Summary
The tympanic membrane (TM) is critical for hearing and requires continuous clearing of cellular debris, but little is known about homeostatic mechanisms in TM epidermis. Using single-cell RNA sequencing, lineage tracing, whole-organ explant, and live-cell imaging, we show that homeostatic TM epidermis is distinct from other epidermal sites and has discrete proliferative zones with a three-dimensional hierarchy of multiple keratinocyte populations. TM stem cells reside in a discrete location of the superior TM and generate long-lived clones and committed progenitors (CPs). CP clones exhibit lateral migration and their proliferative capacity is supported by Pdgfra+ fibroblasts, generating migratory but non-proliferative progeny. Single-cell sequencing of human TM revealed similar cell types and transcriptional programming. Thus, during homeostasis, TM keratinocytes transit through a proliferative CP state and exhibit directional lateral migration. This work forms a foundation for understanding TM disorders and modeling keratinocyte biology.
eTOC blurb
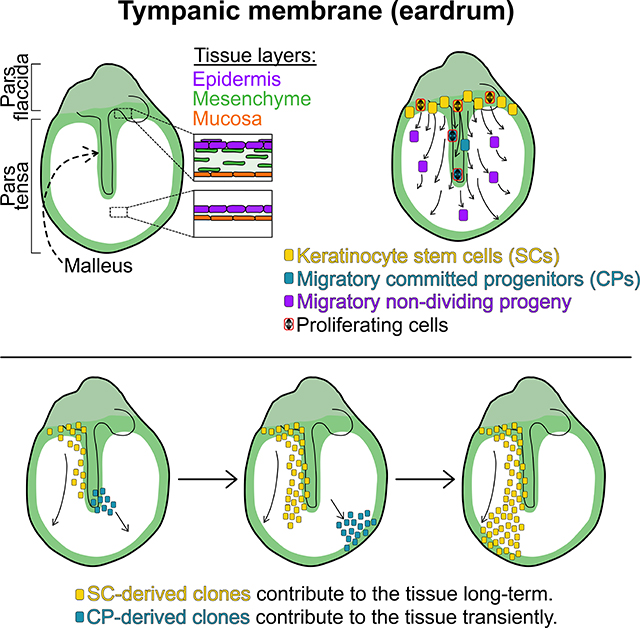
Tward and colleagues show that the tympanic membrane (TM) epidermis has distinct stem cell and committed progenitor regions. In homeostasis, keratinocytes migrate across the TM to maintain a thin vibratory surface, and they proliferate only when in proximity to supporting mesenchyme.
Graphical Abstract
Introduction
Disorders of the tympanic membrane (TM), also known as the eardrum, and middle ear space are common and morbid. In the United States, over 750,000 surgical procedures are performed on the TM each year (Hall et al., 2017), and in 2006 there were 8,000,000 physicians visits by children for ear infections (Soni, 2008). TM disorders including perforation, retraction, cholesteatoma, tympanosclerosis, bullous myringitis, and keratosis obturans can lead to decreased hearing, chronic infections, vertigo, and, in severe cases, meningitis or death. Despite its importance, little is known about how TM cellular homeostasis is maintained.
The TM is part of the conductive apparatus of the ear (Figure 1A). It transmits sound vibrations from the external auditory canal to the middle ear ossicles, which transfer those vibrations to the cochlea. The TM is comprised of two anatomic regions (Figure 1B). The pars tensa vibrates in response to sound waves, and the long process of the first ossicle, the malleus, is embedded within its mesenchymal layer (Figure 1C). The pars flaccida is believed to contribute less to sound transmission. The outer layer of the TM is epidermis continuous with that of the canal. The TM is an unusual site of epidermis as it lacks adnexal structures such as hair follicles and sweat glands. Thus, it provides a unique site to study epidermal cellular dynamics in the absence of confounding from other skin stem cell populations.
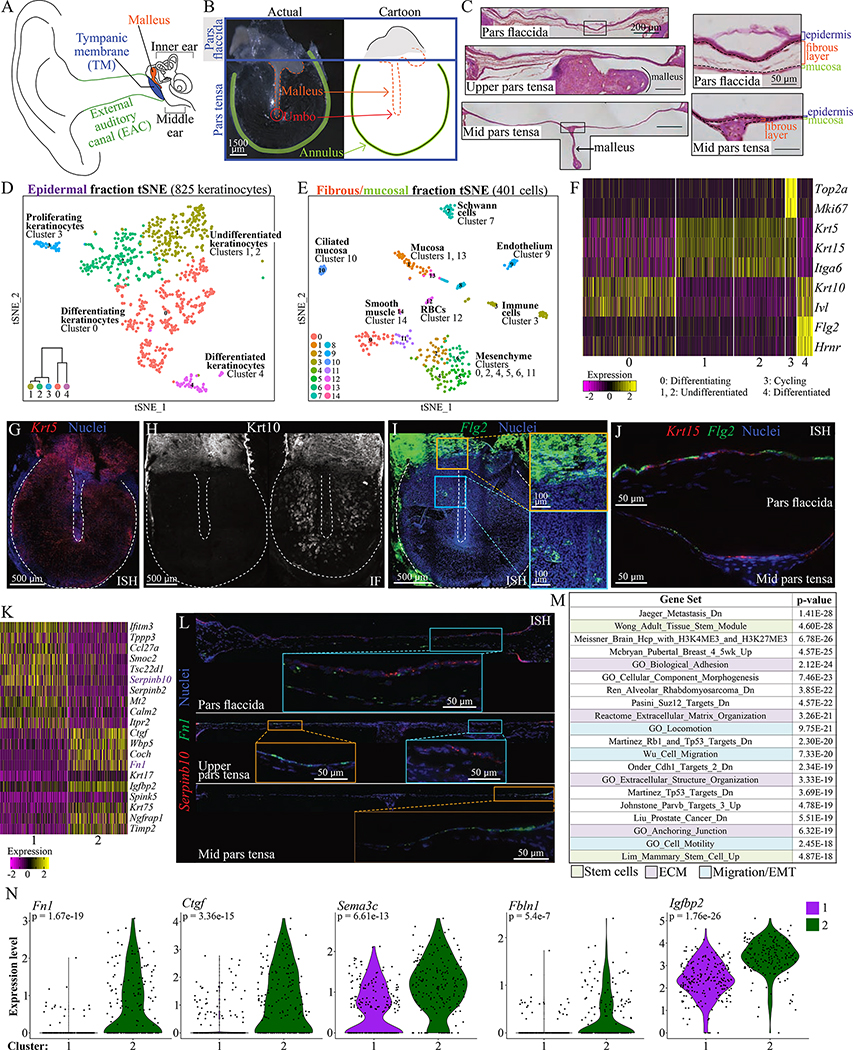
Figure 1: Single-cell RNA sequencing identifies cell types in the murine TM.
(A) Diagram of the ear. (B) A murine TM dissected en bloc and a cartoon representation. (C) TM sections at the pars flaccida (top), upper pars tensa (middle) and mid pars tensa (bottom) with scale bar = 200 μm. Tissue layers are indicated in magnified regions to the right, with scale bar = 50 μm. (D) t-Distributed Stochastic Neighbor Embedding (tSNE) visualization of keratinocyte clusters in the epidermis. The dataset includes 93 cells of mesenchymal, mucosal, or immune types not represented. Dendrogram inset was created with Morpheus and the top 100 differentially upregulated genes in each group (“Morpheus,” n.d.). (E) tSNE of clusters in the fibrous/mucosal fraction. (F) Heat-map showing expression of genes associated with proliferation and differentiation in the keratinocyte clusters. Each column is a cell. (G) ISH for Krt5 in a whole-mount TM. (H) Immunofluorescence (IF) for Krt10 in two whole-mount TMs, illustrating extremes of K10+ cell distribution. (I) ISH for Flg2 in a whole-mount TM. The yellow inset is at the pars flaccida/pars tensa junction; the blue inset is an area of the pars tensa over the malleus. (J) ISH for Flg2 and Krt15 in TM sections. Epidermis is oriented upward. (K) Heat-map showing genes differentially expressed between keratinocyte clusters 1 and 2. (L) ISH for Serpinb10 and Fn1 in TM sections from the pars flaccida (top, ~1500 μm across), upper pars tensa (middle, ~1650 μm across), and mid pars tensa (bottom, ~1800 μm across). Epidermis is oriented upward. (M) Top MSigDB gene sets enriched for genes at least 1.2-fold and significantly upregulated in keratinocyte cluster 2 relative to 1. (N) Violin plots showing expression of Fn1, Ctgf, Sema3c, Fbln1, and Igfbp2 in cells from keratinocyte clusters 1 and 2. Images in C, G-J, and L were acquired in tiles and stitched. Images in G-I are maximum projections of z-stacks. See also Figures S1–3 and Tables S1–2.
In addition to performing the barrier function of any epidermis, the epidermis of the TM and external auditory canal must also prevent accumulation of cellular and exogenous debris via a cleaning mechanism. Since the canal terminates in a blind sac at the TM, if superficial keratinocytes were simply sloughed off, the canal would fill with debris. This would prevent sound transmission, create a fertile environment for infection, and cause erosion of surrounding structures. Variations of this situation occur in cholesteatoma, keratosis obturans, and cerumen impaction (Corbridge et al., 1996; Kuo et al., 2015; Marchisio et al., 2016; Soucek and Michaels, 1993). Previous work demonstrated that ink spots placed on the TMs of mammals, including humans, migrate laterally along the TM and down the canal (Alberti, 1964; Kakoi et al., 1996; Litton, 1963; O’Donoghue, 1984; Stinson, 1936; Tinling and Chole, 2006). Pulse-chase experiments with the thymidine analog BrdU indicated that TM cell proliferation is not uniform, but restricted to a few areas, and that newly proliferated cells are migratory (Kakoi and Anniko, 1997; Koba, 1995). Additional studies demonstrated clonogenic capacity and migratory behavior in vitro of epithelial cells derived from rodent and human TMs (Liew et al., 2018, 2017; Ong et al., 2016; Redmond et al., 2011). Furthermore, following perforation there is a massive cellular response typically leading to rapid healing in all mammals, including humans, investigated thus far (Chari et al., 2019; Lou et al., 2012; Rollin et al., 2011; Santa Maria et al., 2010). Yet, our knowledge of the cellular identities, dynamics, and mechanisms of TM homeostasis is otherwise limited.
At other anatomical sites, epidermis is maintained by a population of basal keratinocyte stem cells (SCs) capable of both self-renewal as well as differentiation, and that exhibit neutral competition kinetics in most studied homeostatic contexts (Clayton et al., 2007; Doupé et al., 2010; Lim et al., 2013; Mascré et al., 2012). Some evidence also exists for a hierarchical differentiation scheme whereby the SCs give rise to a proliferative committed progenitor (CP) or transit amplifying population with decreased propensity for long term tissue maintenance (Jones and Simons, 2008; Mascré et al., 2012; Sada et al., 2016). Part of the challenge in distinguishing between these models is that both the SC and putative CP populations exist in similar locations and express similar markers, and thus the strongest evidence for the existence of keratinocyte CPs comes from mathematical modeling of the fates of clonal progeny (Mascré et al., 2012). Additionally, daughter cells of proliferating skin keratinocytes either remain in the basal layer or commit to differentiation and move apically. Although basal patches arise from individual marked SCs over time, it is believed that, in uninjured skin, basal keratinocytes do not exhibit lateral migration (Rompolas et al., 2016). After wounding, basal (and likely suprabasal) keratinocytes undergo a transcriptional shift and acquire the property of active and directional lateral migration (Grinnell, 1990; Gurtner et al., 2008; Park et al., 2017). The TM epidermis is thus distinct, as the ink dot studies suggest lateral migration occurs in homeostasis and injury. How this impacts tissue maintenance, and whether progenitor populations equivalent to those seen at other sites exist in the TM, has not been evaluated.
In this work, we combine single-cell RNA sequencing (scRNA-Seq), live cell imaging, lineage tracing, and an explant model to explore TM biology. We demonstrate that, under homeostasis, the TM possesses at least three distinct populations of basal keratinocytes: a SC population arising from the junction between the pars tensa and pars flaccida with long-term renewal capability, a CP population near the long process of the malleus in the pars tensa that exhibits proliferative and migratory capacity but lacks long-term renewal potential, and a migratory population throughout the pars tensa that does not proliferate. We also describe a system to maintain murine TMs in culture and implicate Pdgf signaling in the maintenance of regional proliferation of TM keratinocytes.
Results
Identifying cell populations in the murine TM
Prior work described three TM layers—epidermis, fibrous layer, and mucosa—but not the full diversity of cell types (Figure 1C). We sought to define the resident cell populations of the TM via scRNA-Seq using the 10x Genomics Single Cell Solution v2 platform. Thirty-eight adult murine TMs were pooled and enzymatically split into two anatomic fractions, epidermal and fibrous/mucosal, to optimally dissociate different cell types. In a preliminary experiment, we achieved single-cell suspensions of epidermal cells with 79% viability and of fibrous/mucosal cells with 86% viability. Two corresponding datasets were generated and analyzed with Seurat (Figure 1D–E, Tables S1) (Satija et al., 2015).
