Significance
Horizontal gene transfer (HGT)—the transfer of DNA between lineages—is responsible for a large proportion of the genetic variation that contributes to evolution in microbial populations. While HGT can bring beneficial genetic innovation, the transfer of DNA from other species or strains can also have deleterious effects. In this study, we evolve populations of the bacteria Helicobacter pylori and use DNA sequencing to identify over 40,000 genetic variants transferred by HGT. We measure the cost of many of these and find that both strongly beneficial mutations and deleterious mutations are genetic variants transferred by natural transformation. Importantly, we also show how recombination that separates linked beneficial and deleterious mutations resolves the cost of HGT.
Keywords: experimental evolution, antibiotic resistance, evolution, horizontal gene transfer, H. pylori
Abstract
Horizontal gene transfer (HGT) is important for microbial evolution, yet we know little about the fitness effects and dynamics of horizontally transferred genetic variants. In this study, we evolve laboratory populations of Helicobacter pylori, which take up DNA from their environment by natural transformation, and measure the fitness effects of thousands of transferred genetic variants. We find that natural transformation increases the rate of adaptation but comes at the cost of significant genetic load. We show that this cost is circumvented by recombination, which increases the efficiency of selection by decoupling deleterious and beneficial genetic variants. Our results show that adaptation with HGT, pervasive in natural microbial populations, is shaped by a combination of selection, recombination, and genetic drift not accounted for in existing models of evolution.
Horizontal gene transfer (HGT) provides the genetic innovation that underlies major evolutionary transitions (1) and converts benign bacteria into pathogens (2). As soon as whole genomes could be compared, it became evident that HGT is not confined to rare evolutionary leaps, but is pervasive throughout microbial evolution (3–7). Like random mutation, most of the genetic variants served to a population by HGT will be deleterious, and even HGT events that have an overall benefit can incur costs (8). These costs potentially limit the range of genes and genetic variants that can spread by HGT (9, 10), so it is important to understand both the fitness effects of HGT variants that can segregate in an evolving population and the mechanisms that could potentially resolve the costs of HGT.
While most natural microbial populations evolve with genetic variants acquired by HGT, studies of microbial experimental evolution have largely focused on the genetic variants that evolve by spontaneous mutation in clonal populations (11). In nonrecombining, asexual populations, adaptation is characterized by the successive fixation of cohorts of genetic variants driven by beneficial mutations (12, 13). The few studies that have tracked mutations segregating in sexually reproducing microbes have shown how recombination makes natural selection more efficient by decoupling beneficial and deleterious variants, speeding their respective fixation and loss from the population (14–16). In contrast, remarkably little is known about the impact of horizontally transferred genetic variants on the dynamics and outcomes of adaptation. While incorporating DNA from outside of the vertical line of descent has costs (8), comparisons of microbial genomes and microbiomes make it clear that HGT is an important source of genetic innovation (17, 18) and that microbial populations can harbor large amounts of genetic variation (19–22).
Many bacteria, especially human pathogens, can take up DNA that originates from cells of the same or different species directly from their surrounding environment (23). In these conditions, HGT could impact evolving populations of prokaryotes by multiple mechanisms. First, recombination between individuals in the population could decouple linked deleterious and beneficial mutations, increasing the efficiency of natural selection (24). Originally proposed to explain the evolution of recombination in sexual species, the operation of Fisher–Muller mechanisms (24–26) in prokaryotic populations could potentially ameliorate many of the perceived costs of HGT, such as the spread of linked, deleterious genetic variants (27), or facilitate soft sweeps of beneficial mutations (22).
Second, horizontal gene transfer from a distinct donor population can introduce a large amount of novel genetic information into a recipient population. In a previous study, we showed that HGT can provide a directional force that allows neutral and mildly deleterious alleles from the donor to segregate in populations repeatedly subjected to HGT (28). This work suggested that HGT from the donor population could introduce genetic variants with a wide range of fitness effects and potentially contribute to complex evolutionary dynamics in the recipient population. However, the HGT donor strains in that experiment lacked genetic diversity and the fitness effects of only a few genetic variants could be measured. Other experimental studies of evolution have demonstrated clearly that HGT can increase rates of adaptation (29–31) and facilitate adaptation to new niches (32, 33), which is highly repeatable (34). However, these studies have also focused on a few typically beneficial mutations and have been unable to measure the full spectrum of fitness effects available to populations that undergo HGT.
In this study, we evolved populations of antibiotic-sensitive Helicobacter pylori P12 that received HGT from one of two antibiotic-resistant H. pylori clinical “donor” strains (isolates CH426 and CH428) (Fig. 1 and Materials and Methods). The core genomes of the donor strains are ∼3 to 4% diverged from the H. pylori P12 (corresponding to >40,000 variants) and includes genes that provide resistance to clarithromycin (23S ribosomal RNA) and metronidazole (rdxA, CH426; frxA, CH428) (35, 36). In addition to these core genome differences, each donor strain carries around 50 accessory genes that are not found in H. pylori P12. Non-HGT control lines were passaged alongside the HGT treatment populations, but were not inoculated with DNA from the donor strains. While only the HGT treatment populations received gene flow from outside the population, both the HGT and non-HGT treatment populations took up DNA released by other individuals in the population during the course of the experiment (37). We propagated all HGT and non-HGT treatments in four different growth conditions: 1) Media without antibiotics; 2) media supplemented with the antibiotic clarithromycin; 3) media supplemented with the antibiotic metronidazole; and 4) media supplemented with both clarithromycin and metronidazole (double-drug) (SI Appendix, Fig. S1). Since the recipient strain was initially sensitive to both antibiotics, we followed previous studies of antimicrobial resistance evolution and adjusted the growth conditions during the period of the evolution experiment. In particular, we increased the amount of antibiotics supplied to the growth media, and the rate of culture dilution during the course of the experiment, as the populations evolved increased levels of fitness and antibiotic resistance (38–41).
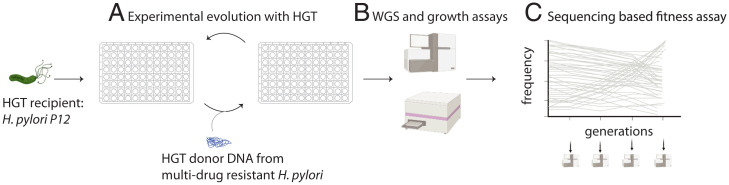
Fig. 1.
Experiment overview: Evolution experiment with HGT from multidrug-resistant strains of H. pylori. The antibiotic-sensitive HGT recipient strain, P12, was propagated with HGT from one of two antibiotic-resistant H. pylori clinical donor strains (CH426 and CH428) in 96-well plates (A). HGT and non-HGT treatment populations were subjected to four different growth conditions: media without antibiotics, media supplemented with the antibiotic clarithromycin, media supplemented with the antibiotic metronidazole, and media supplemented with both clarithromycin and metronidazole. The detailed workflow of the HGT and antibiotic treatments are shown in SI Appendix, Fig. S1. The outcomes of evolution with HGT were analyzed using whole-genome sequencing and growth assays (B). The fitness effects of the genetic variants that evolved in four HGT treatment populations were measured using DNA sequence–based fitness assays (C).
HGT Populations Evolved Higher Fitness but Also Acquired Low-Frequency Deleterious Mutations.
