Abstract
Zanamivir is a highly selective neuraminidase (NA) inhibitor with demonstrated clinical efficacy against influenza A and B virus infections. In phase II clinical efficacy trials (NAIB2005 and NAIB2008), virological substudies showed mean reductions in virus shedding after 24 h of treatment of 1.5 to 2.0 log10 50% tissue culture infective doses compared to a placebo, with no reemergence of virus after the completion of therapy. Paired isolates (n = 41) obtained before and during therapy with zanamivir demonstrated no shifts in susceptibility to zanamivir when measured by NA assays, although for a few isolates NA activity was too low to evaluate. In plaque reduction assays in MDCK cells, the susceptibility of isolates to zanamivir was extremely variable even at baseline and did not correlate with the speed of resolution of virus shedding. Isolates with apparent limited susceptibility to zanamivir by plaque reduction proved highly susceptible in vivo in the ferret model. Further sequence analysis of paired isolates revealed no changes in the hemagglutinin and NA genes in the majority of isolates. The few changes observed were all natural variants. No amino acid changes that had previously been identified in vitro as being involved with reduced susceptibility to zanamivir were observed. These studies highlighted problems associated with monitoring susceptibility to NA inhibitors in the clinic, in that no reliable cell-based assay is available. At present the NA assay is the best available predictor of susceptibility to NA inhibitors in vivo, as measured in the validated ferret model of infection.
Influenza is a respiratory infection that produces significant seasonal morbidity in the general population and excess mortality particularly in high-risk groups, including the elderly and those with underlying chronic disease. Both influenza A and influenza B viruses are responsible for human disease. Antigenic drift in these viruses results from mutations within the genes encoding the two surface glycoproteins, the hemagglutinin (HA) and the neuraminidase (NA) proteins. Control by vaccination requires the yearly prediction of circulating influenza A and B strains and annual vaccination, incorporating current A and B strains. Of the 15 HA and 9 NA subtypes of influenza A, relatively few have produced pandemics in man. However, the potential exists for interspecies transmission and the emergence of further pandemics (30). Additional control measures, including the use of antiviral agents, are therefore important.
Zanamivir (5-acetylamino-2, 6-anhydro-4-guanidino-3, 4, 5-trideoxy-d-glycerol-d-galacto-non-2-enoic acid, also known as GG167) is a potent and selective NA inhibitor that interacts with conserved residues in the active site of the NA enzyme and is effective against all nine NA subtypes of influenza A and B virus NA (10, 28). Zanamivir inhibits a wide range of laboratory and clinical isolates of influenza A and B viruses in vitro and in vivo (24, 25, 31) and has been shown to be effective in human experimental influenza challenge and field studies (11, 13, 19).
Amantadine and rimantadine, the first specific anti-influenza agents, described over 30 years ago, have limited clinical value because they are effective only against influenza A virus and can both cause side effects. Furthermore, drug-resistant viruses readily emerge during therapy with amantadine or rimantadine in up to 30% of subjects with acute influenza infections (20). These resistant strains appear to be fully pathogenic and are transmissible to close contacts (12, 14, 15).
Based on knowledge of the X-ray crystal structure of influenza virus NA, zanamivir was designed to bind only to highly conserved residues within the active site of influenza A and B virus NA (29); as a result, there should be constraints on the development of resistance and the maintenance of NA enzyme function. However, it is important to determine the potential for the development of variants with reduced susceptibility to zanamivir during therapy. From in vitro studies with zanamivir, resistant variants have been isolated in passage experiments after prolonged exposure to the compound, with changes in either the HA or NA gene or both (1, 2, 8, 9, 17, 21, 27). These in vitro studies demonstrated the potential for resistance development and confirmed that neuraminidase variants had reduced stability or enzyme function. The relevance of the in vitro observations to clinical settings is not known. Phase I influenza challenge studies have been reported where influenza virus clinical isolates obtained before and during treatment of experimental influenza virus infection have been evaluated for their susceptibility to zanamivir (13). During 5 days of treatment of acute influenza infection with zanamivir, no influenza virus isolates with reduced susceptibility were reported. However, there has been one report of mutations occurring within the HA and NA genes in a single clinical isolate following delayed and prolonged treatment of influenza B infection in an immunocompromised patient (7). The clinical significance of these mutations has not yet been elucidated, but the virus has significantly reduced infectivity in the ferret model.
As part of the clinical evaluation of zanamivir in acute influenza infection in adults, virological substudies described in this study were undertaken in the phase II clinical trials NAIB2005 and NAIB2008. The clinical details of these two studies have been reported previously (11, 19). Briefly, in NAIB2005, adults with an influenza-like illness of <48-h duration were treated twice daily (BID) for 5 days with zanamivir or a placebo administered by oral inhalation (IH) (10 mg/dose) or by a combination of IH and intranasal (IN) spray (6.4 mg/dose). Hayden et al. (11) reported that with zanamivir treatment (in study NAIAB2005), clinical recovery time was reduced by up to 3 days in those with febrile illness when treatment was initiated early. In study NAIAB2008, adults with influenza-like illness of <48-h duration were treated BID or four times daily (QID) for 5 days with zanamivir or a placebo administered by both the IH and IN routes (19). Similar clinical efficacy in the overall population was observed with either the BID or the QID regimen, with a median reduction in the duration of major symptoms of 1 day in the treated group.
The aim of these virological substudies from the two phase II clinical trials was to evaluate the effects of zanamivir on virus shedding and to monitor the zanamivir susceptibility of influenza clinical isolates obtained before and during therapy. Some isolates were also evaluated in the in vivo ferret model to elucidate the significance of the variability observed in the in vitro assays. Further evaluation of the influenza virus clinical isolates obtained included a comprehensive sequence analysis of both the HA and NA genes. The findings from this detailed analysis of influenza clinical isolates obtained during zanamivir treatment are described below together with recommendations for the most suitable methodology for monitoring the susceptibility of influenza isolates to NA inhibitors.
MATERIALS AND METHODS
Virus.
Influenza virus clinical isolates were obtained from nasal washings taken before, during, and after treatment with either zanamivir or a placebo. All subjects recruited into protocols NAIB2005 and NAIB2008 at the Erasmus University Hospital, Rotterdam, The Netherlands, were evaluated in this virological substudy. The wild-type B/Beijing/1/87 isolate was kindly supplied by M. Zambon (PHLS, Colindale, United Kingdom). The influenza B zanamivir-resistant variant B/Beijing/1/87 isolate P3-12 (containing HA mutations Val105Ala and Leu255Gln and NA mutation Glu116Gly) was isolated by in vitro passage at Glaxo Wellcome Medicines Research Centre, Stevenage, United Kingdom (Barnett et al., submitted for publication).
Cell culture.
Madin-Darby canine kidney (MDCK) cells were purchased from ICN Pharmaceuticals, Ltd., Thame, United Kingdom, and were routinely passaged in Eagle's modified minimal essential medium (MEM) supplemented with 10% fetal calf serum and 1% nonessential amino acids. Tertiary monkey kidney (tMK) cells were obtained from cynomolgus macaques and grown in Earle's minimal essential medium supplemented with 10% fetal bovine serum.
Reagents and compounds.
Indubiose A37 agarose was purchased from IBF Biotechnics, Villeneuve La Garenne, France. Gluteraldehyde and carbol fuchsin (Ziehl-Neelsen strong formulation) were purchased from BDH Laboratory Supplies, Poole, United Kingdom. Tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin was purchased from Worthington Biochemical Corporation, Freehold, N.J. All cell culture media were purchased from the Sigma Chemical Company, Poole, United Kingdom, except for DCCM-1 (defined cell culture medium), which was purchased from Biological Industries, Kibbutz Beth Haemak, Israel. Monoclonal antibodies to influenza A and B virus nucleoprotein antigens were purchased from TCS Biologicals, Ltd., Botolph, Claydon, Buckingham, United Kingdom. Tissue culture plasticware was purchased from GIBCO BRL, Paisley, United Kingdom, and Costar Corporation, Cambridge, Mass. 2-(N-Morpholino)ethanesulphonic acid (MES) and 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUN) were purchased from the Sigma Chemical Company. Black MicroFLUOR 96-well plates were purchased from Dynatech Laboratories, Inc., Chantilly, Va. Zanamivir was synthesized at Glaxo Wellcome Medicines Research Centre.
Virus quantitation.