In the epidermal and fibrous/mucosal fractions, respectively, 918 and 401 cells were captured (Figure 1D–E). In the epidermal fraction, 825 keratinocytes clustered into five populations (0, 1, 2, 3, and 4; Figure 1D). The remaining 93 cells—mesenchymal, mucosal, and immune cells—were removed so as not to confound downstream analyses. Cluster 3 keratinocytes are cycling, marked by Top2a and Mki67 expression (Figure 1F) and enrichment of cell cycle gene sets (Table S2). Almost no cycling cells are seen in the other tissue layers (Figure S1A). When the cell cycle signal was regressed out, cluster 3 cells redistributed amongst clusters 0, 1, and 2 (data not shown). The prominent axis of distinction among the other keratinocyte clusters is differentiation state. Clusters 1 and 2 are relatively undifferentiated; they express basal keratinocytes markers (e.g. Krt5, Krt15, Itga6) and likely represent cells with proliferative potential not dividing at the time of capture (Figure 1F). Cluster 0 is an intermediate differentiation state; cells have lost expression of basal markers and gained expression of differentiation-associated genes (e.g. Krt10, Ivl). Lastly, cluster 4 expresses markers of terminal differentiation (e.g. Flg2, Hrnr). To determine the locations of these populations, we utilized in situ hybridization (ISH) and immunofluorescence (IF). Basal-like Krt5+ cells exist throughout the TM (Figure 1G). The location of more differentiated cells varied; of 34 TMs stained for Krt10, all had expression in the pars flaccida, but expression in the pars tensa was absent in seven (21%), intermediate in 13 (38%), and prominent in 14 (41%), although never seen over the malleus or annulus (Figure 1H). Flg2 is strongly expressed in the pars flaccida, while only occasional cells in the pars tensa express this gene (Figure 1I). In a pars flaccida section, many cells are Flg2+, often overlying less differentiated Krt15+ keratinocytes. In a pars tensa section, Flg2+ cells are observed in the single-cell-thick epidermis (Figure 1J). Thus, the pars flaccida exhibits both differentiation and stratification, whereas the pars tensa exhibits variable (but predominantly basal-like) differentiation with minimal stratification. Overall, the keratinocyte gene expression profiles were similar to those observed in other interfollicular epidermis (Aragona et al., 2017; Cheng et al., 2018; Joost et al., 2018, 2016).
Next, we identified genes differentially expressed between putative progenitor clusters 1 and 2 (Figure 1K). Serpinb10 (cluster 1) and Fn1 (cluster 2) were discriminating markers, confirmed by ISH (Figure 1L). Pars flaccida keratinocytes predominantly express Serpinb10; Fn1 is expressed in this region by fibroblasts. In the pars tensa, a mid-malleus section has only an Fn1+ keratinocyte population, while an upper pars tensa section has separate Fn1+ and Serpinb10+ areas. Regions of adjacent Serpinb10+ and Fn1+ keratinocytes were not observed. In whole-mount TMs, Fn1+ cells are seen near the annulus and over the malleus (Figure S1B). Thus, clusters 1 is Serpinb10+ keratinocytes predominantly in the pars flaccida, and cluster 2 is Fn1+ keratinocytes predominantly in the pars tensa. To further understand the functional distinctions between these clusters, we queried Molecular Signatures Database (MSigDB) for gene sets enriched for genes at least 1.2-fold and significantly (p < 0.05) upregulated in cluster 1 relative to cluster 2 and vice versa (85 and 157 genes, respectively) (Liberzon et al., 2015, 2011; Subramanian et al., 2005). For cluster 1, many enriched terms related to ribosome function and biogenesis (Table S2). For cluster 2, many enriched terms related to cell migration and extracellular matrix (ECM) (Figure 1M, Table S2). The genes upregulated in cluster 2 included Fn1, Ctgf (Kiwanuka et al., 2013), Sema3c (Curreli et al., 2016), Fbln1 (Feitosa et al., 2012), and Igfbp2 (Li et al., 2020), which are characteristic of migratory cells (Figure 1N).
The same approaches were used to define and locate clusters in the fibrous/mucosal fraction. There are multiple Vimentin+ (Vim+) mesenchymal populations, including: Pecam1+ endothelium, Acta2+ smooth muscle, and Mbp+ Schwann cells (Figure S2A). Additional mesenchymal clusters were identified: clusters 0 and 11 express Gpx3 and reside near bony elements (the annulus and malleus) while clusters 2, 4, 5, and 6 express Igfbp3 and are more diffusely spread throughout the fibrous layer (Figure S2B–D). Sox2 is a known marker of mucosa and is expressed in non-ciliated (clusters 1 and 13) and ciliated (cluster 10) mucosal clusters (Figure S3A) (Tucker et al., 2018). Cluster 10 is identifiable as ciliated by expression of motor proteins (Figure S3B). The mucosal clusters express Krt19 while the keratinocyte clusters do not; restriction of Krt19 to the mucosal cells was confirmed by lineage tracing (Figure S3C). Of note, Krt19 has been proposed as a marker of TM epidermal stem cells in human and rat (Kim et al., 2015; Knutsson et al., 2011).
scRNA-Seq of human TM tissue
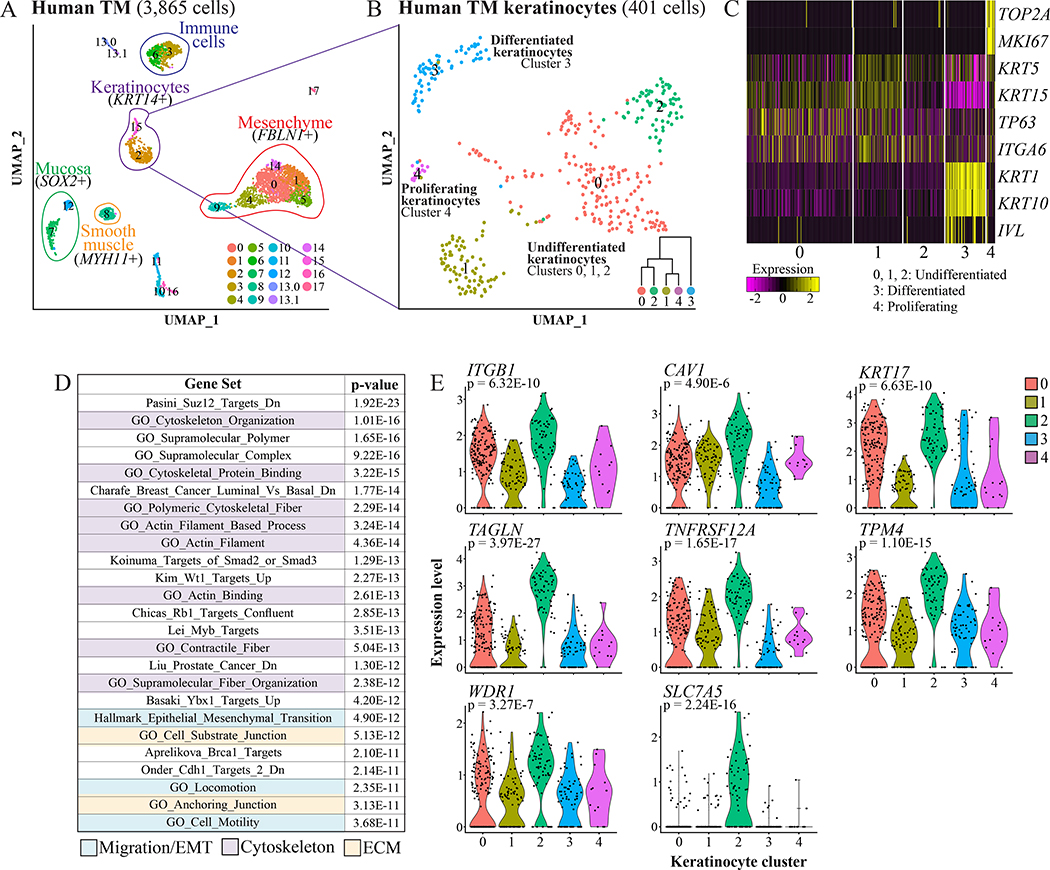
Having cataloged murine TM populations, we addressed whether the human TM has equivalent cell types. Freshly isolated normal TM tissue obtained during a surgical approach for a skull base tumor was mechanically separated into lateral and medial portions, then dissociated and subjected to scRNA-Seq. The resulting datasets were merged and analyzed with the iterative clustering algorithm CellFindR (Yu et al., 2019). Collectively, 3,865 cells were captured and 19 clusters identified, including KRT14+ keratinocytes, FBLN1+ mesenchyme, SOX2+ mucosa, and MYH11+ smooth muscle (Figure 2A, Table S3). The 401 keratinocytes were re-clustered, yielding five groups (Figure 2B, Table S3). As was seen in the murine TM, there is an MKI67+ proliferating cluster (4) and evidence of multiple differentiation states exemplified by differential expression of KRT5, KRT15, TP63, ITGA6, KRT1, and KRT10 (Figure 2C).
Figure 2: scRNA-Seq of the human TM.
(A) UMAPs of clusters identified in a human TM, analyzed with the CellFindR algorithm. (B) UMAPs of the keratinocytes re-clustered independent from other cell types. Dendrogram inset was created with Morpheus and the top 50 significantly differentially upregulated genes in each group (“Morpheus,” n.d.). (C) Heat-map showing expression of genes associated with proliferation and differentiation in the keratinocyte clusters. Each column is a cell. (D) Top MSigDB gene sets enriched for genes at least 1.5-fold and significantly upregulated in human keratinocyte cluster 2 relative to the other keratinocytes. (E) Violin plots of expression of ITGB1, CAV1, KRT17, TAGLN, TNFRSF12A, TPM4, WDR1, and SLC7A5 in the keratinocyte clusters. p values are for expression in cluster 2 relative the rest of the keratinocyte dataset. See also Tables S3–4.
We next asked if any of the basal-like human keratinocyte clusters (0, 1, and 2) upregulated transcriptional programs similar to those of the migratory murine cluster 2. For human keratinocyte clusters 0, 1, and 2, we took the genes at least 1.5-fold and significantly (p < 0.05) upregulated relative to the rest of the keratinocyte dataset (38, 26, and 51 genes, respectively), queried MSigDB, and discovered that cluster 2 had enrichment of gene sets relating to migration, cytoskeleton, and ECM (Figure 2D, Table S4). The genes upregulated in this cluster that have been reported to promote migration include ITGB1 (Tang et al., 2017), CAV1 (Yang et al., 2018), KRT17 (Khanom et al., 2016), TAGLN (Yu et al., 2008), TNFRSF12A (Liu et al., 2019), TPM4 (Zhao et al., 2019), WDR1 (Yuan et al., 2018), and SLC7A5 (Janpipatkul et al., 2014) (Figure 2E).
EdU incorporation demonstrates rapid turnover of the TM epidermis
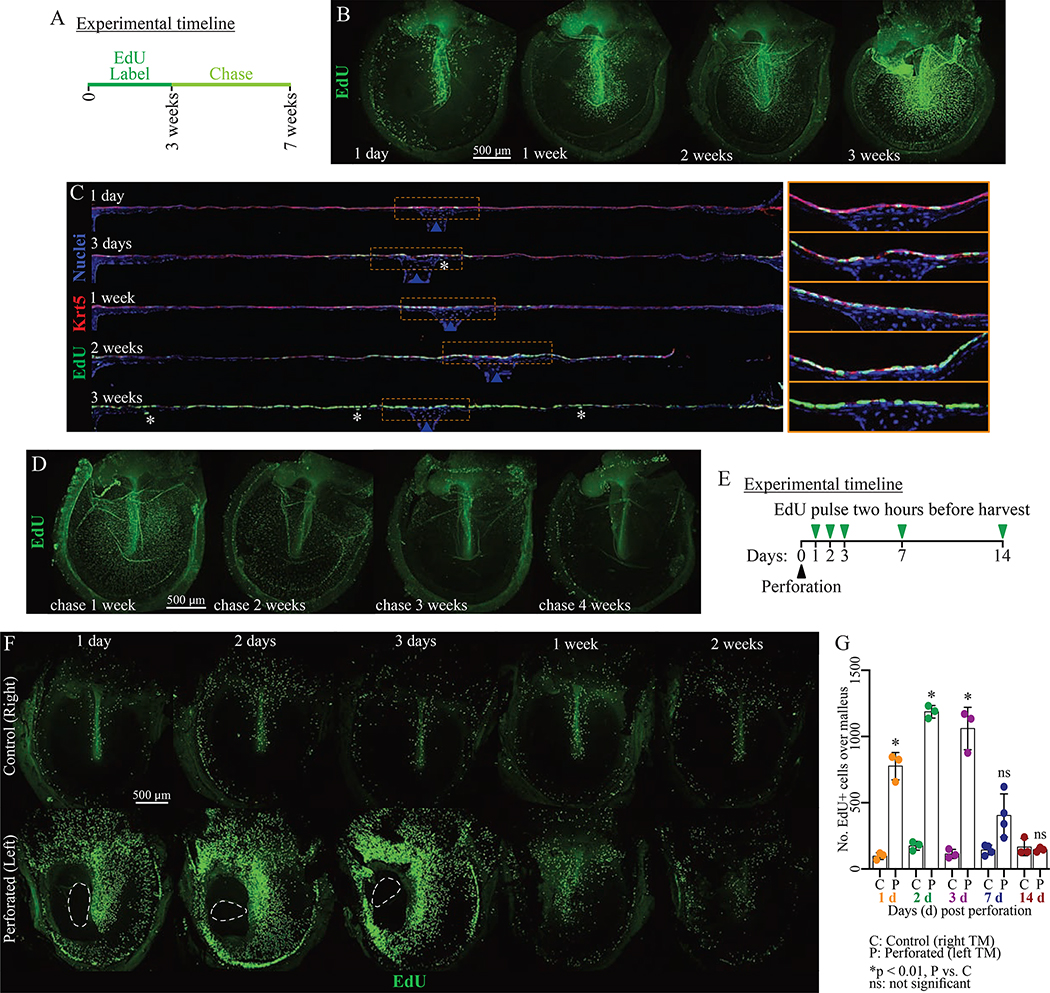
We next characterized cell turnover in the murine TM. We confirmed prior descriptions of proliferation and migration of pars tensa cells with a single EdU injection and chase (Figure S4A) (Kakoi and Anniko, 1997). Cycling cells were seen (1) near the handle of the malleus, (2) near the annulus, (3) in the superior pars tensa, and (4) in the pars flaccida at early timepoints. By later timepoints, labeled cells migrated to intermediate regions of the pars tensa. We next identified which populations were being replenished and how rapidly. We gave mice continuous EdU via their water and observed label accumulation over three weeks, and then removed the EdU and observed label dilution over four weeks (Figure 3A). EdU labeling over the malleus is dense by one week, and the area of EdU+ cells expands outward until nearly all of the keratinocytes have incorporated label by three weeks (Figure 3B). The majority of cells incorporating EdU are Krt5+ keratinocytes (Figure 3C). Rare fibroblasts and mucosal cells also incorporate EdU. Thus, essentially all of the keratinocytes turn over within three weeks and other cell populations are largely quiescent. Rapid turnover of the epidermis was again observed during the four-week chase (Figures 3D).
