Summary
Humans exhibit remarkable interindividual and interpopulation immune response variability upon microbial challenges. Cytokines play a vital role in regulating inflammation and immune responses, but dysregulation of cytokine responses has been implicated in different disease states. Host genetic factors were previously shown to significantly impact cytokine response heterogeneity mainly in European-based studies, but it is unclear whether these findings are transferable to non-European individuals. Here, we aimed to identify genetic variants modulating cytokine responses in healthy adults of East African ancestry from Tanzania. We leveraged both cytokine and genetic data and performed genome-wide cytokine quantitative trait loci (cQTLs) mapping. The results were compared with another cohort of healthy adults of Western European ancestry via direct overlap and functional enrichment analyses. We also performed meta-analyses to identify cQTLs with congruent effect direction in both populations. In the Tanzanians, cQTL mapping identified 80 independent suggestive loci and one genome-wide significant locus (TBC1D22A) at chromosome 22; SNP rs12169244 was associated with IL-1b release after Salmonella enteritidis stimulation. Remarkably, the identified cQTLs varied significantly when compared to the European cohort, and there was a very limited percentage of overlap (1.6% to 1.9%). We further observed ancestry-specific pathways regulating induced cytokine responses, and there was significant enrichment of the interferon pathway specifically in the Tanzanians. Furthermore, contrary to the Europeans, genetic variants in the TLR10-TLR1-TLR6 locus showed no effect on cytokine response. Our data reveal both ancestry-specific effects of genetic variants and pathways on cytokine response heterogeneity, hence arguing for the importance of initiatives to include diverse populations into genomics research.
Keywords: cytokines, genetic variation, Africa, immunogenomics, QTL
Introduction
Cytokines are key regulators of immune response to invading pathogens.1 Variations in cytokine responses determine susceptibility to infectious diseases as well as autoimmune and inflammatory diseases.2 Cytokine responses elicited by different microbial stimuli are known to be highly heterogeneous across individuals and populations.3 Thus, identifying the major factors that determine cytokine responses to microbial and other environmental triggers has the potential to refine our understanding of immune system variation and understand the source of the variability in disease susceptibility between individuals.
The Human Functional Genomics Project (HFGP) was established in 2013 with the aim of identifying genetic as well as non-genetic host factors that determine interindividual variability in immune responses. In cohorts of healthy individuals of Western European ancestry, these functional genomics studies have demonstrated the significant role of genetic as well as environmental and non-genetic (such as age, sex, and seasonality) factors for the interindividual differences in cytokine production capacity upon stimulation.4, 5, 6 Undoubtedly, these studies have broadened our knowledge on the major factors that contribute to cytokine response variability. For example, SNPs at the TLR10-TLR1-TLR6 (MIM: 606270, 601194, 605403) locus showed the strongest effect on production of multiple cytokines.
Prior to this, Wurfel et al. (2008), through candidate gene approach study in individuals recruited from Seattle metropolitan area, had reported a genetic variant, rs5743551, within the TLR1 locus to be strongly associated with induced cytokine production and sepsis outcome.7 Similarly, a previous work on individuals of Caucasian origin demonstrated that variations in the TLR10-TLR1-TLR6 locus drives the most interindividual variation in TLR2-mediated cytokine responses.8 In the same genomic region, previous work on transcriptional response of primary monocytes to viral and bacterial stimuli reported European-specific strong trans-eQTL (expression quantitative trait locus) effect.9 Given that eQTLs effects are context or tissue dependent, this study also focused on self-reported Africans residing in Belgium only. Arguably, the genetic architecture of polymorphisms in the TLR10-TLR1-TLR6 region might have differential effect on various human phenotypes and also varies substantially among populations. Therefore, further exploration of this genomic region by leveraging different molecular phenotypes and individuals from other ethnicities is warranted to draw general conclusions.
Africa is the continent where modern humans evolved and from where they later migrated all around the world. Africa is also extremely diverse culturally, ethnically, and genetically. Burgeoning evidence in the scientific literature shows that Africans have the most genetic variation among all other populations.10,11 Despite this genetic richness, African populations are poorly represented in genetic and functional genomic research, especially compared to individuals of European ancestry.12,13 In the context of induced cytokines responses for example, to the best of our knowledge, only the Hi-Host Phenome Project (H2P2) has incorporated individuals of African ancestry in a study that aimed to identify human genetic variation in pathogen-induced cellular traits. However, this study was conducted in lymphoblast cell lines and also not in Tanzanians.14 The Tanzanian population, among the sub-Saharan African regions, has high prevalence of infectious diseases15 with diverse patterns of causative agents compared to high-income countries.16 Exposure to different environmental conditions including infectious or microorganisms may affect certain genetic markers through positive selection17 with differential effect on modern human populations.18
Therefore, the primary aim of this study was to investigate genetic variants modulating ex vivo microbial-induced cytokine responses in healthy adults of East African ancestry. The other important aim was to assess whether distinct genetic variants and pathways contribute to interpopulation heterogeneity to cytokine responses upon stimulation.
To achieve these aims, we extended the HFGP to include a population of Sub-Saharan Africa: we performed a functional genomics study in healthy adult individuals in the Kilimanjaro region in Northern Tanzania.19 Here, we not only identify significant genetic loci that affect cytokine production in Tanzanian individuals, but we also show considerable differences in the genetic basis for cytokine production between the East Africans from Tanzania and Western European individuals. Finally, we provide evidence for ancestry-associated differences in inflammatory pathways that regulate cytokine production in response to infection.
Subjects and methods
Study design and participants
Data from the 300 Tanzania functional genomics (FG) (300TZFG) were used in this study. Details of this cohort have been described recently.19 In summary, the 300TZFG cohort consists of apparently healthy Tanzanian individuals, aged between 18 and 65 years, living in the Kilimanjaro region in Northern Tanzania. All participants had a negative rapid diagnostic test for malaria and HIV. Exclusion criteria were pregnancy, any acute or chronic disease, use of antibiotics or anti-malaria medication in the 3 months before sampling, tuberculosis in the past year, a blood pressure ≤ 90/60 mmHg or ≥ 140/90 mmHg, or a random blood glucose > 8.0 mmol/L. In this study, a total of 323 and 307 participants had genotype and cytokine data, respectively. After quality control procedures, matched genotype data (SNPs dosages) and cytokine data were available for 271 individuals, consisting of 137 males and 134 females. A general overview of the study samples at various quality control steps are depicted in Figure S1. The general 500FG cohort of individuals of Western European origin consists of 237 males and 296 females with age range of 18 to 75 years.
Ethics approval and consent to participate
The 300TZFG study was approved by the Ethical Committees of the Kilimanjaro Christian Medical University College (CREC) (no. 2443) and the National Institute for Medical Research (NIMR/HQ/R.8a/Vol. IX/2290 and NIMR/HQ/R.8a/Vol.IX/3318) in Tanzania. Written informed consent was obtained from all subjects. The 500FG cohort study was approved by the Ethical Committee of the Radboud University Medical Centre Nijmegen, the Netherlands (NL42561.091.12, 2012/550). Informed consent was provided by all the participants.
Whole blood stimulation experiments
As previously described,19 ex vivo cytokine stimulation experiments were performed at the biotechnology laboratory facility available at Kilimanjaro Clinical Research Institute in Moshi, Tanzania. Whole blood was stimulated with ten stimuli (see Table S16 from Temba et al.19 for details) including bacterial and fungal pathogens as well as TLR3 and TLR4 agonists. 100 mL of heparin blood was added to a 48-wells culture plate and subsequently stimulated with 400 mL of stimulus for 48 h at 37°C and 5% CO2. Stimuli were prepared in RPMI culture medium (Dutch modified, Invitrogen) supplemented with 50 μg/mL gentamicin, 2 mM Glutamax, and 1 mM pyruvate. Supernatants were collected and stored at −80°C until used for ELISA. To minimize variation between measurements, we measured all samples by using kits of the same lot number.
Cytokine measurements
Concentrations of cytokines interleukin (IL)-6, IL-1b, interferon (IFN)-g, tumor necrosis factor (TNF)-, and IL-10 were measured in the stored supernatants via ELISA according to the instructions (given (IL)-6, IL-1b, IL-10, and TNF-: R&D Systems; IFN-g: Sanguin). We excluded IL-10 cytokine responses to S. aureus, C. albicans, and PolyIC, as over 75% of the individuals had values below the detection limit, resulting in 47 cytokine-stimulation combinations used in this analysis.
