Abstract
Rationale
Bedaquiline has been classified as a group A drug for the treatment of multidrug-resistant tuberculosis (MDR-TB) by the World Health Organization; however, globally emerging resistance threatens the effectivity of novel MDR-TB treatment regimens.
Objectives
We analysed pre-existing and emerging bedaquiline resistance in bedaquiline-based MDR-TB therapies, and risk factors associated with treatment failure and death.
Methods
In a cross-sectional cohort study, we employed patient data, whole-genome sequencing (WGS) and phenotyping of Mycobacterium tuberculosis complex (MTBC) isolates. We could retrieve baseline isolates from 30.5% (62 out of 203) of all MDR-TB patients who received bedaquiline between 2016 and 2018 in the Republic of Moldova. This includes 26 patients for whom we could also retrieve a follow-up isolate.
Measurements and main results
At baseline, all MTBC isolates were susceptible to bedaquiline. Among 26 patients with available baseline and follow-up isolates, four (15.3%) patients harboured strains which acquired bedaquiline resistance under therapy, while one (3.8%) patient was re-infected with a second bedaquiline-resistant strain. Treatment failure and death were associated with cavitary disease (p=0.011), and any additional drug prescribed in the bedaquiline-containing regimen with WGS-predicted resistance at baseline (OR 1.92 per unit increase, 95% CI 1.15–3.21; p=0.012).
Conclusions
MDR-TB treatments based on bedaquiline require a functional background regimen to achieve high cure rates and to prevent the evolution of bedaquiline resistance. Novel MDR-TB therapies with bedaquiline require timely and comprehensive drug resistance monitoring.
Short abstract
Bedaquiline resistance emerged in >15% of Mycobacterium tuberculosis complex strains obtained from follow-up isolates of MDR-TB patients. Insufficient backbone regimens and cavitary disease were associated with treatment failure and death. https://bit.ly/2UHoVyG
Introduction
Tuberculosis (TB) is an airborne infectious disease caused by bacteria of the Mycobacterium tuberculosis complex (MTBC). TB remains one of the most challenging health issues worldwide: an estimated 1.4 million people died from this disease in 2019 [1]. Particularly, multidrug-resistant (MDR)-TB, i.e. resistance against at least rifampicin and isoniazid, jeopardises TB control with poor treatment outcomes despite long therapy durations of 9–18 months or more [2, 3].
Based on the result of a systematic review and a meta-analysis, the World Health Organization (WHO) revised the guidelines for the management of patients with MDR-TB by prioritising the fluoroquinolones, bedaquiline and linezolid (all classified as group A agents) in 2019 [2, 4]. In particular, the recently introduced novel anti-TB drug bedaquiline raised great expectations with the potential to reduce death rates [5], shorten MDR-TB treatment durations [6] and decrease treatment failure rates [7–10], and thus has been administered to MDR-TB patients in >50 countries to date [11, 12]. Another important aspect is that MTBC isolates are considered naïve to bedaquiline as it was never administered under programmatic conditions, although recently a study indicated pre-existing bedaquiline resistance pre-dating the introduction of the drug in Southern Africa [13].
Bedaquiline is an adenosine triphosphatase (ATP) synthase inhibitor specifically targeting the protein AtpE, a transmembrane subunit of the ATP synthase. Consequently, mutations in the atpE gene are biologically linked to bedaquiline resistance [14]. Furthermore, mutations in Rv0678 encoding a transcriptional regulator of the MmpS5-MmpL5 efflux pump, and mutations in the putative proline aminopeptidase gene pepQ (Rv3525c) have been shown as secondary resistance mechanisms and, importantly, can lead to cross-resistance against the chemically unrelated WHO group B agent clofazimine [6, 15–18]. In clinical isolates, mutations in Rv0678 seem to be the main resistance-conferring mechanism associated with variable minimum inhibitory concentrations (MICs) and often detected at variable frequencies [19–26]. Of note, Rv0678 mutations often occur in combination with isolates/clones lacking mutations in canonical bedaquiline and clofazimine resistance-associated genes, i.e. hetero-resistance [19–22, 24, 27]. Pre-existing and emerging resistance against bedaquiline in failing treatment regimens have raised concerns to lose this new front-line drug against MDR-TB [4, 26, 28, 29].
We sought to investigate events of bedaquiline resistance acquisition in MDR-TB patients receiving a bedaquiline-based combination therapy and risk factors associated with treatment failure and death. We employed a country-wide cross-sectional cohort study and performed bacterial whole-genome sequencing (WGS), phenotyping and epidemiological analyses to investigate MTBC isolates and MDR-TB patients who received bedaquiline in the Republic of Moldova (a country with one of the highest MDR-TB burdens globally [1]), between 2016 and 2018.
Material and methods
Selection of patients receiving bedaquiline between 2016 and 2018
Using the national TB electronic database (Sistemul Informațional de Monitorizare și Evaluare al Tuberculozei/Tuberculosis Monitoring and Evaluation Information System (SIMETB)) in the Republic of Moldova, we retrospectively identified all MDR-TB patients who started a bedaquiline-containing treatment regimen between 1 January 2016 and 31 December 2018, and who had at least one MTBC isolate (prior to or after initiation of this treatment episode) stored in the biobank of the National Tuberculosis Reference Laboratory in Chişinău, Republic of Moldova. Patient isolates from three other laboratories in the country (Balti, Vorniceni and Bender) are not routinely stored, and were not available for inclusion in this study. Routine phenotypic drug susceptibility test (DST) results for TB and MDR-TB antibiotics, demographic and clinical data including treatment regimens and outcomes were extracted from SIMETB.