Figure 3: The murine TM epidermis turns over rapidly.
(A) Pulse-chase experiment timeline. Mice were exposed to EdU continuously for 3 weeks (pulse), then the label allowed to dilute for 4 weeks (chase). (B, D) Representative whole-mount TMs at indicated (B) pulse and (D) chase timepoints and stained for EdU. (C) Mid-malleus sections (~1700 μm across) at indicated pulse timepoints stained for EdU and Krt5. Images were acquired in tiles and stitched, and TMs were artificially straightened in ImageJ. Asterisks indicate EdU+/Krt5- cells. Blue arrows indicate the malleus. Regions within yellow rectangles are magnified and not straightened to the right. (E) Perforation experiment timeline. (F) Representative whole-mount TMs from perforated (left) and control (right) TMs stained for EdU. On days 1–3, the dashed line indicates the perforation. (G) Number of EdU+ cells in a 400×1200 μm area over the malleus in the perforation time-course. Each dot is 1 TM; 3 animals were analyzed per timepoint. Histogram indicates mean and error bars standard deviation. Results of t-tests for control versus perforated TMs at a single timepoint are indicated. Throughout, scale bars apply to all images in a panel. Images in B, D and F are maximum projections of z-stacks. See also Figure S4.
Wounding alters the proliferation pattern in murine TM
Perforations are a common TM injury. The majority heal without intervention, but some become chronic and morbid (Lou et al., 2012). Further, perforations are sometimes made therapeutically, as in placement of tympanostomy tubes for recurrent otitis media. We previously reported increased proliferation of all three tissue layers in response to perforation (Chari et al., 2019). Here, we aimed to understand how the regional proliferation pattern is affected by wounding. We created perforations in the left TMs of mice, using the right TMs as controls, and pulsed them with EdU two hours prior to harvesting (Figure 3E–G, S4B–C). By 24 hours following injury, there is increased proliferation in all of the zones that proliferate under homeostasis—proximal to the malleus, upper pars tensa, annulus, and pars flaccida—with some extension into the usually non-proliferative pars tensa intermediate zones. On days two and three, there is more proliferation in these intermediate regions, although there is still a region without proliferation on side of the malleus opposite the injury. By one week, the proliferation pattern is reverting to its homeostatic baseline despite the increased tissue volume (Figure S4B). We assessed which cells contribute to healing by staining mid-malleus sections (Figure S4B–C). There is a proliferative response in the mesenchyme (Vim+ cells) and epidermis (superficial to the Vim+ cells). Further, Fn1+ keratinocytes (migratory cluster 2) are seen throughout the epidermis on day three post perforation and more basally on day seven (Figure S4C). There is also Fn1 expression by the reactive mesenchyme in the pars tensa, whereas Fn1+ mesenchyme was restricted to the pars flaccida in homeostasis (Figure 1L).
Live-cell imaging reveals TM keratinocyte migration patterns
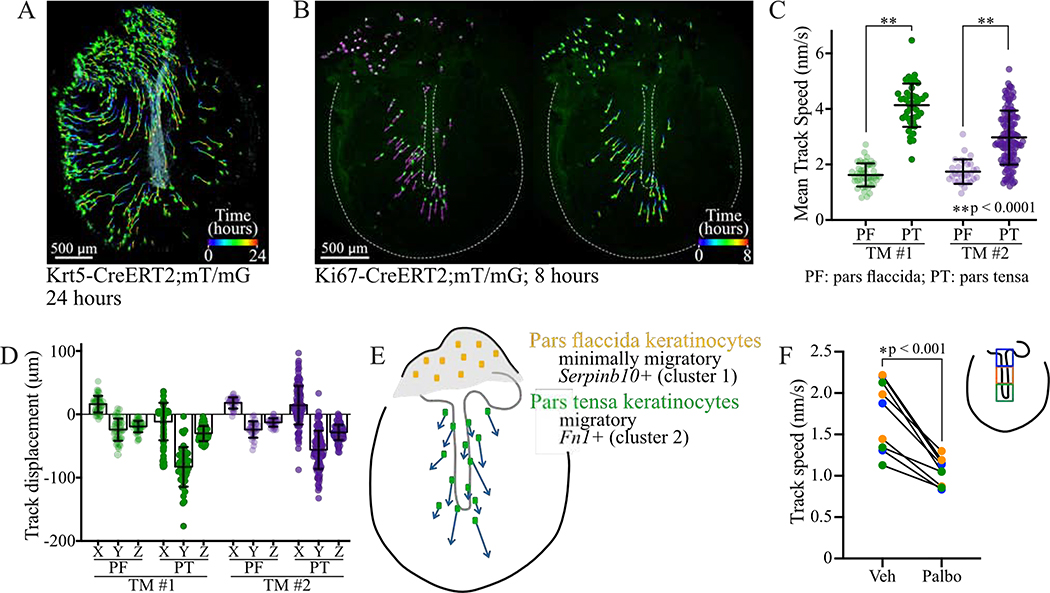
To understand the migratory behavior of TM keratinocytes, we established techniques for time-lapse imaging of TM explants. We employed Krt5-CreERT2 to label cells throughout the TM and Ki67-CreERT2 to target proliferative zones. We used both the mT/mG reporter, in which labeled cells express EGFP, and R26R-Confetti, in which recombination leads to expression of one of four fluorescent proteins (RFP, YFP, mCFP, nGFP) (Basak et al., 2018; Muzumdar et al., 2007; Rock et al., 2009; Snippert et al., 2010). In a representative experiment, a subset of basal-like keratinocytes was labeled in a Krt5-CreERT2;mT/mG mouse. The TM was harvested two days later and imaged hourly over 24 hours. Cells were tracked with Imaris. While radial movement was seen peripherally, the predominant direction of migration over the malleus was superior-to-inferior (Figure 4A, Movie 1).
Figure 4: Live cell imaging of TM explants reveals superior-to-inferior keratinocyte migration.
(A) Krt5-CreERT2;mT/mG TM imaged hourly for 24 hours. Cells were identified and tracked using Imaris. Cells are at their final position, with full tracks traced. (B) Ki67-CreERT2;mT/mG TM imaged hourly for eight hours. Left: cells at their initial positions with vectors showing the directions in which they will move. Right: cells at their final positions with tracks showing their paths. Images in A-B are captured from 3D renderings in Imaris. (C) Mean speed, with standard deviation (SD), of cells in the pars tensa (dark circles) and pars flaccida (light circles) for TMs imaged hourly from independent experiments. Data in green is from the TM in (B). (D) For the same two TMs, X, Y, and Z components of the displacement of cells, with mean and SD. (E) Cartoon summarizing live cell imaging and scRNA-Seq findings. (F) Speed of cell migration in TMs cultured with vehicle or 5 μM palbociclib for 24 hours and imaged every six hours. Results are shown for cells starting in 3 distinct regions over the malleus, depicted in the cartoon inset, and each paired set of points represents speeds during one time interval. Results of unpaired t-tests are indicated. See also Movies 1–2.
To observe recently proliferated cells, we performed imaging on explants from Ki67-CreERT2;mT/mG mice, with labeling predominantly in the pars flaccida and near the malleus. Time-lapse data was collected for 13 Ki67-CreERT2;mT/mG TMs, all with comparable qualitative findings (Figure 4B, Movie 2). Two TMs imaged hourly in independent experiments were analyzed further with Imaris. Pars tensa cells migrated with an average speed of 4.13 and 2.97 nm/sec in the two TMs, while pars flaccida cells migrated with an average speed of 1.63 and 1.74 nm/sec in the same respective TMs (Figure 4C). The average track displacement length was 103.78 and 72.09 μm for pars tensa cells, and 39.21 and 34.17 μm for pars flaccida cells. Breaking the displacement into directional components, the pars tensa cells predominantly move in the negative y-direction (Figure 4D). Thus, pars tensa keratinocytes exhibit directional lateral migration, and pars flaccida keratinocytes do not. These two populations likely correspond to undifferentiated murine clusters 2 (Fn1+) and 1 (Serpinb10+), respectively, as cluster 2 has a transcriptional signature enriched for genes associated with migration (Figures 4E, 1M). Notably, the observed migration is faster than the predicted in vivo speed, which may indicate that dissection and ex vivo culturing is akin to injury and activates migration. However, we hypothesize that the patterns and relative speeds of movement are preserved.
TM keratinocytes ultimately migrate away from proliferative regions to reach the external auditory canal. We determined if cell turnover in the proliferative zones influences migration rates across the TM by culturing explants from Krt5-CreERT2;R26R-Confetti mice in media with and without the Cdk inhibitor palbociclib. TMs were imaged four times over 24 hours, and images were aligned and cell tracks drawn manually. In the area over the malleus, palbociclib slowed the rate of cell movement (Figure 4F). We stained the TMs for cleaved caspase-3 and confirmed that there was no increase in apoptosis with Cdk inhibitor treatment (data not shown). Thus, proliferation and migration appear to be coupled.
Lineage tracing elucidates clonal architecture of TM keratinocytes
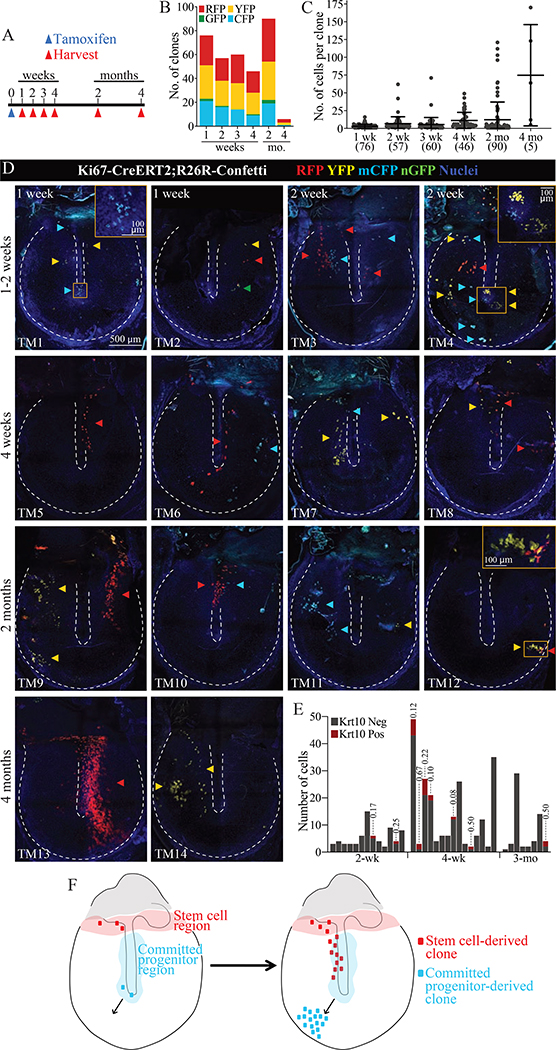
To further understand how proliferation and migration of TM keratinocytes cumulatively maintains the tissue, we determined the shape and location of clonal units of the TM epidermis. To visualize individual clones Ki67-CreERT2;R26R-Confetti mice were given 250 mg/kg tamoxifen to label rare cells and TMs were evaluated two days to four months later (Figures 5). At two days, 0–10 fluorescent cells per pars tensa were seen (data not shown). In 90 TMs evaluated between one week and two months following recombination, 329 pars tensa clones were confidently identified and the number of cells per clone quantified (Figure 5B–C). The majority of clones at every timepoint contained ten or fewer cells. However, the distribution of clone sizes expands at later timepoints. This scaling behavior has been described in other tissues, including testis, interfollicular epidermis, and intestine, and is consistent with neutral drift dynamics in the SC pool (Klein et al., 2010; Lopez-Garcia et al., 2010; Mascré et al., 2012).
Figure 5: Single TM keratinocyte clones arise from distinct stem cell and committed progenitor populations.
(A) Lineage tracing timeline. (B) Number of pars tensa clones expressing each fluorescent proteins. (C) Number of cells per pars tensa clone. Each dot represents a clone, and the error bars show mean and standard deviation. The total number of clones is indicated parenthetically below the x-axis. The large RFP+ clone captured at 4 months in panel (D), TM13, is not included as it could not be accurately quantified. (D) Examples of TMs from the indicated timepoints, with arrowheads annotating individual clones. The annulus and malleus are outlined by dotted lines. The 500 μm scale bar applies to all whole-mount panels. Images were acquired in tiles and stitched and are maximum projections of z-stacks. (E) IF for Krt10 was done on a subset of TMs. Each histogram bar is one clone. For clones with Krt10+ cells, the Krt10+ proportion is indicated above the bar. (F) Model of the location of stem cells and committed progenitors of the murine TM epidermis.
