Abstract
Androgen receptor splice variant 7 (AR-v7), a constitutively active transcription factor, plays a crucial role in the progression of castration-resistant prostate cancer (CRPC). Here, we found that the cleavage and polyadenylation specificity factor 1 (CPSF1) (the largest subunit of the multi-protein cleavage and polyadenylation specificity complex), regulated by the E3 ubiquitin ligases SIAH1, promoted AR-v7 expression. The data from microarray-based analysis and clinical specimen-based analysis showed that SIAH1 expression was decreased in PCa and was negatively correlated with aggressive phenotypes of PCa. SIAH1 repressed PCa cell growth and invasion under castrate conditions. SIAH1 directly interacted with CPSF1 and promoted ubiquitination and degradation of CPSF1. CPSF1 expression was negatively correlated with SIAH1 expression, but positively with PCa progression. CPSF1 overexpression switched the AR splicing pattern and facilitated the generation of the oncogenic isoform (AR-v7) by binding to the AAUAAA polyadenylation signal contained in AR-cryptic exon CE3. Functionally, SIAH1 acted as a tumor suppressor in PCa pathogenesis by repressing CPSF1-mediated AR-v7 generation. Finally, we demonstrated that m6A methylation was concerned with the repression of SIAH1 in PCa. Our results define SIAH1/CPSF1/AR-v7 axis as a regulatory factor of PCa progression, providing a promising target for treating PCa.
Keywords: MT: RNA/DNA Editing, SIAH1, CPSF1, androgen receptor, AR, AR-v7, prostate cancer
Graphical abstract
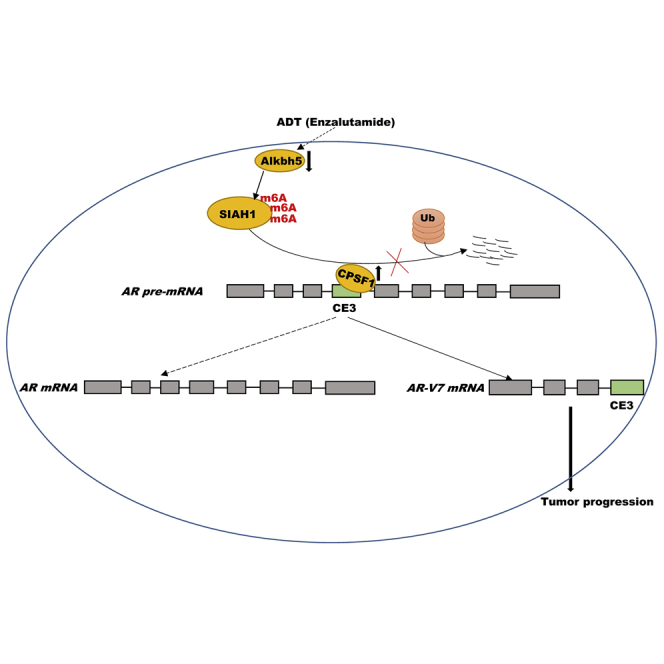
AR-v7, an AR splice variant, plays a crucial role in the progression of castration-resistant PCa. In this study, Sha et al., demonstrated that SIAH1 acted as a tumor suppressor in PCa pathogenesis by repressing CPSF1-mediated AR-v7 generation, and defined SIAH1/CPSF1/AR-v7 axis as a regulatory factor of PCa progression.
Introduction
Prostate cancer (PCa) is one of the most common types of cancer, and is the second most cause of cancer-related mortality in man worldwide.1,2 Although most localized PCa exhibit an indolent form with low tendency to progression, almost all advanced PCa will progress to metastatic phenotype and castration-resistant PCa (CRPC).3,4 Androgen deprivation therapy (ADT) is the mainstay of metastatic PCa treatment and shows a therapeutic benefit for many patients, but patients will inevitably progress to CRPC after ADT.5,6 Enzalutamide (Enza), a recently developed androgen receptor (AR) inhibitor, could effectively repress the CRPC and extend CRPC patient survival. However, Enza treatment causes several adverse effects, such as Enza-resistance and neuroendocrine differentiation of PCa.7,8
Post-transcriptional splicing of pre-mRNA (precursor mRNA) is a key step to enhancing the diversity of the transcriptome and proteome, and exerts a vital role in a wide range of psychopathological processes.9,10 Genome-wide analyses of the transcriptome in human cancer showed that aberrant profiles of pre-mRNA splicing, including exon skipping and intron retention, commonly exist in cancer.11,12 In PCa, the alternative splicing (AS) of AR produce constitutively active forms of AR that lack the ligand-binding domain.13 Among AR splice variants (AR-vs), AR-v7 is the most frequently expressed variant in metastatic CRPC, and AR-v7 overexpression is associated with poor prognosis in PCa.14,15 AR-v7 is produced by the alternative use of the 3′-splice site (ss) next to cryptic exon 3 (CE3) in intron 3 instead of canonical exon 4, leading to the inclusion of CE3, loss of exons 4–8, and generating C-terminally truncated AR.16,17 The molecular mechanisms governing AR-v7 generation are gradually being revealed. Fan et al. showed that histone demethylase JMJD1A facilitates AS of AR-v7 by recruiting HNRNPF to the cryptic exon 3.13 Histone demethylase KDM4B contributes to AR-v7 generation by binding to the splicing factor SF3B3.16 SF3B2 is a crucial determinant of RNA splicing and promotes the expression of AR-v7.9
Polyadenylation signal AAUAAA is one of the core consensus sequences that are correlated with polyadenylation and RNA end cleavage.18, 19, 20 The sequence containing AAUAAA could be identified by CPSF (cleavage and polyadenylation specificity factor), specifically by the CPSF1 subunit.18,21 Emerging studies have demonstrated that CPSF interacts with components of the spliceosome and is correlated with AS of pre-mRNA.22 Kyburz et al. showed that CPSF1 can regulate AS of IL-7 receptor exon 6.22 CPSF1 repression decreases AR-v7 protein expression and enhanced AR-FL protein expression.23 However, the mechanism by which CPSF1 promotes AR-v7 expression is unknown.
SIAH1, an E3 ubiquitin ligase, is involved in PCa and represents a high-value drug target.24 An increasing number of SIAH1-targeting proteins have been identified, such as PHDs, HIPK2, c-MYB, POU2AF1, and β-catenin.25,26 Most SIAH1 targets are correlated with fundamental cellular processes,26 indicating the important role of SIAH1 in intervening and preventing PCa progression. In this study, we demonstrate that m6A methylation was involved the repression of SIAH1 in PCa. SIAH1 directly interacted with CPSF1 and promoted ubiquitination and degradation of CPSF1, and CPSF1 could switch the AR splicing pattern and facilitate the generation of AR-v7.
