Abstract
Microfluidics has proven to be a powerful tool for probing biology at the single-cell level. However, it is only in the past five years that single-cell microfluidics has been used in the field of virology. An array of strategies based on microwells, microvalves and droplets is now available for tracking viral infection dynamics, identifying cell subpopulations with particular phenotypes, as well as high-throughput screening. The insights into the virus-host interactions gained at single-cell level are unprecedented and usually inaccessible by population-based experiments. Therefore, single-cell microfluidics, which opens new avenues for mechanism elucidation and development of antiviral therapeutics, would be a valuable tool for the study of viral pathogenesis.
Keywords: single-cell analysis, microfluidics, virology, heterogeneity, viral infection dynamics, transcriptomic analysis
Dissecting virus-host interplay at the single-cell level
Single-cell biology has accelerated dramatically in the past decade. Many areas of the life sciences and medicine including neuroscience [1], immunology [2], oncology [3], and drug discovery [4] have benefited significantly from single-cell analysis unveiling cell-to-cell heterogeneity and stochasticity in the genome, transcriptome, proteome, and metabolome. Recently, virology has also started to witness the application of single-cell analysis tools [5,6]. Since the lifecycle of viruses relies heavily on the host cell’s machinery, cellular heterogeneity will also be reflected in the unequal outcome of viral infection. Gene expression is fundamentally stochastic [7,8], and the expression of host factors that mediate the viral entry, activate the innate immune response, and shape the intercellular environment for viral replication, is no exception. In addition to the host cellular heterogeneity, individual infectious units delivered to each cell could also be substantially different from each other [9–12]. In particular, for many animal viruses, a plaque forming unit (PFU) may comprise tens to thousands of viral particles [13–15]. Furthermore, RNA viruses evolve with high mutation rates and exist in the form of viral quasispecies (see Glossary), which is a mixture of competent and defective virus variants with distinct genomes and infection abilities [16,17]. Therefore, when a viral infection begins with a low multiplicity of infection (MOI), the introduction of pre-existing viral diversity to individual cells is unavoidable. Together, these multiple layers of heterogeneity predict the intercellular variability in the viral infection outcome. Resolving these differences at the single-cell level would enable the identification of biologically meaningful phenotypes masked by bulk assays, as well as their unique roles during viral infection and/or the emergence of antiviral resistance, and thus represents novel opportunities for mechanism elucidation and development of antiviral therapeutics and vaccines.
Single-cell virology studies have been commonly performed with microwell plates [9,18,19]. Several conventional approaches have been used for single-cell confinement: manual cell micromanipulation and serial dilution are straightforward and low-cost approaches but suffer from laborious procedure; on the other hand, while fluorescence-activated cell sorting (FACS) can send single cells into individual microwells in an automated manner, pre-labeling and large sample volume are required. Additionally, the low throughput of these plate-based approaches has limited their scope of application. For example, to map the variation in the kinetics of viral RNA production, infections initiated by single infectious particles are desired, because the effect of kinetic stochasticity on individual reactions would be averaged and obscured with multiple viral genomes [18]. As direct observation and manipulation of single virions under cell culture conditions remains an unsolved challenge due to their tiny size, experiments of this sort are often implemented by infecting the cells at MOIs <0.1 for random pairing. The sample size, i.e. the number of virus-infected cells, is further reduced under such conditions. Through miniaturizing and integrating experimental workflows, microfluidics provides the capability of overcoming these disadvantages [20–22]. In particular, the small size of microfluidic devices significantly facilitates single-cell biochemical assays and parallel microscopic observation. As thoroughly summarized in several recent reviews [23–25], large-scale capture or patterning of single cells in microfluidic devices has been achieved via a broad range of mechanisms such as dielectrophoresis, optical tweezers, hydrodynamics and acoustic waves. When harmonizing these methods with virological studies, it is important to consider that viruses could be released from infected cells even before cell lysis [26,27]. Unambiguous discrimination between primary and secondary infections in a mixture is technically difficult. To avoid adding one more layer of complexity to the data processing and analysis, single cells and virions ideally should be partitioned into individual compartments where virus-host interactions take place without crosstalk. In consideration of this requirement, most of those relatively practical approaches reported so far could be categorized into microwell-, valve-, and droplet-based strategies (Figure 1, Key Figure). These approaches are different in characteristics such as throughput, spatio-temporal resolution and commercial availability, and therefore offer a resourceful toolbox for diverse applications. In the following sections, we review key challenges in virology that microfluidic single cell technology can address.
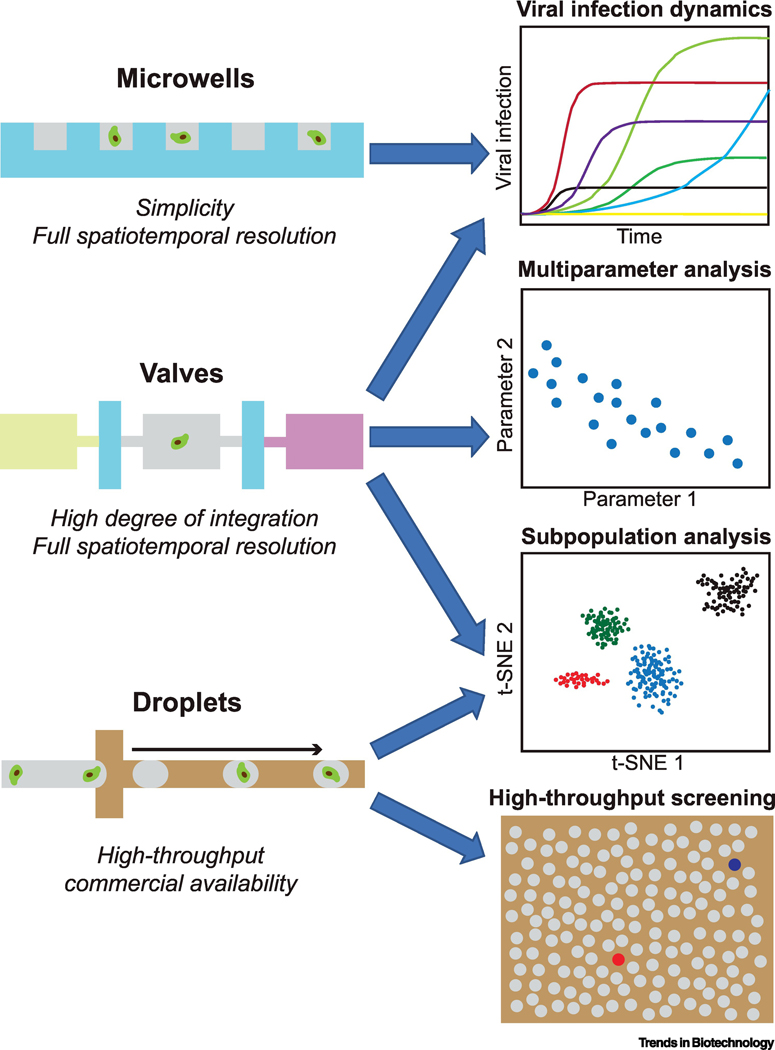
Figure 1, Key Figure.