Quality control of cytokine data
Preprocessing of cytokine data was performed before statistical analysis. Raw cytokine concentrations were first log2-transformed followed by normalization for approximation of standard Gaussian distribution (Figure S2A). Inverse-ranked normalization was performed with the “rntransform” function implemented in the GenABEL R package.20 To examine the presence of outliers, we performed unsupervised hierarchical clustering analysis by using Pearson’s correlation as a measure of similarity. All the samples in the dataset clustered uniformly (Figure S2B).
SNP genotyping, quality control, and imputation
DNA was extracted from whole blood with the DNeasy kit. Genotyping was performed with the Global Screening Array (GSA) SNP chip. We used default settings of Opticall 0.7021 to perform genotype calling. Quality control filters prior to imputation include excluding variants with call rate exceeding 0.1, low minor allele frequencies (MAFs < 0.001), and SNPs deviating from Hardy-Weinberg equilibrium (HWE) with a p value < 1 × 10−4. Next, we excluded 15 samples that were potential genetic outliers through identification by using multi-dimensional scaling plots. A total of 409,261 variants and 308 samples passed the quality control procedures. Strand alignment to a reference panel, 1000 Genomes reference panel dataset, was performed via Genotype harmonizer.22 To improve genome coverage, we performed genotype imputation for all autosomal chromosomes by using the Minimac4 software through the publicly available Michigan Imputation Server.23 The Human Reference Consortium (HCR r.1.1 2016) was used as a reference panel and the dataset was phased with Eagle v2.3. Variants with imputation quality score (R2) < 0.3 were excluded from further analysis. Genotyping and imputation generated a total of 5,271,779 variants from 308 individuals. We further excluded samples because of extreme heterozygosity rates and cryptic relatedness. No sample was removed as a result of incorrect or ambiguous sex information when compared with self-reported sex in the phenotype data. We used a multidimensional scaling approach to examine potential population structure by merging our data with the 1000 Genomes Project data. All individuals in our data clustered homogeneously with individuals of African origin included in the 1000 Genomes dataset (Figure S2C). However, by excluding non-Africans, the Tanzanian data clustered differently from the Africans in the 1000 Genomes Project data (Figure S2D). We considered SNPs with MAFs ≥ 5% and not deviating from HWE with p value > 1 × 10−6, yielding a final dataset of 5,269,992 SNPs that were subsequently used for cytokine QTL analysis.
Cytokine quantitative trait loci (cQTLs) mapping
The cQTL analysis was performed with a linear model implemented in the Matrix-eQTL R package.24 We adjusted the linear model for age, sex, and residency (rural and urban samples) on the inverse ranked normalized cytokine concentrations. We considered the conventional genome-wide significance threshold for statistical significance of p value ≤ 5 × 10−8 to account for multiple testing.
For genome-wide QTL mapping, multiple SNPs are tested throughout the genome and based on linkage disequilibrium (LD) between SNPs, a particular locus might contain multiple cytokine-associated genetic variants. Therefore, we performed LD clumping with the greedy algorithm (SNPs in LD are removed in ascending order of p values) implemented in PLINK25 to identify independent associations. Here, our imputed genotype data were used as a reference dataset for estimating LD, clumps around lead SNPs were formed with a default window size of 250 kb, and r2 threshold greater than 0.1 was applied.
Meta-analysis
We conducted a meta-analysis by using a weighted sum fixed-effect model approach as implemented in METAL software program.26 Meta-analysis was performed by integrating summary statistics of cytokine QTL association results from this study and the 500FG study. To explain briefly, the 500FG is a population-based cohort made up of healthy adult volunteers of Western European origin from the Human Functional Genomics Projects. This cohort consists of 237 males and 296 females with age range of 18 to 75 years. Previously, researchers from our group used this cohort to perform cytokine QTL analyses on pro-inflammatory cytokine responses after bacteria, fungal, and viral stimulations.6 The meta-analysis was restricted to only cytokine-stimulation pairs common in both studies. We calculated heterogeneity statistics based on the chi-square test for all SNPs to estimate heterogeneity of effect sizes in both cohorts. In general, associations with meta p value ≤ 5 × 10−8 were considered genome-wide significant.
Pathway enrichment analysis
The potential biological significance of cQTL was assessed via over-representation analysis (ORA), which performs a hypergeometric test to identify causal pathways. WebGestalt, a freely available online tool and one of the most widely used gene set enrichment analysis tools, was used. We determined statistically significant pathways after correcting for multiple testing (false discovery rate p < 0.05) by using the Benjamini-Hochberg method.
Haplotype and LD block analysis
We extracted common genetic markers that passed quality control after imputation between the 500FG and 300TZFG cohorts to determine haplotype blocks separately in both cohorts by using PLINK (v1.09) software. Default parameters such as pairwise LD calculation based on SNPs within 200 kb window were applied. For each block that was identified per chromosome, we analyzed the number of haplotypes and median haplotype block length between the two populations and carried out Wilcoxon signed-rank non-parametric test to assess statistical significance. Pairwise LD measured as R2 for genetic markers within genomic regions were estimated and visualized with the R package LDheatmap.27
Genetic differentiation or divergence analysis
Interpopulation genetic differentiation analysis was conducted between the two populations with the fixation index (Fst) metric as implemented in PLINK (v1.09) software. Average Fst values were calculated with a step size of 10 kb. The analysis was performed on common SNPs remaining after quality control in each cohort. We used empirical approach by selecting Fst values exceeding 99th percentile per SNP to determine significance.
Statistical analyses
All statistical analyses were performed in R: a free software environment for statistical computing and graphics.28 We used Fligner-Killeen non-parametric test for homogeneity of group variances and robust to dataset with outliers to compared cell culture medium with stimulated cytokine levels. Quality control, preprocessing analysis of genetic data, and LD clumping procedures around any lead SNPs analysis to determine independent loci were performed with the free, open-source whole-genome analysis toolset PLINK 1.70.25 The cytokines association with imputed genome-wide data analyses were performed with a linear model as implemented in the Matrix-eQTL R package.24 The meta-analyses on cQTL summary statistic (p values) were performed with the fixed effects sample-size-weighted analysis method implemented in the METAL software.26
Results
Significant increase in cytokine abundance and interindividual variations upon stimulation
To investigate the contribution of common genetic variations on human induced cytokine responses, we restricted downstream analyses to the 47 cytokine-stimulation combinations passing quality control procedures. Details of these cytokine-stimulation pairs are illustrated in the general overview of the study design and bioinformatics analysis represented in Figure 1. First, we compared the abundance of cytokines from unstimulated state (cell culture medium) and stimulated conditions to establish their potential relationship. Compared to the spontaneous release of cytokines from cells in culture medium and as expected, we observed higher production of cytokines upon stimulation with microbial ligands (Figure 2A). On average, stimulation-induced with S. enteritidis produced the most abundant cytokine response, whereas the least abundance was observed in C. burnetii-stimulated cells. Also, we observed that there is variation in cytokine abundances per stimulation and this is not because of the presence of multiple outliers as depicted in (Figure 2A). Boxplots supporting this observation are presented in (Figure S3A). Moreover, we observed a high level of interindividual variations in cytokine concentrations of samples after stimulation (Figure 2B). To test whether there is a statistically significant difference in the interindividual variation in the production of cytokines upon stimulation compared to unstimulated state, we applied the Fligner-Killeen test for group comparison. We observed more significant increase in interindividual variations in stimulated conditions than in unstimulated states (p value < 2 × 10−16). Computing paired Wilcoxon tests resulted to the same level of significance (Figure S3B). Interestingly, this interindividual variation was highest for IFN-g production under most stimulations, suggesting the major role of host factors (genetic and non-genetic) in determining IFN-g production capacity.