Phenotypic antimicrobial susceptibility testing
For all isolates, we determined growth at 0.5 mg·L−1, 1.0 mg·L−1 (corresponding to the WHO critical concentration) and 2.0 mg·L−1 of each of bedaquiline, clofazimine and linezolid using the Bactec MGIT 960 system (Becton Dickinson, USA). Routine antimicrobial susceptibility testing for other antibiotics were performed in MGIT 960 using WHO recommended critical concentrations and according to manufacturer's instructions and as instructed in the WHO guidelines [30]. Handling details can be found in the supplementary methods.
Whole-genome sequencing
Extracted DNA (see supplementary methods) of 97 M. tuberculosis isolates obtained from 71 MDR-TB patients receiving bedaquiline-containing regimens was subjected to WGS at the Research Center Borstel, Germany with a minimum average genome coverage of ×50, using paired-end DNA libraries and Illumina technology (Nextera-XT and NextSeq500) according to the manufacturer's instructions (Illumina, USA). Fastq files (raw sequencing data) were submitted to the European Nucleotide Archive (accession numbers provided in supplementary table S1) and mapped to the M. tuberculosis H37Rv reference genome (GenBank ID: NC_000962.3) using the MTBseq pipeline [31]. Briefly, we considered mutations (single nucleotide polymorphisms (SNPs), insertions and deletions) in 92 genes implicated in drug resistance [32] and covered by a minimum of one read in both forward and reverse orientation, and one read calling the allele with a phred score of ⩾20. Genotypic resistance prediction was performed on the basis of a curated mutation catalogue employed at the Supranational Reference Laboratory at the Research Center Borstel, based on information available on 3 July 2020. Based on WGS results we classified MTBC isolates as extremely drug resistant (XDR; i.e. MDR plus additional resistance against any fluoroquinolone and at least one injectable drug) according to the WHO classification until December 2020, and pre-XDR (i.e. MDR plus additional resistance against any fluoroquinolone or at least one injectable drug).
Statistics
Predictors for negative treatment outcomes and odds ratios were analysed using univariate logistic regression. Odds ratios for contingency tables with zero cell count were corrected using the Haldane–Anscombe method adding 0.5 to each cell. Means of patient age (non-normal distribution) were compared using Mann–Whitney U-tests; other variables described in supplementary table S2 were compared using Fisher's exact test.
Ethics
No physical interventions took place with the patients and all of the information collected was anonymised at the source. The study protocol was approved by the institutional review board of the Institute of Phthisiopneumology “Chiril Draganiuc”, Chisinau, Republic of Moldova (#1/07.2019).
Results
Study population
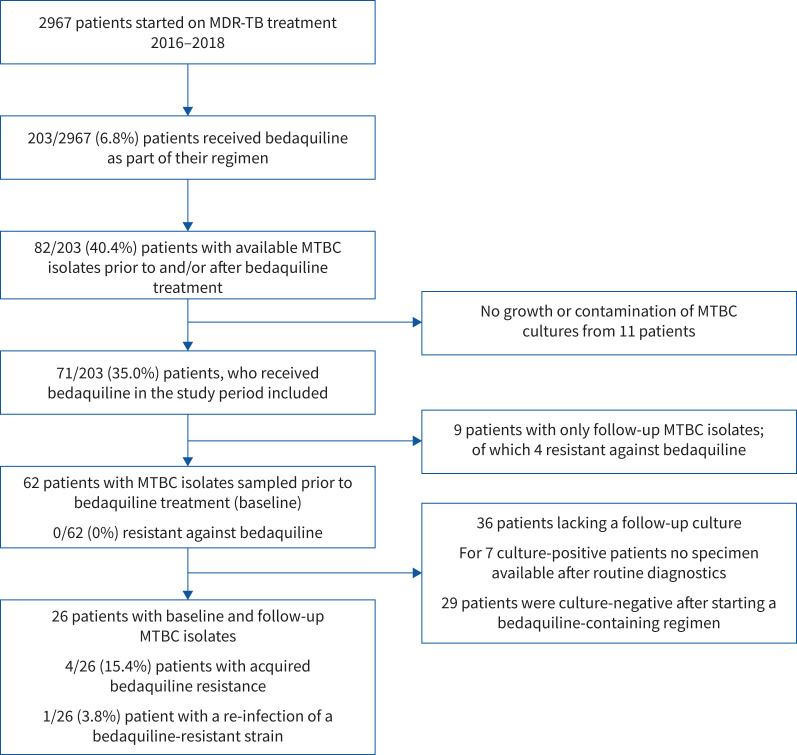
Between 1 January 2016 and 31 December 2018, 2967 patients initiated MDR-TB treatment in the Republic of Moldova (1413 new and 1554 re-treatment cases). In total, 203 (6.8%) of all MDR-TB patients received bedaquiline as part of their anti-TB treatment regimen (figure 1). For this study, we could retrieve MTBC cultures at the National Reference Laboratory for Mycobacteriology in Moldova from 82 (40.4%) out of 203 patients; cultures from the remaining 121 patients were not available after routine diagnostics (figure 1; refer to Material and methods). Furthermore, MTBC cultures from 11 patients failed to grow or were contaminated, and for nine patients we could only receive a follow-up culture, resulting in 62 (30.5%) out of 203 patients with a baseline MTBC isolate, prior to the start of the bedaquiline-containing MDR-TB regimen (figure 1).
FIGURE 1.
Study flow chart. MDR-TB: multidrug-resistant tuberculosis; MTBC: Mycobacterium tuberculosis complex.
This cohort comprised 18 new and 44 re-treatment cases; the median age was 39 years (interquartile range (IQR) 34–45 years). The majority of the patients were HIV-seronegative (54 (87.1%) out of 62), diagnosed with cavitary disease (45, 72.6%) and male (50, 80.6%).
