Summary
Attempts to identify and prioritize functional DNA elements in coding and non-coding regions, particularly through use of in silico functional annotation data, continue to increase in popularity. However, specific functional roles can vary widely from one variant to another, making it challenging to summarize different aspects of variant function with a one-dimensional rating. Here we propose multi-dimensional annotation-class integrative estimation (MACIE), an unsupervised multivariate mixed-model framework capable of integrating annotations of diverse origin to assess multi-dimensional functional roles for both coding and non-coding variants. Unlike existing one-dimensional scoring methods, MACIE views variant functionality as a composite attribute encompassing multiple characteristics and estimates the joint posterior functional probabilities of each genomic position. This estimate offers more comprehensive and interpretable information in the presence of multiple aspects of functionality. Applied to a variety of independent coding and non-coding datasets, MACIE demonstrates powerful and robust performance in discriminating between functional and non-functional variants. We also show an application of MACIE to fine-mapping and heritability enrichment analysis by using the lipids GWAS summary statistics data from the European Network for Genetic and Genomic Epidemiology Consortium.
Key words: functional annotations, multi-dimensional integrated scores, prediction of functional effect, generalized linear mixed model, EM algorithm
Introduction
Ever since the completion of the human genome sequence, substantial effort has been invested into identifying and annotating functional DNA elements. For any given genetic variant, a diverse set of functional annotations is now available. For example, the computational tool PolyPhen1 predicts damaging effects of missense mutations. PhastCons,2 PhyloP,3 and GERP++4 leverage comparative sequence information to identify regions that show evolutionary conservation. The Encyclopedia of DNA Elements (ENCODE) has extensively mapped regions of transcription-factor binding, chromatin structure, and histone modification and has effectively assigned biochemical functions for ∼80% of the genome.5 Other initiatives such as the Roadmap Epigenomics project6 and FANTOM5 project7,8 also provide evidence for potential regulatory variants in the human genome.
Although functional annotations vary considerably with respect to the specific elements they evaluate and the extent of the human genome they annotate, it is well understood that they provide complementary lines of evidence.9 Therefore, if researchers are to obtain a comprehensive understanding of the biological relevance of genomic segments, all of the information provided by different annotations should be jointly synthesized. However, it remains challenging to summarize these diverse functional annotations in an insightful and interpretable manner.
Current algorithmic scoring frameworks utilize a variety of statistical and machine-learning methods to aggregate information from large, diverse sets of individual annotations into single measures of functional importance. Supervised tools such as CADD,10 DANN,11 GWAVA,12 FATHMM-MKL,13 and FATHMM-XF14 build machine-learning classifiers on training sets with pre-labeled functional statuses, e.g., fine-mapped pathogenic or disease-associated variants labeled against benign or neutral variants. Such supervised approaches rely strongly on the quality of labels in the training set. Therefore, they might demonstrate suboptimal performance when inaccurate or biased labels are used. Unsupervised methods such as EIGEN,15 GenoCanyon,16 PINES,17 and FUN-LDA18 do not rely on any labeled training data. They possess advantages in studying non-coding regions, where our current lack of knowledge often precludes gold-standard training data labels. A third group of methods including fitCons19 and LINSIGHT20 use evolution-based approaches that characterize the potential effect of natural selection at each genomic location by using polymorphism and divergence data. Recent reviews provide a more detailed discussion of available functional annotation tools.21, 22, 23
Although existing methods attempt to integrate functional annotations through various approaches, to the best of our knowledge, these methods all summarize the annotation information with a single rating. In doing so, they implicitly assume that variant function can be described along a single axis, whereby variants are more functional at one end of the axis and less functional at the other end. This assumption might be reasonable if interest lies in predicting a specific aspect of variant function (e.g., regulatory behavior) and if all annotations used as input are intended to predict that same aspect. However, if multiple aspects of variant function are simultaneously of interest, then it is unclear how to interpret the one-dimensional consolidation of annotations measuring different aspects of function, especially when these annotations appear to provide orthogonal information, e.g., weak correlation between evolutionary conservation scores and regulatory scores (Figure 1). Therefore, it is of interest to construct multi-dimensional integrative scores capable of capturing multiple facets of variant function simultaneously.
Figure 1.
Heatmap demonstrating the correlation between individual and integrative functional scores for ClinVar pathogenic and benign non-coding variants
In this paper, we propose multi-dimensional annotation-class integrative estimation (MACIE), an unsupervised multivariate mixed-model framework capable of synthesizing multiple categories of annotations and producing interpretable multi-dimensional integrative scores (Figure S1). Instead of a single rating, MACIE explicitly defines variant function as a vector of binary outcomes, each of which captures functionality corresponding to a specific class of annotations. Correlations within and between the different classes of annotations are explicitly modeled, another advancement over existing methods. Using the expectation-maximization algorithm, MACIE calculates the joint posterior probabilities that a genomic position is functional (material and methods).
Because of its multivariate formulation, MACIE is able to provide detailed and nuanced assessments of variant functionality. Output from MACIE is highly interpretable as a result of the specificity allowed by multiple functional classes. Additionally, the MACIE framework allows for considerable versatility to incorporate data in a manner that is most biologically relevant to the scientific question of interest. We apply MACIE to multiple independent coding and non-coding testing sets and show that, compared to existing integrative scores, MACIE scores consistently provide robust and best or near-best performance in discriminating between functional and non-functional variants.
