Significance
Preclinical studies of metastatic melanoma treated with targeted therapeutics have suggested that alternating periods of treatment and withdrawal might delay the onset of resistance. This has been attributed to drug addiction, where cells lose fitness upon drug removal due to the resulting hyperactivation of mitogen-activated protein (MAP) kinase signaling. This study presents evidence that the intermittent treatment response can also be explained by the resensitization of cells following drug removal and enhanced cell loss upon drug rechallenge. Resensitization is accompanied by adaptive transcriptomic switching and occurs despite the sustained expression of resistance genes throughout the intermittent treatment.
Keywords: melanoma, cancer, drug resistance, intermittent treatment, encorafenib
Abstract
Patients with melanoma receiving drugs targeting BRAFV600E and mitogen-activated protein (MAP) kinase kinases 1 and 2 (MEK1/2) invariably develop resistance and face continued progression. Based on preclinical studies, intermittent treatment involving alternating periods of drug withdrawal and rechallenge has been proposed as a method to delay the onset of resistance. The beneficial effect of intermittent treatment has been attributed to drug addiction, where drug withdrawal reduces the viability of resistant cells due to MAP kinase pathway hyperactivation. However, the mechanistic basis of the intermittent effect is incompletely understood. We show that intermittent treatment with the BRAFV600E inhibitor, LGX818/encorafenib, suppresses growth compared with continuous treatment in human melanoma cells engineered to express BRAFV600E, p61-BRAFV600E, or MEK2C125 oncogenes. Analysis of the BRAFV600E-overexpressing cells shows that, while drug addiction clearly occurs, it fails to account for the advantageous effect of intermittent treatment. Instead, growth suppression is best explained by resensitization during periods of drug removal, followed by cell death after drug readdition. Continuous treatment leads to transcriptional responses prominently associated with chemoresistance in melanoma. By contrast, cells treated intermittently reveal a subset of transcripts that reverse expression between successive cycles of drug removal and rechallenge and include mediators of cell invasiveness and the epithelial-to-mesenchymal transition. These transcripts change during periods of drug removal by adaptive switching, rather than selection pressure. Resensitization occurs against a background of sustained expression of melanoma resistance genes, producing a transcriptome distinct from that of the initial drug-naive cell state. We conclude that phenotypic plasticity leading to drug resensitization can underlie the beneficial effect of intermittent treatment.
The discovery that nearly half of melanomas harbor activating mutations in the protein kinase BRAF led to a breakthrough in the treatment of metastatic melanoma (1–3). Inhibitors targeting the most prevalent BRAF mutation, BRAFV600E/K, or its downstream targets, mitogen-activated protein (MAP) kinase kinases 1 and 2 (MEK1/2), show clinical benefit in about 65% of patients with BRAFV600E/K-positive melanoma when administered continuously (4–6). However, resistance invariably develops, limiting median overall survival to ∼2 y (7, 8). Many resistance mechanisms reactivate the MAPK pathway providing a growth advantage in the presence of an inhibitor. Known mechanisms include BRAFV600E amplification, alternative splicing of BRAFV600E, and other oncogenic mutations in the BRAF pathway (e.g., MEK2C125S and NRASQ61K) (7–14). Adaptive resistance also occurs in the absence of genomic alterations and can involve transcriptional changes through epigenetic mechanisms that promote the epithelial-to-mesenchymal transition (EMT), melanocyte dedifferentiation, and neural crest stem cell–like reemergence (15–26).
An emerging body of evidence has suggested that intermittent dosing schedules, in which periods of treatment with targeted therapeutics are interrupted by periods of drug removal, might have advantages over continuous treatment (2, 27, 28). Preclinical studies with patient-derived xenograft (PDX) melanomas or xenografts from established human melanoma cell lines showed that intermittent dosing can delay drug resistance and tumor growth compared with continuous dosing (29–32). Clinical reports and a phase 2 clinical trial have shown dozens of cases where patients with melanoma develop resistance and progress when treated with BRAF or MEK1/2 inhibitors continuously, but then show further response when retreated after a drug holiday period (32–37). By contrast, phase 2 trials of intermittent dosing with BRAF and MEK inhibitor combinations showed worse progression-free survival and no difference in overall survival compared with continuous treatment (38, 39). The reasons for variability in patient responses and trial outcomes are unknown and may reflect an incomplete understanding of mechanisms underlying the response to intermittent treatment.
The current model explaining the beneficial response to intermittent treatment postulates the importance of drug addiction. Here, drug removal allows for the hyperactivation of MAPK signaling, which in turn leads to cell death or cell cycle arrest (2, 29–31, 40–42). Intermittent scheduling is thought to alternate between selection pressure against drug-sensitive cells during periods of drug treatment and selection against drug-resistant cells during periods of drug withdrawal. However, there is limited evidence that patient or xenograft tumors significantly regress when the drug is withdrawn, as predicted by the drug addiction model. Instead, xenograft tumors usually increase in volume with drug withdrawal and decrease volume after drug rechallenge (29, 43, 44). This raises the possibility that other mechanisms besides drug addiction may contribute to improved outcomes seen with intermittent treatment.
Here we use an in vitro strategy to examine cell autonomous responses of metastatic melanoma cells to intermittent treatment with the BRAF inhibitor, LGX818/encorafenib. Like vemurafenib and dabrafenib, LGX818 acts as a type I1/2 BRAF inhibitor (3), but its intermittent scheduling response has not been extensively examined. We report that an intermittent schedule with LGX818 substantially lowers cell viability compared with continuous treatment, in a manner that correlates with the degree of MAPK pathway activation. Both drug addiction following prolonged LGX818 treatment and drug resensitization following withdrawal can be observed over the multicycle time course. However, cell loss is greatest during periods of drug rechallenge, indicating that resensitization is the dominant mechanism underlying the efficacy of the intermittent schedule in this model. Transcriptome profiling through cycles of drug treatment and withdrawal reveals that resensitization is a reversible process that involves adaptive switching between states of drug resistance and drug sensitivity. Importantly, the transcriptome of the resensitized state can be distinguished from that of the initial, drug-sensitive state of naive cells and occurs against a background of sustained elevation of MAPK signaling and known resistance mechanisms. Genes controlling adaptive switching between cell states may be useful targets to delay the onset of resistance in melanoma.
Results
BRAFV600E Amplification Confers Resistance to BRAFV600E and MEK1/2 Inhibitors.
In order to generate cells with amplified BRAF/MAPK signaling, BRAFV600E was overexpressed in a human metastatic melanoma cell line (WM239A) under the control of a cumate-inducible promoter (SI Appendix, Fig. S1A). The oncogene was engineered as a fusion with green fluorescent protein (GFP) and used to confirm expression across the stable cell population after optimizing induction time and cumate concentration (SI Appendix, Fig. S1 B–D). A self-cleaving T2A sequence ensured complete separation of GFP from BRAFV600E following expression (SI Appendix, Fig. S1E).
