Abstract
The APOBEC family of cytidine deaminases is one of the most common endogenous sources of mutations in human cancer. Genomic studies of tumors have found that APOBEC mutational signatures are enriched in the HER2 subtype of breast cancer and are associated with immunotherapy response in diverse cancer types. However, the direct consequences of APOBEC mutagenesis on the tumor immune microenvironment have not been thoroughly investigated. To address this, we developed syngeneic murine mammary tumor models with inducible expression of APOBEC3B. We found that APOBEC activity induced antitumor adaptive immune responses and CD4+ T cell-mediated, antigen-specific tumor growth inhibition. Although polyclonal APOBEC tumors had a moderate growth defect, clonal APOBEC tumors were almost completely rejected, suggesting that APOBEC-mediated genetic heterogeneity limits antitumor adaptive immune responses. Consistent with the observed immune infiltration in APOBEC tumors, APOBEC activity sensitized HER2-driven breast tumors to anti-CTLA-4 checkpoint inhibition and led to a complete response to combination anti-CTLA-4 and anti-HER2 therapy. In human breast cancers, the relationship between APOBEC mutagenesis and immunogenicity varied by breast cancer subtype and the frequency of subclonal mutations. This work provides a mechanistic basis for the sensitivity of APOBEC tumors to checkpoint inhibitors and suggests a rationale for using APOBEC mutational signatures and clonality as biomarkers predicting immunotherapy response in HER2-positive breast cancers.
Keywords: APOBEC, mutational signatures, APOBEC3B, breast cancer, immunotherapy, checkpoint blockade, heterogeneity, T cell, CTLA-4
INTRODUCTION
More than 50 distinct mutational signatures have been identified in cancer genomes (1,2). These signatures are thought to reflect transient or ongoing exogenous and endogenous mutational processes that occur over the lifetime of normal cells and during tumor development. Single-base substitution (SBS) signature 2 is characterized by C-to-T transitions within the trinucleotide motif of TCW (where W represents adenine or thymine), and SBS signature 13 is defined by C-to-G transversions within the same TCW motif. Both signatures are attributed to the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family of cytidine deaminases that catalyze the deamination of cytosine to uracil on single-stranded DNA (2,3). Although APOBEC enzymes have evolutionarily conserved activity in the generation of antibody diversification and restriction of viruses and endogenous retrotransposons, their off-target mutagenic activity on the host somatic genome drives cancer genome instability (4). APOBEC-mediated mutational signatures have been detected in at least 22 different tumor types and are enriched in bladder, head and neck, cervical, and breast cancers (5). Importantly, nearly half of breast cancers exhibit kataegis hypermutation clusters (6). Among breast cancer subtypes, human epidermal growth factor receptor 2-positive (HER2+) breast tumors are reported to have the highest median levels of APOBEC signature enrichment (5).
Somatic mutations in cancer can give rise to unique mutant peptides that serve as immune-reactive neoantigens, allowing cytotoxic T cells to target tumor cells for elimination (7). Thus, work has focused on understanding the role of ongoing mutational processes in contributing to tumor immunogenicity and response to immunotherapies (8,9). Despite the prevalence of APOBEC mutational signatures in breast cancer, these tumors are traditionally thought to be poorly immunogenic or “cold”, with a modest tumor mutation burden (TMB)(2) and low tumor-infiltrating lymphocytes (TILs) relative to more immunogenic cancers (10,11). However, the initial trials of anti-PD-1 and anti-PD-L1 monotherapy in triple-negative breast cancer (TNBC) showed promising objective response rates of up to 19% (12,13). The combination of the PD-L1 antibody atezolizumab and nab-paclitaxel was approved for advanced PD-L1+ TNBC in 2019—the first FDA approval of immunotherapy for breast cancer—based on a slight improvement in progression-free survival (PFS) for all patients and a clinically meaningful improvement in overall survival (OS) for PD-L1+ patients (14). However, there has been conflicting support following this finding. There was no advantage for adding anti-PD-L1 to the more widely available paclitaxel, irrespective of PD-L1 status (15), but in a separate trial, anti-PD-1 and chemotherapy significantly improved PFS for PD-L1+ TNBC patients compared to chemotherapy alone (16). Reflecting the uncertain efficacy of checkpoint inhibitors in breast cancer, the combination of atezolizumab and nab-paclitaxel has been withdrawn for use in TNBC after additional follow-up revealed no improvement in PFS.
Importantly, checkpoint inhibitor clinical trials have been less successful for HER2+ breast cancer patients. In an initial trial for anti-PD-L1 monotherapy in metastatic breast cancer, there were no objective responses in the HER2+ subtype (17). When trastuzumab was combined with anti-PD-1 for HER2+ patients, responses ranged from 0%-15.2% and were dependent on PD-L1 status (18). Finally, the addition of anti-PD-L1 to the antibody-drug conjugate trastuzumab emtansine failed to improve PFS of pretreated HER2+ patients (19). However, APOBEC mutational signatures have yet to be investigated as a specific class of hypermutation that transforms an immunologically “cold” HER2+ breast tumor “hot”, rendering the tumor responsive to checkpoint inhibition.
Work on how mutational signatures impact tumor immunity has revealed several pieces of evidence potentially implicating APOBEC mutagenesis in immunotherapy response. APOBEC signatures are associated with a greater likelihood of response to immune checkpoint inhibition in non-small cell lung cancer (NSCLC)(20), head and neck cancer, bladder cancer (21), and in a small cohort of breast cancer patients (22). In a study using mouse models of TNBC, overexpression of the murine APOBEC3 ortholog sensitized tumors to checkpoint inhibitors (23). Overexpression of human APOBEC3B in a vaccine setting sensitized mouse melanomas to checkpoint inhibition (24). However, the direct consequences of APOBEC mutagenesis on the tumor immune microenvironment and tumor growth in the absence of checkpoint inhibitors have not been thoroughly explored. A mechanistic understanding of how APOBEC mutagenesis alters the tumor immune microenvironment would inform the use of immunotherapies for human tumors with APOBEC mutational signatures. Despite the high enrichment of APOBEC signatures in HER2+ breast cancer, no studies, to our knowledge, have investigated a role for APOBEC mutagenesis in conferring clinical benefit to checkpoint blockade in HER2+ breast cancer.
To address these questions, we developed a syngeneic, immunocompetent murine HER2-driven mammary tumor model with APOBEC activity. Using this model, we examined the consequences of APOBEC activity and genetic heterogeneity on tumor growth, investigated tumor-immune system interactions in APOBEC tumors, and assessed the therapeutic response of these tumors to checkpoint inhibitor therapy. Finally, we examined the relationship between APOBEC mutagenesis and adaptive immune response in human breast tumors.
MATERIALS AND METHODS
Cell line culture, assays, and treatments
All cell lines were grown at 37°C in 5% CO2. SMF cells were provided by Dr. Lewis Chodosh (University of Pennsylvania) and were cultured in Dulbecco’s Modified Eagle Medium (DMEM), 10% fetal bovine serum (FBS), 1% L-glutamine (Gibco 25030-081), 1% penicillin/streptomycin (Gibco 15140-122), and 5 μg/mL insulin (Gemini Bioproducts 700-112P). EMT6 cells were obtained from the American Type Culture Collection (ATCC) via the Duke Cell Culture Facility (CCF) and were cultured in Waymouth’s Medium 752/1, 15% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. NDLUCD cells (25) were a gift from Dr. Alexander Borowsky and were cultured in DMEM, 10% FBS, and 1% penicillin/streptomycin. BT474 cells were obtained from ATCC via the Duke CCF and were cultured in RPMI-1640, 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. SKBR3 cells were obtained ATCC via the Duke CCF and were cultured in DMEM, 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. HEK293T cells were obtained from ATCC via the Duke CCF and were cultured in DMEM, 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. SMF-A3B cells were selected in 1 μg/mL puromycin (Sigma P8833-10MG) and 1 mg/mL neomycin (G418, Sigma, 345810-1GM). EMT6-A3B cells were selected in 4 μg/mL puromycin and 1 mg/mL neomycin. NDLUCD-A3B cells were selected in 0.5 μg/mL puromycin and 1.5 mg/mL neomycin. Doxycycline (Research Products International, D43020-100.0) was added to the cell medium to induce the expression of A3B where specified at concentrations described. Cells were harvested for qRT-PCR, deaminase activity assay, or Western blot analysis. EMT6, BT474, and SKBR3 cell lines were authenticated using STR and tested for mycoplasma contamination by the Duke CCF within one year of use. All cell lines were passaged minimally and for fewer than 20 passages.
Cell viability assays on SMF, EMT6, and NDLUCD cells were performed using CellTiter-Glo (Promega) according to manufacturer instructions. Cells were plated at 2,000 cells per well in an opaque 96-well plate and treated with doxycycline (1 or 2 μg/mL, as indicated) on day 0. Doxycycline in cell medium was refreshed every 3 days.
Colony formation assays on SMF cells were performed by plating cells at 2,000 cells per 10-cm plate and cultured with doxycycline (1 or 2 μg/mL, as indicated) for 14 days. Doxycycline in cell medium was refreshed every 3 days. PBS was used to wash plates, and 0.5% crystal violet in 25% methanal was used to stain cell colonies for 5 minutes. The plates were dried overnight and scanned using a Canon flatbed scanner. Colonies were quantified using ImageJ Fiji.
To isolate single-cell clones from in vitro mutagenized SMF cells, SMF-A3B cells were cultured with 1 μg/mL doxycycline (dox) for 2 weeks and then dox was removed to downregulate A3B. Single-cell clones were derived by limiting dilution by plating APOBEC or control cells onto 96-well plates. Wells were visually inspected to confirm single cells. Wells with growing clones were expanded for subsequent analysis.
For immunofluorescence staining of adherent cells, 5x104 cells per well were plated on coverslips with 0.1% gelatin in a 24-well plate. 1 μg/mL doxycycline was added, and cells were cultured for 3 days prior to fixation in 4% paraformaldehyde. Coverslips were washed in PBS, permeabilized in 0.5% Triton-X 100 (Sigma, 93443), washed in PBS, and blocked in 3% BSA (Sigma, BP1600100) and 10% normal goat serum (Invitrogen, PCN5000) for 1 hour at room temperature. Coverslips were incubated with 1:800 HA-tag rabbit (Cell Signaling 3724S) primary antibody overnight at 4°C, washed, and incubated in with 1:500 goat anti-rabbit AF488 (Life Technologies A1103) secondary antibody for 1 hour at room temperature. Coverslips were then washed in PBS, stained with DAPI for 10 minutes, and mounted on slides with Prolong Gold (Thermo P36930). Slides were imaged Zeiss Axio Imager Widefield fluorescence microscope.
Plasmids and viral transduction
To generate dox-inducible A3B expression in murine cancer cell lines, a 2-vector system was utilized. pLVX-Tet-On Advanced plasmid containing the rtTA cassette was provided by Dr. Ann Marie Pendergast (Duke University). A pLenti-Tet-On-A3B plasmid containing the tetracycline-responsive human APOBEC3B gene (NM_004900.4) that is HA-tagged on the C-terminus was generated by VectorBuilder (Chicago, IL). The APOBEC3B gene contains an in-frame 66 basepair (bp) SV40 T-antigen intron sequence to disrupt transcription of the gene in E. coli for successful cloning, without introducing A3B-catalyzed mutations in the construct sequence. To generate the catalytically inactive mutant of A3B (E255Q), site-directed mutagenesis of the pLenti-Tet-On-A3B plasmid was performed by Genewiz (South Plainfield, NJ). Three million HEK293T cells were transfected with 9 μg psPAX2 and 3 μg pMDG.2 packaging plasmids (gifts from Didier Trono, EPFL, Lausanne, Switzerland; Addgene plasmids 12559 and 12660), 12 μg of the lentiviral expression plasmid, PLUS reagent (Thermo 11514015), and Lipofectamine 2000 (Thermo 11668019). 0.8 mM sodium butyrate was added to cell medium 1 and 2 days post-transfection to prevent epigenetic silencing of the lentiviral vector. Lentivirus was collected in the supernatant and filtered prior to concentrating with Lenti-X™ Concentrator (Clontech 631231), as per the manufacturer’s protocol.
