Abstract
Reduction of dinitrogen by molybdenum nitrogenase relies on complex metalloclusters: the [8Fe:7S] P-cluster and the [7Fe:9S:Mo:C:homocitrate] FeMo-cofactor. Although both clusters bear topological similarities and require the reductive fusion of [4Fe:4S] sub-clusters to achieve their respective assemblies, P-clusters are assembled directly on the NifD2K2 polypeptide prior to the insertion of FeMo-co, which is fully assembled separately from NifD2K2. P-cluster maturation involves the iron protein NifH2 as well as several accessory proteins, whose role has not been elucidated. In the present work, two NifD2K2 species bearing immature P-clusters were isolated from an Azotobacter vinelandii strain in which the genes encoding NifH and the accessory protein NifZ were deleted, and characterized by X-ray absorption spectroscopy and EPR. These analyses showed that both NifD2K2 complexes harbor clusters that are electronically and structurally similar, with each NifDK unit containing two [4Fe:4S]2+/+ clusters. Binding of the accessory protein NifW parallels a decrease in the distance between these clusters, as well as a subtle change in their coordination. These results support a conformational role for NifW in P-cluster biosynthesis, bringing the two [4Fe:4S] precursors closer prior to their fusion, which may be crucial in challenging cellular contexts.
Upon binding of NifW, a subtle conformation change occurs in NifD2K2, decreasing the distance between the two [4Fe:4S] clusters precursors of the P-cluster in nitrogenase.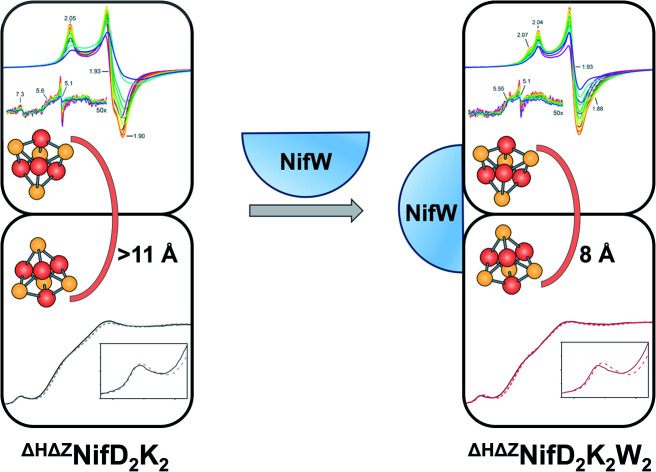
Introduction
Nitrogenases are the catalytic components of biological nitrogen fixation, achieving the nucleotide-dependent reduction of dinitrogen (N2) to ammonia (NH3) at ambient temperature and pressure.1–3 Three structurally and functionally similar but genetically distinct types of nitrogenases have been identified, which are classified according to the heterometal (Mo, V) or lack thereof (Fe) of their cofactors. All microbial nitrogen fixers identified to date produce at least Mo nitrogenase, and certain also produce either or both V- and Fe nitrogenase.4 Mo nitrogenase is composed of catalytic components that include a homodimeric Fe protein, encoded by nifH, and a heterotetrameric MoFe protein, encoded by nifD and nifK. For clarity and consistency, in the present work a nomenclature derived from the corresponding genes will be used to designate the catalytic components as well as certain other proteins involved in maturation of the MoFe protein. For example, Fe protein is designated NifH2 and the MoFe protein is designated NifD2K2 (subscripts indicate subunit organization). NifH2 contains two MgATP-binding sites and a single redox active [4Fe:4S] cluster that sequentially supplies electrons to NifD2K2. NifD2K2 harbors two pairs of complex metalloclusters that participate in catalysis: the [8Fe:7S] P-cluster and the [7Fe:9S:Mo:C:homocitrate] FeMo-cofactor (FeMo-co).3 The P-cluster is located between the subunits of each NifDK catalytic unit and is involved in the inter- and intra-component delivery of electrons from the NifH2 [4Fe:4S] cluster to FeMo-co, which provides the site for substrate binding and reduction.3,5,6 Understanding the biosynthesis of nitrogenase metalloclusters is crucial from the practical perspective of endowing certain eukaryotes the capacity to fix nitrogen by transferring the genetic determinants from diazotrophs, as well as for improving the nitrogen fixing capacity of symbiotic nitrogen fixing microbes associated with crop plants.7–10
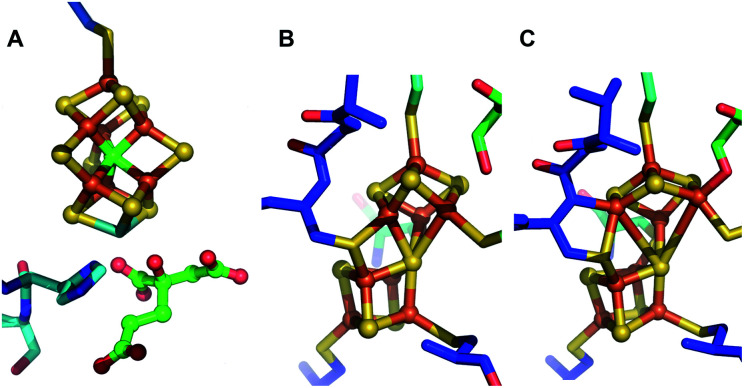
There is some topological similarity between the P-cluster and FeMo-co (Fig. 1) as their [Fe:S] cores can be considered to represent fused or bridged sub-cluster units. Recent refinement of a structural model of NifB, involved in formation of the [Fe:S:C] core common to all three nitrogenase cofactors, suggested a P-cluster-like intermediate as FeMo-co precursor during its formation.11 Nevertheless, there is a fundamental difference between FeMo-co assembly and P-cluster maturation. FeMo-co is separately assembled on a series of scaffolds and subsequently inserted into an immature form of NifD2K2, designated apo-NifD2K2, which already contains mature P-clusters.9,12 In contrast, it appears that an earlier stage of NifD2K2 maturation involves formation of a P-cluster precursor directly within the polypeptide, which is then processed to apo-NifD2K2 replete with mature P-clusters. In the case of Mo nitrogenase of Azotobacter vinelandii, P-cluster maturation involves the interaction of immature forms of NifD2K2 with the dispensable accessory proteins NafH, NifW and NifZ.13,14 P-cluster maturation within NifD2K2 also obligately requires the other nitrogenase catalytic component, NifH2, and MgATP.16–18 The prevailing model for P-cluster maturation is that two [4Fe:4S] clusters are separately formed within opposing NifD and NifK subunits, representing a P-cluster precursor, and then fused in a process involving elimination of a single S atom from one of the [4Fe:4S] units. Although it has been clearly established that each P-cluster precursor contains 8 iron atoms, it is not possible to accurately quantify the S content of P-cluster precursors. It is therefore not possible to distinguish whether P-cluster precursors represent separately paired [4Fe:4S] clusters or an [8Fe:7S] cluster that has already tethered two [4Fe:4S] units together through a shared central S atom. Such a species would be expected to have the same chemical composition as mature P-cluster but differ by some combination of differing redox state, conformation, and/or ligand arrangement.
Fig. 1. Structures of the [Fe:S] clusters of NifD2K2 of A. vinelandii. (A) FeMo-co in DT-reduced NifD2K2 (PDB ID: 3U7Q); (B) P-cluster in the resting all-ferrous PN state (PDB ID: 3MIN); (C) P-cluster in the 2-electron oxidized P2+ state (PDB ID: 2MIN).15.
Besides its key role in P-cluster maturation, NifH2 is also required for FeMo-co insertion into NifD2K2.19,20 Thereby, ΔHNifD2K2§ contains immature P-clusters, but no FeMo-co. Previous work from our laboratories has demonstrated that ΔHNifD2K2 accumulates as a variety of NifD2K2 species, including those having no accessory proteins attached and those bound to either NafH, NifW or NifZ.13 Although the specific functions of these factors are not yet known, biochemical experiments have indicated they sequentially and differentially interact with NifD2K2 during P-cluster maturation in the order NafH, NifW, NifZ. Prior work has also established that, to date, there is no biochemical or physiological defect associated with loss of NafH function,21 whereas inactivation of NifW results in slower diazotrophic growth due to low accumulation of active NifD2K2.22 Inactivation of NifZ also entails slower diazotrophic growth resulting from the accumulation of a mixed population of NifD2K2 species including both fully mature NifD2K2 and a variety of inactive or partially active species.14,21 Among those, some are bound to NifW. These observations have led to the suggestion that NifW serves to stabilize a form of immature NifD2K2 at an early stage in P-cluster maturation and that NifZ could be involved in the dissociation of NifW and further recruitment of NifH2 in the final stage of P-cluster maturation. In the present work, the biophysical properties of cluster species contained in both immature NifD2K2 having no bound accessory protein and immature NifD2K2 with two associated NifW were examined by electron paramagnetic resonance (EPR) and X-ray absorption (XAS) spectroscopies.