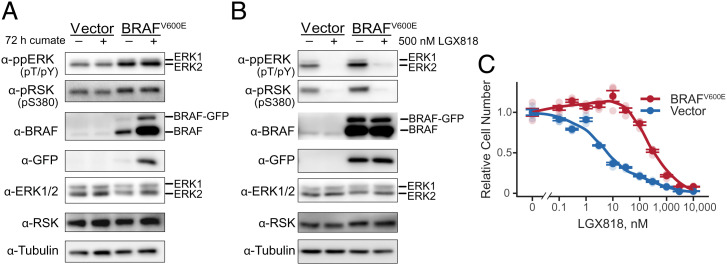
Induction of BRAFV600E increased the levels of active, phosphorylated ERK1/2 (ppERK), as measured by anti-ppERK immunoreactivity (Fig. 1A). Elevated MAPK signaling was confirmed by phosphorylation of the ERK substrate, RPS6KA (ppRSK, Fig. 1A). Both ERK and RSK phosphorylation were strongly blocked by 500 nM LGX818 (Fig. 1B). Partial BRAFV600E expression and pathway activation was apparent even in the absence of cumate, which reflected leakiness of the expression vector following selection (Fig. 1A and SI Appendix, Figs. S1 B and D and S2 A and B).
Fig. 1.
Characterization of melanoma cells with BRAFV600E amplification. (A) WM239A metastatic melanoma cells stably overexpressing BRAFV600E or empty vector were either untreated or induced with cumate for 72 h, monitoring phosphorylated ERK and RSK, total ERK and RSK, and BRAF by Western blotting. GFP and tubulin are controls, respectively, for inducible vector expression and total protein loading. (B) WM239A cells overexpressing BRAFV600E or empty vector were cumate induced for 72 h, then treated with 500 nM LGX818 or dimethylsulfoxide carrier for 2 h, and analyzed by Western blotting as in A. (C) WM239A-BRAFV600E cells were induced with cumate for 72 h, reseeded into 96 wells, and treated for 72 h with varying concentrations of LGX818. Cell numbers were measured using the CellTiter-Glo 2.0 assay, plotting mean ± SEM (n = 4) in dark symbols and individual measurements in light symbols.
Dose–response experiments were used to measure the effect of BRAFV600E overexpression on drug resistance. Cells with amplified BRAFV600E increased the half-maximal inhibitory concentration (IC50) by 50-fold over control cells harboring empty vector (Fig. 1C). Similar increases in IC50 were seen in dose–response measurements with MEK162/binimetinib (SI Appendix, Fig. S2D). Thus, BRAFV600E overexpression strongly increased chemoresistance toward BRAFV600E and MEK inhibitors.
Comparison of Intermittent and Continuous Treatment.
In order to compare intermittent and continuous treatment schedules, cells were seeded in 96-well dishes and cultured for 4 wk, monitoring cell viability at the end of each week (Fig. 2A). Intermittent time courses followed a schedule of 7 d on the drug followed by 7 d off the drug, where cells were treated with 500 nM LGX818 during weeks 1 and 3 and the drug was removed in weeks 2 and 4. In parallel, cells were treated continuously with 500 nM LGX818 over the entire 4-wk period.
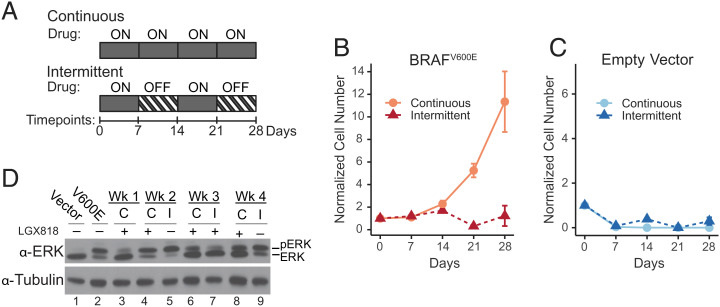
Fig. 2.
Intermittent treatment inhibits cell expansion compared with continuous treatment. (A) Cells were cumate induced for 72 h, reseeded in 96 wells, and cultured for 28 d under continuous or intermittent treatment conditions with 500 nM LGX818. The intermittently treated cells followed a schedule with 7 d on LGX818 and 7 d off, changing media on days 3, 5, and 7 of each week. At the end of each week, cell numbers were quantified using CellTiter-Glo assays and normalized to the initial number of cells seeded. Cell numbers (mean ± SEM) measured at each time point for (B) WM239A-BRAFV600E cells (n = 5 or 6) and (C) WM239A empty vector cells (n = 6) are shown for continuous and intermittent treatments. (D) Western blots of lysates separated by low-bis sodium dodecyl-sulfate polyacrylamide gel electrophoresis show ERK phosphorylation in cells expressing empty vector (lane 1), BRAFV600E (lane 2), and BRAFV600E cells treated continuously “C” or intermittently “I” across the 4 wk with 500 nM LGX818.
Cells treated continuously grew slowly for the first 2 wk after which a drug-resistant population emerged (Fig. 2B). By contrast, cells treated intermittently yielded cell numbers that were initially similar to the continuous experiment for the first 2 wk, but then declined after drug rechallenge in week 3 (Fig. 2B). Thus, intermittent treatment inhibited the cell expansion seen with continuous treatment. Control cells expressing empty vector were strongly suppressed with either treatment schedule (Fig. 2C).
In order to compare responses to BRAFV600E against other oncogenes associated with resistance in melanoma, cells were engineered to individually express MEK2C125S, EGFRL858R, NRASQ61K, or the p61-BRAFV600E splice variant (8–11). Like BRAFV600E, cells expressing p61-BRAFV600E or MEK2C125S substantially increased the phosphorylation of ERK and RSK as well as IC50 with LGX818 or MEK162 (SI Appendix, Fig. S2 A–D). Cells expressing p61-BRAFV600E displayed the greatest resistance to LGX818, with 10-fold higher IC50 than that of BRAFV600E or MEK2C125S. By contrast, EGFRL858R or NRASQ61K only modestly increased ERK or RSK phosphorylation and IC50 (SI Appendix, Fig. S2 A–D). ERK and RSK phosphorylation were suppressed in all cells by 500 nM LGX818, MEK162, or the inhibitor combination (SI Appendix, Fig. S2 E–G).
During continuous treatment with LGX818, cells expressing MEK2C125S or p61-BRAFV600E remained static for the first week, after which a resistant population emerged (SI Appendix, Fig. S3 A and B). Like BRAFV600E, cells expressing MEK2C125S declined with intermittent treatment and remained inhibited for the duration of the time course (SI Appendix, Fig. S3A). Intermittent treatment with either LGX818 or MEK162 only partially inhibited growth of cells expressing p61-BRAFV600E compared with continuous treatment (SI Appendix, Fig. S3 B and C). However, cells were substantially reduced by intermittent treatment with a combination of both LGX818 + MEK162 (SI Appendix, Fig. S3D), indicating that strong inhibition of the MAPK pathway during periods of drug addition is important for maximal efficacy. Cells expressing EGFRL858R or NRASQ61K were strongly repressed by either intermittent or continuous treatment, both of which effectively inhibited expansion over the 4 wk time course (SI Appendix, Fig. S3 E and F). Thus, intermittent treatment showed greater efficacy compared with continuous treatment, but only in cells with the highest levels of ERK activation and the strongest resistance to the inhibitor.
