Highlights
-
•
Toxicogenomics is emerging as a valid approach to characterise the mechanism of action of chemicals.
-
•
Structured pipelines for toxicogenomics increase standardisation and regulatory acceptance.
-
•
We developed the Nextcast software suite for robust analysis and modelling of toxicogenomic data.
-
•
Nextcast offers customisable modular pipelines to tackle multiple biological questions.
Keywords: Nextcast, Toxicogenomics, Pipeline, Software suite, Computational toxicology, Predictive toxicology
Abstract
The recent advancements in toxicogenomics have led to the availability of large omics data sets, representing the starting point for studying the exposure mechanism of action and identifying candidate biomarkers for toxicity prediction. The current lack of standard methods in data generation and analysis hampers the full exploitation of toxicogenomics-based evidence in regulatory risk assessment. Moreover, the pipelines for the preprocessing and downstream analyses of toxicogenomic data sets can be quite challenging to implement. During the years, we have developed a number of software packages to address specific questions related to multiple steps of toxicogenomics data analysis and modelling. In this review we present the Nextcast software collection and discuss how its individual tools can be combined into efficient pipelines to answer specific biological questions. Nextcast components are of great support to the scientific community for analysing and interpreting large data sets for the toxicity evaluation of compounds in an unbiased, straightforward, and reliable manner. The Nextcast software suite is available at: ( https://github.com/fhaive/nextcast).
1. Introduction
Traditional risk assessment strategies provide little understanding of the underlying molecular mechanisms leading to toxic outcomes [1]. It relies on molecular profiling technologies such as genomics, proteomics, and metabolomics to draw comprehensive conclusions on the possible toxicity of a chemical or substance [2], [3], [4]. Toxicogenomics has the potential to widen our understanding of the cascade of events and biological responses to exposure beyond the traditional toxicity endpoints. Toxicogenomics has multiple advantages when applied together with other toxicity testing. It enables predictions of possible long-term effects of exposures, reducing the cost and time of animal testing [5], [6], [7], [8], [9]. Moreover, information derived from toxicogenomics data about key events and their relationships can be used to define adverse outcome pathways (AOP). Finally, toxicogenomics data modelling can be used to derive molecular points of departure (POD) for dose–response assessment [10], [11], [12].
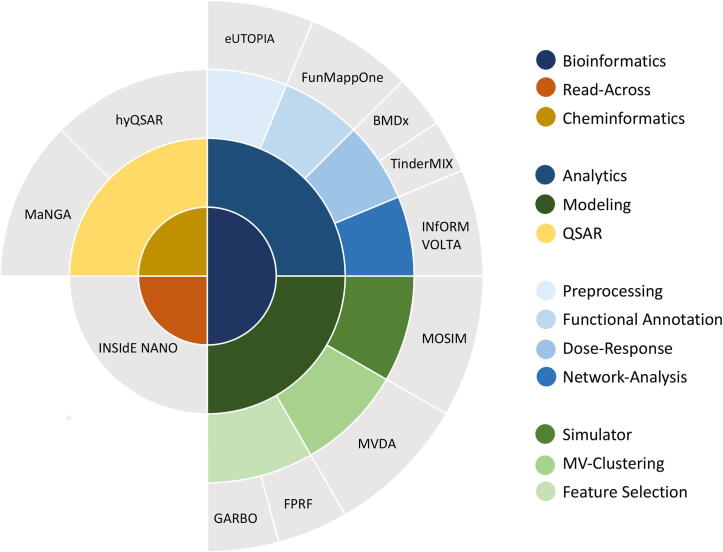
The generation of large amounts of experimental data is increasingly accessible both in academic and industrial research environments. However, standardisation of experimental design, data analysis, and modelling are urgently needed to ensure maximal integration of evidence derived from such data into regulatory safety evaluation. The successful analysis of large omics data sets for the evaluation of adverse effects of chemicals requires simple and straightforward strategies, clear pipelines, and reliable methods. To date, many tools to analyse large data generated with omics- and high-throughput technologies exist [3], but a unified solution addressing all the necessary steps, from the initial data preprocessing to more complex biological questions, is still lacking. Moreover, the change in scientific practices, advocating Open Science principles, requires infrastructures and common strategies supporting the use of FAIR (Findability, Accessibility, Interoperability, and Reusability) principles [13]. The task is not trivial, as the data and tools for data processing are often scattered and inconsistent. To overcome these limitations, we developed during the years multiple software to address specific questions and collected them into an organised software suite that we called Nextcast. Nextcast provides standardised state-of-the-art methods and algorithms to analyse, model, and interpret toxicogenomic and cheminformatic data (Fig. 1).
Fig. 1.
Nextcast is a software suite whose core functionalities allow robust modelling and analysis of bioinformatics (dark blue) and cheminformatics (dark yellow) data as well as read-across analyses (orange). Nextcast components (outer layer in gray) implement methods for omics data analytic such as preprocessing (eUTOPIA), functional annotation (FunMappOne), dose–response (BMDx, TinderMIX), and co-expression network generation and analysis (INfORM, VOLTA). Advanced modelling algorithms are also available (dark green) including data set simulator (MOSIM), multi-view (MV) clustering (MVDA), and feature selection strategies (FPRF, GARBO). Nextcast also includes methods for quantitative structure–activity relationship (QSAR) such as MaNGA and hyQSAR.
Nextcast provides robust pipelines for toxicogenomic data preprocessing and normalisation through the eUTOPIA module [14], which also contains utilities for the identification of statistically significant molecular entities of interest such as genes, transcripts, or CpG sites whose molecular state is differentially represented between sample groups of interest. After obtaining the preprocessed data and a selection of molecular features of interest, depending on the research question, Nextcast offers several tools for downstream analysis, such as FunMappOne, a graphical functional annotation software that allows the simultaneous analysis and comparison of the mechanism of action (MOA) characterising multiple experiments through an easy and interactive grid visualisation [15]. The module INfORM, on the other hand, allows the user to infer gene co-expression networks from differential expression data and uses molecular network inference to highlight biologically meaningful response modules, making them available to the user through several analytical options and high-quality visual outputs [16]. The BMDx and TinderMIX modules allow the user to define molecular points of departure and relevant/optimal doses [10], [12]. see Table 1.
Table 1.
Nextcast components currently utilised and reviewed in the literature.
| Tool | Used in | Cited in | Category |
|---|---|---|---|
| eUTOPIA [14] | Bioinformatics | ||
| https://github.com/Greco-Lab/eUTOPIA | [24], [25], [5], [26], [27], [28], [29], [30] | [31], [2], [3] | Analytics |
| R, Shiny | Preprocessing | ||
| INfORM [16] | Bioinformatics | ||
| https://github.com/Greco-Lab/INfORM | [32], [33], [28], [26], [34] | [31], [2], [3], [4], [35] | Analytics |
| R, Shiny | Network Analysis | ||
| VOLTA [36] | Bioinformatics | ||
| https://github.com/fhaive/VOLTA - | - | Analytics | |
| Python | Network Analysis | ||
| BMDx [10] | Bioinformatics | ||
| https://github.com/Greco-Lab/BMDx | Analytics | ||
| R, Shiny | - | [4] | Dose-Responsive |
| TinderMIX [12] | Bioinformatics | ||
| https://github.com/grecolab/TinderMIX | Analytics | ||
| R | [5], [23] | [4] | Dose-Responsive |
| FunMappOne [15] | Bioinformatics | ||
| https://github.com/Greco-Lab/FunMappOne | [37], [32], [10], [12], [28],[38], [7], [5], [39],[40], [41], [27], [42] | Analytics | |
| R, Shiny | [3], [43] | Functional Annotation | |
| MOSIM [18] | Bioinformatics | ||
| https://doi.org/10.1186/s12859-015-0577-1 | - | [44], [4] | modelling Simulator |
| R | |||
| MVDA [17] | [47], [48], [49], [44],[50], [51], [52], [53] | Bioinformatics | |
| https://github.com/Greco-Lab/MVDA_package | modelling | ||
| R | [45], [46] | [4], [54], [55], [56] | Multi-view clustering |
| FPRF [19] | Bioinformatics | ||
| https://doi.org/10.1371/journal.pone.0107801.s004 | [57], [58] | modelling | |
| R | [59], [60], [50], [61] | [62], [31], [4] | Feature Selection |
| GARBO [20] | Bioinformatics | ||
| https://github.com/Greco-Lab/GARBO | modelling | ||
| Python | [63] | [4] | Feature Selection |
| INSIdE NANO [6] | |||
| http://inano.biobyte.de/ | [64], [65] | [4], [31], [33] | Read-Across |
| MaNGA [21] | |||
| https://github.com/Greco-Lab/MaNGA | |||
| Python | – | [4], [31], [66] | QSAR |
| hyQSAR [22] | – | [4], [31] | QSAR |
Another challenging aspect in toxicogenomics data analysis is the integration of multiple types of omics data. This is considered in the Nextcast software suite through the MVDA methodology for the multi-view clustering or read-across analysis [17]. The MOSIM module is a multi-omics data simulator methodology that is useful in generating synthetic data to test existing or newly developed integrative tools [18]. One of the main needs in computational and predictive toxicology is the identification of models comprising a few predictive features (molecular or intrinsic) of exposure toxicity or susceptibility. The Nextcast suite offers advanced feature selection methodologies for toxicogenomics data, FPRF [19], and Garbo [20]. Moreover, the MaNGA algorithm for feature selection and quantitative structure–activity relationship (QSAR) modelling on chemometric data is provided [21]. Finally, the hyQSAR module is also available as a Nextcast component, allowing integrated hybrid modelling comprising both toxicogenomic and chemoinformatic data [22]. Many of the tools have already been used and reviewed in scientific research (Table 2). Recently, we included the INfORM and TinderMIX modules in an integrative methodology to computationally prioritise drugs that inhibit SARS-CoV-2 infection [23]. Moreover, a systematic review of alternative methods to the Nextcast components has been recently provided in a three-part review mini-series for transcriptomics data in toxicogenomics [2], [3], [4]. Here, we introduce all the components of the Nextcast software suite and we provide comparative analysis against other existing tools. Additionally, we describe how to combine the individual modules to create robust and pipelines for toxicogenomics data analysis. Lastly, we discuss the interoperability of the output of the Nextcast tools with other existing software.
Table 2.