Material and methods
The MACIE generalized linear mixed model (GLMM)
Suppose there are genetic variants in total and we are interested in latent annotation classes, each containing annotation scores. For example, the first class might consist of protein functional scores, and the second class might consist of evolutionary conservation scores. For genetic variant and annotation class , we denote the set of annotations as , such that each variant is described by annotations in total. We are interested in estimating for each variant the vector of binary functional statuses , where is the unobserved latent functional status for class . Continuing our example, would denote membership in the evolutionarily conserved function class, and would denote membership in the regulatory function class. Conditional on and a random effect variable , we assume that the elements of are independent observations, each following a GLMM. That is, for and ,
where additional correlations between elements of are allowed if we assume that
The MACIE score for a given genetic variant is defined by , that is, the posterior probability vector of the unobserved class label , conditional on the observed annotations . Because of thconditional independence of given and (the collections of , , and , respectively, for ), an expectation-maximization (EM) algorithm provides a natural approach.24
The expectation-maximization algorithm
The complete-data log-likelihood of is given by
where are (unknown) model parameters. Because both and are unobserved, we proceed with the following EM algorithm:
-
1.
Initiate reasonable parameter values. Denote the parameter estimates at iteration r by .
-
2.
(E-step a) Compute the posterior distribution via Bayes’ theorem:
-
3.
(E-step b) Compute expected score functions with respect to the posterior distribution of , i.e., , where are the complete data score functions of , respectively.
-
4.
(M-step) Update by solving the expected score equations from step 3.
-
5.
Iterate between steps 2–4 until convergence of all parameters.
To select initial parameter values, for each functional class , we first perform a k-means clustering algorithm with to classify the functional status of each variant . We then fit a linear or logistic regression model for each individual annotation score used in this class and obtained the fitted parameter estimates as initial parameter values. We found that our results are robust to choices of initial values. The EM algorithm proceeds until the relative changes in all estimated parameters are sufficiently small (<10−4) with a maximum of 200 iterations. The final converged estimate (output of step 2) corresponds to the MACIE score for genetic variant . Given the fitted model parameters and the full set of annotation scores for a new genetic variant , the MACIE score of variant is defined as the (predicted) posterior probability vector . It can be calculated with one additional iteration of the EM algorithm. Full details of the EM-algorithm derivation are provided in the supplemental material and methods.
Construction of MACIE training sets
We used the proposed framework to fit the MACIE GLMM models and compute MACIE scores for (1) non-synonymous coding and (2) non-coding and synonymous coding variants separately because the two types of variants are expected to have highly different functional profiles.15 All non-synonymous coding annotations and non-coding and synonymous coding evolutionary conservation annotations were downloaded from EIGEN. The non-coding and synonymous coding epigenetic annotations were downloaded from CADD database10 v1.3.
Non-synonymous coding variants
For the non-synonymous coding training set, we randomly extracted 10% of the variants with a match in the dbNSFP database.25 This database excludes synonymous variants that fall in coding regions but do not alter protein function. Only one unique variant per position was selected, and variants residing on sex chromosomes X and Y were removed so that potential sources of bias or lack of annotations were mitigated. The final set included approximately 2.2 million variants. For each variant in the training set, 4 protein substitution damage scores (SIFT;26 PolyPhenDiv and PolyPhenVar;1 and Mutation Assessor27) and eight evolutionary conservation scores (GERP_NR and GERP_RS;4 PhyloP primate (PhyloPri), placental mammal (PhyloPla), and vertebrate (PhyloVer);3 and PhastCons primate (PhastPri), placental mammal (PhastPla), and vertebrate (PhastVer)2) were extracted from the EIGEN database.15 Thus, we defined the two-class MACIE model for non-synonymous coding variants to assess damaging protein-coding function and evolutionarily conserved function. Full information on the MACIE model for non-synonymous coding variants and the corresponding MACIE GLMM regression parameter estimates from the training set are presented in the supplemental material and methods and Tables S1 and S2. We used this model to compute the MACIE scores for all non-synonymous variants in the human genome.
Non-coding and synonymous coding variants
For the non-coding and synonymous coding training set, we extracted a random sample comprising 10% of the 1000 Genomes Project variants that were located within 500 bp upstream of a gene start site and did not possess a match in dbNSFP. We again removed duplicated variants with multiple alternative alleles and variants on sex chromosomes X and Y to mitigate potential bias. The final training set included 36,431 variants. For each variant in the training set, the same eight evolutionary conservation scores used for coding variants were extracted from the EIGEN database.15 A total of 28 transformed epigenetic scores were additionally extracted from the CADD database10 v.1.3; these included including three histone marks and 12 open chromatin marks from the ENCODE Project;5 three transcription factor binding-site scores; GC content, CpG content; five chromatin-state probabilities derived from the 15-state ChromHMM model;28 a background selection score;29 and two physical-distance metrics.10 Thus, we defined the two-class MACIE model for non-coding and synonymous coding variants to assess evolutionarily conserved function and epigenetic regulatory function. Full information on the MACIE model for non-coding and synonymous coding variants and the corresponding MACIE GLMM regression parameter estimates from the training set are presented in the supplemental material and methods and Tables S1, S3, and S4. We used this model to compute the MACIE scores for all non-coding and synonymous coding variants in the human genome.
