Abstract
Purpose
Glioblastoma (GBM) is characterized by extensive clonal diversity suggesting the presence of tumor cells with varying degrees of treatment sensitivity. Radiotherapy is an integral part of glioblastoma treatment. Whether GBMs are comprised of spatially distinct cellular populations with uniform or varying degrees of radiosensitivity has not been established.
Methods
Spatially distinct regions of three GBMs (J3, J7 and J14) were resected and unique cell lines were derived from each region. DNA from cell lines, corresponding tumor fragments, and patient blood was extracted for whole exome sequencing. Variants, clonal composition, and functional implications were compared and analyzed with superFreq and IPA. Limiting dilution assays were performed on cell lines to measure intrinsic radiosensitivity.
Results
Based on WES, cell lines generated from different regions of the same tumor were more closely correlated with their tumor of origin than the other GBMs. Variant and clonal composition comparisons showed that cell lines from distinct tumors displayed increasing levels of ITH with J3 and J14 having the lowest and highest, respectively. The radiosensitivities of the cell lines generated from the J3 tumor were similar as were those generated from the J7 tumor. However, the radiosensitivities of the 2 cell lines generated from the J14 tumor (J14T3 and J14T6) were significantly different with J14T6 being more sensitive than J14T3.
Conclusion
Data suggest a tumor dependent ITH in radiosensitivity. The existence of ITH in radiosensitivity may impact not only the initial therapeutic response but also the effectiveness of retreatment protocols.
Keywords: Glioblastoma, Intratumor heterogeneity, Radiosensitivity
Introduction
Glioblastoma (GBM) is the most common form of malignant, primary brain cancer [1]. Despite surgical resection and extensive postoperative treatment, which typically includes radiotherapy, GBM inevitably recurs and patients succumb to the disease within two years [2]. Most GBM recurrences are predominately within 2 cm of the initial radiation treatment volume [3–6]. In addition, radiotherapy dose escalation results in similar recurrence patterns and fails to improve local control of tumor growth [7]. Thus, although radiotherapy provides a survival advantage, GBM is considered a radioresistant tumor. Defining the molecules and processes that mediate the radioresistance of GBM cells may lead to the identification of targeted radiosensitizers and the overall improvement of treatment outcomes.
Complicating the development of more effective radiotherapy for GBM is the potential for intratumor heterogeneity (ITH) in radiosensitivity. In terms of genetic, epigenetic and transcriptome measurements [8–11], GBMs have been shown to display ITH, consistent with a high degree of clonal diversity. Based on the analysis of cell lines generated from the same surgical specimen and cultured in serum containing media, GBMs have long been suggested to also be comprised of clones with various intrinsic radiosensitivities [12–15]. However, more recent investigations have shown that GBM cell lines derived using this approach have little in common with the biology of their tumor of origin [16]. In contrast, cell lines generated from GBMs as neurosphere cultures under serum-free conditions have been shown to contain GBM stem-like cells (GSCs) and to better simulate the genetics and gene expression patterns of their original tumors [17–19].
In the study described here, the potential for ITH in intrinsic radiosensitivity was addressed by using neurosphere lines derived from 2 spatially distinct fragments obtained by fluorescence-guided surgical resection of 3 GBMs [20]. The two lines from each tumor were compared in terms of their genomics and radiosensitivity. Whole exome sequencing confirmed that cell lines were most similar to their tumors of origin, while also maintaining heterogeneity between cell lines from the same patient. A significant difference in radiosensitivity was detected between the two cell lines isolated from one of the three tumors, suggesting the potential for the ITH in radiosensitivity in a tumor-dependent manner.
