SUMMARY
Mosquitoes locate and approach humans based on the activity of odorant receptors (ORs) expressed on olfactory receptor neurons (ORNs). Olfactogenetic experiments in Anopheles gambiae mosquitoes revealed that the ectopic expression of an AgOR (AgOR2) in ORNs dampened the activity of the expressing neuron. This contrasts with studies in Drosophila melanogaster in which the ectopic expression of non-native ORs in ORNs confers ectopic neuronal responses without interfering with native olfactory physiology. RNA-seq analyses comparing wild-type antennae to those ectopically expressing AgOR2 in ORNs indicated that nearly all AgOR transcripts were significantly downregulated (except for AgOR2). Additional experiments suggest that AgOR2 protein rather than mRNA mediates this downregulation. Using in situ hybridization, we find that AgOR gene choice is active into adulthood and that AgOR2 expression inhibits AgORs from turning on at this late stage. Our study shows that the ORNs of Anopheles mosquitoes (in contrast to Drosophila) are sensitive to a currently unexplored mechanism of AgOR regulation.
Graphical Abstract

In brief
Maguire et al. discover that the ectopic expression of an olfactory receptor can downregulate the transcription of endogenous odorant receptors in mosquito olfactory neurons. The onset of mosquito odorant-receptor expression by an olfactory neuron continues into adult stages, and is particularly sensitive to exogenous olfactory reception expression.
INTRODUCTION
Malaria is a major cause of human mortality worldwide (Global Malaria Programme, 2019), and it is a global health imperative to prevent the spread of this disease. Malaria is caused by Plasmodium parasites transmitted by the bite of infected Anopheles mosquitoes. To date, antimalarial drugs have been the mainstay of control against malaria, and over the past 15 years, these treatments, along with the distribution of insecticide-treated bed nets, have contributed to an overall reduction of disease transmission. However, the eventual eradication of malaria likely rests on a multidisciplinary approach that integrates our knowledge of host, parasite, and vector biology (Cowman et al., 2016). For example, impairing the ability of the insect vector to bite a human host may further reduce incidences of infection. As such, disrupting the behaviors that bring mosquitoes to humans could dramatically reduce the prevalence of malaria (Beauty and Marquardt, 1996).
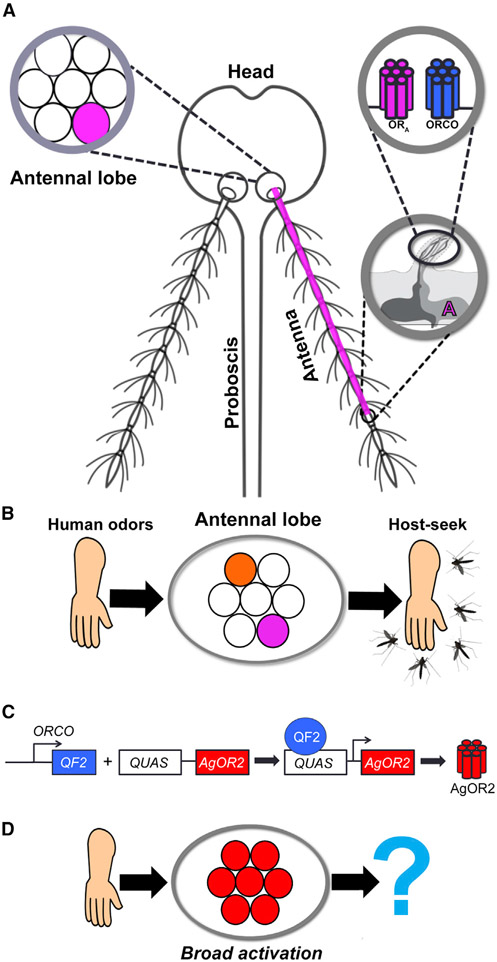
Female Anopheles mosquitoes locate and approach humans (“host seek”) based on specific cues, such as human-derived odors and exhaled CO2, moisture and heat emissions, and body shape (Konopka et al., 2021). The primary way that mosquitoes host seek is through their sense of smell (olfaction). Mosquitoes have evolved a complex repertoire of chemoreceptors that respond to chemical stimuli such as ionotropic receptors (IRs), gustatory receptors (GRs), and the odorant receptors (ORs). Of these, the ORs play a substantial role in mediating how a mosquito responds to human odors (DeGennaro et al., 2013). ORs form heterotetramer complexes with an OR co-receptor (ORCO) (Butterwick et al., 2018). OR-ORCO complexes are expressed on olfactory receptor neurons (ORNs) of the sensory appendages of the mosquito: antennae, maxillary palps, and labella (Riabinina et al., 2016). ORCO+ ORNs are housed in “sensilla,” or the sensory hairs of these appendages. Each sensillum typically contains 1–4 ORNs that each express a unique OR, of which there are 75 in the A. gambiae genome (Giraldo-Calderón et al., 2015). These ORNs are classified and named by the OR gene they express, and each ORCO+ ORN class targets a specific brain region of the mosquito antennal lobe (AL) known as a glomerulus (Couto et al., 2005; Ghaninia et al., 2007). The decision to approach a host is a direct result of activated ORCO+ ORNs targeting a specific combination of glomeruli (Figures 1A and 1B).
Figure 1. Strategy to manipulate the olfactory system of Anopheles mosquitoes.
(A) Anatomy of OR-guided olfaction. Mosquitoes smell odors in the environment using 3 olfactory organs: the proboscis, the maxillary palps (not shown), and the antennae. A single antenna is made up of 13 segments called flagellomeres. Each flagellomere is covered with sensory hairs called sensilla, a single one of which houses up to 4 ORNs. An ORN expresses 1 of 3 chemosensory gene families: the IRs, the GRs, or the ORs. The OR gene family plays an important role in host-seeking behavior. There are 75 different ORs in the Anopheles gambiae genome, each of which is sensitive to specific odorants in the environment. Only 1 OR (ORA) is expressed per ORN (encircled A). At the dendrites of ORN A, ORA couples with ORCO. When odorants bind to ORA-ORCO complexes, ORN A becomes active and sends its excitatory signal down its axon, targeting a discrete brain region of the mosquito AL called a glomerulus (shown as a pink circle).
(B) A human-specific odor code in the mosquito brain. Human odorants bind to specific ORs, activating the ORNs on which they are expressed. Activated ORNs target discrete glomeruli (pink, orange) in the AL to guide host seeking.
(C) Olfactogenetics strategy. Using the Q-system, the ORCO-QF2 transgene (Riabinina et al., 2016) was combined with the effector construct QUAS-AgOR2. The combination of these transgenes causes AgOR2 to be expressed in ORCO+ ORNs.
(D) Test of the olfactogenetics strategy. AgOR2 responds to major components of human and animal odors such as indole and benzaldehyde. We hypothesized that ORCO > AgOR2 mosquitoes would experience broad activation of the majority of the AL in the presence of a human, thereby dysregulating the human-specific odor code in the mosquito brain (Figure 1B). ORCO > AgOR2 mosquitoes would then be evaluated for reduced host-seeking behavior.
Disrupting this specificity by activating all ORCO+ ORNs in the presence of human odors has been hypothesized to prevent host seeking (Clark and Ray, 2016; Jones et al., 2011). Studies in Drosophila, the insect model of olfaction, show that binary systems can be used to express non-native ORs in ORCO+ ORNs to confer ectopic neuronal responses without interfering with native olfactory physiology. Therefore, we examined whether this strategy could be used to disrupt host seeking in A. gambiae mosquitoes. To test this, we expressed A. gambiae OR 2 (AgOR2) in all ORCO+ ORNs. AgOR2 is highly attuned to major components of human and animal odors such as benzaldehyde and indole (Carey et al., 2010), and in the presence of human odors, all ORCO+ neurons expressing AgOR2 should become active. Surprisingly, when we evaluated the olfactory physiology of these experimental mosquitoes, we found that they exhibited reduced responses not only to the cognate ligands of AgOR2 (benzaldehyde and indole) but also to odorants in general. To investigate the molecular basis of this phenotype, we looked for signatures of dysfunction at the level of the transcriptome. Using RNA sequencing (RNA-seq) to compare transcript levels from wild-type antennae to those ectopically expressing AgOR2, we discovered that A. gambiae OR isoforms were significantly downregulated in the experimental line, while the remaining transcripts were largely unchanged. Additional experiments revealed that AgOR2 protein rather than AgOR2 mRNA is needed to reduce native OR levels. We also made the unexpected finding that AgOR gene choice extends into adulthood—a stage in Drosophila when each ORN is thought to have already chosen a distinct OR (McLaughlin et al., 2021)—because AgOR2 suppresses an increase in AgOR-expressing cells observed in wild-type antenna 1–8 days post-eclosion (PE). Overall, our study suggests that an OR-mediated feedback mechanism exists that can regulate A. gambiae OR expression.
RESULTS
Ectopically expressing AgOR2 in ORCO+ neurons impairs olfactory physiology
To activate ORCO+ ORNs in the presence of human odorants, we used olfactogenetics, a technique whereby a specific volatile odorant is used to activate a defined set of OR-expressing neurons (Chin et al., 2018). To accomplish this, an OR with known response properties is ectopically expressed in ORNs of interest through the use of a binary expression system, such as the Q-system (Riabinina et al., 2015) or the GAL4-UAS system. Thus, in the presence of odorants in which the introduced OR normally responds, ORNs with ectopic expression become active.
The recent introduction of the Q-system into Anopheles makes it possible to adapt olfactogenetics in mosquitoes (Riabinina et al., 2016). As a binary expression system, the Q-system works by directing the expression of a specific gene into a specific cell population. This particular system relies on two elements: QF2 and QUAS. The QF2 transcription factor is expressed under the control of a cell-type-specific enhancer/promoter and binds to its upstream activating sequence, QUAS. Once bound by QF2, QUAS initiates the transcription of its effector gene. To ectopically activate ORCO+ ORNs in the presence of human odors, we combined a mosquito line containing ORCO-QF2 (Riabinina et al., 2016), which contains a fusion between the presumptive enhancer and promoter regions of the gene ORCO and the transcription factor QF2, with an effector line containing a QUAS transgene upstream of A. gambiae OR 2, AgOR2 (QUAS-AgOR2). Thus, experimental animals exhibit the ectopic expression of AgOR2 in all ORCO+ ORNs (Figure 1C).
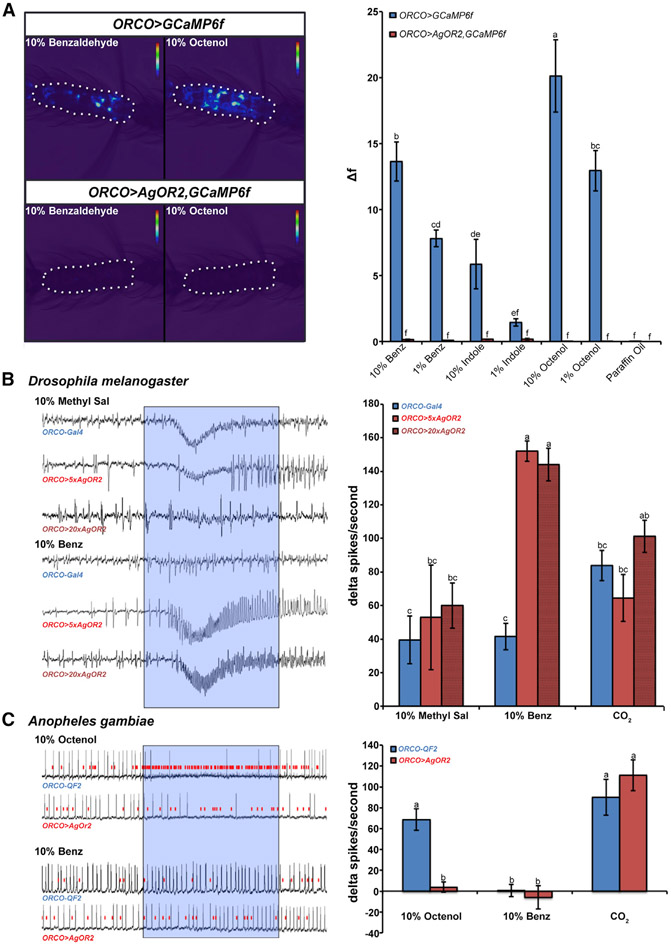
AgOR2 is highly attuned to components of human and animal odors such as benzaldehyde and indole (Carey et al., 2010), and it was expected that ORCO+ ORNs of ORCO >AgOR2 mosquitoes would become active in the presence of these cognate ligands, essentially activating the majority of the olfactory system during host seeking (Figure 1D). To test the functional activity of ORNs ectopically expressing AgOR2 in mosquitoes, we imaged the calcium response of ORCO > AgOR2 antennal segments in a QUAS-GCaMP6f background (ORCO > AgOR2,GCaMP6f). Surprisingly, ORCO > AgOR2,GCaMP6f antennae showed a dampened response not only to various concentrations of benzaldehyde and indole but also to odorants in general (Figure 2A). For example, octenol potentially activates 31 different A. gambiae ORs (Carey et al., 2010), but the ORNs of the experimental mosquitoes did not show a response to this odorant. In Drosophila, a single ORN class can drive behavior at spike rates as low as 10–20 Hz (Bell and Wilson, 2016). When we stimulated ORCO > AgOR2,GCaMP6f with 5 additional odorants known to activate 14–16 different ORN classes at rates higher than 50 spikes/s (Carey et al., 2010), the ORNs still did not exhibit odorant-induced responses (Figure S1).