Results
Benchmarking the performance of MACIE with that of other integrative scoring methods
We compared the predictive performance of MACIE against existing variant classifiers, including CADD,10 FATHMM-XF,14 EIGEN,15 fitCons,19 LINSIGHT,20 and DANN11 over a range of realistic variant-assessment scenarios. Specifically, we assessed the ability of each score to identify clinically significant variants from ClinVar;30,31 loss-of-function variants in the BRCA1 (MIM: 113705) gene uncovered through saturation genome editing (SGE);32 promoters and enhancers from the FANTOM5 project defined by cap analysis of gene expression (CAGE);7,8 and experimentally verified functional variants from massive parallel reporter assays (MPRAs).33,34 Some alternative scoring methods were excluded because of difficulties related to providing a proper comparison of results. For example, LINSIGHT is designed to predict the deleteriousness of non-coding variants, so we did not include it in the comparison for non-synonymous coding variants.
Distribution of posterior probabilities for non-coding and synonymous coding variants in the training set
In Table S5 we provide the posterior probabilities of each functional class averaged across all the non-coding and synonymous coding variants in the training set. The predicted MACIE score for a given variant can be interpreted as the posterior probability that the variant belongs to (0,0), neither conserved nor regulatory classes; (1,0), the conserved but not the regulatory class; (0,1), the regulatory but not the conserved class; and (1,1), both the conserved and the regulatory classes. The four MACIE scores necessarily sum up to 1. A chi-square test comparing observed and expected percentages under independence of evolutionary conservation and regulatory classes gives a significant p value of less than , suggesting that the two classes are correlated. Because the observed percentage of functional variants that belong to (1,1) is statistically significantly greater than the expected percentage under independence (3.18% > 1.92%), we find strong evidence of enrichment of regulatory activity in conserved regions.
ClinVar pathogenic and benign variants
We first validated our method on a testing set consisting of all variants recorded in the ClinVar database.30,31 We extracted variant effect predictor (VEP) information from GENCODE35 and used it to separate non-synonymous coding variants from non-coding and synonymous coding variants in ClinVar. We then applied the two MACIE models described above to the respective partitions. We combined the ClinVar categories “pathogenic” and “likely pathogenic” into a single pathogenic class and treated these variants as the putatively functional class. Similarly, we combined the ClinVar categories “benign” and “likely benign” into a single benign class and treated these variants as the putatively non-functional class. The remaining variants were categorized as having uncertain significance.
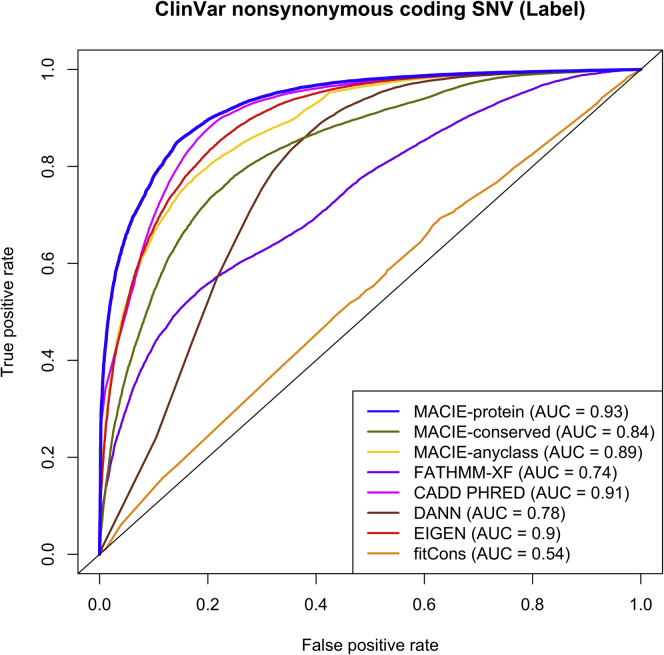
We first tested MACIE’s ability to distinguish pathogenic variants from their benign counterparts among ClinVar non-synonymous variants through two approaches. First, we calculated two marginal MACIE scores: (1) MACIE-damaging protein function score (denoted by MACIE-protein) as the sum of the posterior probabilities of “damaging protein functional/not conserved” and “damaging protein functional/conserved”; and (2) MACIE-conserved score as the sum of the posterior probabilities of “damaging protein functional/conserved” and “not damaging protein functional/conserved.” We also considered the posterior probability of either “damaging protein functional/conserved” (denoted by MACIE-anyclass) by summing the posterior probabilities corresponding to at least one functional class (Table S6). This example illustrates the versatility of MACIE’s posterior probability outputs, which can be summed to form new probability measures with various informative interpretations depending on the specific needs of each analysis.
Figure 2 provides the receiver operating characteristic (ROC) curves and area under the curves (AUC) for the three MACIE approaches and seven one-dimensional scores for ClinVar non-synonymous variants. Of the methods considered, MACIE-damaging protein function score delivered the highest discrimination power (AUC = 0.93), followed by CADD (AUC = 0.91), EIGEN (AUC = 0.90), and MACIE-anyclass (AUC = 0.89). These four methods substantially outperformed the supervised DANN (AUC = 0.78), the supervised FATHMM-XF (AUC = 0.74), and the evolution-based fitCons (AUC = 0.54). We observed similar results when we distinguished between pathogenic missense (as opposed to all non-synonymous) variants and their benign counterparts in ClinVar (Figure S2).
Figure 2.
