact of both chromones on amyloid-β and the cell viability, ROS, and mitochondrial membrane potential were evaluated with human neuroblastoma SH-SY5Y cells. The study indicated that the diaportheones 44 and 45 were able to inhibit the aggregation of amyloid-β by 80% and 74%, respectively, and both compounds showed neuroprotection on SH-SY5Y cells induced by amyloid-β and H2O2. Moreover, Jalili-Baleh et al.107 synthesized multi-functional chromone–lipoic acid conjugates through click reaction and evaluated their neuroprotective, anticholinesterase, anti-amyloid aggregation, antioxidant, and metal-chelation activities. The authors reported that compound 46 (Fig. 9) was the most potent against BuChE with an IC50 value of 8 μM and discussed that it has shown non-competitive mixed-type inhibition of BuChE analyzed through docking and kinetic studies. The study indicated that the compound was also moderately able to inhibit self-mediated amyloid-β aggregation (13%) and selectively chelate with copper ions in a 2 : 1 M ratio. The results highlight the significance of the chromone scaffold in designing multi-target ligands for the treatment of neurodegenerative diseases.
Fig. 9. Chemical structures of compounds 44, 45, and 46.
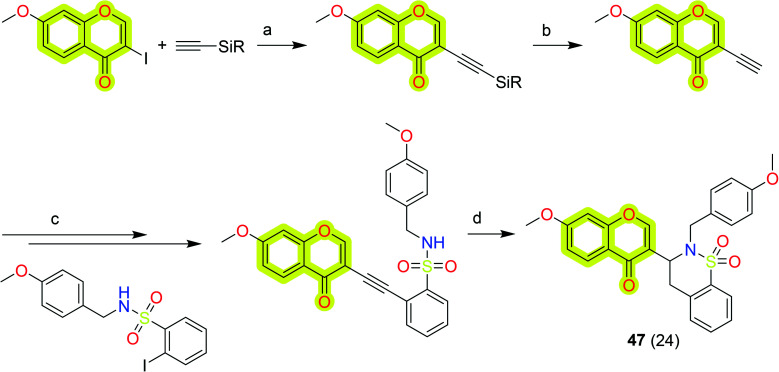
As a molecular hybrid molecule development, the development of a series of isoflavone/benzo-δ-sultam hybrids was reported by Mengheres et al.,108 who analyzed them as potential anti-inflammatory and neuroprotective agents in LPS-activated BV2 microglia. The authors synthesized these hybrid molecules using a two-step reaction by coupling 2-halobenzenesulfonamide derivatives with terminal alkynes, followed by a 6-endo-dig cyclization. The authors performed a biological assay for the inhibition of TNF-α production and nitric oxide (NO) in LPS-stimulated BV2 microglial cells. The results indicated that the most potent hybrid compound, 47, of the study reduced NO production to 41% and TNF-α to 34% at 20 μM, which could be taken for further development. The synthetic route to achieve chromone 47 is shown in Scheme 23.
Scheme 23. Synthetic route for the synthesis of compound 47. Reagents and conditions: (a) (i) [Pd(PPh3)2Cl2] (3 mol%), CuI (20 mol%), THF, TEA, N2, 0 °C; (ii) 3 h, rt, N2; (b) (i) TBAF, CSA, THF, 0 °C; (ii) rt, 3 h; (c) (i) 10% Pd/C (3 mol%), PPh3 (12 mol%), CuI (5 mol%), vacuum/N2; (ii) MeCN, 10 min, N2, 0 °C; (iii) Et3N, N2, 0 °C, 5 min; (iv) 80 °C, overnight; (d) AgNO3 (20 mol%), Et3N, EtOH, N2, 80 °C, 10 min.
3. Concluding remark and future perspective
The evidence presented in this mini-review defines that the treatment of neurodegenerative diseases may be done through actions in several molecular mechanisms, which include inhibition of enzymes such as cholinesterases, BACE1, MAOs, etc., inhibition of amyloid-β formation or disaggregation, and controlling other mechanisms such as oxidative stress, τ-hyperphosphorylation, inflammation, and metal regulation, among others. Currently, due to limitations of available drugs which provide only short-term improvements from a symptomatic perspective, medicinal chemists are continuously working to find new molecules which could be able to seize or cure neurodegenerative diseases completely. Over the past years, the advancement of pharmacologically active molecules highlighted the fruitfulness of the privileged structure concept. These privileged scaffolds exhibit good drug-like properties, possibilities to develop structurally diverse small molecules, and versatile binding properties such as chromone.
The use of the chromone scaffold in medicinal chemistry to develop new anti-neurodegenerative molecules is due to its excellent biological properties and absence of toxic effects. Nevertheless, the structural differences between derivatives tend to direct these compounds toward the desired activities, without taking the risk of off-target effects, in a semi-selective action. Moreover, the potential of chromones for neurodegenerative therapeutics, widely preferred in the last five years, glorified this scaffold for further structural optimizations. The research included in the review indicates that chromones are one of the preferred scaffolds to develop new multi-target molecules for the treatment of neurodegenerative diseases. The chromone derivatives were found potent not only against MAOs, but they were also remarkably potent against AChE, BuChE, BACE1, amyloid-β aggregation, oxidative stress, etc. The optimal length of the linker in designing AChE inhibitors might allow the inhibition of both active sites of the enzyme (CAS and PAS) and a terminal amine group may facilitate the compound–active site interaction. Furthermore, studies indicated that chromones also have the properties of neurogenesis, i.e., the natural chromone was successfully demonstrated by several authors to promote neuron differentiation of neural stem/progenitor cells (NSPCs) and stimulate neurogenesis.
