Significance
Hopanoids are a group of biologically important triterpene scaffolds found in nature, but the discovery of hopane-type triterpene synthases in plants has not been reported. We discovered two types of triterpene synthases synthesizing hopanoid skeletons from monocot and dicot plants and elucidated a mechanism involving the deprotonation at different sites by site-directed mutagenesis experiments and the quantum mechanics and molecular mechanics calculation. Our results provide a genetic element for synthesizing biologically active hopane-type triterpenoids and serve as a foundation for studying the molecular mechanisms of methyl and hydride transfer in the triterpene cyclization mechanism.
Keywords: triterpene skeletons, cyclization, oxidosqualene cyclase, mutants, directed evolution
Abstract
Triterpenoids are biologically active metabolites synthesized from a common linear precursor catalyzed by 2,3-oxidosqualene cyclases (OSCs) to form diverse triterpenoid skeletons. OSCs corresponding to many discovered triterpene alcohols in nature have not been functionally and mechanistically characterized due to the diversity of chemical structures and complexity of the cyclization mechanism. We carried out a genome-wide investigation of OSCs from Avena strigosa and discovered two triterpene synthases, namely, AsHS1 and AsHS2, using a Nicotiana benthamiana expression system. These synthases produce hopenol B and hop-17(21)-en-3β-ol, which are components of surface wax in oat panicles and sheathes, respectively. We demonstrated that substitutions of two to three amino acid residues in AsHS1 with corresponding residues from AsHS2 allowed it to be completely converted into a hop-17(21)-en-3β-ol synthase. AsHS2 mutants with a substitution at site 410 could synthesize hopenol B alone or mixed with a side product isomotiol. The combined quantum mechanics and molecular mechanics calculation demonstrated that the side chain size of the residue at site 410 regulated the relative orientations between the hopyl C22 cation and Phe257, leading to a difference in deprotonation positions through providing or not providing cation–π interaction between the aromatic ring of F257 and the carbocation intermediate. A similar mechanism could be applied to a hopenol B synthase from a dicotyledonous plant Aquilegia. This study provided mechanistic insight into triterpenoid synthesis and discovered key amino acid residues acting on hydride transfer and a deprotonation site to differentiate between hopane-type scaffolds in diverse plant species.
Triterpenoids are a large group of widespread secondary metabolites exhibiting anticoagulant, antiviral, anticancer, antidiabetic, hepatoprotective, hypocholesterolemic, immunomodulatory, neuroprotective, anti-inflammatory, and antioxidant activities (1, 2). Some triterpenoids, such as pentacyclic triterpenes, are components of plant surface wax on the cuticle, which protects the epidermal layer from heat and drought stress. Triterpenoids have many skeleton structures, including tetracyclic and pentacyclic triterpenes with chair–boat–chair (CBC) or chair–chair–chair (CCC) conformation. The tetracyclic skeletons with CBC conformation are lanostane, cycloartane, and cucurbitane, whereas scaffolds with CCC conformation are dammarane, tirucallane, and euphane. Pentacyclic triterpenoids usually have lupane, hopane, ursane, oleanane, taraxaterane, and friedelane skeletons (3, 4). The direct precursor of triterpene aglycone synthesis is 2,3-oxidosqualene, which is catalyzed at the key step of the cyclization reaction by 2,3-oxidosqualene cyclase (OSC), a triterpene synthase (5). The diversity of triterpene skeleton structures is due to divergent evolution of triterpene synthase genes in plants (6–8). With the rapid progress made in genome sequencing technology, the whole-genome sequences of many plants have been released, thereby facilitating OSC gene discovery and cloning. Thirteen and 12 OSC genes exist in the genomes of Arabidopsis thaliana and in Oryza sativa (rice), respectively (9, 10). Using the Saccharomyces cerevisiae and Nicotiana benthamiana heterologous expression systems, the catalytic products of OSC enzymes of Arabidopsis and rice have been mostly identified (11–15). Scientists have identified more than 150 OSCs, including lanosterol synthase (LAS), cucurbitadienol synthase, cycloartenol synthase (CAS), dammarenediol-II synthase, lupeol synthase, β-amyrin synthase, friedelin synthase, and others (4). However, hopane-type triterpene synthases have not yet been identified from any plant.
Plant OSC genes underwent a lineage-dependent evolutionary process to form two distinguished clades, namely, the CAS-derived lineage and the LAS-derived lineage. Most of triterpene synthases from dicotyledonous plants have evolved from the LAS-derived lineage, and those from monocotyledonous plants were from the CAS-derived lineage (7). Dicot OSCs have been extensively studied in many medicinal plants, but novel enzymes are seldom discovered (8, 16). A functional analysis of OSCs from rice and Sorghum bicolor revealed several unprecedented triterpene synthases, such as isoarborinol synthase, fernenol synthase, and simiarenol synthase (7, 12, 17). The discovery of novel OSC enzymes is highly probable through the genome mining of monocotyledonous plants.
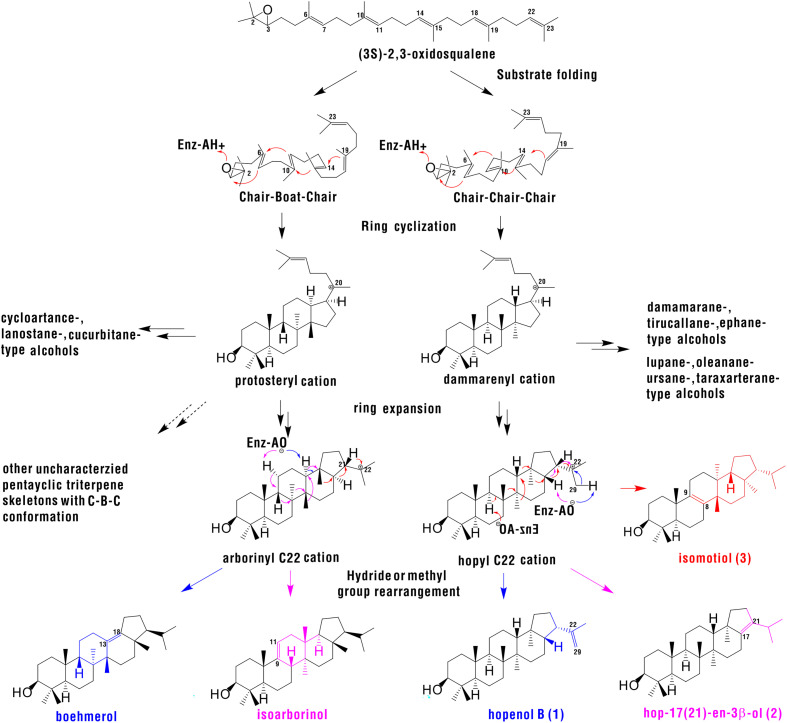
Based on the stereochemistry of OSC catalytic products and the basic principles of chemical reaction, scientists proposed an enzymatic mechanism of triterpene alcohol synthesis as follows: 1) enzymes and substrates form complexes, which initiate prefolding of substrates; 2) protonation of substrate epoxy groups initiates cyclization reactions; 3) ring formations occur; 4) a series of hydride transfer and methyl rearrangement reactions occurs on ring skeletons; and 5) deprotonation or capture of a water molecule forms the final product (18) (Fig. 1). Prefolding of substrates is a very important initial step in the cyclization reaction that determines whether the cyclization product is in a CCC or CBC configuration (19). The diversity of OSC products is determined by the interaction between a series of partially cyclized intermediate cations and catalytic groups in the active center of the enzyme, which form a specific architecture (20). An important foundation for the study of the catalytic mechanism of OSC enzymes is research on human LAS (HsLAS) (21). This human synthase was expressed in Pichia pastoris. Then, the protein was purified by metal chelate chromatography and gel filtration. Finally, a protein crystal with a resolution of 2.1 Å was obtained. Site-directed mutagenesis experiments based on the homologous modeling were widely applied to elucidate the cyclization mechanism of 2,3-oxidosqualene (16). The residues in the hydrophobic active center of various OSCs (e.g., Asp455, Cys456, and Cys533) in S. cerevisiae LAS (ScERG7) provide polar caps, which are beneficial to the formation of the C3 position of hydrogen bonds (22). Cys703, Tyr707, and Tyr99 in ScERG7 can stabilize the C8 and C14 cations during the B ring and the C ring formations (14, 18, 23, 24). Xue et al. identified three key amino acid residues by molecular evolution analysis (14), protein structure modeling, and molecular docking, and these three amino acid residues affected the orientation of the side chain of the fourth amino acid residue Tyr257. Thus, whether the product is a tetracyclic triterpene parkeol in the CBC conformation or a pentacyclic triterpene orysatinol in the abnormal CCC conformation was determined. Phe728 affects the formation of D and E rings in β-amyrin synthase (25), and Phe474 affects substrate folding through π–electron interaction and steric bulk effects (26). The above-described research focused on steps 1 to 3 of the triterpene cyclization process, which determined the types of triterpene skeletons by influencing substrate folding, ring formation, and expansion. However, the mechanisms underlying hydride shift, methyl rearrangement reactions, and deprotonation in the final steps of cyclization need to be elucidated by further research.
