Abstract
Louse flies (Hippoboscidae) are permanent ectoparasites of birds and mammals. They have a cosmopolitan distribution with more than 200 described species. The aim of this study was to reveal host–vector–parasite associations between louse flies, birds, and trypanosomes. A total of 567 louse fly specimens belonging to 7 species were collected from birds at several localities in Czechia, including the rare species Ornithophila metallica and Ornithoica turdi. There was a significant difference in the occurrence of Ornithomya avicularia and Ornithomya fringillina on bird hosts according to their migratory status, O. fringillina being found more frequently on long-distance migrants. Trypanosomes were found in four species, namely, Ornithomya avicularia, O. fringillina, O. biloba, and Ornithoica turdi; the later three species are identified in this paper as natural trypanosome vectors for the first time. The prevalence of trypanosomes ranged between 5 and 19%, the highest being in O. biloba and the lowest being in O. fringillina. Phylogenetic analysis of the SSU rRNA gene revealed that a vast majority of trypanosomes from hippoboscids belong to the avian T. corvi/culicavium group B. Four new lineages were revealed in group B, with louse flies being probable vectors for some of these trypanosome lineages. We also confirmed the transcontinental distribution of several trypanosome lineages. Our results show that hippoboscids of several genera are probable vectors of avian trypanosomes.
Keywords: avian parasite, Trypanosoma, transmission, Hippoboscidae, Ornithomya, Ornithoica, Ornithophila, host specificity
1. Introduction
Louse flies are ectoparasitic insects found on mammals and birds, with both sexes feeding strictly on blood. They belong to the superfamily Hippoboscoidea, along with the medically important tsetse flies (Glossinidae), vectors of African trypanosomes, and bat flies (Nycteribiidae and Streblidae) [1]. The bodies of louse flies show adaptations to the parasitic lifestyle, including dorsoventrally flattened thorax, head and abdomen, claws for better attachment to the host, and modified mouth parts [2]. Louse flies (Hippoboscidae) have a worldwide distribution and contain 213 species [3]; the majority are parasites of birds, while one quarter of them occur on mammals [4]. Eleven avian louse fly species have been found in Czechia, namely: Ornithoica turdi, Ornithomya avicularia, O. chloropus, O. fringillina, O. biloba, Ornithophila metallica, Olfersia fumipennis, Icosta ardeae, Pseudolynchia canariensis, Crataerina pallida, and Stenepteryx hirundinis [5]. Louse fly species differ in their geographic distribution: O. fringillina and O. chloropus are typical for Northern Europe with fewer findings from Central and Southern Europe, whereas O. turdi and O. metallica have been found, rather, in the southern reaches of the northern hemisphere and are typical for Africa [2].
The host specificity of avian louse flies differs among species, ranging from high in O. biloba and S. hirundinis, which are essentially limited to a single host species, to low in Ornithomya avicularia and O. fringillina, which have been found on multiple host genera [6,7,8]. Individual louse flies can even switch their host species when birds are in close contact [9].
Because of their blood-feeding behavior, louse flies are vectors of many pathogens (reviewed in [10]), including trypanosomes. The louse fly O. avicularia was identified as a vector of avian trypanosomes after being fed on a trypanosome-infected rook (Corvus frugilegus), and subsequently transmitting trypanosomes to a canary (Serinus canaria) [11]. Although originally designated as Trypanosoma avium, the species used in these experiments is now considered to be T. corvi [12]. Since then, studies of trypanosomes in avian louse flies have remained scarce [13,14,15]. Trypanosome prevalence in the louse fly Ornithomya avicularia reached 5%, with 90% of the isolates belonging to T. corvi [15].
Avian trypanosomes are polyphyletic, splitting into three groups, each consisting of multiple lineages [16]. T. corvi is closely related to T. culicavium [17], and they both belong to group B, as defined by Zídková et al. [16]. However, trypanosome lineages belonging to the T. avium group C have been isolated from louse flies, as well [16,18]. Apart from Hippoboscidae, avian trypanosomes are transmitted by Nematocera, namely, mosquitoes [14,17,19], black flies [20,21], biting midges [22,23,24], and phlebotomine sandflies [25,26]. A particular trypanosome lineage can be transmitted by multiple vectors [16,26].
In this study, we aimed to (i) describe the occurrence of different avian louse fly species in relationship to the host species, (ii) assess the influence of host migratory status on the species of flies they host, (iii) molecularly identify obtained trypanosomes, using the SSU rRNA gene sequences, and (iv) uncover the associations between different species of hippoboscid flies and the trypanosomes they host and transmit. The louse fly vectorial capacity towards different trypanosome groups is discussed. We present complex data concerning this neglected group of kinetoplastid vectors.
2. Materials and Methods
2.1. Louse Fly Collection, Dissection, and Identification
Louse flies were collected between 2014 and 2019 from adult birds caught in mist nets and from swallow (Hirundo rustica) and swift (Apus apus) nestlings by members of the team and other registered ringers in several Czech localities, mainly Zeměchy (50.231783, 14.272371) and Choteč (49.999069, 14.280239) in Central Bohemia and Milovický forest (48.821274, 16.693175) in South Moravia. Collected louse flies were placed in zip-lock bags and stored in a cooling box before dissection. The flies that did not survive until dissection were stored in 96% ethanol at −20 °C.
