Abstract
Background
Companion animals like dogs play an important role in the lives of many people and are often considered to be members of families, but definitely, any contact with them poses an inherent risk of transmitting zoonotic pathogens. One of these pathogens is the genus Helicobacter which is linked to many disorders in human and animal.
Objectives
The aim of this study was to investigate the presence of some zoonotic species of genus Helicobacter in companion dogs.
Results
Through culturing in a special medium, nine samples (9%) were detected as infected (two pure and seven mixed culture). Based on multiplex‐PCR, 13 samples (13%) were infected by Helicobacter spp. although none of them were infected by H. pylori. Species‐specific PCR indicated that 38.5% or 5/13 of the samples were infected with H. heilmannii, while 15.45% or 2/13 of the samples were infected by H. billis. Multivariate logistic regression analysis showed that the age factor had a significant effect on Helicobacter spp. infection (odds ratio [OR] = 2.42, p = 0.01).
Conclusion
This study revealed the negligible faecal transmission of H. pylori. Moreover, due to the detection of H. Heilmannii and H. billis in feces and their association with human gastric diseases, dog owners should be educated about the risks and transmission modes of zoonotic bacterial infections of dogs.
Keywords: companion dogs, epidemiology, Helicobacter pylori, Helicobacter spp, PCR, zoonotic
The aim of this study was to determine the prevalence of Helicobacter spp. in canine feces by culture and molecular methods. Another objective was to confirm the excretion of H. pylori, H. Heilmannii and H. billis from feces in companion dogs with and without gastrointestinal disorders. This study revealed the negligible faecal transmission of H. pylori. Moreover, due to the detection of H. Heilmannii and H. billis in feces and their association with human gastric diseases, dog owners should be educated about the risks and transmission modes of zoonotic bacterial infections of dogs. The findings of the present study may be useful to clarify the epidemiology of Helicobacter spp. in dogs and humans.
1. INTRODUCTION
Helicobacter pylori, a Gram‐negative and spiral‐shaped bacterium, was discovered in 1982 (Marshall & Warren, 1984). More than half of the population worldwide may have been infected by this bacterium. However, only 5%–10% of them show clinical symptoms (Goh et al., 2011; Mladenova‐Hristova et al., 2017). Having reviewed 263 published articles on the prevalence of H. pylori infection and collecting data from 62 countries, Hooi et al. (2017) showed that Africa has the highest pooled prevalence of H. pylori infection (70.1%), whereas Oceania has the lowest prevalence (24.4%). Among the countries, the prevalence of this infection varies from 18.9% in Switzerland to 87.7% in Nigeria. Based on the regional prevalence estimates, there were approximately 4.4 billion individuals with H. pylori infection worldwide in 2015 (Hooi et al., 2017). At present, the role of H. pylori has been determined in the chronic active gastritis, peptic and duodenal ulcers, gastric adenocarcinoma and gastric mucosa associated lymphoid tissue (MALT) lymphomas. In 1994, the International Agency for Research on Cancer (IARC) categorized H. pylori as a class I carcinogen and reported it in 75% of patients with MALT lymphoma and in 60% of those with an increasing risk of gastric cancer (Malfertheiner et al., 2017; Morgner et al., 2000). Also, H. pylori infection could be related to some important diseases in humans such as the reduction of the ferritin and iron levels in patients with coronary artery disease and changes in lipid profiles and inflammatory factors, such as pre‐eclampsia as a result of impairing the placental development, and glucose homeostasis in patients with type 2 diabetes (Bonfigl, 2016; Di Simone et al., 2017; Fallah et al., 2016). Furthermore, the infection is directly related to the metabolic syndromes such as high values of triglycerides, body mass index and systolic blood pressure and low HDL (Upala et al., 2016). Following the discovery of H. pylori, other spiral bacteria have been observed in many other considered animal species (Dent et al., 1987). In further research employing molecular analysis of the 16rRNA, it was elucidated that these spiral bacteria belonged to the Helicobacter genus (Solnick et al., 1993). At present, this genus, with at least 35 species distributed worldwide, can infect human and animals by colonizing in different anatomical regions of the gastrointestinal system such as oral cavity, stomach, intestine and liver. Depending on the place of colonization, it is also divided into gastric and enterohepatic species (Fox & Wang, 2002; Mladenova‐Hristova et al., 2017; Recordati et al., 2009). Gastric Helicobacter such as Helicobacter heilmannii, Helicobacter felis, Helicobacter salomonis, Helicobacter bizzozeronii, Helicobacter cynogastricus and Helicobacter pylori and enterohepatic Helicobacter such as Helicobacter canis, Helicobacter billis, Helicobacter cinaedi and Helicobacter rappini were isolated from dogs. These species, which are associated with hepatobiliary and gastrointestinal diseases, are transmitted either directly through oral–oral and anal–oral contact or indirectly through water and food in dogs (Buczolits et al., 2003; Ekman et al., 2013; Haesebrouck et al., 2009; Jalava et al., 1997; Jankowski et al., 2016a; Jankowski et al., 2016a; Kubota‐Aizawa et al., 2017; Mladenova‐Hristova et al., 2017; Recordati et al., 2007; Recordati et al., 2009; Rossi et al., 2008; Van den Bulck et al., 2005). Helicobacter spp. infection has been reported in 23%–100% of companion dogs and in 70% and 30% of people in both developing and developed countries, respectively (Agüloğlu et al., 2006; Amorim et al., 2015; Chung et al., 2014; Downsett & Kowolik, 2003; Hong et al., 2015; Hwang et al., 2002; Jankowski et al., 2016a, 2016b; Kubota‐Aizawa et al., 2017; Recordati et al., 2009). Despite the high prevalence of this genus, its transmission mechanism in human–human, animal–human, human–animal and animal–animal paths remains unclear (Ekman et al., 2013; Recordati et al., 2007).
The diagnostic methods include invasive and non‐invasive means; the former with high sensitivity and specificity encompasses gastroscopy and collecting a biopsy sample of the gastric mucosa for rapid urea test, histopathological examination, direct Gram staining, microbiological culture, electron microscopy, fluorescent in situ hybridization and PCR. Furthermore, regardless of the risks of gastroscopy and the associated anaesthesia, it should be noted that many veterinarians and clinics do not have the facilities and ability to perform gastroscopy. Therefore, non‐invasive methods, including serology, culture and PCR on blood, saliva and faecal samples are preferred (Haesebrouck et al., 2009; Hong et al., 2015; Pohl et al., 2019; Shinozaki et al., 2002).
