Abstract
We recently discovered that human neutrophils express immunomodulatory glycoproteins carrying unusual and highly truncated paucimannosidic N-glycans (Man1-3GlcNAc2Fuc0–1), but their biosynthesis remains elusive. Guided by the well-characterized truncation pathway in invertebrates and plants in which the N-acetyl-β-D-hexosaminidase (Hex) isoenzymes catalyze paucimannosidic protein (PMP) formation, we here set out to test if the homologous human Hex α and β subunits encoded by HEXA and HEXB drive a similar truncation pathway in human neutrophils. To this end, we performed quantitative glycomics and glycoproteomics of several CRISPR-Cas9-edited Hex-disrupted neutrophil-like HL-60 mutants (HEXA-KO and HEXB-KO) and matching unedited cell lines. Hex disruption was validated using next-generation sequencing, enzyme-linked immunosorbent assay (ELISA), quantitative proteomics and Hex activity assays. Excitingly, all Hex-disrupted mutants displayed significantly reduced levels of paucimannosylation, particularly Man2-3GlcNAc2Fuc1, relative to unedited HL-60 suggesting that both HEXA and HEXB contribute to PMP formation via a hitherto unexplored truncation pathway in neutrophils. Quantitative N-glycomics indeed demonstrated reduced utilization of a putative noncanonical truncation pathway in favor of the canonical elongation pathway in all Hex-disrupted mutants relative to unedited controls. Quantitative glycoproteomics recapitulated the truncation-to-elongation switch in all Hex-disrupted mutants and showed a greater switch for N-glycoproteins cotrafficking with Hex to the azurophilic granules of neutrophils such as myeloperoxidase. Finally, we supported the Hex-PMP relationship by documenting that primary neutrophils isolated from an early-onset Sandhoff disease patient (HEXB−/−) displayed dramatically reduced paucimannosylation relative to neutrophils from an age-matched unaffected donor. We conclude that both human Hex α and β mediate PMP formation via a putative noncanonical truncation pathway in neutrophils.
Keywords: truncation pathway, paucimannose, N-glycosylation, neutrophil, N-acetyl-β-D-hexosaminidase
Introduction
Neutrophils, an abundant class of granulocytes in the blood circulation, are essential to our innate immune system (Rosales 2018; Liew and Kubes 2019). As first-line responders to pathogenic infections, neutrophils are armed with a plethora of antimicrobial glycoproteins residing in discrete and highly mobile cytosolic granules that may be released via degranulation upon neutrophil activation (Cowland and Borregaard 2016; Ugonotti et al. 2020). Particularly, the azurophilic granules host many potent glycoproteins involved in the neutrophil-mediated innate immune response including myeloperoxidase (MPO), human neutrophil elastase (HNE) and neutrophil cathepsin G (nCG).
We recently discovered that human neutrophils abundantly express paucimannosidic glycans (PMGs, Man1-3GlcNAc2Fuc0–1) (Thaysen-Andersen et al. 2015). PMGs form a peculiar and still poorly understood class of N-glycans in human glycobiology (Loke et al. 2016; Tjondro et al. 2019). We have demonstrated that PMGs are key glyco-features enriched in but not exclusive to the azurophilic granules (Venkatakrishnan et al. 2020), and have shown that they play important biological roles (e.g. modulate enzyme activity, alter protein–protein interactions and facilitate interactions with glycan-binding immunoreceptors) on specific proteins residing in this compartment including MPO, HNE and nCG (Loke et al. 2015; Loke et al. 2017; Tjondro et al. 2021). However, the biosynthesis of paucimannosidic proteins (PMPs) in human neutrophils remains unknown leaving a critical knowledge gap that prevents us from understanding the spatiotemporal expression, regulation and significance of this still understudied class of truncated N-glycoproteins in human glycobiology.
Taking cues from the PMP-rich lower organisms including invertebrates (e.g. flies and worms) and plants, in which the N-acetyl-β-D-hexosaminidase (Hex) isoenzymes are known to catalyze PMP formation via the hydrolysis of N-acetylglucosamine (GlcNAc)-capped glycoprotein substrates in well-described truncation pathways (Zhang et al. 2003; Leonard et al. 2006; Gutternigg et al. 2007; Schachter 2009; Paschinger and Wilson 2019), we have previously proposed that human PMPs may similarly be generated by the action of human Hex isoenzymes (Tjondro et al. 2019). However, the biosynthetic route of PMPs remains, to the best of our knowledge, experimentally unsupported in human neutrophils and other cell types.
