Abstract
During eukaryotic transcription elongation, RNA polymerase II (RNAP2) is regulated by a chorus of factors. Here we identified a common binary interaction module consisting of TFIIS N-terminal domains (TNDs) and natively unstructured TND-interacting motifs (TIMs). This module was conserved among the elongation machinery and linked complexes including TFIIS, Mediator, super elongation complex, elongin, IWS1, SPT6, PP1/PNUTS phosphatase, H3K36me3 readers, and other factors. Using NMR, live-cell microscopy, and mass spectrometry, we revealed the structural basis for these interactions and found that TND-TIM sequences were necessary and sufficient to induce strong and specific co-localization in the crowded nuclear environment. Disruption of a single TIM in IWS1 induced robust changes in gene expression and RNAP2 elongation dynamics, underscoring the functional importance of TND-TIM surfaces for transcription elongation.
Keywords: LEDGF, PSIP1, HRP2, HDGFL2, HDGFRP2, PRO-seq, pausing, release
At the majority of eukaryotic genes, RNA polymerase II (RNAP2) undergoes regulated transcriptional pausing during early elongation, followed by controlled release into productive RNA synthesis (1–3). Disruption of transcription elongation is implicated in cancer (4, 5), neurodegeneration (6), HIV latency (7), deregulation of immune response (8), UV-hypersensitivity disorders (9), and developmental defects (10, 11).
To ensure accurate and efficient transcription elongation, RNAP2 activity is modulated by highly conserved regulators. A number of kinases and phosphatases including P-TEFb/CDK9 (12), CDK7 (13) and PP1/PNUTS (14) regulate phosphorylation patterns on the RNAP2 C-terminal domain (CTD) and therefore control the RNAP2 interactome. TFIIS and elongin promote recovery from transcriptional pauses by rescuing backtracked polymerases using distinct mechanisms (15, 16). Histone chaperones like FACT (17), SPT6 (18) or LEDGF/HRP2 (19) control the structure of nucleosomal DNA during elongation to facilitate progression of transcription elongation. Additionally, super elongation complex (20), P-TEFb (12), PARPs (21), Mediator (22), and topoisomerases (23) are required for efficient transcription elongation.
Protein sequences lacking stable structure are common among eukaryotic transcription regulators (24). While many low-complexity repeat sequences contribute to phase separation (25), the most prevalent unstructured modules mediating specific interactions are relatively small (<15 amino acids) short linear motifs (SLiMs) (24, 26). SLiMs are natively unstructured but adopt defined structures upon docking at their cognate binding sites (24, 26) and often maintain interactions that are subject to regulation by post-translational modification (26, 27). Unlike phase-separated condensates governed by multivalent interaction sites (25, 28), SLiMs display highly selective binding to individual target sites (24, 27). Despite the critical functions disordered sequences like SLiMs play in cells, regulatory networks sustained by such motifs are currently part of the “dark matter” of cell biology.
Our work here revealed that prominent components of the eukaryotic transcription elongation machinery are linked through a network of binary interactions involving a family of unstructured interaction motifs and TFIIS N-terminal domains (TNDs).
Results
TFIIS N-terminal domains (TNDs) are conserved and enriched among transcription elongation factors
We performed domain enrichment analysis on proteins involved in RNAP2 transcription regulatory machinery and found that several structural folds are selectively enriched in transcription initiation, elongation and termination (Fig. 1A, Data S1 and Table S1). For transcription elongation, the most significantly overrepresented domain was the TFIIS N-terminal domain (TND, odds ratio OR≈123, Padj=3.73e-6, hypergeometric test, Fig. 1B). TND-containing proteins are specifically associated with regulation of gene expression, as opposed to other nuclear processes (e.g. DNA damage repair, cell cycle regulation or chromatin organization and others), and constrained to transcription elongation by RNAP2 rather than other polymerases (Fig. 1C and Data S1).
Fig. 1: TFIIS N-terminal domain (TND) is a conserved scaffold enriched among transcription elongation factors.

(A) Magnitude of enrichment of structural folds among transcription initiation, elongation and termination factors (see also Data S1); *** P < 0.001. (B) Domains enriched among transcription elongation factors are ranked based on significance (adjusted P-values). Top hits are highlighted and labeled. (C) Enrichment of TND modules in different nuclear processes, gene expression, and processes involving different RNA polymerases (RNAP). (D) Interaction network of TND-containing proteins with proteins known to participate in transcription elongation from RNAP2 promoters (defined by GO:0006368). Edges represent experimental evidence-based interaction data from the STRING database. Interactions involving TND-containing proteins are highlighted in pink and TND-containing proteins are highlighted in bold. Full labels are provided in Fig. S1. (E) Solution structures of different TNDs determined by NMR spectroscopy. (F) Root-mean-square deviation (RMSD) and sequence identity between each TND pair. (G) Structural multiple sequence alignment generated by PDBeFold. Shading indicates the degree of conservation and fold stabilizing residues are marked by a triangle. The mean fractional solvent accessible surface area (SASA) is also indicated for each position.
Many proteins that contain a TND have documented roles in transcription elongation (Table S2). We plotted experimentally validated interactions between these factors from the STRING database (Fig. 1D and Fig. S1) and found that TND-containing factors operate within a highly interconnected network of elongation complexes that includes RNAP2, the super elongation complex, P-TEFb, FACT, Mediator, TFIIS, elongin, TFIIH, PAF1, NELF, and DSIF complexes. Our analysis does not indicate that TNDs mediate all depicted interactions, however, our results reveal the ubiquitous presence of TNDs across transcription elongation regulators.
We next determined representative NMR solution structures for TNDs from human TFIIS (TCEA1), elongin A (ELOA), the H3K36me3 readers LEDGF (PSIP1) and HRP2 (HDGFRP or HDGFL2), Mediator subunit 26 (MED26), the PP1/PNUTS regulatory subunit 10 (PPP1R10), and IWS1 (Fig. 1E, Fig. S2 and Table S3). The solution structure of each TND showed a compact right-handed bundle of five α-helices, which is characteristic of the TND family (29). Although the sequence identity between some TNDs is less than 10%, the fold is conserved, as the RMSD of the mean Cα coordinates for all pairwise combination of TNDs is <3Å (Fig. 1F). Hydrophobic residues in the helices show the strongest conservation and are buried in the domain core (Fig. 1G). In contrast, most poorly conserved side chains are solvent-exposed and may confer interaction specificity among TNDs.
TNDs mediate selective binding to conserved TIMs in major transcription regulators
While we showed that the TND is a recurrent feature of the elongation machinery, little is known about the interactome of this domain. Structural comparison of the human LEDGF TND in complex with binding partners (27, 30, 31) revealed a striking similarity with Iws1–Spt6 complexes from distantly related species (32, 33) (Fig. S3A–B). The structural conservation of these interfaces led us to hypothesize that TND-containing proteins throughout the elongation machinery may bind their partners through unstructured motifs.