Figure 2. Olfactogenetics impairs Anopheles but not Drosophila ORCO+ ORNs.
(A) ORCO > AgOR2,GCamp6f mosquitoes show impaired olfactory responses to human odorants. The activity of ORNs in antennal segment 11 (outlined with a white dotted line) was detected by calcium imaging of ORCO+ neurons expressing GCaMP6f (Afify et al., 2019). Relative to controls (ORCO > GCaMP6f), mosquito antennal segments with the ectopic expression of AgOR2 and GCaMP6f (ORCO > AgOR2,GCaMP6f) show impaired responses to benzaldehyde (10% and 1%) and indole (10%), the cognate ligands of AgOR2. They also show dampened responses to octenol (10% and 1%). A 2-way repeated measures ANOVA was conducted to test the effect of odorant and genotype on calcium responses (F(6,7) = 75.1, p < 0.001). Groups with different letter values (A–F) are statistically different as determined by the Tukey post hoc honest significant difference (HSD) test. Each sample included in the analysis was taken from a different female mosquito. nORCO>GCaMP6f = 9, nORCO>AgOR2,GCaMP6f = 8.
(B) Ectopic expression of AgOR2 in Drosophila ab1 sensilla does not impair ORN physiology. The cognate ligand of DmOR10a-expressing neurons is methyl salicylate (Methyl Sal). Driving AgOR2 into this neuronal group using the 5xUAS or 20xUAS effector lines does not affect the response of DmOR10a to Methyl Sal. ORCO+ ORNs of ORCO > 5xAgOR2 and ORCO > 20xAgOR2 animals show an ectopic response to benzaldehyde (Benz). The presence of the ORCO− CO2 neuron was used to verify that recordings were taken from the ab1 sensillum. The activity of the CO2 neuron is not affected by the experimental manipulation. Odorant or CO2 stimulus was delivered in the time frame denoted by the blue translucent box. A 2-way repeated measures ANOVA was used to determine the significance of genotype and odorant on delta spikes/s (F(4,28) = 13.7, p < 0.0001). Groups with different letter values (A-C) are statistically different as determined by the Tukey post hoc HSD test. Two to three females per genotype were analyzed. The number of sensilla evaluated for each group was nORCO-GAL4 = 9; nORCO>5xAgOR2 = 4; nORCO>20xAgOR2 = 5.
(C) Olfactogenetics impairs ORCO+ ORN physiology in Anopheles. SSR from the Anopheles maxillary palp cp sensilla. The cognate ligand of AgOR8-expressing neurons is octenol. Driving AgOR2 into this neuron group interferes with the response of AgOR8 to octenol (smallest spiking neurons; red lines indicate AgOR8 activity). The ORNs ectopically expressing AgOR2 do not respond to Benz. The presence of the ORCO− CO2 neuron was used to verify that recordings were taken from a cp sensillum. The activity of the CO2 neuron was not affected by the experimental manipulation. Odorant or CO2 was delivered in the time frame denoted by the blue translucent box. A 2-way repeated measures ANOVA was used to determine that there was a significant effect at odorant and genotype on delta spikes/s (F(4,5) = 8.5, p = 0.01). Groups with different letter values (A and B) are statistically different as determined by the Tukey post doc HSD test. Two to three females per genotype were analyzed. The number of sensilla evaluated for each group was nORCO-QF2 = 6; nORCO>AgOR2 = 5. The error bars in all of the quantified figures represent the standard error of the mean (SEM).
For (B and C) (left), the x axis represents time (ms) and the y axis represents voltage.
One possibility for the observed olfactory defects (Figures 2A and S1) is that the QUAS-AgOR2 transgene inserted into an endogenous olfactory gene and disrupted olfactory function. To date, the genome organization is unavailable for the Anopheles strain used in this study (A. coluzzii N’gousso, ACON), so we used splinkerette mapping (Potter and Luo, 2010) to align the insertion site of the QUAS-AgOR2 element to a related Anopheles strain (A. coluzzii Mali, ACOM). The transgenic line examined in Figures 2A and S1 (line 1) is located in an intergenic region on chromosome 3R between the genes ACOM029303 and ACOM029196. While it is unlikely that this particular insertion site would disrupt the function of an olfactory gene necessary for ORN physiology, we extended our studies to analyze the physiology of two additional QUAS-AgOR2 lines inserted into different regions of the genome. We established line QUAS-AgOR2#2, which maps to an intergenic region between ACOM036217 and ACOM036230, and line QUAS-AgOR2#3, which could not be mapped by splinkerette PCR. When driven into ORCO+ cells, all three lines show similar defects in olfactory physiology when compared to wild type (Figure S2). Furthermore, since these lines were tested as heterozygotes, any recessive mutation in an olfactory gene caused by the QUAS-AgOR2 insertion should be compensated by the wild-type allele. Overall, these data show that the dominant-negative olfactory phenotype (Figures 2A, S1, and S2) is a consequence of ectopic AgOR2 expression rather than the genomic insertion site of the QUAS-AgOR2 element.
Olfactogenetics impairs Anopheles but not Drosophila ORCO+ ORNs
Ectopically expressing an OR in Drosophila ORNs causes the expressing neuron to activate in the presence of the odor ligand of the introduced OR (Carey et al., 2010; Chin et al., 2018; Dobritsa et al., 2003; Hallem et al., 2004; Ray et al., 2007). One possibility as to why ORCO > AgOR2 cells in the mosquito did not respond to benzaldehyde or indole was because the AgOR2 sequence used to create the transgenic QUAS-AgOR2 line was acting in a dominant-negative manner to disrupt ORCO/ORX ion channels. To test this, we ectopically expressed AgOR2 in Drosophila ORCO+ neurons using the GAL4-UAS system and measured the response rate of neurons housed in the ab1 sensilla using single sensillum recordings (SSRs). Ab1 contains 4 ORNs, 3 of which are ORCO+ and express DmOR10a, DmOR42b, and DmOR92a, and one of which is ORCO− and expresses the GR DmGR21, which responds to CO2. To determine whether ectopic expression of AgOR2 impairs native DmOR responses to their cognate ligands, responses of the DmOR10a-expressing neuron to methyl salicylate were measured. We found no difference in how control (ORCO-GAL4) and experimental (ORCO > 5xAgOR2) sensilla responded to methyl salicylate, indicating that AgOR2 expression does not interfere with the olfactory physiology of the neuron in which it is expressed. Furthermore, native responses of DmOR10a were not affected when an even higher dosage of AgOR2 was driven into the neuron (ORCO > 20xAgOR2) (Figure 2B). When we puffed benzaldehyde over the experimental preparation, ORCO+ cells in the Drosophila ab1 sensilla ectopically responded. Interestingly, sensilla that have higher levels of ectopic AgOR2 can still maintain ectopic responses without compromising neuron function. When we used the stronger effector line (20XUAS) to ectopically express AgOR2 in Drosophila ORCO+ neurons, the olfactogenetics approach continued to work and olfactory physiology was not impaired.
In Drosophila, the DmORCO gene turns on relatively late—80 h after puparium formation (APF) during the development of the adult olfactory system—and is approximately coincident with the onset and expression of DmOR genes (Clyne et al., 1999; Elmore et al., 2003; Larsson et al., 2004; Trebels et al., 2021). We next examined whether driving AgOR2 expression earlier in development, before the onset of DmOR expression, would affect odorant responses. Pebbled-Gal4 drives expression in most cells residing in the antenna and maxillary palp during development and can be observed as early as 18 h APF in ORNs (Sweeney et al., 2007), before the detection of the earliest expressing DmOR genes at 24 h APF (McLaughlin et al., 2021). Using pebbled-Gal4 to ectopically drive 5xUAS-AgOR2 or 20xUAS-AgOR2 in ORNs, the DmOR10a-expressing neurons in ab1 sensilla continue to respond normally to methyl salicylate (Figure S3).
The Anopheles capitate peg (cp) sensillum is similar to the ab1 sensillum of Drosophila as it contains ORCO+ ORNs, which express AgOR8 and AgOR28, and one ORCO-independent ORN that responds to CO2. When we ectopically expressed AgOR2 in ORCO+ ORNs in the mosquito and recorded from cp sensilla, we found that the response of the AgOR8-expressing neuron to octenol, its cognate ligand, was eliminated (Figure 2C). In addition, neither AgOR8 nor AgOR28-expressing neurons ectopically respond to benzaldehyde. Similar to Drosophila, the genetic manipulation does not affect the physiology of the ORCO− CO2-responsive neuron (Figure 2C). These data indicate that olfactogenetics affects the physiology of Drosophila and Anopheles ORCO+ ORNs differently: in flies, ectopically expressing ORs does not affect endogenous neuronal function, whereas in Anopheles mosquitoes, the ectopic expression of AgOR2 disrupts the function of the ORNs.
Ectopic AgOR2 protein eliminates olfactory responses in ORCO+ cells
It is possible that driving an AgOR into an Anopheles ORN is cytotoxic, especially if the neuron experiences continual stimulation from the environment. To assess whether ORCO > AgOR2 mosquitoes show ORCO+ cell loss, we (1) compared the number of ORCO+ cells per flagellomere to that of the genetic control and also (2) evaluated whether ORCO > AgOR2 ORN projections to the AL were intact. Using immunochemistry to visualize ORCO+ cells in the periphery and ORN terminals in the AL, we found that ORN cell number and projections were unperturbed in ORCO > AgOR2, indicating that the lack of ORN responses to odors was not due to the death and elimination of neurons ectopically expressing AgOR2 (Figure S4). Next, we tested whether driving any generic transmembrane protein into Anopheles ORNs also hindered olfactory physiology. To evaluate this, we used the ORCO-QF2 driver to ectopically express the transmembrane protein mCD8:GFP into all ORCO+ neurons. There were no differences in the odor-induced responses between control and mCD8:GFP+ ORNs (Figure S5), suggesting that the ectopic expression of another transmembrane protein did not silence ORN activities.
Ectopic AgOR2 expression may inhibit olfactory responses either at the level of AgOR2 mRNA or at the level of AgOR2 protein. To distinguish between these two possibilities, we created a transgenic mosquito line containing a mutated version of AgOR2 (mutAgOR2) that contained a mutation in the start codon of AgOR2 such that QUAS-mutAgOR2 produced mRNA that cannot be translated when combined with ORCO-QF2. We also induced a frameshift mutation at a second in-frame ATG site in mutAgOR2 to eliminate the possibility of having an alternative open reading frame used during translation. We crossed QUAS-mutAgOr2 with ORCO-QF2,QUAS-GCaMP6f and found that the calcium responses were not compromised (Figure 3); only when the wild-type version of the protein was expressed (QUAS-AgOR2) was the odor response impaired (Figures 2A, 2C, S1, and S2). Taken together, these data suggest that the AgOR2 protein itself, and not mRNA, was responsible for the olfactory defect of ORCO+ cells (but see limitations of the study in the discussion).
Figure 3. AgOR2 protein is required for the dominant-negative olfactory phenotype caused by ORCO > AgOR2 expression.