Results
Mature P-clusters in resting state NifD2K2 are diamagnetic and therefore do not exhibit any EPR signature, whereas FeMo-co is paramagnetic and exhibits a characteristic S = 3/2 EPR signal.2 Thus, reduced, fully matured NifD2K2 shows an EPR signature associated with the presence of FeMo-co only. In contrast, ΔHNifD2K2 lacks the S = 3/2 EPR signature associated with FeMo-co but exhibits a complex S = 1/2 signature that has been assigned as a precursor to mature P-cluster formation.16,23 Comparison of EPR analysis of ΔZNifD2K2 or ΔHNifD2K2 suggested that the complex S = 1/2 signature associated with immature P-clusters corresponds to two electronic isomers having close g-tensors of [2.06, 1.93, 1.89] and [2.03, 1.93, 1.86].13,14 Furthermore, fractionation of affinity-purified ΔZNifD2K2 could be resolved into populations that include ΔZNifD2K2 with no associated accessory protein, ΔZNifD2K2 with one NifW bound (ΔZNifD2K2W), and ΔZNifD2K2 with two bound NifW (ΔZNifD2K2W2).14 Initial EPR studies of isolated ΔZNifD2K2W2 revealed a single S = 1/2 species with g = [2.02, 1.93, 1.86]. Nevertheless, analysis of the ΔZNifD2K2 fraction for which no NifW is bound is complicated since inactivation of NifZ does not directly affect FeMo-co formation and only slows, but does not eliminate, P-cluster maturation.14
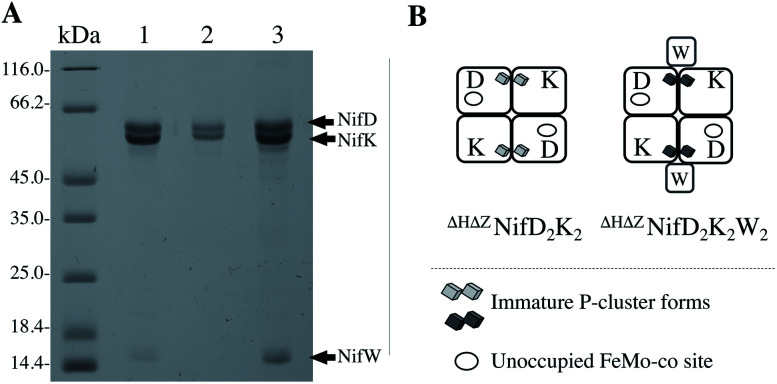
To allow the isolation of a highly enriched fraction of ΔZNifD2K2 containing immature P-clusters and lacking both FeMo-co and bound NifW, a strain was constructed in which both the nifH and nifZ genes are deleted. The Strep-tag placed at the N-terminus of NifD in this construct enables affinity purification of ΔHΔZNifD2K2 fractions that are highly enriched for species uniformly deficient in FeMo-co assembly and P-cluster maturation. ΔHΔZNifD2K2 was further fractionated using anion exchange chromatography allowing isolation of fractions corresponding to ΔHΔZNifD2K2 (no NifW attached to NifD2K2) and ΔHΔZNifD2K2W2 (two NifW attached) (Fig. 2, S1 and S2†).
Fig. 2. NifD2K2 species characterized in this work. (A) SDS-PAGE analysis of proteins isolated by one-step Strep-tag affinity purification (1), and ΔHΔZNifD2K2 (2) and ΔHΔZNifD2K2W2 (3) isolated by ion-exchange chromatography of the sample shown in (1). Molecular weight standards are shown on the left lane. The presence of a small amount of a ΔHΔZNifD2K2NafHX species (less than 8% of the total sample) in (3) was revealed by SDS-PAGE analysis of more concentrated samples as shown in Fig. S1.† (B) Schematic representation of samples shown in (A). Interaction of NifW at the interface of NifD2K2 is only shown for convenience and has not been experimentally established. Corresponding X-band EPR spectra are provided in Fig. S2.†.
The availability of separable forms of NifD2K2 harboring immature P-clusters and having no associated FeMo-co, with or without NifW bound, permitted the detailed biophysical comparison of these species. UV-vis measurements of the isolated samples showed slight differences in their spectra, suggesting subtle structural changes upon binding of NifW (Fig. S3†). To characterize the structural and spectroscopic properties of these clusters, XAS and EPR spectroscopic analyses were performed. For comparative purposes, apo-NifD2K2 was also purified and characterized from a strain in which the FeMo-cofactor anchoring 275Cys residue is substituted by 275Ala (abbreviated as C275ANifD2K2). C275ANifD2K2 contains mature P-clusters, but no FeMo-co.13
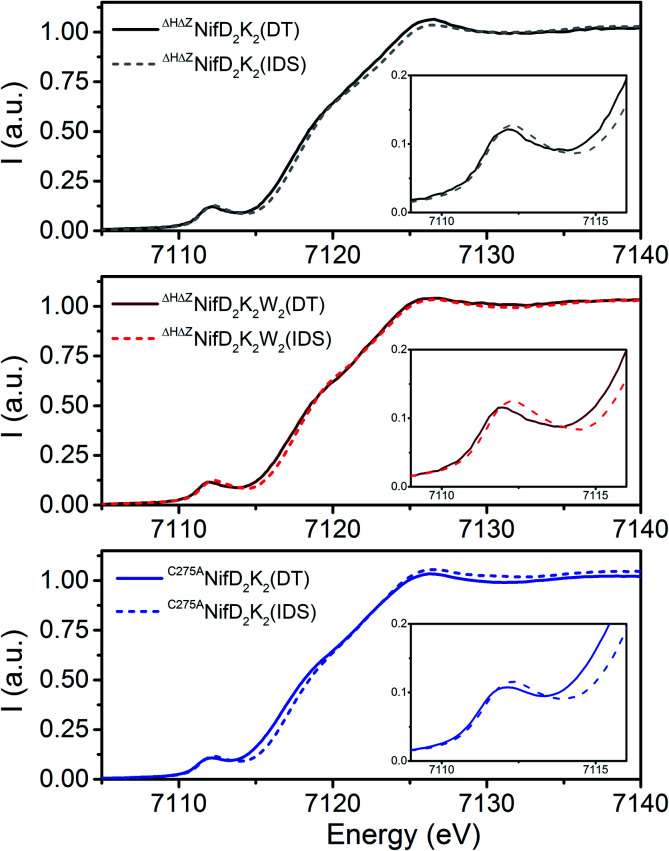
To determine the influence of NifW on immature P-cluster oxidation state and geometry, X-ray absorption spectroscopy (XAS) at the Fe K-edge was performed for ΔHΔZNifD2K2, ΔHΔZNifD2K2W2, and C275ANifD2K2 in both the dithionite (DT)-reduced and indigo disulfonate (IDS)-oxidized states.
As previously noted, C275ANifD2K2 does not contain FeMo-co but contains mature P-clusters, thus providing a useful internal control for comparison of immature and mature P-cluster species.13Fig. 3 compares the DT-reduced and IDS-oxidized forms of each variant under investigation. All spectra appear typical for [Fe:S] clusters, with a single well-defined pre-edge feature ∼7112 eV leading into an edge presenting a shoulder at ∼7118 eV.17,24–27 Upon oxidation, all variants exhibit an increase in both intensity and energetic position of the pre-edge feature, as well as an increase in the energetic position of the rising edge (7115–7119 eV). The position of the pre-edge and rising edge can serve as diagnostics of oxidation state, shifting to higher energies with increased oxidation state. Meanwhile, pre-edge intensity at the Fe K-edge derives from both lowering of centrosymmetry, which induces Fe 3d–4p mixing, and the number of available holes in the valence shell of Fe.28,29 Hence, relative increases in pre-edge intensity can arise from either a decrease in local symmetry, increase in oxidation state, or some combination. Therefore, these changes are all consistent with some degree of Fe-centered oxidation upon exposure to IDS. The observed shift in the rising edge moving from C275ANifD2K2(DT) → C275ANifD2K2(IDS) is +0.7 eV. Previous studies have already established that exposure to IDS oxidizes the mature, all-ferrous P-cluster (PN) to a two-electron oxidized form (P2+).30 Therefore, this +0.7 eV increase in the rising edge can be used as a tentative diagnostic for two-electron oxidation. This is consistent with previous observations for the DT-reduced and IDS-oxidized forms of ΔBNifD2K2, which, like C275ANifD2K2, contains mature P-clusters but no FeMo-co, and displayed a relative shift of +0.6 eV.17 Meanwhile, the observed shifts in the rising edge for both ΔHΔZNifD2K2(DT) → ΔHΔZNifD2K2(IDS) and ΔHΔZNifD2K2W2(DT) → ΔHΔZNifD2K2W2(IDS) are +0.35 eV, half that found for C275ANifD2K2. Therefore, exposure of either immature P-cluster variant to IDS likely results in a one-electron oxidation event.
Fig. 3. Comparison of the normalized Fe K-edge XAS spectra of ΔHΔZNifD2K2 (top), ΔHΔZNifD2K2W2 (middle), and C275ANifD2K2 (bottom) in the DT-reduced (solid) and IDS-oxidized (dashed) forms. Insets show an expanded view of the pre-edge region from 7109–7116 eV.
