Abstract
Despite the widespread use of antiseptics such as chlorhexidine digluconate (CHX) in dental practice and oral care, the risks of potential resistance toward these antimicrobial compounds in oral bacteria have only been highlighted very recently. Since the molecular mechanisms behind antiseptic resistance or adaptation are not entirely clear and the bacterial stress response has not been investigated systematically so far, the aim of the present study was to investigate the transcriptomic stress response in Streptococcus mutans after treatment with CHX using RNA sequencing (RNA-seq). Planktonic cultures of stationary-phase S. mutans were treated with a sublethal dose of CHX (125 µg/mL) for 5 min. After treatment, RNA was extracted, and RNA-seq was performed on an Illumina NextSeq 500. Differentially expressed genes were analyzed and validated by qRT-PCR. Analysis of differential gene expression following pathway analysis revealed a considerable number of genes and pathways significantly up- or downregulated in S. mutans after sublethal treatment with CHX. In summary, the expression of 404 genes was upregulated, and that of 271 genes was downregulated after sublethal CHX treatment. Analysis of differentially expressed genes and significantly regulated pathways showed regulation of genes involved in purine nucleotide synthesis, biofilm formation, transport systems and stress responses. In conclusion, the results show a transcriptomic stress response in S. mutans upon exposure to CHX and offer insight into potential mechanisms that may result in development of resistances.
Keywords: antibacterial, CHX, RNA-seq, Streptococcus mutans, stress response, transcriptomics
1. Introduction
Despite the current COVID-19 pandemic, antimicrobial resistance (AMR) remains one of the greatest challenges for public health in the 21st century [1,2]. For instance, a 2018 report of the Organization for Economic Co-operation and Development (OECD) predicted that 2.4 million people in Europe, North America and Australia will die from infections associated with AMR within the next 30 years, resulting in costs of up to USD 3.5 billion per year for the healthcare services of the 33 countries included in the analysis [3]. Despite many reports about antibiotic resistance, antiseptics or biocides have only recently come into spotlight in the context of AMR [4,5,6,7].
Chlorhexidine is a twofold positively charged bis-biguanide and is mostly used as its digluconate salt (chlorhexidine digluconate, CHX) in clinical practice [4,8]. Since its introduction into dentistry in the late 1960s [9], CHX has come to be considered the gold-standard antiseptic in oral care [4,10]. It is used for plaque control and management of gingivitis [11]; in patients with high caries risk, such as those with fixed orthodontic appliances [12]; or in patients after periodontal or implant surgery [13]. Furthermore, CHX is included in a wide range of over-the-counter oral care products, such as toothpastes or mouthwashes [11,14,15].
Although there is ample evidence that rinsing with CHX can decrease the salivary bacterial load [16] and consequently reduce oral biofilm formation [17,18], the antibacterial efficacy of CHX toward mature oral biofilms may be limited [4,19,20]. For instance, we recently showed that microcosm biofilms from human saliva cultured for 72 h and treated with 0.1% or 0.2% CHX for 1 min resulted in colony-forming-unit reduction rates of less than 1 log10 step [20]. Accordingly, treatment with 0.2% CHX for 1 min only affected the outer layers of biofilms formed in situ for 48 h [19]. This limited efficacy of CHX in mature oral biofilms may be mainly due to the biofilm matrix, which acts as a diffusion barrier for positively charged molecules, such as CHX [21]. Consequently, it is reasonable that bacteria in deeper layers will be exposed to sublethal concentrations of CHX upon application of a CHX-containing mouthwash [4,20]. Several studies from recent years showed that oral, as well as non-oral, bacteria can phenotypically adapt upon multiple exposure to sublethal concentrations in vitro [20,22,23,24,25] and potentially also develop cross resistances toward antibiotics [23,25]. However, the actual molecular mechanisms causing these adaptations, as well as the stress response in bacteria upon exposure toward sublethal concentrations of CHX, are not well understood yet [15].
Therefore, the aim of the present study was to investigate the transcriptomic stress response after sublethal treatment with CHX using an RNA sequencing approach with Streptococcus mutans as model organism due to its key role in biofilm formation and pathogenesis of dental caries [26,27].
2. Materials and Methods
2.1. Chlorhexidine Digluconate
Chlorhexidine digluconate (CHX) was obtained from Sigma-Aldrich (Merck, Darmstadt, Germany) in a concentration of 20% and was dissolved in distilled water to yield the final concentration of 125 µg/mL (0.0125%).
2.2. Bacterial Culture and Treatment
A-type strain of S. mutans (DSM 20523; ATCC 25175) was obtained from the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ; Braunschweig, Germany) to be used in this study. S. mutans was grown in brain heart infusion broth (BHI; Sigma-Aldrich, St. Louis, MO, USA) and on BHI agar plates. Planktonic cultures (5 mL) were grown aerobically overnight at 37 °C. Afterwards, suspensions were harvested by centrifugation (ROTINA 420 R, Hettich Lab Technology, Tuttlingen, Germany) and resuspended in BHI, yielding an optical density (OD) of 2.0 per mL, as measured by means of a spectrophotometer at 600 nm (Ultrospec 3300 pro; Amersham Biosciences, Amersham, UK). Samples were centrifuged, supernatants were discarded and bacterial pellets were resuspended and incubated with 200 μL BHI broth or 200 µL CHX (final concentration: 125 µg/mL) for 5 min.
To evaluate the bacterial ability to replicate, samples were centrifuged, BHI broth or CHX solution was carefully removed, and the bacterial pellet was brought to suspension with 200 μL phosphate-buffered saline (PBS; Biochrom, Berlin, Germany) by frequent pipetting. Tenfold serial dilutions (10−2 to 10−7) were prepared in PBS, and aliquots (20 μL) were plated on BHI agar (both provided by the Institute of Medical Microbiology and Hygiene, University Hospital Regensburg, Germany) according to the method described by Miles et al. [28]. Samples were incubated aerobically for 72 h, and subsequently colony forming units (CFUs) were evaluated (n = 5).
For RNA-Seq, samples were centrifuged after treatment, and distilled water or CHX solution was carefully removed. Pellets were resuspended in 500 μL prewarmed BHI broth and incubated at 37 °C for 30 min. Afterwards, 1 mL of RNAprotect bacteria reagent (Qiagen, Hilden, Germany) was added to the samples and incubated for 5 min at room temperature. Then, samples were centrifuged at 17,035× g for 5 min, supernatants were discarded and pellets were stored at −80 °C until further use (n = 5).