ROC curves comparing the performances of MACIE and other functional scores in discriminating between ClinVar pathogenic and benign non-synonymous coding variants
Next, we identified 40,109 non-coding variants, including 6,551 pathogenic variants and 33,558 benign variants, from the ClinVar database in total. For these non-coding variants, we chose to calculate a marginal MACIE-conserved score because ClinVar pathogenic non-coding variant labels track closely with evolutionary conservation scores (Figure 1). ROC curves and AUCs for discriminating between the pathogenic and benign variants are provided in Figure S3. The MACIE-conserved score showed comparable performance (AUC = 0.95) to the FATHMM-XF score, which showed the highest discrimination power (AUC = 0.97). The outperformance of FATHMM-XF in this specific example should be expected because FATHMM-XF is a supervised machine-learning method trained on labels that bear many similarities to the labels defined in ClinVar, whereas MACIE is an unsupervised method. The MACIE-conserved score substantially outperformed the unsupervised method EIGEN (AUC = 0.84) and the evolution-based method fitCons (AUC = 0.55).
Loss-of-function non-synonymous coding variants in BRCA1
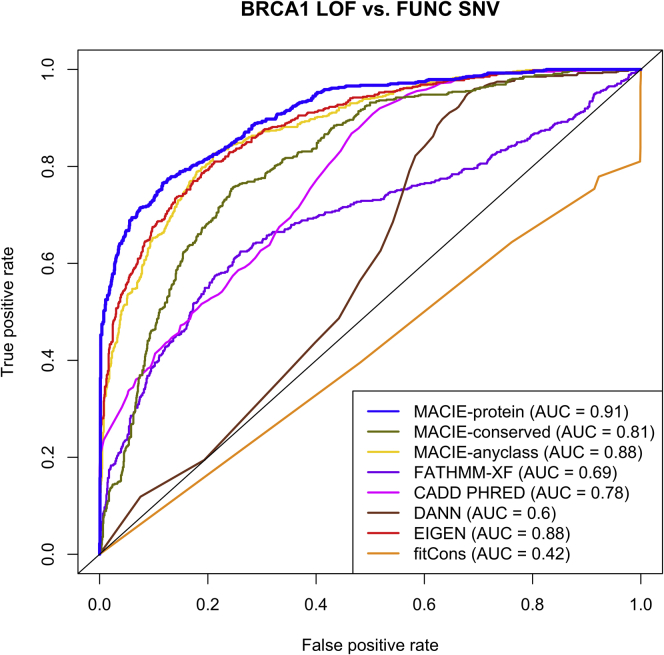
We evaluated MACIE’s performance in predicting the deleteriousness of non-synonymous coding variants located within 13 exons that encode functionally critical domains of BRCA1. We fit based a two-component Gaussian mixture model on the saturation genome editing function scores to classify all BRCA1 variants as loss-of-function (LOF), intermediate (INT), or functional (FUNC) in a decreasing order of severity.32 Thus, FUNC corresponds to benign variants in this experiment. We selected reported LOF non-synonymous coding variants as the putative functional set and designated FUNC non-synonymous coding variants as the putative non-functional set. Among all the methods compared (Figure 3), the MACIE-damaging protein function score showed the highest predictive power (AUC = 0.91), followed by EIGEN (AUC = 0.88) and MACIE-anyclass (AUC = 0.88). The top three scores were much more powerful than CADD (AUC = 0.78), FATHMM-XF (AUC = 0.69), DANN (AUC = 0.60), and fitCons (AUC = 0.42). We observed similar results when distinguishing between BRCA1 LOF non-synonymous coding variants and ClinVar benign non-synonymous coding variants (Figure S4).
Figure 3.
Performance comparing MACIE and other functional scores on BRCA1 non-synonymous coding variants
ROC curves comparing the performances of MACIE and other functional scores in discriminating between loss-of-function (LOF) and functional (FUNC) non-synonymous coding variants within 13 exons that encode functionally critical domains of BRCA1 on the basis of saturation genome editing (SGE) data. Here the LOF class is our putative functional class, and the FUNC class is our putative non-functional class.
FANTOM5 CAGE-defined promoters and enhancers among 1000 genomes non-coding variants
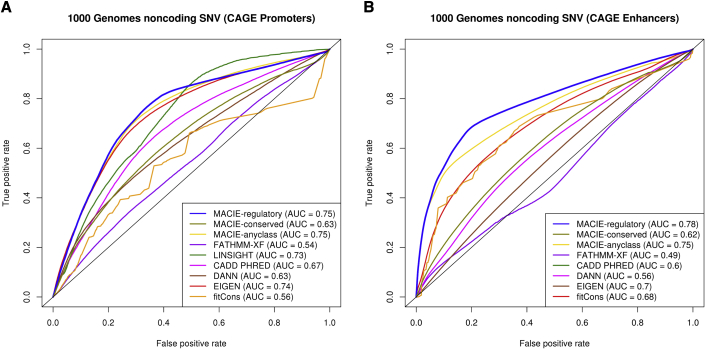
We tested the ability of MACIE to identify promoter regions defined by the FANTOM5 project’s cap analysis of gene expression.7,8 A total of 110,895 out of approximately 80 million non-coding variants from the 1000 Genomes Project phase 3 data36 were mapped to such regions and therefore labeled as CAGE promoters. For each identified CAGE promoter variant, we used the 1000 Genomes Project database to randomly select a matched control variant (non-promoter) that possessed the same minor-allele frequency (MAF) and same minimum distance to any gene transcription start site that was located at least 500 kilobase (kb) away from the promoter variant, yielding a total number of 97,298 variants in the control set (it was not possible to find a matched control for each CAGE variant). Similar to the previous analysis, we calculated a marginal MACIE-regulatory score by summing the two probabilities corresponding to the regulatory class (denoted by MACIE-regulatory). ROC curves and AUCs for discriminating between CAGE promoters and non-promoters are provided in Figure 4A. MACIE-regulatory and MACIE-anyclass scores showed the highest discrimination power (AUC = 0.75), followed by EIGEN with AUC = 0.74. FATHMM-XF (AUC = 0.54) and fitCons (AUC = 0.56) scores performed poorly because these one-dimensional scores are unable to capture epigenetic functionality. We also performed a similar analysis by contrasting CAGE-identified enhancers versus non-enhancers by using non-coding variants from the 1000 Genomes Project. The results were similar, and MACIE-regulatory score displayed the highest predictive power and significantly outperformed all other existing methods (Figure 4B).