To generate SMF-A3B, EMT6-A3B, and NDLUCD-A3B cell lines, SMF, NDLUCD, and EMT6 cells were transduced at 50% confluency in 6-well plates with 1 mL of concentrated lentivirus and 6 μg/mL polybrene (Sigma 107689) at 1000 x g, 33°C for 2 hours. Cells transduced with pLVX-Tet-On Advanced lentivirus were selected in neomycin for at least 10 days. Cells were then transduced with pLenti-Tet-On-A3B lentivirus and selected in puromycin for an additional 14 days.
Animal models
Animal care and animal experiments were performed with the approval of, and in accordance with, guidelines of the Duke University IACUC. Mice were housed under barrier conditions with 12-hour light/12-hour dark cycles. Female FVB mice (FVB/NJ; used with SMF and NDLUCD cells) and female BALB/c mice (BALBc/J; used with EMT6 cells) were obtained from The Jackson Laboratory. Female outbred athymic nude mice (J:NU) and female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from The Jackson Laboratory.
Tumor cell lines were implanted in bilateral 4th inguinal mammary fat pads of 6-8 week old female recipient mice. 2x106 SMF-A3B cells, 5x105 NDLUCD-A3B cells, or 2x104 EMT6-A3B cells in complete cell medium were used for implantation. Tumors were monitored for growth, measured using calipers 2-3 times per week, and sacrificed when tumors reached 10-15 mm in diameter or at experimental endpoint, e.g. when tumors ulcerated or impaired mobility. Tumors and tumor-draining lymph nodes were collected from all mice. Tumor volume was calculated using (L*W*W*π)/6, where L is length of the longer side and W is length of the shorter side. Where indicated, 1 mg/mL of doxycycline supplemented with 5% sucrose was added to mouse drinking water 2 days prior to tumor cell implantation.
For re-challenge experiment, 2x106 cells from clones of in vitro mutagenized SMF-A3B cells were implanted in bilateral 4th inguinal mammary fat pads of 6-8 week old female recipient mice. 30 days later, 2x106 control SMF cells were implanted in the contralateral mammary fat pad as a re-challenge. Tumor onset in re-challenged mice was monitored using calipers 2-3 times per week and compared to control cells implanted in naïve mice.
In vivo depletion antibodies were administered via intraperitoneal injected on day -2 and -1 prior to implantation, then continued twice weekly until endpoint. 300 μg of anti-CD8 (BioXCell BE0117), or 300 μg of anti-IgG2b isotype control (BioXCell BE0090), was used for CD8 depletion alone. 200 μg of anti-CD8 (BioXCell BE0117) and 200 μg of anti-CD4 (BioXCell BE0003-1), or 400 μg of anti-IgG2b isotype control (BioXCell BE0090), was used for CD8/CD4 dual depletion. For anti-PD-1 monotherapy, antibodies were administered when the majority of tumors reached 5 mm in diameter, for a total of 3 doses in one week (day 13, 15, 17) and 3 doses in the next week (day 20, 22, 24). 200 μg of anti-PD-1 (BioXCell BE0146), or 300 μg of anti-IgG2b (BioXCell BE0090), was used for monotherapy. For combination anti-PD-1/anti-CTLA-4 therapy, anti-CTLA-4 monotherapy, and combination anti-CTLA-4/anti-HER2 therapy, antibodies were administered when the majority of tumors reached 5 mm in diameter and continued twice weekly until endpoint. The following concentrations were used for anti-PD-1/anti-CTLA-4 combination therapy: 200 μg of anti-PD-1 (BioXCell BE0146) and 200 μg of anti-CTLA-4 (BioXCell BE0164), or 400 μg of anti-IgG2b isotype control (BioXCell BE0090). The following concentrations were used for anti-CTLA monotherapy and anti-CTLA-4/anti-HER2 combination therapy: 200 μg of anti-CTLA-4 (BioXCell BE0164), 100 μg of anti-HER2/Neu (BioXCell BE0277), or 200 μg of anti-IgG2b isotype control (BioXCell BE0090). Responses to therapy were measured by the percent change in tumor volume from treatment start until endpoint. Only palpable tumors at treatment start were considered. We defined a complete response (CR) as a full tumor regression (−100% change in tumor volume from the treatment start) and a partial response (PR) as any reduction in tumor volume from the treatment start.
Flow cytometry
Bilateral tumors were harvested and aggregated for each mouse, then minced into small chunks. Tumor chunks were digested with warmed digestion buffer (DMEM with 5% FBS and 50 ug/mL gentamicin (Sigma G1397)) containing 300 U/mL collagenase (StemCell 554656) and 100 U/mL hyaluronidase (StemCell 554656) at 37 °C for 1 hour, with vortexing every 15 minutes. Digested tumors were incubated in ACK lysis buffer (150 mM NH4Cl (Sigma 213330), 10 mM KHCO3 (Sigma 60339), 0.1 mM EDTA (Sigma E9884)) for 5 minutes to lysis red blood cells. Tumors were centrifuged, washed in stain buffer (BD Biosciences 554656), decanted, and resuspended in Dispase II (5 mg/mL; StemCell 7913) and DNase I (100 μg/mL; Worthington Biochemical LS002006) for 5 minutes, with mixing. Tumors were then passed through 70 μm strainer, washed in stain buffer, counted, and 1x106 cells in 100 μL of stain buffer were added to 96-well untreated v-bottom plates for staining. Prior to intracellular antigen staining, cells were activated using 2 μL of leukocyte activation cocktail with GolgiPlug (BD Biosciences 550583) for 3 hours at 37°C and 5% CO2.
LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (Thermo L34957) was used to stain dead cells in PBS, according to manufacturer protocol, for 30 minutes at 4°C in the dark. Cells were washed in PBS three times and resuspended in 100 μL of PBS for antibody surface staining. 2 μL of CD16/CD32 Fc Block antibody (BD Biosciences 553141) was added for 10 minutes at 4°C in the dark. Surface antigen antibodies were added at dilutions listed below and incubated for 30 minutes at 4°C in the dark. Cells were washed in PBS and transferred to falcon tubes for analysis.
For intracellular antigen staining, cells were fixed in either Foxp3 Fixation Buffer (BD Biosciences 560409) or BD Cytofix™ Fixation Buffer (BD Biosciences 554655) for 30 minutes at 4°C in the dark. Cells were washed and stored at 4°C in the dark overnight. Cells were permeabilizated in either Foxp3 permeabilization buffer (BD Biosciences 560409) for 30 minutes at 37°C or BD perm/wash buffer for 15 minutes at 4°C. Cells were washed in PBS and resuspended in 100 μL of PBS for intracellular antigen staining using the antibody dilutions listed below and incubated for 25 minutes at room temperature in the dark. Cells were washed in PBS and transferred to falcon tubes for analysis.
Cells were analyzed using a FACSCanto analyzer (BD Biosciences), and data were analyzed using FlowJo software (TreeStar, Ashland, OR). Fluorescence minus one (FMO; all antibodies in the panel, except for one) was used to determine proper gating of individual cell types. Individual cell type compartments were represented as either the percentage of total CD45+ cells or the percentage of total live cells. Regulatory T cell (Treg) and type-2 T helper (Th2) cell compartments were represented as percentage of total CD4+ T cells. The correlation between immune cell frequency and tumor volume was calculated using the mean volume of bilateral tumors at endpoint.
Fluorophore-conjugated antibodies used: PD-L1 (CD274)–BV421 (Clone MIH5, BD, 564716, 1:100); PD-1 (CD279)–AF647 (Clone RMP1-30, BD, 566715, 1:20); CD11c–PECy7 (Clone HL3, BD, 558079, 1:100); MHC-II I-A I-E–AF488 (Clone M5/114.15.2, BD, 562352, 1:100); CD103–PE (Clone M290, BD, 561043, 1:100); EpCAM (CD326)–FITC (Clone G8.8, Biolegend, 118207, 1:500); CD45–PECy5 (Clone 30-F11, BD, 561870, 1:200); CD45–PECy7 (Clone 30-F11, BD, 552848, 1:200); CD45–APC (Clone 30-F11, BD, 561870, 1:200); CD45–V450 (Clone 30-F11, BD, 560501, 1:200); CD45–PE (Clone 30-F11, BD, 561087, 1:500); CD45–PerCP-Cy5.5 (Clone 30-F11, BD, 550994, 1:200); F4/80–AF647 (Clone T45–2342, BD, 565853, 1:50); CD11b-PE (Clone M1/70, BD, 561689, 1:50); CD11b–PECy7 (Clone M1/70, BD, 561098, 1:100); CD11b–APCCy7 (Clone M1/70, BD, 557657, 1:100); NK1.1–APC (Clone PK136, BD, 561117, 1:100); CD3e–PECy7 (Clone 145–2C11, BD, 561100, 1:100); CD3e–PE (Clone 145–2C11, BD, 561824, 1:100); CD3e–PerCP-Cy5.5 (Clone 145–2C11, BD, 561108, 1:100); CD4–APCCy7 (Clone GK1.5, BD, 561830, 1:100); CD8a–APC (Clone 53–6.7, BD, 561093, 1:200); CD8a–AF488 (Clone 53–6.7, BD, 557668, 1:100); Foxp3–AF488 (Clone MF23, BD, 560407, 1:200); Tbet–BV421 (Clone O4–46, BD, 563318, 1:100); Gata3–AF647 (Clone L50–823, BD, 560068, 1:200); IL4–APC (Clone 11B11, Biolegend, 504105, 1:100); granzyme B–FITC (Clone GB11, BD, 515403, 1:500; IFNy–BV421 (Clone XMG1.2, BD, 563376, 1:40); CD16/CD32 Fc Block (Clone 2.4G2, BD, 553141, 1:50).
qRT-PCR and Western blotting
RNA was extracted, cDNA generated, and gene expression level determined by qRT-PCR as previously described in (26). Briefly, 2 μg of RNA was reverse transcribed using ImProm-II Reverse Transcription System (Promega, A3800) with random primers. 2 μL of cDNA was amplified with TaqMan Gene Expression Master Mix (Thermo 4369016) on a Biorad CFX384 instrument in triplicate using the following Taqman probes (Thermo 4331182) were used: APOBEC3B, Hs00358981_m1; ACTB, Hs01060665_g1; Actb, Mm02619580_g1; Gzma, Mm01304452_m1; Tbx21, Mm00450960_m1; Prf1, Mm00812512_m1. mRNA expression was normalized to β-actin expression and presented as the relative fold-change. To compare A3B expression among murine cell lines (SMF-A3B, EMT6-A3B, and NDLUCD-A3B) and human cell lines (BT474), A3B expression was not normalized to account for differences in β-actin expression between mouse and human cells; fold-change of relative Ct value is presented.
To confirm CD8 depletion in SMF tumors, RNA was extracted, cDNA generated, and gene expression was determined by qRT-PCR, as described above, using a Cd8a Taqman probe (Thermo Mm01182107_g1).
For Western blotting of SMF, EMT6, and NDLUCD cells, cells were treated doxycycline as described and harvested. Cells were lysed in RIPA buffer (10 mM Tris-Cl pH 8 (Sigma 252859), 150 mM NaCl (Sigma S9888), 0.1% SDS (Sigma L3771), 0.5% NaDOC (Sigma D6750), 1% NP-40 alternative (Calbiochem 492016)) and 1x Halt Proteinase/Phosphatase Inhibitor (Invitrogen 78444). Protein concentration in the supernatant was determined by Bradford assay (Biorad 5000201). Laemmli Sample Buffer (BioRad 1610747) was added to diluted protein samples and denatured at 95°C for 5 minutes. 20 μg of denatured protein was loaded into wells of 10-15% SDS-PAGE gel and ran at 90-125 V for 1 hour. Gels were transferred to a nitrocellulose membrane (Biorad 1620115) using wet transfer at 90 V for 1 hour. Membranes were incubated with blocking buffer (5% nonfat milk (Biorad) in PBS/T) for 1 hour at room temperature, and then primary antibodies at the dilutions listed below overnight at 4°C. Membranes were washed in PBS-Tween 20 and incubated with secondary antibodies at the dilutions listed below for 1 hour at room temperature in the dark. Membranes were then washed and imaged using a Li-Cor Odyssey infrared imaging system and analyzed in ImageStudio Lite software (Li-Cor Biosciences).
