Summary
Background
In critically ill COVID-19 patients, the initial response to SARS-CoV-2 infection is characterized by major immune dysfunctions. The capacity of these severe patients to mount a robust and persistent SARS-CoV-2 specific T cell response despite the presence of severe immune alterations during the ICU stay is unknown.
Methods
Critically ill COVID-19 patients were sampled five times during the ICU stay and 9 and 13 months afterwards. Immune monitoring included counts of lymphocyte subpopulations, HLA-DR expression on monocytes, plasma IL-6 and IL-10 concentrations, anti-SARS-CoV-2 IgG levels and T cell proliferation in response to three SARS-CoV-2 antigens.
Findings
Despite the presence of major lymphopenia and decreased monocyte HLA-DR expression during the ICU stay, convalescent critically ill COVID-19 patients consistently generated adaptive and humoral immune responses against SARS-CoV-2 maintained for more than one year after hospital discharge. Patients with long hospital stays presented with stronger anti-SARS-CoV-2 specific T cell response but no difference in anti-SARS-CoV2 IgG levels.
Interpretation
Convalescent critically ill COVID-19 patients consistently generated a memory immune response against SARS-CoV-2 maintained for more than one year after hospital discharge. In recovered individuals, the intensity of SARS-CoV-2 specific T cell response was dependent on length of hospital stay.
Funding
This observational study was supported by funds from the Hospices Civils de Lyon, Fondation HCL, Claude Bernard Lyon 1 University and Région Auvergne Rhône-Alpes and by partial funding by REACTing (Research and ACTion targeting emerging infectious diseases) INSERM, France and a donation from Fondation AnBer (http://fondationanber.fr/).
Keywords: Immune memory, T lymphocyte, SARS-CoV-2, Critically ill patients, HLA-DR, Sepsis
Research in context.
Evidence before this study
We searched PubMED for the term ((“COVID-19”) or (“SARS-CoV-2”)) AND ((“memory T cell”) or (“memory T lymphocyte”)) AND ((“critically ill”) or (“ICU”) or (“severe”)) with no limit of publication date. No study was identified evaluating the long-term persistence of SARS-CoV-2 specific T cell response focusing on the subgroup of the most severe critically ill COVID-19 patients.
Added value of this study
We conducted this study to evaluate the presence of SARS-CoV-2 specific T lymphocytes in a cohort of critically ill COVID-19 patients for whom immune dysregulations occurring during ICU stay were characterized. We showed that, despite the presence of major immune alterations during ICU stay, convalescent critically ill COVID-19 patients consistently generated substantial adaptive and humoral immune responses against SARS-CoV-2 maintained for more than one year after hospital discharge. Patients with long hospital stays presented with stronger anti-SARS-CoV-2 specific T cell responses but no difference in anti-SARS-CoV2 IgG levels.
Implications of all the available evidence
Convalescent critically ill COVID-19 patients develop a robust memory immune response against SARS-CoV-2 dependent on length of hospital stay. A better description of the mechanisms underlying the establishment of protective immune memory in recovering individuals is a major concern for public health for predicting and managing potential additional waves of infections in the general population.
Alt-text: Unlabelled box
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is still threatening public health. As of December 2021, over 275 million confirmed cases and more than 5.3 million deaths have been reported since the start of the pandemic (WHO weekly epidemiologic update). COVID-19 is primary a respiratory disease with symptoms ranging from paucisymptomatic or mild infection to severe symptoms requiring hospitalization in intensive care units (ICU).1
The development of specific adaptive immune response is crucial to control and clear viral infections. More specifically, upon infection, virus-specific T and B cells are activated, expand and differentiate into effector cells. Once the infection is cleared, neutralizing antibodies and antigen-specific memory B and T cells persist for a long time after resolution. Such memory immune response plays a central role in the prevention against reinfection and is mobilized during vaccination. It is thus important to characterize in details the magnitude of specific adaptive immune responses in convalescent COVID-19 patients with different severities in order to understand the development and longevity of such protective immunity. A better description of the mechanisms underlying the establishment of protective immune memory in recovering individuals is a major concern for public health for predicting and managing potential additional waves of infections in the general population.
In critically ill COVID-19 patients, the initial response to SARS-CoV-2 infection is characterized by major immune dysfunctions associating a systemic inflammatory response and the development of altered innate and adaptive immune responses.1,2 More specifically, T cell response is markedly affected in critically ill COVID-19 patients and severe lymphopenia, phenotypic and functional T cell alterations have been described in the most severe COVID-19 patients.1, 2, 3 Thus, the capacity of these critically ill patients to mount a robust and persistent SARS-CoV-2 specific T cell response despite the presence of severe immune alterations during the ICU stay is, so far, unknown.
In this context, the goal of the current study was to monitor the immune response including memory T cells specific to SARS-CoV-2 in paired samples collected 9 and 13 months after infection in a cohort of convalescent critically ill COVID-19 patients for whom COVID-19-induced immune dysfunctions during the ICU stay were minutely characterized.
Methods
Clinical study design, patient population and ethics approval
Critically ill patients admitted to two ICUs (Medical and Surgical ICUs of the Edouard Herriot University Hospital) of Lyon university-affiliated hospitals (Hospices Civils de Lyon, Lyon, France) who presented with SARS-CoV-2 pulmonary infection confirmed by RT-PCR were included in the RICO (REA-IMMUNO-COVID) study.2 This study was approved by ethics committee (Comité de Protection des Personnes Ile de France 1 - N°IRB / IORG #: IORG0009918) under agreement number 2020-A01079-30 and was registered at ClinicalTrials.gov (NCT04392401). The committee waived the need for written informed consent because the study was observational, with a low risk to patients, and no specific procedure, other than routine blood sampling, was required. Oral information and agreement to inclusion in the study were mandatory and were systematically obtained before any blood sample was drawn. This was recorded in patients’ clinical files. If a patient was unable to consent directly, the patient's legally authorized representative were contacted and agreement was reconfirmed from the patient at the earliest opportunity. Inclusion criteria were (i): patients aged > 18 years, (ii) hospitalization in ICU for SARS-CoV-2 pneumopathy, (iii) first hospitalization in the ICU for COVID-19, (iv) positive diagnosis of SARS-CoV-2 infection carried out by PCR in at least one respiratory sample, (v) sampling in the first 48 h after admission to the ICU feasible and (vi) patient or next of kin informed of the terms of the study and has not objected to participate.
Upon ICU release, convalescent COVID-19 individuals were requested to participate to the ancillary study: NOSO-COR-IMMUNO. The clinical protocol was approved by ethics committee of Ile de France on October 14 2020 (N° IDRCB: 2020-A02128-31). This study was registered at Clinicaltrials.gov under number NCT04637867. Inclusion criteria were (i) patients aged > 18 years included in NOSO-COR study4 (ii) signed written informed consent (iii) affiliation to social security system. Exclusion criteria were: (i) opposition to participation (ii) breast feeding and pregnancy (iii) currently hospitalized patients (iv) patients under legal assignment. Patients who agreed to participate were invited for two follow-ups medical visits in an ambulatory setting in Edouard Herriot Hospital, Hospices Civils de Lyon, 9 and 13 months after SARS-CoV-2 infection. All participants provided written informed consent and the study was conducted in accordance with Good Clinical Practice.
A cohort of 15 healthy donors either non-infected and non-vaccinated (n = 4), or after full vaccination (n = 4) or after resolution of a non-severe SARS-CoV-2 infection (n = 7) was concomitantly included after informed consent was given. Median age was 48 [29–55] and 10 women and 5 men were included.