We used features shared between the motifs known to bind human LEDGF and IWS1 homologues from other species as a bootstrap to uncover related motifs in humans that bind TNDs, which we refer to as “TND-interacting motifs” (TIMs, Fig. S3). Our proteome-wide search revealed several putative TIMs in different transcriptional regulators (Fig. 2A, Fig. S3, S4 and Data S2). In particular, we discovered three TIMs in IWS1, one of which was a motif known to bind LEDGF/HRP2 (27, 30), as well as TIMs in MED13/MED13L, ELL associated factors 1 and 2 (EAF1–2), and two members of the PAF1 complex: PAF1 and LEO1. Moreover, we identified two conserved TIMs in the N-terminus of SPT6 (SUPT6H, Fig. 2A and Fig. S5). These TIMs are conserved among metazoans and in several cases, the conservation extends to yeast as well. Plotting interactions between these factors based on the STRING database revealed multiple experimentally validated yet uncharacterized interactions between TND- and TIM-containing proteins (Fig. 2B).
Fig. 2: TNDs are selectively recognized by TND-interacting motifs (TIMs) conserved in transcription regulators.

(A) Multiple sequence alignments illustrating conserved TND-recognizing TIMs. (B) Interaction network of proteins containing a TND module or TIM. Edges represent interaction data from the STRING database and are weighted by the confidence score calculated by STRING. TIM-containing proteins are highlighted in bold. (C) Comparison of 2D 15N/1H SOFAST-HMQC NMR spectra of ELOA TND alone (yellow) or with an increasing amount of MED13 TIM (gradient of blue). (D) Quantification of chemical shift perturbation (CSP) in backbone amide signals of the ELOA TND upon titration with MED13 TIM from panel (C). (E) Determination of the dissociation constant (KD) for the ELOA-TND:MED13-TIM complex formation by NMR. Error bars represent the error of the fit for the most perturbed residues (n = 10). (F) Heatmap of the mean CSP of the 10% most affected residues for each TND:TIM binary interaction. (G) Interaction network reconstructed from TND:TIM binary interactions as determined by NMR. Line darkness indicates the strength of each pairwise interaction based on mean CSP values presented in panel (F). (H) Quantification of CSP for each TND:TIM combination at a 1:4 ratio. (I) Examples of CSP values from panel (H) highlighted on the surfaces of different TNDs.
To determine the molecular basis for candidate TIM–TND interactions, we followed the changes in NMR signals of TFIIS, ELOA, LEDGF, HRP2, MED26, PPP1R10 and IWS1 TNDs in the presence of increasing amounts of each of the predicted TIMs (see Methods). The effects of this titration on NMR measurements are exemplified by ELOA TND NMR spectra upon MED13 TIM addition (Fig. 2C), and by analysis of chemical shift perturbations (CSPs) of individual peaks in these spectra (Fig. 2D). This approach allowed us to obtain quantitative measurements of TND-TIM affinities (KD) for the majority of the interactions (Fig. 2E and Fig. S6). Acidic stretches in TIM sequences contain conserved serine and threonine residues, which are amenable to phosphorylation. Measurement of the dissociation constants of selected TNDs with IWS1 phospho- and non-phosphorylated synthetic peptides revealed that phosphorylation consistently increased the affinities of TND interaction partners. Our results confirm that affinities within the TND interactome can be modulated by post-translational phosphorylation (Fig. S7).
Our NMR studies pinpointed the interaction interfaces for a number of transcription elongation regulators and allowed reconstitution of the interaction network mediated by TND-TIM interactions (Fig. 2F–G and Fig. S8). Additionally, visualization of chemical shift perturbations for each TND-TIM pairwise interaction revealed the selectivity of these interactions (Fig. 2H). While most TNDs interact with TIMs through hydrophobic contacts and electrostatic interactions with a positively charged patch on α3, α4, and α5 (Fig. S9), the HRP2 and LEDGF TNDs contain an additional interface created by interhelical loops connecting α1-α2, and α4-α5 (Fig. 2I).
TNDs and TIMs are necessary and sufficient to drive interactions in living cells
Our NMR studies indicated that IWS1 has the highest number of interactions with other TND- and TIM-containing factors of those we tested (Fig. 2G). Intriguingly, IWS1 harbors both a TND and a stretch of TIMs, and could potentially bring multiple TND- and/or TIM-containing factors together at the same time. We therefore focused on IWS1 as a model to establish the selectivity of TND-TIM interactions, as well as to study necessity and sufficiency of these interactions within living cells.
To characterize and validate TND-TIM interactions in the crowded nuclear environment of cells, we employed a fluorescent two-hybrid (F2H)-based interaction assay using U2OS 2–6-3 cells (34). The F2H approach allowed us to assess TIM-mediated co-localization of TagRFP-TNDs at fluorescent foci established by TIM-EGFP-LacI on a lacO array integrated in the U2OS 2–6-3 genome (Fig. 3A and Fig. S10). While TagRFP-tagged TFIIS, ELOA, LEDGF, HRP2 and PPP1R10 TND constructs were re-targeted to the foci with IWS1 TIMs, the IWS1 TND and the TagRFP control did not exhibit significant enrichment at these foci (Fig. 3B and Fig. S11). These results indicate that TIMs are sufficient to recruit TNDs within living cells.
Fig. 3: TND and TIM surfaces are necessary and sufficient for the association of elongation factors in living cells.