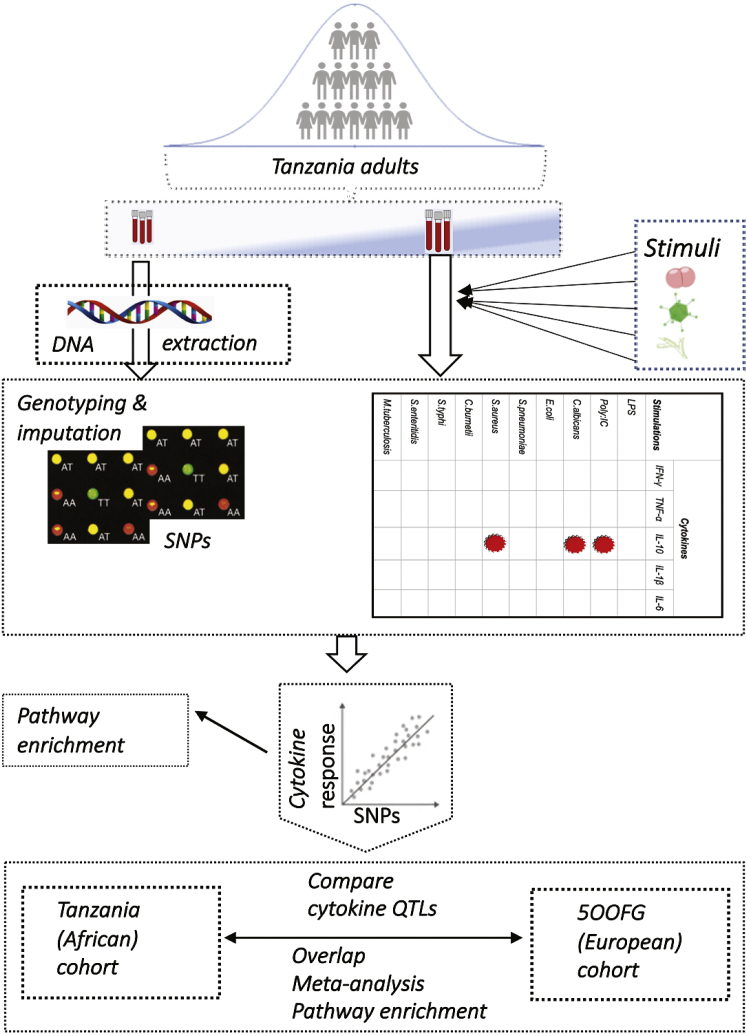
Figure 1.
Schematic diagram of study design and bioinformatics analyses
We utilized samples of a cohort of healthy Tanzanian adults. In vitro stimulation of their blood was performed with ten different stimuli, including bacterial, fungal, and TLR3 and TLR4 agonists followed by measuring five different cytokines in the supernatant. Three (indicated by red ovals) out of the resulting 50 cytokine-stimulation pairs were excluded from downstream analysis after quality control. Genotyping was performed using global screening arrays (GSAs) SNP chip. We performed genome-wide SNP cytokine quantitative trait loci (cQTL) mapping by correlating cytokine abundances and imputed genotype data and subsequently performed pathway enrichment analyses. Finally, we compared the results with another cohort of Western European ancestry individuals by using three different approaches: direct overlap of genetic variants, meta-analysis, and functional enrichment analysis.
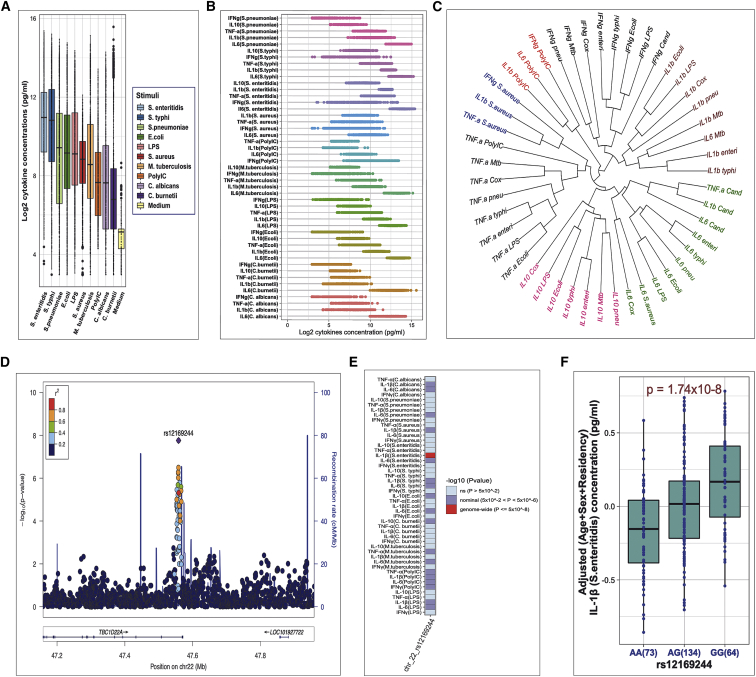
Figure 2.
General overview of cytokine responses architecture and summary of genome-wide significant cQTL results
(A) Boxplot comparing cytokine production capacity of unstimulated state (cell culture medium) and ten different stimuli. Profiles of all cytokines (IFN-g, TNF-, IL-1b, IL-10, and IL-6) were merged per stimuli to compare with unstimulated state.
(B) Dot plot depicting interindividual variability in cytokine response upon stimulation. While the x axis denotes cytokine concentrations after log2 transformation, the y axis represents the total 47 cytokine-stimulation combinations used in the current study. The dot plots have been arranged in ascending order of median values per stimuli.
(C) Dendrogram visualization of unsupervised hierarchical clustering of cytokine-stimulation pairs based on cytokine abundances. The “Euclidean” method in the “hclust” function was used for computing distances.
(D) Regional association plot at the TBC1D22A locus (top SNP rs12169244) associated with IL-1b upon Salmonella enteritidis stimulation. Other SNPs flanking a genomic widow of 400 kb are color-coded according to their linkage disequilibrium (r2) with the top SNP (purple color). The horizontal axis indicates chromosomal positions (NCBI human genome build 37) and the vertical axes represent −log10 p values and recombination rates (cM/Mb) estimated from 1000G (African population) version 3.3.
(E) Heatmap showing the association between the genome-wide significant SNP with all cytokine-stimulation pairs (on the vertical axis). SNP-cytokine associations with p values greater than 5 × 10−2 were considered statistically non-significant (ns).
(F) Boxplot showing distribution of cytokine concentrations (pg/mL) stratified by genotypes of the top SNP (rs12169244).
Induced cytokine responses are clustered in cytokine-dependent manner
The architectures of cytokine response in a population can be dependent on the type of cytokine or the microbial ligands used for the stimulation. Thus, to mine patterns underlying induced cytokine responses, we performed unsupervised hierarchical clustering on the cytokine stimulation combinations. In general, we observed distinct correlations at cytokine level irrespective of pathogens (Figure 2C). Interestingly, all cytokines, including anti-inflammatory cytokine IL-10, showed a very strong cytokine-dependent clustering except for PolyIC stimulation. Many of these stimuli are known to activate TLR-signaling to affect cytokine production. Why PolyIC-induced cytokine production is dependent on the pathogen compared to other stimuli needs to be investigated further.
Identification of genome-wide genetic variants affecting induced cytokine responses
Next, to identify genetic variants significantly associated with cytokine production, we mapped cQTLs at genome-wide scale. We identified one genome-wide significant locus on the basis of the stringent p value threshold of 5 × 10−8, while 80 other loci showed strong suggestive independent associations (p value > 5 × 10−8 to 1 × 10−6) after adjusting for covariates (Table S1). The genome-wide significant hit was SNP rs12169244 on chromosome 22, correlating with IL-1b production upon S. enteritidis stimulation (Figure 2D). SNP rs12169244 is an intronic variant mapping near TBC1D22A (MIM: 616879) gene (TBC1 domain family member 22A), which acts as a GTPase-activating protein for Rab family protein(s). Interestingly, the genome-wide significant SNP also showed nominal association with 18 other cytokine-stimulation pairs, thus suggesting a pleiotropic effect (Figure 2E). At this locus, relative to individuals carrying the AA genotypes, we observed elevated cytokine production in individuals with GG genotypes (Figure 2F). Quantile-quantile (QQ) plot of the association results at this locus is presented in Figure S2E. The genomic inflation factor or lambda(l) was observed to be 1.01, suggesting no evidence of systematic bias in the analysis. We report more detailed information of the top six independent associations in Table 1.
Table 1.
Summary of top six independent SNP-cytokine loci
| Loci | SNPs | Chr | Base pair | Cytokines | Stimulation | p value | Causal genes |
|---|---|---|---|---|---|---|---|
| 1 | rs12169244 | 22 | 47,558,596 | IL-1b | S. enteritidis | 1.74 × 10−8 | TBC1D22Aa |
| 2 | rs9563018 | 13 | 51,656,046 | IL-1b | M. tuberculosis | 7.81 × 10−8 | LINC0037,aGUCY1B2b |
| 3 | rs4474665 | 16 | 17,498,563 | IFN-g | S. enteritidis | 7.99 × 10−8 | XYLT1a |
| 4 | rs74115411 | 1 | 153,331,776 | IL-6 | S. aureus | 8.90 × 10−8 | S100A9a |
| 5 | rs11829172 | 12 | 98,579,287 | IL-1b | S. aureus | 9.63 × 10−8 | MIR4303a |
| 6 | rs10483241 | 12 | 48,630,139 | IFN-g | C. albicans | 9.71 × 10−8 | MIR4303a |
The gene in closest proximity to the cytokine QTL SNPs.