Patient characteristics
For each patient, demographics, comorbidities, time from onset of COVID-19 symptoms to ICU admission, initial presentation of the disease in the ICU including the ratio of the arterial partial pressure of oxygen to the fractional inspired oxygen (PaO2/FiO2 ratio) at admission, antiviral therapy targeting SARS-CoV-2 and organ support, were documented. Organ dysfunctions according to Sequential Organ Failure Assessment (SOFA) score (range 0–24, with higher scores indicating more severe organ failures), and Simplified Acute Physiology Score II (SAPS II; range, 0–164, with higher scores indicating greater severity of illness) were documented. Follow-up included ICU length of stay, in-hospital mortality, day-28 (D28) mortality, day-90 (D90) mortality.
At follow-up visits, the following clinical parameters were recorded: presence and types of persisting symptoms, presence of new symptoms, new hospitalization, contact with an infected person, presence of a positive SARS-CoV-2 PCR test and vaccination status.
Blood samples and cell isolation
Ethylene diamine tetra-acetic acid (EDTA-)-anticoagulated blood was drawn five times after ICU admission: within the first 48 h after admission (Day 0: D0), between 72 h and 96 h after admission (D3), between D7 and D9 (D7), between D12 and D15 (D12), between D20 and D25 (D20) and then twice at follow-up visits (i.e. after 9 and 13 months after hospital discharge). Blood was stored at 4–8 °C and processed within 4 h after withdrawal for flow cytometry analyses and cell culture. Plasma was then frozen at -80 °C within 4 h following blood collection and later used for soluble markers analyses, which were performed by batches after 1 freeze / thaw cycle.
Cytokine measurement
Plasma concentrations of IL-6, TNF-α, IFN-γ and IL-10 were measured by Simpleplex® technology using ELLA instrument (ProteinSimple®, San Jose, CA), following manufacturer's instructions.
Flow cytometry
T lymphocyte immunophenotyping was performed on an automated volumetric flow cytometer from Beckman Coulter (Aquios CL) as previously described.5 The expression of monocyte HLA-DR was determined using the Anti-HLA-DR/Anti-Monocyte Quantibrite assay (BD Biosciences, San Jose, USA). Total number of antibodies bound per cell (AB/C) were quantified using calibration with a standard curve determined with BD Quantibrite phycoerythrin (PE) beads (BD Biosciences) as described elsewhere.6 B and NK lymphocyte immunophenotyping was performed using lyophilized antibody panel from Beckman Coulter (Duraclone kit). Data were acquired on a Navios flow cytometer (Beckman Coulter, Hialeah, FL) and flow data were analyzed using Navios software (Beckman Coulter).
Plasma anti-SARS-CoV-2 antibody concentration measurement
Plasmatic anti-SARS-CoV-2 immunoglobulin measurements were performed using Vidas® SARS-CoV-2 IgG in vitro diagnostic (IVD) assays (bioMérieux, Marcy-l'Étoile, France). Briefly, a solid-phase repository coated with the antigen (recombinant SARS-CoV-2 receptor-binding domain [RBD] of the viral spike [S] protein) served as both solid-phase and pipetting device. After a dilution step, SARS-CoV-2-specific IgG were captured on the coated antigen, and unbound components are washed out. In the second step, human IgG (Vidas® SARS-CoV-2 IgG) were specifically detected by mouse monoclonal antibodies conjugated to alkaline phosphatase and directed against human IgG, respectively. A relative fluorescence value (RFV) was generated (background reading subtracted from the final fluorescence reading). The assay was conducted with a standard solution as well as positive and negative controls. The results were automatically calculated by the instrument, according to standard and an index value (i) was obtained (i = RFVsample/RFVS1). The test is interpreted as negative when i < 1.00 and positive when i ≥ 1.00.
Plasma SARS-CoV-2 viral load measurement
Determination of normalized viral load was performed on plasma samples by RNA extraction on the EMAG® platform (bioMerieux, Marcy-l'Étoile, France). The SARS-CoV-2 load was measured by quantitative RT-PCR, according to a scale of calibrated in-house plasmid, using the RT-PCR RdRp-IP4 developed by the Institut Pasteur (Paris, France).7 The amplification protocol was developed using QuantStudio 5 rtPCR Systems (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The absence of inhibitors in the specimen was checked by using the RNA Internal Control R-GENE® kit (Argene_BioMérieux, Marcy-l'Étoile, France) on each sample. We expressed normalized SARS-CoV-2 load in log10 RNA copies/mL and all viral loads strictly below 4 RNA copies/reaction were considered under the limit of detection and were reported as negative.
Plasma SARS-CoV-2 nucleoprotein concentration measurement
N antigenemia levels were determined with an ELISA microplate assay, COVID-Quantigene® (AAZ, Boulogne-Billancourt, France), according to manufacturer recommendations. Briefly, in each well of 96-wells microplates previously coated with anti-SARS-CoV-2 N-antibodies, 50 μL of a solution containing a biotinylated anti-SARS-CoV-2 N antibodies and 50 μL of sera were added. After incubation at 37 °C for 60 min, plates were washed 5 times with a phosphate buffer solution. Then, 100 μL of a solution containing HRP-conjugated streptavidin were added, followed by incubation for 30 min at 37 °C. Plates were washed 5 times with the phosphate buffer solution, then 50 μL of a solution containing the substrate (3,3’,5,5’-tetramethylbenzidine (TMB)) and 50 μL of a second solution containing urea were added. After 15 min at 37 °C, the colorimetric reaction was stopped by adding 50 μL of H2SO4. Absorbance values were measured at 450 nm, with a reference set at 630 nm. In each plate, standards made of recombinant N antigens were tested, to quantify the N antigenemia levels for each patient's sample. Samples with titers above 180 pg/mL were diluted for precise quantification. Results were expressed as pg/mL.
Anti-SARS-CoV-2 specific T cell proliferation measurement
When sampled at 9- and 13-month follow-up visits, peripheral blood mononuclear cells (PBMCs) from patients and donors were isolated from fresh whole blood by Ficoll-Paque PREMIUM (GE Healthcare) density gradient centrifugation and immediately processed for cell culture. The number of PBMCs per well was adjusted to 1.106 cells/mL re-suspended in complete culture media (RPMI 1640 medium with HEPES, L-glutamine, 10% human AB plasma, 20 mg/mL streptomycin and 2.5 mg/mL fungizone). Cells were then stimulated during 7 days at 37 °C, 5% CO2 with SARS-CoV-2 peptide pools (JPT technology). The peptide pools consisted of 15 amino acid long peptides with 11 amino acid overlap. The following SARS-CoV-2 peptide pools were evaluated: pool of 315 peptides covering the spike (15 mers with 11-aa overlap, delivered in two sub-pools of 158 [S1 pool] and 157 [S2 pool] peptides), pool of 102 peptides covering the nucleoprotein, pool of 53 peptides covering the membrane. Peptides were used at a final concentration of 1.25 µg/mL. All samples were performed in duplicates. EdU click-it reaction was performed using EdU Click-it kit (Thermo Fisher) as previously described.8 CD3+, CD4+ and CD8+ T cell proliferations were analyzed by monitoring EdU-AF488 incorporation into cells. Results were expressed as percentages of cells incorporating EdU.