Therefore, in our opinion, it is expected that the optimization of chromone-type compounds to act as multi-target ligands will have positive outcomes for the treatment of neurodegenerative diseases. Furthermore, computational techniques might be beneficial to synthetic chemists to design and produce new compounds by studying in silico inhibition of each enzyme with promising compounds. The lack of in vivo studies as well as cytotoxicity studies creates limitations in optimization of the most promising compounds; thus these evaluations must be started as the next step for the development of new anti-neurodegenerative agents. In the upcoming years, it is expected that the development of new therapeutic small molecules with the chromone scaffold will disclose further exciting features in the field of neurodegenerative drug discovery.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Material
Acknowledgments
HM wishes to acknowledge the Indian Council of Medical Research (Government of India), New Delhi, India, for providing financial support during the study as a Senior Research Fellowship (Award No. 45/02/2020-Nan/BMS).
References
- Gitler A. D. Dhillon P. Shorter J. Dis. Models Mech. 2017;10:499–502. doi: 10.1242/dmm.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. Dan X. Babbar M. Wei Y. Hasselbalch S. G. Croteau D. L. Bohr V. A. Nat. Rev. Neurol. 2019;15:565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- Fernandopulle M. S. Lippincott-Schwartz J. Ward M. E. Nat. Neurosci. 2021;24:622–632. doi: 10.1038/s41593-020-00785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. Hardy J. Duff K. E. Nat. Neurosci. 2018;21:1350–1358. doi: 10.1038/s41593-018-0221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J. W. Neurochem. Res. 2020;45:144–158. doi: 10.1007/s11064-019-02844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza G. X. Rose S. E. Knupp A. Nicholson D. A. Keene C. D. Young J. E. J. Neurosci. Res. 2021;99:124–140. doi: 10.1002/jnr.24615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun M. A. Lo A. C. Duggan Evans C. Wessels A. M. Ardayfio P. A. Andersen S. W. Shcherbinin S. Sparks J. Sims J. R. Brys M. Apostolova L. G. Salloway S. P. Skovronsky D. M. N. Engl. J. Med. 2021;384:1691–1704. doi: 10.1056/NEJMoa2100708. [DOI] [PubMed] [Google Scholar]
- Trapecar M. Wogram E. Svoboda D. Communal C. Omer A. Lungjangwa T. Sphabmixay P. Velazquez J. Schneider K. Wright C. W. Mildrum S. Hendricks A. Levine S. Muffat J. Lee M. J. Lauffenburger D. A. Trumper D. Jaenisch R. Griffith L. G. Sci. Adv. 2021;7:eabd1707. doi: 10.1126/sciadv.abd1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P. H. Ramamoorthy A. Sahoo B. R. Zheng J. Faller P. Straub J. E. Dominguez L. Shea J.-E. Dokholyan N. V. De Simone A. Ma B. Nussinov R. Najafi S. Ngo S. T. Loquet A. Chiricotto M. Ganguly P. McCarty J. Li M. S. Hall C. Wang Y. Miller Y. Melchionna S. Habenstein B. Timr S. Chen J. Hnath B. Strodel B. Kayed R. Lesné S. Wei G. Sterpone F. Doig A. J. Derreumaux P. Chem. Rev. 2021;121:2545–2647. doi: 10.1021/acs.chemrev.0c01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisiecka D. Kelly H. Jackson J. Disabil. Rehabil. 2021;43:479–488. doi: 10.1080/09638288.2019.1630487. [DOI] [PubMed] [Google Scholar]
