Abstract
Abstract
Objectives
Normal cellular function requires a rate of ATP production sufficient to meet demand. In most neurodegenerative diseases (including Amyotrophic Lateral Sclerosis [ALS]), mitochondrial dysfunction is postulated raising the possibility of impaired ATP production and a need for compensatory maneuvers to sustain the ATP production/demand balance. We investigated intermediary metabolism of neurons expressing familial ALS (fALS) genes and interrogated the functional consequences of glycolysis genes in fitness assays and neuronal survival.
Methods
We created a pure neuronal model system for isotopologue investigations of fuel utilization. In a yeast platform we studied the functional contributions of glycolysis genes in a growth fitness assay iafter expressing of a fALS gene.
Results
We find in our rodent models of fALS, a reduction in neuronal lactate production with maintained or enhanced activity of the neuronal citric acid cycle. This rewiring of metabolism is associated with normal ATP levels, bioenergetics, and redox status, thus supporting the notion that gross mitochondrial function is not compromised in neurons soon after expressing fALS genes. Genetic loss-of-function manipulation of individual steps in the glycolysis and the pentose phosphate pathway blunt the negative phenotypes seen in various fALS models.
Conclusions
We propose that neurons adjust fuel utilization in the setting of neurodegenerative disease-associated alteration in mitochondrial function in a baleful manner and targeting this process can be healthful.
Keywords: Glycolysis, G6PDH, Neuron, ATP, Redox
Highlights
-
•
Our work is the first to perform a comprehensive and quantitative analysis of intermediary metabolism in neurons in the setting of fALS causing gene products.
-
•
Because the cardinal feature of ALS is death of motor neurons, these new studies are directly relevant to the pathogenesis of ALS.
-
•
Our functional interrogations begin to unpack how metabolic re-wiring is induced by fALS genes and it will be very interesting, in the future, to gain insight in amino acid fueling of the TCA cycle.
-
•
We suspect pleiotropic effects of amino acid fueling, and this may lead to very targeted therapeutic interventions.
1. Introduction
Alterations in mitochondrial structure and function are commonly observed in adult-onset neurodegenerative diseases [[1], [2], [3], [4], [5], [6], [7], [8]]. In ALS, mitochondrial dysfunction impairs the efficiency of electron transport chain (ETC) activity and ATP production [[9], [10], [11], [12], [13]] and leads to the accumulation of reactive oxygen and nitrogen species [[14], [15], [16]], abnormal handling of intracellular calcium and cytochrome C release and apoptosis [17]. The extent to which these alterations in mitochondrial function impair cellular operations is unclear. Therapeutic intervention based on combating these mitochondrial abnormalities have displayed variable success in mouse models of ALS and humans [6,18], as reviewed in Vandoorne et al. [19].
Eukaryotic cells monitor their energy economy very carefully to ensure energy production and energy utilization are matched [[20], [21], [22]]. From the energy budget perspective, synaptic transmission, action potentials and the maintenance of the resting membrane potential are considered the most energetically expensive neuronal processes [23]. Even a modest, chronic decrement in ATP production could adversely affect fundamental neuronal biology and consequently impair neuronal functions and survival. In addition, on a vastly different time scale, the activity-driven local ATP synthesis is required for moment-to-moment synaptic function [[24], [25], [26]]. Neuronal glucose oxidative phosphorylation is the prime source of ATP subserving pre- and post-synaptic element cell biology [27]. If neuronal mitochondrial dysfunction in the setting of ALS significantly upset the balance of ATP production with ATP utilization, it would negatively impact all cellular functions.
To achieve cellular homeostasis in the setting of neurodegenerative disease-associated mitochondrial dysfunction, neurons could reduce anabolic processes and/or attempt to re-wire metabolism to maintain adequate ATP levels. Two general strategies could be employed to increase ATP production. First, maneuvers could be deployed to increase NADH production by the tricarboxylic acid (TCA) cycle, forcing more electrons into the ETC. Second, glycolysis might be stimulated. While a less efficient method for ATP production, work in the cancer field establishes that aerobic glycolysis can be a substantial energy source [28]. In either scenario, re-wiring metabolism could have unintended consequences on cellular biochemistry. For example, glycolytic intermediates are the starting point for the pentose phosphate pathway (PPP, which generates NADPH, essential for glutathione and thioredoxin control of reactive oxygen species) [[29], [30], [31]], the hexosamine biosynthetic pathway (HBS, which generates UDP-N-acetylglucosamine, essential for N- and O-glycans within the endoplasmic reticulum) [[32], [33], [34]], and the synthesis of amino acids. Enhanced glycolytic flux, at the expense of PPP or HBS, might adversely impact redox status, ER stress, and protein synthesis. In addition, the source of ATP (that is, glycolysis versus ETC) has cell type-specific effects on signaling pathways. For example, immune cell Akt-FOXO1 signaling is driven by mitochondrial-derived ATP in naïve T cells and by glycolysis in effector T cells [35].
The cardinal feature of ALS is the degeneration of upper and lower motor neurons, and thus, understanding the contribution of metabolic dysfunction of neurons is a critical issue. To understand how fuel is utilized, we undertook glucose isotopologue investigations in cultured rodent cortical neurons expressing two different fALS genes – Cu++/Zn++ superoxide dismutase (SOD1) or TAR DNA binding protein of 43 kDa molecular weight (TDP43). To capture an early time point in fALS pathogenesis, we studied neurons soon after expression of the fALS proteins. Based on our interpretation of these metabolite isotopologue results, we hypothesized that altering the expression of individual glycolytic genes could modify phenotypes induced by the expression of wild type (“Wt”) or disease-causing missense versions (Mutant or “Mt”) of SOD1 or TDP43. We tested this hypothesis in a series of functional studies employing Saccharomyces cerevisiae, Caenorhabditis elegans and Rattus norvegicus spinal cord neuron platforms. These integrated investigations reveal that: 1) there is little evidence of neuronal mitochondrial dysfunction in our models when focusing on outputs such as energy charge or redox state and 2) manipulating fuel utilization by neurons can blunt neuronal dysfunction and death in a variety of ALS models.
2. Results
2.1. Metabolomic investigations of cortical neurons expressing ALS-causing mutant proteins
We began by designing an experimental platform in which heavy atom glucose could be used to monitor how neurons metabolize their major fuel. The following considerations guided our choice of this tissue culture experimental model. First, we needed sufficient material for analysis, and only the embryonic cerebral cortex was a suitably abundant source of neurons to permit our detailed studies. Second, because neuronal metabolism is intimately linked to neuronal connectivity/network activity, it was important to have a mixed neuronal population sufficiently mature that inter-neuronal communication could exist. Our system contrasts with patient-derived induced pluripotent stem cells (iPSC) that are differentiated into motor neurons, where such context-dependent factors are given less emphasis. Third, the use of viruses to express Wt and Mt proteins allows for tight temporal control of gene expression. This allows for a favorable level degree of biochemical synchrony that increases the sensitivity of our measurements. There are two main limitations to our system: 1) Because there are nominally no glial in our cultures, our model does not incorporate the metabolic cross talk between glial cells and neurons. This was a requirement for us because of our goal of biochemically interrogating neuronal metabolism specifically, and 2) Wt and Mt proteins are heterologously expressed and thus potentially introducing non-physiological conditions. To mitigate these concerns, our studies are undertaken at an early time point when a robust metabolic phenotype was evident yet prior to any detectable neuron death [36].
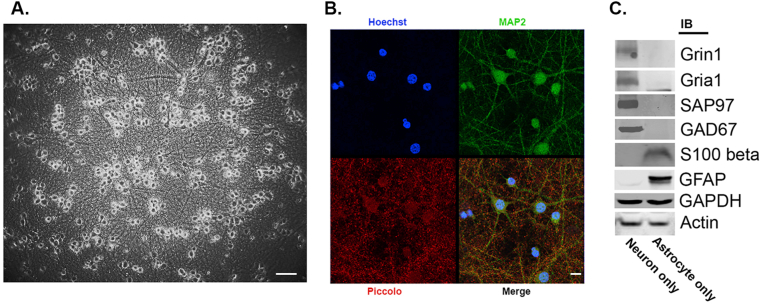
For metabolomic studies, neuronal cultures lacking astrocytes were generated from E17 rat cortex and after 14 days in vitro (DIV) they were infected with recombinant Herpes Simplex Virus (HSV) engineered to express Wt or G86R Mt SOD1 or Wt or M337V mutant TDP43. By phase contrast microscopy, our cultures displayed abundant healthy-appearing neurons with a rich investment of neurites in the intercellular space (Figure 1A). Immunocytochemically, a mesh of MAP2(+) dendrites were decorated with Piccolo-positive pre-synaptic terminals (Figure 1B). Puncta of glutamate receptor subunits Gria1 and Gria2 opposing Piccolo-positive pre-synaptic terminals were abundant as well (Supplementary Figure 1). The co-localization of post synaptic density protein of 95 kDa molecular weight (PSD95) with Gria1 puncta as well as co-localization of Gria1 and GluN1 puncta reinforce the notion that bona fide synaptic structures were assembled in our cultures (Supplementary Figure 1). In western blot assay, our culture system displayed abundant expression of markers of excitatory and inhibitory neurotransmission and little evidence of astrocyte markers (Figure 1C). These observations suggest that our experimental platform captures several network features seen in vivo – this is important because neuronal activity and metabolism are intimately linked [22].
Figure 1.
Characteristics of DIV 14 pure rat cortical neuron cultures. Panel A. Phase contrast image of cultures show abundant phase bright cell bodies with an extensive mat of interstitial neurites. Calibration bar = 85 microns. Panel B. A single confocal slice of cultures immunostained for MAP2 (green) and Piccolo (red); nuclei stained with Hoechst 33342. The merge of individual panels shows abundant pre-synaptic elements opposed to dendrites. Calibration bar = 25 microns Panel C. Western blot of cultures reveals evidence for excitatory (e.g., glutamate ionotropic receptor NMDA type subunit 1, Grin1; glutamate ionotropic receptor AMPA type subunit 1, Gria1; synapse associated protein of 97 kDa, SAP97) and inhibitor synapses (e.g., glutamate decarboxylase 67 kDa, GAD67) in cultures of neurons but not astrocytes (S100 calcium binding protein B, S100beta; glial fibrillary acidic protein, GFAP). Loading control was glyceraldehyde-3-phosphate dehydrogenase (GAPDH) when studying GFAP and actin for all the remaining proteins.
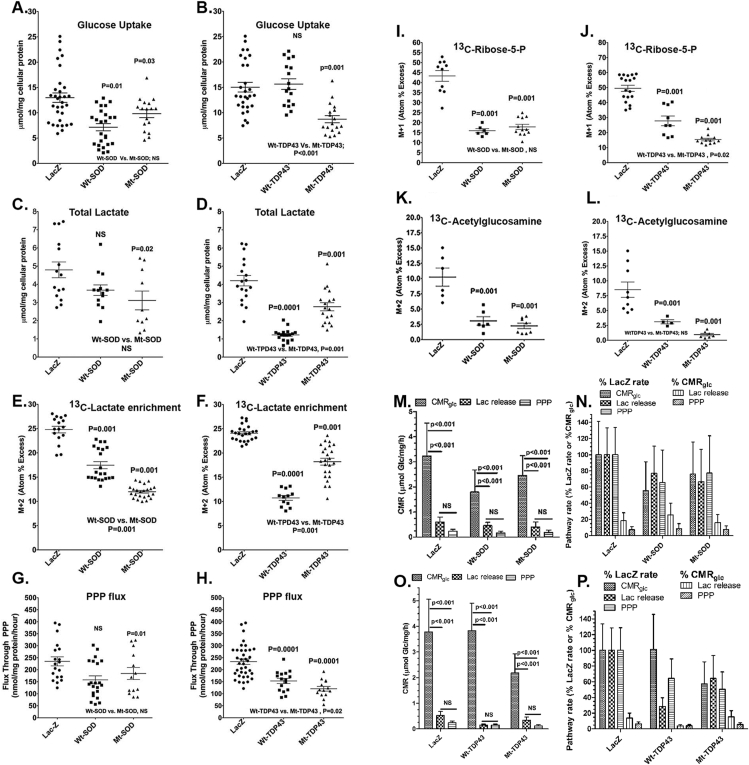
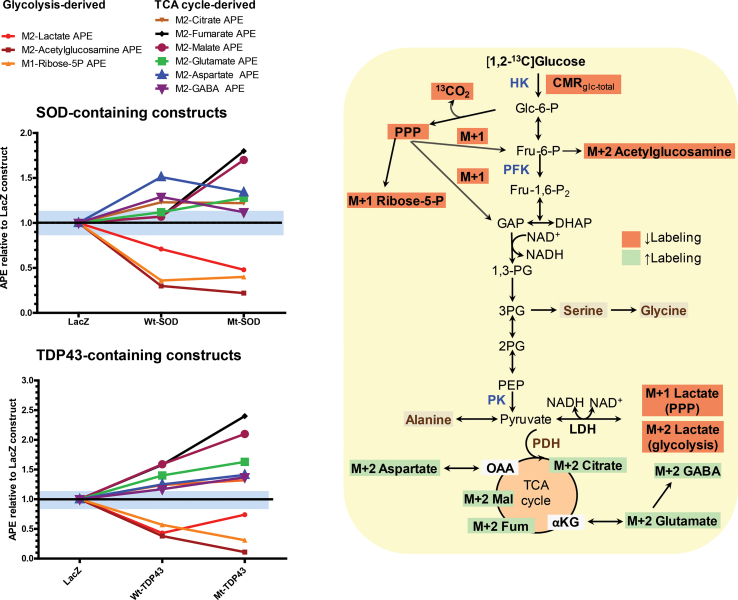
Using [1,2-13C2]-d-glucose to track fuel utilization, we found that trends of metabolic alterations were similar for SOD1 and TDP43 expressing neurons in comparison with the control condition (neurons expressing LacZ). Net glucose uptake (i.e., CMRglc, the cerebral metabolic rate for glucose (glc) or the rate of glucose utilization) from the media (calculated by difference, glucose concentration before minus that after the 4 h incubation) by LacZ expressing neurons was about 3.2–3.8 μmol/mg cellular protein/h, and net uptake was significantly reduced by about 25–45% in Wt SOD1, Mt SOD1 and Mt TDP43 expressing neurons (Figure 2A, B). This was associated with reduced lactate production and release to the medium in SOD1 and TDP43 expressing neurons, although the Wt SOD1 group did not reach statistical significance (Figure 2C, D). For example, extracellular lactate production in LacZ expressing neurons is approximately 1.2 μmol/mg/h while the lactate release from Mt SOD1 expressing neurons was approximately 0.80 μmol/mg/h and from Mt TDP43 expressing neurons approximately 0.68 μmol/mg/h. Isotope enrichment (atom percent excess, APE, see Methods) in lactate is reduced more in Mt SOD1 expressing neurons in comparison with Wt SOD1 expressing neurons. In contrast, total lactate production and isotope enrichment in lactate were reduced more in Wt TDP43 expressing neurons in comparison with Mt TDP43 (although both were lower in values than LacZ expressing neurons) (Figure 2E, F). Taken together, these observations reveal a reduction in glucose utilization, lactate production and release to the medium, and 13C-lactate enrichment in the setting of expression of Wt and Mt SOD and TDP43 constructs in neurons compared with LacZ-expressing neurons (Figure 2A–F and M–P). If we assume lactate and pyruvate are in equilibrium during the 4-hour labeling experiments, the reduction of lactate production and release into the media could reflect either: 1) a reduction of glycolysis (an interpretation aligned with the observed reduced glucose utilization) or 2) enhanced entry of pyruvate into the TCA cycle (see below), perhaps due to changes in regulatory mechanisms (e.g., involving the pyruvate dehydrogenase complex).
Figure 2.
Metabolic interrogation of glycolysis and related pathways in neurons expressing Wt and fALS Mt genes. Pooled data from 3 to 5 experiments comparing neurons expressing LacZ with Wt SOD1/TDP43 with Mt SOD1/TDP43 is presented. Horizontal lines denote means ± SD. ANOVA showed group differences, and values above the scattergram indicate that in post hoc tests the specific group is statistically significantly different from the LacZ group (p value provided) or not different (NS – not significant). Values below the scattergram indicate that the Wt or Mt proteins group are statistically significantly different or not. Panel labels: A, B, Net glucose uptake during the 4 h incubation; C, D, total lactate release to medium; E, F, 13C enrichment of medium lactate; G, H, Flux of glucose through the pentose phosphate pathway (PPP); I, J, 13C enrichment of intracellular ribose-5-phosphate (P); K, L, 13C enrichment of intracellular acetylglucosamine. For the CMRglc summary panels (M and O), data are means and 1 SD, with all rates in units of glucose or its equivalent (i.e., lactate/2). For the pathway rates expressed as percent of the LacZ rate or of CMRglc (Panels N and P), values are mean quotient and vertical bars are CVs for the 100% rates or SDs that account for error propagation for all others. Statistically significant changes are indicated in the panels that illustrate the primary data.
Beyond its role in producing ATP and pyruvate (for entrance via acetyl CoA into the TCA cycle), the metabolism of glucose by glycolysis generates intermediates that serve other cellular functions. The glycolytic intermediate glucose-6-phosphate [Glc-6-P] enters the PPP. The PPP rate is 0.24 μmol/mg/h in LacZ expressing neurons and 0.19 μmol/mg/h for Mt SOD1 and 0.12 μmol/mg/h for Mt TDP43 expressing neurons (Figure 2G, H). When expressed as a fraction of CMRglc, the magnitude of the relative PPP rate (i.e., about 7–9% of CMRglc in LacZ expressing neurons) decreased in proportion to CMRglc in SOD1 neurons, whereas decreased from 6.3% in LacZ neurons to 5.6% in Mt TDP43 expressing neurons (Figure 2M–P). These lower PPP rates were associated with reductions in ribose-5-phosphate levels (a product of PPP) that were disproportionately greater than the respective fall in PPP rate in the Mt SOD1 and TDP43 expressing neurons (Figure 2I, J). The glycolytic intermediate fructose-6-phosphate enters into the HBS pathway. Monitoring acetylglucosamine levels reports on this pathway, and the isotope enrichment in acetylglucosamine in SOD1 and TDP43 expressing neurons was dramatically reduced, more than the decreased in lactate production and release rate, PPP rate, and ribose-5-phosphate enrichment (Figure 2K, L)
By several measures, neurons expressing Mt proteins have more severe deficits than neurons expressing Wt proteins. Net glucose uptake, medium lactate levels, PPP flux, and ribose-5-phosphate enrichment did not differ in the Mt versus Wt SOD1 expressing neurons, but isotope enrichment in lactate was lower in Mt SOD1 vs. Wt SOD1 expressing neurons. Net glucose uptake, PPP flux, and ribose-5-phosphate enrichments were lower in Mt TDP43 vs. Wt TDP43 expressing neurons. We provide a summary of CMRglc for SOD1 and TDP43 expressing neurons as a percentage of LacZ neurons (Figure 2N and P respectively.).
Note that the isotope enrichment of M + 2 lactate would be expected to be half that of [1,2-13C2]glucose (no unlabeled glucose was present in the assay medium) because glycolysis would produce one labeled and one unlabeled pyruvate/lactate causing a 50% dilution. However, the lactate APEs were less than about 25% in all samples (Figure 2E, F) indicating additional dilution by unlabeled lactate. The culture medium contained pyruvate that, along with specific amino acids in the culture medium that can be degraded to produce pyruvate (e.g., alanine, serine, glycine, etc.), can be sources of unlabeled lactate. A likely mechanism is unlabeled pyruvate enters the neuron and is converted to lactate and released, with re-generation of NAD+, more so in the fALS expressing neurons than in LacZ expressing neurons. Metabolism of medium pyruvate to lactate may also facilitate entry of [13C]glucose-derived [13C]pyruvate into the TCA cycle or other pathways and contribute to higher labeling of TCA cycle intermediates and TCA cycle-derived amino acids, but would not alter the conclusion that fALS models have reduced lactate production from glucose and subnormal PPP and HBP pathway fluxes; all cells had the same access to extracellular substrates contained in the medium.