2.3. RNA Extraction, Library Preparation and RNA Sequencing
RNA extraction and sequencing were performed by the NGS Competence Center Tübingen (NCCT), the sequencing partner of the Quantitative Biology Center (QBiC), Tübingen, Germany. RNA of all samples was isolated with the Quick-RNA fungal/bacterial miniprep kit (Zymo Research, Freiburg, Germany). RNA extraction was followed by DNase I treatment in solution using the DNase I recombinant kit (Roche, Basel, Switzerland), as well as purification and concentration of RNA using the RNA Clean & Concentrator-5 kit (Zymo Research, Freiburg, Germany).
Library preparation was performed using an Illumina Stranded Total RNA Prep Ligation with the Ribo-Zero Plus kit (Illumina, San Diego, CA, USA) with 100 ng total RNA. Sequencing was performed on an Illumina NextSeq 500 using a high-output kit (version 2.5; 75 cycles) with 1% PhiX spike-in (Illumina, San Diego, CA, USA).
2.4. RNA Sequencing Data Analysis
Data management and bioinformatic analysis were performed at the Quantitative Biology Center (QBiC), Tübingen, Germany. A Nextflow-based nf-core pipeline nf-core/rnaseq (version 1.4.2; https://github.com/nf-core/rnaseq, accessed on 22 January 2022) was used for the RNA-seq bioinformatic analysis. As part of this workflow, FastQC (version v0.11.8) was used to determine the quality of the FASTQ files [29]. Subsequently, adapter trimming was conducted with Trim Galore (version 0.6.4) [30]. STAR aligner (version 2.6.1, [31]) was used to map the reads that passed quality control against the latest assembly (RefSeq assembly accession GCF_000347875.1) of strain ATCC 25175, downloaded from NCBI. Note that the gtf file for the S. mutans genome had to be modified to be used for the nf-core/rnaseq workflow by removing empty fields for the tag “transcript_id” in the ninth column of that gtf file. RNA-seq data quality control was performed with RSeQC (version 3.0.1) [32] and read counting of the features (e.g., genes) with featureCounts (version 1.6.4) [33]. An aggregation of the quality control for the RNA-seq analysis was performed with MultiQC (version 1.7; http://multiqc.info/, accessed on 22 January 2022) [34].
The analysis of the differential gene expression was performed in R language (version 3.5.1) using DESeq2 (version 1.22) through the Nextflow-based workflow qbic-pipelines/rnadeseq (https://github.com/qbic-pipelines/rnadeseq, accessed on 22 January 2022, version 1.3.2). Genes were considered differentially expressed (DE) when the Benjamini–Hochberg multiple testing adjusted p-value [35] was smaller than 0.05 (padj ≤ 0.05). Multiple testing correction helps to reduce the number of false positives (not real DE genes). In the case of a threshold of 0.05, the proportion of false discoveries in the selected group of DE genes is controlled to be less than the threshold value—in this case, 5%. Genes were further filtered for biological relevance if the log2 fold change (FC) in expression between the two considered groups was above the threshold of 1.0 and less than −1.0. Final reports were produced using the R package rmarkdown (version 2.1) with the knitr (version 1.28) and DT (version 0.13) R packages. The sample similarity heatmaps were created using the edgeR (version 3.26.5) R package. BioCyc database was used for pathway analysis [36]. All DE genes were included, and enrichment was calculated using Fisher’s exact test (p ≤ 0.05).
2.5. Primer Design and qRT-PCR
To validate RNA-Seq data, randomly selected genes were measured by qRT-PCR. The nucleotide sequence of each gene (Table 1) was downloaded from BioCyc [36]. Primers were designed using the Primer3 tool (version 4.1.0, Whitehead Institute for Biomedical Research, Cambridge, MA, USA) and synthesized by Eurofins MWG Synthesis (Ebersberg, Germany). Primer efficiencies were checked by qRT-PCR using different cDNA dilutions. Primers gave a single PCR product, which was verified by gel electrophoresis and melt curves at the end of each run. Total RNA (200 ng) used for RNA-Seq was reverse-transcribed with the Quantitect reverse transcriptase kit (Qiagen, Hilden, Germany) in a reaction volume of 20 μL. Polymerase chain reaction was carried out with 20 pmol of each primer, 5 µL cDNA (0.2 ng) diluted and qPCR master mix with SYBR®Green in a final volume of 20 µL (Applied BiosystemsTM SYBR®Select Master Mix, Thermo Fisher Scientific, Waltham, MA, USA) on the QuantStudio 3 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) following an initial denaturation of the samples at 95 °C for 10 min and 40 cycles of alternating denaturation (95 °C for 1 s), annealing and elongation (60 °C for 20 s).
Table 1.
Primer sequences for qRT-PCR.
| Target Gene | Sequence 5′-3′ (F = Forward; R = Reverse) |
Product Size |
|---|---|---|
| gyrB | F: GCACAAGAGTACGATGCCAGT | 119 bp |
| R: TCCCAAACAAGGTGATGCAGC | ||
| D820_RS03005 | F: CGTGGTTATCAAGTATCGTGTGA | 148 bp |
| R: AAAGAATTGGTCCTGCATCCA | ||
| D820_RS09005 | F: CAGTAGGTGCCGCTCAAACT | 128 bp |
| R: AAGTCCGCCGCCAAACATAT | ||
| glgA | F: GTGCCTTGCCCAAATCCCTT | 145 bp |
| R: ACATATTCACGACGCCAGCC | ||
| ssrS | F: CGGAAGCAACTAAAGTCAGAGCG | 80 bp |
| R: TGGCACCGATGATTCACGTT |
Gene expression was quantified by the comparative threshold cycle (Ct) method (2−∆∆Ct) [37]. First, Ct values of all expressed genes were normalized by the housekeeping gene gyrB. Furthermore, gene expression of treated cells was calculated relative to the untreated control cells (log2 fold change). Median values and quantiles of all experiments were calculated and compared to log2 fold changes obtained from RNA-seq (n = 5).