The morphology of larger clones was further informative. Cells were mostly not contiguous in pars tensa clones, indicating that progeny from multiple progenitors are intermixed as they migrate across the TM (Figure 5D). At one to two weeks, we captured several circular clones near the umbo of the malleus (Figure 5D, TMs 1 and 4), which has been identified as a major site of cell turnover (Knutsson et al., 2011; Liew et al., 2018). At later timepoints, we observed clones near the annulus (e.g., Figure 5D, TM 12) but never a streak radiating exclusively from the malleus to the annulus. These patterns suggest that proliferating cells around the malleus are CPs that migrate away from the umbo towards the external auditory canal (Figure 5F).
Another set of informative clones, observed at four weeks to four months, were elongated along the handle of the malleus or in streaks extending supero-inferiorly over the pars tensa (Figure 5D, TMs 5–10, 13, and 14). The morphology of these clones is consistent with the superior-to-inferior migration pattern observed in the time-lapse studies. Furthermore, most of these clones include cells in the upper pars tensa, near the border with the pars flaccida, which is a known proliferative region. Due to the presence of clones emanating from this region at later timepoints, we posit that the junction of the pars flaccida and pars tensa houses long-term repopulating SCs (Figure 5F). Lastly, we evaluated differentiation of clonal keratinocytes with Krt10 IF. Of 27 TMs stained at two weeks to four months, 12 TMs had 36 clones labeled by the reporter (Figure 5E). Only nine of 36 clones had Krt10+ cells, and these made up a small fraction each clone. Thus, the progeny of individual stem cells remain predominantly basal-like as they migrate across the TM.
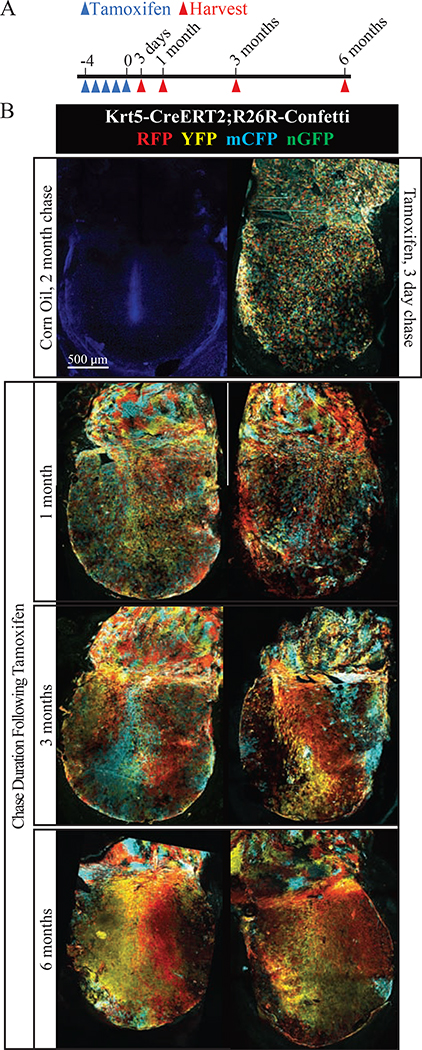
To check for rare contributions from long term repopulating SCs existing along the malleus or at other anatomic locations, we analyzed clonal architecture in densely labeled TMs from Krt5-CreERT2;R26R-Confetti mice (Figures 6, S5). At three days after maximal recombination, keratinocytes throughout the TM are labeled, and cells expressing RFP, YFP, and mCFP are randomly assorted. At six months, the entire TM epidermis remains fluorescent, indicating the long-term repopulating SCs are Krt5+. In the pars flaccida, color blocks emerge rapidly (by one month), indicating less cell mixing than in the pars tensa, consistent with less migratory behavior. This is reminiscent of prior observations in normal interfollicular epidermis.
Figure 6: Clonal drift in densely labeled TMs.
(A) Experimental timeline. (B) Representative whole-mount TMs from indicated timepoints. A TM from a control animal dosed with corn oil is included at the top left. The scale bar applies to all images. Images were acquired in tiles and stitched and are maximum projections of z-stacks. See also Figure S5.
In contrast, color blocks did not emerge at early timepoints in the pars tensa, consistent with active lateral migration and mixing of keratinocytes. However, at timepoints after multiple cycles of epidermal turnover, color streaks formed. Such coarsening is a hallmark of stochastic expansion of individual SC clones (Klein and Simons, 2011). Furthermore, the streaks were oriented supero-inferiorly and connected back to the border with the pars flaccida and never exclusively to the region over the middle or inferior malleus. This is consistent with a model whereby long-term repopulating SCs arise exclusively from the region adjacent to the pars flaccida, and their CP and other non-proliferating progeny migrate over the pars tensa in order to maintain a thin, vibratory surface free of cellular and exogenous debris. To quantify the progression towards tissue-wide oligoclonality, a grid was superimposed over TMs and each box was scored as containing cells of one to three colors (Figure S5). Over 90% of boxes contained mCFP, YFP, and RFP at three days and one month, and this percentage drops sharply at three and six months.
TM fibroblasts provide niche signals that promote keratinocyte turnover
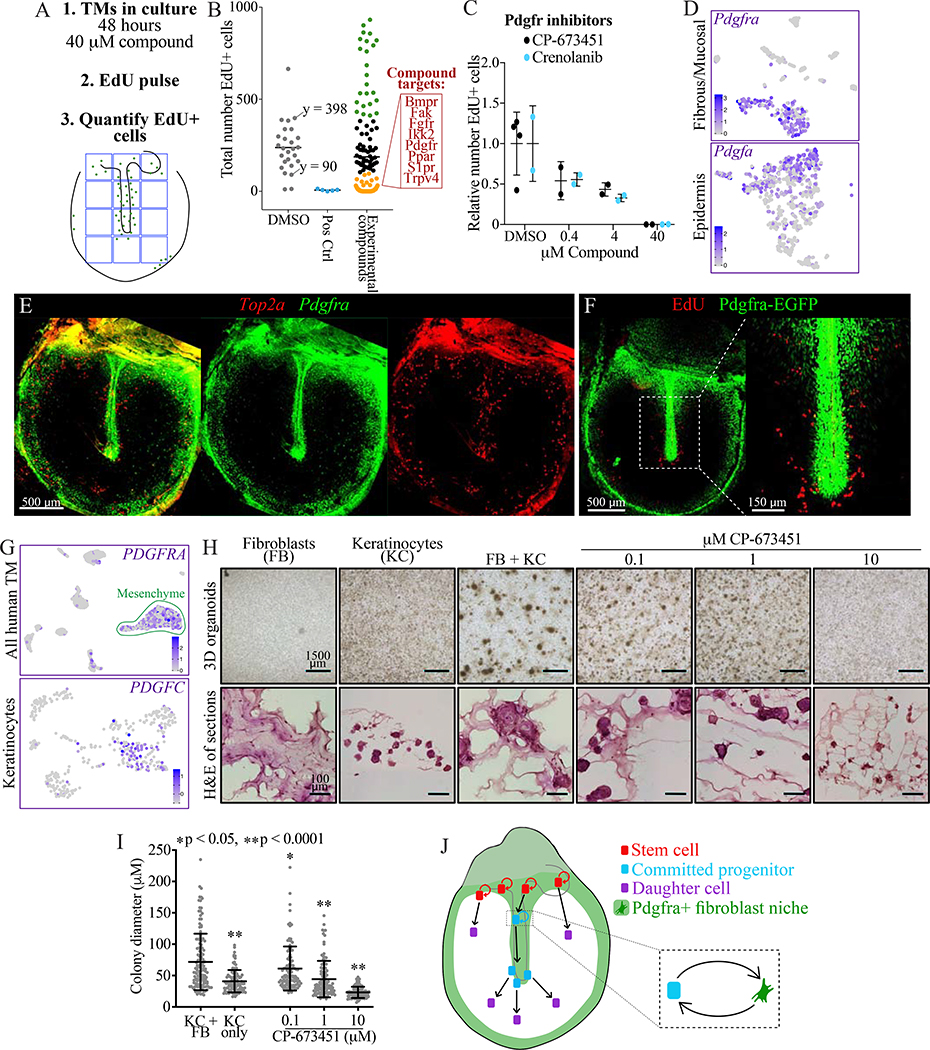
The pattern of proliferation in the TM suggested there were niche signals restricted to the proliferative zones, which we aimed to identify via a small molecule screen. When cultured as explants in non-supplemented media, TMs maintain regionally restricted proliferation. We utilized this system to screen 74 compounds for reduction in EdU incorporation (Table S5). The compound library included only molecular targets whose expression was confirmed in the murine scRNA-Seq data, and predominantly cell surface receptors. TMs were cultured in media containing vehicle (n = 26) or 40 μM of a compound (n = 2 per compound) for 48 hours then pulsed with EdU for two hours. The number of EdU-positive cells in a 1.2 × 1.6 mm region of the pars tensa were counted (Figures 7A–B, S6A). Compounds targeting Cdks or Aurka were positive controls, and they ablated proliferation as expected. Targets for which every compound tested ablated proliferation included: Bmpr, Fak, Fgfr2/3, Ikk2, Pdgfr, Ppar, S1pr/Sphk1, and Trpv4.
Figure 7: Pdgfra signaling in fibroblasts supports turnover of mouse and human TM keratinocytes.
(A) Approach for screening compounds in TM explants. (B) Screen results. The numbers of EdU+ cells in 1,200×1,600 μm areas of the TMs are plotted; each dot is one TM, with a bar indicating mean. DMSO was the negative control and inhibitors of Cdk and Aurka were positive controls. Of TMs treated with one of 71 experimental compounds, data points are colored in black in the DMSO-treated range (90 – 398), in green if >398, and in gold if <90. In the red box are targets for which every compound tested fell in the gold range. (C) Relative number of EdU+ cells in TMs cultured for 48 hours in vehicle or the indicated concentrations of Pdgfra inhibitors. Each dot is a TM and the error bars show mean and standard deviation (SD). (D) tSNEs showing expression of Pdgfra in the murine TM fibrous/mucosal fraction and of Pdgfa in the murine TM epidermis. (E) ISH for Pdgfra and Top2a in a wild-type whole-mount TM. The middle and right panels show each stain independently. (F) Whole-mount TM from a Pdgfra-EGFP mouse injected with EdU and sacrificed 24 hours later. (G) UMAPs showing expression of PDGFRA in the full human TM scRNA-Seq dataset and of PDGFC in the isolated keratinocyte dataset. (H) Organoid cultures of fibroblasts (FB) and keratinocytes (KC) from human TMs, seeded separately or in co-culture and treated with CP-673451. Top: images of 3D cultures. Bottom: H&E stained sections. (I) Quantification of organoid size; each dot is a cell cluster and error bars show mean and SD. Statistical significance of each condition compared to KC+FB, calculated with one-way ANOVA, is indicated. (J) Cartoon summarizing our findings. Fibroblasts create a niche supporting proliferation of keratinocyte stem cells and committed progenitors, while keratinocytes not in proximity to this niche migrate but do not proliferate. Images in E, F, and H (bottom) were acquired in tiles and stitched; images in E and F are maximum projections of z-stacks. See also Figures S6–7 and Table S5.
To characterize these targets, we analyzed their expression in the murine scRNA-Seq data (Figure S7A) and treated explants with multiple concentrations of inhibitors (Figures 7C, S6B). Pdgfra inhibition caused a concentration-dependent decrease in proliferation (Figure 7C). Additionally, Pdgfra is expressed by mesenchyme, while the ligand Pdgfa is expressed by undifferentiated keratinocytes, including cycling cells (Figure 7D). IF for Vim demonstrated that mesenchyme is confined to regions of proliferation (Figure S2A), so communication between keratinocytes and spatially restricted fibroblasts may underlie the proliferative niche. With ISH for Pdgfra and Top2a, as well as a Pdgfra-EGFP reporter mouse injected with EdU 24 hours before TM harvest, we confirmed that Pdgfra+ fibroblasts and cycling keratinocytes colocalize in the tissue (Figure 7E–F) (Hamilton et al., 2003). We next evaluated the human TM. In the scRNA-Seq data, the mesenchyme expresses PDGFRA; the keratinocytes express the ligands PDGFA weakly (not shown) and PDGFC more robustly (Figure 7G). To assess if fibroblasts from a human TM support the growth of human TM keratinocytes, cells were isolated from undiseased surgical TM samples, and 8,000 cells of either a single type or a 50:50 mix of keratinocytes and fibroblasts were plated in an organoid assay (Figure 7H) (Frank et al., 2016). In solitary cultures, the fibroblasts do not form multicellular structures and the keratinocytes form keratin pearls—spherical clusters of keratinocytes. In co-culture, larger colonies comprised of a keratinocyte core and fibroblast periphery are formed, indicating an interaction between the two cell types. Treatment of the co-cultures with the Pdgfr inhibitor CP-673451 reverted the phenotype of these colonies (Figure 7H–I). Therefore, we conclude that the keratinocyte-stromal interaction mediated by Pdgfr signaling is likely conserved in the human TM.