The mutAgOR2 transgene contains an introduced point mutation in the start codon of AgOR2 and a frameshift mutation at a second in-frame ATG site of the gene. Odorant-evoked responses (Δf) were calculated from the 11th segment of the mosquito antennae (outlined with dotted lines). Representative control and experimental calcium imaging responses to 10% Benz and 10% octenol are shown. Antennae from ORCO > mutAgOR2,GCaMP6f mosquitoes show no difference in responses to odorants from control (ORCO > GCaMP6f). A 2-way repeated measures ANOVA was used to determine whether there was a significant effect of odorant and genotype on calcium responses (F(6,9) = 47.3, p < 0.0001). Groups with different letter values (a–d) are statistically different as determined by the Tukey post hoc HSD test. The error bars represent SEMs. Each sample included in the analysis was taken from a different female mosquito. nORCO>GCaMP6f = 11, nORCO>mutAgOR2,GCaMP6f = 5.
Ectopic AgOR2 reduces the transcripts of native AgORs
How might AgOR2 protein impair olfactory responses? We hypothesized this may occur as a result of (1) regulatory mechanisms affecting AgOR transcription, stability, and/or degradation rates or (2) regulatory mechanisms and/or defects in AgOR protein function. These transcriptional or post-translational mechanisms may act together or independently. To distinguish between these two possibilities, we performed isoform-level RNA-seq on antennae isolated from control and ORCO > AgOR2 samples. Of the ~13,000 transcript isoforms detected (Data S1), only 83 were differentially expressed in the ORCO > AgOR2 mosquito antennae. Interestingly, half of these differentially expressed isoforms (41/83) were AgORs, all of which were downregulated in ORCO > AgOR2 antennae, except for AgOR2, which was highly upregulated when compared to the wild type (Figures 4A and S6; Data S2). As shown by the AgOR isoform comparison heatmaps in Figure 4B, control AgOR levels are higher than those in ORCO > AgOR2, with the exception of AgOR2, which is upregulated.
Figure 4. Ectopic AgOR2 protein reduces the transcript levels of AgOR isoforms.
(A) Volcano plot of differentially expressed isoforms. Using Wald tests, we evaluated whether the 13,224 isoforms present in 3 triplicates of control and 3 triplicates of ORCO > AgOR2 antennae (~200 antennae per sample) were differentially expressed. Non-A. gambiae OR and A. gambiae OR isoforms are shown as black or red dots, respectively. Only 0.63% of the transcriptome is differentially regulated in ORCO > AgOR2, in which 49% of those transcripts are AgORs (Figure S6; Data S2). log(q) is the level of significance of β1, which, for each isoform, is defined as TPMcontrol −TPMexperimental. The 41 AgORs found to be significant from the Wald tests are labeled according to their gene annotation in the volcano plot. AgORCO, while not differentially expressed, is also indicated on the plot. All AgORs (with the exception of AgOR2) that are differentially expressed are downregulated in the experimental condition when compared to wild type. Interestingly, AgIR8a and AgIR75k (indicated on the volcano plot) are downregulated in ORCO > AgOR2. The remaining AgIRs and AgGRs are unaffected.
(B and C) Heatmap of the AgOR gene family. A Z score was computed for each cell in the heatmap by subtracting the mean isoform TPM from the TPM of the cell divided by the standard deviation (SD) of the isoform TPM. AgORs are sorted along the x axis according to their significance level (q value) from the Wald tests in (A) Darker orange is most significant and yellow is not significant. Note that ORCO > AgOR2 is listed as O > AgOR2. (C) Ectopic AgOR2 protein is required for the observed downregulation of native AgOR transcripts. Ternary plots were used to visualize the relative ratio of a genotype to the relative abundance level of an isoform using the formula (TPMgenotypex)/(mean TPMcontrol + mean TPMORCO>AgOR2 + mean TPMORCO>mutAgOR2) × 100. There are equal ratios of transcript abundance levels for non-AgOR genes among the 3 genotypes (left). However, the relative contribution to transcript abundance levels of control, ORCO > AgOR2, and ORCO > mutAgOR2 are skewed in the AgOR gene family such that AgOR gene levels in ORCO > mutAgOR2 and control are relatively similar and higher than ORCO > AgOR2 levels, with the exception of AgOR2 itself (whose abundance is similar between ORCO > AgOR2 and ORCO > mutAgOR2) (right).
If AgOR2 ectopic protein was responsible for modulating the steady-state abundance of native AgOR transcripts, then the relative abundance of the A. gambiae OR gene family isoforms in ORCO > mutAgOR2 antennae should not be affected. To test this, we extracted biological triplicates of mosquito antennae from female mosquitoes of the ORCO > mutAgOR2 line and compared the isoform abundance levels of this group to the original RNA-seq dataset. Ternary plots depicting the relative abundance of isoform expression levels in control, ORCO > mutAgOR2, and ORCO > AgOR2 conditions as a position on an equilateral triangle were used to explore how AgOR and other gene sets were differentially expressed. To depict how control, ORCO > mutAgOR2, and ORCO > AgOR2 contribute to relative isoform abundances of non-AgOR genes and AgOR genes, we created 2 discrete ternary plots. As expected, the relative contribution of control, ORCO > mutAgOR2, and ORCO > AgOR2 for all non-A. gambiae OR isoforms in the transcriptome was roughly equal (34.3%:33%:32.6%). However, when removing the contribution of the ectopically induced AgOR2 from the AgOR isoform pool, the relative contribution of the abundance levels of an AgOR was reduced in ORCO > AgOR2 relative to control and ORCO > mutAgOR2 groups: 45.8%:37.8%:16.4% (control:ORCO > mutAgOR2:ORCO > AgOR2) (Figure 4C). Results of the gene set enrichment test and pairwise comparisons of AgOR isoforms can be viewed in Figure S7. The majority of native AgORs are downregulated in ORCO > AgOR2 but not in ORCO > mutAgOR2, with four exceptions: AgOR2, AgOR16, AgOR17, and AgOR33.
AgOR2 feedback occurs during the adult stage of the life cycle of the mosquito
We next examined when, during the life cycle of the mosquito, the downregulation of native AgOR transcripts occurred by ectopic AgOR2. Since ectopic AgOR2 expression is dictated by the ORCO enhancer/promoter region (Figure 1C) (Riabinina et al., 2016), AgOR2-induced olfactory silencing of native AgOR genes should coincide with the AgORCO expression pattern. Holometabolous insects, such as mosquitoes, experience four developmental stages during their lifespan: egg, larva, pupa, and adult stages. Since the lifestyle of adults and larvae differ, the olfactory system is remodeled to take on adult features during pupal development (Mysore et al., 2011, 2013; Trebels et al., 2021), so we performed immunohistochemistry on pupal antennae from ORCO > mCD8:GFP mosquitoes every 4 h starting at the onset of pupal development (0–2 h APF) to just before eclosion (20–22 h APF), which happens at 24 h APF. AgORCO (as reported by mCD8 staining) was expressed immediately after pupal ecdysis at 0–2 h APF (Figures 5A and 5B). Interestingly, ORCO expression turned on gradually in antennal ORNs. Starting at 8–10 h APF (when the first antennal flagellomeres could be observed), each segment gained ~15 ORCO+ cells every 4 h, starting from 37.5 ± 2 cells and ending with 87 ± 20 cells per flagellomere at 20–22 h APF (Figures 5A and 5B). These data suggest that the ectopic expression of AgOR2 in the ORCO > AgOR2 genotype (which coincides with ORCO) also occurred throughout pupal development, and may impinge upon developmental mechanisms that regulate AgOR expression. To see whether this was the case, we used in situ hybridizations to characterize the expression patterns of the 3 AgOR genes most significantly downregulated by ectopic AgOR2: AgOR11, AgOR24, and AgOR41 (Figure 4 and S7; Data S2). In wild-type ORNs, AgOR11+, AgOR24+, and AgOR41+ cells were first detected at 8–10 h APF and continued to increase in number into adulthood up to the last time point sampled: 4–8 days PE (Figures 5C and 5D). This is well past what was typically considered the end of OR gene choice as determined by other insect model organisms of olfaction (Trebels et al., 2021). In ORCO > AgOR2-expressing mosquitoes, AgOR11+, AgOR24+ and AgOR41+ cells were also observed starting at 8–10 h APF and showed no differences in number until 20–22 h APF. From 20 to 22 h APF until the last time point sampled (4–8 days PE), AgOR11-, AgOR24-, and AgOR41-expressing cells did not increase in number as they did in the wild type. We speculate that this may be because at these later stages, AgOR2 mRNA and AgOR2 protein is expressed in every ORN (Figures 5B and 5E) and would prevent ORNs that have not yet chosen an AgOR from expressing transcripts from the AgOR gene family. Overall, these data show that the negative inhibition of AgOR genes by AgOR2 protein occurs during the establishment of AgOR gene choice, which extends into adulthood.
Figure 5. AgOR2-feedback occurs during the adult stage.
(A) AgORCO is expressed at the start of pupal ecdysis. Representative immunohistochemistry images of ORCO-expressing neurons in the ORCO > mCD8:GFP genotype. Pupal antennae were extracted at the indicated time point and stained with anti-mCD8 (green). Cells were scored as ORCO+ based on the presence of mCD8.
(B) ORCO+ cell number increases over pupal development. An ANOVA was used to determine whether the number of cells that express mCD8 change over pupal development. There was a significant effect of time on the number of cells that express mCD8 (F(3,16) = 6.44, p = 0.0046). Groups with different letter values are statistically different as determined by the Tukey post hoc HSD test (A and B).
(C–E) Representative in situ hybridization images of wild-type and ORCO > AgOR2 antennae hybridized to probes against AgOR2 (red) and AgOR11, AgOR24, AgOR41 (green) mRNA. Tissue was sampled every 4 h APF as well as 1–2 days PE and 4–8 days PE. The pink insets (top right) of each panel show AgOR11, AgOR24, AgOR41-expressing cells without AgOR2 co-expression and the blue insets (bottom right) show cells with co-expression. AgOR2 did not co-express with AgOR11, AgOR24, AgOR41 in wild-type ORNs. Independent sample t tests were conducted to compare the number of cells that express AgOR11, AgOR24, and AgOR41 (D) or AgOR2 (E) between wild type and ORCO > AgOR2 for each time point. *p < 0.05, **p < 0.005, ***p < 0.0005. For the results of each statistical test, refer to Table S1. An ANOVA was used to determine whether the number of cells that express AgOR11, AgOR24, and AgOR41 (D) or (E) AgOR2 change within the given genotype over time. There was a significant effect of time on the number of cells that express AgOR11, AgOR24, and AgOR41 in wild type (F(5,24) = 15.8, p < 0.0001) and ORCO > AgOR2 (F(5,22) = 4.54, p = 0.005) flagellomeres and the number of cells that express AgOR2 in ORCO > AgOR2 (F(5,22 = 26.7, p < 0.0001). There was no effect of time of the number of cells that express AgOR2 in wild-type flagellomeres. For both immunochemistry and in situ hybridization experiments, cells were scored once per flagellomere (after 8–10 h APF, inclusive). For each time point, cells from 1 flagellomere per animal in an average of 5 animals per time point were scored. The location of each scored flagellomere was randomized for each antenna.
Flagellomeres in (A and C) are indicated by a dotted white line. The error bars in (C–E) represent SEMs.
Impairing ORCO+ neuron responses inhibits olfactory preference but not host-seeking behavior in Anopheles mosquitoes
In the absence of CO2, ORCO mutant Aedes (DeGennaro et al., 2013; Raji et al., 2019) and Anopheles (Sun et al., 2020) mosquitoes demonstrate reduced host attraction. Under conditions that replicate an environment containing a naturally breathing human, CO2 synergizes with host odors to rescue defects in ORCO mutant attraction (DeGennaro et al., 2013), suggesting that mosquitoes possess redundant mechanisms (such as IR genes) that activate in the presence of CO2 (Raji et al., 2019). The dominant-negative phenotype of ORCO > AgOR2 presented the opportunity to test whether Anopheles mosquitoes with impaired ORCO function also continue to host seek in the presence of CO2. To test this, we used the host-proximity assay (DeGennaro et al., 2013), a population assay that measures the proportion of females that come into olfactory (but not physical) contact with a human arm. As found with ORCO mutant Aedes mosquitoes (DeGennaro et al., 2013; Raji et al., 2019), Anopheles mosquitoes without functional ORCO+ neurons were still attracted to a human host in the presence of CO2 (Figures 6A-6C).
Figure 6. ORCO > AgOR2 mosquitoes remain attracted to a human host.