Figure 4.
Performance comparing MACIE and other functional scores on predicting CAGE promoters and CAGE enhhancers from 1000 Genomes Project phase 3 data
ROC curves comparing the performances of MACIE and other functional scores in discriminating between (A) CAGE-identified promoters and non-promoters and (B) CAGE-identified enhancers and non-enhancers among non-coding variants from 1000 Genomes Project phase 3 data. For CAGE Enhancer predictions, LINSIGHT was excluded because it uses the FANTOM5 enhancer label as one of the genomic features in building the LINSIGHT score.
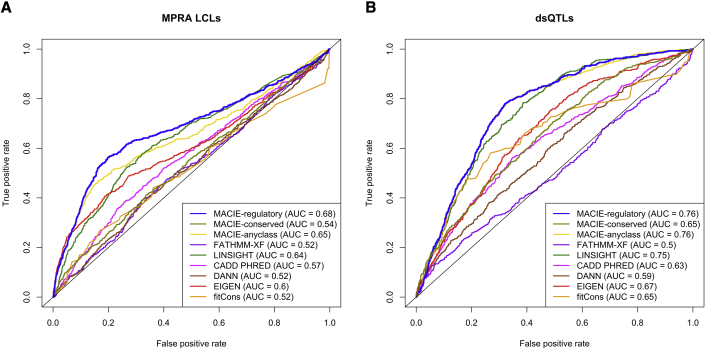
MPRA-validated variants and dsQTLs in lymphoblastoid cell lines
We used test sets from the massively parallel reporter assay to examine the performance of MACIE for predicting cell type- and tissue-specific regulatory variants. The MPRA dataset included validated regulatory variants in lymphoblastoid cell lines (LCLs).33 We paired each positive variant with four control variants from MPRA when neither allele showed significant differential expression at a Bonferroni-corrected p value threshold of 0.1 .37 Figure 5A shows that the MACIE-regulatory score produced the highest discrimination power (AUC = 0.68); the second-best performing method was LINSIGHT (AUC = 0.64).
Figure 5.
Performance comparing MACIE and other functional scores on predicting regulatory variants in LCLs against control variants
ROC curves comparing the performances of MACIE and other functional scores for the prediction of (A) validated regulatory variants in lymphoblastoid cell lines (LCLs) from massively parallel reporter assays (MPRAs) and (B) dsQTLs identified via DNase I sequencing data in LCLs against control variants.
Finally, we evaluated the performance of our proposed method on a collection of dsQTLs that were identified via DNase I sequencing data from human lymphoblastoid cell lines.38 Variants possessing association p values less than and residing within 100 bp of their corresponding DNase I-hypersensitive sites were chosen as the putatively functional set .39 The control set of variants was randomly selected from a larger set of common variants (MAF > 5%) falling in the top 5% of DNase I sensitivity sites used for identifying dsQTLs in the original study . We observed that the MACIE-regulatory score exhibited a larger AUC (AUC = 0.76) than all other methods (Figure 5B). The MACIE-anyclass score also delivered robust performance on MPRA-validated and dsQTL datasets.
In summary, MACIE consistently ranked as one of the most powerful, robust and interpretable functional annotation intergrative score methods across a variety of settings and scientific questions. Given that all the scoring methods in the benchmarking comparisons used a similar set of annotations, the better performance of MACIE across a variety of testing sets shows that whereas one-dimensional scores have gaps in functional profile coverage, a multi-dimensional scoring method offers robust and interpretable predictive performance. The ability of MACIE to interrogate variant functionality from multiple perspectives, at a level that is highly competitive with or better than state-of-the-art methods, is unmatched by existing integrative functional scoring methods.
MACIE prioritizes functional variants by using lipid GWAS data
To illustrate the utility of MACIE scores in identifying plausible functional causal variants in genetic association studies, we applied MACIE to the publicly available lipid GWAS data from the European Network for Genetic and Genomic Epidemiology (ENGAGE) Consortium.40 This dataset consists of lipid GWAS summary statistics for 9.6 million single-nucleotide variants (SNVs) across 62,166 samples (Table S7). We focused on genome-wide significant SNVs associated with low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides (TG), and total cholesterol (TC). In total, for non-synonymous coding SNVs, we found 8, 9, 5, and 10 SNVs that were predicted to belong to the protein-damaging class with probability greater than 0.9 for LDL-C, HDL-C, TG, and TC, respectively. For non-coding and synonymous coding SNVs, 643, 377, 322, and 850 SNVs were predicted to belong to the regulatory class with probability greater than 0.9, and 9, 8, 10, and 12 SNVs were predicted to belong to both the evolutionarily conserved and the regulatory class with probability greater than 0.9 for LDL-C, HDL-C, TG, and TC, respectively. Among all SNVs, 50, 64, 39, and 61 SNVs were predicted to belong to the evolutionarily conserved class with probability greater than 0.9 for LDL-C, HDL-C, TG, and TC, respectively (Tables S8–S15). Compared to the total number of marginally significant SNVs for each trait (Table S7), the MACIE scores reduce the number of SNVs prioritized for follow-up by an order of magnitude, saving much cost and effort in effectively pinpointing SNVs with relevant biological function.