H. pylori is transmitted from experimentally infected dogs to the uninfected ones once being isolated from gastric mucosa and saliva specimens in dogs (Ekman et al., 2013; Jankowski et al., 2016a, 2016b; Lee et al., 1992; Radin et al., 1990). Also, in our previous study, the apparent seroprevalence of H. pylori in related and unrelated individuals with dogs and cats was 72.1% (95% CI: 64.8%–79.4%) and 48.8% (95% CI: 42%–55.6%), respectively. Moreover, the odds of infection in related rather than unrelated individuals was 2.71 (95% CI: 1.73%–4.26%) (Ashrafmodarres et al., 2017).
Therefore, the aim of this study was to determine the prevalence of Helicobacter spp. in canine feces by culture and molecular methods. Another objective was to confirm the excretion of H. pylori from feces in companion dogs with and without gastrointestinal disorders. The results of the present study may be useful to clarify the epidemiology of Helicobacter spp. in dogs and humans.
2. MATERIALS AND METHODS
2.1. Sample collection
The sample studied in this exploratory study included 50 companion dogs with and 50 dogs without gastrointestinal disorders (existence of diarrhoea, vomiting, constipation and loss of appetite) that were referred to the only veterinary hospital in Ahvaz city of Khuzestan province. Also, on the basis of the history and clinical examinations, the dogs infected or suspected of having other body systems diseases were excluded from this study. In addition, there was no history of drug therapy such as antibiotics and proton pump inhibitors (e.g. omeprazole, lansoprazole and pantoprazole) in any of the dogs (at least 2 weeks before sampling). The city of Ahvaz with a population of about 1,300,000 and approximately 4000 domestic dogs is the capital of Khuzestan province (Census, 2016). Companion dogs in this city are often kept only at homes and have no close contact with other dogs and cats (Didehban et al., 2020). In line with the research, the purpose of the study was explained to the dog owners, and informed consent was taken. Sterile cotton swabs were used for scraping the rectal mucosa, and then placed in a 1.5 ml sterile microtube. Swabs were quickly sent to the microbiology lab and cultured in the selective medium. To isolate the Helicobacter spp., a minimum time interval between sampling and culture is crucial due to the possible negative effects on Helicobacter viability. In addition, the age, sex, breed, and habitat (the keeping place) of each dog were recorded.
2.2. Bacterial isolation
To prepare the selective medium, sheep blood agar and several additional antibiotics were required. Therefore, blood agar base (Biolab Diagnostics Laboratory Inc., Hungary) was prepared and autoclaved for 20 min at 121°C and then tempered to 50°C. Then, antibiotics (vancomycin 10 mg/L, trimethoprim 50 mg/L, ceftiufor 50 mg/L, amphotericin B 2–5 mg/L and polymixin B 3500 IU/L) were aseptically added with constant stirring. Finally, the medium was poured into the petri plates.
Before the culturing process, 200 μl of sterile phosphate‐buffered saline (PBS) was added to each microtube, and then the swab was inoculated in the first region of the plate and streaked by loop in another region. The cultured medium was incubated in the microaerophilic condition at 37°C for 3–5 days.
Subsequently, the suspected colonies were detected and purified in the blood agar medium under the previous condition. Meanwhile, the microscopic examination was conducted via preparing the smear and Gram staining. Finally, the suspected isolates inoculated with skim milk were stored at −70°C until being retrieved for further analysis.
2.3. Molecular detection of isolates
2.3.1. DNA extraction
In order for the genomic study of isolates and swab samples, first the DNA extraction was done using the GeneAll DNA kit (GeneAll Biotechnology Co., South Korea) following the manufacturer's instructions. Then, based on genus‐specific and different species‐specific primers, the polymerase chain reaction (PCR) test was carried out for the detection of sample contamination with Helicobacter genus and different species such as H. pylori, H. heilmannii and H. billis. Primer sequences and references of each primer are shown in Table 1. For the detection of Helicobacter genus and pylori species by multiplex PCR, the thermal cycles were set as follows: 1 min at 94°C for denaturation, 2 min at 55°C for annealing and 2 min at 72°C for extension. Thirty‐five repetitions were considered for this stage with an early denaturation at 94°C for 5 min and a final elongation at 72°C for 10 min (Farshad et al., 2004). Each PCR reaction tube contained 25 μl of the reaction mix (consisting of 12.5 μl of Mastermix [Ampliqon, Denmark] with 2 mM MgCl2, 1 μl [10 pmol/μl] of each primer and 5 μl of chromosomal extracted DNA). In order to decrease the negative effects of inhibitory factors, 400 ng/ml of bovine serum albumin (BSA) was added to each reaction (Kreader, 1996).
TABLE 1.
Goal gene, sequence and size of used primers
| Name (gene) | Primer sequence | Size (bp) | Reference |
|---|---|---|---|
| Helicobacter genus (16srRNA) |
5‐GTA AAG GCT CAC CAA GGC TAT‐3 5‐CCA CCT ACC TCT CCC ACA CTC‐3 |
389 | Choi et al. (2001) |
| H. pylori (isocitrate dehydrogenase) |
5‐ATGGCTTACAACCTAAAATTTTACAAAAGCC‐3 5‐TCA CAT GTT TTC AAT CAT CAC GC‐3 |
1200 | Argyros et al. (2000) |
| H. billis (16srRNA) |
5 ‐AGAACTGCATTTGAAACTACTTT‐3 5 ‐GGTATTGCATCTCTTTGTATGT‐3 |
638 | Fox et al. (1995) |
| H. heilmannii (ureB) |
5´‐GGG CGA TAA AGT GCG CTT G‐3´ 5´‐CTG GTC AAT GAG AGC AGG‐3´ |
580 | Neiger et al. (1998) |
The protocols of Fox et al. (1995) and Neiger et al. (1998) were used for the molecular detection of H. billis and H. heilmannii, respectively. The products of PCR were analyzed by electrophoresis in 1.5% agarose gel in TAE (tris‐acetate‐EDTA) buffer, visualized by safe‐staining (SinaGen, Iran), illuminated by a UV transilluminator (Uvitech, Germany), and finally documented by a gel documentation apparatus. The standard strain of H. pylori (Pasteur Institute, Iran) and sterile distilled water were used as positive and negative controls.