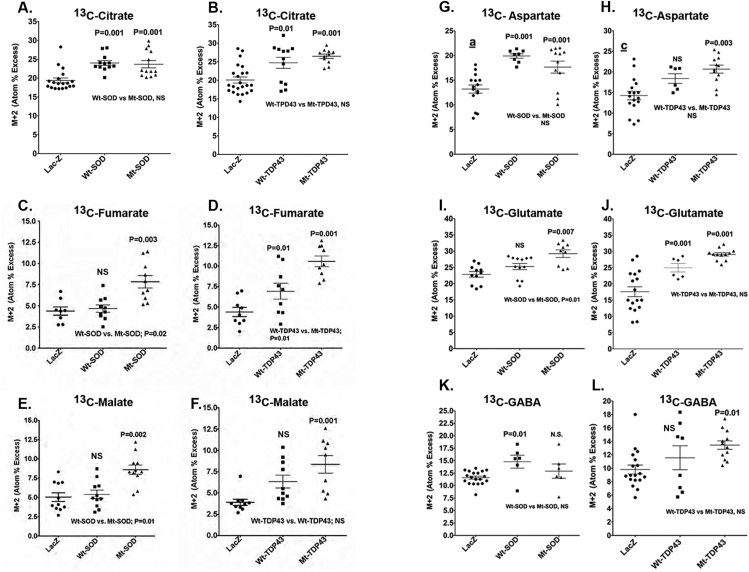
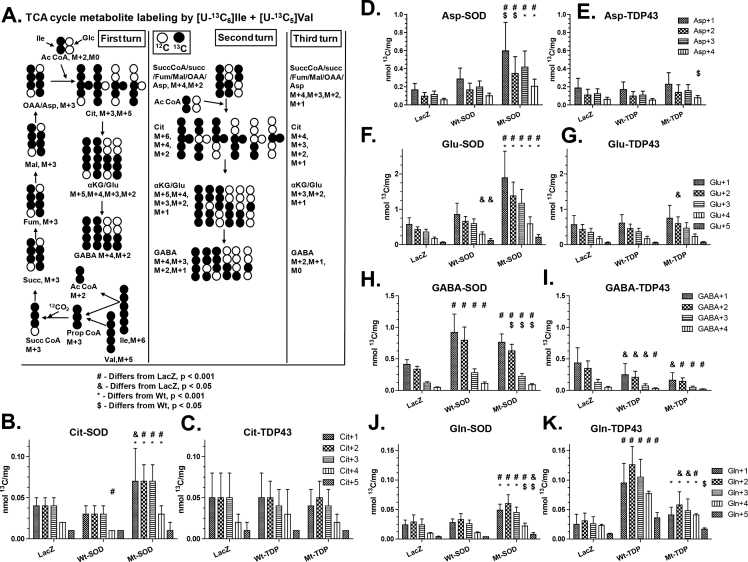
The decrease in lactate labeling, lactate production and release, and PPP flux suggest that reduced glycolysis in SOD1 and TDP43 expressing neurons could impact the operation of the TCA cycle, and therefore, we looked at the labeling of TCA cycle intermediates by [13C]glucose. Isotope enrichment in citrate, fumarate, and malate is maintained or even increased in neurons expressing SOD1 and TDP43 (Figure 3A–F). There is statistically significantly more isotope enrichment in fumarate and in malate in Mt SOD1 or TDP43 expressing samples compared with the Wt versions of these proteins. These observations imply that the entrance of 13C-labeled pyruvate into the TCA cycle is maintained or enhanced despite the Mt protein-evoked reduction in lactate labeling by [13C]glucose, lactate release to the medium, and PPP flux. These results suggest preferential entry of the labeled pyruvate into the TCA cycle compared with its conversion to lactate or use of glucose carbon in the PPP and HBP.
Figure 3.
Metabolic interrogation of TCA cycle and related pathways in neurons expressing Wt and fALS Mt genes. Pooled data from 3 to 5 experiments comparing neurons expressing LacZ with Wt SOD1/TDP43 with Mt SOD1/TDP43 is presented. Horizontal lines denote means ± SD. ANOVA showed group differences and values above the scattergram indicate that in post hoc tests the specific group is statistically significantly different from the LacZ group (p value provided) or not different (NS – not significant). Values below the scattergram indicate whether the Wt or Mt proteins group are statistically significantly different. 13C enrichment of tricarboxylic acid (TCA) cycle constituents (A–F) and TCA cycle-derived amino acids (G–L).
In addition to TCA cycle intermediates, we monitored major amino acids derived from the TCA cycle, and we found significantly increased isotope enrichment in glutamate and aspartate in Mt SOD1 and Mt TDP43 expressing neurons when compared with LacZ expressing neurons (Figure 3G–J). There was statistically significantly higher isotope enrichment in glutamate in Mt SOD1 but not in Mt TDP43 compared with the Wt versions of these proteins. There was a significant increase in isotope enrichment in GABA in Wt SOD1 neurons as well as Mt TDP43 neurons (Figure 3L.). We also measured total amino acid levels and found virtually no differences between the abundance of 19 amino acids in neurons expressing the Wt or Mt TDP43 proteins, although group differences were seen when comparing LacZ to Wt-SOD and Mt-SOD; notably the concentrations of many amino acids, including Glu and Asp levels, were significantly higher in the Mt SOD1 constructs than in the LacZ cultures (Supplemental Table 1). The enhanced labeling of the TCA cycle-derived amino acids by 13C-glucose, indicates that 13C-labeled pyruvate preferentially entered the TCA cycle (as opposed to its conversion to lactate), labeling cycle intermediates including α-ketoglutarate and oxaloacetate that were then transaminated to produce higher levels of labeled glutamate (and secondarily GABA) and aspartate, respectively.
Unexpectedly, the APEs of M + 2 aspartate were higher than those of M + 2 malate and M + 2 fumarate in each of the corresponding LacZ, Wt, and Mt pairs (Figure 3) that are precursors of oxaloacetate and aspartate. This finding appears to violate precursor–product relationships because unlabeled aspartate would dilute the APE to a value below that of the precursor. However, this has been reported previously, and Waagepetersen et al. attributed this to neuronal mitochondrial heterogeneity that results, functionally, in two fumarate/malate/oxaloacetate pools with different turnover rates and sizes [37,38]. The physical association of Mt SOD1 [39,40] and TDP43 [41] with a subpopulation of mitochondria might be a cause for functional heterogeneity. Heterogeneity of mitochondrial enzyme composition and activities and abilities to use certain substrates for nonsynaptic (i.e., somal mitochondria from neurons and glia) compared with synaptic (consisting of at least two sub-populations) mitochondria [42,43] may also contribute to these findings.
The concentrations of many amino acids in the cultures expressing SOD1 constructs were higher than in the control LacZ neuronal cultures (Supplemental Table 1). This is noteworthy because only astrocytes and/or oligodendroglia (but not neurons) express pyruvate carboxylase and thus have the capacity to synthesize oxaloacetate (OAA) from glucose- and medium-derived pyruvate. Since our cultures are nominally devoid of astrocytes and/or oligodendroglia, we hypothesize that there must be other molecules in the culture medium that provide the 4-carbon backbone for synthesis of the observed higher intracellular levels of aspartate (Asp), glutamate (Glu), and γ-aminobutyric acid (GABA). We note that isoleucine (Ile) and valine (Val) are two essential amino acids that are taken up into brain and catabolized to produce propionyl CoA; Ile also produces Ac CoA. Propionyl CoA is metabolized to succinyl CoA and enters the TCA cycle as a potential source of TCA cycle intermediates and TCA cycle-derived amino acids, as illustrated in Supplemental Figure 2A.
To test the hypothesis that amino acids are supplemental oxidative fuel and supply 4-carbon precursors for TCA cycle intermediates and cycle-derived metabolites, the neuronal cultures were incubated with [U–13C6]isoleucine plus [U–13C5]valine for 4 h. Metabolomic analysis revealed extensive labeling of citrate, Asp, Glu, and GABA (Supplemental Figure 2B–I); labeling of alanine, serine, glycine, and lactate was not detected. In addition, low concentrations of 13C-labeled glutamine (Gln) were detected in all samples (Supplemental Figure 2J–K), thus demonstrating that small numbers of non-neuronal cells that contain pyruvate carboxylase and glutamine synthetase must be present. However, Gln labeling was on the order of 10% or less than that of its precursor glutamate (compare ordinate scales in panels F–G with J–K), indicating that these cells would have a minor influence on labeling of metabolites by [1,2-13C2]Glc (compare with main text Figure 2, Figure 3). Metabolite labeling by [13C] (Val + Ile) was seen in both SOD1 and TDP43 culture sets, but only the SOD1 cultures had higher amino acid concentrations (Supplemental Table 1). Notably, the Mt SOD1 constructs had significantly higher amounts of 13C in most isotopologues of citrate, Asp, Glu, and Gln whereas the Wt SOD1 constructs had similar or higher 13C levels in isotopologues of GABA compared with the Mt SOD1 cultures (Supplemental Figure 2B, D, F, H, J). In contrast, most of the TDP43 cultures had small differences, if any, in 13C accumulation in the isotopologues, with the striking exception that Gln that had highest labeling in the Wt TDP43 cultures (Supplemental Figure 2C, E, G, I, K). The M + 1, M + 2, and M + 3 isotopologues had high concentrations compared to other isotopologues. However, M + 1 is not produced in the first turn of the TCA cycle (Supplemental Figure 2A), indicating that the labeled metabolites derived from 13C-labeled Ile and Val remained in the cycle for multiple turns or exited and then re-entered the cycle via exchange reactions. For example, M + 3 and M + 5 citrate are produced in the first turn of the cycle, but all SOD1- and TDP43-expressing cultures have similar levels of M + 1, M + 2, and M + 3 citrate that are formed in the third, second, and first turn of the cycle, respectively. For Asp, Glu, and GABA, the levels of M + 1 isotopologues are also similar to those of M + 2 in most of the SOD1- and TDP43-expressing cultures. Predominant exit of the carbon from the TCA cycle after the first turn would be expected to have the highest concentrations of 13C in the M + 3 for Asp, similar levels for M + 2, M + 3, M + 4, and M + 5 for Glu and M + 2 and M + 4 for GABA, but this is not the case (Supplemental Figure 2A–I). Also of interest, the APEs of Cit, Asp, Glu, and GABA (see legend of Supplemental Figure 2) were considerably lower than those of M + 2 isotopologues derived from metabolism of [1,2-13C2]glucose via the first turn of the TCA cycle indicating much smaller catabolic fluxes of Ile and Val compared with Glc for both the SOD1- and TDP43-expressing cells and their LacZ controls (compare to main text Figure 2, Figure 3; the APEs of precursor 13C-labeled glucose, isoleucine, and valine were similar (>99%) indicating that metabolism, not starting APE, underlies the differences in labeling by Glc and Ile + Val). Taken together, these findings clearly demonstrate that Ile and Val (and other amino acids) in the culture medium are supplemental oxidative fuel for neurons and also supply carbon for synthesis of TCA cycle intermediates and TCA cycle-derived amino acids. In general, metabolite label trapping by catabolism of Ile and Val is higher in the presence of SOD1 constructs than with TDP43.
To summarize, we report that M + 2 labeling by [1,2-13C2]glucose of citrate, fumarate, malate, Asp, and Glu increase, indicating increased flux of M + 2 pyruvate into the TCA cycle and increased labeling of these TCA cycle intermediates and of TCA cycle-derived amino acid pools via transamination exchange reactions. We also found that the total (labeled plus unlabeled) concentrations of these amino acids increased, consistent with the metabolism of unlabeled branched chain amino acids to provide the four-carbon backbone for the synthesis of Asp and Glu, as shown in the supplemental information with labeled valine plus isoleucine. Increased oxidation of non-glucose substrates in the medium (pyruvate and amino acids) via the TCA cycle would reduce demand for glucose (perhaps contributing to reduced CMRglc) and produce ATP and carbon for biosynthetic reactions. Increased oxidation of pyruvate would contribute to reduced lactate levels and labeling, but increased flux of glucose into other pathways, e.g., lipid biosynthesis, could also contribute to reduced lactate levels and labeling.
The alterations in glycolysis and TCA cycle induced by Mt SOD1 and Mt TDP43 expressing neurons (in comparison with LacZ expressing neurons) might be expected to impact cellular energy levels and redox status. We found remarkably minimal effects on the concentrations of adenine nucleotides or the levels of NAD+ and NADH between experimental groups (Supplemental Table 2). ATP and AMP levels were significantly increased in Mt SOD1 expressing neurons and ATP was also elevated in Wt TDP43 expressing neurons. We did not determine the source(s) of this ATP but it is likely that most is derived from the TCA cycle pathways that produce 15 times more ATP than glycolysis. Lactate production is reduced and is a small fraction of glucose utilization (Figure 2C–F; M−P), and glycolysis coupled with TCA cycle activity will produce more ATP via the TCA cycle. We find a statistically significant reduction in NADH and an increase in NADPH in Wt TDP43 expressing neurons (in comparison with LacZ) as well as reductions in NAD+ and NADP+ in Wt SOD1 expressing neurons. Also, some groups showed differences between SOD1 or TDP43 expression and LacZ. If these changes in NADP+ or NADPH levels were a reflection of oxidative stress, we would anticipate corresponding alteration in the ratio of reduced to oxidized glutathione (e.g., the GSH/GSSG ratio.) In fact, we see that expression of Wt or Mt SOD1 or TDP43 has essentially no effects on GSH or GSSG levels. This argues against a significant alteration in redox status at the time point of our interrogations of fALS-expressing neurons. It is possible that fuel utilization by Mt protein-expressing neurons is altered is a way that maintains neuronal redox status and energy charge.
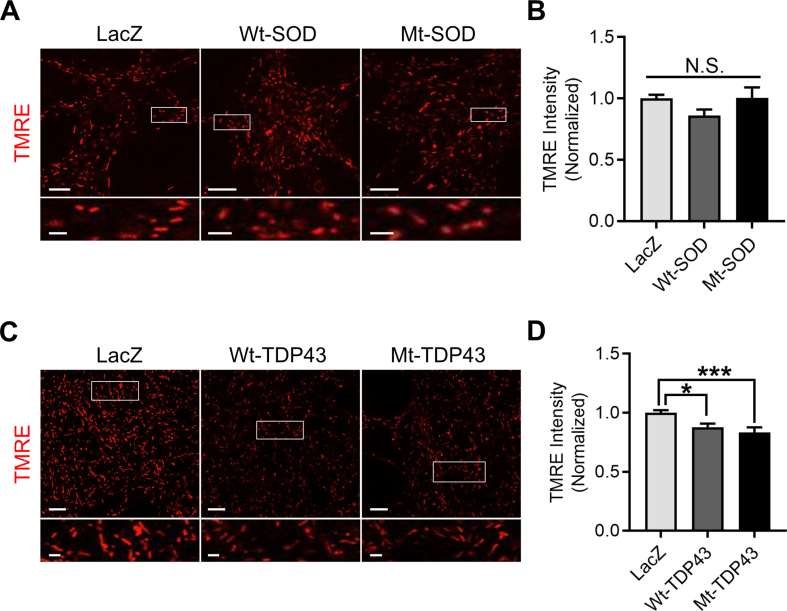
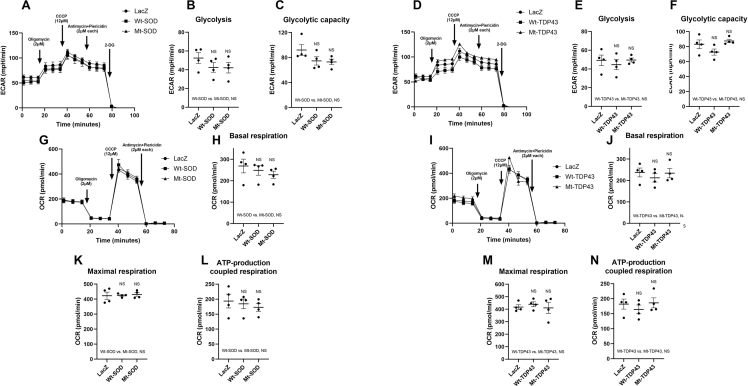
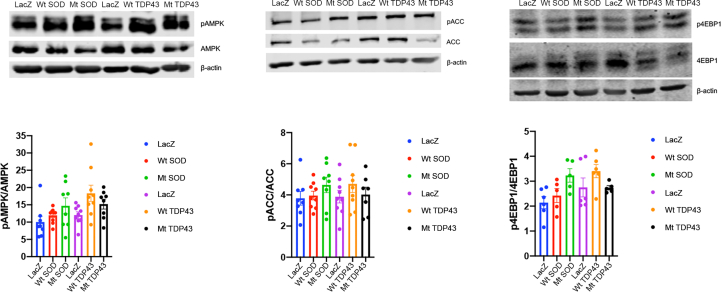
We took several additional approaches to analyze mitochondrial function. First, we analyzed mitochondrial membrane potential in live neurons expressing wild type or mutant SOD1 and TDP43 using tetramethylrhodamine ethyl ester (TMRE) imaging by confocal live cell microscopy. Both Wt and Mt SOD1 did not alter mitochondrial membrane potential as compared to control neurons, as mitochondrial TMRE intensity was not different between conditions (Supplemental Figure 3A, B). However, Wt and Mt TDP43 showed an ∼11% decrease in TMRE intensity as compared to control neurons, suggesting a slight decrease in their mitochondrial membrane potential (Supplemental Figure 3C, D). Second, we studied mitochondrial bioenergetics by monitoring oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) using the Seahorse system. Pure neuronal cultures were established in the specialized Seahorse system plates at the same density used in the metabolic and TMRE experiments, infected with recombinant viruses at DIV12, and subjected to interrogation at DIV14. Typical examples of ECAR data for SOD1 and TDP43 experiments are shown (Supplemental Figure 4A, D). We calculated steady state glycolysis rate and glycolytic capacity and found no statistically significant difference between LacZ expressing neurons in comparison with Wt SOD1 or Mt SOD1 expressing neurons or LacZ expressing neurons, in comparison with Wt TDP43 or Mt TDP43 expressing neurons (Supplemental Figure 4B, C, E, F). The apparent discrepancy between ECAR data and our direct measurements of media lactate (Figure 2D–F) is likely due to the superior sensitivity of isotopologue investigations. Typical examples of OCR data for SOD1 and TDP43 experiments are shown (Supplemental Figure 4G, I). We studied basal and maximal respiration rate and calculated ATP-production coupled respiration and found no statistically significant groups difference between LacZ expressing neurons in comparison with Wt SOD1 or Mt SOD1 expressing neurons, or LacZ expressing neurons in comparison with Wt TDP43 or Mt TDP43 expressing neurons (Supplemental Figure 4H, J–N). Third, we assayed the activation of AMP-dependent protein kinase (AMPK), a master sensor and regulator of intermediary metabolism. In comparison with LacZ expressing neurons, we found neither alterations in AMPK activation nor its downstream targets acetyl-CoA carboxylase (ACC) and factor 4E binding protein (4EBP1) in Wt or Mt SOD1/TDP43 expressing neurons (Supplemental Figure 5). Thus, while we demonstrate substantial re-wiring of intermediary metabolism in our isotopologue studies of neurons expression of fALS-causing proteins (two days post virus infection), a variety of other methods for interrogating mitochondria reveal little evidence for mitochondrial dysfunction. The reduced mitochondrial membrane potential in TDP43 expressing cells is minimal and likely functionally irrelevant. We posit that fALS gene products are leading to plasticity of fuel utilization potentially by an alteration in mitochondrial function that escape detection by certain tools (e.g. TMRE measurements of mitochondrial membrane potential, ECAR and OCR). This is sufficient, at the time point of our studies, to alter fuel utilization such that the production of ATP is adequate to obviate an AMPK-dependent adjustment of anabolism to catabolism. Our studies, at an early time point in fALS gene pathogenesis, identifies specific changes in neuronal metabolism not previously described.