Studies invoking the drug addiction model for intermittent dosing have reported pronounced elevation of ppERK with BRAFV600E amplification (29). Therefore, we characterized the corresponding responses to continuous or intermittent treatment in our BRAFV600E cell system. Western blots showed that ppERK increased with BRAFV600E induction and decreased after the first week in the presence of LGX818, then increased with subsequent weeks of continuous treatment as resistant cell populations emerged (Fig. 2D, lanes 2–4, 6, and 8). By comparison, ppERK was elevated after weeks 2 and 4 of drug removal (lanes 5 and 9), decreasing when the drug was added back during week 3 (Fig. 2D, lane 7). The findings show that continuous treatment with LGX818 maintained levels of ERK activity that supported viability in the presence of the drug, while drug removal during intermittent treatment elevated ERK to levels equal to or greater than seen with the initial induction of BRAFV600E (Fig. 2D, lanes 2, 5, and 9).
Intermittent Treatment Reverses Drug Addiction and Resensitizes Cells to LGX818.
To better understand the effect of the intermittent treatment schedule on drug sensitivity and drug addiction, we characterized the LGX818 dose–response of BRAFV600E cells collected at the end of each week of a continuous or intermittent time course. In this experiment, cells that were cumate induced to express BRAFV600E showed greater drug resistance (IC50 = 130 nM) than cells with empty vector (IC50 = 5 nM) (Fig. 3A). After continuous treatment with 500 nM LGX818 for 7 d, the IC50 increased to 860 nM and remained sustained at 1,000 to 1,200 nM over successive weeks (Fig. 3A). Continuous treatment also resulted in drug addiction, as evidenced by a 30 to 45% reduction in cell numbers at 0 nM LGX818 relative to their maximum levels at 100 nM (Fig. 3A). Thus, drug addiction accompanied resistance to LGX818 in our experimental system, consistent with previous models of resistance to MAPK pathway inhibitors (29, 30, 41–43).
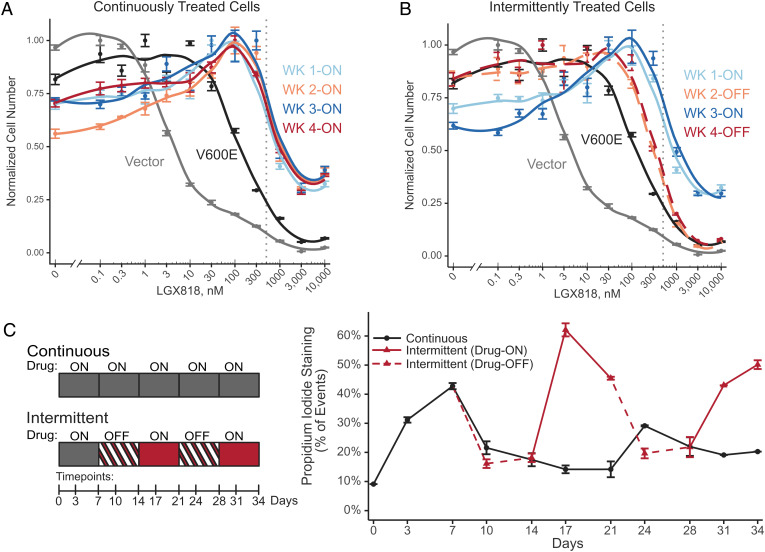
Fig. 3.
Intermittent treatment resensitizes cells to LGX818. WM239A-BRAFV600E cells were cumate induced for 72 h and seeded in 10-cm dishes followed by (A) continuous or (B) intermittent treatments with 500 nM LGX818. At the end of each week, cells were trypsinized, reseeded into 96 wells, and treated for 72 h with varying concentrations of LGX818 (n = 4). Cells with empty vector or BRAFV600E were also assayed for their dose–response to LGX818. Both A and B display the same data for empty vector, BRAFV600E, and week 1 continuous, for ease of comparison. The gray dotted vertical line indicates 500 nM LGX818. (C) Cell death across continuous and intermittent schedules was measured on days 3 and 7 of each week for 5 wk. Adherent and nonadherent cells were collected together, stained with PI, and analyzed by flow cytometry. The media was changed on days 3, 5, and 7 of each week. During the media change on day 5, nonadherent cells were collected by centrifugation of the conditioned media and returned to the dish for flow cytometry analysis on day 7. Measurements report average percentage of PI-positive cells (n = 2 independent experiments), and error bars represent the range.
By contrast, cells treated intermittently varied in their dose–response to LGX818, depending on whether they were collected after periods of drug addition or drug removal. Removing LGX818 in week 2 reduced the IC50 to 300 nM, a threefold decrease from cells treated with the drug in week 1 (Fig. 3B). At the same time, removing LGX818 decreased the extent of drug addiction, as shown by the recovery of cell viability at 0 nM LGX818 (Fig. 3B). Rechallenge with LGX818 in week 3 reversed this behavior, increasing both IC50 and drug addiction back to the levels seen with continuous treatment. Removing LGX818 in week 4 decreased the IC50 and decreased drug addiction back to levels comparable to week 2. Therefore, each cycle of drug removal switched cells to a state with resensitization to the BRAF inhibitor and each cycle of drug addition produced a state of drug addiction.
Cell Loss during Intermittent Treatment Primarily Involves Resensitization after Drug Removal.
Conceivably, either drug addiction or drug resensitization could account for the loss of cell viability observed with intermittent treatment. In order to explore the contribution from each mechanism, we quantified cell death at different times during periods of drug addition or removal. On one hand, if drug addiction were the dominant mechanism, cell loss should be highest during drug-off weeks when ERK hyperactivation would be predicted to promote cell death. On the other hand, if drug resensitization were more important, cell loss should be highest during drug-on weeks when cells would be susceptible to rechallenge with LGX818.
To examine this, cells were seeded in 10-cm dishes and treated with continuous or intermittent schedules over 5 wk. Samples were taken on days 3 and 7 of each week, collecting all adherent and floating cells for analysis of cell viability by propidium iodide (PI) staining using flow cytometry (Fig. 3C). Propidium iodide was used as a marker of cell death, which is caused by parthanatos following drug withdrawal in drug-addicted melanoma cells (30). Continuous treatment led to substantial cell loss during week 1, which fell to lower levels over successive weeks (Fig. 3C) with the expansion of cells able to persist or grow in the presence of LGX818 (Fig. 2B). Parallel flow cytometry measurements showed BRAFV600E expression increased over the first 2 wk of continuous treatment, then maintained over successive weeks (SI Appendix, Fig. S4).
In cells treated intermittently, the percentages of PI-positive cells during drug-off periods (days 10, 14 and 24, 28) were comparable to those in cells treated continuously (days 10 through 28). However, cell death dramatically increased during periods of drug rechallenge (days 17, 21 and 31, 34) where the percentage of PI-positive cells reached levels as high as 60% (Fig. 3C). These results indicate that the reduction in cell fitness with intermittent treatment is best explained by the occurrence of resensitization during periods of drug withdrawal, followed by cell loss when cells are rechallenged with LGX818. This was consistent with the measurements of cell numbers during intermittent treatment, which decreased only when the drug was readded in week 3 (Fig. 2B).
Transcriptomic Responses to Continuous and Intermittent Treatment.