Examples of interoperability of the Nextcast data formats with external tools.
| Nextcast Component | Output | External tool | Description | |
|---|---|---|---|---|
| eUTOPIA | gene expression matrix | MORPHEUS | https://software.broadinstitute.org/morpheus | |
| eUTOPIA | gene expression matrix | t-SNE [82], UMAP [83] | Dimensionality reduction techniques available in R or Python | |
| eUTOPIA | differentially expressed genes | WebGestalt [84], Enrichr [85], PathwAX [86], Ingenuity Pathway Analysis (QIAGEN Inc.,https://digitalinsights.qiagen.com/IPA) | Pathway enrichment analysis | |
| eUTOPIA | differentially expressed genes | STRING [87] | https://string-db.org/ | |
| FunMappOne | enriched GO terms | REVIGO | Tool for summarization and to study of GO terms interactions (available athttp://revigo.irb.hr/) | |
| INfORM | Co-expression networks | Cytoscape [88] and Gephy [89] G | Tools for network visualisation | |
| INfORM | Prioritised genes | WebGestalt [84], Enrichr [85], PathwAX [86], Ingenuity Pathway Analysis (QIAGEN Inc.,https://digitalinsights.qiagen.com/IPA) | Pathway enrichment analysis | |
| INfORM | Prioritised genes | STRING [87] | https://string-db.org/ |
2. Nextcast components
2.1. eUTOPIA: solUTion for Omics data PreprocessIng and Analysis
Preprocessing and statistical analysis are the first steps in any application of omics data. While a wide range of resources is available to perform these tasks, their implementation generally requires advanced knowledge of the statistical methods as well as programming skills. eUTOPIA combines state-of-the-art methods (Table S1) with a user-friendly graphical interface that guides the user through a standardized preprocessing strategy for each specific supported platform [14]. eUTOPIA is able to analyse raw data from multiple platforms, namely Agilent and Affymetrix gene expression microarrays and Illumina DNA methylation microarrays. eUTOPIA allows the raw data to be quality checked, both at the level of individual samples and by comparing all the samples to identify outliers. Moreover, it offers a solution to each step of omics data preprocessing, alongside informative visualisations. A fundamental step in transcriptomics data analysis is to attenuate batch effects while retaining the variation associated with biological variables. Batch effects can be caused by known variables (e.g., dye, RNA quality, experiment date, etc.) or by hidden sources of variation not explained by the known variables. eUTOPIA offers support for the estimation of batch effects and the mitigation of both known and unknown batch effect variables. eUTOPIA further allows the user to statistically evaluate the differences between experimental groups by differential expression or methylation analysis. When performing differential analysis it is important to include in the model all the relevant covariates and any batch variables previously identified and removed. A summary of the methods implemented in each step of the analysis for the different platform can be found in Table S1. Finally, eUTOPIA produces a normalised, batch corrected and annotated expression/methylation data matrix at the desired stage of preprocessing, as well as files with the results of the differential analysis. Furthermore, to ensure reproducibility and transparency, the user can download an analysis report showcasing the steps applied to the data in a visual format. A comparative analysis of the eUTOPIA functionalities against other free analysis tools shows that batch correction and surrogate variable estimation strategies are unavailable in many other tools (Table S2). Moreover, even though eUTOPIA is not the tool with the most functionalities, its features are presented in an easy-to-use workflow that makes the preprocessing task intuitive and less technically challenging for the users.
2.2. FunMappOne: hierarchical organisation and comparison of multiple functional enrichment analysis
FunMappOne is a web-based graphical tool to perform functional annotation of one or multiple toxicogenomic experiments [15]. FunMappOne takes as input a spreadsheet file containing lists of human, mouse or rat genes identifiers. In addition to gene identifiers, gene metrics such as fold-changes or p-values can be provided. FunMappOne allows to query the gProfiler database [67] and compute the enrichment of functional categories from Reactome [68], Kyoto Encyclopedia of Genes and Genomes (KEGG) [69], or Gene Ontology (GO) collections [70]. The over-represented terms or pathways are arranged in a way that easily allows graphical inspection of enriched functional categories over multiple experiments. FunMappOne allows the user to summarise enriched terms by using a three-level hierarchical structure, represented in the form of a directed acyclic graph, that reflects the intrinsic organisation of Reactome, KEGG, and GO annotations. If provided in input, gene metrics can be mapped over enriched terms. The user can upload this information for each experimental condition separately, as well as a set of statistical thresholds and metrics to be associated with the enriched terms. The visual output is an interactive map, which the user can explore in at least three different ways: i) by selecting a subset of experimental conditions; ii) by selecting the level of the hierarchy to visualise or iii) by specifying which categories/terms of interest to be displayed. The samples in the map can be clustered based on the number of shared pathways or on how similar the modifications of the shared pathways are. FunMappOne represents a fast and easy-to-use tool for the final step of most omics-data analyses and allows a clear interpretation of the comparison of multiple experimental conditions with different levels of abstractions. More information on the methods implemented in the FunMappOne tool can be found in Table S3. Many tools are currently available (Table S4) to perform functional and enrichment analysis of omics derived gene lists. To the best of our knowledge, FunMappOne is the only method that summarises the results based on the hierarchical structure of the annotations. Moreover, we are not aware of other publicly available tools that cluster and compare the profiles from multiple experiments.
2.3. INfORM: inference of network response modules
INfORM is an ensemble method for robust gene co-expression network inference and responsive module detection and interpretation [16]. INfORM computes co-expression networks based on multiple correlation and mutual information statistics and multiple network inference algorithms (Table S5). It makes use of the Borda method [71], implemented into the R TopKLists package [72], to integrate all the co-expression networks generated from the ensemble strategy into a final one, ensuring reliable and robust results.
Moreover, INfORM implements widely used community detection algorithms for relevant responsive module identification (Table S5). The quality of responsive modules is assessed by evaluating several characteristics of their nodes and edges, such as their centrality score (computed by several centrality measures such as degree, shortest path among nodes, betweenness), differential log2-fold change, p-value, the median rank of edge weights and number of nodes. These measures are graphically represented in an easy-to-interpret radar chart that also shows the robustness of the modules. INfORM also gives the possibility to perform a functional over-representation analysis of the GO terms over-represented in each responsive module and to compare the similarity between different modules based on the GO terms they enrich. The GO-based module similarity can be visualised as a tile plot to guide the selection of functionally related modules. INfORM, therefore, allows the user to merge statistically significant and biologically relevant modules into an optimised response module. A complete list of methods used in each step of the INfORM analysis is reported in Table S5. We compared INfORM with three publicly available network inference tools (Table S6). Our analysis shows that INfORM is the only one to implement an ensemble strategy. Ensemble methodologies that combine multiple gene co-expression network inference methods give more robust and reliable results [73].
2.4. VOLTA: adVanced mOLecular neTwork Analysis
VOLTA is a network analysis Python package, suited for complex co-expression network analysis [36]. The INfORM and VOLTA tools can be used in combination to compute co-expression networks and to perform advanced network analysis. VOLTA allows the analysis of a single co-expression network, as well as the comparison, clustering and analysis of multiple networks. VOLTA implements several state-of-the-art methodologies for the computation of network similarities and distances, network clustering, community detections, network simplification and common sub-modules identification (Table S7). When compared to other similar software (Table S8), VOLTA offers the widest range of functionalities. VOLTA also allows the comparison of multiple networks and the identification of common sub-structures in different networks. Moreover, VOLTA is a highly flexible tool, allowing users to construct their own custom analysis pipelines, through its individual components. This provides users full control over parameter selection, function selection as well as the combination and re-use of functionalities in different application scenarios. In addition, VOLTA is not only suitable for experienced users but also for novices, as it can be used as a plug-and-play system to suit the individual needs of different users.
2.5. INSIdE NANO: integrated network analysis for nanomaterial characterisation
INSIdE NANO is a network-based web tool ( http://inano.biobyte.de/) for toxicogenomics-based read-across of nanomaterials [6]. The INSIdE NANO network integrates four phenotypic entities in the form of experimental gene expression data for nanomaterial exposures and drug treatments, and prior knowledge between genes known to be associated with chemical exposures or human diseases (Table S9).
In this interaction network, different entities can be compared under the hypothesis that the relatedness of different pairs of exposures can be estimated using the degree of similarity between their specific patterns of the mechanism of action (Table S9). INSIdE NANO can thus be used to contextualize the effects of the nanomaterial exposure on gene regulation by comparing them with those of chemicals and drugs with respect to particular diseases.
The read-across analysis is performed by scanning the network in search of heterogeneous cliques, containing one node for each phenotypic entity category (Table S9). For each clique, the nanomaterial behaviour with respect to a disease can be compared to that of drugs and chemicals. The user can query the database by providing one or more phenotypic entities of interest and a threshold of their similarity score. The output will be a list of cliques containing the entities of interest and other entities strongly connected to them (based on the input threshold). The resulting cliques are prioritised based on the number of known connections that they contain (e.g. drugs used to treat diseases, or chemicals known to cause diseases). Moreover, the INSIdE NANO interface allows investigating which genes underlie the connection.
2.6. BMDx: Benchmark Dose analysis for transcriptomics data
BMDx is a tool for Benchmark Dose (BMD) analysis of omics data developed in R with a shiny graphical interface [10]. The tool analyses transcriptomics data for which multiple doses, at single or multiple time points, are available. It provides a comprehensive survey of dose-dependent transcriptional changes together with dose estimates at which different cellular processes are altered. BMDx can analyse and compare multiple data sets at the same time, making the comparison of different experiments easy.
The steps of the analysis consist of i) filtering the genes by ANOVA or trend test; ii) model fitting and selection. Computation of the BMD (benchmark dose), BMDL, (BMD lower bound), BMDU (BMD upper bound), and IC50 (inhibitory concentration 50) or EC50 (effective concentration 50) for the remaining genes; featuring an interactive visualisation of the fitted model for every gene; iii) functional annotation enrichment of the dose-dependent genes; and iv) comparison of the list of genes/pathways obtained at different time points and experiments. A description of the methods used in the BMDx tool is provided in Table S10. We compared BMDx with other tools for benchmark dose analysis (Table S11). BMDx is one of the few that is able to analyse multiple experiments at the same time. BMDx is designed for the comparative analysis of different toxicogenomics experiments (e.g. multiple chemical exposures) at single or multiple time points. The gene expression data that BMDx accepts as input have to be already preprocessed and normalised. This can be easily achieved with the eUTOPIA module. Moreover, the FunMappOne functionalities are included in the BMDx interface, making it simple to compare different experiments by means of the hierarchical structure of the pathways or GO terms that are enriched by the dose-dependent genes.
2.7. TinderMIX: Time-dose integrated modelling of toxicogenomics data
TinderMIX offers a solution for the simultaneous evaluation of dose-dependent molecular alterations at multiple time points [12]. It provides a tool for the investigation of dynamic dose-dependent alterations improving the interpretation of the kinetics of molecular changes (Table S12). Furthermore, TinderMIX allows the identification of groups of genes with similar sensitivity and kinetics, which can help to identify relevant patterns in biological processes in response to exposures.