Results
Identification of SIAH1 as a biomarker in PCa progression
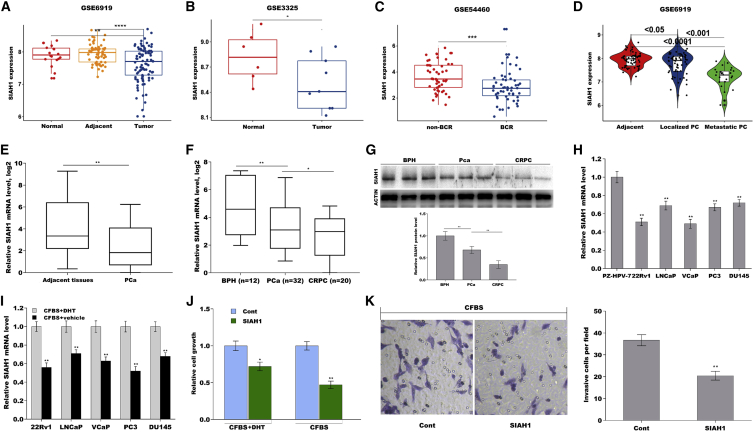
SIAH1, an E3 ubiquitin ligase, is currently being recognized as a critical factor in tumorigenesis and progression of several types of tumors, such as leukemia, breast cancer, and PCa.27,28 To investigate the biological role of SIAH1 in PCa, three sets of data were obtained from Gene Expression Omnibus (GSE6919, GSE3325, and GSE54460). As shown in Figures 1A and 1B, the expression level of SIAH1 was remarkably downregulated in PCa compared with normal tissues. SIAH1 level was also decreased in PCa patients with biochemical recurrence (BCR) after prostatectomy compared with patients without BCR (Figure 1C). SIAH1 level was then analyzed in normal, localized, and metastatic tumor tissues. Figure 1D shows that SIAH1 expression was significantly reduced in localized tumor tissues versus normal tissues (p < 0.05), and was further downregulated in metastatic tumor tissues versus localized tumor tissues (p < 0.001).
Figure 1.
Identification of SIAH1 as a biomarker in PCa progression
SIAH1 expression was analyzed in GSE6919 (A), GSE3325 (B), and GSE54460 datasets (C). (D) SIAH1 expression was analyzed in localized PCa, metastatic PCa tumor tissue, and adjacent normal tissues in GSE6919 datasets. (E) qPCR analysis of SIAH1 expression in PCa tissues and adjacent normal tissues. The 2−ΔΔCt method was used to quantify relative SIAH1 level, and the results were expressed as Log2 (2–ΔΔCt). (F) qPCR analysis of SIAH1 expression in BPH tissues, PCa tissues, and CRPC tissues. (G) Western blot analysis of SIAH1 expression in BPH tissues, PCa tissues, and CRPC tissues. (H) qPCR analysis of SIAH1 expression in PZ-HPV-7, 22Rv1, LNCaP, VCaP, PC3, and DU145 cells. (I) qPCR analysis of SIAH1 expression in PZ-HPV-7, 22Rv1, LNCaP, VCaP, PC3, and DU145 cells under androgen replete conditions or androgen deprivation. (J) Cell growth was detected in 22Rv1 cells with or without SIAH1 overexpression under androgen replete conditions or androgen deprivation using CCK-8 assay. (K) 22Rv1 cells invasion was assessed using transwell invasion assay after SIAH1 overexpression under androgen deprivation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To validate the dysregulated expression of SIAH1 in PCa, 52 matched pairs of normal and PCa tissues were collected. The results from qPCR analysis showed that SIAH1 was significantly decreased in PCa compared with normal tissues (Figure 1E). As expectedly, SIAH1 was reduced in localized PCa, and was further downregulated in CRPC versus localized PCa (Figures 1F and 1G). The association of SIAH1 with clinicopathological parameters is summarized in Table 1. SIAH1 level in PCa tissues was highly correlated with Gleason score (p = 0.026). In vitro, SIAH1 was reduced in PCa cells compared with normal prostate epithelial cells (Figure 1H). Interestingly, androgen deprivation resulted in a decreased expression of SIAH1 in these PCa cells (Figure 1I), indicating that SIAH1 might be involved in castration-resistant progression. Functionally, SIAH1 overexpression resulted in a modest repression on 22Rv1 cells growth under androgen-replete conditions, but SIAH1 caused a markedly repression on 22Rv1 cell growth under androgen deprivation (Figures S1A, S1B, and 1J). Forced expression of SIAH1 also significantly repressed 22Rv1 cell invasion under androgen deprivation (Figure 1K). These data suggest that SIAH1 is inversely correlated with aggressive phenotypes of PCa.
Table 1.
Relationships between SIAH1 expression and clinicopathological parameters in prostate cancer patients
| Variable | n = 52 | SIAH1 level |
p value | |
|---|---|---|---|---|
| Low | high | |||
| Age (years) | ||||
| <65 | 37 | 21 | 16 | 0.614 |
| ≥65 | 15 | 9 | 6 | |
| Prostate-specific antigen | ||||
| <10 | 34 | 22 | 12 | 0.551 |
| ≥ < | 18 | 10 | 8 | |
| Gleason score | ||||
| <7 (3 + 4) | 36 | 19 | 17 | 0.026a |
| ≥7 (4 + 3) | 16 | 11 | 5 | |
| Tumor stage | ||||
| pT2 | 31 | 20 | 11 | 0.472 |
| pT3-4 | 21 | 13 | 8 | |
| Lymph nodes status | ||||
| Negative | 33 | 19 | 14 | 0.098 |
| Positive | 19 | 12 | 7 | |
Statistically significant (χ2 test).
SIAH1 inhibition promoted a switch of AR protein from AR-FL to AR-v7
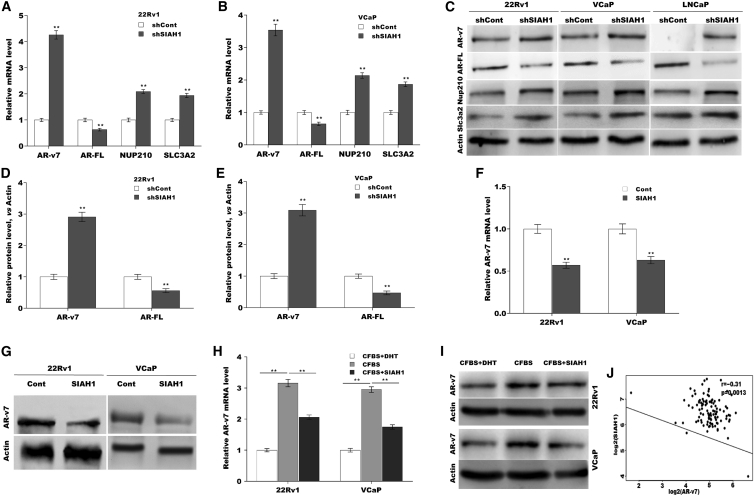
The data from GSE54460 showed that RNA processing is the most highly enriched biologic pathway.29 Given that AR-v7 AS is closely correlated with metastatic CRPC and poor prognosis in PCa, we thus investigated whether SIAH1 regulates AR-v7 generation. 22Rv1 and VCaP cells were used to test the correlation of SIAH1 with AR-v7 because AR-v7 mRNA and protein were constitutively expressed in the two cells.16 As shown in Figures 2A and 2B, SIAH1 knockdown resulted in a switch of the AR splicing from AR-FL to AR-v7 at the mRNA level (Figures S1C–S1F). Similar to the qPCR data, western blot analysis showed that SIAH1 knockdown enhanced AR-v7 protein level and simultaneously reduced AR-FL protein level in 22Rv1 and VCaP cells (Figures 2C–2E and S1G). Previous studies have demonstrated that NUP210 and SLC3A2 are specific AR-V7 target genes.30 To further verify whether SIAH1 knockdown facilitated AR-V7 generation, the expression level of NUP210 and SLC3A2 was assayed in 22Rv1 and VCaP cells after SIAH1 knockdown. Figures 2A–2C showed that SIAH1 knockdown facilitated AR-v7 generation and concurrently upregulated NUP210 and SLC3A2 expression at the mRNA and protein levels (Figure S1G). AR-v7 protein was undetectable in LNCaP cells, but SIAH1 knockdown resulted in the expression of AR-v7 protein (Figure 2C), indicating the critical role of SIAH1 in generating AR-v7 variant.
Figure 2.