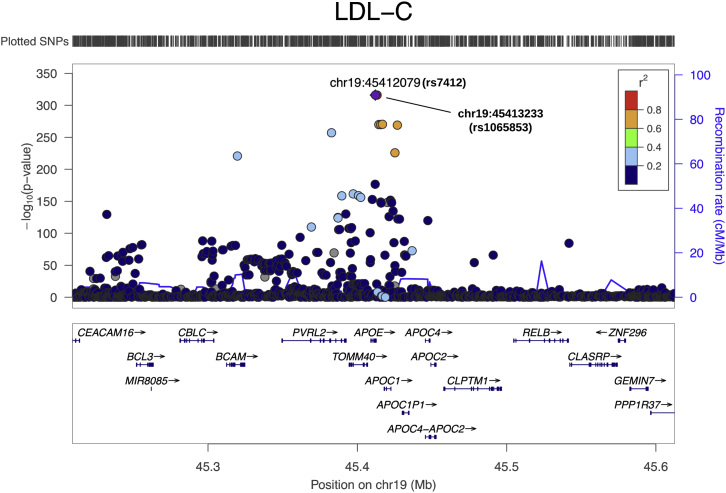
For example, for LDL-C, rs7412 (chr19: 45412079 C/T; ) was the most significant SNV in APOE (MIM: 107741). Because both MACIE-protein and MACIE-conserved scores provided a prediction greater than 0.95, we predicted this known common missense SNV to be functional. These predictions highlight the multiple functional roles of this SNV. It is also worth noting that the second-most-significant SNV rs1065853 (chr19: 45413233 G/T; ) is in extremely high linkage disequilibrium (LD) with the leading SNV rs7412 (Figure 6). MACIE scores indicate that rs1065853 (upstream variant of APOC1 [MIM: 107710]) might possess a regulatory role; its MACIE-regulatory score is greater than 0.99, possibly suggesting that both the missense and regulatory variants can be putatively causal in affecting LDL-C concentrations. Similar results were observed for TC (Figure S5). For HDL-C, although the single most significant SNV was rs17231506 (chr16: 56994528 C/T; ), the MACIE prediction was less than 0.01 for both classes. By scanning across the CETP (MIM: 118470) locus and nearby non-coding regions associated with HDL-C, we found that two SNVs, rs72786786 (chr16: 56985514 G/A; ) and rs12720926 (chr16: 56998918 A/G; ), both in moderate to high LD with the leading SNV rs17231506 (Figure S6), possess a MACIE-regulatory score greater than 0.99; rs72786786 has functional evidence, including binding proteins, motif changes, and in ENCODE DNase and regulatory chromatin states associated with diseases.41 Therefore, these two SNVs might be more functionally important than the leading SNV rs17231506 and might be prioritized for functional follow-up. For TG, there is also a lack of functional evidence for the leading SNV rs964184 (chr11: 116648917 G/C; ) in the APOA1/C3/A4/A5 gene cluster region. However, SNV rs2075290 (chr11: 116653296 C/T; ), which is in moderate LD with rs964184 at this locus, has a MACIE-regulatory score of 0.88 (Figure S7). These examples illustrate how MACIE scores can complement previous literature and provide additional information to aid prioritization of putatively functional causal variants for follow-up.
Figure 6.
LocusZoom plot for GWAS associations of LDL-C at the APOE locus
The lipid GWAS summary statistics were from the European Network for Genetic and Genomic Epidemiology (ENGAGE) Consortium (n = 58,381).40 The MACIE-protein and MACIE-conserved scores for rs7412 are 0.96 and 0.97, respectively. The MACIE-conserved and MACIE-regulatory scores for rs1065853 are <0.01 and >0.99, respectively. LDL-C, low-density lipoprotein cholesterol.
Discussion
As the amount of publicly available annotation data increases and our understanding of variant functional effects continues to grow, describing variant functionality with a flexible yet practically interpretable and intuitive vocabulary will only become more important. Existing one-dimensional integrative scores cannot capture the multi-faceted functional profile of a variant because such ratings necessarily combine diverse, and possibly unrelated, sets of annotations into a single outcome. Oftentimes, they also ignore or do not fully consider the correlation between individual annotations. Current supervised methods further demonstrate performance profiles that are linked strongly to the quality of training-set labels. These supervised scores might lack robustness in the absence of gold-standard training sets.
In this paper, we have proposed MACIE, an unsupervised multivariate mixed-model framework that allows for multiple, possibly correlated, binary functional statuses. This framework offers several fundamental advancements over existing methods. First, MACIE provides multi-dimensional scores that measure functionality across multiple different functional classes. As posterior predictive probabilities, these scores are interpretable and scientifically relevant. They can be further summarized into marginal measures such as “probability of function according to at least one class of annotations” (MACIE-anyclass). The genome-wide average of the MACIE-anyclass score is 0.084, which is consistent with the prediction from previous studies.20,42
Second, the MACIE model accommodates correlations both within and between classes. It has been reported that, although some of the available annotations measure similar notions of functionality, others provide distinct and complementary information.9,22 By flexibly modeling potential, complex correlations across all the annotations, MACIE reflects this underlying biology. In doing so, it is better able to assign each annotation and group of annotations the appropriate amount of influence.