3. Results
3.1. Sublethal Concentration of CHX
The bacterial ability to replicate was investigated for planktonic cultures of S. mutans after treatment with 125 µg/mL CHX by a CFU assay. The chosen concentration of 125 µg/mL was determined by experiments screening multiple concentrations of CHX (data not shown). For the purpose of this study, sublethal doses of CHX were defined as the treatment that resulted in a < 0.5 log10 step reduction in CFU. Untreated controls exhibited 1.0 × 107 to 1.4 × 107 CFU. Samples treated with 125 µg/mL CHX for 5 min showed 4.5 × 106 to 8.5 × 106 CFU, resulting in a reduction in CFU of 0.2 log10 steps, as compared to the untreated control.
3.2. RNA-Seq and Analysis of Differentially Expressed Genes
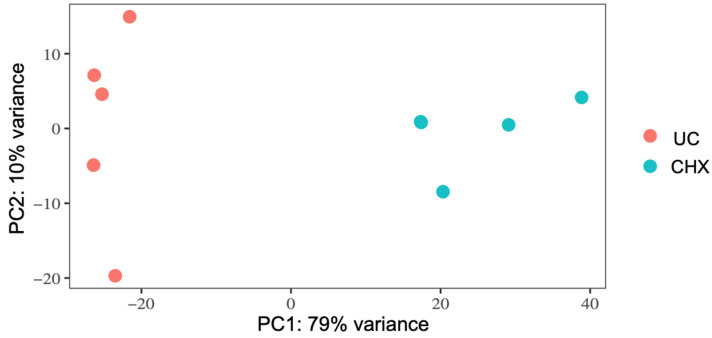
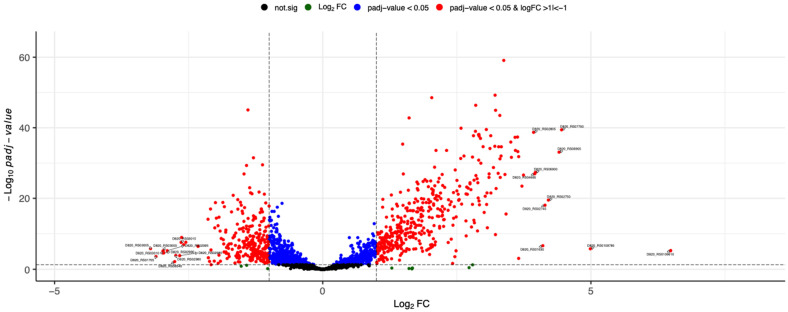
RNA-seq was performed to investigate the molecular stress response of S. mutans upon sublethal treatment with CHX. Five independent experiments were performed on CHX-treated and -untreated bacteria. Principal component analysis (PCA) indicated that all biological replicates grouped together, suggesting that gene expression was significantly influenced by the treatment with CHX compared to untreated samples, explaining 89% of the variance observed in the complete dataset (Figure 1). The distribution of gene expression between the untreated and treated samples is represented by a volcano plot (Figure 2). The genes located outside the borders of the line fold change ≥ 1.0 were considered DE genes. The number of DE genes with an adjusted p-value ≤ 0.05 was 718. Genes situated on the left boundary are downregulated, whereas those on the right are upregulated genes. As compared to the untreated control, 404 genes were upregulated and 271 genes were downregulated in S. mutans after treatment with 125 µg/mL CHX for 5 min. A list of all genes detected by RNA-seq can be found in the Supplementary Material (Table S1).
Figure 1.
Principal component analysis (PCA) of gene expression in S. mutans untreated (UC, orange) or treated with CHX (turquoise). The plot shows the first two principal components (PC1 and PC2), which account for 79% and 10% of the total variation of the data, respectively. UC: untreated control.
Figure 2.
Volcano plot of differential gene expression of S. mutans treated with CHX versus untreated bacteria. Each point represents the average value of one transcript in five replicate experiments. The expression difference is considered significant for a multiple (FDR-based) adjusted p-value of 0.05 (light grey broken horizontal line). The list of DE genes was then further filtered for biological relevance by filtering on a log2 fold change of ≥1 and ≤−1 (red points, outer blue broken vertical lines). The top 20 DE genes based on log2 fold change are labelled.
Table 2 and Figure 2 show the 10 most up- and 10 most downregulated genes. DE genes were selected by their log2 fold change. It was observed that CHX treatment significantly regulated expression of genes involved in transport systems (ABC transporter and PTS system). Table 3 shows selected DE genes involved in oxidative stress (ahpC, ahpF, sod, trxP and tpx), general stress (groEL, groES and D820_RS07050) and acid stress response (aguA and mleP), which were significantly upregulated.
Table 2.
The 10 most up- and downregulated genes in S. mutans upon sublethal CHX treatment. The full list of DE genes can be found in the Supplemental Table S1.
| Gene ID | Gene Name | Product | Log2 FC | p adj |
|---|---|---|---|---|
| D820_RS03005 | NA | PTS fructose transporter subunit IIB |
−3.2 | 1.60 × 10−6 |
| D820_RS01705 | NA | tRNA-Thr | −3.1 | 2.46 × 10−4 |
| D820_RS03010 | NA | PTS mannitol/fructose, IIC component |
−3.0 | 2.26 × 10−5 |
| D820_RS03000 | NA | PTS mannitol transporter subunit IIB |
−3.0 | 6.57 × 10−6 |
| D820_RS02990 | lacC | Tagatose-6-phosphate kinase | −3.0 | 2.40 × 10−5 |
| D820_RS02995 | lacD | Tagatose-bisphosphate aldolase | −2.9 | 6.31 × 10−6 |
| D820_RS08540 | NA | tRNA-Ser | −2.8 | 6.90 × 10−3 |
| D820_RS02980 | lacA | Galactose-6-phosphate isomerase | −2.7 | 1.24 × 10−4 |
| D820_RS02985 | lacB | Galactose-6-phosphate isomerase | −2.7 | 1.43 × 10−4 |
| D820_RS03015 | NA | Lactose-specific phosphotransferase enzyme IIA component |
−2.6 | 2.85 × 10−8 |
| D820_RS02805 | glgA | Glycogen synthase | 3.9 | 0.00 |
| D820_RS04685 | NA | ABC transporter (ATP-binding protein) |
4.0 | 0.00 |
| D820_RS06900 | NA | Hypothetical protein | 4.0 | 0.00 |
| D820_RS01630 | NA | 16S ribosomal RNA | 4.1 | 2.44 × 10−7 |
| D820_RS02745 | NA | ABC transporter (ATP-binding protein) |
4.1 | 0.00 |
| D820_RS02750 | NA | ABC transporter permease | 4.2 | 0.00 |
| D820_RS06905 | lrgB | Antiholin | 4.4 | 0.00 |
| D820_RS07700 | pflB | Formate C-acetyltransferase | 4.5 | 0.00 |
| D820_RS0109785 | NA | 23S ribosomal RNA | 5.0 | 1.79 × 10−6 |
| D820_RS0109610 | NA | 23S ribosomal RNA | 6.5 | 5.73 × 10−6 |
FC: fold change; padj: adjusted p-value; NA: not applicable.