Methods
Glioblastoma collection and derivation of cell lines
Patients underwent 5-ALA fluorescence-guided surgical resection at Addenbrooke’s Hospital. Fluorescence and image guidance were utilized to resect spatially distinct tissue fragments from 5-ALA-fluorescent, non-necrotic tumor mass [20]. Patient and tumor characteristics including preoperative T1-weighted magnetic resonance images are presented in Supplemental Methods and Supplemental Fig. 1. Collected fragments were divided in half for both tissue bank storage (with matched whole blood) and cell line derivation [20]. Informed consent for specimen collection and research use was obtained prior to surgery. Protocols were compliant with UK Human Tissue Act 2004 and approved by Local Regional Ethics Committee (LREC ref. 04/Q0108/60). Resected tissue fragments were minced, enzymatically disaggregated using Accutase and passed through 40 μm sterile filters to generate single cell suspensions. After addition of RBC Lysis buffer and incubation at room temperature for 5 min, suspensions were washed several times with HBSS. Cells were counted and resuspended at 1.0*105 cells/ml in T25 or T75 flasks in serum-free media (Neurobasal A (Invitrogen), 20 mM L-glutamine, 1% PSF solution, 20 ng/ ml hEGF (Sigma), 20 ng/ml hFGF (R&D systems), 2% B27 (Invitrogen), and 1% N2 (Invitrogen)) [21]. After 7–10 days, cells began to form neurospheres, which were disaggregated with Accutase and then plated as single cells on extracellular matrix-coated T75 flasks (Sigma) to form adherent monolayers. Cells were split weekly with Accutase and single cell suspensions were frozen after two successful passages and stored in freezing media (serum-free media with 10% DMSO).
Whole exome sequencing and analysis
DNA was extracted from tissue fragments and cell lines with AllPrep DNA/RNA Mini Kits (Qiagen) according to protocol. DNA was extracted from blood samples with DNeasy Blood and Tissue Kits (Qiagen) according to protocol. Genomic DNA underwent whole exome sequencing (WES) at the Center for Cancer Research Genomics Technology Laboratory, Frederick, Maryland. Library prep was done according to Agilent SureSelect XT (All Exon V5 + UTR) protocol and sequenced on an Illumina HiSeq4000 using paired-end sequencing to an average sequencing depth of > 180x. Alignment and variant calling were performed with the Center for Cancer Research Collaborative Bioinformatics Resource (CCBR) pipeline (https://github.com/CCBR/Pipeliner). After filtering germline and non-protein-altering variants, variants (SNVs and indels) with allelic frequency (VAF) > 5%, were identified for each sample. VAFs for identified SNVs and indels were used to calculate Jaccard Similarity Coefficients for all combinations of samples. Using SNVs and CNAs, SuperFreq software was used to define clonal frequencies in each samples [22]. Ingenuity Pathway Analysis (IPA) software was used to define functional implications.
In vitro radiosensitivity
Cell line radiosensitivity was assessed using the limiting dilution assay. Serial dilutions of single cells were seeded into 96-well plates and 24 h later irradiated (1–3 Gy) using a 320 kV Xray machine (Precision X-Ray Inc., dose rate = 2.3 Gy/min). Plates were incubated for 2–3 weeks and examined by light microscopy to determine the number of positive wells [23]. Positive well refers to any well with 1 or more neurospheres, which are defined as an aggregate of at least 30 cells. Extreme Limiting Dilution Analysis online software (ELDA) was utilized to calculate survival fractions (https://bioinf.wehi.edu.au/software/elda/) [24]. The results are the mean of 3 independent experiments in which 1 plate was used per dose of radiation.