Expression of this gene shows association with cytokine QTL SNP in whole blood.
Functional enrichment analysis of cQTLs identified in Tanzania cohort
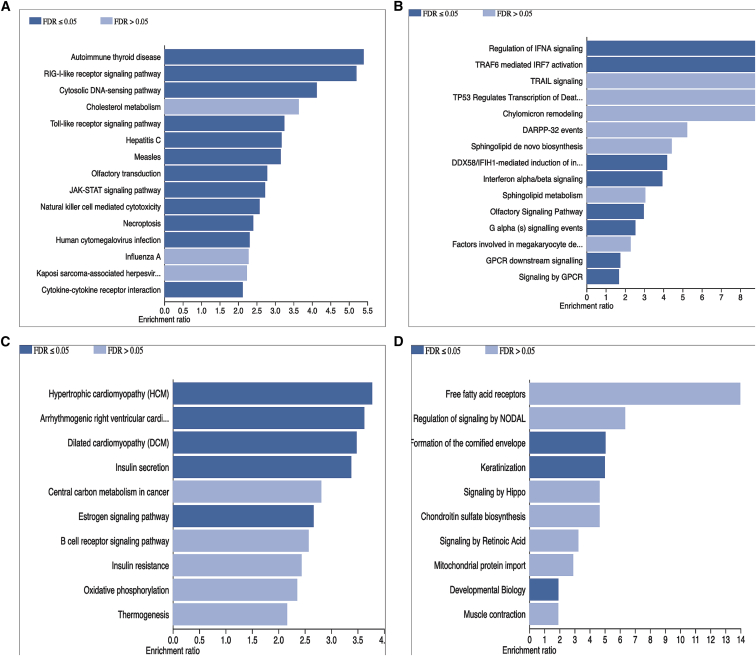
Based on the observation that cytokine-stimulation pairs clustered in a cytokine-dependent manner (Figure 2C), we hypothesized that there are common pathways affecting cytokine production, irrespective of the pathogen that induces the stimulation. Therefore, to identify these pathways, we first prioritized suggestive independent cQTLs (p value < 1 × 10−5) for each of the five cytokines (IL-6, IL-1b, TNF-, IFN-g, and IL-10) and extracted gene sets mapping near these genetic variants by using a window size of 250 kb upstream and downstream of each SNP. Henceforth, we refer to these groups as TNF--based, IL-6-based, IFN-g-based, IL-1b-based, and IL-10-based pathways. As expected, we found significant enrichment of the genes at IFN-g-based cQTLs for various diseases (Figure 3A), and regulation of interferon signaling (Figure 3B), suggesting that cis-genes around IFN-g cQTLs determine IFN-g production by mainly regulating pathogen recognition receptors such as TLRs and DDX58/IFIH1-mediated signaling. Moreover, we observed significantly enriched pathways; for example, estrogen signaling pathway influences the TNF--based category (Figures 3C and 3D). Estrogen is known to regulate cytokine gene expressions in different cell types through estrogen receptor-mediated pathways.29 Bar chart plots showing enrichment results of IL-6-, IL-1b-, IL-10-based pathways, with less significant enriched pathways, are presented in Figures S4A–S4F.
Figure 3.
Bar graphs summarizing IFN-g- and TNF--based pathway enrichment results
(A and B) Top 15 enriched KEGG (A) and reactome (B) pathways for gene sets curated from SNPs associated with IFN-g.
(C and D) Top 10 enriched KEGG (C) and reactome (D) pathways for gene sets curated from SNPs associated with TNF-. Darker blue bars represent statistically significant pathways and lighter blue bars represent non-significant pathways after multiple testing. We set the false discovery rate (FDR)-adjusted p value to 0.05 as significance level.
Limited overlap between cQTLs identified in European and African samples
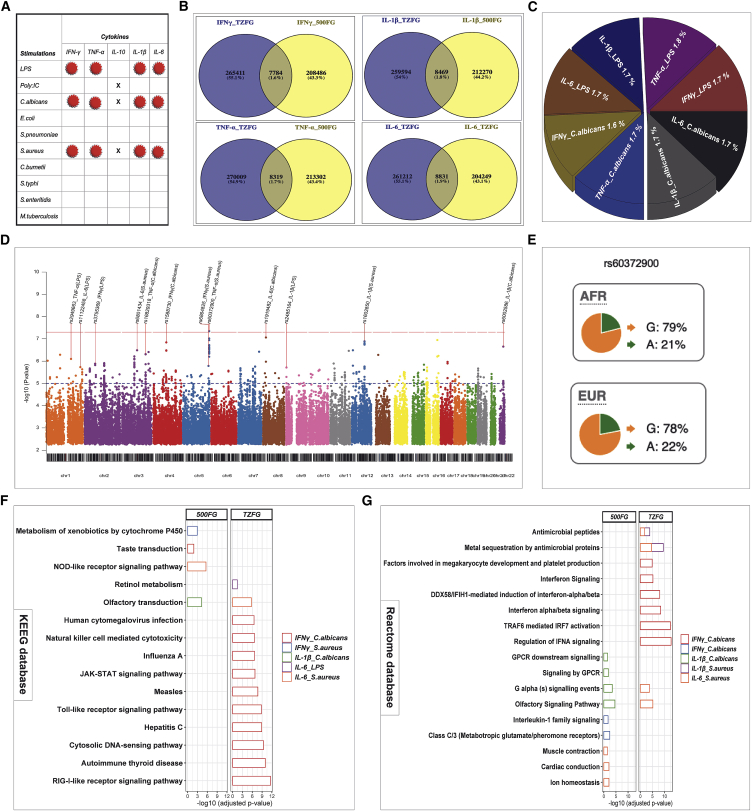
We next assessed the presence of any systematic differences between cQTLs identified via two population-based datasets of African (300TZFG) and European (500FG) individuals. To estimate the degree of shared genetic variants between these two populations, we directly compared the four cytokines (IFN-g, TNF-a, IL-1b, and IL-6) and three stimulations (LPS, Candida albicans, and Staphylococcus aureus) that were common to both cohorts (Figure 4A). Across all the 12 cytokine-stimulation pairs considered, we found only 1.6% to 1.9% of SNPs overlapping between both populations with nominal association threshold (p value < 5 × 10−2). For example, a Venn diagram depicting this observation for S. aureus-cytokines pairs is shown in Figure 4B, while results for LPS- and C. albicans-cytokines are represented in Figure 4C. In addition, no evidence of variant sharing was detected by restricting the analysis to variants showing suggestive association with cytokines (p value < 5 × 10−6), indicating that the identified genetic variants with strong cQTL effect are mostly population-specific variants.
Figure 4.
Comparison of cQTLs between Tanzanian (TZFG) and Western European dataset (500FG)
(A) Illustration of cytokine-stimulation combinations common in both African and European datasets (indicated by red circles). Cells with “x” labels denote cytokine-stimulation pairs excluded from downstream analysis, as more than 75% of samples had values below the detection limit of the assay.
(B) Venn diagrams illustrating S. aureus induced cytokine QTLs overlapping between individuals of African (TZFG) and European (500FG) ancestry.
(C) Pie chart showing the degree of LPS- and C. albicans-induced cytokine QTLs overlapping between individuals of African (TZFG) and European (500FG) ancestry.
(D) Manhattan plot of SNP-cytokines association test results indicating shared cQTL locus between individuals of African and European ancestry. The red horizontal dashed line represents the genome-wide significant threshold (p value < 1 × 10−8) and the blue dashed line denotes the suggestive evidence of association threshold. Nominally significant (p value < 5 × 10−2) cQTLs are plotted.
(E) Bar graphs representation of significantly enriched KEGG pathways after FDR correction.
(F) Bar graphs representation of significantly enriched reactome pathways after FDR correction. The colors of the bars associated with each pathway correspond to cytokine-stimulation pairs as shown by the color legend. Pathway analysis was performed with WebGestalt tool with subsequent visualization with R statistical software.