2.4. Statistical analysis
The statistical analysis of the data was performed using SPSS (version 16.0; SPSS Inc., Chicago, USA). The association between age (year), sex (female or male), breed (Terrier, Doberman, Dachshund, German Shepherd, Spitz, Siberian Husky, Pitbull or Rottweiler), gastrointestinal disorders (yes/no), habitat (yard/apartment) and Helicobacter spp. was analyzed by both χ 2 test and bivariate and multivariate logistic regression. Bivariate logistic regression models were fit to the data for each potential risk factor. Risk factors associated with BoHV‐1 (p ≤ 0.3) in bivariate regression were further analyzed in a multivariate logistic regression model, using a backward, stepwise algorithm. The goodness of fit of the model was determined using the Hosmer and Lemeshow test. The comparison of two diagnostic methods (culture and PCR) was performed by McNemar test and kappa statistic calculation. Differences were considered statistically significant at p ≤ 0.05.
3. RESULTS
3.1. Prevalence of Helicobacter spp
In the total number of 100 samples, there were 56 female and 44 male dogs. The mean score and standard deviation of age were 1.55 and 0.84 years, respectively. The relative frequency of Terrier, Doberman, Dachshund, German Shepherd, Spitz, Siberian Husky, Pitbull and Rottweiler breeds was 25%, 17%, 15%, 13%, 10%, 9%, 8% and 3%, respectively. Fifty‐nine percent of dogs were kept in the apartment and the rest were kept in the yard.
According to the culture and biochemical test, nine of the samples (9%) were positive, but two of them were pure and the rest were a maximum mix of two colonies (Helicobacter and campylobacter). Eighty‐nine percent (eight out of nine positive samples) of samples were collected from dogs with gastrointestinal disorders. The consistency of the results of culture and PCR was demonstrated in nine samples. Four samples shown negative by culture but positive by PCR were isolated from the dogs without gastrointestinal disorders. There was no significant difference between the two diagnostic methods (p > 0.05). Besides, kappa statistic was 0.8 (p < 0.001). The molecular prevalence rate of Helicobacter spp. was 13% (95% CI: 6.4%–19.6%); however, none of them was detected as H. pylori by multiplex‐PCR (Figure 1). In H. heilmannii species‐specific PCR, 38.5% (5/13 samples), and in H. billis species‐specific PCR, 15.4% (2/13 samples) were positive. In all the positive samples, one single Helicobacter species was present. Eighty percent of H. heilmannii positive cases were related to dogs with gastrointestinal disorders, and 100% of H. billis positive cases were related to dogs without gastrointestinal disorders. The results of H. heilmannii and H. billis PCR are displayed in Figures 2 and 3, respectively.
FIGURE 1.
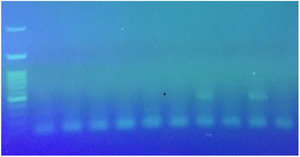
Results of Helicobacter spp. and H. pylori infection by multiplex‐polymerase chain reaction (PCR). Lane 1: 100 bp ladder, lane 2: negative control, lane 3: Helicobacter spp. (389 bp) and H. pylori (1200 bp) positive control, lane 6: Helicobacter spp. positive samples, lanes 4, 5, 7, 8 and 9: negative samples
FIGURE 2.
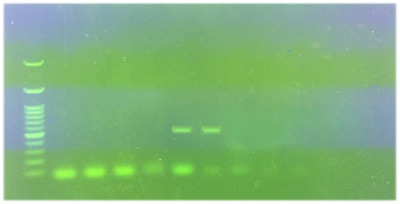
Results of H. heilmannii polymerase chain reaction (PCR) detection. Lane 1: 100 bp ladder, lane 2: negative control, lane 3: positive control (580 bp), lanes 5 and 6: positive sample, lanes 4, 7 and 8: negative samples
FIGURE 3.
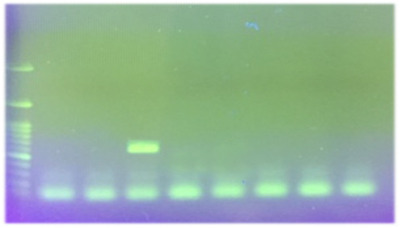
Results of H. billis polymerase chain reaction (PCR) detection. Lane 1: 100 bp ladder, lane 2: positive control (638 bp), lane 3: negative control, lane 4: positive sample, lanes 5, 6, 7 and 8: negative samples
3.2. Risk factors of Helicobacter spp. infections
The statistical analysis showed that the infection was correlated with age orders and increased with aging (p < 0.05). Univariate logistic regression revealed the odds of infection between the age, based on year, and the disease to be 2.42 (95% CI: 1.22%–4.79%); as one year of age increased, the odds of infection increased by 142%. Moreover, 12.5% of fluctuation in the infection was justified by the age factor (Table 2). The prevalence of Helicobacter spp. in male and female dogs was found to be 15.9% and 10.7%, respectively; however, the χ 2 test showed that the difference was not significant (p > 0.05). The odds of infection in males was 1.58 (95% CI: 0.49%–5.08%) compared to those of females. Furthermore, 1.1% of fluctuation in infection was justified by the gender factor (Table 2). The relative frequency of positive cases varied from 0% to 33.3% in the studied breeds, but this difference is not statistically significant (p > 0.05). However, 26.2% of fluctuation of infection was justified by the breed (Table 2). The χ 2 test showed that there was no statistical relationship between the infection and the habitat (p > 0.05). The odds of infection in dogs kept in apartments was 1.13 (95% CI: 0.34%–3.74%) compared to the dogs kept in the yard. Besides, 0.1% of fluctuation in infection was justified by habitat (Table 2). The statistical analysis showed that there was no statistical relationship between the infection and gastrointestinal disorders (p > 0.05). The odds of infection in dogs with gastrointestinal disorders was 1.71 (95% CI: 0.22%–5.66%) compared to dogs without gastrointestinal disorders. That is, 1.5% of fluctuation in infection was justified by gastrointestinal disorders (Table 2).
TABLE 2.