The human Hex isoenzymes, members of the glycoside hydrolase family 20 (GH20), are formed from two catalytically active gene products, the α and β subunits encoded by HEXA and HEXB, respectively (Korneluk et al. 1986). Three dimeric Hex isoenzymes are generated from these two related subunits: the heterodimeric Hex A (αβ) and the homodimeric Hex B (ββ) and Hex S (αα) isoenzymes, Figure 1A. Albeit not a focus of this study, the nucleocytosolic Hex D (Q8WVB3) encoded by HEXDC was recently identified as another member of the GH20 family (Gutternigg et al. 2009). Catalytically, human Hex subunit α and β have been reported to remove both terminal β-linked N-acetylgalactosamine (GalNAc) and GlcNAc residues from various glycoconjugates such as glycolipids, glycoproteins and glycosaminoglycans (Lemieux et al. 2006; Liu et al. 2018). Although the β subunit preferentially hydrolyses N-acetylhexosamine residues (GalNAc/GlcNAc) on neutral substrates, the α subunit acts on both neutral and negatively charged substrates including sialic acid-containing GM2 gangliosides (Kytzia and Sandhoff 1985; Wendeler et al. 2006). Hex A and B are reportedly the major functional Hex isoenzymes in human whereas Hex S is considered a minor unstable form with limited catalytic activity (Hepbildikler et al. 2002).
Fig. 1.
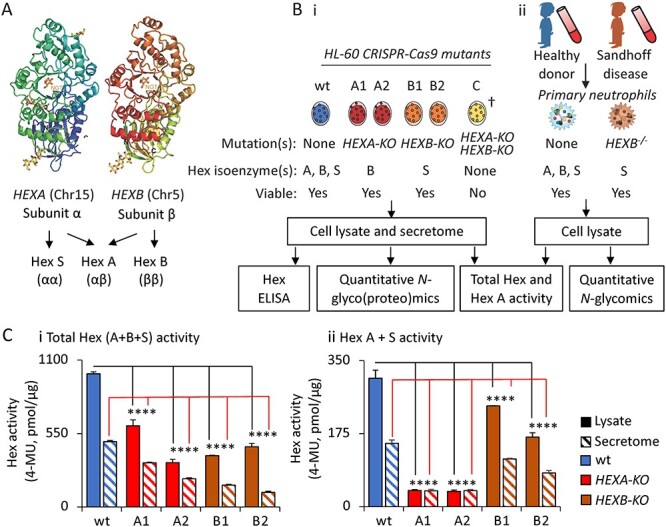
Experimental design and functional validation of Hex-disrupted HL-60 mutants. (A) overview of human Hex genes, subunits and isoenzymes relevant to this study. (B) Experimental workflow for the (i) HL-60 CRISPR-Cas9 mutants and the (ii) Sandhoff disease and healthy donor neutrophils. †Repeat attempts to generate HEXA/HEXB double knockouts were unsuccessful as these mutants displayed severe growth retardation. (C) (i–ii), Determination of the total Hex and Hex A + S enzyme activity in the HL-60 cell lines using MUG(S) assays. Data are plotted as mean + SD, n = 3 technical replicates, ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001 (HL-60 mutant vs wt).
The Hex α and β subunits are recognized to be important in the lysosomal degradation of gangliosides such as GM2 (Breiden and Sandhoff 2019). Unsurprisingly, deleterious mutations in HEXA and HEXB therefore lead to severe neurodegenerative lysosomal storage diseases including Tay-Sachs disease and Sandhoff disease. The Hex deficiency causes an accumulation of glycoconjugates, particularly GM2 in the lysosomes of the ganglioside-rich neurons, leading to neurodegeneration, impaired development and premature death in individuals suffering from Tay-Sachs disease and Sandhoff disease (Breiden and Sandhoff 2019). Although the Hex isoenzymes have been well-studied in the context of GM2 gangliosidosis (Leal et al. 2020), their role in PMP generation in human neutrophils has, so far, only been implicated (Tjondro et al. 2019).
