Abstract
Although obesity has been a longstanding health crisis, the genetic architecture of the disease remains poorly understood. Genome-wide association studies have identified many genomic loci associated with obesity, with genes being enriched in the brain, particularly in the hypothalamus. This points to the role of the central nervous system (CNS) in predisposition to obesity, and we emphasize here several key genes along the satiety signaling pathway involved in genetic susceptibility. Interest has also risen regarding the chronic, low-grade obesity-associated inflammation, with a growing concern towards inflammation in the hypothalamus as a precursor to obesity. Recent studies have found that genetic variation in inflammatory genes play a role in obesity susceptibility, and we highlight here several key genes. Despite the interest in the genetic variants of these pathways individually, there is a lack of research that investigates the relationship between the two. Understanding the interplay between genetic variation in obesity genes enriched in the CNS and inflammation genes will advance our understanding of obesity etiology and heterogeneity, improve genetic risk prediction analyses, and highlight new drug targets for the treatment of obesity. Additionally, this increased knowledge will assist in physician’s ability to develop personalized nutrition and medication strategies for combating the obesity epidemic. Though it often seems to present universally, obesity is a highly individual disease, and there remains a need in the field to develop methods to treat at the individual level.
Keywords: Polygenic obesity, hypothalamus, satiety, inflammation, neuronal genes, precision medicine
Introduction
Obesity is a major public health crisis, on the rise in the United States and worldwide. In 2018 nearly 20% of children and over 40% of adults in the United States were considered obese with the adult rate of obesity predicted to rise to 50% by 2030 [1–3]. Obesity is associated with numerous health, social, and economic consequences, both at an individual and societal level [4–6]. Increased adiposity correlates with an increased risk of total mortality, highlighting the need to better understand the disease to improve treatment development [4].
Obesity is a complex disease resulting from genetic and environmental factors. The heritability of obesity has been studied for a number of years, but much of the genetic architecture underlying the disease continues to elude us [7]. Estimates on the heritability of obesity range from 30% to 90%, with most falling around 70% [8]. Despite the prevalence of obesity, the genetics are still poorly understood—there are more than 900 known low-risk, common genetic variants associated with body mass index (BMI) and over 300 variants associated with waist-to-hip ratio (WHR), but this still only explains a small fraction of variability and many of the underlying causal genes remain unknown [7, 9, 10]. Genome-wide association study (GWAS) findings for obesity are enriched in the central nervous system (CNS), and many of these genes play a role in synaptic function and neurotransmitter signaling [7, 11–13]. Better understanding the genetics of obesity is key for four reasons:
to better understand the biology for obesity development and progression to reveal novel pathways and targets for pharmacotherapy [14, 15]
to advance prediction analyses for genetic risk scores to better highlight and inform at-risk patients and environments [16–18]
to improve pharmacogenetics so patients can be matched with the most effective drug for them [19, 20]
to expand understanding of disease heterogeneity of obesity for researchers, physicians, and the general public to improve patient outcomes and quality of life [21–23].
Many homeostatic processes become dysregulated when fat mass accumulates, but one of the greatest areas of interest is the inflammation associated with increased adiposity [24]. Obesity is associated with a chronic, low-grade inflammation called meta-inflammation [25, 26]. Work has shown that inflammation develops very early in the hypothalamus of the brain when on a high-fat diet (HFD), separate from significant increases in fat and weight [27–29]. Recent findings indicate a role of genetics in modulating inflammatory response, and that this response may play a role in disease progression [30, 31]. For example, a study published in 2020 identified that a central regulator of inflammatory cell function, receptor-interacting serine/threonine-protein kinase 1 (RIPK1), is genetically associated with obesity and a potential therapeutic target for weight loss [32]. This review will focus on genetic contributions of neuronal genes to satiety signaling in obesity and the inflammatory response in the hypothalamus. Better connecting these two areas of research may reveal new pathways and targets for treating obesity and the numerous co-morbidities associated with it.
These are the first steps to developing personalized genomic strategies for treatment, as recent work shows that not everyone responds to a HFD in the same way [22, 23, 33]. More fully understanding the impact of genetic variation on obesity development, progression, and resolution will shed light on a path toward personalized nutrition for treating the obesity epidemic.
Satiety Signaling
The hypothalamus has been a main area of research for the role of the brain in obesity, in part due to its large role in most forms of monogenic obesity and its critical function in satiety and food intake. This region regulates energy homeostasis and systemic metabolism by integrating peripheral anorexigenic (appetite-reducing) and orexigenic (appetite-stimulating) signals to determine appetite [34]. The melanocortin system, consisting of two functionally antagonistic neuronal populations within the arcuate nucleus (ArcN) of the hypothalamus is key to this regulatory function. One subset of neurons expresses the orexigenic neuropeptides: neuropeptide Y (NPY) and Agouti-related peptide (AgRP; NPY/AgRP neurons). The other subset expresses the anorexigenic peptides: pro-opiomelanocortin (POMC) and cocaine and amphetamine regulated transcript (CART; POMC neurons).
As food moves through the gut, signals are sent to the brain via mechanoreceptors, chemoreceptors, and hormones such as cholecystokinin (CCK), glucagon-like peptide (GLP)-1, peptide YY (PYY), and ghrelin (Figure 1) [35–39]. CCK, GLP-1, and PYY are examples of anorexigenic peptides, while ghrelin is a well-known “hunger hormone”, as have been reviewed elsewhere [36–40]. The gut-derived anorexigenic peptides CCK and GLP-1 bind to their respective receptors to stimulate POMC neurons, while PYY binds to its receptors on NPY/AgRP neurons to inhibit signaling (Figure 1) [41–43]. Ghrelin, on the other hand, stimulates NPY/AgRP neurons and modulates expression of adenosine monophosphate-activated protein kinase (AMPK) to promote appetite [44, 45]. Satiety signals can also originate from other tissues, such as leptin from the adipose tissue. Leptin is perhaps the best known satiety signal between the brain and peripheral tissues, and its genetic influence on obesity will be discussed in greater detail later. In the brain, leptin binds leptin receptors on both NPY/AgRP and POMC neurons, triggering the activation of the Janus family of tyrosine kinases (JAK)-signal transducer and activator of transcription (STAT) signaling [46]. In cases of normal signaling, leptin inhibits NPY/AgRP and stimulates POMC neurons, leading to a sense of satiety via the synthesis of POMC and POMC’s subsequent cleavage to the melanocyte-stimulating hormones, α-MSH and β-MSH (Figure 1) [46]. α-MSH can then bind two different melanocortin receptors, melanocortin-3 receptor (MC3R) and melanocortin-4 receptor (MC4R) of the paraventricular nucleus (PVN) of the hypothalamus [47, 48]. Of the two, MC4R is more widely distributed throughout the central nervous system, and has been connected to monogenic and polygenic forms of obesity [13]. Changes in the function of these neuronal populations is known to occur rapidly following HFD feeding, in as little as under a week [49].
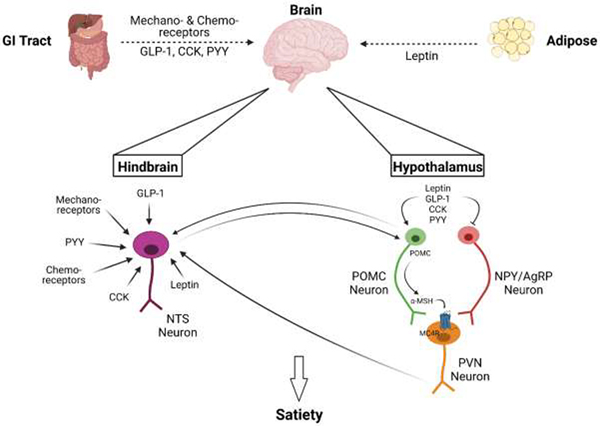
Figure 1. Broad Overview of Satiety Signaling.
The brain incorporates signals from multiple organs, primarily the gut and adipose, to determine appetite (dashed arrow). The gut communicates fullness via mechanoreceptors, chemoreceptors, and hormones including glucagon-like peptide-1 (GLP-1), cholecystokinin (CCK), and peptide YY (PYY). The adipokine leptin is also secreted from white adipose tissue to instigate anorexigenic signals. These signals are integrated in the hypothalamus and the hindbrain. In the hindbrain, neurons of the nucleus of the solitary tract (NTS) receive these signals and can then communicate with neurons in the hypothalamus (solid arrows). In the hypothalamus, there are primarily two neuronal populations: anorexigenic pro-opiomelanocortin (POMC) neurons and orexigenic neuropeptide Y/Agouti-related peptide (NPY/AgRP) neurons. Anorexigenic peptides inhibit NPY/AgRP neurons and stimulate POMC neurons, which then synthesize POMC to be subsequently cleaved to α-melanocyte-stimulating hormone (α-MSH), which then primarily binds to melanocortin-4 receptor (MC4R) of the paraventricular nucleus (PVN) neurons, which then releases downstream signals, including to the NTS, to induce satiety and restrict eating. Created with BioRender.com.
Importantly, a key endpoint of communication in these hypothalamic pathways is the hindbrain, which has been identified as another key hub of energy balance and satiety signaling integration. For example, the PVN neurons described above engage both the nucleus of the solitary tract (NTS) and the central lateral parabrachial nucleus (cLPBN) of the hindbrain [50, 51]. In fact, the NTS likely processes and receives the greatest number of energy status signals, and in turn communicates with the hypothalamus and the parabrachial nucleus [52, 53]. These include the signals from mechanoreceptors in the gut as well as many of the gut-derived hormones, such as CCK, GLP-1, ghrelin, and PYY, plus leptin from adipose (Figure 1) [54–56]. The hindbrain then integrates these multiple signals to control energy balance as reviewed elsewhere [52].
In conjunction to the role of the hypothalamus and hindbrain in homeostatic food intake and satiety, a growing body of evidence also implicates hedonic signaling in the obesity epidemic [12, 57, 58]. For example, genes within human GWAS obesity loci are enriched in the hippocampus [12]. The hippocampus may also play a role in meal size control [59], and hippocampal atrophy has repeatedly been linked to a HFD and obesity, and may alter responses to taste [60, 61]. In addition, the nucleus accumbens can influence food intake pathways, and has been the subject of study for obesity treatment [62–65]. A recent analysis showed strong gene expression enrichment of top obesity/BMI-associated loci in the insula and substantia nigra, regions involved in addiction, motivation, and reward-seeking behavior [66]. Furthermore, the hypothalamus connects with the insula in response to nutrient status and to drive the satiety response [67]. Work has demonstrated a positive relationship between higher BMI and greater hypothalamic gray matter volume and connectivity with the insula [68]. While hedonic signaling is not the focus of this review, it is important to mention its role and dysregulation in the development and maintenance of obesity. Very briefly, excessive dietary fat consumption disrupts the brain hedonic system by altering dopamine signaling to the point of overriding homeostatic mechanisms; more specific details have been described elsewhere [69]. These findings support research that indicates an association between obesity, obesity risk genes, and disordered eating patterns [70]. While further details lie outside the scope of this review, this highlights the necessity of expanding research to other neuron populations and different regions of the brain to determine the role of known genes and to continue identifying genetic factors influencing weight gain.