Antibodies used for immunoblotting: HA-tag Rabbit (Cell Signaling, 3724S, 1:1000); α-Tubulin Mouse (Cell Signaling, 3873, 1:2000); γH2AX (Ser139) Rabbit (Cell Signaling, 2577S, 1:1000); H2A Mouse (Cell Signaling, 3636S, 1:1000); Cleaved PARP (Asp214) Mouse (Cell Signaling, 9544S, 1:1000); Goat anti-Mouse IRDye800 (Li-Cor, 926-32210, 1:5000); Goat anti-Rabbit AF680 (Thermo, A-21076, 1:5000).
Immunohistochemistry (IHC) and immunofluorescence (IF)
SMF tumors were harvested and fixed in 10% normal formalin overnight before paraffin-embedding for immunohistochemistry by Duke Pathology Research Immunohistology Lab (Duke University, Durham, NC). Slides were stained for CD3 or CD45 and imaged at 4 fields-of-view per tumor with Zeiss Axio Imager Widefield fluorescence microscope.
SMF tumors were harvested and frozen in Tissue-Tek OCT Compound (VWR 25608-930) for immunofluorescence staining. Slides were fixed in 4% paraformaldehyde for 10 minutes, washed in PBS, permeabilized in 0.5% Triton-X 100 for 20 minutes, washed in PBS, and blocked in 3% BSA and 10% normal goat serum for 1 hour at room temperature. Slides were incubated with the primary antibodies listed below overnight at 4°C, washed, and incubated in with the secondary antibodies listed below for 1 hour at room temperature. Slides were then washed in PBS, stained with DAPI for 10 minutes, and coverslips were mounted on slides with Prolong Gold (Thermo P36930). For γH2AX foci quantification, 8 fields-of-view were imaged per slide with Leica SP5 Inverted Confocal fluorescence microscope. For assessing expression of HA-tagged A3B in tumors, slides were imaged with Zeiss Axio Imager Widefield fluorescence microscope. Images were analyzed with Image J Fiji. Images were thresholded manually for each stain using Fiji and using positive and negative controls to set the threshold. The same threshold was used for analysis of each image in an experiment. CD8 IHC analysis used the Colour Deconvolution function with H DAB setting, followed by thresholding. Total cells and cells positive for staining were each counted with the Analyze Particles function. The number of positive cells was expressed as a percent of total cells per FOV. For γH2AX IF, nuclei were identified using the DAPI channel and Analyze Particles was used to count total cells. The Find Maxima function was used to count number of foci per FOV, and data were expressed as total number of foci per FOV divided by total cells per FOV.
Antibodies used for histology: CD3 Rabbit (Thermo, RM-9107-S, 1:100 (IHC)); CD45 Rat (BD Biosciences, 550939, 1:50 (IHC)); HA-tag Rabbit (Cell Signaling, 3724S, 1:800 (IF)); γH2AX (Ser139) Rabbit (Cell Signaling, 2577S, 1:800 (IF)); Goat anti-Rabbit AF488 (Life Technologies, A1103, 1:500 (IF)).
Cytidine deaminase activity assay
SMF, EMT6, or NDLUCD cells were treated with doxycycline as described and harvested. Cells were lysed for 10 minutes on ice in 25 mM HEPES (pH 7.4, diluted in molecular grade water, Sigma H3375), 10% glycerol, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM MgCl2, 1 mM ZnCl2, and 1:100 protease inhibitor (Sigma P8340). Protein concentration in supernatants was determined using the DC™ Protein Assay (BioRad) according to the manufacturer’s protocol. 10 μg of protein was incubated for 2 hours at 37°C with 4 pmol of oligonucleotides listed below (Integrated DNA Technologies, Coralville IA), 0.5 μL of uracil DNA glycosylase enzyme (NEB M0280S), 2 μL of 10x uracil DNA glycosylase buffer (NEB M0280S), 2.5 μL RNase A (Thermo EN0531) up to a 20 μL reaction volume with molecular grade H20. Then 10 μL of 1N NaOH was added and heated to 95°C for 10 minutes to break the DNA backbone. 30 μL of 2x RNA loading dye was then added and heated to 95°C for 3 minutes to denature the DNA. 15% Urea-TBE-PAGE gel was made with 3.75 mL of 40% Acryl (29:1), 4.8 g of ultra-pure urea, 1 mL of 10x TBE buffer, 5.25 mL of H20, 99 μL of 10% APS, and 4 μL of TEMED. 15% Urea-TBE-PAGE gel was prewarmed for 1 hour at 150 V. 5 μL of denatured sample was added per well and ran at 150 V for 30-45 minutes. Gels were imaged with Li-Cor Odyssey infrared imaging system and analyzed in ImageStudio Lite software (Li-Cor Biosciences) to quantify the percent of deamination.
Oligonucleotide containing cytosine: /5IRD700/ATTATTATTATTCAAATGGATTTATTTATTTATTTATTTATTT
Positive control oligonucleotide containing uracil: /5IRD700/ATTATTATTATTUAAATGGATTTATTTATTTATTTATTTATTT
RNA-sequencing and analysis
RNA was isolated from SMF tumors using the RNeasy kit (Qiagen). RNA was sequenced using Stranded mRNA-seq libraries and the NovaSeq 6000 S1 sequencing platform with 50 bp paired-end reads by the Duke GCB Sequencing and Genomic Technologies Shared Resource (Duke University, Durham, NC).
RNA-seq data was trimmed with Trim Galore! (Galaxy Version 0.6.3; Krueger, F., Babraham Institute, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and then FastQC (Galaxy Version 0.72+galaxy1; Andrews, S. (n.d.). FastQC A Quality Control tool for High Throughput Sequence Data. Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)) was used to assess quality. Reads were aligned to the GRCm38 reference mouse genome using RNA STAR (Galaxy Version 2.7.5b)(27) and vM25 annotation file downloaded from the Gencode server. Reads were counted with featureCounts (Galaxy Version 1.6.4+galaxy2)(28) and differential gene expression analysis was performed with DESeq2 (Galaxy Version 2.11.40.6+galaxy1)(29).
Gene ontology (GO) analysis was performed using GO Ontology database (DOI: 10.5281/zenodo.4033054 Released 2020-09-10)(30,31) for the log2 fold-change of genes, with a FDR adjusted p-value < 0.05. Gene set enrichment analysis (GSEA) was performed using GSEA v4.1.0(32,33) using preranked gene list and Hallmark v7.2 gene set database at 1,000 permutations.
T cell receptor (TCR)-sequencing and analysis
RNA was isolated from SMF tumors using the RNeasy kit (Qiagen), and TCRβ chain libraries were generated using SMARTer Mouse TCR a/b Profiling Kit (Clontech). Samples were pooled to a final pool concentration of 4 nM and diluted to a final concentration of 13.5 pM, including a 5–10% PhiX Control v3 spike-in. Libraries were sequencing using MiSeq600 v3 300 bp paired-end reads. MiXCR (34) was used to calculate clonotype frequencies with recommended settings and vegan R package (vegan: Community Ecology Package. R package version 2.5-7. https://CRAN.R-project.org/package=vegan) was used to calculate Shannon entropy diversity index. The diversity evenness 50 (DE50) was calculated as the number of clonotypes occupying the top 50% of read counts, divided by the total number of read counts.
Bioinformatics analysis of human breast cancers
We analyzed breast tumors from TCGA for which both whole-exome sequencing (WES) and RNA-sequencing data were available. To estimate immune cell infiltration from RNA-seq data, we quantified the gene expression of individual immune checkpoint genes or immune cell gene signatures (35,36) collapsed into one value for each signature using a PCA-based method (gene list in Supplementary Table S1). Raw gene counts from RNA-seq experiments for TCGA-BRCA (breast carcinoma) patients were first queried from the National Cancer Institute Genomic Data Commons (GDC)(37) using the R package TCGAbiolinks (v2.12.6)(38), and normalized to effective library sizes calculated by the Trimmed Mean of M-values (TMM)(39) method and transformed by the voom method (40) implemented in the R packages edgeR (v3.28.0) (41) and Limma (v3.42.0) (42), respectively. For each gene signature, the first principal component (PC1) of a PCA model was used to summarize the gene expression values of the signature into a single score.
To calculate APOBEC enrichment score from WES data, single nucleotide polymorphisms (SNP) data called by the somatic mutation caller MuTect2 (43) were also queried from the GDC. For each tumor, an APOBEC mutagenesis enrichment score was calculated based on C>T mutations occurring in TCW motifs as described by Roberts et al (5). An APOBEC enrichment score of 2 was used to distinguish tumors with high vs low APOBEC mutagenesis. Quantitative estimates of APOBEC mutagenesis and immune cell infiltration within individual tumors is shown in Supplementary Table S2.
Heatmaps of the relative expression of immune cell gene signatures in APOBEC-high and APOBEC-low tumors were created using R package Morpheus (https://software.broadinstitute.org/morpheus). Samples were grouped by subtype (HER2-enriched or basal-like), and Euclidian hierarchical clustering and cutting the dendrogram was used to identify 2 main immune clusters in the HER2-enriched subtype and 2 main immune clusters in the basal-like subtype (immune cluster 1 and cluster 2).
To measure the correlation of APOBEC enrichment score and immune gene signatures in samples based on subtype, Spearman’s rho was calculated, and the significance was determined by Spearman’s rank correlation test. P-values were adjusted for multiple testing using the Benjamini-Hochberg method to control the false discovery rate. To assess differences in genetic heterogeneity between immune clusters, the number of subclonal mutations per TCGA sample was downloaded from (44).
Statistical analysis
One-way ANOVA and Tukey’s multiple comparisons test were used to assess statistical significance of qRT-PCR gene expression, colony formation assays, and MHC-I expression by flow cytometry. One-way ANOVA and Sidak’s multiple comparisons test were used to assess statistical significance of mouse tumor volume on a single day as indicated, differences in APOBEC enrichment score from human data, and the number of subclonal mutations in immune clusters from human data. Two-way ANOVA and Dunnett’s multiple comparisons test were used to determine the statistical significance of differential cell growth in vitro using CellTiter Glo assay. Two-way repeated-measures ANOVA and Tukey’s multiple comparison were used to measure statistical significance of changes in tumor volume over time in vivo. The adjusted p-values are reported for each. Fisher’s exact test was used to assess differences in response to checkpoint inhibition (CR/PR by percent change in tumor volume from treatment start day). Student’s t-test was used to test the statistical significance of differences in tumor mass at endpoint, flow cytometry, IHC/IF, and TCR-seq diversity measurements. Student’s t-test p-values are reported. Statistical analysis was performed, and graphs were created in R version 4.0.2 or using GraphPad Prism version 8.0.1.
Research reproducibility
Source code to reproduce analyses of APOBEC enrichment and quantification of immune signature gene expression is available at https://github.com/ashleydimarco/alvarezlab-APOBEC. RNA-seq data are available online using National Center for Biotechnology Information’s Short Read Archive (SRA) under project accession number PRJNA701674, and TCR-seq data are under project accession number PRJNA703001.
RESULTS
Expression of APOBEC3B in tumor cells is not lethal and induces cytidine deaminase activity
To induce APOBEC mutagenesis in vivo in an immunocompetent HER2-driven mammary tumor model, we utilized the SMF cell line, which is derived from a mammary tumor arising in the MMTV-Neu/Her2 mouse model on the FVB background (45). We engineered SMF cells to conditionally express the human APOBEC family member, APOBEC3B (A3B), and thereby acquire APOBEC mutational signatures during tumor progression, as expression of A3B is sufficient to induce a kataegis-like pattern (46,47). These SMF-A3B cells allow for titratable and reversible expression of A3B in tumor cells with the administration of doxycycline (dox). To characterize the A3B expression system in vitro, SMF-A3B cells were cultured with increasing concentrations of dox, with or without subsequent removal of dox from the medium. A3B mRNA and protein expression were dose-responsive and reversible (Fig. 1A–B). The A3B protein was constitutively localized to the nucleus in the presence of dox, demonstrating proper subcellular localization (48)(Fig. 1C). In an in vitro cytidine deaminase activity assay, increasing concentrations of dox induced dose-responsive deaminase activity in SMF-A3B cells (Fig. 1D). A3B expression did not affect cell proliferation or survival, as measured by ATP-based cell viability and colony formation assays (Fig. 1E–G). For subsequent experiments, we used a dox concentration (1 μg/mL) that induced A3B expression and deaminase activity comparable to that of the APOBEC-high human HER2+ breast cancer cell line, BT474 (Fig. 1A, D). These data suggest SMF-A3B is a suitable system to induce A3B expression and cytidine deaminase activity in a syngeneic, orthoptic murine tumor model.