No differences were observed for characteristics of patients not included in the subsequent analysis (due to lack of MTBC culture, or only follow-up culture available) with regard to residence, gender, age, microscopy result, case definition, HIV status and treatment outcome (p>0.09; supplementary table S2).
Approximately half of the patients were infected with a MTBC lineage 2 isolate (35 (56.5%) out of 62), while the other patients were infected with a MTBC lineage 4 isolate (27, 43.5%). Isolates of other MTBC lineages were not identified in our MDR-TB cohort (supplementary table S1).
Based on WGS results, 20 (32.3%) out of 62 patients were classified as pre-XDR (supplementary table S1), and 31 (50.0%) were classified as XDR prior to the start of the bedaquiline-containing therapy regimen. Resistance proportions to individual drugs, as predicted by WGS, were as follows: 62 (100%) out of 62 streptomycin, 41 (66.1%) ethambutol, 44 (71.0%) pyrazinamide, 37 (59.7%) fluoroquinolones, 45 (72.6%) kanamycin, 15 (24.2%) amikacin, 14 (22.6%) capreomycin, 47 (75.8%) ethionamide, 16 (25.9%) para-aminosalicylic acid and eight (12.9%) cycloserine (supplementary table S1). Within a personalised therapy regimen of at least five antibiotics, patients received a median of one drug (IQR 0.75–2.0) with WGS-predicted drug resistance at baseline (supplementary table S1).
At the time of analysis, 10 (16.1%) out of 62 patients were either still on treatment or lost to follow-up. Among the remaining 52 patients, 37 (71.2%) were considered as cured (i.e. no clinical or microbiological signs of disease relapse, up to 6 months after treatment completion), three (5.8%) died and 12 (23.1%) patients experienced signs of treatment failure, i.e. no negative culture within 8 months of treatment (supplementary table S1). The proportions of treatment failure (27.0%) and death (8.2%) were comparable among the patients who were excluded from the analysis (p >0.71; supplementary table S2).
In the following section, we first describe bedaquiline resistance-associated mutations and their resulting phenotype. We then investigate in detail events of emerging bedaquiline resistance among 26 patients with baseline and follow-up isolates, and then analyse factors associated with negative treatment outcomes.
Phenotypic and genotypic resistance against bedaquiline
We identified nine MTBC isolates with mutations in the genes atpE and/or Rv0678, out of which one isolate had mutations in atpE only, six isolates had mutations in Rv0678 only and two isolates had mutations in both genes (table 1). Eight MTBC isolates had a bedaquiline MIC of ⩾2.0 mg·L−1 and thus tested resistant in MGIT 960; one isolate (CAR-84) with two mutations in Rv0678 tested susceptible to bedaquiline (MIC 1.0 mg·L−1) (table 1, supplementary figure S1). Notably, we found in seven isolates more than one mutation in atpE and/or Rv0678 at different frequencies, indicating the existence of different subpopulations in these patients. All bedaquiline-resistant isolates were follow-up isolates from patients who were exposed to bedaquiline containing MDR-TB regimens for 77–451 days (table 1).
TABLE 1.
Patients with whole-genome sequencing predicted bedaquiline (BDQ)-resistant Mycobacterium tuberculosis complex isolates in the Republic of Moldova between 2016 and 2018
| Patient (isolate ID), sampling time | Rv0678 (mutation frequency %) | atpE (mutation frequency %) | BDQ MIC MGIT 960 (mg·mL−1) | CFZ MIC MGIT 960 (mg·mL−1) |
| Patient 29 (CAR-13), prior to BDQ | Wild-type | Wild-type | ≤0.5 (S) | ≤0.5 (S) |
| Patient 29 (CAR-38), after BDQ, acquired resistance | Wild-type | I66M# (97%) | >2.0 | ≤0.5 (S) |
| Patient 12 (CAR-52), prior to BDQ | Wild-type | Wild-type | ≤0.5 (S) | ≤0.5 (S) |
| Patient 12 (CAR-61) after BDQ, acquired resistance | 16_del_g (57.4%) 193_del_g# (12.7%) G24D (19.8%) |
Wild-type | 2.0 | 1.0 (S) |
| Patient 2 (CAR-78), prior to BDQ | Wild-type | Wild-type | ≤0.5 (S) | ≤0.5 (S) |
| Patient 2 (CAR-87), after BDQ, acquired resistance | 192insG# (2%) | A63P# (25%) | >2.0 (R) | 2.0 (R) |
| Patient 37 (CAR-10), prior to BDQ | Wild-type | Wild-type | ≤0.5 (S) | ≤0.5 (S) |
| Patient 37 (CAR-18), after BDQ, re-infection | T58P (100%) | Wild-type | 2.0 (R) | 1.0 (S) |
| Patient 57 (CAR-45), prior to BDQ | Wild-type | Wild-type | ≤0.5 (S) | ≤0.5 (S) |
| Patient 57 (CAR-55), after BDQ, acquired resistance | 193_del_g# (44.4%) S63G# (5.5%) |
E61D# (27.5) I66M# (2.6%) |
>2.0 (R) | 1.0 (S) |
| Patient 32 (CAR-84), after BDQ | 192_ins_g# (74.2%) 193_del_g# (5.7%) |
Wild-type | 1.0 (S) | 1.0 (S) |
| Patient 71 (CAR-40), after BDQ | 192ins_g# (23.8%) L142P (64%) |
Wild-type | >2.0 (R) | 2.0 (R) |
| Patient 61 (CAR-1), after BDQ | 136_ins_g (7.1%) 141_ins_c# (69.0%) 195_ins_t# (5.8%) G66W (6.0%) |
Wild-type | >2.0 (R) | 2.0 (R) |
| Patient 33 (CAR-43), after BDQ | 436_ins_t (90.2%) R72W (28.5%) |
Wild-type | >2.0 (R) | >2.0 (R) |
MIC: minimum inhibitory concentration; CFZ: clofazimine; S: susceptible; R: resistant. #: mutation reviewed in [33].