Comparison of the DT-reduced forms of the three variants reveals that ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT) display a nearly superimposable rising edge between 7114–7118 eV, shifted by +0.7 eV relative to C275ANifD2K2(DT) (Fig. S4†). This shift is approximately the same as observed when C275ANifD2K2(DT) and C275ANifD2K2(IDS) (Fig. 3) are compared, supporting an interpretation that ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT) are both two-electron oxidized relative to C275ANifD2K2(DT). Similarly, the rising edges of the IDS-oxidized forms of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 are nearly superimposable and shifted by +0.35 eV relative to C275ANifD2K2(IDS). Interestingly, while the Fe K-edge spectra of ΔHΔZNifD2K2(IDS) and ΔHΔZNifD2K2W2(IDS) are extremely similar, the DT-reduced forms exhibit significant deviations at the pre-edge, around 7112 eV. Namely, while the pre-edge intensities of both ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT) are more intense than C275ANifD2K2(DT), ΔHΔZNifD2K2W2(DT) appears shifted to lower energy by −100 meV relative to either ΔHΔZNifD2K2(DT) or C275ANifD2K2(DT). Besides oxidation state, local symmetry and coordination environment can play a role in energetic position and intensity distribution of the pre-edge, and this shift may be indicative of local structural differences in the Fe environments of ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT). Based on these results proposed Fe oxidation state distributions of the three variants are summarized in Table 1. These distributions are assigned assuming 8 unique Fe per NifDK unit.
Summary of proposed Fe oxidation state distributions of the cluster species found in ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, based on Fe K-edge XAS.
| Sample | DT-reduced | IDS-oxidized |
|---|---|---|
| ΔHΔZNifD2K2 | [6FeII2FeIII] | [5FeII3FeIII] |
| ΔHΔZNifD2K2W2 | [6FeII2FeIII] | [5FeII3FeIII] |
| C275ANifD2K2 | [8FeII] | [6FeII2FeIII] |
The direct overlay of the Fe K-edge XAS spectra of ΔHΔZNifD2K2(DT), ΔHΔZNifD2K2W2(DT), and C275ANifD2K2(IDS) further supports these oxidation state distribution assignments, where the rising edges (7115–7119 eV) of these three samples are superimposable (Fig. S5†). Overlays of the pre-edge regions additionally support small variations in local geometry.
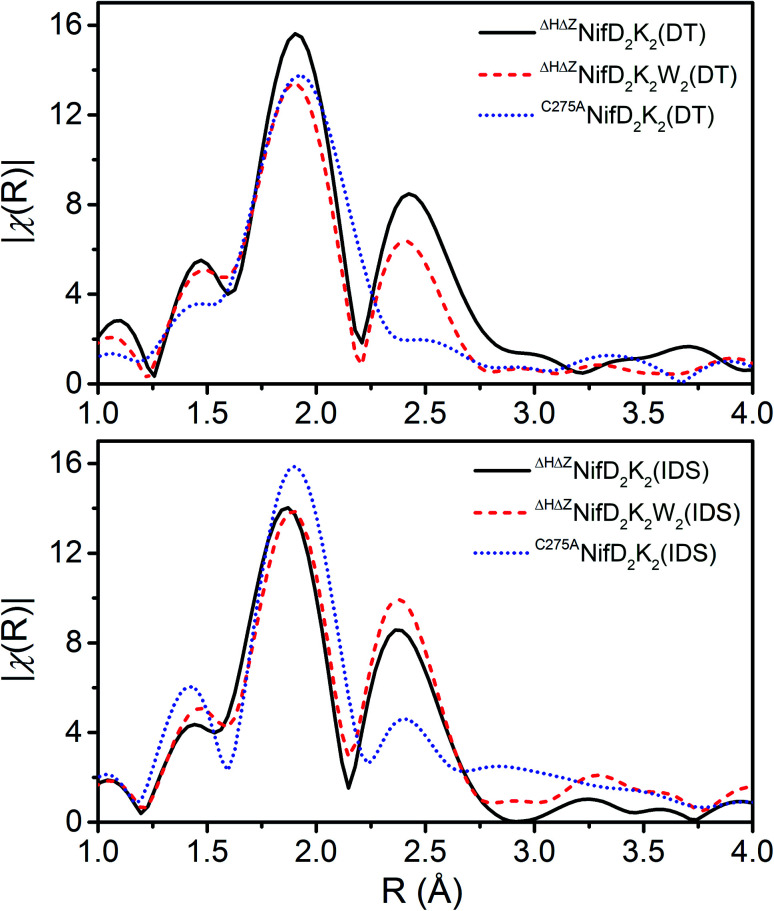
To obtain further information on these species, Fe K-edge EXAFS measurements were performed (Fig. 4 and S6–S8†). As shown in Fig. 4, all spectra exhibit either one or two dominant features around 1.9 and 2.4 Å, which may be attributed to Fe–S and Fe–Fe scatterers, respectively. In C275ANifD2K2(DT), only the dominant Fe–S scatterer is observed due to near complete phase cancellation of the two distinct Fe–Fe scattering paths, Fe–Fe(1) and Fe–Fe(2). This has been previously observed in studies of both mature NifD2K2 and ΔBNifD2K2.17,24,25,31 However, this is not the case for either ΔHΔZNifD2K2(DT) or ΔHΔZNifD2K2W2(DT) which display clear features at 2.4 Å, similar to previous observations for [4Fe:4S] cubane clusters and ΔHNifD2K2.17,25,26 In the IDS-oxidized form of C275ANifD2K2 this feature is apparent, although at a significantly lower intensity in the FTs (Fig. 4 and S6†). This is likely due to the significant structural changes that have been crystallographically observed to accompany the oxidation of mature P-cluster, including light atom coordination of Fe and partial Fe–S bond dissociation.15,32 In the P2+ state, one of the two sub-cubanes assumes an open conformation, with two iron atoms (Fe5 and Fe6) partially dissociated from the bridging sulfide to ligate the backbone amide of a cysteine and the hydroxyl group of a serine, respectively (Fig. 1). A considerable number of other differences are also observed among all three variants for ∼1.9 and ∼2.4 Å scattering features, including variations in total intensity, relative intensity, and small variations in radial distribution. These imply all three variants exhibit structures that are distinct from one another. However, as these spectra involve the complex overlap of at least 3–4 scattering paths, direct interpretation of the absolute values of the R-space spectra are non-diagnostic.
Fig. 4. Comparisons of the Fourier transforms (FTs) of the Fe K-edge EXAFS of the DT-reduced and IDS-oxidized forms of ΔHΔZNifD2K2 (black), ΔHΔZNifD2K2W2 (red), and C275ANifD2K2 (blue) in R-space. Spectra are k3-weighted, and FTs were performed for a k-range of 3–13 Å. No phase-shift was applied.
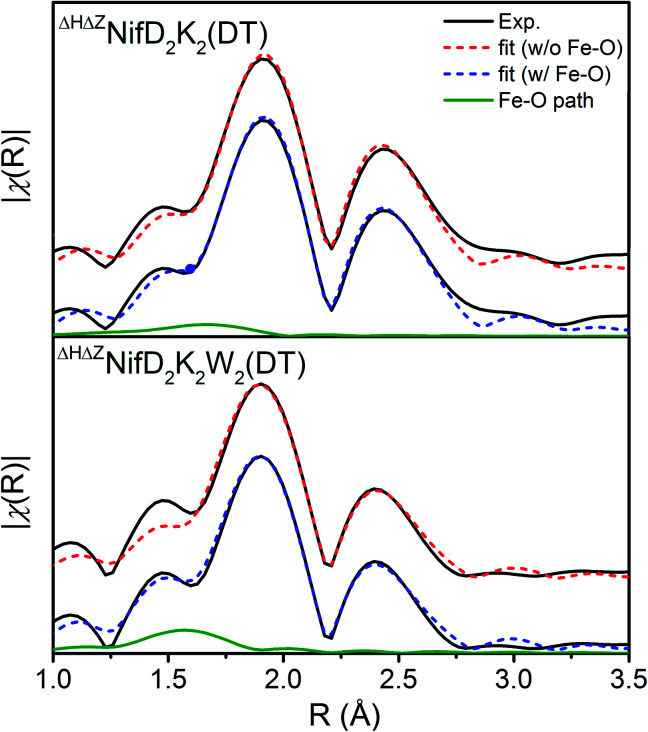
To gain physical insight into these changes, a minimalist model was used to fit the data, involving a single Fe–S scattering path and two unique Fe–Fe scattering paths. The use of two unique Fe–Fe scattering paths is necessitated due to the wide distribution of Fe–Fe distances observed in the mature P-cluster as well as in iron–sulfur cubane clusters.24,33 In addition, the presence of O and N coordination of Fe observed in crystal structures of oxidized mature P-cluster prompted us to test whether such coordination environments exist in the immature variants, using fits that included light atom scatterers.15,32 The resulting best fit parameters for models both with and without an Fe–O scatterer are provided in Table S1.† It is important to note that while an Fe–O scatterer was employed in these models, the similar electron density of O and N make them virtually indistinguishable from one another, and therefore this scattering pathway represents a sum of both possible Fe–O and Fe–N coordination.