Figure 1: SMF-A3B cells express titratable and reversible APOBEC3B without loss of cell viability.
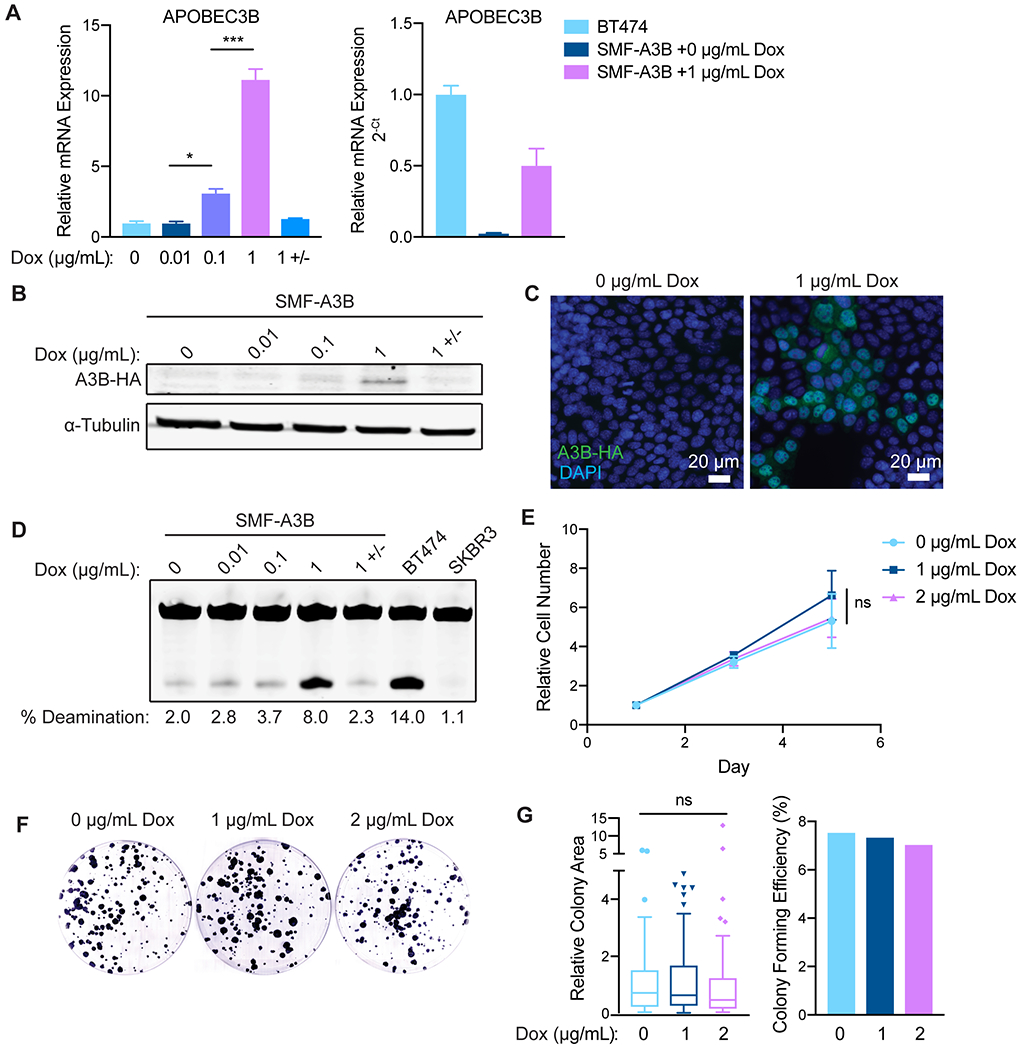
(A) qRT-PCR of A3B gene expression in SMF-A3B cells treated with the indicated concentrations of dox for 2 days. 1 +/− indicates treatment with 1 μg/mL of dox for 2 days, then removal of dox for 2 days. Left: A3B expression relative to 0 μg/mL dox. Right: A3B expression relative to BT474 cells. Data are representative of 2 independent experiments. Results are shown as mean±SD of 3 biological replicates. Significance was determined using a one-way ANOVA and Tukey’s multiple comparisons test. (B) Western blot of HA-tagged A3B in SMF-A3B cells treated with dox as in (A). (C) Immunofluorescence for the HA epitope in SMF-A3B cells treated with dox as in (A), showing nuclear localization of HA-A3B. Blue: DAPI; green: HA. (D) In vitro cytidine deaminase activity assay of SMF-A3B cells treated with dox as in (A). The APOBEC-high human cell line BT474 and the A3B-null human cell line SKBR3 were controls. (E) CellTiter-Glo assay showing growth curves of SMF-A3B cells treated with the indicated concentration of dox. Results are shown as mean±SD of 3 replicates. Statistical significance was determined by two-way ANOVA. (F) Clonogenic assay of SMF-A3B cells cultured with dox for 2 weeks to measure long term survival. Colonies were stained with crystal violet. (G) Quantification of clonogenic assay in (F). Left: Boxplots (Tukey method) depicting the relative colony area. Line shows median value, box shows 25th and 75th percentiles, and whiskers show 1.5-times the interquartile range (IQR); points outside 1.5-times IQR are plotted individually. Statistical significance was determined using a one-way ANOVA. Right: Colony forming efficiency in each condition. ns: not significant, p > 0.05; *p<0.05, **p<0.01, ***p<0.001.
APOBEC activity slows tumor growth and triggers the infiltration of immune cells
Given the evidence that the APOBEC mutational signature is associated with both an immune response and sensitivity to immunotherapy in NSCLC, bladder, and head and neck cancers (20–22), we examined the effects of in vivo APOBEC activity on the tumor immune microenvironment. SMF-A3B cells were orthotopically implanted bilaterally in the mammary gland of syngeneic, immunocompetent wildtype FVB mice. One cohort of mice was administered dox in the drinking water to induce A3B expression and APOBEC activity in the tumor cells throughout tumor growth (“APOBEC tumors”), whereas the control cohort received normal drinking water (Fig. 2A). APOBEC tumors grew significantly slower than control tumors and had a smaller mass at endpoint (Fig. 2B). Immunofluorescence of APOBEC and control tumors for a marker of double-stranded DNA breaks, γH2AX, showed no activation of the DNA damage response in vivo (Supplementary Fig. S1A–B). Similarly, A3B expression for two weeks did not induce γH2AX or cleaved PARP in SMF-A3B cells in vitro (Supplementary Fig. S1C). Taken together with the finding that A3B expression did not affect cell growth in vitro (Fig. 1E–G), this suggests that the growth defect of APOBEC tumors was mediated by a tumor cell-extrinsic mechanism.
Figure 2: A3B expression slows tumor growth and triggers the infiltration of antitumor immune cells into the tumor core.
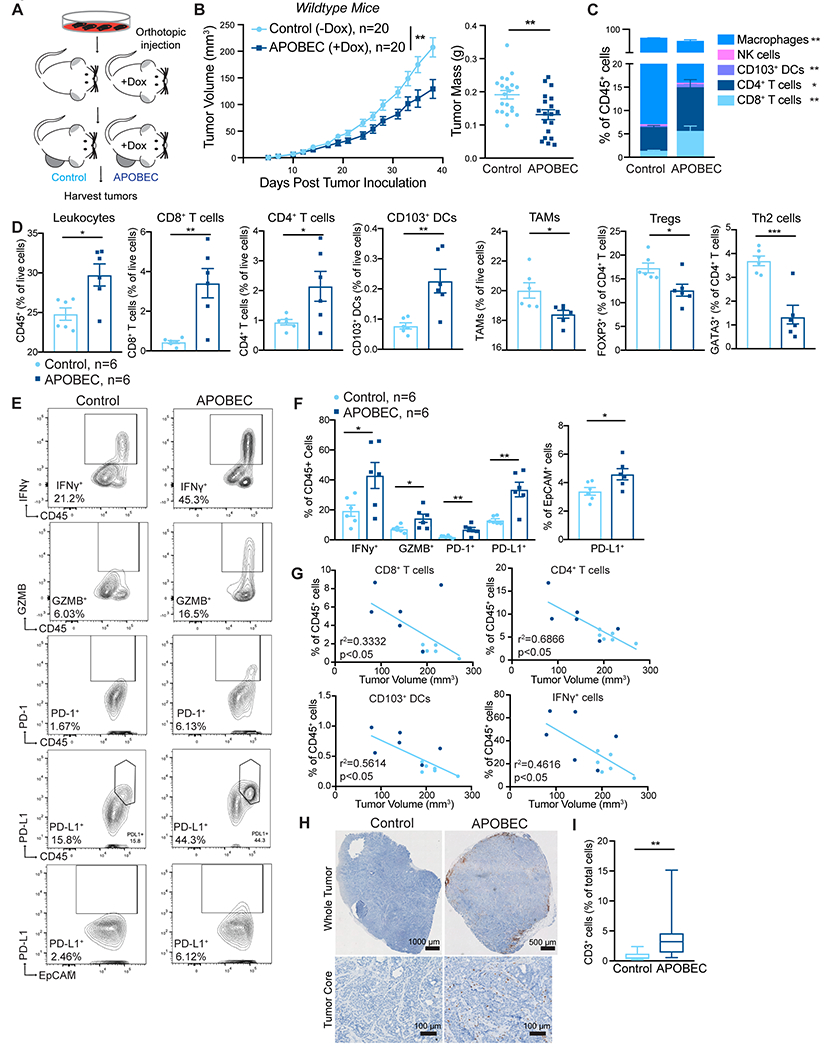
(A) Schematic showing experimental design for tumor growth experiment. SMF-A3B cells were orthotopically implanted in the mammary gland of mice. The APOBEC cohort was administered dox in the drinking water and control cohort was administered normal drinking water until endpoint. (B) Left: tumor volume of control (n=20) and APOBEC tumors (n=20) in wildtype mice. Statistical significance was determined by two-way repeated-measures ANOVA. Right: tumor mass (grams, g) of control and APOBEC tumors at endpoint. Statistical significance was determined by unpaired Student’s t-test. Error bars denote mean±SEM. Data are representative of 2 independent experiments. (C) The frequency of immune cell types, expressed as a percentage of total CD45+ cells, in control (n=6) and APOBEC (n=6) tumors determined by flow cytometry. (D) Flow cytometry quantification of immune cells in control (n=6) and APOBEC (n=6) tumors. Leukocytes, CD8+ T cells, CD4+ T cells, CD103+ DCs, and TAMs are represented as the percentage of total live cells. Tregs and Th2 cells are represented as the percentage of total CD4+ T cells. Statistical significance was determined by unpaired Student’s t-test. Error bars denote mean±SEM. (E-F) Representative flow cytometry plots (E) and quantification (F) for IFNγ, granzyme B, PD-1, and PD-L1 in control (n=6) and APOBEC (n=6) tumors. Statistical significance was determined by unpaired Student’s t-test. Error bars denote mean±SEM. (G) Pearson correlation between immune cell frequency and mean tumor volume (mm3) in control (light blue) and APOBEC (dark blue) tumors. Only significant correlations are shown, R-squared values are indicated. Results in A-G are representative of two independent experiments. (H) IHC CD3 (T cells) in control and APOBEC tumors. Top: tiled scan of the whole tumor. Bottom: representative region in the tumor core. scale bar: 1000 μm (Control), 500 μm (APOBEC) (I) Quantification of CD3 for control (n=4) and APOBEC (n=4) tumors. Four fields-of-view were imaged for each tumor. Boxplots: median percentage of CD3+ cells with minimum and maximum whiskers. Statistical significance was determined by unpaired Student’s t test with Welch’s correction. *p<0.05, **p<0.01, ***p<0.001
To gain insight into how A3B expression altered the tumor microenvironment (TME) of APOBEC tumors, six mice per cohort were randomly selected for immune profiling by flow cytometry (Supplementary Fig. S2 for gating strategy, Supplementary Fig. S3 for representative flow plots). APOBEC tumors showed a substantial infiltration of total leukocytes (CD45+EpCAM−) compared to control tumors (Fig. 2D). CD8+ T cells (CD45+CD3+CD8+), CD4+ T cells (CD45+CD3+CD4+), and CD103+ dendritic cells (DCs; CD45+CD11c+MHC-II+F4/80−CD103+) were expanded in the APOBEC TME, as measured both as the percentage of CD45+ cells (Fig. 2C) and the percentage of total live cells (Fig. 2D). There was no change in the infiltration of natural killer (NK) cells (CD45+NK1.1+CD3−), although immunosuppressive Tregs (CD45+CD3+CD4+FOXP3+), Th2 cells (CD45+CD3+CD4+GATA3+), and tumor-associated macrophages (TAMs; CD45+F4/80+CD11clow) were significantly reduced in the APOBEC tumors (Fig. 2D). APOBEC tumors were comprised of more immune cells producing the proinflammatory cytokine interferon-γ (IFNγ; CD45+IFNγ+) and cytotoxic granule granzyme B (GZMB; CD45+GZMB+)(Fig. 2E–F). Specifically, increased IFNγ on CD8+ T cells and NK cells and increased GZMB on NK cells (Supplementary Fig. S4A) was observed. Finally, PD-1+ immune cells (CD45+PD-1+), PD-L1+ immune cells (CD45+PD-L1+), and PD-L1+ tumor cells (EpCAM+PD-L1+) were elevated in the APOBEC tumors compared to control tumors, suggesting IFNγ signaling and potential feedback mechanisms leading to T-cell dysfunction (49)(Fig. 2E–F). A similar expansion of CD8+ T cells and CD103+ DCs was observed in the tumor-draining lymph nodes from mice with APOBEC tumors (Supplementary Fig. S4B). The defect in APOBEC tumor growth and enhanced immune infiltration was also observed in an independent experiment (Supplementary Fig. S4C–D). Next, we measured the correlation between immune cell abundance and tumor size at endpoint in SMF tumors. CD8+ T cells, CD4+ T cells, CD103+ DCs, and IFNγ+ cells were each negatively correlated with tumor size (Fig. 2G). This suggests that the adaptive immune response may mediate the growth defect observed in APOBEC tumors.