Monogenic Obesity Genes in the Brain
Ordinarily, the satiety signaling pathway is well-controlled. But deleterious mutations in any of the genes along the pathway often causes early onset, severe obesity, which has been extensively described elsewhere [71]. Of those genes expressed in the brain, the two that are perhaps the best studied are the leptin receptor (LEPR) and MC4R. Several single nucleotide variants in LEPR, including Lys109Arg and Gln223Arg are associated with severe obesity [72, 73]. Deficiencies in leptin and leptin receptor are extraordinarily severe but also rare, as is the case with most other mutations along the satiety signaling pathway in genes such as PCSK1, ADCY3, POMC, and SIM1 [74]. The most common gene implicated in monogenic obesity is MC4R: early estimates suggested that approximately 2.5% of severely obese individuals have a deleterious mutation in MC4R, with exact rates varying by populations [75–77]. Most recent analyses demonstrate that approximately 7% of the total population and over 10% of obese patients have a coding variant in MC4R [78]. Nearly 20% of reported single nucleotide variations in the MC4R gene have been predicted to be pathogenic, or likely pathogenic, highlighting the predominance of MC4R in monogenic obesity [71]. New work has uncovered how 19 rare mutations in MC4R disrupt its trafficking and signaling, including plasma membrane localization or interaction with Gαs protein [79]. While deleterious mutations in single genes along the satiety pathway explains a fair number of early onset, severe obesity cases, they do not explain the majority of obesity cases in the clinic, which typically present later in life. For most patients, individually small effects in many genes (polygenic obesity) culminate in an increased risk for obesity, rather than a large effect caused by just one gene. A select number of genes that are both present in the brain and have been implicated in polygenic obesity are discussed below.
Polygenic Obesity Genes in the Brain
As previously mentioned, GWAS has identified hundreds of variants that are associated with BMI and WHR [7, 9]. Although the causal genes for most loci remain unknown, many of the genes at or near these loci are enriched within the CNS, including the hypothalamus and pituitary gland, key sites of satiety regulation [12]. Genes are also enriched in the hippocampus and other areas of the limbic system that are likely to play a role in hedonic signaling of food intake [12, 13, 66]. Gene set enrichment and pathway analysis shows that CNS enriched genes are involved in synaptic function and neurotransmitter signaling [7, 12, 13]. Although to-date many genes identified through human GWAS act through neuronal processes to influence obesity, we have chosen three genes to describe in-depth: FTO, MC4R, and ADCY3.
The first and most significantly obesity-associated gene identified through GWAS is the aptly called fat mass and obesity-associated gene (FTO). A number of variants in FTO have been found in the first intron of the gene and are associated with increased body weight, body fat, energy intake, and other adiposity measures [80]. The best studied variant, rs9939609, has been found to increase risk for obesity 1.7-fold when subjects are homozygous for the obesity-risk “A” allele compared to the low-risk “T” allele [81]. However, the exact mechanism by which this allele increases obesity risk is still poorly understood. Early work demonstrated a relationship between the “obesity-risk” FTO variant and increased food—particularly fat—consumption in children [82, 83]. Research that followed suggested that variants in FTO may alter function of ghrelin, the appetite-promoting gut hormone, where subjects homozygous for the A allele had dysregulated circulating levels of acyl-ghrelin and a diminished postprandial appetite reduction (e.g., reduced satiety response) [84–86]. More recent data indicate that variants in FTO alter eating behavior not through ghrelin itself, but instead through how the central nervous system processes these satiety signals [85–87]. A current leading hypothesis suggests that this altered processing in the hypothalamus may be through connections between FTO and other genes, specifically retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) and the gene Iroquois homeobox 3 (IRX3) [88, 89]. RPGRIP1L is located proximal to FTO and both are regulated by the cut-like homeobox 1 (CUX1) transcription factor [90]. Enhancers in the first intron of FTO—where the most prominent obesity-related SNPs are located—act as long-range regulators for IRX3 gene expression, where presence of these risk alleles in FTO led to increased expression of IRX3 [90]. This connection has been of interest in recent years, and is more extensively reviewed elsewhere [88]. Importantly, it has recently been demonstrated that the FTO genotype may determine the success of weight loss through lifestyle modification, and this may be in part by how these variants allow lifestyle changes to alter expression of FTO and IRX3 [91, 92].
In addition to its association with monogenic obesity, variants in MC4R have been connected to polygenic obesity. As opposed to monogenic variants that truncate protein or severely alter protein folding/function, MC4R variants in polygenic obesity do not completely ablate or disrupt the protein. Indeed, certain polygenic variants within MC4R are beneficial: two coding region variants (Val103Iso and Iso251Leu) are negatively associated with obesity [93, 94]. These variants may affect signaling, as one study found that the presence of the Val103Ile variant led to a twofold decrease in antagonist (hAGRP) potency; other polymorphs were associated with differential responses to the endogenous agonists α-MSH and β-MSH [95]. Additional variants have been detected in GWAS, including the obesity-predisposing rs17782313, rs12970134, and rs476828 [96–98]. Just recently, a large study across 20,000 electronic MEdical Records and GEnomics (eMERGE) network participants and 77,000 independent individuals revealed 125 coding variants, including 30 novel variants [78]. This study confirmed not only the role of MC4R in polygenic obesity, but also heterogeneity in effect of the variants and prevalence in different populations [78]. The specific mutations in MC4R have been shown to influence the effectiveness of behavioral interventions and pharmacotherapy treatment. Patients with non-functional MC4R (monogenic obesity) do not respond well to bypass surgery but can respond to GLP-1 receptor agonism while patients with some functional MC4R (polygenic obesity) do respond to bypass surgery [99–102].
Other players in the melanocortin pathway also contribute to both monogenic and polygenic obesity, including adenylate cyclase isoform 3 (ADCY3), which generates the secondary messenger cAMP [103]. While ADCY3 was best known to participate in olfactory pathways, it has also been linked to adiposity and depression [104–108]. One early study identified a relationship between increased expression of ADCY3 and improved metabolic health in the Goto-Kakizaki rat, where two functional point mutations in the promoter region increased ADCY3 expression that, when combined with forskolin treatment, led to increased cAMP production in the islets, promoting systemic insulin sensitivity [109]. Follow-up studies identified polymorphisms in ADCY3 that confer risk susceptibility to obesity [105, 106, 110]. At the same time, a separate study demonstrated that global ADCY3 knock-out mice became obese due to leptin insensitivity, hyperphagia, and low locomotor activity—suggesting that the activity of ADCY3 in the hypothalamus influenced body weight regulation [111]. Since these initial findings, a number of studies have validated the relationship between ADCY3 function and adiposity: variants that increased expression and/or function of ADCY3 promote decreased adiposity and better metabolic health, while variants that decreased expression and/or function predispose to weight gain [107, 112, 113]. Whether these effects are due to a change in food intake or to peripheral metabolism varies depending on the model used [107, 112, 113]. Our lab recently identified a variant in rat Adcy3 that may be protective against obesity by altering transmembrane packing of the protein, and potentially altering membrane interactions and binding [114, 115]. Other work indicates that impaired signaling between MC4R and ADCY3 in the primary cilia of neurons can increase body weight [116]. Promisingly, it has been shown that genetic variation in ADCY3 alters response to diet. In a randomized trial testing two weight-lowering diets, presence of the G allele at rs10182181 correlated with an increased weight loss response on a low-fat diet, but a diminished response in a protein-rich diet [117]. This particular work highlights the importance an individual’s genetic variation may have not only on developing obesity, but also on weight loss.
Obesity and Hypothalamic Inflammation
We know that genetics can contribute to dysregulated satiety signaling, but could genetic variation also affect the inflammation associated with obesity? It has been previously shown that a subset of the obese population have increased adiposity, but none of the metabolic complications, including reduced inflammation (e.g., metabolically healthy obese) [118, 119]. While adipose tissue is the best studied organ for obesity-associated inflammation, analyses have shown that HFD-induced inflammatory processes develop in the brain well before significant weight gain [27, 28]. There is also evidence that levels of reactive oxygen species (ROS) in the brain increase with diet-induced obesity (DIO) [120]. Hypothalamic inflammation has been connected to obesity since 2005, where the largest group of mRNAs modulated by a HFD was identified as immune-related proteins such as tumor necrosis factor α (TNF-α), interleukin-6 (IL-6), and c-Jun N-terminal kinase 3 (JNK3; also known as mitogen-activated protein kinase (MAPK)-10) [121].
Rodent studies show that a long-term HFD results in hypothalamic inflammation, which in turn induces hypothalamic leptin resistance that perpetuates the development of obesity through increased food intake [27, 122, 123]. High fat diets specifically enriched in saturated fats have been established to have a direct role in inducing this central inflammation [124]. Not all saturated fats increase inflammation equally, either. Rats consuming long-chain saturated fats derived from butter showed overall higher hypothalamic inflammation than rats consuming medium-chain saturated fats derived from coconut oil [124]. It has been postulated that these differences are due to the differential β-oxidation rates of long-chain and medium-chain fatty acids; long-chain saturated fatty acids are less prone to β-oxidation, and may therefore lead to increased lipotoxicity and inflammation [124, 125]. Hypothalamic inflammation has also been shown in human patients, and further positively correlated with serum inflammatory proteins such as IL-6 and C-reactive protein (CRP) [122, 123]. Inappropriate inflammation in the brain can disrupt numerous signaling pathways including the appetite control pathways, disturbing energy homeostasis. This disruption may perpetuate the development and progression of obesity and its sequelae. Furthermore, inflammatory-related hypothalamic abnormalities, such as microstructure and gliosis, are positively associated with obesity, and aberrations in brain volume and connectivity may be predictive of weight loss success [126–131]. To better understand obesity then, we must also consider the role of hypothalamic inflammation.
Inflammation is a key feature of the body’s immune system responding to harmful stimuli, allowing injurious stimuli to be removed and initiating the healing process. Acute inflammation is generally protective, but in the case of obesity, the damaging stimuli cannot be so easily removed, leading to uncontrolled inflammation that becomes chronic and itself harmful. There are, of course, numerous pathways and interactions in inflammatory signaling pathways that have been well-described elsewhere [132]. We describe here only select pathways as they relate to inflammatory genes with known genetic variants and connections to obesity.