In order to explore gene expression changes that accompany drug resensitization, RNA sequencing (RNA-seq) was used to examine cells expressing BRAFV600E after each week of intermittent or continuous treatment with LGX818 (Fig. 4A). Cell viability measurements conducted in parallel matched those observed previously, where resistant cells emerged after 2 wk of continuous treatment with LGX818 and intermittent treatment delayed outgrowth (SI Appendix, Fig. S5). Datasets were also collected on cells with empty vector, BRAFV600E induced for 72 h, and BRAFV600E induced for 72 h, then treated with 500 nM LGX818 for 20 h. These were performed as controls to measure gene expression responses to acute activation or inhibition of the MAPK pathway without long-term selection. Each condition was analyzed in triplicate, except for duplicates of the week 4 continuous condition (Datasets S1 and S2).
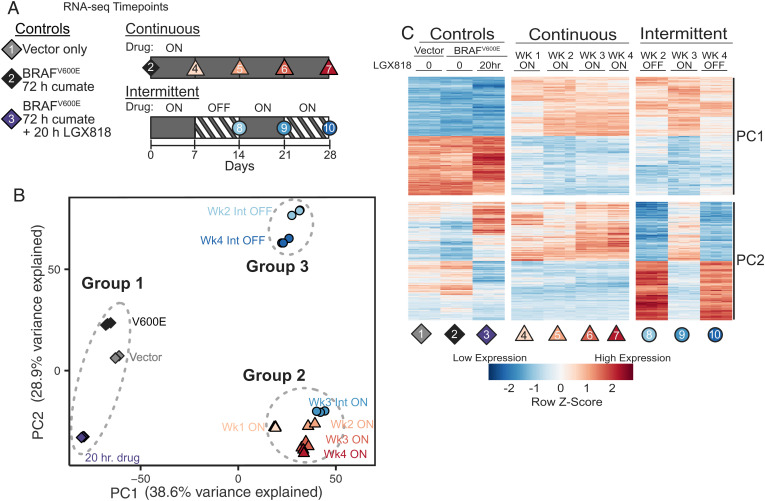
Fig. 4.
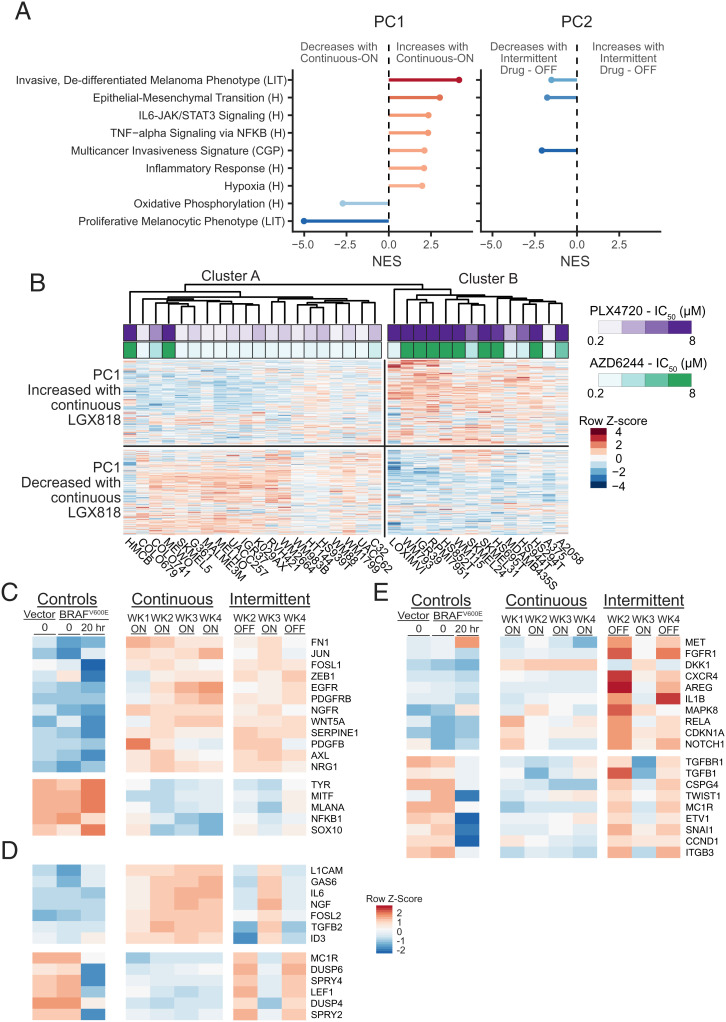
Transcriptomic profiling of continuously and intermittently treated cells. (A) Schematic of RNA samples collected from WM239A-BRAFV600E cells comparing continuous and intermittent drug treatment with 500 nM LGX818. Each condition was collected in biological triplicates, except for week 4 continous (sample 7), which was collected in duplicates, for a total of 29 samples. (B) Principal component analysis (PCA) of the 6,000 highest variance genes. PC1 and PC2 account for almost 70% of the variance in these genes. The 10 different experimental conditions visibly cluster into three separate groups. (C) Heatmap of the 400 genes most positively or negatively associated with PC1 or PC2. Each row is mean centered and scaled to unit variance.
Principal component analysis showed that nearly 70% of the variance across the samples could be accounted for by the first two principal components. Plotting PC1 against PC2 separated the samples into one of three easily identifiable groups (Fig. 4B). One group included cells with empty vector, 72-h induced BRAFV600E, and 72 h BRAFV600E + 20 h LGX818 (Fig. 4B, group 1). A second group contained all cells that were continuously treated (weeks 1 through 4), as well as cells from intermittent week 3, when the drug was reintroduced (Fig. 4B, group 2). The third group included cells treated intermittently in weeks 2 and 4, when the drug was removed (Fig. 4B, group 3). The analysis revealed a striking effect of intermittent treatment, in which cells with the drug removed in weeks 2 and 4 were grouped together and well separated from cells with the drug readded in week 3.
Significantly, the separation between groups 1 and 2 mainly occurred along the PC1 axis, suggesting transcriptomic changes resulting from long-term treatment with LGX818. The separation between cells collected in successive weeks of intermittent treatment occurred primarily along the PC2 axis, corresponding to reversible movement between groups 2 and 3 (Fig. 4B). This suggested that PC2 largely reflects reversible transcriptomic changes accompanying drug removal and drug readdition. A heatmap of the genes highly contributing to either principal component showed that most PC1 gene expression changes after week 1 were sustained in subsequent weeks with either continuous or intermittent treatment (Fig. 4C and Dataset S3). In contrast, PC2 transcripts reversibly switched in expression between the drug-on and drug-off periods during the intermittent time course (Fig. 4C and Dataset S3). Analysis of the complete gene expression datasets showed strong correlations between cells with drug readded during week 3 and those continuously treated throughout weeks 1 through 4 (SI Appendix, Fig. S6). This confirmed that rechallenging cells with LGX818 recapitulates the transcriptomic state of continuously treated cells. Thus, reversible changes in the transcriptome accompany the ability of cells to switch between drug-resistant and -resensitized states during the intermittent time course.
Transcriptome Changes Associated with Drug Resistance.
