Abstract
Simple Summary
Lung cancer is the leading cause of malignancy-related mortality worldwide. AI has the potential to help to treat lung cancer from detection, diagnosis and decision making to prognosis prediction. AI could reduce the labor work of LDCT, CXR, and pathology slides reading. AI as a second reader in LDCT and CXR reading reduces the effort of radiologists and increases the accuracy of nodule detection. Introducing AI to WSI in digital pathology increases the Kappa value of the pathologist and help to predict molecular phenotypes with radiomics and H&E staining. By extracting radiomics from image data and WSI from the histopathology field, clinicians could use AI to predict tumor properties such as gene mutation and PD-L1 expression. Furthermore, AI could help clinicians in decision-making by predicting treatment response, side effects, and prognosis prediction in medical treatment, surgery, and radiotherapy. Integrating AI in the future clinical workflow would be promising.
Abstract
Lung cancer is the leading cause of malignancy-related mortality worldwide due to its heterogeneous features and diagnosis at a late stage. Artificial intelligence (AI) is good at handling a large volume of computational and repeated labor work and is suitable for assisting doctors in analyzing image-dominant diseases like lung cancer. Scientists have shown long-standing efforts to apply AI in lung cancer screening via CXR and chest CT since the 1960s. Several grand challenges were held to find the best AI model. Currently, the FDA have approved several AI programs in CXR and chest CT reading, which enables AI systems to take part in lung cancer detection. Following the success of AI application in the radiology field, AI was applied to digitalized whole slide imaging (WSI) annotation. Integrating with more information, like demographics and clinical data, the AI systems could play a role in decision-making by classifying EGFR mutations and PD-L1 expression. AI systems also help clinicians to estimate the patient’s prognosis by predicting drug response, the tumor recurrence rate after surgery, radiotherapy response, and side effects. Though there are still some obstacles, deploying AI systems in the clinical workflow is vital for the foreseeable future.
Keywords: artificial intelligence, machine learning, lung cancer, radiomics, whole slide imaging, survival prediction
1. Introduction
Lung cancer constitutes the largest portion of malignancy-related deaths worldwide [1]. It is also the leading cause of malignancy-related death in Taiwan [2,3]. The majority of the patients diagnosed with lung cancer are in the late-stage, and therefore have a poor prognosis. In addition to the late stage at diagnosis, the heterogeneity of imaging features and histopathology of lung cancer also makes it a challenge for clinicians to choose the best treatment option.
The imaging features of lung cancer vary from a single tiny nodule to ground-glass opacity, multiple nodules, pleural effusion, lung collapse, and multiple opacities [4]; simple and small lesions are extremely difficult to detect [5]. Histopathological features include adenocarcinoma, squamous cell carcinoma, small cell carcinoma, and many other rare histological types. The histology subtypes vary even more. For example, at least six common subtypes and a total of eleven subtypes of adenocarcinoma were reported in the 2015 World Health Organization classification of lung tumors [6], with more subtypes added to the 2021 version [7]. Treatment options are heavily dependent on the clinical staging, histopathology, and genomic features of the lung cancer. In the era of precision medicine, clinicians need to collect all the features and make a decision to administer chemotherapy, targeted therapy, immunotherapy, and/or combined with surgery or radiotherapy.
Whether to treat or not to treat the disease is always a question in daily practice. Clinicians would like to know the true relationship between the observations and interventions (inputs) and the results (outputs). In other words, to find a model for disease detection, classification, or prediction. Currently, this knowledge is based on clinical trials and the experience of doctors. This exhausts the doctors in reading images and/or pathology slides repeatedly to make an accurate diagnosis. Reviewing charts to determine the best treatment options for patients also consumes a considerable amount of time. A good prediction/classification model would simplify the entire process. Here, artificial intelligence(AI) is introduced.
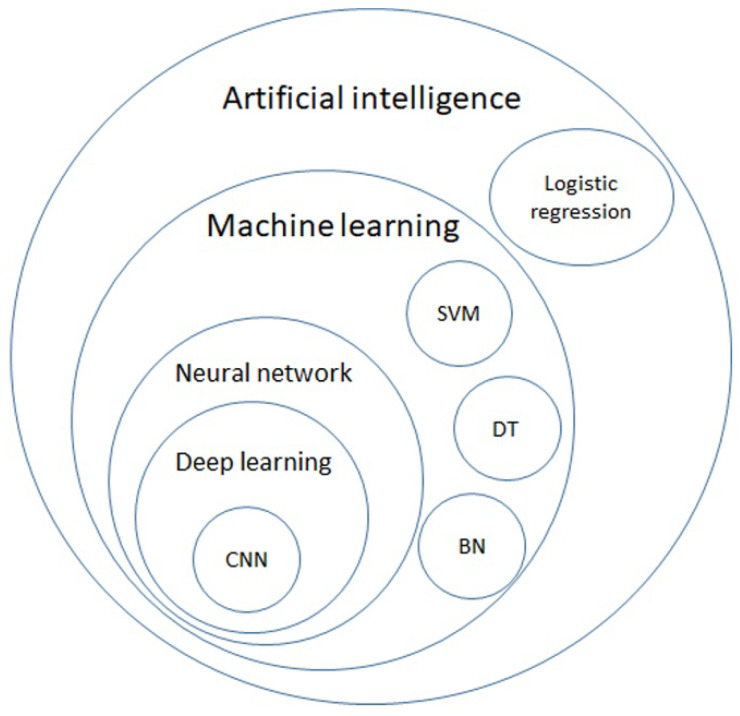
AI is a general term that does not have a strict definition. AI is an algorithm driven by existing data to predict or classify objects [8]. The main components include the dataset used for training, pretreatment method, an algorithm used to generate the prediction model, and the pre-trained model to accelerate the speed of building models and inherit previous experience. Machine learning (ML) is a subclass of AI, and is the science of obtaining algorithms to solve problems without being explicitly programmed, including decision trees (DTs), support vector machines (SVMs), and Bayesian networks (BNs). Deep learning is a further subclass of ML, featured with multiple layered ML, achieving feature selection and model fitting at the same time [9]. The hierarchical relationship between those definitions is displayed in Figure 1.
Figure 1.
Venn diagram of artificial intelligence (AI), machine learning (ML), neural network, deep learning, and further algorithms in each category. AI is a general term for a program that predicts an answer to a certain problem, where one of the conventional methods is logistic regression. ML learns the algorithm through input data without explicit programming. ML includes algorithms such as decision trees (DTs), support vector machines (SVMs), and Bayesian networks (BNs). By using each ML algorithm as a neuron with multiple inputs and a single output, a neural network is a structure that mimics the human brain. Deep learning is formed with multiple layers of neural networks, and convolutional neural network (CNN) is one of the elements of the famous architecture.
However, to develop such a model, a large amount of computation is required. In the past, building a multidimensional algorithm for image analysis has taken hours, or even days, for the human brain. The large computational power required becomes a significant obstacle in creating a sophisticated prediction model. The booming computational power of chip technology and software optimization makes large and sophisticated calculations easier to achieve [10,11]. When a large matrix can be computed in a short time, it is possible to develop models that are much more complex than linear regression or logistic regression. DTs and SVMs were used to build models in the ML era around the year 2000 [12]. By estimating the probability, BNs were used to select treatments by predicting survival [13]. In recent years, deep learning models, including artificial neural networks (ANNs), convolutional networks (CNNs), recurrent neural networks (RCNNs), long-term and short-term memory (LSTM) [14], and generative adversarial networks (GANs) [15], have outperformed most old models and are thus widely used in research and commercial fields [16].
In the 21st century, human life has been largely integrated with AI, and this trend also extends to the medical field. The heterogeneity of lung cancer makes it the best field for AI application. A large number of studies have reported the application in lung nodule detection, diagnostic application in histopathology, disease risk stratification, drug development, and even prognosis prediction. In this article, we present a narrative review of AI applications in lung cancer by introducing AI models first and then reported applications according to the clinical workflow: screening, diagnosis, decision making, and prognosis prediction. Table 1 listed the potential AI application fields in lung cancer.
Table 1.
Summary of AI application fields.
| Screening | Diagnosis | Treatment |
|---|---|---|
| Radiology: CXR [17,18,19,20,21] CXR [17,18,19,20,21] LDCT [22,23,24,25,26,27,28,29,30,31,32,33] Novel tools: Genomics [34] Genomics [34] Proteomics [35,36] Exhaled breath [37,38,39] |
Risk prediction: Radiomics [40,41,42,43,44,45,46] WSI [47,48,49,50,51,52,53] WSI [47,48,49,50,51,52,53] Genomics [50,54] |
Tumor property classification: Drug selection [44,45,46,50,55,56,57] Prognosis prediction: Drug treatment response [58,59,60] Post-Surgery recurrence [54,61,62] Radiotherapy response [63,64] Side effect estimation: Radiation pneumonitis [65,66] |
CXR: Chest X-ray, LDCT: low-dose computed tomography, WSI: whole slide imaging.
2. AI Models
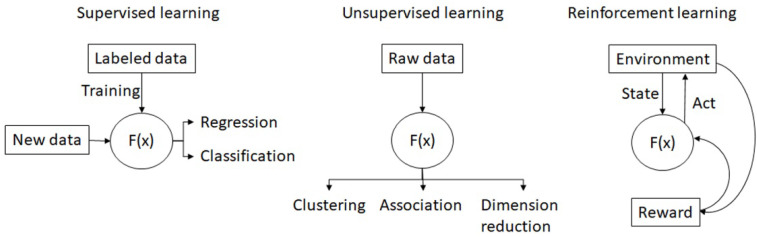
Numerous AI models are constructed with different algorithms are published nowadays. Generally, the AI models can be divided into: supervised learning, unsupervised learning, semi-supervised learning [9], and reinforcement learning (Figure 2).