Statistical analyses
Data are presented as numbers and percentages (qualitative variables) and medians and 25th/75th percentiles (quantitative variables). Fisher's exact test was used for qualitative variables comparison. Quantitative variables were compared with Mann-Whitney U test. For all pairs of immune parameters, Spearman's rho correlation coefficients were estimated and summarized in a correlation matrix. Comparisons between immune parameters measured in patients with short and long hospital length of stays (hospital length of stay below or over 25 days) were limited to values measured at D0, M9 and M13 when all patients from both groups could be sampled. The level of significance was set at 5%. Data were analyzed using Graphpad Prism version 5.03 (Graphpad Software, La Jolla, USA) and R version 3.6.2 (R Core Team).
Role of funding sources
RICO clinical study was supported by funds from the Hospices Civils de Lyon, Fondation HCL and Claude Bernard Lyon 1 University / Région Auvergne Rhône-Alpes.
NOSOCOR clinical study was supported by partial funding by REACTing (Research and ACTion targeting emerging infectious diseases) INSERM, France and a donation from Fondation AnBer (http://fondationanber.fr/).
The funders had no role in study design, data collection and analysis, interpretation, decision to submit, or writing of the manuscript.
Results
Patients characteristics
Sixteen convalescent critically ill COVID-19 patients were included in this study. Demographics and clinical characteristics of patients at ICU admission are presented in Table 1. Patients were predominantly males (75%) with median age of 64 years old. Median duration of symptoms before ICU admission was 9 [7–11] days. These patients were invited for two follow-ups medical visits in an ambulatory setting at respectively 278 [253–301] days and 382 [375–384] days after initial infection. During the first visit (i.e. after 9 months M9), nine patients presented with persistent symptoms such as dyspnea, asthenia, pain or critical illness neuropathy. Two had been re-hospitalized but with no link to the initial COVID-19 infection. At this first visit, none of the patients was vaccinated against SARS-CoV-2. During the second follow-up visit (i.e. after 13 months after initial infection – M13), ten patients had persistent symptoms and one had been re-hospitalized without any link to the COVID-19 infection. At this second visit, three patients had received a first injection and one two injections of anti-COVID-19 vaccine (BNT162b2 vaccine, Pfizer-BioNTech). All participants tested negative for SARS-CoV2 PCR at M9 and M13 follow-up visits.
Table 1.
Clinical characteristics of critically ill patients with COVID-19 at ICU admission. Results are shown as medians and interquartile ranges [Q1-Q3] for continuous variables or numbers and percentages for categorical variables. Patients were separated in two groups based on length of hospital stay. Sepsis-related organ failure assessment (SOFA) and Simplified acute physiology score II (SAPS II) scores were calculated during the first 24 h after intensive care unit (ICU) admission. Data were compared using non-parametric Mann-Whitney test for continuous variables or Fisher's exact test for categorical variables.
| All patients (n = 16) | Short Hospital Stay (n = 7) | Long Hospital Stay (n = 9) | p-value | |
|---|---|---|---|---|
| Demographics | ||||
| Age | 64 [55–68] | 56 [43–72] | 65 [59–67] | 0.6624 |
| Gender (Male) | 12 (75 %) | 4 (57 %) | 8 (89 %) | 0.1457 |
| Body mass index (kg/m2) | 29.5 [26.4–34.3] | 26.5 [25.5–28.7] | 30.5 [28.7–35] | 0.0907 |
| Body mass index > 30 kg/m2 | 8 (50 %) | 1 (14 %) | 7 (78 %) | 0.0117 |
| Comorbidities | ||||
| 0 | 13 (81 %) | 7 (100 %) | 6 (67 %) | 0.0901 |
| ≥ 1 | 3 (19 %) | 0 (0 %) | 3 (33 %) | |
| Charlson score | 0 [0–0] | 0 [0–0] | 0 [0–1] | 0.2125 |
| Admission symptoms | ||||
| Delay between first symptoms (Days) | 9 [7–11] | 9 [8–11] | 9 [6–11] | 0.8143 |
| Fever | 13 (81 %) | 5 (71 %) | 8 (89 %) | |
| Cough | 11 (69 %) | 6 (86 %) | 5 (56 %) | |
| Dyspnea | 9 (56 %) | 2 (29 %) | 7 (78 %) | |
| Diarrhea | 6 (38 %) | 3 (43 %) | 3 (33 %) | |
| Diffuse pain | 3 (19 %) | 1 (14 %) | 2 (22 %) | |
| Altered general status | 9 (56 %) | 5 (71 %) | 4 (44 %) | |
| Other | 4 (25 %) | 1 (14 %) | 3 (33 %) | |
| Severity scores | ||||
| SOFA score | 2 [2–5] | 2 [2–3] | 3 [2–8] | 0.2054 |
| SAPS II score | 28 [22–36] | 27 [22–33] | 29 [20– 40] | 0.8990 |
| PaO2/FiO2 at admission | 162 [89–238] | 238 [78–329] | 147 [90–170] | 0.1893 |
| Organ support | ||||
| Mechanical ventilation) -Noninvasive ventilation) -Invasive ventilation |
16 (100 %) 12 (75 %) 4 (25 %) |
7 (100 %) 7 (100 %) 0 (0 %) |
9 (100 %) 5 (56 %) 4 (44 %) |
|
| 0.0417 | ||||
| Vasoactive drugs | 3 (19 %) | 0 (0 %) | 3 (33 %) | 0.0901 |
| Renal replacement therapy | 2 (13 %) | 0 (0 %) | 2 (22 %) | 0.1824 |
| Follow-up | ||||
| Days in ICU | 8 [3–38] | 4 [1–7] | 30 [14–65] | 0.0031 |
| Days in Hospital | 37 [12–72] | 11 [7–15] | 71 [44–87] | 0.0002 |
| Secondary infections | 8 (50 %) | 0 (0 %) | 8 (89 %) | 0.0004 |
Non-specific immune response to infection
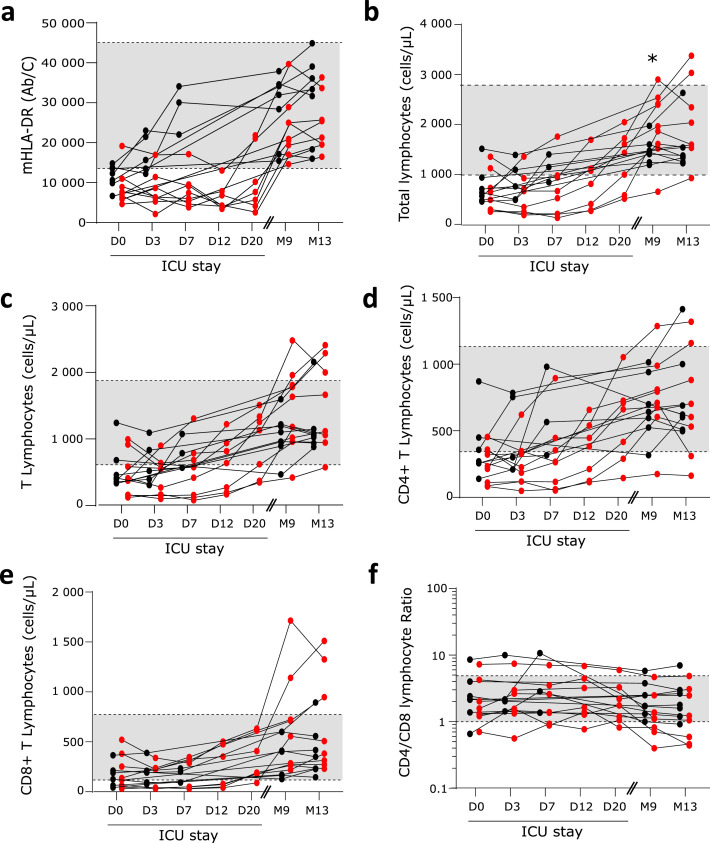
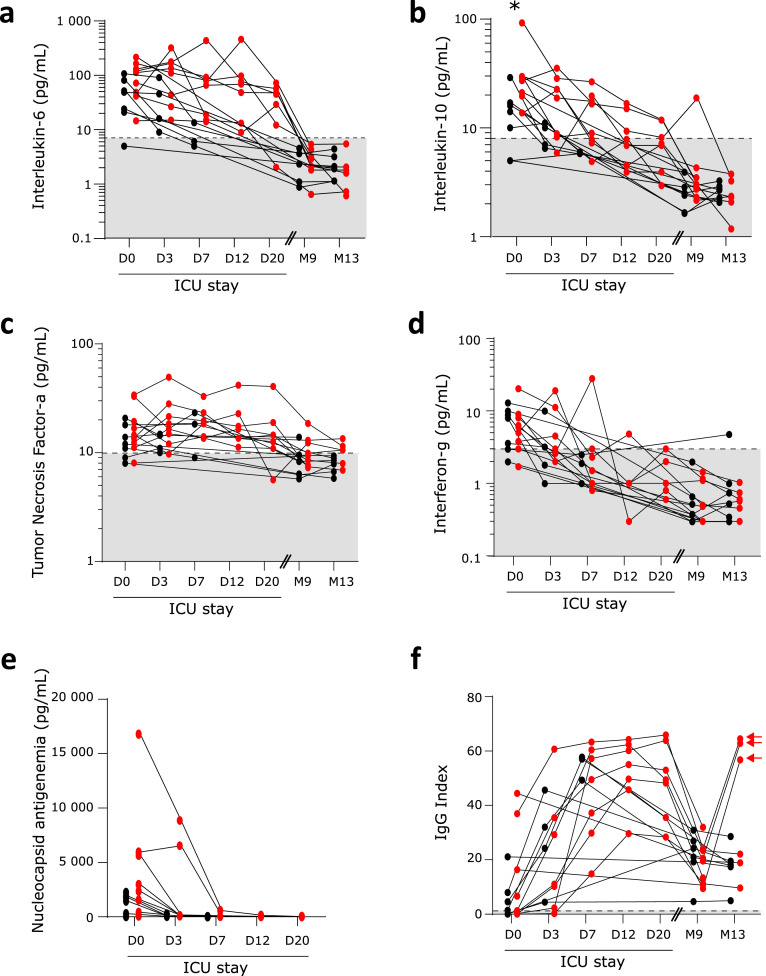