We asked whether the transcripts altered in response to continuous LGX818 treatment might reflect genes that function in cancer drug resistance, using gene set enrichment analysis (45, 46). The PC1 genes that increased expression with prolonged treatment were enriched in molecular signatures associated with resistance to BRAFV600E and MEK inhibitors, including markers of the EMT, NF-κB signaling, inflammatory markers, and hypoxia, while genes that decreased expression were enriched in gene sets associated with a proliferative, melanocytic phenotype (Fig. 5A). These signatures are characteristic of the invasive, dedifferentiated melanoma phenotype associated with melanoma malignancy (15–17, 23). The PC2 genes that reversibly decreased upon drug removal were negatively associated with gene signatures characteristic of the invasive phenotype, suggesting partial reversal of invasion/EMT-like processes.
Fig. 5.
Reversible transcriptome changes are associated with EMT-like responses. (A) Gene set enrichment analysis (GSEA) was performed on genes associated with PC1 and PC2. The gene loadings for each PC were used as the ranks. Shown are the normalized enrichment scores (NESs) for selected gene sets matched to PC1 with significant false discovery rate (FDR)-adjusted P values (q < 0.002). The NESs for the same gene sets matched to PC2 are shown for cases where the FDR-adjusted P value is less than q < 0.05. Gene sets were from the Molecular Signature Database for Hallmark “H,” Chemical and Genetic Perturbations “CGP,” or derived from the literature “LIT.” (B) Clustered heatmap of RNA-seq data of BRAFV600 mutant melanomas in the CCLE. Only the 800 genes identified as PC1 associated in Fig. 4 are shown and used to cluster the cell lines. Cluster A mostly contained cell lines that were sensitive to the BRAF inhibitor, PLX4720, or the MEK inhibitor, AZD6244 (lower IC50). Cluster B mostly contained cell lines with greater resistance to either drug (higher IC50). The two clusters show differential expression of genes that either increased or decreased with continuous treatment in our RNA-seq dataset. (C–E) Heatmaps of RNA-seq data corresponding to literature-curated genes that have been implicated in melanoma drug resistance or sensitivity. (C) Known resistance genes that changed in response to continuous treatment and did not change with drug removal, predicting sustained resistance over the intermittent time course. (D) Resistance genes showing reversible expression between drug-on and drug-off weeks that changed in a manner predicting resensitization upon drug removal. (E) Resistance genes showing reversible expression between drug-on and drug-off weeks, but predicting higher resistance during weeks of drug removal in the intermittent time course.
We next assessed how the expression levels of genes responsive to continuous LGX818 treatment correlated with drug resistance across human melanoma cell lines. RNA-seq data from 34 BRAFV600 melanoma cell lines available from the cancer cell line encyclopedia (CCLE) (47) were used to perform hierarchical clustering based on the expression levels of transcripts highly associated with PC1. These cell lines formed two main clusters, denoted cluster A and cluster B in Fig. 5B. The gene expression patterns of cell lines in cluster A resembled transcript levels in our drug-naive cells (group 1 in Fig. 4B), while cell lines in cluster B showed greater similarly to our cell system after long-term treatment with LGX818 (groups 2 and 3 in Fig. 4B). The degree of resistance among CCLE cell lines, measured by IC50 for the BRAFV600E inhibitor, PLX4720, or the MEK inhibitor, AZD6244, correlated well with cluster membership, largely separating drug-sensitive cells in cluster A from drug-resistant cells in cluster B (Fig. 5B). The analysis reveals that the transcript changes associated with LGX818 resistance in WM239A cells can explain the variance in baseline IC50 across many other BRAFV600 cell lines. Therefore, the transcriptome responses to continuous treatment in our experimental model are consistent with those associated with drug resistance across melanoma cell systems.
It was noteworthy that the transcriptomes of cells in group 3, characterized by drug removal, were distinct from those of cells with empty vector or induced BRAFV600E in group 1 (Fig. 4 B and C). This implies that the resensitized cell state generated by LGX818 removal differs from the initial sensitive state of drug-naive cells. Prominent among the PC2 transcripts that switched reversibly following drug removal and subsequent rechallenge were those that were weakly responsive or in some cases nonresponsive to BRAFV600E induction in drug-naive cells (Fig. 4C). This implies that many PC2 genes normally respond weakly to MAPK signaling but acquire greater responsiveness to this pathway after a week of continuous exposure to the drug. Thus, the resensitized cell state includes new transcriptional responses that only occur after MAPK signaling is activated by drug removal.
We also compared transcript changes corresponding to known resistance genes in melanoma. To do this, we curated a list of 73 genes that were reported to control resistance toward BRAFV600E and/or MEK inhibitors in melanoma cell lines and tumors (genes and references in Dataset S4). These included signaling effectors, transcription factors, and mediators and markers of EMT-like responses, neural crest specification, and melanocyte differentiation. Forty-eight of the curated genes were significantly altered with continuous drug treatment, consistent with the emergence of resistant cell populations. For example, growth factor receptors and ligands, EMT-like and neural crest markers known to promote resistance (e.g., PDGFRB, EGFR, AXL, NGFR, and WNT5A) (12, 21, 42, 48, 49) increased with continuous drug treatment, while genes associated with differentiation to the melanocyte lineage (e.g., MITF, TYR, MLANA, and SOX10) (17, 21) decreased (Fig. 5C).
Among the curated genes associated with drug resistance in melanoma, some transcripts changed reversibly with LGX818 removal in a manner consistent with resensitization (Fig. 5D). For example, growth factors NGF and GAS6 decreased during drug-off weeks, which might predict lower signaling through their respective receptors, NGFR and AXL. Likewise, the transcription factor ID3, which promotes resistance to BRAF inhibitor (50), decreased after drug removal, while LEF1, which associated with greater sensitivity (19, 26, 51), increased. Negative feedback regulators of growth factor signaling, DUSP4/6 and SPRY2/4, reversibly increased after drug removal, consistent with MAPK pathway reactivation (52, 53).
However, the majority of resistance genes from our curated list were inconsistent with resensitization and in fact predicted sustained resistance with intermittent scheduling. These included growth factor receptors/ligands (AXL, NGFR, EGFR, PDGFRB, PDGFB, and WNT5A) and transcription factors (JUN, FOSL1/FRA1, and ZEB1), which remained elevated following drug removal, and differentiation markers (MLANA/MART1, TYR, MITF, and SOX10), which remained repressed (Fig. 5C). Paradoxically, other responsive transcripts predicted increased resistance upon drug removal. For example, FGFR1, MET, AREG, TGFB1, IL1B, ITGB3, and CSPG4 were all up-regulated during drug-off weeks (Fig. 5E), which would be expected to increase receptor signaling and diminish sensitivity to BRAF inhibitors (42, 54–60). Also included in this group were the transcription factors TWIST1, SNAI1, and RELA, which promote EMT-like responses via epigenetic repression of histone marks (61, 62), but increased when the drug was removed (Fig. 5 C and E), and SNAI2, which promotes the dedifferentiated, noninvasive phenotype (61), but increased with drug removal.
Taken together, the gene expression analyses revealed distinct molecular responses to the continuous and intermittent treatment regimens. Many transcripts altered by prolonged drug treatment corresponded to genes implicated in drug resistance in melanoma and were regulated in a manner consistent with the drug-resistant state observed with continuous treatment. Other transcripts switched expression between states of resistance and sensitivity, but only a subset of known resistance genes reversed in a manner consistent with resensitization upon drug removal.
Adaptive Responses to Intermittent Treatment.