- Dewan R. Chia R. Ding J. Hickman R. A. Stein T. D. Abramzon Y. Ahmed S. Sabir M. S. Portley M. K. Tucci A. Ibáñez K. Shankaracharya F. N. U. Keagle P. Rossi G. Caroppo P. Tagliavini F. Waldo M. L. Johansson P. M. Nilsson C. F. Rowe J. B. Benussi L. Binetti G. Ghidoni R. Jabbari E. Viollet C. Glass J. D. Singleton A. B. Silani V. Ross O. A. Ryten M. Torkamani A. Tanaka T. Ferrucci L. Resnick S. M. Pickering-Brown S. Brady C. B. Kowal N. Hardy J. A. Van Deerlin V. Vonsattel J. P. Harms M. B. Morris H. R. Ferrari R. Landers J. E. Chiò A. Gibbs J. R. Dalgard C. L. Scholz S. W. Traynor B. J. Adeleye A. Alba C. Bacikova D. Hupalo D. N. Martinez E. M. Pollard H. B. Sukumar G. Soltis A. R. Tuck M. Zhang X. Wilkerson M. D. Smith B. N. Ticozzi N. Fallini C. Gkazi A. S. Topp S. D. Kost J. Scotter E. L. Kenna K. P. Miller J. W. Tiloca C. Vance C. Danielson E. W. Troakes C. Colombrita C. Al-Sarraj S. Lewis E. A. King A. Calini D. Pensato V. Castellotti B. de Belleroche J. Baas F. ten Asbroek A. L. M. A. Sapp P. C. McKenna-Yasek D. McLaughlin R. L. Polak M. Asress S. Esteban-Pérez J. Muñoz-Blanco J. L. Stevic Z. D'Alfonso S. Mazzini L. Comi G. P. Del Bo R. Ceroni M. Gagliardi S. Querin G. Bertolin C. van Rheenen W. Diekstra F. P. Rademakers R. van Blitterswijk M. Boylan K. B. Lauria G. Duga S. Corti S. Cereda C. Corrado L. Sorarù G. Williams K. L. Nicholson G. A. Blair I. P. Leblond-Manry C. Rouleau G. A. Hardiman O. Morrison K. E. Veldink J. H. van den Berg L. H. Al-Chalabi A. Pall H. Shaw P. J. Turner M. R. Talbot K. Taroni F. García-Redondo A. Wu Z. Gellera C. Ratti A. Brown R. H. Shaw C. E. Ambrose J. C. Arumugam P. Baple E. L. Bleda M. Boardman-Pretty F. Boissiere J. M. Boustred C. R. Brittain H. Caulfield M. J. Chan G. C. Craig C. E. H. Daugherty L. C. de Burca A. Devereau A. Elgar G. Foulger R. E. Fowler T. Furió-Tarí P. Hackett J. M. Halai D. Hamblin A. Henderson S. Holman J. E. Hubbard T. J. P. Jackson R. Jones L. J. Kasperaviciute D. Kayikci M. Lahnstein L. Lawson K. Leigh S. E. A. Leong I. U. S. Lopez J. F. Maleady-Crowe F. Mason J. McDonagh E. M. Moutsianas L. Mueller M. Murugaesu N. Need A. C. Odhams C. A. Patch C. Perez-Gil D. Polychronopoulos D. Pullinger J. Rahim T. Rendon A. Riesgo-Ferreiro P. Rogers T. Savage K. Sawant K. Scott R. H. Siddiq A. Sieghart A. Smedley D. Smith K. R. Sosinsky A. Spooner W. Stevens H. E. Stuckey A. Sultana R. Thomas E. R. A. Thompson S. R. Tregidgo C. Walsh E. Watters S. A. Welland M. J. Williams E. Witkowska K. Wood S. M. Zarowiecki M. Arepalli S. Auluck P. Baloh R. H. Bowser R. Brice A. Broach J. Camu W. Chiò A. Cooper-Knock J. Corcia P. Drepper C. Drory V. E. Dunckley T. L. Faghri F. Farren J. Feldman E. Floeter M. K. Fratta P. Gerhard G. Gibson S. B. Goutman S. A. Heiman-Patterson T. D. Hernandez D. G. Hoover B. Jansson L. Kamel F. Kirby J. Kowall N. W. Laaksovirta H. Landi F. Le Ber I. Lumbroso S. MacGowan D. J. L. Maragakis N. J. Mora G. Mouzat K. Myllykangas L. Nalls M. A. Orrell R. W. Ostrow L. W. Pamphlett R. Pioro E. Pulst S. M. Ravits J. M. Renton A. E. Robberecht W. Robey I. Rogaeva E. Rothstein J. D. Sendtner M. Shaw P. J. Sidle K. C. Simmons Z. Stone D. J. Tienari P. J. Trojanowski J. Q. Troncoso J. C. Valori M. Van Damme P. Van Den Bosch L. Zinman L. Albani D. Borroni B. Padovani A. Bruni A. Clarimon J. Dols-Icardo O. Illán-Gala I. Lleó A. Danek A. Galimberti D. Scarpini E. Serpente M. Graff C. Chiang H.-H. Khoshnood B. Öijerstedt L. Morris C. M. Nacmias B. Sorbi S. Nielsen J. E. Hjermind L. E. Novelli V. Puca A. A. Pastor P. Alvarez I. Diez-Fairen M. Aguilar M. Perneczky R. Diehl-Schimd J. Rogaeva E. Rossi M. Ruiz A. Boada M. Hernández I. Moreno-Grau S. Schlachetzki J. C. Aarsland D. Alba C. Albert M. S. Al-Sarraj S. Attems J. Bacikova D. Barrett M. J. Beach T. G. Bekris L. M. Bennett D. A. Besser L. M. Bigio E. H. Black S. E. Boeve B. F. Bohannan R. C. Brett F. Brice A. Brunetti M. Caraway C. A. Palma J.-A. Calvo A. Canosa A. Clarimon J. Dickson D. Diez-Fairen M. Duyckaerts C. Faber K. Ferman T. Flanagan M. E. Floris G. Foroud T. M. Fortea J. Gan-Or Z. Gentleman S. Ghetti B. Gibbs J. R. Goate A. Goldstein D. González-Aramburu I. Graff-Radford N. R. Hodges A. K. Hu H.