During the ICU stay, immune profile of critically ill COVID-19 patients recapitulated features of immune alterations as described previously.2,3,9 This included decreased expression of monocyte HLA-DR expression (mHLA-DR, Figure 1A), major lymphopenia (Figure 1B) affecting both B, NK, CD4+ and CD8+ T cells (Figure 1 C–E, Supplementary Figure 1A, B), increased plasma concentrations of both pro (Figure 2A, C and D) and anti-inflammatory cytokines (Figure 2B). These alterations were maximal at ICU admission and tended to normalize overtime during the ICU stay.
Figure 1.
Monitoring of COVID-19-induced cellular immune response. Sixteen convalescent critically ill COVID-19 patients were sampled 5 times during ICU stay: within the first 48 h after admission (Day 0: D0, n = 15), between 72 h and 96 h after admission (D3, n = 13), between D7 and D9 (D7, n = 11), between D12 and D15 (D12, n = 7), between D20 and D25 (D20, n = 7) and then twice at follow-up visits: after 9 and 13 months after hospital discharge (M9, n = 16; M13; n = 15). At each sampling time, the following immune parameters were monitored: a- monocytic expression of HLA-DR (mHLA-DR, numbers of antibodies bound per monocytes (Ab/C)), b- absolute counts of circulating lymphocytes (numbers of cells / µL), c- absolute count of CD3+ T lymphocytes (numbers of cells / µL), d- absolute count of CD4+ T cells (numbers of cells / µL), e- absolute count of CD8+ T cells (numbers of cells / µL) and f- ratio of numbers of CD4+ / CD8+ T cells. Results are presented as individual values. Patients with short hospital length of stay (HLS) are represented with black symbols; patients with long HLS with red symbols. Grey zones represent normal values from the routine Immunology Laboratory of Hospices Civils de Lyon for each immune parameter. At D0, M9 and M13, results were compared between patients with short and long HLS using non parametric Mann Whitney test. * p < 0.05.
Figure 2.
Monitoring of COVID-19-induced plasmatic immune response. Sixteen convalescent critically ill COVID-19 patients were sampled 5 times during ICU stay: within the first 48 h after admission (Day 0: D0, n = 15), between 72 h and 96 h after admission (D3, n = 13), between D7 and D9 (D7, n = 11), between D12 and D15 (D12, n = 7), between D20 and D25 (D20, n = 7) and then twice at follow-up visits, i.e. after 9 and 13 months after hospital discharge (M9, n = 16; M13; n = 15). At each sampling time, the following immune parameters were monitored: a- plasmatic concentrations of interleukin-6 (pg/mL), b- plasmatic concentration of interleukin-10 (pg/mL), c- plasmatic concentration of tumor necrosis factor-α (pg/mL), d- plasmatic concentration of interferon-γ (pg/mL), e- plasmatic concentration of nucleoprotein antigen (pg/mL) and f- plasmatic levels of anti-SARS-CoV-2 immunoglobulin G (IgG Index). For this last parameter, results in vaccinated patients are identified by an arrow. To note, nucleoprotein levels were only measured during ICU stay as all patients tested negative for SARS-CoV-2 at M9 and M13 visits. Results are presented as individual values. Patients with short hospital length of stay (HLS) are represented with black symbols; patients with long HLS with red symbols. Grey zones represent normal values from the routine Immunology Laboratory of Hospices Civils de Lyon or from the manufacturer when available. At D0, M9 and M13, results were compared between patients with short and long HLS using non parametric Mann Whitney test. * p < 0.05.
At follow-up visits (i.e. M9 and M13), immune alterations had receded and most patients presented with immune parameters within the range of normal values. In particular, all patients presented with normal mHLA-DR levels (Figure 1A) and no evidence of general persistent lymphopenia (Figure 1B to E, Supplementary Figure 1A, B). Finally, most patients followed up at 9 and 13-month convalescence presented with normal plasmatic concentrations of pro and anti-inflammatory cytokines (Figure 2 A to D) except for plasma TNF-α levels, which remained subnormal in some patients (Figure 2C).
Antigenemia, viremia and anti-SARS-CoV-2 IgG response
Plasma levels of SARS-CoV-2 nucleocapsid were high upon ICU admission but decreased rapidly (Figure 2E) while anti-SARS-CoV-2 IgG concentrations increased over the first week after admission (Figure 2F). At the end of the first week, all critically ill COVID-19 patients presented an elevated concentration of anti-SARS-CoV-2 IgG and all except one tested negative for SARS-CoV-2 antigenemia. Circulating levels of SARS-CoV-2 mRNA were undetectable in plasma for most ICU patients at any time during ICU stay except for four patients who were positive for SARS-CoV-2 PCR in plasma at ICU admission with one maintaining detectable levels of viremia for the first 3 days in ICU (data not shown).
At M9 and M13, all patients remained positive for anti-SARS-CoV-2 IgG in plasma (Figure 2F). However, IgG concentrations had largely decreased compared with levels measured during the ICU stay. Only three patients who received at least one dose of anti-SARS-CoV-2 vaccine at M13 visit presented with a marked elevation of anti-SARS-CoV-2 IgG index (Figure 2F).
