Precision medicine is an emerging field with immense potential for better understanding of diseases and improved treatment outcomes (1). Its focus: patterns of human genetic variation in populations and individuals—and how such patterns influence disease pathology and treatment. The field rejects the “one size fits all” approach to understanding disease, aspiring to develop tailored therapies that optimize treatment efficacy. It’s a promising but fledgling field that faces numerous challenges, both scientific and practical. But one challenge has not been fully appreciated: the lack of genetic diversity in research and clinical studies (2, 3).
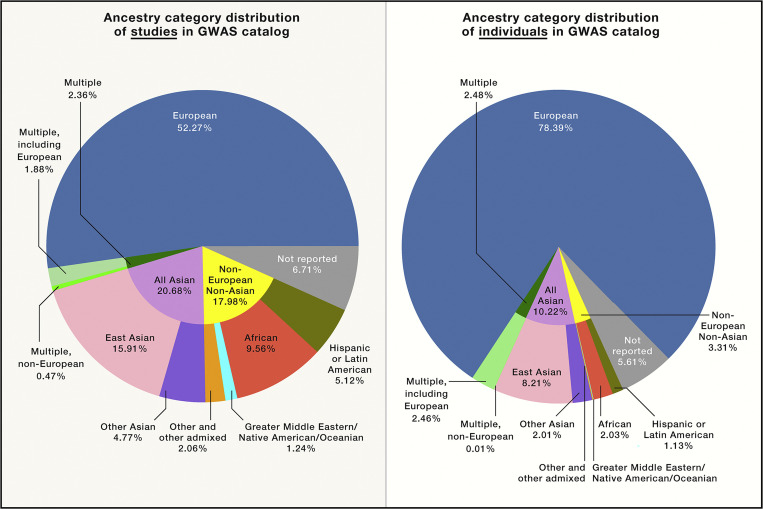
To date, most of the genetic studies that researchers draw on in the context of precision medicine have been conducted on individuals represented by European reference samples (4–9), what is commonly termed “European ancestry.” In 2009, 96% of genome-wide association studies (GWAS) participants were of European descent, with only 0.57% of samples consisting of individuals allied with reference samples from across the African continent [“African ancestry” (10, 11)], although the participation of individuals from non-European regions improved to about 20% in more recent studies, as seen in (Fig. 1) (8). In addition, European populations still receive the most geographically specific descriptions, such as Scandinavian, Finnish, or Northern European (5), whereas the generalized “African ancestry” category remains rather nonspecific and often ambiguously categorized as either “Black” or “African American” (5, 11). Moreover, groups of participating subjects are often defined based on unstandardized and inconsistent criteria, thereby limiting the usefulness of existing datasets (12, 13). However, to include more diverse populations and ancestries in genetic and clinical studies, it is imperative that such inclusion does not conflate race with biological identity.
When including more diverse populations and ancestries in genetic and clinical studies, we need to avoid conflating race with biological identity. Image credit: Shutterstock/tai11.
Race vs Ancestry
Whereas genetic ancestry is a measurable biological parameter, race is a social construct that has often been labeled biological (2, 14–16). Indeed, race is the product of historical, social, and political processes and not a “natural” or biological division of human variation—an understanding that much of the scientific community has started to embrace (13, 17, 18). Nevertheless, race remains commonly reported as a surrogate marker for genetic ancestry and diversity in biomedicine based on the assumption that a socially defined category serves sufficiently as a proxy for genetic ancestry and thus genetic diversity. The inadequacy of the race concept in genetic studies is further heightened by the fact that there is no consensus on the definitions of race categories (19–21).
Race, ethnicity, and genetic ancestry are often used interchangeably with little consensus amongst researchers and clinicians as to how such concepts should be understood and used in clinical genetics practice (21). This confusion is well exemplified by skin color, a trait often used to classify people racially. The racial categories “White, Black, Yellow, Brown, and Red” were introduced in the 18th century by the taxonomist Carl Linnaeus, who incorrectly assumed that variants in skin pigmentation and appearance reflected biological divisions of humanity (22). This categorization persists in common parlance to this day.