As noted above, many transcript changes that were significant after the first 7 d of LGX818 exposure were sustained when the drug was removed in weeks 2 and 4 (Figs. 4C and 5C). Their irreversibility may be due to slow rates of reversal or to an initial selection for drug-resistant cell subpopulations. On the other hand, nearly all genes that changed reversibly during the course of intermittent treatment were found to recover the transcriptome of continuously treated cells during week 3, when the drug was readded (Figs. 4C and 5D). This implies an adaptive mechanism, responsive to epigenetic regulation. However, bulk RNA-seq cannot distinguish between mechanisms involving adaptive transitions in cells switching from drug-resistant to drug-sensitive states, and mechanisms for selection pressure for sensitive cell subpopulations within a resistant majority.
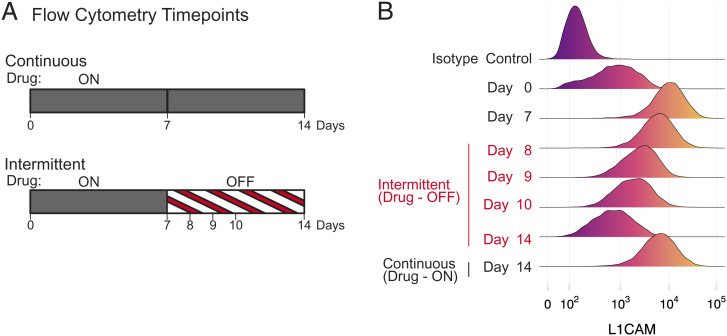
To investigate these possibilities, we examined the dynamics of expression of the neural cell adhesion protein, L1CAM. Bulk transcript levels of L1CAM were elevated in cells treated for prolonged periods with LGX818, consistent with its role in promoting EMT-like responses and as a marker for melanoma drug resistance (18, 63, 64). Its expression decreased following drug removal in a manner that was reversible in subsequent weeks of drug rechallenge and withdrawal (Fig. 5D). The changes in L1CAM protein expression at the single-cell level were measured by flow cytometry at several time points during continuous and intermittent treatment using a fluorescently labeled primary antibody (Fig. 6A). We selected these time points in order to capture the cell population dynamics corresponding to reversal of expression during early stages of drug withdrawal. In drug naive cells (day 0), many cells expressed low levels of L1CAM, some overlapping with the nonspecific isotype control (Fig. 6B). After treating cells with LGX818 for 7 d, the median expression level increased by more than 10-fold over that of drug-naive cells and remained high for 14 d of continuous drug treatment. In the intermittent schedule, L1CAM expression during the first 3 d of drug withdrawal gradually decreased in a unimodal fashion (Fig. 6B). The cells transiently expressed intermediate levels of L1CAM, which then decreased further to match the drug-naive population by the end of the week. The unimodal populations with intermediate L1CAM were inconsistent with selection pressure against a subpopulation of drug-addicted cells, which would have predicted bimodal cell populations shifting from high to low L1CAM. Therefore, the transcript responses following drug withdrawal largely follow an adaptive mechanism for transcript switching, instead of selective depletion of drug-addicted cells.
Fig. 6.
Cell resensitization is accompanied by adaptive gene expression changes in single cells, not by counterselection. (A) WM239A-BRAFV600E cells were cumate induced for 72 h, seeded in 10-cm dishes, and cultured under continuous or intermittent treatment with 500 nM LGX818. At the indicated time points, cells were fixed and stained with a fluorescently labeled primary antibody for L1CAM and then analyzed by flow cytometry. (B) Smoothed histograms of single-cell L1CAM expression by flow cytometry at the different time points. Controls using a fluorescently labeled nonspecific isotype control antibody confirmed specificity of binding to the target protein.
Discussion
Our study demonstrates that adaptive mechanisms for cell switching from drug-resistant to drug-resensitized states can explain the growth suppressive effects of treating melanoma cells intermittently with LGX818. The BRAFV600E-amplified cell model used in our study showed transcripts responsive to the drug that corresponded well with resistance genes known to function across melanoma cell lines and tumors. Importantly, when drug resistance was induced by prolonged treatment, the cells displayed both a drug addiction response after treatment and a drug resensitization response after drug withdrawal. Cell death was highest during periods of rechallenge, showing that cell viability was affected most by the resensitization to kinase inhibitor. Transcriptome analyses showed many gene expression changes that were readily reversible over the intermittent time course, nearly all which returned to levels seen in continuously treated cells when the drug was reintroduced. This indicated that the responses to intermittent treatment were not due to selection pressure, as supported by the expression dynamics of L1CAM in single-cell populations.
Previous studies have shown that drug addiction is caused by resistance mechanisms that maintain MAPK pathway signaling at submaximal levels in the presence of BRAF inhibitor, and hyperactivate MAPK signaling when the drug is removed, thereby triggering cell death or cell cycle arrest (29, 30, 40–42). Thus, it has been theorized that the intermittent treatment effect arises by alternating selection against drug-sensitive cells in the presence of BRAF inhibitor and counterselection against drug-addicted cells in the absence of the inhibitor (2). In our system, MAPK signaling was maximally activated following LGX818 withdrawal, based on increased phospho-ERK1/2 and the expression of downstream pathway targets such as DUSP and SPRY. But while drug addiction was evident, it played a secondary role in the response to intermittent treatment. In cells treated continuously, cell viability was highest at 100 nM LGX818 and at 500 nM fell to levels less than or equal to that in the absence of the drug (Fig. 3A). This may explain why drug addiction did not contribute significantly to growth suppression with intermittent treatment in our experiments, because it would have had its largest effect at drug concentrations lower than we used. While it is hard to extrapolate in vitro cell behavior to clinical outcomes, plasma concentrations of BRAF inhibitors reached in patients (65, 66) may affect the extent to which drug addiction occurs.
Our findings are consistent with a role for phenotypic plasticity in drug resistance, which has been well documented in BRAFV600E melanomas as well as other cancers (15–17). Current models postulate elevated expression of genes in rare cell populations, which enhance resistance in response to BRAF inhibitor by promoting epigenetic pathways for dedifferentiation and transcriptional reprogramming (18–26). Classes of transcripts that promote resistance include drivers of EMT and invasion (AXLhigh, WNT5Ahigh, TGFBhigh, TWISThigh, and SNAI1high) and markers of lineage development from differentiated melanocytes to neural crest stem cells (NGFRhigh, MITFlow, and SOX10low) (20–26). Exploiting the reversibility of these adaptive regulatory events during early, nonmutational phases of drug resistance has been proposed as a treatment strategy for cancer. Our findings concur and further suggest that adaptive mechanisms may underlie beneficial responses to intermittent scheduling.
Significantly, transcriptome profiling of our system revealed many resistance genes that changed with continuous drug treatment but were irreversible over the intermittent time course. These reflect adaptive responses to drug and/or the selection of cell populations able to persist and survive during the first week of LGX818 treatment. This set included most genes characteristic of the invasive, neural crest phenotype and EMT-like responses. This means that resensitization after drug removal occurred despite a large number of resistance genes that did not reverse. Altogether, only a few genes implicated in resistance both displayed reversibility and changed in a direction consistent with resensitization. Leading candidates included LEF1, which is repressed in drug resistance melanoma and strongly associated with phenotype switching to the proliferative, noninvasive state (19, 26, 51), and ID3, which is elevated in resistant cells and whose depletion in resistant cells confers resensitization (50). They did not include genes reported to regulate drug addiction in melanomas, such as JUNB, FOSL1, or CDKN1A (40, 41), consistent with a minimal influence of drug addiction on the intermittent treatment effect.