In sum, expression of ALS-causing Mt proteins SOD1 and TDP43 in cortical neurons leads to reduced lactate, PPP and HBS pathway net labeling by glucose, but this is not associated with deficits in energy and redox status. The TCA cycle operates at a higher-than-normal level with glucose-derived carbon skeletons entering as pyruvate (summarized in Supplemental Figure 6) and likely oxidized medium pyruvate and amino acids in addition to valine and isoleucine (Supplemental Figure 2) resulting in maintenance of ATP levels. While in some prior studies, measures of mitochondrial physiology deviate from normal in ALS [[9], [10], [11], [12], [13]], four reporters of mitochondrial operations in our neuronal models are remarkably normal: 1) ATP/ADP ratio, 2) GSSG/GSH ratio, 3) respiration and 4) the abundance of 13C-labeled TCA cycle intermediates such as citrate, fumarate, and malate, and TCA cycle-derived amino acids (aspartate, glutamate, and GABA). Taken together, these observations suggest that subtle alterations in mitochondrial function exist in our models at the time point of our interrogations that drive re-wiring of metabolism.
2.2. Functional consequences of glycolysis on ALS causing Mt proteins
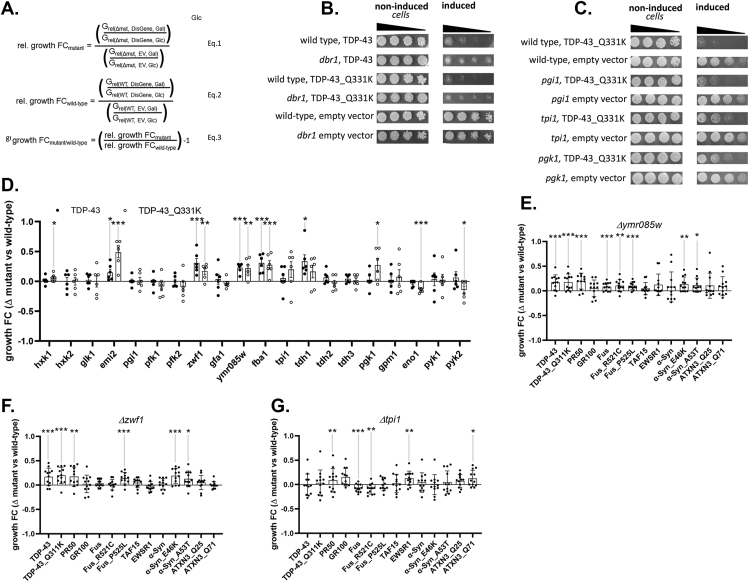
While we have described alterations in intermediary metabolism evoked by disease causing proteins, we do not know whether such rewiring that reduces glucose utilization has consequences for neuronal function and survival. We hypothesized that studying the effects of ablation of individual genes in metabolic pathways (beginning with glycolysis) would be a productive approach to address this rewiring issue (see Methods). To pursue this in an expeditious manner, we undertook a growth fitness assay in S. cerevisiae engineered to inducibly express Wt or Q331K, Mt TDP43 in a wild type background or in the presence of deletions of different glycolysis-associated genes (Supplemental Table 3.). Non-essential genes (e.g., not absolutely required for survival) were studied in a haploid yeast strain with a deletion of the corresponding single allele. Essential genes (e.g., PGI1, GFA1, FBA1, TPI1, PGK1, GPM1 and PYK1) were studied in a diploid yeast strain with a deletion of one of the two alleles. We developed a semi-quantitative assay evaluation that considered three variables affecting cellular growth: 1) the expression of the transgene (Wt TDP-43 or Mt TDP-43), 2) the presence or absence of the gene of interest in glycolysis, and 3) the use of different carbon fuels necessary for either expressing the transgene (galactose, Gal) or inhibiting transgene expression (glucose, Glc) (Figure 4A). The crosstalk between the first and second variables on cellular growth reflects the main signal and the third variable is a source of noise and its magnitude may depend on variable two. Equation (1) reflects the effect of expressing TDP-43 on cellular growth, in the absence of a specific metabolic gene-of-interest. The additional effect of using different carbon fuels (glucose/galactose) is factored in. Equation (2) reflects the effect of expressing TDP-43 on cellular growth, in the presence of the specific metabolic gene-of-interest. Also here, the additional effect of using different carbon fuels (glucose/galactose) is factored in. The ratio of equation #1 to #2 (equation #3) reflects the specific effects of ablating a specific gene-of-interest on TDP-43 impairment of growth fitness. To validate our approach, we examined the effects of a gene previously found in a yeast screen as a suppressor of TDP43 growth fitness [44] (Figure 4B.). In our assay, deletion of this gene, dbr1, promoted growth fitness, thus confirming that our semi-quantitative assay system can be deployed to find modifiers of toxicity.
Figure 4.
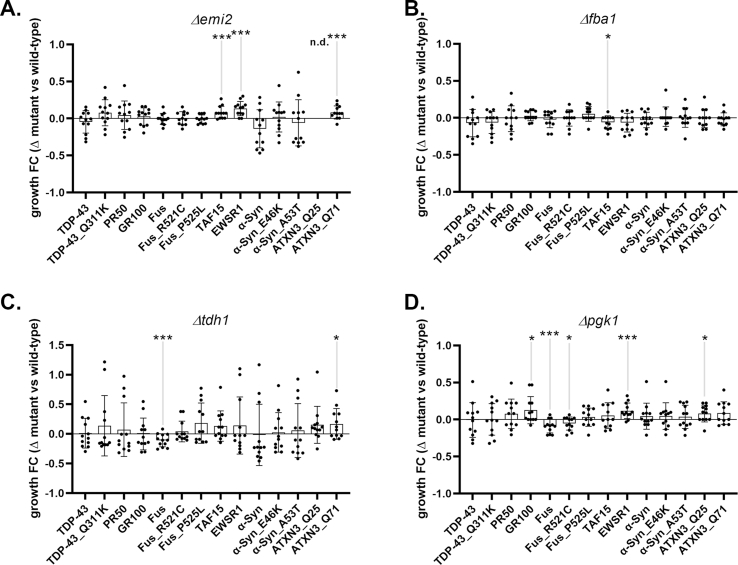
Systematic deletion of individual glycolytic enzymes reveal modifiers of protein toxicity of neurodegenerative disease-associated proteins in yeast. Panel A. Equations used in semi-quantitative growth fitness assay consider three variables: 1) the presence or absence of a specific metabolism gene (noted as Wt or Δmut), 2) the presence of the empty Expression Vector or Disease Gene elaborated by the expression vector (noted as EV or DisGene), and 3) growth in Galactose or Glucose (noted as Gal or Glc). Note, the DisGene is expressed under the control of a galactose inducible promoter, and DisGene expression is inhibited in the presence of glucose. Equation one (Eq. (1)) describes the relative growth fold changes (“rel. growth FC”) in the yeast strain mutant (e. g., lacking) a metabolism gene, expressing the disease gene or not and growing on galactose or glucose. Equation two (Eq. (2)) describes the rel. growth FC of yeast strain WT for the metabolism gene, expressing the disease gene or not and growing on galactose or glucose. Equation (3) describes the specific effect of the metabolism gene on DisGene growth fitness by dividing Eq. (1) by Eq. (2), yielding the growth FCmutant/wild-type. Panel B. As previously published [44], deleting dbr1 partly rescues Wt and Mt (Q331K) TDP43-induced protein toxicity in the corresponding yeast dilution models. Panel C. Representative examples of glycolytic enzyme deletion strains tested for effects on TDP43 toxicity. The strain deleted for pgi1 did not affect protein toxicity, whereas strains deleted for tpi1 or pgk1 reduced Mt TDP-43 toxicity. Panel D. Screening deletion strains for glycolytic enzymes and for branching enzymes into PPP and HBS pathways using the Wt (filled circles) and Mt (open circles) TDP-43 protein toxicity models; n = 6. Growth fitness benefits (group FC > 0) were seen in yeast deleted for hxk1, emi2, zwf1, ymr085w, fba1 tdh1 and pgk1. Growth fitness decrement (growth FC < 0) were seen in yeast deleted for eno1 and pyk2. Panel E. Growth fitness benefits were seen in yeast deleted for ymr085w on a variety of proteotoxic insults relevant to ALS (e.g., TDP43, C9orf72 DPRs, FUS, TAF15, EWSR1) Parkinson's Disease (e.g., α-Syn) and Spinocerebellar Ataxia (e.g., ATXN3); n = 12. Growth fitness benefits were seen in yeast expressing Wt and Mt TDP43, PR50, FUS (Wt and P525L) and Wt and Mt (E46K and A53T) α-syn. Panel F. Growth fitness effects of ablation of zwf1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing Wt and Mt TDP43, PR50, Mt FUS and Mt (E46K and A53T) α-syn. Panel G. Growth fitness effects of ablation of tpi1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing PR50, Wt and Mt FUS, EWSR1 and ATXN3 Q71 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005).
Using this assay platform we assayed 20 non-essential glycolysis associated genes, found that deletion of 5 genes enhanced growth fitness of S. cerevisiae upon induction of Mt TDP43 expression: emi2 (paralog of yeast glucokinase), zwf1 (Glc-6-P dehydrogenase), ymr085w (paralog of yeast glutamine-fructose-6-phosphate amidotransferase), fba1 (fructose 1,6 bisphosphate aldolase), and pgk1 (3-phosphoglycerate kinase) – with enzyme names of mammalian homologs or orthologs in parentheses, respectively (Figure 4C, D). In 3/5 cases, the deletion also improved the growth fitness upon induction of Wt TDP43 expression. In contrast, we found that deletion of 2 genes reduced growth fitness of S. cerevisiae upon induction of Mt (but not Wt) TDP43 expression: eno1 (enolase) and pyk2 (pyruvate kinase).
Next, we assessed whether deletion of the five genes of interest (e.g., emi2, zwf1, ymr085w, fba1 or pgk1) improve the growth fitness of additional yeast-based models of familial ALS or other neurodegenerative diseases. To address this issue, we studied yeast that inducibly-express two different toxic diamino acid peptides (DPRs) derived from the C9ORF72 mutation, Wt or R521C or P525L fused-in-sarcoma (FUS), Wt TAF15, Wt EWSR1, Wt or E46K or A53T alpha synuclein, or ATXN3 with 25 glutamines (Q25) or 71 glutamines (Q71). We found that ymr085w and zwf1 had broad growth fitness phenotypes, showing beneficial actions in 8 and 6 different models, respectively (Figure 4E, F). In contrast, tp1i which did not show a beneficial effect in the original TDP43 screen, had mixed effects (Figure 4G) while emi2, fba1, tdh1 and pgk1 had no consistent effects in these models (Supplemental Figure 7)
In sum, we find that ablation of several yeast genes involved in glycolysis confer growth fitness benefits on TDP43 based models of ALS, and at least two genes (ymr085w and zwf1) confer growth fitness benefits in yeast-based models of several different models of ALS and neurodegeneration in general. zwf1 (G6PDH) is the first step in PPP and ymr085w (GF6PA) is the first step in the HBS pathway. An implication of the removal of these enzymes is that glucose-derived carbon cannot enter the respective branch pathways, thereby altering unidentified processes (e.g., glucose flux into different catabolic routes, actions of regulatory molecules, or unanticipated effects of metabolic shifts) that have favorable outcomes.
2.3. Effects of reducing G6PDH in a C. elegans model of TDP43 proteinopathy
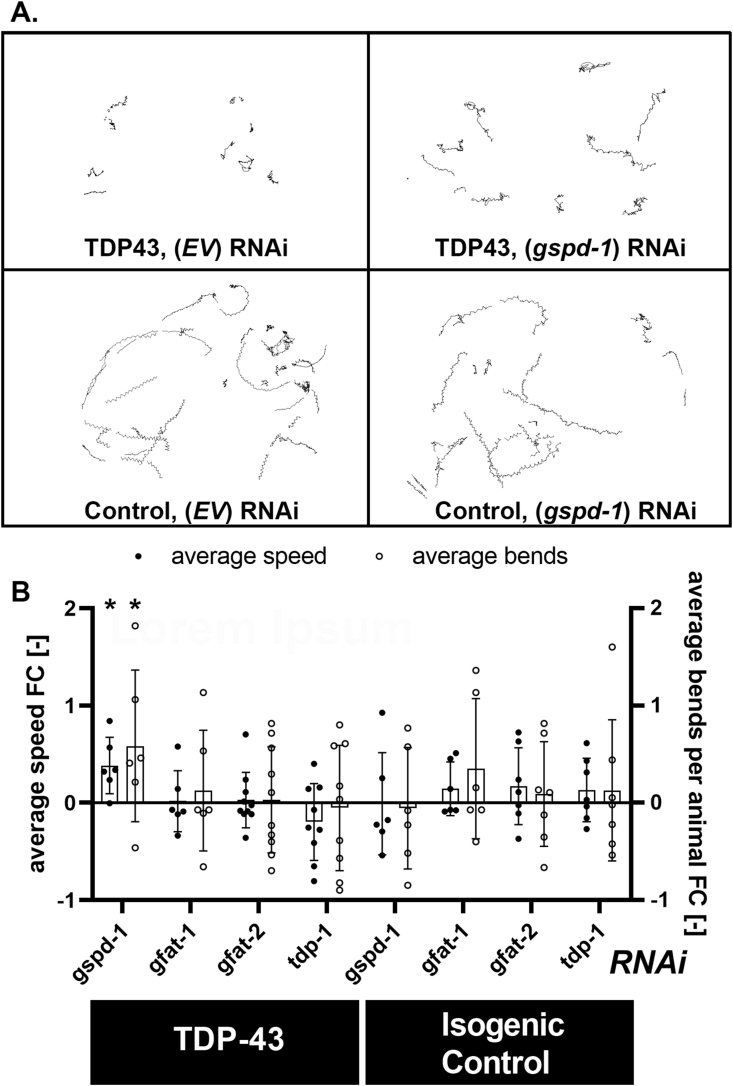
The yeast growth fitness platform is a powerful tool for finding modifiers of mutant gene phenotypes. The extent to which a pleiotropic phenotype like growth fitness reports on neuronal biology requires secondary screens. One experimental platform we next deployed was a C. elegans model of TDP43 proteinopathy. The advantages of this model are that: 1) it is an in vivo system with an intact nervous system and 2) a proven track record of bridging cellular models of biology to mammalian systems [[45], [46], [47], [48]]. We used feeding RNAi to knockdown genes of interest in two different strains (Supplemental Table 3.). CL6303 worms have nervous system-specific expression of human Wt TDP43 and are engineered to have enhanced neuronal RNAi. RK179 worms do not express human TDP43 but have the identical genetic enhancements of neuronal RNAi. After two generations of growth on RNAi bacteria (targeting gspd-1, gfat-1, gfat-2 or tdp-1 – the worm orthologs of G6PDH, GF6PA and TDP43), we studied various locomotor phenotypes. We find that RNAi targeting gspd-1 confers a small but statistically significant locomotor benefit specifically to the human TDP43 expressing animals (Figure 5). Because we are unable to quantify the extent of target knockdown using feeding RNAi in neurons specifically, it is possible the beneficial effects we see are an underestimation. Nonetheless, the beneficial effects on locomotion of reducing G6PDH on TDP43 phenotypes in an in vivo model was an encouraging confirmation that manipulations of intermediary metabolism are worthy of study.
Figure 5.
Knock down of worm G6PDH (gspd-1) suppresses the locomotor defect incurred by nervous system expression of Wt human TDP43. Panel A. Exemplary displacement paths acquired from swimming worms in M9 medium recorded with a high-resolution camera using the ImageJ wrMTrck plugin. Paths were collected from animals expressing TDP43 (CL6303) and isogenic controls (RK179); feeding RNAi targeting gspd-1 or vector controls (EV). Panel B. Quantitative analysis of average speed (pixels/minute, filled circles) and average beats per animal (1/minute, open circles) reveals that average speed and bend rates of TDP43 expressing animals are improved by knock down of gspd-1 but not knock down of gfat-1, gfat-2 or tdp-1 (in comparison with EV control). There is no effect of RNAi knockdown of any of the studied genes in isogenic control animals that do not express TDP43. Thus, the beneficial effect of reducing gspd-1 is specific to animals expressing TDP43. Effects were quantified as average animal speeds and bending rates of 9–15 L4 animals per run; n = 6. (∗∗p < 0.01).
2.4. Effects of reducing G6PDH in mammalian neuron models of ALS
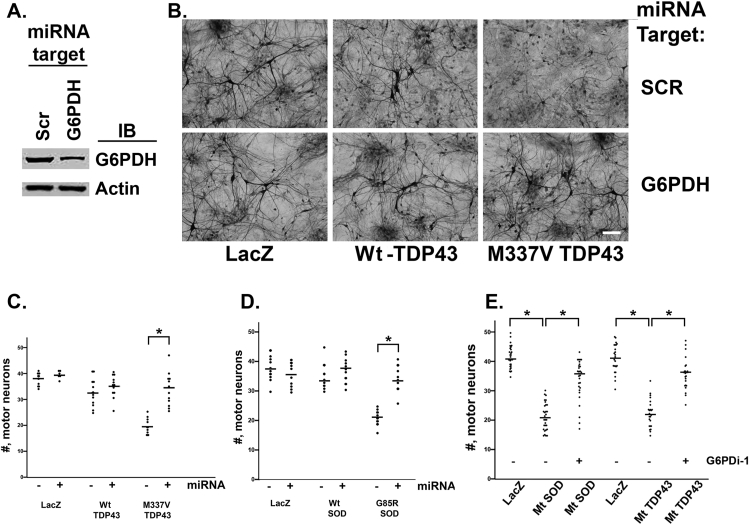
Based on the promising results described above, we assessed whether manipulation of G6PDH in a primary rat neuron model system of TDP43 proteinopathy had beneficial effects. We began by generating a series of microRNAs targeting the rat G6PDH and selected one that achieves a 65% knockdown of the protein (Figure 6A). Next, we generated embryonic rat spinal cord cultures grown on astrocytes and after 14 days in vitro (DIV) they were infected with Herpes Simplex Virus engineered to express LacZ, Wt human TDP43 or human M337V TDP43 ± the miRNA targeting g6pdh or a scrambled targeting sequence (scr) miRNA. In prior work, we showed that infection of cultures with 2 recombinant viruses achieves a >90% co-infection rate of neurons [49]. When we counted motor neurons 5 days later, we found that M337V TDP43 kills ∼50% of motor neurons and knockdown of G6PDH leads to a statistically significant blunting of this noxious effect, whereas G6PDH knockdown had no adverse effect on the survival of LacZ or Wt TDP43 expressing neurons (Figure 6B, C).
Figure 6.