Live louse flies were killed in 96% ethanol, washed twice in 0.9% sterile saline, and dissected on a glass slide under a stereomicroscope. Wings of the louse flies were separated under a stereomicroscope and mounted on glass slides in CMCP-9 mountant (Polysciences, Warrington, PA, USA). Identification was performed using two keys [2,27].
2.2. Trypanosome Cultivation, Microscopy, and Statistics
Trypanosome-positive guts were inoculated on rabbit blood agar, and trypanosomes were cultured as described previously [26]. A part of each positive guts sample was stored in ethanol as a backup for barcoding in case of unsuccessful culture. Thriving trypanosome cultures were frozen in 7% dimethylsulfoxide (final concentration) and stored in liquid nitrogen.
For scanning electron microscopy, a trypanosome-positive gut was treated as described in [26].
Bird migratory status was assigned as described in [28], and the data were processed using R software [29].
2.3. DNA Isolation, Amplification, and Sequencing
Ethanol from samples was evaporated in a thermoshaker at 39 °C. Whole louse fly body samples were crushed in Eppendorf tubes using sterile micropestles. DNA from individual samples was then extracted using the High Pure Template Preparation Kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions.
For trypanosome detection, the SSU rRNA gene was amplified using nested PCR with primers S-762 (5′-GACTTTTGCTTCCTCTAWTG-3′) and S-763 (5′-CATATGCTTGTTTCAAGGAC-3′) [30] in the first step. The first amplification round consisted of 35 cycles and was performed in the final volume of 11 µL of PCR mix (EmeraldAmp GT PCR Master Mix (TaKaRa, Shiga, Japan). The annealing temperature was 55 °C. For the second run, consisting of 35 amplification cycles, 1 µL of the product of the first amplification round was used as a template in 24 µL of PCR mix with TR-F2 (5′-GARTCTGCGCATGGCTCATTACATCAGA-3′) and TR-R2 (5′-CRCAGTTTGATGAGCTGCGCCT-3′) primers [31]. The annealing temperature was 64 °C.
Positive PCR products were purified by ExoSAP-IT™ (Thermo Scientific, Waltham, MA, USA) and sequenced in the Core Facility of the Faculty of Science, Charles University, using the primer 1000R (5′-ATGCCTTCGCTGTAGTTCGTCT-3′) [32], resulting in an approximately 600bp-long sequences. Further sequencing of chosen strains (K51, PAS441, and PAS433) was performed with primers 1000F (5′-AGACGAACTACAGCGAAGGCAT-3′) [32] and kin577F (GCCAGCACCCGCGGT) [16]; the assembly for each of the 3 sequences was carried out in Geneious 9.1.7 and was approximately 1500bp long. Low-quality ends were trimmed from all sequences in BioEdit 7.0.4.1. [33].
2.4. Sequence Analysis
In total, 137 SSU rRNA gene sequences of trypanosomes were used in the phylogenetic analysis, of which 85 were newly determined: 57 were sequences from louse flies, and 18 represented novel genotypes of avian trypanosomes belonging to group B and obtained from avian blood in a parallel study as described in [26].
The sequences were aligned using the MAFFT method [34] on MAFFT 7 online server (https://mafft.cbrc.jp/alignment/server/, accessed on 10 June 2021) with the G-INS-i algorithm and default settings. The final alignment consisted of 1947 characters. The phylogenetic tree was constructed by the maximum likelihood method in RAxML 8.0.0. [35], under GTRGAMMAI model. Statistical support of the topology was assessed by bootstrapping with 1000 pseudoreplicates in RAxML. The tree was then graphically edited in CorelDRAW X8 and Inkscape 2.1.
3. Results
3.1. Collected Louse Flies
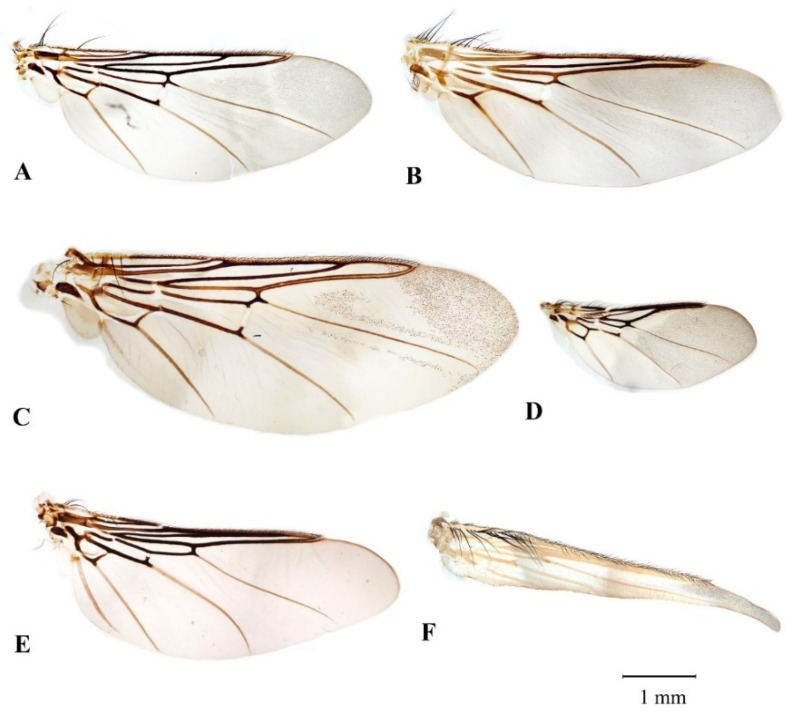
In total, 567 louse flies belonging to 7 species were caught: Ornithomya biloba (306), O. avicularia (133), O. fringillina (78), Ornithoica turdi (14), Stenepteryx hirundinis (2), Ornithophila metallica (1), and Crataerina pallida (33) (Figure 1, Table 1).