Single-cell microfluidics technologies and their typical applications in virology. Microwell-, valve-, and droplet-based strategies are applied in virology studies. Features and typical application opportunities of these methods are presented.
Kinetic analysis of viral infection dynamics
Snapshot determinations of the outcome of viral infection, even at the single-cell resolution, usually quantify the infectious viral particles and nuclei acids. The virus-host interplay itself, to a large extent, remains a black box. Cells have many immune defense functions to restrict or limit viral infection, while viruses have evolved numerous strategies to evade them. Given the same data set, distinct explanations might be plausible. Continuous and non-invasive acquisition of the temporal information of viral infection in single cells provides novel opportunities to correlate specific cellular and molecular features with the fate of the two partners. Combining with live-cell imaging, microfluidic platforms are ideal for monitoring the viral infection dynamics in parallel. One of the first examples was reported in 2015 [28]. Ramji and colleagues used a passive-flow microfluidic channel with 408 to 765 traps to dock single Jurkat T cells in an array and thereupon track the activation dynamics of latent human immunodeficiency virus (HIV) with a green fluorescent protein (GFP) reporter. Noisy HIV reactivation in response to latency reversing agents (LRAs) was observed in this study, indicating a limitation of the effectiveness of the “activate-and-kill” HIV-cure strategy. While this microfluidic approach could be useful under certain circumstances, for unambiguous and long-term monitoring of viral replication in single cells, microwell/valve-based devices are more preferred to prevent crosstalk between infected cells.
The most outstanding characteristic of microwell-based devices is their simplicity. Generally, microwell arrays with each well in a picolitre or nanolitre volume are fabricated from polydimethylsiloxane (PDMS), silicon or glass [29]. For example, the Yin group developed PDMS arrays of 2,500 cubic microwells of 50 μm in length, onto which vesicular stomatitis virus (VSV) infected cells in suspension were directly loaded for random settlement by gravitation (Figure 2A) [30,31]. Theoretically, the seeding cell density could be optimized to maximize the single-cell occupancy (~37%) according to the Poisson distribution, but extra procedures might be required to identify the cell number in each well. Alternately, lower cell densities could be used, leaving most wells empty. Nevertheless, this limit can be overcome by dimension optimization and surface modification of the microwells [32]. Afterwards, the microwells were conveniently sealed to block crosstalk by securing a glass slide lid on the top of the whole device. The researchers also established a dual-color fluorescent reporter system to track the dynamics of virus-host interactions in single cells, where red fluorescent protein (RFP) signals indicated production of infectious virus particles while GFP readouts manifested the activation of cellular innate immunity [33]. With this fluorescent reporter system for live-cell imaging, monitoring of arrayed cells with full spatio-temporal resolution is permitted, making this microwell-based platform superior in tracking viral infection dynamics on the scale of thousands of single cells.
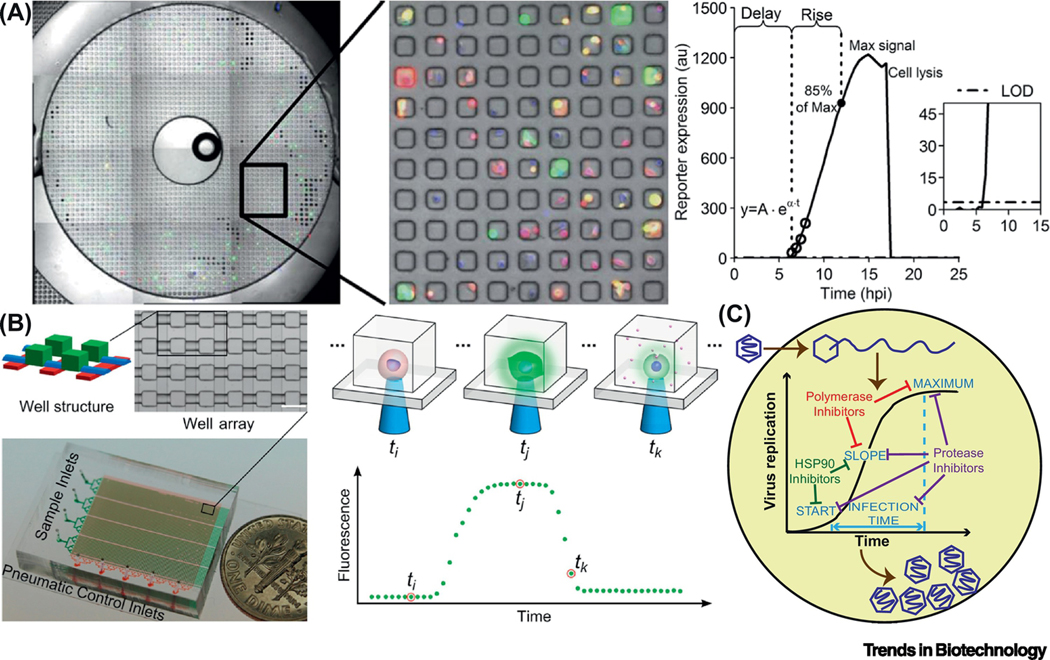
Figure 2.
Microfluidic platforms for kinetic analysis of viral infection dynamics in single cells. (A) One of ten bull’s-eyes on a PDMS microwell device, each containing 2,500 sub-nanoliter volume wells. VSV infected PC3 cells were loaded into the microwell array. They were engineered so that RFP and GFP signals indicated the production of infectious virus particles and the activation of cellular innate immunity, respectively. A model was used to process the fluorescent trajectories from each single cell and derive kinetic parameters. Delay-time: the first data point above the limit of detection (LOD); maximum signal: the highest intensity reached; rise-time: the period between the delay-time and the time when 85% of the maximum signal was reached; production-rate: speed of the production, obtained by fitting the first four data-points above the LOD with an exponential curve. Adapted with permission from [34]. (B) A device composed of 4 groups of 1,600 microchambers for single-cell cultivation, which could be sealed with pneumatic valves. A complete time course of infection by a virus expressing a fluorescent reporter could be recorded. Adapted with permission from [35]. (C) Single-cell analysis of viral infection dynamics could inform mechanism of actions of antiviral drugs [39].