Anti-SARS-CoV-2 T cell response
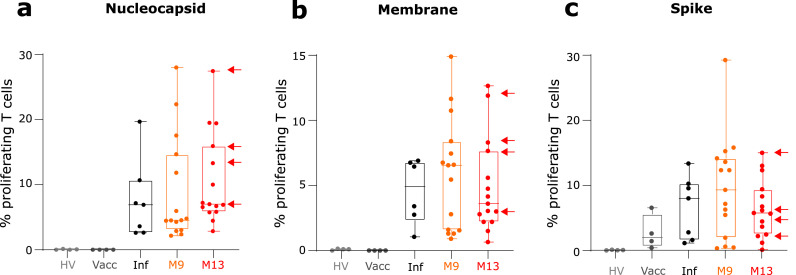
We included a cohort of 4 healthy donors who were not vaccinated against SARS-CoV-2 and had not been in contact with the virus. As expected, in this group of patients, we did not detect any proliferation against any of the SARS-CoV-2 antigens (Figure 3A to C). A group of 4 donors who were fully vaccinated (BNT162b2, Pfizer BioNTech) but had not been in contact with SARS-CoV-2 was also included. In this group, we detected T cell proliferation in all patients but only after T cell stimulation with Spike peptides pool. This shows that this group of donors had developed a memory T cell response against the vaccine antigen (i.e. Spike protein, Figure 3C). Finally, we included a group of 7 donors who had been infected by SARS-CoV-2. In this last group of patients, we detected T cell proliferation in all patients against the 3 SARS-CoV-2 proteins (Figure 3A to C), which illustrated the development of a memory T cell response against several antigens from the virus.
Figure 3.
Monitoring of SARS-CoV-2 specific T cell proliferation. Sixteen convalescent critically ill COVID-19 patients were sampled twice at follow-up visits (i.e. after 9 and 13 months after hospital discharge (M9, n = 16, orange symbols and boxes; M13; n = 15, red symbols and boxes; M13; n = 15). Fifteen healthy donors either non-infected and non-vaccinated (HV, n = 4, light grey symbols), or after full vaccination (Vacc, n = 4, dark grey symbols and boxes) or after resolution of a non-severe SARS-CoV-2 infection (Inf, n = 7, black symbols and boxes) were concomitantly included. At each sampling time, the percentage of T lymphocytes that had proliferated in response to 3 SARS-CoV-2 antigens (Nucleocapsid – Panel a, Membrane – Panel b, Spike – Panel c) was monitored. Results in vaccinated patients are identified by an arrow. Results are expressed as the percentage of proliferating T cells among total T cells and are presented as individual values and as Tukey Box-plots.
At M9 and M13, the presence of anti-SARS-CoV-2 T cells specific for SARS-CoV-2 antigens was detected in 100 % of convalescent critically ill COVID-19 patients (Figure 3A to C). Percentages of SARS-CoV-2 responding T cells in patients were heterogeneous but were mostly equivalent to levels measured in vaccinated controls or healthy individuals after non-severe SARS-CoV-2 infection. T cell proliferation was detected against the three tested SARS-CoV-2 antigens (i.e. nucleocapsid, membrane, spike). Among T cells, proliferation of both T CD4+ and T CD8+ lymphocytes was measured (Supplementary Figure 2A). When comparing the evolution of this specific T cell response overtime, no significant difference between results obtained at M9 and M13 in paired samples was observed (Supplementary Figure 2B).
At M9 and M13, proliferative responses against these 3 antigens were correlated but no correlation was observed between anti-SARS-CoV-2 T cell proliferative response and IgG levels (Supplementary Figure 2C). In addition, T cell response against nucleocapsid antigen was correlated with circulating levels of CD8+ T cells and plasma TNF-α concentration (Supplementary Figure 2C).
Association between hospital length of stay and anti-SARS-CoV-2 specific immune response
As critically ill COVID-19 patients are characterized by long hospital lengths of stay (HLS),10 we stratified this cohort in two groups based on their HLS. The threshold (HLS below or over 25 days) was selected to reflect the bimodal repartition of HLS in this cohort and in accordance with median ICU stay of previously published cohorts of critically ill COVID-19 patients11,12: a group of 7 patients with short HLS (ranging from 7 to 17 days) and a group of 9 patients with long HLS (ranging from 30 to 119 days).
Clinical and demographic characteristics of patients with short and long HLS are presented in Table 1. Critically ill COVID-19 patients with long HLS presented more frequently with obesity (i.e. BMI superior to 30 kg/m²) but were not significantly different in terms of initial severity as evaluated by SAPSII and SOFA scores at ICU admission. Nevertheless, patients with long HLS were more frequently placed under invasive mechanical ventilation (44% vs 0% in patients with short HLS, p = 0.0417). Therefore, this subgroup of patients also more frequently developed secondary / hospital acquired infections (89% vs 0% in patients with short HLS, p = 0.0004).
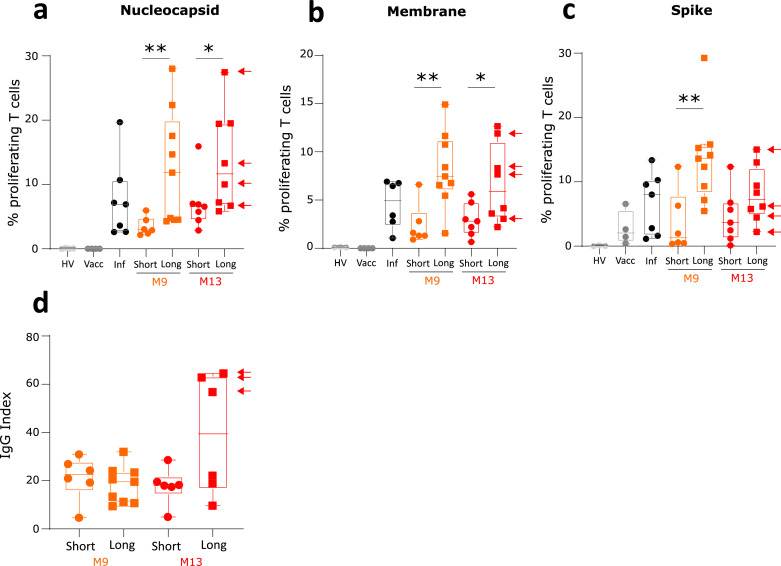
The percentages of anti-SARS-CoV-2 specific proliferating T cells were significantly higher in patients with long HLS (Figure 4A to C). This difference was observed for the three SARS-CoV-2 antigens and was maintained at both M9 and M13 follow-up visits for nucleocapsid and membrane-specific T cell responses. However, no difference regarding plasma anti-SARS-CoV-2 IgG levels was observed between patients with short and long HLS (Figure 4D).
Figure 4.