The localization of T cells in the TME is an important factor that influences tumor immunity and responses to immunotherapy (50). T cells can be excluded from the tumor core and instead localize to the periphery in murine models and human tumors (51,52), and this exclusion may be one mechanism of immune suppression. Therefore, to assess T-cell localization in APOBEC tumors, we performed IHC for CD3 on an independent cohort of SMF tumors. CD3 expression was consistent with flow cytometry analyses and revealed an increase in the total number of T cells in APOBEC tumors. The T cells were most concentrated at the periphery of the APOBEC tumors, although significant T-cell infiltration into the tumor core was seen (Fig. 2H–I, Supplementary Fig. S4E).
To extend these results to additional cell lines, we engineered inducible A3B expression in the HER2-driven mammary tumor cell line NDLUCD (25) and the TNBC cell line EMT6 (Supplementary Fig. S5A–D). As with SMF tumors, EMT6 APOBEC tumors grew more slowly and had increased infiltration of leukocytes when implanted in syngeneic BALB/c mice (Supplementary Fig. S5E–G). A3B expression completely prevented the formation of NDLUCD tumors in syngeneic FVB mice; however, A3B expression did not alter the growth of NDLUCD tumors in vitro (Supplementary Fig. S5H–I). This suggests that APOBEC tumors are immunogenic in multiple breast tumor models.
The growth defect of APOBEC tumors is mediated by the immune system
We next examined whether the growth defect of APOBEC tumors was mediated by immune responses. To address this, SMF-A3B cells were implanted in the mammary fat pad of immunodeficient NSG mice, and mice received either dox or normal drinking water throughout the duration of tumor growth. Control and APOBEC tumors grew at similar rates (Supplementary Fig. S6A), indicating that in the absence of a functional immune system, A3B expression does not affect tumor growth. This is consistent with the finding that A3B expression did not affect the growth or viability of SMF cells in vitro (Fig. 1E–G).
To discern whether the antitumor immune responses in APOBEC tumors were due to the catalytic activity of A3B, and to rule out the possibility that expression of the human A3B protein in mouse cells may be immunogenic, we generated a catalytically inactive A3B mutant by site-directed mutagenesis of one of the A3B catalytic domains (E255Q)., and SMF cells were transduced to generate SMF-A3Binactive cells. Dox treatment led to expression of catalytically dead A3B, and no detectable increase in deaminase activity was observed (Supplementary Fig. S6B–D). SMF-A3Binactive cells were then injected into the mammary glands of immunocompetent wildtype mice on dox water. Tumors expressing catalytically dead A3B (SMF-A3Binactive + dox) grew at similar rates and had similar numbers of total leukocytes (CD45+) and T cells (CD3+) as control tumors (Supplementary Fig. S6E–F). This indicates that the growth defect and immune responses in APOBEC tumors is dependent on A3B catalytic activity and is not the result of expression of the human A3B protein in mouse cells.
To further explore whether the tumor growth defect and immune response in APOBEC tumors was due to A3B-mediated mutagenesis, as opposed to the expression of A3B protein, we utilized the reversibility of the dox-inducible system. After two weeks of dox treatment, dox was removed from SMF-A3B cells to downregulate A3B expression. These in vitro APOBEC-mutagenized cells retain A3B-catalyzed mutations but do not express A3B protein. The proliferation rate of in vitro APOBEC-mutagenized cells was similar to control, non-mutagenized cells (Supplementary Fig. S6G). In contrast, when implanted into the mammary gland of wildtype mice without dox in their drinking water (Supplementary Fig. S6H), the in vitro APOBEC-mutagenized tumors grew more slowly than control tumors and had evidence of an increased adaptive immune response, as measured by qRT-PCR for T cell-specific genes Gzma, Prf-1, Tbx21 (Supplementary Fig. S6I–J). The growth defect of in vitro APOBEC-mutagenized tumors was not evident in NSG mice (Supplementary Fig. S6K), further confirming the role of the adaptive immune response in mediating the growth defect of APOBEC tumors. Together these data reveal that A3B activity promotes an inflamed TME in HER2-driven murine tumors and leads to an immune-dependent growth defect.
APOBEC activity slows tumor growth by stimulating an antigen-specific immune response
To understand the basis of the immune-mediated growth defect of APOBEC tumors, we performed RNA-sequencing on control and APOBEC tumors from either immunocompetent wildtype mice or immunodeficient NSG mice. APOBEC tumors in wildtype mice showed a significant upregulation of adaptive immune response gene ontology (GO) terms, including regulation of T-cell mediated immunity/cytotoxicity/differentiation, antigen processing and presentation, and B-cell activation (Fig. 3A, Supplementary Fig. S7A). The top two pathways enriched in the APOBEC tumors in wildtype mice by GSEA were allograft rejection (Supplementary Fig. S7B) and IFNγ response (Fig. 3B), suggesting an adaptive immune response mechanism of tumor cell killing. In APOBEC tumors harvested from immunodeficient NSG mice, in contrast, the DNA repair pathway was significantly enriched by GSEA (Fig. 3B, Supplementary Fig. S7C), possibly due to the activation of repair pathways following the generation of A3B-catalyzed uracil lesions in the genome.
Figure 3: T cell-dependent antitumor responses in APOBEC tumors are antigen-specific.
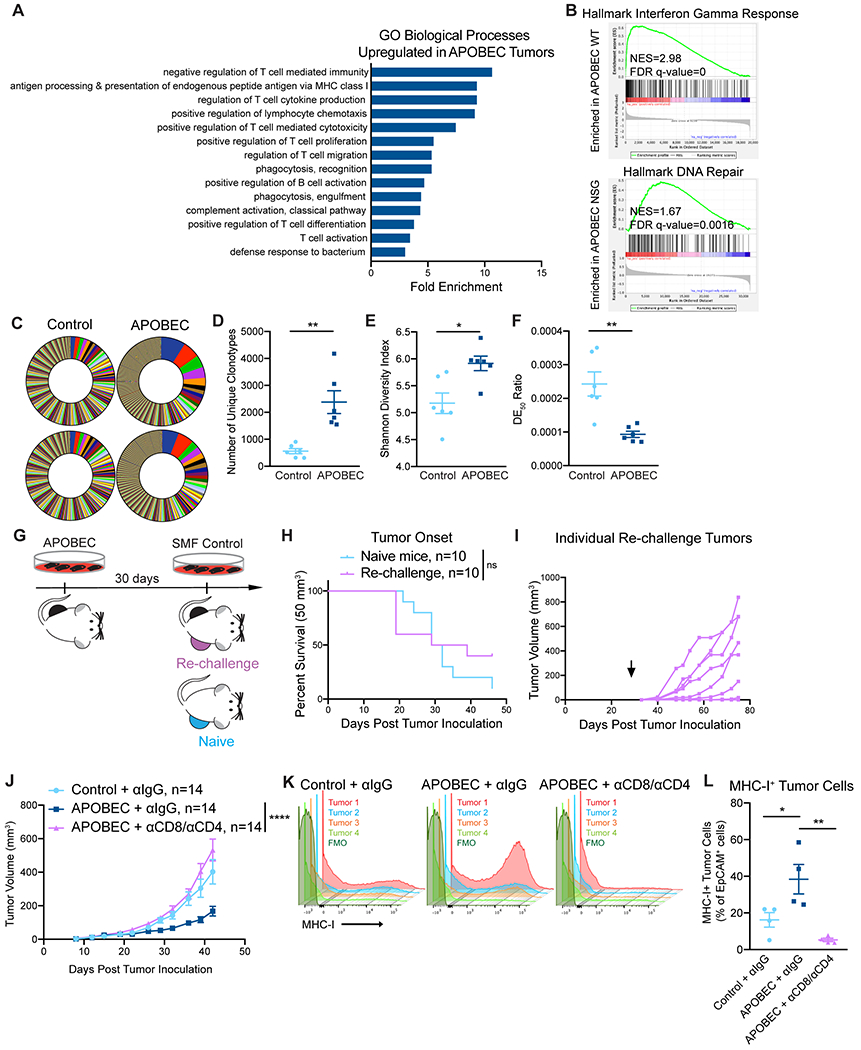
(A) Gene ontology (GO) analysis of differentially expressed genes between control and APOBEC tumors in immunocompetent mice. Fold enrichment of select GO biological processes that were significantly enriched in APOBEC tumors (n=6) compared to control tumors (n=6) are shown; FDR<0.05, Fisher’s test. Also see Supplementary Fig. S6A. (B) GSEA between control and APOBEC tumors. Representative gene sets enriched in APOBEC tumors in immunocompetent mice (top) or immunodeficient mice (bottom) are shown. Normalized Enrichment Scores (NES) and FDR q-values are shown. Also see Supplementary Fig. S6B–C. (C) TCR-sequencing from control (n=6) and APOBEC tumors (n=6) from wildtype mice. Pie charts: unique TCR clonotypes ranked by abundance in two control and two APOBEC tumors. (D) Quantification of the total number of unique clonotypes in control and APOBEC tumors (n=6/cohort). (E) Shannon diversity index of the TCR repertoire in control and APOBEC tumors. (F) TCR DE50 ratios in control and APOBEC tumors. Error bars in (D-F) denote mean±SEM. Statistical significance was determined by unpaired Student’s t test in (E) and unpaired Student’s t test with Welch’s correction in (D, F). (G) Schematic showing experimental design of tumor re-challenge. Clonal APOBEC-mutagenized cells were implanted unilaterally in the mammary fat pad. 30 days later, SMF control cells were implanted in the contralateral mammary fat pad (Re-challenge, n=10). Re-challenge tumors were compared to control cells implanted in naïve mice (Naïve, n=10). (H) Kaplan-Meier curves showing tumor onset (50 mm3) for re-challenge mice and naïve mice from (G). Statistical significance was determined by log-rank test. (I) Tumor growth curves of individual re-challenge tumors from (G). Arrow indicates day of re-challenge. (J) Tumor volume (mm3) over time for control tumors treated with isotype control antibody (n=14) and APOBEC tumors treated with isotype control (n=14) or CD8 and CD4 depletion antibodies (n=14) in wildtype mice. Error bars denote mean±SEM. Statistical significance was determined by two-way repeated-measures ANOVA and Tukey’s multiple comparisons test. (K) Flow cytometry histograms showing MHC-I expression on EpCAM+ tumor cells from tumors in (J). (L) Quantification of MHC-I+ cells, expressed as a percentage of EpCAM+ cells, in tumors (n=4 per cohort) from (J). Results in K and L are from a single experiment. Error bars denote mean±SEM. Statistical significance was determined by one-way ANOVA and Tukey’s multiple comparisons test. ns: not significant, p>0.05; *p<0.05, **p<0.01, ****p<0.0001
Given that antigen presentation pathways were upregulated in APOBEC tumors, we were next interested in studying tumor-specific antigen responses in APOBEC tumors. Therefore, we assessed changes in the T-cell repertoire between control and APOBEC tumors using TCR-sequencing. APOBEC tumors had more unique TCR clonotypes than control tumors (Fig. 3C–D). Using the Shannon entropy diversity index to measure the diversity richness of the clonotypes, we found that APOBEC tumors had a higher clonotype diversity than control tumors (Fig. 3E). Finally, we used the DE50 ratio, a measure of the number of clonotypes making up the top 50% of reads relative to the total number of reads, to assess clonotype evenness. A high DE50 ratio indicates that clonotypes are evenly represented in the population, whereas a low DE50 ratio corresponds to a TCR repertoire that is dominated by specific CDR3 clonotypes. This analysis indicated that APOBEC tumors had a lower DE50 ratio than control tumors (Fig. 3F). Taken together, these analyses indicate that the TCR repertoire of APOBEC tumors exhibit increased diversity richness but decreased diversity evenness; interestingly, this pattern has been associated with productive T-cell responses and successful treatment with immunotherapy (53).