Virus titers were determined for the NAIB2005 samples with both tMK and MDCK cells and for NAIB2008 samples with MDCK cells only. Briefly, confluent MDCK or tMK cells were infected in quadruplicate with 100 μl of a 10-fold dilution series of the nasal washes (ranging from undiluted to 10−5) in serum-free medium containing 1 to 2 μg of trypsin per ml. Virus was adsorbed for 1 h and then the cells were washed four times to remove unadsorbed virus and residual zanamivir. For MDCK cells, cultures were then incubated at 37°C in 5% CO2 for 3 days, and the amount of influenza virus present was then quantified by enzyme-linked immunosorbent assay (ELISA) with mouse monoclonal antibodies to influenza A and B virus nucleoproteins. For tMK cells, cultures were incubated for 6 days and virus was detected by hemagglutination with turkey erythrocytes.
Virus isolation.
Virus stocks of clinical isolates and mutant influenza A virus isolates were prepared by inoculating confluent MDCK cells with virus diluted in serum-free MEM (containing 2 μg of TPCK-treated trypsin per ml). The virus was allowed to adsorb for 1 h at 37°C and then the volume of medium was increased to 5 ml before further incubation at 37°C. Virus stocks were harvested by collecting the supernatant when the cytopathic effect was extensive or after 5 days of incubation. Virus titers were determined by plaque assays, as described below.
Virus plaque assays.
Plaque assays were performed as described previously (31). Confluent monolayers of MDCK cells in six-well tissue culture plates were inoculated with 300 μl of influenza virus diluted in serum-free MEM containing 2 μg of TPCK-treated trypsin per ml. Prior to inoculation, the spent medium was decanted and the cells were washed twice with prewarmed phosphate-buffered saline. After adsorption at room temperature for 1 h, the cell monolayers were overlaid with DCCM-1 containing 0.5% agarose and zanamivir at concentrations ranging from 0.0001 to 100 μg/ml. Two wells of each plate were overlaid with medium alone so that the number of plaques formed in the absence of zanamivir could be determined. Once overlaid, the plates were incubated at 37°C in 5% CO2 for 3 days. After 3 days, the plates were fixed with 5% gluteraldehyde and stained with carbol fuchsin. The plaques were counted and the percentage of plaque reduction for each dilution of zanamivir was determined. The concentration of zanamivir required to reduce plaque numbers by 50% (IC50) was then calculated for each isolate. At least two or three IC50s were determined for each isolate.
NA enzyme inhibition assay.
NA assays (31) use the substrate MUN and are based on the method of Potier et al. (22). Influenza virus NA activity for each virus isolate was first titrated by serial twofold dilutions. The concentration of virus versus NA activity was plotted graphically to determine the virus/enzyme concentration to be used in subsequent inhibition assays. For zanamivir inhibition assays, equal volumes of zanamivir and virus were mixed and preincubated at room temperature for 30 min in Dynatech MicroFLUOR plates. The reaction was initiated by the addition of buffer (32.5 mM MES [pH 6.5], 4 mM CaCl2), containing 100 μM MUN substrate. The reaction mixture was incubated at 37°C for 15 min with shaking, and the reaction was terminated by the addition of 150 μl of 0.014 M NaOH in 83% ethanol. 4-methylumbelliferone was immediately quantified by fluorometric determination with an LS-50B luminescence spectrometer (Perkin-Elmer, Ltd., Beaconsfield, Buckinghamshire, United Kingdom). The excitation wavelength was 365 nm and the emission wavelength was 450 nm. The data were plotted as log zanamivir concentration against fluorescence inhibition, and the IC50s were read from the graph. At least two or three IC50s were determined for each isolate.
Influenza virus infection in ferrets.
Groups of three or four female ferrets (750 to 1,050 g) were infected by IN instillation of 250 μl of a single influenza virus isolate containing either 103.0 PFU/ml (isolate 7249 D1) or 104.0 PFU/ml (isolate 2871 D2 or 7241 D1) while under light anesthesia (isoflurane), as described previously (24). These challenges had been previously determined as the lowest input of virus that would consistently result in infection. Ferrets received two prophylactic doses of zanamivir, at 26 and 2 h prior to infection, and were treated at 5 h after infection and then twice daily for 5 days. Animals were weighed daily for 9 days and IN doses of zanamivir of 1, 0.1, or 0.01 mg/kg of body weight/dose, calculated daily according to animal weight in a volume of 0.25 ml/kg. Virus-infected control animals were sham dosed with pyrogen-free MilliQ phosphate-buffered saline only. Nasal washings were taken on days 1 to 9 following challenge, as described previously (24), and the washings were used in triplicate for the estimation of virus titer by ELISA. The mean nasal-wash virus titer area under the curve (AUC) value for each individual ferret was calculated as the log10 geometric mean of all the ELISA titers determined on days 1 to 9. For each group of ferrets, the log10 geometric mean AUC was also calculated. Statistical analysis was undertaken with a Dunnett's test to evaluate the differences between controls and treated ferret groups. The concentration of zanamivir that inhibits nasal virus titers by 90% (the calculated AUC10) was determined as a measure of susceptibility to zanamivir. Temperature profiles of ferrets were recorded every 10 min with implanted telemetric transmitters (Dataquest; Data Sciences, St. Paul, Minn.), prior to and up to 9 days following infection. Body temperature AUCs were calculated for the period of pyrexic response (0 to 96 h postinfection); AUCs were computed as the area above and below the preinfection mean. Pyrexia was defined as the elevation of core body temperature greater than 2 standard deviations (or more) above the preinfection mean temperature for a period of at least 12 h during the postinfection period. The Duncan's multiple range test was used to analyze temperature values between control and treatment groups. Data were ranked and tested for homogeneity of variance (Levene's test) and normality of distribution (Shapiro-Wilks test).
Sequencing of the HA and NA genes.
Clinical isolates were cultured in MDCK cells and cell supernatant was used to extract viral RNA (vRNA) with the QiaAmp viral RNA extraction kit (Qiagen; Hilden, Germany). The forward primer, p7 (5′-ACTATCATTGCTTTGAGC-3′), was used to prepare cDNA for the HA genes (23) and the primer N2a (5′-GTGAAGATGAATCCAAATCAA-3′) was used for the NA gene of the H3N2 viruses with SuperScript II. Briefly, 1 μg of each forward primer was incubated at 60°C for 10 min with 24 μl of the viral RNA extract. A reaction mixture containing 100 mM deoxynucleoside triphosphate mixture (25 mM [each] dATP, TTP, dCTP, and dGTP), 5 μl of 5 mM dithiothreitol, 5× First Strand buffer (Gibco BRL), and 1 μl SuperScript II reverse transcriptase enzyme (Gibco BRL) was added to each tube and the reactions were incubated at 42°C for 50 min before the reverse transcriptase enzyme was inactivated by incubation at 99°C for 1 min. The forward primer p7 and the reverse primer p1184 (5′-ATGGCTGCTTGAGTGCTT-3′), a vRNA sense primer, were then used to amplify the cDNA obtained. The PCR mixture consisted of 1 μl of 10× PCR buffer, 2.5 mM MgCl2, 0.5 U of Taq polymerase enzyme, 100 mM deoxynucleoside triphosphate mixture (all from Perkin-Elmer Applied Biosystems) and 300 ng (each) of primers p7 and p1184 as described above. Cycling conditions for a single round of PCR with the Perkin-Elmer 9600 thermal cycler were as follows: 94°C for 4 min, 1 cycle; then 94°C for 1 min, 50°C for 1.5 min, and 72°C for 2 min for 35 cycles. Amplicons were visualized by agarose gel electrophoresis and purified with the Bio101 GeneClean II DNA purification kit (Anachem, Luton, United Kingdom). The purified DNA was cycle sequenced with the ABI Terminator BIGDYE cycle sequencing kit and run on an ABI Prism 377 DNA sequencer (Perkin-Elmer, Foster City, Calif.) according to the manufacturer's instructions. The sequence data was analyzed with Lasergene software (DNASTAR, Inc.). For sequencing, three additional internal primers were also used: R792 (5′-CAGTATGTCTCCCGGTTT-3′), R570 (5′-TGGCATAGTCACGTTCAG-3′), and R362 (5′-TAAGGGTAACAGTTGCTG-3′) (23). PCR primers p7 and p1184 were also used for sequencing.