Due to the high degree of correlation between scattering path degeneracy, N, and bond variance, σ2, it is generally difficult to accurately fit both simultaneously without some structural reference to restrict N. To fit the unknown structures of the clusters in ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, the sum of N for Fe–Fe(1) and Fe–Fe(2) was fixed to 3, which is the theoretical Fe–Fe path degeneracy for a [4Fe:4S] cubane cluster, and the ratios of N for either path were allowed to vary. As there are 8 unique Fe absorbers in each variant, the minimal increment in N for Fe–O/N and Fe–S scattering paths was limited to 0.125. Meanwhile, the minimal increment used in variation of the Fe–Fe scattering paths was 0.25, as the loss or addition of a single Fe–Fe scatterer must be observed by both Fe involved. The degeneracies of each pathway for C275ANifD2K2(DT) and C275ANifD2K2(IDS) were determined from equivalent crystal structures of the mature P-cluster in the PN and the P2+ states and fixed during fitting.15,33
As shown in Table S1,† the fit Fe–S, Fe–Fe(1), and Fe–Fe(2) scattering distances all appear to fall within typical ranges for both varying forms of the P-cluster and more generally for homometallic iron–sulfur clusters.34–38 A comparison of the fit parameters of the respective DT-reduced and IDS-oxidized forms reveals only relatively minor contractions for ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2. These changes are considerably more significant when comparing C275ANifD2K2(DT) and C275ANifD2K2(IDS), which exhibit contractions of ∼0.06 Å and ∼0.10 Å in Fe–Fe(1) and Fe–Fe(2), respectively. However, based on the Fe oxidation state distributions for the DT-reduced and IDS-oxidized variants, these larger contractions are not surprising due to C275ANifD2K2(IDS) being two-electron oxidized relative to C275ANifD2K2(DT). Notably, the structural parameters of ΔHΔZNifD2K2(DT), ΔHΔZNifD2K2W2(DT), and C275ANifD2K2(IDS) appear quite similar, further supporting the oxidation state distribution assignments shown in Table 1 (Fig. S8†).
Inclusion of an Fe–O/N scattering pathway provides an improved fit for all variants in both DT-reduced and IDS-oxidized forms in terms of χ2 and R-factors. This is not unexpected, as increasing the number of free parameters in a model will typically provide a better fit in terms of mean square deviation. However, fits performed for ΔHΔZNifD2K2W2 involving an Fe–O scatterer show significantly greater improvement in these parameters than for either ΔHΔZNifD2K2 or C275ANifD2K2. This is also clearly visualized in Fig. 5, where the inclusion of the Fe–O scattering contribution to the fit provides only a slight improvement between 1.25–1.75 Å for ΔHΔZNifD2K2(DT), but a significant improvement over this same R-range for C275ANifD2K2(DT).
Fig. 5. Comparison of the Fe K-edge EXAFS of DT-reduced ΔHΔZNifD2K2 (top) and ΔHΔZNifD2K2W2 (bottom) experimental spectra (black) and respective best fit models with (blue, dashed) and without (red, dashed) inclusion of an Fe–O/N scattering path contribution (green, solid). Corresponding fit parameters are provided in Table S1.† Spectra are k3-weighted, and FTs were performed for a k-range of 3–13 Å. Fits were performed using k3-weighting over ranges of k = 3–13 Å−1 and R = 1–3 Å.
To evidence the statistical improvement gained in fits of ΔHΔZNifD2K2W2 by inclusion of an Fe–O scattering path, we have included the reduced χ2 statistic in Table S1.† This parameter is calculated by dividing the root mean-square deviation, χ2, by the difference between the total number of degrees of freedom and the free parameters used in the fit. The number of degrees of freedom (χind) is calculated by eqn (1).
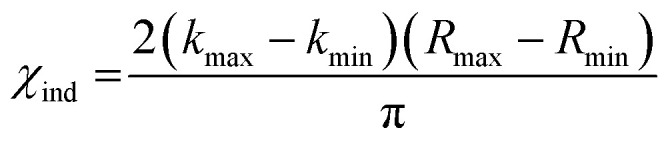 |
1 |
For the present spectra χind equates to 12.4. When including an Fe–O scattering path to the EXAFS model, two additional free parameters are necessary, raising the total number of free parameters from 7 to 9 while the total number of degrees of freedom remains the same. This reduces the unused number of degrees of freedom from 5.4 to 3.4, meaning a >1.6-fold improvement in χ2 must be achieved to improve the reduced χ2 statistic. Given the weak scattering properties of O compared to heavier atoms such as S and Fe, the presence of an Fe–O scatterer is not expected to make particularly large spectral contributions. In turn, reduced χ2 serves as a particularly harsh measure of statistical significance. Nevertheless, the reduced χ2 statistic is improved for both ΔHΔZNifD2K2W2(DT) and ΔHΔZNifD2K2W2(IDS) by inclusion of an Fe–O scatterer with N = 0.5. These statistical improvements are nearly invariant for a range of 0.25–0.5, equating to the coordination of 2–4 light atoms (O or N) per immature P-cluster of ΔHΔZNifD2K2W2.
While the presence of Fe–O/N coordination in ΔHΔZNifD2K2W2 is well supported by the observed statistical improvements, inclusion of this scattering path to fits of either ΔHΔZNifD2K2 or C275ANifD2K2 results in a poorer reduced χ2 statistic, with increases of 40–80%. However, this lack of improvement in reduced χ2 does not exclude the possibility that light atom scatterers are present in the Fe coordination environments of these samples. Namely, the overall contribution of such a light atom scatterer to the EXAFS is expected to be small, especially considering that heavier scatterers such as S and Fe are present. Additionally, variation in bond distances between Fe and O or N can greatly affect whether these scattering paths will accumulate into some significant spectral contribution. For example, the P2+ state of the mature P-cluster displays Fe–O coordination from the NifK Ser188 residue at 1.90 Å, and Fe–N from the NifD Cys88 residue at 2.15 Å.15
Due to the large difference in distances of these two bonds (0.25 Å), attempts to account for both with a single scattering path would require a very large bond variance σ2, which would further diminish the contributions of these already weak scattering paths to the total EXAFS spectrum. With such small contributions it is little surprise that inclusion of an Fe–O/N scatterer to C275ANifD2K2(IDS), which contains mature P2+, does not significantly improve reduced χ2.
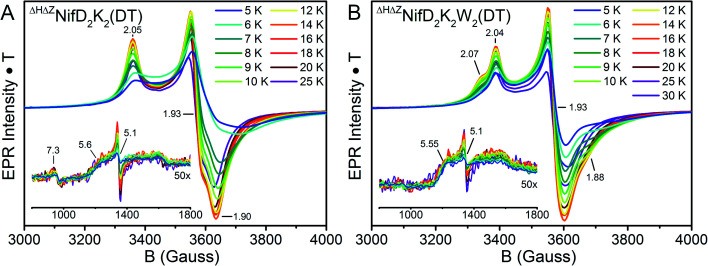
The oxidation state assignments from our XAS studies (Table 1) imply that the IDS-oxidized forms of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 should be paramagnetic, and possibly the DT-reduced forms as well. Therefore, to further investigate the electronic structures of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, X-band EPR measurements of these systems were performed. The EPR spectra of DT-reduced ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 (Fig. 6 and S2†) are dominated by broad S = 1/2 signals similar to those previously observed in ΔZNifD2K2.14ΔHΔZNifD2K2 displays a slightly rhombically distorted axial signal, with clear inflections at g ≈ 2.05, 1.93, and 1.90. Meanwhile, ΔHΔZNifD2K2W2 is dominated by an axial signal with inflections at g ≈ 2.04 and 1.93, and additional shoulders appearing at g ≈ 2.07 and 1.88. Additionally, both ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 samples exhibit weak signals at g ≈ 5.60 and 5.11, consistent with transitions anticipated for an S = 3/2 system with negative zero-field splitting. These signals had previously been detected in ΔHNifD2K2 and ΔZNifD2K2, but are not present in ΔBNifD2K2, implying they are associated with an immature P-cluster species.14 The appearance of a very minor signal g ≈ 7.3 is also observed in ΔHΔZNifD2K2, possibly corresponding to the Ms = ±1/2 manifold of an S = 5/2 signal. SDS-PAGE analysis of the ΔHΔZNifD2K2W2 sample (Fig. S1†) reveals the presence of a low amount of NafH, which could suggest that elements of the spectrum might originate from a low level of NifD2K2 with bound NafH. However, the normalized intensities of this signal are invariant between the protein isolated by one-step Strep-tag affinity purification and the further-purified ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 fractions, supporting that this signal is independent of NafH (Fig. S2†). Alternatively, a similar S = 3/2 signal has also been observed in the X-band EPR spectrum of NifH2, which contains a subunit-bridging [4Fe:4S] cluster, as well as other [Fe:S] clusters having dominant S = 1/2 signals.39–42 The S = 3/2 signal observed here therefore may belong to an S = 1/2, 3/2 spin admixture, originating from a cluster species with an exchange-coupled system.43,44
Fig. 6. Temperature-dependent X-band EPR of DT-reduced (A) ΔHΔZNifD2K2 and (B) ΔHΔZNifD2K2W2. Spectra were collected using 1 mW power at 9.65 GHz. All spectra are normalized for power, gain, collection time, and temperature.
Variable temperature measurements were performed to further characterize and quantify the observed EPR signals (Fig. 6). The obtained spectra reveal similar saturation behavior in DT-reduced ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, with saturation observed below 12 K and temperature dependent broadening occurring above 20 K. This narrow temperature range of unsaturated signal intensity has been commonly observed in S = 1/2 [4Fe:4S]+ clusters.45 Meanwhile, the temperature-corrected intensities of the respective S = 3/2 signals are relatively invariant across the measured temperature range. Based on the Asaa–Vanngard corrected spin-integration against a CuSO4 standard, the S = 1/2 signal of DT-reduced samples accounts for ∼3.75 spins/ΔHΔZNifD2K2 and ∼3.5 spins/ΔHΔZNifD2K2W2, respectively.46 By comparison, the minor S = 3/2 signal accounts for approximately 0.3 spins in either DT-reduced ΔHΔZNifD2K2 or ΔHΔZNifD2K2W2; we note the broadness of this signal precludes precise quantification. These results support ∼2 spins per NifDK(W) unit, indicating the spectra presented in Fig. 6 represent a convolution of at least two unique S = 1/2 species having similar spectral properties. When combined with the described XAS measurements, these results support the presence of two unique [4Fe:4S]+ clusters in both ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2.