Reduction of G6PDH abundance or activity protects against the motor neuron toxicity of Mt SOD1 and Mt TDP43. Panel A. Lysates from pure neuronal cultures infected with HSV engineered to express a miRNA targeting G6PDH or a scramble (scr) sequence miRNA were immunoblotted for G6PDH and actin. The miRNA targeting G6PDH reduces neuronal G6PDH expression by ∼60% of the control. Panel B. Representative photomicrographs of SMI32 immunostained DIV14 spinal cord cultures expressing LacZ, Wt-TDP43 or M337V Mt TDP43 and either the miRNA targeting G6PDH or the scramble sequence miRNA. Neurons with cell bodies larger than 25 microns are motor neurons. Fewer motor neurons are evident in the M337V mutant TDP43 + miRNA scr in comparison with the M337V Mt TDP43 + miRNA G6PDH. Scale bar = 75 microns. Panel C. There is no statistical difference in the number (denoted by #) of motor neurons expressing LacZ with or with knockdown of G6PDH. Similarly, expression of Wt TDP43 does not lead to motor neuron death with or without knockdown of G6PDH. Expression of M337V Mt TDP43 does lead to a significant loss of motor neurons and knockdown of G6PDH leads to a statistically significant blunting of the Mt TDP43 toxicity. Results from ≥4 experiments, ∗p < 0.05. Panel D. There is no statistical difference in the number of motor neurons expressing LacZ with or with knockdown of G6PDH. Similarly, expression of Wt SOD1 does not lead to motor neuron death with or without knockdown of G6PDH. Expression of G85R Mt SOD1 does lead to a significant loss of motor neurons and knockdown of G6PDH leads to a statistically significant blunting of the Mt SOD1 toxicity. Results from ≥4 experiments, ∗p < 0.05. Panel E. A three-way comparison of number of motor neurons expressing LacZ, Mt SOD1 − G6PDi-1 and Mt SOD1 + G6PDi-1. Statistically significant group differences were seen by ANOVA and post hoc test reveal that Mt SOD1 − G6PDi-1 is significant different from LacZ and from Mt SOD1 + G6PDi-1. Results from ≥4 experiments, ∗p < 0.05. A three-way comparison of number of motor neurons expressing LacZ, Mt TDP43 − G6PDi-1 and Mt TDP43 + G6PDi-1. Statistically significant group differences were seen by ANOVA and post hoc test reveal that Mt TDP43 − G6PDi-1 is significant different from LacZ and from Mt TDP43 + G6PDi-1. Results from ≥4 experiments, ∗p < 0.05.
Encouraged by these observations and noting the similarity of metabolic rewiring evoked by mutant forms of TDP43 and SOD1, we studied the effects of G6PDH reduction in cultures expressing Wt SOD1 or G85R SOD1. We found that G85R SOD1 kills ∼50% of motor neurons and knockdown of G6PDH leads to a statistically significant blunting of this noxious effect (Figure 6D). Again, G6PDH knockdown had no adverse effects on LacZ or Wt SOD1 expressing neurons.
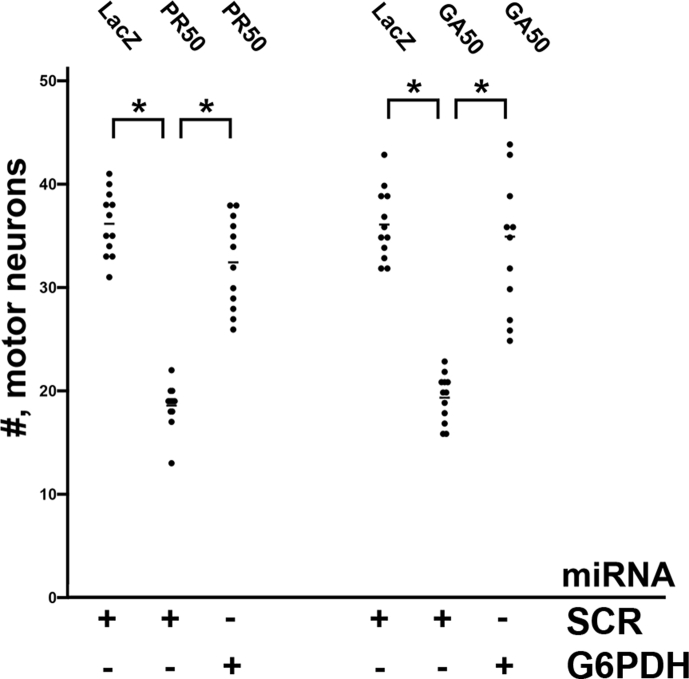
Finally we looked at two other fALS models involving the expression of GA50 or PR50, noxious DPRs that are generated from the hexanucleotide expanded mutant version of C9ORF72 [[50], [51], [52], [53]]. We find that GA50 or PR50 kills ∼50% of motor neurons and knockdown of G6PDH leads to a statistically significant blunting of this noxious effect (Supplemental Figure 8).
The primary goal of research into neurodegenerative diseases is to find targets for manipulation by small molecules, i.e., drugs. In light of our observations on the contribution of G6PDH to neuronal dysfunction in ALS models, we wondered whether this was a druggable target. Several different G6PDH inhibitors have been described but their specificity is open to question. A new, highly selective small molecule G6PDH inhibitor was reported last year, “G6PDi-1” [54], and we tested it in our system. We found that G6PDi-1 has no effects on the survival of motor neurons expressing no transgene, LacZ or Wt SOD1 or Wt TDP43. In contrast, motor neuron death induced by the expression of Mt SOD1 or Mt TDP43 is robustly suppressed by administration of G6PDi-1 (Figure 6E).
Together these results suggest that reducing the abundance or inhibiting the activity of G6PDH in neurons confers a broad beneficial effect on cellular phenotypes evoked by the expression of fALS genes.
2.5. Metabolomic effects of G6PDH knockdown in neurons expressing fALS proteins.
The conversion of Glc-6-P to 6-phosphogluconolactone by G6PDH is the first and rate-limiting enzyme in PPP. The ratio of Glc-6-P that enters glycolysis (via metabolism to fructose-6-phosphate) to PPP is ∼10:1 and we considered the possibility that reducing G6PDH levels could make this ratio even larger. If this were true, it might enhance glycolytic flux into lactate and the PPP and HBS pathways and potentially help mend the demonstrated defects we find in fALS expressing neurons (Figure 2). To examine this issue, we undertook glucose isotopologue interrogations of pure neuron cultures expressing Mt fALS genes ±65% knockdown of G6PDH that blunts motor neuron death (Supplemental Figure 9). In comparison with a scrambled sequence miRNA (control), the miRNA targeting G6PDH in the mutant constructs had no effects compared with controls on glucose uptake, total lactate, isotope enrichment in lactate, flux through PPP, isotope enrichment in R-5-P or isotope enrichment in acetylglucosamine in neurons expressing Mt SOD1 or Mt TDP43. In terms of TCA cycle intermediates, reduction of G6PDH that enhanced motor neuron survival led to increased isotope enrichment in malate only in Mt TDP43, but not SOD1, expressing neurons. In terms of amino acids, reduction of G6PDH reduced isotope enrichment in aspartate and glutamate only in the Mt SOD1 expressing neurons. There are differences in the magnitudes of glucose and lactate levels and of metabolite APEs for the control miRNA-treated neurons in Supplemental Figure 9 compared with data in Figure 2, Figure 3, probably due to miRNA treatment. However, the APEs of aspartate remained higher than fumarate and the glutamate APEs exceeded those of GABA, whereas the glutamate APEs in the miRNA-treated constructs were about twice those of citrate, contrasting miRNA-untreated neurons. Together these observations lead to two conclusions: 1) knockdown of G6PDH has no effect of the flux of glucose through glycolysis in neurons expressing fALS proteins, and 2) the beneficial effects of manipulation of G6PDH in the presence of Mt SOD1 or Mt TDP43 are metabolically distinct and thus potentially mechanistically distinct.
3. Discussion
Here, we have investigated the intermediary metabolism of neurons that express fALS genes. We find that neurons expressing two different fALS Mt proteins display a reduction of net utilization of glucose via lactate production, PPP and HBS pathways. At the same time, the fuel entrance into TCA cycle is maintained or increased. This may be due to enhanced entry of pyruvate, as well as carbon skeletons from growth medium amino acids. While mitochondrial dysfunction has been described in some ALS disease models, we find that in our in vitro model of early stage disease (48 h of fALS gene expression) neuronal mitochondrial operations are sufficiently robust to maintain ATP levels and healthy redox status. It appears that the fALS genes induce quantitative (and potentially qualitative) alterations to metabolic pathways potentially as a mechanism to maintain ATP and redox status. It is conceivable that the metabolic abnormalities we detect are responsible for the described elevated α-ketoglutarate, oxaloacetate and 2-hydroxybutyrate levels in the plasma from ALS patients [55]. The case that this is pathophysiologically significant derives from the observation that loss-of-function genetic maneuvers at specific steps of the glycolysis pathway suppress toxic phenotypes. Null or partial reduction in G6PDH has broad beneficial effect in three experimental platforms (e.g., S. cerevisiae, C. elegans and rodent spinal cord neurons). This is unlikely to be due to specific effects on glycolysis and TCA cycle metabolism, and the mechanisms by which reducing G6PDH blunts the toxicity of Mt SOD1 versus Mt TDP43 may be distinct.
Alterations in the energy economy in ALS-based experimental models have been recognized for many years. The wide variation in experimental platforms makes it difficult to generate a single, unified model of ALS-associated metabolic perturbation. Even within a single experimental platform such as human cells re-programmed to adopt neuronal fate, there are significant differences in differentiation protocols, culture conditions and age at interrogation that make establishment of a coherent assessment of data problematic (c.f., [[56], [57], [58], [59], [60], [61]]). The variety of metabolic abnormalities observed in ALS models may reflect the evolution of the disease process over time to which each platform is specifically tuned to probe.
Mitochondrial abnormalities (such as sequestration of toxic proteins and perturbed morphology and function) have been described in SOD1, TDP43, VCP, CHCHD10 and C9ORF72 models of ALS [9,[39], [40], [41],[62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73]]. This is associated with alterations in glycolysis [[74], [75], [76]], glutaminolysis [74], PPP [[75], [76], [77]], amino acid depletion [74,78] and impaired glucose utilization [76]. The above noted observations are derived from non-neuronal cells (i.e., NSC-34 cells, fibroblasts, muscle cells or HeLa cells) or tissues composed of multiple cells types (i.e., fly third instar larvae, mouse cerebral cortex or spinal cord or human autopsy brain). Because the routes by which fuel is metabolized to generate ATP varies widely between cell types, the above cited work provides no insight into intermediary metabolism of neurons. Motor neurons differentiated from patient derived induced pluripotent stem cell (iPSC) have undergone metabolic interrogations (c.f., [66]) but this platform has two, intertwined limitations – these cultures lack neuronal cell type diversity (typically, motor neurons account for ∼70% of cells in such cultures [79,80]) and they lack synaptic transmission. Since synaptic transmission is the quantitatively greatest driver of energy consumption, iPSC-based studies have exiguous utility for certain types of metabolic investigations. We provide an in depth and quantitative analysis of fuel utilization, redox status and ATP/ADP/AMP abundance in a population of interconnected vertebrate neurons. Because glucose is the prime fuel for neuronal ATP production, our work highlights the fundamental alterations in neuronal intermediary metabolism incurred by fALS genes.
In vivo, the metabolism of non-neuronal cells is coupled to that of neurons, and lactate may be a glial cell-derived fuel transferred to neurons in disease models [73,74]. The concept of the “astrocyte-neuron lactate shuttle” as proposed by Pellerin and Magistretti [81] has engendered significant debate for many years [22,82,83] but strong in vivo evidence indicates that lactate shuttle activity, if any, is minor during activation of normal brain [22]{Rothman:2022hn}. We are open to the possibility that in vivo, neuronal glucose metabolism is supplemented by molecules shared by neighboring astrocytes and oligodendrocytes, with the requirement that rigorous, quantitative studies establish fuel sharing.
We found a consistent reduction in net labeling of glycolytic products by [1,2-13C2]-d-glucose in neurons expressing Mt SOD1 or Mt TDP43, an observation which complements work by the Borges and Zarensca laboratories in multi-cell type fALS models [[76], [77], [78]]. Glycolysis is regulated at a number of steps – classically, glucose uptake from the extracellular milieu and allosteric regulation of enzymes that engage in essentially irreversible reactions (e.g., hexokinase, phosphofructokinase and pyruvate kinase). Allosterically active small molecules include ATP, AMP, cellular pH, fructose 2,6 bisphosphate (F-2,6-BP), F-1,6-BP, and alanine. In addition, glycolysis responds to mechanical extracellular cues, in particular the structure of the actomyosin cytoskeleton [84]. At present, we do not know which of these points of control account for our finding of reduced glycolytic flux in neurons expressing Mt SOD1 or Mt TDP43.
One simple hypothesis is that reduction in glycolytic flux to produce lactate and feed the PPP and HBP pathways in neurons expressing Mt SOD1 or Mt TDP43 is the primary pathophysiological driver. If so, neurons might enact metabolic re-wiring to maintain ATP production and several observations are consistent with this notion. First, substrate flux through PPP and HBS pathways are reduced and the TCA cycle operates normally or even at supra-normal levels. Our data suggest enhanced utilization of pyruvate and other carbon skeleton sources including the culture medium amino acids valine and isoleucine. Second, the expression of Mt SOD1 or Mt TDP43 in neurons has minimal effects on adenylate nucleotide levels and redox status. Several observations, however, suggest that the simple hypothesis is incomplete. First, in our yeast screen, we found that ablation of multiple, specific glycolytic enzymes conferred benefits in the growth fitness assay and this would be expected to reduce glycolytic flux. Second, knockdown of G6PDH was beneficial in multiple additional experimental platforms but does not, in fact, normalize lactate formation and the fluxes of branch pathways that diverge from glycolysis (PPP and HBP). A simple explanation is that the beneficial metabolic consequences of ablation or knockdown of specific glycolysis-associated enzymes occurs by routing substrates for utilization into pathways we are not monitoring. We suspect, however, that this is not the complete story.
Our current model does not include consideration of glycolytic enzyme spatial distribution. For example, glycolytic enzymes such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and phosphoglycerate kinase are located on transport vesicles and their local, “on-board” ATP production is required for fast axonal transport [85]. Additionally, glycolytic enzymes have the capacity to undergo phase separation to form membraneless condensates that enhance substrate flux [86]. Beyond subcellular localization, metabolic enzymes can have cell biological functions beyond catabolism. GAPDH interacts with ADP-ribosylation factor 1 GTP'ase activating protein to regulate coat protein I-mediated vesicle fission and this occurs independent of GAPDH's catalytic activity [87]. A similar phenomenon has been described for aldehyde dehydrogenase (ALDH) family member ALDH7A1 [88]. GAPDH is also sensitive to oxidative stress and S-nitrosylation that causes its translocation to the nucleus where it can trigger apoptosis [89]. Thus, along with effects on intermediary metabolism, consideration of metabolic enzyme subcellular distribution (and dynamics) and non-canonical protein–protein interactions will be required to truly understand the interface between intermediary metabolism machinery with neurodegeneration.
Oxidative stress is a cardinal feature of neurodegenerative disease in general [90,91] and ALS specifically [92] and direct targeting of reactive oxygen species (ROS) generation systems has been repeatedly shown to confer benefits in model systems [15,93]. The potent antioxidant Edaravone is one of two Food and Drug Administration approved medications for ALS [94]. Endogenous antioxidants (i.e., the glutathione system, catalase, and SOD1) depend on the production of NADPH [30]. Because G6PDH is one of the major producers of NADPH, it is perhaps counterintuitive that reducing its abundance or activity would be beneficial as we found. In addition to providing reducing power to antioxidant systems, G6PDH also has pro-oxidant activities – for example, both nitric oxide synthase (NOS) and NADPH oxidase use NADPH to generate oxidants (nitric oxide and ROS, respectively) [[95], [96], [97]]. A variety of studies suggest that reducing the function of NADPH oxidase [[98], [99], [100]] or NOS [16,101,102] confers benefits in ALS models. In addition, the dysregulation of G6PDH has been show to drive reductive stress, protein aggregation and cardiomyopathy in ∝B-Crystallin mutant mice [103]. The opposing pro- and anti-oxidant activities of G6PDH may be linked to its dynamic subcellular localization [30]. Because G6PDH deficiency can cause a number of different conditions such as hemolytic anemia or jaundice [104,105] it seems unlikely G6PDH is druggable. While the metabolic effects of G6PDH in the setting of fALS mutant proteins is clearly not straightforward, we believe that deconstructing the role of G6PDH and pathological rewired metabolism in general will reveal novel targets for therapeutic intervention.
3.1. Study limitations
In this study, we provided [1,2-13C2]-d-glucose to neurons and monitored M + 2 lactate production to interrogate glycolysis and PPP. Several factors impact total and M + 2 lactate levels. For example, our measurements do not account for lactate generated from non-glycolytic sources (such as pyruvate in the medium, pyruvate recycling or oxidation of unlabeled amino acids) which would lower lactate APE. In addition, lactate formation and release to the medium occurs via concentration gradient-driven reactions, and the enormous extracellular compared with intracellular volume will ‘pull’ lactate into the medium due to dilution, thereby increasing the apparent lactate production and glycolytic rates. In addition, M + 1 lactate production and calculated PPP rate would be underestimated when PPP intermediates are exported for nucleotide biosynthesis.
While neurons incubated with [1,2-13C2]-d-glucose for 4 h are very likely to have achieved isotopic steady state for glycolytic intermediates [106,107], longer incubation times would be required to reach isotopic steady state of TCA cycle-derived amino acids (owing to their higher concentrations and longer half lives in brain in vivo). For comparison to M + 2 lactate, we report M + 2-labeled TCA cycle intermediates and TCA cycle-derived amino acids that are derived from the first turn of the TCA cycle. Subsequent turns of the cycle would produce additional isotopologues. Potential isotopologue labeling differences among experimental groups can then arise from different TCA cycle rates and differences in metabolite retention within the cycle before label is incorporated into the amino acids. These possibilities were not examined in the present study.
Finally, our neuronal cultures are bathed in an abundance of glucose and additional potential fuels and in the nominal absence of glial cells. These conditions clearly deviate from the situation in vivo. First, the levels of fuels in the neuronal microenvironment in situ have not been defined and so it is not possible to compare to the composition of culture media we used. Second, the sharing of fuels by glial cells with neurons (i.e., glutamine, etc.) and vice-versa (e.g., neuronal release glutamate and GABA neurotransmitters that are known to be oxidized by astrocytes, and lactate [e.g., Figure 2] that can be available to astrocytes for metabolism, or release for re-uptake by neurons, and lactate release to blood) is minimized or not occurring in our cultures. [13C]Glutamine in cultures incubated with [U–13C]-labeled valine plus isoleucine demonstrates the presence of small numbers of astrocytes and/or oligodendrocytes, and their interactions with neurons remains to be evaluated. The extent to which observations made in our simplified system can be extended to in vivo models awaits further work.
4. Methods
4.1. Neuronal cultures
Pure neuronal cultures were prepared using E17 rat cortex (meninges removed), and dissociated cells were grown on tissue culture plasticware coated with poly-l-lysine (50 nM) and laminin (3 μg/mL) in NeuroBasal + B27. Fifty percent of media was refreshed 3× a week.
Spinal cord cultures were prepared as previously described [47,49,53]. In brief, astrocyte monolayers were generated from rat P1-3 rat cortex by dissociation and plated on German glass coverslips previously acid washed and coated with poly-l-lysine (50 nM) and laminin (3 μg/mL). When approximately ∼70% confluent, E15 rat spinal cord was dissociated and plated on top of the astrocytes in Minimal Essential Media with Earle's salts (NEM) + 10% Horse serum (HS) with growth factors at 2 μg/mL (CT-1, CNTF and GDNF). NEM + 10% HS is conditioned over astrocytes for one day prior to use. After 2 days, cytosine arabinoside (AraC) was added (5 μM) to arrest the proliferation of astrocytes. Fifty percent of the growth media was replaced with fresh media three times a week and leading to progressive dilution of the AraC. For biochemical studies the same general approach was used except that astrocytes and neuron were grown on poly-l-lysine/laminin coated tissue coated plastic dishes.