Figure 2.
The concept map of supervised learning, unsupervised learning and reinforcement learning.
2.1. Supervised Learning
In supervised learning, researchers need to prepare the labeled dataset with both inputs and desired outputs (answers) to train the algorithm. It is suitable to solve prediction problems, such as classification and regression. The architecture of the algorithms varies. Researchers can use multiple binary nodes to create DTs as a classifier, or find a plane in a multidimensional space as a SVM classifier. Bayesian classifiers used input data to calculate the probability of correct classification. With the probability calculated from the above-mentioned algorithm, researchers can turn the answer into a continuous variable to solve regression problems and vice versa. Most AI applications predicting survival [13,59], cancer risk [34,35,36,37,38,39], nodule detection [22,23], and nodule characteristics [33] are based on supervised learning.
2.2. Unsupervised Learning
In unsupervised learning, the algorithm divides the samples according to the inputs by itself. Labeled data are not necessary. It is suitable to do clustering, to find associations between samples, and to do dimensionality reduction. For example, cluster analysis was used to find oncogenes in lung cancer [67,68].
2.3. Semi-Supervised Learning
Though supervised learning provides a more accurate algorithm, the labeled data are relatively rare, and the labeling process is labor intensive. Unsupervised learning can adopt unlabeled data but the algorithm is less accurate. Therefore, semi-supervised learning could have both of the advantages when using supervised learning to generate a labeling tool and use supervised learning to generate a large scaled labeled dataset for further training [52].
2.4. Reinforcement Learning
Reinforcement learning is a reward-based system. The algorithm evolves as it interacts with the environment (dataset). A reward function is used to adjust the algorithm or the network. This type of AI is famous for playing chess, shogi, and Go through self-play [69] or generating data with GANs [70]. With this technique, researchers can develop a self-evolving AI for nodule hunting on CT images and achieve better accuracy [15,71,72].
In conclusion, there is no best method to build AI models for all. The best method should be tailored according to the clinical question and the dataset used for training.
3. Screening
Approximately 7% of patients diagnosed with lung cancer are asymptomatic [73], and more than half of the patients who underwent lung cancer resection were asymptomatic [74]. Several attempts at screening have been made, including imaging, sputum cytology [75,76], blood test screening [77,78], and breath test [79,80]. However, only image screening is able to provide the relevant clues. Although chest X-rays (CXRs) are widely used clinically, low-dose computed tomography (LDCT) is the only method that has been proven to diagnose lung cancer earlier and extend the survival of lung cancer patients [81,82].
The reading workflow of repetitive imaging provides room for AI to participate, because human eyes become sore and images start to blur after reading images for a long time. Furthermore, mistakes in reading CXR or LDCT images occur, and it constitutes a large number of malpractice law suits [83]. Though experts were shown to detect more pulmonary nodules on CXRs [84], approximately 20% of lung nodules <3 cm are missed by radiologists [85]. In the 21st century, the prediction accuracy of pulmonary nodules on CXRs has improved with the computer-aided diagnosis systems or AI-based programs. The sensitivity of radiologists improves from 65.1% to 70.3% with the assistance of AI and the false negative rate decreases from 0.2 to 0.18, changing the diagnosis in 6.7% of the cases [17]. In CT images, the sensitivity of lung nodules were more than 90% spotted by AI-based programs [23]. Integrating AI into lung cancer screening protocol is an ongoing event.
3.1. DICOM Format
Digital imaging and communications in medicine (DICOM) is the standard format for image restoration and transfer to enable communication between different servers, manufacturers, and hospitals [86]. The DICOM not only carries pixel data of the image file but also a patient identification number, image type, machine-related parameters, and other information in a format managed by the Medical Imaging and Technology Alliance, a division of the National Electrical Manufacturers Association. After its first publication in 1993, DICOM changed the workflow of radiology, allowing image data to be transmitted quickly and to be analyzed by computers. Later, huge datasets were established for data sharing, model training, or as a benchmark for model testing, and are shown in Table 2.
Table 2.
Summary of frequently used datasets for model training.
| Database | Year | Material | Volume | Features |
|---|---|---|---|---|
| JSRT [87] | 1998 | CXR | 154 | Contains 100 CXRs with malignant nodule, 54 CXRs with benigh nodule, and 93 normal CXRs |
| Shenzhen CXR set [88] | 2012 | CXR | 662 | Contains 326 normal CXRs, and 336 CXRs with tuberculosis. Ribs were labeled. |
| Montgomery CXR set [88] | 2014 | CXR | 138 | Contains 80 normal CXRs, and 58 CXRs with tuberculosis. Ribs were labeled. |
| ChestXray8 [89] | 1992–2015 | CXR | 108,948 | Classified into 8 features: atelectasis, cardiomegaly, effusion, infiltration, mass, nodule, normal, pneumonia, and pneumothorax |
| ChestXray14 [89] | 1992–2015 | CXR | Classified into 14 features: atelectasis, cardiomegaly, consolidation, edema, effusion, emphysema, fibrosis, hernia, infiltration, mass, nodule, pleural thickening, pneumonia, pneumothorax. | |
| PadChest [90] | 2009–2017 | CXR | >160,000 | Labeled with 174 different radiographic findings, 19 differential diagnoses and 104 anatomic locations |
| LIDC [91] | 2011 | LDCT | 1018 | Nodules were annotated and labeled with nodule sizes |
| LUNA16 [23] | 2016 | LDCT | 888 | Adapted from LIDC, with additional nodules found during model training. 1186 lung nodules annotated in 888 CT scans |
| MIMIC-CXR [92] | 2011–2016 | CXR | 377,110 | Classified into 14 labels derived from two natural language processing tools. |
| ChestXpert [93] | 2019 | CXR | 224,316 | Labeled with 14 features: no finding, enlarged cardiom, cardiomegaly, lung opacity, lung lesion, edema, consolidation, pneumonia, atelectasis, pneumothorax, pleural effusion, pleural other, fracture, support devices |
| VinDr-RibCXR [94] | 2020 | CXR | 18,000 | Rib suppression images |
| RadGraph [95] | 2021 | CXR | 500 | Inference dataset of MMIC-CXR and reports |
| REFLACX [96] | 2021 | CXR | 3032 | Labeled by 5 radiologists and synchronized sets of eye-tracking data and timestamped report transcriptions |
CXR: chest CX-ray set, JSRT: Japanese Society of Radiological Technology, LIDC: Lung Image Database Consortium, LUNA: LUng Nodule Analysis, REFLACX: Reports and Eye-Tracking Data for Localization of Abnormalities in Chest X-rays.
3.2. CXR
CXRs are the most frequently used imaging modality in the medical field. With 0.1 mSv radiation exposure, similar to 10 days of natural background radiation, CXR provides a good examination of the patient’s thorax. Far before digital imaging, the computer-aided diagnosis(CAD) system for CXR has been developed since the 1960s [97]. Image features, such as shape, size, intensity, and texture, must be manually labeled before being sent for further analysis. In the digital era, computers can directly analyze images. By computing the image pixel-by-pixel, radiomics expands the definition of image features from a computer perspective. By computing the image texture and density using different mathematical techniques, the region of interest area can be converted to higher dimension data and expressed as a huge matrix. Because the principle of radiomics is math, the various image qualities of CXR give the computer another task. To obtain accurate radiomics data, image augmentation is an important procedure before nodule detection [98], including pre-processing, lung segmentation [88], and rib suppression [94].
Further malignancy/benign classification was performed using a different algorithm. DT-based algorithms were widely used to analyze these features before 2011. Later, deep-learning-based algorithms demonstrated their power in image analysis. CheXNet, a radiologist-level deep learning algorithm trained on Chest-Xray14, one of the largest CXR databases in the world, exceeds radiologist performance in the detection of 14 pulmonary diseases, including lung nodules and lung masses with an area under the receiver operating curve (AUROC, AUC) 0.78 and 0.87, respectively [18]. Further deep learning models pushed the sensitivity to 0.83 at a false-positive rate of 0.2 per CXR [19]. Currently, several software programs have been approved by the FDA [17,20,21].
3.3. Chest CT
CT technology provides a noninvasive method to explore the 3-dimensional structure of the thorax. As the technology advanced, the radiation exposure has reduced from 7 mSv (conventional chest CT) to 1.6 mSv (LDCT). Screening with LDCT showed an approximately 20% mortality reduction in two large randomized control trials: the National Lung Screening Trial (NLST) [81] and the Dutch-Belgian Randomized Lung Cancer Screening Trial (Dutch acronym: NELSON study) [82]. The Multicentric Italian Lung Detection (MILD) trial showed that prolonged LDCT screening for more than five years reduced lung cancer mortality and overall mortality at ten years. These trials boosted the demand for image reading. The application of AI in LDCT reading can help radiologists reduce laborious work, minimize reader variability, and improve screening efficiency [99,100]. The main task for AI application in LDCT reading is the same as in CXR: nodule detection and classification/malignancy prediction. However, unlike CXR, the radiodensity of LDCT is based on an international standard scale with a Hounsfield unit (HU) and fixed resolution. The preprocessing of the CT images focused on denoising, resizing, and lung segmentation.