Figure 1.
Wings of hippoboscid species caught during the study (photos by AS). Wing shape, length, and hair distribution are used for species determination: (A)—Ornithomya fringillina, (B)—O. biloba, (C)—O. avicularia, (D)—Ornithoica turdi, (E)—Ornithophila metallica, (F)—Stenepteryx hirundinis.
Table 1.
Avian hosts of hippoboscids according to the migratory status of the hosts (M): L—long-distance migrants, S—short-distance migrants, R—residents; Louse fly species: OA—Ornithomya avicularia, OF—Ornithomya fringillina, OB—Ornithomya biloba, OT—Ornithoica turdi, OM—Ornithophila metallica, SH—Stenepteryx hirundinis, CP—Crataerina pallida.
| Avian Host | M | Louse Fly Species | ||||||
|---|---|---|---|---|---|---|---|---|
| OA | OF | OB | OT | OM | SH | CP | ||
| Acrocephalus arundinaceus | L | 1 | ||||||
| Acrocephalus palustris | L | 2 | 8 | |||||
| Acrocephalus schoenobaenus | L | 1 | ||||||
| Acrocephalus scirpaceus | L | 4 | 21 | |||||
| Anthus trivialis | L | 1 | ||||||
| Apus apus | L | 33 | ||||||
| Carduelis cannabina | S | 2 | ||||||
| Coccothraustes coccothraustes | S | 5 | 4 | 1 | ||||
| Cyanistes caeruleus | R | 1 | 3 | 1 | ||||
| Delichon urbicum | L | 1 | ||||||
| Dendrocopos major | R | 3 | ||||||
| Dendrocopos medius | R | 6 | ||||||
| Emberiza calandra | S | 1 | 2 | |||||
| Emberiza citrinella | R | 18 | 6 | 4 | ||||
| Emberiza schoeniclus | S | 1 | 1 | |||||
| Erithacus rubecula | S | 1 | ||||||
| Ficedula albicollis | L | 4 | ||||||
| Fringilla coelebs | S | 5 | ||||||
| Hirundo rustica | L | 1 | 2 | 293 | 1 | |||
| Jynx torquilla | L | 1 | 1 | |||||
| Lanius collurio | L | 2 | ||||||
| Locustella luscinioides | L | 2 | ||||||
| Luscinia svecica | L | 1 | ||||||
| Motacilla alba | S | 2 | ||||||
| Panurus biarmicus | R | 1 | ||||||
| Parus major | S | 11 | 3 | |||||
| Passer montanus | R | 2 | 1 | 3 | ||||
| Phylloscopus collybita | S | 1 | 3 | |||||
| Picus viridis | R | 2 | ||||||
| Prunella modularis | S | 7 | 3 | |||||
| Riparia riparia | L | 3 | 9 | |||||
| Sitta europaea | R | 3 | 1 | |||||
| Sturnus vulgaris | S | 2 | ||||||
| Sylvia atricapilla | S | 10 | ||||||
| Sylvia borin | L | 1 | 3 | |||||
| Sylvia communis | L | 2 | 3 | |||||
| Sylvia curruca | L | 3 | ||||||
| Turdus merula | R | 15 | ||||||
| Turdus philomelos | S | 23 | 5 | |||||
| Total | 133 | 78 | 306 | 14 | 1 | 2 | 33 | |
3.2. Host Specificity of Hippoboscids
In total, 39 avian species belonging to 28 genera were found to host louse flies; of those, 17 species were long-distance migrants, 13 were short-distance migrants, and 9 were residents. Ornithomya avicularia was found on 32 species from 23 genera, O. fringillina on 19 species from 12 genera, O. biloba on 4 species and genera, and Ornithoica turdi was found on 4 species and genera (namely, Coccothraustes coccothraustes, Emberiza citrinella, Sitta europaea, Turdus philomelos). A single specimen of Ornithophila metallica was found on C. coccothraustes. Stenepteryx hirundinis was found on two species from two genera of Hirundinidae, and Crataerina pallida was found on its specific host species, Apus apus (Table 1).