We showed that MACIE delivered powerful and robust performance in discriminating between functional and non-functional variants in multiple independent testing sets, including (1) validated, clinically relevant variants in ClinVar, (2) SGE-verified loss-of-function variants in BRCA1, and (3) experimentally validated functional variants in FANTOM5 and MPRA. Using lipid GWAS summary statistics from the ENGAGE Consortium, we illustrated that MACIE offers an effective tool for fine-mapping studies to prioritize putative functional variants for experimental follow-up. MACIE is also informative in prioritizing lipid-associated variants through a stratified LD-score regression-heritability enrichment analysis (supplemental materials and methods).23,43 MACIE scores have already been used, for example, in the identification and characterization of inflammation and immune-related risk variants in squamous cell lung cancer.44 Finally, the proposed MACIE scores can be used as a weighting scheme to further empower variant-set analyses of rare variants.45
Our proposed MACIE framework provides a multi-dimensional functional-class extension of several existing unsupervised single scoring frameworks, such as EIGEN.15 MACIE fits a mixed model to the set of annotations for several latent functional classes and outputs the corresponding posterior component probabilities, which are highly interpretable. If we assume that there exists a single latent dichotomous variable summarizing functional status and that all annotations are independent on condition of the univariate functional status, then MACIE reduces to the GenoCanyon framework (Figure S1B).16 MACIE produces multi-dimensional functional scores and has advantages over GenoCanyon, which predicts the regulatory potential of genomic locations by using a single-dimensional score. The MACIE-damaging protein function score (AUC = 0.93) substantially outperformed GenoCanyon (AUC = 0.69) in the ClinVar coding testing set, and the MACIE-conserved score (AUC = 0.95) substantially outperformed GenoCanyon (AUC = 0.78) in the ClinVar non-coding testing set. In addition, the performance of the MACIE-regulatory score (AUC = 0.76) is comparable to that of GenoCanyon (AUC = 0.73) in predicting regulatory function, for example, in the dsQTL testing set.
The versatility of the MACIE approach does introduce additional decisions that investigators need to make. First, researchers need to decide which set of annotations to include and how to assign the annotations to different classes; both the quality of the annotations as well as the accuracy of their assignment can impact MACIE’s performance. In this paper, we assigned these annotation scores on the basis of biological knowledge. Second, the exponential family assumption in the model might also require a proper transformation of some individual annotation scores before model fitting. Third, users need to consider the trade-offs between a more complex model and computation time; these trade-offs include the number of classes, the number of functional scores in each class, and the number of variants used for training. Such issues will become more relevant when the MACIE framework is extended to integrate cell-type-specific, tissue-specific, species-specific, or phenotype-related annotations and to include more variants in the training set, for example from the Trans-Omics for Precision Medicine Program.17,18,37,46 Nevertheless, these choices again highlight the flexibility of the MACIE approach. Unlike other one-dimensional algorithms that rely on assumptions more likely to be satisfied when the number of annotations is small, the MACIE statistical model scales well with increasing annotation data. Thus, MACIE can be expected to provide more meaningful predictions as the availability of variants and annotation scores continues to expand and the quality of these data improves.
A final important consideration in practical analysis concerns the differences between supervised and unsupervised methods. The performance of unsupervised scores might lag behind that of supervised methods when training datasets with relevant, high-quality labels are available. We observed this behavior when comparing MACIE to FATHMM-XF for ClinVar non-coding variants. Future extensions of interest include development of tools capable of integrating both supervised and unsupervised methods to further improve prediction accuracy.37
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants R35-CA197449, U19-CA203654, U01-HG009088, U01HG012064 and R01-HL113338 to X. Lin and by the NIH grants R01-AG072272, R01-MH095797, and R01-MH106910 to I.I.L.
Declaration of interests
X. Lin is a consultant of AbbVie Pharmaceuticals and Verify Life Sciences.
Published: February 24, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.01.017.
Contributor Information
Iuliana Ionita-Laza, Email: ii2135@columbia.edu.
Xihong Lin, Email: xlin@hsph.harvard.edu.
Data and code availability
The code for training MACIE models and MACIE scores used in all benchmarking examples are available for download at https://github.com/xihaoli/MACIE. Precomputed MACIE scores for all possible variants in the human genome are available for download at https://zenodo.org/record/5755656; https://zenodo.org/record/5756449; https://zenodo.org/record/5756479; and https://zenodo.org/record/5756563 (DOIs: 10.5281/zenodo.5755656; 10.5281/zenodo.5756449; 10.5281/zenodo.5756479; and 10.5281/zenodo.5756563). All genomic coordinates are given in NCBI Build 37/UCSC hg19.