Results
Whole exome sequencing (WES)
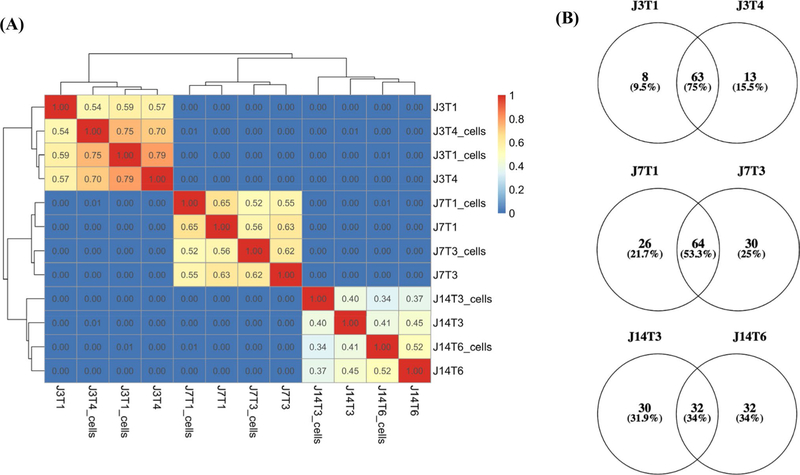
Because the clonal diversity within a tumor (i.e., intratumor heterogeneity) is typically established according to genetic alterations, initial analyses involved WES with a focus on neurosphere cell lines established from the same GBM. Specifically, two spatially distinct specimens were resected from 3 patients with the diagnosis of glioblastoma. Each surgical specimen was then divided into two sections: half was submitted for WES and the other half used to derive a neurosphere cell line, which was subsequently submitted for WES. Initially, variant profiles generated from cell lines and tumor fragments were compared according to Jaccard similarity coefficients, which grade similarity from 0–1.00. As shown in Fig. 1a, cell lines and tumor fragments isolated from the same tumor (J3, J7 or J14) were similar to each other and not to samples from the other 2 tumors. This lack of similarity is consistent with the expected inter-tumor heterogeneity. Cell lines compared to their respective tumor fragments showed varying degrees of similarity, suggesting that, in general, the cell lines were representative of the in situ tumor. The similarity coefficients for the 2 cell lines derived from the same tumor were 34% for J14 cell lines, 52% for J7s and 75% for J3s, suggesting that the paired cell lines reflect different levels of intra-tumor heterogeneity. As an additional assessment of the relative amount of heterogeneity reflected by the cell lines derived from spatially distinct regions, mutation burdens were compared (Fig. 1b). As shown by Venn diagrams (Fig. 1b), the percentage of mutations in common between cell lines from a given tumor also suggested that the level of ITH reflected by paired cell lines could be ranked as J14 > J7 > J3.
Fig. 1.
Correlation between samples and variant burden. a Jaccard similarity coefficients (Jaccard Index) were based on variant allele frequencies (vaf) of all J3, J7, and J14 samples sequenced. Germline variants, variants with vaf < 0.05, and non-protein altering variants were removed for this analysis. b Venn diagrams display the number (and %) of unique and shared variants (SNVs and indels) between cell lines for J3T1 and J3T4, J7T1 and J7T3, and J14T3 and J14T6. Non-synonymous, protein-altering mutations with vaf > 0.05 were included in the analysis
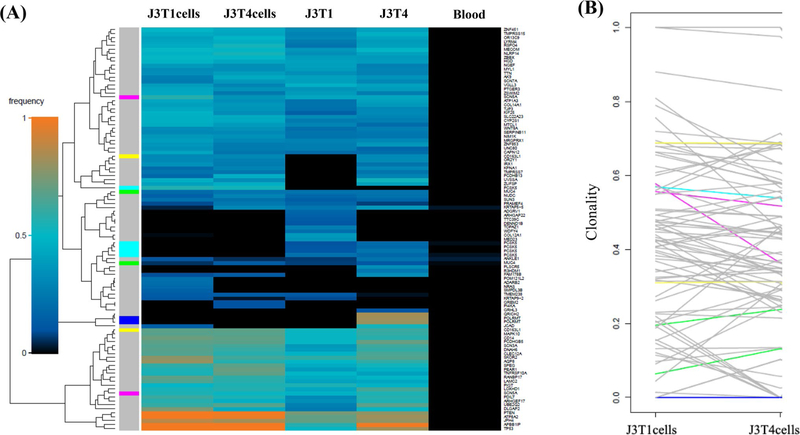
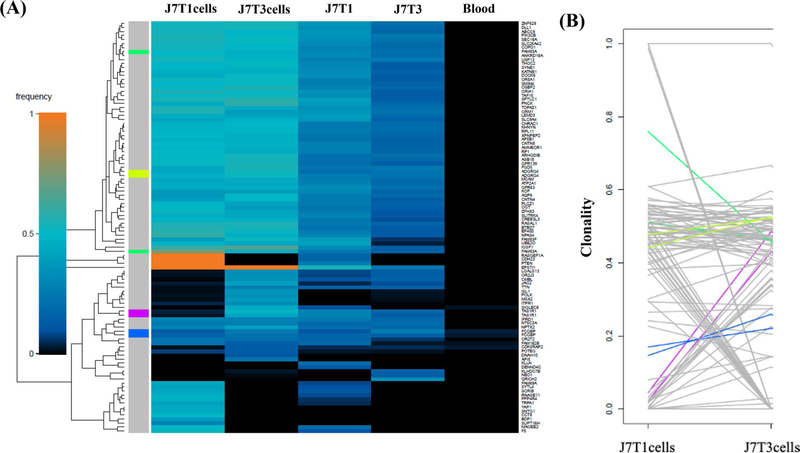
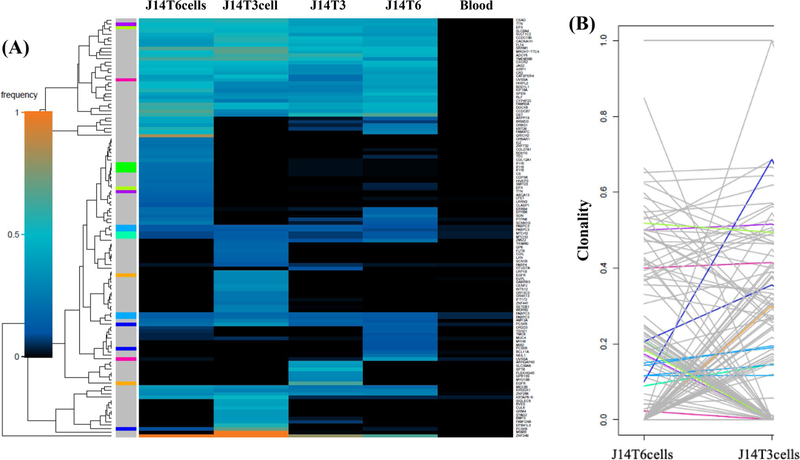