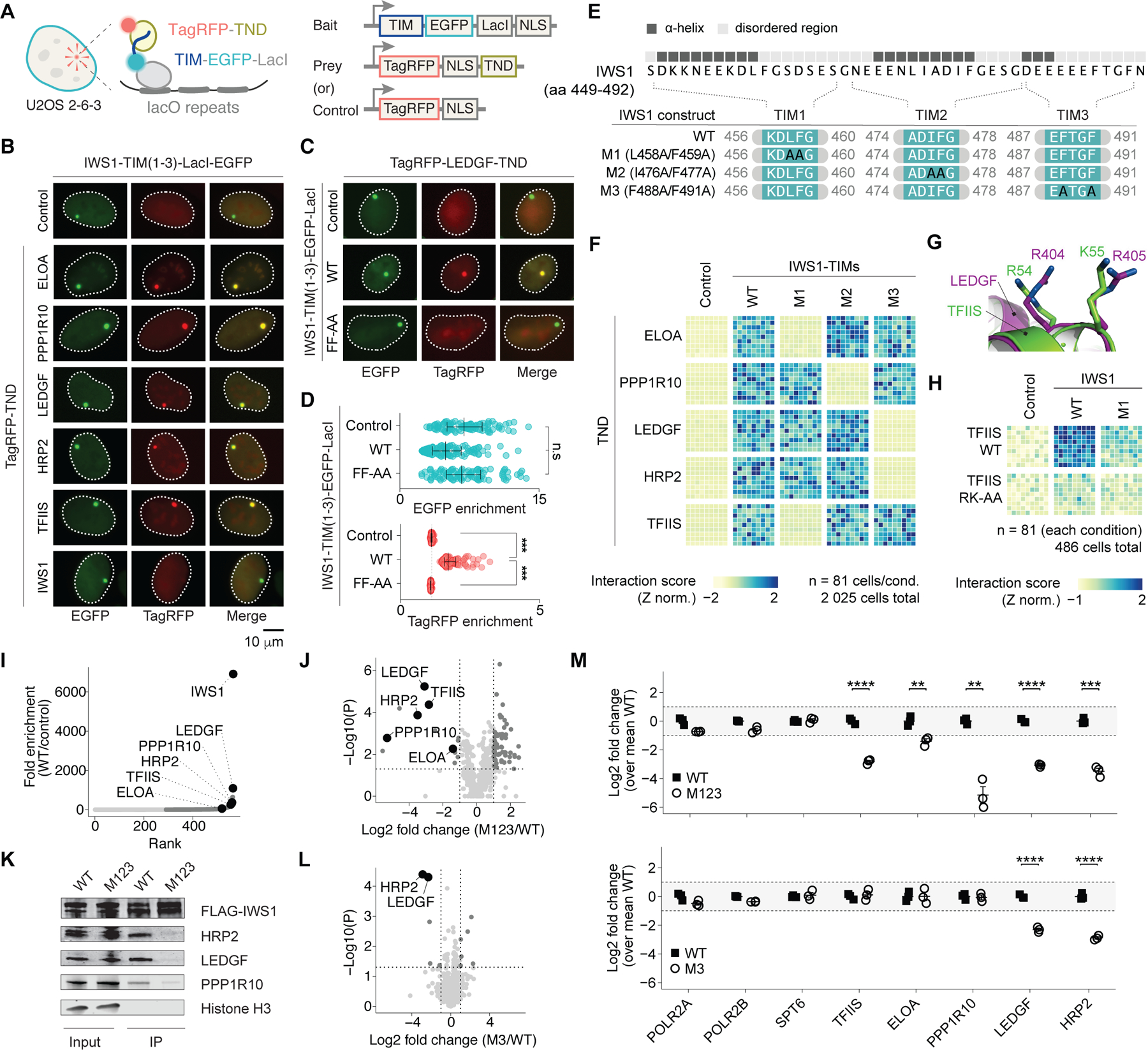
(A) Fluorescent two-hybrid (F2H)-based protein-protein interaction assay and construct design. TND-containing proteins are fused to TagRFP and TIM-containing factors to EGFP-LacI. (B) Fluorescence images of U2OS 2–6-3 cells expressing IWS1-TIM(1–3)-EGFP-LacI bait and a TagRFP-TND or control. See also Fig. S11. (C) Images of cells expressing TagRFP-LEDGF-TND and EGFP-LacI fused to WT or F488A/F491A TIM(1–3) IWS1. (D) Quantification of enrichment at fluorescent foci (n = 100) from panel (C); *** P < 0.001. (E) IWS1 mutants affecting TIMs employed in this study. Helical propensity determined by TALOS+ is indicated. (F) Single-cell interaction scores for EGFP-LacI control, WT or mutant (M1, 2 and 3) IWS1-TIM-EGFP-LacI fusion with TagRFP-TNDs (n = 81 each). See also Fig. S12 and S13. (G) LEDGF and TFIIS protein structures with residues important for interaction with TIMs labeled. (H) Single-cell interaction score for full-length WT and M1 mutant IWS1 fused to EGFP-LacI or control with TagRFP-TFIIS WT or RK-AA mutant. See also Fig. S14. (I) Enrichment of proteins in the IWS1 interactome measured by pull-down mass spectrometry. (J) Volcano plot of differentially enriched proteomic interaction partners in M123 FLAG-IWS1 compared to WT. (K) Western blot analysis of co-IP of WT and M123 FLAG-IWS1 protein variants. (L) Volcano plot of differentially enriched proteomic interaction partners in M3 FLAG-IWS1 compared to WT. (M) Fold change enrichment of selected proteins from mass spectrometry study presented in panels (J) and (L). **** P < 0.0001, *** P < 0.001, ** P < 0.01; Student’s t-test.
Fluorescence recovery after photobleaching (FRAP) revealed that binary TND-TIM interactions have a short-lived character. Specifically, FRAP half-lives for TagRFP-IWS1-TND (11.7 ± 3.6 s) and TagRFP-PPP1R10-TND (14.9 ± 9 s) were increased compared to the TagRFP control (4.5 ± 1.0 s), which reflects diffusion only (Fig. S10). Our measurements indicate that the interaction half-lives of TIM-TND interactions in cells were on the order of seconds.
We interrogated the selectivity of these interactions by introducing a structure-guided mutation of a single TIM (F488A/F491A, FF-AA) into the F2H bait containing all three IWS1 TIMs. This mutation abolished binding of TagRFP-LEDGF-TND (Fig. 3C–D), indicating that TND-TIM binary interactions depend on specific sequence-encoded molecular features of TIMs. We mutated conserved bulky hydrophobic residues in three distinct TIMs of IWS1 (Fig. 3E) and observed that mutant M1 selectively abolished the interaction with TFIIS and ELOA TND, while M2 abolished interaction with PPP1R10, and M3 abolished interaction with LEDGF and HRP2 (Fig. 3F and Fig. S12, S13). Thus, each TIM of IWS1 mediates a selective interaction with cognate TNDs.
To address whether TND-TIM interfaces are necessary for association of factors, we examined the interaction of full-length IWS1 with full-length WT and mutant TFIIS. We mutated two surface residues in the TFIIS TND (R54A, K55A; RK-AA) that are homologous to residues that abolish interactions of LEDGF TND with the MLL TIM (Fig. 3G) (27); mutation of either the TND or TIM compromised TagRFP enrichment of full-length TFIIS at the fluorescent foci (Fig. 3H, Fig. S14).
To evaluate the IWS1 interactome, we performed mass spectrometry following pull-down using FLAG-IWS1 as a bait or empty control. IWS1 binding partners enriched using this approach included the TND-containing factors LEDGF, PPP1R10, ELOA, TFIIS and HRP2, as well as subunits of RNAP2, chromatin remodeling complexes, splicing factors, and chromatin modifiers (Fig. 3I, and Fig. S15A–B). TND-containing interaction partners were selectively depleted in the combined “M123 IWS1” mutant containing disruptions to all three TIMs (Fig. 3J and Data S3, all mutations as indicated in Fig. 3E). In addition, analysis of the underlying structural folds revealed the TND as the most significantly depleted structural fold from the M123 IWS1 interactome (Fig. S15C). As orthogonal validation, we performed co-immunoprecipitation (co-IP) experiments with FLAG-tagged WT or M123 IWS1 mutant. Western blot analysis of co-IP samples confirmed depletion of TND-containing factors from the IWS1 interactome in M123 samples (Fig. 3K, additional pull-down validation in Fig. S15D). We further observed selective loss of TND-containing proteins by mutating a single TIM. Consistent with our F2H interaction data, mutation of IWS1 TIM3 (“M3”) resulted in selective depletion of LEDGF and HRP2 while the interactions of the other TND-containing proteins were preserved (Fig. 3L). Moreover, disrupting IWS1 TIMs impaired association with TND-containing proteins, but not association with RNAP2 and SPT6 (Fig. 3M), as expected for interactions mediated through other portions of the protein. Taken together, our work confirmed that TND-TIM surfaces are necessary and sufficient to mediate selective association of elongation factors in cells.