Utilizing a set of Hex-disrupted CRISPR-Cas9 HL-60 mutants and primary neutrophils isolated from Sandhoff disease and healthy donors and powered by advances in quantitative glycomics and glycoproteomics (Thaysen-Andersen et al. 2021), we document herein that both Hex α and β mediate the formation of PMPs and suggest that these trimming processes are part of a putative noncanonical truncation pathway in human neutrophils.
Results and discussion
Homology between human Hex and Hex from PMP-rich organisms
We firstly tested for homology between human Hex and Hex produced by well-studied PMP-rich model organisms including Caenorhabditis elegans, Arabidopsis thaliana and Drosophila melanogaster. The human Hex α (UniProtKB, P06865) and β (P07686) subunits showed homology to several nonhuman Hex known to mediate PMP formation including A. thaliana HEXO3 (Q8L7S6, ~49% sequence similarity) and D. melanogaster Hex fdl (Q8WSF3, ~40% sequence similarity) (Leonard et al. 2006; Gutternigg et al. 2007), Supplementary Figure S1A and B. The nucleocytosolic human Hex D (Gutternigg et al. 2009; Alteen et al. 2016) neither showed homology to Hex α and β (~17% sequence similarity), nor clustered with most other PMP-producing nonhuman Hex. This approach however does not account for conserved domains that may localize to different polypeptide regions, a feature shown by some GH20 enzymes. Reportedly, Hex D preferentially cleaves GalNAc- over GlcNAc-terminating substrates (Gutternigg et al. 2009, Alteen et al. 2016). Thus, Hex D was considered irrelevant in the context of PMP formation and not explored further. Similarly, human O-GlcNAcase (Hex C, O60502), another nucleocytosolic β-hexosaminidase belonging to the GH84 family (Hart et al. 2007), was also considered irrelevant for further exploration due to its subcellular localization that presumably renders this enzyme unable to act on luminal substrates in the glycosylation pathways.
Both the human Hex α and β subunits, but not Hex D, were found to feature a key tryptophan residue (W329, numbering based on human Hex α) conserved across the PMP-producing nonhuman Hex variants, Supplementary Figure S1C. This strategically located tryptophan residue was found to be important for efficient docking of the N-glycan core of N-glycoprotein substrates into the active site for hydrolysis of terminal nonreducing end GlcNAc residues, as demonstrated for the Streptococcus pneumoniae Hex homolog StrH (P49610) (Pluvinage et al. 2011). Although more refined structural biology is required to elucidate how N-glycoprotein substrates interact with human Hex, our sequence analysis supports the involvement of the human Hex α and β subunits in PMP formation.
Experimental design to test the involvement of human hex α and β subunits in PMP formation
Based on the initial sequence analysis, we decided to generate a set of CRISPR-Cas9-edited HL-60 knockout mutants (HEXA-KO and HEXB-KO), Figure 1B. Two independent clones were generated for each of the knockout variants herein termed A1 and A2 for the HEXA-KO and B1 and B2 for the HEXB-KO. HL-60 is an immortalized promyelocytic cell line widely recognized as a suitable in vitro model system in neutrophil research (Babatunde et al. 2021). Further, we have previously demonstrated, via comprehensive profiling of the expression and activity of a collection of glycosyltransferases, similarity between the glycosylation machinery of HL-60 and primary neutrophils (Marathe et al. 2008), supporting the use of HL-60 in this study.
The mutants and relevant unedited wildtype (wt) controls were carefully validated using next-generation sequencing (NGS) and Hex-centric enzyme-linked immunosorbent assay (ELISA), quantitative proteomics and activity assays, and were then glycoprofiled using state-of-the-art quantitative glycomics and glycoproteomics. Finally, in vivo observations were made by profiling primary neutrophils from a Sandhoff disease patient (HEXB−/−) and an age-paired healthy donor using quantitative glycomics.