Monitoring of SARS-CoV-2 specific immune response in patients depending on hospital length of stay. Sixteen convalescent critically ill COVID-19 patients were sampled twice at follow-up visits (i.e. after 9 (M9, n = 16, orange symbols and boxes) and 13 months after hospital discharge (M13; n = 15, red symbols and boxes) and stratified according to length of hospital stay. Results obtained in patients with length of hospital stay inferior to 30 days (n = 7 patients, short, symbols = circles) and in patients with length of hospital stay superior to 30 days (n = 9 patients, Long, symbols = squares) are shown. Fifteen healthy donors either non-infected and non-vaccinated (HV, n = 4, light grey symbols), or after full vaccination (Vacc, n = 4, dark grey symbols and boxes) or after resolution of a non-severe SARS-CoV-2 infection (Inf, n = 7, black symbols and boxes) were concomitantly included. The percentages of T lymphocytes that had proliferated in response to 3 SARS-CoV-2 antigens (Nucleocapsid – Panel a, Membrane – Panel b, Spike –Panel c, expressed as percentages of proliferating T cells among total T cells) and the plasmatic concentrations of anti-SARS-CoV-2 IgG (IgG index – Panel d) were monitored. Results in vaccinated patients are identified by an arrow. Results are presented as individual values and as Tukey Box-plots. Comparison between the two groups of patients at each time point were performed using non parametric Mann Whitney test. * p < 0.05 ** p < 0.01.
Convalescent patients with short and long HLS did not differ for any immune parameter when measured at M9 and M13 except for the numbers of circulating total lymphocytes (Figure 1B) and of NK cells (Supplementary Figure 1A) which were higher in patients with long HLS.
Upon ICU admission, patients with short and long HLS did not significantly differ in the measurement immune parameters except for IL-10 concentration, which was increased at D0 in patients with long HLS (Figure 2B). However, patients with long HLS tended to present with more profound immune alterations such as decreased mHLA-DR (Figure 1A), T cell lymphopenia (Figure 1C), higher inflammatory response (Figure 2A), higher plasmatic antigenemia (Figure 2E) and more frequently detectable plasmatic viral mRNA (0 % in patients with short HLS at D0 versus 50 % in patients with long HLS) compared with patients with short HLS. In addition, the decreased MHC class II expression on monocytes (mHLA-DR) observed at D0 in patients with long HLS tended to persist overtime when compared with normal values (Figure 1A).
Discussion
In this study, we assessed the longevity of anti-SARS-CoV-2 specific immune response in convalescent critically ill COVID-19 patients longitudinally monitored during the acute phase of the disease and 9 and 13 months after symptom onset. We showed that, despite the presence of profound immune alterations during the ICU stay (systemic inflammation, lymphopenia and decreased MHC class II expression), convalescent critically ill COVID-19 patients consistently generated a good memory T cell response against SARS-CoV-2 that was maintained for more than one year after hospital discharge. Patients with long hospital and ICU stays presented with stronger anti-SARS-CoV-2 specific T cell response but no difference in anti-SARS-CoV-2 antibody levels.
Severe COVID-19 patients develop altered immune response upon SARS-CoV-2 infection, which associates a systemic inflammatory response and innate and adaptive immune alterations.1,3 More specifically, T lymphocyte response is profoundly altered in severe COVID-19 patients and the presence of an extensive lymphopenia has repeatedly been described as a prominent feature of critically ill COVID-19 patients.1,13 COVID-19-induced lymphopenia affect all lymphocyte subpopulations, develop upon ICU admission and persist during ICU stay.2,14 Decreased circulating numbers of lymphocytes is also associated with functional and phenotypic alterations with many similarities with T lymphocyte alterations described in patients with bacterial sepsis.3,11,15 Finally, some studies associated the intensity of lymphocyte diminution and deleterious outcome.16 Antigen-presentation capacity is also altered in critically ill COVID-19 patients as illustrated by the decreased expression of the MHC class II molecule HLA-DR on circulating monocytes.1,2,9,15 Whether such initial alterations of immune response might persist over time upon ICU discharge and could alter the development of SARS-CoV-2 specific adaptive and humoral immune responses in the most severe survivors critically ill individuals was, so far, unknown.
Observations from the current study confirmed that critically ill COVID-19 patients developed COVID-19-induced immune alterations including increased plasma cytokine concentrations and decreased T, B and NK cell numbers in association with markedly reduced mHLA-DR during ICU stay when compared with normal values. These alterations persisted over the first week after admission with a tendency to improve over time. We completed these data by monitoring these immune parameters in the same patients 9 and 13 months after hospital discharge. We showed that COVID-19 induced immune alterations did not persist in convalescent individuals upon ICU discharge. Thus, COVID-19-induced immune alterations appear as a transitory phenomenon following acute infection. As described in other cohorts of critically ill patients, such initial response may be considered as part of the physiologic host response to stress initiated to control the overwhelming inflammatory response to infection to prevent its deleterious consequences for the patient.11
When assessing SARS-CoV-2 specific humoral immune response, we observed a strong anti-SARS-CoV-2 antibody production during ICU the stay despite the parallel presence of immune alterations. This is in agreement with previous studies and suggests that severe disease manifestations are not caused by a lack of humoral anti-SARS-CoV-2 immune response.17, 18, 19, 20
Upon infection resolution, previous studies in non-severe COVID-19 patients described the persistence of SARS-CoV-2 neutralizing antibodies up to 6 to 8 months in 90% of recovered individuals.21,22 We showed that all convalescent critically ill patients from the current cohort had detectable anti-SARS-CoV-2 antibody titers 9 and 13 months after infection. However, we observed a substantial decline of antibody levels over time. Thus, even in patients with initially elevated anti-SARS-CoV-2 antibody titers, a marked decline in IgG concentrations was observed.
To note, in this study, four patients received at least one vaccine injection before the second follow-up visit. This was associated with a strong humoral response with elevated anti-SARS-CoV-2 IgG levels at M13. This highlights the good serological response to vaccine of these patients because, despite the substantial decline of anti-SARS-CoV-2 levels over time, humoral immune response could be re-stimulated by vaccination in convalescent critically ill COVID-19 patients.
Different read-outs are currently used in the literature to identify SARS-CoV-2 specific T cells, including expression of activation markers, IFN-γ production by ELISPOT, IFN-γ release assay or intracellular cytokine staining after stimulation with SARS-CoV-2 peptides.23,24 For example, anti-SARS-CoV-2 specific T cell response was evaluated either through Fluorospot assay (detection of IL-2 and or IFN-γ T cells) and through intracellular cytokine staining after stimulation with SARS-CoV-2 peptide pools in cohorts of mild to critical COVID-19 patients.25,26 Both articles described the persistence of anti-SARS-CoV-2 specific T cells on a long-term basis in recovered COVID-19 patients. These techniques, with short stimulation duration, are highly efficient in monitoring SARS-CoV-2 specific T lymphocyte effector response. However, they may lack sensitivity in detecting the persistence of memory T cells when measured at distance of infection or vaccination.27 On the contrary, T cell proliferation measured after one week of antigen stimulation is a sensitive technique to evaluate memory T cell responses and has been identified by experts as the reference technique for the follow-up of patients with primary immune deficiencies.28,29
Using this technique, we observed that convalescent critically ill COVID-19 patients developed a persistent memory response against SARS-CoV-2 when sampled after 9 and 13 months. This completes the data from the literature, that reported detectable SARS-CoV2 specific T cells upon disease resolution in convalescent donors.30, 31, 32, 33 However, previous studies mostly included patients after non-severe SARS-CoV-2 infections and none used T cell proliferation to evaluate memory T cell response. Thus, to our knowledge, this is the first report of the persistence of a strong memory T cell response after more than one year in convalescent critically ill COVID-19 patients using a T cell proliferation assay. Both CD4+ and CD8+ T cells specific responses to SARS-CoV-2 were detected and responses against SARS-CoV-2 antigens were correlated showing that a broad immune response towards SARS-CoV-2 was generated.