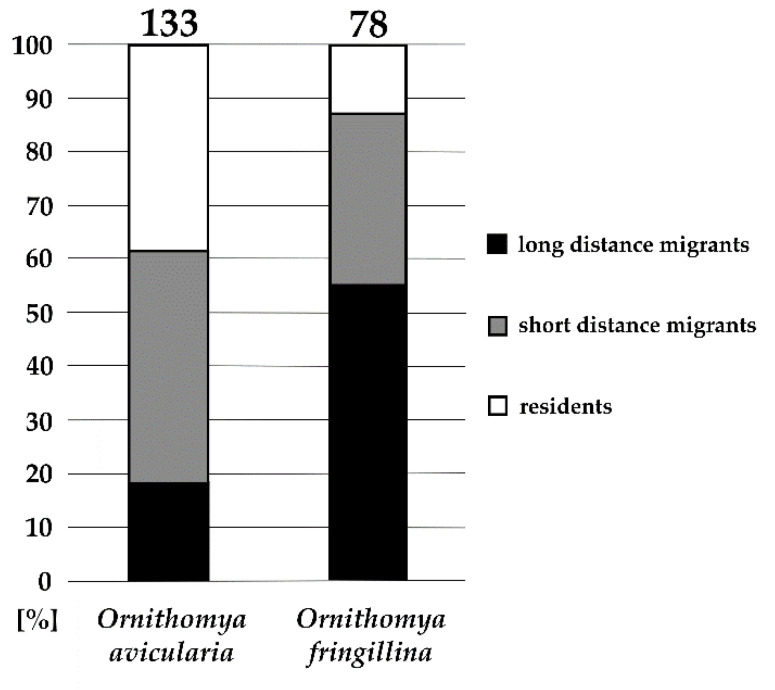
We evaluated the differences in host preferences of the two opportunistic louse fly species, O. avicularia and O. fringillina, according to the migratory status of their bird hosts. The difference between these preferences is significant (Pearson’s Chi-squared test, X = 34.042, df = 2, p < 0.001). Although in different ratios, both species occurred on residents and short- and long-distance migrants (Figure 2).
Figure 2.
The occurrence of Ornithomya spp. on bird hosts in relation to host migratory status; numbers of collected specimens are given above columns.
3.3. Prevalence of Trypanosomes in Louse Flies
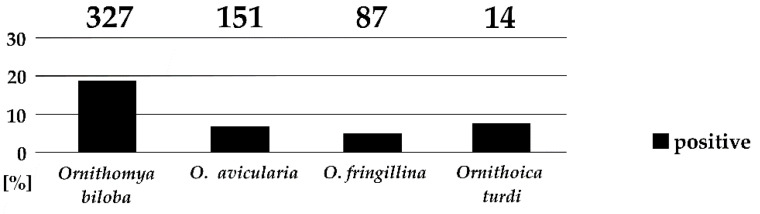
Four of the seven collected louse fly species harbored trypanosomes in their gut, namely, Ornithomya biloba, O. avicularia, O. fringillina, and Ornithoica turdi. Trypanosome infections were mature, localized in the hindguts, and putative metacyclic forms were observed in the vast majority of cases. The prevalence of trypanosomes differed significantly, with the highest prevalence in O. biloba (18.7%), followed by O. turdi (7.1%), O. avicularia (6.6%), and O. fringillina (4.6%; Figure 3). The prevalence of trypanosomes significantly differs among the four louse fly species (Fisher’s Exact Test, p < 0.001). No trypanosomes were found in 33 specimens of Crataerina pallida, which is specific for swifts.
Figure 3.
Prevalence of trypanosomes in Ornithomya spp. and Ornithoica turdi. Numbers of examined specimens are given above the columns.
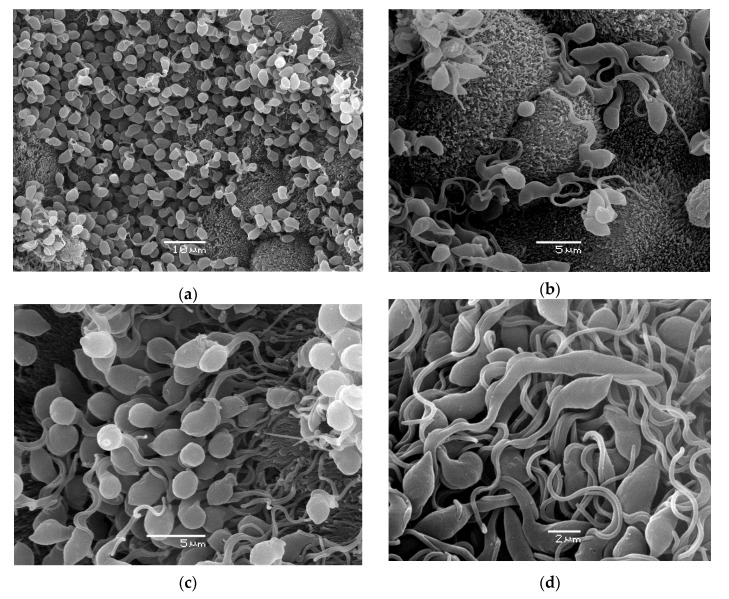
In the vast majority of cases, trypanosomes massively colonized hippoboscid guts (Figure 4).
Figure 4.
Trypanosomes in O. avicularia gut, natural infection. Cultivation of these resulted in a new strain OA23 identified as belonging to the new lineage B13. The female louse fly was caught on a juvenile thrush (T. philomelos) in Central Bohemia. Epimastigotes were attached to the gut epithelium (a–c) or were free, probably metacyclic forms (d).