Validation of Hex disruption in the HL-60 mutants
Both the Hex genotype and phenotype of the CRISPR-Cas9 HL-60 mutants were investigated relative to unedited wt HL-60 cells to validate Hex disruption. NGS of the polymerase-chain reaction (PCR) amplified region around the CRISPR-Cas9 target site confirmed various insertion–deletions (indels) in the target Hex genes in all HL-60 mutants, Supplementary Figure S2A. The first and second exon of HEXA and HEXB were targeted, respectively. A one-base insertion occurred in the exon of allele 1 of A1 and in A2. The second allele of A1 was found to have seven mutated nucleotides. A four-nucleotide insertion and four-nucleotide mutation occurred in B1 and B2. The predicted protein sequences of the Hex mutants showed that the various indels caused a termination of translation in all mutants except allele 2 of A1, Supplementary Figure S2B. The Hex-disrupted HL-60 mutants showed similar or only mildly perturbed growth characteristics relative to unedited wt HL-60 indicating that the generated Hex-disrupted cells remained viable under the assayed conditions, Supplementary Figure S3. Interestingly, despite several attempts, we were unable to generate a HEXA/HEXB double knockout line due to insufficient cellular growth of this variant, suggesting that a single functional Hex gene may be essential to maintain viability of HL-60 under the tested conditions.
ELISA and tandem mass tag (TMT) label-assisted quantitative proteomics were used to profile the protein expression level of the Hex α and β subunits in both the lysate and secretome of the HL-60 mutants, Supplementary Figure S4i–iv and Supplementary Table I. Both the ELISA and the proteomics data showed that Hex α and β were significantly reduced albeit still present in the lysate and secretome of all Hex-disrupted mutants compared to wt HL-60. Although all mutants featured indels (see above), it is possible that the indels did not terminate protein translation possibly explaining the residual Hex protein in the Hex-disrupted mutants. Hex α and β were found in the low expression range in the proteomics data across all samples. Hex α was identified via four peptides in the secretome whereas Hex β was only identified by a single peptide in the lysate fraction. The low Hex sequence coverage and the inconsistent identification of the Hex subunits across the two cellular fractions is likely a result of the high complexity of the HL-60 protein mixture and the abundant structural and metabolic proteins masking less abundant proteins such as the Hex subunits. For that reason, we were unable to confirm by proteomics the exact mutations of the Hex α and β polypeptides still expressed by the Hex-disrupted mutants. Sensitive proteomics performed on enriched or isolated Hex α and β is therefore required to establish accurate levels of Hex α and β and unpick how the indels in HEXA and HEXB translate into mutant Hex protein. Contrary to Hex α and β, the nucleocytoplasmic O-GlcNAcase and Hex D were absent or present below the level of detection in the HL-60 proteome.
Importantly, and in line with the Hex-centric ELISA and proteomics data, we demonstrated significant reduction of the Hex enzyme activity in both the secretome and lysate of all Hex-disrupted mutants compared to wt HL-60 utilizing well-established fluorometric Hex activity assays, Figure 1Ci–ii. Taking together, despite exhibiting some residual Hex activity, the generated HL-60 Hex α and β mutants were therefore found to be a suitable model system to probe the relationship between the Hex isoenzymes and PMP generation in human neutrophils.
Disruption of either Hex α or β impedes PMP generation in HL-60
Utilizing our established porous graphitized carbon liquid chromatography tandem mass spectrometry (PGC-LC–MS/MS)-based glycomics platform (Jensen et al. 2012; Hinneburg et al. 2019), we then quantitatively profiled the N-glycome of both the lysate and secretome of the Hex-disrupted mutants and wt HL-60, Figure 2A, Supplementary Table II and Supplementary Data. The oligomannosidic M6 (17.7%) and paucimannosidic M2F (16.0%) N-glycans were found to be very abundant structures in wt HL-60 in congruence with the N-glycan structures prominently expressed by human neutrophils and in accordance with N-glycans known to be carried by proteins residing in the azurophilic granules of neutrophils (Reiding et al. 2019; Venkatakrishnan et al. 2020; Tjondro et al. 2021). Importantly, the total paucimannosylation levels were significantly reduced in the lysates of all Hex α and β mutants (17.1%–22.2%) compared to wt HL-60 (29.8% ± 1.7%, P < 0.01). Although the secretome, as expected, showed less paucimannosylation, the secreted proteins generally displayed a similar reduction in the PMG levels upon Hex disruption as observed for the lysate. The level of reduction varied between mutants in line with the variable Hex disruption efficiency across clones as noted above (see Figure 1C). Taken together, this illustrates that PMPs are not only produced and stored in cytosolic granules, but may also be secreted from HL-60 cells in line with the presence of Hex in the HL-60 secretome (see above) and our previous observations on HL-60 (Thaysen-Andersen et al. 2015).