TinderMIX fits multiple models of the molecular alteration (measured as fold-changes) as a function of dose and time. Then, it selects the best fitting model for each gene and represents it as a 2D contour plot. This results in an integrated time- and dose–effect map, where a responsive area is identified based on the user-selected threshold. The responsive area consists of the area in which a monotonic alteration can be observed with respect to the doses for a subset of the time points. Each gene showing dynamic dose-dependent response is then labelled according to the integrated point of departure that considers both the time and the dose, giving insight into the sensitivity and kinetics of the molecular alterations. Finally, the dynamic dose–response as a whole can be investigated by grouping the genes by the assigned labels and identifying over-represented pathways for each group.
A few other time- and dose/concentration integrative analysis have been suggested for the modelling of gene expression data. [74] To the best of our knowledge, TinderMIX is currently the only method that gives an estimation of the dynamic point of departure of the molecular alterations.
2.8. FPRF: A robust and accurate method for feature selection and prioritisation from multi-class omics data
FPRF (Fuzzy Pattern Random Forest) implements a feature selection algorithm for multi-omics data. The tool is optimised for the detection of highly relevant patterns associated with predictive variables (Table S13) [19]. Feature relevance determination is a fundamental step for the discovery of biomarkers (e.g. genes able to discriminate with high precision in different clinical conditions) together with the development of predictive models based on these features. The most commonly used approaches to feature selection are univariate and wrapper methods. Despite their diffusion, a common problem of these and other approaches is the stability of relevant features.
FPRF is based on the Random Forests algorithm [75] and a robust feature selection mechanism based on a data transformation process called fuzzy patterns. Before model training, data is discretised into fuzzy patterns employing a set of membership functions, assigning to each feature (a gene or transcript) a fuzzy level of activity (low, low-middle, middle, middle-high, high). After this process, the fuzzy patterns are used to build a predictive model based on random forests, which is in turn used to prioritise the fuzzy patterns using permutation-based feature relevance scores. FPRF produces a predictive model based on the fuzzy patterns, together with a list of prioritised features based on their relevance in the learning phase. When compared to other tools, FPRF is one of the few to combine fuzzy pattern generation over the data set and random forest learning models (Table S14)
2.9. GARBO: Genetic AlgoRithm for biomarker selection in high-dimensional Omics
Genetic AlgoRithm for biomarker selection in high-dimensional Omics (GARBO) is a multi-island-based genetic algorithm for the concurrent optimisation of model accuracy and the number of features used in predictive tasks [20]. The optimisation strategy implemented in GARBO is based on variable length chromosome, dynamic genetic operators, migration of optimal individuals in the populations and a random forest based fitness evaluation (Table S15). Given a classification task, GARBO explores the space of feature sets by evaluating the accuracy related to random forest classifiers built upon these sets to find the best-performing/minimum-sized set.
GARBO has been validated on the classification of cancer patients and the prediction of drug sensitivity using omics data from The Cancer Genome Atlas (TCGA), The Cancer Cell Line Encyclopedia (CCLE), and the Genomics of Drug Sensitivity in Cancer (GDSC). Compared to six other state-of-the-art algorithms, GARBO demonstrated good performances in optimising both accuracy and number of features [20]. A comparative analysis between GARBO and other tools is present in Table S16.
2.10. MaNGA: A multi-niche/multi-objective genetic algorithm for QSAR modelling
MaNGA is a multi-niche/multi-objective genetic algorithm for quantitative structure–activity relationship (QSAR) modelling that simultaneously enables stable feature selection as well as robust and validated regression models with maximised applicability domain (Table S17) [21].
Starting from chemical descriptors and a continuously measured endpoint for a given set of compounds, MaNGA builds predictive models that are both internally and externally validated. The models are optimised for high predictivity and reliable applicability domain. MaNGA strategy starts with creating multiple niches with an independent training-test split of the data set. While the population in each niche evolves independently towards the optimal solution, the niches are also communicating between each other and migrating their optimal solutions. When compared with other QSAR tools, MaNGA is one of the few to perform multi-objective feature selection (Table S18). Indeed, the selected models are ranked according to i) their number of selected molecular descriptors, ii) their predictive performances, iii) applicability domain and iv) their stability across the different niches. The top-ranked model is returned as the final solution.
2.11. hyQSAR: Hybrid quantitative structure–activity relationship modelling
hyQSAR is a suite of instruments for training and analysing data-driven QSAR models [22]. Its models can be fed with structural data of chemical compounds (e.g. molecular descriptors or substructure fingerprints), transcriptomic data (e.g., gene expression values or fold changes), or both, and applied to predict a numerical activity/property of interest. hyQSAR predictions are based on linear models, and during training, the least absolute shrinkage and selection operator (LASSO) is used to improve generalisation and feature selection (Table S19). The user can choose between several transformations to be applied separately to the structural and the transcriptional components of the input. The hyper-parameters are the penalisation factor of LASSO and, optionally, the exponents of the transformations for the structural and the transcriptomic inputs. They are chosen by grid search, using random splits to improve generalisability. hyQSAR allows internal and external model validation according to the Organisation for Economic Co-operation and Development (OECD) requirements. To the best of our knowledge, hyQSAR is one of the few strategy that generate QSAR models with mixed omics and cheminformatics features (Table S18).
2.12. MVDA: A multi-view clustering approach
The MVDA (Multi-View Data Analysis) is a tool for clustering samples in a multi-omics data set. MVDA implements a multi-view late integration strategy that combines dimensionality reduction, unsupervised learning clustering, and matrix factorisation [17].
MVDA analyses multi-omics data for the same set of samples and, if available, an initial samples stratification, and produces a multi-view clustering computed by taking into account: i) the sample stratification over all omics data layers, ii) the influence of the omics layer on each cluster and iii) the relevant omics features characterising each cluster. The first step of the MVDA analysis consists of reducing the dimensionality of the omics layers by clustering the features and extracting a representative prototype, such as the cluster centroid, for each group. These prototypes are used to cluster the samples in each omic layer. Eventually, a matrix-factorisation approach is used to combine the single view grouping into a multi-view clustering. If an initial sample stratification is available, a feature selection step on the prototype or a semi-supervised matrix factorisation can be also performed. A description of the steps and methods implemented in the MVDA methodology, and its comparison to other similar tools, are available in Tables S20 and S21.
2.13. MOSIM: Multi-omics data simulator
The ability of multi-view learning algorithms to take into account different omics data layers allows this class of algorithms to build more robust models of the biological system under study. To ease the development and debugging of new algorithms, it is important to rely on perfectly known ground-truth benchmark data. In the case of biological systems, this is not always possible, and to this purpose, MOSIM (Multi-Omics Simulator) has been proposed as a generator of synthetic multi-omics data based on graph theory and ordinary differential equations (Table S22) [18].
MOSIM can reproduce key characteristics of transcriptional and post-transcriptional regulatory networks topology, such as hierarchical modularity and the scale-free property of many real-life network systems. Moreover, the rate of concentration of transcripts is explicitly modelled. The strength of MOSIM is derived by the integration of these two aspects, specifically, the complex interaction patterns described by the modules in the network are reflected in the model of activity of each entity (gene or miRNA) which can produce complex behaviours such as cooperation, competition, and inhibition of regulatory entities acting on each node of the network. To the best of our knowledge, MOSIM is one of the few tools able to model multi-view entities such as mRNA, miRNA and transcription factors (Table S23).
3. Use of the Nextcast components
Toxicogenomics aims at linking the safety assessment of chemicals to the underlying biological mechanisms. However, this can pose multiple challenges, such as the identification of the best experimental design, a standardised way for data preprocessing, identification of the modelling methodologies that can be used for omics data, as well as concerns related to the robustness and quality of the results and their interpretation. Nextcast offers a flexible solution for tackling these problems. The modular structure allows the use of the tools independently or in combination to produce more complex pipelines that can turn raw data into scientific knowledge. Here, we provide examples of Nextcast pipelines able to answer specific biological questions.
3.1. Characterisation of the MOA of a compound
One of the key aspects addressed by toxicogenomics investigation is the characterisation of the mechanism of action (MOA) of a compound. The MOA comprises all the molecular alterations induced by a specific exposure. The characterisation of the MOA can be performed by comparing transcriptomics or epigenomics data between the sample groups and identifying the differences induced by the exposure.
In Fig. 2, we provide some possible approaches available in Nextcast for the investigation of the MOA. To ensure a robust and reproducible analysis the raw transcriptomics data need to be systematically preprocessed. This can be achieved through a well-established pipeline implemented in the eUTOPIA tool [14]. After an evaluation (visual and statistical) of the normalisation, batch effect removal, and quality control procedures, an annotated expression matrix can be generated. Moreover, pairwise comparisons between treatments or different conditions can be performed (e.g. treatment vs. control), generating a list of differentially expressed genes (DEGs).
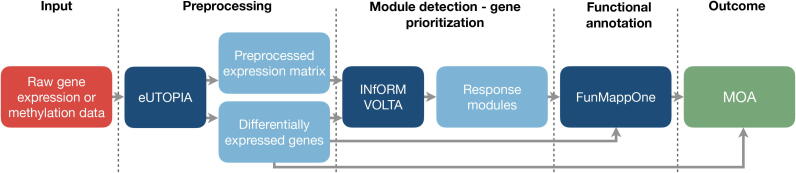
Fig. 2.
Nextcast pipeline for the characterisation of the MOA of a compound. Raw omics data is preprocessed with eUTOPIA. The output of the tool includes a matrix with normalised (and batch corrected) expression values and a list of differentially expressed genes. This data can be fed to INfORM to identify a set of responsive gene modules. VOLTA can be further used to analyse networks built with INfORM. Alternatively, differentially expressed genes can be directly provided as the input for the FunMappOne tool to perform enrichment analysis and identify the underlying biological processes. The result is a list of regulated genes and corresponding enriched pathways or regulated genes in co-expressed modules and their corresponding pathways. The red box represents the input for the pipeline while the green box describes the outcome of the pipeline. The dark blue boxes correspond to the individual Nextcast components of the “Analytics” category, and the light blue boxes indicate the intermediate outputs/inputs.
To grasp the systemic effects in the biological system, the biological activities and the molecular responses triggered by the chemical exposure should be investigated (e.g., immune system activation, changes in the metabolism, effects on the cell cycle, triggered apoptotic pathways). An easy-to-do characterisation of the MOA can be achieved by running FunMappOne [15], either directly with the set of DEGs, or after an intermediate step of prioritising gene modules with INfORM and VOLTA [16], [36]. Eventually, the enriched terms obtained from FunMappOne allow characterising the functional effects of the compound on a more systemic level. Furthermore, it is possible to investigate the specific key genes and their activation patterns (up-regulation/down-regulation) in the biological functions to further explore the MOA.