Following the same streamlined pipeline, it is possible to quantitatively characterize the dynamic kinetics of viral and host gene expression, as well as how they contribute to the outcome of VSV infection [34]. As reflected by Pearson correlation coefficients, viral protein production was correlated with innate immune response on onset and increase time, but not on magnitude. Higher yields of virus multiplication were facilitated by earlier viral gene expression and slower anti-viral responses. Moreover, single cells in which a viral stress response was activated were likely to produce more viral proteins and lyse earlier. It was thus speculated that anti-viral cytokines secretion from these cells was enhanced to “warn” more neighboring cells, as a potential mechanism to suppress the spread of viral infection. Overall, this work underscores the superiority of single-cell analysis of viral infection in identification and quantification of sub-populations and rare events over conventional bulk assays.
Guo and colleagues reported a valve-based device that was assembled from multiple PDMS layers and composed of 4 groups of 1,600 microchambers for cultivation of single, virus-infected cells (Figure 2B). These chambers were connected to channels that could be blocked by deformation of specific channels in the control layer [35]. These pneumatic valves could be reversibly switched by alternating the pressure applied on the control layer(s), owing to the elastic nature of PDMS [36,37]. However, this was essentially a “microwells-like” platform, because the valves were merely used to isolate virus-infected cells infused into the microfluidic channels, playing a role similar to the glass slide lid on microwells. Employing poliovirus-GFP (PV-GFP) as the model, viral infection with a variety of MOIs, cell cycle stages and viral genotypes were performed with live-cell imaging for a 24 h period. Fluorescence intensities detected from single cells over time were processed in an automatic manner with a customized R package [38]. The kinetic parameters were compared in order to elucidate origins of the observed between-cell variability of infection outcomes. Independent contributions of viral and host factors, which are normally masked by population-based experiments, were deconvoluted: the yield and speed of viral replication were virus-dependent whereas the onset was host-dependent.
For the microwell- or valve-based devices mentioned above, practically only 10~30% of the microwells accommodate a single cell. To more adequately exploit the device capacity, Liu and colleagues recently added a trapping microstructure to each microchamber and improved the single-cell occupancy to 86% [39]. Using this device with 5,700 (5×1,140) microchambers, single-cell analysis of antiviral therapeutics was enabled, because even under a condition that the fraction of infected cells was reduced to 10%, the number of infected cells in this group (~100) would still be enough to perform statistical tests. When evaluating different classes of antivirals on the dynamics of PV infection, single-cell analysis provided information on the mechanism of action and synergy of antiviral cocktails (Figure 2C). The resolution is unprecedented as conventional population-based approaches only provide a measure of efficacy. Therefore, single-cell analysis holds the promise to have a transformative impact on the drug development process.
Transcriptome analysis of virus-host interactions
Any phenotypic cell-to-cell variation in the outcome of viral infection is essentially the manifestation of the heterogeneity in mRNA and proteins. In recent years, there has been a growing interest in exploiting transcriptome analysis of virus-host interactions to explore viral diversity and evolution, map cellular heterogeneity in response to viral infection, and particularly, identify cell subpopulations displaying specific phenotypes (Figure 3A) [5,6]. As the core element, single-cell RNA sequencing (scRNA-seq) has been realized on disparate platforms. However, plate-based methods, which are highly dependent on manual cell picking or FACS for single-cell isolation, suffer from complicated workflows involving large numbers of assay plates [40]. Microfluidic-generated nanoliter-scale droplets have emerged as advantageous reactors for scRNA-seq with high throughput and improved sensitivity [20,41,42].
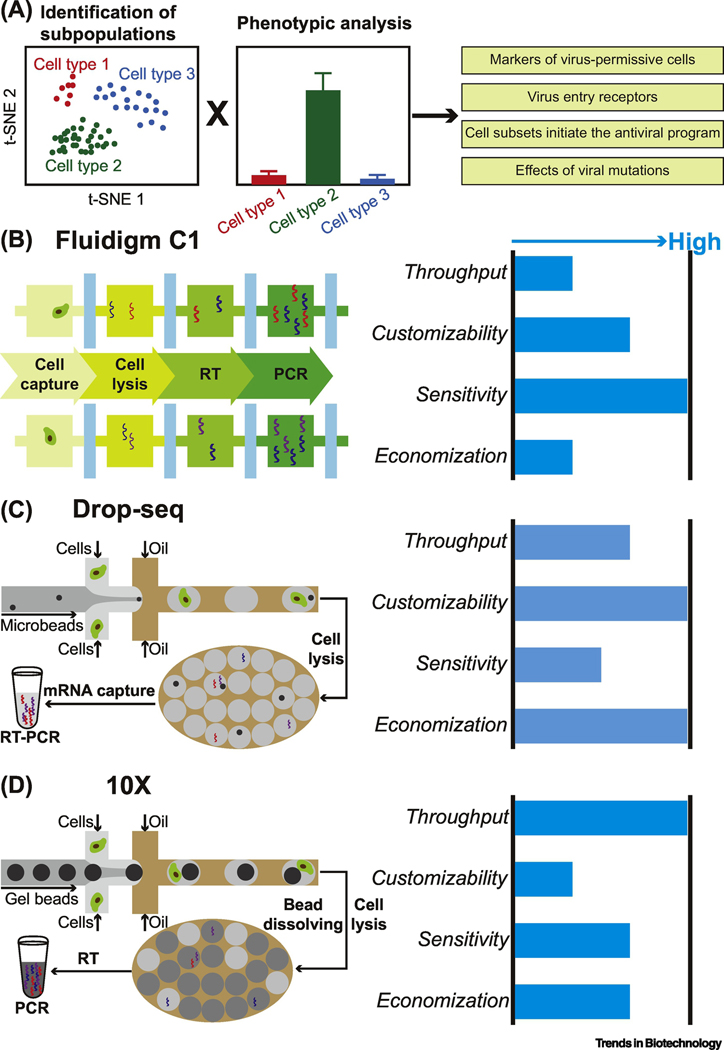
Figure 3.
Transcriptome analysis of virus-host interactions at the single-cell level. (A) ScRNA-seq reveals distinct cell subpopulations. Combined with phenotypical analysis, single-cell transcriptome analysis could be used to investigate viral diversity and identify cell subpopulations with specific phenotypes. (B-D) Characteristics of main scRNA-seq technologies (Fluidigm C1, Drop-seq and 10X) applied in the field of virology.