Emerging bedaquiline resistance
Among 26 patients with available MTBC isolates prior and after administration of bedaquiline, baseline and follow-up isolate differed by a maximum of four SNPs, while four (15.4%) patients were probably re-infected with a second isolate with 26–1126 SNPs difference compared to the respective baseline isolate (supplementary table S1).
In total, four (15.4%) out of 26 patient isolates acquired bedaquiline resistance following 90, 159, 348 and 451 days of bedaquiline administration (table 1). One follow-up isolate (patient 29) with the mutation atpE p.I66M (97% frequency) was phenotypically bedaquiline-resistant, but clofazimine-susceptible (table 1). A second follow-up isolate (patient 12) carried three mutations in the gene Rv0678 with variable frequencies (p.D5fs (57%), p.G24D (19.8%) and p.S64fs (13%)). Another two follow-up isolates acquired the mutations atpE p.A63P (25%) in combination with Rv0678 p.S64fs (2%) (patient 2), and the combination atpE p.E61D (28%), atpE p.I66M (3%), Rv0678 p.S63fs (5%) and p.S64fs (44%) (patient 57; table 1).
No additional drug resistances emerged under bedaquiline-containing treatment regimens, except patient 56, who had a follow-up isolate which acquired the mutation rrs g.1484 g/t in the 16S rRNA gene mediating cross-resistance against the second-line injectable drugs kanamycin, capreomycin and amikacin (supplementary table S1). However, we found one isolate that virtually lost phenotypic resistance against all second-line injectable drugs as well as one fluoroquinolone resistance mediating mutation (patient 2; supplementary table S1). The baseline isolate from patient 2 harboured the mutations rrs 1401 a/g (63%) and gyrA p.D94G (61%) in combination with gyrA p.S91P (44%). In the follow-up isolate that tested susceptible to all second-line injectable drugs, the mutations rrs g.1401 a/g and gyrA p.D94G were reduced to a frequency of 4% and 0.5%, respectively. In contrast, the mutation gyrA p.S91P increased to a frequency of 96% (supplementary table S1).
Among the four patients re-infected with a different MTBC strain, patient 37 was re-infected with a bedaquiline-resistant strain carrying the mutation Rv0678 p.T58P at a frequency of 100%. Although the second isolate had the same genotype (lineage 2.2.1/Europe/Russian W148 outbreak), a genetic distance of 26 SNPs compared to the baseline isolate clearly pointed towards a re-infection (table 1, supplementary table S1).
Risk factors for failure of bedaquiline-based MDR-TB therapies
To determine risk factors of bedaquiline-based therapies, we analysed predictors for a negative outcome, i.e. death or treatment failure, for 52 (83.9%) out of 62 patients with available treatment outcome data. In a univariate logistic regression analysis, we included the following factors: MTBC lineage, gender, case definition, XDR, presence of cavities in chest radiographs, HIV status, age and number of drugs with predicted resistance (in the following also referred to as “inactive drugs”) as part of the bedaquiline containing MDR-TB therapy regimen (table 2). We found cavitary disease (p=0.011) associated with a negative treatment outcome. Furthermore, an increasing number of inactive drugs increased the odds of treatment failure (OR 1.92 per unit increase, 95% CI 1.15–3.21; p=0.012) (table 2, figure 2). Likewise, most regimens included antibiotics with WGS-predicted resistance at baseline and patients with a negative outcome had more inactive drugs (on average 2.33) included in their bedaquiline-containing regimen compared to patients with positive treatment outcomes (on average 1.27 inactive drugs; p=0.018, Mann–Whitney U-test).
TABLE 2.
Univariate analysis of factors associated with negative treatment outcomes of bedaquiline-based multidrug-resistant tuberculosis therapies
| Negative outcome | Positive outcome | OR (95% CI) | p-value | |
| Patients | 15 | 37 | ||
| Lineage | ||||
| L2 | 8 (53.3) | 20 (48.6) | Reference | |
| L4 | 7 (46.7) | 17 (51.4) | 1.03 (0.31–3.43) | 1.0 |
| Gender | ||||
| Female | 4 (26.7) | 6 (19.4) | Reference | |
| Male | 11 (73.3) | 31 (83.8) | 0.53 (0.13–2.25) | 0.448 |
| Case | ||||
| New case | 4 (26.7) | 13 (32.4) | Reference | |
| Previously treated | 11 (73.3) | 24 (67.6) | 1.49 (0.39–5.63) | 0.747 |
| Resistance category | ||||
| Not XDR# | 6 (40.0) | 18 (35.1) | Reference | |
| XDR# | 9 (53.3) | 19 (45.9) | 1.42 (0.42–4.80) | 0.760 |
| Cavitary | ||||
| No | 0+ (0.0) | 13+ (32.4) | Reference | |
| Yes | 15+ (100.0) | 24+ (67.6) | 17.08 (0.95–308.6) | 0.011 |
| HIV status | ||||
| Negative | 11 (73.3) | 34 (91.9) | Reference | |
| Positive | 4 (26.7) | 3 (8.1) | 4.12 (0.80–21.34) | 0.173 |
| Inactive drugs ¶ | 2 | 1 | 1.92 (1.15–3.21) | 0.012 |
| Age¶ years | 35 | 41 | 0.97 (0.90–1.04) | 0.427 |
Data are presented as n or n (%), unless otherwise stated. Bold type represents statistical significance (p<0.05). XDR: extensively drug resistant. #: World Health Organization classification until 2020, i.e. resistance against one fluoroquinolone and one second-line injectable drug; ¶: median; +: Haldane–Anscombe correction adding 0.5 to each cell to allow calculation of odds ratio.