Many studies have used AI algorithms to detect lung nodules in chest CT images [101,102]. Because they used different models on different datasets and evaluated the models with different benchmarks, such as sensitivity, specificity, AUC, and accuracy, it was difficult to evaluate the models scientifically. A series of grand challenges, such as the Automated Nodule Detection 2009 (ANODE09) study [22] and the Lung Nodule Analysis 2016 (LUNA16) challenge [23] were conducted to find the benchmark model of nodule detection on CT. The best algorithm achieved a sensitivity of 97.2% at one false-positive rate per scan. AI has also been proven to help radiologists increase the sensitivity of nodule detection [24,25,26] and reduce interpretation time. AI has proven to be a good concurrent reader or a second reader. It was noticeable that consumer AI was not approved as the first reader, in case that the radiologists may not have the chance to access to the AI-missed nodules.
Lung nodule classification and malignancy prediction are important tasks in nodule detection. Nodules are classified according to their texture as solid, part-solid, or non-solid, and their size. An AI model trained on the MILD trial [103] dataset and externally validated on the Danish Lung Cancer Screening Trial (DLCST) [104] showed that AI performed equivalently to a human expert on differential six textures (sold, part-solid, non-solid, calcified, perifissural, and speculated) [27]. The classification was then used to predict the malignancy probability as recommended by the Lung CT Screening Reporting and Data System (LUNG-RADS) [105] and the Fleischner guideline [106]. Traditionally, researchers have used AI to classify lung nodules as a feature extraction step, and then go for malignancy prediction. Later, researchers substituted the classification step with radiomics’ feature extraction to increase prediction accuracy [28,29,30].
Similar to nodule detection on CT, challenges were conducted to compare the prediction models. In 2015, the LUNGx Challenge for computerized lung nodule classification was established. Ten teams sent their reports with AUC between 0.50 and 0.68, and only three of them performed statistically better than a random guess, while radiologists performed with AUC between 0.70–0.85. The technology advanced rapidly in the ISBI 2018 Lung Nodule Malignancy Prediction Challenge, and 11 participants completed the challenge with an AUC between 0.70–0.91. The top five participants used deep learning models with AUC between 0.87–0.91 without significant differences from each other [31]. The accuracy was 93% with a sensitivity of 82% and precision of 84% based on the weighted voting method of the autoencoder, ResNet, and handicraft features [32], and 96% with deep convolutional network learning (DCN) [33].
3.4. Novel Screening Tests
Genomics [107], proteomics, microbiomes [108], and exhaled breath [109] are novel screening tools for lung cancer [110]. Although these screening methods yield a large set of signals for each patient, an advanced algorithm would elevate the diagnostic yield.
Genomics is one of the most popular topics in oncology. With the polymerase chain reaction (PCR) amplification method and related technology, scientists can now analyze the whole genome [111], exome, transcriptome [112], and epigenome of cancers and produce large sets of information about patients and their tumors. By analyzing whole-genome and whole-transcriptome sequencing data from treatment-naïve patients in The Cancer Genome Atlas [113] (TCGA), the machine learning model successfully discriminated cancer-free healthy controls from patients with cancer [34].
Proteins and other metabolites acquired from plasma and urine samples are relatively easy to obtain and have been studied as a screening tool for lung cancer for decades. To handle the extremely large number of variables produced by proteomics, researchers have used machine learning methods to reduce dimensionality and feature selection [35,36]. In 2003, the machine learning methods were applied to analyze 1676 original and 124 prescreened mass spectra data from 24 diseased and 17 healthy specimens, and researchers successfully built predictive models to discriminate lung cancer specimens from healthy specimens. However, the most accurate predictions were obtained using less interpretable models [35]. Following this idea, the urine proteome combined with machine learning analysis successfully established models that can discriminate lung cancer samples not only from healthy ones, but also from samples from other cancers [36].
Exhaled breath is composed of volatile organic compounds (VOCs) and exhaled breath condensates [79,109]. To date, more than 3000 VOCs have been identified to be related to lung cancer [114], however, not a single VOC could be accurate enough for diagnosis. Therefore, a composite prediction model is a way to solve this problem, in addition to escalating sensor technology. In 2018, a logistic regression model that was able to discriminate patients with lung cancer in both smokers (sensitivity, 95.8%; specificity, 92.3%), and non-smokers (sensitivity, 96.2%; specificity, 90.6%) [37]. Using this, an SVM model [38] and an ANN model [39] were created.
4. Diagnosis
When a nodule is detected, clinicians must know the properties of the lung nodule. The gold standard is to acquire tissue samples via either biopsy or surgery. The image features provide a way to guess the properties of the lung nodule by radiomics as mentioned in the previous section. Aside from imaging features, the histopathological features also affect further treatment. Following the path of digital radiology, whole slide imaging (WSI) has opened the trend of digital histopathology. With digitalized WSI data, AI can help pathologists with daily tasks and beyond, ranging from tumor cell recognition and segmentation [47], histological subtype classification [48,49,50,51], PD-L1 scoring [52], to tumor-infiltrating lymphocyte (TIL) count [53].
4.1. Radiomics
Following the idea of radiomics in nodule detection and malignancy risk stratification, radiomics was applied to predict the histopathological features of lung nodules/masses [40]. Researchers used logistic regression of radiomics and clinical features to distinguish small cell lung cancer from non-small cell lung cancer with an AUC of 0.94 and an accuracy of 86.2% [41]. The LASSO logistic regression model was used to classify adenocarcinomas and squamous cell carcinomas in the NSCLC group [42]. Further molecular features such as Ki-67 [43], epidermal growth factor receptor (EGFR) [44], anaplastic lymphoma kinase (ALK) [45], and programmed cell death 1 ligand, (PD-L1) [46] were also shown to be predictable with AI-analyzed radiomics, a non-invasive and simple method.
4.2. WSI
The emergence of WSI is a landmark in modern digital pathology. The WSI depends on a slide scanner that can transform glass slides into digital images with the desired resolution. Once the images are stored on the server, pathologists can view them on their personal computers or handheld devices. Similar to DICOM in diagnostic radiology, in 2017, the FDA approved two vendors for the WSI system for primary diagnosis [115,116]. Meanwhile, the DICOM also planned support for WSI in the PACS systems to facilitate the adaption of digital pathology in hospitals and further information exchange [117,118]. These features enable the building of a digital pathology network to share expertise for consultations and make education across the country possible [119].
Each WSI digital slide is a large image. It may contain more than 4 billion pixels and may exceed 15 GB when scanned with a resolution of 0.25 micrometers/pixel, referred to as 40× magnification [118,120]. With recent advances in AI and DL in image classification, segmentation, and transformation, digitalized WSI provides another broad field to play. There are many applications for deep learning in cytopathology.
4.3. Histopathology
Detecting cancerous regions is the most basic and essential task of deep learning in pathology. Some models combine the detection, segmentation, and histological subtyping together [47,48,49]. Accuracy depends on the data quality, quantity, and abundance of the malignant cell differentiation status. It is difficult to perform histological subtyping of lung cancer without special immunohistochemistry (IHC) staining. This causes inter-observer disagreement when reading H&E staining. While the agreement between pathologists came to a Kappa value of 0.485, a trained AI model can achieve a Kappa value of up to 0.525 when compared with a pathologist [48]. In the detection of lymph node metastasis, a well-trained AI model can help reduce human workload and prevent errors [121]. It obviously performs better than a pathologist in a limited time and has a greater detection rate of single-cell metastasis or micro-metastasis [121].
Although WSI with H&E-stained slides is designed to view the morphology of tissues, with the aid of AI, researchers have designed methods to predict specific gene mutations, PD-L1 expression level, treatment response, and even the prognosis of patients. Focusing on lung adenocarcinoma, Coudray et al. developed an AI application using Inception-V3 for the prediction of frequently appearing gene mutations including STK11, EGFR, FAT1, SETBP1, KRAS, and TP53 [50]. The AUC of this prediction reached 0.754 for EGFR and 0.814 for KRAS which can be treated with effective targeted agents. Sha et al., used ResNet-18 as the backbone to predict the PD-L1 status in NSCLC [55]. Their model showed an AUC between 0.67 and 0.81, while different PD-L1 cutoff levels were chosen. They believed that the morphological features may be related to PD-L1 expression level.
Next-generation sequencing (NGS) plays an important role in modern lung cancer treatment [122]. Successful NGS testing depends on a sufficient number of tumor cells and tumor DNA. AI can assist in determining tumor cellularity [123,124]. In addition, a trained AI can help count the immune cells, while the tissue specimen is adequately stained for special surface markers [53]. Since the PD-L1 expression level is the key predictor for immunotherapy in lung cancer, AI has been trained to count the proportion score for PD-L1 expression [52,125]. When properly stained, computer-aided PD-L1 scoring and quantitative tumor microenvironment analysis may meet the requests of pathologists, and eliminate inter-observer variations and achieve precise lung cancer treatment [126].
However, there are several barriers to the translation of AI applications into clinical services. First, AI applications may not work well when applied to other pathology laboratories, scanners, or diverse protocols [127]. Second, most AIs are designed for their own unique functions. Users are requested to launch several applications for different purposes and spend a lot of time transferring the data. Medical devices powered by AI applications require approval by regulations. Most articles and works were in-house studies and laboratory-developed tests. All of these barriers may restrict the deployment of trained AI models in daily clinical practice [119].