And yet, scientifically speaking, skin color serves exactly as an example of how genetic variation does not fall along racial lines. In geographical areas with high frequency of sunlight, such as sub-Saharan Africa, Southern India, Southeast Asia, and Australia, dark skin has served human populations as protective mechanism in areas of intense UV light exposure. Long-term existence by human populations in these areas, characteristic of our species for much of human evolutionary history, led to adaptive melanin distributions and concordant dark pigmentation. In populations that moved into areas with lower incident UV strength, such as Northern Europe and Siberia, light skin has emerged over recent human evolution as a result of the need to obtain sufficient UV radiation and facilitate the production of Vitamin D (difficult to achieve for dark skin in low UV intensity northern regions). This so-called “Vitamin D hypothesis” illustrates how variation in skin color—an externally obvious example of genetic variation in humans—is a product of natural selection in varying environments (23).
The distribution of skin color variation globally makes evident the way in which ancestral influences on pigmentation do not fall along racial lines but rather along geographical ones. Genetic studies in multiple populations across the African continent, as well as non-African populations, reveal differential contributions of genetic variants to skin pigmentation. For example, variants in the MC1R gene likely do not contribute to variation in skin pigmentation within some, but not all, African populations (24–26). On the other hand, MC1R variants contribute greatly to pigmentation variation within many, but not all, European populations (27–29).
Attempts to understand skin pigmentation variation along racial lines would lead one to believe that all Africans, which fall under the race category “Black,” exhibit the same genetic determinants of skin pigmentation. In fact, there is significant diversity in the frequency of skin pigmentation genes between populations within the African continent and across the world (25). However, to date ancestral groups from the African continent make up only 2% of the individuals in the reference genome catalog used for such studies (8), and a large number of other populations from multiple geographies across the globe are not present at all. This is problematic given the large degree of variation in skin color, as well as genetic diversity, on the African continent (30).
Fig. 1.
Distribution of ancestral categories used in genetics studies by percentage, as of 2019. European ancestry predominates, and there is a lack of sufficient representation of other ancestral groups. Reprinted from ref. 8, with permission from Elsevier.
Racial Categorization, not Genetic Ancestry
There is no genetic homogeneity in social “race” groups. However, some genetic markers of ancestry should contribute to understanding patterns associated with human health. The preferential usage of genetic ancestry markers is the key point for discarding race as a category when assessing genetic targets in precision medicine. Indeed, using race instead of actual genetic ancestry in genetic and clinical studies may result in a variety of consequences, such as genetic misdiagnosis and adverse drug reactions.
To best illustrate such unintended consequence, we offer two examples from neurology and cardiovascular medicine. Consider first epidemiological studies that have suggested individuals who self-reported as Black exhibited lower risk of Parkinson’s Disease (PD) compared with individuals who self-reported as White (31–33). More specifically, variants in MC1R that contribute to lighter pigmentation were associated with a nearly threefold greater risk of PD than those with MC1R variants that correspond to darker pigmentation (33, 34). As we noted above, although MC1R variants have differential functions across multiple geographic areas and human populations, these variants are not mapped according to social race. Hence, it is not sufficient to categorize subjects based on self-identified race or even just skin color to determine the risk of PD, despite what previous epidemiological data have suggested. Rather, it is essential to establish the presence or absence of specific genetic risk variants (e.g., in MC1R) that mediate such risk. If researchers develop potential treatments that target variants, such as those within MC1R, it would be impossible to properly stratify patients just based on their skin color or assigned race.
The diversity of reference samples, biobanks, and datasets must be expanded to better represent the high degree of genetic diversity across populations. In doing so, however, we must make a distinction between race and genetic ancestral diversity.
Similarly, therapeutic approaches that are designed based on “racial categorization” may lead to adverse drug reactions. For example, many adverse drug reactions are caused by polymorphisms in the individual’s genome that affect drug metabolism, with the frequency of such polymorphisms having been shown to vary among different populations (35). This is well illustrated by the rs2242480 polymorphism within the CYP3A4 gene. It is present at a frequency of 7% in individuals with English and Scottish ancestry but at a frequency of 84% in the Yoruba population in Ibadan, Nigeria (35). The rs2242480 variant is associated with increased metabolism of warfarin, an anticoagulant (35). Populations that exhibit greater frequencies of this variant require greater doses of warfarin, because they metabolize it faster. Thus, it is wise to avoid a “one size fits all” approach in warfarin prescription; the presence or absence of rs2242480 variant should be evaluated. And the presence of such genetic variants cannot be assumed based on just “racial” categorization, as English and Scottish ancestry do not reflect a baseline for all European populations and the Ibadan Yoruba do not in any way reflect the patterns and diversity across the entire African continent.