FIGURE 2.
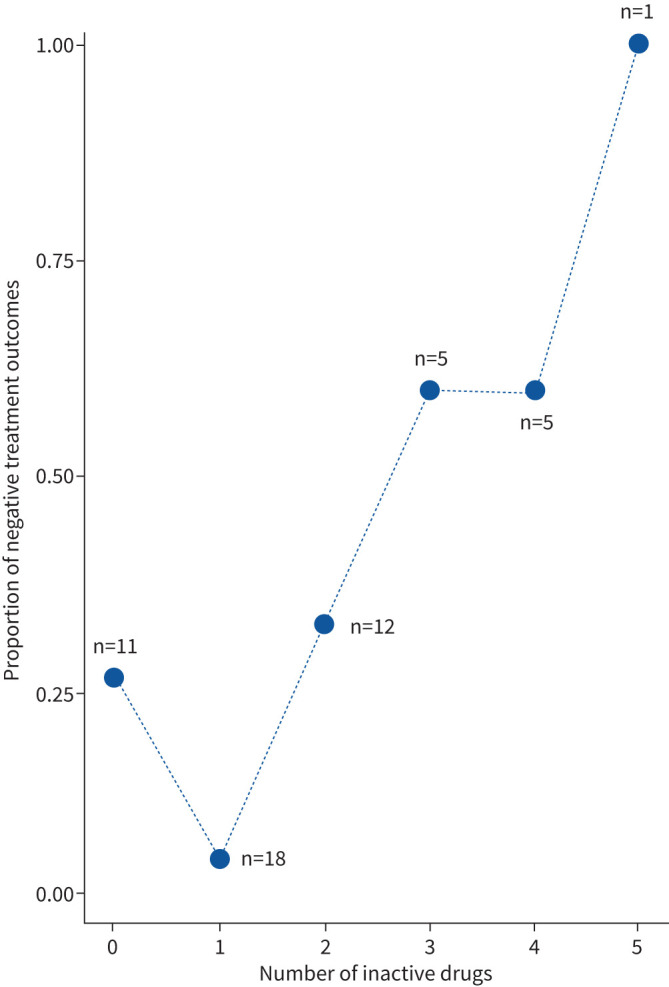
Proportions of negative treatment outcomes, relative to the number of drugs used despite predicted resistance at baseline, for patients in the Republic of Moldova who received bedaquiline and at least four additional drugs as part of their multidrug-resistant tuberculosis regimen (2016–2018). Numbers indicate patients per category.
Discussion
In this cross-sectional cohort study, we report bedaquiline resistance acquisition in >15% of all MDR-TB patients who received bedaquiline as part of their treatment regimen, and with available follow-up cultures. The odds for negative treatment outcomes increased with the presence of cavities and the number of ineffective drugs included in a regimen. Nevertheless, despite high numbers of re-treatment cases and extended drug resistance profiles, bedaquiline-based regimens achieved high cure rates (71%) among our patient cohort, which is also reflected by the overall low number of positive follow-up cultures.
Emerging resistance against bedaquiline has been described in patients previously [19–25, 28, 29]. In addition, high MICs among patient isolates who were never exposed to bedaquiline (and/or clofazimine) may potentially jeopardise the effectiveness of currently endorsed MDR-TB treatment regimens [13, 19–25, 28, 29]. New information from this study, in which all available baseline isolates were bedaquiline-susceptible, indicates that the success of bedaquiline-containing therapies relies on a functional background regimen excluding underling resistances to the administered antibiotics [4]. Another important factor associated with treatment outcomes is the presence of pulmonary cavities, which further promote the emergence of drug resistance most likely due to higher bacterial loads [34–37]. Larger cavities may hinder drugs in reaching the central lesion that contain the highest bacterial load [37]. For example, clofazimine is unable to reach the necrotic centre of caseous lesions, and bedaquiline accumulates rather in cellular regions of a granuloma, while linezolid and moxifloxacin effectively penetrate all lesion types [36, 38, 39]. Thus, cavities provide a micro-environment for the infecting MTBC strain with variable drug concentrations, and especially bedaquiline levels might temporarily fall below effective concentrations. This changing micro-environment may select different subpopulations of bedaquiline-resistant clones, but susceptible bacteria remain which lack any mutations in the canonical resistance genes (Rv0678, atpE, pepQ), as seen in our study and observed previously [19–22, 24, 27].
The complex pathology of granulomas, pharmacokinetics and pharmacodynamics can usually not be considered in routine clinical practice. However, rapid and comprehensive DST is key for personalised MDR-TB treatment [40]. Resistance to pyrazinamide, for instance, is not routinely performed in many high-burden MDR-TB countries, and fluoroquinolones may be still given to patients despite proven resistance when no other antibiotics are available (personal communication; Aliona David, Chiril Draganiuc Phthisiopneumology Institute, Chisinau, Republic of Moldova). Nevertheless, we show the immediate risk of resistance development when bedaquiline is administered in partially ineffective background regimens. This is aggravated by the fact that the diagnostic work-up of rifampicin-resistant TB and MDR-TB is currently hampered by a lack of rapid (i.e. sputum-based) genotypic tests to rule out resistance to the WHO group A medicines bedaquiline and linezolid [18]. New approaches such as early targeted next-generation sequencing, e.g. using the Deeplex assay [41], or even WGS of the MTBC genome directly from patient specimens [42] may complement confirmatory phenotypic DST to further improve management of MDR-TB patients in high-burden countries such as the Republic of Moldova.