SIAH1 inhibition promoted a switch of AR protein from AR-FL to AR-v7
qPCR analysis of AR-FL and AR-v7 mRNA level in 22Rv1 (A) and VCaP (B) cells after SIAN1 knockdown. (C–E) Western blot analysis for AR-FL, AR-v7, Nup210, and Slc3a2 protein in 22Rv1, VCaP, and LNCaP cells with or without SIAN1 knockdown. (F) qPCR analysis of AR-v7 mRNA level in 22Rv1 and VCaP cells with or without SIAN1 overexpression. (G) Western blot analysis for AR-v7 protein level in 22Rv1 and VCaP cells with or without SIAN1 overexpression. qPCR (H) and western blot analysis (I) of AR-v7 expression in 22Rv1 and VCaP cells in the presence or absence of SIAH1 overexpression under androgen replete or deprivation. (J) The negative association between SIAH1 level and AR-v7 level in GSE54460 datasets. ∗p < 0.05, ∗∗p < 0.01.
On the other hand, forced expression of SIAH1 significantly reduced the mRNA and protein level of AR-v7 in 22Rv1 and VCaP cells (Figures 2F and 2G). Furthermore, androgen deprivation resulted in a marked increase of AR-v7 expression, but the effect was blocked by SIAH1 overexpression (Figures 2H and 2I). SIAH1 expression was inversely correlated with AR-V7 expression in GSE54460 datasets (Figure 2J). As an E3 ubiquitin ligase, SIAH1 regulates target protein stability by promoting ubiquitin-dependent proteasome degradation. We next investigated the interaction between SIAH1 and AR-v7 in 2c2Rv1 cells via co-immunoprecipitation (co-IP) of SIAH1, and data showed that SIAH1 did not directly bind to AR-v7 (Figure S2A). A cycloheximide (CHX) chase assay showed that SIAH1 did not affect the stability of AR-v7 in SIAH1-overexpressing 22Rv1 cells (Figures S2B and S2C), indicating that SIAH1 regulates AR-v7 expression in an indirect way.
CPSF1 level was inversely correlated with SIAH1 level in PCa
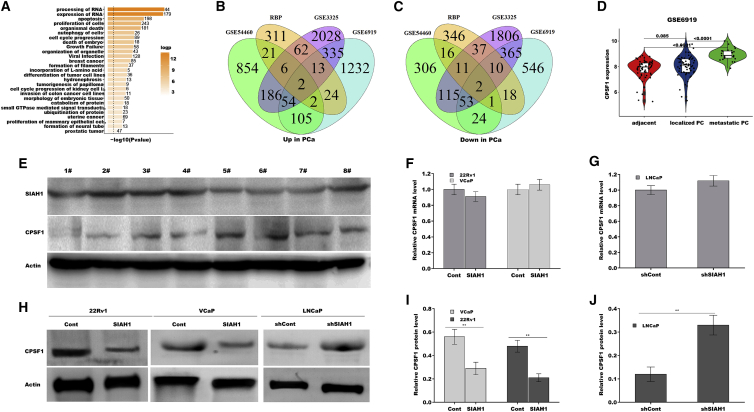
SIAH1 is downregulated in PCa patients with BCR (GSE54460, Figure 1C), and KEGG pathway analysis from GSE54460 datasets showed that processing of RNA is the most highly enriched biologic pathway (Figure 3A).29 Therefore, we speculated whether downregulated SIAH1 promotes AR-v7 generation by regulating splicing factors (or RNA binding protein [RBP]). Four hundred and forty-one genes encoding RBP and splicing factors were selected by gene ontology terms in AmiGO (Table S2).31 Venn diagram analysis showed that two splicing factors were concurrently upregulated in GSE54460, GSE3325, and GSE6919 datasets (Figure 3B), and two splicing factors were concurrently downregulated in these datasets (Figure 3C; Table S3). Of four aberrant splicing factors, cleavage and polyadenylation specificity factor 1 (CPSF1) level was positively correlated with PCa progression (Figure 3D). Especially, PCa tissues highly expressing SIAH1 exhibited a relatively lower CPSF1 protein level in eight randomly selected samples (Figure 3E). The functional correlation of SIAH1 with CPSF1 was then investigated in PCa cells. SIAH1 overexpression or knockdown did not affect the mRNA expression of CPSF1 in 22Rv1, VCaP, and LNCaP cells (Figures 3F and 3G), but SIAH1 overexpression reduced the protein level of CPSF1 in 22Rv1 and VCaP cells (Figures 3H and 3I). SIAH1 knockdown enhanced CPSF1 protein level in LNCaP cells (Figures 3H, 3J, and S2D).
Figure 3.
CPSF1 level was inversely correlated with SIAH1 level in PCa
(A) The most highly enriched biologic pathway was shown using KEGG pathway analysis in GSE54460 datasets. (B and C) Venn diagram analysis were used to predict abnormally expressed splicing factors in GSE54460, GSE3325, and GSE6919 datasets. (D) CPSF1 expression was analyzed in localized PCa, metastatic PCa tumor tissue, and adjacent normal tissues in GSE6919 datasets. (E) Western blot analysis of SIAH1 and CPSF1 expression in eight randomly selected samples. qPCR analysis of CPSF1 expression in 22Rv1 and VCaP cells after SIAH1 overexpression (F) and in LNCaP cells after SIAH1 knockdown (G). (H–J) Western blot and quantitative analysis for CPSF1 protein in 22Rv1 and VCaP cells after SIAN1 overexpression (H and I) and in LNCaP cells after SIAH1 knockdown (H and J). ∗p < 0.05, ∗∗p < 0.01.
SIAH1 promoted ubiquitination and proteasomal degradation of CPSF1
We next investigated whether SIAH1 reduces CPSF1 protein level by promoting its ubiquitination and proteasomal degradation. To this end, we first assayed the interaction between SIAH1 and CPSF1 through reciprocal co-IP, and data verified the direct interaction between SIAH1 and CPSF1 in 22Rv1 cells (Figures 4A and 4B) and in VCaP cells (Figures S2E and S2F) by co-IP assay. A CHX chase assay showed that SIAH1 overexpression significantly reduced the stability of CPSF1 protein (Figures 4C and 4D), whereas SIAH1 knockdown increased the stability of CPSF1 protein (Figures 4E, 4F, S2G, and S2H). The ubiquitination of CPSF1 was assayed via CPSF1 IP and subsequent western blot analysis for ubiquitin. As shown in Figure 4G, SIAH1 overexpression remarkably enhanced CPSF1 ubiquitination in 22Rv1 cells. Furthermore, MG132, a proteasome inhibitor, was used to treat SIAH1-overexpressing 22Rv1 cells and assessed whether SIAH1-mediated CPSF1 degradation could be rescued by MG132. As expected, the decreased expression of CPSF1 was prevented after MG132 treatment (Figure 4H). Proximity ligation assay (PLA) assay further verified the interaction of SIAH1-CPSF1 (Figure 4I). These data suggest that SIAH1 binds to CPSF1 and results in the decrease of CPSF1 protein level through promoting ubiquitination and proteasomal degradation.
Figure 4.
SIAH1 promoted ubiquitination and proteasomal degradation of CPSF1
(A and B) The interaction between SIAH1 and CPSF1 in 22Rv1 cells was determined by reciprocal co-IP assay. (C and D) The protein level of CPSF1 in 22Rv1 cells was assessed by CHX pulse-chase assay in the presence or absence of SIAH1 overexpression. CHX (50 μg/mL) was used for inhibiting the de novo protein synthesis and then relative protein density of CPSF1 was assessed at different time points (0, 2, 3, and 6 h). (E and F) CHX (50 μg/mL) was used for inhibiting the de novo protein synthesis and then relative protein density of CPSF1 was assessed at different time points (0, 3, and 6 h) in LNCaP cells. (G) The ubiquitination of CPSF1 was assessed by CPSF1 IP followed by western blot with ubiquitin antibody. (H) Western blot analysis for CPSF1 protein in 22Rv1 cells after SIAH1 overexpression in the presence or absence of MG132 (20 μM). (I) The interaction of SIAH1-CPSF1 was verified by PLA assay in 22Rv1 cells.