4.2. Metabolomic interrogations of primary neuron cultures
To study glucose utilization, metabolomic studies involved replacing standard media with an otherwise identical media in which all glucose was [1,2-13C2]-d-glucose (APE >99%) and collecting media and cells 4 h later for subsequent interrogations. We found that combining cells from two 60 mm dishes was compatible with our interrogations and a single experiment typically involved twenty-four 60 mm dishes. To study amino acid utilization, metabolomic studies involved replacing standard media with an otherwise identical media in which all valine and isoleucine was [U–13C6]isoleucine plus [U–13C5]valine (APE > 99%) and collecting media and cells 4 h later for subsequent interrogations.
4.3. Biochemistry
Sample preparation for immune blotting was as previously described [47,49,53]. Briefly, cells were lysed in RIPA buffer (NaCl 150 mM, NP-40 1%, NaDeoxycholate 0.5%, SDS 0.1%, Tris pH 7.4 25 mM), sonicated (15% duty cycle for 10 s on ice), allowed to incubate for 30 min on a rotating drum at 4 °C before centrifugation at 16,000g in a microcentrifuge for 10 min. The supernatant was recovered, protein determined, and western blot was performed using denaturing SDS-PAGE gel.
To determine the efficacy of HSV engineered to express the miRNA targeting.
G6PDH, pure neuronal cultures were infected with recombinant virus and 48 h later immunoblotted as described above.
4.4. Immunocytochemistry
Pure neuronal cultures grown on glass coverslips were fixed with 4% paraformaldehyde for 10–20 min and then washed extensively with PBS before incubating with primary antibodies overnight at room temperature (diluent was Dulbecco's Modified Eagle Medium (DMEM) + 10% fetal bovine serum (FBS) + 1% triton X-100). The following day, coverslips were washed twice with PBS, incubated with species-specific Alexa fluor secondary antibodies (Invitrogen) for 4–6 h as room temperature before washing in PBS twice, then water and mounting on slide with “Fluoro-Gel (with Tris buffer)”, Electron Microscopy Supplies. Hoechst 33342 (Thermo Scientific, diluted 1:1000 from manufacturers stock) was included with both primary and secondary antibodies. Images were captures on a Leica DM14000 inverted confocal microscope with a 63×, 1.3 NA oil objective. Lasers and photomultipliers were set to non-saturating levels and single z-stack images are presented as typical examples of immunocytochemistry.
4.5. Mitochondrial TMRE imaging by confocal microscopy
Live neurons were cultured on glass-bottomed culture dishes (MatTek; P35G-1.5-14-C) for imaging. Neurons were incubated for 45 min in 0.1 μM Tetramethylrhodamine ethyl ester (TMRE, red; Fisher Scientific, #T669) two days after transduction. After TMRE incubation and two washes in neuronal media, neurons were subjected to live cell confocal microscopy with a Zeiss LSM 980 confocal microscope with GaAsp detectors using an α Plan-Apochromat 100× 1.46 Oil DIC immersion objective (Zeiss) with Zen Blue Software (Zeiss). Live neurons were imaged in a temperature-controlled chamber (37 °C) at 5% CO2. For analysis, the maximum TMRE intensity per neuron was measured using ImageJ 1.51j8 (NIH), and the TMRE intensity for each neuron was normalized to the average TMRE intensity in neurons from the control (LacZ) plate of the corresponding experiment. TMRE intensities between conditions were analyzed using one-way ANOVA with Tukey's post hoc test. Data presented are means ± s.e.m and were analyzed from n > 35 neurons (see figure legend for details) from N = 3 independent experiments per condition. Statistics and graphing were performed using Prism 7 (GraphPad) software. All images were assembled using ImageJ 1.51j8 (NIH). All final figures were assembled in Affinity Designer.
4.6. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements
OCR and ECAR were measured in a XFe96 extracellular flux analyzer (Seahorse Bioscience). Cortical neurons were seeded in Agilent Seahorse XF96 cell culture microplate coated poly-l-lysine/laminin and incubated in a CO2 incubator at 37 °C for 12 days before infection with recombinant HSV. After two days, 1 h prior to the assay, growth medium was replaced with XF assay medium containing Seahorse XF DMEM Medium (pH 7.4) (Agilent 103575-100), 10 mM Seahorse XF Glucose (Agilent 103577-100), 1 mM Seahorse XF Pyruvate (Agilent 103578-100), and 1× GlutaMAX (Gibco 35050-061) and the place was placed in a 37 °C incubator without CO2. Basal respiration was calculated by subtracting the average OCR obtained after the third and fourth injections (2 μM Antimycin A+ 2 μM Piericidin A, and 50 mM 2-deoxy-d-glucose (2-DG), respectively) from the baseline OCR. Maximal respiration capacity was calculated by subtracting the average OCR obtained after the third and fourth injections from OCR obtained after carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (12 μM) injection. ATP-production coupled respiration was calculated by subtracting the OCR obtained after injecting 2 μM oligomycin A from the basal OCR. Basal ECAR was calculated by subtracting the average non-glycolytic ECAR obtained after 2-DG injection from the baseline ECAR. Finally, the glycolytic capacity was calculated by subtracting the average non-glycolytic ECAR from ECAR obtained after injecting 2 μM oligomycin A.
4.7. Measurement of 13C-isotopogues
Measurement was performed in the Metabolomics Core facility of the Children Hospital of Philadelphia using either: (1) Liquid Chromatography–Mass Spectrometer (LC–MS) system, an Agilent Triple Quad 6410 MS combined with an Agilent LC 1260 Infinity: (2) Gas-Chromatograph–Mass Spectrometer (GC–MS) system, a Hewlett-Packard 5971 Mass Selective Detector (MSD), coupled with a 5890 HP-GC, and (3) GC–MS, an Agilent System 6890 GC-5973 MSD.
13C enrichment in the TCA cycle intermediates and/or in amino acids were performed as previously described [[109], [110], [111]]. Briefly, cells were washed twice with PBS and then metabolites extracted with 4% perchloric acid (PCA). Cell extracts were neutralized with KOH. The neutralized extracts were subjected to either AG-1 column (100–200 mesh, 0.5 × 2.5 cm, Bio-Rad) for enriching the organic acids, or AG-50 (100–200 mesh, 0.5 × 2.5 cm, Bio-Rad) for enriching amino acids. The collected samples were then converted to t-butyldimethylsilyl derivatives for GC–MS analysis. Enrichment in 13C glutamate isotopologues was monitored using ions at m/z 432, 433, 434, 435, 436 and 437 for M0, M + 1, M + 2, M + 3, M + 4 or M + 5 (containing 1 to 5 13C atoms above M0, the natural abundance), respectively. Isotopic enrichment in 13C GABA was monitored using ions at m/z 274, 275, 276, 277, and 278 for M0, M + 1, M + 2, M + 3, or M + 4 (containing 1 to 4 13C atoms above M0, the natural abundance). Isotopic enrichment in 13C aspartate isotopologues was monitored using ions at m/z 418, 419, 420, 421 and 422 for M0, M + 1, M + 2, M + 3 and M + 4 (containing 1 to 4 13C atoms above M0, the natural abundance), respectively. Isotopic enrichment in 13C lactate was monitored using ions at m/z 261, 262, 263 and 264 for M0, M1, M2 and M3 (containing 1 to 3 13C atoms above natural abundance), respectively. Isotopic enrichment in 13C citrate isotopologues was assayed using ions at m/z 459, 460, 461, 462, 463, 464 and 465 for M0, M + 1, M + 2, M + 3, M + 4, M5 and M + 6 (containing 1 to 6 13C atoms above natural abundance), respectively. Isotopic enrichment in 13C malate isotopologues was determined using ions at m/z 419, 420, 421, 422 and 423 for M0, M + 1, M + 2, M + 3 and M + 4 (containing 1 to 4 13C atoms above natural abundance), respectively. Isotopic enrichment in 13C fumarate isotopologues was monitored using ions at m/z 287, 288, 289, 290 and 291 for M0, M + 1, M + 2, M + 3 and M + 4 (containing 1 to 4 13C atoms above natural abundance), respectively [[109], [110], [111]].
The LC–MS system was used to determine isotopic enrichment in 13C-labeled ribose-5-P, and 13C-labeled acetylglucosamine. Protein was precipitated with PCA. The level of protein in 100ul was normalized to 1 ml for calculation of the relevant metabolite as amount/mg protein. Then, sample was derivatized by adding, 50 μl 0.3 N NaOH, 50ul 0.5 M 1-phenyl-3-methyl-5-pyrazolone (PMP), and placed into heater at 70 °C for 30 min. After sample cooled, 50 μl of 0.3 N HCL was added to the vial and mixed well. Then, 0.5 ml ultra-pure H2O and 2 ml ethyl acetate were added. Samples were vortexed for 1 min, centrifuged for 1 min at 2500 RPM, the upper layer was removed and discarded, and the water phase was evaporated completely. The dry sample was reconstituted with 100 μl 0.1% formate in water (Solution A), vortexed, transferred into injection vials, and 3 μl was injected into LC–MS. Chromatographic conditions were as follows: Separation was performed on Agilent Poroshell 120 EC-C18 column. The mobile phase consists of Solution A and Solution B (0.1% formate in acetonitrile mixed with 0.005% trifluroacetate). LC flow was directed into waste for the first 1.5 min, and then, diverted into MS for next 6 min and back to waste at 7.5 min to flush the column, and stop time of 10 min. MS condition were: Capillary voltage, 4000 V, nebulizer 25 p.s.i, and dry gas temperature at 350 °C. Fragmentor and collision energy voltage were established for each compound by MassHunter Optimizer software. Metabolites were monitored using multiple-reaction monitoring (MRM) in positive mode. For ribose-5-P, at retention time (RT) 3.7 min, we monitored M0, M + 1 and M + 2 using 561-175,562-175 and 563-175 MRM, respectively. For acetylglucosamine, at RT 4.9 min, we monitored M0, M + 1 and M + 2 using 552-175,553-175 and 554-175 MRM, respectively. For glucosamine, we monitored M0, M + 1 and M + 2 using 510-175, 511-175 and 512-175 MRM, respectively. Glucosamine, if present in the sample, was below the detection limit, and, therefore, not reported in the Results Section.
4.8. Measurement of metabolite levels
The levels of media glucose and its net uptake during the incubation were determined as described [109,110,112] and the results were expressed as μmol/mg cellular protein. The levels of ATP, ADP and AMP were measured as in [109,110,112]. The levels of NADP+ and NADPH were determined according to the manufacturer's protocol using the BioVision Colorimetric Kit (Catalog#K347). The levels of NAD+ and NADH were determined according to the manufacture's protocol using the BioVision Colorimetric Kit (Catalog#K337). The concentrations of amino acids were determined with an Agilent 1260 Infinity HPLC system, utilizing pre-column derivatization with O-phthalaldehyde [109,112]. Lactate levels were determined enzymatically as given in [109,110,112]. Concentrations of representative of the TCA cycle intermediates (fumarate, citrate and succinate) in cell extracts were determined by isotope dilution approach using GC–MS system as described [113]. Briefly, an aliquot of the sample (100 μL) was spiked with a mixture of 13C-labeled organic acids of known concentrations. Then, GC–MS measurement of 13C isotopic abundance in each sample was performed (GC–MS parameters and measurements as described above), and the concentrations in the sample were calculated as described [113].
Concentrations of GSH and GSSG were determined by isotope dilution approach using N-ethylmaleimide (NEM) derivatization and LC–MS system. Method details are as in [114]. Briefly, cell pellets were spiked with a known concentration of [13C2,15N]-GSH (Sigma) and GSH-ethylester (GSH-EE, Sigma) which were used as an internal standard. Then, sample was subjected to three freeze/thaw cycles in 50 μL of 240 mM NEM in15% Methanol, vortexed vigorously, and incubated for 1 h at room temperature. Incubation was followed by addition of 50 μL of precipitating solution that contained 240 mM NEM, 10% sulfosalicylic acid, 2 mM EDTA and 15% methanol. Samples were vortexed vigorously and centrifuged for 5 min at 14000 rpm, and the supernatant was transferred to LC–MS injection vials, and samples injected to LC–MS; the conditions of LC and MS were similar to above. For the RT of GSH 3.9 min and GSSG 3.6 min, we used 436-307 MRM to monitor 13C–15N-GSH, 461-332 MRM to monitor GSH-EE, 433-304 MRM to monitor GSH and 613-355 MRM to monitor GSSG. Calculation of the concentrations was determined as in [113]. All measurements described above were validated by standard curves derived from pure metabolites. Cellular protein levels were determined with Coomassie blue (Thermo Fisher Scientific) [109,112].
4.9. Calculations
13C-Enrichment in 13C-labeled mass isotopomers is expressed by Atom Percent Excess (APE), which is the fraction (%) of analyte containing 13C atoms above natural abundance. The amount of 13C-labeled mass isotopomer was calculated by the product of (APE/100) times concentration (nmoles x mg−1 cellular protein) and is expressed as nmoles 13C metabolite per mg cellular protein. The flux of the relative pentose phosphate (RPP) was quantified by the ratio of M + 1 to M + 2 in 13C-labeled lactate and overall flux through pentose phosphate pathway (PPP) was calculated as described [115]. CMRglc[(M + 1)lactate/(M + 1 + M + 2)lactate], where M + 1 and M + 2 denote the 13C contents of lactate: (M + 1)lactate represents lactate derived from the PPP due to decarboxylation of 13C-labeled carbon one of [1,2-13C2]glucose-6-phosphate, and (M + 2)lactate represents lactate derived from the glycolytic pathway.
Overall rates of glucose utilization (CMRglc) were calculated by dividing net glucose uptake by the 4 h incubation interval. Overall rate of lactate release was calculated as net increase in medium lactate concentration divided by the 4 h experimental interval, and the rate was converted to glucose release equivalents by dividing by 2. The PPP was converted to the same units as CMRglc (i.e., nmol to μmol), and all rates are expressed as μmol glucose or its equivalent/mg/h so that they can be compared directly. For data summaries, rates were expressed either % of CMRglc or as % of the respective rate in LacZ constructs (Figure 2M–P). For the 100% values for LacZ cells, the error bars for the rate calculations are coefficients of variation cv = 100×SD/mean. For all other values, the SDs were calculated as using a method that accounts for error propagation and expressed as percentages.
4.10. Yeast strains
The rationale for selecting yeast strains for these studies was guided by three considerations. First, there are often multiple Isoforms (encoded by separate genes) of particular glycolytic enzymes that differ in intrinsic activity, localization and responsiveness to environmental conditions [85,[116], [117], [118], [119]]. Second, in addition to energy production per se, metabolic intermediates can play crucial roles in a variety of cell biological processes (i.e., senolysis [120], TOR modulation [121] and chromatin regulation [122]). Third metabolic enzymes can participate in protein–protein interactions that modify cellular processes independent of their enzymatic activity [87,110]. For these reasons, it may be difficult to predict the effect of ablation of a specific gene on both substrate flux and molecular cell biology in general. Thus, it was important to adopt an approach that was not constrained by prior understanding of the function of specific metabolism enzyme.
All yeast strains in this study are taken from the publicly available yeast deletion library (distributed by Horizon Discovery) (Supplemental Table 3). In case of deletion mutants of essential genes, we used BY4743 (MATa/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; met15Δ0/MET15; LYS2/lys2Δ0; ura3Δ0/ura3Δ0) based strains whereas we used BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) based deletion strains for non-essential genes [123]. All yeast strains were checked by colony PCR for corresponding deletions before use.
4.11. Yeast plasmids
All vectors used in this study were generous gifts from James Shorter. cDNAs of ATXN3 Q25 and ATXN3 Q71 were generous gifts of Erich E. Wancker. In the plasmids the expression of proteins of interest (TDP-43, TDP-43 Q331K, TAF15, EWSR1, Fus, Fus R521C, Fus P525L, α-Syn, α-Syn E46K α-Syn A53T, ATXN3 Q25, ATXN3 Q71) is under the control of a galactose inducible promoter. Vectors (pRS423Gal, pRS413Gal) were originally modified from redesigned pRS vectors [124].
4.12. Yeast transformation
Yeast strains were transformed using the lithium-acetate method modified from Ito et al. [125]. Briefly, 1 ml of yeast overnight cultures in YPD define was used for inoculating 28 ml of fresh YPD in a 250 ml Erlenmeyer flask. Cells were harvested for 5 min at 1,000 g after about 4 h after reaching an OD600 of 0.6–0.8. Cells were washed in 10 ml of 1× TE buffer, pH8. Cells were made competent by suspending them in 1 ml of TE based 0.1 M lithium acetate solution and incubating them for 10 min at room temperature. On wet ice, 0.5–2 μg plasmid DNA was mixed with 5 μl of carrier DNA (Clontech Order No.: 630440) and 12 μl of competent yeast cells. 60 μl of PEG-solution (0.1 M lithium acetate and 60% PEG 8,000 in 1× TE buffer) were added and mixed gently. The transformation reaction was incubated at 30 °C for 30 min before adding 8 μl of DMSO. After that, the cells were heat shocked for 7 min at 42 °C. Cells were applied to selective agar plates (SD–His) and checked for transgenic colonies after 4–7 days at 30 °C. Finally, single colonies were selected and verified by colony PCR.
4.13. Yeast disease gene toxicity modifier assay
The assay was performed as described earlier [44,126] with modifications. Briefly, yeast cells were grown for 3 d at 30 °C in liquid medium with glucose (SD –His). After that, cells were grown for 17 h at 30 °C in liquid medium containing raffinose (SRaf –His). Cultures were harvested and re-suspended in YNB without amino acids adjusted to an OD600 of 100. Cells of different samples were spotted in identical serial dilutions on plates containing either galactose (SGal–His) for disease protein expression or glucose (SD–His) as a non-expression control. Cells on plates were grown for 2 d before photos were taken for data analysis. Spots were evaluated and ratios were calculated for number of spots with growth with disease protein expression in relation to number of dilution steps with growth without disease protein expression. After that, growth ratios with and without a deletion for a metabolic gene were compared to each other (Figure 4A.). Experiments were performed done in 2–4 biological replicates with 3 technical replicates each.
4.14. C. elegans strains
CL6303 (eri-1(mg366) IV; uls60[P-unc-119::YFP + P-unc-119::sid-1]; dvIs62(snb-1::TDP-43)X, CL6186 – eri-1(mg366) IV), and CL6264 (uls60[P-unc-119::YFP + P-unc-119::sid-1]) were a generous gift of Christopher D. Link. Strain RK179 (eri-1(mg366) IV; uls60[P-unc-119::YFP + P-unc-119::sid-1]) was constructed and genotyped using CL6186 and CL6264 as parental strains.
4.15. Worm cultivation
C. elegans were cultivated at 20 °C on nematode growth medium (NGM) agar surface, unless otherwise stated, using standard techniques [127]. NGM plates were seeded with 50 μl of the E. coli strain OP50 as the food source. E. coli cells in general were grown for 1–2 d and formed a lawn before the worms were placed on plates.
4.16. Knock-down of genes of interest in worms:
For feeding RNAi experiments NGM plates were seeded with E. coli (HT115 (F-, mcrA, mcrB, IN(rrnD-rrnE)1, rnc14::Tn10(DE3 lysogen: lacUV5 promoter-T7 polymerase)) expressing empty vector (L4440) or different RNAi-encoding plasmids (Supplemental Table 4).For feeding RNAi experiments NGM plates were seeded with E. coli (HT115 (F-, mcrA, mcrB, IN(rrnD-rrnE)1, rnc14::Tn10(DE3 lysogen: lacUV5 promoter-T7 polymerase)) expressing empty vector (L4440) or different RNAi-encoding plasmids (Supplemental Table 4). We tested the worm strain CL6303 and RK179 strains, enhanced for neuronal import of dsRNA from the bacterial source and RNAi-dependent silencing of the target gene, to achieve knockdown of the genes of interest specifically in neurons. L4-staged worms were placed on either empty vector or gene of interest RNAi plates (NGM with 25 μg/mL Carbenicillin and 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG)) for 2–4 generations to ensure successful knockdown of the gene of interest. Subsequently, L4 staged and young adult worms were used in the worm fitness assay.