As an additional approach to defining the genetic diversity between the paired cell lines, WES results were subjected to superFreq analysis. superFreq utilizes somatic SNVs and CNAs to cluster genes according to similar coverage and VAF; clonalities are calculated with clones sorted by predicted size [22]. Figure 2a shows a superFreq-generated SNV heatmap for the two tumor fragments and two neurosphere lines derived from tumor J3. From this clustering it appears that the two cell lines are more similar than the two tumor fragments. The clonality of the individual variants in the J3T1 and J3T4 cell lines were directly compared in Fig. 2b. Each line represents a single variant; the end of each line corresponds to the clonality of each variant in the individual cell lines. As shown, most variants seem to be shared between cell lines with minor differences in clonality, suggesting that J3T1 and J3T4 lines exhibit a relatively low amount of ITH. The superFreq heatmap for the J7 tumor also showed that the two cell lines cluster together as do the two tumor fragments (Fig. 3a). However, as shown in Fig. 3b, whereas many of the individual variants are similar between J7T1 and J7T3 cell lines, there are also a number of variants that are present in one of the cell lines but not in the other indicative of a greater degree of diversity in variant clonality than the J3 cell lines. Finally, superFreq analyses of J14 tumor components are shown in Fig. 4. As for the other tumors, the J14 cell lines are more similar to each other than the tumor fragments (Fig. 4a). In the heat map there are extensive differences in SNV presence and frequency between J14T3 cells and J14T6 cells, a number of which are mirrored in their respective tumor fragments. The differences in J14T3 and J14T6 clonality is clearly evident in Fig. 4b, which of the 3 pairs of tumor cell lines evaluated shows the largest number of variants detected in one cell line and absent in the other. Consistent with Fig. 1, superFreq analyses estimate that the greatest degree of ITH is reflected by the cell lines generated from the J14 tumor followed by J7 with the least detected in the J3 cell lines.
Fig. 2.
J3 superFreq analysis. a Heatmaps comparing presence and frequencies of protein-altering SNVs that change significantly between J3 samples. Genes are assigned unique colors if they are repeatedly mutated (more than one SNV is detected for that gene). b Variant shift analysis charts the differences in relative clonality, or cellular fraction of the sample, across J3 cell lines for each variant detected in the corresponding heatmap. Each variant is represented by an individual line
Fig. 3.