(A) Representative images of wild-type, ORCO-QF2, QUAS-AgOR2, and ORCO > AgOR2 mosquitoes (circled) in the host-proximity assay. Mosquitoes attracted to an arm (2.5 cm from the cage) that land on the net are counted.
(B) Results of the host-proximity assay. A 1-way ANOVA between subjects was conducted to compare the effect of genotype on percentage of attraction. There was no effect of genotype on percentage of attraction at the p < 0.05 level for the 4 groups (F(3) = 1.08, p = 0.37). A total of 20–30 female mosquitoes were tested per trial. The number of trials per genotype: nwild-type = 5; nORCO-QF2 = 5; nQUAS-AgOR2 = 7; nORCO>AgOR2 = 7.
(C) Time course of mosquito attraction toward a human host by genotype. Over the course of 3 min, there was no difference in the percentage of mosquitoes attracted to a human host.
(D) Results of the olfactory-based oviposition preference assay. Paired-samples t tests were conducted to compare the total number of eggs laid in solvent control double cups and in lemongrass oil (0.1%) double cups for each genotype (wild type, ORCO-QF2, QUAS-AgOR2, and ORCO > AgOR2). Control genotypes (wild type, ORCO-QF2, and QUAS-AgOR2) laid fewer eggs in lemongrass oil than the solvent control double cups (wild type: solvent control [mean = 632.4, SD = 208.5] and lemongrass oil [mean = 213, SD = 207.7], t(6) = 2.8, p < 0.05; ORCO-QF2: solvent control [mean = 560.1, SD = 394.9] and lemongrass oil [mean = 116.1, SD = 151.9], t(6) = 2.8, p < 0.05; QUAS-AgOR2: solvent control (mean = 384.3, SD = 246.4] and lemongrass oil [mean = 47.7, SD = 31.6], t(6) = 3.4, p < 0.05]). ORCO > AgOR2 mosquitoes laid the same number of eggs in solvent (mean = 612, SD = 374) and lemongrass oil (mean = 535.7,SD = 376.8), t(6) = 0.34, p = 0.74 double cups. ns, not significant, *p < 0.05. The number of trials per genotype: nwild-type = 7; nORCO-QF2 = 7; nQUAS-AgOR2 = 7; nORCO>AgOR2 = 7. The error bars in (B–D) represent SEMs.
While Anopheles mosquitoes continue to show strong attraction to host odors in the presence of CO2, it is likely that a loss of functional ORCO-expressing neurons impairs the ability of mosquitoes to discriminate between odorants (DeGennaro et al., 2013). Oviposition preference by gravid females is in part mediated by their olfactory system (Montell and Zwiebel, 2016). Sun et al. (2020) demonstrated that Anopheles mosquitoes without ORCO function cannot discriminate between substrates emanating control versus attractive odors, but it is unknown whether defects in ORCO function also cause mosquitoes to become insensitive to repellent odors. Lemongrass oil is a known mosquito repellent that activates a subset of ORCO+ ORNs (Afify et al., 2019). When we evaluated repellent odorant discrimination using an oviposition-based assay, we found that control genotypes laid fewer eggs on substrates emanating 0.1% lemongrass oil odor. In contrast, ORCO > AgOR2 mosquitoes laid the same number of eggs on oviposition substrates emanating 0.1% lemongrass oil or control odorants, indicating that an impairment in ORCO function causes a mosquito to lose sensitivity to repellent odors (Figure 6D).
DISCUSSION
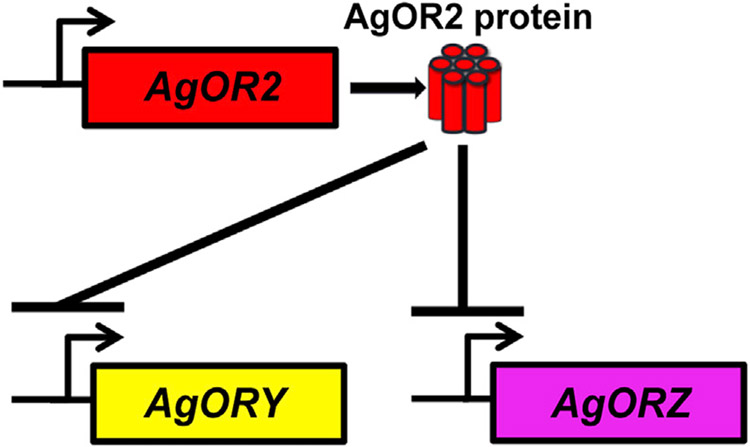
We report that a mechanism may exist to regulate AgOR gene stability in insects. A main finding of our study—that AgOR protein downregulates the expression of native AgORs (Figure 7)—is a pattern more closely resembling OR-regulatory processes in mice than those of Drosophila. This study demonstrates that driving ectopic expression of AgOR2 in non-native ORCO+ ORNs does not disrupt Drosophila neuron function, whereas in Anopheles it does (Figures 2 and S1-S3). Furthermore, ectopically expressing a DmOR gene in Drosophila ORNs does not affect the gene expression of native DmORs (Ray et al., 2007), whereas a similar manipulation in Anopheles leads to robust changes in gene expression (Figures 4 and S7; Data S2).
Figure 7. Summary model.
Ectopic AgOR2 negatively regulates the expression of AgOR transcripts. Our data implicate a mechanism of negative regulation of most of the AgOR transcripts (e.g., AgORY, AgORZ) by ectopic AgOR2 protein.
From these experiments, we hypothesize that unexpected similarities of AgOR gene regulation may exist between Anopheles and mice. For example, both Anopheles and mice contain a negative feedback loop by which OR protein inhibits the expression of the remaining ORs. In Anopheles mosquitoes, as was the case in mice, this feedback pathway likely requires intact OR protein (Figures 3 and 4; with 3 exceptions: see Figure S7) since expressing mutant OR genes lacking either the entire coding sequence or the start codon permits a second OR gene to be expressed (Feinstein et al., 2004; Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). Frameshift mutations also allow for the co-expression of functional OR genes (Serizawa et al., 2003). It remains to be determined whether mice and Anopheles mosquitoes use similar cellular machinery to regulate OR stability (Dalton et al., 2013).
Our data demonstrate that AgOR2 feedback occurs during the development of the olfactory system in Anopheles mosquitoes (Figure 5). Why might OR feedback be important for olfactory development? One consequence of OR feedback is that it increases the likelihood that a single ORN expresses only 1 OR, which is an important developmental mechanism in mice because ORs play an instructive role in guiding ORN axonal projections to the olfactory region of the brain (Mombaerts et al., 1996; Vassar et al., 1994; Wang et al., 1998). In experiments analogous to those presented here, when multiple ORs are genetically engineered to co-express in a single ORN in mice, the topographic map of projections to the olfactory center in the brain is perturbed (Clowney et al., 2012). While we do not know when ORNs target the AL in Anopheles, our methods expressed AgOR2 in ORCO+ neurons as early as 0–2 h APF (Figure 5). If ORNs have not yet targeted the AL at this early stage, then our data suggest that AgORs in Anopheles are likely not involved in axon guidance, as we did not detect obvious deformations of ORN targeting or AL structure in the adult brain (Figure S4). Why else might AgOR feedback be important during olfactory development? OR feedback (at least in mice) serves to reduce the expression of the remaining OR genes in an ORN, ensuring the singular expression of an OR by stabilizing the gene choice. We show that AgOR gene choice in ORNs continues into the adult stage (Figures 5C and 5D), which is traditionally thought to end at the late pupal stage (McLaughlin et al., 2021; Trebels et al., 2021). It is an intriguing possibility that the process of AgOR gene choice and stabilization remains active during adult stages as a way to allow the mosquito to synergize the physiology of ORNs with biological needs. Anopheles mosquitoes rely heavily on their sense of olfaction to integrate ecologically relevant stimuli that change over the course of their adult lifespan. When females first eclose, they are uninterested in host odors (Omondi et al., 2019) and instead actively search for sugar-rich resources from plants to supplement their nutrient reserves. After a period of ~4 days PE, they develop an attraction to host odorants (Foster and Takken, 2004; Omondi et al., 2019) and following a blood meal will experience a refractory period to host odorants until after oviposition. These changes in behavior have been correlated with changes in chemosensory gene transcript abundance both in Anopheles (Omondi et al., 2019) and Aedes aegypti (Tallon et al., 2019). More recently, Hill et al. (2021) demonstrated that Aedes aegypti mosquitoes show age- and state-dependent regulation of AeOR gene expression during adulthood. It would be interesting to determine whether this dynamic regulation observed in bulk antennal tissue is a consequence of mosquito OR gene levels changing within their native cells or whether new ORs are being chosen and stabilized via OR feedback.
AgORs can be co-expressed within the same ORN when transcribed as polycistronic mRNA (Karner et al., 2015). Polycistronic AgOR mRNA is observed in cases when AgOR genes are clustered tightly together within the genome. Such clustering of OR genes is commonplace in mosquito species such as A. gambiae (Hill et al., 2002) and Aedes aegypti (Bohbot et al., 2007), as well as in mice (Sullivan et al., 1996; Zhang et al., 2004); however, to our knowledge, polycistronic OR mRNA has not been observed in rodent olfactory systems. It is possible that polycistronic OR expression avoids the negative feedback mechanism of OR regulation, enabling the neuron to co-express multiple ORs.
While ectopically expressing AgOR2 downregulates native AgOR genes, it was surprising that ORCO > AgOR2 neurons did not show responses to the cognate ligands of AgOR2 (benzaldehyde and indole). From our RNA expression data, we estimate that a single wild-type ORN contributes 0.006 transcripts per million (TPM) of a given AgOR to a single antennal sample (calculations in Data S3). We estimate that ectopically expressing AgOR2 increases the AgOR expression level of a single cell to 0.02 TPM (2.5×). If all of the expressed AgORs are being translated in a cell, this would elevate the amount of AgOR protein that is normally observed in wild-type ORNs. We hypothesize that elevated AgOR protein levels impair olfactory physiology by disrupting the stoichiometry of the AgOR-ORCO complexes, rendering them non-functional. While research has shown that AgORs form stable heteromeric complexes with AgORCO (Benton et al., 2006; German et al., 2013; Neuhaus et al., 2005; Tsitoura et al., 2010), the stoichiometry underlying AgOR-ORCO channels is unknown. RNA-seq data from this study (Data S3) and from two independent studies (Athrey et al., 2017; Pitts et al., 2011) show that in wild-type conditions, there is a conserved ~1:1 relationship between total AgOR transcripts and AgORCO in bulk antennal tissue. Interestingly, we see that ectopic AgOR2 expression does not change AgORCO expression (Figure 4B), skewing the estimated ratio of AgORs:AgORCO in a single cell from 1.2:1 in wild-type to 3:1 in ORCO > AgOR2 (Data S3). Since ORCO is required to traffic OR:ORCO complexes to the dendritic surfaces (Benton et al., 2006), increasing AgOR2 expression may interfere with this process. Alternatively, changes to the OR:ORCO stoichiometry may result in a functional defect in the channel itself, whereby the malformed complexes cannot respond to odors even if they have been successfully trafficked to the membrane. Finally, mosquito-specific defects caused by the ectopic expression of AgOR2 may depend on its partnering with AgORCO, which was not present in the Drosophila experiments. Future studies will be required to examine these possibilities.
This study adds to the mounting evidence that the disruption of a single sensory modality is insufficient to eliminate host-seeking behavior toward a naturally breathing host. As first demonstrated in Aedes aegypti, mosquitoes with a mutation in the ORCO gene remain attracted to humans in the presence of CO2 (DeGennaro et al., 2013); under these conditions, we also find that Anopheles mosquitoes with impaired ORCO neuronal function continue to host seek (Figures 6A-6C). In addition to odors, mosquitos are attracted to a wide variety of human-derived cues, including heat, CO2, visual stimuli, and moisture, and so one sensory modality is likely able to compensate for the loss of another (DeGennaro et al., 2013; Greppi et al., 2020; McMeniman et al., 2014; Raji et al., 2019). While the loss of ORCO function does not impair host-seeking behavior in the presence of CO2, Sun et al., 2020 (Sun et al., 2020) demonstrated that Anopheles ORCO mutant mosquitoes lose their preference for attractive odorants. Interestingly, loss of ORCO function also prevents Anopheles mosquitoes from discriminating between repellent and control odorants (Figure 6D). These data have important consequences for vector control strategies aimed at manipulating ORCO function, as mosquitoes defective in ORCO+ ORN activities will not only continue to host-seek but may also have reduced sensitivity to select repellents.