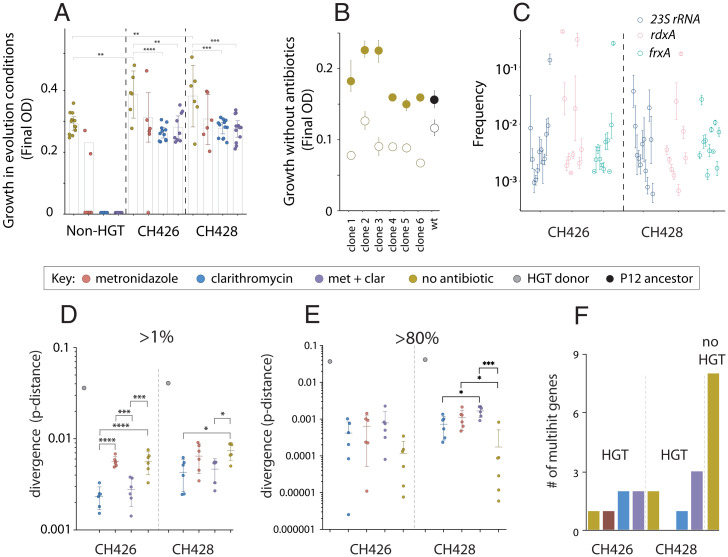
After 192 generations of evolution we measured the change in carrying capacity (final optical density [OD]600) of all evolved populations. As expected (28, 30), we found that HGT treatment populations adapted more quickly than the non-HGT treatments to growth media that did not contain antibiotics (one-way ANOVA, post hoc t test, CH426, t = 2.953, P = 0.0046; CH428, t = 2.795, P = 0.0102) (Fig. 2A) and that HGT treatment populations were able to survive and adapt to antibiotic treatments that drove non-HGT treatment populations extinct. Previous studies have shown that clarithromycin resistance mutations come with a fitness cost in growth conditions without antibiotics (42). We confirmed this by engineering a 23S rRNA allele (A2143G) from one of the donor strains, known to cause clarithromycin resistance (42), into clones isolated from six HGT populations that evolved without clarithromycin and also into the H. pylori P12 ancestor. In each case, this 23S rRNA allele causes a significant fitness defect, confirming that clarithromycin resistance is costly, and that the magnitude of the cost is dependent on the genetic background (Fig. 2B, one-way ANOVA, F(6,14) = 7.68, P = 0.0009).
Fig. 2.
HGT increases the rate of phenotypic and molecular evolution. The maximum carrying capacity of each evolved population after 192 generations, measured in the same growth conditions as the evolution experiment. For example, the non-HGT treatment populations evolved and were measured in media without antibiotic (gold circles), while the non-HGT populations evolved in metronidazole were measured in media containing metronidazole (red circles). The non-HGT clarithromycin (blue circles)- and double-drug (purple circles)-treated populations went extinct and could not be measured (A). Growth experiments were carried out in media without clarithromycin to measure the cost of mutations in the 23S rRNA gene (B). The wild-type (black filled circle) and evolved clones (gold filled circles) were engineered with the A2143G substitution in 23S rRNA, that confers resistance to clarithromycin (open circles). The average frequency of variants in genes known to cause resistance to clarithromycin (23S rRNA), and metronidazole (rdxA and frxA) in populations that had evolved with HGT but in media without antibiotic, after 192 generations of evolution (C). The core genome dissimilarity (pairwise nucleotide p-distance) between the donor strains (CH426 and CH428) and the H. pylori P12 recipient strain (gray circles), and core genome dissimilarity between individual evolved populations and the H. pylori P12 recipient strain. In the case of the evolved populations, dissimilarity was calculated based on the number of variants that attained a frequency >1% or higher (D) or >80% (E). The number of multihit genes that evolved de novo mutations in parallel more often than expected in the six sequenced populations from each treatment (F). The no-HGT treatment populations evolved significantly more multihit genes by de novo mutation than the HGT treatment populations (one-sample Z test, z = 3.83, P = 6E−5). Asterisks indicate significance by Tukey's multiple comparison test: *P≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001.
To identify the HGT and de novo mutations that evolved during the experiment, we carried out high coverage (∼600-fold), whole-population, whole-genome sequencing of six replicate populations from each of the eight HGT experimental treatments, six replicates from the non-HGT, no-antibiotic treatment and two non-HGT treatment populations that survived the metronidazole treatment (n = 56). Due to the significant divergence of the donor strains from the recipient strains, we developed (Materials and Methods and SI Appendix, Fig. S2) and validated (SI Appendix) a method for combining donor and recipient genomes into an extended HGT reference genome so that we could readily identify possible HGT events in whole-population sequence data. In previous work, we have shown that HGT from a metronidazole-resistant donor strain facilitated the spread of mildly deleterious metronidazole resistance alleles into populations evolving in growth media without metronidazole (28). We found that resistance mutations for clarithromycin and metronidazole were segregating in populations not treated with these drugs, at frequencies from around 0.01 to 10% (Fig. 2C). This confirmed that not only the mildly deleterious metronidazole alleles, but also the strongly deleterious clarithromycin resistance alleles can establish at low to intermediate frequencies in populations evolving with HGT even in the absence of selection for those antibiotics.
HGT from Diverged Strains Causes an Inundation of Low-Frequency Genetic Variants with Only a Few Variants Promoted by Selection.
In addition to the known resistance alleles in the 23S rRNA, rdxA and frxA genes, we discovered tens of thousands of genetic variants segregating in evolved populations. To quantify how much genetic variation had been transferred by HGT, we computed the core genome dissimilarity (nucleotide p-distance) of each evolved replicate population with the P12 recipient strain, based on the numbers of genetic variants that were detected at a frequency greater than 1%. We found a high degree of polymorphism corresponding to up to 1% divergence from the P12 ancestor strain, with significant variation across treatments, (one-way ANOVA, CH426: F(3,20) = 19.42, P < 0.0001; CH428: F(3,20) = 4.58, P = 0.0134) (Fig. 2D and SI Appendix, Fig. S3). Compared with the P12 ancestor strain, the no-antibiotic treatment populations showed significantly greater core genome dissimilarity than the clarithromycin-treated HGT populations (Tukey’s multiple comparisons test, P ≤ 0.05; Fig. 2D). However, when we calculated p-distance based on high-frequency polymorphisms (>80%) (Fig. 2E), the difference between no-antibiotic and antibiotic treatment was reduced, and for some of CH428 treatment populations, had reversed (Fig. 2E). These results are consistent with natural transformation contributing large amounts of genetic variation that establishes at low or intermediate frequencies, with a small proportion of variants driven to high frequencies by natural selection.
Reduced Signal of De Novo Parallel Evolution in HGT Treatment Lines.