The NA gene of the H3N2 virus was amplified and sequenced with the same conditions except for the primer sets (N2a, 5′-GTGAAGATGAATCCAAATCAA-3′ [forward], and N2b, 5′-GCGAAAGCTTATATAGGC-3′ [reverse]). Internal sequencing primers for the NA gene were as follows: N2c (5′-GGTAACTACTGTAACATTGCA-3′), N2d (5′-GAGAACCTTATGTGTCATGCG-3′), N2e (5′-CAGGAGTCGGAATCGGTTTGT-3′), N2f (5′-GTCAGGAAGTGCTCAGCA-3′), and N2g (5′-AGTGAAAGGCTGGGCCTT-3′). All primers were synthesized by PE Applied Biosystems and purified by high-performance liquid chromatography. As for the HA sequence, the PCR primers were also used for the sequencing reactions. Primer sets for the H1N1 viruses and the influenza B viruses were recommended by Alan Hay (NIMR, London, United Kingdom). Primers underlined indicate those used for the PCR; these primers were also used for sequencing. For the influenza A (H1N1) NA gene, the primers were as follows: H1N1F1 (5′-AGCAAAAGCAGGAGTTTAAAATGA-3′), H1N1F2 (5′-TTCTCACTTGGAATGCAGAACCTT-3′), H1N1F3 (5′-ATAATGACCGATGGCCCGAGTAAT-3′), H1N1F4 (5′-AGGACTAAAAGTAACAGACTCAGA-3′), H1N1R1 (3′-CAACTTTTTTGAGGAACAAAGAT-5′), H1N1R2 (3′-TCCTGATTTTCATTGTCTGGAGTCT-5′), H1N1R3 (3′-TATTACTGGCTACCGGGCTAATTA-5′), and H1N1R4 (3′-AAGAGTGAACCTTACGTCTTGGAA-5′). For the influenza A (H1N1) HA gene, the primers were as follows: H1A1F1 (5′-CAACCAAATGAAAGCAAAACTAC-3′), H1A1F2 (5′-CCCTGAGAATGGAACATGTTACCC-3′), H1A1F3 (5′-GGTATGCTTTCGCACTGAGTAGAG-3′), H1A2F4 (5′-ACATTCCATCCATTCAATCCAGAG-3′), H1A2R1 (3′-CCCAGAAACGTCACGTCTTATACG-5′), H1A1R1 (3′-AACCTCGGTAACGGCCAAAGTAAC-5′), H1A1R2 (3′-CCATACGAAAGCGTGACTCATCTC-5′), H1A1R3 (3′-TCTTTAAACGATACCGACTGCCTC-5), and H1A1R4 (3′-GGGACTCTTACCTTGTACAATGGG-5′). For the influenza B virus NA gene, the primers were as follows: BNAF1 (5′-AGGCCAAAAATGAACAATGCTAC-3′), BNAF2 (5′-GCACTCCTAATTAGCCCTCATAGA-3′), BNAF3 (5′-ACAAGAAAGTGCCTGCAATTGCAT-3′), BNAF4 (5′-GCATCAAGGGAGGATTTGTTCATC-3′), BNAR1 (3′-GGAATGACCTGAATTAACAAAGAC-5′), BNAR2 (3′-CGTAGTTCCCTCCTAAACAAGTAG-5′), BNAR3 (3′-TGTTCTTTCACGGACGTTAACGTA-5′), and BNAR4 (3′-CGTGAGGATTAATCGGGAGTATCT-5′). For the influenza B virus HA gene, the primers were as follows: BHA1F1 (5′-ATTGTACTACTCATGGTAGTAACA-3′), BHA1F2 (5′-ATGCTTTCCTATAATGCACGACAG-3′), BHA1F3 (5′-TGCGCGAGTGGCAGGAGCAAAGTA-3′), BHA2F1, (5′-ATGGAACCAAATATAGACCTCCTG-3′), BHA2R1 (3′-ACGAACAAACGAACAATGGTAATG-5′), BHA1R1 (3′-TCTTCCTCCTACCCTTCCTTACTA-5′), BHA1R2 (3′-ACGCGCTCACCGTCCTCGTTTCAT-5′), and BHA1R4 (3′-TACGAAAGGATATTACGTGCTGTC-5′).
RESULTS
Quantitation of virus shedding.
In NAIB2005, a total of 51 subjects were included in the virological subset from which 60% of viruses were typed as influenza B by hemagglutination inhibition or by sequencing, and the remainder were typed as influenza A (H3N2). For NAIB2008, 41 subjects were included in the subset, of which 73% of viruses were typed as influenza A (H3N2) virus and the remainder were typed as influenza A (H1N1). Virus titrations were determined for the NAIB2005 samples with both tMK cells and MDCK cells. Both cell lines gave statistically comparable results. In NAIB2008, virus titrations were undertaken in MDCK cells. All virus titrations were performed blinded and titers were calculated with the Spearman-Karber equation (5). For comparison of the virological data from these two studies, graphs of median virus titers have been reproduced in Fig. 1 and 2. The results from both studies show similar trends, with reductions in virus shedding in all the IN plus IH groups compared to the placebo group. In the BID and QID treatment arms of NAIB2008, there was a statistically significant reduction in virus shedding from the placebo group (P = 0.005 and P = 0.021, respectively, for AUC determinations). In addition, the duration of virus shedding was reduced below the limit of detection in the IN plus IH groups by 1 to 2 days compared to the placebo groups. From this comparison, it is apparent that the greatest differences in virus titers were observed at day 2 within 24 h of initiating therapy with zanamivir. This appears to be due mainly to the fact that placebo virus titers have not dropped significantly at day 2 but by day 3, there are considerable reductions in virus shedding in the placebo group. Although virus shedding continues to drop in the treated groups, this is partly masked by the reductions in titers in the placebo group. No reemergence of virus after cessation of treatment was observed. Overall the reductions in virus titers in the two studies show a 1.6 to 2.1 log10 reduction in mean virus shedding and a 1.5 to 3.75 median log10 reduction in virus shedding at day 2. This contrasts with the results obtained for the IH-only group, where no significant reductions in virus titers compared with placebo were observed. These results indicated that with IH treatment alone, insufficient compound was reaching the nose to affect virus growth, and this has been confirmed with γ-scintigraphy studies (unpublished data). Despite this, clinical efficacy was comparable with IN plus IH administration of the compound and with IH administration alone, suggesting that viral replication in the nose has little clinical significance compared to the rest of the respiratory tract.
FIG. 1.
Clinical study NAIB2005. Median viral titer versus day of study is shown. ⧫, placebo (n = 16); ■, IH zanamivir (n = 19); ▴, IH plus IN zanamivir (n = 16). P = 0.623, IH versus placebo; P = 0.3, IH plus placebo, by AUC analysis. Limit of detection, 1.5 log10 TCID50s/ml.
FIG. 2.
Clinical study NAIB2008. Median viral titer versus day of study is shown. ⧫, placebo (n = 14); ▴, IH plus IN zanamivir BID (n = 13); ●, IH plus IN zanamivir QID (n = 13). P = 0.005, BID versus placebo; P = 0.021, QID versus placebo, by AUC analysis. Limit of detection, 1.5 log10 TCID50s/ml.
Susceptibility of influenza virus clinical isolates to zanamivir in NA enzyme inhibition assays.
A total of 15 (43%) therapy isolates from NAIB2005 and 12 (46%) therapy isolates from NAIB2008 were successfully grown together with baseline isolates for susceptibility monitoring. Isolates with the longest exposure to zanamivir were chosen for analysis and some matched placebo controls were included. These matched pairs of clinical isolates were evaluated for their susceptibility to zanamivir in the NA enzyme inhibition assay. Twenty matched pairs of clinical isolates from NAIB2005 were evaluated for NA enzyme susceptibility, after first carefully determining virus enzyme input (Table 1). All 20 paired clinical isolates were equally sensitive to zanamivir, with IC50s falling in the range of 0.8 to 2.7 ng/ml. These values fall within the range previously observed for clinical isolates (31). The in vitro control influenza B zanamivir-resistant variant B/Beijing/1/87 isolate P3-12 and the wild-type B/Beijing/1/87 isolate gave IC50s of 2,080 to 2,326 ng/ml and 4.3 to 14.3 ng/ml, respectively.
TABLE 1.