4.17. Worm swimming assay
Worms were grown for 2–4 generations either on HT115 carrying either L4440 empty vector or L4440 expressing the RNAi for a gene of interest. After that, 9–15 L4 stage and young adult worms were transferred onto a RNAi plate without E. coli cells and suspended in a drop M9 buffer. After that, the animals were tested immediately in a worm swimming assay as described earlier using the ImageJ plugin wrMTrck [128]. Briefly, 1-minute movies were taken using bright-field microscopy with a high-resolution CCD camera. We obtained 6–14 movies of each knock-down tested. Using the wrMTrck plugin readout we determined the average speed, fold change (FC), the average bending rate FC, average path length, and the average distance traveled by an animal per minute. FC's were calculated as values when a metabolism gene was knocked down in relation to the empty vector control.
4.18. miRNA design for the knock-down of glycolysis genes in rat primary neurons
All miR RNA's in this paper were designed using the BLOCK iT™ RNA designer (https://rnaidesigner.thermofisher.com/rnaiexpress/). Briefly, miR RNA was selected as target design option, the rat (R. norvegicus) open reading frame (ORF) was used for miR RNA design, minimum G/C content of 35% and maximum of 55% were selected. The three top ranking sequences selected by the design tool that do not overlap in their position in the ORF were selected. The tool designs a top strand as well as a bottom strand. For cloning the miR RNA constructs into the p1006 (+) empty vector using the FseI and BspEI restriction sites, we added 5′ and 3′ flanking sequences to the top strands (5′: CCTGGAGGCTTGCTGAAGGCTGTA; 3′: CAGGACACAAGGCCTGTTACTAGCACTCACATGGAACAAATGGCCT), as well as 5′ and 3′ flanking sequences to the bottom strands (5′: CCGGAGGCCATTTGTTCCATGTGAGTGCTAGTAACAGGCCTTGTGTCCTG; 3′: AGCATACAGCCTTCAGCAAGCCTCCAGGCCGG). The final top and bottom oligonucleotides were purchased from IDT and dissolved in nuclease free water at 200 μM. 4 nmol of top and bottom oligonucleotides each were mixed in annealing buffer (10 mM Tris–HCl, pH8.0, 1 mM EDTA, 100 mM NaCl) and heated to 95 °C for 4 min, followed by cooling for 10 min at room temperature. The annealed double-stranded oligonucleotide was diluted to 10 nM in annealing buffer. In parallel the HSV p1006 (+) vector was digested using FseI and BspEI (both NEB) and the linearized vector was gel purified. The FseI/BspEI linearized HSV p1006 (+) (2.5 ng/μl) and the double-stranded miR RNA construct (4 nM) were ligated using the T4 fragment as recommended by the manufacturer for 30 min at room temperature and transformed into chemically competent E. coli cells (TOP10; Thermo). Finally, single colonies were picked, purified (Qiagen), sequence verified (NU sequencing facility) and tested for knock-down efficiency of heterologous expressed Flag-tagged rat G6PDH in HEK293 cells in Lipofectamine 2000 (Thermo) transfections as recommended by the manufacturer. Constructs showing efficient target construct knockdown were packed in HSV viral vectors at the Massachusetts General Hospital Gene Delivery Technology Core on a fee for service basis.
4.19. Statistical analysis
Data were analyzed using Prism (GraphPad Software, La Jolla, CA). Significant differences between two groups were determined using paired Student's t-test (two-tailed). Significant differences within groups greater than two were determined using one-way ANOVA followed by Tukey's test for multiple comparisons. Data are presented as mean ± SD or SE where noted. For all tests, the significance threshold was set to p < 0.05.
Inclusion and diversity statement
We worked to ensure sex balance in the selection of subjects and experimental samples throughout this project. T.B.B. self-identifies as an underrepresented minority, and is funded by an NIH Diversity Research Supplement from the National Institute of Neurological Disorders and Stroke (attached to R00NS109252 (Y.C.W.). None of the other authors of this manuscript self-identify as an underrepresented ethnic minority, a member of the LGBTQ+ community, living with a disability or received support from a program designed to increase minority representation in science. We actively worked to promote gender balance in our reference list.
Data and Image processing
Data processing was kept to a minimum and conform to the guidelines provided to authors on the Molecular Metabolism website.
Studies involving human and animals
No human samples were used in this study and the use of vertebrate animals was covered by our approved Animal Care and Use Committee Protocol.
Resource availability
All yeast and worm strains used in this study are freely available to qualified investigators by simple request. In addition, plasmids used for the creation of viruses (amplicons) are similarly freely available to qualified investigators by simple request.
Data and code availability
All primary data collected in the course of these experiments is freely available to qualified investigators by simple request.
Author contributions
S.-P.R. performed all the S. cerevisiae, C. elegans studies, analyzed data and created figures; J. M.-P. created all the mammalian neuron cultures and performed toxicity studies; M.G. performed C. elegans studies; T.B.B. and Y.C.W. conducted confocal live cell microscopy and analyzed data; R.P.C. conducted the Seahorse studies; V.M. and C.D. performed biochemical studies; G.D., N.S.C. and R.G.K. analyzed and interpreted metabolomic data; J.M.-P. and R.G.K. performed all the metabolomic studies; R.G.K. supervised and integrated the project. All authors contributed to the writing and editing of the manuscript.
Key resources table.
| Reagent or Resource | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-MAP2 | Abcam | Cat.No. ab5392 RRID AB_2138153 |
| Mouse monoclonal anti-Piccolo | Synaptic Systems | Cat.No. 142111, clone 68G2, RRID AB_2619829 |
| Rabbit polyclonal anti-Glia1 | Synaptic Systems | Cat.No. 182003 RRID AB_2113441 |
| Guinea pig polyclonal anti-Glia2 | Synaptic Systems | Cat.No. 182105 RRID AB_2619875 |
| Mouse monoclonal anti-GluN1 | Synaptic Systems | Cat.No. 114011 RRID AB_887750 |
| Mouse monoclonal anti-PSD95 | NeuroMab (UCDavis) | Clone K28/43, RRID:AB_2292909 |
| Rabbit polyclonal anti-S100 beta | LSBio | Cat.No. LS-C268728 RRID:AB_11143806 |
| Mouse monoclonal anti-GFAP | Synaptic Systems | Cat.No.173211 RRID:AB_2232308 |
| Mouse monoclonal anti-neurofilament H, non-phosphorylated clone SMI 32 | BioLegend | Cat.No. 801701 RRID AB_2314912 |
| Mouse monoclonal anti-glial fibrillary protein | Synaptic Systems | Cat.No. 173211, clone 186C6, RRID AB_2721090 |
| Polyclonal rabbit anti-glucose-6-phosphate dehydrogenase | Abcam | Cat. No. ab993 RRID AB_296714 |
| Rabbit monoclonal anti-AMPK | Cell Signaling | Cat.No.2532S RRID AB_10624867 |
| Rabbit monoclonal anti-phosphoAMPK | Cell Signaling | Cat.No.2535S RRID AB_331250 |
| Rabbit monoclonal anti-ACC | Cell Signaling | Cat.No.3676SAB RRID AB_2219397 |
| Rabbit monoclonal anti-phosphoACC | Cell Signaling | Cat.No.3661S RRID AB_330337 |
| Rabbit monoclonal anti-4EBP1 | Cell Signaling | Cat.No.9452 RRID AB_331692 |
| Rabbit monoclonal anti-phospho4EBP1 | Cell Signaling | Cat.No.2855S RRID AB_560835 |
| Mouse monoclonal anti-HA (clone 16B12) | BioLegend | Cat.No. 901514 RRID:AB_2563417 |
| Rabbit polyclonal anti-FLAG | Sigma Aldrich | Cat.No. 60-031 RRID:AB_10718939 |
| Polyclonal goat anti-guinea Pig IgG (H + L), Alexa Fluor 488 | Life Technology | Cat.No. A-11073 RRID:AB_2534117 |
| Polyclonal goat anti-mouse IgG (H + L), Alexa Fluor 594 | Life Technology | Cat.No. A11032 RRID:AB_2534091 |
| Polyclonal goat anti-rabbit IgG (H + L), Alexa Fluor 488 | Life Technology | Cat.No.A11034 RRID:AB_2576217 |
| Polyclonal goat anti-chicken IgG (H + L), Alexa Fluor 488 | Life Technology | Cat.No.A11039 RRID AB_2534069 |
| Bacterial and virus strains | ||
| HSV-WT-superoxide dismutase (SOD1) | Lim et al., 2012 | |
| HSV-G86R SOD1 | Lim et al., 2012 | |
| HSV-WT-TAR DNA binding protein 43 (TDP43) | Jablonski et al., 2015 | |
| HSV-M337V TDP43 | Jablonski et al., 2015 | |
| HSV-miRNA to G6PDH | This study | |
| HSV-miRNA, scrambled (control) sequence | This study | |
| E. coli (HT115 (F-, mcrA, mcrB, IN(rrnD-rrnE)1, rnc14::Tn10(DE3 lysogen: lacUV5 promoter -T7 polymerase)) expressing empty vector (L4440) or different RNAi-encoding plasmids (C. elegans RNAi Collection (Ahringer)) | Source BioScience | SCR_017064 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| [1,2-13C2]-d-glucose | Sigma–Aldrich | Cat.No. 453188 |
| [U–13C6]isoleucine | Sigma–Aldrich | Cat.No. 604798 |
| [U–13C5]valine | Sigma–Aldrich | Cat.No. 758159 |
| [13C2,15N]-GSH | Sigma–Aldrich | Cat.No. 683620 |
| Tetramethylrhodamine ethyl ester (TMRE) | Fisher Scientific | Cat. No. T669 |
| Antimycin A | Sigma | Cat. No. A8674 |
| Piericidin A | Cayman | Cat. No. 15379 |
| 2-deoxy-d-glucose (2-DG) | Cayman 14325 | Cat. No. 14325 |
| carbonyl cyanide m-chlorophenyl hydrazone (CCCP) | Sigma | Cat. No. C2759 |
| Oligomycin A | Sigma | Cat. No. 75351 |
| Critical commercial assays | ||
| NAD+/NADH | BioVision | #K337 |
| NADP+/NADPH | BioVision | #K347 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Time-pregnant female rats (Sprague–Dawley) – Charles River | ||
| C. elegans strain: (eri-1(mg366) IV; uls60[P-unc-119::YFP + P-unc-119::sid-1]; dvIs62(snb-1::TDP-43)X) | Christopher LInk | CL6306 |
| C. elegans strain: eri-1(mg366) IV) | Christopher LInk | CL6186 |
| C. elegans strain: (uls60[P-unc-119::YFP + P-unc-119::sid-1]) | Christopher LInk | CL6264 |
| C. elegans strain: (eri-1(mg366) IV; uls60[P-unc-119::YFP + P-unc-119::sid-1]) | This study | RK179 |
| BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) | Horizon Discovery | YSC1048 |
| BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xxxΔ0) | Horizon Discovery | YSC1053 |
| BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0) | Horizon Discovery | YSC1050 |
| BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 xxx/xxxΔ0) | Horizon Discovery | YSC1057 |
| Oligonucleotides | ||
| G6pd_3_top: CCTGGAGGCTTGCTGAAGGCTGTATGCTGAGGAGTTACGGGCAAAGAACTGTTTTGGCCACTGACTGACAGTTCTTTCCGTAACTCCTCAGGACACAAGGCCTGTTACTAGCACTCACATGGAACAAATGGCCT | IDT | This study |
| G6pd_3_bottom: CCGGAGGCCATTTGTTCCATGTGAGTGCTAGTAACAGGCCTTGTGTCCTGAGGAGTTACGGAAAGAACTGTCAGTCAGTGGCCAAAACAGTTCTTTGCCCGTAACTCCTCAGCATACAGCCTTCAGCAAGCCTCCAGGCCGG | IDT | This study |
| Recombinant DNA | ||
| Rat G6pd (NM_017006.2) in pcDNA3.1+/C-(K)-DYK | Genscript | This study |
| Software and algorithms | ||
| ImageJ plugin wrMTrck | J Vis Exp 52321 | http://www.phage.dk/plugins/wrmtrck.html |
| Other | ||
Acknowledgments:
This work was supported by the National Public Health Service (NS05225 and NS087077, R.G.K.; R00NS109252, Y.C.W.; CA197532 and AG049665 to N.S.C.), the Heather Koster Family Charitable Fund and the Les Turner ALS Foundation. R.P.C. was supported by a Northwestern University Pulmonary and Critical Care Department Cugell predoctoral fellowship. The 13C-enrichments (APEs), PPP rates, and metabolite profiles were determined by the Director and members of the Metabolomics Core Facility at the Children's Hospital of Philadelphia, and we thank Ilana Nissim, Yevgeny Daikhin and Oksana Horyn for their expert contributions to these studies; Christopher Link for sharing several strains of C. elegans, Joshua Rabinowitz for the gift of G6PDi-1, Richard Kibbey for expert advice and members of the Kalb lab for fruitful discussions during the performance of these studies.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101468.
Conflict of interest
None.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Total intracellular amino acid levels from neuronal cultures engineered to express LacZ, Wt or Mt versions of SOD1 or TDP43. Mean, standard error and ANOVA with post hoc analysis are presented.
. Adenylate nucleotide levels and redox status in neurons expressing Wt or familial Mt ALS genes. Mean, standard deviation and Student's t-test values from analysis of ATP, ADP, AMP, NAD+, NADH, NADP+, NADPH, GSSG and GSH are presented. Comparisons are from neurons expressing LacZ versus Wt-SOD1 versus Mt-SOD1 or LacZ versus Wt-TDP43 versus Mt-TDP43.
Yeast deletion strains used in these studies.
Feeding RNAi bacteria studied in C. elegans model of fALS.
Supplemental Figure 1.
Immunocytochemically-defined pre- and post-synaptic elements indicate that there is synaptic connectivity in cortical neuron cultures. Panel A. DIV 14 cortical neuron cultures underwent immunocytochemical staining for Hoechst staining of nuclei, the Gria1 subunit of AMPA receptors (green) and the presynaptic protein, Piccolo (red). Merge image of a single confocal slice reveals pre- and post-synaptic apposition of puncta reflecting the existence of AMPA'ergic synapses. Calibration bar, 30 microns. Panel B. DIV 14 cortical neuron cultures underwent immunocytochemical staining for Hoechst staining of nuclei, the Gria2 subunit of AMPA receptors (green) and the presynaptic protein, Piccolo (red). Merge image of a single confocal slice reveals pre- and post-synaptic apposition of puncta reflecting the existence of AMPA'ergic synapses. Calibration bar, 30 microns. Panel C. DIV 14 cortical neuron cultures underwent immunocytochemical staining for Hoechst staining of nuclei, Gria1 subunit of AMPA receptors (green) and the postsynaptic density protein, PSD95 (red). Merge image of a single confocal slice reveals Gria1 puncta adjacent to PSD95 reflecting the existence of AMPA'ergic synapses. Calibration bar, 30 microns. Panel D. DIV 14 cortical neuron cultures underwent immunocytochemical staining for Hoechst staining of nuclei, the Gria1 subunit of AMPA receptors (green) and GluN1 subunit of NMDA receptors (red). Merge image of a single confocal slice reveals Gria1 puncta co-localizing with GluN1 puncta reflecting the existence of synapses containing both AMPA and NMDA receptors. Calibration bar, 30 microns.
Supplemental Figure 2.
Labeling of TCA cycle-derived metabolites by [U–13C6]isoleucine (Ile) plus [U–13C5]valine. Cultured neurons were incubated for 4 h with labeled Ile plus Val exactly as described for [1,2-13C2]glucose (Glc) except that the unlabeled Ile and Val were replaced with labeled Ile plus Val at the same concentrations as in the culture medium (See Methods). Panel A. Catabolism of Val and Ile produces propionyl (Prop) CoA, and Ile also produces acetyl (Ac) CoA. Prop CoA enters the TCA cycle at the succinyl (Succ) CoA step and is successively converted to succinate (Succ), fumarate (Fum), malate (Mal), and oxaloacetate (OAA) that can be transaminated to aspartate (Asp). OAA can combine with 13C-labeled Ac CoA derived from Ile or unlabeled Ac CoA derived predominantly from unlabeled Glc (and other unlabeled compounds in the medium, including pyruvate and amino acids) to generate citrate (Cit) and α-ketoglutarate that can transaminated to glutamate (Glu). Glu can be decarboxylated to produce γ-aminobutyric acid (GABA) or converted to glutamine (Gln). Labeling patterns arising from catabolism of labeled Val plus Ile are illustrated in panel A (filled symbols represent 13C) to produce isotopologues M + i, where i denotes the additional mass due to 13C atoms. For simplicity, the numbers in legends reflect M + i above unlabeled (M0) compounds. Symmetry at the Succ/Fum steps causes randomization of the label. The second and third turns of the cycle produce larger numbers of isotopologues than the first turn. Due to space constraints, the labeling patterns are not illustrated for the third turn and only M + i are indicated for each compound. Note that additional isotopologues (not shown) can arise from unlabeled endogenous compounds (e.g., Glc, Asp, Glu, GABA, etc.) entering the TCA cycle. The values for atom percent excess (APE) for each isotopologue for each compound among the three experimental groups were not statistically significantly different (not shown). The mean total APEs for each isotopologue (i.e., the sum of all isotopologue APEs for each compound) ranged from about 4.7 to 7.5% for citrate, 0.9–3% for Asp, 0.3–3.8% for Glu, 0.3–3.7% for GABA, and 0.4–2.8% for Gln. (The APEs for some isotopologues are quite low (<0.4%) and close to the level of detection, i.e., M + 5 and M + 6 for citrate, M + 5 for Glu, and M + 4 for GABA). Note that most of these total APE values are much lower than the M + 1 and M + 2 APEs for compounds derived from [1,2-13C2]Glc (compare to main text Figure 2, Figure 3), indicating lower catabolic fluxes of Ile plus Val into the TCA cycle-derived compounds compared with glucose. Because the total concentrations of many amino acids were significantly increased in the SOD1 constructs (See Supplemental Table 1), the amount of 13C for each isotopologue was calculated as follows: (APE/100)∗concentration = nmol 13C/mg protein and used for between-group comparisons. Mean 13C concentrations and SDs (vertical bars) for 8–12 samples from 3 independent cultures are plotted in panels B–K for SOD1 and TDP43 constructs that express LacZ, Wt and Mt proteins; isotopologues not shown had negligible labeling (e.g., Citrate M + 6), and those with the low APEs noted above have minor relevance. Significant differences among these three groups for each isotopologue of each compound were identified by ANOVA and Tukey's post hoc test.
Supplemental Figure 3.
Analysis of mitochondrial membrane potential in neurons expressing fALS genes. Panel A. Representative confocal microscopy images (top) and corresponding insets (bottom) of TMRE intensity as a measure of mitochondrial membrane potential in live neurons expressing control (LacZ), wild type SOD1 (Wt-SOD) or mutant SOD1 (Mt-SOD). Scale bar: 5 μm (top); 1 μm (bottom insets). Panel B. Graph of TMRE intensity (normalized to control LacZ condition) in live neurons expressing control (LacZ), wild type SOD1 (Wt-SOD) or mutant SOD1 (Mt-SOD). n = 71 neurons (control), 40 neurons (wild type SOD1), 41 neurons (mutant SOD1) from N = 3 independent experiments/condition. Data are means ± s.e.m. ANOVA with Tukey's post hoc test. N.S. (Not Significant); P = 0.1393 (control vs wild type); P = 0.997 (control vs mutant); P = 0.1853 (wild type vs mutant). Panel C. Representative confocal microscopy images (top) and corresponding insets (bottom) of TMRE intensity as a measure of mitochondrial membrane potential in live neurons expressing control (LacZ), wild type TDP43 (Wt-TDP43) or mutant TDP43 (Mt-TDP43). Scale bar: 5 μm (top); 1 μm (bottom insets). Panel D. Graph of TMRE intensity (normalized to control LacZ condition) in live neurons expressing control (LacZ), wild type TDP43 (Wt-TDP43) or mutant TDP43 (Mt-TDP43). n = 44 neurons (control), 40 neurons (wild type TDP43), 37 neurons (mutant TDP43) from N = 3 independent experiments/condition. Data are means ± s.e.m. ANOVA with Tukey's post hoc test. ∗P = 0.0197 (control vs wild type); ∗∗∗P = 0.001 (control vs mutant); P = 0.5847 (wild type vs mutant).