The inflammatory response can be broadly summarized into four key steps: 1) stimuli recognition by cell surface pattern receptors, 2) signaling activation, 3) inflammatory marker release, and 4) recruitment of inflammatory cells. We will distinguish where in each of these steps the obesity-related genes discussed below are found.
Stimuli Recognition.
(Figure 2). Microbial structures (pathogen-associated molecular patterns, PAMPs), endogenous signals such as danger-associated molecular patterns (DAMPs), or cytokines trigger the inflammatory response by activating pattern-recognition receptors (PRRs). PRRs are expressed on immune and nonimmune cells alike, and one of the most highly conserved and best-studied families are the Toll-like receptors (TLRs). TLRs activate an intracellular signaling cascade that leads to nuclear translocation of transcription factors such as nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). Increased adiposity contributes to increased TLR4 signaling, which is considered one of the main triggers for meta-inflammation [133].
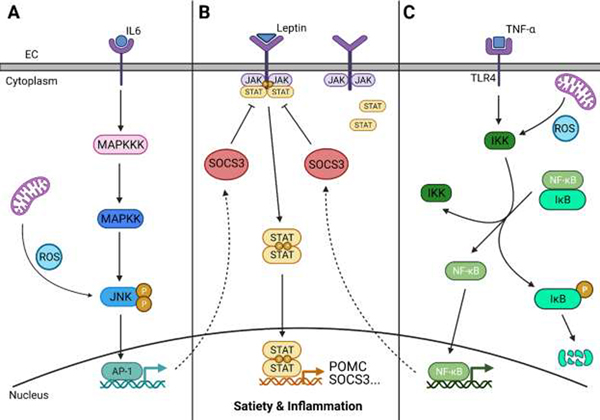
Figure 2. Broad Overview of Relevant Inflammatory Pathways: JNK, JAK-STAT, and NF-κB Signaling.
Pattern recognition receptors (PRRs), expressed on immune and non-immune cells, can be activated by many different signals, including cytokines. A) c-Jun N-terminal kinase 3 (JNK3) also known as mitogen-activated protein kinase (MAPK)-10, is activated when inflammatory cytokines begin a kinase signaling cascade via a MAPK kinase (MAPKK) and MAPKK kinase (MAPKKK) (solid line). JNK may also be activated by reactive oxygen species (ROS) produced by mitochondria. Activated JNK phosphorylates transcription factors, including activator protein-1 (AP-1), which then regulate the inflammatory response. AP-1 also has a response element in the promoter region of suppressor of cytokine signaling 3 (SOCS3) (dashed line). B) When a ligand, such as leptin, binds to the receptor, two receptor-associated Janus family of tyrosine kinases (JAK) proteins are brought in close contact and transphosphorylate, creating a docking site for the cytoplasmic JAK-signal transducer and activator of transcription (STAT) proteins. The JAK proteins then phosphorylate the STAT proteins, which dimerize and translocate to the nucleus to regulate transcription for satiety signaling pathways (e.g., pro-opiomelanocortin, POMC) and the inflammatory response. SOCS3 inhibits the JAK-STAT pathway and down-regulates leptin signaling. C) The PRR for NF-κB, typically Toll-like receptor 4 (TLR4), activates IκB kinase (IKK), a protein complex that phosphorylates the protein IκB, releasing it from the NF-κB complex. ROS may also activate IKK. NF-κB then translocates to the nucleus to activate gene transcription of many pro-inflammatory cytokines. Increased activation of NF-κB in POMC neurons has also been associated with increased expression of SOCS3 (dashed line). Created with BioRender.com.
Signaling Activation.
(Figure 2). MAPK. This family of serine/threonine protein kinases direct cellular responses to stimuli including the inflammatory cytokines TNF-α and IL-6. JNK is one of three mammalian MAPKs, and is activated by inflammatory stimuli (including ROS) and stress causing a signaling cascade through a MAPK kinase (MAPKK) and MAPKK kinase (MAPKKK). Activated JNK can phosphorylate the transcription factor activator protein (AP)-1 to result in regulation of the inflammatory response. JAK-STAT. The JAK-STAT pathway acts in inflammatory signaling as well as satiety signaling, responding to diverse signals including cytokines and leptin. Two receptor-associated JAKs are activated by a ligand and transphosphorylate, creating a docking site for two latent transcription factors, the cytoplasmic STATs. Once the STATs are phosphorylated, they dimerize and can translocate to the nucleus to regulate transcription of inflammatory genes, primarily cytokines. NF-κB. Following stimulation, PRRs activate IκB kinase (IKK), which regulates the NF-κB pathway by phosphorylating the inhibitory protein IκB. ROS have also been suggested to activate the NF-κB pathway through alternative IκB phosphorylation [134]. Phosphorylated IκB is degraded by the proteasome, subsequently releasing NF-κB for nuclear translocation. NF-κB can then activate gene transcription of pro-inflammatory cytokines.
Inflammatory Marker Release.
Inflammatory markers include inflammatory cytokines and chemokines, inflammatory proteins and enzymes, and ROS. Key pro-inflammatory cytokines include IL-1β, IL-6, and TNF-α. Perhaps the most notable inflammatory protein is CRP. These markers then go on to recruit inflammatory cells to the site of interest.
Inflammatory Cell Recruitment.
While the inflammatory response involves a highly coordinated network of several cell types, most immune cells associated with obesity-induced inflammation are macrophages, which are capable of phagocytosis, antigen presentation, and immune response modulation. Microglia are the brain’s resident macrophages, with some participation by astrocytes in neuroprotection [135, 136].
Mechanistically, hypothalamic inflammation appears to be characterized by reactive gliosis, an increase in microglia infiltration and astrocyte proliferation, and has been found in rodent models, nonhuman primate models, and human models alike [122, 123]. Reactive gliosis observed in the hypothalamus may initially be protective in order to limit inflammation and neuron loss, and appearance of reactive gliosis corresponds with pro-inflammatory markers of the ArcN returning to basal levels [122]. As over-nutrition becomes chronic, the capacity of the supportive glial cells to control damage is overwhelmed and inflammation reoccurs [122]. Recent work has identified the mitochondrial uncoupling protein 2 (UCP2) as being a master regulator of microglial activation [137, 138]. A HFD induces UCP2-dependent microglial mitochondrial changes in dynamics and functions, resulting in ROS production and inflammatory activation [137, 138]. Separately, work has shown that activation of IKKβ/NF-κB signaling pathways in astrocytes of the hypothalamus are key in the development of hypothalamic inflammation, HFD-induced hyperphagia, and DIO susceptibility [139]. As hypothalamic inflammation and gliosis are evident in animals before the development of other metabolic symptoms, including weight gain, it has been suggested that hypothalamic inflammation is causative in the pathophysiology of obesity [123].
Research suggests that a HFD determines microglial presence and activity in the ArcN independent of body weight [122, 140, 141]. Enteric gavage in mice with long-chain saturated fatty acids increased hypothalamic inflammatory markers and accumulation of hypothalamic microglia in a way that isocaloric gavages with medium-chain saturated fats or unsaturated fats did not [142]. Importantly, hypothalamic microglia have been identified as sensors of dietary saturated fatty acids that then control the intensity of the resulting inflammation [142]. Microglial content determines the intensity of the inflammatory response, mediate neuronal stress, and regulate satiety-signaling neurons when saturated fatty acid content is high to maintain consistent daily food intake [142]. Better understanding the response of microglia under different diet conditions in promoting and regulating hypothalamic inflammation may further reveal pathways to target for obesity treatment.
Genetic Variations in Inflammatory-related Genes Impact Obesity
In addition to identifying genes that are enriched in the CNS, human GWAS have also found variants within inflammation genes, including TLR4 and MAPK3 [12]. A recent study also identified inflammatory genes such as Src homology 2 adaptor protein 3 (SH2B3) and diacylglycerol lipase beta (DAGLB) within genetic loci that are associated with increased adiposity and lower cardiometabolic risk, further suggesting a role of inflammation genes in metabolically healthy obese individuals [143]. In addition, as previously mentioned, variants near the central regulator of inflammatory cell function RIPK1 have been connected to obesity [32]. Although genetic variation of inflammation genes and how they contribute to the obesity epidemic is not well understood, we discuss three genes in-depth below: SOCS3, JNK, and UCP2.
The JAK-STAT pathway plays a prominent role in inflammatory signaling as well as satiety signaling. Activation of the JAK-STAT-SOCS3 signaling pathway by leptin has been highlighted in obesity research [144–146]. Binding and activation of STAT3 mediates the effects of leptin on energy homeostasis and neuroendocrine function and activates the negative feedback regulator, suppressor of cytokine signaling 3 (SOCS3), to limit leptin receptor action and reduce downstream MC4R activation [146]. Increased SOCS3 expression is associated with increased leptin resistance, and SOCS3 over-activation has been linked to the development of insulin resistance in the brain and in peripheral tissues [146–148]. There is some evidence that demonstrates genetic variation in or near the SOCS3 gene is associated with obesity traits, where decreased SOCS3 expression is associated with improved leptin sensitivity and limited diet-induced weight gain [148–152]. Other studies have identified tentative interactions between variants in SOCS3, IKBKB, and NFKB1 as affecting BMI and waist circumference [153]. This is particularly interesting as other research has shown that over-nutrition inappropriately activates the pro-inflammatory master switch IKKβ/NF-κB in hypothalamic neurons and promotes expression of SOCS3, further perpetuating dysregulation in leptin and melanocortin signaling [154]. Both JAK-STAT-SOCS3 and IKKβ/NF-κB pathways have shown some relation to TLR4 signaling [155–158]. While current research on the genetic variants of these genes is limited, this offers a promising avenue of research for hypothalamic inflammation under the context of obesity.
In addition to SOCS3, one candidate gene approach among obese patients found a common SNP in the JNK gene (rs1062225) that was associated with hypothalamic inflammation [123]. JNK codes for a MAPK that responds to inflammatory cytokines and may also regulate activation of the PI3K/Akt signaling pathway [159, 160]. Work has shown that constitutive JNK activation in NPY/AgRP neurons leads to adiposity via leptin resistance [161]. This may be through connections with SOCS3, as there is evidence that the downstream target of JNK, AP-1, has a response element in the SOCS3 promoter region and can enhance SOCS3 expression [162, 163]. The potential importance of the JNK gene for the inflammatory response in the hypothalamus is validated by the fact that inhibition of the JNK protein restored hypothalamic insulin signaling in HFD-fed rats and improved metabolic parameters [121]. Further evidence validating JNK signaling as being important for hypothalamic health is a recent study that confirmed dual specificity phosphatase 8 (DUSP8), which governs JNK signaling, as a novel hypothalamic factor for type 2 diabetes (T2D) [164]. Genetic variants along the JNK signaling pathway have not been as well studied as previously mentioned genes under an obesity context, but these findings do demonstrate the importance of this pathway for hypothalamic and global metabolic health.