Fig. 1.
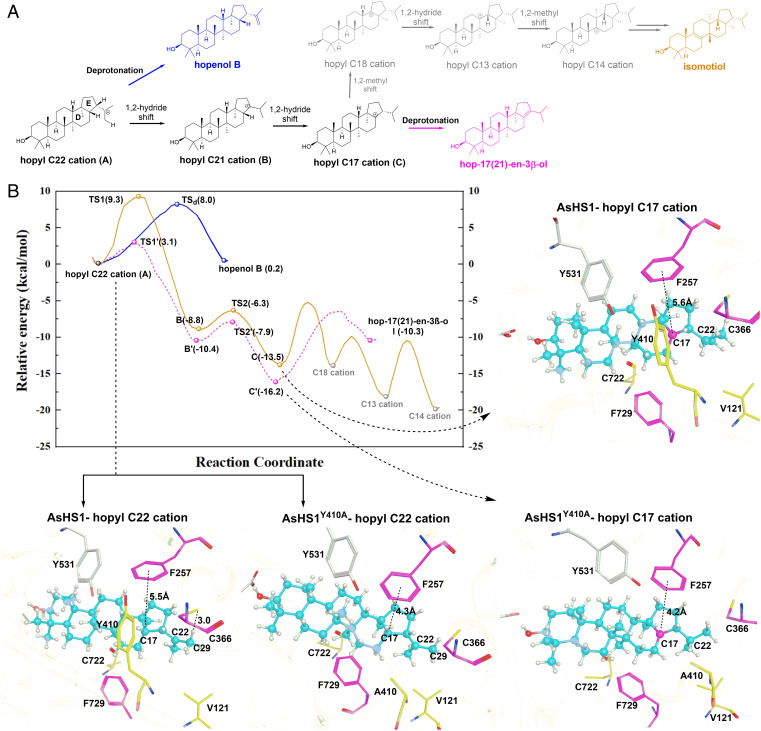
Biosynthetic pathway of triterpene formation. The precursor of triterpene, (3S)-2,3-oxidosqualene, is folded into a CBC or CCC conformation, followed by ring expansion to form protosteryl and dammarenyl cation, which can be deprotonated indirectly or after hydride and methyl shifting to produce tetracyclic CBC or CCC triterpene alcohols. Tetracyclic carbocations can undergo ring expansion to generate arborinyl C22 or hopyl C22 cations, which can give rise to pentacyclic triterpenes, such as boehmerol, isoarborinol, hopenol B (1), hop-17(21)-en-3β-ol (2), and isomotiol (3) through hydride and methyl group rearrangements and deprotonation at different sites.
Avenacins (also known as saponins) are antibacterial substances that accumulate in oat roots. β-amyrin synthase is a key enzyme involved in the synthesis of these saponins, but the origin of this enzyme differs from that of β-amyrin synthases in dicotyledonous plants (27). So far, it is the only single functional β-amyrin synthase that has been identified from monocotyledonous plants (16). Finding out how this OSC originated and evolved would be interesting. Thus, we identified 13 OSC genes from the oat genome, among which four genes formed an independent branch that included the Sad1 gene. Two OSC enzymes involved in the synthesis of hopenol B and hop-17(21)-en-3β-ol, respectively, were identified by transient expression in leaves of N. benthamiana. The quantum mechanics and molecular mechanics (QM/MM) molecular dynamics (MD) simulations and site-directed mutation revealed that hopane-type triterpene synthases produced different triterpenes by adjusting the distances between key amino acid residue(s) and the intermediate cation. Mutation of the equivalent amino acid residues of AcHS1, an Aquilegia coerulea triterpene synthase that also cyclizes oxidosqualene to hopenol B, similarly generated hop-17(21)-en-3β-ol as a side product. These results indicated that the mechanism underlying the formation of different hopane-type triterpenes is conserved in plants. Our results provide a genetic element for synthesizing hopane-type triterpenoids and present another important foundation for studying the molecular mechanisms underlying methyl and hydride transfer in the triterpene skeleton cyclization mechanism.
Results
Identification and Phylogenetic Analysis of OSCs from Avena strigosa.
Thirteen putative OSC genes, namely, AsOSC1–AsOSC13 (SI Appendix, Table S2), were returned after searching the oat genome sequence with rice OSC sequences. AsOSC10 and AsOSC11 only contained pseudogenes encoding partial OSC proteins, but the other 11 OSCs contained 18 exons that encoded complete OSC proteins of 751 to 766 amino acids. The AsOSC3 gene is the sad1 gene identified in previous studies (28) that encodes a β-amyrin synthase and participates in the biosynthesis of avenacin in oat. According to transcriptome sequencing analysis, AsOSC1 and AsOSC5 were expressed in all tissues. AsOSC2 was expressed in leaves, panicles, and spikelets. AsOSC3 and AsOSC4 were specifically expressed in roots. AsOSC4 was not expressed in root tips. AsOSC6 and AsOSC13 were expressed in all tissues except root tips. AsOSC7 was highly expressed in panicles, whereas AsOSC8 was expressed in shoots and spikelets. AsOSC9 and AsOSC10 were specifically expressed in panicles and leaves, and AsOSC12 was expressed in leaves, sheathes, and seedlings (SI Appendix, Table S2).
With the exception of AsOSC11, the other 12 OSC genes from oat were phylogenetically analyzed by constructing of a maximum likelihood tree. According to the functional study of rice OSC, oat OSCs can be divided into six subgroups. AsOSC1 and AsOSC2 were grouped with rice CAS, and AsOSC9 and AsOSC12 were grouped with cycloartenol synthase. These two subgroups are tetracyclic triterpene synthases with CBC conformation. AsOSC5, AsOSC6, and AsOSC11 were grouped with rice α-β-amyrin and achilleol B synthases. AsOSC10 was grouped with rice isoarborinol synthase, which is a pentacyclic triterpene synthase with CBC conformation. Expression and phylogenetic analysis indicated that AsOSC13 and rice poaceatapetol synthase had similar conserved functions. Interestingly, AsOSC3, AsOSC4, AsOSC7, and AsOSC8 formed a discrete branch from rice OSCs. Functional analysis of OSCs in this discrete branch might reveal triterpene synthases functionally different from rice OSCs; one of them, namely, AsOSC3/SAD1, has already been characterized as β-amyrin synthase (28) (SI Appendix, Fig. S1).