Interestingly, many transcripts that responded reversibly with each cycle of drug removal were elevated to levels well above those seen in drug-naive cells. In fact, many reversible transcripts showed little or no response to short-term induction of BRAFV600E. This suggests that genes that were not normally downstream of MAPK signaling became pathway targets after 7 d of drug treatment. Conceivably, drug treatment may have enriched a subpopulation of cells with an expanded range of transcriptional responses. Alternatively, new targets may have been triggered by the hyperactivated MAPK signaling that followed drug withdrawal. Further definition of the cellular mechanisms involved in reversible resensitization may lead to novel targets and the potential contribution of MAPK signaling thresholds to the resensitized cell state.
The partial reversal of resistance gene expression upon drug removal helped explain the partial resensitization response of only a threefold decrease in IC50. Therefore, the drug resistant state in our hands appears to represent an intermediate-to-late stage in transcriptome reprogramming, with a few rapidly reversible and many slowly reversible resistance genes (21). Conceivably, a longer period of drug withdrawal might have eventually returned cells to the initial transcriptome and sensitivity level of drug-naive cells, as observed in other studies of transcriptome dynamics (25). However, in order for intermittent treatment to block cell expansion, the amount of time needed for resensitization must be balanced against the faster cell doubling time during the period of drug removal. Therefore, partial resensitization may be a practical condition needed for a successful intermittent treatment schedule.
So far, intermittent treatment of melanoma has been unproven, with clinical outcomes arguing for and against its potential effectiveness. On one hand, two recent phase II trials revealed worse progression-free survival in patients with melanoma treated intermittently with dabrafenib + trametinib or vemurafenib + cobimetinib compared with those treated with continuous therapy, with no difference in overall survival (38, 39). On the other hand, retrospective and prospective studies of dozens of patients who progress on BRAF or MEK inhibitor have reported that more than one-third show a second clinical response after a drug holiday period (27, 33–37). Preclinical studies using xenografts have been mixed as well. While some have reported delayed emergence or complete suppression of resistant tumors using intermittent scheduling (29–32), others have observed more rapid outgrowth of tumors (43, 44). Factors that could affect whether beneficial responses are seen with intermittent treatment, in vitro or in animals, may include drug concentration relative to IC50, doubling times for resistant vs. sensitive cells, and cell specificity in signaling pathway activation. A notable report showed a substantial benefit of intermittent treatment in xenografts from a drug-resistant melanoma cell line, but only when the dosing schedule was tailored individually for each mouse (32). Optimal scheduling was established using a predator–prey model for adaptive drug therapy, which postulates expansion and loss of sensitive cells during drug-off and drug-on periods, respectively (67, 68). Early stage clinical trials are ongoing to test these intriguing concepts for melanoma, breast, and prostate cancer, and the possibility that personalized scheduling may be optimized by dynamic measurements of cancer blood markers (68–70).
In summary, our findings show that intermittent treatment can suppress cell growth and delay the emergence of drug resistance by transitioning from a drug-resistant state to a more sensitive state through adaptive transcriptional mechanisms. Significantly, this can occur against a background of many resistance genes that either fail to reverse when the drug is removed or change in a manner that would paradoxically predict increased resistance. Thus, we propose that intermittent treatment generates a distinct cellular state that accompanies resensitization, which may include transcripts not normally regulated by MAPK signaling and are triggered by higher signaling thresholds. Genes controlling resensitization may be useful targets to augment or improve the response duration of current treatment strategies for melanoma.
Materials and Methods
The human metastatic melanoma cell line, WM239A, was a kind gift from Meenhard Herlyn, Wistar Institute, Philadelphia, PA. LGX818/encorafenib and MEK162/binimetinib were obtained from Selleck Chemicals. The CellTiter-Glo 2.0 assay (Promega) was used to determine cell numbers. Detailed methods for construction of cell lines, biochemical- and cell-based assays, and RNA-seq measurements are described in detail in SI Appendix, which includes SI Appendix, SI Materials and Methods, Figs. S1–S6, and Datasets S1–S4.
Supplementary Material
Acknowledgments
We are indebted to Theresa Nahreini, Joseph Dragavon, and Dan Timmons for assistance from the Biochemistry Cell Culture Facility, the BioFrontiers Advanced Light Microscopy Core, and the BioFrontiers Computing Core at the University of Colorado, Boulder, which are supported by NIH Grants S10OD025072, S10OD021601, and S10OD012300, and P30CA046934. We also thank the Genomics Shared Resource at the University of Colorado Cancer Center, supported by NIH Grant P30CA046934. Support was provided by an ALSAM Foundation Therapeutic Innovation Grant from the University of Colorado Skaggs School of Pharmacy and NSF Integrative Graduate Education and Research Traineeship Grant 1144807 to the Interdisciplinary Quantitative Biology Program (A.J.K.), a Sponsored Research Award from Array BioPharma (S.A.S.), NIH Grant T32GM142607 (K.R.H.), and NIH Grant R35GM136392 (N.G.A.).
Footnotes
Reviewers: M.C., University of Texas Southwestern Medical Center; M.M., Huntsman Cancer Institute.
Competing interest statement: B.B. is a former employee of Array BioPharma Inc., which developed the inhibitor compounds used in this research (encorafenib and binimetinib) and provided seed grant funding for this study.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113535119/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in NCBI Gene Expression Omnibus (GSE117123). A runnable and editable version of the code used in this study can be found on code ocean (https://codeocean.com/capsule/9070543/tree/v1), and a code repository is available on GitHub at https://github.com/andykavran/Intermittent_Drug_Treatment. All other study data are included in the article and/or supporting information.