UCP2 is a mitochondrial protein known to be involved in energy homeostasis, that protects against ROS, promotes fatty acid oxidation, and is a master regulator of microglial activation [137, 138, 165–167]. Variants in the UCP2 gene and promoter region have been connected to obesity, serum lipids, insulin resistance, and metabolic efficiency [168–170]. UCP2 is connected to ROS regulation, where decreased expression of UCP2 has been related to increased ROS accumulation and oxidative stress [171, 172]. UCP2 is also required for ghrelin to act on NPY/AgRP neurons, driven partially by the role of UCP2 in ROS scavenging [173]. But researchers have also shown that HFD-induced microglial activation in the hypothalamus is associated with increased expression of UCP2 in the microglia, where ablating expression of UCP2 in the brain microglia prevented DIO and improved metabolic parameters in male mice [137]. Separately, increased UCP2 expression has been correlated with decreased risk for chronic inflammatory diseases such as rheumatoid arthritis and Crohn’s disease, but increased risk for sepsis and inflammatory bowel disease [174–176]. Whether these inconsistencies are due to genotype differences, tissue-specific actions, or pathway-specific roles remains to be determined. Taken together, these data show that the relationship between UCP2 and obesity must not only consider its haplotype, but also the tissue of study.
Connections between satiety signaling and inflammatory pathways
Very little work has been done to connect genetic variants in satiety signaling pathways to hypothalamic inflammation, and/or variants in inflammatory pathways to satiety signaling. Given the contributions of dysregulation in both pathways to the development and maintenance of obesity, this presents a large gap in knowledge. Described below are the known links between FTO, MC4R, and ADCY3 and inflammation, as well as the connections between SOCS3, JNK, and UCP2 and satiety (Figure 3).
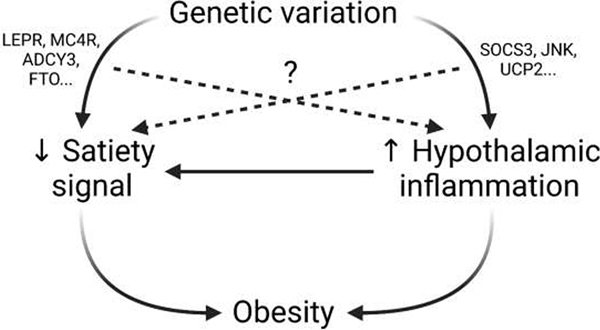
Figure 3. Graphical Abstract.
It is known that genetic variation can alter satiety signaling and hypothalamic inflammation, and that dysfunction in both of these pathways contributes to the development and progression of obesity (solid line). It is also known that hypothalamic inflammation dysregulates satiety signaling (solid line), further contributing to weight gain. What remains to be determined is the extent to which genetic variations in satiety-signaling genes (LEPR, MC4R, ADCY3, FTO, etc) contribute to development of inflammation, and how variations in inflammatory-related genes (SOCS3, JNK, UCP2, etc) might alter satiety signaling (dashed line). Better understanding the connections between these two pathways will reveal new drug targets for obesity treatment, and will allow physicians to gain a wider perspective on how genetic variants will affect metabolic health. Created with BioRender.com.
Genes known to participate in satiety signaling have limited available evidence of also being connected to inflammation. The exact relationship between FTO and inflammation remains unclear. Early studies found that presence of the obesity-risk rs9939609 variant was associated with increased plasma CRP levels, independent of adiposity, as well as a predisposition in T2D patients towards inflammatory obesity [177, 178]. Another study found pleiotropic associations for a different FTO variant, rs1558902, with obesity measures and CRP [30]. However, subsequent studies investigating the relationship between FTO and inflammatory markers do not show significant associations [179, 180]. While at the time of this article there is no publicly available data on how FTO expression and activity are related to or affected by hypothalamic inflammation, research has found a connection between FTO and NF-κB activation in pancreatic β-cells, which may indicate a connection in the hypothalamus that has not yet been elucidated [181]. MC4R activation has been linked to anti-inflammatory and neuroprotective effects in vitro, and activating MC4R ameliorated the reactive phenotype in in vitro astrocytes [182–185]. Given the connection between astrocytes and the development of hypothalamic inflammation under HFD conditions, this also opens up the possibility that dysfunctional MC4R mutations may predispose or protect against hypothalamic inflammation as well as obesity. While much focus has naturally been on the role of ADCY3 in the hypothalamus, there is some evidence indicating importance to its signaling in peripheral tissues as well [113]. Of particular note, haploinsufficiency of Adcy3 in mice negatively altered fatty acid oxidation genes in white adipose tissue while separate work suggests that enhancing fatty acid oxidation in the hypothalamus reduced local inflammatory responses [113, 186]. Further work is needed to identify if changes in expression of ADCY3 in the hypothalamus alter fatty acid oxidation, but this relationship may provide a link between ADCY3 expression, hypothalamic inflammation, and adiposity. Taken together, this indicates a need to better understand how genetic variants in known obesity-causing genes may also promote hypothalamic inflammation. The relationship between hypothalamic inflammation and obesity development cannot be ignored, and elucidating connections here may reveal new targets in known pathways.
There are also links between hypothalamic inflammation and satiety signaling that still need to be explored (Figure 3). Current evidence shows that hypothalamic inflammation is responsive to a HFD, and leads to obesity-associated problems [187]. Hypothalamic inflammation has been reported to disrupt hypothalamic hormonal (leptin, insulin, etc) signaling, leading to dysfunction of the central energy balance [154]. As has been reviewed elsewhere, hypothalamic inflammation has been shown to disrupt the hypothalamic endocrine systems to promote obesity, and may also activate the endocannabinoid system to promote overeating [29]. Expression changes in SOCS3, regulator of the JAK-STAT pathway, are well known to be instrumental to the development of leptin and insulin resistance and dysregulation of satiety signaling [29, 148, 188–191]. HFD feeding has also been connected to hypothalamic JNK activation, and a JNK-inhibitory peptide was once considered as a possible diabetes therapeutic [29, 161, 192–194]. Constitutive activation of JNK in NPY/AgRP neurons has been shown to induce hyperphagia and obesity in mice, and other studies have demonstrated a role of the JNK isoforms in feeding [161, 194]. Research has also shown that UCP2 modulates POMC and NPY/AgRP neuronal function, inhibiting glucose-induced activation in the former and promoting firing of the latter through its ROS regulators [195]. This work shows that hypothalamic inflammation affects satiety signaling and may explain its potentially causal role in DIO. It is therefore important to also consider genetic variants in these inflammatory-related genes in the context of obesity, as they may also explain a portion of a patient’s predisposition to gaining weight. Better understanding the relationship between expression of inflammatory-related genes and dysfunction in these signaling pathways will illuminate our understanding of the connections between these two pathways as well as revealing potential drug targets.
Other Influences on Hypothalamic Inflammation
It is important to note that the gut-brain axis communicates not only satiety signals, but may also connect inflammation between the two tissues. Current research suggests that gut inflammation is another early consequence of a HFD and may have a causative role in obesity [28, 196]. The microbiome may be essential for mediating the relationship between nutritional components of a HFD and neuronal inflammation, and these changes in the gut-brain axis might induce the appetite and satiety dysregulation that promote the development of obesity [123]. As previously discussed, saturated fats have been well-connected to hypothalamic obesity [124, 125, 142]. Fatty acids also impact the gut microbiome. Human studies and other work have shown that a HFD alters gut microbiota, including reducing microbial richness and diminishing gut barrier integrity, both of which are ultimately associated with increased adiposity and worsened metabolic health [24, 28]. Studies in mice have investigated how different levels of saturation in fats affect the gut microbiome, and determined that diets richer in saturated fats have worsened outcomes [197, 198]. A study in humans also revealed that saturated fat content has a greater effect on microbe diversity compared to protein source [199]. Early changes in the gut microbiota influence inflammation in the intestines, including an increased endotoxin production, and alter the permeability of the intestinal barrier that leads to endotoxin release and TLR4 activation elsewhere in the body [28, 133, 200]. Recent work has also shown that a switch to a fat-rich diet leads to a rapid and profound change in gut microbiota diversity and oxidative stress in the hypothalamus [201]. This body of work strongly suggests a connection between gut microbiota and the hypothalamus in the adaptation to HFD and regulating energy homeostasis, with the effect strongly driven by the content of saturated fatty acids in the diet. While not the focus of this review, fully understanding the connections between the gut and the brain is a key area of obesity research important for understanding and treating the disease.
Another key influence on hypothalamic inflammation is the blood-brain barrier (BBB), which controls the passage of most blood components into the central nervous system [202]. Damage to this barrier can lead to leakage of unwanted substances into the brain that can propagate inflammation. Studies have demonstrated that a HFD typically leads to increased permeability of the BBB [203–205]. It has been shown that this effect differed between DIO and diet-resistant rats, indicating a potential impact of genetic variation [205]. The gut microbiome has been shown to affect BBB permeability [206, 207], and DIO-associated changes in the gut may then affect the BBB to propagate hypothalamic inflammation. Further connecting back to nutrients in the diet, a recent study showed that omega-3 polyunsaturated fatty acids were positively associated with BBB integrity and better brain health [208]. When investigating the role of hypothalamic inflammation on obesity, it is important to consider the many different factors that can determine the onset and severity of inflammation, whether they be genetic, environmental, or a mixture of both.
Conclusion
Concern for obesity and its prevalence is only rising: current trends suggest that an average of 50% of adults in the United States will be considered obese within ten years [1, 2]. It is well-known that much of what drives the obesity epidemic is the increase of calorie-rich foods high in saturated fats coupled with reduced physical activity [209, 210]. But obesity is a self-propagating cycle. Dietary fat causes an inflammatory response in the brain very quickly and in turn disrupts normal homeostatic processes [121, 211]. But not everyone responds to a HFD or obesity in the same way. Genetic variants in genes along both the satiety signaling and the inflammatory response pathways may explain individual susceptibility to gaining weight and/or suffering metabolic consequences. As new variants in brain genes are identified, it will be important to assess how these genes contribute to obesity. While hypothalamic inflammation is well-known to contribute to downstream events in the progression of obesity, many currently identified obesity-related genes that are involved in satiety signaling (FTO, IRX3, MC4R, ADCY3, etc) have not been evaluated for their role in the development and maintenance of inflammation, or how this inflammation affects their activity in turn. Further, genetic research on inflammatory-related genes (SOCS3, JNK, UCP2, etc) is limited. Connections between these pathways present the possibility of variants in multiple genes acting in tandem to predispose to or protect against obesity. Merging our knowledge of these fields will improve our understanding on how obesity first develops under a HFD and how dysregulation of inflammatory pathways helps persist and worsen the disease.