Characterization of Hopenol B and Hop-17(21)-en-3β-ol Synthases in A. strigosa.
To study the catalytic functions of AsOSC4, AsOSC7, and AsOSC8, we cloned coding sequences of AsOSC4, AsOSC7, and AsOSC8 genes from the complementary DNA (cDNA) of oat roots, panicles, and seedlings. The sequencing results showed that they were completely consistent with the sequences annotated by transcriptome and genome data. Each one of the OSCs was subcloned with the expression vector pEAQ-HT, and the metabolites in N. benthamiana leaves infected by Agrobacterium tumefaciens harboring one of the three OSCs were extracted and analyzed by gas chromatography–mass spectrometry (GC-MS). However, AsOSC4 has not been cloned and expressed successfully. The results showed that compared with the control without transient expression of any foreign OSC, the leaves transiently expressing AsOSC7 generated a new peak at 21.5 min, whereas the extract of N. benthamiana leaves transiently expressing AsOSC8 solely produced a new peak at 19.3 min in GC-MS analysis (Fig. 2A). Their mass spectra were inconsistent with those of α-amyrin, β-amyrin, or lupeol, thereby indicating that AsOSC7 and AsOSC8 produced an unknown triterpene alcohol. These results indicated that the catalytic functions of OSCs in this oat-specific group are likely different from one another.
Fig. 2.
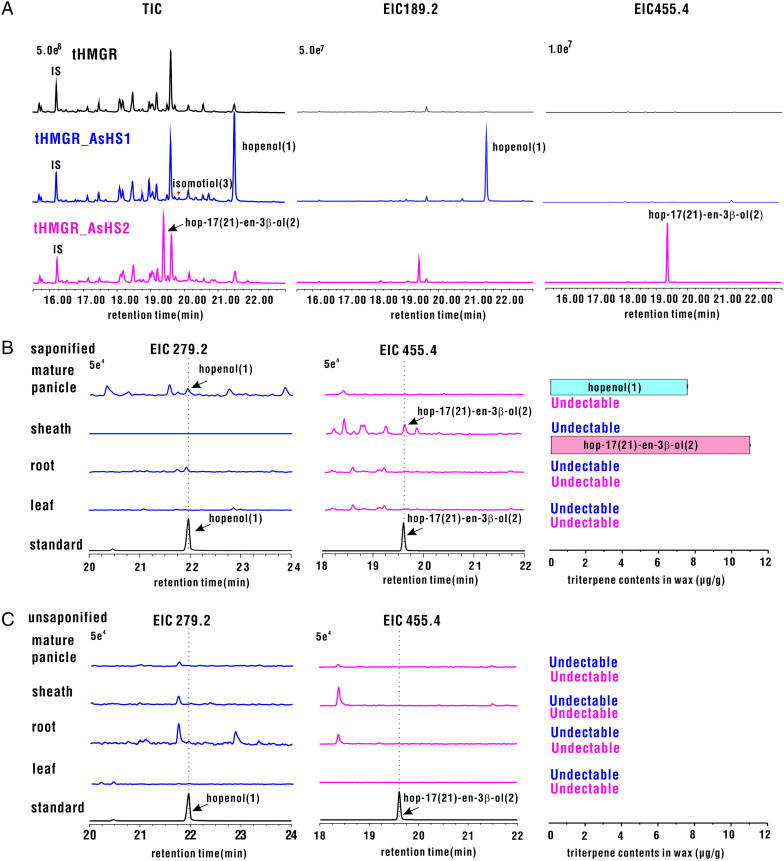
Divergence of catalytic function and amino acid residues in the active site between two OSCs from oat. (A) Total ion chromatogram (TIC) and extracted ion chromatogram (EIC) at m/z 189.2 (EIC 189.2) and m/z 455.4 (EIC 455.4) of N. benthamiana leaves expressing AsHS1 and AsHS2 coexpressed with tHMGR from oat (44). (B and C) EIC and absolute amounts of hopenol B and hop-17(21)-en-3β-ol were quantified by GC-MS analysis of extracts from saponified (B) and unsaponified (C) surface wax of four tissues using the standard curve method. To avoid the interference of other compounds in oat tissues, EIC 279.2 was used to indicate hopenol B for samples of extracted oat tissues. EIC 279.2 and EIC 455.4, EIC at m/z 279.2 and m/z 455.4, respectively. All values are from three biological replicates; 1, hopenol B; 2, hop-17(21)-en-3β-ol; 3, isomotiol.
We carried out a large-scale product extraction to determine the structure of AsOSC7 and AsOSC8 products; we obtained compound 1 (3.8 mg) and compound 2 (2.4 mg) from each of the pure products for NMR analysis. Compounds 1 and 2 were obtained as colorless crystals. The 1H NMR, 13C NMR, and heteronuclear single quantum coherence (HSQC) data of compound 1 exhibited one double bond (δC 148.7/110.1) and one hydroxyl (δH 3.19, δC 79.0) and seven methyl groups assigned as the following: δH 0.97 and δC 28.0, δH 0.76 and δC 15.4, δH 0.82 and δC 15.9, δH 0.96 and δC 16.6, δH 0.93 and δC 16.7, δH 0.72 and δC 16.1, and δH 1.75 and δC 25.0. The 1H NMR and 13C NMR spectra of compound 1 were determined to be hopenol-type triterpene. The heteronuclear multiple-bond correlation (HMBC) spectrum showed that the olefinic protons δH 4.78 (2H, br, s) were correlated with δC 46.5 and δC 25.0, which resulted in the determination of compound 1 as hopenol B (29) (Fig. 1 and Dataset S1). The 1H NMR and 13C NMR spectra of compound 2 were similar to those of 1, except for the fact that no olefinic protons and two methyl groups, namely, δH 0.92 and δC 21.3 and δH 0.97 and δC 21.9, were correlated with the olefinic carbons (δC 139.9/136.1) in the HMBC spectrum. By comparing the 1H NMR data of compound 2 with the reported data, compound 2 was identified as hop-(17)21-en-3β-ol (30) (Fig. 1 and Dataset S2). All the above results showed that the products of AsOSC7 and AsOSC8 were hopenol B and hop-17(21)-en-3β-ol, respectively. Thus, AsOSC7 and AsOSC8 were named AsHS1 and AsHS2, respectively. Hopenol B and hop-17(21)-en-3β-ol are both pentacyclic triterpenes with CCC conformation. The molecule of 2,3-oxidosqualene passed through dammarane cation and underwent ring expansion to form the hopyl C22 cation, which was directly deprotonated at C29 to form hopenol B, whereas the hopyl C22 cation underwent proton shift and deprotonation at C17 to generate hop-17(21)-en-3β-ol (Fig. 1).