References
- 1.Davies H., et al. , Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Holderfield M., Deuker M. M., McCormick F., McMahon M., Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 14, 455–467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karoulia Z., Gavathiotis E., Poulikakos P. I., New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer 17, 676–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giugliano F., et al. , First line treatment of BRAF mutated advanced melanoma: Does one size fit all? Cancer Treat. Rev. 99, 102253 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Dummer R., et al. , Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF -mutant melanoma (COLUMBUS): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2045, 1–13 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Halle B. R., Johnson D. B., Defining and targeting BRAF mutations in solid tumors. Curr. Treat. Options Oncol. 22, 30 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Tangella L. P., Clark M. E., Gray E. S., Resistance mechanisms to targeted therapy in BRAF-mutant melanoma - A mini review. Biochim. Biophys. Acta, Gen. Subj. 1865, 129736 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Proietti I., et al. , Mechanisms of acquired BRAF inhibitor resistance in melanoma: A systematic review. Cancers (Basel) 12, 2801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prahallad A., et al. , Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Wagle N., et al. , Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, 3085–3096 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulikakos P. I., et al. , RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480, 387–390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazarian R., et al. , Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H., et al. , Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 3, 724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corcoran R. B., et al. , BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal. 3, ra84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arozarena I., Wellbrock C., Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 19, 377–391 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Boumahdi S., de Sauvage F. J., The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 19, 39–56 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Rambow F., Marine J. C., Goding C. R., Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 33, 1295–1318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambow F., et al. , Toward minimal residual disease-directed therapy in melanoma. Cell 174, 843–855.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Hugo W., et al. , Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell 162, 1271–1285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y., et al. , Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. U.S.A. 114, 13679–13684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaffer S. M., et al. , Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Widmer D. S., et al. , Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 25, 343–353 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Verfaillie A., et al. , Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 6, 6683 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song C., et al. , Recurrent tumor cell-intrinsic and -extrinsic alterations during MAPKi-induced melanoma regression and early adaptation. Cancer Discov. 7, 1248–1265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fallahi-Sichani M., et al. , Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol. Syst. Biol. 13, 905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsdale R., et al. , The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci. Signal. 8, ra82 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Dooley A. J., Gupta A., Bhattacharyya M., Middleton M. R., Intermittent dosing with vemurafenib in BRAF V600E-mutant melanoma: Review of a case series. Ther. Adv. Med. Oncol. 6, 262–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tétu P., et al. , Mitogen-activated protein kinase blockade in melanoma: Intermittent versus continuous therapy, from preclinical to clinical data. Curr. Opin. Oncol. 33, 127–132 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Das Thakur M., et al. , Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494, 251–255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong A., et al. , Exploiting drug addiction mechanisms to select against MAPKi-resistant melanoma. Cancer Discov. 8, 74–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue Y., et al. , An approach to suppress the evolution of resistance in BRAFV600E-mutant cancer. Nat. Med. 23, 929–937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smalley I., et al. , Leveraging transcriptional dynamics to improve BRAF inhibitor responses in melanoma. EBioMedicine 48, 178–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreuer M., et al. , Combination of dabrafenib plus trametinib for BRAF and MEK inhibitor pretreated patients with advanced BRAFV600-mutant melanoma: An open-label, single arm, dual-centre, phase 2 clinical trial. Lancet Oncol. 18, 464–472 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Valpione S., et al. , Rechallenge with BRAF-directed treatment in metastatic melanoma: A multi-institutional retrospective study. Eur. J. Cancer 91, 116–124 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Tietze J. K., et al. , The efficacy of re-challenge with BRAF inhibitors after previous progression to BRAF inhibitors in melanoma: A retrospective multicenter study. Oncotarget 9, 34336–34346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stagno A., et al. , Case Report: Rechallenge with BRAF and MEK inhibitors in metastatic melanoma: A further therapeutic option in salvage setting? Front. Oncol. 11, 645008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matter A. V., Micaletto S., Urner-Bloch U., Dummer R., Goldinger S. M., Long-term response to intermittent binimetinib in patients with NRAS-mutant melanoma. Oncologist 25, e1593–e1597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Algazi A. P., et al. , Continuous versus intermittent BRAF and MEK inhibition in patients with BRAF-mutated melanoma: A randomized phase 2 trial. Nat. Med. 26, 1564–1568 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez-Cao M., et al. , Intermittent BRAF inhibition in advanced BRAF mutated melanoma results of a phase II randomized trial. Nat. Commun. 12, 7008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong X., et al. , Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 550, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriceau G., et al. , Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 27, 240–256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C., et al. , Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Reger de Moura C., et al. , Intermittent versus continuous dosing of MAPK inhibitors in the treatment of BRAF-mutated melanoma. Transl. Oncol. 13, 275–286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez I. M., et al. , In Vivo ERK1/2 reporter predictively models response and resistance to combined BRAF and MEK inhibitors in melanoma. Mol. Cancer Ther. 18, 1637–1648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Barretina J., et al. , The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller J., et al. , Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 5, 5712 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anastas J. N., et al. , WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J. Clin. Invest. 124, 2877–2890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachindra L. L., et al. , New role of ID3 in melanoma adaptive drug-resistance. Oncotarget 8, 110166–110175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichhoff O. M., et al. , Differential LEF1 and TCF4 expression is involved in melanoma cell phenotype switching. Pigment Cell Melanoma Res. 24, 631–642 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Lito P., et al. , Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lake D., Corrêa S. A., Müller J., Negative feedback regulation of the ERK1/2 MAPK pathway. Cell. Mol. Life Sci. 73, 4397–4413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metzner T., et al. , Fibroblast growth factor receptors as therapeutic targets in human melanoma: Synergism with BRAF inhibition. J. Invest. Dermatol. 131, 2087–2095 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Straussman R., et al. , Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng Y. K., et al. , Pan-erbB inhibition potentiates BRAF inhibitors for melanoma treatment. Melanoma Res. 24, 207–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young H. L., et al. , An adaptive signaling network in melanoma inflammatory niches confers tolerance to MAPK signaling inhibition. J. Exp. Med. 214, 1691–1710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu L., et al. , The CSPG4-specific monoclonal antibody enhances and prolongs the effects of the BRAF inhibitor in melanoma cells. Immunol. Res. 50, 294–302 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Woods D., et al. , Induction of beta3-integrin gene expression by sustained activation of the Ras-regulated Raf-MEK-extracellular signal-regulated kinase signaling pathway. Mol. Cell. Biol. 21, 3192–3205 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu X., Tao X., Lu W., Ding Y., Tang Y., Blockade of integrin β3 signals to reverse the stem-like phenotype and drug resistance in melanoma. Cancer Chemother. Pharmacol. 83, 615–624 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Caramel J., et al. , A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Konieczkowski D. J., et al. , A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 4, 816–827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ernst A. K., et al. , Knockdown of L1CAM significantly reduces metastasis in a xenograft model of human melanoma: L1CAM is a potential target for anti-melanoma therapy. PLoS One 13, e0192525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiefel H., et al. , EMT-associated up-regulation of L1CAM provides insights into L1CAM-mediated integrin signalling and NF-κB activation. Carcinogenesis 33, 1919–1929 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Delord J. P., et al. , Phase I dose-escalation and -expansion study of the BRAF inhibitor encorafenib (LGX818) in metastatic BRAF-mutant melanoma. Clin. Cancer Res. 23, 5339–5348 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Grippo J. F., et al. , A phase I, randomized, open-label study of the multiple-dose pharmacokinetics of vemurafenib in patients with BRAF V600E mutation-positive metastatic melanoma. Cancer Chemother. Pharmacol. 73, 103–111 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Silva A. S., et al. , Evolutionary approaches to prolong progression-free survival in breast cancer. Cancer Res. 72, 6362–6370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J., Cunningham J. J., Brown J. S., Gatenby R. A., Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat. Commun. 8, 1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park D. S., et al. , Searching for Goldilocks: How evolution and ecology can help uncover more effective patient-specific chemotherapies. Cancer Res. 80, 5147–5154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.West J., et al., Towards Multidrug Adaptive Therapy. Cancer Res. 80, 1578–1589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. J. Kavran et al., Intermittent treatment of BRAF-V600E melanoma cells delays resistance by adaptive resensitization to drug rechallenge. Gene Expression Omnibus. https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse117123. Deposited 15 July 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in NCBI Gene Expression Omnibus (GSE117123). A runnable and editable version of the code used in this study can be found on code ocean (https://codeocean.com/capsule/9070543/tree/v1), and a code repository is available on GitHub at https://github.com/andykavran/Intermittent_Drug_Treatment. All other study data are included in the article and/or supporting information.