In doing so, we can identify new potential targets for treating obesity. There are several main benefits to doing so. The first is that current treatments for obesity, which typically target satiety, are limited in number and efficacy and so developing new strategies is of high importance [212–214]. It has been shown in mice that reducing expression of inflammatory-related genes, including TLR4 and TNF-α, can protect against DIO and associated comorbidities, opening up new pathways to target [215–218]. Better understanding the connection between satiety and inflammation will ensure that future treatment options are even more effective than those currently available. The second benefit is that studies have demonstrated that genetic variation can impact drug response to lipid lowering statins or sulfonylureas for treating T2D—this is likely to also be true for anti-obesity medication [19, 20]. Incorporating a patient’s genetic variation into healthcare decisions will improve treatment and patient outcome. Genetic studies will also elucidate novel biological pathways and connections to help us understand underlying molecular mechanisms, enable polygenic risk scores to identify individuals most at risk, and inform pharmacogenetic analysis to determine which specific medications an individual will respond to best.
Each person is unique, with their bodies responding to a HFD and inflammatory signaling in different ways; we must strive to develop treatments that can ensure the best possible weight loss success for every individual. Using genetic information to advise patients is still a developing field [219, 220]. Although work is still needed to optimize the use of genetic information, increasing our understanding of the genetic variants and the biological pathways of obesity will help inform physicians of how best to use a patient’s genetic information to help them reach their metabolic health and weight loss goals. Because obesity is so extraordinarily complex, the more we understand the connections between pathways and the roles individual variants may play, the better we can treat each individual patient.
Highlights.
GWAS has identified numerous obesity-related genes as being enriched in the brain
Genetic variants in genes along the satiety signaling pathway are known to contribute to obesity
Recent studies have also identified genetic variants in inflammatory genes that contribute to obesity
Connections between variants in satiety signaling and inflammation should be explored
Personalized nutrition and medicine approaches improve obesity patient outcomes
Sources of Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases grants R01DK120667 (LSW) and K01DK117069 (CCK), and National Institute on Drug Abuse of the National Institutes of Health grant T32DA041349 (AMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ward ZJ, et al. , Projected U.S. State-Level Prevalence of Adult Obesity and Severe Obesity. N Engl J Med, 2019. 381(25): p. 2440–2450. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein EA, et al. , Obesity and severe obesity forecasts through 2030. Am J Prev Med, 2012. 42(6): p. 563–70. [DOI] [PubMed] [Google Scholar]
- 3.Hales CM, et al. , Prevalence of Obesity and Severe Obesity Among Adults: United States, 2017–2018. NCHS Data Brief, 2020(360): p. 1–8. [PubMed] [Google Scholar]
- 4.Hruby A and Hu FB, The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics, 2015. 33(7): p. 673–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hruby A, et al. , Determinants and Consequences of Obesity. Am J Public Health, 2016. 106(9): p. 1656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apovian CM, Obesity: definition, comorbidities, causes, and burden. Am J Manag Care, 2016. 22(7 Suppl): p. s176–85. [PubMed] [Google Scholar]
- 7.Yengo L, et al. , Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet, 2018. 27(20): p. 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maes HH, Neale MC, and Eaves LJ, Genetic and environmental factors in relative body weight and human adiposity. Behav Genet, 1997. 27(4): p. 325–51. [DOI] [PubMed] [Google Scholar]
- 9.Pulit SL, et al. , Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet, 2019. 28(1): p. 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohde K, et al. , Genetics and epigenetics in obesity. Metabolism,2019. 92: p.37–50. [DOI] [PubMed] [Google Scholar]
- 11.Loos RJ, The genetics of adiposity. Curr Opin Genet Dev, 2018. 50: p.86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke AE, et al. , Genetic studies of body mass index yield new insights for obesity biology. Nature, 2015. 518(7538): p. 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turcot V, et al. , Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet, 2018. 50(1): p. 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouni-Berthold I and Berthold HK, Current Options for the Pharmacotherapy of Obesity. Curr Pharm Des, 2019. 25(18): p. 2019–2032. [DOI] [PubMed] [Google Scholar]
- 15.Ruban A, et al. , Current treatments for obesity. Clin Med (Lond),2019.19(3):p.205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viljakainen H, et al. , Genetic risk score predicts risk for overweight and obesity in Finnish preadolescents. Clin Obes, 2019. 9(6): p. e12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason KE, et al. , Genetic risk of obesity as a modifier of associations between neighbourhood environment and body mass index: an observational study of 335 046 UK Biobank participants. BMJ Nutr Prev Health, 2020. 3(2): p. 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dashti HS, et al. , Polygenic risk score for obesity and the quality, quantity, and timing of workplace food purchases: A secondary analysis from the ChooseWell 365 randomized trial. PLoS Med, 2020. 17(7): p. e1003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Iruela C, et al. , KIF6 gene as a pharmacogenetic marker for lipid -lowering effect in statin treatment. PLoS One, 2018. 13(10): p. e0205430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J, et al. , KCNJ 11, ABCC8 and TCF7L2 polymorphisms and the response to sulfonylurea treatment in patients with type 2 diabetes: a bioinformatics assessment. BMC Med Genet, 2017. 18(1): p. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seral-Cortes M, et al. , Development of a Genetic Risk Score to predict the risk of overweight and obesity in European adolescents from the HELENA study. Sci Rep, 2021. 11(1): p. 3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.deToro-Martin J, et al. , Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients, 2017. 9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrington WT, et al. , Improving Metabolic Health Through Precision Dietetics in Mice. Genetics, 2018. 208(1): p. 399–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saltiel AR and Olefsky JM, Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest, 2017. 127(1): p. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hotamisligil GS, et al. , Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest, 1995. 95(5): p. 2409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotamisligil GS, et al. , IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science, 1996. 271(5249): p. 665–8. [DOI] [PubMed] [Google Scholar]
- 27.Jais A and Bruning JC, Hypothalamic inflammation in obesity and metabolic disease. J Clin Invest, 2017. 127(1): p. 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KA, et al. , High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One, 2012. 7(10): p. e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai D and Liu T, Hypothalamic inflammation: a double-edged sword to nutritional diseases. Ann N Y Acad Sci, 2011. 1243: p. E1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraja AT, et al. , Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab, 2014. 112(4): p. 317–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galmes S, et al. , A Genetic Score of Predisposition to Low-Grade Inflammation Associated with Obesity May Contribute to Discern Population at Risk for Metabolic Syndrome. Nutrients, 2019. 11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karunakaran D, et al. , RIPK1 gene variants associate with obesity in humans and can be therapeutically silenced to reduce obesity in mice. Nat Metab, 2020. 2(10): p. 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeevi D, et al. , Personalized Nutrition by Prediction of Glycemic Responses. Cell, 2015. 163(5): p. 1079–1094. [DOI] [PubMed] [Google Scholar]
- 34.Ahima RS and Antwi DA, Brain regulation of appetite and satiety. Endocrinol Metab Clin North Am, 2008. 37(4): p. 811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips RJ and Powley TL, Gastric volume rather than nutrient content inhibits food intake. Am J Physiol, 1996. 271(3 Pt 2): p. R766–9. [DOI] [PubMed] [Google Scholar]
- 36.Kojima M, et al. , Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature, 1999. 402(6762): p. 656–60. [DOI] [PubMed] [Google Scholar]
- 37.Batterham RL, et al. , Gut hormone PYY(3–36) physiologically inhibits food intake. Nature, 2002. 418(6898): p. 650–4. [DOI] [PubMed] [Google Scholar]
- 38.Gibbs J, Young RC, and Smith GP, Cholecystokinin decreases food intake in rats. J Comp Physiol Psychol, 1973. 84(3): p. 488–95. [DOI] [PubMed] [Google Scholar]
- 39.Turton MD, et al. , A role for glucagon-like peptide-1 in the central regulation of feeding. Nature, 1996. 379(6560): p. 69–72. [DOI] [PubMed] [Google Scholar]
- 40.Liebling DS, et al. , Intestinal satiety in rats. J Comp Physiol Psychol,1975.89(8): p. 955–65. [DOI] [PubMed] [Google Scholar]
- 41.Shah M and Vella A, Effects of GLP-1 on appetite and weight. Rev Endocr Metab Disord, 2014. 15(3): p. 181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan W, et al. , Cholecystokinin-mediated suppression of feeding involves the brainstem melanocortin system. Nat Neurosci, 2004. 7(4): p. 335–6. [DOI] [PubMed] [Google Scholar]
- 43.Holzer P, Reichmann F, and Farzi A, Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides, 2012. 46(6): p. 261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibrahim Abdalla MM, Ghrelin - Physiological Functions and Regulation. Eur Endocrinol, 2015. 11(2): p. 90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kola B, et al. , The orexigenic effect of ghrelin is mediated through central activation of the endogenous cannabinoid system. PLoS One, 2008. 3(3): p. e1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campfield LA, et al. , Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science, 1995. 269(5223): p. 546–9. [DOI] [PubMed] [Google Scholar]
- 47.Ellacott KL and Cone RD, The role of the central melanocortin system in the regulation of food intake and energy homeostasis: lessons from mouse models. Philos Trans R Soc Lond B Biol Sci, 2006. 361(1471): p. 1265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balthasar N, et al. , Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell, 2005. 123(3): p. 493–505. [DOI] [PubMed] [Google Scholar]