-C. Hupalo D. Infante J. Iranzo A. Kaiser S. M. Kaufmann H. Keith J. Kim R. C. Klein G. Krüger R. Kukull W. Kuzma A. Lage C. Lesage S. Lleó A. Leverenz J. B. Logroscino G. Lopez G. Love S. Mao Q. Marti M. J. Martinez-McGrath E. Masellis M. Masliah E. May P. McKeith I. Mesulam M.-M. Monuki E. S. Morris C. M. Newell K. L. Norcliffe-Kaufmann L. Palmer L. Pastor P. Perkins M. Pletnikova O. Molina-Porcel L. Renton A. E. Reynolds R. H. Rodríguez-Rodríguez E. Rogaeva E. Rohrer J. D. Sanchez-Juan P. Scherzer C. R. Serrano G. E. Shakkottai V. Sidransky E. Tayebi N. Thomas A. J. Tilley B. S. Troakes C. Troncoso J. C. Walton R. L. Woltjer R. Wszolek Z. K. Xiromerisiou G. Zecca C. Phatnani H. Kwan J. Sareen D. Broach J. R. Simmons Z. Arcila-Londono X. Lee E. B. Shneider N. A. Fraenkel E. Ostrow L. W. Baas F. Zaitlen N. Berry J. D. Malaspina A. Fratta P. Cox G. A. Thompson L. M. Finkbeiner S. Dardiotis E. Miller T. M. Chandran S. Pal S. Hornstein E. MacGowan D. J. Heiman-Patterson T. Hammell M. G. Patsopoulos N. A. Butovsky O. Dubnau J. Nath A. Bowser R. Harms M. Aronica E. Poss M. Phillips-Cremins J. Crary J. Atassi N. Lange D. J. Adams D. J. Stefanis L. Gotkine M. Baloh R. H. Babu S. Raj T. Paganoni S. Shalem O. Smith C. Zhang B. Harris B. Broce I. Drory V. Ravits J. McMillan C. Menon V. Wu L. Altschuler S. Amar K. Archibald N. Bandmann O. Capps E. Church A. Coebergh J. Costantini A. Critchley P. Ghosh B. C. P. Hu M. T. M. Kobylecki C. Leigh P. N. Mann C. Massey L. A. Morris H. R. Nath U. Pavese N. Paviour D. Sharma J. Vaughan J. Neuron. 2021;109:448–460.e4. doi: 10.1016/j.neuron.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber J. Schaprian T. Berkan K. Reetz K. França M. C. Rezende T. J. R. Hong J. Liao W. Warrenburg B. Gaalen J. Durr A. Mochel F. Giunti P. Garcia-Moreno H. Schoels L. Hengel H. Synofzik M. Bender B. Oz G. Joers J. Vries J. J. Kang J. Timmann-Braun D. Jacobi H. Infante J. Joules R. Romanzetti S. Diedrichsen J. Schmid M. Wolz R. Klockgether T. Mov. Disord. 2021;36(10):2273–2281. doi: 10.1002/mds.28610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, Dementia Fact-Sheet, https://www.who.int/news-room/fact-sheets/detail/dementia [Google Scholar]
- Durães F. Pinto M. Sousa E. Pharmaceuticals. 2018;11:44. doi: 10.3390/ph11020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adan G. Mitchell J. W. Ziso B. Larner A. J. Curr. Treat. Options Neurol. 2021;23:1. doi: 10.1007/s11940-020-00656-y. [DOI] [Google Scholar]
- Rohman M. A. Baruah P. Bhatta A. Mitra S. J. Mol. Liq. 2019;290:111210. doi: 10.1016/j.molliq.2019.111210. [DOI] [Google Scholar]
- Manzoor S. Prajapati S. K. Majumdar S. Raza K. Gabr M. T. Kumar S. Pal K. Rashid H. Kumar S. Krishnamurthy S. Hoda N. Eur. J. Med. Chem. 2021;215:113224. doi: 10.1016/j.ejmech.2021.113224. [DOI] [PubMed] [Google Scholar]
- Li Q. Chen Y. Xing S. Liao Q. Xiong B. Wang Y. Lu W. He S. Feng F. Liu W. Chen Y. Sun H. J. Med. Chem. 2021;64:6856–6876. doi: 10.1021/acs.jmedchem.1c00167. [DOI] [PubMed] [Google Scholar]
- Maqbool M. Mobashir M. Hoda N. Eur. J. Med. Chem. 2016;107:63–81. doi: 10.1016/j.ejmech.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Manzoor S. Hoda N. Eur. J. Med. Chem. 2020;206:112787. doi: 10.1016/j.ejmech.2020.112787. [DOI] [PubMed] [Google Scholar]
- Hyun D.-H. Arch. Pharmacal Res. 2019;42:436–445. doi: 10.1007/s12272-019-01147-8. [DOI] [PubMed] [Google Scholar]
- Ross C. A. Poirier M. A. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Woerman A. L. Acta Neuropathol. 2021;142:1–3. doi: 10.1007/s00401-021-02311-5. [DOI] [PubMed] [Google Scholar]