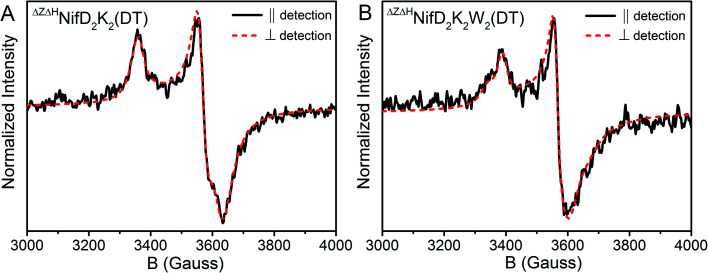
In the context of the P-cluster, the potential presence of two unique [4Fe:4S]+ per NifDK unit of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 is not surprising. However, if these clusters were in relatively close spatial proximity, as would be required for formation of the mature P-cluster, some significant degree of spin–spin coupling would be anticipated.47 Additionally, simulations of ΔHΔZNifD2K2(DT) or ΔHΔZNifD2K2W2(DT) using either a single S = 1/2 component or two uncoupled S = 1/2 components in a 1 : 1 stoichiometry cannot reasonably account for either the level of broadening or additional inflections around g = 2 observed in these spectra. Therefore, to investigate these species further, parallel mode X-band measurements of DT-reduced ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 were performed (Fig. 7).
Fig. 7. Comparison of perpendicular (black, solid) and parallel mode (red, dashed) X-band EPR spectra of DT-reduced (A) ΔHΔZNifD2K2 and (B) ΔHΔZNifD2K2W2. Spectra were measured at 16 K, 1 mW, and 9.65 GHz.
The transition selection rule for parallel mode detection is ΔmS = 0, and thus requires mixing of mS levels to gain intensity. Such mixing occurs in a limited number of circumstances. Examples include (i) Kramer's systems with half-integer S > ½ and very weak zero-field splitting (on the order of the Zeeman effect), (ii) non-Kramer's systems with energetically close lying integer spin levels, and (iii) spin-coupled systems in which the coupling strength competes with the Zeeman effect.48 Parallel mode intensity is observed in spectra of both DT-reduced ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, indicating the observed signals could originate from weak spin-coupling. On this basis, spectra of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 were each fit using two spin-coupled S = 1/2 systems (Fig. S9 and Table S2†). Due to the large number of parameters involved in fitting such a system, a definitive, unique fit was not possible. Spectra of ΔHΔZNifD2K2 could be reasonably fit using two overlapping S = 1/2 systems with equal contributions in the absence of spin-coupling. Inclusion of spin-coupling resulted in an estimate of J = 1.2 × 10−3 cm−1 between two MS = 1/2 components a and b with ga = [2.06, 1.91, 1.91] and gb = [2.05, 1.93, 1.86], corresponding to a lower limit on the e−–e− distance of approximately 11 Å based on the point-dipole approximation. Meanwhile, spectra of ΔHΔZNifD2K2W2 required the use of spin coupling to be simulated, with a larger axial e−–e− coupling of J = 3.3 × 10−3 cm−1 between ga = [2.05, 1.93, 1.90] and gb = [2.06, 1.93, 1.87], resulting in a reduced e−–e− distance of ∼8 Å. The dependence of the simulated spectra on the magnitude of the principal values of the e−–e− coupling interaction matrix is provided in Fig. S10.† Although the point-dipole approximation has been shown to overestimate e−–e− distances in spin-delocalized systems, the relative inter-cluster distances are expected to remain consistent given the similarity of ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT). These results support that ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT) consist of two unique, separate [4Fe:4S]+ clusters, and that the distance between these clusters is decreased in the NifW-bound form.
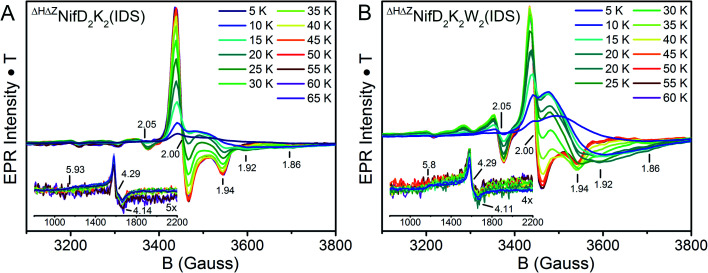
Further EPR measurements were performed on IDS-oxidized samples. Mature P-cluster in the P2+ state exhibits a low-field parallel mode signal (appearing at g ≈ 12.0 for A. vinelandii NifD2K2, g = 15.6 for Xanthobacter autotrophicus NifD2K2, and g = 16.0 for Gluconacetobacter diazotrophicus NifD2K2) associated with an integer spin system.48,49 This is not the case for IDS-oxidized ΔHΔZNifD2K2 or ΔHΔZNifD2K2W2, in agreement with previous reports for ΔHNifD2K2.16 Given the EPR features observed for ΔHΔZNifD2K2(DT) and ΔHΔZNifD2K2W2(DT) combined with the XAS-based oxidation state assignments summarized in Table 1, IDS-oxidized samples of both species were anticipated to be non-integer spin. Indeed, IDS oxidation of either ΔHΔZNifD2K2 or ΔHΔZNifD2K2W2 resulted in the complete disappearance of the spectra observed for the dithionite-reduced species, and the appearance of a new, complicated set of signals in the CW X-band EPR (Fig. 8). Importantly, re-reduction of these samples with sodium dithionite after desalting resulted in spectra identical to the reduced states presented in Fig. 6 (see Fig. S11†). Therefore, IDS-oxidation of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 is a reversible process, and none of the features observed in these spectra appear to originate from cluster damage.
Fig. 8. Temperature-dependent X-band EPR of IDS-oxidized (A) ΔHΔZNifD2K2 and (B) ΔHΔZNifD2K2W2. Spectra were collected using 1 mW power at 9.65 GHz. All spectra are normalized for power, gain, collection time, and temperature. The signals appearing in the low-field region are provided as insets.
In the IDS-oxidized forms of both ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, low-field inflections are clearly observed starting at g = 5.8 and 4.3 (Fig. 8, insets). Temperature-dependent measurements reveal an increase in intensity of the g = 4.3 inflection between 5–20 K, followed by a decrease in intensity above 20 K. This result is consistent with the presence of a rhombic S = 5/2 species, with the g = 4.3 inflection arising from population of the Ms = 3/2 manifold as a function of temperature. Meanwhile, the presence of an inflection at g = 5.8 at low temperatures is consistent with the presence of an S = 3/2 species with negative zero-field splitting, like that observed in the equivalent DT-reduced species (Fig. S12†).
At intermediate fields (3200-3800 G), the spectra of IDS-oxidized ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 become complex and strongly temperature dependent. At low temperatures both display inflections at g ∼2.00 (sharp) and 1.94 (broad). The extremely broad nature of the signal centered around 1.94 is consistent with rapid spin-lattice relaxation. At temperatures above 20 K, this broad signal quickly disappears and is replaced by a well-defined axial S = 1/2 signal having inflections at g = 2.00, 2.00 and 1.94, which take full form at 40 K (ΔHΔZNifD2K2) and 45 K (ΔHΔZNifD2K2W2). These signals continue to persist at higher temperatures without any significant decrease in intensity, similar to previous observations for [2Fe:2S] and [3Fe:4S] clusters.45,50 Additionally, both species display further minor inflections at g = 2.05, 1.92 and 1.86, and possibly at 2.15 and 2.09 as well. A similarly shaped S = 1/2 spectrum has been observed for the minor component of the OX state of the open-cubane active site of the hybrid cluster protein (Hcp), but differs significantly in both temperature dependence and observed g-values (1.971, 1.951, 1.898).51–53 While the observed inflections in the IDS-oxidized ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 spectra display very similar g-values, the intensity ratios of these components and their temperature dependence are distinct (Fig. 8). These additional inflections are generally indicative of a spin–spin coupled system with non-integer S > 1/2. The presence of spin–spin coupling is consistent with our observations of the DT-reduced species.