- 49.Wei W, et al. , Diet composition, not calorie intake, rapidly alters intrinsic excitability of hypothalamic AgRP/NPY neurons in mice. Sci Rep, 2015. 5: p. 16810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li MM, et al. , The Paraventricular Hypothalamus Regulates Satiety and Prevents Obesity via Two Genetically Distinct Circuits. Neuron, 2019. 102(3): p. 653–667 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng H, et al. , A potential role for hypothalamomedullary POMC projections in leptin-induced suppression of food intake. Am J Physiol Regul Integr Comp Physiol, 2010. 298(3): p. R720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grill HJ and Hayes MR, Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell Metab, 2012. 16(3): p. 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herbert H, Moga MM, and Saper CB, Connections of the parabrachial nucleus with the nucleus of the solitary tract and the medullary reticular formation in the rat. J Comp Neurol, 1990. 293(4): p. 540–80. [DOI] [PubMed] [Google Scholar]
- 54.Daniels D and Mietlicki-Baase EG, Glucagon-Like Peptide 1 in the Brain: Where Is It Coming From, Where Is It Going? Diabetes, 2019. 68(1): p. 15–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campos CA, et al. , CCK-induced reduction of food intake and hindbrain MAPK signaling are mediated by NMDA receptor activation. Endocrinology, 2012. 153(6): p. 2633–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faulconbridge LF, et al. , Hyperphagic effects of brainstem ghrelin administration. Diabetes, 2003. 52(9): p. 2260–5. [DOI] [PubMed] [Google Scholar]
- 57.Berthoud HR, Munzberg H, and Morrison CD, Blaming the Brain for Obesity: Integration of Hedonic and Homeostatic Mechanisms. Gastroenterology, 2017. 152(7): p. 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alonso-Alonso M, et al. , Food reward system: current perspectives and future research needs. Nutr Rev, 2015. 73(5): p. 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suarez AN, et al. , Ghrelin and Orexin Interact to Increase Meal Size Through a Descending Hippocampus to Hindbrain Signaling Pathway. Biol Psychiatry, 2020. 87(11): p. 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee TH and Yau SY, From Obesity to Hippocampal Neurodegeneration: Pathogenesis and Non-Pharmacological Interventions. Int J Mol Sci, 2020. 22(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mestre ZL, et al. , Hippocampal atrophy and altered brain responses to pleasant tastes among obese compared with healthy weight children. Int J Obes (Lond), 2017. 41(10): p. 1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Domingo-Rodriguez L, et al. , A specific prelimbic-nucleus accumbens pathway controls resilience versus vulnerability to food addiction. Nat Commun, 2020. 11(1): p. 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Will MJ, Franzblau EB, and Kelley AE, Nucleus accumbens mu-opioids regulate intake of a high-fat diet via activation of a distributed brain network. J Neurosci, 2003. 23(7): p. 2882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casquero-Veiga M, et al. , Stimulating the nucleus accumbens in obesity: A positron emission tomography study after deep brain stimulation in a rodent model. PLoS One, 2018. 13(9): p. e0204740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Formolo DA, et al. , Deep Brain Stimulation for Obesity: A Review and Future Directions. Front Neurosci, 2019. 13: p. 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ndiaye FK, et al. , The expression of genes in top obesity-associated loci is enriched in insula and substantia nigra brain regions involved in addiction and reward. Int J Obes (Lond), 2020. 44(2): p. 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright H, et al. , Differential effects of hunger and satiety on insular cortex and hypothalamic functional connectivity. Eur J Neurosci, 2016. 43(9): p. 1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le TM, et al. , The interrelationship of body mass index with gray matter volume and resting-state functional connectivity of the hypothalamus. Int J Obes (Lond), 2020. 44(5): p. 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coccurello R and Maccarrone M, Hedonic Eating and the "Delicious Circle": From Lipid-Derived Mediators to Brain Dopamine and Back. Front Neurosci, 2018. 12: p. 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Micali N, et al. , Are obesity risk genes associated with binge eating in adolescence? Obesity (Silver Spring), 2015. 23(8): p. 1729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fairbrother U, et al. , Genetics of Severe Obesity. Curr Diab Rep, 2018. 18(10): p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Considine RV, et al. , The hypothalamic leptin receptor in humans: identification of incidental sequence polymorphisms and absence of the db/db mouse and fa/fa rat mutations. Diabetes, 1996. 45(7): p. 992–4. [DOI] [PubMed] [Google Scholar]
- 73.Matsuoka N, et al. , Human leptin receptor gene in obese Japanese subjects: evidence against either obesity-causing mutations or association of sequence variants with obesity. Diabetologia, 1997. 40(10): p. 1204–10. [DOI] [PubMed] [Google Scholar]
- 74.Ranadive SA and Vaisse C, Lessons from extreme humano besity: monogenic disorders. Endocrinol Metab Clin North Am, 2008. 37(3): p. 733–51, x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farooqi IS, et al. , Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med, 2003. 348(12): p. 1085–95. [DOI] [PubMed] [Google Scholar]
- 76.Wang CL, et al. , Several mutations in the melanocortin 4 receptor gene are associated with obesity in Chinese children and adolescents. J Endocrinol Invest, 2006. 29(10): p. 894–8. [DOI] [PubMed] [Google Scholar]
- 77.Polak E, et al. , The prevalence of melanocortin-4 receptor gene mutations in Slovak obese children and adolescents. J Pediatr Endocrinol Metab, 2016. 29(1): p. 55–61. [DOI] [PubMed] [Google Scholar]
- 78.Namjou B, et al. , Evaluation of the MC4R gene across eMERGE network identifies many unreported obesity-associated variants. Int J Obes (Lond), 2021. 45(1): p. 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brouwers B, et al. , Human MC4R variants affect endocytosis, trafficking and dimerization revealing multiple cellular mechanisms involved in weight regulation. Cell Rep, 2021. 34(12): p. 108862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Q, et al. , Complex Relationship between Obesity and the Fat Mass and Obesity Locus. Int J Biol Sci, 2017. 13(5): p. 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frayling TM, et al. , A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science, 2007. 316(5826): p. 889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wardle J, et al. , The FTO gene and measured food intake in children. Int J Obes (Lond), 2009. 33(1): p. 42–5. [DOI] [PubMed] [Google Scholar]
- 83.Tanofsky-Kraff M, et al. , The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am J Clin Nutr, 2009. 90(6): p. 1483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karra E, et al. , A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest, 2013. 123(8): p. 3539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Magno F, et al. , Influence of FTO rs9939609 polymorphism on appetite, ghrelin, leptin, IL6, TNFalpha levels, and food intake of women with morbid obesity. Diabetes Metab Syndr Obes, 2018. 11: p. 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Melhorn SJ, et al. , FTO genotype impacts food intake and corticolimbic activation. Am J Clin Nutr, 2018. 107(2): p. 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Danaher J, Stathis CG, and Cooke MB, Similarities in Metabolic Flexibility and Hunger Hormone Ghrelin Exist between FTO Gene Variants in Response to an Acute Dietary Challenge. Nutrients, 2019. 11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.deAraujo TM and Velloso LA, Hypothalamic IRX3: A New Player in the Development of Obesity. Trends Endocrinol Metab, 2020. 31(5): p. 368–377. [DOI] [PubMed] [Google Scholar]
- 89.Stratigopoulos G, et al. , Cut-lik ehomeobo x1(CUX1) regulate sexpressio no fth efat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J Biol Chem, 2011. 286(3): p. 2155–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smemo S, et al. , Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature, 2014. 507(7492): p. 371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doaei S, et al. , Changes in FTO and IRX3 gene expression in obese and overweight male adolescents undergoing an intensive lifestyle intervention and the role of FTO genotype in this interaction. J Transl Med, 2019. 17(1): p. 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiang L, et al. , FTO genotype and weight loss in diet and lifestyle interventions : a systematic review and meta-analysis. Am J Clin Nutr, 2016. 103(4): p. 1162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Young EH, et al. , The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond), 2007. 31(9): p. 1437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang D, et al. , Association of the MC4R V103I polymorphism with obesity: a Chinese case-control study and meta-analysis in 55,195 individuals. Obesity (Silver Spring), 2010. 18(3): p. 573–9. [DOI] [PubMed] [Google Scholar]
- 95.Xiang Z, et al. , Pharmacological characterization of 40 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelanocortin-derived agonists and the agouti-related protein (AGRP) antagonist. Biochemistry, 2006. 45(23): p. 7277–88. [DOI] [PubMed] [Google Scholar]
- 96.Willer CJ, et al. , Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet, 2009. 41(1): p. 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zobel DP, et al. , Variants near MC4R are associated with obesity and influence obesity-related quantitative traits in a population of middle-aged people: studies of 14,940 Danes. Diabetes, 2009. 58(3): p. 757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thorleifsson G, et al. , Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet, 2009. 41(1): p. 18–24. [DOI] [PubMed] [Google Scholar]
- 99.Iepsen EW, et al. , GLP-1 Receptor Agonist Treatment in Morbid Obesity and Type 2 Diabetes Due to Pathogenic Homozygous Melanocortin-4 Receptor Mutation: A Case Report. Cell Rep Med, 2020. 1(1): p. 100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iepsen EW, et al. , Patients with Obesity Caused by Melanocortin-4 Receptor Mutations Can Be Treated with a Glucagon-like Peptide-1 Receptor Agonist. Cell Metab, 2018. 28(1): p. 23–32 e3. [DOI] [PubMed] [Google Scholar]
- 101.Hatoum IJ, et al. , Melanocortin-4 receptor signaling is required for weight loss after gastric bypass surgery. J Clin Endocrinol Metab, 2012. 97(6): p. E1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Holzapfel C, et al. , Association between Single Nucleotide Polymorphisms and Weight Reduction in Behavioural Interventions-A Pooled Analysis. Nutrients, 2021. 13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang H and Storm DR, Calmodulin-regulated adenylyl cyclases: cross-talk and plasticity in the central nervous system. Mol Pharmacol, 2003. 63(3): p. 463–8. [DOI] [PubMed] [Google Scholar]
- 104.Redei EE, et al. , Pilot validation of blood-based biomarkers during pregnancy and postpartum in women with prior or current depression. Transl Psychiatry, 2021. 11(1): p. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nordman S, et al. , Genetic variation of the adenylyl cyclase 3(AC3) locus and its influence on type 2 diabetes and obesity susceptibility in Swedish men. Int J Obes (Lond), 2008. 32(3): p. 407–12. [DOI] [PubMed] [Google Scholar]