IWS1 is a central hub maintaining interactions with multiple factors through TND-TIM surfaces
We obtained solution structures of IWS1 TIMs in complex with TFIIS and PPP1R10 TNDs and compared them to our previous LEDGF-IWS1 structure (Fig. 4A) (27). While some structural features of these interfaces are conserved, each of the TNDs preferentially interacts with a different portion of IWS1 TIMs underscoring the selectivity of TND-TIM interactions (Fig. 4A and Fig. S16A–C). Our solution structures of TND-TIM modular interactions enabled coarse classification of TIMs based on recurrent sequence-encoded features (Fig. S17).
Fig. 4: IWS1 is a central hub organizing transcription regulators.

(A) Solution structures of TFIIS, PPP1R10 and LEDGF TND in complex with IWS1 TIMs. Residues contributing to the interaction with each TND are indicated, as are TIM1, 2 and 3 interfaces mutated in Fig. 3F. (B) Competition curves of TFIIS-IWS1 (left) or ELOA-IWS1 (right) with unlabelled ELOA, TFIIS or LEDGF TND domain measured via NMR. (C) Competition curves of FLAG-IWS1 bound to GST-LEDGF TND (left) or GST-HRP2 TND (right) with increasing amount of untagged HRP2 or LEDGF TND domain using AlphaScreen assay. (D) Schematic of competitive interactions of IWS1. (E) Quantification of chemical shift perturbation (CSP) in backbone amide signals of the IWS1 TIMs upon addition of unlabeled TFIIS-TND, PPP1R10-TND and LEDGF-TND individually and simultaneously measured in 3D-HNCO experiments. IWS1 residues selectively responding to addition of TFIIS-TND are highlighted in green, PPP1R10 in yellow and LEDGF in violet. (F) Sum of CSPs for IWS1 residues selectively responding to addition of TFIIS, PPP1R10 and LEDGF TNDs for conditions presented in panel (E). (G) Comparison of the interaction surface area, number of hydrogen bonds and salt bridges at the PPP1R10:TIM2, LEDGF:TIM2 and LEDGF:TIM3 interfaces. (H) Detail of the IWS1-PPP1R10-LEDGF interface. Residues important to support the interaction with each TND are presented as sticks and labeled. (I) Hybrid structure of the IWS1-SPT6-TFIIS-PPP1R10-LEDGF quinary complex.
While PPP1R10 showed a unique mechanism of IWS1 TIM recognition, the interaction profile of HRP2 is similar to LEDGF and the ELOA profile is similar to TFIIS (Fig. S16D). Competition studies confirmed binding competition between TFIIS and ELOA, and between LEDGF and HRP2 for interactions mediated by an individual TIM on IWS1 (Fig. 4B–D). Notably, the residues responsible for distinct interactions with TIMs are highly conserved between LEDGF and HRP2 as well as between ELOA and TFIIS TND (Fig. S16E), indicating that interaction specificity is encoded not only within the TIM sequence, but also by structural features of each TND.
As TFIIS, LEDGF, and PPP1R10 TND each recognize distinct IWS1 TIMs, we hypothesized that binding of these domains might not be mutually exclusive. Chemical shift perturbations of IWS1 TIM signals upon addition of each of the domains separately show a degree of overlap, however, perturbations distinct for each of the interactions can be defined (Fig. 4E–F). Measurement of IWS1 TIM spectra upon concurrent addition of all three TNDs revealed their ability to simultaneously bind to IWS1-TIMs (Fig. 4E–F).
To understand how LEDGF and PPP1R10 bind simultaneously to IWS1, we analyzed the molecular contacts with IWS1 at the interfaces of both solution structures. The LEDGF-IWS1 interaction is supported primarily through the TIM3 interface, which has higher interaction area and stabilizing contacts compared to the LEDGF-TIM2 interface (Fig. 4G). This is consistent with our mutagenesis data, as mutation of TIM3 and not TIM2 abolished the interaction between the LEDGF TND and IWS1 TIMs (Fig. 3F). We calculated a hybrid structure of the IWS1-SPT6-TFIIS-PPP1R10-LEDGF quinary complex without steric clashes, explaining the non-mutually exclusive nature of these interactions (Fig. 4H–I). Our structure showed that IWS1 is capable of inducing the co-association of all five factors based solely on TIM-TND modules and serves as a hub that maintains association of these transcription elongation regulators.
IWS1-TIM disruption alters transcription elongation dynamics of RNAP2
Because of its association with RNAP2, IWS1 represents a defining building block for elongation complex composition (35–37). We therefore hypothesized that IWS1 TIMs may contribute to regulation of transcription elongation. We focused on mutation of TIM3 (M3), which impairs the interaction of IWS1 with LEDGF and HRP2, while preserving the interactions with SPT6 and subunits of RNAP2 (Fig. 3M).
To determine the contribution of this single TIM to transcription elongation, we sought to re-express IWS1 in a cell line lacking endogenous IWS1. The deep deletion of IWS1 in KELLY neuroblastoma cells (Fig. 5A) leads to undetectable expression compared to IMR-32 neuroblastoma cells (Fig. 5B). We therefore expressed either wild-type or M3 FLAG-IWS1 in KELLY cells (Fig. 5B–C) and compared expression profiles using RNA-seq. We found 430 transcripts with decreased expression and 709 transcripts with increased expression in M3 cells (Fig. 5D, statistical criteria defined in the Methods section), as well as altered patterns of RNA splicing (Fig. S18A–B). Gene set enrichment analysis indicated that a diverse set of transcripts was affected in M3 cells, consistent with a broadly altered transcriptional landscape (Fig. S18C).
Fig. 5: Mutation of IWS1 TIMs alters elongation dynamics of RNAP2.

(A) IWS1 expression levels in different cell lines with deep deletion, amplification or diploid cells from cBioPortal. (B) Western blot analysis of IWS1 expression in IMR-32 and KELLY cells. Cell lines with endogenous expression (endo) and cells re-expressing IWS1 wild type (WT) and mutant (M3) were analyzed. (C) Construct design for IWS1 re-expression in KELLY cells. (D) Changes in mRNA levels of KELLY cells expressing IWS1 mutant M3 compared to WT IWS1 determined by RNA-seq. Selected differentially expressed transcripts are labeled. (E) Genome-wide distribution of IWS1 WT and M3 as determined by ChIP-seq. (F) Example browser tracks of IWS1 WT and M3 localization at gene bodies. (G) Correlation of RNA-seq expression changes induced by IWS1 M3 compared to WT, with IWS1 M3 ChIP-seq density. (H) Metagene plot of average IWS1 WT and M3 ChIP-seq density at gene bodies. Average ChIP-seq signal at promoter proximal region presented in inset. (I) Density of IWS1 M3 ChIP-seq signal at decreased, increased, and unaffected genes based on RNA-seq data from panel (D). (J) Metagene plot comparing difference in average PRO-seq signal between IWS1 M3 and WT at decreased, increased, and unaffected genes. The difference in PRO-seq signal near promoters is presented in inset. Solid lines are running means. (K) Example PRO-seq browser tracks at a decreased gene. Additional examples are provided in Fig. S19D–F. (L) Transcript-level changes in pausing index between WT and M3 IWS1 determined by PRO-seq.