Fig. 2.
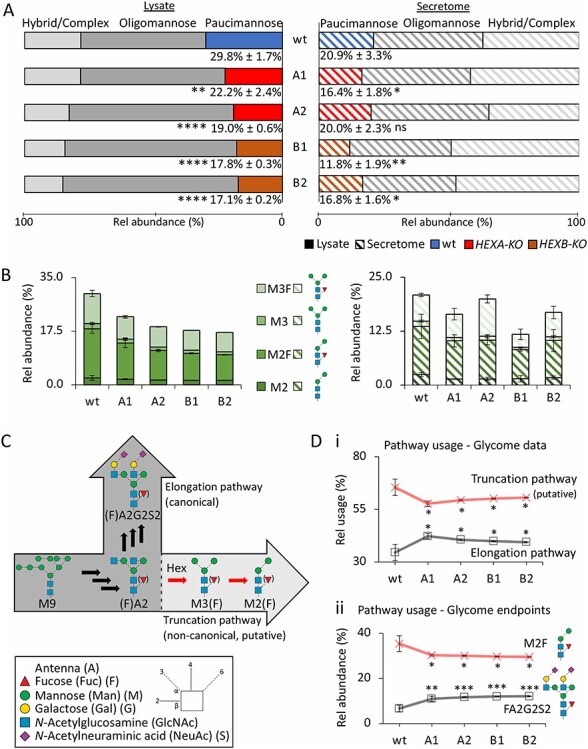
Hex disruption reduces paucimannosylation in the HL-60 N-glycome. Distribution of (A), N-glycan types and (B), PMG species (M2, M2F, M3, M3F) of the lysate (full bars, left) and secretome (striped bars, right) of the Hex-disrupted mutants (A1, A2, B1, B2) and wt HL-60. The total paucimannose levels are indicated as mean ± SD. (C), Graphical representation of the canonical elongation pathway and the proposed noncanonical truncation pathway mediated by human Hex. (D), Relative utilization of the proposed truncation and elongation pathway based on quantitative glycomics data when considering (i) all observed N-glycans mapping to the two pathways and (ii) the endpoint structures of the two pathways (M2F and FA2G2S2). For all panels, data have been plotted as mean ± SD, n = 3 technical replicates, ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.005 (HL-60 mutant vs wt). Glycan visualization and linkage representation as per the symbol nomenclature for glycans (Neelamegham et al. 2019).
Notably, in the lysate, Hex disruption caused a consistent increase in oligomannosylation compared to wt HL-60, an observation recapitulated in primary neutrophils (see below). Albeit unexpected, we could speculate that the accumulation of underprocessed oligomannosidic glycoproteins in truncation-deficient cells results from a less efficient glycosylation machinery unable to keep up with the early processing of trafficking glycoprotein intermediates in these cells, however, the underpinning mechanisms for this observation require further investigation. The fucosylated PMGs (M3F and M2F) appeared more sensitive to Hex disruption relative to the nonfucosylated species (M3 and M2), Figure 2B. Importantly, the reductions in the PMG levels in the HL-60 mutants accurately reflected the magnitude of the Hex disruption in those mutants as determined via established Hex activity assays (Figure 1C).