Table 3.
Differentially expressed genes in S. mutans upon CHX treatment related to stress response.
| Gene ID | Gene Name | Product | Log2 FC | p adj |
|---|---|---|---|---|
| D820_RS06120 | ahpC | Peroxiredoxin | 1.1 | 1.50 × 10−6 |
| D820_RS06115 | ahpF | Alkyl hydroperoxide reductase subunit F | 1.8 | 6.00 × 10−17 |
| D820_RS06685 | sod | Superoxide dismutase | 1.6 | 1.71 × 10−6 |
| D820_RS07435 | trxB | Thioredoxin disulfide reductase | 1.1 | 5.15 × 10−4 |
| D820_RS05415 | tpx | 2-Cys-peroxiredoxine | 1.0 | 1.02 × 10−2 |
| D820_RS07050 | NA | DNA protection protein | 1.9 | 2.41 × 10−12 |
| D820_RS00990 | groES | Co-Chaperone GroES | 1.1 | 8.15 × 10−6 |
| D820_RS00995 | groEL | Chaperone GroEL | 1.5 | 0.00 |
| D820_RS05270 | NA | ATP-dependent Clp protease ATP-binding subunit | 1.1 | 1.04 × 10−7 |
| D820_RS03310 | clpB | Clp proteinase ATP-binding subunit ClpB | 2.4 | 4.29 × 10−15 |
| D820_RS06955 | NA | ATP-dependent Clp protease ATP-binding subunit | 1.4 | 3.43 × 10−10 |
| D820_RS08310 | aguA | Agmatine deiminase | 1.3 | 1.27 × 10−14 |
| D820_RS08960 | mleP | Malate permease | 3.7 | 0.00 |
FC: fold change; padj: adjusted p-value; NA: not applicable.
3.3. Pathway Enrichment Analysis with Differentially Expressed Genes
Pathway enrichment analysis was performed using the BioCyc database, including all upregulated DE genes or all downregulated DE genes after CHX treatment. Significantly enriched pathways that were upregulated were composed of genes involved in carboxylate degradation, glycan biosynthesis, purine nucleotide biosynthesis, L-histidine biosynthesis and L-ascorbate degradation (Table 4). Lactose and galactose degradation, protein modification and L-phenylalanine biosynthesis were found to be downregulated after CHX treatment (Table 4).
Table 4.
Significantly up- and downregulated pathways in S. mutans upon CHX treatment.
| Pathway | Genes Involved in Pathway (Ensemble IDs) |
p-Value | |
|---|---|---|---|
| Upregulated | |||
| Carboxylate degradation | D820_RS00885, D820_RS07700, D820_RS07305, D820_RS08215, D820_RS08265, D820_RS08275, D820_RS08270, D820_RS04975, D820_RS04980, D820_RS08965 | 5.00 × 10−5 | |
| 5-Aminoimidazole ribonucleotide biosynthesis | D820_RS09340, D820_RS09355, D820_RS09335, D820_RS09300, D820_RS09345 | 2.00 × 10−3 | |
| Glycan pathway | D820_RS07040, D820_RS02810, D820_RS02685, D820_RS02800, D820_RS02795, D820_RS02790, D820_RS02805 | 3.00 × 10−3 | |
| L-ascorbate degradation | D820_RS08215, D820_RS08265, D820_RS08275, D820_RS08270 | 6.00 × 10−3 | |
| L-histidine biosynthesis | D820_RS03965, D820_RS03970, D820_RS03955, D820_RS03950, D820_RS03960, D820_RS03940, D820_RS03930 | 7.00 × 10−3 | |
| Purine nucleotide biosynthesis | D820_RS04105, D820_RS04110, D820_RS04100, D820_RS09285, D820_RS09290, D820_RS09325, D820_RS09250, D820_RS08295, D820_RS09340, D820_RS09355, D820_RS09335, D820_RS09300, D820_RS09345 | 1.00 × 10−2 | |
| Inosine-5’-phosphate biosynthesis | D820_RS09285, D820_RS09290, D820_RS09325, D820_RS09250 | 2.00 × 10−2 | |
| Downregulated | |||
| Lactose degradation | D820_RS02995, D820_RS02980, D820_RS02985, D820_RS03025, D820_RS02990 | 4.00 × 10−6 | |
| Galactose degradation | D820_RS02995, D820_RS02980, D820_RS02985, D820_RS03025, D820_RS02990 | 4.00 × 10−6 | |
| Protein modification | D820_RS05715, D820_RS02255, D820_RS08010 | 2.00 × 10−2 | |
| L-phenylalanine biosynthesis | D820_RS03740, D820_RS07095 | 2.00 × 10−2 |
3.4. Validation of RNA-Seq Data Using qRT-PCR
To validate RNA-Seq data, randomly selected genes were measured by qRT-PCR to evaluate the transcription levels. Overall, the expression levels of all selected genes were similar to those of the RNA-seq, with a Spearman correlation coefficient of 0.8. The results showed that the D820_RS03003 expression levels were downregulated in CHX-treated bacteria compared to the untreated control (Table 5). In comparison, expression levels were upregulated in D820_RS09005, ssrA and glgA in CHX-treated bacteria compared to the untreated control (Table 5), which is in line with the DE results from the RNA-Seq experiment.
Table 5.
Validation of differentially expressed genes using qRT-PCR. Transcript levels of selected genes (Table 1) were corrected to gyrB. Each value (log2 fold change) is the median of five replicate PCR reactions.
| Gene | RNA-Seq | qRT-PCR |
|---|---|---|
| D820_RS03005 | −3.2 | −2.4 |
| D820_RS09005 | 3.3 | 3.4 |
| ssrS | 3.4 | 2.5 |
| glgA | 3.9 | 4.0 |
4. Discussion
Despite the widespread use of antiseptics such as CHX in dental practice and oral care, the risks of potential resistance toward these antimicrobial compounds in oral bacteria have only been highlighted very recently [4,5,20,22,24]. Since the molecular mechanisms behind antiseptic resistance or adaptation are not entirely clear yet and the bacterial stress response upon sublethal exposure toward CHX has not been investigated systematically so far [4], the aim of the present study was to investigate the transcriptomic stress response in S. mutans after sublethal treatment with CHX.