To dissect how IWS1 TIMs influence transcription dynamics within cells, we assessed genomic localization of IWS1 with ChIP-seq, and transcription dynamics using PRO-seq (38). Analysis of anti-FLAG ChIP-seq signals uncovered that both WT and M3 IWS1 are strongly enriched at gene bodies and depleted from intergenic regions (Fig. 5E–F). The genes most strongly downregulated upon TIM3 mutation were those with high IWS1 density (Spearman R=–0.53, P<2.2e-16; Fig. 5G–I). We therefore conclude that genes with increased expression likely do not reflect direct IWS1 activity, but instead represent compensatory effects. IWS1 showed a mean profile spanning the entire gene body with peak enrichment observed ~150 bp downstream of the transcription start site (TSS, Fig. 5H). We did not detect changes in IWS1 localization or abundance between WT and M3 (Fig. 5E–F and H), indicating that TIM3 does not significantly contribute to IWS1 gene targeting.
PRO-seq transcripts were classified as either decreased (n = 412), increased (n = 675), or unchanged (n = 7,767) using the RNA-seq data and criteria described in the Methods section. Analysis of differences between PRO-seq metagene profiles revealed that compared to WT, M3 causes significant redistribution of RNAP2 density specifically at transcripts with decreased expression (Fig. 5J–K and Fig. S19). In particular, M3 caused an accumulation of RNAP2 occupancy within the promoter-proximal region of decreased genes (Fig. S19A–B), consistent with a defect in the release of paused RNAP2 into productive elongation. In agreement with this, M3 led to broadly lower RNAP2 density across the bodies of decreased genes (Mann-Whitney P<2.2e-16, Fig. S19C), and an elevated pausing index compared to transcripts with increased or unchanged expression levels (Mann-Whitney, P<2.2e-16, Fig. 5L). Together our results confirmed that disruption of a single TIM in IWS1 induces widespread elongation defects at native genomic loci, demonstrating a functional role for TND-TIM interactions during transcription.
Discussion
The organizational principles that permit diverse complexes to coordinate transcription elongation and enable efficient gene expression remain a major frontier. Here we uncovered a ubiquitous feature of transcription elongation complexes that mediates their interconnectivity (Fig. 6). In particular, we demonstrated that the TND module mediates interactions with intrinsically disordered sequences, which we refer to as “TIMs”. This binary interaction module facilitates ties between different families of transcription elongation factors and influences transcription elongation dynamics.
Fig. 6: Schematic of the TND-TIM module-driven interaction network.
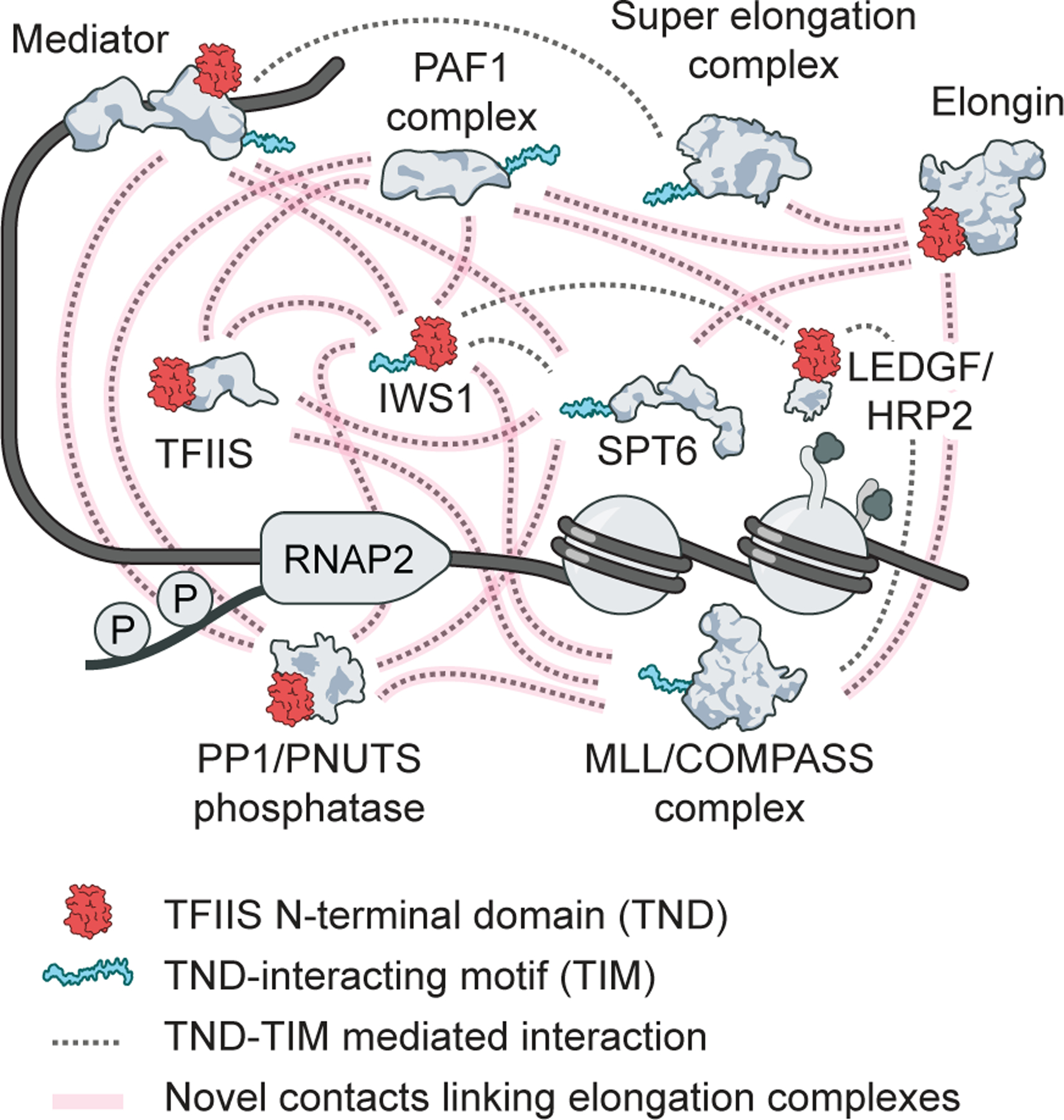
All TND-TIM interactions are represented by grey lines, previously unknown TND-TIM interaction interfaces in the elongation machinery are highlighted in pink.