Web resources
1000 Genomes, http://www.1000genomes.org
BRCA1 SGE, https://sge.gs.washington.edu/BRCA1
ChromHMM, http://compbio.mit.edu/ChromHMM
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar
ENGAGE Consortium, http://diagram-consortium.org/2015_ENGAGE_1KG
FANTOM5 CAGE, https://fantom.gsc.riken.jp/5/data
FATHMM-XF, http://fathmm.biocompute.org.uk/fathmm-xf
fitCons, http://compgen.cshl.edu/fitCons
GENCODE, https://www.gencodegenes.org
LocusZoom, http://locuszoom.org
MACIE, https://github.com/xihaoli/MACIE; https://zenodo.org/record/5755656; https://zenodo.org/record/5756449; https://zenodo.org/record/5756479; https://zenodo.org/record/5756563
OMIM, http://www.omim.org
Supplemental information
References
- 1.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., et al. FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J., et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie G.R., Dunham I., Zeggini E., Flicek P. Functional annotation of noncoding sequence variants. Nat. Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shihab H.A., Rogers M.F., Gough J., Mort M., Cooper D.N., Day I.N., Gaunt T.R., Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34:511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Q., Hu Y., Sun J., Cheng Y., Cheung K.-H., Zhao H. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci. Rep. 2015;5:10576. doi: 10.1038/srep10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodea C.A., Mitchell A.A., Bloemendal A., Day-Williams A.G., Runz H., Sunyaev S.R. PINES: phenotype-informed tissue weighting improves prediction of pathogenic noncoding variants. Genome Biol. 2018;19:173. doi: 10.1186/s13059-018-1546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backenroth D., He Z., Kiryluk K., Boeva V., Pethukova L., Khurana E., Christiano A., Buxbaum J.D., Ionita-Laza I. FUN-LDA: A latent Dirichlet allocation model for predicting tissue-specific functional effects of noncoding variation: methods and applications. Am. J. Hum. Genet. 2018;102:920–942. doi: 10.1016/j.ajhg.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulko B., Hubisz M.J., Gronau I., Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y.-F., Gulko B., Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 2017;49:618–624. doi: 10.1038/ng.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H., Thomas P.D. Tools for predicting the functional impact of nonsynonymous genetic variation. Genetics. 2016;203:635–647. doi: 10.1534/genetics.116.190033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee P.H., Lee C., Li X., Wee B., Dwivedi T., Daly M. Principles and methods of in-silico prioritization of non-coding regulatory variants. Hum. Genet. 2018;137:15–30. doi: 10.1007/s00439-017-1861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B., Lu Q., Zhao H. An evaluation of noncoding genome annotation tools through enrichment analysis of 15 genome-wide association studies. Brief. Bioinform. 2019;20:995–1003. doi: 10.1093/bib/bbx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dempster A.P., Laird N.M., Rubin D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B. 1977;39:1–38. [Google Scholar]
- 25.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr407. e118–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst J., Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McVicker G., Gordon D., Davis C., Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kheradpour P., Ernst J., Melnikov A., Rogov P., Wang L., Zhang X., Alston J., Mikkelsen T.S., Kellis M. Systematic dissection of regulatory motifs in 2,000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Z., Liu L., Wang K., Ionita-Laza I. A semi-supervised approach for predicting cell-type specific functional consequences of non-coding variation using MPRAs. Nat. Commun. 2018;9:5199. doi: 10.1038/s41467-018-07349-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degner J.F., Pai A.A., Pique-Regi R., Veyrieras J.-B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., Crawford G.E., et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M.J., Pan Z., Liu Z., Wu J., Wang P., Zhu Y., Xu F., Xia Z., Sham P.C., Kocher J.-P.A., et al. Predicting regulatory variants with composite statistic. Bioinformatics. 2016;32:2729–2736. doi: 10.1093/bioinformatics/btw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surakka I., Horikoshi M., Mägi R., Sarin A.-P., Mahajan A., Lagou V., Marullo L., Ferreira T., Miraglio B., Timonen S., et al. ENGAGE Consortium The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feitosa M.F., Wojczynski M.K., Straka R., Kammerer C.M., Lee J.H., Kraja A.T., Christensen K., Newman A.B., Province M.A., Borecki I.B. Genetic analysis of long-lived families reveals novel variants influencing high density-lipoprotein cholesterol. Front. Genet. 2014;5:159. doi: 10.3389/fgene.2014.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rands C.M., Meader S., Ponting C.P., Lunter G. 8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014;10:e1004525. doi: 10.1371/journal.pgen.1004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., et al. ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun R., Xu M., Li X., Gaynor S., Zhou H., Li Z., Bossé Y., Lam S., Tsao M.-S., Tardon A., et al. Integration of multiomic annotation data to prioritize and characterize inflammation and immune-related risk variants in squamous cell lung cancer. Genet. Epidemiol. 2021;45:99–114. doi: 10.1002/gepi.22358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Li Z., Zhou H., Gaynor S.M., Liu Y., Chen H., Sun R., Dey R., Arnett D.K., Aslibekyan S., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. TOPMed Lipids Working Group Dynamic incorporation of multiple in silico functional annotations empowers rare variant association analysis of large whole-genome sequencing studies at scale. Nat. Genet. 2020;52:969–983. doi: 10.1038/s41588-020-0676-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for training MACIE models and MACIE scores used in all benchmarking examples are available for download at https://github.com/xihaoli/MACIE. Precomputed MACIE scores for all possible variants in the human genome are available for download at https://zenodo.org/record/5755656; https://zenodo.org/record/5756449; https://zenodo.org/record/5756479; and https://zenodo.org/record/5756563 (DOIs: 10.5281/zenodo.5755656; 10.5281/zenodo.5756449; 10.5281/zenodo.5756479; and 10.5281/zenodo.5756563). All genomic coordinates are given in NCBI Build 37/UCSC hg19.