Zanamivir susceptibility of paired influenza virus clinical isolates from NAIB2005
| Treatment group | Virus subtype | Subject | Sample day | Plaque reduction IC50 (μg/ml)a | NA IC50 (ng/ml)ab |
|---|---|---|---|---|---|
| IN + IH | B | 2849 | 1 | 0.0003 ± 0.0002 | 0.9 ± 0.14 |
| 4 | 0.0002 | 3.5 ± 3.11 | |||
| B | 2853 | 1 | 1.38 ± 0.04 | 1.1 ± 0.57 | |
| 2 | 0.69 ± 0.66 | 1.35 ± 0.64 | |||
| B | 2867 | 1 | 27.80 ± 24.75c | 2.7 ± 1.56 | |
| 2 | 4.89 ± 2.48 | 2.5 ± 1.41 | |||
| B | 2868 | 1 | 0.004 ± 0.006 | 0.8 ± 0.28 | |
| 2 | 0.012 ± 0.021 | 0.9 ± 0.00 | |||
| B | 2885 | 1 | 35.25 ± 56.13c | 2.3 ± 0.71 | |
| 2 | 33.73 ± 57.39c | 2.65 ± 0.92 | |||
| IN | B | 2861 | 1 | 0.25 ± 0.10 | 2.45 ± 0.78 |
| 2 | NT | 2.05 ± 0.49 | |||
| 4 | 0.14 ± 0.22 | NT | |||
| B | 2871 | 1 | 0.003 ± 0.001 | 2.25 ± 1.48 | |
| 2 | 5.75 ± 6.63 | 2.1 ± 0.85 | |||
| B | 2878 | 1 | 0.74 ± 0.83 | 1.5 ± 0.71 | |
| 4 | 0.4 ± 0.35 | 1.55 ± 0.78 | |||
| B | 2881 | 1 | 0.61 ± 0.59 | 1.6 ± 0.71 | |
| 5 | 0.46 ± 0.36 | 1.0 | |||
| B | 2884 | 1 | 0.84 ± 0.63 | 1.5 ± 0.71 | |
| 2 | 0.28 ± 0.13 | 1.4 ± 0.85 | |||
| B | 2895 | 1 | 13.88 ± 19.54 | 1.75 ± 0.21 | |
| 2 | 15.87 ± 22.25 | 2.3 ± 0.57 | |||
| B | 2897 | 1 | NT | 1.35 ± 0.64 | |
| 2 | 11.78 ± 16.29 | 1.7 | |||
| 3 | 8.21 ± 11.15 | 1.5 | |||
| H3N2 | 2873 | 1 | 0.002 ± 0.002 | 1.1 ± 0.14 | |
| 2 | 0.011 ± 0.010 | 0.95 ± 0.21 | |||
| B | 2909 | 1 | 33.42 ± 57.66c | 0.9 ± 0.28 | |
| 2 | 0.39 ± 0.43 | 1.1 ± 0.42 | |||
| B | 2910 | 1 | 1.40 ± 1.97 | 1.6 ± 0.14 | |
| 4 | 15.80 ± 22.34 | 0.9 ± 0.57 | |||
| Placebo | B | 2857 | 1 | 0.99 ± 0.1 | 2.4 ± 0.57 |
| 4 | 1.5 ± 0.22 | 2.55 ± 1.06 | |||
| B | 2862 | 1 | 0.17 ± 0.15 | 1.25 ± 0.07 | |
| 4 | 0.04 ± 0.06 | 1.3 ± 0.85 | |||
| B | 2877 | 1 | 50.09 ± 70.59c | 2.35 ± 0.92 | |
| 2 | 37.12 ± 52.02 | 2.7 ± 1.13 | |||
| B | 2882 | 1 | 0.22 ± 0.15 | NT | |
| 4 | 0.1 ± 0.05 | NT | |||
| B | 2865 | 1 | 1.04 ± 1.16 | 2.3 ± 0.57 | |
| 2 | 1.09 ± 1.33 | 2.25 ± 0.49 | |||
| Negative control | B/Beijing/1/87 | 0.025 | 4.3 | ||
| Positive control | B/Beijing/Zrd | >100 | 2080 |
Values represent the means of at least two assays. NT, not tested.
Results obtained in replicate assays varied at most by two- to threefold.
One replicate had an IC50 of >100 μM.
Zanamivir-resistant influenza B/Beijing/1/87 (HA, Val105-Ala, Leu225→Gln; NA, Glu116→Gly).
Out of a total of 19 pairs of clinical isolates from NAIB2008, four pairs from the BID and QID treatment arms contained insufficient levels of NA enzyme activity even after concentration to enable an assessment of zanamivir susceptibility to be made. For the remaining 15 matched pairs where NA enzyme susceptibility to zanamivir was measured, 13 showed no reproducible difference between samples taken before and during treatment, with IC50s ranging from 0.4 to 3.1 ng/ml (Table 2). Again, these data are consistent with previously published data (31). Two pairs of isolates collected from subjects 7257 and 7259 did demonstrate a small apparent reduction in susceptibility to zanamivir during treatment. For isolates from subject 7259, a shift in susceptibility of two- to fourfold (mean, 3.6-fold) was observed. For isolates from subject 7257, initial experiments showed a shift in sensitivity of 5.6- to 11.3-fold (mean, 7.8-fold); however, this was not reproducible in freshly grown material (Table 2), where only one assay gave a shift in susceptibility. The absolute values obtained for each isolate from subjects 7257 and 7259 still fall within the range of those quoted previously as being highly sensitive to zanamivir. The matched isolates from subject 7257 were also sensitive to zanamivir in yield reduction assays with no shift in susceptibility during treatment (data not shown).
TABLE 2.
Zanamivir susceptibility of paired influenza virus clinical isolates from NAIB2008
| Treatment group | Virus subtype | Subject | Sample day | Plaque reduction IC50 (μg/ml)a | NA IC50 (ng/ml)a |
|---|---|---|---|---|---|
| QID | H3N2 | 7222 | 1 | 0.04 ± 0.02 | C |
| 2 | 0.05 ± 0.06 | C | |||
| H3N2 | 7232 | 1 | 0.04 ± 0.02 | 0.9 ± 0.57 | |
| 3 | 0.05 ± 0.06 | 1.65 ± 0.35 | |||
| H3N2 | 7241 | 1 | >100 | C | |
| 4 | >100 | C | |||
| H3N2 | 7228 | 1 | 0.04 ± 0.05 | 0.75 ± 0.35 | |
| 3 | 0.09 ± 0.05 | 0.95 ± 0.78 | |||
| H3N2 | 7249 | 1 | >100 | 0.9 ± 0.14 | |
| 4 | >100 | 0.75 ± 0.21 | |||
| H3N2 | 7257 | 1 | >100 | 0.4 ± 0.14 | |
| 2 | >100 | 3.1 ± 0.42 | |||
| BID | H3N2 | 7227 | 1 | >100 | C |
| 2 | >100 | C | |||
| H3N2 | 7244 | 1 | >100 | 0.8 ± 0.14 | |
| 2 | >100 | 0.6 ± 0.28 | |||
| H3N2 | 7256 | 1 | >100 | 1.1 ± 0.28 | |
| 3 | >100 | 2.75 ± 1.63 | |||
| H3N2 | 7230 | 1 | 0.22 ± 0.12 | C | |
| 2 | 50.29 ± 49.72b | C | |||
| H3N2 | 7259 | 1 | NT | 0.8 ± 0.14 | |
| 3 | NT | 2.85 ± 2.05 | |||
| H1N1 | 7239 | 1 | >100 | 10 ± 00 | |
| 2 | >100 | 0.95 ± 0.07 | |||
| Placebo | H3N2 | 7229 | 1 | >100 | 0.945 ± 0.36 |
| 3 | >100 | 1.25 ± 0.35 | |||
| H3N2 | 7237 | 1 | >100 | 1.35 ± 0.07 | |
| 3 | >100 | 1.4 ± 0.71 | |||
| H3N2 | 7248 | 1 | NT | 1.05 ± 0.49 | |
| 4 | NT | 1.05 ± 0.35 | |||
| H1N1 | 7231 | 1 | NT | 0.5 ± 0.42 | |
| 3 | NT | 0.5 ± 0.42 | |||
| H1N1 | 7242 | 1 | >100 | 0.85 ± 0.35 | |
| 4 | >100 | 0.9 ± 0.28 | |||
| H3N2 | 7250 | 1 | NT | 1.3 ± 0.57 | |
| 5 | NT | 0.9 ± 0.57 | |||
| H3N2 | 7258 | 1 | NT | 1.2 ± 0.57 | |
| 3 | NT | 1.05 ± 0.35 | |||
| Negative control | B/Beijing/1/87 | 0.012 | 14.3 | ||
| Positive control | B/Beijing/ Zrc | >100 | 2,326 |
Values represent the means of at least two assays. C, insufficient NA activity following concentration; 76% of isolates were influenza A (H3N2); the remainder were influenza A (H1N1); NT, not tested.