J7 superFreq analysis. a Heatmaps comparing presence and frequencies of protein-altering SNVs that change significantly between J7 samples. Genes are assigned unique colors if they are repeatedly mutated (more than one SNV is detected for that gene). b Variant shift analysis charts the differences in relative clonality, or cellular fraction of the sample, across J7 cell lines for each variant detected in the corresponding heatmap. Each variant is represented by an individual line
Fig. 4.
J14 superFreq analysis. a Heatmaps comparing presence and frequencies of protein-altering SNVs that change significantly between J14 samples. Genes are assigned unique colors if they are repeatedly mutated (more than one SNV is detected for that gene). b Variant shift analysis charts the differences in relative clonality, or cellular fraction of the sample, across J14 cell lines for each variant detected in the corresponding heatmap. Each variant is represented by an individual line
Radiosensitivity
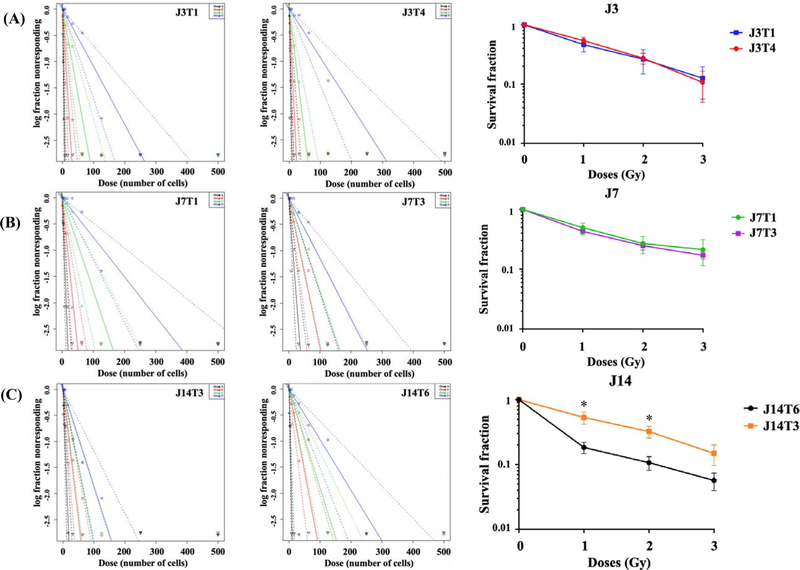
The limiting dilution assay (LDA) was then used to determine the radiosensitivity of the tumor cell lines. Serial dilutions of each cell line were seeded to 96-well plates; 24 h later individual plates were irradiated and returned to the incubator. After 2–3 weeks, the wells were scored as positive or negative, based on the presence of a neurosphere; survival curves were constructed using Extreme Limiting Dilution Assay software [24]. For each cell line, representative neurosphere frequency results plotted as a function of cells seeded are shown for each radiation dose in Fig. 5a. Based on results from 3 independent experiments, radiation survival curves were then generated (Fig. 5b). For the cell lines derived from the J3 and J7 tumors, no difference in radiosensitivity was detected. However, for the J14 cell lines, which displayed the greatest level of ITH, there was a difference in radiosensitivity between the J14 cell lines with J14T3 significantly more resistant than J14T6. To determine whether specific mutations may account for the difference in radiosensitivity detected in the J14 cell lines, the unique variants (see Fig. 1b) in J14T3 and J14T6 cells were subjected to IPA (supplemental Fig. 1). Among the top molecular and cellular processes associated with the mutations detected in J14T3 cells, but not in J14T6 cells were cell cycle and DNA replication, recombination, and repair.
Fig. 5.
Limiting dilution assays. Stem cell frequency estimates (with confidence intervals) generated through ELDA and survival fraction as a function of radiation dose (last column) for a J3T1 and J3T4, b J7T1 and J7T3, and c J14T3 and J14T6. Black, red, green, and blue lines correspond to 0, 1, 2, and 3 Gy, respectively. Dotted lines represent confidence intervals. Survival fractions were calculated by dividing inverse of stem cell frequency estimate by inverse of stem cell frequency estimate of the control group. *p < 0.05 by 2-way ANOVA
Conclusions
Although characterized by extensive clonal diversity, whether the radioresistance of GBMs is the result of a uniform response among the various clones or the presence of a resistant subpopulation remains unclear. As an approach to begin to address this issue, we used GSC cultures initiated from spatially distinct sections of the same GBM. GSCs are a clonogenic subpopulation considered critical to the development and maintenance of GBMs. Moreover, GSCs, as compared to bulk tumor cells, are thought to be responsible for GBM radioresistance [25]. Thus, GSCs appear to provide a clinically relevant in vitro model for investigating not only the basic biology of GBMs, but also, specifically, their radioresponse.