We reasoned that the growth suppression of APOBEC tumors could be mediated by either an innate or an adaptive immune response to specific antigens. To distinguish between these possibilities, we performed a re-challenge experiment. We isolated clones of in vitro APOBEC-mutagenized cells and implanted unilaterally in the mammary fat pad. 30 days later, control SMF cells were implanted in the contralateral mammary fat pad as a re-challenge (Fig. 3G). Tumors in formed with similar kinetics as in naïve mice, and 7 out of 10 re-challenge tumors grew progressively (Fig. 3H–I). This suggests that APOBEC-mediated mutagenesis likely leads to the generation of tumor-specific antigens which are targeted by T cells to induce tumor growth inhibition.
CD4+ T cells are required for the tumor growth defect of APOBEC tumors
To explore the requirement for T cells in mediating antitumor immune responses against APOBEC tumors, we depleted CD8+ T cells in APOBEC tumor-bearing mice. We confirmed that CD8+ T cells were completely depleted in the peripheral blood using flow cytometry, and in the tumor at endpoint using qRT-PCR (Supplementary Fig. S8A–C). The growth defect of APOBEC tumors was not rescued upon CD8+ T-cell depletion alone (Supplementary Fig. S8D). We next depleted CD8+ T cells and CD4+ T cells simultaneously (Supplementary Fig. S8E–G). In the absence of both CD4+ and CD8+ T cells, the APOBEC tumor growth defect was completely rescued, and APOBEC tumors grew similarly to the control tumors (Fig. 3J). Control or CD4/CD8-depleted tumors were harvested to assess MHC-I expression on tumor cells. In the presence of T cells, APOBEC tumor cells had higher expression of MHC-I compared to control tumors. In contrast, when T cells were depleted, MHC-I expression on tumor cells was abrogated (Fig. 3K–L). Together, these results revealed that T cells are required for MHC-I upregulation and slowed tumor growth in APOBEC tumors.
APOBEC activity sensitizes breast tumors to immune checkpoint inhibition
Because we found that A3B expression stimulated T cell-mediated antitumor immune responses, we next asked if APOBEC activity rendered the tumors responsive to checkpoint inhibition. Control tumors did not benefit from checkpoint inhibition, consistent with the clinical observation that checkpoint inhibition is not effective in HER2+ breast cancer patients (17–19). In contrast, APOBEC tumor growth was significantly blunted upon treatment with anti-PD-1/anti-CTLA4 therapy (Fig. 4A). Checkpoint inhibitor treatment led to a PR in only 1 of the 13 control tumors (Fig. 4B, Supplementary Fig. S9A–B). In contrast, 7 out of 11 APOBEC tumors exhibited a CR or PR with combination checkpoint inhibition (Fig. 4B, Supplementary Fig. S9A–B). Neither control nor APOBEC tumors responded significantly to anti-PD-1 monotherapy (Fig. 4C–D, Supplementary Fig. S9C–D). However, APOBEC tumors benefited from anti-CTLA-4 monotherapy, with 6/9 tumors exhibiting a CR/PR, whereas no control tumors responded (Fig. 4E–F, Supplementary Fig. S9E, G). Monoclonal antibodies targeting HER2 are commonly combined with checkpoint inhibitors in clinical trials for HER2+ breast cancer patients (11,54). Thus, we examined the response of control and APOBEC tumors to anti-CTLA-4 and anti-HER2 combination therapy. Six of 9 control tumors had a PR to combination therapy, whereas all 10 APOBEC tumors experienced a CR (Fig. 4E–F, Supplementary Fig. S9F–G). These results show that APOBEC activity sensitizes HER2-driven murine breast cancers to anti-CTLA-4 checkpoint inhibition, and the addition of HER2-directed therapy leads to a complete response in all APOBEC tumors.
Figure 4: APOBEC tumors are sensitive to combination anti-PD-1/anti-CTLA4 immune checkpoint blockade.
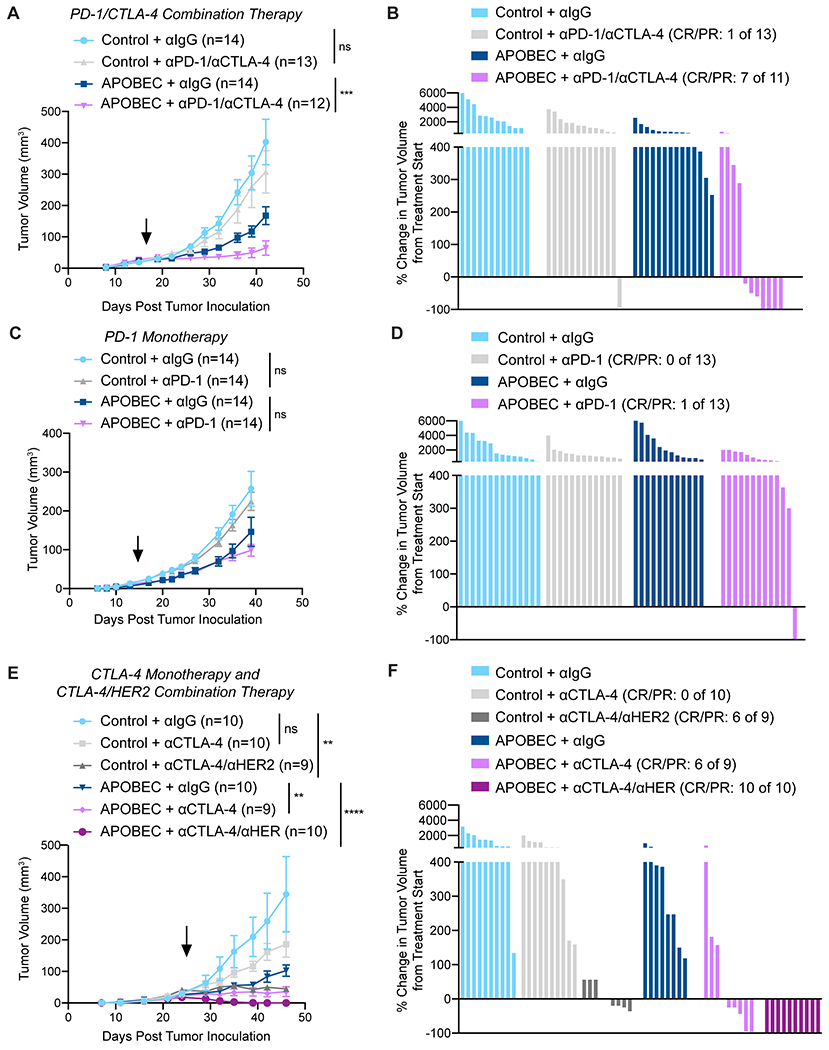
(A) Tumor volume (mm3) over time for control and APOBEC tumors treated with isotype control or anti-PD-1/anti-CTLA-4. Arrow: treatment start. Error bars denote mean±SEM. Statistical significance was determined by two-way repeated-measures ANOVA and Tukey’s multiple comparisons test. (B) Percent-change in tumor volume from treatment start for palpable tumors from (A) until endpoint. Bars denote individual tumors. (C) Tumor volume (mm3) over time for control and APOBEC tumors treated with isotype control or anti-PD-1 monotherapy. Arrow: treatment start. Error bars denote mean±SEM. Statistical significance was determined by two-way repeated-measures ANOVA and Tukey’s multiple comparisons test. (D) Percent-change in tumor volume from treatment start for palpable tumors from (C) until endpoint. Bars denote individual tumors. (E) Tumor volume (mm3) over time for control and APOBEC tumors treated with isotype control, anti-CTLA-4 monotherapy, or anti-CTLA-4/anti-HER2. Arrow: treatment start. Error bars denote mean±SEM. Statistical significance was determined by two-way repeated-measures ANOVA and Tukey’s multiple comparisons test. (F) Percent-change in tumor volume from treatment start for palpable tumors from (E) until endpoint. Bars denote individual tumors. CR, complete response; PR, partial response. ns: not significant, p>0.05; **p<0.01, ***p<0.001, ****p<0.0001
Clonal APOBEC tumors are maintained in cancer-immune equilibrium
Genomic studies of human cancer suggest that episodic APOBEC mutagenesis may fuel cancer heterogeneity and evolution (55). In melanoma and NSCLC, mutational and neoantigen heterogeneity reduces antitumor immunity (56,57) and response to checkpoint inhibitor therapy. For instance, lung tumors with more clonal neoantigens are better controlled by neoantigen-specific T cells and have improved responses to checkpoint inhibitors (58). Thus, we were interested in understanding the consequences of APOBEC-mediated genetic heterogeneity on antitumor immunity and mammary tumor growth in immunocompetent mice. To assess differences between heterogenous and clonal APOBEC tumors, SMF-A3B cells were cultured with dox for 2 weeks and then dox was removed to downregulate A3B (referred to as “parental APOBEC”; control, non-mutagenized cells referred to as “parental control”). We next derived single-cell clones by limiting dilution from the parental APOBEC and parental control populations. We identified two control clones (clone 2 and 3) and two APOBEC clones (clone 2 and 3) that grew at the same rate as parental cells in vitro (Supplementary Figure S10A–B) and formed tumors similarly to their parental counterparts in immunocompromised, athymic nude mice (Fig. 5A–B). However, when these clones were injected into immunocompetent, wildtype mice, only the control clones formed progressively growing tumors. In contrast, both APOBEC clones gave rise to very small tumors that remained in a cancer-immune equilibrium until the animals were sacrificed (Fig. 5C–D). Comparing the size of tumors formed in the presence or absence of the adaptive immune response revealed that clonal APOBEC tumors were significantly smaller than control tumors in wildtype mice but not in athymic nude mice (Fig. 5E–F). Thus, APOBEC-mediated heterogeneity may limit the potential of a fully productive immune response against hypermutated breast tumors. In contrast, clonal APOBEC tumor growth may be controlled in cancer-immune equilibrium.
Figure 5: Complete immune-mediated suppression of clonal APOBEC tumor growth.