Discussion
The nitrogenase MoFe protein from A. vinelandii, designated here as NifD2K2, contains two complex metalloclusters: the P-cluster and the FeMo-cofactor. Although both clusters share similar topological features, their assembly pathways differ significantly. The biosynthesis of FeMo-co does not occur on NifD2K2; instead, it is maturated on other scaffolds, and once complete inserted into an apo-form of NifD2K2 containing mature P-clusters.9 In contrast, maturation of individual P-clusters is proposed to occur in situ, via the fusion of two distinct [4Fe:4S] clusters already contained within apposing NifDK subunits through a process involving the NifD2K2 catalytic partner, NifH2, and MgATP.18 In A. vinelandii, NifD2K2 sequentially binds the accessory proteins NafH, NifW and NifZ prior to the final maturation step completed by NifH2.13 Support for the [4Fe:4S] cluster fusion model has come from a variety of spectroscopic characterizations of NifD2K2 produced in the absence of NifH2 (ΔHNifD2K2), which contain immature P-clusters but lack FeMo-co.16–18,23,54
Previous X-band perpendicular mode EPR characterization of the immature P-clusters contained in ΔHNifD2K2 revealed a mixture of two S = 1/2 electronic isomers with similar g-tensors (g ∼ [2.06, 1.93, 1.89] and [2.03, 1.93, 1.86]).13,14,16 A similar signature was also observed for NifD2K2 produced in the absence of NifZ (ΔZNifD2K2).14 Recent biochemical studies established that both ΔHNifD2K2 and ΔZNifD2K2 isolated by single-step affinity purification represent mixed populations of NifD2K2.13,14 Specifically, ΔHNifD2K2 was found to be separately bound to either NafH, NifW, or NifZ, or unbound. Meanwhile, ΔZNifD2K2 was resolved into three fractions, namely unbound, or with one or two associated NifW (ΔZNifD2K2W and ΔZNifD2K2W2, respectively). Importantly, the ΔZNifD2K2W2 fraction was found to be highly enriched in one of the S = 1/2 signals (g ∼ [2.03, 1.93, 1.86]), and did not exhibit the S = 3/2 signal associated with FeMo-co. This result indicated that the two S = 1/2 electronic isomers observed in ΔHNifD2K2 and ΔZNifD2K2 represented a mixture of two different populations of immature P-cluster.
In the present work, we have shown that a strain with both nifH and nifZ gene deletions has enabled the isolation of sub-populations of immature ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, each harboring immature P-clusters which are differentiable by their respective EPR signatures. As described below, isolation of these species permitted the detailed spectroscopic analysis of the immature P-cluster states associated with the corresponding samples.
Perpendicular-mode X-band EPR measurements demonstrated that a combination of the individual immature P-cluster species respectively contained in ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 species recapitulates the complex EPR signature found in mixed species contained in either ΔHNifD2K2 and ΔZNifD2K2 samples prior to their fractionation (Fig. S1†). Thus, this complex signature represents at least two distinct populations having two different states of immature P-clusters, rather than arising from individual sub-clusters or oscillating conformations contained within a single immature P-cluster species. Measurements of the Fe K-edge XAS permitted assignment of redox states of the clusters observed in each species. Both reduced samples of ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 displayed a [6FeII2FeIII] oxidation state, consistent with the presence of two [4Fe:4S]+ cubanes as evidenced by EPR. Oxidation with IDS lead to the one-electron oxidation of either species, which suggests that only one subcluster within an individual P-cluster precursor is oxidized to [4Fe:4S]2+ while the other maintains its valency, indicating different redox properties for each subcluster. In contrast, IDS treatment of mature, all-ferrous P-cluster (PN) results in a two-electron oxidation to form P2+, shifting from an [8Fe:7S]0 state to a [8Fe:7S]2+ state, consistent with previous measurements.25
EPR simulations of reduced ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2, combined with spin quantification, suggest their corresponding spectra represent two unique S = 1/2 species having similar electronic properties arising from two distinct [4Fe:4S] clusters. Weak spin-coupling detected via parallel-mode X-band EPR further supports that these two species are spatially close enough to display spin–spin coupling. This coupling is significantly increased in the ΔHΔZNifD2K2W2 sample when compared to ΔHΔZNifD2K2, suggesting a decrease in distance between the two [4Fe:4S]+ cubanes in the NifW bound state, and hence an associated conformational change in ΔHΔZNifD2K2 upon binding of NifW. Spin–spin coupling is also observed in equivalent oxidized samples, again differing between ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2. The unique spectra of these oxidized samples are reminiscent of open [4Fe:4S] clusters, albeit with significantly different temperature-dependent behavior.51–53,55
Despite having similar electronic properties, the immature P-clusters contained in ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 display significant structural differences, as indicated in our XAS analyses by both shifts in the intensity and energetic position of the pre-edge feature and by variations in the EXAFS region. Remarkably, EXAFS analysis of ΔHΔZNifD2K2W2 suggests additional Fe-coordination of 1–2 light atom(s) (N or O) per [4Fe:4S] subcluster when compared to the ΔHΔZNifD2K2 species, even when regarding stringent considerations of fitting statistics. This observation suggests a transition or stabilization of the coordination environment for one or, perhaps, both sub-clusters upon NifW binding.
It should be noted that although this work has revealed that NifD2K2 from cells deleted for the nifH and nifZ genes represents a pool of two distinct ΔHΔZNifD2K2 and ΔHΔZNifD2K2W2 species, this does not necessarily indicate that either species accumulates significantly under normal physiological conditions. The ΔHΔZNifD2K2 species lacking bound NifW may accumulate due to interruption of the maturation process in the absence of NifZ and NifH2. Both transcriptomic and proteomic studies have revealed that NifD and NifK are expressed and accumulate in vivo at much higher levels than the NafH, NifW, and NifZ assembly factors.56,57 Consequently, when the maturation process is interrupted by the inactivation of NifZ and NifH2, the available NifW would become sequestered into a NifD2K2W2 complex, and excess immature NifD2K2 lacking NifW would be forced to accumulate. In contrast, under physiological conditions maturation must occur more rapidly than assembly intermediates can accumulate. Hence, deleting genes involved in P-cluster maturation offers biochemical snapshots of this process. In this context, ΔHΔZNifD2K2W2 allows the characterization of the NifDKW complex occurring in physiological conditions, and ΔHΔZNifD2K2 represents an immature NifD2K2 species prior to interaction with NifW.
Conclusions
Taken together, our results shed light on the structure of the individual immature P-cluster states and on the possible roles of NifW. Upon binding of NifW to ΔHΔZNifD2K2, a decrease in the distance between both [4Fe:4S]+ clusters occurs along with the coordination of 2–4 light atoms (O or N) per [8Fe:8S] unit. The combination of these observations supports a NifW-instigated conformational change in the NifD2K2 at the NifD/NifK subunit interface to assist in cubane fusion. A conformational role for NifW in optimizing a configuration of NifD and NifK to promote efficient P-cluster maturation is consistent with the observation that loss of NifW function only results in the lower accumulation of active NifD2K2 that displays the characteristic EPR signature associated with FeMo-co.22 The possible participation of assembly factors such as NafH and NifW in promoting subunit interaction has previously been indicated by studies that have demonstrated both NifD and NifK can accumulate in the absence of each other, and that crude extracts harbouring separately produced NifD or NifK can be mixed to achieve only a very low level of activity.58 Nevertheless, NifD- and NifK-subunits produced in the absence of each other have yet to be purified and characterized. In our current working model, NifW conformationally assists the fusion of the two subclusters in a pathway that also involves NifZ, NifH2 and MgATP. In this provisional model, NifZ is proposed to assist dissociation of NifW from the NifD2K2W2 complex and, possibly recruit NifH2 to support sub-cluster fusion. In support of this model an interaction between NifW and NifZ, based on yeast-two-hybrid studies has been suggested.59
An analogous example of [4Fe:4S] cluster fusion following slight structural changes has also been recently reported for formation of a stable [8Fe:8S] precursor of FeMo-co, the K-cluster.11 Strikingly, the ligand environment of the K-cluster appears identical to that of the P-cluster, underlining a structural convergence for [4Fe:4S] cluster fusion. Formation of P-clusters directly within an immature form of NifD2K2 represents a fundamentally different pathway than completion of FeMo-co formation prior to its insertion into an apo-form of NifD2K2 that already contains intact P-clusters. Nevertheless, P-clusters and FeMo-co share striking topological similarities and a common mechanistic feature in their respective assemblies involving reductant-dependent fusion of sub-clusters. Further studies are required to fully elucidate the conformational changes which occur upon binding of NifW to NifD2K2 – namely, structural resolution via crystallography could determine the structural changes occurring around the clusters.
Data availability
Additional data and parameters regarding spectral fitting are available in the ESI file.† Further data may be furnished upon request.
Author contributions
C. V. S. and E. J. V. contributed equally to this work. C. V. S., E. J.-V., L. D. and D. R. D. conceived and supervised the project. C. V. S., E. J.-V., A. P.-G. and Z.-Y. Y. acquired and analysed the data. C. V. S. and L. D. wrote the draft. All authors participated in manuscript preparation and draft revision before the final submission.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
C. V. S., S. D., and L. D. would like to thank the Max-Planck Society for funding. S. D. and C. V. S. acknowledge the DFG SPP 1927 “Iron–Sulfur for Life” (project DE 1877/1-1) for funding. L. D. thanks the Peter und Traudl Engelhorn Stiftung for funding. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. C. V. S., S. D., and L. D. gratefully acknowledge Matthew Latimer for his technical assistance during XAS measurements at beamline 9-3. George E. Cutsail III, Justin H. Henthorn, and Patricia Rodríguez Macía are also thanked for their assistance in XAS data collection. E. J.-V. and A. P. G. were supported by Bill and Melinda Gates Foundation grants BNF Cereals Phase II (OPP1143172) and BNF Cereals Phase III (INV-005889). Work in the laboratories of L. C. S and D. R. D. is supported by grants from the U.S. Department of Energy, Office of Science, Basic Energy Science, DE-SC0010834 and DE-SC0010867, respectively. This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation (OPP1143172, INV-005889). Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission.
Electronic supplementary information (ESI) available: Experimental procedures, supplementary figures and tables. See DOI: 10.1039/d1sc06418e
Footnotes
NifD2K2 isolated from a particular genetic background in which gene-encoding components involved in maturation have been deleted are indicated by a superscript. For example, NifD2K2 isolated from a strain deleted for the gene encoding NifH is indicated as ΔHNifD2K2.