In the present cohort, SARS-CoV-2-specific T cells could be detected up to 13 months after initial infection and no decline was observed between results obtained after 9 and 13 months. At these late time-points, when focusing on anti-SARS-CoV-2 specific immune response, no correlations were observed between antigen-specific T cell response and anti-SARS-CoV-2 IgG levels and no increase in SARS-CoV-2 specific T cell response was observed in vaccinated patients. Thus, while anti-SARS-CoV-2 antibody levels seem to decay upon disease resolution,34, 35, 36 SARS-CoV-2-specific memory T cells persist for longer time in convalescent individuals. In the present context of consecutive COVID-19 waves, the capacity of this persistent memory T cell response to prevent reinfection should be investigated.
In convalescent individuals, T cell proliferation to SARS-CoV-2 nucleocapsid was correlated with the number of circulating CD8+ T cells. This is not unexpected as cytotoxic CD8+ T lymphocytes are central to the antiviral effector response.27 A significant correlation was also observed with plasma TNF-α levels. While TNF-α can have divergent effects on T cell subpopulation functions and survival37 and as this cytokine is the sole to maintain subnormal levels in convalescent individuals, deciphering its role in COVID-19 pathophysiology may be of major importance.
We observed that patients with complicated clinical courses leading to long hospital stays developed significantly better memory T cell response than patients with short clinical courses. The intensity of memory T cell response did not correlate with acute COVID-19 severity as evaluated by severity score at ICU admission (SAPS II, SOFA). However, patients with long HLS tended to present with more important COVID-19 induced immune alterations upon ICU admission and higher plasmatic antigenemia and viremia. Accordingly; in the study by Vibholm et al., the magnitude of CD8+ T cell response correlated with SARS-CoV-2 copies per swab.38 They also showed that persistently positive individuals had increased breath and magnitude of CD8+ specific response compared to PCR negative individuals. This is in accordance with our data, as patients with long hospital stays presented with higher and more persistent antigenemia and more frequently detectable SARS-CoV-2 mRNA in plasma compared with patients with uncomplicated clinical courses. This suggests that viral persistence resulted in an ongoing immune stimulation. Prolonged disease course and consequent larger exposure to virus experienced in hospitalized patients may provide a timeframe in which enhanced specific adaptive immune cells stimulation and maturation take place compared to shorter course and mild infections.
In addition, patients with better T cell response and long hospital stays presented with deep and persistent inflammation during the first week after admission; in addition with profound lymphopenia and decreased MHC class II expression. In this context, deciphering the mechanisms through which initial immune response to acute infection may influence the development of specific T cell response might be important to delineate the beneficial versus deleterious effects of this initial response to stress.
Interestingly, no differences were observed in humoral immune response to SARS-CoV-2 in patients with long and short hospital stays. This suggests that the mechanisms leading to humoral versus adaptive memory immune responses might differ in critically ill patients. In particular, while T follicular helpers might be fully efficient in generating SARS-CoV-2 specific B cells and neutralizing antibodies; immediate cytotoxic CD8+ T lymphocyte response might be insufficient to clear the virus. In the absence of effective antiviral therapies, immunoadjuvant treatments targeting such T cell alterations might represent innovative therapies in the most severe COVID-19 patients.39, 40, 41, 42, 43
A first limitation of this work is the limited number of patients included. This is especially true for the analysis of patients by subgroups depending on HLS and for adjustment for potential relevant clinical confounding factors. In addition, because of the low number of patients in each group, statistical testing could only detect the most profound differences. Finally, because they left the ICU, patients with short HLS could not be monitored overtime during their ICU stays. Thus, results from this purely observational study remain preliminary and should be confirmed prospectively on a larger cohort of patients. In line, the relatively small size of the cohort did not allow us to identify patients’ characteristics that would predict the maintenance of the anti-SARS-CoV-2 immune memory. In addition, as we restricted our cohort to individuals who recovered from severe COVID-19, these data did not describe the immune memory response after asymptomatic or mild COVID-19. Finally, protective efficacy of the anti-SARS-CoV-2 immune response analyzed in our study remains unclear as the exact magnitude of adaptive immune responses to SARS-CoV-2 infection required for protection from reinfection is not known so far. However, as recent longitudinal studies on larger cohorts suggest that the presence of cellular and humoral immunity was highly associated with prevention from reinfection with SARS-CoV-2,44,45 we can extrapolate that convalescent critically ill COVID-19 patients may have a strong protective humoral and adaptive immunity against SARS-CoV-2 reinfection. Again, this should be confirmed in a larger dedicated study.
In conclusion, results of the present study showed that, despite the presence of major lymphopenia and decrease in MHC class II expression on monocytes during ICU stay, convalescent critically ill COVID-19 patients consistently generated a good memory immune response against SARS-CoV-2, associating adaptive and humoral responses. This response was maintained for more than one year after hospital discharge. Patients with long hospital length of stay presented with stronger anti-SARS-CoV-2 specific T cell response but no difference in anti-SARS-CoV2 IgG levels.
Contributors
FV, MG, GM, FB, RC, RP, LG, MB and MO were involved in the design, implementation, and day-to-day management of the study. ACL, TR, LA, MC, MSE, LH, PV included participants in the study. MBD, FMS, JSC were responsible for the virological analyses. FV, MG, RC, FB, CT, KBP, VC, MAC, MO, RP and LG were responsible for the immunological analyses. MB, FB and FV were involved in the statistical analyses. FB, GM and FV wrote the original draft of the manuscript, which was reviewed and edited by MG, RC, ACL, RP, TR, MSE, LH and PV. All authors have read and approved the manuscript. All authors had full access to all the data and accept responsibility for the decision to submit for publication.
Data sharing statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of interests
MB, CT, KBG, VC and MAC are bioMérieux's employees. This private company had no role in the study design, result analysis and decision to publish this study. PV received consulting fees and payment for a literature review from Pfizer and Astellas. All other authors have declared no conflicts of interest.
Acknowledgments
The authors would like to thank the clinical teams from all ICUs in HCL who were involved in this project while dedicated to their clinical duties during COVID-19 pandemic as well as patients and their families who agreed to participate to this clinical study. The authors would also like to thank Valérie Cerro, Laurie Bignet, Marion Provent, Lauredana Baboi, Jessica Rousson, Florence Raynal, Laure Giambra (Services de Réanimation, Hospices Civils de Lyon), Sabrina Bennia (Service Hygiène, Epidémiologie, Infectiovigilance et Prévention, Hospices Civils de Lyon), Anne Termoz et Julie Haesebaert (Pôle de santé publique, Hospices Civils de Lyon), Camille Mena, Catherine Planckaert, Nadège Trehet-Mandez, Nathalie Paquin, Pr Roland Chapurlat (Prévention des Maladies Osseuses, Hospices Civils de Lyon) for clinical trial management. Last, the authors would like to thank Elisabeth Cerrato for technical support.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103967.