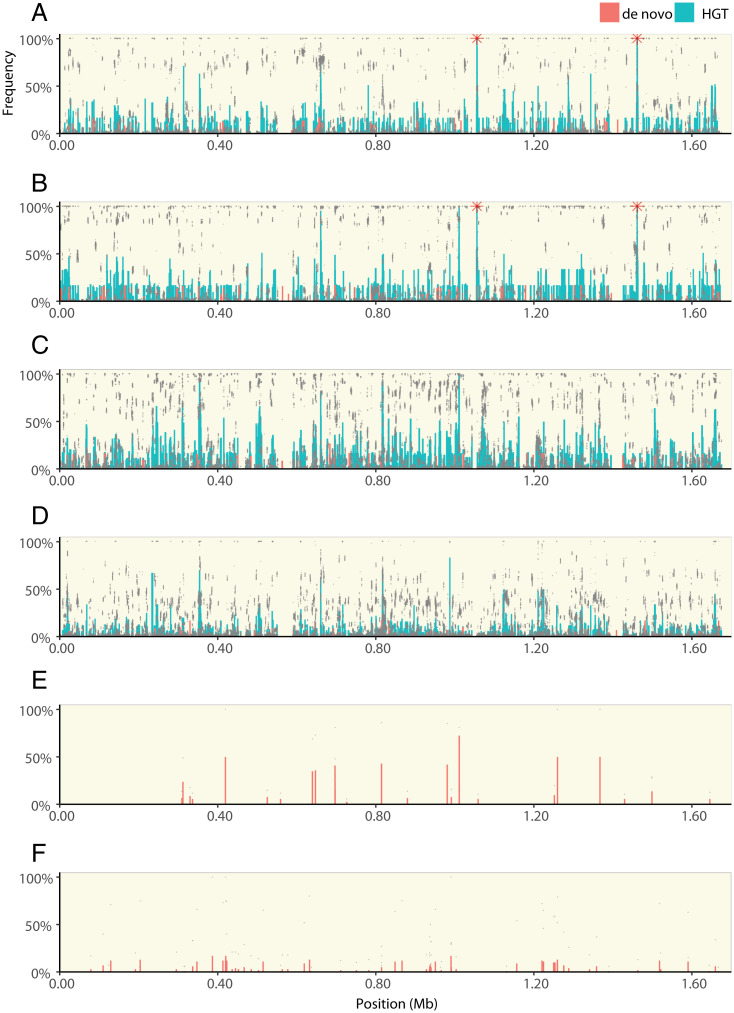
To understand whether these de novo variants contributed to adaptation in HGT populations, we looked for examples of parallel evolution—the evolution of high-frequency genetic variants in the same gene across multiple independently evolving populations—a frequently used measure of selection in natural and experimental populations (13, 43–45). We first looked at those mutations that evolved during the experiment, i.e., de novo mutations that did not originate by HGT from the donor strains. To do this, we used a model that compares the number of parallel, multihit genes in evolved populations with a null model of neutral evolution (15) (Materials and Methods). We found significantly fewer multihit genes in the HGT treatment populations than non-HGT populations (μ = 2.25, σ = 1.9, one-sample Z test, z = 3.83, P = 6E−5, Fig. 2F), supporting that HGT treatment populations are less dependent on spontaneously evolving genetic variants for adaptation. To confirm that the HGT-treated populations also had access to de novo genetic variations, we compared the total number of variants across all treatments. Although >99% of the variation in HGT treatment populations originated from HGT, there was significantly more de novo variants in HGT populations than the non-HGT populations (Fig. 3 and SI Appendix, Fig. S4) (Welsh’s test, two tailed, P = 0.006).
Fig. 3.
The distribution of HGT and de novo variants across the H. pylori genome. Each panel shows the frequency of genetic variants along the length of the H. pylori genome. Populations evolving with HGT from the CH428 donor in growth media with clarithromycin and metronidazole (A), clarithromycin (B), metronidazole (C), or in growth media without antibiotic (D). Populations evolving without HGT, including the two populations that survived treatment with metronidazole (E) and populations that evolved in growth media without antibiotic (F). Each panel (except E) shows sequencing data from six replicate populations, including data points from each individual population (gray dots), the average frequency of each HGT (green bars), and de novo (red bars) genetic variant. Red stars indicate 23S rRNA mutations. Data from populations that evolved with HGT from CH426 are shown in SI Appendix, Fig. S3.
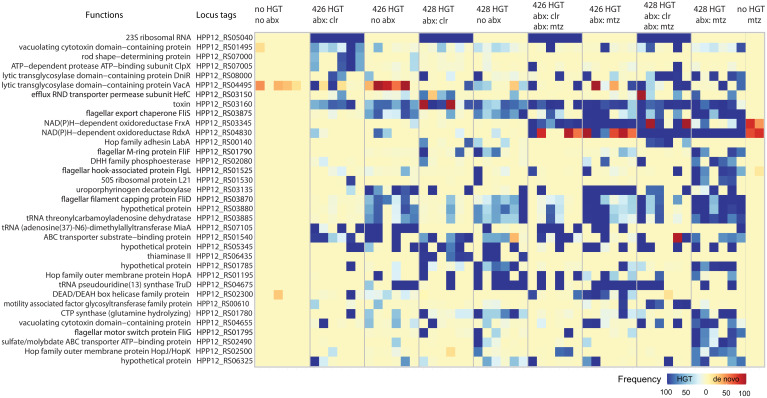
The parallel evolution analysis described above for de novo variants compares the observed number of multihit genes with a null model based on neutral, spontaneously occurring mutations. The genetic variants that originate by HGT from the donor strain should not be compared with this model since horizontally transferred genetic variation does not arise by a random mutational process. Instead, HGT variants are limited to the discrete set of variants that are available for transfer from the donor strains. To find the HGT genes most enriched by selection, we identified multihit genes that had attained a population frequency above 50% in at least four of the six sequenced replicates in at least one of the treatment conditions (Fig. 4). These selection criteria recovered 35 out of 1,469 genes, including the known causes of clarithromycin resistance (23S rRNA) and metronidazole resistance (rdxA and frxA). In addition to other suspected contributors to antibiotic resistance such as the efflux pump regulator hefC (HPP12_RS03150) (46), our data revealed a number of genes not previously attributed to antibiotic resistance. These may be compensatory mutations that do not directly block antibiotic action, but increase fitness by amelioration of the costs of antibiotic resistance (47–52).
Fig. 4.
Multihit genes. Multihit genes were defined as genes that evolved high-frequency variants (>0.5) in four or more populations within at least a single treatment (e.g., 428 HGT abx: mtz, are populations that evolved with HGT from donor CH428 and in growth media supplemented with metronidazole). Each treatment category has six sequenced populations. Blue squares correspond to horizontally transferred variants, while red squares are non-HGT genetic variants, those that evolved de novo, or may have been very low frequency standing genetic variation in the recipient H. pylori P12 ancestor population.
The Distribution of Fitness Effects of Horizontally Transferred Genetic Variants.
The growth assays of the reconstructed 23S rRNA mutant (Fig. 2A) and parallel evolution analysis of the evolved populations (Figs. 2E and 4) confirm that both deleterious and beneficial mutations can be introduced into the population by horizontal gene transfer. We wanted to carry out a more comprehensive measurement of the fitness effects of horizontally transferred variants, but it is difficult to reconstruct large numbers of mutations in H. pylori without using antibiotic resistance markers that carry their own costs. To circumvent this limitation and measure the full spectrum of fitness effects of the genetic variants transferred by HGT, we carried out sequencing-based fitness assays. To do this, we sampled evolved populations that carried large amounts of genetic diversity and then propagated them for five transfers (Materials and Methods and SI Appendix, Figs. S5 and S6). By sequencing whole population samples at multiple time points during the fitness assay we could track thousands of variants per population.
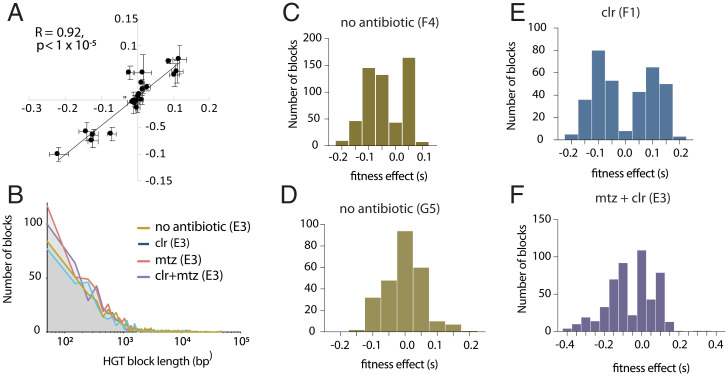
In a well-recombined population, each allele will be present on a large number of genetic backgrounds, and assuming that epistasis is negligible, many deleterious and beneficial alleles can independently segregate in a population (Materials and Methods). We checked the repeatability of our fitness assay by comparing two measurements for a set of variants that disrupted gene function and found a high degree of repeatability across assays (Pearson’s R = 0.92, P = 1 × 10−5, Fig. 5A and SI Appendix, Fig. S7). Due to the large number of variants segregating in the populations, we found that, even with high recombination rates, there were blocks of linked alleles. These linked blocks of genetic variants could be identified since they each had similar selection coefficients and were physically close on the genome (Materials and Methods and SI Appendix, Fig. S6). We treated these blocks of linked variants as a single allele for further analysis. We found that the range and distributions of block sizes were similar across different treatments and populations (Fig. 5B and Dataset S1) and consistent with HGT experiments carried out in other species (34, 53, 54). We plotted the distribution of fitness effects for those “blocks” of HGT genetic variants that evolved in populations without (Fig. 5 C and D) and with antibiotics (Fig. 5 E and F). These data further support that after 192 generations of evolution, large numbers of HGT-derived beneficial and deleterious mutations continue to segregate in all populations, showing that the adaptive genetic variation introduced by HGT came at the cost of significant genetic load (Fig. 5 C–F).