References
- Seefeldt L. C. Yang Z. Y. Lukoyanov D. A. Harris D. F. Dean D. R. Raugei S. Hoffman B. M. Reduction of Substrates by Nitrogenases. Chem. Rev. 2020;120(12):5082. doi: 10.1021/acs.chemrev.9b00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Stappen C. Decamps L. Cutsail 3rd G. E. Bjornsson R. Henthorn J. T. Birrell J. A. DeBeer S. The Spectroscopy of Nitrogenases. Chem. Rev. 2020;120(12):5005. doi: 10.1021/acs.chemrev.9b00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einsle O. Rees D. C. Structural Enzymology of Nitrogenase Enzymes. Chem. Rev. 2020;120(12):4969. doi: 10.1021/acs.chemrev.0c00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo M. A. Dos Santos P. C. Distribution of Nitrogen-Fixation Genes in Prokaryotes Containing Alternative Nitrogenases. ChemBioChem. 2020;21(12):1749. doi: 10.1002/cbic.202000022. [DOI] [PubMed] [Google Scholar]
- Peters J. W. Fisher K. Newton W. E. Dean D. R. Involvement of the P-Cluster in Intramolecular Electron-Transfer within the Nitrogenase MoFe Protein. J. Biol. Chem. 1995;270(45):27007. doi: 10.1074/jbc.270.45.27007. [DOI] [PubMed] [Google Scholar]
- Chan J. M. Christiansen J. Dean D. R. Seefeldt L. C. Spectroscopic evidence for changes in the redox state of the nitrogenase P-cluster during turnover. Biochemistry. 1999;38(18):5779. doi: 10.1021/bi982866b. [DOI] [PubMed] [Google Scholar]
- Xiang N. Guo C. Y. Liu J. W. Xu H. Dixon R. Yang J. G. Wang Y. P. Using synthetic biology to overcome barriers to stable expression of nitrogenase in eukaryotic organelles. Proc. Natl. Acad. Sci. U. S. A. 2020;117(28):16537. doi: 10.1073/pnas.2002307117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S. Gregg C. M. Allen R. S. Menon A. Hussain D. Gillespie V. Johnston E. Byrne K. Colgrave M. L. Wood C. C. A Synthetic Biology Workflow Reveals Variation in Processing and Solubility of Nitrogenase Proteins Targeted to Plant Mitochondria, and Differing Tolerance of Targeting Sequences in a Bacterial Nitrogenase Assay. Front. Plant Sci. 2020;11:552160. doi: 10.3389/fpls.2020.552160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buren S. Jimenez-Vicente E. Echavarri-Erasun C. Rubio L. M. Biosynthesis of Nitrogenase Cofactors. Chem. Rev. 2020;120(12):4921. doi: 10.1021/acs.chemrev.9b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Torrejon G. Buren S. Veldhuizen M. Rubio L. M. Biosynthesis of cofactor-activatable iron-only nitrogenase in Saccharomyces cerevisiae. Microb. Biotechnol. 2021;14(3):1073. doi: 10.1111/1751-7915.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L. P. Cherrier M. V. Amara P. Rubio L. M. Nicolet Y. An unexpected P-cluster like intermediate en route to the nitrogenase FeMo-co. Chem. Sci. 2021;12:5269–5274. doi: 10.1039/D1SC00289A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J. Goodwin P. J. Lanzilotta W. N. Seefeldt L. C. Dean D. R. Catalytic and biophysical properties of a nitrogenase Apo-MoFe protein produced by a nifB-deletion mutant of Azotobacter vinelandii. Biochemistry. 1998;37(36):12611. doi: 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
- Jimenez-Vicente E. Yang Z. Y. Ray W. K. Echavarri-Erasun C. Cash V. L. Rubio L. M. Seefeldt L. C. Dean D. R. Sequential and differential interaction of assembly factors during nitrogenase MoFe protein maturation. J. Biol. Chem. 2018;293(25):9812. doi: 10.1074/jbc.RA118.002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vicente E. Yang Z. Y. Martin Del Campo J. S. Cash V. L. Seefeldt L. C. Dean D. R. The NifZ accessory protein has an equivalent function in maturation of both nitrogenase MoFe protein P-clusters. J. Biol. Chem. 2019;294(16):6204. doi: 10.1074/jbc.RA119.007905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. W. Stowell M. H. Soltis S. M. Finnegan M. G. Johnson M. K. Rees D. C. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry. 1997;36(6):1181. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- Ribbe M. W. Hu Y. Guo M. Schmid B. Burgess B. K. The FeMoco-deficient MoFe protein produced by a nifH deletion strain of Azotobacter vinelandii shows unusual P-cluster features. J. Biol. Chem. 2002;277(26):23469. doi: 10.1074/jbc.M202061200. [DOI] [PubMed] [Google Scholar]
- Corbett M. C. Hu Y. Naderi F. Ribbe M. W. Hedman B. Hodgson K. O. Comparison of iron–molybdenum cofactor-deficient nitrogenase MoFe proteins by X-ray absorption spectroscopy: implications for P-cluster biosynthesis. J. Biol. Chem. 2004;279(27):28276. doi: 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
- Hu Y. Fay A. W. Lee C. C. Ribbe M. W. P-cluster maturation on nitrogenase MoFe protein. Proc. Natl. Acad. Sci. U. S. A. 2007;104(25):10424. doi: 10.1073/pnas.0704297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C. Dean D. R. Burgess B. K. Iron–molybdenum cofactor biosynthesis in Azotobacter vinelandii requires the iron protein of nitrogenase. J. Biol. Chem. 1987;262(29):14327. doi: 10.1016/S0021-9258(18)47941-0. [DOI] [PubMed] [Google Scholar]
- Robinson A. C. Chun T. W. Li J. G. Burgess B. K. Iron–Molybdenum Cofactor Insertion into the Apo-MoFe Protein of Nitrogenase Involves the Iron Protein-MgATP Complex. J. Biol. Chem. 1989;264(17):10088. doi: 10.1016/S0021-9258(18)81770-7. [DOI] [PubMed] [Google Scholar]
- Jacobson M. R. Cash V. L. Weiss M. C. Laird N. F. Newton W. E. Dean D. R. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Gen. Genet. 1989;219(1–2):49. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- Kim S. Burgess B. K. Evidence for the direct interaction of the nifW gene product with the MoFe protein. J. Biol. Chem. 1996;271(16):9764. doi: 10.1074/jbc.271.16.9764. [DOI] [PubMed] [Google Scholar]
- Gavini N. Ma L. Watt G. Burgess B. K. Purification and characterization of a FeMo cofactor-deficient MoFe protein. Biochemistry. 1994;33(39):11842. doi: 10.1021/bi00205a021. [DOI] [PubMed] [Google Scholar]
- Van Stappen C. Thorhallsson A. T. Decamps L. Bjornsson R. DeBeer S. Resolving the structure of the E1 state of Mo nitrogenase through Mo and Fe K-edge EXAFS and QM/MM calculations. Chem. Sci. 2019;10(42):9807. doi: 10.1039/C9SC02187F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrave K. B. Liu H. I. Ma L. Burgess B. K. Watt G. Hedman B. Hodgson K. O. EXAFS studies on the PN and POX states of the P-clusters in nitrogenase. JBIC, J. Biol. Inorg. Chem. 1998;3(4):344. doi: 10.1007/s007750050243. [DOI] [Google Scholar]
- Musgrave K. B. Angove H. C. Burgess B. K. Hedman B. Hodgson K. O. All-ferrous titanium(iii) citrate reduced Fe protein of nitrogenase: An XAS study of electronic and metrical structure. J. Am. Chem. Soc. 1998;120(21):5325. doi: 10.1021/ja980598z. [DOI] [Google Scholar]
- Kowalska J. K. Hahn A. W. Albers A. Schiewer C. E. Bjornsson R. Lima F. A. Meyer F. DeBeer S. X-ray Absorption and Emission Spectroscopic Studies of [L2Fe2S2](n) Model Complexes: Implications for the Experimental Evaluation of Redox States in Iron–Sulfur Clusters. Inorg. Chem. 2016;55(9):4485. doi: 10.1021/acs.inorgchem.6b00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman G. R. Yafet Y. Eisenberger P. Blumberg W. E. Observations and interpretation of X-ray absorption edges in iron compounds and proteins. Proc. Natl. Acad. Sci. U. S. A. 1976;73(5):1384. doi: 10.1073/pnas.73.5.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson H. P. Knight J. R. X-Ray Absorption Edges of Transition Metal Salts. Phys. Rev. 1956;102(3):632. doi: 10.1103/PhysRev.102.632. [DOI] [Google Scholar]
- Pierik A. J. Wassink H. Haaker H. Hagen W. R. Redox properties and EPR spectroscopy of the P clusters of Azotobacter vinelandii MoFe protein. Eur. J. Biochem. 1993;212(1):51. doi: 10.1111/j.1432-1033.1993.tb17632.x. [DOI] [PubMed] [Google Scholar]
- Christiansen J. Tittsworth R. C. Hales B. J. Cramer S. P. Fe and Mo EXAFS of Azotobacter vinelandii Nitrogenase in Partially Oxidized and Singly Reduced Forms. J. Am. Chem. Soc. 1995;117(40):10017. doi: 10.1021/ja00145a012. [DOI] [Google Scholar]