It remains to be determined if other culture conditions or differentiation of HL-60 into mature neutrophil-like cells will alter, and possibly exacerbate, the Hex- and PMG-deficient phenotype in the generated HL-60 Hex α and β mutants. Unfortunately, our attempts to generate HL-60 cells with a more complete reduction in Hex (thus completely devoid of PMGs) through the disruption of both HEXA and HEXB in the same cell line proved unsuccessful. Access to double knockout HL-60 cells presumably displaying a stronger Hex- and PMG-deficient phenotype would be valuable to confirm the contribution of the Hex α and β subunits to the PMP generation, rule out the involvement of other β-hexosaminidases (i.e. Hex D, O-GlcNAcase or other yet-to-be-identified human Hex variants), and create neutrophil-like cells devoid of paucimannose for functional studies. Interestingly, HEXA−/-HEXB−/− mice have successfully been produced (Sango et al. 1996; Hepbildikler et al. 2002). In accordance with our observations, biochemical characterization of urine from these double knockout mice showed an accumulation of free GlcNAc-capped biantennary N-glycans. Similar N-glycan fragments have been identified in the urine of a patient with Sandhoff disease (Strecker et al. 1977) further supporting the mechanistic link between Hex α and β and PMPs reported herein.
Quantitative glycomics suggests a putative noncanonical truncation pathway in HL-60
We then sought to explore if human Hex α and β mediate PMP generation via a noncanonical truncation pathway proposed to coexist with the canonical elongation pathway in the secretory pathway of neutrophils, Figure 2C. To test this hypothesis, we firstly used the quantitative glycomics data to assess the relative usage of the canonical elongation and the putative noncanonical truncation pathways. For this purpose, we only considered the identified N-glycans mapping to either of these two pathways; thus, the underprocessed N-glycan intermediates upstream of the elongation and truncation pathways including all oligomannosidic- and hybrid-type N-glycans were excluded in the analysis. Supporting our hypothesis, the putative truncation pathway was consistently underutilized in all Hex-disrupted mutants compared to wt HL-60 in favor of an increased utilization of the elongation pathway (all P < 0.05), Figure 2Di and Supplementary Table III. This observation, which points to a truncation-to-elongation switch in Hex-disrupted HL-60, was also reflected in the analysis of abundant N-glycans mapping to the endpoints of the two competing N-glycan processing pathways (M2F and FA2G2S2, respectively) (all P < 0.05), Figure 2Dii.
Glycoproteomics recapitulates the truncation-to-elongation switch in Hex-disrupted HL-60
Next, we utilized quantitative glycoproteomics to further investigate the putative truncation pathway in the Hex-disrupted HL-60 mutants, Figure 3 and Supplementary Table IV–VIII. When considering the entire HL-60 N-glycoproteome dataset, we observed a significant underutilization of the putative truncation pathway in all Hex-disrupted mutants compared to wt HL-60 (all P < 0.001, average of 0.8× fold reduction in mutants relative to wt), Figure 3Ai and Supplementary Table VI-A. The underutilization of the proposed truncation pathway was slightly more pronounced when considering only neutrophil proteins known to cotraffic and colocalize with Hex in the azurophilic granules (all P < 0.001, 0.7× fold reduction) (Rorvig et al. 2013), Figure 3Aii and Supplementary Table VI-B. These observations indicate that proteins destined for the azurophilic granules have a greater propensity for entering the proposed Hex-mediated truncation pathway, an intuitive relationship given the enhanced potential of such glycoproteins to interact with the cotrafficking Hex isoenzymes. However, given that HL-60 is an immortalized promyelocytic cell type in which most protein cargo expressed at that stage of neutrophil maturation including Hex reportedly traffic to the azurophilic granules as elegantly described by the targeted-by-timing model (Rorvig et al. 2013; Cowland and Borregaard 2016) and supported by our observation that glycoproteins known to reside in the azurophilic granules constituted nearly half of the HL-60 glycoproteome, the HL-60 cell may not be an ideal model system to decipher which glycoprotein subsets (if any) are destined to enter the truncation pathway. Investigations monitoring the dynamic glycosylation of maturing neutrophils isolated from the bone marrow of Hex-deficient patients and healthy donors are warranted to expand on these findings.
Fig. 3.