One replicate had an IC50 of >100 μM.
Zanamivir-resistant influenza B/Beijing/1/87 (HA, Val105-Ala, Leu225-Gln; NA, Glu116-Gly).
Susceptibility of influenza virus clinical isolates to zanamivir in plaque reduction assays.
As previously observed (31), data from plaque reduction assays were highly variable, limiting the assay's value in susceptibility monitoring. Data was pooled from individual experiments to generate a geometric mean value for each isolate (Tables 1 and 2). The in vitro controls, influenza B zanamivir-resistant variant B/Beijing/1/87 isolate P3-12 and the wild-type B/Beijing/1/87 isolate, gave IC50s of >100 and 0.012 to 0.025 μg/ml, respectively.
The susceptibility to zanamivir of the 20 paired clinical isolates from NAIB2005 ranged from means of 0.002 to 50 μg/ml (Table 1). Considerable variability was observed between replicate assays for some isolates. For four isolates, at least one replicate experiment gave a value of >100 μg/ml, the highest concentration tested. Analysis of the data from paired isolates showed that there was a consistent shift in sensitivity between only two pairs of isolates: isolates from day 1 and day 2 from patient 2871, a patient that had received IH zanamivir, and isolates from day 1 and day 4 from patient 2862, a patient that had received a placebo. The day 2 isolate from patient 2871 was approximately 2,000 times less sensitive to zanamivir than the day 1 isolate (before treatment). For the isolates from patient 2862, the day 4 isolate showed a fourfold higher sensitivity to zanamivir than the day 1 isolate. No corresponding change in NA susceptibility was observed for any isolate from these two subjects. All of the paired isolates from the NAIB2005 study plaqued reasonably well in tissue culture.
The paired clinical isolates from NAIB2008 were tested for in vitro susceptibility to zanamivir by plaque reduction (Table 2). The isolates tested plaqued poorly in MDCK cells, forming large but very diffuse plaques. Of the 15 pairs of isolates tested, 12 were apparently insensitive to zanamivir inhibition at the highest concentration tested, 100 μg/ml. This included representatives from all three treatment groups, including the placebo group. Only four pairs of isolates were susceptible to zanamivir by plaque reduction, with a mean range of susceptibility from 0.04 to 50 μg/ml. Although differences in susceptibility were observed between pre- and intratreatment samples for these isolates, the differences observed were not reproducible between assays. In addition to the matched pairs of clinical isolates, six day 1 virus isolates from subjects 7240, 7261, 7233, 7246, 7247, and 7245, who had rapidly resolved shedding virus (<30 h), were also tested for in vitro susceptibility to zanamivir by plaque reduction. These isolates appeared to be insensitive to zanamivir by plaque reduction, with IC50s of >100 μg/ml, suggesting that plaque reduction data were not predictive of in vivo efficacy. In addition, no correlation between plaque reduction susceptibility, baseline virus titer, and time to resolution of virus shedding was observed in isolates from the QID treatment arm (Table 3). In the NAIB2005 study, in addition to obtaining daily nasal-wash samples from a subset of subjects, throat swabs were taken on day 6 from the majority of subjects. There was no evidence, from the culture of day 6 swabs taken from 256 subjects treated with zanamivir, that influenza virus shedding rebounded after treatment had ceased.
TABLE 3.
Lack of correlation of antiviral efficacy in MDCK cells and response to zanamivir therapy in humansa
| Subject and day (D) | Plaque reduction IC50 (μg/ml) | Pretreatment titer (log10 TCID50/ml) | Day virus undetectable |
|---|---|---|---|
| 7222 D1 | 0.04 | 5.8 | 3 |
| 7228 D1 | 0.04 | 5.8 | 4 |
| 7232 D1 | 0.04 | 5.05 | 4 |
| 7240 D1 | >100 | 6.05 | 2 |
| 7241 D1 | >100 | 4.05 | 2 |
| 7249 D1 | >100 | 4.3 | 5 |
| 7257 D1 | >100 | 4.05 | 3 |
| 7261 D1 | >100 | 4.8 | 2 |
Subjects enrolled in the NAIB2008 clinical study were placed in a treatment group receiving IH plus IN zanamivir QID.
In vivo susceptibility of isolates from subjects 2871 D2, 7241 D1, and 7249 D1.
Ferrets were infected with the above influenza clinical isolates and treated with zanamivir at 0.01, 0.1, and 1.0 mg/kg. The progress of influenza infection in the animal model was monitored by measurement of core body temperature, nasal-wash viral titer, nasal turbidity, and comparison of these parameters in zanamivir-treated animals with sham-treated animals (Tables 4 to 6). The concentration of zanamivir that inhibits nasal virus titers by 90% (AUC10) was determined as a measure of susceptibility to zanamivir for comparison with the in vitro data. The in vitro susceptibility in the plaque reduction and NA assays (IC50s) and the AUC10s determined in vivo together with influenza A and B controls have been tabulated to allow comparisons to be made between the three assay systems used in this study (Table 7).
TABLE 4.
Efficacy of IN zanamivir in ferrets following infection with day 2 isolate from subject 2871 (virus 2871 D2)a
| Treatment dose (mg/kg) | No. of animals with pyrexia/no. treated | Mean body temperature AUC (% control) | Mean nasal-wash virus AUC (TCID50) | Mean nasal-wash virus titer (% of control)b | Mean nasal-wash turbidity (% of control) |
|---|---|---|---|---|---|
| 1.0 | 0/4 | 3.3* | 4.93 | 10.57** | 65.4 |
| 0.1 | 0/4 | 6.8* | 5.17 | 18.62*** | 80.4 |
| 0.01 | 4/4 | 52.0* | 5.79 | 77.65 | 103.7 |
| Vehicle | 4/4 | 100 | 5.90 | 100 | 100 |
*, significantly different from untreated control (P = 0.01 by Duncan's multiple range test); **, significantly different from untreated control (P ≤ 0.01 by Dunnett's test); ***, significantly different from untreated control (P ≤ 0.05 by Dunnett's test).
Virus titrated in triplicate for each time point for individual ferrets.
TABLE 6.
Efficacy of IN zanamivir in ferrets following infection with day 2 isolate from subject 7249 (virus 7249 D2)a
| Treatment dose (mg/kg) | No. of animals with pyrexia/no. treated | Mean body temperature AUC (% control) | Mean nasal-wash virus AUC (TCID50) | Mean nasal-wash virus titer (% of control)b | Mean nasal-wash turbidity (% of control) |
|---|---|---|---|---|---|
| 1.0 | 0/4 | 2.9* | 5.50 | 7.0** | 57.4 |
| 0.1 | 0/4 | 14.4* | 5.48 | 6.8** | 93.8 |
| 0.01 | 3/3 | 50.7 | 5.99 | 21.7** | 82.8 |
| Vehicle | 3/3 | 100 | 6.65 | 100 | 100 |
*, significantly different from untreated control (P = 0.05 by Duncan's multiple range test); **, significantly different from untreated control (P ≤ 0.01 by Dunnett's test); ***, significantly different from untreated control (P ≤ 0.05 by Dunnett's test).
Virus titrated in triplicate for each time point for individual ferrets.
TABLE 7.
Comparison of in vitro and in vivo susceptibility data for influenza virus clinical isolates
| Virus | NA assay IC50 (ng/ml)a | Plaque assay IC50 (μg/ml) | Ferret nasal-wash virus titers (EDAUC10 mg/kg)b |
|---|---|---|---|
| 2871 D2 (influenza B) | 2.1 ± 0.85 | 5.7 ± 6.63 | 0.82 |
| 7241 D1 (influenza A/H3N2) | Cc | >100 | 0.02 |
| 7249 D1 (influenza A/H3N2) | 0.9 ± 0.14 | >100 | 0.1 |
| A/Mississippi/1/85 (H3N2) | 1.3 | 0.0047 | 0.32 |
| B/Victoria/102/85 | 1.3 | 0.0017 | 0.59 |
Although numbers are small, a rank correlation coefficient of 0.95 (P = 0.0513) was observed on comparison of the NA assay and the in vivo assay.
AUC10, the concentration of zanamivir that inhibits nasal virus titers by 90% (calculated as 10% of the AUC), was determined as a measure of susceptibility to zanamivir. ED, effective dose.
NA activity was too low to assay.