4.4. Cytology
The WSI for cytology differs from pathology. Cytology slides are not even sliced flat layers. Instead, they have an entire cell on the glass and would be multiple cell layers. Cytologists tend to use the focus function and look into the cells. While digitalizing the cytology glass slide, the focus function was simulated through the Z-stack function and multiple layers of different focus [128,129]. This method yields a larger WSI file, approximately 10 times that of a typical histological case. Multiple image layers also increase complexity and pose challenges to AI applications.
Few articles have discussed cytology, especially those focusing on lung cancer. For thyroid cancer, Lin et al. proposed a DL method for thyroid fine-needle aspiration (FNA) samples and ThinPrep (TP) cytological slides for detecting papillary thyroid carcinoma [130]. The authors did not claim the ability to detect other cell types of thyroid cancer using their method. AI can be performed for various cytology samples from lung cancer patients, including pleural effusion, lymph node aspiration, tissue aspiration samples, and endobronchial ultrasound-guided fine-needle aspiration (EBUS-TBNA) of mediastinal lymph nodes.
5. Decision Making and Prognosis Prediction
Oncologists prefer to deploy this technique to its limits. There are many exciting possibilities for the use of the AI technique. By predicting treatment response, including survival and adverse events, AI was proven to have the potential to play a role in clinical decision making [13], to help surgeons choose the specific groups of patients to receive surgery, and to aid radiotherapists in planning the radiation zone.
5.1. Medication Selection
In late-stage lung cancer, the identification of driver mutations, PD-L1 expression, and tumor oncogenes affects most the treatment of choice. Using WSI and radiomics, AI could help to identify EGFR mutations [44,50], ALK [45], and PD-L1 expression [46,55,56]. EGFR mutation subtypes have also been classified using radiomic features [57].
Another research point is the use of radiomics, WSI, and clinical data to directly predict cancer treatment response or survival [131]. Dercle et al. retrospectively analyzed the data from prospective clinical trials and found that the AI model based on the random forest algorithm and CT-based radiomic features predicted the treatment sensitivity of nivolumab with an AUC of 0.77, docetaxel with an AUC of 0.67, and gefitinib with an AUC of 0.82 [58]. CT-based radiomics models have also been reported to predict the overall survival of lung cancer [59,60].
One patent application publication declared that using radiomics features of segmented cell nuclei of lung cancer can predict responses to immunotherapy with an AUC up to 0.65 in the validation dataset [132]. Although there is no specific survival prediction model for lung cancer, Ellery et al. developed a risk prediction model using the TCGA Pan-Cancer WSI database including lung cancer [133]. However, the DL algorithm did not provide acceptable prediction power for lung adenocarcinoma or lung squamous cell carcinoma.
5.2. Surgery
The gold standard for the treatment of early-stage lung cancer is surgical resection. AI was applied to pre-surgical evaluation [61,62], and prognosis prediction after surgery, and could help identify patients who are suitable to receive adjuvant chemotherapy after surgery [54].
In pre-surgical evaluation, radiologist-level AI could help predict visceral pleural invasion [62], and identify early stage lung adenocarcinomas suitable for sub-lobar resection [61]. After surgery, AI could play a role in predicting prognosis. The model based on radiomic feature nomograms could identify high-risk groups whose postsurgical tumor recurrence risk is 16-fold higher than that of low-risk group [134]. The CNN model pre-trained with the radiotherapy dataset successfully predict a 2-year overall survival after surgery [135]. The model integrating genomic and clinicopathological features was able to identify patients at risk for recurrence and who were suitable to receive adjuvant therapy [54].
5.3. Radiotherapy
SBRT is currently the standard of care to treat early-stage lung cancer and/or provide local control for patients who are medically inoperable or refuse surgery. Radiomics-based models have been reported to successfully predict 1-year tumor recurrence via CT scans performed after 3 and 6 months of SBRT [63]. Lewis and Kemp also developed a model trained on TCGA dataset to predict cancer resistance to radiation [64]. As a well-known side effect of radiotherapy, radiation pneumonitis can be lethal, and clinicians would like to prevent this situation. The AI model based on pretreatment CT radiomics was superior to the traditional model using dosimetric and clinical predictors in predicting radiation pneumonitis [65]. Another ANN algorithm trained with radiomics extracted from a 3D dose map of radiotherapy has been shown to predict the acute and late pulmonary toxicities with an accuracy of 0.69 [66]. A well-designed prediction model for radiation pneumonitis may help to prevent radiation pneumonitis in the future.
6. Future Development
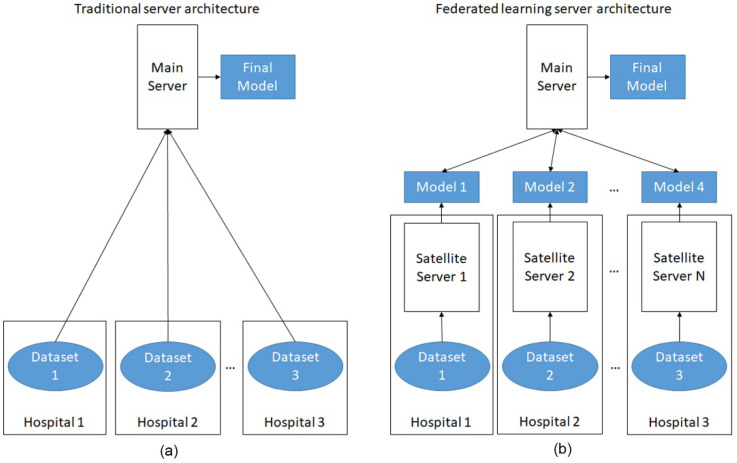
The future of AI applications in lung cancer could focus on integration and applications. First, because AI is a data-driven technology, scientist can integrate small datasets to create large data sets for training. However, regulations regarding data sharing are a huge obstacle for researchers. Federated learning, a method that shares the trained parameters rather than sharing the data, is a simple solution [136,137]. In federated learning, the models were trained at each different hospitals separately and only the trained models were sent to the main server, so that the main server does not touch the raw data directly. The final model was then reported back to individual hospitals (Figure 3).
Figure 3.
The comparison of traditional AI server architecture and federated learning server architecture. (a) In traditional server architecture, the main server processes all the raw data at the same site, leading to concerns about privacy; (b) In federated learning, the datasets are processed at each individual site and only the trained models are shared with the main server. Privacy of each dataset is protected.
Second, most previous researches were conducted by separate specialists and focused on separated fields such as radiology, pathology, surgery, or clinical oncology. However, integrating all aspects such as radiology, pathology, demographics and clinical data, and both old and new technologies could better reflect reality. The combination of different features also helps researchers build predictive models [138,139]. This brings about the idea of multi-omics [140,141] or “Medomics” [40]. Similar to multidisciplinary teams in clinical lung cancer treatment [142,143], the combination of different domain knowledge and multidisciplinary integration is worth pursuing in the future.
Apart from improvement in model accuracy by increasing the training sample size and multidisciplinary integration, another issue is the application of AI programs. Although the studies above all showed the promising results of applying AI in lung cancer and some products were approved by the FDA [17,20,21,116,144], real implementation of the clinical workflow is rare. The user interface, speed of data analysis, expanse of the AI program, internet bandwidth, and resources consumed by the AI program are all barriers to real-world applications. More infrastructure needs to be constructed before we can enter the AI-assisted world.
Acknowledgments
Thanks to the Ministry of Science and Technology, Taiwan, for funding.
Author Contributions
Conceptualization, H.-Y.C.; methodology, H.-Y.C. and H.-S.C.; writing—original draft preparation, H.-Y.C. and H.-S.C.; writing—review and editing, H.-S.C. and Y.-M.C.; supervision, Y.-M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Technology, Taiwan, grant number MOST 110-2321-B-075-001.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cancer. [(accessed on 29 November 2021)]. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer.
- 2.Luo Y.H., Chiu C.H., Scott Kuo C.H., Chou T.Y., Yeh Y.C., Hsu H.S., Yen S.H., Wu Y.H., Yang J.C., Liao B.C., et al. Lung Cancer in Republic of China. J. Thorac. Oncol. 2021;16:519–527. doi: 10.1016/j.jtho.2020.10.155. [DOI] [PubMed] [Google Scholar]
- 3.Cause of Death Statistics. [(accessed on 1 October 2021)]; Available online: https://www.mohw.gov.tw/lp-4650-2.html.