Meta-analysis revealed shared genetic locus related to cytokine response in African and European cohorts
We further sought to identify cQTLs that show effect on cytokines in the same allelic direction in both populations by meta-analyzing summary statistics of each marker across the two cohorts. To accomplish this, we extracted the common markers showing nominally significant association (p values < 5 × 10−2) in each independent study. Interestingly, among all the 12 cytokine-stimulation pairs analyzed separately, we identified one genome-wide significant locus at chromosome 5, the lead SNP rs60372900 correlating with TNF- levels after S. aureus stimulation. This intronic variant is mapped near a protein-coding gene ZNF354A. Remarkably, data from 1000 Genomes Project Phase 3 shows nearly equal minor allele frequencies of 21% and 22% in Europeans and Africans, respectively, of rs60372900. (Figure 4E). This observation underscores the regulatory potential of this variant on induced cytokine levels in both populations. Meta-analysis performed by integrating the African and the European data also identified several suggestive (p value < 1 × 10−5) associations (Figure 4D). Summary statistics of meta-analysis results are presented in Table 2.
Table 2.
Summary of top loci identified by meta-analysis of TZFG and 500FG dataset
| Loci | SNPs | Chr | Base pair | Cytokines | Stimulation | Z score | Meta p value | TZFG p value | 500FG p value | Allelic direction | Causal genes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs60372900 | 5 | 178,220,798 | TNF-a | S. aureus | 5.47 | 4.50 × 10−8 | 6.56 × 10−3 | 1.24 × 10−6 | ++ | AACSP1a |
| 2 | rs1919482 | 8 | 19,754,718 | IL-6 | C. albicans | 5.353 | 8.69 × 10−8 | 2.31 × 10−5 | 5.78 × 10−4 | ++ | LPL,a,bINT,bCSGALNCb |
| 3 | rs1602850 | 12 | 86,657,959 | IL-1b | S. aureus | 5.281 | 1.29 × 10−7 | 1.13 × 10−4 | 2.54 × 10−4 | ++ | MGAT4Ca |
| 4 | rs1588730 | 4 | 89,955,623 | IFN-g | C. albicans | 5.26 | 1.44 × 10−7 | 2.70 × 10−5 | 8.05 × 10−4 | ++ | FAM13A,a,bTIGD2b |
| 5 | rs6002586 | 22 | 42,441,412 | IL-1b | C. albicans | 5.18 | 2.22 × 10−7 | 9.63 × 10−4 | 6.57 × 10−5 | ++ | WBP2NL,aSMDT1,bCYP2D6,bMEI1,bNAGA,bPHETA2b |
| 6 | rs6801434 | 3 | 100,809,977 | IL-6 | S. aureus | 5.108 | 3.25 × 10−7 | 2.22 × 10−4 | 3.51 × 10−4 | ++ | ABI3BP,aTFGb |
| 7 | rs16829318 | 3 | 158,418,720 | TNF-a | C. albicans | 5.032 | 4.87 × 10−7 | 4.08 × 10−4 | 3.12 × 10−4 | ++ | RARRES1,aLXN,bGFM1,bMLF1,bRSRC1b |
| 8 | rs2949663 | 1 | 167,391,498 | TNF-a | LPS | −4.935 | 8.00 × 10−7 | 6.17 × 10−4 | 3.55 × 10−4 | – | POU2F1,aCD247,bAL359962.1b |
| 9 | rs3755369 | 2 | 70,942,964 | IFN-g | LPS | 4.84 | 1.30 × 10−6 | 1.30 × 10−3 | 2.93 × 10−4 | ++ | ADD2,a,bCLEC4Fb |
| 10 | rs6864825 | 5 | 173,659,256 | IFN-g | S. aureus | −4.794 | 1.64 × 10−6 | 1.40 × 10−4 | 2.16 × 10−3 | – | HMP19a |
| 11 | rs11122458 | 1 | 230,308,855 | IL-6 | LPS | 4.768 | 1.86 × 10−6 | 2.51 × 10−3 | 2.25 × 10−4 | ++ | GALNT2a |
| 12 | rs2485164 | 9 | 1,706,565 | IL-1b | LPS | 4.761 | 1.93 × 10−6 | 1.00 × 10−3 | 5.48 × 10−4 | ++ | SMARCA2a |
The gene in closest proximity to the cytokine QTL SNPs.
Expression of this gene shows association with cytokine QTL SNP in whole blood.
Population-specific cQTLs converge on distinct pathways to regulate cytokine levels
As we observed a few overlaps of cQTLs between the African and the European cohorts (Figures 4B and 4C), we tested whether these cQTLs affect cytokine production capacity through distinct pathways. We first identified independent suggestive associated cQTLs (p value < 1 × 10−5) for all cytokine-stimulation pairs separately for each population, and then we extracted genes around these cQTLs and performed overrepresentation analysis as described in the subjects and methods section. Intriguingly, we observed significant enrichment of genes for different pathways in different populations (Figure 4F). For example, in the Tanzanian cohort, the top-ranked KEGG pathways were mostly related to human viral infectious diseases such as hepatitis C, measles, influenza A, and human cytomegalovirus infection. Notably, these pathways were distinct for both populations. In support of this finding, using Reactome database, we found enrichment for regulation of type I interferon signaling pathways (Figure 4G). In contrast, we observed moderate enrichment for IL-1 signaling pathway and olfactory signaling pathway in the European cohort.
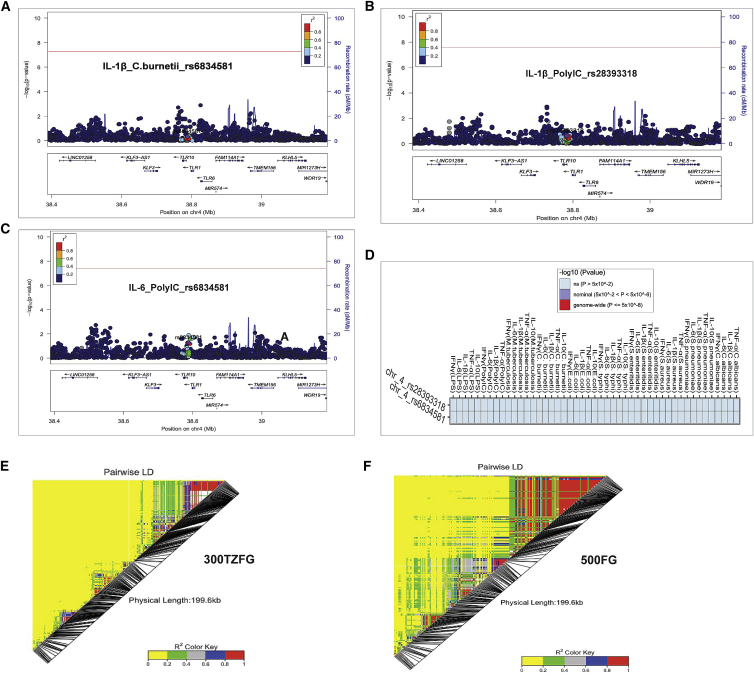
Exploring the Toll-like receptor (TLR) locus in the Tanzania cohort
TLRs are pathogen recognition receptors through which our innate immune system “senses” foreign antigens and activates host protective inflammatory responses.30 It has already been shown that the TLR10-TLR1-TLR6 locus is under strong positive selection in European populations.31 Our previous cQTL mapping study in the European population identified 17 independent loci, and among them, the SNPs at the TLR10-TLR1-TLR6 locus on chromosome 4 showed the strongest association (p = 3.9 × 10−25) with cytokine production.6 Notably, a SNP, rs28393318, was strongly associated with PolyIC-induced IL-1b production and another SNP, rs6834581, was associated with both PolyIC-induced IL-6 and C. burnettii-induced IL-1b levels. Given the availability of these cytokine-stimulation pairs in this study cohort, we attempted to interrogate the transferability of the TLR10-TLR1-TLR6 locus findings in the context of cytokine responses to an African population. Mapping cQTLs revealed that, while these polymorphisms (rs6834581 and rs28393318) significantly affect cytokines (IL-6 and IL-1b) in European samples, we found no association (p value > 0.05) in the African cohort (Figures 5A–5C). Testing the effect of these SNPs on other cytokine-stimulation pairs also showed no association (Figure 5D). Interestingly, the lead cQTL SNPs (with smallest p values) at these loci in the African cohort are not in LD with the lead SNPs in the European cohort (see Figures S5A–S5C for regional association plots and Figures S5D–S5F for heatmaps depicting the lack of correlation).