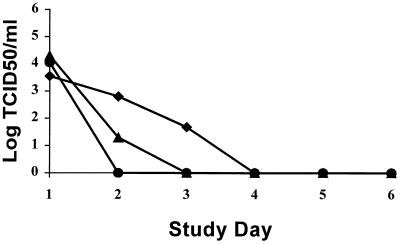
The in vivo susceptibility of influenza virus isolate 2871 D2 to zanamivir is summarized in Table 4. In ferrets infected with isolate 2871 D2, the pyrexic response was completely inhibited in all animals treated with 1 or 0.1 mg of zanamivir per kg, respectively. All animals treated with 0.01 mg of zanamivir per kg had pyrexia between days 2 to 6 postinfection, but temperatures were statistically lower than those of control animals (P ≤ 0.01), which all had a marked pyrexic response 2 to 3 days after infection, with elevated temperatures recorded up to day 6. Treatment with 1.0 and 0.1 mg of zanamivir per kg lowered nasal-wash viral titers, compared with control values (P < 0.01 and P < 0.05, respectively). The dose required to reduce the nasal-wash virus titer AUC to 10% of the sham-treated control value was calculated as 0.82 mg/kg, similar to the value previously calculated for the laboratory influenza strain B/Victoria/102/85 (Table 7). Nasal-wash turbidity was reduced to 35 and 20% of control values. The virus isolate 2871 D2 was sensitive to inhibition by zanamivir in the NA assay, and this was confirmed by a full response to therapy in the ferret model. The results obtained in the plaque reduction assay were, therefore, not predictive of results obtained in either NA or in vivo assays. (Table 7).
In ferrets infected with influenza virus isolate 7241 D1, the pyrexic response was completely inhibited in animals treated with 1 mg of zanamivir per kg (Table 5). Ferrets treated with 0.1 to 0.01 mg of zanamivir per kg had elevated temperatures 2 to 6 days postinfection, although these were statistically lower than those of control animals at both doses (P ≤ 0.01). Treatment with 1.0, 0.1, and 0.01 mg of zanamivir per kg lowered nasal-wash viral titers significantly compared with control animals giving percent AUCs of 1.83, 3.99, and 15.63, respectively. An AUC10 of 0.02 mg/kg for reduction in nasal-wash viral titer was calculated and this compares well with the zanamivir sensitivity of the control virus, influenza A/Mississippi/1/85, in this animal model system (Table 7). Nasal-wash turbidities were reduced to 73.81, 79.76, and 92.26% of control values at doses of 1, 0.1, and 0.01 mg/kg, respectively. The virus isolate 7241 D1 was shown to be highly sensitive to zanamivir in the ferret model. Low levels of NA activity in influenza clinical isolate 7241 are therefore not predictive of poor efficacy of zanamivir in vivo.
TABLE 5.
Efficacy of IN zanamivir in ferrets following infection with day 1 isolate from subject 7241 (virus 7241 D1)a
| Treatment dose (mg/kg) | No. of animals with pyrexia/no. treated | Mean body temperature AUC (% control) | Mean nasal-wash virus AUC (TCID50) | Mean nasal-wash virus titer (% of control)b | Mean nasal-wash turbidity (% of control) |
|---|---|---|---|---|---|
| 1.0 | 0/4 | 12.01* | 2.97 | 1.83** | 73.81 |
| 0.1 | 4/4 | 33.49* | 3.43 | 3.99** | 79.76 |
| 0.01 | 4/4 | 65.09* | 3.64 | 15.63** | 92.26 |
| Vehicle | 4/4 | 100 | 4.91 | 100 | 100 |
*, significantly different from untreated control (P = 0.01 by Duncan's multiple range test); **, highly significantly different from untreated control (P ≤ 0.01 by Dunnett's test).
Virus titrated in triplicate for each time point for individual ferrets.
The numbers of animals with a pyrexic response and the magnitude of response following infection with influenza virus isolate 7249 D1 are shown in Table 6. All sham-treated control animals infected with influenza virus isolate 7249 D1 had a pyrexic response within 2 to 4 days postinfection. In contrast, the temperature responses of zanamivir-treated ferrets were completely suppressed in all animals at doses of 1 or 0.1 mg/kg and significantly reduced at 0.01 mg/kg (P ≤ 0.01). Significant reductions in nasal-wash virus titer were observed on days 1 to 6 postinfection at all doses tested, with reductions to 7.0, 6.8, and 21.7% of control values (AUCs, days 1 to 6) at doses of 1, 0.1, and 0.01 mg/kg, respectively. From these values, the AUC10 was calculated as 0.10 mg/kg, which is comparable to that of other influenza strains (Table 7). Clinical isolates 7241 D1 and 7249 D1 were both apparently not susceptible to zanamivir in the plaque reduction assay. However, both of these isolates were highly susceptible to zanamivir in vivo, indicating that the plaque reduction assay is not predictive of in vivo susceptibility to zanamivir.
Sequences of the NA and HA genes.
Sequences of the NA and HA genes from matched isolates gathered before and during treatment were compared to identify possible treatment-induced changes. A total of 28 matched isolates from the treated groups, including 7 influenza B isolates and 6 placebo matched pairs, were sequenced. The nucleotide and predicted amino acid sequences obtained from the clinical isolates were also compared with the sequence data for prototype strains predicted to be circulating during the 1994 to 1995 and 1995 to 1996 seasons. For these seasons, the prototype H3N2 strain used for reference was A/Wuhan/359/95 (H3N2). Table 8 gives a summary of the predicted amino acid changes observed in the HA and NA genes during treatment. The majority of the matched isolates sequenced (79%) showed no coding changes, including the two influenza B matched isolates from NAIB2005, which showed apparent shifts in susceptibility to zanamivir by plaque reduction. Only four H3N2 isolates from NAIB2008 had sequence changes in the HA and/or NA gene. Isolates from subjects 7228 and 7249 had a predicted HA coding change at residue 275Gly→Asp. However, this mutation is not in a conserved region and has been observed in other virus variants. Samples from subject 7230 showed a predicted coding change in HA1 at residue 135Arg→Lys. Although this mutation is close to the primary sialic acid binding site, the change is conservative and a known natural variant and was present in several isolates at baseline in this trial. Isolates with the 135Lys residue showed a range of susceptibility to zanamivir in vitro, and two isolates (7241 D1 and 7249 D1) were highly susceptible to zanamivir in vivo in the ferret model. Isolates from subject 7228 showed a predicted coding change in NA at residue 20Ile→Met. This forms part of the transmembrane region of the NA gene and is not conserved. A limited search showed no other published variant sequences that had a methionine at this location; however, in our study we identified one other clinical isolate that had a methionine at this location at baseline. Isolates from subject 7241 showed a predicted NA coding change at residue 29Ile→Pro that is also located within the transmembrane region of the NA, which is not highly conserved. No changes were observed in NA residues previously shown to be involved with in vitro susceptibility to zanamivir.
TABLE 8.
Summary of susceptibility data in vitro and coding changesa
| Study/treatment group | Subject | Sample day | Plaque reduction assay IC50 (μg/ml) | NA assay IC50 (ng/ml) | HA coding changesb | NA coding changesb |
|---|---|---|---|---|---|---|
| NAIB2005/IH | 2871 | 1 | 0.003 ± 0.001 | 2.25 ± 0.49 | ||
| 2 | 5.75 ± 6.63 | 2.1 ± 1.48 | None | None | ||
| NAIB2008/QID | 7228 | 1 | 0.04 ± 0.05 | 0.75 ± 0.35 | ||
| 3 | 0.09 ± 0.05 | 0.95 ± 0.78 | G275D* | None | ||
| NAIB2008/QID | 7241 | 1 | >100 | C | ||
| 2 | >100 | C | None | L29P† | ||
| NAIB2008/QID | 7249 | 1 | >100 | 0.9 ± 0.14 | ||
| 4 | >100 | 0.75 ± 0.21 | G275D* | None | ||
| NAIB2008/BID | 7230 | 1 | 0.22 ± 0.12 | C | ||
| 2 | 50.3 ± 49.72 | C | R135K*‡ | Y22F* |
All isolates with coding changes were influenza A (H3N2) isolates. Changes were observed in influenza virus clinical isolates during treatment with zanamivir in studies NAIB2005 and NAIB2008.
Results indicate a comparison between pre- and intratreatment samples. *, natural variation; †, transmembrane region; ‡, close to the primary sialic acid binding site.