To study the biochemical functions and biological significances of AsHS1 and AsHS2 in plants, we verified the expression of AsHS1 and AsHS2 genes by qRT-PCR using 11 kinds of tissues and organs at different developmental stages. The AsHS1 gene was expressed in both young and mature panicles, but the expression level was highest in mature panicles and was about 20 times that of a young panicle (SI Appendix, Fig. S2A). Besides shoots, AsHS2 was very strongly expressed in sheaths, and the expression level was approximately ten times more than that in shoots (SI Appendix, Fig. S2A). The above expression results by qRT-PCR were consistent with the RNA-sequencing data. Therefore, we chose four tissues, namely, root, leaf, leaf sheath, and mature panicles, to analyze the triterpene composition and to elucidate the biological roles of AsHS1 and AsHS2 in plants. Many hopane-type triterpenoids are components of epidermal wax. Thus, GC-MS was used to analyze the wax on the surface of saponified plant tissues. Trace amounts of hopenol B and hop-17(21)-en-3β-ol (8 and 11 μg/g) were detected in mature panicles and sheaths, respectively (Fig. 2B and SI Appendix, Fig. S3 A and B). However, they were not detected in nonsaponin waxy extracts (Fig. 2C). Similarly, hopenol B and hop-17(21)-en-3β-ol were not detected in saponified and unsaponifiable extracts of the whole cells of the above tissues and organs (SI Appendix, Fig. S3 A and B). Coexpression analysis of AsHS1 and AsHS2 with candidate metabolic synthetic genes, such as cytochrome P450s (CYPs), acyltransferases (ACTs), and UDP-glucosyltransferases (UGTs), showed that more CYPs and ACTs have higher Pearson correlation coefficients (>0.9) than UGTs (SI Appendix, Fig. S2B). The above results indicate that AsHS1 and AsHS2 are involved in the biosynthesis of wax on the surface of panicles and sheaths, respectively, and probably existed in the form of triterpene esters.
Identification of Key Amino Acids Responsible for AsHS1 and AsHS2 Product Specificities.
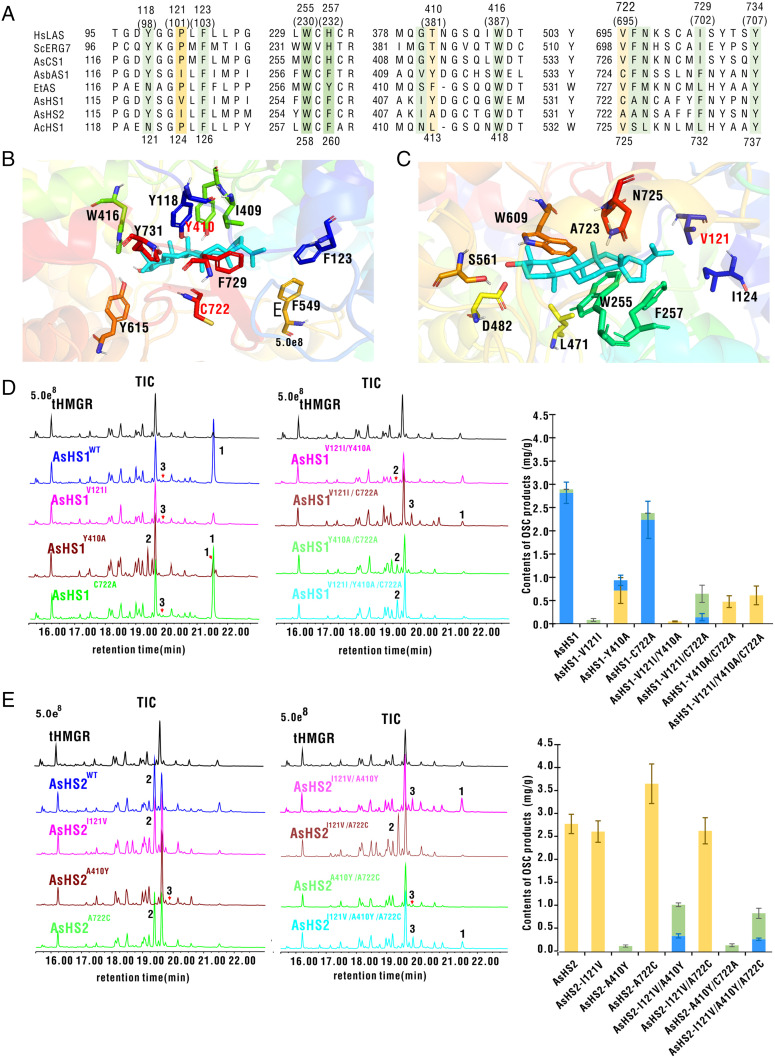
To study the potential molecular mechanism underlying the functional divergence of AsHS1 and AsHS2, we compared their differences in amino acid sequence and found 199 divergent sites that had different amino acid residues between AsHS1 and AsHS2 (SI Appendix, Fig. S4). Homologous modeling and molecular docking showed that three sites were distributed in the active center of enzymes within 4 Å from the hopyl C22 cation. They were V121, Y410, and C722 in AsHS1 and I121, A410, and A722 in AsHS2 (Fig. 3A). However, other amino acid residues within an active center, such as Y118, F123, W387, D482, F729, and Y707, did not differ between the two enzymes (Fig. 3 B and C).
Fig. 3.
Identification of key amino acid residues in the active site between AsHS1 and AsHS2 and validation of effects of single, double, or triple replacements of three different amino acid residues on AsHS product specificity. (A) Selected regions of five functionally characterized OSCs from diverse organisms and three OSCs characterized in this study were aligned. Residues within a 4-Å distance to hopyl C22 cation are shown in colored background. Among those sites, different residues between AsHS1 (hopenol B synthase) and AsHS2 (hop-17[21]-en-3β-ol synthase) are shown in light yellow background; HsLAS, a human lanosterol synthase; ScERG7, Saccharomyces cerevisiae lanosterol synthase; AsCS1, Avena strigose CAS; AsbAS1, A. strigose β-amyrin synthase; EtAS, Euphorbia tirucalli β-amyrin synthase; AcHS1, Aquilegia coerulea hopenol B synthase. The numbers above the alignment refer to sites of AsHS1 and AsHS2, and corresponding sites in HsLAS are included in parentheses. The numbers below the alignment refer to sites of AcHS1. (B and C) All residues of AsHS1 within a 4-Å distance to hopyl C22 cation are shown. The amino acid residues of AsHS1 differing to AsHS2 are highlighted in red font. (D) Extracts from leaves of N. benthamiana WT and mutant plants with single-, double-, and triple-site mutations of AsHS1 and analyzed by GC-MS. (E) Extracts from leaves of N. benthamiana WT plants and mutants with single-, double-, and triple-site mutations of AsHS2 analyzed by GC-MS. The quantities of hopenol B (blue), hop-17(21)-en-3β-ol (yellow), and isomotiol (green) were determined by integrating peak areas from GC-MS analysis using coprostanol as an internal standard. The expression levels of OSC mRNA were quantified by qRT-PCR analysis (SI Appendix, Fig. S10). The activities of WT and mutants are presented as means ± SE; n = 3 (contents of products mg/g); tHMGR from oat (44); 1, hopenol B; 2, hop-17(21)-en-3β-ol; 3, isomotiol.
The effects of three different amino acid residues on product specificity were verified by constructing a mutagenesis library with single or combinational substitutions on the background of wild-type (WT) AsHS1. GC-MS analysis results showed that the V121I substitution resulted in the absence of the hopenol B. In the latter case, AsHS1Y410A retained the ability to produce a small amount of hopenol B (0.22 ± 0.11 mg/g) and gained the function to produce hop-17(21)-en-3β-ol (0.71 ± 0.28 mg/g). The substitution of C722A only resulted in a slight decrease of hopenol B yield (Fig. 3D and SI Appendix, Fig. S5).