The suggested strategy has been successfully utilised in a wide range of applications ranging from the study of nickel-induced allergic contact dermatitis [29], copper oxide nanoparticles induced asthma [24], and the characterisation of the effects of ten carbon nanomaterials in three cell lines [76]. Moreover, the eUTOPIA pipeline has been widely applied to create harmonised transcriptomics data collections [28], [25]. FunMappOne, on the other hand, has proven to be an effective tool for comparing the pathway enrichment of different experimental conditions in multiple studies [37], [7]. The Nextcast components have also been used jointly to characterise the transcriptomic signature underlying atopic dermatitis [32]. Two sets of relevant genes involved in the disease were identified and functionally characterised and compared employing the FunMappOne visualisation, while INfORM was used to study the co-expression network and the corresponding modules of differentially expressed genes between lesional and non-lesional samples. Furthermore, in a recent study by Kinaret et al., eUTOPIA and FunMappOne have been successfully utilised to characterise the mechanism of toxicity of 28 distinct nanomaterials by interpreting the varying effects observed in mouse airways [27].
3.2. Using toxicogenomics in estimating relevant doses for a compound
The study of the dose–response relationship is one of the cornerstones of toxicology. It is used to observe the relationship of exposures and apical endpoints to determine safe, hazardous, beneficial and/or effective exposure levels of chemicals, drugs, and compounds. BMD analysis is a relevant tool in health risk assessment to identify the effective doses of compounds to trigger particular biological responses [10], [11], [77]. Furthermore, it is relevant to distinguish between the patterns of molecular alteration that are a direct consequence of the exposure from secondary effects resulting from genomic regulatory loops. The BMDx tool can be used to identify genes with expression patterns showing dose–response behaviour and estimate their active concentrations or benchmark doses [10]. In the case of experiments where multiple time-points are available, the TinderMIX tool can be instrumental in identifying genes showing a dynamic-dose dependent effect and estimate their PODs [12].
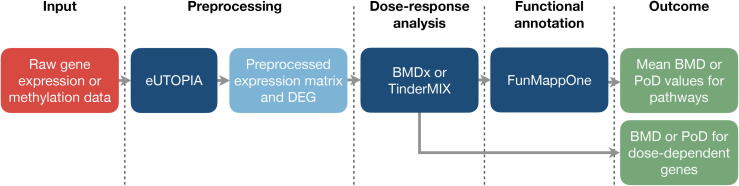
Fig. 3 provides a suggested pipeline for the dose–response analysis of toxicogenomics data using Nextcast. The combination of the tools allows a flexible approach from preprocessing to functional annotation of the dose-dependent features. BMDx can be particularly useful for gaining BMD values for each gene and mean BMD values for biological pathways [10], as well as for comparing multiple exposures. TinderMIX, on the other hand, can be used to obtain dynamic-dose dependent PODs for each gene [12]. Eventually, genes showing a relevant (time-) dose-dependency can be functionally annotated by FunMappOne, helping to understand the impact of a chemical [15].
Fig. 3.
Nextcast pipeline for the estimation of relevant doses of chemical exposure. Raw omics data can be preprocessed with eUTOPIA to obtain a matrix with normalised (and batch corrected) expression values and a list of differentially expressed genes. These data can be given in input to BMDx for a benchmark dose analysis or to TinderMIX to identify dynamic-dose responsive genes. Eventually, enrichment analysis can be conducted for the set of dose-dependent genes to identify the affected biological processes. The red box indicates the input for the pipeline, while the green boxes mark the output. The dark blue boxes are the individual Nextcast components of the ”Analytics” category, and the light blue box shows the intermediate output/input.
The strategy was recently applied for the systematic comparison of the gene expression and DNA methylation dynamic dose–response in a macrophage model after multi-walled carbon nanotube (MWCNT) exposure [5]. Gene expression and DNA methylation data were preprocessed and analysed by using eUTOPIA, while TinderMIX was used to identify dynamic dose-dependent features whose functionality was annotated and compared using FunMappOne.
3.3. Toxicogenomics and structural predictors
Early assessment of adverse effects induced by drugs or chemical exposures in humans is critical to avoid potential long-lasting harm. Moreover, the identification of valuable biomarkers from toxicogenomics data plays a central role in toxicity assessment, since they can be detected earlier than histopathological or clinical phenotypes. To this end, Nextcast provides multiple customisable pipelines (Fig. 4). The eUTOPIA tool supports the preprocessing of the raw data and produces an expression matrix and a ranked list of significantly altered genes between the exposed and control samples [14].
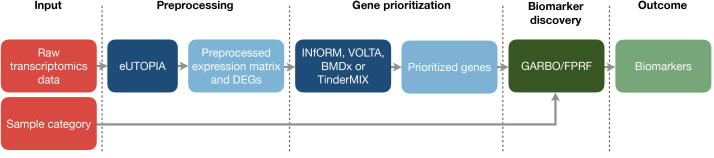
Fig. 4.
Nextcast pipeline for biomarker identification from toxicogenomics data. Raw omics data can be preprocessed with eUTOPIA. Preprocessed transcriptomics data can be provided as input to INfORM, VOLTA (after INfORM), BMDx, or TinderMIX to identify a set of biomarkers in a univariate way. The whole list of genes or only the prioritised set can be provided to the feature selection algorithm (GARBO or FPRF) to identify the smallest predictive set of biomarkers. The red boxes represent the input for the pipeline. The sample category is the variable of interest for the biomarker discovery phase. The lighter green box marks the output of the pipeline, dark blue and dark green boxes indicate the individual Nextcast components belonging to the ”Analytics” and ”modelling” categories, respectively. The light blue boxes represent the intermediate outputs/inputs.
These genes can be already considered markers of exposure since they represent the whole set of molecular alterations induced in the biological system. Alternatively, the most central genes involved in the processes can be identified in a gene co-expression network by using INfORM [16]. Alternatively, genes can be prioritised based on dose-dependency by the means of the BMDx or TinderMIX tools. To take into account the non-linear dependencies among expression levels, the univariate analysis of individual genes should be complemented by multivariate feature selection. The goal of feature selection is to express high-dimensional data with a low number of features to reveal significant underlying information and to identify a set of biomarkers for a particular phenotype. Nextcast has two feature selection methods available that can be used in this pipeline. One is FPRF, which is a random forest-based method that produces a ranking of the genes based on their discriminative power [19]. The other one is GARBO, which implements more advanced modelling based on a genetic algorithm that allows the modelling of non-linear correlation between candidate biomarkers and the phenotype of interest [20]. Both methods can be implemented to derive a reduced set of responsive genes, taking into account the predictivity with respect to the level of a toxic response. FPRF and GARBO can be run on the whole set of genes available in the data set or, to reduce their computational cost, they can be run on a prioritised set of genes that can be represented by: i) the differentially expressed genes identified with eUTOPIA, ii) the genes involved into relevant co-expression modules identified with INfORM or iii) the dynamic dose-dependent genes identified with BMDx or TinderMIX. The INfORM and GARBO methodologies were recently applied to identify candidate biomarkers to distinguish between irritant and allergic contact dermatitis [63]. INfORM was used to infer and compare co-expression networks of the two kinds of dermatitis. The GARBO methodology was then applied to optimise the number of relevant features to use when testing the accuracy of omics-based biomarker panels.
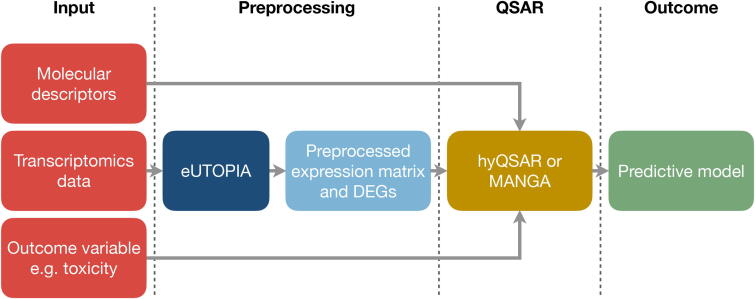
Another important aspect tackled down by toxicogenomics is the modelling of an outcome of interest, for example, chemical toxicity, starting from transcriptomics data from exposure experiments and chemical characteristics of the compounds, such as the PubChem CACTVS fingerprints, molecular descriptors and so on. This can be streamlined in Nextcast by combining the eUTOPIA and the hyQSAR or MaNGA modules. hyQSAR and MaNGA are two algorithms for QSAR modelling [21], [22]. The transcriptomics data is first fed to eUTOPIA producing an expression matrix (Fig. 5). hyQSAR and MaNGA are modules that can then be used to train predictive models for a variable of interest, such as chemical toxicity, by integrating toxicogenomics and cheminformatics data. Several aspects can dictate the choice of the predictive module (i.e. MaNGA or hyQSAR). Based on the dimensionality of the data set, hyQSAR may be preferred over MaNGA when the sample size is relatively small (e.g. less than 100 samples) since it learns a linear model and the only other hyper-parameter to estimate is the amount of regularisation. On the other hand, MaNGA may be preferred when the sample size is high since it is possible to learn more flexible models like Random Forests and SVMs, that usually require a higher amount of samples to reliably capture non-linear relationships and account for feature interactions at the expense of extensive hyper-parameters tuning and higher computational demands. Both approaches generate predictive models that are internally and externally validated according to the QSAR standards [21], [22].
Fig. 5.
Nextcast pipeline for biomarkers identification and QSAR models development from toxicogenomics and cheminformatics data. Raw omics data can be preprocessed with eUTOPIA. Then, the preprocessed transcriptomics data, chemical representation data, and the outcome variable can be provided to hyQSAR or MaNGA to identify the optimal predictive model. The red boxes indicate the input for the pipeline while the green box is the output. The dark blue and the yellow box are the individual Nextcast components, and the light blue box represents the intermediate output/input.
A similar strategy was used in a recent publication, where the hyQSAR tool was applied to build hybrid QSAR models for the prediction of the binding affinity to human serum albumin from transcriptomics data and molecular descriptors for a set of 57 drugs [22]. The developed model was compared with those identified only using the molecular descriptors, as in classical QSAR analysis. The results showed that the hybrid model had overall better predictive performances. Moreover, the model was also shown to be able to provide new avenues for the interpretation of chemical-biological interactions.
3.4. Multi-view clustering for chemical read-across
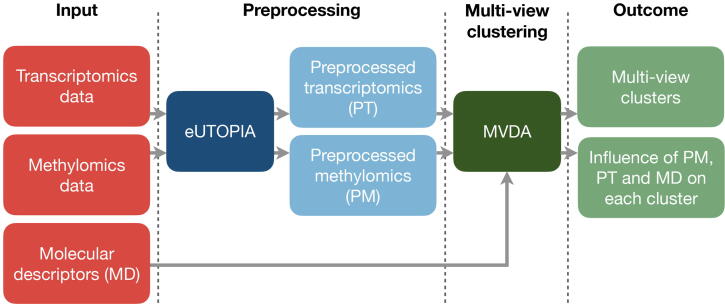
Multi-view learning and data integration strategies have become well-established methodologies in biomedical research where more comprehensive knowledge can be derived from the joint analysis of multiple data layers [78], [52], [79]. Multi-view learning, and in particular multi-view unsupervised clustering, is available in Nextcast through the use of the MVDA pipeline [17] (Fig. 6).