This pattern can be seen across the medical landscape, with similar problematic outcomes; in particular, using race in medical algorithms remains highly debated. For example, the usage of race corrections in estimated glomerular filtration rate (eGFR) equation to predict risk of end-stage kidney disease has been effectively challenged. Recent research demonstrates that race adjustments do not improve the accuracy of the eGFR test and that they may in fact disadvantage patient groups (36, 37). Corrections that mistake race for a biological category in this case may undermine the proportion of Black patients who meet the diagnostic threshold for kidney disease (37). Additionally, calibration for race as a biological unit shifts risk estimates for Black patients to a higher risk category, consequently shifting all non-Black patients to lower risk groups, which may not necessarily be accurate (36). Thus, the correction for race carries direct clinical implications and risk of misdiagnosis and is now being widely revised and reconsidered as a practice (38, 39).
Capturing Genetic Diversity
We need treatments based on actual and not assumed genetic variation. That means assessing the patterns of diversity that reflect the distribution of human genetic variation across the globe. To this end, genetic ancestry should be understood as a continuum that it is not categorized in such a way that serves as a surrogate for race (40). Contemporary usage of continental ancestry categories (e.g., European, Middle Eastern, South Asian, Oceanic, East Asian, American, and African) serves as an example of how presumed “ancestral” geographies are assumed as equivalent to biological categories and serve as a false proxy for race. Such groupings correspond to Western racial categorizations and assume genetic homogeneity based on geographical separation, but these groupings misrepresent the actual distribution of genetic variants and neglect continuous movement of people and the resulting degree of mixture across global populations.
As precision medicine moves to the forefront of biomedical science, the scientific and medical communities must acknowledge the challenges of a lack of genetic diversity in our research pools and reference samples and prioritize efforts to reflect the global distribution of human genetic variation more accurately. The diversity of reference samples, biobanks, and datasets must be expanded to better represent the high degree of genetic diversity across populations. In doing so, however, we must make a distinction between race and genetic ancestral diversity. Otherwise, we risk mistakenly attributing biological factors to a socially constructed reality. Failure to do so would impede an effective understanding of the genetics and genomics in human disease.
Footnotes
The authors declare no competing interest.
Any opinions, findings, conclusions, or recommendations expressed in this work are those of the authors and have not been endorsed by the National Academy of Sciences.
Change History
April 5, 2022: The article text has been updated.
References
- 1.Ashley E. A., Towards precision medicine. Nat. Rev. Genet. 17, 507–522 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Adigbli G., Race, science and (im)precision medicine. Nat. Med. 26, 1675–1676 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Sirugo G., Tishkoff S. A., Williams S. M., The quagmire of race, genetic ancestry, and health disparities. J. Clin. Invest. 131, e150255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustamante C. D., Burchard E. G., De la Vega F. M., Genomics for the world. Nature 475, 163–165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fullerton S. M., Yu J. H., Crouch J., Fryer-Edwards K., Burke W., Population description and its role in the interpretation of genetic association. Hum. Genet. 127, 563–572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landry L. G., Ali N., Williams D. R., Rehm H. L., Bonham V. L., Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Aff. (Millwood) 37, 780–785 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Ramos E., Callier S. L., Rotimi C. N., Why personalized medicine will fail if we stay the course. Per. Med. 9, 839–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirugo G., Williams S. M., Tishkoff S. A., The missing diversity in human genetic studies. Cell 177, 1080 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., De T., Zhong Y., Perera M. A., The advantages and challenges of diversity in pharmacogenomics: Can minority populations bring us closer to implementation? Clin. Pharmacol. Ther. 106, 338–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Need A. C., Goldstein D. B., Next generation disparities in human genomics: Concerns and remedies. Trends Genet. 25, 489–494 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Popejoy A. B., Fullerton S. M., Genomics is failing on diversity. Nature 538, 161–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonham V. L., Green E. D., Pérez-Stable E. J., Examining how race, ethnicity, and ancestry data are used in biomedical research. JAMA 320, 1533–1534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman C. W., Brett A. S., Race and pharmacogenomics-personalized medicine or misguided practice? JAMA 325, 625–626 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Benn Torres J., Anthropological perspectives on genomic data, genetic ancestry, and race. Am. J. Phys. Anthropol. 171 (suppl. 70), 74–86 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Birney E., et al., The language of race, ethnicity, and ancestry in human genetic research. arXiv [Preprint] (2021). 10.48550/arXiv.2106.10041 (Accessed 6 July 2021). [Google Scholar]