CPSF1 promoted AS of AR-v7
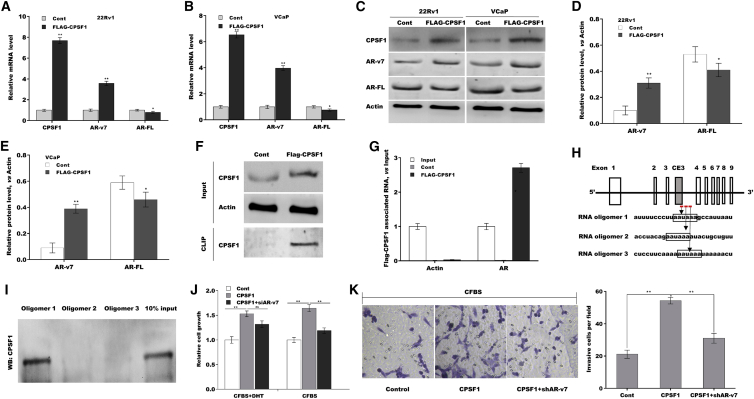
Then we investigated whether CPSF1 regulates AR-v7 generation. To this end, CPSF1 was overexpressed in 22Rv1 and VCaP cells, and then the mRNA and protein expression of AR-v7 was assessed. As shown in Figures 5A and 5B, CPSF1 overexpression resulted in a switch of the AR splicing from AR-FL to AR-v7 at the mRNA level. CPSF1 also increased AR-v7 protein level and simultaneously reduced AR-FL protein level in 22Rv1 and VCaP cells (Figures 5C–5E). An RNA-CLIP assay of RNA-protein complexes was carried out to assess the binding of CPSF1 to endogenous AR pre-mRNA. As shown in Figures 5F and 5G, when CPSF1 protein labeled with FLAG in 22Rv1 cells was immunoprecipitated with anti-FLAG antibody (Figure 5F) and then co-immunoprecipitated RNA was assessed using qPCR amplifying the region containing CE3, FLAG-CPSF1 protein directly bound to the RNA region (Figure 5G).
Figure 5.
CPSF1 promoted AS of AR-v7
qPCR analysis of CPSF1, AR-v7, and AR-FL expressions in 22Rv1 (A) and VCaP (B) cells with or without CPSF1 overexpression. (C–E) Western blot and quantitative analysis for AR-v7 and AR-FL protein levels in 22Rv1 and VCaP cells with or without CPSF1 overexpression. (F and G) RNA-CLIP assay was used to identify the binding of CPSF1 to AR. FLAG-tagged CPSF1 was overexpressed in 22Rv1 cells, and then IP was carried out using anti-FLAG antibody in cell lysates after UV-crosslinking. Cell lysates before (Input) and after IP (CLIP-product) were assessed using western blot with anti-CPSF1 antibody (F), and total RNA was acquired from input and CLIP-product and subject to qPCR analysis for AR (G). (H) Diagram of putative CPSF1-binding sites in CE3 and subsequent intron. (I) Biotin-labeled RNA oligomers were synthesized and RNA pull-down assay was performed using three RNA oligomers. (J) Under androgen deprivation or replete conditions, 22Rv1 cells growth was assessed after CPSF1 overexpression in the presence or absence of AR-v7 knockdown. (K) Under androgen deprivation, 22Rv1 cells invasion was assessed after CPSF1 overexpression in the presence or absence of AR-v7 knockdown. ∗p < 0.05, ∗∗p < 0.01.
Polyadenylation signal AAUAAA is a core consensus sequence that is associated with polyadenylation and RNA end cleavage. CPSF1 is involved in the regulation of AS of several genes by binding to the region containing AAUAAA.18 Here, five regions containing AAUAAA were identified in CE3 and subsequent intron, and RNA pull-down was performed using three biotin-tagged RNA oligomers derived from CE3 or subsequent introns (Figure 5H). Figure 5I shows that RNA oligomer 1 located in CE3 specifically pulled down CPSF1 protein from nuclear extracts of 22Rv1 cells, indicating that CPSF1 might bind to the AAUAAA polyadenylation signal located in CE3, and thus promote the cleavage downstream of AAUAAA. Functionally, CPSF1 overexpression promoted 22Rv1 cells growth under androgen-replete or deprivation conditions, but the effect was blocked by AR-v7 inhibition (Figures 5J, S3A, S3B, and S4A–S4C). Forced expression of CPSF1 also facilitated 22Rv1 cell invasion under androgen deprivation, whereas AR-v7 partially blocked the effect (Figure 5K). These data suggest that CPSF1 contributes to PCa progression by promoting AR-v7 generation.
SIAH1 inhibited PCa cell growth and invasion by regulating CPSF1-mediated AR-v7 generation
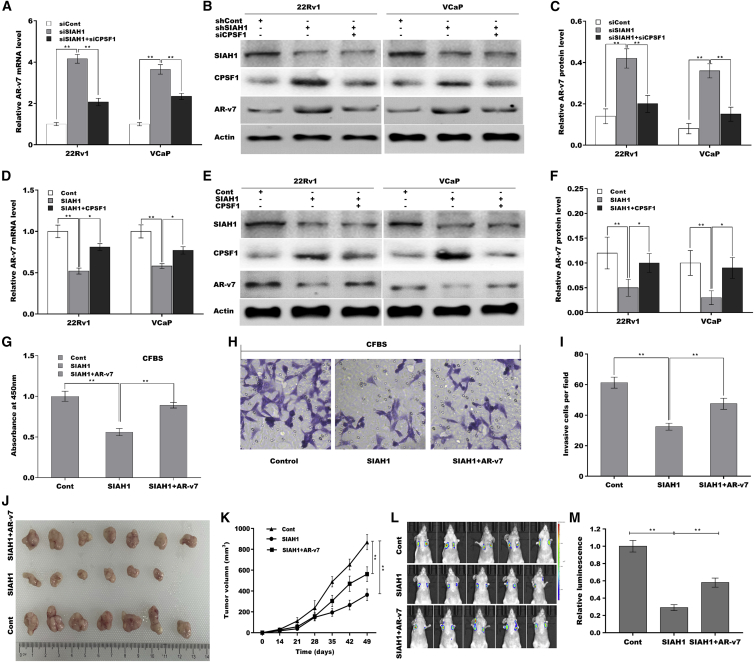
Given that SIAH1 represses CPSF1 expression, and that CPSF1 increases AR-v7 generation, we next investigated whether SIAH1 reduces AR-v7 splicing by regulating CPSF1. As shown in Figures 6A–6C, SIAH1 knockdown increased AR-v7 mRNA and protein expression of AR-v7 in 22Rv1 and VCaP cells, whereas CPSF1 silencing (Figures S3C and S3D) partially inhibited the effect. In addition, SIAH1 overexpression reduced AR-v7 mRNA (Figure 6D) and protein expression (Figures 6E and 6F), whereas CPSF1 overexpression weakened the effect.
Figure 6.