Figure 5.
Regional association plots and heatmaps of variants in the Toll-like receptor (TLR) locus
(A–C) Regional association plots of SNPs flanking genomic window of 400 kb around SNPs rs28393318 and rs6834581 associated with IL-6 and IL-1b concentrations at the TLR10-TLR1-TLR6 locus. These polymorphisms reached the genome-wide significant threshold (red lines) but showed non-significant association in the Tanzania cohort.
(D) Heatmap showing the association between SNPs (rs28393318 and rs6834581) with all cytokine-stimulation pairs (on the vertical axis). SNP-cytokine associations with p values greater than 5 × 10−2 were considered non-significant (ns).
(E and F) Linkage disequilibrium (LD) patterns of genetic variants in the TLR locus for the Tanzanians (E) and Europeans (F). Red color depicts areas of strong LD and yellow represents sections of weak LD. The black lines show the location of each marker in this genomic region.
In addition, through publicly available eQTL database searches, there was no evidence of the lead cQTL SNPs from the African cohort showing effect on expression levels of TLR10-TLR1-TLR6 genes (data not shown). Furthermore, we observed varying LD patterns between the populations in this genomic region. While we observed some isolated markers in smaller LD blocks in the Tanzanians (Figure 5E), areas of stronger LD corresponding to uniform haplotype blocks were seen in the Europeans (Figure 5F). This observation points to the role of different haplotypes at this locus in the African population than in the European population contributing to the cytokine regulation in response to pathogens.
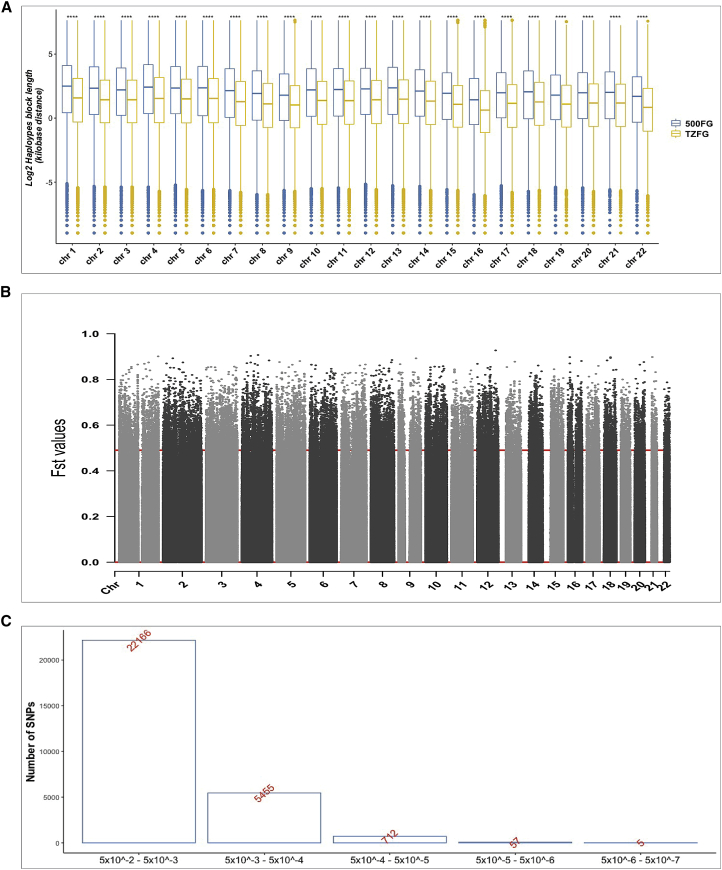
Haplotype block lengths varied between the African and European cohorts
To get some insights into the substantial lack of cQTLs shared between both populations, we tested whether average length of haplotype blocks vary significantly between two populations. Across the entire autosomes, we observed significantly higher median haplotype block length in the 500FG samples contrary to the observed haplotype block length in the Tanzanians (Figure 6A). Also, compared to the 500FG samples, we observed higher number of blocks in the Tanzanians. Details of the results supporting Figure 6A are depicted in Table S2. These observations indicate differential genetic architecture underlying both populations, which could partly explain the limited concordance we observed with cQTLs between two populations.
Figure 6.
Haplotype analysis and candidate markers under positive selection
(A) Boxplot of median haplotype blocks length (y axis) per chromosome (x axis) for the Tanzanians (yellow) and Europeans (steel blue). The asterisks (∗) show the degree of significance difference between both populations based on Wilcoxon signed-rank non-parametric test.
(B) Manhattan Fst plot of SNPs (in grays) across the genome. The red horizontal line represents the 99th percentile, threshold of significance.
(C) Bar plot of the distribution of significant genetic markers across different p value categories, ranging from nominal to suggestive associations.
Limited evidence for cQTLs in Tanzanians to be under positive selection
To test whether some of cQTLs identified in the Tanzanians are under positive selection, we first computed genome-wide Fst values for each SNP to uncover differential markers or signals of positive selection between both populations. The genome-wide Fst averaged 0.10. The top 1% Fst values, corresponding to a threshold of 0.535, were identified as markers (n = 37,867) showing significant differentiation (Figure 6B). We then tested how many of the nominally associated cQTLs (p < 0.05) were also showing a very high Fst value. We found less than 0.75% of the cQTLs (p < 0.05) showed >0.535 of Fst value, indicating the possibility that only a few cQTLs were under positive selection (Table S3). Interestingly, there were five strongly associated cQTLs (p < 10−6) with high Fst value, while the majority of the cQTLs were nominal associations (Figure 6C). Although this observation suggests lack of strong enrichment of cQTLs to be under natural selection, some of these loci could be interesting candidates to understand their function in the context of evolutionary genetics.
Discussion
This study identified genetic loci in a Tanzanian population that affect cytokine responses of primary immune cells to diverse pathogenic stimuli. We also show that the genetic loci that regulate cytokine responses differ between populations of African ancestry and Western European ancestry. Notably, there is less than 2% overlap among cQTLs between the two populations, including lack of association at the TLR10-TLR1-TLR6 locus in the Tanzanian population.
One of the important observations of our study is the pathogen-independent clustering of cytokine responses. The pattern with which an individual elicits cytokine response to pathogens could be either cytokine and/or stimulus dependent. While previous studies in the European population reported pathogen-dependent cytokine responses,5,6 we observed the contrary in our Tanzanian cohort in which certain cytokine-stimulation pairs, particularly IL-10, clustered at the cytokine level, which suggests that anti-inflammatory responses are organized to respond to different pathogens through a specific cytokine pathway. Unfortunately, IL-10 measurement was not available from the previous 500FG study for exploration of whether this pattern is specific to anti-inflammatory cytokines. Nevertheless, it will be interesting to explore further through future studies whether ancestry could drive the differences at the patterns of cytokine clusters.
Our study also identified a genome-wide significant locus exerting a strong effect on cytokine levels in response to pathogens. The most significant SNP, rs12169244, associated with IL-1b levels resides near the cis-region of TBC1D22A rather than IL1B. In addition, by conducting a meta-analysis between the 500FG and 300TZFG cohorts, we found another genome-wide significant cQTL for TNF- with the top SNP, rs60372900, residing near ZNF354A (MIM: 602444). We made a similar observation in our previous studies using European-based cohorts,5,6 where all the significant cQTLs were located in trans, suggesting the significant role of trans-regulatory pathways in modulating cytokine responses upon stimulations. We were unable to establish any correlation between these top SNPs and potential causal genes by using available eQTL datasets. Thus, it is difficult to speculate the mechanistic role of any cis-genes, and we therefore emphasize the need for relevant eQTL datasets from the African population.
Comparative analysis of genetic variants associated with cytokine response upon stimulation revealed that most of the genetic variants exert their effect on cytokines response in a population-specific manner, as a very limited percentage (1.6% to 1.9%) of shared effect was observed. This result may not come as a surprise considering the distinct genetic architecture that is known to exist between populations on different continents, most likely due to different infectious pressure.32 These findings highlight the potential role of common variants to the remarkable interpopulation diversity in immune responses and susceptibility to infectious diseases.33,34 In this context, by performing simulation analyses, studies have shown that the accuracy of Fst estimation directly depends on the number of genetic polymorphisms,35 suggesting the intrinsic role of genetic markers on population differentiation estimates. Although our Fst analysis provided some indication for genetic differentiation between the two populations, including a large number of samples and whole-genome sequence data to represent the true population diversity, particularly from the African population, will help in accurately estimating the genetic differentiation. Evidently, non-genetic factors such as gut microbiome,36 seasonality,4 age and sex,37,38 and urban or rural living also contribute to interindividual differences in immune responses. This observation again highlights the fact that host genetics is shaped by strong selective pressure from environmental factors, mainly pathogens.