Appendix. Supplementary materials
References
- 1.Osuchowski M.F., Winkler M.S., Skirecki T., et al. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med. 2021;9(6):622–642. doi: 10.1016/S2213-2600(21)00218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venet F., Cour M., Rimmele T., et al. Longitudinal assessment of IFN-I activity and immune profile in critically ill COVID-19 patients with acute respiratory distress syndrome. Crit Care. 2021;25(1):140. doi: 10.1186/s13054-021-03558-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remy K.E., Mazer M., Striker D.A., et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5(17) doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saadatian-Elahi M., Picot V., Henaff L., et al. Protocol for a prospective, observational, hospital-based multicentre study of nosocomial SARS-CoV-2 transmission: NOSO-COR Project. BMJ Open. 2020;10(10) doi: 10.1136/bmjopen-2020-039088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gossez M., Malcus C., Demaret J., Frater J., Poitevin-Later F., Monneret G. Evaluation of a novel automated volumetric flow cytometer for absolute CD4+ T lymphocyte quantitation. Cytom B Clin Cytom. 2017;92(6):456–464. doi: 10.1002/cyto.b.21360. [DOI] [PubMed] [Google Scholar]
- 6.Demaret J., Walencik A., Jacob M.C., et al. Inter-laboratory assessment of flow cytometric monocyte HLA-DR expression in clinical samples. Cytom B Clin Cytom. 2013;84(1):59–62. doi: 10.1002/cyto.b.21043. [DOI] [PubMed] [Google Scholar]
- 7.Etievant S., Bal A., Escuret V., et al. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J Clin Med. 2020;9(6) doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poujol F., Monneret G., Friggeri A., et al. Flow cytometric evaluation of lymphocyte transformation test based on 5-ethynyl-2′deoxyuridine incorporation as a clinical alternative to tritiated thymidine uptake measurement. J Immunol Methods. 2014;415:71–79. doi: 10.1016/j.jim.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet B., Cosme J., Dupuis C., et al. Severe COVID-19 is characterized by the co-occurrence of moderate cytokine inflammation and severe monocyte dysregulation. EBioMedicine. 2021;73 doi: 10.1016/j.ebiom.2021.103622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karagiannidis C., Mostert C., Hentschker C., et al. Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Respir Med. 2020;8(9):853–862. doi: 10.1016/S2213-2600(20)30316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venet F., Textoris J., Blein S., et al. Immune profiling demonstrates a common immune signature of delayed acquired immunodeficiency in patients with various etiologies of severe injury. Crit Care Med. 2021 doi: 10.1097/CCM.0000000000005270. [DOI] [PubMed] [Google Scholar]
- 12.COVID-ICU Group on behalf of the REVA Network and the COVID-ICU Investigators Clinical characteristics and day-90 outcomes of 4244 critically ill adults with COVID-19: a prospective cohort study. Intensive Care Med. 2021;47(1):60–73. doi: 10.1007/s00134-020-06294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monneret G., Cour M., Viel S., Venet F., Argaud L. Coronavirus disease 2019 as a particular sepsis: a 2-week follow-up of standard immunological parameters in critically ill patients. Intensive Care Med. 2020;46(9):1764–1765. doi: 10.1007/s00134-020-06123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeannet R., Daix T., Formento R., Feuillard J., Francois B. Severe COVID-19 is associated with deep and sustained multifaceted cellular immunosuppression. Intensive Care Med. 2020;46(9):1769–1771. doi: 10.1007/s00134-020-06127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J Intensive Care. 2020;8:36. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roltgen K., Powell A.E., Wirz O.F., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54) doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisberg S.P., Connors T.J., Zhu Y., et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat Immunol. 2021;22(1):25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seow J., Graham C., Merrick B., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conti F., Oriol G., Cheynet V., et al. Seroconversion in septic ICU patients presenting with COVID-19: necessary but not sufficient. Arch Med Res. 2021;52(8):850–857. doi: 10.1016/j.arcmed.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dan J.M., Mateus J., Kato Y., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529) doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherina N., Piralla A., Du L., et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med (N Y) 2021;2(3):281–295. doi: 10.1016/j.medj.2021.02.001. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bercovici N., Duffour M.T., Agrawal S., Salcedo M., Abastado J.P. New methods for assessing T-cell responses. Clin Diagn Lab Immunol. 2000;7(6):859–864. doi: 10.1128/cdli.7.6.859-864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogbe A., Kronsteiner B., Skelly D.T., et al. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat Commun. 2021;12(1):2055. doi: 10.1038/s41467-021-21856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcotte H., Piralla A., Zuo F., et al. Immunity to SARS-CoV-2 up to 15 months after infection. iScience. 2022;25(2) doi: 10.1016/j.isci.2022.103743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen K.W., Linderman S.L., Moodie Z., et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. 2021;2(7) doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson H.L., Amara RR. T cell vaccines for microbial infections. Nat Med. 2005;11(4 Suppl):S25–S32. doi: 10.1038/nm1212. [DOI] [PubMed] [Google Scholar]
- 28.Notarangelo LD. Primary immunodeficiencies. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S182–S194. doi: 10.1016/j.jaci.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 29.McCusker C., Upton J., Warrington R. Primary immunodeficiency. Allergy Asthma Clin Immunol. 2018;14(Suppl 2):61. doi: 10.1186/s13223-018-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilich T., Nelde A., Heitmann J.S., et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci Transl Med. 2021;13(590) doi: 10.1126/scitranslmed.abf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Potti A., Lange J., Buggert M. Deciphering the ins and outs of SARS-CoV-2-specific T cells. Nat Immunol. 2021;22(1):8–9. doi: 10.1038/s41590-020-00838-5. [DOI] [PubMed] [Google Scholar]
- 32.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung J.H., Rha M.S., Sa M., et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat Commun. 2021;12(1):4043. doi: 10.1038/s41467-021-24377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cromer D., Juno J.A., Khoury D., et al. Prospects for durable immune control of SARS-CoV-2 and prevention of reinfection. Nat Rev Immunol. 2021;21(6):395–404. doi: 10.1038/s41577-021-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marot S., Malet I., Leducq V., et al. Rapid decline of neutralizing antibodies against SARS-CoV-2 among infected healthcare workers. Nat Commun. 2021;12(1):844. doi: 10.1038/s41467-021-21111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamayoshi S., Yasuhara A., Ito M., et al. Antibody titers against SARS-CoV-2 decline, but do not disappear for several months. EClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2021.100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta A.K., Gracias D.T., Croft M. TNF activity and T cells. Cytokine. 2018;101:14–18. doi: 10.1016/j.cyto.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vibholm L.K., Nielsen S.S.F., Pahus M.H., et al. SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses. EBioMedicine. 2021;64 doi: 10.1016/j.ebiom.2021.103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monneret G., de Marignan D., Coudereau R., et al. Immune monitoring of interleukin-7 compassionate use in a critically ill COVID-19 patient. Cell Mol Immunol. 2020;17(9):1001–1003. doi: 10.1038/s41423-020-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukaszewicz A.C., Venet F., Faure A., Vignot E., Monneret G. Immunostimulation with interferon-gamma in protracted SARS-CoV-2 pneumonia. J Med Virol. 2021;93(10):5710–5711. doi: 10.1002/jmv.27172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Laarhoven A., Kurver L., Overheul G.J., et al. Interferon gamma immunotherapy in five critically ill COVID-19 patients with impaired cellular immunity: a case series. Med (N Y) 2021;2(10):1163–1170. doi: 10.1016/j.medj.2021.09.003. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazer M.B., Turnbull I.R., Miles S., et al. Interleukin-7 reverses lymphopenia and improves T-cell function in coronavirus disease 2019 patient with inborn error of toll-like receptor 3: a case report. Crit Care Explor. 2021;3(7):e0500. doi: 10.1097/CCE.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Veerdonk F.L., Giamarellos-Bourboulis E., Pickkers P., et al. A guide to immunotherapy for COVID-19. Nat Med. 2022;28(1):39–50. doi: 10.1038/s41591-021-01643-9. [DOI] [PubMed] [Google Scholar]
- 44.Lumley S.F., O'Donnell D., Stoesser N.E., et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384(6):533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanrath A.T., Payne B.A.I., Duncan C.J.A. Prior SARS-CoV-2 infection is associated with protection against symptomatic reinfection. J Infect. 2021;82(4):e29–e30. doi: 10.1016/j.jinf.2020.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.