Fig. 5.
The distribution of fitness effects of HGT variants. Regression of fitness values for a set of indel mutations identified in double-drug–selected population E3 and measured in growth media with clarithromycin (y axis) and growth media with clarithromycin and metronidazole (x axis) (A) (Materials and Methods and Dataset S2). The distribution of HGT block length for four different evolved populations (E3, F1, G5, and F4) measured growth media without antibiotics (B). The distribution of fitness effects of blocks of HGT variants, for HGT treatment populations measured in the same conditions as their evolution treatment; growth media without antibiotic (C and D), growth media with clarithromycin (E), or clarithromycin and metronidazole (F).
Linkage Disequilibrium Decays with Distance from the Clarithromycin Resistance Allele in 23s rRNA.
Without recombination, the fixation of a strongly selected variant would result in a hard selective sweep and purge genetic variation at nonlinked sites (55). However, we found that high- and low-frequency variants are interspersed along the length of the genome in all HGT treatment populations (Fig. 3). This supports the idea that, across the scale of H. pylori’s single chromosome, many genetic variants introduced by HGT segregate independently (Fig. 3 and SI Appendix, Fig. S4). To understand the scale of recombination and linkage, we looked at 18 HGT variants distributed across the 2.9 kb 23S rRNA gene. The 23S rRNA gene carries the single SNP required for clarithromycin resistance. We expected that the other variant alleles in the 23S rRNA gene (6 alleles from donor 426 and 14 alleles from donor 428), would fix together with the resistance allele. However, only the known resistance alleles from each donor strain fixed (CH426 = A2142G; CH428 = A2143G). We regressed the frequency of the 23S rRNA variants introduced by HGT against their genetic distance from the selected, resistance allele. We found that the frequencies of HGT variants were negatively correlated to their genetic distance from the resistance locus (Fig. 6A) (Pearson’s correlation coefficient, CH426, R = −0.98, P < 0.0001; CH428, R = −0.75, P = 0.0021). This decay of linkage disequilibrium within a few kilobases of a strongly selected locus suggests that in addition to an initial HGT event that introduces a block of 23s rRNA alleles from the donor, there was subsequent “backcrossing” between strains that carried the wild-type version of 23S RNA gene.
Fig. 6.
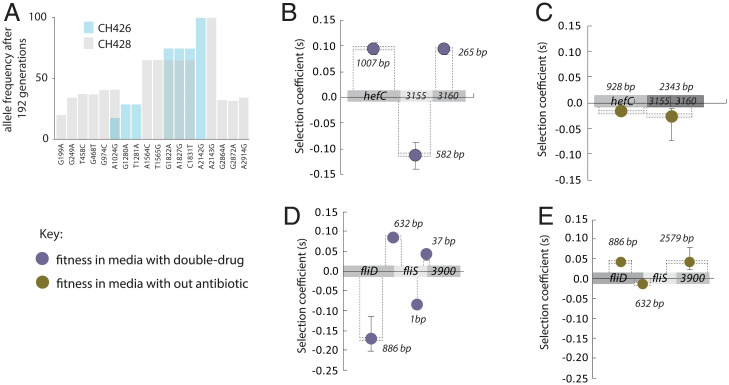
Recombination and selection. The frequency of HGT alleles in the 23s rRNA gene after 192 generations of selection in clarithromycin (A). The average frequency for horizontally transferred 23S rRNA gene variants in clarithromycin HGT treatment populations with the CH426 donor (blue bars) and the CH428 donor (gray bars). A single substitution in the 23S rRNA gene is sufficient for clarithromycin resistance and was fixed in conditions with clarithromycin (CH426 = A2142G; CH428 = A2143G). Each bar shows the mean of six populations. The frequencies of the other 23S rRNA alleles are negatively correlated with genetic distance to the fixed mutation (Pearson’s correlation coefficient, CH426, R = −0.98, P < 0.0001; CH428, R = −0.75, P = 0.0021). The fitness effects of HGT blocks in multihit genes measured in two environments, growth media with clarithromycin and metronidazole (purple circles B and D) and growth media without antibiotics (gold circles, C and E). The size of the selected blocks is indicated by the number of bases shown, and dashed lines show the approximate position of the blocks on the genes. For example, B shows that in the double-drug conditions, the 582-bp fragment is deleterious, while the 265-bp fragment is beneficial. However, in no-antibiotic conditions these two blocks segregate together as a weakly deleterious block. R is Pearson’s correlation coefficient.
Recombination Separates Deleterious and Beneficial Mutations.
So far, our results are consistent with the well-established natural competence of H. pylori (56). The evolving recipient populations of H. pylori frequently take up DNA from their environment, including DNA from the lysis of other recipient cells, as well as the donor DNA added to the growth media. If an HGT-originating fragment recombines with a wild-type–originating fragment of DNA from the same locus, this could result in a new recombinant individual that contains a beneficial variant but not the previously linked deleterious variant. The net effect of HGT variants on many different genetic backgrounds across the population is that the beneficial variant will spread, while the frequency of the deleterious allele will decrease (26). To identify examples of physically close deleterious and beneficial mutations segregating in opposite directions, we looked at the multihit genes that evolved in the E3 population (Fig. 4). The E3 population evolved in double-drug growth conditions, and the sequencing-based fitness assay for this population was carried out in four environmental conditions (no antibiotic, metronidazole, clarithromycin, and double-drug). Looking at this population, we focused on the subset of blocks that contained multihit genes. As expected, we found that nearly all of the multihit genes contained at least one block with strong beneficial effects in double-drug conditions (Dataset S1). Two examples are the multihit gene regions (hefC-RSO3155-RSO3160, Fig. 6B and fliD-fliS-RSO3900, Fig. 6D), where the deleterious blocks are flanked by beneficial blocks. In the double-drug environment, these alleles are segregating as strongly deleterious alleles, but when measured in growth media without antibiotic, the same loci have smaller fitness effects and segregate as part of larger linked blocks of variants (Fig. 6 C and E).
In H. pylori, the size, distribution, and complexity of HGT events are well characterized (56). Complex recombinant blocks can be generated in a single HGT event since the transformation of a long fragment of donor DNA can result in multiple breakpoints, and “clusters” of DNA interspersed with wild-type and donor alleles (56, 57). Additionally, a donor fragment of DNA that had been integrated into the genome could later backcross by recombination with a wild-type version of the fragment taken up from other individuals in the population (i.e., between-individual recombination) (37). Both processes are well established in H. pylori (31, 58), although it is difficult to determine the relative contribution of these two processes in our experiment. Altogether, these data show that recombination has resulted in multiple combinations of alleles segregating in each population, and show how selection can efficiently purge or promote deleterious and beneficial alleles, respectively, depending on the environmental conditions (SI Appendix, Fig. S8). A range of theoretical (24, 26, 59, 60) and empirical (14–16) studies have shown how recombination increases the efficiency of natural selection by decoupling deleterious and beneficial mutations so that each can be selected on their individual fitness effects. While most theory has focused on Fisher–Muller mechanisms in the context of meiosis, our data agree with theoretical results that show that recombination could prevent the fixation of deleterious mutations in microbial populations (27, 61). It is important to note that estimates of the fitness effects of blocks assumes that epistasis is negligible. We think that this is reasonable because the strongest epistatic effects would be between the primary resistance alleles and other compensatory mutations that increase fitness. We measured the fitness effects of segregating blocks in populations where these primary resistance mutations had fixed or were almost entirely absent, so these strong epistatic effects should not affect our results, although we cannot rule out epistatic effects at other loci.