Prevalence of Helicobacter spp. in dogs in the southwest of Iran based on age, sex, breed, keeping place and gastrointestinal disorders
| Category | Groups | Prevalence | OR | 95% CI for OR | p‐Value |
|---|---|---|---|---|---|
| Age (year) | <2 b | 5.3% (3/57) | |||
| ≥2a | 23.3% (10/43) | 2.42 | 1.22–4.79 | <0.05 | |
| Sex | Femalea | 10.7% (6/56) | 1 | – | – |
| Malea | 15.9% (7/44) | 1.58 | 0.49–5.08 | >0.05 | |
| Breed | Dobermana | 0% (0/17) | – | – | – |
| Spitza | 0% (0/10) | – | – | – | |
| Pitbulla | 0% (0/8) | – | – | – | |
| German Shepherd a | 7.7% (1/13) | 1 | – | – | |
| Siberian Husky a | 11.1% (1/9) | 1.5 | 0.08–27.61 | >0.05 | |
| Terrier a | 24% (6/25) | 3.79 | 0.41–35.49 | >0.05 | |
| Dachshund a | 26.7% (4/15) | 4.36 | 0.42–45.26 | >0.05 | |
| Rottweiler a | 33.3% (1/3) | 6 | 0.26–140.05 | >0.05 | |
| Habitat | Yarda | 12.2% (5/41) | 1 | – | – |
| Apartmenta | 13.6% (8/59) | 1.13 | 0.34–3.74 | >0.05 | |
| Gastrointestinal disorders | Noa | 10% (5/50) | 1 | – | – |
| Yesa | 16% (8/50) | 1.71 | 0.52–5.66 | >0.05 |
Note: The different lowercase letters in each variable represent a significant difference.
Abbreviations: CI, confidence interval; OR, odds ratio.
Multivariate logistic regression showed that 43.7% of the fluctuation of infection was justified by the factors such as age, sex, breed, gastrointestinal disorders and habitat. However, in backward stepwise logistic regression, only the age factor was significantly related to the infection (p = 0.012) (Hosmer and Lemeshow test: χ 2 = 4.11, df = 4, p = 0.39).
4. DISCUSSION
This pioneering epidemiological survey, in addition to determining Helicobacter spp. in feces, identified some of the factors associated with its occurrence in companion dogs in the southwest of Iran. Understanding the epidemiology of this organism is definitely an essential key to establishing appropriate prevention strategies.
In this study, using culture and multiplex‐PCR methods to assess faecal samples, Helicobacter spp. was identified in 9% and 13% of the dogs, respectively; however, this observed difference was not statistically significant. According to other studies, prevalence of Helicobacter spp. DNA in feces was reported to be 100% by Ekman et al. (2013), 62.5% by Hong et al. (2015) and 23.3% by Jankowski et al. (2016a). In gastrointestinal biopsies of dogs with gastritis by PCR, the prevalence of Helicobacter spp. in Japan and Poland was reported as 34.7% and 100%, respectively (Jankowski et al., 2016a; Kubota‐Aizawa et al., 2017). The quantitative polymerase chain reaction (qPCR), histological, histochemical and immunohistochemical evaluations on gastric samples revealed that Helicobacter spp. was present in 47.8%, 65.2%, 75.4% and 82.6% of dogs, respectively (Amorim et al., 2015). The relative frequency of Helicobacter spp. in oral cavity (dental plaque and saliva) samples was 71.1%–100% (Ekman et al., 2013; Jankowski et al., 2016a; Recordati et al., 2007a). In Brazil, Helicobacter spp. infection has been reported as 94.7% and 100% by rapid urease test and histological analysis (Okubo et al., 2017). The large discrepancy in the frequency percentage of Helicobacter spp. may depend on the diagnostic method, specimen type, sample size and management and environmental factors (Chung et al., 2014; Falsafi et al., 2009; Hong et al., 2015; Kabir, 2001; Shinozaki et al., 2002; Smith et al., 2012). The detection of Helicobacter spp. in faecal samples is performed by microbiological culture, determination of anti‐Helicobacter antibodies and bacterial DNA using PCR (Falsafi et al., 2009; Mishra et al., 2008; Smith et al., 2012). Microbiological cultures are not performed routinely to diagnose Helicobacter spp. in faecal samples since the sensitivity of this method is low, and H. pylori can be difficult to culture. Furthermore, faecal samples have a high concentration of other microorganisms and a low concentration of gastric Helicobacter spp.; in addition to this, the long‐time passage of bacteria through the gastrointestinal tract has harmful effects on its viability (Falsafi et al., 2009; Smith et al., 2012). The PCR can be used to detect Helicobacter spp. in faecal samples. The sensitivity and specificity of this method are between 69% and 94% and 97.1% and 100%, respectively. However, factors such as the small amount of gastric Helicobacter spp. in feces, a degradation of bacterial DNA in the large intestine, the presence of polymerase inhibitors such as complex polysaccharides, the DNA extraction method and type of PCR used affect its validity. Moreover, the PCR does not require a high concentration of bacteria and/or the presence of live bacteria. Furthermore, the use of a semi‐nested or nested‐PCR increases the accuracy of this method compared to the classical PCR (Mishra et al., 2008; Smith et al., 2012; Tonkic et al., 2012). According to Prachasilpchai et al. (2007), the percentage frequency of Helicobacter spp. detected by haematoxylin and eosin stain (H&E), Warthin Starry stain (WSS), immunohistochemistry (IHC) and PCR in canine stomach turned out to stand at 17.3%, 46.7%, 30.7% and 10.7%, respectively. Although the detection of Helicobacter spp. revealed a statistically meaningful difference between H&E and WSS, IHC and H&E and PCR and H&E, the result of PCR was not in stark contrast with that of WSS and IHC. Despite the fact that IHC was proved to be much more sensitive in pinpointing infection compared to H&E and WSS, it is relatively costly and demanding in terms of time and experience; thus, it is not being deployed for routine tests. However, H&E staining may have low sensitivity when few bacteria are present, so the use of special stains such as WSS facilitates histological identification of bacteria (Dunn et al., 1997). The recognition of the particular species or strains of Helicobacter is feasible via PCR due to being a straightforward, precise, time‐saving, automatic and highly efficient method. PCR is applicable to a wide array of samples ranging from gastric biopsies, gastric juice and dental plaque to feces. The sensitivity of this method is considerably higher than that of histology, bacterial culture and urease evaluation although the type of primer used can alter its sensitivity (Prachasilpchai et al., 2007; Sabbagh et al., 2019; Simpson et al., 1999). Finally, not a single ‘gold‐standard’ method exists for the detection of the infection induced by Helicobacter spp.; thus, its confirmation requires the joint application of at least two tests (Jankowski et al., 2017; Patel et al., 2014). Ekman et al. (2013) and Hong et al. (2015) used laboratory dogs that were kept together in the same kennel, thus facilitating the transmission of the Helicobacter spp. compared to the dogs living alone. In addition, the large percentage of Helicobacter spp. may be due to the small sample size. For example, the sample size in the surveys of Ekman et al. (2013), Hong et al. (2015) and Jankowski et al. (2016a) was 14, 8 and 30 dogs, respectively, which is much smaller than the sample size in the present study (100 dogs). Specimen type is also influential. For example, Jankowski et al. (2016a) showed that the frequency of Helicobacter spp. in saliva and gastric biopsy specimens was 76.6% and 100%, respectively. Also, Recordati et al. (2007) detected Helicobacter spp. DNA by nested PCR in 94.7% of gastric biopsies, 44.7% of dental plaque and 50% of saliva samples in dogs. The high prevalence shown in the studies by Hong et al. (2015) and Jankowski et al. (2016a) may be related to the fact that all dogs under the experiment had gastritis, whereas in the present study, only 50% of dogs had gastrointestinal disorders. In addition, the low prevalence of Helicobacter spp. in this study may reflect the regional differences such as management and environmental factors between Iran and other countries because the dogs in this study, with no previous antibiotic therapy background, were kept only at homes and did not have any close contact with other dogs and cats.