Fig. 6.
Nextcast pipeline with multi-view clustering for chemical read-across. Raw omics data can be preprocessed with eUTOPIA. The preprocessed multi-view data for the same samples and/or chemical structure data (e.g. molecular descriptors) can be fed to MVDA to obtain the multi-view cluster assignment of each sample and the influence of each view on the clustering. Red boxes indicate the input while the lighter green boxes mark the output of the pipeline. The dark blue and dark green boxes are the individual Nextcast components, and the light blue boxes correspond to the intermediate output/input.
An example of the application of MVDA is the read-across analysis of compounds based on their toxicogenomics and chemical characterisation. The use of computational strategies for hazard assessment is essential to reduce the time and costs of the safety assessment of compounds. Classical read-across-based approaches are based on the assumption that structurally similar compounds also have similar toxicokinetic and toxicodynamic properties [80]. Thus one can hypothesise that compounds with unknown properties will most likely behave in a manner that resembles the most structurally similar ones. A complementary approach can be based on the grouping of compounds based on toxicogenomics data where compounds inducing similar molecular alterations would be clustered together. More interestingly, intrinsic properties and toxicogenomics data can be integrated to obtain a more comprehensive clustering. This integrative clustering analysis can be performed with our MVDA tool, by using toxicogenomics (e.g. gene expression profiles, methylation data, etc.) signatures and structural data of chemical agents (e.g. binary fingerprints, molecular descriptors, etc.) as input.
If the user has omics data available in a raw data format, the eUTOPIA tool can be used to obtain their robust and effective preprocessing. Otherwise, the preprocessed omics data can be fed directly into the MVDA pipeline. The results of the analysis will be a grouping of the compounds based on both intrinsic properties and molecular alteration information and a score of the influence of each view on each final group.
MVDA was originally developed as a tool for patient subtyping from multi-omics data [17]. However, it is a general-purpose tool that can be used in different domains of applications. For example, Li et al. [46] applied it to perform a multi-view clustering of patients from medical imaging data by integrating histogram features from multi-parametric magnetic resonance imaging.
3.5. Interoperability of Nextcast data formats
Nextcast uses data representations that comply with well-accepted standardised formats [81] and offers a high degree of interoperability of its outputs with other external software (Table 2 and supplementary methods). As for the interoperability between the Nextcast components, some of the analytics tools require the expression data and the metadata table, describing the samples, to be manipulated and stored as spreadsheet files. Automatic conversion of the eUTOPIA outputs in a ready-to-use format for BMDx, INfORM and FunMappOne is provided in the eUTOPIA interface. In particular, the spreadsheet file required as input for the FunMappOne module can be generated by specifying which of the comparisons performed during the analysis should be included and how they are grouped. The gene expression matrix and the list of genes with log2-fold changes and p-values, required by INfORM for the generation of the networks, can be exported from eUTOPIA for each one of the comparisons. The user can choose to include all the genes present in the experimental data or to filter them by using only the genes that are differentially expressed in each comparison. Lastly, if preprocessing data with an experimental setup containing multiple doses and/or multiple time points, the data can be directly exported in a format ready for the BMDx tool. Other kind of data filtering, splitting or merging with external data sets needs to be manipulated either manually or through the use of customised scripts outside the Nextcast environment.
3.6. Example application of the Nextcast pipelines on real data
Toxicogenomics aims at linking the safety assessment of chemicals to the underlying biological mechanisms by means of omics data analysis [2], [3], [4]. In the last years, many datasets have been generated to characterise the molecular mechanism of action (MOA) of chemical exposure by transcriptomics profiling the exposed system. The FAIRness of the data sharing and reusing is a topic currently discussed by the scientific community [90], [91], [92]. The availability of well-reported standardised pipelines in Nextcast also support and increase the FAIRness of the data [91]. Analysis of toxicogenomic data generally consists in elucidating the MOA of exposure and to identifying related biomarkers. The most common approach is to characterise the MOA as the molecules that are significantly altered between the exposed and the control samples as shown in Fig. 2. More recently, particular relevance has been given to the dose dependent analysis of toxicogenomic data for the identification transcriptomic alterations with a monotonic patter with respect to increasing doses or concentrations. It could be speculated that these alterations can be used to dissect the direct effects of the exposure from other secondary regulatory circuits happening in the cells. Moreover, benchmark dose analysis allow to identify the reference doses at which particular cellular processes are altered [93]. This type of analysis can be easily performed in Nextcast as shown in Fig. 3. In the last decade, it has become clear that complex phenotypes are the results of the interactions of different molecules. Thus, biological network analysis has been successfully increasingly applied in toxicogenomic studies [94]. Markers of exposures can be identified by studying the gene co-expression network starting from transcriptomics data [4], [95]. For example, Nextcast offers the possibility to identify key genes associated to the exposures as those more central to the co-expression networks in terms of different topological properties (Fig. 2). In the following sections we showcase how the theoretical pipelines described in Figs. 2 can be applied to address the aforementioned points. We used toxicogenomics data derived from a dose-time exposure series of multi-walled carbon nanotubes (MWCNT) on THP-1 macrophages (data previously published in Saarimäki & Kinaret et al. [5], available on the NCBI Gene Expression Omnibus (GEO) database under the series accession number GSE146710). Detailed information on the analyses can be found in the supplementary methods.
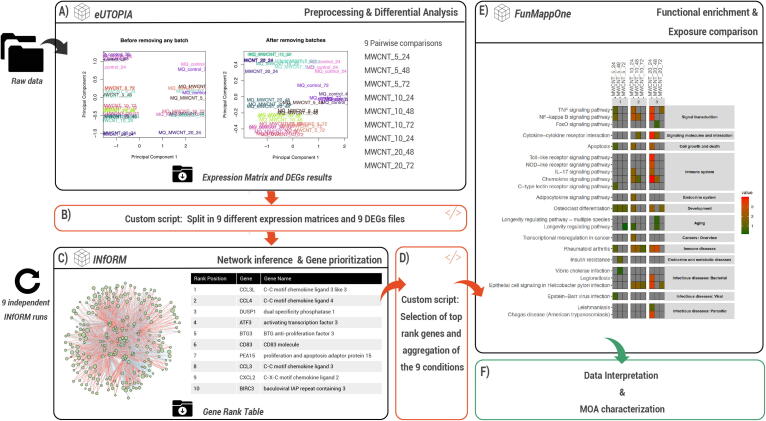
3.6.1. Characterisation of the MOA of MWCNT
Prioritising the most significant molecular perturbations is an effective way to characterise the MOA of a compound [95]. Here we showcase an example of MOA characterisation of MWCNT that first uses network based metrics to prioritise relevant genes and than characterise them by means of functional annotation (Fig. 2). The alternative strategy that performs directly functional annotation of the differentially expressed genes is shown in Figure S1. The pipelines start with the preprocessing of the data and the identification of the differentially expressed genes using eUTOPIA (Fig. 7A). After co-expression network inference, INfORM is able to prioritise the genes in the network based on both a consensus of centrality measures and the level of deregulation of the gene expression (Fig. 7C). Fig. 7C reports an example of gene rank obtained from the high dose and early time point MWCNT exposure. The data reported in the table highlights the prominent role of the immune response in the adaptation response, as well as the control of cell cycle and apoptosis. FunMappOne is able to summarise the functions of the relevant genes as an heatmap (Fig. 7E). As expected, the FunMappone output always presents the highest values of deregulation at 24 h, regardless of the dose, while the system gradually turned back towards homeostasis at 48 and 72 h, respectively. In detail, low and intermediate doses after 3 days of exposure, virtually showed the complete resolution of the inflammatory response as compared to day 1. Furthermore, the amplitude of the adaptation response increased with the dose. As expected, both inflammatory and pro-fibrotic pathways were up-regulated one day after all the exposures: TNF, NFB and IL-17, among the others, showed a consistent up-regulation that increased with the dose. NFB role in MWCNT molecular mechanism of toxicity has been extensively studied and is well accepted [96]. Similarly, IL-17 mediates protective innate immunity mechanisms against a plethora of pathogens, and is nowadays regarded a potential pivotal therapeutical target in inflammation pathogenesis [97], [98], [99], [100], [101], [102].
Fig. 7.
Example application of the characterisation of the MWCNT MOA employing INfORM. (A) eUTOPIA was used to preprocess input raw data and to perform differential analysis. The normalised expression matrix, as well as the lists of differentially expressed genes, were exported. (B) A custom script was used to select the most frequently deregulated 1,000 genes across the exposures and to produce inputs for INfORM. (C) INfORM was used to infer the gene co-expression networks and to rank the genes according to their topological properties. (D) The first 200 positions of each list were selected and combined in a format compatible with the FunMappOne input. (E) FunMappOne was used to perform enrichment analysis of the KEGG human pathways. (F) The output was interpreted for MOA characterisation of MWCNT exposures at different doses and time points.
3.6.2. Characterisation of the dose–response to MWCNT and identification of effective doses
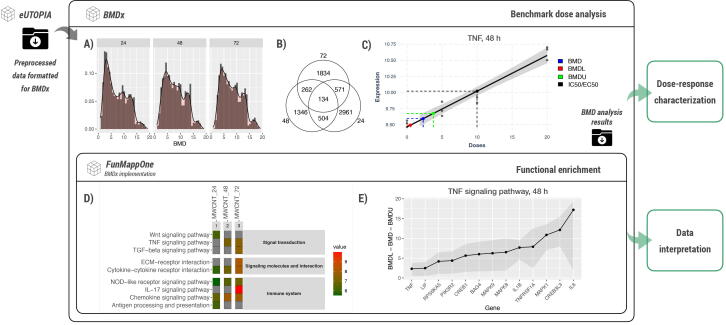
Benchmark dose analysis can help to distinguish the direct effects of an exposure from the indirect ones, as they are likely to show dose-dependent alteration. At the same time, understanding the point of departure, i.e. the dose at which the expression of a gene diverges from the steady state, can help in the estimation of safe or effective doses of controlled exposures. Here we showcase how the pipeline shown in Fig. 3 can elucidating the dose-dependent effects of MWCNT exposure. After preprocessing the data with eUTOPIA, the bechmark dose dependent analysis was performed by means of BMDx. As a result, distinct sets of dose-dependent genes were obtained for each time point (Fig. 8, Fig. 8B). Specifically, 4170, 2246 and 2801 genes were considered altered in a dose-dependent manner at 24 h, 48 h and 72 h, respectively (Fig. 8B). The results can be investigated through various visualisations, both at the level of individual genes as well as at the level of the gene sets at each time point with comparisons between them. Here, we showcase the distribution of the calculated BMD values at each time point (Fig. 8A), how these gene sets overlap (Fig. 8B) as well as the representation of the model fit on the gene TNF at 48 h (Fig. 8C). These results suggest that more genes are showing dose-dependent changes in their expression at 24 h as compared to later time points. Furthermore, the BMD values are generally lower at 24 h as compared to 48 and 72 h. The higher BMD values at later time points recapitulate the mechanisms observed in the previous network based example. At lower exposure doses, the system generally adapts and reaches homeostasis faster than at higher doses. Hence, the doses at which significant changes can be observed still at 48 h and 72 are higher than those at 24 h and before. The dose-responsive genes can be characterised by means of functional enrichment. A small selection of the enriched pathways is shown here for the purpose of clarity (Fig. 8D). For instance, the heatmap shows that the KEGG term “Cytokine-cytokine receptor interaction” is enriched at all instances with increasing mean BMD value at each time point. This value can be used as an estimation for the dose at which significant changes related to the biological function can be observed. Finally, the BMDL, BMD and BMDU values for the genes in a specific pathway (e.g., TNF signalling pathway in Fig. 8E) can be investigated.