A few microwell-based devices reported for RNA-seq of virus-infected single-cells still require complex fluid manipulations to separately load different reagents into each well [43]. In contrast, a unique characteristic of valve-based microfluidics is its capability to accomplish a scRNA-seq workflow step-by-step. By programming the operations of the in-line microvalves, reagents can be directed to prescribed locations in a stepwise manner [44]. Such high-density microfluidic chips are called integrated fluidic circuits (IFCs)” A commercially available platform is Fluidigm C1 (Figure 3B) [45], which can process up to 800 cells in parallel. Precise control of the IFCs enables automatic cell capture and lysis, reverse transcription, and PCR amplification of cDNA in nanoliter reaction volumes. Single-cell reaction products are then recovered and quantified. Using this tool, the phenotypic heterogeneity in viral gene expression among single cells can be uncovered, as demonstrated with a host-virus model system, Emiliania huxleyi and its specific virus EhV-201 [46]. Based on this heterogeneity, cells at different infection states could be discerned and an unrecognized host response was revealed. Besides, more investigations were conducted from the perspective of the cells by resolving the heterogeneity in cellular transcriptome in response to viral infection. By examining the expression of receptors mediating flavivirus entry into single cells in the developing brain, Nowakowski and colleagues revealed a plausible candidate responsible for Zika virus (ZIKV) infection of neural stem cells [47]. In another ZIKV infection study, Onorati and colleagues performed single-cell RNA-sequencing on neocortical and spinal cord neuroepithelial stem cells to model ZIKV-related neuropathogenesis [48]. The Ciuffi group respectively identified biomarkers of HIV permissiveness and a 134-gene-specific transcriptional signature of the HIV-inducible cell [49,50]. Collectively, these studies show the attractive power of microfluidics-based scRNA-seq in revealing cellular and molecular mechanisms during viral pathogenesis.
Cell subpopulations with particular phenotypes could significantly affect the overall population behavior during viral infection. Identifying these subpopulations, which might be very rare, is challenging for valve-based systems, as they require relatively large physical space for each single-cell unit and addition peripherals to provide actuation. Under such circumstances, droplet microfluidics that permits several orders of magnitude higher throughputs is preferred. In general, monodisperse water-in-oil droplets are generated in microfluidic channels with a flow-focusing [51] or T-junction [52] design at a frequency as high as thousands per second. With certain configurations of sampling channels, single cells and reagents could be encapsulated together into individual droplets. Millions of such independent micro-reactors can be obtained and stabilized in oil. Barcoding strategies are used during the cell encapsulation process to distinguish reaction products in individual droplets from each other, so the droplets can be broken later for analysis in their entirety [53]. For example, Macosko and colleagues reported an open-source Drop-seq pipeline (Figure 3C) [54]: first, microbeads functionalized with barcoded oligonucleotides are encapsulated together with cells and reagents in droplets using a microfluidic flow-focusing geometry; then, single cells are subsequently lysed in individual droplets and the polyadenylated mRNAs released are captured by the microbeads; and finally, the barcoded transcriptomes are recovered from the droplets for the following amplification and sequencing. Utilizing this platform for transcriptional profiling of HSV-1-infected single cells, Drayman and colleagues identified a rare subset of abortively infected cells that initiate the “anti-viral program” [55]. Moreover, developmental pathways, for example the WNT/β-catenin pathway, were significantly upregulated in highly infected cells. Similarly, using the same technique, Wyler and colleagues found that in a subset of HSV-1-infected cells, the transcription factor NRF2 was activated, which was correlated to an anti-viral program [56]. In both studies, single-cell microfluidics paved the way for identification of key cell subpopulations with phenotypes that would not be manifested when evaluated with bulk assays.
In Drop-seq, both the microbeads and the cells for loading are usually diluted to minimize their respective multiplet rates in droplets. However, following a double-Poisson distribution, a natural byproduct is the low cell capture rate (number of cells sequenced/number of cells loaded; <5%), which becomes an obstacle when limited samples are available. To achieve a super-Poissonian distribution, close-packing of deformable particles has been demonstrated feasible [57]. Utilizing this strategy, the GemCode technology has been developed and commercialized by 10X Genomics to encapsulate single gel beads into droplets at ~80% fill rate, resulting in cell capture rates higher than 40% (Figure 3D) [58]. Tens of thousands of single cells in droplets could be generated and collected per ~6-min run. Despite higher experimental cost, this platform exhibits better molecular sensitivity and precision than Drop-seq [59], and hence has been welcomed by the virology community. Several research groups have used it to quantify gene expression during the infection of ZIKV [60], HIV [61], influenza virus [62–67], cytomegalovirus (CMV) [68–72], and human papillomavirus (HPV) [73], respectively. For instance, the Bloom group unveiled extremely wide cell-to-cell variation during influenza A virus (IAV) infection [62], which was later re-observed by other groups [63–65]. Quantitatively, Gini coefficients over 0.64 were calculated from the distributions of viral mRNA amount in infected cells. While this heterogeneity was partially ascribed to the absence of some viral genes in some infected cells, contributions from other viral and host factors were not neglectable. On this basis, the group developed a method to combine transcriptome analysis and full-length PacBio sequencing of all viral genes in single cells [70]. Thereupon, viral genetic variation of IAV, well known for its rapid mutation, was assessed as a source of heterogeneity in innate immune activation among virus-infected cells. Even though diverse immunostimulatory defects were identified in most virions, viral genotype-independent stochastic outcome of infection still existed. It was thus concluded that innate immune response to influenza virus infection could not be deterministically induced solely by interferon-inducing viral genetic defects, once again suggesting the complexity but also the necessity of studying viral infection at the single-cell resolution.
A limitation of both Drop-seq and 10X Genomics technologies is that only A-tailed mRNA transcripts would be quantified. Recently reported droplet-assisted RNA targeting by single-cell sequencing (DART-seq), which was a simple alteration of the Drop-seq, promises multiplexed RNA amplicon sequencing and transcriptome profiling simultaneously in single cells [74]. This was accomplished by attaching custom primers with different sequences to a subset of the oligo dT sequences on Drop-seq beads. Using reovirus as a model, the researches demonstrated the capability of DART-seq in capturing multiple non-polyadenylated transcripts, in addition to transcriptomes in single cells.