In this study, the relative frequency of H. heilmannii, H. billis and H. pylori was detected to be 38.5%, 15.4% and 0%, respectively. H. billis is grouped in enterohepatic Helicobacter; thus, its isolation was expected from feces but H. pylori and H. heilmannii are grouped in gastric helicobacter, so their isolation may not be expected. Also, in all the positive dogs, one single Helicobacter species was present. Similarly, the frequency percentage of H. canis and H. billis in faecal samples of 14 clinically normal Beagles held for educational purposes was 100% and 14.3%, respectively, but that of H. pylori, H. felis, H. salomonis and H. bizzozeronii was 0% (Ekman et al., 2013). Moreover, the prevalence of H. heilmannii and H. salomonis in faecal samples of 30 dogs with gastritis was 71.4% and 28.6%, respectively, but that of H. felis, H. pylori and H. bizzozeronii was 0% (Jankowski et al., 2016a). Hong et al. (2015) reported that the frequency of H. heilmannii and H. felis was 37.5% and 25%, respectively. The prevalence of H. heilmannii, H. salomonis, H. bizzozeronii, H. pylori and H. felis in saliva of 30 dogs with gastrointestinal disorders has been reported to be 95.7%, 17.4%, 13%, 8.7% and 4.4%, respectively (Jankowski et al., 2016a). H. pylori infection in dogs is rare. For example, in one study, H. pylori infection was found in 144 dogs with gastrointestinal diseases (Kubota‐Aizawa et al., 2017). In addition, Abdel‐Raouf et al. (2014) reported 41.1%, 42.9% and 50% infection caused by H. pylori in stool, saliva and stomach juice samples of dogs by microbiological culture, respectively; in the same vein, Elhariri et al. (2017) reported 37.2% infection caused by this bacterium in serology. However, the failure of the serological methods in differentiating between the current infection and the previous exposure is regarded as the main shortcoming due to resulting in misinterpretation (Sabbagh et al., 2019). In addition, another source of misinterpretation is the cross‐reacting antigens, especially flagellar proteins, residing between H. pylori and campylobacters (Mégraud & Lehours, 2007).
In the present study, age unlike gender, breed, gastrointestinal disorders and habitat was significantly related to the infection caused by Helicobacter spp. Regarding the age, the higher infection rate in older dogs was probably due to a greater exposure to the Helicobacter spp. over time. In the present study, the relative frequency of positive samples in dogs with gastrointestinal disorders was not statistically higher than those without gastrointestinal disorders, thus proving that Helicobacter infection could be asymptomatic in dogs. In several studies, no significant relationship was detected between infection and age, gender, gastrointestinal disorders and domestic habitat (Ekman et al., 2013; Elhariri et al., 2017; Okubo et al., 2017; Recordati et al., 2007). However, Kubota‐Aizawa et al. (2017) showed that dogs positive for Helicobacter spp. had a significantly higher frequency of chronic diarrhoea than the negative dogs. A clear relationship between the presence of Helicobacter spp. and both mild to moderate epithelial injury and mild to moderate intraepithelial lymphocyte infiltration of the canine stomach has been reported by Amorim et al. (2015). Hwang et al. (2002) showed that the total positive rate in clinically abnormal dogs is significantly higher compared to the clinically normal dogs in the urease test and PCR. It should be kept in mind that although the veterinary hospital in Ahvaz is the most important and the largest veterinary centre in southwest of Iran, the findings might not reflect the whole dog population in Ahvaz. Also, due to the significant size of the sample in this study (100 dogs and bacterial culture as well as direct PCR on their feces) and limited financial resources and time, only the prevalence of three zoonotic species of Helicobacter spp. was evaluated, and thus other species such as H. bizzozeronii, H. salomonis, H. canis and H. felis will be investigated in the upcoming research.
5. CONCLUSION
The rate of Helicobacter spp. infection in feces of companion dogs with and without gastrointestinal disorders is scant but noticeable, especially in zoonotic species such as H. billis and H. heilmannii. Canine feces can be a potential source of Helicobacter spp.; therefore, dog owners should be educated about the risks and transmission modes of bacteria by veterinarians in the southwest of Iran. In addition, the present study indicated that the transmission risk of H. pylori through feces of dogs appears to be negligible.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was ‘observational study’ and the research protocol was reviewed and approved by research committee of the Faculty of Veterinary Medicine, Shahid Chamran University of Ahvaz and documented by the number 915824. Before beginning work on the present study, the dog owners were briefed on the purpose of the research and written informed consent was obtained from the owners for the participation of their animals in this study. During collecting rectal swabs, these animals were not disturbed.
AUTHOR CONTRIBUTIONS
Data curation (equal), investigation (equal), methodology (equal), validation (equal), visualization (equal) and writing—review and editing (equal): Mahdi Fatemi Khader. Conceptualization (equal), data curation (equal), formal analysis (lead), investigation (equal), methodology (equal), project administration (lead), resources (equal), software (equal), supervision (lead), validation (equal), visualization (equal), writing—original draft preparation (equal) and writing—review and editing (equal): Mahdi Pourmahdi Borujeni. Conceptualization (equal), data curation (equal), investigation (equal), methodology (equal), resources (equal), software (equal), validation (equal), visualization (equal), writing—original draft preparation (equal) and writing—review and editing (equal): Naghmeh Moori Bakhtiari. Data curation (equal), investigation (equal), methodology (equal), resources (equal) and writing—review and editing (equal): Reza Avizeh.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.765.
ACKNOWLEDGEMENT
The authors would like to express their sincere terms of gratitude to the staff of the Veterinary Hospital of Shahid Chamran University of Ahvaz and all the dog owners for their kind cooperation in sampling.