SIAH1 inhibited PCa growth and metastasis through regulating CPSF1-mediated AR-v7 generation
(A) qPCR analysis of AR-v7 expression in 22Rv1 and VCaP cells after SIAH1 knockdown in the presence or absence of CPSF1 knockdown. (B and C) Western blot and quantitative analysis for AR-v7 protein in 22Rv1 and VCaP cells after SIAH1 knockdown in the presence or absence of CPSF1 knockdown. (D) qPCR analysis of AR-v7 expression in 22Rv1 and VCaP cells after SIAH1 overexpression in the presence or absence of CPSF1 overexpression. (E and F) Western blot and quantitative analysis for AR-v7 expression in 22Rv1 and VCaP cells after SIAH1 overexpression in the presence or absence of CPSF1 overexpression. (G) Under androgen deprivation conditions, 22Rv1 cell growth was detected after SIAH1 overexpression in the presence or absence of AR-v7 overexpression. (H and I) Under androgen deprivation conditions, 22Rv1 cell invasion was assessed after SIAH1 overexpression in the presence or absence of AR-v7 overexpression. (J and K) 22Rv1 cells stably expressed SIAH1 and AR-v7 were subcutaneously injected into the flanks of BALB/c mice (n = 7) and tumor volume was calculated. (L and M) Cancer metastasis was assayed using a small living animal fluorescence imaging system after SIAH1 and AR-v7 overexpression (n = 5). ∗p < 0.05, ∗∗p < 0.01.
Functionally, SIAH1 overexpression inhibited 22Rv1 cell growth under androgen-deprivation conditions, but the effect was blocked by AR-v7 overexpression (Figures S3E, S3F, and 6G). Forced expression of SIAH1 facilitated 22Rv1 cell invasion under androgen deprivation, whereas AR-v7 also partially blocked the effect (Figures 6H and 6I). The effect of the SIAH1/AR-v7 axis on regulating tumor growth and metastasis was further investigated in vivo. Figures 6J and 6K showed that SIAH1 overexpression in 22Rv1 cells significantly repressed tumor growth in nude mice, whereas AR-v7 partially blocked the effect. The results from in vivo imaging showed that forced expression of SIAH1 repressed cancer metastasis, whereas AR-v7 overexpression also blocked the effect (Figures 6L and 6M), indicating that SIAH1 inhibits PCa cell growth and invasion in vitro and in vivo by repressing AR-v7 generation.
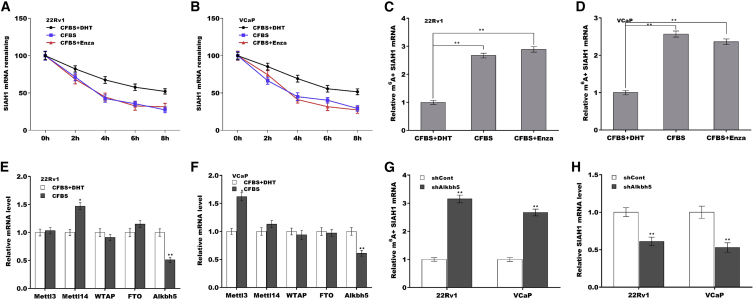
m6A modification was associated with the repression of SIAH1 in PCa cells
mRNA decay is an important step in regulating gene expression and is usually regulated by N6-methyladenosine (m6A) modification. Previous studies have demonstrated that SIAH1 mRNA contains internal m6A modifications.32 We thus investigated the correlation of SIAH1 expression with m6A modifications under androgen deprivation. As shown in Figures 7A and 7B, androgen deprivation remarkably reduced the mRNA stability of SIAH1 in 22Rv1 and VCaP cells after treatment with actinomycin D to terminate transcription. The results from gene-specific m6A qPCR assays showed that m6A levels in the SIAH1 mRNA were significantly increased under androgen deprivation (Figures 7C and 7D). The m6A modification is regulated by m6A methylase “writers” (Mettl3, Mettl14, and WTAP) and demethylases “erasers” (FTO and Alkbh5).33 As shown in Figures 7E and 7F, Alkbh5 level was concurrently decreased in 22Rv1 and VCaP cells under androgen deprivation (Figure S3G), indicating that Alkbh5 might regulate SIAH1 mRNA levels. As expected, Alkbh5 knockdown increased m6A levels in SIAH1 mRNA (Figure 7G), reduced SIAH1 mRNA (Figure 7H) and protein (Figures S3H and S3I) expression, and increased AR-v7 expression (Figures S3J and S3K) in 22Rv1 and VCaP cells. Taken together, these data demonstrate that Alkbh5-dependent m6A demethylation of SIAH1 mRNA promotes AR-v7 generation by increasing CPSF1 expression under androgen deprivation.
Figure 7.
m6A modification was associated with the repression of SIAH1 in PCa cells
(A and B) Under androgen deprivation or replete conditions, the mRNA stability of SIAH1 in 22Rv1 and VCaP cells was assessed at the indicated time points after treatment with actinomycin D. (C and D) Gene-specific m6A qPCR assays of m6A+ SIAH1 expressions in 22Rv1 and VCaP cells under androgen deprivation or replete conditions. (E and F) qPCR analysis of Mettl3, Mettl14, WTAP, FTO, and Alkbh5 expressions in 22Rv1 and VCaP cells under androgen deprivation or replete conditions. (G) Gene-specific m6A qPCR assays of m6A+ SIAH1 expression in 22Rv1 and VCaP cells after Alkbh5 knockdown. (H) qPCR assays of SIAH1 expression in 22Rv1 and VCaP cells after Alkbh5 knockdown. ∗p < 0.05, ∗∗p < 0.01.
Discussion
ADT is the mainstay of treatment to locally advanced or metastatic PCa, but patients will inevitably progress to CRPC after therapy.5,6,34 Enza could effectively repress the CRPC and extend overall patient survival. However, Enza treatment promotes Enza resistance and neuroendocrine differentiation of PCa.7,8 Therefore, it is of urgency to uncover underlying mechanisms involved in PCa progression. In this study, we demonstrate that (1) Alkbh5 downregulation increases m6A level in SIAH1 mRNA and results in subsequent SIAH repression under androgen deprivation, (2) SIAH1 repression promotes AR-v7 generation, (3) SIAH1 level is negatively correlated with CPSF1 level in PCa, (4) SIAH1 reduces CPSF1 expression by promoting its ubiquitination and proteasomal degradation, and (5) CPSF1 promotes a switch of AR protein from AR-FL to AR-v7. These data verified the important role of SIAH1/CPSF1/AR-v7 axis in PCa progression, providing a promising target for treating PCa.
SIAHs (SIAH1 and SIAH2) regulate cell survival and oncogenesis by promoting ubiquitin-dependent proteasomal degradation of target proteins.28 Nemani et al. demonstrated for the first time that human SIAHs are correlated with tumor progression.35 SIAHs are activated during intestinal epithelium apoptosis, and SIAH mRNA expression is increased in cancer cells selected for repression of tumorigenic phenotype. The biological role of SIAH2 in PCa has been widely verified. SIAH2 regulates prostate-specific antigen gene expression under low and physiologic concentrations of androgens and is thus correlated with PCa recurrence.36 SIAH2−/− mice show less progression to neuroendocrine PCa.37 It is well known that SIAH1 is a target for activation by p53 and plays an important role in p53-dependent cell-cycle arrest, functioning as a tumor suppressor.38,39 However, the role of SIAH1 in PCa progression remains unclear. In this study, we demonstrated that the expression level of SIAH1 is downregulated in PCa tissues versus normal tissues. The data from microarray-based analysis also showed that SIAH1 expression is decreased in PCa and is negatively correlated with aggressive phenotypes of PCa. Moreover, SIAH1 overexpression suppresses PCa cell growth and invasion under castration conditions.