To further investigate the potential molecular mechanism underlying the functional conversion of AsHS1, we analyzed mutants with the combinations of two- and three-site amino acid substitutions. As expected, all combinations with the Y410A substitution produced hop-17(21)-en-3β-ol as a single product (Fig. 3D and SI Appendix, Fig. S5), and the triple mutant attained a hop-17(21)-en-3β-ol yield of 0.61 ± 0.2 mg/g (Fig. 3D and SI Appendix, Fig. S5). Interestingly, AsHS1V121I/C722A continued to produce 0.14 ± 0.18 mg/g hopenol B; it also produced an additional triterpene alcohol, which was the same as the minor product of WT AsHS1. The yield of this product increased to 0.5 ± 0.18 mg/g (Fig. 3D and SI Appendix, Fig. S5). MS and NMR analysis results indicated that the structure of this product was isomotiol (Dataset S3). These results indicated that the combined substitutions of Y410A, V121I, and C722A allowed the conversion of AsHS1 into an OSC producing hop-17(21)-en-3β-ol as a single product.
We next examined the effects of three amino acid sites on the functional maintenance of AsHS2. The results of amino acid substitution at a single site showed that the substitution of I121V had little effect on the yield of hop-17(21)-en-3β-ol (decreased by 6%), and the mutation of A722C resulted in an ∼30% increase of the product (Fig. 3E and SI Appendix, Fig. S6). The substitution of A410Y allowed AsHS2 to synthesize a small amount of isomotiol (Fig. 3E and SI Appendix, Fig. S6). Mutant analysis of two- and three-site combinational substitutions showed that the AsHS2I121V/A722C produced the same amount of hop-17(21)-en-3β-ol as WT AsHS2 (Fig. 3E and SI Appendix, Fig. S6). The mutants AsHS2I121V/A410Y and AsHS2I121V/A410Y/A722C lost their ability to synthesize hop-17(21)-en-3β-ol but produced hopenol B and isomotiol (Fig. 3E and SI Appendix, Fig. S6). Mutant AsHS2A410Y/A722C produced a low level (0.15 ± 0.03 mg/g) of isomotiol as a single product (Fig. 3E and SI Appendix, Fig. S6). The above results indicated that the combined mutations at two or three sites failed to completely convert AsHS2 into a single functional hopenol B synthase but generated isomotiol synthase (AsHS2A410Y and AsHS2A410Y/A722C) or an OSC that synthesized both isomotiol and hopenol B (AsHS2I121V/A410Y and AsHS2I121V/A410Y/A722C), as shown in Fig. 3E and SI Appendix, Fig. S6.
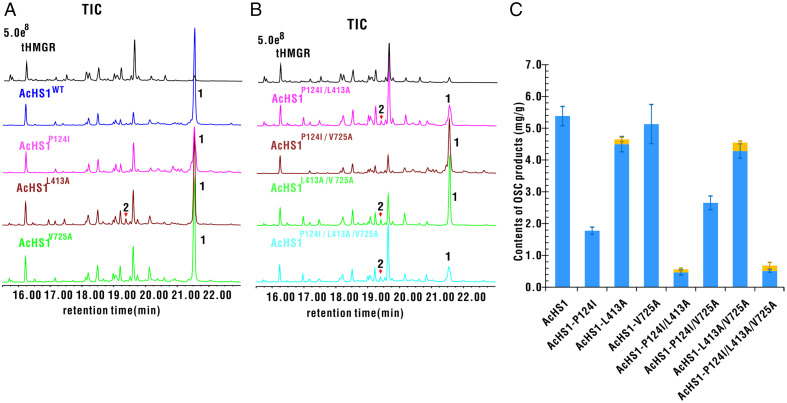
To verify the universal roles of the three amino acid residues at sites 121, 410, and 722 in hopane skeleton cyclization, we carried out mutagenesis experiments on AcHS1, which is also a hopenol B synthase identified from a basal dicot plant A. coerulea. AcHS1 had three different amino acid residues (Phe, Lue, and Val) at sites 124, 413, and 725, which corresponded to sites 121, 410, and 722, respectively, in AsHS1/AsHS2. We replaced those three amino acids residues with those at the corresponding sites of AsHS2 to generate a mutant library. Although amino acid substitution at a single site did not lead to the loss of ability to synthesize hopenol B, the L413A substitution led to the production of a small amount of hop-17(21)-en-3β-ol (0.15 ± 0.06 mg/g) (Fig. 4 and SI Appendix, Fig. S7). In fact, hop-17(21)-en-3β-ol was synthesized by all double and triple mutants with a L413A substitution. Mutations of P124I/L413A and P124I/L413A/V725A dramatically decreased the production of hopenol B (Fig. 4 and SI Appendix, Fig. S7). The result suggested that the residue at site 413 was crucial and conserved for the synthesis of hop-17(21)-en-3β-ol in diverse plants.
Fig. 4.
TICs and contents of OSC products of N. benthamiana leaf extracts expressing the WT and mutants of Aquilegia hopenol B synthase. (A and B) To validate the effects of amino acid substitutions at the corresponding positions of Aquilegia hopenol B synthase, mutagenesis of three amino acid residues of AcHS1 was carried out. Single- (A), double-, and triple-site mutations (B) of AcHS1 were analyzed by GC-MS; 1, hopenol B; 2, hop-17(21)-en-3β-ol. (C) The quantities of hopenol B (blue) and hop-17(21)-en-3β-ol (yellow) were determined by integrating peak areas from GC-MS analysis using coprostanol as an internal standard. The expression levels of OSC mRNA were quantified by qRT-PCR analysis (SI Appendix, Fig. S10). The activities of WT and mutants are presented as means ± SE; n = 3 (contents of products mg/g); tHMGR, from oat (44); 1, hopenol B; 2, hop-17(21)-en-3β-ol.
The above results suggested that sites 121, 410, and 722 are responsible for controlling the hydride shift and deprotonation. However, these sites are not conserved (16) (Fig. 3A), and saturation mutagenesis was carried out to address whether there are some rules. For hopenol B synthase AsHS1 and AcHS1, mutation of the amino acid at site 121 into a negatively charged amino acid lysine or an aromatic amino acid tyrosine depleted their function. Aliphatic amino acid (121S, 121N, and 121A) substitution usually reduces enzymatic activity. The relatively conservative 410 mutations (410S, 410N, and 410A) enable it to synthesize hop-17(21)-en-3β-ol. In contrast, the less conservative 722 mutations led to little change in the product spectrum and productivity (SI Appendix, Fig. S9). Mutations at site 410 (410K, 410D, 410S, 410N, and 410Y) of AsHS2 yielded isomotiol, and mutations at sites 722 and 121 mostly reduced enzymatic activity. Interestingly, the I121N mutant afforded isomotiol, suggesting that site 121 is more important than site 722 for hop-17(21)-en-3β-ol synthase (SI Appendix, Fig. S9).
Conserved Molecular Mechanism of Hopenol B and Hop-17(21)-en-3β-ol Formation through Deprotonation at Different Sites.
To decipher the formation mechanism of hopenol B or hop-17(21)-en-3β-ol, the hybrid QM/ MM calculation (SI Appendix, Supplementary Experimental Methods) was performed to investigate the three plausible reaction pathways (direct deprotonation pathway of hopenol B and rearrangement pathways of hop-17(21)-en-3β-ol and isomotiol, respectively) (Fig. 5A) on AsHS1/AsHS2 and their mutants. Based on the QM/MM optimized active pocket of enzyme–ligand complex bound with the hopyl C22 cation (hereafter referred to as A state), the aromatic residues F257 and F729 attracted our attention since they may facilitate the rearrangement reactions, especially F257, which is closed to the E ring of carbocation intermediate. Additionally, there is no other appropriate general base residue except C366, which is closed to the C29 site of hopyl C22 cation in AsHS1 (Fig. 5B). Thus, the interactions between hopyl C22 cation and F257/C366/Y410 were exhaustively detected along the reaction pathway by means of a QM/MM scan driven from suitable reaction coordinates (SI Appendix, Supplementary Experimental Methods).
Fig. 5.