Limitations of the study
Our study uncovers the existence of a mechanism of AgOR regulation in insects whereby ectopic expression of an AgOR results in the downregulation of other native AgOR gene isoforms (Figure 4). A major limitation of this study is that only a single insect OR was examined in Anopheles mosquitoes. It remains to be determined whether ectopic expression of other insect ORs besides AgOR2, such as Drosophila ORs or other mosquito ORs, can similarly trigger AgOR-expression regulation or whether the effects observed are specific to AgOR2. In addition, the mutations introduced to eliminate AgOR2 protein expression while retaining mRNA levels (mutAgOR2; data shown in Figures 3 and 4) may trigger nonsense-mediated decay; this could reduce mutAgOR2 abundance. Finally, while Anopheles mosquito olfactory neurons are affected by ectopic AgOR2 expression, it remains to be determined to what extent this mechanism is used during normal olfactory neuron development. Nonetheless, this work lays the technical and conceptual foundations to explore cellular mechanisms used by mosquito ORNs to regulate AgOR expression.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Christopher J. Potter (cpotter@jhmi.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
Raw and analyzed RNA-seq datafiles generated during this study have been deposited on NCBI (dataview.ncbi.nlm.nih.gov/) and are publicly available as of the date of publication. The BioProject accession number is listed on the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Apocrypta bakeri ORCO | (Butterwick et al., 2018) | #15B2 |
| Rat anti-CD8 | Invitrogen | Cat#MCD0800; RRID:AB_10392843 |
| Mouse anti-nc82 | Developmental Studies Hybridoma Bank | RRID: AB_2392664 |
| Alexa-488 goat anti-rat | Invitrogen | Cat#A110066 |
| Chemicals, peptides, and recombinant proteins | ||
| Benzaldehyde | Millipore Sigma | B1334; CAS: 100-52-7 |
| Methyl Salicylate | Millipore Sigma | M6752; CAS: 119-36-8 |
| Lemongrass oil | Millipore Sigma | W262440; CAS: 8007-02-01 |
| Critical commercial assays | ||
| In-Fusion HD Cloning System | Clontech | 639645 |
| Midiprep kit | Qiagen | 12145 |
| Deposited data | ||
| Raw and analyzed RNA-seq data | This paper | BioProject: PRJNA771697 |
| Experimental models: Organisms/strains | ||
| An. coluzzii: ORCO-QF2 | (Riabinina et al., 2016) | BEI Resources:MRA-1300 |
| An. coluzzii: QUAS-mCD8::GFP | (Riabinina et al., 2016) | BEI Resources:MRA-1301 |
| An. coluzzii: QUAS-GCaMP6f | (Afify et al., 2019) | N/A |
| An. coluzzii: Wild-type M-form strain Ngousso | Insect Transformation Facility (Rockville, MD) | N/A |
| D. melanogaster: ORCO-GAL4; P{Orco-GAL4.W}11.17 | Bloomington Drosophila Stock Center | BDSC: 26818 |
| D. melanogaster: 5xUAS-AgOR2; w[*];Df(2L)dp-79b Dp(2;2)dpp[d21]/cyo; P{w[+mC]=UAS-Agam\Or2}3/TM3,Sb[1] | Bloomington Drosophila Stock Center | BDSC: 58828 |
| D. melanogaster: Pebbled-GAL4; w[*]P{w[+m*]=GAL4}peb | Bloomington Drosophila Stock Center | BDSC: 80570 |
| An. coluzzii: QUAS-AgOR2 | This paper | N/A |
| An. coluzzii: QUAS-mutAgOR2 | This paper | N/A |
| D. melanogaster: 20xUAS-AgOR2 | This paper | N/A |
| Oligonucleotides | ||
| Aga_OR2_F: ATTCGTTAACAGATCTAT GCTGATCGAAGAGTGTCCGA | This paper | N/A |
| Aga_OR2_R: CCTTCACAAAGATCGAC GTCTTAGTTGTACACTCGGCGCAGC | This paper | N/A |
| InfuMUTAgOr2_for: ATTCGTTAACAGATCTTTTC TGATCGAAGAGTGTCCGATAATTG | This paper | N/A |
| InfuMUTAGOr2_rev: CGTCATTTTTCTCGAGTA GAGAGCGTACTCGGCGGC | This paper | N/A |
| UAS-AgOr2-FOR: TTACTTCAGGCGGCC GCAAA ATGCTGATCGAAGAGTGTCCG | This paper | N/A |
| UAS-AgOR2-REV: ACAAAGATCCTCTAGA TTAGTTGTACACTCGGCGCAG | This paper | N/A |
| gcamp6f_for2: ATGGTATGGCTAGCATGACTG | This paper | N/A |
| gcamp6f_rev: GTAGTTTACCTGACCATCCCC | This paper | N/A |
| AgOr2_for1: TAATTGGTGTCAATGTGCGAG | This paper | N/A |
| AgOr2_rev2: TTATCGGCTCCTCAAAGTCTG | This paper | N/A |
| Recombinant DNA | ||
| pXL-BacII-15xQUAS-TATA-AgOR2-Sv40 | This paper | N/A |
| pXL-BacII-15xQUAS_TATA-Sv40 | (Riabinina et al., 2016) | Addgene Plasmid #104875 |
| pXL-BacII-15xQUAS-TATA-mutAgOR2-Sv40 | This paper | N/A |
| pJFRC-20xUAS-AgOR2 | This paper | N/A |
| pJFRC-20xUAS-IVS-CD8GFP | (Pfeiffer et al., 2010) | Addgene Plasmid #26220 |
| pXL-BACII-DsRed-OR7_9kbProm-QF2-hsp70 | (Riabinina et al., 2016) | Addgene Plasmid #104877 |
| Software and algorithms | ||
| MacVector v17.0.10 | MacVector, Inc. | Macvector.com |
| Fiji software | (Schindelin et al., 2012) | imagej.net |
| JMP Version 9 | SAS Institute, Inc | jmp.com |
| AUTOSPIKE software | USB-IDAC System; Syntech | ockenfels-syntech.com |
| Kallisto v0.46.0 | (Bray et al., 2016) | N/A |
| R | (The R Development Core Team and Computing, 2018) | r-project.org |
| Sleuth v0.30.0 | (Pimentel et al., 2017) | rdocumentation.org |
| Ggtern R package | (Hamilton and Ferry, 2018) | cran.r-project.org |
| Other | ||
| Company used for D. melanogaster transformation | Rainbow Transgenics | rainbowgene.com |
| Company used for An. coluzzii transformation | The Insect Transformation Facility | ibbr.umd.edu/facilities/itf |
| Company used for RNA-seq library preparation and sequencing | Genewiz, Inc. | genewiz.com |
| Company used to construct RNA probes | Molecular instruments | molecularinstruments.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Anopheles gambiae M-form strain Ngousso (the M-form of An. gambiae is now referred to as Anopheles coluzzii) were grown at 28°C, 70-75% relative humidity and 14hr light/10hr dark cycle. Larvae were reared at low densities (175 larvae/1L dH2O) to ensure large adult size. They were provided with TetraMin Tropical Flakes and Purina Cat Chow Indoor pellets ad libitum. Pupae were hand collected and allowed to eclose in small cages, where they were provided with 10% sucrose continuously. Almost all pupae eclosed the day after collection. Adult males and females were kept together in the same cage for 7-10 days, after which they were fed mouse blood from anaesthetized mice according to Johns Hopkins University Animal Care and Use Committee (ACUC) approved protocol #M019M483. Eggs were collected from the resulting gravid females by providing them with a cup of water containing wet filter paper on which to deposit their eggs as an oviposition substrate. Each generation was screened for the presence of the eye specific marker encoded by the inserted plasmid cassette.
Drosophila melanogaster: Flies were reared at 25°C and 70% humidity on a standard cornmeal diet.
METHOD DETAILS
Insect stocks
Mosquitoes: ORCO-QF2 and QUAS-mCD8::GFP transgenic mosquito stocks were generated as described in Riabinina et al. 2016 (Riabinina et al., 2016). QUAS-GCaMP6f was generated as described in Afify et al. 2019 (Afify et al., 2019). Wild-type Ngousso mosquitoes were a gift from the Insect Transformation Facility (Rockville, MD). D. melanogaster: ORCO-GAL4; P{Orco-GAL4.W} 11.17 (BDSC: 26818), 5xUAS-AgOR2; w[*];Df(2L)dp-79b Dp(2;2)dpp[d21]/cyo;P{w[+mC]=UAS-Agam\Or2}3/TM3,Sb[1] (BDSC: 58828), and Pebbled-GAL4; w[*]P{w[+m*]=GAL4}peb (BDSC: 80570) lines were obtained from the Bloomington Drosophila Stock Center.
Recombinant DNA construction
Plasmids were constructed by enzyme digestions, PCR, subcloning and the In-Fusion HD Cloning System (Clontech, 639645). Plasmid inserts were verified by restriction enzyme digests and DNA sequencing. Insertions of each plasmid into the Anopheles genome (QUAS-AgOR2, QUAS-mutAgOR2) or the Drosophila genome (20xUAS-AgOR2) were verified by sequencing the vector-specific cassette within the transgenic animal. To create the pXL-BacII-15xQUAS-TATA-AgOR2-Sv40 plasmid, we linearized the pXL-BacII-15xQUAS_TATA-Sv40 (Riabinina et al., 2016) vector with Xhol. The cDNA of AgOR2 was amplified from Bloomington Drosophila Stock Center 58828 using the oligos Aga_OR2_F (5′-ATTCGTTAACAGATCTATGCTGATCGAAGAGTGTCCGA-3′) and Aga_OR2_R (5′-CCTTCACAAAGATCGACGTCTTAGTTGTACACTCGGCGCAGC-3′). The resultant PCR product was then infusion-subcloned back into the construct. To create the pXL-BacII-15xQUAS-TATA-mutAgOR2-Sv40 reporter line, a double digest of pXL-BacII-15xQUAS-TATA-AgOR2-Sv40 with BgIII and XhoI was performed. AgOR2 was amplified from pXL-BacII-15xQUAS-TATA-AgOr2-Sv40 using a forward primer that mutated the start codon: ATG→TTT (InfuMUTAgOr2_for: 5′- ATTCGTTAACA GATCTTTTCTGATCGAAGAGTGTCCGATAATTG-3′). The reverse primer was engineered to create a frameshift mutation at a second in-frame ATG site located between the first and second transmembrane domains of AgOR2 (InfuMUTAGOr2_rev: 5′- CGT CATTTTTCTCGAGTAGAGAGCGTACTCGGCGGC-3′). The pJFRC-20xUAS-AgOR2 reporter was created in Drosophila to test whether increasing the dosage of AgOR2 affects olfactory physiology. The construct was made by digesting pJFRC-20xUAS-IVS-CD8GFP (Pfeiffer et al., 2010) with NotI and XbaI and isolating the linearized 8.1kb vector. AgOR2 was PCR amplified from pXL-BacII-15xQUAS-TATA-AgOR2-Sv40 using the primers UAS-AgOr2-FOR (5′-TTACTTCAGGCGGCC GCAAA ATGCTGATC GAAGAGTGTCCG-3′) and UAS-AgOR2-REV (5′-ACAAAGATCCTCTAGA TTAGTTGTACACTCGGCGCAG-3′). The PCR product was infusion cloned into the digested pJFRC-20xUAS vector. Upon sequence confirmation, the plasmid was isolated using a Midiprep kit (Qiagen 12145) and sent to Rainbow Transgenics (rainbowgene.com) for injection into the attP2 site (RFT # 8622). The pXL-BACII-DsRed-OR7_9kbProm-QF2-hsp70 construct was used to generate the ORCO-QF2 driver line in this study. Construction of this plasmid is described in Riabinina et al. 2016 (Riabinina et al., 2016). All plasmids were constructed using MacVector v17.0.10 (MacVector, Inc.; Macvector.com).