Fatemi Khader, M. , Pourmahdi Borujeni, M. , Moori Bakhtiari, N. , & Avizeh, R. (2022). An exploratory study on the presence of Helicobacter heilmannii and Helicobacter billis in the feces of companion dogs. Veterinary Medicine and Science, 8, 537–545. 10.1002/vms3.765
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abdel‐Raouf, M. , Abdel‐Gleel, Y. , & Enab, A. (2014). Study on the role of pet animals for Helicobacter pylori transmission. Journal of American Science, 10, 20–28. [Google Scholar]
- Agüloğlu, S. , Turhanoğlu, M. , Eskimez, S. , & Tacir, I. (2006). Detection of Helicobacter pylori colonization in human dental plaques and saliva of patients with chronic gastritis. Biotechnology and Biotechnological Equipment, 20, 173–178. [Google Scholar]
- Amorim, I. , Smet, A. , Alves, O. , Teixeira, S. , Saraiva, A. L. , Taulescu, M. , Reis, C. , Haesebrouck, F. , & Gärtner, F. (2015). Presence and significance of Helicobacter spp. in the gastric mucosa of Portuguese dogs. Gut Pathogens, 7, 12. 10.1186/s13099-015-0057-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros, F. C. , Ghosh, M. , Huang, L. , Masubuchi, N. , Cave, D. R. , & Grubel, P. (2000). Evaluation of a PCR primer based on the isocitrate dehydrogenase gene for detection of Helicobacter pylori in feces. Journal of Clinical Microbiology, 38, 3755–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafmodarres, F. , Pourmahdi Borujeni, M. , Avizeh, R. , Gharibi, D. , & Hashemi, S. J. (2017). Seroepidemiological study of Helicobacter pylori in related and non‐related people with dogs and cats in Ahvaz (2014–2015). Journal of Veterinary Research, 72, 137–145. 10.22059/jvr.2017.62598 [DOI] [Google Scholar]
- Bonfigli, A. R. , Boemi, M. , Festa, R. , Bonazzi, P. , Brandoni, G. , Spazzafumo, L. , Olivieri, F. , Ceriello, A. , Genovese, S. , & Testa, R. (2016). Randomized, double‐blind, placebo controlled trial to evaluate the effect of Helicobacter pylori eradication on glucose homeostasis in type 2 diabetic patients. Nutrition, Metabolism & Cardiovascular Diseases, 26, 893–898. 10.1016/j.numecd.2016.06.012 [DOI] [PubMed] [Google Scholar]
- Buczolits, S. , Hirt, R. , Rosengarten, R. , & Busse, H. J. (2003). PCR‐based genetic evidence for occurrence of Helicobacter pylori and novel Helicobacter species in the canine gastric mucosa. Veterinary Microbiology, 95, 259–270. 10.1016/s0378-1135(03)00182-2 [DOI] [PubMed] [Google Scholar]
- Census (2016). Census of the Islamic Republic of Iran, Statistical Centre of Iran. https://www.amar.org.ir/english/Population‐and‐Housing‐Censuses
- Choi, Y. K. , Han, J. H. , & Joo, H. S. (2001). Identification of novel Helicobacter species in pig stomachs by PCR and partial sequencing. Journal of Clinical Microbiology, 39, 3311–3315. 10.1128/JCM.39.9.3311-3315.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, T. H. , Kim, H. D. , Lee, Y. S. , & Hwang, C. Y. (2014). Determination of the prevalence of Helicobacter heilmannii‐like organisms Type 2 (HHLO‐2) infection in humans and dogs using non‐invasive genus/species‐specific PCR in Korea. Journal of Veterinary Medical Science, 76, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, J. C. , McNulty, C. A. , Uff, J. C. , Wilkinson, S. P. , & Gear, M. W. (1987). Spiral organisms in the gastric antrum. Lancet (London, England), 2(8550), 96. 10.1016/s0140-6736(87)92754-1 [DOI] [PubMed] [Google Scholar]
- Didehban, N. , Pourmahdi Borujeni, M. , Avizeh, R. , & Mosallanejad, B. (2020). Problematic behaviors in companion dogs: A survey of their prevalence and associated factors. Journal of Veterinary Behavior, 39, 6–13. 10.1016/j.jveb.2020.06.003 [DOI] [Google Scholar]
- Di Simone, N. , Tersigni, C. , Cardaropoli, S. , Franceschi, F. , Di Nicuolo, F. , Castellani, R. , Bugli, F. , de Waure, C. , Cavaliere, A. F. , Gasbarrini, A. , Sanguinetti, M. , Scambia, G. , & Todros, T. (2017). Helicobacter pylori infection contributes to placental impairment in preeclampsia: Basic and clinical evidences. Helicobacter, 22, e12347. 10.1111/hel.12347 [DOI] [PubMed] [Google Scholar]
- Dowsett, S. A. , & Kowolik, M. J. (2003). Oral Helicobacter pylori: Can we stomach it? Critical Reviews in Oral Biology & Medicine, 14, 226–233. [DOI] [PubMed] [Google Scholar]
- Dunn, B. E. , Cohen, H. , & Blaser, M. J. (1997). Helicobacter pylori. Clinical Microbiology Reviews, 10, 720–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman, E. , Fredriksson, M. , & Trowald‐Wigh, G. (2013). Helicobacter spp. in the saliva, stomach, duodenum and faeces of colony dogs. The Veterinary Journal, 195, 127–129. 10.1016/j.tvjl.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Elhariri, M. , Elhelw, R. , Hamza, D. , & El‐Mahallawy, H. S. (2017). Serologic evidence and risk factors for Helicobacter pylori infection in animals and humans. Journal of Infection in Developing Countries, 11, 414–419. 10.3855/jidc.9339 [DOI] [PubMed] [Google Scholar]
- Fallah, S. , Ahmadi, R. , Moradi, N. , Fadaei, R. , Sezavar, S. H. , & Seifi, M. (2016). Helicobacter pylori infection and iron deficiency in patients with coronary artery disease. Cellular and Molecular Biology, 62, 8–14. [PubMed] [Google Scholar]
- Falsafi, T. , Favaedi, R. , Mahjoub, F. , & Najafi, M. (2009). Application of Stool‐PCR test for diagnosis Helicobacter pylori infection in children. World Journal of Gastroenterology, 15, 484–488. 10.3748/wjg.15.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshad, S. , Rasouli, M. , & Alborzi, A. (2004). Simultaneous detection of Helicobacter genus and Helicobacter pylori species using a multiplex PCR method. Iranian Biomedical Journal, 8, 205–209. [Google Scholar]