- 16.Mathieson I., Scally A., What is ancestry? PLoS Genet. 16, e1008624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Fuentes et al., AAPA statement on race and racism. Am. J. Phys. Anthropol. 169, 400–402 (2019). [DOI] [PubMed]
- 18.Graves J. L. Jr, Goodman A. H., Racism, Not Race: Answers to Frequently Asked Questions (Columbia University Press, New York, 2021). [Google Scholar]
- 19.Tatonetti N. P., Elhadad N., Fine-scale genetic ancestry as a potential new tool for precision medicine. Nat. Med. 27, 1152–1153 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Belbin G. M., et al. , Toward a fine-scale population health monitoring system. Cell 184, 2068–2083.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Popejoy A. B., et al. ; Clinical Genome Resource (ClinGen) Ancestry and Diversity Working Group, Clinical genetics lacks standard definitions and protocols for the collection and use of diversity measures. Am. J. Hum. Genet. 107, 72–82 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutherford A., How to Argue with a Racist: What Our Genes Do (and Don’t) Say about Human Difference (The Experiment, New York, 2020). [Google Scholar]
- 23.Clemens T. L., Adams J. S., Henderson S. L., Holick M. F., Increased skin pigment reduces the capacity of skin to synthesise vitamin D3. Lancet 1, 74–76 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Feng Y., McQuillan M. A., Tishkoff S. A., Evolutionary genetics of skin pigmentation in African populations. Hum. Mol. Genet. 30 (R1), R88–R97 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford N. G., et al. ; NISC Comparative Sequencing Program, Loci associated with skin pigmentation identified in African populations. Science 358, eaan8433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding R. M., et al. , Evidence for variable selective pressures at MC1R. Am. J. Hum. Genet. 66, 1351–1361 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J., et al. , A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 4, e1000074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nan H., et al. , Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 129, 2250–2257 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sulem P., et al. , Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 39, 1443–1452 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Tishkoff S. A., et al. , The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey M., Anderson S., Hall D. A., Parkinson’s disease in African Americans: A review of the current literature. J. Parkinsons Dis. 10, 831–841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao X., Simon K. C., Han J., Schwarzschild M. A., Ascherio A., Genetic determinants of hair color and Parkinson’s disease risk. Ann. Neurol. 65, 76–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao X., Simon K. C., Han J., Schwarzschild M. A., Ascherio A., Family history of melanoma and Parkinson disease risk. Neurology 73, 1286–1291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tell-Marti G., et al. , The MC1R melanoma risk variant p.R160W is associated with Parkinson disease. Ann. Neurol. 77, 889–894 (2015). [DOI] [PubMed] [Google Scholar]
- 35.De T., Park C. S., Perera M. A., Cardiovascular pharmacogenomics: Does it matter if you’re Black or White? Annu. Rev. Pharmacol. Toxicol. 59, 577–603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bundy J. D., et al. ; CRIC Study Investigators, Prediction of end-stage kidney disease using estimated glomerular filtration rate with and without race: A prospective cohort study. Ann. Intern. Med., 10.7326/M21-2928 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai J. W., et al. , Evaluating the impact and rationale of race-specific estimations of kidney function: Estimations from U.S. NHANES 2015–2018. EClinicalMedicine 42, 101197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diao J. A., et al. , Clinical implications of removing race from estimates of kidney function. JAMA 325, 184–186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts D. E., Abolish race correction. Lancet 397, 17–18 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Lewis A. C. F., et al. , Getting genetic ancestry right for science and society. arXiv [Preprint] (2021) 10.48550/arXiv.2110.05987. [DOI] [PMC free article] [PubMed]