- McMurray L. Macdonald J. A. Ramakrishnan N. K. Zhao Y. Williamson D. W. Tietz O. Zhou X. Kealey S. Fagan S. G. Smolek T. Cubinkova V. Žilka N. Spillantini M. G. Tolkovsky A. M. Goedert M. Aigbirhio F. I. ACS Chem. Neurosci. 2021;12(11):1885–1893. doi: 10.1021/acschemneuro.0c00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teravskis P. J. Ashe K. H. Liao D. Neuroscientist. 2020;26:503–520. doi: 10.1177/1073858420916696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone M. Solfrizzi V. D'Urso F. Di Gioia I. Sardone R. Dibello V. Stallone R. Liguori A. Ciritella C. Daniele A. Bellomo A. Seripa D. Panza F. Expert Opin. Emerging Drugs. 2020;25:319–335. doi: 10.1080/14728214.2020.1808621. [DOI] [PubMed] [Google Scholar]
- Li D. Liu C. Nat. Chem. Biol. 2021;17:237–245. doi: 10.1038/s41589-020-00708-z. [DOI] [PubMed] [Google Scholar]
- Prasad A. Bharathi V. Sivalingam V. Girdhar A. Patel B. K. Front. Mol. Neurosci. 2019;12(25) doi: 10.3389/fnmol.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool M. Gadhavi J. Singh A. Hivare P. Gupta S. Hoda N. Org. Biomol. Chem. 2021;19:1589–1603. doi: 10.1039/D0OB02226H. [DOI] [PubMed] [Google Scholar]
- Maqbool M. Gadhavi J. Hivare P. Gupta S. Hoda N. Eur. J. Med. Chem. 2020;207:112705. doi: 10.1016/j.ejmech.2020.112705. [DOI] [PubMed] [Google Scholar]
- Oukoloff K. Nzou G. Varricchio C. Lucero B. Alle T. Kovalevich J. Monti L. Cornec A.-S. Yao Y. James M. J. Trojanowski J. Q. Lee V. M.-Y. Smith A. B. Brancale A. Brunden K. R. Ballatore C. J. Med. Chem. 2021;64:1073–1102. doi: 10.1021/acs.jmedchem.0c01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhav H. Hoda N. Eur. J. Med. Chem. 2021;210:112955. doi: 10.1016/j.ejmech.2020.112955. [DOI] [PubMed] [Google Scholar]
- Yao H. Uras G. Zhang P. Xu S. Yin Y. Liu J. Qin S. Li X. Allen S. Bai R. Gong Q. Zhang H. Zhu Z. Xu J. J. Med. Chem. 2021;64(11):7483–7506. doi: 10.1021/acs.jmedchem.1c00160. [DOI] [PubMed] [Google Scholar]
- Kumar J. Gill A. Shaikh M. Singh A. Shandilya A. Jameel E. Sharma N. Mrinal N. Hoda N. Jayaram B. ChemistrySelect. 2018;3:736–747. doi: 10.1002/slct.201702599. [DOI] [Google Scholar]
- Jameel E. Naz H. Khan P. Tarique M. Kumar J. Mumtazuddin S. Ahamad S. Islam A. Ahmad F. Hoda N. Hassan M. I. Chem. Biol. Drug Des. 2017;89:741–754. doi: 10.1111/cbdd.12898. [DOI] [PubMed] [Google Scholar]
- Jameel E. Meena P. Maqbool M. Kumar J. Ahmed W. Mumtazuddin S. Tiwari M. Hoda N. Jayaram B. Eur. J. Med. Chem. 2017;136:36–51. doi: 10.1016/j.ejmech.2017.04.064. [DOI] [PubMed] [Google Scholar]
- Jameel E. Umar T. Kumar J. Hoda N. Chem. Biol. Drug Des. 2016;87:21–38. doi: 10.1111/cbdd.12629. [DOI] [PubMed] [Google Scholar]
- Ellis G. P., Chromenes, Chromanones, and Chromones-Introduction, Chemistry of Heterocyclic Compounds: A Series Of Monographs, 2008, pp. 1–10, 10.1002/9780470187012.ch1 [DOI] [Google Scholar]
- Tomer N. Goel A. Ghule V. D. Malhotra R. J. Mol. Struct. 2021;1227:129549. doi: 10.1016/j.molstruc.2020.129549. [DOI] [Google Scholar]
- Gaspar A. Matos M. J. Garrido J. Uriarte E. Borges F. Chem. Rev. 2014;114:4960–4992. doi: 10.1021/cr400265z. [DOI] [PubMed] [Google Scholar]
- Reis J. Gaspar A. Milhazes N. Borges F. J. Med. Chem. 2017;60:7941–7957. doi: 10.1021/acs.jmedchem.6b01720. [DOI] [PubMed] [Google Scholar]
- Joolakanti H. B. Battu S. Kamepalli R. Kolanupaka H. R. Bobbili H. R. Chem. Data Collect. 2021;32:100651. doi: 10.1016/j.cdc.2021.100651. [DOI] [Google Scholar]
- Sestili P. Stocchi V. Front. Pharmacol. 2020;11:854. doi: 10.3389/fphar.2020.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Chen H. Wang P. Li Y. Wold E. A. Leonard P. G. Joseph S. Brasier A. R. Tian B. Zhou J. J. Med. Chem. 2020;63:5242–5256. doi: 10.1021/acs.jmedchem.0c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y. J. Abhijit K. Russ. J. Gen. Chem. 2020;90:1074–1082. doi: 10.1134/S1070363220060225. [DOI] [Google Scholar]