Fig. 8.
Example application of the characterisation of the dose–response to MWCNT with BMDx. The preprocessed data were downloaded from eUTOPIA in a format compatible with the BMDx input. After completing the benchmark dose analysis, the results can be explored via various visual presentations. For example, (A) the distributions of the computed BMD values were compared between the time points. The BMD values computed at 24 h of exposure exhibit a higher peak at low doses compared to the later time points. (B) the Venn diagram indicates a larger number of dose-dependent genes at 24 h than at 48 and 72 h. (C) The best model for TNF with the computed BMD (blue), BMDL (red), BMDU (green) and IC/EC50 (green) values. (D) Selected pathways enriched in the functional enrichment indicate that the mean BMD values for distinct biological functions increase at later time points. The colour of the cell represents the mean BMD values of the genes enriching the pathway. (E) Line graph representing the genes enriching TNF signalling pathway at 48 h with their BMD, BMDL and BMDU values plotted.
4. Conclusions
Currently, a large amount of toxicogenomics data is available to the scientific community [103], [104], [25]. This data is used to answer different questions such as mechanism of action reconstruction, biomarker selection, evaluation of dose dependent alteration, inference of molecular co-alteration, which require complex and specific analytical strategies. Many modular and heterogeneous components may be strung together in novel ways to answer these research questions on an ever-growing size of experimental and simulated data sets. Abstracting the software from the underlying programming languages and execution environments improves both user’s experience and the scalability of workflows. It also allows integration of new workflow steps and even existing web services. Therefore, we developed the Nextcast software suite, which contains a wide variety of tools for comprehensive, easy-to-perform toxicogenomic data analysis. As scientific workflows usually involve multiple actors with different levels of involvement and technical expertise, Nextcast aims at catering to these actors with multiple entry points to the development of the data pipelines, and it guides users with diverse backgrounds in the evaluation of the workflows and their results. Nextcast is further designed to allow high flexibility in any type of analysis that needs to be performed while providing standardised pipelines and ensuring the compatibility between the provided tools. While these standardised pipelines compiled using the state-of-the-art methods are a step towards more robust and reproducible toxicogenomics, the importance of documentation of the decisions taken during the analytical steps should not be overlooked. Solely reporting the methods and parameters is often not enough to obtain full reproducibility. Instead, complete documentation and scientific justification of choices made during the experiment and data analysis is crucial for gaining trust in toxicogenomics derived evidence. In conclusion, Nextcast provides the needed, user-friendly infrastructure to make comparable, systematic toxicogenomic analysis, and thus it will be of great support to the scientific community, regulators, and stakeholders.
Funding
This research was funded by the EU H2020 projects NanoSolveIT (Grant No. 814572) and NanoinformaTIX (grant agreement No 814426), Academy of Finland (Grant No. 322761), and Novo Nordisk Foundation.
CRediT authorship contribution statement
Angela Serra: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Project administration. Laura Aliisa Saarimäki: Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Alisa Pavel: Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Giusy del Giudice: Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Michele Fratello: Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Luca Cattelani: Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft. Antonio Federico: Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Omar Laurino: Writing - original draft. Veer Singh Marwah: Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Vittorio Fortino: Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Giovanni Scala: Methodology, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Pia Anneli Sofia Kinaret: Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Dario Greco: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to Troy Faithfull for his critical comments on the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.csbj.2022.03.014.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Chepelev N.L., Moffat I.D., Labib S., Bourdon-Lacombe J., Kuo B., Buick J.K., Lemieux F., Malik A.I., Halappanavar S., Williams A., et al. Integrating toxicogenomics into human health risk assessment: lessons learned from the benzo [a] pyrene case study. Crit Rev Toxicol. 2015;45:44–52. doi: 10.3109/10408444.2014.973935. [DOI] [PubMed] [Google Scholar]
- 2.Kinaret P.A.S., Serra A., Federico A., Kohonen P., Nymark P., Liampa I., Ha M.K., Choi J.-S., Jagiello K., Sanabria N., et al. Transcriptomics in toxicogenomics, part i: experimental design, technologies, publicly available data, and regulatory aspects. Nanomaterials. 2020;10:750. doi: 10.3390/nano10040750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Federico A., Serra A., Ha M.K., Kohonen P., Choi J.-S., Liampa I., Nymark P., Sanabria N., Cattelani L., Fratello M., et al. Transcriptomics in toxicogenomics, part ii: preprocessing and differential expression analysis for high quality data. Nanomaterials. 2020;10:903. doi: 10.3390/nano10050903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serra A., Fratello M., Cattelani L., Liampa I., Melagraki G., Kohonen P., Nymark P., Federico A., Kinaret P.A.S., Jagiello K., et al. Transcriptomics in toxicogenomics, part iii: data modelling for risk assessment. Nanomaterials. 2020;10:708. doi: 10.3390/nano10040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saarimäki L.A., Kinaret P.A., Scala G., del Giudice G., Federico A., Serra A., Greco D. Toxicogenomics analysis of dynamic dose-response in macrophages highlights molecular alterations relevant for multi-walled carbon nanotube-induced lung fibrosis. NanoImpact. 2020;20 [Google Scholar]
- 6.Serra A., Letunic I., Fortino V., Handy R.D., Fadeel B., Tagliaferri R., Greco D. INSIdE NANO: a systems biology framework to contextualize the mechanism-of-action of engineered nanomaterials. Sci Rep. 2019;9:179. doi: 10.1038/s41598-018-37411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavel A., Del Giudice G., Federico A., Di Lieto A., Kinaret P.A., Serra A., Greco D. Integrated network analysis reveals new genes suggesting covid-19 chronic effects and treatment. Brief Bioinform. 2021;22:1430–1441. doi: 10.1093/bib/bbaa417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scala G., Marwah V., Kinaret P., Sund J., Fortino V., Greco D. Integration of genome-wide mRNA and miRNA expression, and DNA methylation data of three cell lines exposed to ten carbon nanomaterials. Data in Brief. 2018;19:1046–1057. doi: 10.1016/j.dib.2018.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinaret P.A.S., Del Giudice G., Greco D. Covid-19 acute responses and possible long term consequences: What nanotoxicology can teach us. Nano Today. 2020;35 doi: 10.1016/j.nantod.2020.100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serra A., Saarimäki L.A., Fratello M., Marwah V.S., Greco D. Bmdx: a graphical shiny application to perform benchmark dose analysis for transcriptomics data. Bioinformatics. 2020;36:2932–2933. doi: 10.1093/bioinformatics/btaa030. [DOI] [PubMed] [Google Scholar]
- 11.Phillips J.R., Svoboda D.L., Tandon A., Patel S., Sedykh A., Mav D., Kuo B., Yauk C.L., Yang L., Thomas R.S., Gift J.S., Davis J.A., Olszyk L., Merrick B.A., Paules R.S., Parham F., Saddler T., Shah R.R., Auerbach S.S. BMDExpress 2: enhanced transcriptomic dose-response analysis workflow. Bioinformatics. 2019;35:1780–1782. doi: 10.1093/bioinformatics/bty878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serra A., Fratello M., Del Giudice G., Saarimäki L.A., Paci M., Federico A., Greco D. Tindermix: Time-dose integrated modelling of toxicogenomics data. GigaScience. 2020;9:giaa055. doi: 10.1093/gigascience/giaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., Bouwman J., Brookes A.J., Clark T., Crosas M., Dillo I., Dumon O., Edmunds S., Evelo C.T., Finkers R., Gonzalez-Beltran A., Gray A.J.G., Groth P., Goble C., Grethe J.S., Heringa J., ’t Hoen P.A.C., Hooft R., Kuhn T., Kok R., Kok J., Lusher S.J., Martone M.E., Mons A., Packer A.L., Persson B., Rocca-Serra P., Roos M., van Schaik R., Sansone S.-A., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M.A., Thompson M., van der Lei J., van Mulligen E., Velterop J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., Mons B., et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marwah V.S., Scala G., Kinaret P.A.S., Serra A., Alenius H., Fortino V., Greco D. eutopia: solution for omics data preprocessing and analysis. Source Code Biol Med. 2019;14:1–12. doi: 10.1186/s13029-019-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scala G., Serra A., Marwah V.S., Saarimäki L.A., Greco D. Funmappone: a tool to hierarchically organize and visually navigate functional gene annotations in multiple experiments. BMC Bioinform. 2019;20:1–7. doi: 10.1186/s12859-019-2639-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marwah V.S., Kinaret P.A.S., Serra A., Scala G., Lauerma A., Fortino V., Greco D. Inform: Inference of network response modules. Bioinformatics. 2018;34:2136–2138. doi: 10.1093/bioinformatics/bty063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serra A., Fratello M., Fortino V., Raiconi G., Tagliaferri R., Greco D. Mvda: a multi-view genomic data integration methodology. BMC Bioinform. 2015;16:1–13. doi: 10.1186/s12859-015-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fratello M., Serra A., Fortino V., Raiconi G., Tagliaferri R., Greco D. A multi-view genomic data simulator. BMC Bioinform. 2015;16:1–15. doi: 10.1186/s12859-015-0577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortino V., Kinaret P., Fyhrquist N., Alenius H., Greco D. A robust and accurate method for feature selection and prioritization from multi-class omics data. PloS ONE. 2014;9 doi: 10.1371/journal.pone.0107801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortino V., Scala G., Greco D. Feature set optimization in biomarker discovery from genome-scale data. Bioinformatics. 2020;36:3393–3400. doi: 10.1093/bioinformatics/btaa144. [DOI] [PubMed] [Google Scholar]