SIAH1 expression is regulated by a lot of epigenetic mechanisms, such as DNA methylation, histone lysine methylation, and acetylation.28 Histone-lysine N-methyltransferase EHMT2 directly binds to SIAH1 promoter and represses its transcription in lung cancer cells.40 Consequently, EHMT2 inhibition increases SIAH1 expression and inhibits cancer cells growth. Trichostatin-A (a pan-HDAC inhibitor) treatment results in an increased of SIAH1 expression in human kidney-derived cells.41 SIAH1 mRNA contains internal m6A modifications, and m6A acts as a negative regulator of gene expression.32 We thus investigated whether aberrant m6A in SIAH1 mRNA is correlated with SIAH1 repression under androgen deprivation. The current data showed that m6A level in the SIAH1 mRNA is increased in 22Rv1 and VCaP cells under androgen deprivation, resulting in a subsequent repression of SIAH1. The m6A modification is regulated by m6A methylases (Mettl3, Mettl14, and WTAP) and demethylases (FTO and Alkbh5).33 We further revealed that m6A demethylase Alkbh5 is inhibited in 22Rv1 and VCaP cells under androgen deprivation. More important, Alkbh5 knockdown increases m6A levels in SIAH1 and results in a decreased SIAH1 mRNA level.
As an E3 ubiquitin ligase, SIAH1 directly interacts with CPSF1 and promotes ubiquitination and degradation of CPSF1. CPSF1 expression is negatively correlated with SIAH1 expression, but positively correlated with PCa progression. Functionally, forced expression of CPSF1 switches the AR splicing pattern and promotes the generation of the oncogenic isoform, AR-v7, by binding to the AAUAAA polyadenylation signal contained in the AR-cryptic exon CE3. In a previous study, Van Etten et al. demonstrated that the generation of AR variants AR-v1, AR-v7, and AR-v9 is facilitated by CPSF1.23 Given the negative regulation of SIAH1 in CPSF1 expression, here we also investigated whether SIAH1 decreases the generation AR-v9 as well as AR-v7. Indeed, SIAH1 knockdown results in significant increase of AR-v9 levels, whereas SIAH1 overexpression represses AR-v9 expression (Figures S5A and S5B). The mRNA and protein expression of AR-v1 could not be assessed due to lack of specific antibody and qPCR primer. In conclusion, these data demonstrate that Alkbh5-dependent m6A demethylation of SIAH1 mRNA promotes AR-v7 generation by increasing CPSF1 expression under androgen deprivation.
Materials and methods
Clinical sample collection
Prostate cancer (PCa) tissues and the adjacent non-tumor tissues were obtained from Renji hospital affiliated to Shanghai Jiaotong University School of Medicine. The clinicopathological parameters in prostate cancer patients were available in Table 1. The protocols used in the study were approved by the Hospital’s Protection of Human Subjects Committee and all patients provided written informed consent.
Cell culture
LNCaP, 22Rv1, PC3, DU145, and VCaP cells were obtained from ATCC (Rockville, MD, USA) and maintained in RPMI-1640 supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO, USA) in a humidified incubator containing 5% CO2 at 37°C. For hormone-deficient treatment, ADT, we used phenol red-free media supplemented with 10% charcoal dextran-treated serum.
Overexpression and RNA interference
The recombinant lentivirus encoding full-length human SIAH1 cDNA (Lv-SIAH1), CPSF1 cDNA (Lv-CPSF1), AR-v7 cDNA (Lv-AR-v7), and control (Lv-cont) were purchased from Genechem (Shanghai, China) to overexpress SIAH, CPSF1, and AR-v7, respectively. Retroviral infection was carried out as described previously.42
Oligomers for shSIAH1 or shAR-v7 were synthesized from Sangon Biotech (Shanghai, Chain) and cloned into a pGE1 vector. siRNAs targeting cellular mRNA (CPSF1 and Alkbh5) were designed and purchased from Genechem (Shanghai, China). siRNAs were transfected using Lipofectamine RNAiMAX reagents (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer protocols. The gene expression efficiency after treatment with siRNA, shRNA, or recombinant lentivirus was assessed using qPCR or western blot. Sequences of siRNA and shRNA were provided in Table S1.
Western blot analysis
22Rv1, VCaP, and LNCaP cells were lysed in RIPA Lysis and Extraction buffer (Thermo Fisher Scientific), and protein concentration was measured using the BCA protein assay kit (Pierce, Rockford, IL, USA). Fifty or 10 μg of protein was loaded onto 10% SDS-PAGE gels, and then transferred to PVDF membranes (Roche, Basel, Switzerland). After blocking in 5% skimmed milk at room temperature for 1 h, the membranes were incubated with primary antibodies against SIAH1 (1:1,000; Abcam, ab2237), CPSF1 (1:1,000; Abcam, ab81552), AR (1:2,000; Abcam, ab133273), AR-V7 (1:1,000; Abcam, ab198394), and β-actin (1:1,000; Abcam, ab8226) overnight at 4°C, followed by incubation with secondary antibody (goat anti-rabbit IgG H&L [HRP]; 1:1,000; Abcam, ab205718) for 1 h. Immunoblots were visualized using a chemiluminescence detection system (GE Healthcare, Chicago, IL, USA), and luminescent images were scanned into a computer and analyzed using ImageJ software (National Institute of Health, USA).
Extraction of total RNAs and quantitative real-time PCR
Total RNA was isolated from 22Rv1, VCaP, and LNCaP cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and first-strand cDNA was synthesized using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qPCR was carried out in triplicate using SYBR Green qPCR Master Mix (MedChemExpress) on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) based on the manufacturer’s recommendations. β-Actin was used as an internal reference and the gene expressions were quantified using the 2−ΔΔCT method. All primers were purchased from Sangon Biotech (Shanghai, China) and primer sequences for qPCR analysis are listed in Table S1.
Transwell invasion assay
The 24-well transwell chambers (BD Biosciences, San Jose, CA, USA) with 8-μm pores coated with Matrigel (BD Biosciences) were used to assess the invasiveness of 22Rv1 cells. The transfected 22Rv1 cells (1 × 104 cells/well) in serum-free DMEM/F12 medium were seeded into the upper chamber. The lower chamber was fulfilled with complete growth medium as a chemoattractant. After culturing for 36 h at 37°C, noninvasive cells inside the upper chamber were scraped off using cotton swabs, and invasive cells on the lower membrane surface were fixed and then stained with 0.1% crystal violet (Sigma-Aldrich) for 15 min. The invasive cells were photographed and counted under a microscope (Nikon, Tokyo, Japan).
PLA
The SIAH1-CPSF1 interaction was detected by PLA using anti-SIAH1 and anti-CPSF1 antibodies and PLA probes in 22Rv1 cells. The experiments were carried out as previously mentioned.43 In brief, 22Rv1 cells were grown on glass coverslips, fixed with methanol or 4% PFA, permeabilized with acetone or 0.2% PBS-Tween, and then incubated with SIAH1 and CPSF1 antibodies (Olink Biosciences, Uppsala, Sweden) at 4°C overnight. PLA minus and PLA plus probes were added and incubated for 1 h at 37°C. Further oligonucleotides were added, and allowed to hybridize to the PLA probes, and a ligase joined the two hybridized oligonucleotides. The fluorescence images were captured using an Olympus Fluoview FV10i Laser confocal microscope (Olympus, Tokyo, Japan). Images were collected sequentially on a confocal laser scanning microscope (Olympus UPLSAPO 60XO, NA 1.35) and analyzed using Olympus FV10-ASW version 3.0 Software.