Anopheles gambiae transgenics
Anopheles gambiae M-form strain Ngousso (the M-form of An. gambiae is now referred to as Anopheles coluzzii) mosquitoes were grown at 28°C, 70-75% relative humidity, 12h light/dark cycle. Freshly deposited eggs were collected by providing mated, gravid females with wet filter paper as an oviposition substrate for 15-20min, after which the eggs were collected and systematically arranged side-by-side on a double-sided tape fixed to a coverslip. Aligned embryos were covered with halocarbon oil and injected at their posterior pole with an injection cocktail between 30-40min after egg laying. Injection cocktails consisted of a mixture of two plasmids, one with a piggyBac vector carrying the transgene of interest with a dominant visible marker gene – enhanced cyan fluorescent protein (ECFP) – under the regulatory control of the 3xP3 promoter, and a piggyBac transposase-expressing plasmid consisting of the transposase open reading frame under the regulatory control of the promoter from the An. stephensi vasa gene. Vector concentrations were at 150 ng/uL and the transposase-expressing plasmid was at 300 ng/uL in 5mM potassium chloride, 0.1mM sodium phosphate pH 6.8. Halocarbon oil was immediately removed and coverslips with injected embryos were placed in trays of water at 28°C, where the first instar larvae hatched ~24hr later. The Insect Transformation Facility (https://www.ibbr.umd.edu/facilities/itf) within the University of Maryland College Park’s Institute for Bioscience and Biotechnology Research performed all embryo microinjections. Adults developing from injected embryos were separated by sex at the pupal stage before mating, and small groups of 5-10 injected adult males or females were crossed to wild-type Ngousso adults of the opposite sex. The progeny from these matings were screened during the third or fourth larval instar for the presence of vector-specific marker gene expression. Transgenic larvae were saved and adults from these larvae were outcrossed to wild-type for a total of 5 generations.
Calcium imaging
Preparation: In vivo preparation of mosquitoes (ages 3-10 days) and optical imaging of odor-evoked calcium responses are described in Afify et al. 2019 (Afify et al., 2019). Genotyping mosquitoes: After the recordings were made for each sample, the bodies of all mosquitoes were frozen for subsequent genomic DNA extraction and genotyping. At the time of the experiment, our transgenic lines were not homozygous. Because all QUAS effector lines (QUAS-AgOR2, QUAS-mutAgOR2, QUAS-GCaMP6f) are marked with the dominant eye marker, ECFP, we had to determine – for each sample – whether the mosquito contained a single copy of QUAS-AgOR2, a single copy of QUAS-GCaMP6f, or both QUAS-AgOR2 and QUAS-GCaMP6f transgenes (for experiments in Figures 2A, S1, and S2). For the experiment in Figure 3, we had to determine – for each sample – whether the mosquito contained a single copy of QUAS-mutAgOR2, a single copy of QUAS-GCaMP6f, or both QUAS-mutAgOR2 and QUAS-GCaMP6f transgenes. To genotype QUAS-GCaMP6f, we used the primers gcamp6f_for2 (5′-ATGGTATGGCTAGCATGACTG-3′) and gcamp6f_rev (5′- GTAGTTTACCT GACCATCCCC-3′). Females that did not have any amplification of GCaMP6f were discarded from the analysis. To genotype QUAS-AgOR2 or QUAS-mutAgOR2, we used the following primers: AgOr2_for1 (5′- TAATTGGTGTCAATGTGCGAG-3′) and AgOr2_rev2 (5′- TTATCGGCTCCTCAAAGTCTG-3′). The PCR was designed so that both the wild-type AgOR2 (1542bp) and the transgenic AgOR2 (966bp) (or mutAgOR2; 968bp) – if present – would amplify. For each female, we determined whether she contained the wild-type and transgenic copy of AgOR2 (or mutAgOR2) or only the wild-type AgOR2. Scoring of all calcium imaging files was done blind to genotype. Analysis: To make the heatmaps (ΔF), Fiji software (Schindelin et al., 2012) was used with a custom-built macro. This Macro uses the “Image stabilizer” plug-in to correct for movements in the recording, followed by the “Z project” function to calculate the mean baseline fluorescence (mean intensity in the first 9s of recording, before stimulus delivery). The “Image calculator” function was used to subtract the mean baseline fluorescence from the image of maximum fluorescence after odorant delivery (this image was manually chosen). Afterward, this ΔF image was used to produce heatmaps. To quantify the ΔF value for each segment to each odorant, the “ROI manager” tool in Fiji was used to manually select an ROI. For each sample, we manually drew the ‘antennal ROI’ around the 11th antennal segment from an epifluorescent image taken for each sample prior to the calcium imaging. We also drew a ‘background ROI’ outside of the tissue using the same surface area as the ‘antennal ROI’ to control for any background signal. A “task manager” was used to store the location of the antennal ROI and the background, and the mean intensity across the antennal segment (or background control) for each odorant was stored for each ROI. The final ΔF value was taken for each antennal segment and each odorant as the mean intensity of the ‘antennal ROI’ minus the ‘background ROI.’ Of note, the imaging software was upgraded for Figure 3, which explains why the (ΔF) values in Figure 3 are on a different scale than Figures 2A, S1, and S2. Data were analyzed using JMP Version 9, SAS Institute Inc.; jmp.com.
SSR
Drosophila preparation. Flies were housed on standard food in groups of a maximum of 10. Analysis was done on ab1 sensilla from 6-10 day old male flies. Sensilla of targeted ORNs were prepared and identified using methods described in Lin & Potter 2015 (Lin and Potter, 2015). Briefly, ab1 sensilla were identified by their response to CO2. Ab1 is the only sensillar group that houses the CO2-responsive neuron, and so a CO2-response was indicative that the sensillum recording from was ab1. Signals were amplified 100X (USB-IDAC System; Syntech, Hilversum, The Netherlands), inputted into a computer via a 16-bit analog-digital converter and analyzed off-line with AUTOSPIKE software (USB-IDAC System; Syntech; ockenfels-syntech.com). The low cutoff filter setting was 50Hz, and the high cutoff was 5kHz. To deliver odorants or CO2 to ab1, a constant air stream was guided through a serological pipette with a tip placed 1cm from the antenna. The chemical cartridge was laterally inserted into this airflow. Stimuli consisted of 1000ms air pulses passed over odorant sources, which were various odorants diluted in paraffin oil (30μL on a filter paper of 1x2cm). For the benzaldehyde (Millipore Sigma, B1334; CAS: 100-52-7) analysis, every spike was counted (neuron subtypes were not distinguished). For the methyl salicylate (Millipore Sigma, M6752; CAS:119-36-8) analysis, only responses from the DmOR10a-positive neuron were included. Delta spikes/second were calculated by manually counting the number of spikes in a 0.5s window at stimulus delivery (200ms after stimulus onset to account for the delay due to the air path) and then subtracting the number of spontaneous spikes in a 0.5s window before stimulation, multiplied by 2 to obtain delta spikes per second. Mosquito preparation. Mated females were 5-12 days old and not bloodfed. Extracellular recordings of the cps on the maxillary palps were made using the same equipment as for Drosophila SSR (see above). Cp sensilla were also identified by their response to CO2. Cp is the only sensillum that houses the CO2-responsive neuron, and so a CO2-response was indicative that the sensillum we were recording from was cp. SSR data were analyzed using JMP Version 9, SAS Institute Inc.; jmp.com.
RNA-seq
To generate experimental pools of ORCO>AgOR2 antennae, mosquitoes of the genotype QUAS-AgOR2 were crossed to ORCO-QF2. Simultaneously, we generated control samples by crossing ORCO>AgOR2 or ORCO-QF2 strains to wild-type (2 crosses). All 3 crosses were conducted in large breeding cages that consisted of ~75 males and ~75 females. Crosses were blood-fed a total of 4 times and progeny from each cross were screened for the eye-specific fluorescent markers. Larvae generated from experimental crosses were screened for the presence of ECFP and DsRed, markers for the QUAS and the ORCO-QF2 transgenes, respectively. Larvae that did not contain both markers were discarded. For the control larvae progeny, animals that contained a single eye marker (either DsRed or ECFP, respectively) were kept in control pools that consisted either of ORCO-QF2 or QUAS-AgOR2 alone. All mosquitoes used in this study were heterozygous for a given transgene. ORCO>mutAgOR2 and QUAS-mutAgOR2 library preparations were made using the procedure described above with the exception that the antennae were extracted at a different time. To prepare antennal RNA-seq libraries, we isolated RNA from approximately 200 antennae from age-matched cohorts (11 total samples: 3 experimental samples containing ORCO>AgOR2 antennae (ORCO-QF2 + QUAS-AgOR2), 5 control samples consisting of 2 samples from ORCO-QF2 antennae, 2 samples from QUAS-AgOR2 antennae, and 1 sample from QUAS-mutAgOR2 antennae, and 3 experimental samples containing ORCO>mutAgOR2 antennae). These mated females were within their fertile period (5-20 days old) and did not receive a bloodmeal. To create the antennal RNA-seq libraries, we removed the whole antenna from the base of the pedicel and isolated total RNA using TriZol purification methods. The tissues were disrupted and homogenized using a power pestle with disposable RNase free pestles. Total RNA samples were stored at −80°C and shipped to Genewiz, Inc. (genewiz.com), where they were first assessed for quantity (Qubit Quantification) and quality (Agilent 2100 Bioanalyzer). RNA library preparation with polyA selection was then carried out using the Illumina HiSeq with a 2x150bp configuration. Paired end reads were pseudoaligned to the AgamP4.12 reference transcriptome (Giraldo-Calderón et al., 2015) and isoform-level abundances were quantified using kallisto v0.46.0 (Bray et al., 2016) using default parameters with the following exceptions: −t4 and −bl00. Per-sample abundances were aggregated and normalized in R (R Core Team, 2018) using sleuth v0.30.0 (Pimentel et al., 2017). For each pairwise comparison (AgOR2 v control, mutAgOR2 v control, and mutAgOR2 v AgOR2), we fit a full sleuth model using condition as an explanatory variable. Differentially expressed isoforms were identified using the Wald test (q≤0.01, Benjamini-Hochberg corrected) for each model fit. Ternary plots were constructed using the ggtern R package (Hamilton and Ferry, 2018). Gene ontology (GO) annotations for the differentially expressed isoforms were assigned by Vectorbase.org (Giraldo-Calderón et al., 2015). Raw and analyzed RNA-seq data were deposited on NCBI, BioProject: PRJNA771697.
Immunohistochemistry
Adult Antennae: Antennae of 5+ day old ORCO>AgOR2 and QUAS-AgOR2 were extracted and stained as described in Basrur et al. 2020 (Basrur et al., 2020), with one exception. Female heads were incubated for 1 hour 20 minutes in chitinase-chymotrypsin solution before fixation and subsequent antennal removal. Mouse anti-Apocrypta bakeri ORCO monoclonal antibody #15B2 (Butterwick et al., 2018) (1:50 dilution, gift of Joel Butterwick and Vanessa Ruta) was used to visualize ORCO-positive cells. Brains: Brains of female mosquitoes of ORCO>mCD8::GFP and ORCO>AgOR2,mCD8::GFP genotypes were extracted and stained as described previously (Riabinina et al., 2016). Once genotyped via a PCR designed to amplify the transgenic and wild-type copies of AgOR2 (see ‘calcium imaging section’ above), experimental and control brains were separated and stained in two different groups. Rat anti-CD8 (Invitrogen Cat#MCD0800, 1:100) was used to visualize the ORN projections to the ALs and mouse nc82 (Developmental Studies Hybridoma Bank, 1:50) was added to visualize the structure of the brain. Pupal Antennae: ORCO>mCD8::GFP larvae were collected in 2 trays, each of which contained ~60 larvae. At the given timepoint APF, the cephalothorax and abdomen of the pupae were extracted and the antennae, oculus, compound eye, and rudimentary appendages were placed in 4% paraformaldehyde (PFA) with 0.1% triton phosphate buffer solution (PBS-T). Antennae were then dissected out and washed 3x in 1X PBS-T (0.1%). Antennae were then blocked for 30min at 4°C with 5% normal goat serum (NGS). To visualize mCD8::GFP expression, we added rat anti-CD8(Invitrogen, Cat#MCD0800, 1:100), which was left to incubate on a rotator overnight at 4°C. The next day, antennae were washed 3x in 1X PBS-T (0.1%) and Alexa-488 goat anti-rat (Invitrogen Cat#A110066, 1:200) was added to the solution. After 1.5hr at 25°C, antennae were washed 2X in 1X PBS-T (0.1%) and the last wash in 1xPBS to remove the triton. Antennae were placed on slides and mounted in SlowFade Gold Antifade Mountant.