- Silva C. F. M. Pinto D. C. G. A. Silva A. M. S. Expert Opin. Drug Discovery. 2018;13:1141–1151. doi: 10.1080/17460441.2018.1543267. [DOI] [PubMed] [Google Scholar]
- Alhadrami H. A. Sayed A. M. Al-Khatabi H. Alhakamy N. A. Rateb M. E. Pharmaceuticals. 2021;14:541. doi: 10.3390/ph14060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittisrisopit S. Bunbamrung N. Thawai C. Tadtong S. Niemhom N. Komwijit S. Rachtawee P. Pittayakhajonwut P. Nat. Prod. Res. 2019:1–6. doi: 10.1080/14786419.2019.1679135. [DOI] [PubMed] [Google Scholar]
- Ayaz M. Sadiq A. Junaid M. Ullah F. Ovais M. Ullah I. Ahmed J. Shahid M. Front. Aging Neurosci. 2019;11:155. doi: 10.3389/fnagi.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista F. I. Henriques A. G. Silva A. M. S. Wiltfang J. da Cruz e Silva O. A. B. ACS Chem. Neurosci. 2014;5:83–92. doi: 10.1021/cn400213r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang P.-W. Cui G.-Z. Zhang Y.-J. Zhang M.-X. Guo H. Zhang J.-B. Lu Z.-Q. Isaiah A.-O. Lin Y.-X. CNS Neurosci. Ther. 2013;19:154–162. doi: 10.1111/cns.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowndhararajan K. Deepa P. Kim M. Park S. Kim S. Brain Sci. 2018;8:104. doi: 10.3390/brainsci8060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. S. Perfilieva E. Björk-Eriksson T. Alborn A.-M. Nordborg C. Peterson D. A. Gage F. H. Nat. Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- Marlier Q. Verteneuil S. Vandenbosch R. Malgrange B. Front. Neurosci. 2015;9:458. doi: 10.3389/fnins.2015.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbán N. Guillemot F. Front. Cell. Neurosci. 2014;8:396. doi: 10.3389/fncel.2014.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. Iacovitti L. Brain Res. 2015;1628:327–342. doi: 10.1016/j.brainres.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Zhao F. Tao W. Shang Z. Zhang W. Ruan J. Zhang C. Zhou L. Aiello H. Lai H. Qu R. Front. Pharmacol. 2020;11:556845. doi: 10.3389/fphar.2020.556845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. He M. Wang F. Zhang H. Li Y. Yang J. Wu C. Front. Neurosci. 2019;13:834. doi: 10.3389/fnins.2019.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. B. Park H. R. Jang Y. J. Choi S. Y. Son T. G. Lee J. Br. J. Pharmacol. 2013;168:421–431. doi: 10.1111/j.1476-5381.2012.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang P.-W. Cui G.-Z. Zhang Y.-J. Zhang M.-X. Guo H. Zhang J.-B. Lu Z.-Q. Isaiah A.-O. Lin Y.-X. CNS Neurosci. Ther. 2013;19:154–162. doi: 10.1111/cns.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Nascimento B. O. da Silva Neto O. C. Teodoro M. T. F. de Oliveira Silva E. Guedes M. L. S. David J. M. Phytochem. Lett. 2020;39:124–127. doi: 10.1016/j.phytol.2020.08.002. [DOI] [Google Scholar]
- He Q. Hu D.-B. Zhang L. Xia M.-Y. Yan H. Li X.-N. Luo J.-F. Wang Y.-S. Yang J.-H. Wang Y.-H. Phytochemistry. 2021;181:112554. doi: 10.1016/j.phytochem.2020.112554. [DOI] [PubMed] [Google Scholar]
- Kim J. Lim J. Kang B. Y. Jung K. Choi H. J. Neurochem. Int. 2017;105:11–20. doi: 10.1016/j.neuint.2017.01.018. [DOI] [PubMed] [Google Scholar]
- Hiep N. T. Kwon J. Kim D.-W. Hwang B. Y. Lee H.-J. Mar W. Lee D. Phytochemistry. 2015;111:141–148. doi: 10.1016/j.phytochem.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Lu Q. Harmalkar D. S. Quan G. Kwon H. Cho J. Choi Y. Lee D. Lee K. J. Nat. Prod. 2021;84:1359–1365. doi: 10.1021/acs.jnatprod.1c00121. [DOI] [PubMed] [Google Scholar]
- Dhiman P. Malik N. Sobarzo-Sánchez E. Uriarte E. Khatkar A. Molecules. 2019;24:418. doi: 10.3390/molecules24030418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat I. U. H. Bhat R. Biology. 2021;10:586. doi: 10.3390/biology10070586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Zhuang P. Shen B. Zhang Y. Shen J. Brain Res. 2012;1429:36–42. doi: 10.1016/j.brainres.2011.10.030. [DOI] [PubMed] [Google Scholar]