Conclusions
In natural populations of microbes, DNA can spread horizontally by a range of mechanisms. For example, plasmid conjugation and bacteriophage selfishly promote their own spread and can prevail despite significant costs (62). In contrast, for other mediums of HGT—nanotubes, vesicles, and natural transformation—the integration of DNA sequences by HGT is random with respect to its fitness effect in a given environment. Here, we study HGT by natural transformation, which provides an opportunity to separate the evolutionary forces that shape the dynamics of HGT variants from the confounding effects of infectious mechanisms of HGT. Our results confirm two important roles of recombination in populations evolving with HGT. First, HGT from diverged populations can introduce a vast amount of genetic variation into the evolving population, even without selection for those variants. The scale of genetic variation in HGT compared with non-HGT treatment populations (Fig. 3 and SI Appendix, Fig. S3) shows how the genome content of a local donor population could significantly influence the evolution of a naturally competent recipient population. Second, our data show that Fisher–Muller mechanisms function in recombining populations of microbes and can ameliorate the costs of the horizontal import of deleterious genetic variants in adapting populations. Although H. pylori has an exceptionally high recombination rate (63), the observation of gene-specific sweeps in natural microbiomes (21, 64, 65) suggests that the two impacts of recombination observed in this study—the introduction and reshuffling of novel genetic variation—are likely to be common in natural microbial populations. Future experimental and theoretical studies need to take horizontal gene flow and recombination into account when considering adaptation in natural and clinical microbial communities.
Materials and Methods
Bacterial Strains and Antibiotic Resistance Profile.
The ancestral, recipient strain for experimental evolution was isogenic with H. pylori P12 wild-type (vacA m1s1, metronidazole sensitive: minimal inhibitory concentration [MIC] 0.19 mg/L; clarithromycin sensitive: MIC 0.023 mg/L). The two strains that provided donor genomic DNA (gDNA) for the evolution experiment are H. pylori CH426 and H. pylori CH428. These strains were isolated by gastric biopsy and determined by E tests in a clinical laboratory to be resistant to amoxicillin (CH426: >256 mg/L, CH428: >256 mg/L), metronidazole (CH426: >256 mg/L, CH428: >256 mg/L) and clarithromycin (CH426: 12 mg/L, CH428: >256 mg/L). We carried out follow-up testing for clarithromycin and metronidazole to determine the minimal concentrations required to inhibit the growth of the recipient H. pylori P12, but that did not inhibit the donor strains CH426 and CH428. This was carried in the same conditions as the evolution experiment: 132 μL of brain heart infusion (BHI) growth media were supplemented with antibiotic, in 96-well plates, incubated at 37 °C in a 5% CO2 incubator and the minimal inhibitory concentrations of clarithromycin and metronidazole determined were 0.03 mg/L and 0.8 mg/L, respectively.
Growth Conditions.
H. pylori strains were routinely grown on GC (gonococcus) agar-based medium (Oxoid) supplemented with protease peptone 1.5 g/L, 10% vol/vol horse serum (HS), 1% vol/vol of vitamin mix, 10 mg/L of vancomycin, and 20 mg/L of trimethoprim. Culture plates were incubated at 37 °C in an anaerobic jar with microaerobic conditions generated by BD GasPak EZ Campy sachets (Becton Dickinson). For liquid cultures, the strains were inoculated in BHI broth (Oxoid) supplemented with 10% vol/vol of fetal bovine serum (Serena), 1% vol/vol of vitamin mix, and 10 mg/L of vancomycin. Cultures were incubated in a 5% CO2 incubator at 37 °C with either shaking at 120 rpm or without shaking for the evolution experiment. Agar plates and liquid cultures were supplemented with additional antibiotics as needed for selection and routine culture.
Experimental Evolution.
We propagated populations of antibiotic-sensitive H. pylori in growth media with or without antibiotic, and with or without HGT from antibiotic-resistant clinical strains. To allow for the evolving populations to incorporate DNA via HGT, we increased the concentration of antibiotic incrementally and did not add metronidazole to the metronidazole and double-drug treatments until week 2 (SI Appendix, Fig. S1). The evolution experiment was established with 48 replicate populations of H. pylori P12 wild-type in 96-well microplates with 132 μL of growth medium per well with starting cell density of ∼2 × 104 colony-forming units (CFU). Thirty-six of these 48 populations were “HGT treatment populations” exposed to gDNA extracted from one of the two clinical donors, CH426 and CH428. For the HGT treatment, ∼300 ng of gDNA was added to the growth medium for each population; this was carried out 5 to 6 h after the dilution into fresh medium to allow for physiological adaptation to new conditions. Antibiotic concentrations and dilutions were as follows: At the beginning of the experiment, H. pylori P12 cultures could tolerate very low concentrations of antibiotic and slow dilution rates, but we anticipated that HGT populations would rapidly adapt to these conditions. In order to maintain strong selection on the evolving populations, we changed the dilution rates and antibiotic concentrations as the experimental populations adapted. For the first transfer, 24 of the 36 HGT populations (12 for each of the two donors) were cultured for 18 h before adding clarithromycin at a concentration of 0.03 mg/L. This was to give the antibiotic-sensitive H. pylori P12 cells a chance to take up the DNA, so that it could be integrated and expressed before the media was supplemented with antibiotic. This concentration of antibiotic used was significantly less than our measured MIC for the CH426 (256 mg/L) and CH428 (12 mg/L) donor strains, but was lethal to the H. pylori P12 populations that had not received HGT. The 36 HGT populations with and without clarithromycin were transferred to fresh growth media every 72 h with a 1:16 dilution, corresponding to four generations per transfer. After seven transfers (28 generations), all replicate populations were split into two independent evolution experiments. We continued the 48 populations described above (24 HGT treatment populations with clarithromycin, 12 HGT treatment populations without clarithromycin, and 12 non-HGT controls without clarithromycin) except that the concentration of clarithromycin was increased to 0.4 mg/L to select for higher resistance. The second set of 48 populations was exposed to the second antibiotic, metronidazole (0.8 mg/L) as well as the higher concentration of clarithromycin (0.4 mg/L). All HGT populations in both evolutionary experiments were reexposed to gDNA extracted from donors at this point (generation 28) and at generations 56, 137, and 186. We monitored the growth of all populations and found that after 72 generations, all cultures were attaining high densities after ∼36 h, so the dilutions were carried out every 48 h (instead of every 72 h), and the clarithromycin concentration was increased to 2 mg/L for both single- and double-drug treatments (SI Appendix, Fig. S1). After 64 generations, the dilution for transfer in the clarithromycin experiment was changed from 1:16 to 1:32 (5 generations per transfer). These treatments were kept constant until the end of the experimental evolution (∼192 generations). The growth of the evolving populations was measured before every next transfer by BioTek microplate reader at 37 °C with OD600. Glycerol stocks of evolved populations were prepared every 20 generations by mixing with glycerol to 25% and stored at −80 °C.
Construction of Clarithromycin-Resistant Mutants.