Multi-parameter measurements
Recently, efforts have been made towards acquiring multi-dimensional data for more comprehensive analysis on virus-host interactions. For example, in order to extract biological meaningful information from the scRNA-seq data, the association between a specific phenotype of interest and a specific transcriptional signature should be established. Usually, the cell phenotype is assessed by protein expression and activity at the population level. A more straightforward strategy would be characterizing the phenotype of interest and the transcriptome on each single cell. To execute such workflows for multiple omics analyses, valve-based microfluidic systems shows attractive promise because of their capability of high-degree integration of complex experimental procedures from cell loading, biochemical stimuli, washing, cell lysis, to lysate retrieval in a highly controllable and hands-off manner. The Tay group developed an assay for ultra-sensitive quantification of protein and mRNA in up to 144 single cells compartmentalized in microfluidic chambers (Figure 4) [75]. As an application, the authors explored herpes simplex virus 1 (HSV-1) infection at the single-cell resolution. A549 cells were infected by an HSV-1 strain expressing infected-cell polypeptide 4-yellow fluorescent protein (ICP4-YFP), loaded onto the device, monitored by time-lapse imaging, and lysed to determine HSV-1 viral proteins. Unsurprisingly, an evidently higher abundance of viral proteins was found in those YFP-positive cells, due to the essential role of ICP4 in the regulation of viral gene expression. However, the perceived YFP intensity was only partially correlated with viral protein counts, suggesting the existence of abortive infection.
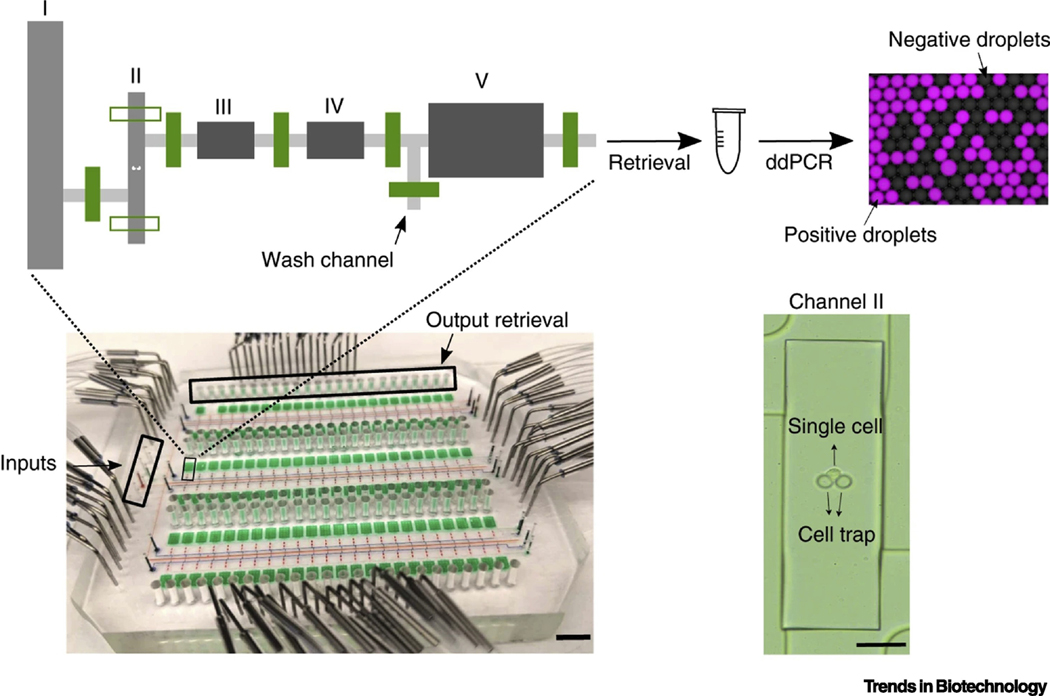
Figure 4.
An integrated valve-based microfluidic system for multi-parameter measurements. A device containing 144 assay chambers and the schematic of one unit are shown. Inset shows a single cell is trapped in chamber II. By switching the valves, on-chip cell loading, lysis, proximity ligation and product retrieval could be accomplished stepwisely. Droplet digital PCR (ddPCR) was then used for quantification of proteins or mRNA. Adapted from [75], under a Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
High-throughput screening
As suggested by viral quasispecies theory [16,17], infection experiments implemented in bulk should be dominated by major alleles. The only way to study those minor alleles would be isolating thousands or even millions of single cells infected by single virions without competition. Well-known for high-throughput single-cell analysis and screening applications, droplet-microfluidic platforms provide possibility to discover exceedingly rare biomarkers [76,77]. As such, Fischer and colleagues reported a flow focusing system to co-encapsulate murine norovirus (MNV-1) and single RAW 264.7 cells in droplets with volumes of ~65 pL [78]. The viral particles were diluted to infect the cells at an MOI of 0.01 in the presence of neutralizing monoclonal antibody. Infectious particles detected after 24 hours was about half of genome copy number, which was a much higher fraction than those obtained with standard bulk experiments. Therefore, under such a condition, virus variants able to escape evolutionary stressors could be effectively isolated. This platform represents a high-throughput and cost-effective tool for isolation and analysis of minor alleles in viral quasispecies, which might be extended to screening applications.
As mentioned earlier, using a passive-flow microfluidic device, noisy HIV reactivation was observed in response to LRAs [28]. With the single cells docked in the same microchannel, expression of HIV-GFP in activated cells could be tracked, but it was impractical to quantify the amount of HIV RNA associated to each cell or sort cells out for downstream in-depth characterization. To address this issue, Yucha and colleagues used a T-junction microfluidic device for encapsulation of lymphocytes latently infected by HIV [79]. In contrast to the commercial Fluidigm and 10X Genomics platforms designed for polyadenylated mRNA analysis, this relatively cost-effective approach also allowed fluorescence detection of unspliced RNA and multiply spliced RNA, with the use of intra-droplet PCR amplification of HIV RNA. As a result, HIV transcriptionally reactivated cells could be screened for characteristics of human gDNA and mRNA. Application of the methodology to CD4+ T cells isolated from participants on an antiretroviral therapy revealed that when evaluating HIV reactivation at the single-cell level, LRAs may either enhance transcription from active cells or increase the number of transcriptionally active cells. Overall, this heterogeneity highlighted the potential of single-cell analysis to promote the development of HIV reactivation strategies.
Concluding remarks and future perspectives
Single-cell virology has been largely an uncharted territory until recently. We have summarized the key ideas and specific examples of using single-cell microfluidics as a transformative tool to improve our understandings in the molecular and cellular basis of virus-host interactions (Table 1), which are practically inaccessible through traditional population-scale experiments. Most of these exploratory studies were accomplished in the past five years. In addition to microfluidics, the availability of computational biology methods [80] and single-cell sequencing techniques [20] has fueled a rapid progress in this field. The modern era of single-cell virology is just beginning and will substantially contribute to the war against viral infectious diseases.