- 4.Panunzio A., Sartori P. Lung Cancer and Radiological Imaging. Curr. Radiopharm. 2020;13:238–242. doi: 10.2174/1874471013666200523161849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migliore M., Palmucci S., Nardini M., Basile A. Imaging patterns of early stage lung cancer for the thoracic surgeon. J. Thorac. Dis. 2020;12:3349–3356. doi: 10.21037/jtd.2020.02.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M.B., Chirieac L.R., Dacic S., Duhig E., Flieder D.B., et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson A.G., Tsao M.S., Beasley M.B., Borczuk A.C., Brambilla E., Cooper W.A., Dacic S., Jain D., Kerr K.M., Lantuejoul S., et al. The 2021 WHO Classification of Lung Tumors: Impact of advances since 2015. J. Thorac. Oncol. 2021;17:362–387. doi: 10.1016/j.jtho.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Klang E. Deep learning and medical imaging. J. Thorac. Dis. 2018;10:1325–1328. doi: 10.21037/jtd.2018.02.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson C.E., Marti J.M., Radivojevic T., Jonnalagadda S.V.R., Gentz R., Hillson N.J., Peisert S., Kim J., Simmons B.A., Petzold C.J., et al. Machine learning for metabolic engineering: A review. Metab. Eng. 2021;63:34–60. doi: 10.1016/j.ymben.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Leiserson C.E., Thompson N.C., Emer J.S., Kuszmaul B.C., Lampson B.W., Sanchez D., Schardl T.B. There’s plenty of room at the Top: What will drive computer performance after Moore’s law? Science. 2020;368:eaam9744. doi: 10.1126/science.aam9744. [DOI] [PubMed] [Google Scholar]
- 11.Shalf J. The future of computing beyond Moore’s Law. Philos. Trans. A Math. Phys. Eng. Sci. 2020;378:20190061. doi: 10.1098/rsta.2019.0061. [DOI] [PubMed] [Google Scholar]
- 12.Somvanshi M., Chavan P., Tambade S., Shinde S. A review of machine learning techniques using decision tree and support vector machine; Proceedings of the 2016 International Conference on Computing Communication Control and Automation (ICCUBEA); Pune, India. 12–13 August 2016; pp. 1–7. [Google Scholar]
- 13.Sesen M.B., Nicholson A.E., Banares-Alcantara R., Kadir T., Brady M. Bayesian networks for clinical decision support in lung cancer care. PLoS ONE. 2013;8:e82349. doi: 10.1371/journal.pone.0082349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao R., Huo Y., Bao S., Tang Y., Antic S.L., Epstein E.S., Balar A.B., Deppen S., Paulson A.B., Sandler K.L. International Workshop on Machine Learning in Medical Imaging. Springer; New York, NY, USA: 2019. Distanced LSTM: Time-distanced gates in long short-term memory models for lung cancer detection; pp. 310–318. [PMC free article] [PubMed] [Google Scholar]
- 15.Onishi Y., Teramoto A., Tsujimoto M., Tsukamoto T., Saito K., Toyama H., Imaizumi K., Fujita H. Automated pulmonary nodule classification in computed tomography images using a deep convolutional neural network trained by generative adversarial networks. BioMed Res. Int. 2019;2019:6051939. doi: 10.1155/2019/6051939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kourou K., Exarchos T.P., Exarchos K.P., Karamouzis M.V., Fotiadis D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sim Y., Chung M.J., Kotter E., Yune S., Kim M., Do S., Han K., Kim H., Yang S., Lee D.J., et al. Deep Convolutional Neural Network-based Software Improves Radiologist Detection of Malignant Lung Nodules on Chest Radiographs. Radiology. 2020;294:199–209. doi: 10.1148/radiol.2019182465. [DOI] [PubMed] [Google Scholar]
- 18.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., Ding D., Bagul A., Langlotz C., Shpanskaya K. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv. 20171711.05225 [Google Scholar]
- 19.Kim Y.-G., Cho Y., Wu C.-J., Park S., Jung K.-H., Seo J.B., Lee H.J., Hwang H.J., Lee S.M., Kim N. Short-term reproducibility of pulmonary nodule and mass detection in chest radiographs: Comparison among radiologists and four different computer-aided detections with convolutional neural net. Sci. Rep. 2019;9:18738. doi: 10.1038/s41598-019-55373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tam M., Dyer T., Dissez G., Morgan T.N., Hughes M., Illes J., Rasalingham R., Rasalingham S. Augmenting lung cancer diagnosis on chest radiographs: Positioning artificial intelligence to improve radiologist performance. Clin. Radiol. 2021;76:607–614. doi: 10.1016/j.crad.2021.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Kim J.H., Han S.G., Cho A., Shin H.J., Baek S.-E. Effect of deep learning-based assistive technology use on chest radiograph interpretation by emergency department physicians: A prospective interventional simulation-based study. BMC Med. Inform. Decis. Mak. 2021;21:311. doi: 10.1186/s12911-021-01679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Ginneken B., Armato S.G., III, de Hoop B., van Amelsvoort-van de Vorst S., Duindam T., Niemeijer M., Murphy K., Schilham A., Retico A., Fantacci M.E. Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: The ANODE09 study. Med. Image Anal. 2010;14:707–722. doi: 10.1016/j.media.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Setio A.A.A., Traverso A., De Bel T., Berens M.S., Van Den Bogaard C., Cerello P., Chen H., Dou Q., Fantacci M.E., Geurts B. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017;42:1–13. doi: 10.1016/j.media.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Roos J.E., Paik D., Olsen D., Liu E.G., Chow L.C., Leung A.N., Mindelzun R., Choudhury K.R., Naidich D.P., Napel S. Computer-aided detection (CAD) of lung nodules in CT scans: Radiologist performance and reading time with incremental CAD assistance. Eur. Radiol. 2010;20:549–557. doi: 10.1007/s00330-009-1596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo S.B., Freedman M.T., Gillis L.B., White C.S., Mun S.K. JOURNAL CLUB: Computer-aided detection of lung nodules on CT with a computerized pulmonary vessel suppressed function. Am. J. Roentgenol. 2018;210:480–488. doi: 10.2214/AJR.17.18718. [DOI] [PubMed] [Google Scholar]
- 26.Liang M., Tang W., Xu D.M., Jirapatnakul A.C., Reeves A.P., Henschke C.I., Yankelevitz D. Low-dose CT screening for lung cancer: Computer-aided detection of missed lung cancers. Radiology. 2016;281:279–288. doi: 10.1148/radiol.2016150063. [DOI] [PubMed] [Google Scholar]
- 27.Ciompi F., Chung K., Van Riel S.J., Setio A.A.A., Gerke P.K., Jacobs C., Scholten E.T., Schaefer-Prokop C., Wille M.M., Marchiano A. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci. Rep. 2017;7:46479. doi: 10.1038/srep46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y., Li C., Jin L., Gao P., Zhao W., Ma W., Tan M., Wu W., Duan S., Shan Y. Radiomics for lung adenocarcinoma manifesting as pure ground-glass nodules: Invasive prediction. Eur. Radiol. 2020;30:3650. doi: 10.1007/s00330-020-06776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins S., Wang H., Liu Y., Garcia A., Stringfield O., Krewer H., Li Q., Cherezov D., Gatenby R.A., Balagurunathan Y. Predicting malignant nodules from screening CT scans. J. Thorac. Oncol. 2016;11:2120–2128. doi: 10.1016/j.jtho.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu S.-J., Wang C.-W., Pan K.-T., Wu Y.-C., Wu C.-T. Localized thin-section CT with radiomics feature extraction and machine learning to classify early-detected pulmonary nodules from lung cancer screening. Phys. Med. Biol. 2018;63:065005. doi: 10.1088/1361-6560/aaafab. [DOI] [PubMed] [Google Scholar]
- 31.Balagurunathan Y., Beers A., Mcnitt-Gray M., Hadjiiski L., Napel S., Goldgof D., Perez G., Arbelaez P., Mehrtash A., Kapur T. Lung Nodule Malignancy Prediction in Sequential CT Scans: Summary of ISBI 2018 Challenge. IEEE Trans. Med. Imaging. 2021;40:3748–3761. doi: 10.1109/TMI.2021.3097665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao N., Qiang Y., Bilal Zia M., Wang S., Lian J. Ensemble classification for predicting the malignancy level of pulmonary nodules on chest computed tomography images. Oncol. Lett. 2020;20:401–408. doi: 10.3892/ol.2020.11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lv E., Liu W., Wen P., Kang X. Classification of Benign and Malignant Lung Nodules Based on Deep Convolutional Network Feature Extraction. J. Healthc. Eng. 2021;2021:8769652. doi: 10.1155/2021/8769652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poore G.D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Hilario M., Kalousis A., Müller M., Pellegrini C. Machine learning approaches to lung cancer prediction from mass spectra. Proteomics. 2003;3:1716–1719. doi: 10.1002/pmic.200300523. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C., Leng W., Sun C., Lu T., Chen Z., Men X., Wang Y., Wang G., Zhen B., Qin J. Urine proteome profiling predicts lung cancer from control cases and other tumors. EBioMedicine. 2018;30:120–128. doi: 10.1016/j.ebiom.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tirzïte M., Bukovskis M., Strazda G., Jurka N., Taivans I. Detection of lung cancer with electronic nose and logistic regression analysis. J. Breath Res. 2018;13:016006. doi: 10.1088/1752-7163/aae1b8. [DOI] [PubMed] [Google Scholar]