Several lines of evidence have demonstrated that TLRs have undergone different selection pressures across populations and have identified the TLR10-TLR1-TLR6 locus as a target of positive selection in contemporary European populations.39,40 Indeed, it has been hypothesized that this locus may have been selected by the severe ancient and medieval plague epidemics in Europe, as this locus modulates the immune response to Yersinia pestis.41 Our analysis evaluating LD patterns in this locus is consistent with these previous findings. We observed much longer blocks of LD in the Europeans as compared to the Africans, a typical characteristic of a region undergoing natural selection.42 Another typical example is TLR6, and variants in this gene have been shown to exhibit differential allele frequencies between European populations and other populations.43 Interestingly, the variation in the TLR10-TLR1-TLR6 locus has been reported to be strongly influenced in Europeans by introgression from Neanderthals, thus showing the impact that archaic humans had on immune responses of modern human populations.40 As practically no introgression of Neanderthals and Denisovans is found in African populations, it becomes less surprising the lack of impact of variation in this locus on the immune response of African populations. Our observation of lack of impact of the TLR10-TLR1-TLR6 locus on cytokine responses in the African population strengthens the possibility that this locus has putatively undergone adaptive evolution exclusively in Europe.
In the same line of comparison of molecular phenotypes across diverse ancestries, a study systematically assessing allelic variants in cytokine genes showed that African American women were more likely to harbor allelic variants known to upregulate pro-inflammatory cytokines compared to women of European ancestry.44 Also, on the basis of genotype-expression data, 9.3% of genes expressed in macrophages exhibit ancestry-related variances in the regulatory response to infection, and African ancestry typically predicts stronger inflammation than European ancestry.9 Our pathway enrichment analyses based on genes mapped to cQTLs also showed similar results: contrary to European samples, cQTL genes from the Tanzanian cohort were significantly enriched in the interferon pathway. This is interesting because, using the blood transcriptional data from the same cohort, we recently observed significant upregulation of interferon genes among urban residents of Tanzania.19 In line with this, Manry et al. (2011) have also shown significant differences in the number of polymorphisms within the population between the genes encoding the various IFNs and their receptors, in which the highest levels of nucleotide diversity within interferon genes were observed in African populations.45 Interestingly, Quach et al. (2016) have shown that virus-specific eQTLs in Europeans are mostly population specific and Neanderthals introduced regulatory variants into European genomes.33 Given the lack of Neanderthal variants in African genomes and the possibility that people in Tanzania are much more exposed to Mycobacterium tuberculosis46 than the people living in the Netherlands, it is tempting to speculate that the natural selection had independently led to an increased interferon response in populations of Africa and is partly driven by host genetic variations. Whether this enhanced interferon response in the Tanzanian population influences risk for COVID-19 remains to be tested.
We also acknowledge some limitations with our study. First, using only 300 samples in our discovery cohort, we are unable to recruit a sufficiently large independent cohort of Tanzanian volunteers for replication analysis. Second, contrary to the Tanzania cohort, immune cell counts data used for correcting cytokines abundance prior to cQTL mapping were only available for the European cohort (500FG). Therefore, it is possible that some of the cQTLs are false positive associations and further validation is required. Third, a number of recent studies have identified the inability of the currently available SNP arrays to capture or assign non-European variants and haplotypes accurately.47,48 We therefore reasoned that some critical variants in the Tanzanians might be missed, especially untagged variants with different frequencies and rare variants. Therefore, whole-genome sequence data from diverse populations are very much needed to capture population-specific and rare variants to study their impact of immune function.
Nevertheless, this study offers unique contribution to studies comprehensively investigating the genetics of cytokines response variability in non-European ancestry, most importantly leveraging a large variety of pathogens for inducing cytokine responses. Follow-up studies in larger cohorts are required to validate our findings and also to discover more genetic variants with less strong effects.
Conclusions
Our study shows that at both SNPs and immunological pathway levels, the cytokine production capacity upon diverse stimulations is modulated in a population-specific manner. We also demonstrate that the TLR1-TLR6-TLR10 locus significantly affects cytokine production capacity only in individuals of European descent. Given the dissimilarity of genetic determinants regulating induced cytokine responses in both populations, we argue for the inclusion of under-represented populations in genomics research to make new discoveries in health and disease disparities seen among individuals and populations.
Acknowledgments
This study was funded by the following grants: the European Union’s Horizon 2020 Research and Innovation Program under the ERA-Net Cofund action no. 727565; the Joint Programming Initiative, A Healthy Diet for a Healthy Life (JPI-HDHL; project 529051018) awarded to M.G.N., Q.d.M., and A.V.; ZonMw (the Netherlands Organization for Health Research and Development) awarded to M.G.N., Q.d.M., and A.V.; and Radboud Revolving Research Funds (3R-Fund) awarded to G.S.T. Spinoza Prize (NWO SPI94-212) and ERC Advanced grant (no. 833247) awarded to M.G.N. Hypatia tenure track grant from Radboud UMC to V.K. The authors are grateful to all volunteers from the 300TZFG and 500FG cohorts without whom this research would not have been possible for participation in the studies.
Declaration of interests
The authors declare no competing interests.
Published: February 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.01.014.
Data and code availability
Summary statistics of the association results have been deposited to the EBI GWAS Catalog under the accession number GCST90093065. All other data and code underlying the findings are also available on reasonable request from the corresponding author.
Web resources
Ensembl, https://www.ensembl.org
HaploReg, http://pubs.broadinstitute.org/
Human Functional Genomics Project, http://www.humanfunctionalgenomics.org/site/
LDlink, https://ldlink.nci.nih.gov
LocusZoom, http://locuszoom.org
Online Mendelian Inheritance in Man, https://www.omim.org/
Supplemental information
References
- 1.Akdis M., Burgler S., Crameri R., Eiwegger T., Fujita H., Gomez E., Klunker S., Meyer N., O’Mahony L., Palomares O., et al. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2011;127 doi: 10.1016/j.jaci.2010.11.050. 701–21.e1, 70. [DOI] [PubMed] [Google Scholar]
- 2.Vignali D.A.A., Kuchroo V.K. IL-12 family cytokines: immunological playmakers. Nat. Immunol. 2012;13:722–728. doi: 10.1038/ni.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labuda L.A., de Jong S.E., Meurs L., Amoah A.S., Mbow M., Ateba-Ngoa U., van der Ham A.J., Knulst A.C., Yazdanbakhsh M., Adegnika A.A. Differences in innate cytokine responses between European and African children. PLoS ONE. 2014;9:e95241. doi: 10.1371/journal.pone.0095241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ter Horst R., Jaeger M., Smeekens S.P., Oosting M., Swertz M.A., Li Y., Kumar V., Diavatopoulos D.A., Jansen A.F.M., Lemmers H., et al. Host and Environmental Factors Influencing Individual Human Cytokine Responses. Cell. 2016;167:1111–1124.e13. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Oosting M., Deelen P., Ricaño-Ponce I., Smeekens S., Jaeger M., Matzaraki V., Swertz M.A., Xavier R.J., Franke L., et al. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat. Med. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Oosting M., Smeekens S.P., Jaeger M., Aguirre-Gamboa R., Le K.T.T., Deelen P., Ricaño-Ponce I., Schoffelen T., Jansen A.F.M., et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016;167:1099–1110.e14. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Wurfel M.M., Gordon A.C., Holden T.D., Radella F., Strout J., Kajikawa O., Ruzinski J.T., Rona G., Black R.A., Stratton S., et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am. J. Respir. Crit. Care Med. 2008;178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikacenic C., Reiner A.P., Holden T.D., Nickerson D.A., Wurfel M.M. Variation in the TLR10/TLR1/TLR6 locus is the major genetic determinant of interindividual difference in TLR1/2-mediated responses. Genes Immun. 