To engineer clarithromycin-resistant mutants, we first obtained clarithromycin-sensitive strains: The H. pylori P12 wild-type and six clone isolates, one from each of the six HGT populations (CH426 donor) that evolved in growth media without antibiotic. Briefly, the PCR products containing single resistance mutation at position 2143 in the 23S rRNA gene were amplified from a clone selected from the evolved CH426 HGT-treated population selected on clarithromycin that evolved in well A1, Ev426ClrA1. According to whole genome sequencing, population Ev426ClrA1 fixed a single resistance mutation in 23S rRNA, so that this is the sole variant that distinguishes this allele from the wild-type 23S rRNA. There are no other mutations in this gene. The sequence of the target site in the 23S rRNA gene of the isolate was confirmed by Sanger sequencing, oligo sequences: 23S_rRNA.F: CGACTTTCGTCTCTGCTTGA and 23S_rRNA.R: AACACAGCACTTTGCCAACT, and products were used to generate mutants through transformation. The successful transformants were selected on GC plates supplemented with clarithromycin (2 mg/L).
For transformation, freshly grown cells were patched (1- to 1.5-cm size) on new GC agar plates and incubated for 5 to 6 h at 37 °C in microaerobic conditions. Approximately 1 μg of PCR product was pipetted onto the patch of bacteria and the plates were incubated overnight. Cells were harvested and resuspended in 1 mL of BHI liquid media. After that, 100 μL of the resuspension was spread on GC plates supplemented with clarithromycin to select for transformants.
Sequencing and Assembly of Genomes.
DNA sequencing.
Whole-population whole-genome DNA from the donors, ancestor, and evolved populations was prepared with the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich) following the standard protocol recommended by the manufacturer. Samples were sent to GENEWIZ (Suzhou, China) for sequencing on the Illumina MiSeq next-generation platform with paired-end and 150-bp read length. GENEWIZ returned 2 GB of data, with an average Phred quality score (Q score) of 30 for greater than or equal to 80% of the Illumina paired-end short reads. Adaptor trimming was performed at GENEWIZ, but reads were further filtered and trimmed based on quality scores using the BBDuk package (https://jgi.doe.gov/data-and-tools/bbtools/). DNA from the same donor samples was also prepared for long-read sequencing with the MinION long-read sequencing platform by Oxford Nanopore Technology (66). Long reads were demultiplexed and base-called according to Wick et al. (67). The Unicycler and Bandage software packages were used for hybrid assembly and visualization of the short and long reads of donor DNA (66).
The ancestral population was derived from H. pylori P12, which as a complete reference sequence available from the National Center for Biotechnology Information (NCBI) Genome Database so resequencing was sufficient to produce a complete genome, chromosome: NC_011498, plasmid: NC_011499 (68). Although donors and the ancestor were each derived from a single isolated cell, the resequencing and variant-calling software package breseq was used in polymorphic mode with an absolute read count threshold of 2 for all short-read samples to detect any low-level variants that might have arisen during the cell culture (69).
Discovery of HGT variants through resequencing.
Donor short reads were mapped to the reference sequence and unmapped short reads were binned by breseq. Evolved sequences were also mapped to the reference sequence and the variants that were identified were compared with those of the relevant donor with the breseq suite of tools for variant analysis, gdtools. Any of these overlapping variants that further overlapped with the ancestor or the non-HGT controls were removed, and the remainder were presumed to represent putative HGT events. This initial set of HGT events formed the “core HGT reference” dataset (SI Appendix, Fig. S2, “path 1”). For each evolved HGT population, all HGT events of any frequency were applied to the P12 reference genome to produce a set of core putative HGT genome FASTA files.
Identification of HGT variants by alignment with the HGT-hybrid reference.
The binned reads from the alignment of the donors to the reference genome were assembled into scaffolds via the de novo assembly software package SPAdes (70). All scaffolds of at least 300 bp were collected and locally blasted against the relevant donor genome and the P12 chromosomal and plasmid sequences using the BLAST+ executables (71). Only those scaffolds that had less than 100% BLASTN identity to the P12 sequences and 100% BLASTN identity to the donor sequence were retained. These scaffolds were then concatenated with the H. pylori P12 sequences into an extended, combined genome. Breseq was used to map and variant call each of the evolved read sets against this combined (HGT-hybrid) reference genome. The regions of the donor nodes for which there was read coverage were extracted based on the most conservative estimates for the start and end positions of the regions of coverage. All variants present at any frequency were applied to the extracted coverage regions. Coverage regions were blasted against the H. pylori P12 sequences to filter out donor coverage regions caused by mismapping of H. pylori P12 reads to the donor scaffolds. The process was also performed for the controls to ensure that the dataset reflected actual HGT events in the HGT populations. Donor coverage sequences were aligned to the putative HGT genomes produced from the application of the core HGT dataset via long-read mapping with BBMap (https://jgi.doe.gov/data-and-tools/bbtools/) and consensus sequences generated with Samtools (72). For each population, the resulting consensus sequence was extracted, producing the genomic sequence that represented the total putative variation (regardless of proposed frequency) derived from HGT—the “extended HGT reference genomes” (SI Appendix, Fig. S2, “path 2”).
Refinement of HGT-derived variant frequencies.
Each extended HGT reference genome was aligned to the H. pylori P12 chromosomal sequence, and the genome alignment program “progressiveMauve” was used to collect the reciprocal sequence differences in terms of single nucleotide polymorphisms (SNPs) or indels of any length (gaps) (73). A final breseq run was performed for each of the evolved short reads against a reference that included the H. pylori P12 sequences and the extended HGT reference genome for that population. For each HGT population, the bam file obtained from the breseq run was analyzed at the sites specified by the progressiveMauve SNPs and gap files for reference and HGT allele coverage for both the H. pylori P12 and extended HGT reference sequences with the use of a custom script. Final HGT frequencies were calculated for each of the predicted HGT events from the extended HGT reference genome and written to a gd file compatible with the breseq gdtools for variant comparison. Coverage information was written to a CSV file for downstream analysis.
Analysis of patterns of HGT uptake.
For each evolved population, variants with frequencies at or above each of three threshold values (1%, 50%, and 80%) were applied to the H. pylori P12 reference genome. Gene annotation of all HGT, donor, and ancestral genomes was performed with Prokka (74) for consistent open reading frame (ORF) identification. For each frequency cutoff, the genome comparison software package “Roary” was used to identify and cluster orthologous groups across the donor, ancestral, and corresponding HGT genome sequences that represent the core genome using a conservative definition of novelty, with a BLASTP identity cutoff of 90%, above which ORFs would be clustered together and no splitting of paralogs (default = 95% and allow splitting; developer discourages BLASTP cutoffs below 90%) (75). Core genomes for each of the HGT, ancestral, and donor sequences were produced as part of the Roary output, and nucleotide p-distance was calculated using Mega (76, 77). The presence/absence (binary) data for accessory genes identified and clustered across the donor, ancestral, and HGT genomes of a given frequency threshold by Roary were converted into pairwise Euclidean distances using the R package “proxy” (78). The package “Scoary” was used to assess patterns of novel gene uptake (79). For both the core genome dissimilarity given by (pairwise nucleotide p-distance) and unshared accessory gene (pairwise Euclidean distance) data, within-group mean distances and groupwise mean distances to H. pylori P12, and the SDs thereof, were calculated. Distances to H. pylori P12 and to one another were also calculated for each of the donors as above. HGT genome data were split according to DNA donor and one-way ANOVAs followed up with Tukey’s multiple comparisons tests performed in GraphPad Prism version 7.0.0 for Mac OS X (GraphPad Software, https://www.graphpad.com:443/). Statistical significance was indicated using asterisks: >0.05 (ns), ≤0.05 (*), ≤0.01 (**), ≤0.001 (***), <0.0001 (****).
Identifying multihit genes (parallel evolution) in de novo mutations in evolved populations.