3.4. Phylogenetic Analysis
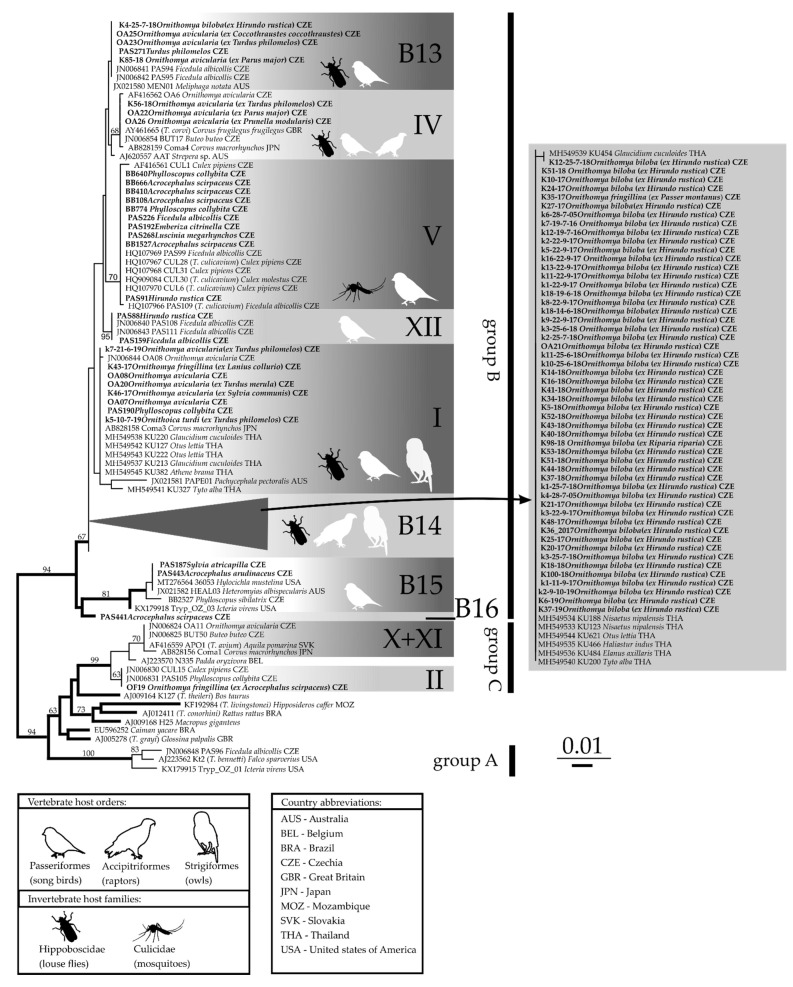
Trypanosome lineages obtained from louse flies belonged to groups B (T. corvi/culicavium) and C (T. avium/thomasbancrofti). The majority (69) of these trypanosome sequences belonged to group B, whereas only two belonged to group C (Figure 5). Some obtained Trypanosoma sequences from louse flies belonged to lineages described previously, while 61 of them belonged to new lineages. Newly determined unique sequences are available in GenBank under accession numbers OM509725-OM509732.
Figure 5.
Maximum likelihood unrooted phylogenetic tree of avian trypanosome SSU rRNA gene sequences. The three groups of avian trypanosomes (A—T. bennetti group; B—T. corvi/culicavium group, and C—T. avium/thomasbancrofti group) along with previously described lineages (I–XII) are labelled after [16]; new lineages in the group B are labelled B13–B16. Original sequences are described in bold. The sequence description contains identifier, trypanosome host, and country of collection. Bootstrap support higher than 50 is shown at the branches.
The well-known species transmitted by louse flies is T. corvi (lineage IV). Three newly obtained Trypanosoma sequences from O. avicularia belonged to this lineage, together with previously sequenced trypanosomes from O. avicularia and birds (buzzard Buteo buteo, rook Corvus frugilegus, large-billed crow C. macrorhynchos, and currawong Strepera sp.) from Europe, Asia, and Australia.
The second described Trypanosoma species from the B group is T. culicavium (lineage V). In addition to the originally identified avian host is the collared flycatcher (Ficedula albicollis). We have found this lineage in several new hosts, namely, the Eurasian reed warbler (Acrocephalus scirpaceus), swallow (Hirundo rustica), chiffchaff (Phylloscopus collybita), yellowhammer (Emberiza citrinella), and nightingale (Luscinia megarhynchos).
Two new trypanosome sequences were isolated from birds clustered with lineage XII; this lineage consists of only trypanosome sequences from birds; vectors of these trypanosomes remain unknown.
Our phylogenetic analysis revealed four new lineages in the group B, which we designate B13 to B16 (Figure 5).
The lineage B13 consisted of isolates from hippoboscids (O. avicularia and O. biloba) and passerines. In addition to a song thrush (Turdus philomelos) isolate and two isolates from collared flycatcher (Ficedula albicollis), this lineage contained a sequence from an Australian passerine yellow spotted honeyeater (Meliphaga notata). The lineage B13 was previously identified and is as of yet unnamed [16]. Trypanosome sequences from louse flies clustering with this lineage indicate hippoboscids as newly identified vectors of the lineage B13.
The lineage B14 consisted of trypanosome isolates from hippoboscids (all except one from O. biloba, one from O. fringillina caught on a sparrow (Passer montanus)) and trypanosomes of raptors and owls from Thailand. This lineage differed from the lineage I, its closest relative, by four nucleotides in the sequenced region.
B15 and B16 were ancestral lineages at the base of group B and consisted of only trypanosome sequences originating from Passerines; interestingly, B15 contained our new sequences from European passerines blackcap (S. atricapilla), great reed warbler A. arundinaceus, and wood warbler P. sibilatrix, together with American and Australian birds: wood thrush (Hylocichla mustelina), yellow-breasted chat (Icteria virens), and ashy robin (Heteromyias albispecularis). B16 consisted of a single sequence from the reed warbler A. scirpaceus. Vectors of these two lineages remain unknown. The two new lineages B15 and B16 differed in 52 and 23 nucleotides, respectively, from lineage I.
A single new trypanosome sequence obtained from the hippoboscid Ornithomya fringillina (OF19 [18]) was placed by our phylogenetic analysis to the group C, lineage II (Trypanosoma thomasbancrofti).