2013;14:52–57. doi: 10.1038/gene.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nédélec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.C., Freiman A., Sams A.J., Hebert S., et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Retshabile G., Mlotshwa B.C., Williams L., Mwesigwa S., Mboowa G., Huang Z., Rustagi N., Swaminathan S., Katagirya E., Kyobe S., et al. Whole-Exome Sequencing Reveals Uncaptured Variation and Distinct Ancestry in the Southern African Population of Botswana. Am. J. Hum. Genet. 2018;102:731–743. doi: 10.1016/j.ajhg.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magalhães T.R., Casey J.P., Conroy J., Regan R., Fitzpatrick D.J., Shah N., Sobral J., Ennis S. HGDP and HapMap analysis by Ancestry Mapper reveals local and global population relationships. PLoS ONE. 2012;7:e49438. doi: 10.1371/journal.pone.0049438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley A.R., Callier S.L., Rotimi C.N. Evaluating the promise of inclusion of African ancestry populations in genomics. NPJ Genom. Med. 2020;5:5. doi: 10.1038/s41525-019-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Pittman K.J., Barker J.R., Salinas R.E., Stanaway I.B., Williams G.D., Carroll R.J., Balmat T., Ingham A., Gopalakrishnan A.M., et al. An Atlas of Genetic Variation Linking Pathogen-Induced Cellular Traits to Human Disease. Cell Host Microbe. 2018;24:308–323.e6. doi: 10.1016/j.chom.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karimuribo E.D., Mboera L.E., Mbugi E., Simba A., Kivaria F.M., Mmbuji P., Rweyemamu M.M. Are we prepared for emerging and re-emerging diseases? Experience and lessons from epidemics that occurred in Tanzania during the last five decades. Tanzan. J. Health Res. 2011;13(5, Suppl 1):387–398. doi: 10.4314/thrb.v13i5.8. [DOI] [PubMed] [Google Scholar]
- 16.Kumburu H.H., Sonda T., Mmbaga B.T., Alifrangis M., Lund O., Kibiki G., Aarestrup F.M. Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Trop. Med. Int. Heal. 2017;22:454–464. doi: 10.1111/tmi.12836. [DOI] [PubMed] [Google Scholar]
- 17.Netea M.G., Wijmenga C., O’Neill L.A.J. Genetic variation in Toll-like receptors and disease susceptibility. Nat. Immunol. 2012;13:535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson E.K., Kwiatkowski D.P., Sabeti P.C. Natural selection and infectious disease in human populations. Nat. Rev. Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temba G.S., Kullaya V., Pecht T., Mmbaga B.T., Aschenbrenner A.C., Ulas T., Kibiki G., Lyamuya F., Boahen C.K., Kumar V., et al. Urban living in healthy Tanzanians is associated with an inflammatory status driven by dietary and metabolic changes. Nat. Immunol. 2021;22:287–300. doi: 10.1038/s41590-021-00867-8. [DOI] [PubMed] [Google Scholar]
- 20.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 21.Shah T.S., Liu J.Z., Floyd J.A., Morris J.A., Wirth N., Barrett J.C., Anderson C.A. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–1603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deelen P., Bonder M.J., van der Velde K.J., Westra H.J., Winder E., Hendriksen D., Franke L., Swertz M.A. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res. Notes. 2014;7:901. doi: 10.1186/1756-0500-7-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin J.H., Blay S., McNeney B., Graham J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006;16:1–9. [Google Scholar]
- 28.Team R.C.R. R Foundation for Statistical Computing; 2016. A Language and Environment for Statistical Computing. [Google Scholar]
- 29.Cerillo G., Rees A., Manchanda N., Reilly C., Brogan I., White A., Needham M. The oestrogen receptor regulates NFkappaB and AP-1 activity in a cell-specific manner. J. Steroid Biochem. Mol. Biol. 1998;67:79–88. doi: 10.1016/S0960-0760(98)00078-8. [DOI] [PubMed] [Google Scholar]
- 30.Hug H., Mohajeri M.H., La Fata G. Toll-like receptors: Regulators of the immune response in the human gut. Nutrients. 2018;10:E203. doi: 10.3390/nu10020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Areal H., Abrantes J., Esteves P.J. Signatures of positive selection in Toll-like receptor (TLR) genes in mammals. BMC Evol. Biol. 2011;11:368. doi: 10.1186/1471-2148-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang L., Smith J.A., Zhao W., Kho M., Turner S.T., Mosley T.H., Kardia S.L.R., Zhou X. Genetic Architecture of Gene Expression in European and African Americans: An eQTL Mapping Study in GENOA. Am. J. Hum. Genet. 2020;106:496–512. doi: 10.1016/j.ajhg.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quach H., Rotival M., Pothlichet J., Loh Y.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M., et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwok A.J., Mentzer A., Knight J.C. Host genetics and infectious disease: new tools, insights and translational opportunities. Nat. Rev. Genet. 2020;22:137–153. doi: 10.1038/s41576-020-00297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willing E.M., Dreyer C., van Oosterhout C. Estimates of genetic differentiation measured by F(ST) do not necessarily require large sample sizes when using many SNP markers. PLoS ONE. 2012;7:e42649. doi: 10.1371/journal.pone.0042649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Ter Horst R., Jansen T., Jacobs L., Bonder M.J., et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167:1125–1136.e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patin E., Hasan M., Bergstedt J., Rouilly V., Libri V., Urrutia A., Alanio C., Scepanovic P., Hammer C., Jönsson F., et al. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol. 2018;19:302–314. doi: 10.1038/s41590-018-0049-7. [DOI] [PubMed] [Google Scholar]
- 38.Bakker O.B., Aguirre-Gamboa R., Sanna S., Oosting M., Smeekens S.P., Jaeger M., Zorro M., Võsa U., Withoff S., Netea-Maier R.T., et al. Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nat. Immunol. 2018;19:776–786. doi: 10.1038/s41590-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barreiro L.B., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J.K., Bouchier C., Tichit M., Neyrolles O., Gicquel B., et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5:e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dannemann M., Andrés A.M., Kelso J. Introgression of Neandertal- and Denisovan-like Haplotypes Contributes to Adaptive Variation in Human Toll-like Receptors. Am. J. Hum. Genet. 2016;98:22–33. doi: 10.1016/j.ajhg.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laayouni H., Oosting M., Luisi P., Ioana M., Alonso S., Ricaño-Ponce I., Trynka G., Zhernakova A., Plantinga T.S., Cheng S.C., et al. Convergent evolution in European and Rroma populations reveals pressure exerted by plague on Toll-like receptors. Proc. Natl. Acad. Sci. USA. 2014;111:2668–2673. doi: 10.1073/pnas.1317723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucek K., Willi Y. Drivers of linkage disequilibrium across a species’ geographic range. PLoS Genet. 2021;17:e1009477. doi: 10.1371/journal.pgen.1009477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickrell J.K., Coop G., Novembre J., Kudaravalli S., Li J.Z., Absher D., Srinivasan B.S., Barsh G.S., Myers R.M., Feldman M.W., Pritchard J.K. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ness R.B., Haggerty C.L., Harger G., Ferrell R. Differential distribution of allelic variants in cytokine genes among African Americans and White Americans. Am. J. Epidemiol. 2004;160:1033–1038. doi: 10.1093/aje/kwh325. [DOI] [PubMed] [Google Scholar]
- 45.Manry J., Laval G., Patin E., Fornarino S., Itan Y., Fumagalli M., Sironi M., Tichit M., Bouchier C., Casanova J.L., et al. Evolutionary genetic dissection of human interferons. J. Exp. Med. 2011;208:2747–2759. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheriff F.G., Manji K.P., Manji M.P., Chagani M.M., Mpembeni R.M., Jusabani A.M., Alwani Z.R., Karimjee T.S. Latent tuberculosis among pregnant mothers in a resource poor setting in Northern Tanzania: a cross-sectional study. BMC Infect. Dis. 2010;10:52. doi: 10.1186/1471-2334-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lankheet I., Vicente M., Barbieri C., Schlebusch C. The performance of common SNP arrays in assigning African mitochondrial haplogroups. BMC Genom. Data. 2021;22:43. doi: 10.1186/s12863-021-01000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnston H.R., Hu Y.J., Gao J., O’Connor T.D., Abecasis G.R., Wojcik G.L., Gignoux C.R., Gourraud P.A., Lizee A., Hansen M., et al. CAAPA Consortium Identifying tagging SNPs for African specific genetic variation from the African Diaspora Genome. Sci. Rep. 2017;7:46398. doi: 10.1038/srep46398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics of the association results have been deposited to the EBI GWAS Catalog under the accession number GCST90093065. All other data and code underlying the findings are also available on reasonable request from the corresponding author.