All of the variants discovered in the evolved populations were compared against the genomes of the donor strains and the recipient strain. If a variant was determined not to match that of the donor or the recipient strains, this was designated as de novo, or arising by spontaneous mutation during the period of the evolution experiment. Only de novo variants that reached a population frequency of at least 5% were included in the set of “called” variants for the purposes of the parallel evolution analysis. Parallel evolution is the evolution of the same variant across multiple, independently evolving, populations and provides evidence for the action of natural selection acting on each population due to similar environmental conditions across replicates. Gene hits—the number of populations in which a de novo mutation occurs for each gene—were tabulated and compared with a null model of evolution without selection. To do this, we employed an in-house script that simulated evolution by drawing of mutations from the H. pylori genome using a uniform probability distribution weighted by gene size. Simulations were carried out with 10,000 repetitions.
Sequence-based fitness assay.
Four of the evolved populations were selected: Populations 426F4 and 428G5 evolved with HGT in media without antibiotic, 428F1 evolved in media with clarithromycin, and population 428E3 evolved in media with clarithromycin and metronidazole. All populations were measured in growth media without antibiotic. The F1 population was also measured in media with clarithromycin, and the E3 population was also measured in growth media with clarithromycin and metronidazole. To start, the cryopreserved stocks of evolved populations were defrosted at room temperature. At least 20 μL of culture was transferred to fresh media supplemented with appropriate antibiotics in a 96-well microplate and incubated at 37 °C in 5% CO2 incubator without shaking to recover the populations. After 48 h, the populations were expanded to 5-mL cultures with the same antibiotic conditions. These 5-mL cultures served as the ancestral populations for the fitness assay (T0). For the 428F1 population (a clarithromycin-adapted population), the first transfer was initiated after 48 h by separating the ancestral population into two independent replicate transfers with 1:32 dilution. One replicate continued being exposed to clarithromycin, while the other was switched to antibiotic-free conditions. This transfer pattern was repeated for five transfers (∼20 generations) for every 48 h. A similar process was applied for the 426F4 and 428G5 populations but since these populations evolved in media without antibiotics, they were only measured in growth media without antibiotics. For the E3 population, the first transfer was commenced after 48 h by separating the ancestral population into four independent replicate transfers with 1:16 dilution. These replicate populations were exposed to either metronidazole, clarithromycin, double-drug, or antibiotic-free conditions. Portions of each time point were cryopreserved and DNA was extracted for whole-population whole-genome sequencing.
Calculation of Selection Coefficients Assignment of Linked Variants into Blocks.
DNA was prepared and paired-end short reads were processed as above. Reads were aligned to the H. pylori P12 reference sequences both with and without the relevant extended HGT reference genome (428E3, 428F1, 426F4, or 428G5) with breseq, and precise frequencies for previously identified HGT events were found as described above. Ancestral and de novo variants previously identified in the population were detected in the time-course data and coverage values calculated using a bam file generated by aligning reads to the H. pylori P12 reference sequences only. Remaining variants identified in the time-course sequencing sets not previously identified as belonging to any of the other categories were classified as “miscellaneous.” The coverage values for all variants of interest for all time points for each of the eight populations were collated together for input to a batch script for calculation of selection coefficients using a logistic link function as in McDonald et al. (14). We found that some variants were fixed or lost during the fitness assay, and some instances were present for only one time point. The trajectories of such variants were interpreted by the selection calculation algorithm as strong beneficial or deleterious variants, but we were concerned that some might have been fixed or lost by chance, since we only had evidence for an intermediate frequency at a single time point. Therefore, to gather a more conservative estimate of the distribution of fitness effects and reduce the possibility of false extreme values, we removed variants that could not be detected in the zeroth time point. In addition, time points where a variant attained a frequency of 0 or 1 during the course of the experiment were removed. If the removal of such a time point resulted in there being fewer than four time points for the selection coefficient calculation, then this variant was removed from the analysis.
If two physically close alleles have significantly different selection coefficients, this corresponds to these two alleles having different evolutionary trajectories as they segregate over evolutionary time. For this to happen, these two alleles must have become decoupled by recombination. We developed a program to group continuous runs of physically linked variants that had very similar selection coefficients. Using an in-house script, selection coefficients for which P < 0.05 were organized by genomic position and blocked according to whether, for a given selection coefficient, the adjacent selection coefficient was within the bounds of its confidence interval. If so, it was added to the growing block; if not, a new block was started. This process identified groups of linked blocks that had very similar selection coefficients. Note that only contiguous runs of physically linked alleles that have similar selection coefficients could be blocked together. The frequency distributions for blocks longer than the length of a codon (4 bp and above) of each population were produced, and Kruskal–Wallis and Kolmogorov–Smirnov tests were carried out using GraphPad Prism version 7.0.0 for Mac OS X.
Selection of Block Subsets from Double-Drug–Evolved Population 428E3.
Population E3 had evolved in double-drug conditions and could be measured in four different growth conditions: No antibiotic, metronidazole, clarithromycin, and clarithromycin + metronidazole. We wanted to track the performance of genetic variants in these different environments to measure the repeatability of the experimental measurements of selection coefficients, and to identify environment-dependent fitness effects. However, the high rates of recombination generated variation in the size and number of blocks, even during the course of the transfers for the fitness assay. This precluded an automated, large-scale comparison of the performance blocks between treatments for several reasons. First, a single block in one treatment might have broken into several smaller blocks in another treatment, and differences in the selection coefficients calculated for the blocks could be due to different genetic variants or environment-specific effects of the same variants. Second, blocks may contain multiple genes, with HGT variants in each of those genes and it would be difficult determine that the same variant is driving a different fitness outcome for the block across different environments. Third, while a block or particular genetic variant could have a selection coefficient calculated in one treatment, it might not be present in another treatment, due to strong selection pressures causing its early fixation or loss. As described above, only variants present at each time point at a frequency greater than 0 or less than 1 were included in the set of variants for which selection coefficients were calculated. To overcome the limitations, we focused on subsets of the blocks where we were confident that we could identify the variant that drove the observed fitness coefficient. To check the replicability of our measurements across different experiments, we correlated the calculated fitness values for individual genetic variants for different fitness assays of the same population (SI Appendix, Fig. S7). This revealed a high level of repeatability (population E3, R = 0.82, P < 2.2e−16; population F1, R = 0.96, P < 2.2e−16). However, we thought that this correlation, considering each HGT variant, would overestimate the strength of the relationship between the two measurements since many loci are linked into blocks. The ideal comparison would be to compare average “s” values for the same blocks in two repeat measurements of the same population; however, blocks vary across repeats, and it could be that having a single different variant in a block could alter its fitness trajectory. To overcome this problem, we found a conservative set of blocks that were each likely to have the same genetic variant driving fitness. To do this we identified 22 blocks that each had a loss-of-function mutation—either a frameshift or early stop codon in the reading frame of a gene (Dataset S2). We assumed that the fitness trajectory of these blocks would be dominated by the loss of function of this gene, and we plotted the correlation of the fitness effects of these mutants as a more conservative estimate of the repeatability of the measurement of blocks across replicates.
Supplementary Material
Acknowledgments
This work was supported by Australia Research Council Future Fellowship FT170100441 (M.J.M.) and National Health Medical Research Council Ideas Grant APP1186140 (M.J.M. and T.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. I.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119010119/-/DCSupplemental.
Data Availability
Raw sequencing reads used to generate the data in Figs. 2 and 3 have been deposited in National Center for Biotechnology Information under the Bioproject ID PRJNA720176. Genetic variant blocks are provided as Datasets S1–S3. Custom scripts used for bespoke analyses are available at GitHub (https://github.com/woodlaur189).