- 38.Huang C.-H., Zeng C., Wang Y.-C., Peng H.-Y., Lin C.-S., Chang C.-J., Yang H.-Y. A study of diagnostic accuracy using a chemical sensor array and a machine learning technique to detect lung cancer. Sensors. 2018;18:2845. doi: 10.3390/s18092845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kort S., Brusse-Keizer M., Gerritsen J.W., Schouwink H., Citgez E., de Jongh F., van der Maten J., Samii S., van den Bogart M., van der Palen J. Improving lung cancer diagnosis by combining exhaled-breath data and clinical parameters. ERJ Open Res. 2020;6:00221–02019. doi: 10.1183/23120541.00221-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu G., Jochems A., Ibrahim A., Yan C., Sanduleanu S., Woodruff H.C., Lambin P. Structural and functional radiomics for lung cancer. Eur. J. Nucl. Med. Mol. Imaging. 2021;48:3961–3974. doi: 10.1007/s00259-021-05242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S., Liu S., Zhang C., Yu H., Liu X., Hu Y., Xu W., Tang X., Fu Q. Exploratory study of a CT Radiomics model for the classification of small cell lung cancer and non-small-cell lung cancer. Front. Oncol. 2020;10:1268. doi: 10.3389/fonc.2020.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu X., Dong D., Chen Z., Fang M., Zhang L., Song J., Yu D., Zang Y., Liu Z., Shi J. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur. Radiol. 2018;28:2772–2778. doi: 10.1007/s00330-017-5221-1. [DOI] [PubMed] [Google Scholar]
- 43.Gu Q., Feng Z., Liang Q., Li M., Deng J., Ma M., Wang W., Liu J., Liu P., Rong P. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radiol. 2019;118:32–37. doi: 10.1016/j.ejrad.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 44.Wang S., Shi J., Ye Z., Dong D., Yu D., Zhou M., Liu Y., Gevaert O., Wang K., Zhu Y. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019;53:1800986. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song L., Zhu Z., Mao L., Li X., Han W., Du H., Wu H., Song W., Jin Z. Clinical, conventional CT and radiomic feature-based machine learning models for predicting ALK rearrangement status in lung adenocarcinoma patients. Front. Oncol. 2020;10:369. doi: 10.3389/fonc.2020.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y., Liu Y.-L., Feng Y., Yang X.-Y., Zhang J., Chang D.-D., Wu X., Tian X., Tang K.-J., Xie C.-M. A CT-derived deep neural network predicts for programmed death ligand-1 expression status in advanced lung adenocarcinomas. Ann. Transl. Med. 2020;8:930. doi: 10.21037/atm-19-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Šarić M., Russo M., Stella M., Sikora M. CNN-based method for lung cancer detection in whole slide histopathology images; Proceedings of the 2019 4th International Conference on Smart and Sustainable Technologies (SpliTech); Split, Croatia. 18–21 June 2019; pp. 1–4. [Google Scholar]
- 48.Wei J.W., Tafe L.J., Linnik Y.A., Vaickus L.J., Tomita N., Hassanpour S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 2019;9:3358. doi: 10.1038/s41598-019-40041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gertych A., Swiderska-Chadaj Z., Ma Z., Ing N., Markiewicz T., Cierniak S., Salemi H., Guzman S., Walts A.E., Knudsen B.S. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 2019;9:1483. doi: 10.1038/s41598-018-37638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D., Moreira A.L., Razavian N., Tsirigos A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Chen H., Gan C., Lin H., Dou Q., Tsougenis E., Huang Q., Cai M., Heng P.-A. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 2019;50:3950–3962. doi: 10.1109/TCYB.2019.2935141. [DOI] [PubMed] [Google Scholar]
- 52.Kapil A., Meier A., Zuraw A., Steele K.E., Rebelatto M.C., Schmidt G., Brieu N. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies. Sci. Rep. 2018;8:17343. doi: 10.1038/s41598-018-35501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aprupe L., Litjens G., Brinker T.J., van der Laak J., Grabe N. Robust and accurate quantification of biomarkers of immune cells in lung cancer micro-environment using deep convolutional neural networks. PeerJ. 2019;7:e6335. doi: 10.7717/peerj.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones G.D., Brandt W.S., Shen R., Sanchez-Vega F., Tan K.S., Martin A., Zhou J., Berger M., Solit D.B., Schultz N. A genomic-pathologic annotated risk model to predict recurrence in early-stage lung adenocarcinoma. JAMA Surg. 2021;156:e205601. doi: 10.1001/jamasurg.2020.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sha L., Osinski B.L., Ho I.Y., Tan T.L., Willis C., Weiss H., Beaubier N., Mahon B.M., Taxter T.J., Yip S.S. Multi-field-of-view deep learning model predicts nonsmall cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J. Pathol. Inform. 2019;10:24. doi: 10.4103/jpi.jpi_24_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang M., Sun D., Guo Y., Guo Y., Xiao J., Wang L., Yao X. Assessing PD-L1 expression level by radiomic features from PET/CT in nonsmall cell lung cancer patients: An initial result. Acad. Radiol. 2020;27:171–179. doi: 10.1016/j.acra.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 57.Li S., Ding C., Zhang H., Song J., Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med. Phys. 2019;46:4545–4552. doi: 10.1002/mp.13747. [DOI] [PubMed] [Google Scholar]
- 58.Dercle L., Fronheiser M., Lu L., Du S., Hayes W., Leung D.K., Roy A., Wilkerson J., Guo P., Fojo A.T. Identification of non–small cell lung cancer sensitive to systemic cancer therapies using radiomics. Clin. Cancer Res. 2020;26:2151–2162. doi: 10.1158/1078-0432.CCR-19-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le V.-H., Kha Q.-H., Hung T.N.K., Le N.Q.K. Risk score generated from CT-based radiomics signatures for overall survival prediction in non-small cell lung cancer. Cancers. 2021;13:3616. doi: 10.3390/cancers13143616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun F., Chen Y., Chen X., Sun X., Xing L. CT-based radiomics for predicting brain metastases as the first failure in patients with curatively resected locally advanced non-small cell lung cancer. Eur. J. Radiol. 2021;134:109411. doi: 10.1016/j.ejrad.2020.109411. [DOI] [PubMed] [Google Scholar]
- 61.Yoshiyasu N., Kojima F., Hayashi K., Bando T. Radiomics technology for identifying early-stage lung adenocarcinomas suitable for sublobar resection. J. Thorac. Cardiovasc. Surg. 2021;162:477–485.e1. doi: 10.1016/j.jtcvs.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Choi H., Kim H., Hong W., Park J., Hwang E.J., Park C.M., Kim Y.T., Goo J.M. Prediction of visceral pleural invasion in lung cancer on CT: Deep learning model achieves a radiologist-level performance with adaptive sensitivity and specificity to clinical needs. Eur. Radiol. 2021;31:2866–2876. doi: 10.1007/s00330-020-07431-2. [DOI] [PubMed] [Google Scholar]
- 63.Mattonen S.A., Palma D.A., Haasbeek C.J., Senan S., Ward A.D. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Med. Phys. 2014;41:033502. doi: 10.1118/1.4866219. [DOI] [PubMed] [Google Scholar]
- 64.Lewis J.E., Kemp M.L. Integration of machine learning and genome-scale metabolic modeling identifies multi-omics biomarkers for radiation resistance. Nat. Commun. 2021;12:2700. doi: 10.1038/s41467-021-22989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krafft S.P., Rao A., Stingo F., Briere T.M., Court L.E., Liao Z., Martel M.K. The utility of quantitative CT radiomics features for improved prediction of radiation pneumonitis. Med. Phys. 2018;45:5317–5324. doi: 10.1002/mp.13150. [DOI] [PubMed] [Google Scholar]
- 66.Bourbonne V., Da-Ano R., Jaouen V., Lucia F., Dissaux G., Bert J., Pradier O., Visvikis D., Hatt M., Schick U. Radiomics analysis of 3D dose distributions to predict toxicity of radiotherapy for lung cancer. Radiother. Oncol. 2021;155:144–150. doi: 10.1016/j.radonc.2020.10.040. [DOI] [PubMed] [Google Scholar]
- 67.Girard L., Zochbauer-Muller S., Virmani A.K., Gazdar A.F., Minna J.D. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 68.Shen R., Olshen A.B., Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–2912. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silver D., Hubert T., Schrittwieser J., Antonoglou I., Lai M., Guez A., Lanctot M., Sifre L., Kumaran D., Graepel T. A general reinforcement learning algorithm that masters chess, shogi, and Go through self-play. Science. 2018;362:1140–1144. doi: 10.1126/science.aar6404. [DOI] [PubMed] [Google Scholar]
- 70.Shi H., Lu J., Zhou Q. A novel data augmentation method using style-based GAN for robust pulmonary nodule segmentation; Proceedings of the 2020 Chinese Control and Decision Conference (CCDC); Hefei, China. 22–24 August 2020; pp. 2486–2491. [Google Scholar]
- 71.Ali I., Hart G.R., Gunabushanam G., Liang Y., Muhammad W., Nartowt B., Kane M., Ma X., Deng J. Lung Nodule Detection via Deep Reinforcement Learning. Front. Oncol. 2018;8:108. doi: 10.3389/fonc.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Capizzi G., Sciuto G.L., Napoli C., Połap D., Woźniak M. Small lung nodules detection based on fuzzy-logic and probabilistic neural network with bioinspired reinforcement learning. IEEE Trans. Fuzzy Syst. 2019;28:1178–1189. doi: 10.1109/TFUZZ.2019.2952831. [DOI] [Google Scholar]