Mechanism of hopane-type triterpene cyclization. (A) The reaction pathways starting from the hopyl C22 cation (also called A state herein) to produce hopenol B (blue), hop-17(21)-en-3β-ol (magenta), and isomotiol (orange) in AsHS1/AsHS2. The blue pathway is named the “direct protonation” pathway, while another pathway is named the “rearrangement” pathway, which could be further divided into two pathways from a branching point hopyl C17 cation (C state). (B) The relative energy profile for WT AsHS1 hopenol B (blue), isomotiol (orange), and AsHS1Y410A mutant hop-17(21)-en-3β-ol pathways. The representative structures of A and C states in WT AsHS1 and AsHS1Y410A mutant are shown.
In WT AsHS1, for the reaction energetics, direct deprotonation of A state to produce the hopenol B is feasible with a reaction barrier of 8.0 kcal/mol (Fig. 5B), by the help of C366 serving as a general base. The catalytic role of C366 could be verified by experimental mutations that AsHS1C366D still retains the native activity (yields hopenol B), while C366G mutant is inactive (SI Appendix, Fig. S8). Considering that the rearrangement pathway involved a 1,2-hydride shift to form hopyl C21 cation (B state) with a higher barrier (9.3 kcal/mol), the direct deprotonation of A state is dynamically preferred, leading to the major product hopenol B. Nevertheless, the cascade reactions of A state along the rearrangement pathway are competitive in view of the notably exothermal thermodynamic properties, as shown in Fig. 5B. For the enzyme–ligand interaction, it is obvious that the stabilization of F257 is relatively weak for A state of WT AsHS1, since the aromatic ring of F257 is not close to the E ring of carbocation (the distance to C17 is 5.5 Å). This is attributed to the sterically hindered effects arising from the large side chain of Y410, which anchors the relative orientations between the intermediate and F257 (SI Appendix, Fig. S11). Differently, subsequent hydride and methyl shifts are stabilized by F729 since it is close to the D ring of intermediates (the distance to C14 is 4.6 Å) and thus facilitates the ultimate formation of the isomotiol as byproduct.
In the AsHS1Y410A mutant, the hindered effect is lacking, with a much smaller side chain of A410 instead of Y410; accordingly, the conformation of A state in the AsHS1Y410A mutant becomes flexible and prefers an optimal bind pose distinct from that in WT AsHS1. As shown in Fig. 5B, the F257 of AsHS1Y410A is close to the E ring of A state (with a 4.3-Å distance to C17). As a result, F257 significantly promotes the hydride shifts by mean of stabilizing the carbocation intermediates and transition state (TS1′ and TS2′) through cation–π interactions. This is proven by the reaction energetics, only a very low barrier (3.1 kcal/mol) for the 1,2-hydride shifts (from A state to B′ state), and also tiny barrier (2.5 kcal/mol) to C′ state. Subsequent deprotonation from C21 of C′ state (C366 as a general base) conquers the 9.8 kcal/mol barrier and produces the final product hop-17(21)-en-3β-ol.
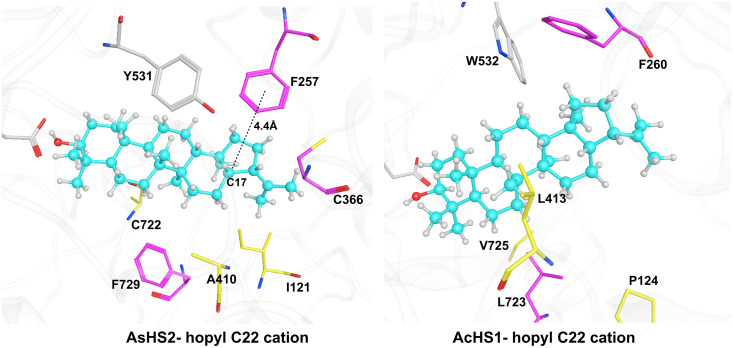
Similar to the AsHS1Y410A mutant, it is also A410 with a small side chain in WT AsHS2; thus, the cation–π interactions between the F257 and the hopyl C22 cation intermediate are also strong (Fig. 6) and promote the hydride shifts. However, the experimentally expressed A410Y single mutant did not produce the hopenol B as in AsHS1. This is probably due to the pocket contour difference between AsHS1 and AsHS2. As mentioned above, besides the key residue Y410/A410 that is highly related to hopenol B/hop-17(21)-en-3β-ol production, the pocket contour residues, especially the V121 and C722 in AsHS1, will also affect the conformations of the intermediates. This is consistent with the experimental results that the I121V/A410Y/A722C triple mutant of AsHS2 turned the hop-17(21)-en-3β-ol activity into hopenol B activity, while the I121V/A722C mutant of AsHS2 did not change the activity. Although isomotiol is also largely produced in the I121V/A410Y/A722C mutant, it could be expected if considering the high feasibility of hydride shifts and subsequent rearrangements similar as AsHS1 WT pathways.
Fig. 6.
The representative structures of the hopyl C22 cation in (Left) AsHS2 and (Right) AcHS1.
In AcHS1, although the Y410 in AsHS1 is changed to L413, we speculate that the large aliphatic side chain of L413 still has a similar function as AsHS1 Y410 (as shown in Fig. 6) and thus produces the hopenol B as main product (SI Appendix, Fig. S8). We have also attempted to verify the function of leucine by the AsHS1Y410L mutant. Interestingly, the AsHS1Y410L mutant still retained the activity to produce hopenol B (SI Appendix, Fig. S8), which further proved the similar function of tyrosine and leucine. Combining the computational results and the experimentally expressed AsHS1 Y410A/Y410L mutants together, it could be concluded that the different cation–π interaction arising from the sterically hindered effects due to the side chain volume of position 410 is a key factor for the production of hopenol B and hop-17(21)-en-3β-ol.
Saturation mutagenesis of sites 257 and 729 suggested that F257 replacement inactivates AsHS1, reducing the activity of AsHS2 and AcHS1. Mutation of F729 into aliphatic amino acids mostly reduces yield, and F729Y has nearly no effect on enzyme activity (SI Appendix, Fig. S9).
Discussion
Hopanoids are the main components of bacterial sterols and are widely distributed in nonvascular and higher plants (31–33). Hopenol B from fossil dammar resin of early Paleogene time (65 Mya) was proposed to be derived from the Dipterocarpaceae family (34). Bacteria-derived hopanoids are formed by the cyclization of squalene and catalyzed by squalene-hopene cyclases, while plant-derived hopanoids originated from cyclization of 2,3-oxidosqualene by OSCs (35). To date, the discovery of hopane-type triterpene synthases in higher plants has not been reported. Our phylogenetic tree showed that oat hopane-type triterpene synthases (AsHS1 and AsHS2) were formed after the subfamily Pooideae diverged from the Oryzoideae (65 Mya), which is close to the early Paleogene time (36). Our study provided an alternative source of hopanoids in the geological sediments.