Whole mount antennal RNA in situ hybridization
RNA was detected in whole mount antennae using the hybridization chain reaction (HCR) technique as described in (Younger et al., 2020) with few modifications. Probes were designed against AgOR2, AgOR11, AgOR24, and AgOR41. The 488 and 546 amplifiers, probe hybridization buffer, amplification buffer, and probe wash buffer were purchased from Molecular Instruments (molecularinstruments.com/). AgOR11, AgOR24, and AgOR41 probes were designed to be conjugated with Alexa 488 and probes for AgOR2 were designed to be conjugated with Alexa 546. Heads from female wild-type and ORCO>AgOR2 pupae (0-2hr APF, 4-6hr APF, 8-10hr APF, 12-14hr APF, 16-18hr APF, and 20-22hr APF) and adults (1 day PE, 4-8 days PE) were digested in a chitinase-chymotrypsin solution for 25 minutes and fixed overnight at 4°C. The following day, the heads were dehydrated with a graded series of methanol/0.1% PBS-Tween-10 at 4°C. Tissues were incubated in 100% methanol for 2-3 days and were subsequently rehydrated with a series of graded methanol/0.1% PBS-Tween-20 at 4°C. Heads were then washed in 0.1% PBS-Tween-20 and pupal or adult antennae were dissected. Adult (but not pupal) antennae were digested in 20 ug/mL Recombinant Proteinase-K Solution in 0.1% PBS-Tween for 30 minutes at room temperature, washed in 0.1% PBS-Tween-20, and fixed in 4% paraformaldehyde in 0.1% PBS-Tween-20 for 20 minutes and then washed 3x in 0.1% PBS-Tween-20. Probe hybridization, wash, and amplification steps were then followed as described in Younger et al. 2020 (Younger et al., 2020).
Confocal imaging and analyses
Brains and antennae were imaged on an LSM 700 Zeiss confocal microscope with a 40x immersion-corrected objective at a resolution of 512x512 pixel resolution with 0.9uM Z-steps. For illustration purposes, confocal images were processed using Fiji (Schindelin et al., 2012) to collapse the maximum intensity projection of Z-stacks into a single image. For the immunohistochemistry experiment, we used cells that were immunoreactive to mCD8 as a proxy for ORCO expression. Cells per flagellomere were counted manually in pupal and adult antennae.
Host-proximity assay
The design and methods of the host-proximity assay are modified from DeGennaro et al. 2013 (DeGennaro et al., 2013). Briefly, for each trial, 20-30 adult female Anopheles mosquitoes (5+ days PE, mated but not blood-fed) were sorted under cold anesthesia (4°C), placed in 24-oz deli containers (https://www.amazon.com/gp/product/B00NB9WCEO/ref=oh_aui_detailpage_o06_s00?ie=UTF8&psc=1), and fasted with access to water for 16-24hr prior to assaying. Pre-fasting and behavior experiments took place at 27°C and 70-80% relative humidity. Experiments took placed after zeitgeber time 0 (ZT0) and continued until ZT8. 5 minutes before the start of the assay, mosquitoes were released into a modified BugDorm (https://shop.bugdorm.com/bugdorm-1-insect-rearing-cage-p-1.html). After 5 minutes had elapsed, pure CO2 and synthetic air (air flow rate: 0.1L/min, CO2 flowrate 145MM) were mixed in an adaptor before being pulsed (3 sec on: 3 sec off) into a flypad (8.1 x 11.6 cm catalogue #59-114; Flystuff.com, San Diego, CA), which was placed at the bottom of the BugDorm. Mosquitoes were then presented with a single volunteer’s arm, which was placed 2.5 cm away from one side of the BugDorm so that mosquitoes could not come into direct contact with the arm. To control the distance from the arm to the cage, a Q-Snap needlework frame (https://www.amazon.com/dp/B00013MV30/ref=twister_B07CQQJKL2?_encoding=UTF8&psc=1) was placed flush against the cage and the arm was pressed against the vials. The arm was elevated 2.7 cm by placing it on a plastic microcentrifuge test-tube rack. An HDR-CX260V camera (Sony) was positioned to take images of mosquitoes responding to the human arm. Trials ran for 3 minutes. To quantify mosquito responses that came into ‘close proximity’ to the human arm, we counted the number of mosquitoes resting on the screen. We did not include mosquitoes that had landed on the white area surrounding the screen nor the mosquitoes that were in flight. For the % attraction figure, we scored the number of mosquitoes that came into close proximity of the arm every 10 seconds from minute 1 to minute 3 and then divided this number by the total number of mosquitoes in the trial. Data were analyzed using JMP Version 9, SAS Institute Inc.; jmp.com.
Olfactory-based oviposition preference assay
Olfactory-based oviposition preference was evaluated in groups of 10, 10-12 day old gravid females for the following genotypes: wild-type, ORCO-QF2, QUAS-AgOR2, and ORCO>AgOR2. Three days prior to preference assay, blood-fed females were placed in aluminum mosquito cages (BioQuip, Cat#1450A) and allowed to acclimate to a 12L:12D light:dark cycle maintained at 27°C, 75% RH. On day four, control (paraffin oil) and repellent (0.1% lemongrass oil (Millipore Sigma; W262440; CAS:8007-02-01; diluted in paraffin oil) ‘double cups’ (Mwingira et al., 2019) were placed in opposite corners of the cage. Double cups separate the influence of physical and olfactory cues on oviposition site selection by gravid females. A double cup consists of a small cup (4.3 cm top diameter x 2.9 cm bottom diameter x 3.2 cm height) filled with 10 mL of deionized water placed inside a larger cup (5.7 cm top diameter x 3.8 cm bottom diameter x 6.3 cm height) filled with 1 mL control or repellent solution. To induce egg laying, filter paper, a mosquito oviposition substrate, was placed in the small cup and left for 24 hrs. The small cup prevented contamination of the filter paper with the control or repellent solution. The following day, the number of eggs laid on the control and repellent double cup filter papers were scored manually for each genotype. Egg laying for each genotype was assessed over the course of 7 trials for each genotype and the position of control and repellent double cups were randomized across all trials. For each genotype, a paired-samples t-test was used to assess if there were differences in egg number between the control and repellent double cups. Statistical analyses were performed in R (R Core Team, 2018).
QUANTIFICATION AND STATISTICAL ANALYSIS
Information regarding the quantification and statistical analyses performed on all data are described in the method details section or in the figure legends. All statistical tests were run in JMP Version 9 (SAS Institute, Inc.; jmp.com) or R (R Core Team, 2018; r-project.org).
Supplementary Material
Highlights.
Ectopic OR expression disrupts mosquito olfactory neuron activity
Expression of an OR in mosquitoes reduces endogenous OR expression
Onset of mosquito OR expression extends into adult stages
Mosquitoes host seek despite the lack of OR neuron function
ACKNOWLEDGMENTS
This research benefited from the limitless enthusiasm and support of Potter Lab members, both past and present. In particular, we would like to thank Kateline Robinson Shaw and Liz Marr for assisting with molecular biology. Darya Task and Joanna Konopka helped rear the transgenic mosquitoes and improved the writing of the manuscript. We thank Robert Johnston and Liz Urban for help with in situs. We also thank Greg Artiushin for the graphic designs created for the article. Studies outlined in this document were supported by grants from the National Institutes of Health, to C.J.P. (NIAID R01Al137078); the Department of Defense, to C.J.P. (W81XWH-17-PRMRP); and a Johns Hopkins Malaria Research Institute postdoctoral fellowship to S.E.M. We thank the Johns Hopkins Malaria Research Institute and Bloomberg Philanthropies for their support.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110494.
REFERENCES
- Afify A, Betz J, Riabinina O, Lahondère C, and Potter C (2019). Commonly used insect repellents hide human odors from Anopheles mosquitoes. Curr. Biol 29, 3669–3680.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athrey G, Cosme L, Popkin-Hall Z, Pathikonda W, and Slotman M (2017). Chemosensory gene expression in olfactory organs of the anthropophilic Anopheles coluzzii and zoophilic Anopheles quadriannulatus. BMC Genomics 18, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrur N, De Obaldia M, Morita T, Herre M, von Heynitz R, Tsitohay Y, and Vosshall L (2020). Fruitless mutant male mosquitoes gain attraction to human odor. Elife 9, e63982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauty B, and Marquardt W (1996). The Biology of Disease Vectors, First Edition (University Press of Colorado; ). [Google Scholar]
- Bell J, and Wilson R (2016). Behavior reveals selective summation and max pooling among olfactory processing channels. Neuron 91, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick S, and Vosshall L (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot J, Pitts R, Kwon H-W, Rützler M, Robertson H, and Zwiebel L (2007). Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol. Biol 16, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Pimentel H, Melsted P, and Pachter L (2016). Near-optimzal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Butterwick J, Mármol J, Kim K, Kahlson M, Rogow J, Walz T, and Ruta V (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey A, Wang G, Su C-Y, Zwiebel L, and Carlson J (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin S, Maguire S, Huoviala P, Jefferis G, and Potter C (2018). Olfactory neurons and brain centers directing oviposition decisions in Drosophila. Cell Rep. 24, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, and Ray A (2016). Olfactory mechanisms for discovery of odorants to reduce insect-host contact. J. Chem. Ecol 42, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney E, LeGros M, Mosley C, Clowney F, Markenskoff-Papadimitriou C, Myllys M, Barnea G, Larabell C, and Lomvardas S (2012). Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151, 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P, Warr C, Freeman M, Lessing D, Kim J, and Carlson J (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, and Dickson B (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol 15, 1535–1547. [DOI] [PubMed] [Google Scholar]
- Cowman A, Healer J, Marapana D, and Marsh K (2016). Malaria: biology and disease. Cell 167, 610–624. [DOI] [PubMed] [Google Scholar]
- Dalton R, Lyons D, and Lomvardas S (2013). Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell 155, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro M, McBride C, Seeholzer L, Nakagawa T, Dennis E, Goldman C, Jasinskiene N, James A, and Vosshall L (2013). Orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A, van der Goes van Naters W, Warr C, Steinbrecht A, and Carlson J (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. [DOI] [PubMed] [Google Scholar]
- Elmore T, Ignell R, Carlson J, and Smith D (2003). Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. J. Neurosci 23, 9906–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, and Mombaerts P (2004). Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell 117, 833–846. [DOI] [PubMed] [Google Scholar]
- Foster W, and Takken W (2004). Nectar-related vs. human-related volatiles: behavioural response and choice by female and male Anopheles gambiae (Diptera:Culicidae) between emergence and first feeding. Bull. Entomol. Res 94, 145–157. [DOI] [PubMed] [Google Scholar]
- German P, van der Poel S, Carraher C, Kralicek A, and Newcomb R (2013). Insights into subunit interactions within the insect olfactory receptor complex using FRET. Insect Biochem. Mol. Biol 43, 138–145. [DOI] [PubMed] [Google Scholar]
- Ghaninia M, Hansson B, and Ignell R (2007). The antennal lobe of the African malaria mosquito, Anopheles gambiae - innervation and three-dimensional reconstruction. Arthropod Struct. Development 36, 23–39. [DOI] [PubMed] [Google Scholar]
- Giraldo-Calderón G, Emrich S, MacCallum R, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, Consortium V, Madey G, et al. (2015). Vector-Base: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Malaria Programme (2019). World Malaria Report 2019 (WHO; ), p. 232. [Google Scholar]