In the study described here, genomic analyses suggested that GSC cell lines derived from the same GBM corresponded to different degrees of clonal diversity. Cell lines established from the J3 tumor appeared similar with respect to genomic alterations and thus not consistent with presence of ITH. In contrast, cell lines generated from the J7 tumor displayed a detectable level of clonal diversity with substantially more detected in the J14 cell lines. Whereas the radiosensitivity of cell lines from the J3 tumor did not differ nor did those from the J7, the J14 cell lines were significantly different in terms of radiosensitivity. Based on the analysis of only 2 fragments from each tumor, it is not possible to conclude whether the results from the J3 and J7 tumor cell lines were due to the absence of ITH in radiosensitivity or simply an inadequate number of tumor fragments evaluated. However, the significant difference in radiosensitivity between the cell lines derived for the J14 tumor illustrates the potential for ITH in radiosensitivity. The specific mechanism responsible for the difference in radiosensitivity of the J14 cell lines was not apparent from functional analyses (IPA) of their genetic alterations, which is consistent with mutation profiles typically not being predictive of tumor radiosensitivity. However, a quantitative comparison of the genomic profiles showed that the cell lines generated from the J14 tumor had a greater discrepancy in the number of mutations than the cell lines from J3 and J7 tumors, which had similar radiosensitivities. These results suggest that the more disparate tumor clones are in terms of mutational burden, the higher the possibility of a difference in their radioresponse and ITH in radiosensitivity.
The limitations of this study include the use of only 3 tumors and that only 2 fragments from each tumor were evaluated, which may underestimate the presence of ITH in radiosensitivity. The existence of ITH in GBM radiosensitivity is likely to have the most proximate clinical impact on the treatment for recurrent tumors. Whereas molecular differences have been detected between newly diagnosed and recurrent GBM, whether these affect radiosensitivity and the response to re-irradiation remain to be determined. Although some GBMs respond better than others to radiotherapy, the vast majority recur and require additional therapy. If ITH in clonal radiosensitivity was present at the time of the initial treatment, recurrence is likely to involve the selection and emergence of the radioresistant subpopulations, which would portend a poor response to a re-irradiation protocol (e.g. the J14 tumor evaluated herein). However, because about 50% of recurrent GBMs respond to a second course of radiotherapy (43% 6-month PFS) [26], analyses of the initial surgical specimen for ITH and tumor cell radiosensitivity may aid in selecting patients for re-irradiation protocols.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11060-020-03643-0) contains supplementary material, which is available to authorized users.