DISCUSSION
Zanamivir was designed to bind to the highly conserved residues within the active site of influenza A and B virus NA. Previous in vitro studies have shown that passage of various influenza A and B strains in the presence of zanamivir results in the selection of virus variants that have reduced susceptibility in vitro to zanamivir. Biological studies and sequence analysis of the viruses isolated from such passage experiments have shown that reduced susceptibility to zanamivir in vitro can result from amino acid sequence changes in either the viral HA or NA or in both proteins (1, 2, 4, 8, 9, 16, 17, 18, 21, 26, 27). The key mutation in the NA gene associated with the resistance phenotype which produces a 30- to 40-fold shift in susceptibility is 119Glu→Gly (influenza A) or 116Glu→Gly (influenza B), which also reduces enzyme stability (4, 26). Several mutations in the HA have been identified, including those at residues K68R and G75E in HA2 and G135D, N145S, N150S, T155A, S165N, S186F, K222T, V223I, R229I/S in HA1, some of which appear to contribute to in vitro resistance and in some cases to drug dependence (1, 2, 9, 17, 18, 21, 27). Interestingly, one HA variant which caused high-level resistance in vitro showed no change in susceptibility to zanamivir in vivo, indicating that the HA mutation may not contribute to resistance in vivo (21). Furthermore, a double HA-plus-NA mutant possessed significantly less resistance in vivo (10-fold) than in vitro (100-fold) (1). These studies suggest that HA mutations identified in vitro may not be important in conferring reduced susceptibility to zanamivir in vivo and that it may be NA mutations alone that are critical. At present, however, the relationship between HA and NA mutations and zanamivir susceptibility in vivo is not clear. It is therefore important to monitor virus susceptibility to different uses of the compound in the clinic. In one clinical study of an immunocompromised patient given 14 days of treatment with zanamivir, an influenza B mutant with changes in both the HA (198Thr→Ile) and NA (152Arg→Lys) regions was isolated, suggesting that HA mutations may play a role in vivo. The mutation in NA reduced the affinity of the enzyme for zanamivir and also reduced its catalytic activity. This double mutant also had reduced infectivity when evaluated in the ferret model. This clinical study and the previous in vitro studies have confirmed that mutations selected by zanamivir in the NA compromise enzyme stability or function, leading to less-fit viruses. Interestingly, the mutant virus was found to be sensitive to zanamivir in cell culture systems, probably due to changes in receptor binding specificity. This study highlighted problems associated with cell culture systems, such as the MDCK plaque reduction assay, for monitoring in vivo resistance (7).
In the clinical studies described here, susceptibility monitoring was undertaken with both enzyme NA assays and plaque reduction assays in MDCK cells. Here again as reported previously (31), the plaque reduction assay was found to be highly variable and unreliable in monitoring resistance in clinical isolates, whereas the NA assay was highly reproducible (Tables 1 and 2). This variability is probably explainable in part by the observation that influenza viruses have a widely varying dependence on NA activity for replication in cell culture and virus may spread from cell to cell directly, bypassing the NA function. In contrast to the isolate obtained from an immunocompromised patient where the in vivo resistant isolate in cell culture was highly sensitive, here some isolates were highly sensitive in the NA assay but appeared resistant in the MDCK cell plaque assay. Despite the variations in plaque reduction observed in NAIB2005, only one matched pair of isolates showed a consistent reduction in susceptibility to zanamivir. However, many isolates from NAIB2008 appeared insensitive by plaque reduction, even at baseline. The isolates from NAIB2008 all formed large diffuse plaques, which made some plaque reduction assays difficult to interpret and may have contributed to the susceptibility profile seen in tissue culture. All the isolates studied failed to agglutinate chick erythrocytes but agglutinated guinea pig erythrocytes efficiently (unpublished data), indicating changes in receptor specificity relative to the viruses that circulated in the previous influenza outbreak. In addition, a proportion of the isolates from NAIB2008 contained insufficient NA activity to enable susceptibility testing in the NA enzyme assay. This variability in plaque reduction susceptibility profiles and levels of NA activity required further investigation, and key isolates were evaluated in vivo for a definitive answer regarding zanamivir susceptibility. Of the three isolates examined, one (2871 D2) developed an apparent reduction in susceptibility, as measured by plaque reduction, during treatment; one (7241 D1) was apparently insensitive by plaque reduction at baseline; and one (7249) had undetectable neuraminidase activity. All these isolates were highly susceptible to zanamivir in the ferret model (assessed by reduction of viral shedding over time), showing that the plaque reduction assay was not predictive of zanamivir susceptibility in vivo. Furthermore, there was no apparent correlation between susceptibility of virus isolates by plaque reduction at baseline and the time to clearance of virus (Table 3). This again indicates the limitations of the plaque reduction assay as a predictor of the susceptibility of virus to zanamavir in the clinic. Due to the acute nature of influenza infection and the efficacy of zanamivir, the number of paired pre- and intratreatment isolates is relatively small, which precludes the use of statistical analysis to determine the significance of this observation. In addition, low-level NA activity did not influence susceptibility to zanamivir in vivo. Data from this study indicate that the NA enzyme assay is currently the only in vitro assay that is predictive of in vivo susceptibility to zanamivir (rank correlation coefficient, 0.95; P = 0.0513). Furthermore, it should be stressed that in all susceptibility assays, including assays of enzyme activity, in vitro virus assays, and in vivo assays, the IC50s determined will be influenced by the level of input virus used. It is therefore essential to quantify virus and to standardize the level of virus used for susceptibility assays to reduce variability and to avoid incorrect reporting of resistance.
These results emphasize problems associated with the assays currently used for monitoring susceptibility to NA inhibitors. To address some of these problems, further assay development is in progress. Firstly, a more sensitive chemiluminescent neuraminidase assay is being evaluated to allow monitoring of NA susceptibility in viruses with low NA activity. Secondly, alternate respiratory cell lines with α-2-6 sialic acid receptor specificity are being tested for sensitivity to influenza viruses, in an attempt to find a cell system which is more representative of the respiratory tract. Thirdly, other methods of evaluating the antiviral activity of NA inhibitors in influenza virus-infected MDCK cells have been investigated. Initial experiments involved an ELISA-based assay that detects influenza A and B nucleoprotein antigens to assess virus yield using a relatively high input of virus. This assay was also found to be too variable, depending on input, and not to be sensitive enough to accurately evaluate zanamivir susceptibility with clinical isolates. In addition, we evaluated the susceptibility of a small number of isolates to zanamivir in MDCK cells in a 24-h yield reduction assay where virus was quantified by plaque titration. The input of virus used was found to be absolutely critical. Using an input of 0.01 PFU/cell isolates that were sensitive in the NA enzyme assay but not by plaque reduction were also sensitive by yield reduction (data not shown). This yield reduction assay, however, is very labor intensive and is not applicable to the testing of large numbers of clinical isolates. In addition, many influenza virus clinical isolates do not plaque well without cell passage, and assessment of virus input and resulting yield will be necessary by other techniques.
Further characterization of matched clinical isolates by sequencing the HA genes, both HA1 and HA2, and the NA gene was undertaken. The majority of isolates revealed no amino acid changes between the matched pairs, although there were changes from the baseline sequence of the prototype strain A/Wuhan/359/95 (H3N2). This is attributable to the known variability within the HA and NA genes of influenza clinical isolates. Amino acid changes in either HA1 or in the transmembrane region of NA were observed in isolates for four subjects (7228, 7230, 7241, and 7249) on zanamivir therapy. However, all amino acid changes were conservative and resulted in changes that are present in natural variants. None of the amino acid changes identified from in vitro studies to be involved with reduced susceptibility to zanamivir were observed in these clinical studies.
In conclusion, there was consistency observed in results between susceptibility monitoring by the neuraminidase assay and the in vivo ferret model plus the sequencing results. These results indicated, in these two clinical substudies, that no zanamivir resistance had emerged during 5 days of therapy of acute infection. In addition to the analysis of paired isolates, samples in NAIB2005 were also taken from the majority of subjects on day 6 for culture to check that virus had not reemerged 24 h after treatment had stopped. No evidence of an increase in virus shedding at day 6 was observed in a total of 256 treated subjects. The NA assay has proved to be a reliable, rapid system for monitoring the susceptibility of clinical isolates to zanamivir and in conjunction with sequencing studies, is currently the best method for surveillance studies. Evaluation of influenza virus isolates from zanamivir phase III treatment and prophylaxis studies are in progress to continue monitoring the susceptibility of clinical isolates to zanamivir.