- Greppi C, Laursen W, Budelli G, Chang E, Daniels A, van Giesen L, Smidler A, Catteruccia F, and Garrity P (2020). Mosquito heat seeking is driven by an ancestral cooling receptor. Science 367, 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E, Ho M, and Carlson J (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. [DOI] [PubMed] [Google Scholar]
- Hamilton N, and Ferry M (2018). ggtern: ternary diagrams using ggplot2. J. Stat. Softw. Code Snippets 87, 1–17. [Google Scholar]
- Hill C, Fox A, Pitts R, Kent L, Tan P, Chrystal M, Cravchik A, Collins F, Robertson H, and Zwiebel L (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178. [DOI] [PubMed] [Google Scholar]
- Hill S, Taparia T, and Ignell R (2021). Regulation of the antennal transcritome of the dengue vector, Aedes aegyti, during the first gonotropic cycle. BMC Genomics 22, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Pask G, Rinker D, and Zwiebel L (2011). Functional agonism of insect odorant receptor ion channels. Proc. Natl. Acad. Sci. USA 108, 8821–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner T, Kellner I, Schultze A, Breer H, and Krieger J (2015). Co-expression of six tightly clustered odorant receptor genes in the antenna of the malaria mosquito Anopheles gambiae. Front. Ecol. Evol 3. 10.3389/fevo.2015.00026. [DOI] [Google Scholar]
- Konopka J, Task D, Afify A, Raji J, Deibel K, Maguire S, Lawrence R, and Potter C (2021). Olfaction in Anopheles mosquitoes. Chem. Senses 46, bjab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson M, Domingos A, Jones W, Chiappe M, Amrein H, and Vosshall L (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. [DOI] [PubMed] [Google Scholar]
- Lewcock J, and Reed R (2004). A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl. Acad. Sci. USA 101, 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-C, and Potter C (2015). Re-classifcation of Drosophila melanogaster trichoid and intermediate sensilla using fluorescence-guided single sensillum recording. PLoS One 10, e0139675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin C, Brbić M, Xiw Q, Li T, Horns F, Kolluru S, Kebschull J, Vacek D, Xie A, Li J, et al. (2021). Single-cell transcriptomes of developing and adult olfacotry receptor neurons in Drosophila. eLife 10, e63856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C, Corfas R, Matthews B, Ritchie S, and Vosshall L (2014). Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 156, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao S, Nemes A, Mendelsohn M, Edmondson J, and Axel R (1996). Visualizing an olfactory sensory map. Cell 87, 675–686. [DOI] [PubMed] [Google Scholar]
- Montell C, and Zwiebel L (2016). Chapter ten - mosquito sensory systems. Adv. Insect Physiol 51, 293–328. [Google Scholar]
- Mwingira V, Spitzen J, Mboera L, Torres-Estrada J, and Takken W (2019). The influence of larval stage and density on oviposition site-selection behavior of the Afrotropical malaria mosquito Anopeles coluzzii (Diptera: Culicidae). J. Med. Entomol 57, 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K, Flannery E, Tomchaney M, Severson D, and Duman-Scheel M (2013). Disruption of Aedes aegypti olfactory system development through Chitosan/siRNA nanoparticle targeting of semaphorin-1a. PLoS Negl. Trop. Dis 7, e2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K, Flister S, Müller P, Rodrigues V, and Reichert H (2011). Brain development in the yellow fever mosquito Aedes aegypti: a comparative immunocytochemical analysis using cross-reacting antibodies from Drosophila melanogaster. Developmental Genes Evol. 221, 281–296. [DOI] [PubMed] [Google Scholar]
- Neuhaus E, Gisselmann G, Zhang W, Dooley R, Störtkuhl K, and Hatt H (2005). Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci 8, 15–17. [DOI] [PubMed] [Google Scholar]
- Omondi A, Ghaninia M, Dawit M, Svensson T, and Ignell R (2019). Age-dependent regulation of host seeking in Anopheles coluzzii. Scientific Rep. 9, 9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B, Ngo T-T, Hibbard K, Murphy C, Jenett A, Truman J, and Rubin G (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray N, Puente S, Melsted P, and Pachter L (2017). Differential analysis of RNA-Seq incorporating quantification uncertainty. Nat. Methods 14, 687–690. [DOI] [PubMed] [Google Scholar]
- Pitts R, Rinker D, Jones P, Rokas A, and Zwiebel L (2011). Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics 12, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C, and Luo L (2010). Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One 5, e10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Raji J, Melo N, Castillo J, Gonzalez S, Saldana V, Stensmyr M, and DeGennaro M (2019). Aedes aegypti mosquitoes detect acidic volatiles found in human odor using the IR8a pathway. Curr. Biol 29, 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van der Goes van Naters W, Shiraiwa T, and Carlson J (2007). Mechanisms of odor receptor gene choice in Drosophila. Neuron 53, 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu M, Luo L, and Potter C (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12, 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Task D, Marr E, Lin C-C, Alford R, O’Brochta D, and Potter C (2016). Organization of olfactory centers in the malaria mosquito Anopheles gambiae. Nat. Commun 7, 13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, and Sakano H (2003). Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302, 2088–2094. [DOI] [PubMed] [Google Scholar]
- Shykind B, Rohani S, O’Donnell S, Nemes A, Mendelsohn M, Sun A, Axel R, and Barnea G (2004). Gene switching and the stability of odorant receptor gene choice. Cell 117, 801–815. [DOI] [PubMed] [Google Scholar]
- Sullivan S, Adamson M, Ressler K, Kozak C, and Buck L (1996). The chromosomal distribution of mouse odorant receptor genes. Proc. Natl. Acad. Sci. USA 93, 884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Liu F, Ye Z, Baker A, and Zwiebel L (2020). Mutagenesis of the orco odorant receptor co-receptor impairs olfactory function in the malaria vector Anopheles coluzzii. Insect Biochem. Mol. Biol 127, 103497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney L, Couto A, Chou Y, Berdnik D, Dickson B, Luo L, and Komiyama T (2007). Temporal target restriction of olfactory receptor neurons by Semaphorin-1a/PlexinA-mediated axon-axon interactions. Neuron 53, 185–200. [DOI] [PubMed] [Google Scholar]
- Tallon A, Hill S, and Ignell R (2019). Sex and age modulate antennal chemosensory-related genes linked to the onset of host seeking in the yellow-fever mosquito, Aedes aegypti. Scientific Rep. 9, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Development Core Team (2018). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Trebels B, Dippel S, Goetz B, Graebner M, Hofmann C, Hofmann F, Schmid F-R, Uhl M, YVuong M-P, Weber V, et al. (2021). Metamorphic development of the olfactory system in the red flour beetle (Trobolium castaneum, HERBST. BMC Biol. 19, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitoura P, Andronopoulou E, Tsikou D, Agalou A, Papakonstantinou M, Kotzia G, Labropoulou V, Swevers L, Georgoussi Z, and Iatrou K (2010). Expression and membrane topology of Anopheles gambiae odorant receptors in Lepidopteran Insect Cells. PLoS One 5, e15428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Chao S, Sitcheran R, Nuñez J, Vosshall L, and Axel R (1994). Topographic organization of sensory projections to the olfactory bulb. Cell 79, 981–991. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, and Axel R (1998). Odorant receptors govern the formation of a precise topographic map. Cell 93, 47–60. [DOI] [PubMed] [Google Scholar]
- Younger M, Herre M, Ehrlich A, Gong Z, Gilbert Z, Rahiel S, Matthews B, and Vosshall L (2020). Non-canonical odor coding ensures unbreakable mosquito attraction to humans. Preprint at bioRxiv. 10.1101/2020.11.07.368720. [DOI] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, and Firestein S (2004). Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics 83, 802–811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and analyzed RNA-seq datafiles generated during this study have been deposited on NCBI (dataview.ncbi.nlm.nih.gov/) and are publicly available as of the date of publication. The BioProject accession number is listed on the key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Apocrypta bakeri ORCO | (Butterwick et al., 2018) | #15B2 |
| Rat anti-CD8 | Invitrogen | Cat#MCD0800; RRID:AB_10392843 |
| Mouse anti-nc82 | Developmental Studies Hybridoma Bank | RRID: AB_2392664 |
| Alexa-488 goat anti-rat | Invitrogen | Cat#A110066 |
| Chemicals, peptides, and recombinant proteins | ||
| Benzaldehyde | Millipore Sigma | B1334; CAS: 100-52-7 |
| Methyl Salicylate | Millipore Sigma | M6752; CAS: 119-36-8 |
| Lemongrass oil | Millipore Sigma | W262440; CAS: 8007-02-01 |
| Critical commercial assays | ||
| In-Fusion HD Cloning System | Clontech | 639645 |
| Midiprep kit | Qiagen | 12145 |
| Deposited data | ||
| Raw and analyzed RNA-seq data | This paper | BioProject: PRJNA771697 |
| Experimental models: Organisms/strains | ||
| An. coluzzii: ORCO-QF2 | (Riabinina et al., 2016) | BEI Resources:MRA-1300 |
| An. coluzzii: QUAS-mCD8::GFP | (Riabinina et al., 2016) | BEI Resources:MRA-1301 |
| An. coluzzii: QUAS-GCaMP6f | (Afify et al., 2019) | N/A |
| An. coluzzii: Wild-type M-form strain Ngousso | Insect Transformation Facility (Rockville, MD) | N/A |
| D. melanogaster: ORCO-GAL4; P{Orco-GAL4.W}11.17 | Bloomington Drosophila Stock Center | BDSC: 26818 |
| D. melanogaster: 5xUAS-AgOR2; w[*];Df(2L)dp-79b Dp(2;2)dpp[d21]/cyo; P{w[+mC]=UAS-Agam\Or2}3/TM3,Sb[1] | Bloomington Drosophila Stock Center | BDSC: 58828 |
| D. melanogaster: Pebbled-GAL4; w[*]P{w[+m*]=GAL4}peb | Bloomington Drosophila Stock Center | BDSC: 80570 |
| An. coluzzii: QUAS-AgOR2 | This paper | N/A |
| An. coluzzii: QUAS-mutAgOR2 | This paper | N/A |
| D. melanogaster: 20xUAS-AgOR2 | This paper | N/A |
| Oligonucleotides | ||
| Aga_OR2_F: ATTCGTTAACAGATCTAT GCTGATCGAAGAGTGTCCGA | This paper | N/A |
| Aga_OR2_R: CCTTCACAAAGATCGAC GTCTTAGTTGTACACTCGGCGCAGC | This paper | N/A |
| InfuMUTAgOr2_for: ATTCGTTAACAGATCTTTTC TGATCGAAGAGTGTCCGATAATTG | This paper | N/A |
| InfuMUTAGOr2_rev: CGTCATTTTTCTCGAGTA GAGAGCGTACTCGGCGGC | This paper | N/A |
| UAS-AgOr2-FOR: TTACTTCAGGCGGCC GCAAA ATGCTGATCGAAGAGTGTCCG | This paper | N/A |
| UAS-AgOR2-REV: ACAAAGATCCTCTAGA TTAGTTGTACACTCGGCGCAG | This paper | N/A |
| gcamp6f_for2: ATGGTATGGCTAGCATGACTG | This paper | N/A |
| gcamp6f_rev: GTAGTTTACCTGACCATCCCC | This paper | N/A |
| AgOr2_for1: TAATTGGTGTCAATGTGCGAG | This paper | N/A |
| AgOr2_rev2: TTATCGGCTCCTCAAAGTCTG | This paper | N/A |
| Recombinant DNA | ||
| pXL-BacII-15xQUAS-TATA-AgOR2-Sv40 | This paper | N/A |
| pXL-BacII-15xQUAS_TATA-Sv40 | (Riabinina et al., 2016) | Addgene Plasmid #104875 |
| pXL-BacII-15xQUAS-TATA-mutAgOR2-Sv40 | This paper | N/A |
| pJFRC-20xUAS-AgOR2 | This paper | N/A |
| pJFRC-20xUAS-IVS-CD8GFP | (Pfeiffer et al., 2010) | Addgene Plasmid #26220 |
| pXL-BACII-DsRed-OR7_9kbProm-QF2-hsp70 | (Riabinina et al., 2016) | Addgene Plasmid #104877 |
| Software and algorithms | ||
| MacVector v17.0.10 | MacVector, Inc. | Macvector.com |
| Fiji software | (Schindelin et al., 2012) | imagej.net |
| JMP Version 9 | SAS Institute, Inc | jmp.com |
| AUTOSPIKE software | USB-IDAC System; Syntech | ockenfels-syntech.com |
| Kallisto v0.46.0 | (Bray et al., 2016) | N/A |
| R | (The R Development Core Team and Computing, 2018) | r-project.org |
| Sleuth v0.30.0 | (Pimentel et al., 2017) | rdocumentation.org |
| Ggtern R package | (Hamilton and Ferry, 2018) | cran.r-project.org |
| Other | ||
| Company used for D. melanogaster transformation | Rainbow Transgenics | rainbowgene.com |
| Company used for An. coluzzii transformation | The Insect Transformation Facility | ibbr.umd.edu/facilities/itf |
| Company used for RNA-seq library preparation and sequencing | Genewiz, Inc. | genewiz.com |
| Company used to construct RNA probes | Molecular instruments | molecularinstruments.com |