Lastly, we attempted to identify a pro-proliferative signal sent from the Pdgfra+ fibroblasts to keratinocytes. We utilized a curated list of receptor-ligand pairs, CellPhoneDB (Vento-Tormo et al., 2018), and identified ones with the ligand expressed exclusively by Pdgfra+ fibroblasts and the receptor expressed by keratinocytes (although not necessarily exclusively). Among the ligand-receptor pairs with the requisite pattern were: Tgfb3-Tgfbr1/2; Fgf7-Fgfr2/3; and Igf1/2-Igf1r (Figure S7A). Fgfr and Igfr inhibition ablated proliferation in the compound screen, and the activity of high-dose inhibition was validated in the concentration-response experiment (Figures 7B, S8). Tgfbr inhibition did not impact proliferation (Figure S6B). We also tested inhibition of these targets in the organoid assay, and three Fgfr inhibitors had a significant effect (Figure S7B–D). In the human TM, as in the murine TM, the mesenchyme expresses FGF7, and the keratinocytes express FGF receptors (Figure S7E). Thus, Fgf may be a component of a signaling axis from the fibroblasts to the keratinocytes.
Discussion
The TM is a unique epidermal site
Our study elucidates the proliferative, migratory, and clonal dynamics of TM keratinocytes. The TM conducts sound from the external auditory canal to the ossicular chain, which transmits it to the inner ear where it is converted to neural signals. The TM epidermis is in many ways similar to epidermis at other anatomical sites. But, its position at the blind end of the ear canal necessitates an additional function: clearing the canal of squamous and foreign debris. To achieve this, zones of proliferation are in the central and superior regions of the pars tensa, confirmed here by EdU labeling, and keratinocytes migrate inferiorly and radially outward, as we visualized with live cell imaging. This arrangement is similar to re-epithelialization following wounding, where a proliferative region exists behind a migrating front (Park et al., 2017).
Epidermal thickness is a key property that differs by anatomic site. Across the murine TM alone, the epidermis varies in thickness from a one to roughly five cells. The pars flaccida epidermis is more stratified than the pars tensa epidermis. In our scRNA-Seq data, we captured cells spanning the differentiation spectrum. As expected, staining for Flg2 showed markedly more differentiation in the pars flaccida. In much of the pars tensa, there is no apparent cell stratification under homeostasis, and concomitantly few terminally differentiated keratinocytes. However, there were occasional Flg2+ cells in the epidermal monolayer, in contrast to the obligate coupling of differentiation and stratification seen in skin elsewhere (Clayton et al., 2007; Lechler and Fuchs, 2005). How differentiation is induced independent of exit from the basal layer, and whether it is linked to the lateral migration instead of to stratification, are questions for further study.
Clonal dynamics in the TM
The atypical arrangement and migratory behavior of the TM epidermis reveal clonal dynamics of keratinocytes. Regarding the TM epidermal stem cell (SC) pool, both Ki67-CreERT2 and Krt5-CreERT2 lineage traces are qualitatively consistent with a model of population asymmetry and neutral drift, in which some SCs are lost over time and others divide symmetrically to maintain the SC pool (Klein and Simons, 2011). Quantification of single Ki67-CreERT2;R26R-Confetti clones demonstrated widening of the distribution of clone sizes, also known as scaling behavior, over time. In Krt5-CreERT2;R26R-Confetti samples, the emergence of more distinctive and larger color blocks over time also supports this model.
A challenge in interpreting these data is that the TM SC niche is not well defined. We know generally where the proliferative zones are, but also demonstrated that most TM keratinocytes become proliferative following injury. Live imaging and lineage tracing support the hypothesis that the cells maintaining the TM epidermis long-term reside in the superior pars tensa or potentially the pars flaccida. In the live imaging, the predominant direction of migration is superior-to-inferior. In the Ki67-CreERT2 studies, none of the larger clones emanated from the umbo, but many involved the superior pars tensa. Finally, in the Krt5-CreERT2 experiments, the predominant orientation of color blocks is vertical. These findings are consistent with a stable progenitor niche in the superior pars tensa. The proliferating cells over the malleus behave more like committed progenitors (CPs), making a limited contribution to the tissue before migrating into the ear canal.
Previous data from other areas of mouse epidermis has yielded conflicting results depending on the methodology used to distinguish between neutrally competing SCs and a hierarchical model (Fuchs, 2016). The spatial separation of keratinocyte SCs and CPs within the TM presents the clearest evidence we know yet of the capacity of keratinocytes to exist in a clonal hierarchy. The fact that this arrangement exists within the TM does not necessarily argue for its existence in other areas of the epidermis, but it does argue that given the proper signals such an arrangement may arise.
Lateral keratinocyte migration establishes the clonal hierarchy
Although it appears that in uninjured TM the majority of keratinocytes do not have long-term repopulation activity, they may act as facultative SCs in response to injury. We and others have demonstrated a massive proliferative response following perforation (Chari et al., 2019; Santa Maria et al., 2010). One possibility is that the majority of pars tensa keratinocytes are capable of long-term repopulation activity but lack the niche signals to execute that program. This model may explain the pattern of proliferation observed in the TM under homeostasis. If the niche signals to support proliferation come predominantly from stromal cells (perhaps Pdgfra+ cells), then, as the progeny of the long-term repopulating cells migrate inferiorly, if they are in proximity to these niche signals—such as over the malleus—they will continue to proliferate. If they are not in proximity to these signals—as in over the body of the pars tensa—then they may continue to migrate but cease proliferation. Because the proliferative cells over the malleus inexorably move away from the malleus, they would be removed from proximity to the niche signals and stop proliferating. In this way, the migration itself may confer the hierarchical behavior, rather than an enforced cell-autonomous differentiation program.
One outstanding question is how the direction of migration is enforced. Although we do not yet have direct evidence, we suspect it may be related to inherent properties of the pars tensa. Previous work has shown that asymmetries in the tension of structures on which keratinocytes are grown may confer directional migration (Lü et al., 2016, 2013). Under this model, when progeny of TM SCs transition inferiorly onto the body of the pars tensa, they experience vectors of force not present in the uppermost pars tensa or pars flaccida and begin to migrate. An additional possibility is that collagen fibers in the pars tensa permits directional migration in a way that the pars flaccida ECM does not. Deep to the basement membrane there is a highly organized network of radial collagen fibers that may approximate some of the migratory patterns of the keratinocytes (Fay et al., 2006). These hypotheses bear further experimental validation.
Niche signals maintain keratinocyte proliferation
The stromal signals required for keratinocyte proliferation in uninjured epidermis are not well defined. To maintain primary keratinocytes in culture requires a mix of growth factors that may replicate aspects of wound healing rather than physiologic maintenance. Further, organotypic models of stratified squamous epithelia almost invariably require fibroblasts as “feeders” to maintain the keratinocytes in a healthy state (Oh et al., 2013). Despite the well-established necessity of stroma in supporting keratinocytes, the exact mechanisms remain cryptic. We propose Pdgf signaling as one component of this crosstalk.
Because basal keratinocytes of the TM express Pdgf and mesenchymal cells express Pdgfr, we hypothesize that keratinocytes send trophic signals to the mesenchyme. Given that inhibition of Pdgfr ablates proliferation of keratinocytes, this argues for signals being sent back from the mesenchyme to the keratinocytes. We have not yet definitively identified these signals, but they may be diffusible factors, membrane bound factors, ECM components, or even mechanical forces. Our data suggests Fgf7 may be one of these signals, and this is further supported by published findings that FGF improves healing of TM perforations and Fgfr inhibition slows healing (Hakuba et al., 2003; Kaftan et al., 2012; Kato and Jackler, 1996). Further studies will clarify the mechanisms supporting TM keratinocyte turnover. Because of the limited zones of proliferation within the pars tensa, the TM may be an ideal model system to better understand keratinocyte-stromal interactions, and we may then extend these findings to epidermis of other parts of the body, as well as other squamous epithelia.
Implications for human disease
Lastly, TM physiology is important to understand because it is deranged in pathological conditions. Abnormal migration has been documented in keratosis obturans, in which desquamated keratin builds up in the ear canal, and myringitis granulosa, a chronic inflammatory condition of the lateral TM (Corbridge et al., 1996; Makino et al., 1988; Soucek and Michaels, 1993). TM cell proliferation and migration have also been implicated in closure of perforations, which occurs spontaneously in ~90% of cases (Chari et al., 2019; Lou, 2012; Lou et al., 2012). In the remaining cases, chronic perforations can result in hearing impairment, infections, and drainage. In all of these pathologies, advances in care have been hampered by lack of understanding of normal physiology.
Another example is the common and morbid condition cholesteatoma, an abnormal growth of TM keratinocytes that can invade the middle ear space and destroy structures in and surrounding the ear (Kuo et al., 2015). The underlying pathogenesis is not well understood, but this study suggests a potential mechanism. The majority of acquired cholesteatomas occur as a consequence of retraction of the pars flaccida or superior pars tensa. If there is minimal lateral migratory activity of the pars flaccida, then, following retraction, progeny of proliferating cells may simply stratify and accumulate within the retraction pocket. In contrast, because there is minimal stratification and rapid lateral migration of the keratinocytes of the pars tensa, retraction in that area would not give rise to the accumulation of squamous debris. The findings herein thus provide a basis for deepening understanding of cholesteatoma and other conditions of the TM, as well as basic epidermal biology.
Limitations of the study
In this paper, we identify markers of distinct murine keratinocyte populations (particularly Fn1 and Serpinb10), but do not validate the long-term contributions of these populations to the tissue with lineage tracing in homeostasis or injury. Strategies to label specific populations or areas of the tissue are needed to better characterize and validate their contributions to the organ. Even in our Ki67 lineage trace studies, we could not fully isolate pars tensa from pars flaccida contributions, as the border cannot be robustly identified. Further, our conclusions about migration, clonal dynamics, and molecular mechanisms are based on ex vivo analysis, terminally harvested tissue, and an explant culture system, respectively. In vivo time-lapse and mechanistic analyses, particularly of the role of Pdgfr, are needed for greater insight into the cellular dynamics and underlying signaling. Lastly, our conclusions from the human scRNA-Seq dataset were constrained by its small size and lack of samples for validation.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Aaron D. Tward (aaron.tward@ucsf.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All scRNA-Seq datasets generated during this study have been deposited in NCBI’s Gene Expression Omnibus (GEO; accession number: GSE128892).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mice were maintained and experiments conducted following the guidelines of the Institutional Animal Care and Use Committee at the University of California, San Francisco (approval number AN165495). All experiments using wild type mice were done with strain FVB/NJ. All Cre driver lines (Ki67-CreERT2 (Basak et al., 2018), Krt5CreERT2 (Rock et al., 2009), and Krt19-CreERT (Means et al., 2008)) and floxed reporter lines (R26R-Confetti (Snippert et al., 2010) and mT/mG (Muzumdar et al., 2007)) were received and maintained on a C57BL/6 background. Pdgfra-EGFP mice (Hamilton et al., 2003) were received and maintained on a FVB/NJ background. All strains are available from commercial vendors. For most studies, we used adult animals—both male and female—between 6 and 12 weeks of age at the initiation of experiments. For whole-mount ISH, we used mice between 3 and 5 weeks of age to facilitate dissection of the TM in the bony ring without requiring decalcification. Additional details are included in the Methods Details section below.
Primary human tissue samples
Normal TM tissue was obtained from surgical procedures at University of California, San Francisco under approval of the Institutional Review Board (approval number 16–19811) and with informed consent from the patients. For scRNA-Seq, TM tissue was removed from a 34-year-old female as part of a surgical approach for unrelated tumor adjacent to the brain. Fibroblasts and keratinocytes used in the organoid assay were derived from undiseased TM tissue from patients undergoing surgery for cholesteatoma in the pars flaccida region of the TM. Fibroblasts were derived from tissue from a 55-year-old male, and keratinocytes were derived from tissue from a 27-year-old female.
METHODS DETAILS
TM dissociations
Murine
TMs were dissected from 19 6-week-old FVB/NJ mice (10 female, 9 male). They were stored in phosphate buffered saline (PBS) on ice until all tissue had been collected. They were incubated in dispase (Corning) at 37°C for five minutes and then mechanically separated into epidermal and fibrous/mucosal fractions. The epidermal tissue was dissociated in TrypLE (Life Technologies) and the fibrous/mucosal tissue was dissociated in 0.2 mg/mL collagenase P (Sigma-Aldrich) and 5 μg/mL DNAse. Both dissociations were done at 37°C for 10 minutes, with trituration every five minutes for the fibrous/mucosal tissue and only at the 10-minute endpoint for the epidermis. Cells were passed through 40 μm strainers and collected by centrifugation. Dead cells were removed with the Dead Cell Removal Kit (Miltenyi Biotec) per the manufacturer instructions and again collected by centrifugation. They were resuspended at 1,000 cells/μL in 0.04% BSA in PBS, and 25,000 cells were loaded for single cell capture.
Human
Prior to dissociation, the TM was mechanically separated into an outer epidermal portion and an inner fibrous/mucosal portion. A scalpel was used to cut the tissue into pieces less than 2 mm. The outer TM was dissociated in 1 mg/mL collagenase P and 5 μg/mL DNAse at 37°C for one hour, with trituration every 20 minutes. The inner TM was incubated in dispase at 37°C for 30 minutes and then transferred to collagenase P and DNAse for an additional hour, with trituration every 20 minutes. Cells were passed through 40 μm strainers and collected by centrifugation. Red blood cells were lysed (Alfa Aesar J62990), and then dead cells were removed (Dead Cell Removal Kit; Miltenyi Biotec) following the manufacturers’ instructions. Cells were resuspended at 1,000 cells/μL in 0.04% BSA in PBS, and 25,000 cells were loaded for single cell capture.
scRNA-Seq analysis
Isolated cells were run on the Chromium Controller (10X Genomics) with Single Cell 3’ Reagent Kit v2 and the generated libraries were sequenced on Illumina HiSeq 4000. Human data was aligned to hg19 and mouse to mm10. Data was run through CellRanger 2.0.0 (10x Genomics) and then analyzed via R primarily through single cell analysis package Seurat version 3 (R Core Team, 2018; Satija et al., 2015). Standard processing steps were conducted to limit unwanted sources of variation through removal of cells expressing less than 200 genes and genes expressed in less than 3 cells. The matrices of data were log normalized in a sparse data matrix and principal component analysis was performed and the first 10 components were used to cluster the cells by Louvain clustering implemented in Seurat. tSNE or UMAP plots were independently generated to aid in 2D representation of multidimensional data. On the murine keratinocytes, we ran cell cycle analysis by assigning cells, based on expression of genes, to either S, G1 or G2 phase and then conducting linear regression based on these characteristics. For the human TM, datasets were merged via Seurat objects in R. The read count matrices were combined together and were regressed via batches in the latest Seurat implementation in 3.1.1. The merged dataset was analyzed with CellFindR, an iterative clustering algorithm based on Seurat (Yu et al., 2019). Once the keratinocytes were identified, we isolated and re-clustered only the keratinocytes after rescaling the data.