4. Discussion
Although hippoboscids were already identified as vectors of avian trypanosomes by the middle of the 20th century [11], their vectorial role has scarcely been studied since. The finding of trypanosomes in several louse fly species in our study, as well as the transcontinental distribution of group B trypanosomes transmitted by the hippoboscids, indicate a high potential for louse flies as vectors of avian trypanosomes in other continents as well.
4.1. Findings of Avian Hippoboscids
While three common species belonging to the genus Ornithomya were present in all catching sites, Ornithophila metallica and Ornithoica turdi occurred only in the southernmost site of Milovický forest. O. turdi was reported from South Moravia previously but was considered non-breeding [2,36]. However, the relatively frequent occurrence, and occurrence on resident birds suggest this species completes its life cycle in Czechia. In fact, we recorded many more sightings of specimens that eluded capture due to their high agility and small size.
O. metallica has been reported in Czechia only twice in 1956 and 1973 in South Moravia [2,5], and this species is considered a southern element.
4.2. Host Specificity of Avian Louse Flies
The host specificity of avian louse flies differed among louse fly genera and species. Some species are considered strictly host specific; Crataerina pallida for the swift, Stenepteryx hirundinis for the common house martin Delichon urbicum, and swallow Hirundo rustica [2,6], as supported by our findings. Similarly, Ornithomya biloba is host specific toward Hirundinidae, with the vast majority of specimens found on swallows [2,6]. Nevertheless, we also document occasional findings of O. biloba on other avian hosts, namely, blue tit (Cyanistes caeruleus) and sparrow (Passer montanus). The louse fly species O. avicularia, O. fringillina, and Ornithoica turdi displayed very low host specificity and have been found on hosts belonging to several passerine families; this corresponds to previous studies [2,6].
4.3. Migration Status of Louse Fly Hosts
The geographical distribution of Ornithomya fringillina is holarctic, the species being found mainly in Northern Europe, with scarce findings in Central and Southern Europe, presumably on migrants [2]. We compared the host species composition of O. avicularia and O. fringillina according to the bird migratory status. O. fringillina was found most frequently on long-distance migrants, whereas O. avicularia was found mostly on residents and short-distance migrants. Although the difference was significant, it was not clear-cut; the question persists whether specimens of O. fringillina found on resident birds emerged locally or switched hosts from northern migrants [9]. Interestingly, a recent study in Finland did not find any difference between three Ornithomya species abundances in relation to host migratory status [7], which might support the hypothesis that O. fringillina found in Czechia originated from northern breeding grounds, given that emerging louse flies attach to the first available avian host specimen, which then brings it to the south during its fall migration.
4.4. Trypanosomes in Avian Hippoboscids
To date, the only hippoboscid species found to be naturally infected with trypanosomes was Ornithomya avicularia. This species was used for the experimental transmission of trypanosomes [11,13], and trypanosomes were readily found in specimens captured on raptor nestlings [15]. Trypanosomes were found in O. fringillina experimentally fed on infected birds and were, in one case, transmitted to the same host species by intraperitoneal injection [20]. In this study, we have found O. avicularia, O. biloba, O. fringillina, and Ornithoica turdi naturally infected with avian trypanosomes. Trypanosome infections were mature, localized in the hindguts, and putative metacyclic forms were observed. Therefore, we consider these hippoboscid species as competent vectors. The prevalence of trypanosomes in different louse fly species varied form 18.7% in O. biloba to 4.6% in O. fringillina. The prevalence of 6.6% in O. avicularia is only slightly higher than in previous screening of this hippoboscid species caught on raptors [15].
Although the permanent presence of host blood in the louse fly intestine enables trypanosomes to thrive regardless of the vector capacity of the louse fly, the massive infections of hippoboscid guts indicate a true ability of these insects to transmit avian trypanosomes. Apart from being true vectors, hippoboscids can serve as unspecific hosts for group C trypanosomes transmitted by mosquitoes [18] or blackflies [16], but their potential to transmit them remains to be confirmed.
4.5. Phylogeny of Avian Trypanosomes and Host–Parasite Associations
Our phylogenetic analysis was unable to resolve the relationships between all particular lineages within the group C, which is in agreement with previous studies [16].
The lineage B14 is frequently found in hippoboscids, namely, in Ornithomya biloba from swallows; the only exception out of 53 isolates from louse flies was a single specimen from O. fringillina caught on a sparrow. Interestingly, the trypanosome clade found in O. biloba, which exhibited the highest prevalence in louse flies, could not be detected in its avian hosts (59 barn swallows sampled, see [18]). In Thailand, trypanosome sequences clustering to the lineage B14 were obtained from blood samples from owls and raptors [36]. Given the strict host specificity of O. biloba and its documented geographic distribution, it is likely that the B14 trypanosome lineage is transmitted to raptors and owls by other hippoboscid species in Thailand.
The lineages in the group B of avian trypanosomes indicate specificity toward the louse fly host species O. biloba (lineage B14) and O. avicularia (lineage B13, IV and I). The trypanosome sequences isolated from O. fringillina were scattered among different lineages (B14, I and lineage II from the group C).
A trypanosome from the only positive Ornithoica turdi clusters within the lineage I, which originally contained only two trypanosome sequences from O. avicularia [16] but now, in addition to louse fly sequences, also contains lineages from passerines and owls [37], suggesting low host specificity toward both avian and hippoboscid hosts.