- Fox, J. G. , & Wang, T. C. (2002). Helicobacter pylori infection: Pathogenesis. Current Opinion in Gastroenterology, 18, 15–25. [DOI] [PubMed] [Google Scholar]
- Fox, J. G. , Yan, L. L. , Dewhirst, F. E. , Paster, B. J. , Shames, B. , Murphy, J. C. , Hayward, A. , Belcher, J. C. , & Mendes, E. N. (1995). Helicobacter bilis sp. nov., a novel Helicobacter isolated from bile, livers, and intestines of aged, inbred mouse strains. Journal of Clinical Microbiology, 33, 445–454. 10.1128/jcm.33.2.445-454.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, K. L. , Chan, W. K. , Shiota, S. , & Yamaoka, Y. (2011). Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter, 16, 1–9. 10.1111/j.1523-5378.2011.00874.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesebrouck, F. , Pasmans, F. , Flahou, B. , Chiers, K. , Baele, M. , Meyns, T. , Decostere, A. , & Ducatelle, R. (2009). Gastric Helicobacters in domestic animals and nonhuman primates and their significance for human health. Clinical Microbiology Reviews, 22, 202–223. 10.1128/CMR.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S. , Chung, Y. , Kang, W. G. , Choi, Y. S. , & Kim, O. (2015). Comparison of three diagnostic assays for the identification of Helicobacter spp. in laboratory dogs. Laboratory Animal Research, 31, 86–92. 10.5625/lar.2015.31.2.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooi, J. K. Y. , Lai, W. Y. , Ng, W. K. , Suen, M. M. Y. , Underwood, F. E. , Tanyingoh, D. , Malfertheiner, P. , Graham, D. Y. , Wong, V. W. S. , Wu, J. C. Y. , Chan, F. K. L. , Sung, J. J. Y. , Kaplan, G. G. , & Ng, S. C. (2017). Global prevalence of Helicobacter pylori infection: Systematic review and meta‐analysis. Gastroenterology, 153, 420–429. 10.1053/j.gastro.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Hwang, C. Y. , Han, H. R. , & Youn, H. Y. (2002). Prevalence and clinical characterization of gastric Helicobacter species infection of dogs and cats in Korea. Journal of Veterinary Science, 3, 123–133. [PubMed] [Google Scholar]
- Jalava, K. , Kaartinen, M. , Utriainen, M. , Happonen, I. , & Hanninen, M. L. (1997). Helicobacter salomonis sp. nov., a canine gastric Helicobacter sp. related to Helicobacter felis and Helicobacter bizzozeronii . International Journal of Systematic Bacteriology, 47, 975–982. [DOI] [PubMed] [Google Scholar]
- Jankowski, M. , Spużak, J. , Kubiak, K. , Glińska‐Suchocka, K. , & Biernat, M. (2016a). Detection of Helicobacter spp. in the saliva of dogs with gastritis. Polish Journal of Veterinary Sciences, 19, 133–140. 10.1515/pjvs-2016-0017 [DOI] [PubMed] [Google Scholar]
- Jankowski, M. , Spużak, J. , Kubiak, K. , Glińska‐Suchocka, K. , & Biernat, M. (2016b). Detection of gastric Helicobacter spp. in stool samples of dogs with gastritis. Polish Journal of Veterinary Sciences, 19, 237–243. 10.1515/pjvs-2016-0030 [DOI] [PubMed] [Google Scholar]
- Jankowski, M. , Spużak, J. , Kubiak, K. , Glińska‐Suchocka, K. , & Biernat, M. (2017). An evaluation of the usefulness of invasive and non‐invasive methods used to diagnose Helicobacter spp. infections in dogs. Polish Journal of Veterinary Sciences, 20, 491–499. 10.1515/pjvs-2017-0059 [DOI] [PubMed] [Google Scholar]
- Kabir, S. (2001). Detection of Helicobacter pylori in faeces by culture, PCR and enzyme immunoassay. Journal of Medical Microbiology, 50, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kreader, C. A. (1996). Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Applied and Environmental Microbiology, 62, 1102–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota‐Aizawa, S. , Ohno, K. , Fukushima, K. , Kanemoto, H. , Nakashima, K. , Uchida, K. , Chambers, J. K. , Goto‐Koshino, Y. , Watanabe, T. , Sekizaki, T. , Mimuro, H. , & Tsujimoto, H. (2017). Epidemiological study of gastric Helicobacter spp. in dogs with gastrointestinal disease in Japan and diversity of Helicobacter heilmannii sensu stricto. Veterinary Journal, 225, 56–62. 10.1016/j.tvjl.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Lee, A. , Krakowka, S. , Fox, J. G. , Otto, G. , Eaton, K. A. , & Murphy, J. C. (1992). Role of Helicobacter felis in chronic canine gastritis. Veterinary Pathology, 29, 487–494. [DOI] [PubMed] [Google Scholar]
- Malfertheiner, P. , Megraud, F. , O'Morain, C. A. , Gisbert, J. P. , Kuipers, E. J. , Axon, A. T. , Bazzoli, . F. , Gasbarrini, A. , Atherton, J. , Graham, D. Y. , Hunt, R. , Moayyedi, P. , Rokkas, T. , Rugge, M. , Selgrad, M. , Suerbaum, S. , Sugano, K. , & El‐Omar, E. M. (2017). European Helicobacter and Microbiota study group and consensus panel. Management of Helicobacter pylori infection‐the Maastricht V/Florence consensus report. Gut, 66, 6–30. 10.1136/gutjnl-2016-312288 [DOI] [PubMed] [Google Scholar]
- Marshall, B. J. , & Warren, J. R. (1984). Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet (London, England), 1(8390), 1311–1315. 10.1016/s0140-6736(84)91816-6 [DOI] [PubMed] [Google Scholar]
- Mégraud, F. , & Lehours, P. (2007). Helicobacter pylori detection and antimicrobial susceptibility testing. Clinical Microbiology Reviews, 20, 280–322. 10.1128/CMR.00033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, S. , Singh, V. , Rao, G. R. , Jain, A. K. , Dixit, V. K. , Gulati, A. K. , & Nath, N. (2008). Detection of Helicobacter pylori in stool specimens: Comparative evaluation of nested PCR and antigen detection. The Journal of Infection in Developing Countries, 2, 206–210. 10.3855/jidc.264 [DOI] [PubMed] [Google Scholar]