Table 1.
Applications of single-cell microfluidics in virology
| Application | Microfluidic technology | Throughput (No. of cells) | Virus of interest | Description | Reference(s) |
|---|---|---|---|---|---|
| Viral infection dynamics | Trap array, customized | Up to 700 | HIV | Drug-specific patterns of noisy HIV activation dynamics were revealed |
[28] |
| Microwells, customized | 2000~3000 | VSV | Interferential effects of defective interfering particles (DIPs) on viral infection were characterized |
[30,31] | |
| Innate immune activation by VSV infection was quantitative profiled |
[34] | ||||
| Valves, customized | 1000~2000 | PV | Viral and host factors independently contributed to the cell-to-cell variation of infection outcomes | [35] | |
| ~5000 | PV | Single-cell analysis of antiviral therapeutics revealed more than efficacy | [39] | ||
| Transcriptome analysis | Microwells, customized | 600~700 | HPV | Diversity of HPV-18 expression and splicing at the single-cell level was characterized |
[43] |
| Valves, Fluidigm C1 | Up to 800 | EhV201 | Heterogeneity in viral gene expression was unmasked to map cells into their infection state | [46] | |
| ZIKV | Expression analysis revealed a candidate ZIKV entry receptor in neural stem cells | [47] | |||
| Neuroepithelial stem cells were characterized to model ZIKV-induced pathogenesis |
[48] | ||||
| HIV | Cellular markers of HIV permissiveness were revealed by single-cell RNA-sequencing |
[49] | |||
| A 134-gene-specific transcriptional signature for the HIV-inducible cell was identified |
[50] | ||||
| Droplets, Drop-seq (open-source) | Thousands | HSV-1 | Activation of anti-viral and developmental programs were revealed in distinct sub-populations | [55] | |
| In a subpopulation of infected-cell, NRF2 was activated, which restrict HSV-1 infection |
[56] | ||||
| Droplets, 10X Genomics | Tens of thousands | ZIKV | An immunocompetent mouse model of ZIKV infection was established |
[60] | |
| HIV | Host transcriptional factors associated with HIV downregulation were identified | [61] | |||
| IAV | Extremely wide cell-to-cell variation in viral gene expression was observed | [62] | |||
| Innate immune response to IAV in the respiratory epithelium was investigated at the single-cell level | [63] | ||||
| A common pattern of IAV gene expression IAV heterogeneity that governs the antiviral response was revealed | [64] | ||||
| Impact of defective virus genomes on host cell responses was evaluated | [65] | ||||
| PacBio sequencing was used to assess how viral mutations contribute to the heterogeneity in immune activation |
[66] | ||||
| IBV | A cell population that can survive IBV infection was found to maintain epithelial barrier function |
[67] | |||
| Human CMV | Transcriptional landscape during human CMV latent infection was defined | [68] | |||
| Latently infected cells were driven towards a weaker immune-responsive monocyte state | [69] | ||||
| A subpopulation of myeloid cells permissive to human CMV replication was identified | [70] | ||||
| CRISPR/Cas9-based screening was used for high-resolution scanning of functional elements in the human CMV genome | [71] | ||||
| Murine CMV | Ly49R activation receptor was needed for natural killer cell immunity against murine CMV infection |
[72] | |||
| HPV | HPV expression differed between hyperplastic skin lesions | [73] | |||
| Droplets, DART-seq | Tens of thousands | Reovirus | DART-seq enabled amplicon sequ transcriptome profiling technology multiplexed encing and | [74] | |
| Multi-parameter measurements | Valves, customized | Up to 144 | HSV-1 | A platform was developed for ultra-sensitive quantification of protein and mRNA |
[75] |
| High-throughput screening | Droplets, customized | Thousands to millions | MNV-1 | A platform was developed for isolation and analysis of minor alleles in viral quasispecies | [78] |
| Hundreds of thousands | HIV | HIV transcriptionally reactivated cells were screened for downstream in-depth characterization |
[79] |
Kinetic analysis of viral infections is performed through un-interrupted microfluidic cultivation and imaging of single cells. The more components are tracked, the deeper the molecular network of virus-host interactions would be dissected. Future integration of novel fluorescence labeling techniques [81,82] with the platforms should allow joint analysis of viral and cellular heterogeneity with high spatio-temporal details. On the other hand, in transcriptome analyses, the impact of batch effects on the observed cell-to-cell expression variability should not be underestimated, which is a common challenge faced by all scRNA-seq technologies [83]. A solution to reduce this impact, from the technical perspective, would be increasing the number of samples processed in one run (currently 1 for Fluidigm C1 and drop-seq; 8 for 10X).
Ideal microfluidic technologies would also allow both high-throughput analysis and multi-parameter measurement, two capabilities which unfortunately usually exist in tension with each other. Because performing multiplexed assays targeting different analytes of interest, let alone detection at multiple time points, would inevitably drive the overall design, the integration of functional elements, and the automated manipulation of the microfluidic devices towards a more sophisticated direction. As a result, the number of single cells that can be accommodated within a given device footprint would be reduced. Therefore, while realizing the ultimate aim asks for continuous and incremental efforts from engineers and scientists with diverse expertise (see Outstanding Questions), near-future development is likely to innovate and further develop at least one of the two capabilities. As such, systems that demand minimal prior experience in operation and meanwhile promise high-throughput transcriptomic analysis have been commercialized. Because these “plug-and-play” tools are user-friendly, we have already started to witness the increasing utilization by virologists to gain the next phase of insights into virus-host interactions. The other focus, “complex” systems promising multi-parameter measurement, will require tighter collaborations between engineers and virologists to identify specific analytic targets and determine the roadmap for assay development accordingly. Although the development and operation of these highly individualized microfluidic devices are typically carried out by skilled users, which somehow hinders widespread popularization in virology laboratories, they will confer more impressive opportunities to answer frontier questions hanging over the field.
Outstanding questions:
Other than single cells, how can single virions, which are much smaller in size, be manipulated on microfluidic platforms?
How to find the right balance between the depth to which the single cells are analyzed and the number of cells?
RNA-sequencing is the most popular omic analytical method applied in single-cell virology. How to integrate other omic analysis tools, for example mass spectrometry for proteomic analysis, into the microfluidic platforms for multi-omics analysis?