Supplemental Figure 4.
Expression of Wt-SOD, Mt-SOD, Wt-TDP43, or Mt-TDP43 in neurons does not significantly alter mitochondrial respiration and extracellular acidification rates. Panel A. ECAR profile of LacZ, Wt-SOD, and Mt-SOD expressing neurons monitored under basal condition and upon the sequential injections of oligomycin A, CCCP, Antimycin A/Piericidin A, and 2-DG (injections are indicated by arrows). Panel B. Basal ECARs of LacZ, Wt-SOD, and Mt-SOD expressing neurons. Panel C. ECARs of LacZ, Wt-SOD, and Mt-SOD expressing neurons upon the injection of oligomycin A. Panel D. ECAR profile of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons monitored under basal condition and upon the sequential injections of oligomycin A, CCCP, Antimycin A/Piericidin A, and 2-DG (injections are indicated by arrows). Panel E. Basal ECAR of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons. Panel F. ECARs of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons upon the injection of oligomycin A. Panel G. OCR profile of LacZ, Wt-SOD, and Mt-SOD expressing neurons monitored under basal condition and upon the sequential injections of oligomycin A, CCCP, Antimycin A/Piericidin A, and 2-DG (injections are indicated by arrows; injection of 2-DG not shown). Panel H. Basal OCRs of LacZ, Wt-SOD, and Mt-SOD expressing neurons. Panel I. OCR profile of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons monitored under basal condition and upon the sequential injections of oligomycin A, CCCP, and Antimycin A/Piericidin A, and 2-DG (injections are indicated by arrows; injection of 2-DG not shown). Panel J. Basal OCRs of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons. Panel K. Maximal OCRs of LacZ, Wt-SOD, and Mt-SOD expressing neurons upon the injection of CCCP. Panel L. ATP-production coupled OCRs of LacZ, Wt-SOD, and Mt-SOD expressing neurons. Panel M. Maximal OCRs of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons upon the injection of CCCP. Panel N. ATP-production coupled OCRs of LacZ, Wt-TDP43, and Mt-TDP43 expressing neurons. Panels A, D, G, and I are representatives of 4 biologically independent experiments with an n = 6 technical replicates. For B, C, E, F, H, and J-N, n = 4 biologically independent experiments. Data are mean ± s.e.m. (A-N). One-way ANOVA with Dunnett's T3 multiple comparisons tests (α = 0.05) were used for determining statistical significance.
Supplemental Figure 5.
AMPK is not activated in neurons expressing fALS genes. DIV14 pure neuronal cultures were infected with HSV engineered to express LacZ, Wt or Mt form of SOD1 or TDP43 and two days later Western blots were performed on lysates. AMPK activation is monitored by blotting for phosphorylation of threonine 172 (e.g., pAMPK) and the state of phosphorylation of downstream targets ACC (phosphorylated serine 79, pACC) and 4EBP1 (phosphorylated threonine 37/46, p4EBP1). Representative blots are displayed and quantification of pAMPK/AMPK, pACC/ACC and p4EBP1/4EBP1 presented below. There was no statistically significant difference between the ratio of phosphorylated to non-phosphorylated species for any of these measures of AMPK activation by neurons expressing Wt or Mt fALS proteins versus LacZ. These results were derived from 6 to 9 independent pure neuron cultures.
Supplemental Figure 6.
Experimental ALS causes upregulation of flux of [1,2-13C2]glucose into oxidative pathways and downregulation of glycolysis and its branch pathways. Mean values for the atom percent excess (APE, i.e., the fraction of the analyte containing 13C atoms above natural abundance; see Methods) for each metabolite were normalized to the respective values for the LacZ constructs and plotted for each experimental group. The individual and mean values for each experimental group and statistical differences among groups are presented in Figure 2, Figure 3 of the main text. The blue shaded area represents ± mean 95% confidence interval (CI) that was calculated from the 95% CIs for all compounds in the LacZ constructs. Metabolites produced by (i.e., lactate) or derived from branch pathways (i.e., ribose-5-phosphate (P) and acetylglucosamine) of the glycolytic pathway (pathways are illustrated in the right panel) have reduced enrichment from [1,2-13C2]glucose in the Wt and Mt Cu++/Zn++ superoxide dismutase (SOD1) (top left panel) and TAR DNA binding protein of 43 kDa molecular weight (TDP43) (bottom left panel) constructs compared with LacZ constructs. In contrast, metabolites that are components of the tricarboxylic acid (TCA) cycle (fumarate (Fum), malate (Mal), and citrate (Cit)) and amino acids derived from the TCA cycle (aspartate, glutamate, and γ-aminobutyric acid (GABA)) have increased enrichment compared with the LacZ construct. Other abbreviations: M + 1 or M + 2, mass of the compound containing 1 or 2 13C atoms derived from [1,2-13C2]glucose, respectively; HK, hexokinase; Glc-6-P, glucose-6-P; PPP, pentose phosphate pathway; Fru-6-P, fructose-6-P; PFK, phosphofructo-1-kinase; Fru-1,6-P2, fructose-1,6-bisphosphate; GAP, glyceraldehyde-3-P; DHAP, dihydroxyacetone-P; 1,3-PG, 1,3-diphosphoglycerate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; PK, pyruvate kinase; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; OAA, oxaloacetate; ∝- KG, ∝-ketoglutarate.
Supplemental Figure 7.
Deletion of individual glycolytic enzymes reveal modifiers of protein toxicity of neurodegenerative disease-associated proteins in yeast Panel A. Growth fitness effects of ablation of emi2 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing TAF15, EWSR1 and ATXN3 Q71. Panel B. Growth fitness effects of ablation of fba1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing TAF15. Panel C. Growth fitness effects of ablation of tdh1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing ATXN3 Q71. Growth fitness detriment was seen in yeast expressing FUS. Panel D. Growth fitness effects of ablation of pgk1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing GR100, EWSR1, ATXN3 Q25. Growth fitness detriment was seen in yeast expressing FUS and FUS R521C (∗p < 0.05, ∗∗∗p < 0.005).
Supplemental Figure 8.
Reduction of G6PDH abundance protects against the motor neuron toxicity of PR50 and GA50. Panel A. A three-way comparison of number of motor neurons expressing LacZ + scr, PR50 + scr miRNA and PR50 + G6PDH miRNA. Statistically significant group differences were seen by ANOVA and post hoc test reveal that PR50 + scr miRNA is significant different from LacZ + scr and from PR50 + G6PDH miRNA. Results from ≥4 experiments, ∗, p < 0.05. A three-way comparison of number of motor neurons expressing LacZ + scr, GA50 + scr miRNA and GA50 + G6PDH miRNA. Statistically significant group differences were seen by ANOVA and post hoc test reveal that GA50 + scr miRNA is significant different from LacZ + scr and from GA50 + G6PDH miRNA. Results from ≥4 experiments, ∗, p < 0.05.
Supplemental Figure 9.
Metabolic interrogation of glycolysis, the TCA cycle and related pathways in neurons expressing fALS genes with or without knockdown of G6PDH. Scattergrams of data from neurons expressing Mt SOD1 or Mt TDP43 with miRNA mediated knockdown of G6PDH or expression of a control miRNA (”Cont.”). P values from Student's t-test are presented in every panel. The only statistically significant differences were: 1) higher levels of M + 2-malate in Mt TDP43 expressing neurons with G6PDH knockdown versus control, 2) lower levels of M + 2-asparate in Mt SOD1 expressing neurons with G6PDH knockdown versus controls and 3) lower levels of M + 2-glutamate in Mt SOD1 expressing neurons with G6PDH knockdown versus controls (compare to Figure 2, Figure 3 in main text).
References
- 1.Archer S.L. Mitochondrial dynamics–mitochondrial fission and fusion in human diseases. The New England Journal of Medicine. 2013;369(23):2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 2.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 3.Casajus Pelegay E., Puzzo F., Yilmazer A., Cagin U. Targeting mitochondrial defects to increase longevity in animal models of neurodegenerative diseases. Advances in Experimental Medicine & Biology. 2019;1134(Chapter 5):89–110. doi: 10.1007/978-3-030-12668-1_5. [DOI] [PubMed] [Google Scholar]
- 4.Area-Gomez E., Guardia-Laguarta C., Schon E.A., Przedborski S. Mitochondria, OxPhos, and neurodegeneration: cells are not just running out of gas. The Journal of Clinical Investigation. 2019;129(1):34–45. doi: 10.1172/JCI120848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J., Wang L., Yan T., Perry G., Wang X. TDP-43 proteinopathy and mitochondrial abnormalities in neurodegeneration. Molecular and Cellular Neurosciences. 2019;100:103396. doi: 10.1016/j.mcn.2019.103396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y., Chen M., Jiang J. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion. 2019;49:35–45. doi: 10.1016/j.mito.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Rangaraju V., Lewis T.L., Hirabayashi Y., Bergami M., Motori E., Cartoni R., et al. Pleiotropic mitochondria: the influence of mitochondria on neuronal development and disease. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2019;39(42):8200–8208. doi: 10.1523/JNEUROSCI.1157-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan K., Anichtchik O., Luo S. Mitochondrial integrity in neurodegeneration. CNS Neuroscience and Therapeutics. 2019;25(7):825–836. doi: 10.1111/cns.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattiazzi M., D'Aurelio M., Gajewski C.D., Martushova K., Kiaei M., Beal M.F., et al. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. The Journal of Biological Chemistry. 2002;277(33):29626–29633. doi: 10.1074/jbc.M203065200. [DOI] [PubMed] [Google Scholar]
- 10.Dupuis L., Oudart H., René F., Gonzalez de Aguilar J.-L., Loeffler J.-P. Evidence for defective energy homeostasis in amyotrophic lateral sclerosis: benefit of a high-energy diet in a transgenic mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(30):11159–11164. doi: 10.1073/pnas.0402026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magrané J., Sahawneh M.A., Przedborski S., Estévez Á.G., Manfredi G. Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2012;32(1):229–242. doi: 10.1523/JNEUROSCI.1233-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung C., Higgins C.M.J., Xu Z. Mitochondrial electron transport chain complex dysfunction in a transgenic mouse model for amyotrophic lateral sclerosis. Journal of Neurochemistry. 2002;83(3):535–545. doi: 10.1046/j.1471-4159.2002.01112.x. [DOI] [PubMed] [Google Scholar]
- 13.Bowling A.C., Schulz J.B., Brown R.H., Beal M.F. Superoxide dismutase activity, oxidative damage, and mitochondrial energy metabolism in familial and sporadic amyotrophic lateral sclerosis. Journal of Neurochemistry. 1993;61(6):2322–2325. doi: 10.1111/j.1471-4159.1993.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim G.H., Kim J.E., Rhie S.J., Yoon S. The role of oxidative stress in neurodegenerative diseases. Experimental Neurobiology. 2015;24(4):325–340. doi: 10.5607/en.2015.24.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greco V., Longone P., Spalloni A., Pieroni L., Urbani A. Crosstalk between oxidative stress and mitochondrial damage: focus on amyotrophic lateral sclerosis. Advances in Experimental Medicine & Biology. 2019;1158:71–82. doi: 10.1007/978-981-13-8367-0_5. [DOI] [PubMed] [Google Scholar]
- 16.Pirie E., Oh C.-K., Zhang X., Han X., Cieplak P., Scott H.R., et al. S-nitrosylated TDP-43 triggers aggregation, cell-to-cell spread, and neurotoxicity in hiPSCs and in vivo models of ALS/FTD. Proceedings of the National Academy of Sciences of the United States of America. 2021;118(11) doi: 10.1073/pnas.2021368118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith E.F., Shaw P.J., De Vos K.J. The role of mitochondria in amyotrophic lateral sclerosis. Neuroscience Letters. 2019;710:132933. doi: 10.1016/j.neulet.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 18.Marques E.P., Wyse A.T.S. Creatine as a neuroprotector: an actor that can play many parts. Neurotoxicity Research. 2019;36(2):411–423. doi: 10.1007/s12640-019-00053-7. [DOI] [PubMed] [Google Scholar]
- 19.Vandoorne T., De Bock K., Van Den Bosch L. Energy metabolism in ALS: an underappreciated opportunity? Acta Neuropathologica. 2018;135(4):489–509. doi: 10.1007/s00401-018-1835-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature Cell Biology. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dienel G.A. Brain glucose metabolism: integration of energetics with function. Physiological Reviews. 2019;99(1):949–1045. doi: 10.1152/physrev.00062.2017. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y., Herman P., Rothman D.L., Agarwal D., Hyder F. Evaluating the gray and white matter energy budgets of human brain function. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2018;38(8):1339–1353. doi: 10.1177/0271678X17708691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Licznerski P., Park H.-A., Rolyan H., Chen R., Mnatsakanyan N., Miranda P., et al. ATP synthase c-subunit leak causes aberrant cellular metabolism in fragile X syndrome. Cell. 2020;182(5):1170–1179. doi: 10.1016/j.cell.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangaraju V., Calloway N., Ryan T.A. Activity-driven local ATP synthesis is required for synaptic function. Cell. 2014;156(4):825–835. doi: 10.1016/j.cell.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann K., Deny S., Ganguli S., Clandinin T.R. Coupling of activity, metabolism and behaviour across the Drosophila brain. Nature. 2021;593(7858):244–248. doi: 10.1038/s41586-021-03497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall C.N., Klein-Flügge M.C., Howarth C., Attwell D. Oxidative phosphorylation, not glycolysis, powers presynaptic and postsynaptic mechanisms underlying brain information processing. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2012;32(26):8940–8951. doi: 10.1523/JNEUROSCI.0026-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Science Advances. 2016;2(5) doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood T. Physiological functions of the pentose phosphate pathway. Cell Biochemistry and Function. 1986;4(4):241–247. doi: 10.1002/cbf.290040403. [DOI] [PubMed] [Google Scholar]
- 30.Stanton R.C. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life. 2012;64(5):362–369. doi: 10.1002/iub.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kletzien R.F., Harris P.K., Foellmi L.A. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. Federation of American Societies for Experimental Biology Journal: Official Publication of the Federation of American Societies for Experimental Biology. 1994;8(2):174–181. doi: 10.1096/fasebj.8.2.8119488. [DOI] [PubMed] [Google Scholar]
- 32.Denzel M.S., Storm N.J., Gutschmidt A., Baddi R., Hinze Y., Jarosch E., et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156(6):1167–1178. doi: 10.1016/j.cell.2014.01.061. [DOI] [PubMed] [Google Scholar]
- 33.Slawson C., Hart G.W. O-GlcNAc signalling: implications for cancer cell biology. Nature Reviews Cancer. 2011;11(9):678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z.V., Deng Y., Gao N., Pedrozo Z., Li D.L., Morales C.R., et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156(6):1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu K., Yin N., Peng M., Stamatiades E.G., Shyu A., Li P., et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science (New York, N.Y.) 2021;371(6527):405–410. doi: 10.1126/science.abb2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mojsilovic-Petrovic J., Jeong G.-B., Crocker A., Arneja A., David S., Russell D.S., et al. Protecting motor neurons from toxic insult by antagonism of adenosine A2a and Trk receptors. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2006;26(36):9250–9263. doi: 10.1523/JNEUROSCI.1856-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waagepetersen H.S., Sonnewald U., Larsson O.M., Schousboe A. Compartmentation of TCA cycle metabolism in cultured neocortical neurons revealed by 13C MR spectroscopy. Neurochemistry International. 2000;36(4–5):349–358. doi: 10.1016/s0197-0186(99)00143-6. [DOI] [PubMed] [Google Scholar]
- 38.Sonnewald U., Hertz L., Schousboe A. Mitochondrial heterogeneity in the brain at the cellular level. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 1998;18(3):231–237. doi: 10.1097/00004647-199803000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Israelson A., Arbel N., Da Cruz S., Ilieva H., Yamanaka K., Shoshan-Barmatz V., et al. Misfolded mutant SOD1 directly inhibits VDAC1 conductance in a mouse model of inherited ALS. Neuron. 2010;67(4):575–587. doi: 10.1016/j.neuron.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasinelli P., Belford M.E., Lennon N., Bacskai B.J., Hyman B.T., Trotti D., et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43(1):19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Wang W., Wang L., Lu J., Siedlak S.L., Fujioka H., Liang J., et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nature Medicine. 2016;22(8):869–878. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai J.C., Walsh J.M., Dennis S.C., Clark J.B. Synaptic and non-synaptic mitochondria from rat brain: isolation and characterization. Journal of Neurochemistry. 1977;28(3):625–631. doi: 10.1111/j.1471-4159.1977.tb10434.x. [DOI] [PubMed] [Google Scholar]
- 43.Leong S.F., Lai J.C., Lim L., Clark J.B. The activities of some energy-metabolising enzymes in nonsynaptic (free) and synaptic mitochondria derived from selected brain regions. Journal of Neurochemistry. 1984;42(5):1306–1312. doi: 10.1111/j.1471-4159.1984.tb02788.x. [DOI] [PubMed] [Google Scholar]
- 44.Armakola M., Higgins M.J., Figley M.D., Barmada S.J., Scarborough E.A., Diaz Z., et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nature Genetics. 2012;44(12):1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim M.A., Selak M.A., Xiang Z., Krainc D., Neve R.L., Kraemer B.C., et al. Reduced activity of AMP-activated protein kinase protects against genetic models of motor neuron disease. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2012;32(3):1123–1141. doi: 10.1523/JNEUROSCI.6554-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Periz G., Lu J., Zhang T., Kankel M.W., Jablonski A.M., Kalb R., et al. Regulation of protein quality control by UBE4B and LSD1 through p53-mediated transcription. PLoS Biology. 2015;13(4) doi: 10.1371/journal.pbio.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhai J., Zhang L., Mojsilovic-Petrovic J., Jian X., Thomas J., Homma K., et al. Inhibition of cytohesins protects against genetic models of motor neuron disease. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2015;35(24):9088–9105. doi: 10.1523/JNEUROSCI.5032-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jablonski A.M., Lamitina T., Liachko N.F., Sabatella M., Lu J., Zhang L., et al. Loss of RAD-23 protects against models of motor neuron disease by enhancing mutant protein clearance. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2015;35(42):14286–14306. doi: 10.1523/JNEUROSCI.0642-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mojsilovic-Petrovic J., Nedelsky N., Boccitto M., Mano I., Georgiades S.N., Zhou W., et al. FOXO3a is broadly neuroprotective in vitro and in vivo against insults implicated in motor neuron diseases. Journal of Neuroscience. 2009;29(25):8236–8247. doi: 10.1523/JNEUROSCI.1805-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen X., Tan W., Westergard T., Krishnamurthy K., Markandaiah S.S., Shi Y., et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84(6):1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ash P.E.A., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.-L., DeJesus-Hernandez M., et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizielinska S., Grönke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science (New York, N.Y.) 2014;345(6201):1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta R., Lan M., Mojsilovic-Petrovic J., Choi W.H., Safren N., Barmada S., et al. The proline/arginine dipeptide from hexanucleotide repeat ExpandedC9ORF72Inhibits the proteasome. eNeuro. 2017;4(1) doi: 10.1523/ENEURO.0249-16.2017. ENEURO.0249–16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghergurovich J.M., García-Cañaveras J.C., Wang J., Schmidt E., Zhang Z., TeSlaa T., et al. A small molecule G6PD inhibitor reveals immune dependence on pentose phosphate pathway. Nature Chemical Biology. 2020;16(7):731–739. doi: 10.1038/s41589-020-0533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawton K.A., Cudkowicz M.E., Brown M.V., Alexander D., Caffrey R., Wulff J.E., et al. Biochemical alterations associated with ALS. Null. 2012;13(1):110–118. doi: 10.3109/17482968.2011.619197. [DOI] [PubMed] [Google Scholar]