S. mutans was chosen as a model organism for the present study due to its key role with regard to biofilm formation and pathogenesis of dental caries [26,27]. A type strain was used in order to ensure that the transcriptomic stress response of a sublethal treatment with CHX was investigated on a strain not pre-adapted to CHX. For this purpose, it was necessary to choose “sublethal” treatment stress conditions but not kill the bacteria in order to allow for transcriptomic adaptations. Thus, sublethal conditions were defined as conditions that result in a reduction in bacterial ability to replicate in <0.5 log10 step as measured by CFU, which means that less than 50% of the bacteria are influenced in their ability to replicate. Bacteria were treated with CHX at a concentration of 125 µg/mL for 5 min, which resulted in a CFU reduction of 0.2 log10 steps. After treatment, bacteria were cultured in fresh nutrient broth for 30 min to allow for regulation of transcriptomic stress response. Afterwards, RNA was extracted and used for RNA-seq. RNA-seq data were validated using qRT-PCR to evaluate the transcription levels of randomly selected genes.
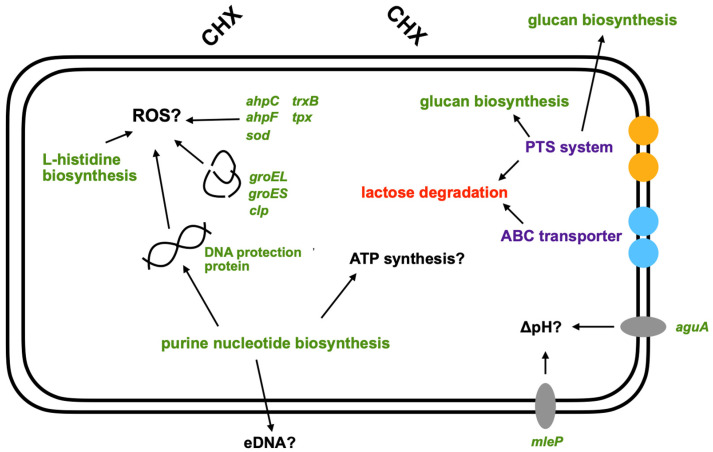
Sublethal treatment of S. mutans with CHX showed a strong treatment effect that led to a clear separation of samples into the two experimental groups, thereby explaining 89% of the variance in the dataset. The reason for this is most likely the differential regulation of gene expression, with 404 genes upregulated and 271 genes downregulated. Further analysis of DE genes and significantly regulated pathways showed regulation of genes involved in purine nucleotide synthesis, biofilm formation, transport systems and stress responses (Figure 3).
Figure 3.
Schematic representation of a possible stress response in S. mutans to CHX treatment found in this study. Shown are the mechanisms described in the discussion in response to CHX treatment. Green: upregulated; red: downregulated; purple: up- and downregulated. Black arrows represent the links between different mechanisms. ROS: reactive oxygen species; PTS: phosphotransferase system; ABC: ATP-binding cassette; eDNA: extracellular DNA.
Amongst the DE genes and significantly regulated pathways, an upregulation in the expression of genes involved in purine nucleotide synthesis was detected. For the bacterial cell, purines primarily contribute to cell division and synthesis of necessary energy and cofactors [38]. For biofilm formation, purines are required for the production of extracellular DNA [39,40], as well as synthesis of secondary messengers from guanosine monophosphate (GMP) and adenosine monophosphate (AMP), which also play a role in the overall stress response [41]. Purine nucleotide biosynthesis has been shown to be the most regulated pathway in biofilm formation in Gram-positive bacteria [42]. Further studies have found a link between increased purine nucleotide biosynthesis and the formation of “persister” bacteria and the emergence of antibiotic resistance [43,44]. This is explained by the fact that the increase in purines leads to an increase in ATP concentration, which is needed for a stronger formation of polymers, such as for the construction of the peptidoglycan layer [43].
After treatment with CHX, regulation of ATP-binding cassette (ABC) transporters occurred. These are transport systems responsible for the uptake of nutrients, amino acids, ions or peptides, as well as the release of hydrophobic substances or toxins [45]. In addition, ABC transporters play a role in DNA repair and translation of mRNA [46,47]. Regulation of ABC transporters upon exposure to CHX has been discussed and linked to resistance formation in other studies [48,49,50,51]. Treatment of Acinetobacter baumannii with CHX resulted in the upregulation of the efflux system AdeAB and AceI, which were responsible for increased tolerances [50]. A study by Liu et. al. also demonstrated that deletion of the ABC transporter LmrB resulted in enhanced biofilm formation and a higher resistance to acid, H2O2, CHX, penicillin and erythromycin [52]. Another group of transporters constitutes the phosphotransferase system (PTS), members of which were also significantly regulated in S. mutans after treatment with CHX in the present work. The role of these transporters is mainly the uptake and phosphorylation of sugar derivatives [53,54]. Accordingly, regulation of PTS may be responsible for reduced lactose and galactose degradation. In addition, it may be related to the upregulation of glycan synthesis [55,56]. The accumulation of intracellular polysaccharides (IPS) can be enhanced by nutrient deficiency, thus helping the cell to survive by promoting the emergence of “persister” bacteria [57]. Glucan synthesis outside the cell indicates formation of extracellular polymeric substances (EPS), hinting to increased biofilm formation [58,59]. The increased biofilm formation after treatment of S. mutans with CHX was also demonstrated in a study by Dong et al. [60].
Sublethal CHX treatment also resulted in increased expression of specific genes encoding for the synthesis of enzymes, such as superoxide dismutase, thioredoxin, peroxiredoxin, and alkyl hydroperoxide reductases, indicating intracellular oxidative stress [61]. Oxidative stress may lead to damage of proteins or DNA [62,63]. This is confirmed by the regulation of genes encoding for chaperones (groEL and groES) and a DNA protective protein after treatment with CHX. In addition, increased expression of Clp proteases was observed, which play a major role in cell survival after stressful events, such as heat shock, acid stress or oxidative stress [64,65]. In a study by Deng et al., it was shown that deletion of the protease ClpP in S. mutans leads to a higher sensitivity to CHX [66]. In addition, measurement of metabolic activity showed increased tolerance to toxic H2O2 and CHX after pre-incubation with sublethal levels of the corresponding compounds in the wild-type strain but not in the mutant [66]. Accordingly, the increase in regulation of Clp proteases observed in the present work may be related to oxidative stress.