Immunofluorescence (IF)
Whole-mount TMs were either dissected en bloc, or the auditory bulla was isolated and the TM dissected out following fixation and decalcification. Following dissection, TMs were fixed in 4% paraformaldehyde (PFA) at 4°C for two hours (en bloc) or overnight (auditory bullae). They were decalcified in 5% EDTA overnight (en bloc) or for one to three days with daily solution changes (auditory bullae). TMs were permeabilized in PBS with 1% Triton X-100 for two hours at room temperature (RT), blocked in PBS with 1% Triton X-100 and 10% fetal bovine serum (FBS; blocking buffer) for two hours at RT, and incubated in primary antibody diluted in blocking buffer at 4°C overnight. The primary antibodies (and dilutions) used were: rabbit anti-Cytokeratin 10 (1:100, Abcam, ab76318), rabbit anti-Vimentin (1:200, Abcam, ab92547), rabbit anti-Acta2 (1:100, Cell Signaling Technology, 19245), rat anti-CD31 (1:100, BD Biosciences, 553370), rabbit anti-MBP (1:50, Cell Signaling Technology, 78896), and goat anti-Sox2 (1:100, Neuromics, GT15098). They were washed and then incubated in Alexafluor-coupled secondary antibody in blocking buffer (1:250, Thermo Fisher Scientific) for one hour at RT. They were again washed and, if nuclear stain was desired, incubated in Hoechst dye diluted 1:2,000 in PBS for 30 minutes. They underwent final washes and were then mounted. Whole-mount TMs were imaged on a Nikon AZ100 macro confocal microscope as either single-frame or tiled acquisitions. Images were processed using Fiji (ImageJ) software (Schindelin et al., 2012). Whole-mounts are shown as maximum projections of z-stacks.
To prepare paraffin sections, auditory bullae were fixed and decalcified as above, dehydrated and embedded in paraffin, and then sectioned at 7-μm thickness. Staining was done as previously described (Hu et al., 2017). Briefly, paraffin was dissolved in Histo-Clear II (Electron Microscopy Sciences) and then the tissue was rehydrated. Slides were sub-boiled in citrate buffer (10 mM citric acid, 2 mM EDTA, 0.05% Tween-20, pH 6.2) for ten minutes in a microwave to retrieve antigens. Samples were permeabilized and blocked at RT for one hour in Animal-Free Blocker (Vector Labs) supplemented with 1% Triton X-100 and 2% normal goat serum (Cell Signaling Technology). They were incubated in primary antibody diluted in blocking buffer at 4°C overnight. The primary antibodies (and dilutions) used were: rabbit anti-Keratin 5 (1:1000, Biolegend, 905501), rabbit anti-Vimentin (1:200, Abcam, ab92547), and chicken anti-GFP (1:500, Abcam, ab13970). The following day slides were washed and then incubated in Alexafluor-coupled secondary antibody (1:250, Thermo Fisher Scientific) at RT for one hour. They were again washed and mounted with ProLong Gold Antifade Mountant with DAPI (Life Technologies). Images were acquired in tiles and stitched on a Leica DM6 B microscope, and merged files further processed using Fiji (ImageJ) software.
In situ hybridization (ISH)
Prior to dissection of TMs to be used for in situ hybridization (ISH), mice were euthanized with CO2 and thoracotomy, and then their vasculature was perfused with RNase-free PBS followed by 10% normal buffered formalin (NBF; sections) or 4% PFA (whole-mounts) to fix the tissue in situ. For sections, auditory bullae were then isolated, incubated in NBF at RT for 24 hours, then decalcified, paraffin embedded, and sectioned as described for IF. The RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics) protocol was followed, with the following specifications: manual target retrieval was done for 15 minutes and digestion with Protease Plus for 30 minutes. Images were acquired in tiles and stitched on a Leica DM6 B microscope, and merged files further processed using Fiji (ImageJ) software.
For whole-mount TMs, mice age 3 to 5 weeks old were used so that the TM could be dissected out in a bony ring without the need for decalcification. A protocol for staining of whole-mount zebrafish embryos was followed (Gross-Thebing et al., 2014). Briefly, the TMs were fixed in 4% PFA diluted in RNase-free PBS for 6 hours at RT, then washed in PBT (RNase-free PBS with 0.1% Tween 20), dehydrated in increasing concentrations of methanol, then stored in 100% methanol at −20°C overnight or for several days. The methanol was removed and the TMs air-dried, then incubated in Protease III at RT for 20 minutes, washed, and probes hybridized at 50°C overnight. The following day, the TMs were washed in 0.2X SSCT, fixed in 4% PFA at RT for 10 minutes, and again washed. The signal was then amplified and developed following the steps in the RNAscope Multiplex Fluorescent Reagent Kit v2 protocol. Images were acquired on a Nikon AZ100 macro confocal microscope and processed with Fiji (ImageJ). They were acquired in tiles and z-stacks, and stitched composites are shown as maximum projections.
EdU administration and detection
For injection, EdU (Carbosynth Limited) was resuspended at 5 mg/mL in saline and passed through a 0.2 μm filter. One mg was given by intraperitoneal (IP) injection. For continuous administration, mice were given an IP injection at the start of the experiment and then provided with EdU in the water, resuspended at 0.5 mg/mL with 1% sucrose and passed through a 0.2 μm filter. Water bottles were shielded from light and the water supply changed every three days. Samples were dissected and processed as for IF. For EdU detection, the Click-iT EdU Alexa Fluor 488 Imaging Kit (ThermoFisher Scientific) was used. For combined EdU and protein detection, EdU detection was completed first and then the IF protocol was followed beginning at the blocking step. Whole-mounts in which EdU was detected were acquired in a single frame with z-stack on the Nikon AZ100 macro confocal microscope and processed, including with maximum projection, with Fiji (ImageJ). Sections were imaged as described above.
Perforations
Perforations were created in mouse TMs as previously described (Chari et al., 2019). Briefly, animals were anesthetized with isoflurane, the left TM visualized with a dissection microscope, and a perforation created in the anterior pars tensa using a 25-gauge needle. In each mouse, the right TM served as a control and was not perforated. Two hours before sacrifice, mice received a single dose of EdU as described above. For each timepoint, whole-mounts from three mice were analyzed.
Live-cell imaging
To label TMs for live cell imaging, recombination was induced in Ki67-CreERT2;mT/mG or Krt5-CreERT2;mT/mG mice with a single IP injection of 250 mg/kg or 1 mg tamoxifen, respectively. TMs were harvested en bloc and cultured floating on Advanced DMEM (Life Technologies 12491023) supplemented with 1% penicillin/streptomycin (Fisher Scientific). Live-cell imaging was done on a Nikon AZ100 macro confocal microscope in an incubated chamber maintained at 37°C with 5% CO2 and 95% humidity. Z-stacks were acquired approximately every hour. Cells were identified and tracked using Imaris software. The images and movies presented were captured from 3D renderings in Imaris. For the palbociclib experiment, recombination was induced in Krt5-CreERT2;R26R-Confetti mice. TMs were harvested as above, cultured in media with either vehicle or 5 μM palbociclib (Selleckchem), and imaged every six hours for 24 hours on a Leica M205 FA microscope. Cells were manually annotated and tracked using Fiji (ImageJ) software.
Lineage tracing with the R26R-Confetti reporter
For minimal labeling, Ki67-CreERT2;R26R-Confetti mice were given a single IP injection of 250 mg/kg tamoxifen. For maximal labeling, Krt5-CreERT2;R26R-Confetti mice were given 2 mg tamoxifen per day for 5 consecutive days. At the specified timepoints, auditory bullae were harvested, fixed in 4% PFA at 4°C for 4 hours, decalcified in 5% EDTA at 4°C overnight, and then the bone was resected to leave the TM in a bony ring. Krt5-CreERT2;R26R-Confetti TMs were directly mounted for imaging. Prior to mounting, Ki67-CreERT2;R26R-Confetti TMs were permeabilized in 0.5% Triton X-100 in PBS at RT for 2 hours, and nuclei were stained with Hoechst as above. Some Ki67-CreERT2;R26R-Confetti TMs were also stained for Krt10 following the IF protocol. TMs were imaged on a Leica TCS SP8 confocal microscope. CFP, YFP, GFP, RFP, and Hoechst were detected using the following excitation/emission wavelengths, respectively: 458/463–490, 514/540–570, 488/490–510, 555/580–640, 405/410–500. Images were acquired in tiles and stitched, and merged files further processed with Fiji (ImageJ), including the creation of maximum projections of z-stacks. For the Ki67-CreERT2;R26R-Confetti lineage trace, images were manually analyzed for discernible clones, and numbers of cells per clone quantified. For each timepoint, the total number of TMs imaged and the total number with discernible clones are as follows, respectively, in parentheses: 1 week (14, 9), 2 weeks (18, 12), 3 weeks (11, 10), 4 weeks (18, 13), 2 months (29, 22), 3 months (39, 12), and 4 months (19, 5). For the Krt5-CreERT2;R26R-Confetti experiment, a subset of samples was quantified by imposing a grid of 5,000 μm2 boxes over the maximum projections and manually counting the number of fluorescent proteins expressed in each box. The total number of TMs imaged and the total number quantified are as follows, respectively, in parentheses: 3 days (5, 2), 1 month (4, 3), 3 months (13, 3), and 6 months (11, 5). Representative TMs are shown in the figures.
TM explants and compound screen
For explant culture, murine TMs were floated in Advanced DMEM supplemented with 1% penicillin-streptomycin. They were maintained in an incubator at 37°C with 5% CO2. For drug exposures, the TMs were cultured in media with drug for 48 hours and then pulsed with 10 μM EdU for two hours. Whole-mounts were then processed as described above. Compounds tested in the screen are listed in Table S5 (Selleckchem). Crenolanib used after the initial screen was from MedChem Express, and CP-673451 used after the initial screen was from Tocris. We used additional small molecule inhibitors to treat TM explants and organoids in concentration response: BGJ398 (Cayman Chemical), PRN1371 (MedChem Express), HC-067047 (MedChem Express), GW788388 (Selleckchem), and SB-525334 (Cayman Chemical).
Human TM cell organoid cultures
Keratinocytes and fibroblasts were isolated from human TM tissue using procedures similar to ones previously described for isolation of cells from foreskin (Ridky et al., 2010). Briefly, the tissue was incubated in dispase at 4°C overnight, and then the epidermis and underlying tissue separated if possible. For keratinocyte isolation, the epidermis or bulk TM tissue was minced, then incubated in 0.5% trypsin (Life Technologies) at 37°C for 3 minutes. The enzyme was quenched with soybean trypsin inhibitor (Thermo Fisher Scientific), then the cells and tissue pelleted and subsequently plated. Keratinocytes were cultured in a 50/50 mix of Keratinocyte-SFM with supplements (EGF and BPE) and Medium 154 with HKGS (Thermo Fisher Scientific), plus 1% penicillin-streptomycin. For fibroblast isolation, the tissue was incubated in 1 mg/mL collagenase P in a 1:1:1 ratio of HBSS to dispase to fibroblast media (Advanced DMEM with 5% FBS and 1% penicillin-streptomycin) at 37°C for 1 hour. The tissue was then transferred to 0.5% trypsin and incubated at 37°C for 5 minutes. The digestion was quenched with fibroblast media, then both enzyme solutions were centrifuged and the cell pellets plated in fibroblast media.
The 3D organoid cultures were setup similarly to an assay previously described (Frank et al., 2016). We used KGM, a media previously employed for growth of organotypic skin cultures, which consists of a 3:1 mix of DMEM and Ham’s F12 supplemented with 10% FBS, 0.18 mM adenine (Sigma A9795), 0.4 μg/mL hydrocortisone (EMD Millipore 386698), 5 μg/mL insulin (Sigma I1882), 0.1 nM cholera toxin (EMD Millipore 227036), 10 ng/mL EGF (Life Technologies PHG0315), 0.01 mg/mL ciprofloxacin hydrochloride (Sigma PHR1044), 1.36 ng/mL triiodo-L-thyronine (Sigma T2877), 1% antibiotic-antimycotic, and 1% penicillin-streptomycin (Ridky et al., 2010). Per replicate, 8,000 cells (all keratinocytes, all fibroblasts, or a 50:50 mixture of the two) were resuspended in 45 μL KGM media, then 45 μL Matrigel (Corning 354234) was added and the resulting 90 μL plated in a transwell insert in a 24-well plate. The culture was allowed to set at 37°C for 20 minutes, then 600 μL of KGM media with 1 μM of the ROCK inhibitor thiazovivin (Selleckchem) was added to the well. After 24 hours, the media was changed to KGM without the ROCK inhibitor; for inhibitor studies, the compound was added at this point. Media was changed every 2–3 days. After 14 days, the cultures were fixed with 4% PFA at 4°C overnight, then washed with PBS. Images of the whole 3D cultures were captured on a Leica M205 FA microscope. The cultures were then equilibrated in 30% sucrose, embedded in OCT, and sectioned at 10 μm thickness for hematoxylin and eosin (H&E) staining. H&E images were acquired in tiles and stitched on a Leica DM6 B microscope, and merged files further processed using Fiji (ImageJ) software. Organoid size was quantified by measuring the greatest diameter of the cell clusters in H&E sections.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analyses were done using Seurat functions for scRNA-seq data and GraphPad Prism for all other data. For scRNA-Seq data, Wilcoxon rank sum test with Bonferroni corrections was used. For other data, statistical significance was determined by t-test when comparing two groups and by one-way ANOVA when comparing more than two unmatched groups. Statistical results and sample numbers can be found within the figure panels and figure legends.