5. Summary
Avian trypanosomes are transmitted by louse flies belonging to several species and genera. Ornithomya fringillina is probably brought from the north on migrating birds, while Ornithoica turdi breeds in Central Europe. Avian trypanosomes transmitted by louse flies have a transcontinental distribution.
Acknowledgments
We thank František Novák, Oldřich Myška, Karel Pithart, and others for their help with louse fly collection. We thank Miroslav Hyliš for help with processing a sample for electron microscopy, Jan Holeček for the graphical assembly of louse fly wing pictures, and Magdalena Fialová for help with PCR. We also thank William A. Bourland for carefully reading and editing the manuscript.
Author Contributions
Conceptualization, M.S.; formal analysis, I.Č., A.S., and M.S.; investigation, A.S., J.B., and M.S.; writing—original draft preparation, A.S.; writing—review and editing, M.S., J.B., I.Č., and A.S.; supervision, M.S.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received Charles University institutional funding (PROGRES Q43, Cooperatio Biology, UNCE/SCI/012-204072/2018, SVV 260432/2018).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences obtained in this study have been deposited in GenBank under the accession numbers OM509725-OM509732.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Petersen F.T., Meier R., Kutty S.N., Wiegmann B.M. The phylogeny and evolution of host choice in the Hippoboscoidea (Diptera) as reconstructed using four molecular markers. Mol. Phylogenetics Evol. 2007;45:111–122. doi: 10.1016/j.ympev.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 2.Chalupský J. Čeleď Hippoboscidae–Klošovití. In: Chvála M.K., editor. Fauna ČSR, Svazek 22, Krevsající Mouchy a Střečci. Nakladatelství Československé Akademie Věd; Prague, Czech Republic: 1980. pp. 475–478. [Google Scholar]
- 3.Dick C.W. Checklist of world Hippoboscidae (Diptera: Hippoboscoidea) Department of Zoology, Field Museum of Natural History; Chicago, IL, USA: 2006. pp. 1–7. [Google Scholar]
- 4.Maa T. A revised checklist and concise host index of Hippoboscidae (Diptera) Pacific Insects Monogr. 1969;20:261–299. [Google Scholar]
- 5.Sychra O. Hippoboscidae Samouelle, 1819. In: Jedlička L., Kúdela M., Stloukalová V., editors. Checklist of Diptera of the Czech Republic and Slovakia. Electronic Version 2. 2009. [(accessed on 14 May 2021)]. Available online: http://zoology.fns.uniba.sk/diptera2009>+CD-ROM. [Google Scholar]
- 6.Oboňa J., Sychra O., Greš S., Heřman P., Manko P., Roháček J., Šestáková A., Šlapák J., Hromada M. A revised annotated checklist of louse flies (Diptera, Hippoboscidae) from Slovakia. ZooKeys. 2019;862:129–152. doi: 10.3897/zookeys.862.25992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehikoinen A., Pohjola P., Valkama J., Mutanen M., Jaakko L. Promiscuous specialists: Host specificity patterns among generalist louse flies. PLoS ONE. 2021;16:e0247698. doi: 10.1371/journal.pone.0247698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomás A., da Fonseca I.P., Valkenburg T., Rebelo M.T. Louse flies in Azorean and mainland populations of four Passeriformes species: A new perspective to parasite island syndromes. Int. J. Parasitol. Parasites Wildl. 2020;14:33–40. doi: 10.1016/j.ijppaw.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbet G.B. The life-history and host-relations of a hippoboscid fly Ornithomyia fringillina Curtis. J. Anim. Ecol. 1956;25:403–420. doi: 10.2307/1934. [DOI] [Google Scholar]
- 10.Bezerra-Santos M.A., Otranto D. Keds, the enigmatic flies and their role as vectors of pathogens. Acta Trop. 2020;209:105521. doi: 10.1016/j.actatropica.2020.105521. [DOI] [PubMed] [Google Scholar]
- 11.Baker J.R. Studies on Trypanosoma avium Danilewsky 1885.2. Transmission by Ornithomyia avicularia. Parasitology. 1956;46:321–334. doi: 10.1017/s0031182000026536. [DOI] [PubMed] [Google Scholar]
- 12.Baker J.R. Biology of the trypanosomes of birds. In: Lumsden W.H.R., Evans D.A., editors. Biology of the Kinetoplastida. Academic Press; London, UK: New York, NY, USA: San Francisco, CA, USA: 1976. pp. 131–174. [Google Scholar]
- 13.Mungomba L.M., Molyneux D.H., Wallbanks K.R. Host-parasite relationship of Trypanosoma corvi in Ornithomyia avicularia. Parasitol. Res. 1989;75:167–174. doi: 10.1007/BF00931269. [DOI] [PubMed] [Google Scholar]
- 14.Votýpka J., Oborník M., Volf P., Svobodová M., Lukeš J. Trypanosoma avium of raptors (Falconiformes): Phylogeny and identification of vectors. Parasitology. 2002;12:253–263. doi: 10.1017/S0031182002002093. [DOI] [PubMed] [Google Scholar]
- 15.Svobodová M., Volf P., Votýpka J. Trypanosomatids in ornithophilic bloodsucking Diptera. Med. Vet. Entomol. 2015;29:444–447. doi: 10.1111/mve.12130. [DOI] [PubMed] [Google Scholar]