References
- 1.Timmis J. N., Ayliffe M. A., Huang C. Y., Martin W., Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Gal-Mor O., Finlay B. B., Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell. Microbiol. 8, 1707–1719 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Lawrence J. G., Ochman H., Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. U.S.A. 95, 9413–9417 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochman H., Lawrence J. G., Groisman E. A., Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Koonin E. V., Makarova K. S., Aravind L., Horizontal gene transfer in prokaryotes: Quantification and classification. Annu. Rev. Microbiol. 55, 709–742 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin E. V., Horizontal gene transfer: Essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000 Res. 5, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher Y., et al. , Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37, 283–328 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Baltrus D. A., Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Sorek R., et al. , Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449–1452 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Cohen O., Gophna U., Pupko T., The complexity hypothesis revisited: Connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481–1489 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Cvijović I., Nguyen Ba A. N., Desai M. M., Experimental studies of evolutionary dynamics in microbes. Trends Genet. 34, 693–703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang G. I., et al. , Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good B. H., McDonald M. J., Barrick J. E., Lenski R. E., Desai M. M., The dynamics of molecular evolution over 60,000 generations. Nature 551, 45–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald M. J., Rice D. P., Desai M. M., Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531, 233–236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leu J. Y., Chang S. L., Chao J. C., Woods L. C., McDonald M. J., Sex alters molecular evolution in diploid experimental populations of S. cerevisiae. Nat. Ecol. Evol. 4, 453–460 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Cooper T. F., Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biol. 5, e225 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smillie C. S., et al. , Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Hehemann J. H., et al. , Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Schloissnig S., et al. , Genomic variation landscape of the human gut microbiome. Nature 493, 45–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garud N. R., Good B. H., Hallatschek O., Pollard K. S., Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol. 17, e3000102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendall M. L., et al. , Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 10, 1589–1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barroso-Batista J., et al. , The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 10, e1004182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitz P., Blokesch M., Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 37, 336–363 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J., The evolutionary advantage of recombination. Genetics 78, 737–756 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller H. J., The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 (1964). [DOI] [PubMed] [Google Scholar]
- 26.Fisher R. A., The Genetical Theory of Natural Selection (Oxford University Press, Oxford, UK, 1930). [Google Scholar]
- 27.Takeuchi N., Kaneko K., Koonin E. V., Horizontal gene transfer can rescue prokaryotes from Muller’s ratchet: Benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 4, 325–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods L. C., et al. , Horizontal gene transfer potentiates adaptation by reducing selective constraints on the spread of genetic variation. Proc. Natl. Acad. Sci. U.S.A. 117, 26868–26875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slomka S., et al. , Experimental evolution of Bacillus subtilis reveals the evolutionary dynamics of horizontal gene transfer and suggests adaptive and neutral effects. Genetics 216, 543–558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baltrus D. A., Guillemin K., Phillips P. C., Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62, 39–49 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Utnes A. L., et al. , Growth phase-specific evolutionary benefits of natural transformation in Acinetobacter baylyi. ISME J. 9, 2221–2231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu H. Y., Sprouffske K., Wagner A., Assessing the benefits of horizontal gene transfer by laboratory evolution and genome sequencing. BMC Evol. Biol. 18, 54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peabody V G. L., Li H., Kao K. C., Sexual recombination and increased mutation rate expedite evolution of Escherichia coli in varied fitness landscapes. Nat. Commun. 8, 2112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Power J. J., et al. , Adaptive evolution of hybrid bacteria by horizontal gene transfer. Proc. Natl. Acad. Sci. U.S.A. 118, e2007873118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodwin A., et al. , Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28, 383–393 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Jeong J. Y., et al. , Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182, 5082–5090 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baltrus D. A., Guillemin K., Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol. Lett. 255, 148–155 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Toprak E., et al. , Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 44, 101–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chevereau G., et al. , Quantifying the determinants of evolutionary dynamics leading to drug resistance. PLoS Biol. 13, e1002299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perron G. G., Zasloff M., Bell G., Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci. 273, 251–256 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spagnolo F., Rinaldi C., Sajorda D. R., Dykhuizen D. E., Evolution of resistance to continuously increasing streptomycin concentrations in populations of Escherichia coli. Antimicrob. Agents Chemother. 60, 1336–1342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Björkholm B., et al. , Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 98, 14607–14612 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichman H. A., Badgett M. R., Scott L. A., Boulianne C. M., Bull J. J., Different trajectories of parallel evolution during viral adaptation. Science 285, 422–424 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Colosimo P. F., et al. , Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Tenaillon O., et al. , The molecular diversity of adaptive convergence. Science 335, 457–461 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Hirata K., et al. , Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J. Gastroenterol. Hepatol. 25 (suppl. 1), S75–S79 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Barrick J. E., Kauth M. R., Strelioff C. C., Lenski R. E., Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27, 1338–1347 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loftie-Eaton W., et al. , Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 1, 1354–1363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bottery M. J., Wood A. J., Brockhurst M. A., Adaptive modulation of antibiotic resistance through intragenomic coevolution. Nat. Ecol. Evol. 1, 1364–1369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Björkman J., Nagaev I., Berg O. G., Hughes D., Andersson D. I., Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287, 1479–1482 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Nagaev I., Björkman J., Andersson D. I., Hughes D., Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40, 433–439 (2001). [DOI] [PubMed] [Google Scholar]
- 52.San Millan A., et al. , Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croucher N. J., Harris S. R., Barquist L., Parkhill J., Bentley S. D., A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 8, e1002745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mell J. C., Lee J. Y., Firme M., Sinha S., Redfield R. J., Extensive cotransformation of natural variation into chromosomes of naturally competent Haemophilus influenzae. G3 (Bethesda) 4, 717–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Messer P. W., Petrov D. A., Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28, 659–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bubendorfer S., et al. , Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat. Commun. 7, 11995 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin E. A., et al. , Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5, e1000337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuipers E. J., Israel D. A., Kusters J. G., Blaser M. J., Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J. Bacteriol. 180, 2901–2905 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller H., Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932). [Google Scholar]
- 60.Kondrashov A. S., Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440 (1988). [DOI] [PubMed] [Google Scholar]
- 61.Redfield R. J., Evolution of bacterial transformation: Is sex with dead cells ever better than no sex at all? Genetics 119, 213–221 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koonin E. V., Makarova K. S., Wolf Y. I., Krupovic M., Evolutionary entanglement of mobile genetic elements and host defence systems: Guns for hire. Nat. Rev. Genet. 21, 119–131 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Sakoparnig T., Field C., van Nimwegen E., Whole genome phylogenies reflect the distributions of recombination rates for many bacterial species. eLife 10, e65366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shapiro B. J., et al. , Population genomics of early events in the ecological differentiation of bacteria. Science 336, 48–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeuchi N., Cordero O. X., Koonin E. V., Kaneko K., Gene-specific selective sweeps in bacteria and archaea caused by negative frequency-dependent selection. BMC Biol. 13, 20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wick R. R., Judd L. M., Holt K. E., Deepbinner: Demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLOS Comput. Biol. 14, e1006583 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fischer W., et al. , Strain-specific genes of Helicobacter pylori: Genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 38, 6089–6101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Darling A. E., Mau B., Perna N. T., progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5, e11147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Page A. J., et al. , Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stecher G., Tamura K., Kumar S., Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyer D., Buchta C., Proxy: Distance and similarity measures. https://cran.r-project.org/web/packages/proxy/proxy.pdf (Accessed 8 June 2021).
- 79.Brynildsrud O., Bohlin J., Scheffer L., Eldholm V., Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 17, 238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads used to generate the data in Figs. 2 and 3 have been deposited in National Center for Biotechnology Information under the Bioproject ID PRJNA720176. Genetic variant blocks are provided as Datasets S1–S3. Custom scripts used for bespoke analyses are available at GitHub (https://github.com/woodlaur189).