Current sequencing methods only quantify the transcriptome profiles at a specific time point, usually post viral infection. Would future technologies achieve analysis both before and after infection?
How to study the cell-to-cell spread of viral infection at the single-cell level?
Another challenge for widespread implementation of these platforms, in our opinion, lies not in the microfluidic technologies, but instead the data processing and analysis methods. With unique virology questions in mind, single-cell data sets, usually huge in size, have been obtained with differences in quality and/or format, and are therefore processed by diverse customized computational models and tools. This trend seems likely to continue as the data acquisition capability being pushed toward multimodal omics [84]. Adopting a flexible strategy at this early, exploratory phase of investigation is fairly reasonable, but it also increases the difficulty to compare and integrate cross-experiment data. The emergency of relatively unified but versatile pipelines will hallmark the maturation of this research field.
Highlights:
Recent development in single-cell microfluidics is about to unlock a new era of virology, by providing additional dimensions of our understanding in virus-host interactions and antiviral therapeutic mechanisms.
Microwell- and valve-based technologies have been developed for resolving viral infection dynamics at the single-cell resolution. Novel insights and perspectives masked by conventional population-based experiments have been revealed.
Transcriptome analysis of virus-host interactions at the single-cell level has entered the age of mass production along with the commercialization of several valve- and droplet-based microfluidic platforms for single-cell sequencing.
Single-cell microfluidics technologies with higher degrees of integration and more comprehensive analytical capability are under way, which might revolutionize the study of viral pathogenesis.
Acknowledgement
This work was supported by NIH/NIAID 7R01AI134911 awarded to S.-Y. Z.
Glossary
- Defective interfering particles
virus-like particles with large deletions in the infectious genome, spontaneously formed during virus growth
- Fluorescence-activated cell sorting (FACS)
a specialized form of flow cytometry that sorts cells into sub-populations based on fluorescent characteristics of each cell
- Gini coefficient
a measurement of the degree of inequality in a distribution. This coefficient ranges from 0 (complete equality) to 1 (complete inequality)
- Latency reversing agents (LRAs)
pharmacological compounds that induce HIV out of latency, allowing its recognition by the immune system
- Multiplicity of infection (MOI)
the number of virions that infect a cell
- PacBio sequencing
a single-molecule real-time DNA sequencing method provides long sequencing reads, developed by Pacific BioSciences
- Polydimethylsiloxane (PDMS)
a common material used for the fabrication of microfluidic devices
- Viral quasispecies
a population structure of viruses with an extremely large number of variant genomes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Poulin J. et al. (2016) Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay PK et al. (2014) Single-cell technologies for monitoring immune systems. Nat. Immunol 15, 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baslan T. and Hicks J. (2017) Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat. Rev. Cancer 17, 557–569. [DOI] [PubMed] [Google Scholar]
- 4.Heath JR et al. (2016) Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov 15, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rato S. et al. (2017) Exploring viral infection using single-cell sequencing. Virus Res. 239, 55–68. [DOI] [PubMed] [Google Scholar]
- 6.Cristinelli S. and Ciuffi A. (2018) The use of single-cell RNA-Seq to understand virus–host interactions. Curr. Opin. Virol 29, 39–50. [DOI] [PubMed] [Google Scholar]
- 7.Raj A. and van Oudenaarden A. (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elowitz MB (2002) Stochastic gene expression in a single cell. Science 297, 1183–1186. [DOI] [PubMed] [Google Scholar]
- 9.Combe M. et al. (2015) Single-cell analysis of RNA virus infection identifies multiple genetically diverse viral genomes within single infectious units. Cell Host Microbe 18, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vignuzzi M. et al. (2006) Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renner DW et al. (2018) Impacts of genome-wide analyses on our understanding of human herpesvirus diversity and evolution. J. Virol 92, e00908–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue KS et al. (2018) Within-host evolution of human influenza virus. Trends Microbiol. 26, 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korboukh VK et al. (2014) RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem 289, 29531–29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfson KJ et al. (2015) Particle-to-PFU ratio of Ebola virus influences disease course and survival in cynomolgus macaques. J. Virol 89, 6773–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chouljenko DV et al. (2012) Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J. Virol 86, 4262–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andino R. and Domingo E. (2015) Viral quasispecies. Virology 479–480, 46–51. [DOI] [PMC free article] [PubMed]
- 17.Lauring AS and Andino R. (2010) Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 6, e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte MB and Andino R. (2014) Single-cell analysis uncovers extensive biological noise in poliovirus replication. J. Virol 88, 6205–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heldt FS et al. (2015) Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat. Commun 6, 8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prakadan SM et al. (2017) Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat. Rev. Genet 18, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett MR and Hasty J. (2009) Microfluidic devices for measuring gene network dynamics in single cells. Nat. Rev. Genet 10, 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reece A. et al. (2016) Microfluidic techniques for high throughput single cell analysis. Curr. Opin. Biotech 40, 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosic S. et al. (2015) Microfluidic sample preparation for single cell analysis. Anal. Chem 88, 354–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin L. et al. (2018) Micro/nanofluidics-enabled single-cell biochemical analysis. TrAC-Trend. Anal. Chem 99, 66–74. [Google Scholar]
- 25.Wu J. and Lin J. (2019) Microfluidic technology for single-cell capture and isolation. In Microfluidics for Single-Cell Analysis (Lin J, ed), 27–51, Springer Singapore. [Google Scholar]
- 26.Sattentau Q. (2008) Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol 6, 815–826. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y. et al. (2015) Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramji R. et al. (2015) A passive-flow microfluidic device for imaging latent HIV activation dynamics in single T cells. Integr. Biol 7, 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindstrom S. and Andersson-Svahn H. (2011) Miniaturization of biological assays - overview on microwell devices for single-cell analyses. Biochim. Biophys. Acta-Gen. Subj 1810, 308–316. [DOI] [PubMed] [Google Scholar]
- 30.Warrick JW et al. (2016) Tools for single-cell kinetic analysis of virus-host interactions. PLoS One 11, e0145081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akpinar F. et al. (2016) High-throughput single-cell kinetics of virus infections in the presence of defective interfering particles. J. Virol 90, 1599–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Revzin A. et al. (2005) Development of a microfabricated cytometry platform for characterization and sorting of individual leukocytes. Lab Chip 5, 30–37. [DOI] [PubMed] [Google Scholar]