Universal DST is an important measure, but not the only way to reduce the risk of drug resistance development under therapy. The current WHO recommendations for the management of MDR-TB give advice for situations with underlying drug resistances or intolerance against certain anti-TB drugs, and provide recommendations for surgery as adjunctive therapy option [43]. Unfortunately, in the Moldovian setting, the design of second-line treatment regimens is driven by the availability of drugs. Surgery is performed in accordance with WHO recommendations and mostly on the basis of individual decisions, especially in patients with cavitary diseases. Furthermore, poor treatment adherence and treatment interruption as well as patients refusing medication probably contribute to the selection of bedaquiline-resistant strains. Thus, increasing patient awareness about the importance of medication adherence and regular drug supply is crucial to design effective MDR-TB therapies and reduce the risk of resistance evolution.
However, high costs of bedaquiline, lack of evidence on drug safety, limited experience with regard to side-effects, and sometimes bureaucratic barriers further complicate the programmatic implementation of bedaquiline in many high-burden MDR-TB settings [44]. These factors were the reasons only 203 patients were actually treated with bedaquiline in the Republic of Moldova during the study period 2016–2018, as compared to all eligible MDR-TB patients in that timeframe. Following the WHO endorsement of bedaquiline as a group A MDR-TB drug in August 2018 [45], bedaquiline became available for the majority of all MDR-TB patients.
This cross-sectional cohort study has some limitations. We could only retrieve MTBC isolates from 35% of all patients who received bedaquiline between 2016 and 2018 in the Republic of Moldova. This emphasises the immediate benefit of universal culture and DST for future studies; in particular, systematic biobanking of MDR MTBC strains will provide crucial information for drug resistance surveys. However, sampling of MTBC isolates in this study occurred prior to phenotypic and genotypic investigations and was not biased towards bedaquiline resistance. Due to overall rapid culture conversion times under bedaquiline treatment, and thus a lack of follow-up cultures for many patients, acquired bedaquiline resistance is mainly observed among patients with longer culture conversion times. Furthermore, cavitary disease is associated with negative treatment outcomes, but is probably also contributing to the emergence of antibiotic resistance during therapy, and needs to be considered as confounding factor for any association between resistance and treatment outcome.
In conclusion, we show that bedaquiline resistance emerged among >15% of MTBC strains from MDR-TB patients with available bacterial isolates prior and during bedaquiline therapy. MDR-TB therapy with an insufficient number of active drugs and cavitary disease were considered risk factors for treatment failure and death in this cohort. Availability of adequate treatment regimens based on information provided by comprehensive and timely genotypic and phenotypic DSTs will be key to improve treatment outcomes for patients with MDR-TB and to avoid the evolution of drug resistance in circulating MDR-MTBC strains.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods erj-00621-2021.supplemeny (89.3KB, pdf)
Table S1 erj-00621-2021.supplement_tableS1 (609.5KB, pdf)
Table S2 erj-00621-2021.supplement_tableS2 (137.8KB, pdf)
Shareable PDF
Acknowledgements
We thank Fenja Boysen, Tanja Niemann, Vanessa Mohr, Carina Hahn, Tanja Struve-Sonnenschein, Anja Lüdemann and Darshaalini Nadarajan (Research Center Borstel, Borstel, Germany) for excellent technical assistance.
Footnotes
This article has supplementary material available from erj.ersjournals.com
This article has an editorial commentary: https://doi.org/10.1183/13993003.00149-2022
Author contributions: J. Heyckendorf and M. Merker conceived the study. E. Chesov performed minimum inhibitory concentration determination. F.P. Maurer and S. Andres supported implementation of drug susceptibility testing in Moldova and provided reference diagnostics. S. Niemann, C. Utpatel and I. Barilar performed whole genome sequencing and molecular drug resistance prediction. E. Chesov and M. Merker analysed whole genome sequencing data. E. Chesov, M. Merker and M. Reimann performed statistical analysis. E. Chesov, D. Chesov, J. Heyckendorf and M. Merker drafted the manuscript. All authors made substantial intellectual contribution, revised the manuscript and gave final approval.
Conflict of interest: E. Chesov has nothing to disclose.
Conflict of interest: D. Chesov has nothing to disclose.
Conflict of interest: F.P. Maurer has nothing to disclose.
Conflict of interest: S. Andres has nothing to disclose.
Conflict of interest: C. Utpatel has nothing to disclose.
Conflict of interest: I. Barilar has nothing to disclose.
Conflict of interest: A. Donica has nothing to disclose.
Conflict of interest: M. Reimann has nothing to disclose.
Conflict of interest: S. Niemann reports grants from EXC 2167 Precision Medicine in Inflammation, grants from Leibniz Science Campus Evolutionary Medicine of the LUNG, grants from German Center for Infection Research, during the conduct of the study.
Conflict of interest: C. Lange reports personal fees from Chiesi, Gilead, Janssen, Novartis, Oxfordimmunotec and Insmed, outside the submitted work.
Conflict of interest: V. Crudu has nothing to disclose.
Conflict of interest: J. Heyckendorf has nothing to disclose.
Conflict of interest: M. Merker has nothing to disclose.
Support statement: This work has been funded by the CARE consortium (Common Action against HIV/TB/HCV across the Regions of Europe), European Union's Horizon 2020 Research and Innovation programme, grant number 825673, the German Excellence Cluster Precision Medicine in Chronic Inflammation (EXC 2167), and the German Center for Infection Research.