References
- 1.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS (2018) CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-Oncol 20:iv1–iv86. 10.1093/neuonc/noy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, Groups EOfRaToCBTaR, Group NCIoCCT (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 3.Hochberg FH, Pruitt A (1980) Assumptions in the radiotherapy of glioblastoma. Neurology 30:907–911. 10.1212/wnl.30.9.907 [DOI] [PubMed] [Google Scholar]
- 4.Yamahara T, Numa Y, Oishi T, Kawaguchi T, Seno T, Asai A, Kawamoto K (2010) Morphological and flow cytometric analysis of cell infiltration in glioblastoma: a comparison of autopsy brain and neuroimaging. Brain Tumor Pathol 27:81–87. 10.1007/s10014-010-0275-7 [DOI] [PubMed] [Google Scholar]
- 5.Lee SW, Fraass BA, Marsh LH, Herbort K, Gebarski SS, Martel MK, Radany EH, Lichter AS, Sandler HM (1999) Patterns of failure following high-dose 3-D conformal radiotherapy for high-grade astrocytomas: a quantitative dosimetric study. Int J Radiat Oncol Biol Phys 43:79–88. 10.1016/s0360-3016(98)00266-1 [DOI] [PubMed] [Google Scholar]
- 6.Oppitz U, Maessen D, Zunterer H, Richter S, Flentje M (1999) 3D-recurrence-patterns of glioblastomas after CT-planned postoperative irradiation. Radiother Oncol 53:53–57 [DOI] [PubMed] [Google Scholar]
- 7.Chan JL, Lee SW, Fraass BA, Normolle DP, Greenberg HS, Junck LR, Gebarski SS, Sandler HM (2002) Survival and failure patterns of high-grade gliomas after three-dimensional conformal radiotherapy. J Clin Oncol 20:1635–1642. 10.1200/JCO.2002.20.6.1635 [DOI] [PubMed] [Google Scholar]
- 8.Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA 110:4009–4014. 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahlokozera T, Vellimana AK, Li T, Mao DD, Zohny ZS, Kim DH, Tran DD, Marcus DS, Fouke SJ, Campian JL, Dunn GP, Miller CA, Kim AH (2018) Biological and therapeutic implications of multisector sequencing in newly diagnosed glioblastoma. Neuro-Oncol 20:472–483. 10.1093/neuonc/nox232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker NR, Hudson AL, Khong P, Parkinson JF, Dwight T, Ikin RJ, Zhu Y, Cheng ZJ, Vafaee F, Chen J, Wheeler HR, Howell VM (2016) Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci Rep 6:22477. 10.1038/srep22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (New York, NY) 344:1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allalunis-Turner MJ, Barron GM, Day RS III, Dobler KD, Mirzayans R (1993) Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat Res 134:349–354 [PubMed] [Google Scholar]
- 13.Ramsay J, Ward R, Bleehen NM (1992) Radiosensitivity testing of human malignant gliomas. Int J Radiat Oncol Biol Phys 24:675–680. 10.1016/0360-3016(92)90713-r [DOI] [PubMed] [Google Scholar]
- 14.Taghian A, Suit H, Pardo F, Gioioso D, Tomkinson K, DuBois W, Gerweck L (1992) In vitro intrinsic radiation sensitivity of glioblastoma multiforme. Int J Radiat Oncol Biol Phys 23:55–62. 10.1016/0360-3016(92)90543-q [DOI] [PubMed] [Google Scholar]
- 15.Ke C, Tran K, Chen Y, Di Donato AT, Yu L, Hu Y, Linskey ME, Wang PH, Limoli CL, Zhou YH (2014) Linking differential radiation responses to glioma heterogeneity. Oncotarget 5:1657–1665. 10.18632/oncotarget.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9:391–403. 10.1016/j.ccr.2006.03.030 [DOI] [PubMed] [Google Scholar]
- 17.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64:7011–7021. 10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed] [Google Scholar]
- 18.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- 19.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN (2015) Cancer stem cells in glioblastoma. Genes Dev 29:1203–1217. 10.1101/gad.261982.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccirillo SGM, Dietz S, Madhu B, Griffiths J, Price SJ, Collins VP, Watts C (2012) Fluorescence-guided surgical sampling of glioblastoma identifies phenotypically distinct tumour-initiating cell populations in the tumour mass and margin. Br J Cancer 107:462–468. 10.1038/bjc.2012.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fael Al-Mayhani TM, Ball SLR, Zhao J-W, Fawcett J, Ichimura K, Collins PV, Watts C (2009) An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods 176:192–199. 10.1016/j.jneumeth.2008.07.022 [DOI] [PubMed] [Google Scholar]
- 22.Flensburg C, Sargeant T, Oshlack A, Majewski I (2018) Super-Freq: integrated mutation detection and clonal tracking in cancer. bioRxiv. 10.1101/380097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenman R, Burk D, Virolainen E, Buick RN, Church J, Schwartz DR, Carey TE (1989) Clonogenic cell assay for anchorage-dependent squamous carcinoma cell lines using limiting dilution. Int J Cancer 44:131–136 [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Smyth GK (2009) ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- 25.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 26.Kazmi F, Soon YY, Leong YH, Koh WY, Vellayappan B (2019) Re-irradiation for recurrent glioblastoma (GBM): a systematic review and meta-analysis. J Neurooncol 142:79–90. 10.1007/s11060-018-03064-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.