Because TND-TIM surfaces are necessary and sufficient to induce colocalization (Fig. 3), it is tempting to speculate that these interactions act as a collective “molecular glue” to maintain and coordinate all members of the transcription elongation machinery at transcriptional hubs in the crowded nuclear environment. Indeed, several proteins in our network (e.g. IWS1, SPT6, and LEO1) harbor multiple TIMs. This opens the possibility that TND- and TIM-containing factors may participate in dynamic higher-order structures such as oligomers, or nucleate higher concentrations of their binding partners at biologically active sites. However, each TIM enables binding to only one TND at a time, and therefore TNDs compete for access to each TIM. Because individual TND-TIM interactions do not display the high multivalency of other unstructured sequences that drive liquid-liquid phase separation, the interactions between TNDs and TIMs are resistant to 1,6-hexanediol treatment (Fig. S20). As a result, the nature of the interaction mediated by this structural module is more akin to transient protein complexes than to membraneless organelles.
Given the largely binary character of TND-TIM interactions, TND-containing factors recruited to an individual TIM are likely to have temporally non-overlapping activities. But like the C-terminal domain of RNAP2, the modularity and sensitivity to phosphorylation of TIMs may also permit them to be employed differently in distinct epochs of the transcription cycle. Additionally, features that alter the relative affinities of TND-TIM interactions offer a potential avenue for rapid regulation. Indeed, we observe that phosphorylation of serine and threonine residues in the acidic stretch generally increases TND-TIM affinity. Such a switch may enable regulation of responses to intrinsic and extrinsic signaling cues, and would allow efficient tuning of RNAP2 pausing, processivity and transitions from distinct states during transcription elongation.
Mutation of a single IWS1 TIM (TIM3) induced extensive transcription elongation defects. Affected genes displayed promoter-proximal pile-up of RNAP2 (Fig. 5J), and exhibited a significantly increased pausing index (Fig. 5L). These results indicate that TIM3-interacting proteins contribute to pause release and the transition to productive elongation near the barrier imposed by the +1 nucleosome (39, 40). TIM3 mutation did not affect interaction of IWS1 with RNAP2, but selectively disrupted interaction with HRP2 and LEDGF (Fig. 3). Notably, genetic depletion of these two chromatin readers induces similar accumulation of paused RNAP2 and defects early in transcription elongation (19). This supports our finding that the TIM interaction platform of IWS1 coordinates the activities needed for efficient transcription elongation.
Supplementary Material
One-Sentence Summary:
The transcription elongation machinery is linked via a network of conserved unstructured motifs and TFIIS N-terminal domains.
Acknowledgments:
We would like to thank David L. Spector (Cold Spring Harbor Laboratory) for the generous gift of U2OS 2–6-3 cells, and Charles Lin (Baylor College of Medicine) for the generous gift of KELLY cells. The authors would also like to thank the Nascent Transcriptomics Core at Harvard Medical School for assistance with PRO-seq. HCH is a CPRIT Scholar. We also thank Alena Krenkova, Martin Hubalek, Pavel Talacko and Karel Harant from the IOCB and BIOCEV MS core facilities for their support in proteomics data acquisition and analysis. We thank the BCM Integrated Microscopy core facility and Nikon USA for assistance with FRAP.
Funding:
GACR grant GA19–14360S (VV)
Ministry of Education of the Czech Republic grant LO1304 (VV)
European Regional Development Fund OP RDE: Chemical Biology for Drugging Undruggable Targets (ChemBioDrug) grant CZ.02.1.01/0.0/0.0/16_019/0000729 (VV)
National Institutes of Health grant R35GM137996 (HCH)
Cancer Prevention and Research Institute of Texas grant RR170036 (HCH)
Gabrielle’s Angel Foundation Medical Research Award (HCH)
V Foundation grant V2018–003 (HCH)
Mark Foundation for Cancer Research grant 20–024-ASP (HCH)
FWO postdoctoral funding (JD)
FWO grant G099018 (ZD)
KU Leuven Research Fund (ZD)
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: KA is a consultant for Syros Pharmaceuticals, and is a stockholder and scientific advisory board member of CAMP4 Therapeutics.
Data and materials availability: The structures, NMR restraints and resonance assignments were deposited in the Protein Data Bank (PDB, accession codes: 6ZUY, 6ZUZ, 6ZV0, 6ZV1, 6ZV2, 6ZV3 and 6ZV4) and Biological Magnetic Resonance Bank (BMRB, accession codes: 34535, 34536, 34537, 34538, 34539, 34540 and 34541). All high-throughput sequencing data used in this project have been deposited in the Gene Expression Omnibus (GEO) database with SuperSeries accession number GSE179917. All analyses were performed using publicly or commercially available software. Scripts for data analysis and visualization are available at Zenodo (41–43).
References and Notes
- 1.Core LJ, Waterfall JJ, Lis JT, Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science (80-. ) (2008), doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed]
- 2.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science (80-. ) (2009), doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed]
- 3.Mayer A, Di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS, Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell (2015), doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed]
- 4.Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY, Morrow JJ, Wu Q, Mack SC, Hubert CG, Gillespie SM, Flavahan WA, Hoffmann T, Thummalapalli R, Hemann MT, Paddison PJ, Horbinski CM, Zuber J, Scacheri PC, Bernstein BE, Tesar PJ, Rich JN, Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature (2017), doi: 10.1038/nature23000. [DOI] [PMC free article] [PubMed]
- 5.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A, AFF4, a Component of the ELL/P-TEFb Elongation Complex and a Shared Subunit of MLL Chimeras, Can Link Transcription Elongation to Leukemia. Mol. Cell (2010), doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed]
- 6.Yuva-Aydemir Y, Almeida S, Krishnan G, Gendron TF, Gao FB, Transcription elongation factor AFF2/FMR2 regulates expression of expanded GGGGCC repeat-containing C9ORF72 allele in ALS/FTD. Nat. Commun (2019), doi: 10.1038/s41467-019-13477-8. [DOI] [PMC free article] [PubMed]
- 7.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK, HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med (2018), doi: 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed]
- 8.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I, Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. U. S. A (2009), doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed]
- 9.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LHF, Cockayne Syndrome A and B Proteins Differentially Regulate Recruitment of Chromatin Remodeling and Repair Factors to Stalled RNA Polymerase II In Vivo. Mol. Cell (2006), doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed]
- 10.Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald DJ, Lin S, Cantor AB, Orkin SH, Zon LI, TIF1γ Controls Erythroid Cell Fate by Regulating Transcription Elongation. Cell (2010), doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed]