- Rutledge H. L. Rittle J. Williamson L. M. Xu W. A. Gagnon D. M. Tezcan F. A. Redox-Dependent Metastability of the Nitrogenase P-Cluster. J. Am. Chem. Soc. 2019;141(25):10091. doi: 10.1021/jacs.9b04555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatzal T. Aksoyoglu M. Zhang L. Andrade S. L. Schleicher E. Weber S. Rees D. C. Einsle O. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science. 2011;334(6058):940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Hodgson K. O. Holm R. H. Crystal-Structure of [(C2H5)4n]3[Fe4S4(Sch2ph)4], a Reduced Ferredoxin Site Analog with a Nontetragonal Fe4S4 Core Structure in the Solid-State. J. Am. Chem. Soc. 1979;101(16):4586. doi: 10.1021/ja00510a025. [DOI] [Google Scholar]
- Sharp C. R. Duncan J. S. Lee S. C. [Fe(4)S(4)](q) cubane clusters (q = 4+, 3+, 2+) with terminal amide ligands. Inorg. Chem. 2010;49(14):6697. doi: 10.1021/ic100742c. [DOI] [PubMed] [Google Scholar]
- Ohki Y. Tanifuji K. Yamada N. Imada M. Tajima T. Tatsumi K. Synthetic analogues of [Fe4S4(Cys)3(His)] in hydrogenases and [Fe4S4(Cys)4] in HiPIP derived from all-ferric [Fe4S4{N(SiMe3)2}4] Proc. Natl. Acad. Sci. U. S. A. 2011;108(31):12635. doi: 10.1073/pnas.1106472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moula G. Matsumoto T. Miehlich M. E. Meyer K. Tatsumi K. Synthesis of an All-Ferric Cuboidal Iron–Sulfur Cluster [Fe(iii)4S4(SAr)4] Angew. Chem., Int. Ed. Engl. 2018;57(36):11594. doi: 10.1002/anie.201803679. [DOI] [PubMed] [Google Scholar]
- Wenke B. B. Spatzal T. Rees D. C. Site-Specific Oxidation State Assignments of the Iron Atoms in the [4Fe:4S](2+/1+/0) States of the Nitrogenase Fe–Protein. Angew. Chem., Int. Ed. Engl. 2019;58(12):3894. doi: 10.1002/anie.201813966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P. A. Day E. P. Kent T. A. Orme-Johnson W. H. Munck E. M. EPR, and magnetization studies of the Azotobacter vinelandii Fe protein. Evidence for a [4Fe–4S]1+ cluster with spin S = 3/2. J. Biol. Chem. 1985;260(20):11160. doi: 10.1016/S0021-9258(17)39160-3. [DOI] [PubMed] [Google Scholar]
- Carney M. J. Papaefthymiou G. C. Spartalian K. Frankel R. B. Holm R. H. Ground spin state variability in [Fe4S4(SR)4]3−. Synthetic analogs of the reduced clusters in ferredoxins and other iron–sulfur proteins: cases of extreme sensitivity of electronic state and structure to extrinsic factors. J. Am. Chem. Soc. 1988;110(18):6084. doi: 10.1021/ja00226a025. [DOI] [PubMed] [Google Scholar]
- Flint D. H. Emptage M. H. Guest J. R. Fumarase-a from Escherichia coli – Purification and Characterization as an Iron–Sulfur Cluster Containing Enzyme. Biochemistry. 1992;31(42):10331. doi: 10.1021/bi00157a022. [DOI] [PubMed] [Google Scholar]
- Duin E. C. Lafferty M. E. Crouse B. R. Allen R. M. Sanyal I. Flint D. H. Johnson M. K. [2Fe–2S] to [4Fe–4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry. 1997;36(39):11811. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- Hagen W. R. Eady R. R. Dunham W. R. Haaker H. A novel S = 3/2 EPR signal associated with native Fe-proteins of nitrogenase. FEBS Lett. 1985;189(2):250. doi: 10.1016/0014-5793(85)81033-4. [DOI] [PubMed] [Google Scholar]
- Hans M. Buckel W. Bill E. The iron–sulfur clusters in 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Biochemical and spectroscopic investigations. Eur. J. Biochem. 2000;267(24):7082. doi: 10.1046/j.1432-1327.2000.01809.x. [DOI] [PubMed] [Google Scholar]
- Rupp H. Cammack R. Rao K. K. Hall D. O. Electron-Spin Relaxation of Iron–Sulfur Proteins Studied by Microwave-Power Saturation. Biochim. Biophys. Acta. 1978;537(2):255. doi: 10.1016/0005-2795(78)90509-3. [DOI] [PubMed] [Google Scholar]
- Aasa R. Vanngard T. Epr Signal Intensity and Powder Shapes – Re-Examination. J. Magn. Reson. 1975;19(3):308. [Google Scholar]
- Riplinger C. Kao J. P. Rosen G. M. Kathirvelu V. Eaton G. R. Eaton S. S. Kutateladze A. Neese F. Interaction of radical pairs through-bond and through-space: scope and limitations of the point-dipole approximation in electron paramagnetic resonance spectroscopy. J. Am. Chem. Soc. 2009;131(29):10092. doi: 10.1021/ja901150j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surerus K. K. Hendrich M. P. Christie P. D. Rottgardt D. Ormejohnson W. H. Munck E. Mossbauer and Integer-Spin Epr of the Oxidized P-Clusters of Nitrogenase – Pox Is a Non-Kramers System with a Nearly Degenerate Ground Doublet. J. Am. Chem. Soc. 1992;114(22):8579. doi: 10.1021/ja00048a034. [DOI] [Google Scholar]
- Owens C. P. Katz F. E. Carter C. H. Oswald V. F. Tezcan F. A. Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer. J. Am. Chem. Soc. 2016;138(32):10124. doi: 10.1021/jacs.6b06783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen W. R. Dunham W. R. Johnson M. K. Fee J. A. Quarter field resonance and integer-spin/half-spin interaction in the EPR of Thermus thermophilus ferrodoxin. Possible new fingerprints for three iron clusters. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1985;828(3):369. doi: 10.1016/0167-4838(85)90318-8. [DOI] [PubMed] [Google Scholar]
- Hagen W. R. Pierik A. J. Veeger C. Novel electron paramagnetic resonance signals from an Fe/S protein containing six iron atoms. J. Chem. Soc., Faraday Trans. 1. 1989;85(12):4083. doi: 10.1039/F19898504083. [DOI] [Google Scholar]
- Pierik A. J. Hagen W. R. Dunham W. R. Sands R. H. Multi-frequency EPR and high-resolution Mossbauer spectroscopy of a putative [6Fe–6S] prismane-cluster-containing protein from Desulfovibrio vulgaris (Hildenborough). Characterization of a supercluster and superspin model protein. Eur. J. Biochem. 1992;206(3):705. doi: 10.1111/j.1432-1033.1992.tb16977.x. [DOI] [PubMed] [Google Scholar]
- Hagen W. R. Structure and function of the hybrid cluster protein. Coord. Chem. Rev. 2022;457:214405. doi: 10.1016/j.ccr.2021.214405. [DOI] [Google Scholar]
- Broach R. B. Rupnik K. Hu Y. L. Fay A. W. Cotton M. Ribbe M. W. Hales B. J. Variable-temperature, variable-field magnetic circular dichroism spectroscopic study of the metal clusters in the Delta nifB and Delta nifH MoFe proteins of nitrogenase from Azotobacter vinelandii. Biochemistry. 2006;45(50):15039. doi: 10.1021/bi061697p. [DOI] [PubMed] [Google Scholar]
- Roessler M. M. Evans R. M. Davies R. A. Harmer J. Armstrong F. A. EPR spectroscopic studies of the Fe–S clusters in the O2-tolerant [NiFe]-hydrogenase Hyd-1 from Escherichia coli and characterization of the unique [4Fe–3S] cluster by HYSCORE. J. Am. Chem. Soc. 2012;134(37):15581. doi: 10.1021/ja307117y. [DOI] [PubMed] [Google Scholar]
- Hamilton T. L. Jacobson M. Ludwig M. Boyd E. S. Bryant D. A. Dean D. R. Peters J. W. Differential accumulation of nif structural gene mRNA in Azotobacter vinelandii. J. Bacteriol. 2011;193(17):4534. doi: 10.1128/JB.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poza-Carrion C. Jimenez-Vicente E. Navarro-Rodriguez M. Echavarri-Erasun C. Rubio L. M. Kinetics of Nif gene expression in a nitrogen-fixing bacterium. J. Bacteriol. 2014;196(3):595. doi: 10.1128/JB.00942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C. Burgess B. K. Dean D. R. Activity, Reconstitution, and Accumulation of Nitrogenase Components in Azotobacter vinelandii Mutant Strains Containing Defined Deletions within the Nitrogenase Structural Gene-Cluster. J. Bacteriol. 1986;166(1):180. doi: 10.1128/jb.166.1.180-186.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. Pulakat L. Parker K. C. Gavini N. Genetic analysis on the NifW by utilizing the yeast two-hybrid system revealed that the NifW of Azotobacter vinelandii interacts with the NifZ to form higher-order complexes. Biochem. Biophys. Res. Commun. 1998;244:498. doi: 10.1006/bbrc.1998.8119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional data and parameters regarding spectral fitting are available in the ESI file.† Further data may be furnished upon request.