- 16.Zídková L., Cepicka I., Szabová J., Svobodová M. Biodiversity of avian trypanosomes. Infect. Genet. Evol. 2012;12:102–112. doi: 10.1016/j.meegid.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Votýpka J., Szabová J., Rádrová J., Zídková L., Svobodová M. Trypanosoma culicavium sp. nov., an avian trypanosome transmitted by Culex mosquitoes. Int. J. Syst. Evol. Microbiol. 2012;62:745–754. doi: 10.1099/ijs.0.032110-0. [DOI] [PubMed] [Google Scholar]
- 18.Fialová M., Santolíková A., Brontáková A., Brzoňová J., Svobodová M. Complete life cycle of Trypanosoma thomasbancrofti, an avian trypanosome transmitted by culicine mosquitoes. Microorganisms. 2021;9:2101. doi: 10.3390/microorganisms9102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett G.F. Development of trypanosomes of the T. avium complex in the invertebrate host. Can. J. Zool. 1970;48:945–957. doi: 10.1139/z70-169. [DOI] [PubMed] [Google Scholar]
- 20.Bennett G.F. On the specificity and transmission of some avian trypanosomes. Can. J. Zool. 1961;39:17–33. doi: 10.1139/z61-003. [DOI] [Google Scholar]
- 21.Votýpka J., Svobodová M. Trypanosoma avium: Experimental transmission from black flies to canaries. Parasitol. Res. 2004;92:147–151. doi: 10.1007/s00436-003-1034-z. [DOI] [PubMed] [Google Scholar]
- 22.Miltgen F., Landau I. Culicoides nubeculosus, an experimental vector of a new trypanosome from psittaciform Trypanosoma bakeri n. sp. Ann. Parasitol. Hum. Comp. 1982;57:423–428. doi: 10.1051/parasite/1982575423. [DOI] [PubMed] [Google Scholar]
- 23.Chandenier J., Landau I., Baccam D. Experimental transmission of passeriform trypanosomes by Culicoides. Ann. Parasitol. Hum. Comp. 1991;66:9–13. doi: 10.1051/parasite/19916619. [DOI] [Google Scholar]
- 24.Svobodová M., Dolnik O.V., Čepička I., Rádrová J. Biting midges (Ceratopogonidae) as vectors of avian trypanosomes. Parasites Vectors. 2017;10:224. doi: 10.1186/s13071-017-2158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato H., Gomez E.A., Cáceres A.G., Vargas F., Mimori T., Yamamoto K., Iwata H., Korenaga M., Velez L., Hashiguchi Y. Natural Infections of Man-Biting Sand Flies by Leishmania and Trypanosoma Species in the Northern Peruvian Andes. Vector-Borne Zoonotic Dis. 2011;11:515–521. doi: 10.1089/vbz.2010.0138. [DOI] [PubMed] [Google Scholar]
- 26.Svobodová M., Rádrová J. Phlebotomine sandflies—potential vectors of avian trypanosomes. Acta Protozool. 2018;57:53–59. doi: 10.4467/16890027AP.18.005.8399. [DOI] [Google Scholar]
- 27.Hutson A.M. Keds, flat-flies and bat-flies. Diptera, Hippoboscidae and Nycteribiidae. Handbk Ident. Br. Insects. 1984;10:1–40. [Google Scholar]
- 28.Reif J., Vermouzek Z., Voříšek P., Šťastný K., Bejček V., Flousek J. Population changes in Czech passerines are predicted by their life-history and ecological traits. IBIS Int. J. Avian Sci. 2010;152:610–621. doi: 10.1111/j.1474-919X.2010.01036.x. [DOI] [Google Scholar]
- 29.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. [(accessed on 29 August 2021)]. Available online: https://www.R-project.org/ [Google Scholar]
- 30.Maslov D.A., Lukeš J., Jirků M., Simpson L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: Implications for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 1996;75:197–205. doi: 10.1016/0166-6851(95)02526-X. [DOI] [PubMed] [Google Scholar]
- 31.Votýpka J., Rádrová J., Skalický T., Jirků M., Jirsová D., Mihalca A.D., D’Amico G., Petrželková K.J., Modrý D., Lukeš J. A tsetse and tabanid fly survey of African great apes habitats reveals the presence of a novel trypanosome lineage but the absence of Trypanosoma brucei. Int. J. Parasitol. 2015;45:741–748. doi: 10.1016/j.ijpara.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Brotánková A., Čepička I., Brzoňová J., Svobodová M. Trypanosomes of the Trypanosoma theileri group: Phylogeny and new potential vectors. Microorganisms. 2022;10:294. doi: 10.3390/microorganisms10020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Simp. Ser. 1999;41:95–98. doi: 10.14601/PHYTOPATHOL_MEDITERR-14998U1.29. [DOI] [Google Scholar]
- 34.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Šochová E., Husník F., Nováková E., Halajian A., Hypša V. Arsenophonus and Sodalis replacements shape evolution of symbiosis in louse flies. PeerJ. 2017;5:e4099. doi: 10.7717/peerj.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pornpanom P., Salakij C., Prasopsom P., Lertwatcharasarakul P., Kasorndorkbua C., Santavakul M. Morphological and molecular characterization of avian trypanosomes in raptors from Thailand. Parasitol. Res. 2019;118:2419–2429. doi: 10.1007/s00436-019-06379-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences obtained in this study have been deposited in GenBank under the accession numbers OM509725-OM509732.