- 73.In K.-H., Kwon Y.-S., Oh I.-J., Kim K.-S., Jung M.-H., Lee K.-H., Kim S.-Y., Ryu J.-S., Lee S.-Y., Jeong E.-T. Lung cancer patients who are asymptomatic at diagnosis show favorable prognosis: A Korean Lung Cancer Registry Study. Lung. Cancer. 2009;64:232–237. doi: 10.1016/j.lungcan.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 74.Quadrelli S., Lyons G., Colt H., Chimondeguy D., Buero A. Clinical characteristics and prognosis of incidentally detected lung cancers. Int. J. Surg. Oncol. 2015;2015:287604. doi: 10.1155/2015/287604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melamed M.R., Flehinger B.J., Zaman M.B., Heelan R.T., Perchick W.A., Martini N. Screening for early lung cancer. Results of the Memorial Sloan-Kettering study in New York. Chest. 1984;86:44–53. doi: 10.1378/chest.86.1.44. [DOI] [PubMed] [Google Scholar]
- 76.Hocking W.G., Hu P., Oken M.M., Winslow S.D., Kvale P.A., Prorok P.C., Ragard L.R., Commins J., Lynch D.A., Andriole G.L., et al. Lung cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. J. Natl. Cancer Inst. 2010;102:722–731. doi: 10.1093/jnci/djq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu G.C.W., Lazare K., Sullivan F. Serum and blood based biomarkers for lung cancer screening: A systematic review. BMC Cancer. 2018;18:181. doi: 10.1186/s12885-018-4024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montani F., Marzi M.J., Dezi F., Dama E., Carletti R.M., Bonizzi G., Bertolotti R., Bellomi M., Rampinelli C., Maisonneuve P., et al. miR-Test: A blood test for lung cancer early detection. J. Natl. Cancer Inst. 2015;107:djv063. doi: 10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- 79.Campanella A., De Summa S., Tommasi S. Exhaled breath condensate biomarkers for lung cancer. J. Breath Res. 2019;13:044002. doi: 10.1088/1752-7163/ab2f9f. [DOI] [PubMed] [Google Scholar]
- 80.Lopez-Sanchez L.M., Jurado-Gamez B., Feu-Collado N., Valverde A., Canas A., Fernandez-Rueda J.L., Aranda E., Rodriguez-Ariza A. Exhaled breath condensate biomarkers for the early diagnosis of lung cancer using proteomics. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313:L664–L676. doi: 10.1152/ajplung.00119.2017. [DOI] [PubMed] [Google Scholar]
- 81.National Lung Screening Trial Research T., Aberle D.R., Adams A.M., Berg C.D., Black W.C., Clapp J.D., Fagerstrom R.M., Gareen I.F., Gatsonis C., Marcus P.M., et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Koning H.J., van der Aalst C.M., de Jong P.A., Scholten E.T., Nackaerts K., Heuvelmans M.A., Lammers J.J., Weenink C., Yousaf-Khan U., Horeweg N., et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020;382:503–513. doi: 10.1056/NEJMoa1911793. [DOI] [PubMed] [Google Scholar]
- 83.Baker S.R., Patel R.H., Yang L., Lelkes V.M., Castro A., 3rd Malpractice suits in chest radiology: An evaluation of the histories of 8265 radiologists. J. Thorac. Imaging. 2013;28:388–391. doi: 10.1097/RTI.0b013e3182a21be2. [DOI] [PubMed] [Google Scholar]
- 84.Sakai M., Kato A., Kobayashi N., Nakamura R., Okawa S., Sato Y. Improved Lung Cancer Detection in Cardiovascular Outpatients by the Pulmonologist-based Interpretation of Chest Radiographs. Intern. Med. 2015;54:2991–2997. doi: 10.2169/internalmedicine.54.4171. [DOI] [PubMed] [Google Scholar]
- 85.White C.S., Salis A.I., Meyer C.A. Missed lung cancer on chest radiography and computed tomography: Imaging and medicolegal issues. J. Thorac. Imaging. 1999;14:63–68. doi: 10.1097/00005382-199901000-00006. [DOI] [PubMed] [Google Scholar]
- 86.About DICOM: Overview. [(accessed on 3 December 2021)]. Available online: https://www.dicomstandard.org/about.
- 87.Shiraishi J., Katsuragawa S., Ikezoe J., Matsumoto T., Kobayashi T., Komatsu K.-I., Matsui M., Fujita H., Kodera Y., Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 2000;174:71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 88.Jaeger S., Candemir S., Antani S., Wang Y.X., Lu P.X., Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014;4:475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Honolulu, HI, USA. 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- 90.Bustos A., Pertusa A., Salinas J.-M., de la Iglesia-Vayá M. Padchest: A large chest x-ray image dataset with multi-label annotated reports. Med. Image Anal. 2020;66:101797. doi: 10.1016/j.media.2020.101797. [DOI] [PubMed] [Google Scholar]
- 91.Armato S.G., III, McLennan G., Bidaut L., McNitt-Gray M.F., Meyer C.R., Reeves A.P., Zhao B., Aberle D.R., Henschke C.I., Hoffman E.A. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson A.E., Pollard T.J., Greenbaum N.R., Lungren M.P., Deng C.-Y., Peng Y., Lu Z., Mark R.G., Berkowitz S.J., Horng S. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv. 2019 doi: 10.1038/s41597-019-0322-0.1901.07042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison; Proceedings of the AAAI Conference on Artificial Intelligence; Honolulu, HI, USA. 27 January–1 February 2019; pp. 590–597. [Google Scholar]
- 94.Nguyen H.C., Le T.T., Pham H., Nguyen H.Q. VinDr-RibCXR: A Benchmark Dataset for Automatic Segmentation and Labeling of Individual Ribs on Chest X-rays. arXiv. 20212107.01327 [Google Scholar]
- 95.Jain S., Agrawal A., Saporta A., Truong S.Q., Bui T., Chambon P., Zhang Y., Lungren M.P., Ng A.Y., Langlotz C. RadGraph: Extracting Clinical Entities and Relations from Radiology Reports. arXiv. 20212106.14463 [Google Scholar]
- 96.Lanfredi R.B., Zhang M., Auffermann W.F., Chan J., Duong P.-A.T., Srikumar V., Drew T., Schroeder J.D., Tasdizen T. REFLACX, a dataset of reports and eye-tracking data for localization of abnormalities in chest X-rays. arXiv. 2021 doi: 10.1038/s41597-022-01441-z.2109.14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lodwick G.S., Keats T.E., Dorst J.P. The Coding of Roentgen Images for Computer Analysis as Applied to Lung Cancer. Radiology. 1963;81:185–200. doi: 10.1148/81.2.185. [DOI] [PubMed] [Google Scholar]
- 98.Munir K., Elahi H., Ayub A., Frezza F., Rizzi A. Cancer diagnosis using deep learning: A bibliographic review. Cancers. 2019;11:1235. doi: 10.3390/cancers11091235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Van Riel S.J., Jacobs C., Scholten E.T., Wittenberg R., Wille M.M.W., de Hoop B., Sprengers R., Mets O.M., Geurts B., Prokop M. Observer variability for Lung-RADS categorisation of lung cancer screening CTs: Impact on patient management. Eur. Radiol. 2019;29:924–931. doi: 10.1007/s00330-018-5599-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schreuder A., Scholten E.T., van Ginneken B., Jacobs C. Artificial intelligence for detection and characterization of pulmonary nodules in lung cancer CT screening: Ready for practice? Transl. Lung Cancer Res. 2021;10:2378. doi: 10.21037/tlcr-2020-lcs-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu B., Chi W., Li X., Li P., Liang W., Liu H., Wang W., He J. Evolving the pulmonary nodules diagnosis from classical approaches to deep learning-aided decision support: Three decades’ development course and future prospect. J. Cancer Res. Clin. Oncol. 2020;146:153–185. doi: 10.1007/s00432-019-03098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li D., Mikela Vilmun B., Frederik Carlsen J., Albrecht-Beste E., Ammitzbøl Lauridsen C., Bachmann Nielsen M., Lindskov Hansen K. The performance of deep learning algorithms on automatic pulmonary nodule detection and classification tested on different datasets that are not derived from LIDC-IDRI: A systematic review. Diagnostics. 2019;9:207. doi: 10.3390/diagnostics9040207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pastorino U., Rossi M., Rosato V., Marchianò A., Sverzellati N., Morosi C., Fabbri A., Galeone C., Negri E., Sozzi G. Annual or biennial CT screening versus observation in heavy smokers: 5-year results of the MILD trial. Eur. J. Cancer Prev. 2012;21:308–315. doi: 10.1097/CEJ.0b013e328351e1b6. [DOI] [PubMed] [Google Scholar]
- 104.Pedersen J.H., Ashraf H., Dirksen A., Bach K., Hansen H., Toennesen P., Thorsen H., Brodersen J., Skov B.G., Døssing M. The Danish randomized lung cancer CT screening trial—Overall design and results of the prevalence round. J. Thorac. Oncol. 2009;4:608–614. doi: 10.1097/JTO.0b013e3181a0d98f. [DOI] [PubMed] [Google Scholar]
- 105.Martin M.D., Kanne J.P., Broderick L.S., Kazerooni E.A., Meyer C.A. Lung-RADS: Pushing the limits. Radiographics. 2017;37:1975–1993. doi: 10.1148/rg.2017170051. [DOI] [PubMed] [Google Scholar]
- 106.MacMahon H., Naidich D.P., Goo J.M., Lee K.S., Leung A.N., Mayo J.R., Mehta A.C., Ohno Y., Powell C.A., Prokop M. Guidelines for management of incidental pulmonary nodules detected on CT images: From the Fleischner Society 2017. Radiology. 2017;284:228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 107.Chabon J.J., Hamilton E.G., Kurtz D.M., Esfahani M.S., Moding E.J., Stehr H., Schroers-Martin J., Nabet B.Y., Chen B., Chaudhuri A.A. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–251. doi: 10.1038/s41586-020-2140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cammarota G., Ianiro G., Ahern A., Carbone C., Temko A., Claesson M.J., Gasbarrini A., Tortora G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:635–648. doi: 10.1038/s41575-020-0327-3. [DOI] [PubMed] [Google Scholar]