- 106.Speliotes EK, et al. , Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet, 2010. 42(11): p. 937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grarup N, et al. , Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat Genet, 2018. 50(2): p. 172–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wray NR, et al. , Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry, 2012. 17(1): p. 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Abdel-Halim SM, et al. , Mutations in the promoter of adenylyl cyclase (AC)-III gene, overexpression of AC-III mRNA, and enhanced cAMP generation in islets from the spontaneously diabetic GK rat model of type 2 diabetes. Diabetes, 1998. 47(3): p. 498–504. [DOI] [PubMed] [Google Scholar]
- 110.Wen W, et al. , Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet, 2012. 44(3): p. 307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang Z, et al. , Adult type 3 adenylyl cyclase-deficient mice are obese. PLoS One, 2009. 4(9): p. e6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pitman JL, et al. , A gain-of-function mutation in adenylate cyclase 3 protects mice from diet-induced obesity. PLoS One, 2014. 9(10): p. e110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tong T, et al. , Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci Rep, 2016. 6: p. 34179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keele GR, et al. , Genetic Fine-Mapping and Identification of Candidate Genes and Variants for Adiposity Traits in Outbred Rats. Obesity (Silver Spring), 2018. 26(1): p. 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chitre AS, et al. , Genome-Wide Association Study in 3,173 Outbred Rats Identifies Multiple Loci for Body Weight, Adiposity, and Fasting Glucose. Obesity (Silver Spring), 2020. 28(10): p. 1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Siljee JE, et al. , Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet, 2018. 50(2): p. 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goni L, et al. , Interaction between an ADCY3 Genetic Variant and Two Weight-Lowering Diets Affecting Body Fatness and Body Composition Outcomes Depending on Macronutrient Distribution: A Randomized Trial. Nutrients, 2018. 10(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Navarro E, et al. , Can metabolically healthy obesity be explained by diet, genetics, and inflammation? Mol Nutr Food Res, 2015. 59(1): p. 75–93. [DOI] [PubMed] [Google Scholar]
- 119.Bluher M, Metabolically Healthy Obesity. Endocr Rev, 2020. 41(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Freeman LR, et al. , Obesity increases cerebrocortical reactive oxygen species and impairs brain function. Free Radic Biol Med, 2013. 56: p. 226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.DeSouza CT, et al. , Consumption of a fat-rich diet activates a proinflammatory response and induces insulin resistance in the hypothalamus. Endocrinology, 2005. 146(10): p. 4192–9. [DOI] [PubMed] [Google Scholar]
- 122.Thaler JP, et al. , Obesity is associated with hypothalamic injury in rodents and humans. J Clin Invest, 2012. 122(1): p. 153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kreutzer C, et al. , Hypothalamic Inflammation in Human Obesity Is Mediated by Environmental and Genetic Factors. Diabetes, 2017. 66(9): p. 2407–2415. [DOI] [PubMed] [Google Scholar]
- 124.Maric T,Woodside B, and Luheshi GN, The effects of dietary saturated fat on basal hypothalamic neuroinflammation in rats. Brain Behav Immun, 2014. 36: p. 35–45. [DOI] [PubMed] [Google Scholar]
- 125.Sergi D and Williams LM, Potential relationship between dietary long-chain saturated fatty acids and hypothalamic dysfunction in obesity. Nutr Rev, 2020. 78(4): p. 261–277. [DOI] [PubMed] [Google Scholar]
- 126.Thomas K, et al. , Higher body mass index is linked to altered hypothalamic microstructure. Sci Rep, 2019. 9(1): p. 17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szabo-Reed AN, et al. , Modeling interactions between brain function, diet adherence behaviors, and weight loss success. Obes Sci Pract, 2020. 6(3): p. 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Contreras-Rodriguez O, et al. , Altered cross-talk between the hypothalamus and non-homeostatic regions linked to obesity and difficulty to lose weight. Sci Rep, 2017. 7(1): p. 9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Riederer JW, et al. , Understanding Neuronal Architecture in Obesity through Analysis of White Matter Connection Strength. Front Hum Neurosci, 2016. 10: p. 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sewaybricker LE, et al. , Initial evidence for hypothalamic gliosis in children with obesity by quantitative T2 MRI and implications for blood oxygen-level dependent response to glucose ingestion. Pediatr Obes, 2019. 14(2): p. e12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Syan SK, et al. , Dysregulated resting state functional connectivity and obesity: a systematic review. Neurosci Biobehav Rev, 2021. [DOI] [PubMed] [Google Scholar]
- 132.Chen L, et al. , Inflammatory responses and inflammation-associated diseases in organs. Oncotarget, 2018. 9(6): p. 7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rogero MM and Calder PC, Obesity, Inflammation, Toll-Like Receptor 4 and Fatty Acids. Nutrients, 2018. 10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morgan MJ and Liu ZG, Crosstalk of reactive oxygen species and NF-kappa B signaling. Cell Res, 2011. 21(1): p. 103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mendes NF, et al. , Hypothalamic Microglial Activation in Obesity: A Mini-Review. Front Neurosci, 2018. 12: p. 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rahman MH, et al. , Interglial Crosstalk in Obesity-Induced Hypothalamic Inflammation. Front Neurosci, 2018. 12: p. 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim JD, et al. , Microglial UCP2 Mediates Inflammation and Obesity Induced by High-Fat Feeding. Cell Metab, 2019. 30(5): p. 952–962 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.DeSimone R, et al. , The mitochondrial uncoupling protein-2 is a master regulator of both M1 and M2 microglial responses. J Neurochem, 2015. 135(1): p. 147–56. [DOI] [PubMed] [Google Scholar]
- 139.Douglass JD, et al. , Astrocyte IKKbeta/NF-kappaB signaling is required for diet-induced obesity and hypothalamic inflammation. Mol Metab, 2017. 6(4): p. 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gutierrez-Martos M, et al. , Cafeteria diet induces neuroplastic modifications in the nucleus accumbens mediated by microglia activation. Addict Biol, 2018. 23(2): p. 735–749. [DOI] [PubMed] [Google Scholar]
- 141.Gao Y, et al. , Hormones and diet, but not body weight, control hypothalami cmicroglial activity. Glia, 2014. 62(1): p. 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Valdearcos M, et al. , Microglia dictate the impact of saturated fat consumptionon hypothalamic inflammation and neuronal function. Cell Rep, 2014. 9(6): p. 2124–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang LO, et al. , Genome-wide discovery of genetic loci that uncouple excess adiposity from its comorbidities. Nat Metab, 2021. 3(2): p. 228–243. [DOI] [PubMed] [Google Scholar]
- 144.Bjorbaek C, et al. , Identification of SOCS-3 as a potential mediator of central leptin resistance. Mol Cell, 1998. 1(4): p. 619–25. [DOI] [PubMed] [Google Scholar]
- 145.Bjorbaek C, et al. , The role of SOCS-3 in leptin signaling and leptin resistance. J Biol Chem, 1999. 274(42): p. 30059–65. [DOI] [PubMed] [Google Scholar]
- 146.Wunderlich CM, Hovelmeyer N, and Wunderlich FT, Mechanisms of chronic JAK-STAT3-SOCS3 signaling in obesity. JAKSTAT, 2013. 2(2): p. e23878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang Z, et al. , Regulation of insulin and leptin signaling by muscle suppressor of cytokine signaling 3 (SOCS3). PLoS One, 2012. 7(10): p. e47493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pedroso JA, et al. , Inactivation of SOCS3 in leptin receptor-expressing cells protects mice from diet-induced insulin resistance but does not prevent obesity. Mol Metab, 2014. 3(6): p. 608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Talbert ME, et al. , Polymorphisms near SOCS3 are associated with obesity and glucose homeostasis traits in Hispanic Americans from the Insulin Resistance Atherosclerosis Family Study. Hum Genet, 2009. 125(2): p. 153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tang W, et al. , Association of two polymorphisms within and near SOCS3 gene with obesity in three nationalities in Xinjiang province of China. Acta Pharmacol Sin, 2011. 32(11): p. 1381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Howard JK, et al. , Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat Med, 2004. 10(7): p. 734–8. [DOI] [PubMed] [Google Scholar]
- 152.Briancon N, et al. , Combined neural inactivation of suppressor of cytokine signaling-3 and protein-tyrosine phosphatase-1B reveals additive, synergistic, and factor-specific roles in the regulation of body energy balance. Diabetes, 2010. 59(12): p. 3074–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tessier F, et al. , Investigating Gene-Gene and Gene-Environment Interactions in the Association Between Overnutrition and Obesity-Related Phenotypes. Front Genet, 2019. 10: p. 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang X, et al. , Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell, 2008. 135(1): p. 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fu XQ, et al. , Activation of STAT3 is a key event in TLR4 signaling-mediated melanoma progression. Cell Death Dis, 2020. 11(4): p. 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu X, et al. , SOCS3 promotes TLR4 response in macrophages by feedback inhibiting TGF-beta1/Smad3 signaling. Mol Immunol, 2008. 45(5): p. 1405–13. [DOI] [PubMed] [Google Scholar]
- 157.Wolters TLC, et al. , IGF1 potentiates the pro-inflammatory response in human peripheral blood mononuclear cells via MAPK. J Mol Endocrinol, 2017. 59(2): p. 129–139. [DOI] [PubMed] [Google Scholar]
- 158.Wang JW, et al. , Loganin alleviates LPS-activated intestinal epithelial inflammation by regulating TLR4/NF-kappaB and JAK/STAT3 signaling pathways. Kaohsiung J Med Sci, 2020. 36(4): p. 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Junyent F, et al. , Gene expression profile in JNK3 null mice: a novel specific activation of the PI3K/AKT pathway. J Neurochem, 2011. 117(2): p. 244–52. [DOI] [PubMed] [Google Scholar]
- 160.Nakano R, Nakayama T, and Sugiya H, Biological Properties of JNK3 and Its Function in Neurons, Astrocytes, Pancreatic beta-Cells and Cardiovascular Cells. Cells, 2020. 9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tsaousidou E, et al. , Distinct Roles for JNK and IKK Activation in Agouti-Related Peptide Neurons in the Development of Obesity and Insulin Resistance. Cell Rep, 2014. 9(4): p. 1495–506. [DOI] [PubMed] [Google Scholar]
- 162.Barclay,J.L.,et al. , Characterization of the SOCS3 promoter response to prostaglandin E2 in T47D cells. Mol Endocrinol, 2007. 21(10): p. 2516–28. [DOI] [PubMed] [Google Scholar]
- 163.Gao S, Howard S, and LoGrasso PV, Pharmacological Inhibition of c-Jun N-terminal Kinase Reduces Food Intake and Sensitizes Leptin's Anorectic Signaling Actions. Sci Rep, 2017. 7: p. 41795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schriever SC, et al. , Type 2 diabetes risk gene Dusp8 regulates hypothalamic Jnk signaling and insulin sensitivity. J Clin Invest, 2020. 130(11): p. 6093–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krauss S,Zhang CY, and Lowell BB, The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol, 2005. 6(3): p. 248–61. [DOI] [PubMed] [Google Scholar]