- Li Y.-C. Shen J.-D. Li J. Wang R. Jiao S. Yi L.-T. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2013;40:138–143. doi: 10.1016/j.pnpbp.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Zhang K. Pan X. Wang F. Ma J. Su G. Dong Y. Yang J. Wu C. Sci. Rep. 2016;6:30951. doi: 10.1038/srep30951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C. Du Q. Li W. Deng R. Wang Q. Xu A. Shen J. Mol. Neurobiol. 2018;55:9334–9348. doi: 10.1007/s12035-018-1042-8. [DOI] [PubMed] [Google Scholar]
- Zhang C.-Y.-Y. Zeng M.-J. Zhou L.-P. Li Y.-Q. Zhao F. Shang Z.-Y. Deng X.-Y. Ma Z.-Q. Fu Q. Ma S.-P. Qu R. Int. Immunopharmacol. 2018;64:175–182. doi: 10.1016/j.intimp.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang R. Ma Z. Liu K. Li Y. Liu D. Xu L. Deng X. Qu R. Ma Z. Ma S. Life Sci. 2019;221:241–248. doi: 10.1016/j.lfs.2019.02.033. [DOI] [PubMed] [Google Scholar]
- Fang A. Li Y. Wu X. Wu B. Zhang Y. Metab. Brain Dis. 2020;35:1085–1093. doi: 10.1007/s11011-020-00599-y. [DOI] [PubMed] [Google Scholar]
- Jia Z. Yang J. Cao Z. Zhao J. Zhang J. Lu Y. Chu L. Zhang S. Chen Y. Pei L. Behav. Brain Res. 2021;414:113463. doi: 10.1016/j.bbr.2021.113463. [DOI] [PubMed] [Google Scholar]
- Xiao Z. Cao Z. Yang J. Jia Z. Du Y. Sun G. Lu Y. Pei L. Biochem. Pharmacol. 2021;190:114594. doi: 10.1016/j.bcp.2021.114594. [DOI] [PubMed] [Google Scholar]
- Yang J. Jia Z. Xiao Z. Zhao J. Lu Y. Chu L. Shao H. Pei L. Zhang S. Chen Y. Drug Des., Dev. Ther. 2021;15:3163–3180. doi: 10.2147/DDDT.S314076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. Zhang S. Cheng Y. Lu Y. Jia Z. Yang X. Zhang S. Guo W. Pei L. Brain Res. 2022;1774:147723. doi: 10.1016/j.brainres.2021.147723. [DOI] [PubMed] [Google Scholar]
- Takao K. Saito T. Chikuda D. Sugita Y. Chem. Pharm. Bull. 2016;64:1499–1504. doi: 10.1248/cpb.c16-00527. [DOI] [PubMed] [Google Scholar]
- Wang Z. Wu J. Yang X. Cai P. Liu Q. Wang K. D. G. Kong L. Wang X. Bioorg. Med. Chem. 2016;24:5929–5940. doi: 10.1016/j.bmc.2016.09.050. [DOI] [PubMed] [Google Scholar]
- Reis J. Cagide F. Chavarria D. Silva T. Fernandes C. Gaspar A. Uriarte E. Remião F. Alcaro S. Ortuso F. Borges F. J. Med. Chem. 2016;59:5879–5893. doi: 10.1021/acs.jmedchem.6b00527. [DOI] [PubMed] [Google Scholar]
- Jia W. Z. Cheng F. Zhang Y. J. Ge J. Y. Yao S. Q. Zhu Q. Chem. Biol. Drug Des. 2017;89:141–151. doi: 10.1111/cbdd.12841. [DOI] [PubMed] [Google Scholar]
- Li F. Wu J.-J. Wang J. Yang X.-L. Cai P. Liu Q.-H. Kong L.-Y. Wang X.-B. Bioorg. Med. Chem. 2017;25:3815–3826. doi: 10.1016/j.bmc.2017.05.027. [DOI] [PubMed] [Google Scholar]
- Nesi G. Chen Q. Sestito S. Digiacomo M. Yang X. Wang S. Pi R. Rapposelli S. Eur. J. Med. Chem. 2017;141:232–239. doi: 10.1016/j.ejmech.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Reis J. Manzella N. Cagide F. Mialet-Perez J. Uriarte E. Parini A. Borges F. Binda C. J. Med. Chem. 2018;61:4203–4212. doi: 10.1021/acs.jmedchem.8b00357. [DOI] [PubMed] [Google Scholar]
- Reis J. Cagide F. Valencia M. E. Teixeira J. Bagetta D. Pérez C. Uriarte E. Oliveira P. J. Ortuso F. Alcaro S. Rodríguez-Franco M. I. Borges F. Eur. J. Med. Chem. 2018;158:781–800. doi: 10.1016/j.ejmech.2018.07.056. [DOI] [PubMed] [Google Scholar]
- Valencia M. E. Herrera-Arozamena C. de Andrés L. Pérez C. Morales-García J. A. Pérez-Castillo A. Ramos E. Romero A. Viña D. Yáñez M. Laurini E. Pricl S. Rodríguez-Franco M. I. Eur. J. Med. Chem. 2018;156:534–553. doi: 10.1016/j.ejmech.2018.07.026. [DOI] [PubMed] [Google Scholar]
- Makhaeva G. F. Boltneva N. P. Lushchekina S. V. Rudakova E. V. Serebryakova O. G. Kulikova L. N. Beloglazkin A. A. Borisov R. S. Richardson R. J. Bioorg. Med. Chem. 2018;26:4716–4725. doi: 10.1016/j.bmc.2018.08.010. [DOI] [PubMed] [Google Scholar]