- 11.Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, Rajagopalan R, Venditti CP, Gripp K, Samanich J, Zackai EH, Deardorff MA, Clark D, Allen JL, Dorsett D, Misulovin Z, Komata M, Bando M, Kaur M, Katou Y, Shirahige K, Krantz ID, Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet (2015), doi: 10.1038/ng.3229. [DOI] [PMC free article] [PubMed]
- 12.Peterlin BM, Price DH, Controlling the Elongation Phase of Transcription with P-TEFb. Mol. Cell (2006), doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed]
- 13.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL, TFIIH-Associated Cdk7 Kinase Functions in Phosphorylation of C-Terminal Domain Ser7 Residues, Promoter-Proximal Pausing, and Termination by RNA Polymerase II. Mol. Cell. Biol (2009), doi: 10.1128/mcb.00637-09. [DOI] [PMC free article] [PubMed]
- 14.Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover-Cutter K, Brannan K, Bentley DL, Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a “Sitting Duck Torpedo” Mechanism. Mol. Cell (2019), doi: 10.1016/j.molcel.2019.09.031. [DOI] [PMC free article] [PubMed]
- 15.Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C, Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature (2007), doi: 10.1038/nature05701. [DOI] [PubMed]
- 16.Aso T, Lane WS, Conaway JW, Conaway RC, Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science (80-. ) (1995), doi: 10.1126/science.7660129. [DOI] [PubMed]
- 17.Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, Zhang Z, Carragher B, Potter CS, D’Arcy S, Luger K, FACT caught in the act of manipulating the nucleosome. Nature (2020), doi: 10.1038/s41586-019-1820-0. [DOI] [PMC free article] [PubMed]
- 18.Bortvin A, Winston F, Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science (80-. ) (1996), doi: 10.1126/science.272.5267.1473. [DOI] [PubMed]
- 19.LeRoy G, Oksuz O, Descostes N, Aoi Y, Ganai RA, Kara HO, Yu JR, Lee CH, Stafford J, Shilatifard A, Reinberg D, LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Sci. Adv (2019), doi: 10.1126/sciadv.aay3068. [DOI] [PMC free article] [PubMed]
- 20.Luo Z, Lin C, Shilatifard A, The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol (2012), doi: 10.1038/nrm3417. [DOI] [PubMed]
- 21.Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, Kraus WL, Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science (80-. ) (2016), doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed]
- 22.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CAS, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, Florens L, Seidel CW, Lin C, Smith ER, Shilatifard A, Conaway RC, Conaway JW, Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell (2011), doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed]
- 23.Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, Guha R, Wilson K, Zhang X, Zhang H, Piotrowski J, Thomas CJ, Singer DS, Pugh BF, Pommier Y, Przytycka TM, Kouzine F, Lewis BA, Zhao K, Levens D, RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell (2016), doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed]
- 24.Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ, Davey NE, Short linear motifs: Ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev (2014), doi: 10.1021/cr400585q. [DOI] [PubMed]
- 25.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA, Coactivator condensation at super-enhancers links phase separation and gene control. Science (80-. ) (2018), doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed]
- 26.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ, Attributes of short linear motifs. Mol. Biosyst (2012), doi: 10.1039/c1mb05231d. [DOI] [PubMed]
- 27.Sharma S, Čermáková K, De Rijck J, Demeulemeester J, Fábry M, El Ashkar S, Van Belle S, Lepšík M, Tesina P, Duchoslav V, Novák P, Hubálek M, Srb P, Christ F, Řezáčová P, Hodges HC, Debyser Z, Veverka V, Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation. Proc. Natl. Acad. Sci (2018), doi: 10.1073/pnas.1803909115. [DOI] [PMC free article] [PubMed]
- 28.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK, Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell (2019), doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed]
- 29.Booth V, Koth CM, Edwards AM, Arrowsmith CH, Structure of a conserved domain common to the transcription factors TFIIS, Elongin A, and CRSP70. J. Biol. Chem (2000), doi: 10.1074/jbc.M002595200. [DOI] [PubMed]
- 30.Tesina P, Cermáková K, Horejší M, Procházková K, Fábry M, Sharma S, Christ F, Demeulemeester J, Debyser Z, De Rijck J, Veverka V, Rezácová P, Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat. Commun (2015), doi: 10.1038/ncomms8968. [DOI] [PubMed]
- 31.Čermaková K, Tesina P, Demeulemeester J, El Ashkar S, Méreau H, Schwaller J, Řezáčová P, Veverka V, De Rijck J, Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res (2014), doi: 10.1158/0008-5472.CAN-13-3602. [DOI] [PubMed]
- 32.Diebold ML, Koch M, Loeliger E, Cura V, Winston F, Cavarelli J, Romier C, The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J (2010), doi: 10.1038/emboj.2010.272. [DOI] [PMC free article] [PubMed]
- 33.McDonald SM, Close D, Xin H, Formosa T, Hill CP, Structure and Biological Importance of the Spn1-Spt6 Interaction, and Its Regulatory Role in Nucleosome Binding. Mol. Cell (2010), doi: 10.1016/j.molcel.2010.11.014. [DOI] [PMC free article] [PubMed]
- 34.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL, From silencing to gene expression: Real-time analysis in single cells. Cell (2004), doi: 10.1016/S0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed]
- 35.Reim NI, Chuang J, Jain D, Alver BH, Park PJ, Winston F, The conserved elongation factor Spn1 is required for normal transcription, histone modifications, and splicing in Saccharomyces cerevisiae. Nucleic Acids Res (2020), doi: 10.1093/nar/gkaa745. [DOI] [PMC free article] [PubMed]
- 36.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA, The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev (2007), doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed]
- 37.Dronamraju R, Kerschner JL, Peck SA, Hepperla AJ, Adams AT, Hughes KD, Aslam S, Yoblinski AR, Davis IJ, Mosley AL, Strahl BD, Casein Kinase II Phosphorylation of Spt6 Enforces Transcriptional Fidelity by Maintaining Spn1-Spt6 Interaction. Cell Rep (2018), doi: 10.1016/j.celrep.2018.11.089. [DOI] [PMC free article] [PubMed]
- 38.Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT, Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc (2016), doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed]
- 39.Core L, Adelman K, Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev (2019), doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed]
- 40.Weber CM, Ramachandran S, Henikoff S, Nucleosomes are context-specific, H2A.Z-Modulated barriers to RNA polymerase. Mol. Cell (2014), doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed]
- 41.Hodges HC, Code: “hodgeslab/workflows”, Version 20210915. Zenodo (2021), doi: 10.5281/zenodo.5511049. [DOI]
- 42.Adelman lab, Code: “AdelmanLab/NIH_scripts”, Version 1.0. Zenodo (2021), doi: 10.5281/zenodo.5519915. [DOI]
- 43.Adelman lab, Code: “AdelmanLab/GetGeneAnnotation_GGA”, Version 1.0. Zenodo (2021), doi: 10.5281/zenodo.5519928. [DOI]
- 44.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD, The Pfam protein families database in 2019. Nucleic Acids Res (2019), doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed]