References
- 1.World Health Organization (WHO) . Global Tuberculosis Report 2020. Available from: www.who.int/tb/publications/global_report/en/ Date last accessed: 23 October 2020.
- 2.World Health Organization (WHO) . WHO Consolidated Guidelines on Tuberculosis. Module 4: Treatment – Drug-Resistant Tuberculosis Treatment. https://apps.who.int/iris/bitstream/handle/10665/332397/9789240007048-eng.pdf Date last accessed: 7 July 2021. [PubMed]
- 3.Lange C, Dheda K, Chesov D, et al. . Management of drug-resistant tuberculosis. Lancet 2019; 394: 953–966. doi: 10.1016/S0140-6736(19)31882-3 [DOI] [PubMed] [Google Scholar]
- 4.Ahmad N, Ahuja SD, Akkerman OW, et al. . Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet 2018; 392: 821–834. doi: 10.1016/S0140-6736(18)31644-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnippel K, Ndjeka N, Maartens G, et al. . Effect of bedaquiline on mortality in South African patients with drug-resistant tuberculosis: a retrospective cohort study. Lancet Respir Med 2018; 6: 699–706. doi: 10.1016/S2213-2600(18)30235-2 [DOI] [PubMed] [Google Scholar]
- 6.Diacon AH, Pym A, Grobusch MP, et al. . Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med 2014; 371: 723–732. doi: 10.1056/NEJMoa1313865 [DOI] [PubMed] [Google Scholar]
- 7.Ndjeka N, Schnippel K, Master I, et al. . High treatment success rate for multidrug-resistant and extensively drug-resistant tuberculosis using a bedaquiline-containing treatment regimen. Eur Respir J 2018; 52: 1801528. doi: 10.1183/13993003.01528-2018 [DOI] [PubMed] [Google Scholar]
- 8.Olayanju O, Limberis J, Esmail A, et al. . Long-term bedaquiline-related treatment outcomes in patients with extensively drug-resistant tuberculosis from South Africa. Eur Respir J 2018; 51: 1800544. doi: 10.1183/13993003.00544-2018 [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Fox T, Manning K, et al. . Improved treatment outcomes with bedaquiline when substituted for second-line injectable agents in multidrug-resistant tuberculosis: a retrospective cohort study. Clin Infect Dis 2019; 68: 1522–1529. doi: 10.1093/cid/ciy727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesov D, Heyckendorf J, Alexandru S, et al. . Impact of bedaquiline on treatment outcomes of multidrug-resistant tuberculosis in a high-burden country. Eur Respir J 2021; 57: 2002544. doi: 10.1183/13993003.02544-2020 [DOI] [PubMed] [Google Scholar]
- 11.Andries K, Verhasselt P, Guillemont J, et al. . A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005; 307: 223–227. doi: 10.1126/science.1106753 [DOI] [PubMed] [Google Scholar]
- 12.Veziris N, Bernard C, Guglielmetti L, et al. . Rapid emergence of Mycobacterium tuberculosis bedaquiline resistance: lessons to avoid repeating past errors. Eur Respir J 2017; 49: 1601719. doi: 10.1183/13993003.01719-2016 [DOI] [PubMed] [Google Scholar]
- 13.Beckert P, Sanchez-Padilla E, Merker M, et al. . MDR M. tuberculosis outbreak clone in Eswatini missed by Xpert has elevated bedaquiline resistance dated to the pre-treatment era. Genome Med 2020; 12: 104. doi: 10.1186/s13073-020-00793-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huitric E, Verhasselt P, Koul A, et al. . Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother 2010; 54: 1022–1028. doi: 10.1128/AAC.01611-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2014; 58: 2979–2981. doi: 10.1128/AAC.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andries K, Villellas C, Coeck N, et al. . Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 2014; 9: e102135. doi: 10.1371/journal.pone.0102135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almeida D, Ioerger T, Tyagi S, et al. . Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2016; 60: 4590–4599. doi: 10.1128/AAC.00753-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kranzer K, Kalsdorf B, Heyckendorf J, et al. . New World Health Organization treatment recommendations for multidrug-resistant tuberculosis: are we well enough prepared? Am J Respir Crit Care Med 2019; 200: 514–515. doi: 10.1164/rccm.201902-0260LE [DOI] [PubMed] [Google Scholar]
- 19.Bloemberg GV, Keller PM, Stucki D, et al. . Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 2015; 373: 1986–1988. doi: 10.1056/NEJMc1505196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann H, Kohl TA, Hofmann-Thiel S, et al. . Delamanid and bedaquiline resistance in Mycobacterium tuberculosis ancestral Beijing genotype causing extensively drug-resistant tuberculosis in a Tibetan refugee. Am J Respir Crit Care Med 2016; 193: 337–340. doi: 10.1164/rccm.201502-0372LE [DOI] [PubMed] [Google Scholar]
- 21.Andres S, Merker M, Heyckendorf J, et al. . Bedaquiline-resistant tuberculosis: dark clouds on the horizon. Am J Respir Crit Care Med 2020; 201: 1564–1568. doi: 10.1164/rccm.201909-1819LE [DOI] [PubMed] [Google Scholar]
- 22.Polsfuss S, Hofmann-Thiel S, Merker M, et al. . Emergence of low-level delamanid and bedaquiline resistance during extremely drug-resistant tuberculosis treatment. Clin Infect Dis 2019; 69: 1229–1231. doi: 10.1093/cid/ciz074 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Gao M, Du J, et al. . Reduced susceptibility of Mycobacterium tuberculosis to bedaquiline during antituberculosis treatment and its correlation with clinical outcomes in China. Clin Infect Dis 2020; ciaa1002. doi: 10.1093/cid/ciaa1002 [DOI] [PubMed] [Google Scholar]
- 24.Nimmo C, Brien K, Millard J, et al. . Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine 2020; 55: 102747. doi: 10.1016/j.ebiom.2020.102747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Wang B, Hu M, et al. . Primary clofazimine and bedaquiline resistance among isolates from patients with multidrug-resistant tuberculosis. Antimicrob Agents Chemother 2017; 61: e00239-e00317. doi: 10.1128/AAC.00239-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimmo C, Millard J, van Dorp L, et al. . Population-level emergence of bedaquiline and clofazimine resistance-associated variants among patients with drug-resistant tuberculosis in southern Africa: a phenotypic and phylogenetic analysis. Lancet Microbe 2020; 1: e165–e174. doi: 10.1016/S2666-5247(20)30031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vos M, Ley SD, Wiggins KB, et al. . Bedaquiline microheteroresistance after cessation of tuberculosis treatment. N Engl J Med 2019; 380: 2178–2180. doi: 10.1056/NEJMc1815121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villellas C, Coeck N, Meehan CJ, et al. . Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J Antimicrob Chemother 2017; 72: 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen TVA, Anthony RM, Bañuls A-L, et al. . Bedaquiline resistance: its emergence, mechanism, and prevention. Clin Infect Dis 2018; 66: 1625–1630. doi: 10.1093/cid/cix992 [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization (WHO) . Technical Manual for Drug Susceptibility Testing of Medicines Used in the Treatment of Tuberculosis. https://www.who.int/publications/i/item/9789241514842 Date last accessed: 28 October 2020.