- 56.Singh T., Jiao Y., Ferrando L.M., Yablonska S., Li F., Horoszko E.C., et al. Neuronal mitochondrial dysfunction in sporadic amyotrophic lateral sclerosis is developmentally regulated. Scientific Reports. 2021;11(1) doi: 10.1038/s41598-021-97928-7. 18916–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang T., Liu H., Itoh K., Oh S., Zhao L., Murata D., et al. C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metabolism. 2021;33(3):531–539. doi: 10.1016/j.cmet.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alves C.J., Dariolli R., Jorge F.M., Monteiro M.R., Maximino J.R., Martins R.S., et al. Gene expression profiling for human iPS-derived motor neurons from sporadic ALS patients reveals a strong association between mitochondrial functions and neurodegeneration. Frontiers in Cellular Neuroscience. 2015;9:289. doi: 10.3389/fncel.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Genin E.C., Madji Hounoum B., Bannwarth S., Fragaki K., Lacas-Gervais S., Mauri-Crouzet A., et al. Mitochondrial defect in muscle precedes neuromuscular junction degeneration and motor neuron death in CHCHD10S59L/+ mouse. Acta Neuropathologica. 2019;138(1):123–145. doi: 10.1007/s00401-019-01988-z. [DOI] [PubMed] [Google Scholar]
- 60.Kiskinis E., Sandoe J., Williams L.A., Boulting G.L., Moccia R., Wainger B.J., et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14(6):781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee H., Lee J.J., Park N.Y., Dubey S.K., Kim T., Ruan K., et al. Multi-omic analysis of selectively vulnerable motor neuron subtypes implicates altered lipid metabolism in ALS. Nature Neuroscience. 2021;24(12):1673–1685. doi: 10.1038/s41593-021-00944-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson C.J., Bredvik K., Burstein S.R., Davis C., Meadows S.M., Dash J., et al. ALS/FTD mutant CHCHD10 mice reveal a tissue-specific toxic gain-of-function and mitochondrial stress response. Acta Neuropathologica. 2019;138(1):103–121. doi: 10.1007/s00401-019-01989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang P., Deng J., Dong J., Liu J., Bigio E.H., Mesulam M., et al. TDP-43 induces mitochondrial damage and activates the mitochondrial unfolded protein response. PLoS Genetics. 2019;15(5) doi: 10.1371/journal.pgen.1007947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Straub I.R., Janer A., Weraarpachai W., Zinman L., Robertson J., Rogaeva E., et al. Loss of CHCHD10-CHCHD2 complexes required for respiration underlies the pathogenicity of a CHCHD10 mutation in ALS. Human Molecular Genetics. 2018;27(1):178–189. doi: 10.1093/hmg/ddx393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baek M., Choe Y.-J., Bannwarth S., Kim J., Maitra S., Dorn G.W., et al. TDP-43 and PINK1 mediate CHCHD10S59L mutation-induced defects in Drosophila and in vitro. Nature Communications. 2021;12(1):1924–2020. doi: 10.1038/s41467-021-22145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu C.-H., Davidson S., Harapas C.R., Hilton J.B., Mlodzianoski M.J., Laohamonthonkul P., et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183(3):636–649.e18. doi: 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parone P.A., Da Cruz S., Han J.S., McAlonis-Downes M., Vetto A.P., Lee S.K., et al. Enhancing mitochondrial calcium buffering capacity reduces aggregation of misfolded SOD1 and motor neuron cell death without extending survival in mouse models of inherited amyotrophic lateral sclerosis. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2013;33(11):4657–4671. doi: 10.1523/JNEUROSCI.1119-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan W., Naniche N., Bogush A., Pedrini S., Trotti D., Pasinelli P. Small peptides against the mutant SOD1/Bcl-2 toxic mitochondrial complex restore mitochondrial function and cell viability in mutant SOD1-mediated ALS. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2013;33(28):11588–11598. doi: 10.1523/JNEUROSCI.5385-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi S.Y., Lopez-Gonzalez R., Krishnan G., Phillips H.L., Li A.N., Seeley W.W., et al. C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nature Neuroscience. 2019;22(6):851–862. doi: 10.1038/s41593-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang Y.-C., Hung W.-T., Chang Y.-C., Chang H.C., Wu C.-L., Chiang A.-S., et al. Pathogenic VCP/TER94 alleles are dominant actives and contribute to neurodegeneration by altering cellular ATP level in a Drosophila IBMPFD model. PLoS Genetics. 2011;7(2) doi: 10.1371/journal.pgen.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim N.C., Tresse E., Kolaitis R.-M., Molliex A., Thomas R.E., Alami N.H., et al. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78(1):65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartolome F., Wu H.-C., Burchell V.S., Preza E., Wray S., Mahoney C.J., et al. Pathogenic VCP mutations induce mitochondrial uncoupling and reduced ATP levels. Neuron. 2013;78(1):57–64. doi: 10.1016/j.neuron.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S., Wu Z., Li Y., Tantray I., De Stefani D., Mattarei A., et al. Altered MICOS morphology and mitochondrial ion homeostasis contribute to poly(GR) toxicity associated with C9-ALS/FTD. Cell Reports. 2020;32(5):107989. doi: 10.1016/j.celrep.2020.107989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valbuena G.N., Rizzardini M., Cimini S., Siskos A.P., Bendotti C., Cantoni L., et al. Metabolomic analysis reveals increased aerobic glycolysis and amino acid deficit in a cellular model of amyotrophic lateral sclerosis. Molecular Neurobiology. 2016;53(4):2222–2240. doi: 10.1007/s12035-015-9165-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allen S.P., Rajan S., Duffy L., Mortiboys H., Higginbottom A., Grierson A.J., et al. Superoxide dismutase 1 mutation in a cellular model of amyotrophic lateral sclerosis shifts energy generation from oxidative phosphorylation to glycolysis. Neurobiology of Aging. 2014;35(6):1499–1509. doi: 10.1016/j.neurobiolaging.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 76.Manzo E., Lorenzini I., Barrameda D., O'Conner A.G., Barrows J.M., Starr A., et al. Glycolysis upregulation is neuroprotective as a compensatory mechanism in ALS. eLife. 2019;8:606. doi: 10.7554/eLife.45114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tefera T.W., Bartlett K., Tran S.S., Hodson M.P., Borges K. Impaired pentose phosphate pathway in the spinal cord of the hSOD1G93A mouse model of amyotrophic lateral sclerosis. Molecular Neurobiology. 2019;56(8):5844–5855. doi: 10.1007/s12035-019-1485-6. [DOI] [PubMed] [Google Scholar]
- 78.Tefera T.W., Borges K. Neuronal glucose metabolism is impaired while astrocytic TCA cycling is unaffected at symptomatic stages in the hSOD1G93A mouse model of amyotrophic lateral sclerosis. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2019;39(9):1710–1724. doi: 10.1177/0271678X18764775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziller M.J., Ortega J.A., Quinlan K.A., Santos D.P., Gu H., Martin E.J., et al. Dissecting the functional consequences of De Novo DNA methylation dynamics in human motor neuron differentiation and physiology. Cell Stem Cell. 2018 doi: 10.1016/j.stem.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ortega J.A., Daley E.L., Kour S., Samani M., Tellez L., Smith H.S., et al. Nucleocytoplasmic proteomic analysis uncovers eRF1 and nonsense-mediated decay as modifiers of ALS/FTD C9orf72 toxicity. Neuron. 2020;106(1):90–107.e13. doi: 10.1016/j.neuron.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pellerin L., Magistretti P.J. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(22):10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jolivet R., Allaman I., Pellerin L., Magistretti P.J., Weber B. Comment on recent modeling studies of astrocyte-neuron metabolic interactions. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2010;30(12):1982–1986. doi: 10.1038/jcbfm.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mangia S., DiNuzzo M., Giove F., Carruthers A., Simpson I.A., Vannucci S.J. Response to “comment on recent modeling studies of astrocyte-neuron metabolic interactions”: much ado about nothing. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31(6):1346–1353. doi: 10.1038/jcbfm.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park J.S., Burckhardt C.J., Lazcano R., Solis L.M., Isogai T., Li L., et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature. 2020;578(7796):621–626. doi: 10.1038/s41586-020-1998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zala D., Hinckelmann M.-V., Yu H., Lyra da Cunha M.M., Liot G., Cordelières F.P., et al. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013;152(3):479–491. doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 86.Jin M., Fuller G.G., Han T., Yao Y., Alessi A.F., Freeberg M.A., et al. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Reports. 2017;20(4):895–908. doi: 10.1016/j.celrep.2017.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang J.-S., Hsu J.-W., Park S.-Y., Li J., Oldham W.M., Beznoussenko G.V., et al. GAPDH inhibits intracellular pathways during starvation for cellular energy homeostasis. Nature. 2018;561(7722):263–267. doi: 10.1038/s41586-018-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang J.-S., Hsu J.-W., Park S.-Y., Lee S.Y., Li J., Bai M., et al. ALDH7A1 inhibits the intracellular transport pathways during hypoxia and starvation to promote cellular energy homeostasis. Nature Communications. 2019;10(1):1–15. doi: 10.1038/s41467-019-11932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tristan C., Shahani N., Sedlak T.W., Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cellular Signalling. 2011;23(2):317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee K.H., Cha M., Lee B.H. Neuroprotective effect of antioxidants in the brain. International Journal of Molecular Sciences. 2020;21(19):7152. doi: 10.3390/ijms21197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fransen M., Revenco I., Li H., Costa C.F., Lismont C., Van Veldhoven P.P. Peroxisomal dysfunction and oxidative stress in neurodegenerative disease: a bidirectional crosstalk. Advances in Experimental Medicine & Biology. 2020;1299:19–30. doi: 10.1007/978-3-030-60204-8_2. [DOI] [PubMed] [Google Scholar]
- 92.Barber S.C., Mead R.J., Shaw P.J. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochimica Et Biophysica Acta. 2006;1762(11–12):1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 93.Sorce S., Stocker R., Seredenina T., Holmdahl R., Aguzzi A., Chiò A., et al. NADPH oxidases as drug targets and biomarkers in neurodegenerative diseases: what is the evidence? Free Radical Biology & Medicine. 2017;112:387–396. doi: 10.1016/j.freeradbiomed.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 94.Abe K., Itoyama Y., Sobue G., Tsuji S., Aoki M., Doyu M., et al. Confirmatory double-blind, parallel-group, placebo-controlled study of efficacy and safety of edaravone (MCI-186) in amyotrophic lateral sclerosis patients. Amyotrophic Lateral Sclerosis & Frontotemporal Degeneration. 2014;15(7–8):610–617. doi: 10.3109/21678421.2014.959024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brennan A.M., Suh S.W., Won S.J., Narasimhan P., Kauppinen T.M., Lee H., et al. NADPH oxidase is the primary source of superoxide induced by NMDA receptor activation. Nature Neuroscience. 2009;12(7):857–863. doi: 10.1038/nn.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leopold J.A., Walker J., Scribner A.W., Voetsch B., Zhang Y.-Y., Loscalzo A.J., et al. Glucose-6-phosphate dehydrogenase modulates vascular endothelial growth factor-mediated angiogenesis. The Journal of Biological Chemistry. 2003;278(34):32100–32106. doi: 10.1074/jbc.M301293200. [DOI] [PubMed] [Google Scholar]
- 97.Leopold J.A., Zhang Y.-Y., Scribner A.W., Stanton R.C., Loscalzo J. Glucose-6-phosphate dehydrogenase overexpression decreases endothelial cell oxidant stress and increases bioavailable nitric oxide. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003;23(3):411–417. doi: 10.1161/01.ATV.0000056744.26901.BA. [DOI] [PubMed] [Google Scholar]
- 98.Marrali G., Casale F., Salamone P., Fuda G., Caorsi C., Amoroso A., et al. NADPH oxidase (NOX2) activity is a modifier of survival in ALS. Journal of Neurology. 2014;261(11):2178–2183. doi: 10.1007/s00415-014-7470-0. [DOI] [PubMed] [Google Scholar]
- 99.Seredenina T., Nayernia Z., Sorce S., Maghzal G.J., Filippova A., Ling S.-C., et al. Evaluation of NADPH oxidases as drug targets in a mouse model of familial amyotrophic lateral sclerosis. Free Radical Biology & Medicine. 2016;97:95–108. doi: 10.1016/j.freeradbiomed.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 100.Chiot A., Zaïdi S., Iltis C., Ribon M., Berriat F., Schiaffino L., et al. Modifying macrophages at the periphery has the capacity to change microglial reactivity and to extend ALS survival. Nature Neuroscience. 2020;23(11):1339–1351. doi: 10.1038/s41593-020-00718-z. [DOI] [PubMed] [Google Scholar]
- 101.Drechsel D.A., Estévez Á.G., Barbeito L., Beckman J.S. Nitric oxide-mediated oxidative damage and the progressive demise of motor neurons in ALS. Neurotoxicity Research. 2012;22(4):251–264. doi: 10.1007/s12640-012-9322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X., Zhang X., Li C., Guan T., Shang H., Cui L., et al. S-nitrosylated protein disulfide isomerase contributes to mutant SOD1 aggregates in amyotrophic lateral sclerosis. Journal of Neurochemistry. 2013;124(1):45–58. doi: 10.1111/jnc.12046. [DOI] [PubMed] [Google Scholar]
- 103.Rajasekaran N.S., Connell P., Christians E.S., Yan L.-J., Taylor R.P., Orosz A., et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130(3):427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luzzatto L., Nannelli C., Notaro R. Glucose-6-phosphate dehydrogenase deficiency. Hematology/Oncology Clinics of North America. 2016;30(2):373–393. doi: 10.1016/j.hoc.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 105.Garcia A.A., Koperniku A., Ferreira J.C.B., Mochly-Rosen D. Treatment strategies for glucose-6-phosphate dehydrogenase deficiency: past and future perspectives. Trends in Pharmacological Sciences. 2021;42(10):829–844. doi: 10.1016/j.tips.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Savaki H.E., Davidsen L., Smith C., Sokoloff L. Measurement of free glucose turnover in brain. Journal of Neurochemistry. 1980;35(2):495–502. doi: 10.1111/j.1471-4159.1980.tb06293.x. [DOI] [PubMed] [Google Scholar]
- 107.Veech R.L., Harris R.L., Veloso D., Veech E.H. Freeze-blowing: a new technique for the study of brain in vivo. Journal of Neurochemistry. 1973;20(1):183–188. doi: 10.1111/j.1471-4159.1973.tb12115.x. [DOI] [PubMed] [Google Scholar]
- 109.Nissim I., Horyn O., Nissim I., Daikhin Y., Wehrli S.L., Yudkoff M., et al. Effects of a glucokinase activator on hepatic intermediary metabolism: study with 13C-isotopomer-based metabolomics. The Biochemical Journal. 2012;444(3):537–551. doi: 10.1042/BJ20120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li B., Qiu B., Lee D.S.M., Walton Z.E., Ochocki J.D., Mathew L.K., et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li C., Nissim I., Chen P., Buettger C., Najafi H., Daikhin Y., et al. Elimination of KATP channels in mouse islets results in elevated [U-13C]glucose metabolism, glutaminolysis, and pyruvate cycling but a decreased gamma-aminobutyric acid shunt. The Journal of Biological Chemistry. 2008;283(25):17238–17249. doi: 10.1074/jbc.M709235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nissim I., Horyn O., Daikhin Y., Chen P., Li C., Wehrli S.L., et al. The molecular and metabolic influence of long term agmatine consumption. The Journal of Biological Chemistry. 2014;289(14):9710–9729. doi: 10.1074/jbc.M113.544726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weinberg J.M., Venkatachalam M.A., Roeser N.F., Nissim I. Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(6):2826–2831. doi: 10.1073/pnas.97.6.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moore T., Le A., Niemi A.-K., Kwan T., Cusmano-Ozog K., Enns G.M., et al. A new LC-MS/MS method for the clinical determination of reduced and oxidized glutathione from whole blood. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 2013;929:51–55. doi: 10.1016/j.jchromb.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 115.Lee W.N., Boros L.G., Puigjaner J., Bassilian S., Lim S., Cascante M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2-13C2]glucose. The American Journal of Physiology. 1998;274(5):E843–E851. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- 116.van Geersdaele L.K., Stead M.A., Harrison C.M., Carr S.B., Close H.J., Rosbrook G.O., et al. Structural basis of high-order oligomerization of the cullin-3 adaptor SPOP. Acta Crystallographica. Section D, Biological Crystallography. 2013;69(Pt 9):1677–1684. doi: 10.1107/S0907444913012687. [DOI] [PubMed] [Google Scholar]
- 117.Jang S., Nelson J.C., Bend E.G., Rodríguez-Laureano L., Tueros F.G., Cartagenova L., et al. Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron. 2016;90(2):278–291. doi: 10.1016/j.neuron.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li S., Xiong G.-J., Huang N., Sheng Z.-H. The cross-talk of energy sensing and mitochondrial anchoring sustains synaptic efficacy by maintaining presynaptic metabolism. Nature Metabolism. 2020;2(10):1077–1095. doi: 10.1038/s42255-020-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Johmura Y., Yamanaka T., Omori S., Wang T.-W., Sugiura Y., Matsumoto M., et al. Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science (New York, N.Y.) 2021;371(6526):265–270. doi: 10.1126/science.abb5916. [DOI] [PubMed] [Google Scholar]
- 121.Chin R.M., Fu X., Pai M.Y., Vergnes L., Hwang H., Deng G., et al. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510(7505):397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gut P., Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502(7472):489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 123.Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast (Chichester, England) 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-132. [DOI] [PubMed] [Google Scholar]
- 124.Chee M.K., Haase S.B. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomycescerevisiae. G3: Genes – Genomes – Genetics. 2012;2(5):515–526. doi: 10.1534/g3.111.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. Journal of Bacteriology. 1983;153(1):163–168. doi: 10.1128/JB.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., et al. A yeast functional screen predicts new candidate ALS disease genes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(52):20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nussbaum-Krammer C.I., Neto M.F., Brielmann R.M., Pedersen J.S., Morimoto R.I. Investigating the spreading and toxicity of prion-like proteins using the metazoan model organism C. elegans. Journal of Visualized Experiments: Journal of Visualized Experiments. 2015;(95):52321. doi: 10.3791/52321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total intracellular amino acid levels from neuronal cultures engineered to express LacZ, Wt or Mt versions of SOD1 or TDP43. Mean, standard error and ANOVA with post hoc analysis are presented.
. Adenylate nucleotide levels and redox status in neurons expressing Wt or familial Mt ALS genes. Mean, standard deviation and Student's t-test values from analysis of ATP, ADP, AMP, NAD+, NADH, NADP+, NADPH, GSSG and GSH are presented. Comparisons are from neurons expressing LacZ versus Wt-SOD1 versus Mt-SOD1 or LacZ versus Wt-TDP43 versus Mt-TDP43.
Yeast deletion strains used in these studies.
Feeding RNAi bacteria studied in C. elegans model of fALS.
Data Availability Statement
All primary data collected in the course of these experiments is freely available to qualified investigators by simple request.