In addition to the suggestion that the regulatory changes in gene expression occur due to oxidative stress, there is also evidence of intracellular acid stress in S. mutans after treatment with CHX, which may be explained by the acidic pH of CHX (pH 5.5). This assumption can be supported by a study of Svensäter et al. which showed that exposure of planktonic growing S. mutans to pH values between 6.0 and 3.5 results in the induction of an acid tolerance response [67]. To protect themselves from low pH, oral bacteria attempt to maintain intracellular pH by alkalinization [68]. In the present work, malate permease (mleP) was found to be upregulated, which plays a role in malolactic fermentation. This involves the conversion of L-malate to L-lactate with the acquisition of ATP and CO2, leading to alkalinization of the environment [69,70]. Furthermore, an increased expression of agmatine deiminase (aguA) was found in this study. Agmatine is a decarboxylated derivative of arginine that is taken up by the cell and degraded by agmatine deiminase. The degradation produces ATP, CO2 and NH3, which eventually leads to an increase in intracellular pH [71,72].
5. Conclusions
In conclusion, we have identified and analyzed a considerable number of genes and pathways significantly regulated in S. mutans after sublethal treatment with CHX (Figure 3). RNA-seq showed increased expression of genes related to oxidative stress. Furthermore, regulation of transporters and increased biofilm formation were found, which could support the risk of development of resistance. Due to the acidic pH of CHX, acid stress, in addition to oxidative stress, occurred. This is the first attempt to assess the transcriptomic stress response following sublethal treatment of S. mutans with CHX. Further investigations should focus on the transcriptomic stress response in clinical isolates or strains exhibiting phenotypic adaptation toward CHX in order to assess potential resistance mechanisms. Furthermore, whereas the present study investigated the transcriptomic stress response in planktonic bacteria, future studies should include bacteria grown in biofilms, which may exhibit different stress responses.
Acknowledgments
Andreas Rosendahl is gratefully acknowledged for his excellent support in qRT-PCR experiments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10030561/s1, Table S1: List of all genes of S. mutans detected by RNA-Seq.
Author Contributions
Conceptualization, D.M., F.C., A.A.-A., S.C., J.G., T.M. and K.-A.H.; methodology, D.M., S.C., J.G., C.E. and M.W.; validation, D.M., S.C., J.G., M.W., W.B., T.M. and K.-A.H.; formal analysis, D.M., S.C., K.-A.H., T.M. and F.C.; investigation, D.M., S.C., J.G., C.E. and M.W.; data curation, D.M., S.C. and K.-A.H.; writing—original draft preparation, D.M., X.M. and F.C.; writing—review and editing, F.C., A.A.-A., T.M., K.-A.H., M.W., S.C., J.G. and W.B.; visualization, D.M. and S.C.; supervision, F.C.; project administration, F.C. and D.M.; funding acquisition, F.C. and A.A.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation; grants CI 263/3-1 and AL 1179/4-1).
Data Availability Statement
All data supporting the reported results are available upon request from the corresponding author. The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [73] and are accessible through GEO Series accession number GSE197633 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197633, accessed on 22 January 2022).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tacconelli E., Pezzani M.D. Public health burden of antimicrobial resistance in Europe. Lancet Infect. Dis. 2019;19:4–6. doi: 10.1016/S1473-3099(18)30648-0. [DOI] [PubMed] [Google Scholar]
- 2.Knight G.M., Glover R.E., McQuaid C.F., Olaru I.D., Gallandat K., Leclerc O.J., Fuller N.M., Willcocks S.J., Hasan R., van Kleef E., et al. Antimicrobial resistance and COVID-19: Intersections and implications. Elife. 2021;10:e64139. doi: 10.7554/eLife.64139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.OECD . Stemming the Superbug Tide. OECD; Paris, France: 2018. [Google Scholar]
- 4.Cieplik F., Jakubovics N.S., Buchalla W., Maisch T., Hellwig E., Al-Ahmad A. Resistance Toward Chlorhexidine in Oral Bacteria-Is There Cause for Concern? Front. Microbiol. 2019;10:587. doi: 10.3389/fmicb.2019.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao X., Auer D.L., Buchalla W., Hiller K.A., Maisch T., Hellwig E., Al-Ahmad A., Cieplik F. Cetylpyridinium Chloride: Mechanism of Action, Antimicrobial Efficacy in Biofilms, and Potential Risks of Resistance. Antimicrob. Agents Chemother. 2020;64:e00576-20. doi: 10.1128/AAC.00576-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kampf G. Acquired resistance to chlorhexidine-is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 2016;94:213–227. doi: 10.1016/j.jhin.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 7.Venter H., Henningsen M.L., Begg S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017;61:1–10. doi: 10.1042/EBC20160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies G.E., Francis J., Martin A.R., Rose F.L., Swain G. 1:6-Di-4′-chlorophenyldiguanidohexane (hibitane); laboratory investigation of a new antibacterial agent of high potency. Br. J. Pharmacol. Chemother. 1954;9:192–196. doi: 10.1111/j.1476-5381.1954.tb00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjermo P. Chlorhexidine in dental practice. J. Clin. Periodontol. 1974;1:143–152. doi: 10.1111/j.1600-051X.1974.tb01250.x. [DOI] [PubMed] [Google Scholar]
- 10.Jones C.G. Chlorhexidine: Is it still the gold standard? Periodontol 2000. 1997;15:55–62. doi: 10.1111/j.1600-0757.1997.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 11.Van der Weijden F.A., Van der Sluijs E., Ciancio S.G., Slot D.E. Can Chemical Mouthwash Agents Achieve Plaque/Gingivitis Control? Dent. Clin. N. Am. 2015;59:799–829. doi: 10.1016/j.cden.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Pithon M.M., Sant’Anna L.I., Baião F.C., dos Santos R.L., Coqueiro Rda S., Maia L.C. Assessment of the effectiveness of mouthwashes in reducing cariogenic biofilm in orthodontic patients: A systematic review. J. Dent. 2015;43:297–308. doi: 10.1016/j.jdent.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Solderer A., Kaufmann M., Hofer D., Wiedemeier D., Attin T., Schmidlin P.R. Efficacy of chlorhexidine rinses after periodontal or implant surgery: A systematic review. Clin. Oral Investig. 2019;23:21–32. doi: 10.1007/s00784-018-2761-y. [DOI] [PubMed] [Google Scholar]