- 33.Swick A. et al. (2014) Visualizing infection spread: dual-color fluorescent reporting of virus-host interactions. Biotechnol. Bioeng 111, 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timm AC et al. (2017) Quantitative profiling of innate immune activation by viral infection in single cells. Integr. Biol 9, 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo F. et al. (2017) Single-cell virology: on-chip investigation of viral infection dynamics. Cell Rep. 21, 1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unger MA (2000) Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116. [DOI] [PubMed] [Google Scholar]
- 37.Hong JW and Quake SR (2003) Integrated nanoliter systems. Nat. Biotechnol 21, 1179–1183. [DOI] [PubMed] [Google Scholar]
- 38.Caglar MU et al. (2018) Sicegar: R package for sigmoidal and double-sigmoidal curve fitting. PeerJ 6, e4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W. et al. (2019) More than efficacy revealed by single-cell analysis of antiviral therapeutics. Sci. Adv 5, eaax4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaitin DA et al. (2014) Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Streets AM et al. (2014) Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. U. S. A 111, 7048–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu AR et al. (2014) Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 11, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L. et al. (2015) Full-length single-cell RNA-seq applied to a viral human cancer: applications to HPV expression and splicing analysis in HeLa S3 cells. GigaScience 4, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kellogg RA et al. (2014) High-throughput microfluidic single-cell analysis pipeline for studies of signaling dynamics. Nat. Protoc 9, 1713–1726. [DOI] [PubMed] [Google Scholar]
- 45.Pollen AA et al. (2014) Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol 32, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenwasser S. et al. (2019) Unmasking cellular response of a bloom-forming alga to viral infection by resolving expression profiles at a single-cell level. PLoS Pathog. 15, e1007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowakowski TJ et al. (2016) Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell 18, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onorati M. et al. (2016) Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rato S. et al. (2017) Single-cell analysis identifies cellular markers of the HIV permissive cell. PLoS Pathog. 13, e1006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golumbeanu M. et al. (2018) Single-cell RNA-seq reveals transcriptional heterogeneity in latent and reactivated HIV-infected cells. Cell Rep. 23, 942–950. [DOI] [PubMed] [Google Scholar]
- 51.Anna SL et al. (2003) Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett 82, 364–366. [Google Scholar]
- 52.Nisisako T. et al. (2002) Droplet formation in a microchannel network. Lab Chip 2, 24–26. [DOI] [PubMed] [Google Scholar]
- 53.Klein AM et al. (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macosko EZ et al. (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drayman N. et al. (2019) HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations. eLife 8, e46339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyler E. et al. (2019) Single-cell RNA-sequencing of herpes simplex virus 1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun 10, 4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abate AR et al. (2009) Beating Poisson encapsulation statistics using close-packed ordering. Lab Chip 9, 2628–2631. [DOI] [PubMed] [Google Scholar]
- 58.Zheng GXY et al. (2017) Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X. et al. (2019) Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell 73, 130–142.e5. [DOI] [PubMed] [Google Scholar]
- 60.Gorman MJ et al. (2018) An immunocompetent mouse model of Zika virus infection. Cell Host Microbe 23, 672–685.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradley T. et al. (2018) Single-cell analysis of quiescent HIV infection reveals host transcriptional profiles that regulate proviral latency. Cell Rep. 25, 107–117.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell AB et al. (2018) Extreme heterogeneity of influenza virus infection in single cells. eLife 7, e32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramos I. et al. (2019) Innate immune response to influenza virus at single-cell resolution in human epithelial cells revealed paracrine induction of interferon lambda 1. J. Virol 93, e00559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vera JC et al. (2019) A common pattern of influenza A virus single cell gene expression heterogeneity governs the innate antiviral response to infection. bioRxiv, 858373.
- 65.Wang C. et al. (2020) Cell-to-cell variation in defective virus expression and effects on host responses during influenza virus infection. mBio 11, e02880–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell AB et al. (2019) Single-cell virus sequencing of influenza infections that trigger innate immunity. J. Virol 93, e00500–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dumm RE et al. (2019) Non-lytic clearance of influenza B virus from infected cells preserves epithelial barrier function. Nat. Commun 10, 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shnayder M. et al. (2018) Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. mBio 9, e00013–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shnayder M. et al. (2020) Single cell analysis reveals human cytomegalovirus drives latently infected cells towards an anergic-like monocyte state. eLife 9, e52168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galinato M. et al. (2019) Single-cell transcriptome analysis of CD34+ stem cell-derived myeloid cells infected with human cytomegalovirus. Front. Microbiol 10, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hein MY and Weissman JS (2019) Functional single-cell genomics of human cytomegalovirus infection. bioRxiv, 775080. [DOI] [PubMed]
- 72.Gamache A. et al. (2019) Ly49R activation receptor drives self-MHC–educated NK cell immunity against cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A 116, 26768–26778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devitt K. et al. (2019) Single-cell RNA sequencing reveals cell type-specific HPV expression in hyperplastic skin lesions. Virology 537, 14–19. [DOI] [PubMed] [Google Scholar]
- 74.Saikia M. et al. (2019) Simultaneous multiplexed amplicon sequencing and transcriptome profiling in single cells. Nat. Methods 16, 59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin J. et al. (2019) Ultra-sensitive digital quantification of proteins and mRNA in single cells. Nat. Commun 10, 3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joensson HN and Svahn HA (2012) Droplet microfluidics-a tool for single-cell analysis. Angew. Chem. Int. Edit 51, 12176–12192. [DOI] [PubMed] [Google Scholar]
- 77.Neun S. et al. (2019) Single-cell activity screening in microfluidic droplets. In Methods in Enzymology (Allbritton NL, and Kovarik ML, eds), 95–112, Academic Press. [DOI] [PubMed] [Google Scholar]
- 78.Fischer AE et al. (2015) A high-throughput drop microfluidic system for virus culture and analysis. J. Virol. Methods 213, 111–117. [DOI] [PubMed] [Google Scholar]
- 79.Yucha RW et al. (2017) High-throughput characterization of HIV-1 reservoir reactivation using a single-cell-in-droplet PCR assay. EBioMedicine 20, 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riordon J. et al. (2019) Deep learning with microfluidics for biotechnology. Trends Biotechnol. 37, 310–324. [DOI] [PubMed] [Google Scholar]
- 81.Hicks SC et al. (2018) Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 19, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dean KM, and Palmer AE (2014) Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol 10, 512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Z. et al. (2015) Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell 58, 644–659. [DOI] [PubMed] [Google Scholar]
- 84.Ma A. et al. (2020) Integrative methods and practical challenges for single-cell multi-omics. Trends Biotechnol. DOI: 10.1016/j.tibtech.2020.02.013. [DOI] [PMC free article] [PubMed]