- 21.Serra A., Önlü S., Festa P., Fortino V., Greco D. Manga: a novel multi-niche multi-objective genetic algorithm for qsar modelling. Bioinformatics. 2020;36:145–153. doi: 10.1093/bioinformatics/btz521. [DOI] [PubMed] [Google Scholar]
- 22.Serra A., Önlü S., Coretto P., Greco D. An integrated quantitative structure and mechanism of action-activity relationship model of human serum albumin binding. J Cheminformatics. 2019;11:38. doi: 10.1186/s13321-019-0359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serra A., Fratello M., Federico A., Ojha R., Provenzani R., Tasnadi E., Cattelani L., Del Giudice G., Kinaret P.A., Saarimäki L.A., et al. Computationally prioritized drugs inhibit sars-cov-2 infection and syncytia formation. Briefings Bioinform. 2022;23:bbab507. doi: 10.1093/bib/bbab507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kooter I., Ilves M., Gröllers-Mulderij M., Duistermaat E., Tromp P.C., Kuper F., Kinaret P., Savolainen K., Greco D., Karisola P., et al. Molecular signature of asthma-enhanced sensitivity to cuo nanoparticle aerosols from 3d cell model. ACS Nano. 2019;13:6932–6946. doi: 10.1021/acsnano.9b01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saarimäki L.A., Federico A., Lynch I., Papadiamantis A.G., Tsoumanis A., Melagraki G., Afantitis A., Serra A., Greco D. Manually curated transcriptomics data collection for toxicogenomic assessment of engineered nanomaterials. Sci Data. 2021;8:1–10. doi: 10.1038/s41597-021-00808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottman N., Barrientos-Somarribas M., Fyhrquist N., Alexander H., Wisgrill L., Olah P., Tsoka S., Greco D., Levi-Schaffer F., Soumelis V., et al. Microbial and transcriptional differences elucidate atopic dermatitis heterogeneity across skin sites. Allergy. 2021;76:1173–1187. doi: 10.1111/all.14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinaret P.A., Ndika J., Ilves M., Wolff H., Vales G., Norppa H., Savolainen K., Skoog T., Kere J., Moya S., et al. Toxicogenomic profiling of 28 nanomaterials in mouse airways. Adv Sci. 2021;8:2004588. doi: 10.1002/advs.202004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Federico A., Hautanen V., Christian N., Kremer A., Serra A., Greco D. Manually curated and harmonised transcriptomics datasets of psoriasis and atopic dermatitis patients. Sci Data. 2020;7:1–6. doi: 10.1038/s41597-020-00696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisgrill L., Werner P., Jalonen E., Berger A., Lauerma A., Alenius H., Fyhrquist N. Integrative transcriptome analysis deciphers mechanisms of nickel contact dermatitis. Allergy. 2021;76:804–815. doi: 10.1111/all.14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ndika J., Ilves M., Kooter I.M., Gröllers-Mulderij M., Duistermaat E., Tromp P.C., Kuper F., Kinaret P., Greco D., Karisola P., et al. Mechanistic similarities between 3d human bronchial epithelium and mice lung, exposed to copper oxide nanoparticles, support non-animal methods for hazard assessment. Small. 2020;16:2000527. doi: 10.1002/smll.202000527. [DOI] [PubMed] [Google Scholar]
- 31.Afantitis A., Melagraki G., Isigonis P., Tsoumanis A., Varsou D.D., Valsami-Jones E., Papadiamantis A., Ellis L.-J.A., Sarimveis H., Doganis P., et al. Nanosolveit project: Driving nanoinformatics research to develop innovative and integrated tools for in silico nanosafety assessment. Computat Struct Biotechnol J. 2020;18:583–602. doi: 10.1016/j.csbj.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Möbus L., Rodriguez E., Harder I., Stölzl D., Boraczynski N., Gerdes S., Kleinheinz A., Abraham S., Heratizadeh A., Handrick C., et al. Atopic dermatitis displays stable and dynamic skin transcriptome signatures. J Allergy Clini Immunol. 2021;147:213–223. doi: 10.1016/j.jaci.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Nymark P., Bakker M., Dekkers S., Franken R., Fransman W., García-Bilbao A., Greco D., Gulumian M., Hadrup N., Halappanavar S., et al. Toward rigorous materials production: new approach methodologies have extensive potential to improve current safety assessment practices. Small. 2020;16:1904749. doi: 10.1002/smll.201904749. [DOI] [PubMed] [Google Scholar]
- 34.Suojalehto H., Ndika J., Lindström I., Airaksinen L., Karvala K., Kauppi P., Lauerma A., Toppila-Salmi S., Karisola P., Alenius H. Transcriptomic profiling of adult-onset asthma related to damp and moldy buildings and idiopathic environmental intolerance. Int J Mol Sci. 2021;22:10679. doi: 10.3390/ijms221910679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma X., Meng Y., Wang P., Tang Z., Wang H., Xie T. Bioinformatics-assisted, integrated omics studies on medicinal plants. Briefings Bioinform. 2020;21:1857–1874. doi: 10.1093/bib/bbz132. [DOI] [PubMed] [Google Scholar]
- 36.Pavel A., Federico A., Del Giudice G., Serra A., Greco D. Volta: adVanced mOLecular neTwork analysis. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marttila S., Chatsirisupachai K., Palmer D., de Magalhães J.P. Ageing-associated changes in the expression of lncrnas in human tissues reflect a transcriptional modulation in ageing pathways. Mech Ageing Develop. 2020;185 doi: 10.1016/j.mad.2019.111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallud A., Delaval M., Kinaret P., Marwah V.S., Fortino V., Ytterberg J., Zubarev R., Skoog T., Kere J., Correia M., et al. Multiparametric profiling of engineered nanomaterials: Unmasking the surface coating effect. Adv Sci. 2020;7:2002221. doi: 10.1002/advs.202002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das S., Tamang J.P. Changes in microbial communities and their predictive functionalities during fermentation of toddy, an alcoholic beverage of india. Microbiol Res. 2021;248 doi: 10.1016/j.micres.2021.126769. [DOI] [PubMed] [Google Scholar]
- 40.Scala G., Delaval M.N., Mukherjee S.P., Federico A., Khaliullin T.O., Yanamala N., Fatkhutdinova L.M., Kisin E.R., Greco D., Fadeel B., et al. Multi-walled carbon nanotubes elicit concordant changes in dna methylation and gene expression following long-term pulmonary exposure in mice. Carbon. 2021;178:563–572. doi: 10.1016/j.carbon.2021.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhutia M.O., Thapa N., Shangpliang H.N.J., Tamang J.P. Metataxonomic profiling of bacterial communities and their predictive functional profiles in traditionally preserved meat products of sikkim state in india. Food Res Int. 2021;140 doi: 10.1016/j.foodres.2020.110002. [DOI] [PubMed] [Google Scholar]
- 42.Kharnaior P., Tamang J.P. Bacterial and fungal communities and their predictive functional profiles in kinema, a naturally fermented soybean food of india, nepal and bhutan. Food Res Int. 2021;140 doi: 10.1016/j.foodres.2020.110055. [DOI] [PubMed] [Google Scholar]
- 43.Ejigu G.F., Jung J. Review on the computational genome annotation of sequences obtained by next-generation sequencing. Biology. 2020;9:295. doi: 10.3390/biology9090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng I.S.L., Lumley T. Review of statistical learning methods in integrated omics studies (an integrated information science) Bioinform Biol Insights. 2018;12 doi: 10.1177/1177932218759292. 1177932218759292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galdi P, Napolitano F, Tagliaferri R. Consensus clustering in gene expression, in: International meeting on computational intelligence methods for bioinformatics and biostatistics, Springer; 2014. p. 57–67.
- 46.Li C., Wang S., Serra A., Torheim T., Yan J.-L., Boonzaier N.R., Huang Y., Matys T., McLean M.A., Markowetz F., et al. Multi-parametric and multi-regional histogram analysis of mri: modality integration reveals imaging phenotypes of glioblastoma. European Radiol. 2019;29:4718–4729. doi: 10.1007/s00330-018-5984-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad A., Fröhlich H. Integrating heterogeneous omics data via statistical inference and learning techniques. Genom Comput Biol. 2016;2:e32. [Google Scholar]
- 48.Ray B., Ghedin E., Chunara R. Network inference from multimodal data: a review of approaches from infectious disease transmission. J Biomed Inform. 2016;64:44–54. doi: 10.1016/j.jbi.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parimbelli E., Marini S., Sacchi L., Bellazzi R. Patient similarity for precision medicine: A systematic review. J Biomed Inform. 2018;83:87–96. doi: 10.1016/j.jbi.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Serra A., Galdi P., Tagliaferri R. Machine learning for bioinformatics and neuroimaging. Wiley Interdisc Rev Data Min Knowl Disc. 2018;8 [Google Scholar]
- 51.Mallik S, Maulik U, Tomar N, Bhadra T, Mukhopadhyay A, Mukherji A. Machine learning and rule mining techniques in the study of gene inactivation and rna interference, Modulating Gene Expression-Abridging the RNAi and CRISPR-Cas9 Technologies; 2019.
- 52.Serra A, Galdi P, Tagliaferri R. Multiview learning in biomedical applications, in: Artificial Intelligence in the Age of Neural Networks and Brain Computing, Elsevier; 2019. p. 265–280.