Supplementary Material
Movie 1: Time-lapse imaging of a Krt5-CreERT2;mT/mG TM. Related to Figure 4. A Krt5-CreERT2;mT/mG mouse was given a single dose of 1 mg tamoxifen and a TM harvested two days later. It was maintained in Advanced DMEM in an incubator surrounding an AZ100 macro confocal microscope and imaged every hour for 24 hours. The resulting video loops four times. The first and last loops are not annotated, the second includes dragontails indicating where the cells have recently been, and the third includes the full tracks for each cell. Cells were tracked using the Spots function in Imaris.
Movie 2: Time-lapse imaging of a Ki67-CreERT2;mT/mG TM. Related to Figure 4. A Ki67-CreERT2;mT/mG mouse was given a single dose of 250 mg/kg tamoxifen and a TM harvested two days later. It was maintained in Advanced DMEM in an incubator surrounding an AZ100 macro confocal microscope and imaged every hour for eight hours. The resulting video loops four times. The first and last loops are not annotated, the second includes dragontails indicating where the cells have recently been, and the third includes the full tracks for each cell. Cells were tracked using the Spots function in Imaris.
Table S1: ScRNA-Seq gene expression matrices for the murine TM epidermal fraction (keratinocytes) and fibrous/mucosal fraction. Related to Figure 1. For each cluster, the average expression of each gene and the average differential expression relative to all of the other clusters are included.
Table S2: Gene sets from the Molecular Signatures Database (MSigDB) enriched in murine keratinocyte clusters. Related to Figure 1. For cluster 3, genes with at least 1.5-fold increased expression relative to the remaining keratinocytes in the dataset (clusters 0, 1, 2, and 4) and that met a significance threshold of p < 0.05 were used to compute overlaps in MSigDB. For clusters 1 and 2, genes with at least 1.2-fold increased expression in cluster 1 versus 2 or vice versa and that met a significance threshold of p < 0.05 were used to compute overlaps in MSigDB. For all, the top 50 gene sets are shown.
Table S3: ScRNA-Seq gene expression matrices for the human TM dataset. Related to Figure 2. For each cluster, the average expression of each gene and the average differential expression relative to all of the other clusters are included.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Cytokeratin 10; 1:100 | Abcam | Cat# ab76318, RRID: AB_1523465 |
| Rabbit polyclonal anti-Keratin 5; 1:1000 | Biolegend | Cat# 905501, RRID: AB_2565050 |
| Rabbit monoclonal anti-Vimentin; 1:200 | Abcam | Cat# ab92547, RRID: AB_10562134 |
| Rabbit monoclonal anti-α-Smooth Muscle Actin (Acta2); 1:100 | Cell Signaling Technology | Cat# 19245, RRID: 2734735 |
| Rat monoclonal anti-CD31 (PECAM); 1:100 | BD Biosciences | Cat# 553370, RRID: AB_394816 |
| Rabbit monoclonal anti-Myelin basic protein (MBP); 1:50 | Cell Signaling Technology | Cat# 78896 |
| Goat polyclonal anti-Sox2; 1:100 | Neuromics | Cat# GT15098, RRID: AB_2195800 |
| Chicken polyclonal anti-GFP; 1:500 | Abcam | Cat# ab13970, RRID: AB_300798 |
| Goat anti-Chicken IgY (H+L), Alexa Fluor 488; 1:250 | Thermo Fisher Scientific | Cat# A-11039, RRID: AB_2534096 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed, Alexa Fluor 555; 1:250 | Thermo Fisher Scientific | Cat# A-21428, RRID: AB_2535849 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed, Alexa Fluor 488; 1:250 | Thermo Fisher Scientific | Cat# A-11006, RRID: AB_2534074 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed; 1:250 | Thermo Fisher Scientific | Cat# A-11055, RRID: AB_2534102 |
| Biological Samples | ||
| Healthy tympanic membrane tissue for scRNA-Seq, from 34-year-old female | University of California, San Francisco | Under IRB approval |
| Undiseased tympanic membrane tissue used to derive fibroblasts for organoid experiments, from 55-year-old male | University of California, San Francisco | Under IRB approval |
| Undiseased tympanic membrane tissue used to derive keratinocytes for organoid experiments, from 27-year-old female | University of California, San Francisco | Under IRB approval |
| Chemicals | ||
| 5-Ethynyl-2’-deoxyuridine (EdU) | Carbosynth Limited | NE08701 |
| Crenolanib | MedChem Express | Cat# HY-13223; CAS No: 670220–88–9 |
| CP-673451 | Tocris | Cat# 5993; CAS No: 343787–29–1 |
| Palbociclib | MedChem Express | Cat# HY-50767; CAS No: 571190–30–2 |
| Thiazovivin | Selleckchem | Cat# S1459; CAS No: 1226056–71–8 |
| Compound library | Selleckchem | Details in Table S9 |
| BGJ398 | Cayman Chemical | Cat# 19157; CAS No: 872511–34–7 |
| PRN1371 | MedChem Express | Cat# HY-101768; CAS No: 1802929–43–6 |
| HC-067047 | MedChem Express | Cat# HY-100208; CAS No: 883031–03–6 |
| SB-525334 | Cayman Chemical | Cat# 16281; Cas No: 356559–20–1 |
| GW788388 | Selleckchem | Cat# S2750; Cas No: 452342–67–5 |
| Critical Commercial Assays | ||
| Dead Cell Removal Kit | Miltenyi Biotec | Cat# 130–090–101 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostics | Cat# 323100 |
| Click-iT EdU Alexa Fluor 488 Imaging Kit | Thermo Fisher Scientific | Cat# C10337 |
| Deposited Data | ||
| Raw scRNA-Seq datasets (4) | This paper | GEO: GSE128892 |
| Experimental Models: Organisms/Strains | ||
| Mouse: FVB/NJ | From: The Jackson Laboratory | Cat# 001800, RRID: IMSR_JAX:001800 |
| Mouse: Ki67-CreERT2; Mki67tm2.1(cre/ERT2)Cle/J | From: The Jackson Laboratory | Cat# 029803, RRID: IMSR_JAX:029803 |
| Mouse: Krt5-CreERT2; Tg(KRT5-cre/ERT2)1Blh | From: Sarah Knox | MGI: 4358332 |
| Mouse: R26R-Confetti; Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J | From: Ophir Klein | RRID: IMSR_JAX:013731 |
| Mouse: mT/mG; Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | From: Ophir Klein | RRID: IMSR_JAX:007676 |
| Mouse: Krt19-CreERT; Krt19tm1(cre/ERT)Ggu/J | From: Julie Sneddon | MGI: 3797107, RRID: IMSR_JAX:026925 |
| Mouse: Pdgfra-EGFP; Pdgfratm11(EGFP)Sor/J | From: Tien Peng | RRID: IMSR_JAX007669 |
| Software and Algorithms | ||
| Cell Ranger 2.0.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Seurat | https://satijalab.org/seurat/ | |
| CellFindR | https://github.com/kevyu27/CellFindR | |
| R | https://www.r-project.org/ | |
| Fiji (ImageJ) | https://fiji.sc/ | |
| Imaris | Bitplane | http://www.bitplane.com/ |
Highlights.
Single-cell sequencing of murine and human tympanic membrane (TM) cells.
TM keratinocyte stem cells reside in a specific region of the tissue.
Keratinocytes migrate over the tissue to maintain homeostasis.
Spatially restricted mesenchyme supports proliferation of keratinocyte progenitors.
Acknowledgements
We thank Ophir D. Klein, Diana J. Laird, and Tien Peng for helpful discussions and reading of the manuscript (O.D.K. and D.J.L.); Bruce Wang and Licia Selleri for use of equipment; the Biological Imaging Development Center, Broad Center Microscopy Core, and Institute for Human Genetics for their expertise and equipment. Also, we thank our funding sources: American Otological Society (S.M.F. and A.D.T.), the ARCS foundation (L.E.B.), Hearing Research Inc. (A.D.T.), the National Institute of Deafness and Communication Disorders R01DC018076 (A.D.T.), the National Institute of Dental and Craniofacial Research R01DE029890 (A.D.T.), and the National Institute of Diabetes and Digestive and Kidney Diseases R01DK118421 (J.B.S.).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti PW, 1964. Epithelial migration on the tympanic membrane. J Laryngol Otol 78, 808–830. [DOI] [PubMed] [Google Scholar]
- Aragona M, Dekoninck S, Rulands S, Lenglez S, Mascré G, Simons BD, Blanpain C, 2017. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat Commun 8, 14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak O, Krieger TG, Muraro MJ, Wiebrands K, Stange DE, Frias-Aldeguer J, Rivron NC, van de Wetering M, van Es JH, van Oudenaarden A, Simons BD, Clevers H, 2018. Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc Natl Acad Sci U S A 115, E610–E619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari D, Frumm S, Akil O, Tward A, 2019. Cellular Dynamics in Early Healing of Mouse Tympanic Membranes. Otol Neurotol 40, E160–E166. [DOI] [PubMed] [Google Scholar]
- Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, Fassett MS, Golovato J, Gray M, Ghadially R, Liao W, White BEP, Mauro TM, Mully T, Kim EA, Sbitany H, Neuhaus IM, Grekin RC, Yu SS, Gray JW, Purdom E, Paus R, Vaske CJ, Benz SC, Song JS, Cho RJ, 2018. Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep 25, 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH, 2007. A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189. [DOI] [PubMed] [Google Scholar]
- Corbridge RJ, Michaels L, Wright T, 1996. Epithelial migration in keratosis obturans. Am J Otolaryngol 17, 411–414. [DOI] [PubMed] [Google Scholar]
- Curreli S, Wong BS, Latinovic O, Konstantopoulos K, Stamatos NM, 2016. Class 3 semaphorins induce F-actin reorganization in human dendritic cells: Role in cell migration. J Leukoc Biol 100, 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé DP, Klein AM, Simons BD, Jones PH, 2010. The Ordered Architecture of Murine Ear Epidermis Is Maintained by Progenitor Cells with Random Fate. Dev Cell 18, 317–323. [DOI] [PubMed] [Google Scholar]
- Fay JP, Puria S, Steele CR, 2006. The discordant eardrum. Proc Natl Acad Sci U S A 103, 19743–19748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitosa NM, Zhang J, Carney TJ, Metzger M, Korzh V, Bloch W, Hammerschmidt M, 2012. Hemicentin 2 and Fibulin 1 are required for epidermal-dermal junction formation and fin mesenchymal cell migration during zebrafish development. Dev Biol 369, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Peng T, Zepp J, Snitow M, Vincent T, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, Lu MM, Morrisey EE, 2016. Emergence of a wave of Wnt signaling that regulates lung alveologenesis through controlling epithelial self-renewal and differentiation. Cell Rep 17, 2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, 2016. Epithelial Skin Biology: Three Decades of Developmental Biology, a Hundred Questions Answered and a Thousand New Ones to Address. Curr Top Dev Biol 116, 357–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell F, 1990. The activated keratinocyte: up regulation of cell adhesion and migration during wound healing. J Trauma 30, S144–149. [PubMed] [Google Scholar]
- Gross-Thebing T, Paksa A, Raz E, 2014. Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol 12, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT, 2008. Wound repair and regeneration. Nature 453, 314–321. [DOI] [PubMed] [Google Scholar]
- Hakuba N, Taniguchi M, Shimizu Y, Sugimoto A, Shinomori Y, Gyo K, 2003. A new method for closing tympanic membrane perforations using basic fibroblast growth factor. Laryngoscope 113, 1352–1355. [DOI] [PubMed] [Google Scholar]
- Hall MJ, Schwartzman A, Zhang J, Liu X, 2017. Ambulatory Surgery Data From Hospitals and Ambulatory Surgery Centers: United States, 2010. Natl Health Stat Report 1–15. [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P, 2003. Evolutionary Divergence of Platelet-Derived Growth Factor Alpha Receptor Signaling Mechanisms. Mol Cell Biol 23, 4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK-H, Du W, Shelton SJ, Oldham MC, DiPersio CM, Klein OD, 2017. A FAK-YAP-mTOR signaling axis regulates stem cell-based tissue renewal in mice. Cell Stem Cell 21, 91–106.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janpipatkul K, Suksen K, Borwornpinyo S, Jearawiriyapaisarn N, Hongeng S, Piyachaturawat P, Chairoungdua A, 2014. Downregulation of LAT1 expression suppresses cholangiocarcinoma cell invasion and migration. Cell Signal 26, 1668–1679. [DOI] [PubMed] [Google Scholar]
- Jones P, Simons BD, 2008. Epidermal homeostasis: do committed progenitors work while stem cells sleep? Nat Rev Mol Cell Biol 9, 82–88. [DOI] [PubMed] [Google Scholar]
- Joost S, Jacob T, Sun X, Annusver K, La Manno G, Sur I, Kasper M, 2018. Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell Rep 25, 585–597.e7. [DOI] [PubMed] [Google Scholar]
- Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, Kasper M, 2016. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 3, 221–237.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaftan H, Reuther L, Miehe B, Hosemann W, Beule A, 2012. Inhibition of fibroblast growth factor receptor 1: influence on tympanic membrane wound healing in rats. Eur Arch Otorhinolaryngol 269, 87–92. [DOI] [PubMed] [Google Scholar]
- Kakoi H, Anniko M, 1997. Auditory Epithelial Migration III: An Immunohistologic Study Using Anti-Brdu Antibody on Tympanic Membrane in Mouse. Ann Otol Rhinol Laryngol 106, 414–421. [DOI] [PubMed] [Google Scholar]