- Makhaeva G. F. Boltneva N. P. Lushchekina S. V. Rudakova E. V. Serebryakova O. G. Kulikova L. N. Beloglazkin A. A. Borisov R. S. Richardson R. J. Bioorg. Med. Chem. 2018;26:4716–4725. doi: 10.1016/j.bmc.2018.08.010. [DOI] [PubMed] [Google Scholar]
- Mphahlele M. Agbo E. Gildenhuys S. Int. J. Mol. Sci. 2018;19:4112. doi: 10.3390/ijms19124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mphahlele M. J. Agbo E. N. Gildenhuys S. Setshedi I. B. Biomolecules. 2019;9:736. doi: 10.3390/biom9110736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mphahlele M. J. Gildenhuys S. Agbo E. N. Int. J. Mol. Sci. 2019;20:5451. doi: 10.3390/ijms20215451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke C. Christmann J. Yin J. Alonso J. M. Serrano E. Chioua M. Ismaili L. Martínez-Grau M. A. Beadle C. D. Vetman T. Dato F. M. Bartz U. Elsinghorst P. W. Pietsch M. Müller C. E. Iriepa I. Wille T. Marco-Contelles J. Gütschow M. ACS Omega. 2019;4:22161–22168. doi: 10.1021/acsomega.9b03409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuther-Conrad W. Diez-Iriepa D. Iriepa I. López-Muñoz F. Martínez-Grau M. A. Gütschow M. Marco-Contelles J. RSC Med. Chem. 2021;12:1000–1004. doi: 10.1039/D1MD00105A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao K. Endo S. Nagai J. Kamauchi H. Takemura Y. Uesawa Y. Sugita Y. Bioorg. Chem. 2019;92:103285. doi: 10.1016/j.bioorg.2019.103285. [DOI] [PubMed] [Google Scholar]
- Takao K. Shiori U. Kamauchi H. Sugita Y. Bioorg. Chem. 2019;87:594–600. doi: 10.1016/j.bioorg.2019.03.042. [DOI] [PubMed] [Google Scholar]
- Takao K. Sakatsume T. Kamauchi H. Sugita Y. Chem. Pharm. Bull. 2020;68:1082–1089. doi: 10.1248/cpb.c20-00579. [DOI] [PubMed] [Google Scholar]
- Pachón-Angona I. Refouvelet B. Andrýs R. Martin H. Luzet V. Iriepa I. Moraleda I. Diez-Iriepa D. Oset-Gasque M.-J. Marco-Contelles J. Musilek K. Ismaili L. J. Enzyme Inhib. Med. Chem. 2019;34:479–489. doi: 10.1080/14756366.2018.1545766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpitimpiti A. N. Petzer J. P. Petzer A. Jordaan J. H. L. Lourens A. C. U. Mol. Diversity. 2019;23:897–913. doi: 10.1007/s11030-019-09917-8. [DOI] [PubMed] [Google Scholar]
- Wang X.-B. Yin F.-C. Huang M. Jiang N. Lan J.-S. Kong L.-Y. RSC Med. Chem. 2020;11:225–233. doi: 10.1039/C9MD00441F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanhom P. Nualnoi T. Khongkow P. Lee V. S. Lomlim L. Med. Chem. Res. 2020;29:564–574. doi: 10.1007/s00044-020-02508-5. [DOI] [Google Scholar]
- Shaikh S. Dhavan P. Ramana M. M. V. Jadhav B. L. Mol. Diversity. 2021;25:811–825. doi: 10.1007/s11030-020-10060-y. [DOI] [PubMed] [Google Scholar]
- Rao Y. J. Abhijit K. Mallikarjun G. Hemasri Y. Chem. Pap. 2021;75:703–716. doi: 10.1007/s11696-020-01332-w. [DOI] [Google Scholar]
- Malafaia D. Oliveira A. Fernandes P. A. Ramos M. J. Albuquerque H. M. T. Silva A. M. S. Int. J. Mol. Sci. 2021;22:4145. doi: 10.3390/ijms22084145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdpour S. Jalili-Baleh L. Nadri H. Forootanfar H. Bukhari S. N. A. Ramazani A. Ebrahimi S. E. S. Foroumadi A. Khoobi M. Bioorg. Chem. 2021;110:104750. doi: 10.1016/j.bioorg.2021.104750. [DOI] [PubMed] [Google Scholar]
- Tan M. A. Zakharova E. An S. S. A. Biology. 2021;10:199. doi: 10.3390/biology10030199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili-Baleh L. Nadri H. Forootanfar H. Küçükkılınç T. T. Ayazgök B. Sharifzadeh M. Rahimifard M. Baeeri M. Abdollahi M. Foroumadi A. Khoobi M. Daru, J. Pharm. Sci. 2021;29:23–38. doi: 10.1007/s40199-020-00378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengheres G. Rice C. R. Olajide O. A. Hemming K. Bioorg. Med. Chem. Lett. 2021;34:127761. doi: 10.1016/j.bmcl.2020.127761. [DOI] [PubMed] [Google Scholar]