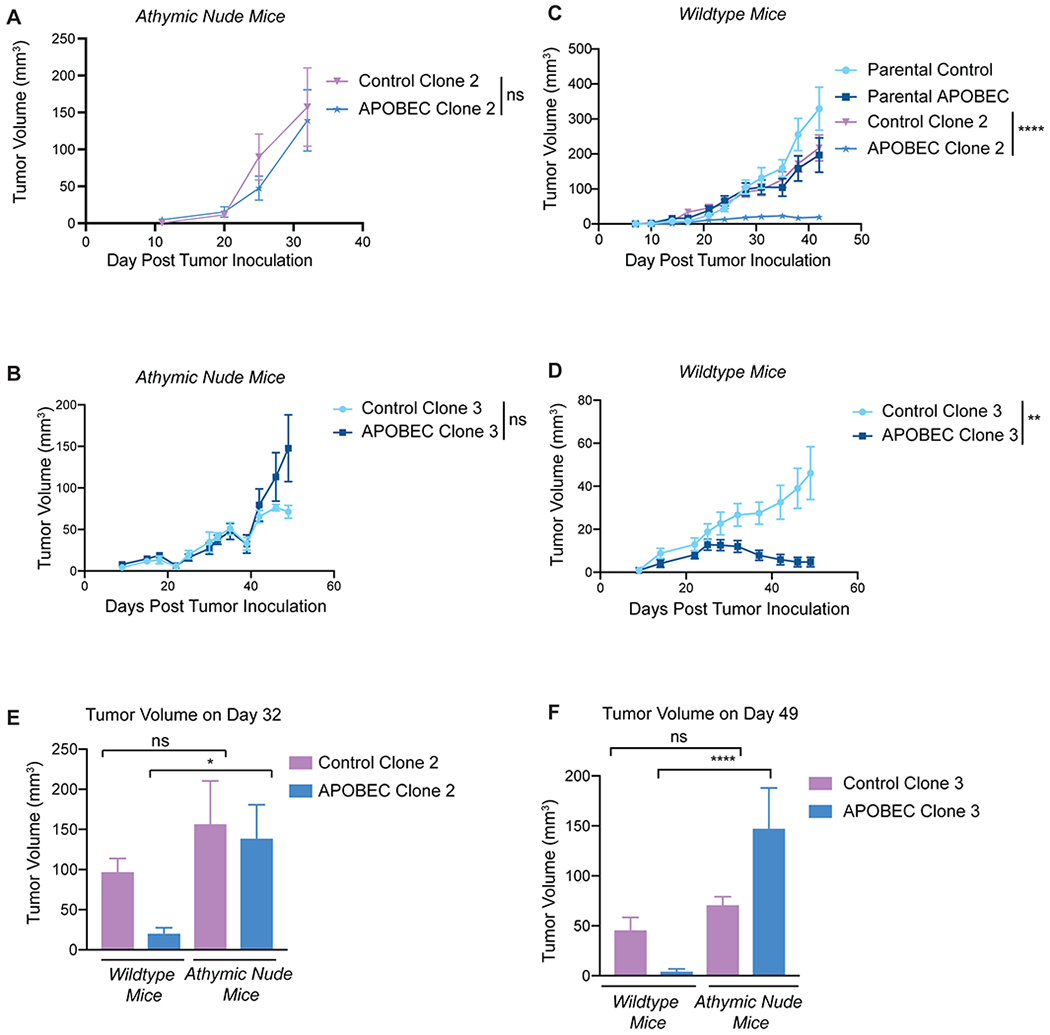
(A) Tumor volume (mm3) over time for control clone 2 and APOBEC clone 2 injected in the mammary gland of athymic nude mice. N= 4 mice per clone. (B) Tumor volume (mm3) over time for control clone 3 and APOBEC clone 3 injected in the mammary gland of athymic nude mice. N=6 mice per clone. (C) Tumor volume (mm3) over time for control and APOBEC clones, as well as the corresponding polyclonal parental populations, injected in the mammary gland of immunocompetent wildtype mice. N=14 mice per cell line. (D) Tumor volume (mm3) over time for control clone 3 and APOBEC clone 3 injected in the mammary gland of wildtype mice. N=10 mice per clone. For (A-D), statistical significance was determined by two-way repeated-measures ANOVA; error bars denote mean±SEM. (E) Comparison of tumor volume on day 32 between control clone 2 and APOBEC clone 2 grown in immunocompromised nude mice from (A) and immunocompetent wildtype mice from (C). (F) Comparison of tumor volume on day 49 between control clone 3 and APOBEC clone 3 grown in immunocompromised nude mice from (B) or immunocompetent wildtype mice from (D). For (E-F), statistical significance was determined by one-way ANOVA and Sidak’s multiple comparisons test; error bars denote mean±SEM; ns: not significant, p>0.05; *p<0.05, ****p<0.0001.
Association between APOBEC signatures and adaptive immune responses in breast cancer
We next determined whether human breast tumors with APOBEC mutagenesis have evidence of an increased adaptive immune response. To do this, we analyzed breast tumors from TCGA. Similar to previous reports (5), we found that the HER2-enriched subtype had the highest median APOBEC enrichment scores and the largest proportion of tumors with enrichment scores >2 (Fig. 6A). The relationship between APOBEC mutagenesis and the expression of immune signatures in HER2-enriched and basal-like breast cancers was determined using the PAM50 subtype. The basal-like category includes most TNBCs and is considered the most immunologically active breast cancer subtype (59). We segregated basal-like and HER2-enriched tumors into APOBEC-high (Fig. 6B) or APOBEC-low groups (Supplementary Fig. S11A) using an APOBEC enrichment score cutoff of 2. Hierarchical clustering of tumors based on immune cell signatures revealed two main clusters in each subtype. Tumors in cluster 1 had high expression of immune signatures that were reflective of an antitumor adaptive immune response, including Th1 cells, activated DCs (aDCs), CD8+ T cells, cytotoxic cells, interferon signaling pathway (IFN), major histocompatibility complex class II antigen presentation pathway (MHC-II), and checkpoint genes such as LAG3, PD1, PDL1, PDL2, CTLA4, and TIM3. Tumors in cluster 2 had low expression of antitumor immune response signatures and high expression of several immunosuppressive gene signatures, such as macrophages and neutrophils.
Figure 6: The TME phenotype of APOBEC-high human breast cancers is dependent on molecular subtype and the number of subclonal mutations.
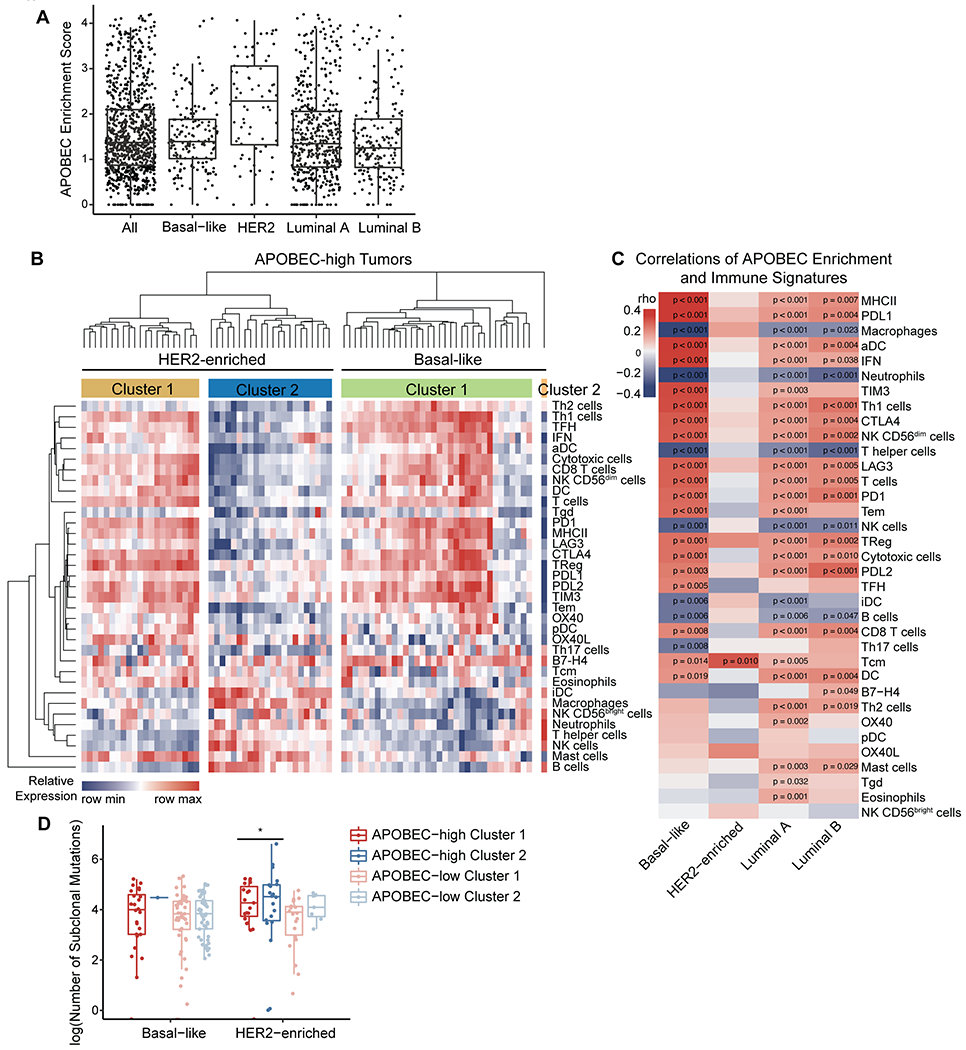
(A) APOBEC enrichment score calculated from WES data for all TCGA breast cancer samples. APOBEC enrichment score of 2 or higher delineates APOBEC-high tumors. Boxplots: Line shows median value, box shows 25th and 75th percentiles, and whiskers show 1.5-times the IQR. (B) Heatmap showing the relative expression of immune cell gene signatures from TCGA RNA-seq data in APOBEC-high tumors, grouped by breast cancer subtype. (C) Heatmap showing correlation Spearman’s rho values between APOBEC enrichment score and immune gene signatures for each molecular subtype of breast cancer. Red, immune signature positively correlated with APOBEC enrichment score; blue, immune signature negatively correlated with APOBEC enrichment score. (D) The number of subclonal mutations (from (44)) in APOBEC-high clusters depicted in (B) and APOBEC-low clusters depicted in Supplementary Figure S9. Boxplots: Line shows median value, box shows 25th and 75th percentiles, and whiskers show 1.5-times the IQR. Statistical significance was determined one-way ANOVA and Sidak’s multiple comparisons test. *p<0.05.
Most APOBEC-high basal-like tumors fell within cluster 1, reflective of an antitumor adaptive immune response (Fig. 6B). These results are consistent with the well-defined hot TME of TNBC and their response to immune checkpoint inhibition. Within cluster 1, seven basal-like patient tumors exhibited an intermediate antitumor immune gene expression, and only one patient tumor fell within cluster 2. In contrast to basal-like tumors, half of the APOBEC-high HER2-enriched tumors fell within cluster 1 and half within cluster 2 (Fig. 6B).
To further explore the differences in immune cell gene expression signatures between basal-like, HER2-enriched, and luminal A/B tumors, we analyzed the correlation between the APOBEC signature enrichment score and each immune cell gene signature, as measured by a quantitative score (Supplementary Table S3). In the basal-like subtype, APOBEC signature enrichment score positively correlated with numerous adaptive immune response gene signatures (e.g. MHC-II, aDCs, IFN, Th1 cells) and checkpoint genes (e.g. PDL1, TIM3, CTLA4), and negatively correlated with known immunosuppressive cell types (macrophages, neutrophils) (Fig. 6C). Luminal A/B subtypes showed similar patterns of correlation between APOBEC enrichment and immune signatures (Fig. 6C). In contrast, all but one of the immune signatures (Tcm, central memory T cells), did not significantly correlate with the APOBEC enrichment score in the HER2-enriched subtype, despite this subtype possessing the highest median APOBEC enrichment scores (Fig. 6C). In summary, the APOBEC mutational signature associated with antitumor adaptive immunity gene expression in basal-like breast cancer patients, but there was no evidence of association in HER2-enriched patients.
Association between subclonal mutations and immune signatures in APOBEC-high tumors
We next defined the fraction of HER2-enriched tumors with high APOBEC mutational enrichment and a favorable TME. 58.1% of all HER2-enriched tumors analyzed were APOBEC-high (43 of 74; Fig. 6B, Supplementary Fig. S11A). Of APOBEC-high, HER2-enriched tumors, 48.8% fell within TME cluster 1 (21 of 43; Fig. 6A–B). Thus, 28.4% of all HER2-enriched tumors had high APOBEC enrichment and a favorable TME (21 of 74). To understand the differences in immune infiltration between APOBEC-high, HER2-enriched tumors in clusters 1 and 2, we examined clinical features of tumors from each cluster. There were no statistically significant differences in estrogen receptor status, p53 status by IHC, node positivity, risk of recurrence, or pathological stage between cluster 1 and 2 of APOBEC-high HER2 tumors.