- 14.Sanz M., Serrano J., Iniesta M., Santa Cruz I., Herrera D. Antiplaque and antigingivitis toothpastes. Monogr. Oral Sci. 2013;23:27–44. doi: 10.1159/000350465. [DOI] [PubMed] [Google Scholar]
- 15.Cieplik F., Kara E., Muehler D., Enax J., Hiller K.A., Maisch T., Buchalla W. Antimicrobial efficacy of alternative compounds for use in oral care toward biofilms from caries-associated bacteria in vitro. Microbiologyopen. 2019;8:e00695. doi: 10.1002/mbo3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marui V.C., Souto M.L.S., Rovai E.S., Romito G.A., Chambrone L., Pannuti C.M. Efficacy of preprocedural mouthrinses in the reduction of microorganisms in aerosol: A systematic review. J. Am. Dent. Assoc. 2019;150:1015–1026.e1. doi: 10.1016/j.adaj.2019.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Arweiler N.B., Boehnke N., Sculean A., Hellwig E., Auschill T.M. Differences in efficacy of two commercial 0.2% chlorhexidine mouthrinse solutions: A 4-day plaque re-growth study. J. Clin. Periodontol. 2006;33:334–339. doi: 10.1111/j.1600-051X.2006.00917.x. [DOI] [PubMed] [Google Scholar]
- 18.Auschill T.M., Hein N., Hellwig E., Follo M., Sculean A., Arweiler N.B. Effect of two antimicrobial agents on early in situ biofilm formation. J. Clin. Periodontol. 2005;32:147–152. doi: 10.1111/j.1600-051X.2005.00650.x. [DOI] [PubMed] [Google Scholar]
- 19.Zaura-Arite E., van Marle J., ten Cate J.M. Confocal Microscopy Study of Undisturbed and Chlorhexidine-treated Dental Biofilm. J. Dent. Res. 2001;80:1436–1440. doi: 10.1177/00220345010800051001. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz S.R., Hirsch S., Hiergeist A., Kirschneck C., Muehler D., Hiller K.A., Maisch T., Al-Ahmad A., Gessner A., Buchalla W., et al. Limited antimicrobial efficacy of oral care antiseptics in microcosm biofilms and phenotypic adaptation of bacteria upon repeated exposure. Clin. Oral Investig. 2021;25:2939–2950. doi: 10.1007/s00784-020-03613-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubovics N.S., Goodman S.D., Mashburn-Warren L., Stafford G.P., Cieplik F. The dental plaque biofilm matrix. Periodontol. 2000. 2021;86:32–56. doi: 10.1111/prd.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagawa H., Izutani N., Kitagawa R., Maezono H., Yamaguchi M., Imazato S. Evolution of resistance to cationic biocides in Streptococcus mutans and Enterococcus faecalis. J. Dent. 2016;47:18–22. doi: 10.1016/j.jdent.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Wand M.E., Bock L.J., Bonney L.C., Sutton J.M. Mechanisms of Increased Resistance to Chlorhexidine and Cross-Resistance to Colistin following Exposure of Klebsiella pneumoniae Clinical Isolates to Chlorhexidine. Antimicrob. Agents Chemother. 2017;61:e01162-16. doi: 10.1128/AAC.01162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verspecht T., Rodriguez Herrero E., Khodaparast L., Khodaparast L., Boon N., Bernaerts K., Quirynen M., Teughels W. Development of antiseptic adaptation and cross-adapatation in selected oral pathogens in vitro. Sci. Rep. 2019;9:8326. doi: 10.1038/s41598-019-44822-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laumen J.G.E., Van Dijck C., Manoharan-Basil S.S., Abdellati S., De Baetselier I., Cuylaerts V., De Block T., Van den Bossche D., Xavier B.B., Malhotra-Kumar S., et al. Sub-Inhibitory Concentrations of Chlorhexidine Induce Resistance to Chlorhexidine and Decrease Antibiotic Susceptibility in Neisseria gonorrhoeae. Front. Microbiol. 2021;12:776909. doi: 10.3389/fmicb.2021.776909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemos J.A., Palmer S.R., Zeng L., Wen Z.T., Kajfasz J.K., Freires I.A., Abranches J., Brady L.J. The Biology of Streptococcus mutans. Microbiol. Spectr. 2019;7:GPP3-0051-2018. doi: 10.1128/microbiolspec.GPP3-0051-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemos J.A., Burne R.A. A model of efficiency: Stress tolerance by Streptococcus mutans. Pt 11Microbiology. 2008;154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miles A.A., Misra S.S., Irwin J.O. The estimation of the bactericidal power of the blood. J. Hyg. 1938;38:732–749. doi: 10.1017/S002217240001158X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. FastQC. A Quality Control Tool for High Throughput Sequence Data. 2010. [(accessed on 22 January 2022)]. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 30.Krueger F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files, with Some Extra Functionality for MspI-Digested RRBS-Type (Reduced Representation Bisufite-Seq) Libraries. 2012. [(accessed on 22 January 2022)]. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 31.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Wang S., Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 33.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 34.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 36.Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q., et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019;20:1085–1093. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Pedley A.M., Benkovic S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas V.C., Thurlow L.R., Boyle D., Hancock L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008;190:5690–5698. doi: 10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dengler V., Foulston L., DeFrancesco A.S., Losick R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2015;197:3779–3787. doi: 10.1128/JB.00726-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahmi T., Faozia S., Port G.C., Cho K.H. The second messenger c-di-AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall homeostasis, SpeB expression, and virulence in Streptococcus pyogenes. Infect. Immun. 2019;87:e00147-19. doi: 10.1128/IAI.00147-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gélinas M., Museau L., Milot A., Beauregard P.B. Cellular adaptation and the importance of the purine biosynthesis pathway during biofilm formation in Gram-positive pathogens. bioRxiv. 2020;2020 doi: 10.1101/2020.12.11.422287. [DOI] [Google Scholar]