- Kakoi H, Anniko M, Pettersson CÅ-V, 1996. Auditory Epithelial Migration: I. Macroscopic Evidence of Migration and Pathways in Rat. Acta Oto-Laryngol 116, 435–438. [DOI] [PubMed] [Google Scholar]
- Kato M, Jackler RK, 1996. Repair of chronic tympanic membrane perforations with fibroblast growth factor. Otolaryngol Head Neck Surg 115, 538–547. [DOI] [PubMed] [Google Scholar]
- Khanom R, Nguyen CTK, Kayamori K, Zhao X, Morita K, Miki Y, Katsube K-I, Yamaguchi A, Sakamoto K, 2016. Keratin 17 Is Induced in Oral Cancer and Facilitates Tumor Growth. PLoS ONE 11, e0161163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Kim J, Seonwoo H, Jang K-J, Kim YJ, Lim HJ, Lim K-T, Tian C, Chung JH, Choung Y-H, 2015. Latent progenitor cells as potential regulators for tympanic membrane regeneration. Sci Rep 5, 11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwanuka E, Hackl F, Caterson EJ, Nowinski D, Junker JPE, Gerdin B, Eriksson E, 2013. CCN2 is transiently expressed by keratinocytes during re-epithelialization and regulates keratinocyte migration in vitro by the ras-MEK-ERK signaling pathway. J Surg Res 185, e109–e119. [DOI] [PubMed] [Google Scholar]
- Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD, 2010. Mouse Germ Line Stem Cells Undergo Rapid and Stochastic Turnover. Cell Stem Cell 7, 214–224. [DOI] [PubMed] [Google Scholar]
- Klein AM, Simons BD, 2011. Universal patterns of stem cell fate in cycling adult tissues. Development 138, 3103–3111. [DOI] [PubMed] [Google Scholar]
- Knutsson J, Unge M. von, Rask-Andersen H, 2011. Localization of Progenitor/Stem Cells in the Human Tympanic Membrane. Audiol Neurootol 16, 263–269. [DOI] [PubMed] [Google Scholar]
- Koba R, 1995. Epidermal Cell Migration and Healing of the Tympanic Membrane: An Immunohistochemical Study of Cell Proliferation Using Bromodeoxyuridine Labeling. Ann Otol Rhinol Laryngol 104, 218–225. [DOI] [PubMed] [Google Scholar]
- Kuo C-L, Shiao A-S, Yung M, Sakagami M, Sudhoff H, Wang C-H, Hsu C-H, Lien C-F, 2015. Updates and Knowledge Gaps in Cholesteatoma Research. Biomed Res Int 2015, 854024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E, 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Forbes ME, Fuller GN, Li J, Yang X, Zhang W, 2020. IGFBP2: integrative hub of developmental and oncogenic signaling network. Oncogene 39, 2243–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP, 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew LJ, Chen LQ, Wang AY, von Unge M, Atlas MD, Dilley RJ, 2018. Tympanic Membrane Derived Stem Cell-Like Cultures for Tissue Regeneration. Stem Cells Dev 27, 649–657. [DOI] [PubMed] [Google Scholar]
- Liew LJ, Day RM, Dilley RJ, 2017. Tympanic membrane organ culture using cell culture well inserts engrafted with tympanic membrane tissue explants. BioTechniques 62, 109–114. [DOI] [PubMed] [Google Scholar]
- Lim X, Tan SH, Koh WLC, Chau RMW, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R, 2013. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science 342, 1226–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litton WB, 1963. Epithelial migration over tympanic membrane and external canal. Arch Otolaryngol 77, 254–257. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu Y, Peng L, Li J, Wu K, Xia L, Wu J, Wang S, Wang X, Liu Q, Zeng W, Xia Y, 2019. TWEAK/Fn14 Signals Mediate Burn Wound Repair. J Invest Dermatol 139, 224–234. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ, 2010. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825. [DOI] [PubMed] [Google Scholar]
- Lou ZC, 2012. Spontaneous Healing of Traumatic Eardrum Perforation: Outward Epithelial Cell Migration and Clinical Outcome. Otolaryngol Head Neck Surg 147, 1114–1119. [DOI] [PubMed] [Google Scholar]
- Lou Z-C, Lou Z-H, Zhang Q-P, 2012. Traumatic tympanic membrane perforations: a study of etiology and factors affecting outcome. Am J Otolaryngol 33, 549–555. [DOI] [PubMed] [Google Scholar]
- Lü D, Li Z, Gao Y, Luo C, Zhang F, Zheng L, Wang J, Sun S, Long M, 2016. β1 integrin signaling in asymmetric migration of keratinocytes under mechanical stretch in a co-cultured wound repair model. Biomed Eng Online 15, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü D, Liu X, Gao Y, Huo B, Kang Y, Chen J, Sun S, Chen L, Luo X, Long M, 2013. Asymmetric Migration of Human Keratinocytes under Mechanical Stretch and Cocultured Fibroblasts in a Wound Repair Model. PLoS One 8, e74563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K, Amatsu M, Kinishi M, Mohri M, 1988. The clinical features and pathogenesis of myringitis granulosa. Arch Otorhinolaryngol 245, 224–229. [DOI] [PubMed] [Google Scholar]
- Marchisio P, Pipolo C, Landi M, Consonni D, Mansi N, Di Mauro G, Salvatici E, Di Pietro P, Esposito S, Felisati G, Principi N, 2016. Cerumen: A fundamental but neglected problem by pediatricians. Int J Pediatr Otorhinolaryngol 87, 55–60. [DOI] [PubMed] [Google Scholar]
- Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohée S, Sotiropoulou PA, Simons BD, Blanpain C, 2012. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257–262. [DOI] [PubMed] [Google Scholar]
- Means AL, Xu Y, Zhao A, Ray KC, Gu G, 2008. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis 46, 318–323. Morpheus, https://software.broadinstitute.org/morpheus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L, 2007. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- O’Donoghue GM, 1983. Epithelial migration on the tympanic membrane of children. Int J Pediatr Otorhinolaryngol 6, 119–125. [DOI] [PubMed] [Google Scholar]
- Oh JW, Hsi T-C, Guerrero-Juarez CF, Ramos R, Plikus MV, 2013. Organotypic Skin Culture. J Invest Dermatol 133, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong HT, Redmond SL, Marano RJ, Atlas MD, von Unge M, Aabel P, Dilley RJ, 2017. Paracrine Activity from Adipose-Derived Stem Cells on In Vitro Wound Healing in Human Tympanic Membrane Keratinocytes. Stem Cells Dev 26, 405–418. [DOI] [PubMed] [Google Scholar]
- Park S, Gonzalez DG, Guirao B, Boucher JD, Cockburn K, Marsh ED, Mesa KR, Brown S, Rompolas P, Haberman AM, Bellaïche Y, Greco V, 2017. Tissue-scale coordination of cellular behavior promotes epidermal wound repair in live mice. Nat Cell Biol 19, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2018. R: A language and environment for statistical computing.
- Redmond SL, Levin B, Heel KA, Atlas MD, Marano RJ, 2011. Phenotypic and genotypic profile of human tympanic membrane derived cultured cells. J Mol Histol 42, 15–25. [DOI] [PubMed] [Google Scholar]
- Ridky TW, Chow JM, Wong DJ, Khavari PA, 2010. Invasive 3-Dimensional Organotypic Neoplasia from Multiple Normal Human Epithelia. Nat Med 16, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BLM, 2009. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollin M, Rogers P, Robinson P, 2011. Natural History of Pediatric Tympanic Membrane Perforation. Otol Neurotol 32, 246–251. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V, 2016. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, Tumbar T, 2016. Defining the cellular lineage hierarchy in the inter-follicular epidermis of adult skin. Nat Cell Biol 18, 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Maria PL, Redmond SL, Atlas MD, Ghassemifar R, 2010. Histology of the healing tympanic membrane following perforation in rats. Laryngoscope 120, 2061–2070. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A, 2015. Spatial reconstruction of single-cell gene expression. Nat Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji - an Open Source platform for biological image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H, 2010. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- Soni A, 2008. Statistical Brief #228: Ear Infections (Otitis Media) in Children (0–17): Use and Expenditures, 2006. Agency for Healthcare Research and Quality. [Google Scholar]
- Soucek S, Michaels L, 1993. Keratosis of the tympanic membrane and deep external auditory canal. A defect of auditory epithelial migration. Eur Arch Otorhinolaryngol 250, 140–142. [DOI] [PubMed] [Google Scholar]
- Stinson WD, 1936. Reparative processes in the membrana tympani: some interesting manifestations. Arch Otolaryngol 24, 600–605. [Google Scholar]
- Subramanian A., Tamayo P., Mootha VK., Mukherjee S., Ebert BL., Gillette MA., Paulovich A., Pomeroy SL., Golub TR., Lander ES., Mesirov JP., 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yan T, Zhang J, Jiang X, Zhang D, Huang Y, 2017. Notch1 Signaling Contributes to Hypoxia-induced High Expression of Integrin β1 in Keratinocyte Migration. Sci Rep 7, 43926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinling SP, Chole RA, 2006. Gerbilline cholesteatoma development Part I: Epithelial migration pattern and rate on the gerbil tympanic membrane: comparisons with human and guinea pig. Otolaryngol Head Neck Surg 134, 788–793. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Dyer CJ, Fons Romero JM, Teshima THN, Fuchs JC, Thompson H, 2018. Mapping the distribution of stem/progenitor cells across the mouse middle ear during homeostasis and inflammation. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA, 2018. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xu T, Qiu P, Xu G, 2018. Caveolin-1 promotes pituitary adenoma cells migration and invasion by regulating the interaction between EGR1 and KLF5. Exp Cell Res 367, 7–14. [DOI] [PubMed] [Google Scholar]
- Yu H, Königshoff M, Jayachandran A, Handley D, Seeger W, Kaminski N, Eickelberg O, 2008. Transgelin is a direct target of TGF-beta/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB J 22, 1778–1789. [DOI] [PubMed] [Google Scholar]
- Yu KS, Frumm SM, Park JS, Lee K, Wong DM, Byrnes L, Knox SM, Sneddon JB, Tward AD, 2019. Development of the Mouse and Human Cochlea at Single Cell Resolution. bioRxiv 739680. [Google Scholar]
- Yuan B, Zhang R, Hu J, Liu Z, Yang C, Zhang T, Zhang C, 2018. WDR1 Promotes Cell Growth and Migration and Contributes to Malignant Phenotypes of Non-small Cell Lung Cancer through ADF/cofilin-mediated Actin Dynamics. Int J Biol Sci 14, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jiang M, Wang Z, 2019. TPM4 promotes cell migration by modulating F-actin formation in lung cancer. Onco Targets Ther 12, 4055–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1: Time-lapse imaging of a Krt5-CreERT2;mT/mG TM. Related to Figure 4. A Krt5-CreERT2;mT/mG mouse was given a single dose of 1 mg tamoxifen and a TM harvested two days later. It was maintained in Advanced DMEM in an incubator surrounding an AZ100 macro confocal microscope and imaged every hour for 24 hours. The resulting video loops four times. The first and last loops are not annotated, the second includes dragontails indicating where the cells have recently been, and the third includes the full tracks for each cell. Cells were tracked using the Spots function in Imaris.
Movie 2: Time-lapse imaging of a Ki67-CreERT2;mT/mG TM. Related to Figure 4. A Ki67-CreERT2;mT/mG mouse was given a single dose of 250 mg/kg tamoxifen and a TM harvested two days later. It was maintained in Advanced DMEM in an incubator surrounding an AZ100 macro confocal microscope and imaged every hour for eight hours. The resulting video loops four times. The first and last loops are not annotated, the second includes dragontails indicating where the cells have recently been, and the third includes the full tracks for each cell. Cells were tracked using the Spots function in Imaris.
Table S1: ScRNA-Seq gene expression matrices for the murine TM epidermal fraction (keratinocytes) and fibrous/mucosal fraction. Related to Figure 1. For each cluster, the average expression of each gene and the average differential expression relative to all of the other clusters are included.
Table S2: Gene sets from the Molecular Signatures Database (MSigDB) enriched in murine keratinocyte clusters. Related to Figure 1. For cluster 3, genes with at least 1.5-fold increased expression relative to the remaining keratinocytes in the dataset (clusters 0, 1, 2, and 4) and that met a significance threshold of p < 0.05 were used to compute overlaps in MSigDB. For clusters 1 and 2, genes with at least 1.2-fold increased expression in cluster 1 versus 2 or vice versa and that met a significance threshold of p < 0.05 were used to compute overlaps in MSigDB. For all, the top 50 gene sets are shown.
Table S3: ScRNA-Seq gene expression matrices for the human TM dataset. Related to Figure 2. For each cluster, the average expression of each gene and the average differential expression relative to all of the other clusters are included.
Data Availability Statement
All scRNA-Seq datasets generated during this study have been deposited in NCBI’s Gene Expression Omnibus (GEO; accession number: GSE128892).