CHX pulse-chase assay
Cells were treated with CHX to inhibit translation allowing the rates of CPSF1 protein degradation to be evaluated in the presence or absence of SIAH1 overexpression. In brief, 22Rv1 cells were transfected with Lv-SIAH1 and 24 h post transfection was treated with 100 μg/mL CHX (GlpBio, Montclair, CA, USA). Protein lysates were collected following 0, 3, or 6 h exposure to CHX. Western blot analysis was performed as described above.
RNA pull-down assay
Oligomer 1, 2, 3 were biotin labeled with the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Roche), and purified using an RNeasy Mini Kit (QIAGEN, Duesseldorf, Germany). RNA pull-down assay was performed as previously mentioned.44 After pull-down with streptavidin beads, the proteins were run in 10% SDS-PAGE and transferred to PVDF membranes (Millipore), and western blot analysis was done using anti-CPSF1 antibody as described above.
Co-IP
22Rv1 cells grown in 10-cm dishes at a confluency of 70%–80% were lysed with NP40 buffer. Then, 22Rv1 cell lysates were used for IP with CPSF1 (2 μg/mg; Abcam, ab81552) or SIAH1 (2 μg/mg; Abcam, ab2237) antibody overnight at 4°C. Protein A/G beads (Thermo Fisher Scientific) were added to the IP reactions and left rotating overnight at 4°C. After washing with cold PBS twice, the beads were pelleted by centrifugation at 13,000 rpm at 4°C for 5 min, re-suspended in 10 μL NUPAGE loading buffer, and centrifuged for 2 min at 13,000 rpm before loading onto an SDS gel for western blot analysis.
Cell growth assay
Cell growth was measured using a cell counting kit-8 (CCK-8) assay as per the manufacturer’s instructions. The 22Rv1 cells were seeded in 96-well plates (4 × 103 cells/well) after treatment with indicated reagents for 72 h under androgen replete conditions or androgen deprivation conditions, and then CCK-8 solution (10 μL, Dojindo, Japan) was added and the cells were incubated for another 2 h at 37°C. Absorbance was measured at a wavelength of 450 nm.
mRNA stability
22Rv1 and VCaP cells were treated with actinomycin D (5 μg/mL; Sigma) in the presence or absence of androgen deprivation or Enza (10 μM; Selleck Chemicals, Houston, TX, USA) and collected at 0, 2, 4, 6, or 8 h. Then, RNA was extracted with TRIzol Reagent for qPCR as described above. The results were normalized to GAPDH before the half-lives were calculated.
Crosslinking-immunoprecipitation
The Human CPSF1 coding sequence with an N-terminal FLAG was synthesized and cloned into pcDNA 3.1 to overexpress CPSF1. The cosslinking-immunoprecipitation (CLIP) assay performed in this study has been described previously.45 In brief, 2 × 106 22Rv1 cells, transfected with FLAG-CPSF1-pcDNA 3.1 or empty vector pcDNA 3.1, were crosslinked by UV at 480 mJ for 48 h and then harvested with 500 μL RIPA buffer (Roche). Then IP was carried out with anti-FLAG affinity gel (Sangon Biotech) and IP products were treated with Proteinase K for 0.5 h at 37°C. After IP of RNA-protein adducts, beads were washed and treated sequentially with calf intestinal alkaline phosphatase, and then polynucleotide kinase and γ-32P-ATP. RNA was extracted using a phenol-chloroform mixture and qPCR was carried out as described above.
Gene-specific m6A qPCR
The Magna MeRIP m6A Kit (Merck Millipore, Billerica, MA, USA) was used to examine m6A modifications on individual genes following the manufacturer’s instructions. In brief, total RNA was fragmented to about 100 nt in length and purified RNA fragments were incubated with m6A antibody-conjugated protein A/G magnetic beads in 1× IP buffer, which contained RNase inhibitors, at 4°C overnight. Methylated RNAs were eluted from beads with free m6A, and recovered using the RNeasy kit (QIAGEN). Fragmented RNA was analyzed by qPCR.
Xenograft mouse model
The BALB/c mice (6–8 weeks, SPF level, 20–22 g) were purchased from Charles River (Beijing, China) and maintained in specific, pathogen-free facilities. The animal experiments involved in this study were approved by the Experimental Animal Committee of Renji hospital affiliated to Shanghai Jiaotong University School of Medicine and we confirm that all experiments conformed to all relevant regulatory standards. 22Rv1 cells stably expressing SIAH1 and AR-v7 were constructed. 22Rv1 cells (2.5 × 106 cells/100 μL PBS) were subcutaneously injected into the flanks of mice (n = 7). The development of s.c. tumors was monitored using a digital Vernier caliper at the specified times. The volume of the tumor was calculated from the formula length × width2 × 0.5, approximating the volume of the elliptical entity.
In vivo metastasis assay
BALB/c nude mice (male, 6–8 weeks, SPF level, 20–22 g), purchased from Charles River, were applied for in vivo metastasis assay and the construction of tumor metastatic model was approved by the Experimental Animal Committee of Renji hospital affiliated to Shanghai Jiaotong University School of Medicine. 22Rv1 cells stably expressing SIAH1 and AR-v7 (2.5 × 106/200 μL PBS) were tail vein injected into the mice (n = 5). In vivo imaging of lung metastasis at 10 weeks after tail vein inoculation was observed using a small living animal fluorescence imaging system (INDEC BioSystems, USA).
Statistical analysis
Data are presented as mean ± standard deviation from three independent experiments and analyzed using SPSS 19.0 software (IBM, NY, USA). The difference between two groups was compared using two-tailed Student’s t test or one-way analysis of variance, followed by the Scheffé test. p value < 0.05 was considered statistically significant.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81772742, 81702840, 81702542, and 81901747), the Science and Technology Commission of Shanghai Municipality (19ZR143100 and 19411967400), the Shanghai Shenkang Hospital Development Center (SHDC12015125 and 16CR3049A), the Shanghai Municipal Commission of Health and Family Planning (201640247), and the Shanghai Jiao Tong University (YG2017MS47 and YG2017MS52).
Author contributions
J.S. and Weiliang X. participated in the conception and design of the study. L.X., Q.H., and X.D. performed the analysis and interpretation of the data. Y.Z., J.P., B.D., and W.X. contributed to drafting the article. All authors read and approved the final submitted manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.03.008.
Contributor Information
Wei Xue, Email: uroxuewei@163.com.
Jianjun Sha, Email: shajianjunrj@126.com.