- 109.Peled N., Fuchs V., Kestenbaum E.H., Oscar E., Bitran R. An Update on the Use of Exhaled Breath Analysis for the Early Detection of Lung Cancer. Lung Cancer Targets Ther. 2021;12:81. doi: 10.2147/LCTT.S320493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiang D., Zhang B., Doll D., Shen K., Kloecker G., Freter C. Lung cancer screening: From imaging to biomarker. Biomark. Res. 2013;1:4. doi: 10.1186/2050-7771-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Daniels M., Goh F., Wright C.M., Sriram K.B., Relan V., Clarke B.E., Duhig E.E., Bowman R.V., Yang I.A., Fong K.M. Whole genome sequencing for lung cancer. J. Thorac. Dis. 2012;4:155. doi: 10.3978/j.issn.2072-1439.2012.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi Y., Qu J., Wu S., Hao Y., Zhang J., Ning J., Yang X., Lofaro L., Pankratz D.G., Babiarz J. Improving lung cancer risk stratification leveraging whole transcriptome RNA sequencing and machine learning across multiple cohorts. BMC Med. Genom. 2020;13:151. doi: 10.1186/s12920-020-00782-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chang K., Creighton C., Davis C., Donehower L. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Phillips M., Gleeson K., Hughes J.M.B., Greenberg J., Cataneo R.N., Baker L., McVay W.P. Volatile organic compounds in breath as markers of lung cancer: A cross-sectional study. Lancet. 1999;353:1930–1933. doi: 10.1016/S0140-6736(98)07552-7. [DOI] [PubMed] [Google Scholar]
- 115.Evans A.J., Bauer T.W., Bui M.M., Cornish T.C., Duncan H., Glassy E.F., Hipp J., McGee R.S., Murphy D., Myers C. US Food and Drug Administration approval of whole slide imaging for primary diagnosis: A key milestone is reached and new questions are raised. Arch. Pathol. Lab. Med. 2018;142:1383–1387. doi: 10.5858/arpa.2017-0496-CP. [DOI] [PubMed] [Google Scholar]
- 116.Abels E., Pantanowitz L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J. Pathol. Inform. 2017;8:23. doi: 10.4103/jpi.jpi_11_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niazi M.K.K., Parwani A.V., Gurcan M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–e261. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.DICOM Whole Slide Imaging (WSI) [(accessed on 29 November 2021)]. Available online: https://dicom.nema.org/Dicom/DICOMWSI/
- 119.Sakamoto T., Furukawa T., Lami K., Pham H.H.N., Uegami W., Kuroda K., Kawai M., Sakanashi H., Cooper L.A.D., Bychkov A. A narrative review of digital pathology and artificial intelligence: Focusing on lung cancer. Transl. Lung Cancer Res. 2020;9:2255. doi: 10.21037/tlcr-20-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Giovagnoli M.R., Giansanti D. Artificial Intelligence in Digital Pathology: What Is the Future? Part 1: From the Digital Slide Onwards. Healthc. Multidiscip. Digit. Publ. Inst. 2021;9:858. doi: 10.3390/healthcare9070858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bejnordi B.E., Veta M., Van Diest P.J., Van Ginneken B., Karssemeijer N., Litjens G., Van Der Laak J.A., Hermsen M., Manson Q.F., Balkenhol M. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Biermann J., Adkins D., Agulnik M., Benjamin R., Brigman B., Butrynski J., Cheong D., Chow W., Curry W., Frassica D. National comprehensive cancer network. Bone cancer. J. Natl. Compr. Cancer Netw. 2013;11:688–723. doi: 10.6004/jnccn.2013.0088. [DOI] [PubMed] [Google Scholar]
- 123.Furukawa T., Kuroda K., Bychkov A., Pham H., Kashima Y., Fukuoka J. Verification of Deep Learning Model to Measure Tumor Cellularity in Transbronchial Biopsies of Lung Adenocarcinoma. Laboratory Investigation, Nature Publishing Group; New York, NY, USA: 2019. [Google Scholar]
- 124.Sakamoto T., Furukawa T., Pham H.H., Kuroda K., Tabata K., Kashima Y., Okoshi E.N., Morimoto S., Bychkov A., Fukuoka J. Collaborative workflow between pathologists and deep learning for evaluation of tumor cellularity in lung adenocarcinoma. bioRxiv. 2022 doi: 10.1101/2022.01.11.475587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hondelink L.M., Hüyük M., Postmus P.E., Smit V.T., Blom S., von der Thüsen J.H., Cohen D. Development and validation of a supervised deep learning algorithm for automated whole-slide programmed death-ligand 1 tumour proportion score assessment in non-small cell lung cancer. Histopathology. 2021;80:635–647. doi: 10.1111/his.14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu J., Lin D. A Review of Artificial Intelligence in Precise Assessment of Programmed Cell Death-ligand 1 and Tumor-infiltrating Lymphocytes in Non− Small Cell Lung Cancer. Adv. Anat. Pathol. 2021;28:439–445. doi: 10.1097/PAP.0000000000000322. [DOI] [PubMed] [Google Scholar]
- 127.Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Silva V.W.K., Busam K.J., Brogi E., Reuter V.E., Klimstra D.S., Fuchs T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Giansanti D., Grigioni M., D’Avenio G., Morelli S., Maccioni G., Bondi A., Giovagnoli M.R. Virtual microscopy and digital cytology: State of the art. Ann. Dell’istituto Super. Di Sanità. 2010;46:115–122. doi: 10.4415/ANN_10_02_03. [DOI] [PubMed] [Google Scholar]
- 129.Boschetto A., Pochini M., Bottini L., Giovagnoli M.R., Giansanti D. The focus emulation and image enhancement in digital cytology: An experience using the software Mathematica. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2015;3:110–116. doi: 10.1080/21681163.2014.885852. [DOI] [Google Scholar]
- 130.Lin Y.-J., Chao T.-K., Khalil M.-A., Lee Y.-C., Hong D.-Z., Wu J.-J., Wang C.-W. Deep Learning Fast Screening Approach on Cytological Whole Slides for Thyroid Cancer Diagnosis. Cancers. 2021;13:3891. doi: 10.3390/cancers13153891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Echle A., Rindtorff N.T., Brinker T.J., Luedde T., Pearson A.T., Kather J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer. 2021;124:686–696. doi: 10.1038/s41416-020-01122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Predicting Response to Immunotherapy Using Computer Extracted Featuresof Cancer Nuclei from Hematoxylin and Eosin (H&E) Stained Images of Non-Small Cell Lung Cancer (NSCLC) [(accessed on 6 March 2022)]; Available online: https://patft.uspto.gov/netacgi/nph-Parser?Sect1=PTO2&Sect2=HITOFF&p=1&u=%2Fnetahtml%2FPTO%2Fsearchbool.html&r=1&f=G&l=50&co1=AND&d=PTXT&s1=11,055,844.PN.&OS=PN/11,055,844&RS=PN/11,055,844.
- 133.Wulczyn E., Steiner D.F., Xu Z., Sadhwani A., Wang H., Flament-Auvigne I., Mermel C.H., Chen P.-H.C., Liu Y., Stumpe M.C. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE. 2020;15:e0233678. doi: 10.1371/journal.pone.0233678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.D’Antonoli T.A., Farchione A., Lenkowicz J., Chiappetta M., Cicchetti G., Martino A., Ottavianelli A., Manfredi R., Margaritora S., Bonomo L. CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad. Radiol. 2020;27:497–507. doi: 10.1016/j.acra.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 135.Hosny A., Parmar C., Coroller T.P., Grossmann P., Zeleznik R., Kumar A., Bussink J., Gillies R.J., Mak R.H., Aerts H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jochems A., Deist T.M., Van Soest J., Eble M., Bulens P., Coucke P., Dries W., Lambin P., Dekker A. Distributed learning: Developing a predictive model based on data from multiple hospitals without data leaving the hospital–a real life proof of concept. Radiother. Oncol. 2016;121:459–467. doi: 10.1016/j.radonc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 137.Jochems A., Deist T.M., El Naqa I., Kessler M., Mayo C., Reeves J., Jolly S., Matuszak M., Ten Haken R., van Soest J. Developing and validating a survival prediction model for NSCLC patients through distributed learning across 3 countries. Int. J. Radiat. Oncol. Biol. Phys. 2017;99:344–352. doi: 10.1016/j.ijrobp.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang D.D., Zhou W., Yan H., Wong M., Lee V. Personalized prediction of EGFR mutation-induced drug resistance in lung cancer. Sci. Rep. 2013;3:2855. doi: 10.1038/srep02855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Giang T.-T., Nguyen T.-P., Tran D.-H. Stratifying patients using fast multiple kernel learning framework: Case studies of Alzheimer’s disease and cancers. BMC Med. Inform. Decis. Mak. 2020;20:108. doi: 10.1186/s12911-020-01140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gao Y., Zhou R., Lyu Q. Multiomics and machine learning in lung cancer prognosis. J. Thorac. Dis. 2020;12:4531. doi: 10.21037/jtd-2019-itm-013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wissel D., Rowson D., Boeva V. Hierarchical autoencoder-based integration improves performance in multi-omics cancer survival models through soft modality selection. bioRxiv. 2022 doi: 10.1101/2021.09.16.460589. [DOI] [Google Scholar]
- 142.Coory M., Gkolia P., Yang I.A., Bowman R.V., Fong K.M. Systematic review of multidisciplinary teams in the management of lung cancer. Lung Cancer. 2008;60:14–21. doi: 10.1016/j.lungcan.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 143.Denton E., Conron M. Improving outcomes in lung cancer: The value of the multidisciplinary health care team. J. Multidiscip. Healthc. 2016;9:137–144. doi: 10.2147/JMDH.S76762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wichmann J.L., Willemink M.J., De Cecco C.N. Artificial Intelligence and Machine Learning in Radiology: Current State and Considerations for Routine Clinical Implementation. Investig. Radiol. 2020;55:619–627. doi: 10.1097/RLI.0000000000000673. [DOI] [PubMed] [Google Scholar]