- Mladenova‐Hristova, I. , Grekova, O. , & Patel, A. (2017). Zoonotic potential of Helicobacter spp. Journal of Microbiology, Immunology and Infection, 50, 265–269. 10.1016/j.jmii.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Morgner, A. , lehn, N. , Andersen, L. P. , Thiede, C. , Bennedsen, M. , Trebesius, K. , Neubauer, B. , Neubauer, A. , Stolte, M. , & Bayerdörfferet, E. (2000). Helicobacter heilmannii‐associated primary gastric low‐grade MALT lymphoma: Complete remission after curing the infection. Gastroenterology, 118, 821–828. [DOI] [PubMed] [Google Scholar]
- Neiger, R. , Dieterich, C. , Burnens, A. , Waldvogel, A. , Corthésy‐Theulaz, I. , Halter, F. , Lauterburg, B. , & Schmassmann, A. (1998). Detection and prevalence of Helicobacter infection in pet cats. Journal of Clinical Microbiology, 36, 634–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo, B. M. , Ricci‐Azevedo, R. , Zobiole, N. N. , Buccini, D. F. , & Moreno, S. E. (2017). Prevalence of Helicobacter spp. in dogs from Campo Grande‐Ms. Ciência Animal Brasileira, 18, e17286. 10.1590/1089-6891v18e-17286 [DOI] [Google Scholar]
- Patel, S. K. , Pratap, C. B. , Jain, A. K. , Gulati, A. K. , & Nath, G. (2014). Diagnosis of Helicobacter pylori: What should be the gold standard? World Journal of Gastroenterology, 20, 12847–12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachasilpchai, W. , Nuanualsuwan, S. , Chatsuwan, T. , Techangamsuwan, S. , Wangnaitham, S. , & Sailasuta, A. (2007). Diagnosis of Helicobacter spp. infection in canine stomach. Journal of Veterinary Science, 8, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl, D. , Keller, P. M. , & Bordier, V. (2019). Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World Journal of Gastroenterology, 25, 4629–4660. 10.3748/wjg.v25.i32.4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin, M. J. , Eaton, K. A. , Krakowka, S. , Morgan, D. R. , Lee, A. , Otto, G. , & Fox, J. G. (1990). Helicobacter pylori gastric infection in gnotobiotic beagle dogs. Infection and Immunity, 58, 2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recordati, C. , Gualdi, V. , Craven, M. , Sala, L. , Luini, M. , Lanzoni, A. , Rishniw, M. , Simpson, K. W. , & Scanziani, E. (2009). Spatial distribution of Helicobacter spp. in the gastrointestinal tract of dogs. Helicobacter, 14, 180–191. 10.1111/j.1523-5378.2009.00674.x [DOI] [PubMed] [Google Scholar]
- Recordati, C. , Gualdi, V. , Tosi, S. , Facchini, R. V. , Pengo, G. , Luini, M. , Simpson, K. W. , & Scanziani, E. (2007). Detection of Helicobacter spp. DNA in the oral cavity of dogs. Veterinary Microbiology, 119, 346–351. [DOI] [PubMed] [Google Scholar]
- Rossi, M. , Hänninen, M. L. , Revez, J. , Hannula, M. , & Zanoni, R. G. (2008). Occurrence and species level diagnostics of Campylobacter spp., enteric Helicobacter spp. and Anaerobiospirillum spp. in healthy and diarrheic dogs and cats. Veterinary Microbiology, 129, 304–314. 10.1016/j.vetmic.2007.11.014 [DOI] [PubMed] [Google Scholar]
- Sabbagh, P. , Mohammadnia‐Afrouzi, M. , Javanian, M. , Babazadeh, A. , Koppolu, V. , Vasigala, V. R. , Nouri, H. R. , & Ebrahimpour, S. (2019). Diagnostic methods for Helicobacter pylori infection: Ideals, options, and limitations. European Journal of Clinical Microbiology & Infectious Diseases, 38, 55–66. 10.1007/s10096-018-3414-4 [DOI] [PubMed] [Google Scholar]
- Shinozaki, J. K. , Sellon, R. K. , Cantor, G. H. , Besser, T. E. , Mealey, K. L. , & Vaden, S. L. (2002). Fecal polymerase chain reaction with 16S ribosomal RNA primers can detect the presence of gastrointestinal Helicobacter in dogs. Journal of Veterinary Internal Medicine, 16, 426–432. [DOI] [PubMed] [Google Scholar]
- Simpson, K. W. , Strauss‐Ayali, D. , McDonough, P. L. , Chang, Y. F. , & Valentine, B. A. (1999). Gastric function in dogs with naturally acquired gastric Helicobacter spp. infection. Journal of Veterinary Internal Medicine, 13, 507–515. [DOI] [PubMed] [Google Scholar]
- Smith, S. , Fowora, M. A. , Lesi, O. A. , Agbebaku, E. , Odeigah, P. , Abdulkareem, F. B. , Onyekwere, C. A. , Agomo, C. A. , & Contreras, M. (2012). Application of stool‐PCR for the diagnosis of Helicobacter pylori from stool in Nigeria—A pilot study. Springerplus, 1, 78. 10.1186/2193-1801-1-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick, J. V. , O'Rourke, J. , Lee, A. , Paster, B. J. , Dewhirst, F. E. , & Tompkins, L. S. (1993). An uncultured gastric spiral organism is a newly identified Helicobacter in humans. The Journal of Infectious Diseases, 168, 379–385. 10.1093/infdis/168.2.379 [DOI] [PubMed] [Google Scholar]
- Tonkic, A. , Tonkic, M. , Lehours, P. , & Mégraud, F. (2012). Epidemiology and diagnosis of Helicobacter pylori infection. Helicobacter, 17, 1–8. 10.1111/j.1523-5378.2012.00975.x [DOI] [PubMed] [Google Scholar]
- Upala, S. , Jaruvongvanich, V. , Riangwiwat, T. , Jaruvongvanich, S. , & Sanguankeo, A. (2016). Association between Helicobacter pylori infection and metabolic syndrome: A systematic review and meta‐analysis. Journal of Digestive Diseases, 17, 433–440. 10.1111/1751-2980.12367 [DOI] [PubMed] [Google Scholar]
- Van den Bulck, K. , Decostere, A. , Baele, M. , Driessen, A. , Debongnie, J. C. , Burette, A. , Stolte, M. , Ducatelle, R. , & Haesebroucket, F. (2005). Identification of non‐Helicobacter pylori spiral organisms in gastric samples from humans, dogs, and cats. Journal of Clinical Microbiology, 43, 2256–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