AsHS1 and AsHS2 commonly use hopyl C22 cation to produce hopenol B and hop-17(21)-en-3β-ol, but they differ in the hydride rearrangement and deprotonation site during cyclization reactions. MD simulations and site-directed mutagenesis experiments showed that the key amino acid residues of oat hopane-type triterpene synthases that determine the hydride rearrangement and deprotonation process were two phenylalanines at sites 257 and 729, respectively. Mutagenesis experiments of an A. coerulea hopenol B synthase also showed the important role of phenylalanine at corresponding site 260. AcHS1 was independently evolved in a basal dicot (SI Appendix, Fig. S2). The distance between F257/260 and hopyl C22 cation is therefore critical for the determination of hopenol B and hop-17(21)-en-3β-ol formation, and this mechanism is very conserved in both monocot and dicot hopane-type synthases. Site 257 corresponds to site 232 in HsLAS, which has a histidine residue. H232 acts with the C9 carbocation of lanosteryl cation and accepts protons to terminate cyclization reactions, thereby forming lanosterol (21). β-amyrin synthase has a conserved amino acid residue (tyrosine or phenylalanine) at this site, which has been proposed to stabilize baccharenyl cations via cation–π interactions during β-amyrin formation (16). Our previous studies on functional interconversion between rice parkeol synthase and orysatinol synthase (OsOS) also showed that the side chain direction of Y257 affected product formation (14). Therefore, we deduced that H232/Y(F)257 plays important but distinct roles in diverse triterpene cyclization reactions through the interaction with different intermediate carbocations.
According to our evolutionary analysis, oat OSCs can be divided into five subgroups. One subgroup includes a characterized enzyme OsOSC6 (α/β-amyrin synthase) from rice (13) and three OSCs from oat, AsOSC5, AsOSC6, and AsOSC11. AsOSC11 is considered as a pseudogene, and its expression was detected in all investigated tissues. The expression patterns of AsOSC5 and AsOSC6 are similar to those of rice OsOSC6. Therefore, we proposed that the functions of AsOSC5 and AsOSC6 might be similar to those of OsOSC6 (13). Four OSCs, AsOSC3/SAD1, AsOSC4, AsHS1, and AsHS2 formed an oat-specific subgroup. Gene expression pattern analysis showed the following: sad1 and AsOSC4 were specifically expressed in roots, AsHS1 was specifically expressed in panicle, and AsHS2 was specifically expressed in shoots and sheathes. Such results indicated that the expression pattern of OSC genes in this oat-specific branch had stronger tissue specificity than its adjacent OsOSC6 branch. Furthermore, the tissue-specific expression pattern of metabolic biosynthetic genes always suggested functional specialization. Recent research showed that sorghum OSCs participate in the biosynthesis of the surface wax (17). Hopenol B and hop-17(21)-en-3β-ol were only detected in the wax extracts and not in the whole-cell extracts of oat panicles and sheathes, respectively. Coexpression analysis also discovered more ester synthesis related genes than saponin synthesis related genes. We predicted that AsHS1 and AsHS2 participate in a major way in the biosynthesis of surface wax in oat panicles and sheathes, which may facilitate survival in extreme environments, such as under drought or cold stress.
Materials and Methods
Gene Annotation.
To identify OSC genes, the A. strigosa S75 genome sequence (version 8.0) (37) was searched by tblastn to find sequences of OSCs from A. strigosa using rice OSCs as a bait (10). A. coerulea OSCs were predicted using the Phytozome portal’s database A. coerulea genome, version 3.1 (https://phytozome-next.jgi.doe.gov/info/Acoerulea_v3_1) (38).
Phylogenetic Analysis.
Multiple alignments of OSC protein sequences were performed (Dataset S4), and a codon matrix was produced using the MUSCLE alignment package in MEGA 7 (39). Functionally characterized plant OSCs and other OSCs from lower organisms as outgroup sequences were derived from a review paper (8). The evolutionary history was inferred using the maximum likelihood method based on the Jones–Taylor–Thornton matrix-based model (40). The bootstrap consensus tree inferred from 1,000 replicates was taken to represent the evolutionary history of the taxa analyzed.
RNA Extraction and cDNA Synthesis.
Total RNA was extracted from A. strigosa panicles and sheathes using TRI Reagent (Invitrogen). The cDNA was synthesized from RNA using SuperScript III (Invitrogen) according to the manufacturer’s instructions.
Cloning and Expression of OSCs in N. benthamiana.
The AsHS1 (AsOSC7) and AsHS2 (AsOSC8) coding sequences were amplified from the cDNA of A. strigosa S75 panicles and sheathes using Phusion polymerase (New England Biolabs). AcHS1 was predicted using the Phytozome database A. coerulea genome version 3.1 and was synthesized by GENEWIZ. Each sequence was cloned into the pDONR207 vector (Invitrogen) and transferred into pEAQ-HT-DEST1 using the att L × att R reaction clonase II enzyme (Invitrogen). The expression constructs were transformed into chemically competent A. tumefaciens strain GV3101 by flash-freezing in liquid nitrogen. Five-week-old N. benthamiana were infiltrated with three leaves per plant as replicates. A. tumefaciens strain carrying the truncated 3-hydroxy-3-methyl glutaryl-coenzyme A (HMG-CoA) reductase gene (tHMGR) from oat was coinfiltrated with strains carrying OSC genes. Leaves were harvested 5 d after infiltration and lyophilized.
Metabolite Extraction and GC-MS Analysis.
Triterpenes were extracted from 10 mg of dry materials according to the triterpene extraction method and derivatized using 50 μL of trimethylsilyl imidazole/pyridine reagent (Sigma-Aldrich, 92718). Samples were analyzed using a GC–triple quadrupole–MS instrument (Agilent Technologies). Full details appear in the SI Appendix, Supplementary Experimental Methods.
Triterpene Purification and Structural Identification.
The dried leaves (30 g) were saponified and extracted with hexane (three times). The hexane extract was concentrated under reduced pressure and partitioned with a silica gel column using sequential gradient elution of hexane/ethyl acetate (100:0 → 0:100) to obtain 20 fractions (Fr1 to Fr20). Compound 1 (3.8 mg, retention time [tR] = 15.0 min) was purified from Fr5 by semipreparative high-performance liquid chromatography using a semipreparative column (Thermo Hypersil GOLD C18 column, 250 mm × 10 mm, 5 μm) with water/methanol as the eluent, respectively. Using the same methodology, hop-17(21)-en-3β-ol (2.4 mg, tR = 14.0 min) and isomotiol (3.4 mg, tR = 15.0 min) were purified. The purified compounds were dissolved with CDCl3 for NMR analysis, including 1H-NMR, 13C-NMR, HMBC, HSQC, 1H-1H correlation spectroscopy, and rotating frame Overhauser effect spectroscopy. Chemical shifts were recorded in parts per million and referenced to the residual solvent peak or to an internal tetramethylsilane standard. Full details appear in the SI Appendix, Supplementary Experimental Methods.
Extraction of Wax Compositions of A. strigosa Tissues.
The panicle, sheath, whole root, and leaf tissues were collected from 2-mo-old oats, and tissue samples were lyophilized. Wax was obtained by immersing the dry materials (1 g) in chloroform (100 mL) for 1 min. Then, the plant materials were removed by filtration immediately, and the solvent was evaporated, leaving the whole tissue extraction behind as a solid residue. The unsaponified and saponified wax were analyzed using a GC-MS instrument. Full details appear in the SI Appendix, Supplementary Experimental Methods.
Modeling and Identification of Key Residues.
Three-dimensional models of the AsHS1, AsHS2, and AcHS1 proteins were generated by modeling with the HsLAS (Protein Data Bank ID: 1w6k/1w6j) template using SWISS-MODEL software (21, 41) and also by AlphaFold2 (42). The three-dimensional structure of hopyl cations were obtained by Chemdraw 15.0. Docking searches were performed using the Lamarckian genetic algorithm, with a maximum of 25,000,000 energy evaluations and other options set as default in Autodocking suite (43). Potential models were returned ranked on the basis of binding energy, and the top-ranked model was assumed to be the most likely of actual structure. The models were graphically rendered using PyMOL software (version 2.4). Furthermore, classical MD and QM/MM modeling were performed to decipher the atomic-level catalytic mechanism and identify the key residues responsible for regulating the reaction pathways; more details are provided in the SI Appendix, Supplementary Experimental Methods.