- 166.Brand MD and Esteves TC, Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab, 2005. 2(2): p. 85–93. [DOI] [PubMed] [Google Scholar]
- 167.Graier WF, Trenker M, and Malli R, Mitochondrial Ca2+, the secret behind the function of uncoupling proteins 2 and 3? Cell Calcium, 2008. 44(1): p. 36–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Le Fur S, et al. , The common -866 G/A polymorphism in the promoter of uncoupling protein 2 is associated with increased carbohydrate and decreased lipid oxidation in juvenile obesity. Diabetes, 2004. 53(1): p. 235–9. [DOI] [PubMed] [Google Scholar]
- 169.Bulotta A, et al. , The common-866G/A polymorphism in the promoter region of the UCP-2 gene is associated with reduced risk of type 2 diabetes in Caucasians from Italy. J Clin Endocrinol Metab, 2005. 90(2): p. 1176–80. [DOI] [PubMed] [Google Scholar]
- 17 0.Su M, et al. , UCP2 and UCP3 variants and gene-environment interaction associated with prediabetes and T2DM in a rural population: a case control study in China. BMC Med Genet, 2018. 19(1): p. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pierelli G, et al. , Uncoupling Protein 2: A Key Player and a Potential Therapeutic Target in Vascular Diseases. Oxid Med Cell Longev, 2017. 2017: p. 7348372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arsenijevic D, et al. , Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet, 2000. 26(4): p. 435–9. [DOI] [PubMed] [Google Scholar]
- 173.Andrews ZB, et al. , UCP2 mediates ghrelin's action on NPY/AgRP neurons by lowering free radicals. Nature, 2008. 454(7206): p. 846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yu X, et al. , Association of UCP2 –866 G/A polymorphism with chronic inflammatory diseases. Genes Immun, 2009. 10(6): p. 601–5. [DOI] [PubMed] [Google Scholar]
- 175.Jin X, et al. , miRNA-133a-UCP2 pathway regulates inflammatory bowel disease progress by influencing inflammation, oxidative stress and energy metabolism. World J Gastroenterol, 2017. 23(1): p. 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moon JS, et al. , UCP2-induced fatty acid synthase promotes NLRP3 inflammasome activation during sepsis. J Clin Invest, 2015. 125(2): p. 665–80. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 177.Fisher E, et al. , Association of the FTO rs9939609 single nucleotide polymorphism with C-reactive protein levels. Obesity (Silver Spring), 2009. 17(2): p. 330–4. [DOI] [PubMed] [Google Scholar]
- 178.Alipour M, Rostami H, and Parastouei K, Association between inflammatory obesity phenotypes, FTO-rs9939609, and cardiovascular risk factors in patients with type 2 diabetes. J Res Med Sci, 2020. 25: p. 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Olza J, et al. , Influence of FTO variants on obesity, inflammation and cardiovascular disease risk biomarkers in Spanish children: a case-control multicentre study. BMC Med Genet, 2013. 14: p. 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zimmermann E, et al. , Influences of the common FTO rs9939609 variant on inflammatory markers throughout a broad range of body mass index. PLoS One, 2011. 6(1): p. e15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fan HQ, et al. , FTO Inhibits Insulin Secretion and Promotes NF-kappaB Activation through Positively Regulating ROS Production in Pancreatic beta cells. PLoS One, 2015. 10(5): p. e0127705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kamermans A, et al. , Setmelanotide, a Novel, Selective Melanocortin Receptor-4 Agonist Exerts Anti-inflammatory Actions in Astrocytes and Promotes an Anti-inflammatory Macrophage Phenotype. Front Immunol, 2019. 10: p. 2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Carniglia L, et al. , Effect of NDP-alpha-MSH on PPAR-gamma and -beta expression and anti-inflammatory cytokine release in rat astrocytes and microglia. PLoS One, 2013. 8(2): p. e57313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Caruso C, et al. , Melanocortin 4 receptor activation induces brain-derived neurotrophic factor expression in rat astrocytes through cyclic AMP-protein kinase A pathway. Mol Cell Endocrinol, 2012. 348(1): p. 47–54. [DOI] [PubMed] [Google Scholar]
- 185.Caruso C, et al. , Activation of melanocortin 4 receptors reduces the inflammatory response and prevents apoptosis induced by lipopolysaccharide and interferon-gamma in astrocytes. Endocrinology, 2007. 148(10): p. 4918–26. [DOI] [PubMed] [Google Scholar]
- 186.McFadden JW, et al. , Increasing fatty acid oxidation remodels the hypothalamic neurometabolome to mitigate stress and inflammation. PLoS One, 2014. 9(12): p. e115642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Valdearcos M, Xu AW, and Koliwad SK, Hypothalamic inflammation in the control of metabolic function. Annu Rev Physiol, 2015. 77: p. 131–60. [DOI] [PubMed] [Google Scholar]
- 188.Matarazzo V, et al. , Inactivation of Socs3 in the hypothalamus enhances the hindbrain response to endogenous satiety signals via oxytocin signaling. J Neurosci, 2012. 32(48): p. 17097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Heldsinger A, et al. , Ghrelin induces leptin resistance by activation of suppressor of cytokine signaling 3 expression in male rats: implications in satiety regulation. Endocrinology, 2014. 155(10): p. 3956–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pedroso JA, et al. , Changes in Leptin Signaling by SOCS3 Modulate Fasting-Induced Hyperphagia and Weight Regain in Mice. Endocrinology, 2016. 157(10): p. 3901–3914. [DOI] [PubMed] [Google Scholar]
- 191.Timp er K andBruni ng JC,Hypothala miccircu itsregulat ingappetite andenergy homeostasis: pathways to obesity. Dis Model Mech, 2017. 10(6): p. 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kaneto H, et al. , Possible novel therapy for diabetes with cell-permeable JNK-inhibitory peptide. Nat Med, 2004. 10(10): p. 1128–32. [DOI] [PubMed] [Google Scholar]
- 193.Unger EK, et al. , Functional role of c-Jun-N-terminal kinase in feeding regulation. Endocrinology, 2010. 151(2): p. 671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nogueiras R and Sabio G, Brain JNK and metabolic disease. Diabetologia, 2021. 64(2): p. 265–274. [DOI] [PubMed] [Google Scholar]
- 195.Diano S and Horvath TL, Mitochondrial uncoupling protein 2(UCP2) in glucose and lipid metabolism. Trends Mol Med, 2012. 18(1): p. 52–8. [DOI] [PubMed] [Google Scholar]
- 196.Tilg H, et al. , The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol, 2020. 20(1): p. 40–54. [DOI] [PubMed] [Google Scholar]
- 197.de Wit N, et al. , Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am J Physiol Gastrointest Liver Physiol, 2012. 303(5): p. G589–99. [DOI] [PubMed] [Google Scholar]
- 198.Patterson E, et al. , Impact of dietary fatty acids on metabolic activity and host intestinal microbiota composition in C57BL/6J mice. Br J Nutr, 2014. 111(11): p. 1905–17. [DOI] [PubMed] [Google Scholar]
- 199.Lang JM, et al. , Impact of Individual Traits, Saturated Fat, and Protein Source on the Gut Microbiome. mBio, 2018. 9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.de La Serre CB, et al. , Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol, 2010. 299(2): p. G440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fouesnard M, et al. , Dietary switch to Western diet induces hypothalamic adaptation associated with gut microbiota dysbiosis in rats. Int J Obes (Lond), 2021. [DOI] [PubMed] [Google Scholar]
- 202.O'Brown NM, Pfau SJ, and Gu C, Bridging barriers: a comparative look at the blood-brain barrier across organisms. Genes Dev, 2018. 32(7–8): p. 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kanoski SE, et al. , The effects of a high-energy diet on hippocampal function and blood-brain barrier integrity in the rat. J Alzheimers Dis, 2010. 21(1): p. 207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kanoski SE and Davidson TL, Western diet consumption and cognitive impairment: links to hippocampal dysfunction and obesity. Physiol Behav, 2011. 103(1): p. 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Davidson TL, et al. , The effects of a high-energy diet on hippocampal-dependent discrimination performance and blood-brain barrier integrity differ for diet-induced obese and diet-resistant rats. Physiol Behav, 2012. 107(1): p. 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Braniste V, et al. , The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med, 2014. 6(263): p. 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tang W, et al. , The Impact of Gut Microbiota Disorders on the Blood-Brain Barrier. Infect Drug Resist, 2020. 13: p. 3351–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Barnes S, et al. , Omega-3 fatty acids are associated with blood-brain barrier integrity in a healthy aging population. Brain Behav, 2021. 11(8): p. e2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rakhra V, et al. , Obesity and the Western Diet: How We Got Here. Mo Med, 2020. 117(6): p. 536–538. [PMC free article] [PubMed] [Google Scholar]
- 210.Kopp W, How Western Diet And Lifestyle Drive The Pandemic Of Obesity And Civilization Diseases. Diabetes Metab Syndr Obes, 2019. 12: p. 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Clegg DJ, et al. , Consumption of a high-fat diet induces central insulin resistance independent of adiposity. Physiol Behav, 2011. 103(1): p. 10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mark AL, Dietary therapy for obesity is a failure and pharmacotherapy is the future: a point of view. Clin Exp Pharmacol Physiol, 2006. 33(9): p. 857–62. [DOI] [PubMed] [Google Scholar]
- 213.Saunders KH, et al. , Obesity Pharmacotherapy. Med Clin North Am, 2018. 102(1): p. 135–148. [DOI] [PubMed] [Google Scholar]
- 214.Tak YJ and Lee SY, Long-Term Efficacy and Safety of Anti-Obesity Treatment: Where Do We Stand? Curr Obes Rep, 2021. 10(1): p. 14–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Uysal KT, et al. , Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature, 1997. 389(6651): p. 610–4. [DOI] [PubMed] [Google Scholar]
- 216.Tsukumo DM, et al. , Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes, 2007. 56(8): p. 1986–98. [DOI] [PubMed] [Google Scholar]
- 217.Mendes NF, et al. , TGF-beta 1 down-regulation in the mediobasal hypothalamus attenuates hypothalamic inflammation and protects against diet-induced obesity. Metabolism-Clinical and Experimental, 2018. 85: p. 171–182. [DOI] [PubMed] [Google Scholar]
- 218.Romanatto T, et al. , Deletion of tumor necrosis factor-alpha receptor 1(TNFR1) protects against diet-induced obesity by means of increased thermogenesis. J Biol Chem, 2009. 284(52): p. 36213–36222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 219.Turnwald BP, et al. , Learning one's genetic risk changes physiology independent of actual genetic risk. Nat HumBehav, 2019. 3(1): p. 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ahn WK and Lebowitz MS, An experiment assessing effects of personalized feedback about genetic susceptibility to obesity on attitudes towards diet and exercise. Appetite, 2018. 120: p.23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]