- 53.Mallik S., Zhao Z. Graph-and rule-based learning algorithms: a comprehensive review of their applications for cancer type classification and prognosis using genomic data. Briefings Bioinform. 2020;21:368–394. doi: 10.1093/bib/bby120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T., Shao W., Huang Z., Tang H., Zhang J., Ding Z., Huang K. Mogonet integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nature Commun. 2021;12:1–13. doi: 10.1038/s41467-021-23774-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu M., Yi H., Ma S. Vertical integration methods for gene expression data analysis. Briefings Bioinform. 2021;22:bbaa169. doi: 10.1093/bib/bbaa169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park S., Xu H., Zhao H. Integrating multidimensional data for clustering analysis with applications to cancer patient data. J Am Stat Assoc. 2021;116:14–26. doi: 10.1080/01621459.2020.1730853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaur H., Huggins D.R., Rupp R.A., Abatzoglou J.T., Stöckle C.O., Reganold J.P. Agro-ecological class stability decreases in response to climate change projections for the pacific northwest, usa. Front Ecol Evol. 2017;5:74. [Google Scholar]
- 58.Crabtree N.M., Moore J.H., Bowyer J.F., George N.I. Multi-class computational evolution: development, benchmark evaluation and application to rna-seq biomarker discovery. BioData Mining. 2017;10:1–18. doi: 10.1186/s13040-017-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa P.M., Fadeel B. Emerging systems biology approaches in nanotoxicology: Towards a mechanism-based understanding of nanomaterial hazard and risk. Toxicol Appl Pharmacol. 2016;299:101–111. doi: 10.1016/j.taap.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 60.Vijayakumar S., Conway M., Lió P., Angione C. Seeing the wood for the trees: a forest of methods for optimization and omic-network integration in metabolic modelling. Briefings Bioinform. 2018;19:1218–1235. doi: 10.1093/bib/bbx053. [DOI] [PubMed] [Google Scholar]
- 61.Mirza B., Wang W., Wang J., Choi H., Chung N.C., Ping P. Machine learning and integrative analysis of biomedical big data. Genes. 2019;10:87. doi: 10.3390/genes10020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scala G., Federico A., Fortino V., Greco D., Majello B. Knowledge generation with rule induction in cancer omics. Int J Mol Sci. 2020;21:18. doi: 10.3390/ijms21010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fortino V., Wisgrill L., Werner P., Suomela S., Linder N., Jalonen E., Suomalainen A., Marwah V., Kero M., Pesonen M., et al. Machine-learning–driven biomarker discovery for the discrimination between allergic and irritant contact dermatitis. Proc Nat Acad Sci. 2020;117:33474–33485. doi: 10.1073/pnas.2009192117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He J., Zhu G., Wang G., Zhang F. Oxidative stress and neuroinflammation potentiate each other to promote progression of dopamine neurodegeneration. Oxidative Med Cellul Longev. 2020;2020 doi: 10.1155/2020/6137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta G., Gliga A., Hedberg J., Serra A., Greco D., Odnevall Wallinder I., Fadeel B. Cobalt nanoparticles trigger ferroptosis-like cell death (oxytosis) in neuronal cells: Potential implications for neurodegenerative disease. FASEB J. 2020;34:5262–5281. doi: 10.1096/fj.201902191RR. [DOI] [PubMed] [Google Scholar]
- 66.Lambrinidis G., Tsantili-Kakoulidou A. Multi-objective optimization methods in novel drug design. Expert Opin Drug Discov. 2021;16:647–658. doi: 10.1080/17460441.2021.1867095. [DOI] [PubMed] [Google Scholar]
- 67.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g: Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucl Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., Matthews L., May B., Milacic M., Rothfels K., Shamovsky V., Webber M., Weiser J., Williams M., Wu G., Stein L., Hermjakob H., D’Eustachio P. The reactome pathway knowledgebase. Nucl Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucl Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schimek M.G., Budinská E., Kugler K.G., Švendová V., Ding J., Lin S. Topklists: a comprehensive r package for statistical inference, stochastic aggregation, and visualization of multiple omics ranked lists. Stat Appl Genet Mol Biol. 2015;14:311–316. doi: 10.1515/sagmb-2014-0093. [DOI] [PubMed] [Google Scholar]
- 72.Schimek M., Budinska E., Kugler K., Svendova V., Ding J., Lin S. Topklists: a comprehensive r package for statistical inference, stochastic aggregation, and visualization of multiple omics ranked lists. Stat Appl Genet Mol Biol. 2015:311–316. doi: 10.1515/sagmb-2014-0093. [DOI] [PubMed] [Google Scholar]
- 73.Marbach D., Costello J.C., Küffner R., Vega N.M., Prill R.J., Camacho D.M., Allison K.R., Kellis M., Collins J.J., Stolovitzky G. Wisdom of crowds for robust gene network inference. Nature Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schüttler A., Altenburger R., Ammar M., Bader-Blukott M., Jakobs G., Knapp J., Krüger J., Reiche K., Wu G.-M., Busch W. Map and model–moving from observation to prediction in toxicogenomics. GigaScience. 2019;8:giz057. doi: 10.1093/gigascience/giz057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 76.Scala G., Kinaret P., Marwah V., Sund J., Fortino V., Greco D. Multi-omics analysis of ten carbon nanomaterials effects highlights cell type specific patterns of molecular regulation and adaptation. NanoImpact. 2018;11:99–108. doi: 10.1016/j.impact.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ewald J., Soufan O., Xia J., Basu N. Fastbmd: an online tool for rapid benchmark dose–response analysis of transcriptomics data. Bioinformatics. 2021;37:1035–1036. doi: 10.1093/bioinformatics/btaa700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Serra A., Fratello M., Greco D., Tagliaferri R. Data integration in genomics and systems biology. 2016 IEEE Congress on Evolutionary Computation (CEC), IEEE. 2016;2016:1272–1279. [Google Scholar]
- 79.Nguyen N.D., Wang D. Multiview learning for understanding functional multiomics. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamon L., Aschberger K., Asturiol D., Richarz A., Worth A. Grouping of nanomaterials to read-across hazard endpoints: a review. Nanotoxicology. 2019;13:100–118. doi: 10.1080/17435390.2018.1506060. [DOI] [PubMed] [Google Scholar]
- 81.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. Minimum information about a microarray experiment (miame)–toward standards for microarray data. Nature Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 82.Van der Maaten L., Hinton G. Visualizing data using t-sne. J Mach Learn Res. 2008;9 [Google Scholar]
- 83.McInnes L, Healy J, Melville J. Umap: Uniform manifold approximation and projection for dimension reduction, arXiv preprint arXiv:1802.03426; 2018.
- 84.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(2019):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14 doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogris C., Helleday T., Sonnhammer E.L. PathwAX: a web server for network crosstalk based pathway annotation. Nucleic Acids Res. 2016;44:W105–W109. doi: 10.1093/nar/gkw356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., Jensen L.J., von Mering C. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucl Acids Res. 2020;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bastian M, Heymann S, Jacomy M. Gephi: An open source software for exploring and manipulating networks; 2009.
- 90.Jeliazkova N., Apostolova M.D., Andreoli C., Barone F., Barrick A., Battistelli C., Bossa C., Botea-Petcu A., Châtel A., Angelis I.D., Dusinska M., Yamani N.E., Gheorghe D., Giusti A., Gómez-Fernández P., Grafström R., Gromelski M., Jacobsen N.R., Jeliazkov V., Jensen K.A., Kochev N., Kohonen P., Manier N., Mariussen E., Mech A., Navas J.M., Paskaleva V., Precupas A., Puzyn T., Rasmussen K., Ritchie P., Llopis I.R., Rundén-Pran E., Sandu R., Shandilya N., Tanasescu S., Haase A., Nymark P. Towards FAIR nanosafety data. Nat Nanotechnol. 2021;16:644–654. doi: 10.1038/s41565-021-00911-6. [DOI] [PubMed] [Google Scholar]
- 91.Saarimäki L.A., Melagraki G., Afantitis A., Lynch I., Greco D. Prospects and challenges for FAIR toxicogenomics data. Nat Nanotechnol. 2021;17:17–18. doi: 10.1038/s41565-021-01049-1. [DOI] [PubMed] [Google Scholar]
- 92.Grafström R., Haase A., Kohonen P., Jeliazkova N., Nymark P. Reply to: Prospects and challenges for FAIR toxicogenomics data. Nat Nanotechnol. 2021;17:19–20. doi: 10.1038/s41565-021-01050-8. [DOI] [PubMed] [Google Scholar]
- 93.Thomas R.S., Allen B.C., Nong A., Yang L., Bermudez E., Clewell H.J., Andersen M.E. A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci. 2007;98:240–248. doi: 10.1093/toxsci/kfm092. [DOI] [PubMed] [Google Scholar]
- 94.Pavel A, Serra A, Cattelani L, Federico A, Greco D. Network analysis of microarray data, in: Methods in Molecular Biology, Springer, US, 2021, pp. 161–186. URL: 10.1007/978-1-0716-1839-4_11. doi: 10.1007/978-1-0716-1839-4_11. [DOI] [PubMed]
- 95.Kinaret P., Marwah V., Fortino V., Ilves M., Wolff H., Ruokolainen L., Auvinen P., Savolainen K., Alenius H., Greco D. Network analysis reveals similar transcriptomic responses to intrinsic properties of carbon nanomaterials in vitro and in vivo. ACS Nano. 2017;11:3786–3796. doi: 10.1021/acsnano.6b08650. [DOI] [PubMed] [Google Scholar]
- 96.He X., Young S.-H., Schwegler-Berry D., Chisholm W.P., Fernback J.E., Ma Q. Multiwalled carbon nanotubes induce a fibrogenic response by stimulating reactive oxygen species production, activating nf-kb signaling, and promoting fibroblast-to-myofibroblast transformation. Chem Res Toxicol. 2011;24:2237–2248. doi: 10.1021/tx200351d. [DOI] [PubMed] [Google Scholar]
- 97.Zenobia C., Hajishengallis G. Basic biology and role of interleukin-17 in immunity and inflammation. Periodontology. 2015;2000(69):142–159. doi: 10.1111/prd.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor E.S., Wylie A.G., Mossman B.T., Lower S.K. Repetitive dissociation from crocidolite asbestos acts as persistent signal for epidermal growth factor receptor. Langmuir. 2013;29:6323–6330. doi: 10.1021/la400561t. [DOI] [PubMed] [Google Scholar]
- 99.Wang X., Xia T., Ntim S.A., Ji Z., Lin S., Meng H., Chung C.-H., George S., Zhang H., Wang M., Li N., Yang Y., Castranova V., Mitra S., Bonner J.C., Nel A.E. Dispersal state of multiwalled carbon nanotubes elicits profibrogenic cellular responses that correlate with fibrogenesis biomarkers and fibrosis in the murine lung. ACS Nano. 2011;5:9772–9787. doi: 10.1021/nn2033055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palomäki J., Välimäki E., Sund J., Vippola M., Clausen P.A., Jensen K.A., Savolainen K., Matikainen S., Alenius H. Long, needle-like carbon nanotubes and asbestos activate the NLRP3 inflammasome through a similar mechanism. ACS Nano. 2011;5:6861–6870. doi: 10.1021/nn200595c. [DOI] [PubMed] [Google Scholar]
- 101.Meunier E., Coste A., Olagnier D., Authier H., Lefèvre L., Dardenne C., Bernad J., Béraud M., Flahaut E., Pipy B. Double-walled carbon nanotubes trigger IL-1b release in human monocytes through nlrp3 inflammasome activation. Nanomed: Nanotechnol Biol Med. 2012;8:987–995. doi: 10.1016/j.nano.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 102.Li R., Wang X., Ji Z., Sun B., Zhang H., Chang C.H., Lin S., Meng H., Liao Y.-P., Wang M., Li Z., Hwang A.A., Song T.-B., Xu R., Yang Y., Zink J.I., Nel A.E., Xia T. Surface charge and cellular processing of covalently functionalized multiwall carbon nanotubes determine pulmonary toxicity. ACS Nano. 2013;7:2352–2368. doi: 10.1021/nn305567s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Igarashi Y., Nakatsu N., Yamashita T., Ono A., Ohno Y., Urushidani T., Yamada H. Open tg-gates: a large-scale toxicogenomics database. Nucl Acids Res. 2015;43:D921–D927. doi: 10.1093/nar/gku955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ganter B, Snyder RD, Halbert DN, Lee MD. Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class-dependent effects using the drugmatrix database; 2006. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.