Supplemental information
References
- 1.Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M., Jemal A., Kramer J.L., Siegel R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M., Gavin A., Visser O., Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Al-Salama Z.T. Apalutamide: a review in non-metastatic castration-resistant prostate cancer. Drugs. 2019;79:1591–1598. doi: 10.1007/s40265-019-01194-x. [DOI] [PubMed] [Google Scholar]
- 6.Scher H.I., Fizazi K., Saad F., Taplin M.E., Sternberg C.N., Miller K., Wit R., Mulders P., Chi K.N., Shore N.D., et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.Lin T.H., Izumi K., Lee S.O., Lin W.J., Yeh S., Chang C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis. 2013;4:e764. doi: 10.1038/cddis.2013.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucci M., Zichi C., Buttigliero C., Vignani F., Scagliotti G.V., Di Maio M. Enzalutamide-resistant castration-resistant prostate cancer: challenges and solutions. OncoTargets Ther. 2018;11:7353–7368. doi: 10.2147/OTT.S153764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamura N., Nimura K., Saga K., Ishibashi A., Kitamura K., Nagano H., Yoshikawa Y., Ishida K., Nonomura N., Arisawa M., et al. SF3B2-Mediated RNA splicing drives human prostate cancer progression. Cancer Res. 2019;79:5204–5217. doi: 10.1158/0008-5472.CAN-18-3965. [DOI] [PubMed] [Google Scholar]
- 10.Sharp A., Coleman I., Yuan W., Sprenger C., Dolling D., Rodrigues D.N., Russo J.W., Figueiredo I., Bertan C., Seed G., et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Invest. 2019;129:192–208. doi: 10.1172/JCI122819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Manley J.L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan L., Zhang F., Xu S., Cui X., Hussain A., Fazli L., Gleave M., Dong X., Qi J. Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4584–E4593. doi: 10.1073/pnas.1802415115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C., Chen Y., Mohammad T.A., Chen Y., Fedor H.L., et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scher H.I., Lu D., Schreiber N.A., Louw J., Graf R.P., Vargas H.A., Johnson A., Jendrisak A., Bambury R., Danila D., et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–1449. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan L., Chen Z., Lu J., Liang Y., Wang M., Roggero C.M., Zhang Q.J., Gao J., Fang Y., Cao J., et al. Histone lysine demethylase KDM4B regulates the alternative splicing of the androgen receptor in response to androgen deprivation. Nucleic Acids Res. 2019;47:11623–11636. doi: 10.1093/nar/gkz1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L.L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evsyukova I., Bradrick S.S., Gregory S.G., Garcia-Blanco M.A. Cleavage and polyadenylation specificity factor 1 (CPSF1) regulates alternative splicing of interleukin 7 receptor (IL7R) exon 6. RNA. 2013;19:103–115. doi: 10.1261/rna.035410.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutz C.S., Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:23–31. doi: 10.1002/wrna.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller W., Bienroth S., Lang K.M., Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3' processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyburz A., Friedlein A., Langen H., Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3' end processing and splicing. Mol. cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 23.Van Etten J.L., Nyquist M., Li Y., Yang R., Ho Y., Johnson R., Ondigi O., Voytas D.F., Henzler C., Dehm S.M. Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Cancer Res. 2017;77:5228–5235. doi: 10.1158/0008-5472.CAN-17-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y., Sessions E.H., Zhang F., Ban F., Placencio-Hickok V., Ma C.T., Zeng F.Y., Pass I., Terry D.B., Cadwell G., et al. Identification and characterization of small molecule inhibitors of the ubiquitin ligases Siah1/2 in melanoma and prostate cancer cells. Cancer Lett. 2019;449:145–162. doi: 10.1016/j.canlet.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Q., Hou J., Feng L., Lv A., Ke X., Liang H., Wang F., Zhang K., Chen K., Cui H. PHF19 promotes the proliferation, migration, and chemosensitivity of glioblastoma to doxorubicin through modulation of the SIAH1/beta-catenin axis. Cell Death Dis. 2018;9:1049. doi: 10.1038/s41419-018-1082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko H.R., Jin E.J., Lee S.B., Kim C.K., Yun T., Cho S.W., Park K.W., Ahn J.Y. SIAH1 ubiquitin ligase mediates ubiquitination and degradation of Akt3 in neural development. J. Biol. Chem. 2019;294:15435–15445. doi: 10.1074/jbc.RA119.009618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer O.H., Stauber R.H., Bug G., Hartkamp J., Knauer S.K. SIAH proteins: critical roles in leukemogenesis. Leukemia. 2013;27:792–802. doi: 10.1038/leu.2012.284. [DOI] [PubMed] [Google Scholar]
- 28.Knauer S.K., Mahendrarajah N., Roos W.P., Kramer O.H. The inducible E3 ubiquitin ligases SIAH1 and SIAH2 perform critical roles in breast and prostate cancers. Cytokine Growth Factor Rev. 2015;26:405–413. doi: 10.1016/j.cytogfr.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Long Q., Xu J., Osunkoya A.O., Sannigrahi S., Johnson B.A., Zhou W., Gillespie T., Park J.Y., Nam R.K., Sugar L., et al. Global transcriptome analysis of formalin-fixed prostate cancer specimens identifies biomarkers of disease recurrence. Cancer Res. 2014;74:3228–3237. doi: 10.1158/0008-5472.CAN-13-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugiura M., Sato H., Okabe A., Fukuyo M., Mano Y., Shinohara K.I., Rahmutulla B., Higuchi K., Maimaiti M., Kanesaka M., et al. Identification of AR-V7 downstream genes commonly targeted by AR/AR-V7 and specifically targeted by AR-V7 in castration resistant prostate cancer. Transl. Oncol. 2021;14:100915. doi: 10.1016/j.tranon.2020.100915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 34.Chen D.Y., See L.C., Liu J.R., Chuang C.K., Pang S.T., Hsieh I.C., Wen M.S., Chen T.H., Lin Y.C., Liaw C.C., et al. Risk of cardiovascular ischemic events after surgical castration and gonadotropin-releasing hormone agonist therapy for prostate cancer: a nationwide cohort study. J. Clin. Oncol. 2017;35:3697–3705. doi: 10.1200/JCO.2016.71.4204. [DOI] [PubMed] [Google Scholar]
- 35.Nemani M., Linares-Cruz G., Bruzzoni-Giovanelli H., Roperch J.P., Tuynder M., Bougueleret L., Cherif D., Medhioub M., Pasturaud P., Alvaro V., et al. Activation of the human homologue of the Drosophila sina gene in apoptosis and tumor suppression. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi J., Tripathi M., Mishra R., Sahgal N., Fazli L., Ettinger S., Placzek W.J., Claps G., Chung L.W., Bowtell D., et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer cell. 2013;23:332–346. doi: 10.1016/j.ccr.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi J., Pellecchia M., Ronai Z.A. The Siah2-HIF-FoxA2 axis in prostate cancer - new markers and therapeutic opportunities. Oncotarget. 2010;1:379–385. doi: 10.18632/oncotarget.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuzawa S., Takayama S., Froesch B.A., Zapata J.M., Reed J.C. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roperch J.P., Lethrone F., Prieur S., Piouffre L., Israeli D., Tuynder M., Nemani M., Pasturaud P., Gendron M.C., Dausset J., et al. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21(Waf1) Proc. Natl. Acad. Sci. U. S. A. 1999;96:8070–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho H.S., Kelly J.D., Hayami S., Toyokawa G., Takawa M., Yoshimatsu M., Tsunoda T., Field H.I., Neal D.E., Ponder B.A., et al. Enhanced expression of EHMT2 is involved in the proliferation of cancer cells through negative regulation of SIAH1. Neoplasia. 2011;13:676–684. doi: 10.1593/neo.11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer O.H., Muller S., Buchwald M., Reichardt S., Heinzel T. Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. FASEB J. 2008;22:1369–1379. doi: 10.1096/fj.06-8050com. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Xu H., Jiao H., Wang S., Xiao Z., Zhao Y., Bi J., Wei W., Liu S., Qiu J., et al. STX2 promotes colorectal cancer metastasis through a positive feedback loop that activates the NF-kappaB pathway. Cell Death Dis. 2018;9:664. doi: 10.1038/s41419-018-0675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu D.S., Wang H.J., Tai S.K., Chou C.H., Hsieh C.H., Chiu P.H., Chen N.J., Yang M.H. Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell. 2014;26:534–548. doi: 10.1016/j.ccell.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Casas-Vila N., Sayols S., Perez-Martinez L., Scheibe M., Butter F. The RNA fold interactome of evolutionary conserved RNA structures in S. cerevisiae. Nat. Commun. 2020;11:2789. doi: 10.1038/s41467-020-16555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutluay S.B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., Bieniasz P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.