- 45.Bateman A, Martin MJ, O’Donovan C, Magrane M, Alpi E, Antunes R, Bely B, Bingley M, Bonilla C, Britto R, Bursteinas B, Bye-AJee H, Cowley A, Da Silva A, De Giorgi M, Dogan T, Fazzini F, Castro LG, Figueira L, Garmiri P, Georghiou G, Gonzalez D, Hatton-Ellis E, Li W, Liu W, Lopez R, Luo J, Lussi Y, MacDougall A, Nightingale A, Palka B, Pichler K, Poggioli D, Pundir S, Pureza L, Qi G, Rosanoff S, Saidi R, Sawford T, Shypitsyna A, Speretta E, Turner E, Tyagi N, Volynkin V, Wardell T, Warner K, Watkins X, Zaru R, Zellner H, Xenarios I, Bougueleret L, Bridge A, Poux S, Redaschi N, Aimo L, ArgoudPuy G, Auchincloss A, Axelsen K, Bansal P, Baratin D, Blatter MC, Boeckmann B, Bolleman J, Boutet E, Breuza L, Casal-Casas C, De Castro E, Coudert E, Cuche B, Doche M, Dornevil D, Duvaud S, Estreicher A, Famiglietti L, Feuermann M, Gasteiger E, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Jungo F, Keller G, Lara V, Lemercier P, Lieberherr D, Lombardot T, Martin X, Masson P, Morgat A, Neto T, Nouspikel N, Paesano S, Pedruzzi I, Pilbout S, Pozzato M, Pruess M, Rivoire C, Roechert B, Schneider M, Sigrist C, Sonesson K, Staehli S, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey AL, Wu CH, Arighi CN, Arminski L, Chen C, Chen Y, Garavelli JS, Huang H, Laiho K, McGarvey P, Natale DA, Ross K, Vinayaka CR, Wang Q, Wang Y, Yeh LS, Zhang J, UniProt: The universal protein knowledgebase. Nucleic Acids Res (2017), doi: 10.1093/nar/gkw1099. [DOI]
- 46.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, Von Mering C, STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res (2015), doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed]
- 47.Cherepanov P, Sun ZYJ, Rahman S, Maertens G, Wagner G, Engelman A, Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol (2005), doi: 10.1038/nsmb937. [DOI] [PubMed]
- 48.Krystkowiak I, Davey NE, SLiMSearch: A framework for proteome-wide discovery and annotation of functional modules in intrinsically disordered regions. Nucleic Acids Res (2017), doi: 10.1093/nar/gkx238. [DOI] [PMC free article] [PubMed]
- 49.Davey NE, Cowan JL, Shields DC, Gibson TJ, Coldwell MJ, Edwards RJ, SLiMPrints: Conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Res (2012), doi: 10.1093/nar/gks854. [DOI] [PMC free article] [PubMed]
- 50.Mészáros B, Erdös G, Dosztányi Z, IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res (2018), doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed]
- 51.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN, PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta - Proteins Proteomics (2010), doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed]
- 52.Söding J, Biegert A, Lupas AN, The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res (2005), doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed]
- 53.Chiu J, March PE, Lee R, Tillett D, Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res (2004), doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed]
- 54.Renshaw PS, Veverka V, Kelly G, Frenkiel TA, Williamson RA, Gordon SV, Hewinson RG, Carr MD, Sequence-specific assignment and secondary structure determination of the 195-residue complex formed by the Mycobacterium tuberculosis protein CFP-10 and ESAT-6. J. Biomol. NMR (2004),, doi: 10.1023/B:JNMR.0000048852.40853.5c. [DOI] [PubMed]
- 55.Veverka V, Lennie G, Crabbe T, Bird I, Taylor RJ, Carr MD, NMR assignment of the mTOR domain responsible for rapamycin binding [3]. J. Biomol. NMR (2006),, doi: 10.1007/s10858-005-4324-1. [DOI] [PubMed]
- 56.Veverka V, Crabbe T, Bird I, Lennie G, Muskett FW, Taylor RJ, Carr MD, Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: Compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene (2008), doi: 10.1038/sj.onc.1210693. [DOI] [PubMed]
- 57.Herrmann T, Güntert P, Wüthrich K, Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol (2002), doi: 10.1016/S0022-2836(02)00241-3. [DOI] [PubMed]
- 58.Shen Y, Delaglio F, Cornilescu G, Bax A, TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR (2009), doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed]
- 59.Harjes E, Harjes S, Wohlgemuth S, Müller KH, Krieger E, Herrmann C, Bayer P, GTP-Ras Disrupts the Intramolecular Complex of C1 and RA Domains of Nore1. Structure (2006), doi: 10.1016/j.str.2006.03.008. [DOI] [PubMed]
- 60.Peruzzini R, Lens Z, Verger A, Dewitte F, Ferreira E, Baert JL, Villeret V, Landrieu I, Cantrelle FX, 1H, 15N and 13C assignments of the N-terminal domain of the Mediator complex subunit MED26. Biomol. NMR Assign (2016), doi: 10.1007/s12104-016-9673-z. [DOI] [PubMed] [Google Scholar]
- 61.Lens Z, Cantrelle FX, Peruzzini R, Hanoulle X, Dewitte F, Ferreira E, Baert JL, Monté D, Aumercier M, Villeret V, Verger A, Landrieu I, Solution Structure of the N-Terminal Domain of Mediator Subunit MED26 and Molecular Characterization of Its Interaction with EAF1 and TAF7. J. Mol. Biol (2017), doi: 10.1016/j.jmb.2017.09.001. [DOI] [PubMed]
- 62.Dominguez C, Boelens R, Bonvin AMJJ, HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc (2003), doi: 10.1021/ja026939x. [DOI] [PubMed]
- 63.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov (2012), doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed]
- 64.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal (2013), doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed]
- 65.Erde J, Loo RRO, Loo JA, Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J. Proteome Res (2014), doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed]
- 66.Langerová H, Lubyová B, Zábranský A, Hubálek M, Glendová K, Aillot L, Hodek J, Strunin D, Janovec V, Hirsch I, Weber J, Hepatitis B Core Protein Is Post-Translationally Modified through K29-Linked Ubiquitination. Cells (2020), doi: 10.3390/cells9122547. [DOI] [PMC free article] [PubMed]
- 67.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J, The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods (2016), doi: 10.1038/nmeth.3901. [DOI] [PubMed]
- 68.Hodges HC, Stanton BZ, Cermakova K, Chang C-YY, Miller EL, Kirkland JG, Ku WL, Veverka V, Zhao K, Crabtree GR, Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol 25, 61–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y, rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A (2014), doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed]
- 70.Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y, MATS: A Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res (2012), doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed]
- 71.Park JW, Tokheim C, Shen S, Xing Y, Identifying differential alternative splicing events from RNA sequencing data using RNASeq-MATS. Methods Mol. Biol (2013), doi: 10.1007/978-1-62703-514-9_10. [DOI] [PubMed]
- 72.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol (2009), doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed]
- 73.Pohl A, Beato M, bwtool: A tool for bigWig files. Bioinformatics (2014), doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed]
- 74.Reimer KA, Mimoso CA, Adelman K, Neugebauer KM, Co-transcriptional splicing regulates 3′ end cleavage during mammalian erythropoiesis. Mol. Cell (2021), doi: 10.1016/j.molcel.2020.12.018. [DOI] [PMC free article] [PubMed]
- 75.Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Grüning B, Ramírez F, Manke T, pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics (2021), doi: 10.1093/bioinformatics/btaa692. [DOI] [PMC free article] [PubMed]
- 76.Cermakova K, Courtney Hodges H, Next-generation drugs and probes for chromatin biology: From targeted protein degradation to phase separation. Molecules (2018), doi: 10.3390/molecules23081958. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