- 31.Kohl TA, Utpatel C, Schleusener V, et al. . MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 2018; 6: e5895. doi: 10.7717/peerj.5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merker M, Kohl TA, Barilar I, et al. . Phylogenetically informative mutations in genes implicated in antibiotic resistance in Mycobacterium tuberculosis complex. Genome Med 2020; 12: 27. doi: 10.1186/s13073-020-00726-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadura S, King N, Nakhoul M,et al. . Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 2020; 75: 2031–2043. doi: 10.1093/jac/dkaa136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbanowski ME, Ordonez AA, Ruiz-Bedoya CA, et al. . Cavitary tuberculosis: the gateway of disease transmission. Lancet Infect Dis 2020; 20: e117–e128. doi: 10.1016/S1473-3099(20)30148-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mbuagbaw L, Guglielmetti L, Hewison C, et al. . Outcomes of bedaquiline treatment in patients with multidrug-resistant tuberculosis. Emerg Infect Dis 2019; 25: 936–943. doi: 10.3201/eid2505.181823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strydom N, Gupta SV, Fox WS, et al. . Tuberculosis drugs’ distribution and emergence of resistance in patient's lung lesions: a mechanistic model and tool for regimen and dose optimization. PLoS Med 2019; 16: e1002773. doi: 10.1371/journal.pmed.1002773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong CWM, Elkington PT, Friedland JS. Tuberculosis, pulmonary cavitation, and matrix metalloproteinases. Am J Respir Crit Care Med 2014; 190: 9–18. doi: 10.1164/rccm.201311-2106PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irwin SM, Prideaux B, Lyon ER, et al. . Bedaquiline and pyrazinamide treatment responses are affected by pulmonary lesion heterogeneity in Mycobacterium tuberculosis infected C3HeB/FeJ mice. ACS Infect Dis 2016; 2: 251–267. doi: 10.1021/acsinfecdis.5b00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarathy J, Blanc L, Alvarez-Cabrera N, et al. . Fluoroquinolone efficacy against tuberculosis is driven by penetration into lesions and activity against resident bacterial populations. Antimicrob Agents Chemother 2019; 63: e02516-e02518. doi: 10.1128/AAC.02516-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gröschel MI, Walker TM, van der Werf TS, et al. . Pathogen-based precision medicine for drug-resistant tuberculosis. PLoS Pathog 2018; 14: e1007297. doi: 10.1371/journal.ppat.1007297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feuerriegel S, Kohl TA, Utpatel C, et al. . Rapid genomic first- and second-line drug resistance prediction from clinical Mycobacterium tuberculosis specimens using Deeplex-MycTB. Eur Respir J 2021; 57: 2001796. doi: 10.1183/13993003.01796-2020 [DOI] [PubMed] [Google Scholar]
- 42.Goig GA, Cancino-Muñoz I, Torres-Puente M, et al. . Whole-genome sequencing of Mycobacterium tuberculosis directly from clinical samples for high-resolution genomic epidemiology and drug resistance surveillance: an observational study. Lancet Microbe 2020; 1: e175–e183. doi: 10.1016/S2666-5247(20)30060-4 [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization (WHO) . WHO Operational Handbook on Tuberculosis. Module 4: Treatment – Drug-Resistant Tuberculosis Treatment. www.who.int/publications/i/item/9789240006997 Date last accessed: 12 July 2021.
- 44.Lienhardt C, Raviglione M, Spigelman M, et al. . New drugs for the treatment of tuberculosis: needs, challenges, promise, and prospects for the future. J Infect Dis 2012; 205: Suppl. 2, S241–S249. doi: 10.1093/infdis/jis034 [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization (WHO) . Rapid Communication: Key Changes to Treatment of Multidrug- and Rifampicin-Resistant Tuberculosis (MDR/RR-TB). www.who.int/tb/publications/2018/WHO_RapidCommunicationMDRTB.pdf Date last accessed: 12 July 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods erj-00621-2021.supplemeny (89.3KB, pdf)
Table S1 erj-00621-2021.supplement_tableS1 (609.5KB, pdf)
Table S2 erj-00621-2021.supplement_tableS2 (137.8KB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00621-2021.Shareable (389.3KB, pdf)