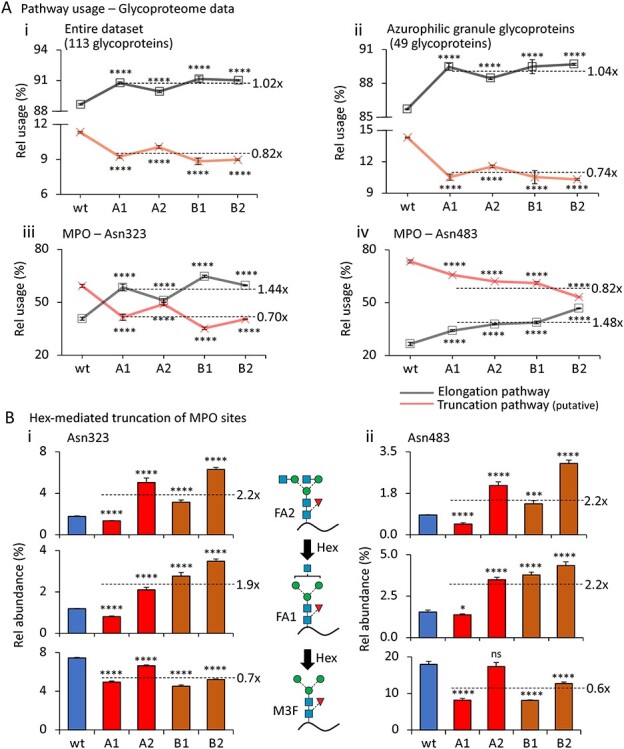
Hex disruption reduces paucimannosylation of azurophilic granule-resident proteins in the HL-60 N-glycoproteome. (A), Relative utilization of the proposed truncation and elongation pathway (see Figure 2C for schematics) based on all observed N-glycopeptides carrying N-glycans mapping to the two pathways arising from (i) the entire HL-60 glycoproteome, (ii) HL-60 source proteins known to traffic to and reside in the azurophilic granule (broken line represents average fold change relative to wt) (Rorvig et al. 2013), and (iii) Asn323 and (iv) Asn483 of MPO, a prominent azurophilic granule-resident glycoprotein. (B), Relative abundance of the GlcNAc-capped Hex substrates (FA1 and FA2, top graphs) and immediate Hex product (M3F, bottom graphs) decorating (i) Asn323 and (ii) Asn483 of MPO. The mean relative abundance for all HL-60 mutants (A1, A2, B1, B2, broken lines) and the fold change relative to the unedited wt are provided for each of the three N-glycan structures quantified across the two sites. For all panels, data have been plotted as mean ± SD, n = 3 technical replicates, ns, not significant, *P < 0.05, ***P < 0.005, ****P < 0.001 (mutant vs wt HL-60). See Figure 2 for glycan symbol and linkage representation (Neelamegham et al. 2019).
Underutilization of the putative truncation pathway was also observed when performing the pathway analysis on the truncation-prone N-glycosylation sites (i.e. Asn323 and Asn483) of the azurophilic granule-resident MPO (all P < 0.001, average of 0.7–0.8× fold reduction in mutants relative to wt) (Reiding et al. 2019; Tjondro et al. 2021), Figure 3Aiii and iv and Supplementary Table VI-C and Table VI-D. Upon further quantitative interrogation of the N-glycans decorating Asn323 and Asn483 of MPO, we observed that Hex disruption, as expected, resulted in an elevation of the GlcNAc-capped Hex substrates (FA1 and FA2, 1.9–2.2× fold elevation), except for the A1 mutant, which, however, still displayed a higher elongation efficiency compared to the wt and the other Hex mutants (see Figure 2Di). Nonetheless, this observation again illustrates the different glycophenotypes arising from the variable degree of Hex disruption achieved with the investigated mutants. Importantly, the immediate Hex product (M3F, 0.6–0.7× fold reduction) was reduced in all Hex mutants relative to wt HL-60 although only a minor decrease was observed for the A2 mutant, Figure 3Bi and ii and Supplementary Table VII. Collectively, the glycoproteomics and Hex activity data (see Figure 1C) support that both Hex α and β are involved in PMP formation via a putative truncation pathway in HL-60. Although our data show that glycoprotein products of the truncation pathway may also be secreted from HL-60 cells (see Figure 2A), the data indicate that luminal glycoproteins cotrafficking with Hex α and β to the azurophilic granules such as MPO are particularly prone to enter the trimming cascade in neutrophil-like cells.