Next, in light of previous findings that tumors with more subclonal mutations have a less productive immune response (57,58), we postulated that genetic heterogeneity may underly the TME differences between APOBEC-high basal-like and HER2-enriched tumors. To test this, we used the clonal phylogenies of TCGA breast cancers generated by Raynaud and colleagues (44) to explore the relationship between subclonal mutations and immunogenicity in human breast cancer. We compared the number of subclonal mutations between tumors with a hot TME (cluster 1) and tumors with a cold TME (cluster 2). In HER2-enriched tumors, APOBEC-high tumors in cluster 2 had more subclonal mutations than APOBEC-high tumors in cluster 1 (Fig. 6D), despite the fact that the APOBEC enrichment scores were similar between these two groups (Supplementary Fig. S11B). This suggests that the immunogenicity of APOBEC-high tumors between breast cancer subtypes may be due to intratumor genetic diversity. Basal-like tumors are less heterogenous and have lower APOBEC enrichment scores on average than HER2-enriched tumors. Conversely, HER2-enriched tumors with high APOBEC enrichment scores and high genetic heterogeneity may undergo immune escape and acquire a cold TME.
DISCUSSION
APOBEC mutational signatures have been identified in more than 22 different cancer types (5), but the functional consequences of APOBEC activity on the tumor immune microenvironment have not been explored. Here, we showed that APOBEC activity promoted an immunologically hot tumor microenvironment, leading to slowed tumor growth. We found that the slowed growth of APOBEC tumors was due to an antigen-specific adaptive immune-mediated mechanism that required the activity of CD4+ T cells. APOBEC tumors exhibited a T cell-dependent upregulation of MHC-I expression, which associated with increased TCR diversity within tumors. Consistent with increased immune cell infiltration, APOBEC tumors were sensitive to checkpoint inhibitors. Although other studies have examined how APOBEC mutagenesis sensitizes tumors to checkpoint inhibitors (23,24), this is the first study, to our knowledge, to comprehensively define the direct consequences of APOBEC activity on the tumor immune microenvironment in the absence of therapy. Most of our findings were made using the SMF transgenic mammary tumor cell line, which expresses HER2/Neu, a potential foreign antigen for wildtype mice. EMT6 cells are immunogenic, suggesting that they may also express a strong tumor antigen. It is therefore possible that APOBEC activity must act in the context of a universally expressed tumor antigen in order to elicit a robust anti-tumor immune response. If a foreign antigen such as HER2 is required for APOBEC-catalyzed immunogenicity in breast cancer patients, these findings have clinical significance, considering the ongoing trials focusing on HER2 as an antigen in breast cancer (54).
The role of CD4+ T cells in the APOBEC-dependent antitumor immune response is intriguing and opens up the possibility for CD4+ T cell-directed therapies, such as CTLA-4 inhibitors or CD4+ T-cell adoptive transfer, to treat APOBEC-high patients. Our work showed that A3B activity sensitized HER2-driven mammary tumors to anti-CTLA-4 monotherapy and anti-CTLA-4/anti-PD-1 combination therapy, whereas anti-PD-1 monotherapy alone was ineffective. Similarly, Hollern and colleagues found that single-agent anti-PD-1 is inferior to the combination therapy for TNBC (23). Thus, although the majority of immunotherapy trials focus on re-invigorating CD8+ cytotoxic T cells, our findings and others suggest that harnessing the activity of CD4+ helper T cells may be more beneficial for breast tumors with APOBEC mutational signatures. We found that CD8+ T cells were not required for growth defect of APOBEC tumors, suggesting some redundancy in the cytotoxic cells that mediate growth suppression of APOBEC tumors.
At the same time that APOBEC-catalyzed mutations may promote immunogenicity, APOBEC activity can also generate genetic heterogeneity and fuel tumor evolution (4). Although there is growing interest in understanding how intratumor genetic diversity impacts productive immune responses, little is known about the effects of APOBEC-catalyzed subclonal diversification on tumor immunogenicity. When we examined the relationship between APOBEC mutagenesis and immunogenicity in human breast cancers, we observed a correlation between APOBEC enrichment scores and immune cell gene signatures in basal-like tumors, consistent with findings in other tumor types (20,21). In contrast, there was no correlation between APOBEC enrichment and immune cell signatures in HER2-enriched breast cancers. In fact, half of HER2-enriched tumors with high APOBEC enrichment scores (cluster 2) had low expression of adaptive immune signatures. In light of our finding that APOBEC activity promoted immune infiltration in HER2-driven mouse mammary tumors, this suggested that cluster 2 tumors may have evolved immune-suppressive mechanisms that limit antitumor adaptive immune responses. Although the details of such mechanisms remain unknown, initial insight came from examining the frequency of subclonal mutations in these tumors. Among APOBEC-high tumors, immune-suppressed (cluster 2) tumors had a higher number of subclonal mutations than immune-infiltrated (cluster 1) tumors. These results are reminiscent of the finding that breast tumors with high heterogeneity have less infiltration of antitumor immune cells, including CD8+ and CD4+ T cells, lower expression of PD-L1, and lower expression of cytolytic enzymes, granzyme A and perforin-1 (60). These results suggest that a subset of APOBEC-high HER2 tumors with a high frequency of subclonal mutations can evade immune activation, and that APOBEC mutational signatures and mutational clonality should be considered together as biomarkers for predicting response to immunotherapy in women with breast cancer. These results mirror our findings in mouse tumors, where clonal APOBEC tumors are controlled by the immune system more profoundly than polyclonal APOBEC tumors. We propose a model (Supplementary Fig. S11C), whereby APOBEC mutagenesis leads to immune infiltration and immunotherapy benefit in both mouse models and human breast tumors yet can also foster subclonal diversification to promote evasion of immune responses.
Supplementary Material
SYNOPSIS:
APOBEC mutagenic activity is shown to sensitize HER2-driven mammary tumors to immune checkpoint inhibition, an effect dependent on T-helper cells. Data highlight potential biomarkers that could be used to predict immunotherapy responses in patients with HER2+ breast cancer.
Acknowledgements
We thank Dr. Lewis Chodosh (University of Pennsylvania) for providing the SMF cell line and Dr. Alexander Borowsky for providing NDLUCD cells. We thank Dr. Michael Plebanek and Dr. Nicolas Devito (Duke University) for technical advice with flow cytometry and Dr. Andrea Walens (University of North Carolina at Chapel Hill) for reviewing the manuscript. This work was funded by the National Cancer Institute under award R01CA208042 (to J.V.A.) and T32-CA009111 (to A.V.D.), as well as the American Cancer Society under award 132556-RSG-18-130-CCG (to J.V.A.) and by startup funds from the Duke Cancer Institute, the Duke University School of Medicine, the Whitehead Foundation (to J.V.A.), and the National Institutes of Health under T32-GM007184 (to A.V.D.).
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emens LA. Breast cancer immunotherapy: facts and hopes. Clin Cancer Res. 2018;24:511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams S, Gatti-Mays ME, Kalinsky K, Korde LA, Sharon E, Amiri-Kordestani L, et al. Current landscape of immunotherapy in breast cancer: A review. JAMA Oncol. 2019;5:1205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nanda R, Chow LQM, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34:2460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emens LA, Cruz C, Eder JP, Braiteh F, Chung C, Tolaney SM, et al. Long-term Clinical Outcomes and Biomarker Analyses of Atezolizumab Therapy for Patients With Metastatic Triple-Negative Breast Cancer: A Phase 1 Study. JAMA Oncol. 2019;5:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emens LA, Adams S, Barrios CH, Diéras V, Iwata H, Loi S, et al. First-line atezolizumab plus nab-paclitaxel for unresectable, locally advanced, or metastatic triple-negative breast cancer: IMpassion130 final overall survival analysis. Ann Oncol. 2021;32:983–93. [DOI] [PubMed] [Google Scholar]
- 15.Miles D, Gligorov J, André F, Cameron D, Schneeweiss A, Barrios C, et al. Primary results from IMpassion131, a double-blind, placebo-controlled, randomised phase III trial of first-line paclitaxel with or without atezolizumab for unresectable locally advanced/metastatic triple-negative breast cancer. Ann Oncol. 2021; [DOI] [PubMed] [Google Scholar]
- 16.Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im S-A, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–28. [DOI] [PubMed] [Google Scholar]
- 17.Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau H-T, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat. 2018;167:671–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loi S, Giobbie-Hurder A, Gombos A, Bachelot T, Hui R, Curigliano G, et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b-2 trial. Lancet Oncol. 2019;20:371–82. [DOI] [PubMed] [Google Scholar]
- 19.Emens LA, Esteva FJ, Beresford M, Saura C, De Laurentiis M, Kim S-B, et al. Trastuzumab emtansine plus atezolizumab versus trastuzumab emtansine plus placebo in previously treated, HER2-positive advanced breast cancer (KATE2): a phase 2, multicentre, randomised, double-blind trial. Lancet Oncol. 2020;21:1283–95. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Jia M, He Z, Liu X-S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018;37:3924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barroso-Sousa R, Jain E, Cohen O, Kim D, Buendia-Buendia J, Winer E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol. 2020;31:387–94. [DOI] [PubMed] [Google Scholar]
- 23.Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia-Recio S, Mott KR, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179:1191–1206.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Driscoll CB, Schuelke MR, Kottke T, Thompson JM, Wongthida P, Tonne JM, et al. APOBEC3B-mediated corruption of the tumor cell immunopeptidome induces heteroclitic neoepitopes for cancer immunotherapy. Nat Commun. 2020;11:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pénzváltó Z, Chen JQ, Tepper CG, Davis RR, Silvestrini MT, Umeh-Garcia M, et al. A syngeneic erbb2 mammary cancer model for preclinical immunotherapy trials. J Mammary Gland Biol Neoplasia. 2019;24:149–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mabe NW, Fox DB, Lupo R, Decker AE, Phelps SN, Thompson JW, et al. Epigenetic silencing of tumor suppressor Par-4 promotes chemoresistance in recurrent breast cancer. J Clin Invest. 2018;128:4413–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 34.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–1. [DOI] [PubMed] [Google Scholar]
- 35.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 36.Faruki H, Mayhew GM, Serody JS, Hayes DN, Perou CM, Lai-Goldman M. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. J Thorac Oncol. 2017;12:943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamin DI, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. Calling Somatic SNVs and Indels with Mutect2. BioRxiv. 2019; [Google Scholar]
- 44.Raynaud F, Mina M, Tavernari D, Ciriello G. Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet. 2018;14:e1007669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elson A, Leder P. Protein-tyrosine phosphatase epsilon. An isoform specifically expressed in mouse mammary tumors initiated by v-Ha-ras OR neu. J Biol Chem. 1995;270:26116–22. [DOI] [PubMed] [Google Scholar]
- 46.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikkilä J, Kumar R, Campbell J, Brandsma I, Pemberton HN, Wallberg F, et al. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br J Cancer. 2017;117:113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017;19:1189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beatty GL, Winograd R, Evans RA, Long KB, Luque SL, Lee JW, et al. Exclusion of T cells from pancreatic carcinomas in mice is regulated by ly6c(low) F4/80(+) extratumoral macrophages. Gastroenterology. 2015;149:201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosoi A, Takeda K, Nagaoka K, Iino T, Matsushita H, Ueha S, et al. Increased diversity with reduced “diversity evenness” of tumor infiltrating T-cells for the successful cancer immunotherapy. Sci Rep. 2018;8:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa RLB, Czerniecki BJ. Clinical development of immunotherapies for HER2+ breast cancer: a review of HER2-directed monoclonal antibodies and beyond. NPJ Breast Cancer. 2020;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
- 56.Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9:5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell. 2019;179:219–235.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–41. [DOI] [PubMed] [Google Scholar]
- 60.McDonald K-A, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, et al. Tumor Heterogeneity Correlates with Less Immune Response and Worse Survival in Breast Cancer Patients. Ann Surg Oncol. 2019;26:2191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