REFERENCES
- 1.Blick T J, Sahasrabudhe A, McDonald M, Owens I J, Morley P J, Fenton R J, McKimm-Breschkin J L. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-Neu5Ac2en. Virology. 1998;246:95–103. doi: 10.1006/viro.1998.9194. [DOI] [PubMed] [Google Scholar]
- 2.Blick T J, Tiong T, Sahasrabudhe A, Varghese J N, Colman P M, Hart G J, Bethell R C, McKimm-Breschkin J L. Generation and characterization of an influenza virus neuraminidase variant with decreased susceptibility to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology. 1995;214:475–484. doi: 10.1006/viro.1995.0058. [DOI] [PubMed] [Google Scholar]
- 3.Campion K, Silagy C, Cooper C, Bolton P, Watts R, Liaw T, Narayan K, Delooze F the MIST Study Group. Efficacy and safety of inhaled zanamivir in the treatment of influenza A and B virus infections. Lancet. 1998;352:1877–1881. [PubMed] [Google Scholar]
- 4.Colacino J M, Chirgadze N Y, Garman E, Murti K G, Loncharich R J, Baxter A J, Staschke K A, Laver W G. A single sequence change destabilizes the influenza virus neuraminidase tetramer. Virology. 1997;236:66–75. doi: 10.1006/viro.1997.8709. [DOI] [PubMed] [Google Scholar]
- 5.Doherty R M. Animal virus titration techniques. In: Harris R J C, editor. Techniques in experimental virology. New York, N.Y: Academic Press; 1964. pp. 169–223. [Google Scholar]
- 6.Goto H, Bethell R C, Kawaoka Y. Mutations affecting the sensitivity of the influenza virus neuraminidase to 4-guanidino-Neu5Ac2en. Virology. 1997;238:265–272. doi: 10.1006/viro.1997.8810. [DOI] [PubMed] [Google Scholar]
- 7.Gubareva L V, Matrosovitch M N, Brenner M D, Bethell R C, Webster R G. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis. 1998;178:1257–1262. doi: 10.1086/314440. [DOI] [PubMed] [Google Scholar]
- 8.Gubareva L V, Robinson M J, Bethell R C, Webster R G. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol. 1997;71:3385–3390. doi: 10.1128/jvi.71.5.3385-3390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubareva L V, Bethell R, Hart G J, Murti K G, Penn C R, Webster R G. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J Virol. 1996;70:1818–1827. doi: 10.1128/jvi.70.3.1818-1827.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubareva L V, Penn C R, Webster R G. Inhibition of replication of avian influenza viruses by the neuraminidase inhibitor 4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid. Virology. 1995;212:323–330. doi: 10.1006/viro.1995.1489. [DOI] [PubMed] [Google Scholar]
- 11.Hayden F G, Osterhaus A D M E, Treanor J J, Fleming D M, Aoki F Y, Nicholson K G, Bohnen A M, Hirst H M, Keene O, Wightman K. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza virus infections. N Engl J Med. 1997;337:874–880. doi: 10.1056/NEJM199709253371302. [DOI] [PubMed] [Google Scholar]
- 12.Hayden F G. Amantadine and rimantadine—clinical aspects. In: Richman D D, editor. Antiviral drug resistance. New York, N.Y: John Wiley & Sons Ltd.; 1996. pp. 59–77. [Google Scholar]
- 13.Hayden F G, Treanor J J, Betts R F, Lobo M, Esinhart J D, Hussey E K. Safety and efficacy of the neuraminidase inhibitor GG167 in experimental human influenza. JAMA. 1996;275:295–299. [PubMed] [Google Scholar]
- 14.Hayden F G, Belshe R B, Clover R D, Hay A J, Oakes M G, Soo W. Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. N Engl J Med. 1989;321:1696–1702. doi: 10.1056/NEJM198912213212502. [DOI] [PubMed] [Google Scholar]
- 15.Houck P, Hemphill M, LaCroix S, Hirsh D, Cox N. Amantadine-resistant influenza A in nursing homes. Identification of a resistant virus prior to drug use. Arch Intern Med. 1995;155:533–537. [PubMed] [Google Scholar]
- 16.McKimm-Breschkin J L, Sahasrabudhe A, Blick T J, McDonald M, Colman P M, Hart G J, Bethell R C, Varghese J N. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–2462. doi: 10.1128/jvi.72.3.2456-2462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKimm-Breschkin J L, Blick T J, Sahasrabudhe A, Tiong T, Marshall D, Hart G J, Bethell R C, Penn C R. Generation and characterization of variants of NWS/G7OC influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob Agents Chemother. 1996;40:40–46. doi: 10.1128/aac.40.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKimm-Breschkin J L, McDonald M, Blick T J, Colman P M. Mutation in the influenza virus neuraminidase gene resulting in decreased susceptibility to the neuramindiase inhibitor 4-guanidino-Neu5Ac2en leads to instability of the enzyme. Virology. 1996;225:240–242. doi: 10.1006/viro.1996.0595. [DOI] [PubMed] [Google Scholar]
- 19.Monto, A. S., D. P. Robinson, L. Herlocher, J. M. Hinson, M. Elliott, and O. Keene. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza A and B virus infections. J. Infect. Dis., in press. [DOI] [PubMed]
- 20.Nicholson K G. Use of antivirals in influenza in the elderly: prophylaxis and therapy. Gerontology. 1996;42:280–289. doi: 10.1159/000213804. [DOI] [PubMed] [Google Scholar]
- 21.Penn C R, Barnett J M, Bethell R C, Fenton R, Gearing K L, Healy N, Jowett A J. Selection of influenza virus with reduced susceptibility in vitro to the neuraminidase inhibitor GG167 (4-guanidino-Neu5Ac2en): changes in the haemagglutinin may compensate for loss of neuraminidase activity. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza III. Amsterdam, The Netherlands: Elsevier Science B. V.; 1996. pp. 735–740. [Google Scholar]
- 22.Potier M, Mameli L, Belisle M, Dallaire L, Melancon S B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 23.Rocha E, Xu X, Hall H E, Allen J R, Regnery H L, Cox N J. Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of cell- and egg-grown viruses. J Gen Virol. 1993;74:2513–2518. doi: 10.1099/0022-1317-74-11-2513. [DOI] [PubMed] [Google Scholar]
- 24.Ryan D M, Ticehurst J, Dempsey M H. GG167 (4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid) is a potent inhibitor of influenza virus in ferrets. Antimicrob Agents Chemother. 1995;39:2583–2584. doi: 10.1128/aac.39.11.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan D, Ticehurst J, Dempsey M H, Penn C R. Inhibition of influenza virus replication in mice by GG167 is consistent with extracellular activity of viral neuraminidase (sialidase) Antimicrob Agents Chemother. 1994;38:2270–2275. doi: 10.1128/aac.38.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahasrabudhe A, Lawrence L, Epa V C, Varghese J N, Colman P M, McKimm-Breschkin J L. Substrate, inhibitor, or antibody stabilizes the Glu 119 Gly mutant influenza virus neuraminidase. Virology. 1998;247:14–21. doi: 10.1006/viro.1998.9222. [DOI] [PubMed] [Google Scholar]
- 27.Staschke K A, Colacino J M, Baxter A J, Air G M, Bansal A, Hornback W J, Munroe J E, Laver G W. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology. 1995;214:642–646. doi: 10.1006/viro.1995.0078. [DOI] [PubMed] [Google Scholar]
- 28.Thomas G P, Forsyth M, Penn C R, McCauley J W. Inhibition of the growth of influenza viruses in vitro by 4-guanidino-2,4-dideoxy-2,3-N-acetylneuraminic acid. Antivir Res. 1994;24:352–356. doi: 10.1016/0166-3542(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 29.von Itzstein M, Wu W Y, Kok G B, Pegg M S, Dyason J C, Jin B, Van Phan T, Smythe M L, White H F, Oliver S W, Colman P M, Varghese J N, Ryan D M, Woods J M, Bethell R C, Hotham V J, Cameron J M, Penn C R. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 30.Webster R G, Shortridge K F, Kawaoka Y. Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol Med Microbiol. 1997;18:275–279. doi: 10.1111/j.1574-695X.1997.tb01056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods J M, Bethell R C, Coates J A V, Healy N, Hiscox S A, Pearson B A, Ryan D M, Ticehurst J, Tilling J, Walcott S M, Penn C R. 4-Guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid is a highly effective inhibitor both of the sialidase (neuraminidase) and of growth of a wide range of influenza A and B viruses in vitro. Antimicrob Agents Chemother. 1993;37:1473–1479. doi: 10.1128/aac.37.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]