- 43.Mongodin E., Finan J., Climo M.W., Rosato A., Gill S., Archer G.L. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 2003;185:4638–4643. doi: 10.1128/JB.185.15.4638-4643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yee R., Cui P., Shi W., Feng J., Zhang Y. Genetic Screen Reveals the Role of Purine Metabolism in Staphylococcus aureus Persistence to Rifampicin. Antibiotics. 2015;4:627–642. doi: 10.3390/antibiotics4040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davidson A.L., Dassa E., Orelle C., Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraburtty K. Translational regulation by ABC systems. Res. Microbiol. 2001;152:391–399. doi: 10.1016/S0923-2508(01)01210-4. [DOI] [PubMed] [Google Scholar]
- 47.Goosen N., Moolenaar G.F. Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair. Res. Microbiol. 2001;152:401–409. doi: 10.1016/S0923-2508(01)01211-6. [DOI] [PubMed] [Google Scholar]
- 48.Hassan K.A., Jackson S.M., Penesyan A., Patching S.G., Tetu S.G., Eijkelkamp B.A., Brown M.H., Henderson P.J.F., Paulsen I.T. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA. 2013;110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J., Zhang J., Guo L., Zhao W., Hu X., Wei X. Inactivation of a putative efflux pump (LmrB) in Streptococcus mutans results in altered biofilm structure and increased exopolysaccharide synthesis: Implications for biofilm resistance. Biofouling. 2017;33:481–493. doi: 10.1080/08927014.2017.1323206. [DOI] [PubMed] [Google Scholar]
- 50.Hassan K.A., Liu Q., Henderson P.J.F., Paulsen I.T. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio. 2015;6:e01982-14. doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mombeshora M., Mukanganyama S. Development of an accumulation assay and evaluation of the effects of efflux pump inhibitors on the retention of chlorhexidine digluconate in Pseudomonas aeruginosa and Staphylococcus aureus. BMC Res. Notes. 2017;10:328. doi: 10.1186/s13104-017-2637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Guo L., Lui J., Zhang J., Zeng H., Ning Y., Wei Y. Identification of an Efflux Transporter LmrB Regulating Stress Response and Extracellular Polysaccharide Synthesis in Streptococcus mutans. Front. Microbiol. 2017;8:962. doi: 10.3389/fmicb.2017.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vadeboncoeur C., Pelletier M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 54.Deutscher J., Francke C., Postma P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nobre dos Santos M., Melo dos Santos L., Francisco S.B., Cury J.A. Relationship among dental plaque composition, daily sugar exposure and caries in the primary dentition. Caries Res. 2002;36:347–352. doi: 10.1159/000065959. [DOI] [PubMed] [Google Scholar]
- 56.Liu C., Niu Y., Zhou X., Zheng X., Wang S., Guo Q., Li Y., Li M., Li J., Yang Y., et al. Streptococcus mutans copes with heat stress by multiple transcriptional regulons modulating virulence and energy metabolism. Sci. Rep. 2015;5:12929. doi: 10.1038/srep12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Busuioc M., Mackiewicz K., Buttaro B.A., Piggot P.J. Role of intracellular polysaccharide in persistence of Streptococcus mutans. J. Bacteriol. 2009;191:7315–7322. doi: 10.1128/JB.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colby S.M., Russell R.R. Sugar metabolism by mutans streptococci. Soc. Appl. Bacteriol. Symp. Ser. 1997;26:80s–88s. doi: 10.1046/j.1365-2672.83.s1.9.x. [DOI] [PubMed] [Google Scholar]
- 59.Rainey K., Michalek S.M., Wen Z.T., Wu H. Glycosyltransferase-Mediated Biofilm Matrix Dynamics and Virulence of Streptococcus mutans. Appl. Environ. Microbiol. 2019;85:e02247-18. doi: 10.1128/AEM.02247-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong L., Tong Z., Linghu D., Lin Y., Tao R., Liu J., Tian Y., Ni L. Effects of sub-minimum inhibitory concentrations of antimicrobial agents on Streptococcus mutans biofilm formation. Int. J. Antimicrob. Agents. 2012;39:390–395. doi: 10.1016/j.ijantimicag.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 61.Kajfasz J.K., Ganguly T., Hardin E.L., Abranches J., Lemos J.A. Transcriptome responses of Streptococcus mutans to peroxide stress: Identification of novel antioxidant pathways regulated by Spx. Sci. Rep. 2017;7:16018. doi: 10.1038/s41598-017-16367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cabiscol E., Tamarit J., Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- 63.Imlay J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemos J.A., Burne R.A. Regulation and Physiological Significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 2002;184:6357–6366. doi: 10.1128/JB.184.22.6357-6366.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou X.H., Zhang J.Q., Song X.Y., Ma X.B., Zhang S.Y. Contribution of ClpP to stress tolerance and virulence properties of Streptococcus mutans. J. Basic Microbiol. 2014;54:1222–1232. doi: 10.1002/jobm.201300747. [DOI] [PubMed] [Google Scholar]
- 66.Deng D.M., Ten Cate J.M., Crielaard W. The adaptive response of Streptococcus mutans towards oral care products: Involvement of the ClpP serine protease. Eur. J. Oral Sci. 2007;115:363–370. doi: 10.1111/j.1600-0722.2007.00477.x. [DOI] [PubMed] [Google Scholar]
- 67.Svensäter G., Larsson U.B., Greif E.C., Cvitkovitch D.G., Hamilton I.R. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 1997;12:266–273. doi: 10.1111/j.1399-302X.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 68.Burne R.A., Marquis R.E. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 69.Sheng J., Baldeck J.D., Nguyen P.T.M., Quivey R.G., Jr., Marquis R.E. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can. J. Microbiol. 2010;56:539–547. doi: 10.1139/W10-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheng J., Marquis R.E. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 2007;272:196–201. doi: 10.1111/j.1574-6968.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- 71.Simon J.P., Stalon V. Enzymes of agmatine degradation and the control of their synthesis in Streptococcus faecalis. J. Bacteriol. 1982;152:676–681. doi: 10.1128/jb.152.2.676-681.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griswold A.R., Jameson-Lee M., Burne R.A. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 2006;188:834–841. doi: 10.1128/JB.188.3.834-841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the reported results are available upon request from the corresponding author. The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [73] and are accessible through GEO Series accession number GSE197633 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197633, accessed on 22 January 2022).