Mutagenesis Experiments.
Mutagenesis was performed using the QuikChange site-directed mutagenesis method (Stratagene, Agilent). The expression plasmids for AsHS1, AsHS2, and AcHS1 were used as templates for PCR-based site-directed mutagenesis. The primers used for site-directed mutagenesis are listed in SI Appendix, Table S1, with the substitutions underlined. The PCR system and procedure are shown in SI Appendix, Supplementary Experimental Methods. Functional characterization of the mutants was conducted similar to that for the AsHS1 and AsHS2 mentioned above.
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (grants 31970314 to Z.X., U21A20243 to Z.X., and 21773313 to R.W.) and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team). We thank Yaonan Wang (Capital University of Medicine) and Dr. Shengnan Tan (Analysis and Test Center, Northeast Forestry University) for assistance with NMR analysis and Michael J. Stephenson (John Innes Centre) and Shihong Luo (Shenyang Agricultural University) for NMR assignments of OSC products. We thank James Reed (John Innes Centre) in A. coerulea OSC identification and Anne Osbourn (John Innes Centre) for the critical comments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118709119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Raw and assembled RNA-seq and genomic DNA data can be found in a previously published paper (37). DNA squences of 13 AsOSC genes in SI Appendix, Table S2 and AcHS1 can also be found in GenBank (accession nos. OM401318–OM401331).
References
- 1.Liu J., Henkel T., Traditional Chinese medicine (TCM): Are polyphenols and saponins the key ingredients triggering biological activities? Curr. Med. Chem. 9, 1483–1485 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Moses T., Papadopoulou K. K., Osbourn A., Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 49, 439–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincken J. P., Heng L., de Groot A., Gruppen H., Saponins, classification and occurrence in the plant kingdom. Phytochemistry 68, 275–297 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Wang J., et al. , Diverse triterpene skeletons are derived from the expansion and divergent evolution of 2,3-oxidosqualene cyclases in plants. Crit. Rev. Biochem. Mol. Biol. 10.1080/10409238.2021.1979458 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Abe I., Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 24, 1311–1331 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Phillips D. R., Rasbery J. M., Bartel B., Matsuda S. P., Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Xue Z., et al. , Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 193, 1022–1038 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Thimmappa R., Geisler K., Louveau T., O’Maille P., Osbourn A., Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65, 225–257 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Husselstein-Muller T., Schaller H., Benveniste P., Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol. Biol. 45, 75–92 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Inagaki Y. S., et al. , Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering. New Phytol. 191, 432–448 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Morlacchi P., et al. , Product profile of PEN3: The last unexamined oxidosqualene cyclase in Arabidopsis thaliana. Org. Lett. 11, 2627–2630 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Ito R., et al. , Triterpene cyclases from Oryza sativa L.: Cycloartenol, parkeol and achilleol B synthases. Org. Lett. 13, 2678–2681 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Sun J., Xu X., Xue Z., Snyder J. H., Qi X., Functional analysis of a rice oxidosqualene cyclase through total gene synthesis. Mol. Plant 6, 1726–1729 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Xue Z., et al. , Identification of key amino acid residues determining product specificity of 2,3-oxidosqualene cyclase in Oryza species. New Phytol. 218, 1076–1088 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Xue Z., et al. , Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat. Commun. 9, 604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K., Zhang M., Ye M., Qiao X., Site-directed mutagenesis and substrate compatibility to reveal the structure-function relationships of plant oxidosqualene cyclases. Nat. Prod. Rep. 38, 2261–2275 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Busta L., Schmitz E., Kosma D. K., Schnable J. C., Cahoon E. B., A co-opted steroid synthesis gene, maintained in sorghum but not maize, is associated with a divergence in leaf wax chemistry. Proc. Natl. Acad. Sci. U.S.A. 118, e2022982118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu T. K., Chang C. H., Liu Y. T., Wang T. T., Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase: A chemistry-biology interdisciplinary study of the protein’s structure-function-reaction mechanism relationships. Chem. Rec. 8, 302–325 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Ma Y. S., et al. , New insights into substrate folding preference of plant OSCs. Sci. Bull. (Beijing) 61, 1407–1412 (2016). [Google Scholar]
- 20.Wu S., et al. , An unexpected oxidosqualene cyclase active site architecture in the Iris tectorum multifunctional α-amyrin synthase. ACS Catal. 10, 9515–9520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thoma R., et al. , Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432, 118–122 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Wendt K. U., Enzyme mechanisms for triterpene cyclization: New pieces of the puzzle. Angew. Chem. Int. Ed. Engl. 44, 3966–3971 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Chang C. H., et al. , The cysteine 703 to isoleucine or histidine mutation of the oxidosqualene-lanosterol cyclase from Saccharomyces cerevisiae generates an iridal-type triterpenoid. Biochimie 94, 2376–2381 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Wu T. K., et al. , Differential stereocontrolled formation of tricyclic triterpenes by mutation of tyrosine 99 of the oxidosqualene-lanosterol cyclase from Saccharomyces cerevisiae. Eur. J. Org. Chem. 2009, 5731–5737 (2009). [Google Scholar]
- 25.Salmon M., et al. , A conserved amino acid residue critical for product and substrate specificity in plant triterpene synthases. Proc. Natl. Acad. Sci. U.S.A. 113, E4407–E4414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito R., et al. , β-amyrin synthase from Euphorbia tirucalli. Steric bulk, not the π-electrons of Phe, at position 474 has a key role in affording the correct folding of the substrate to complete the normal polycyclization cascade. Org. Biomol. Chem. 12, 3836–3846 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Qi X., et al. , A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haralampidis K., et al. , A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. U.S.A. 98, 13431–13436 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsunaga S., Morita R., Hopenol-B, a triterpene alcohol from Euphorbia supina. Phytochemistry 22, 605–606 (1983). [Google Scholar]
- 30.Kakuda R., Iijima T., Yaoita Y., Machida K., Kikuchi M., Triterpenoids from Gentiana scabra. Phytochemistry 59, 791–794 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Shinozaki J., Shibuya M., Masuda K., Ebizuka Y., Squalene cyclase and oxidosqualene cyclase from a fern. FEBS Lett. 582, 310–318 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Ma K., et al. , Characterization and biosynthesis of a rare fungal hopane-type triterpenoid glycoside involved in the antistress property of Aspergillus fumigatus. Org. Lett. 21, 3252–3256 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Inayama S., et al. , Isolation of a hopane-type triterpenoid, zeorin, from a higher plant, Tripterygium regelii. Chem. Pharm. Bull. (Tokyo) 37, 2836–2837 (1989). [Google Scholar]
- 34.Dang T., Prestwich G. D., Site-directed mutagenesis of squalene-hopene cyclase: Altered substrate specificity and product distribution. Chem. Biol. 7, 643–649 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Paul S., Dutta S., Preservation of hopenol-B in Eocene dammar resins: Insights into the evolution of biochemical synthesis of angiosperm metabolites. Geobios 50, 453–457 (2017). [Google Scholar]
- 36.Ma P. F., et al. , The Pharus latifolius genome bridges the gap of early grass evolution. Plant Cell 33, 846–864 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., et al. , Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 12, 2563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filiault D. L., et al. , The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife 7, e36426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones D. T., Taylor W. R., Thornton J. M., The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forli S., et al. , Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed J., et al. , A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab. Eng. 42, 185–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Raw and assembled RNA-seq and genomic DNA data can be found in a previously published paper (37). DNA squences of 13 AsOSC genes in SI Appendix, Table S2 and AcHS1 can also be found in GenBank (accession nos. OM401318–OM401331).