Supporting further the relationship between human Hex and PMPs, we have previously used immunocytochemistry to show that human Hex A (αβ) colocalizes with PMG epitopes in the azurophilic granules of HL-60 (Thaysen-Andersen et al. 2015). In that study we also demonstrated using MALDI-MS that human Hex A in vitro can generate the paucimannosidic M3F from GlcNAc-capped FA2 substrates (Thaysen-Andersen et al. 2015) providing important in vitro support for the involvement of Hex in PMP generation. In another study we demonstrated in silico that sites modified with PMGs are highly solvent accessible in line with the notion that truncated N-glycan structures arise from excessive trimming by glycan processing enzymes (Thaysen-Andersen and Packer 2012; Lee et al. 2014). Taken together, these observations support the link between Hex α and β and PMP generation through a putative truncation pathway that appears to coexist and compete with the canonical elongation pathway in HL-60 cells and which proposedly favors glycoproteins trafficking to the azurophilic granules of human neutrophils such as MPO and other microbicidal glycoproteins.
Hex mediates PMP formation in primary neutrophils
We then sought to validate the relationship between human Hex and PMP generation in primary blood neutrophils. For this purpose, we analyzed neutrophils from a clinically diagnosed Sandhoff disease patient (HEXB−/−, IVS11 + 5G > A genotype) and a healthy age-matched control individual, Figure 4 and Supplementary Table VIII. As expected, the Sandhoff disease neutrophils displayed very low total Hex and Hex A + S activity whereas the β-galactosidase activity remained similar to the levels in unaffected neutrophils. In line with the data from the Hex-disrupted HL-60 mutants, quantitative N-glycomics showed a considerable reduction in the PMG levels in the Sandhoff disease neutrophils (21.2% ± 0.9%) compared to the PMG levels in unaffected neutrophils (44.0% ± 1.7%). The truncation-to-elongation switch was also recapitulated in the Sandhoff disease neutrophils as demonstrated by a lower utilization of the putative truncation pathway (31.9% ± 1.4%) relative to the usage of this proposed pathway in unaffected neutrophils (49.4% ± 6.6%).
Fig. 4.
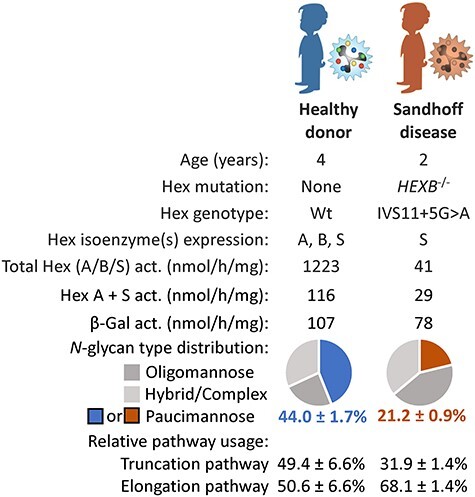
Sandhoff disease neutrophils display reduced levels of paucimannosylation. Donor metadata including age, Hex genotype, Hex and β-galactosidase activity and quantitative N-glycomics data showing the relative level of paucimannosylation and the relative utilization of the putative truncation pathway in primary blood neutrophils isolated from a Sandhoff disease patient (HEXB−/−, brown) and an age-paired healthy donor (blue).
Although the Hex-PMP relationship was confirmed in primary neutrophils from a single Sandhoff disease patient, due to the extremely limited access to biologically relevant specimens from individuals suffering from such rare disorders and unfortunately often do not live beyond infancy, these data add valuable support for the notion that the Hex isoenzymes mediate PMP generation via a putative noncanonical truncation pathway in human neutrophils. There is, however, a need to confirm these in vivo observations on larger patient cohorts ideally spanning individuals affected by both Tay-Sachs disease (HEXA−/−) and Sandhoff disease (HEXB−/−) to assess the relative impact of the Hex α and β subunits on PMP formation. Longitudinally monitoring of neutrophils and other cells and tissues of interest from late-onset patients suffering from Hex deficiency could also provide value observations of the dynamics of the Hex-PMP relationship before and during the disease course. Studies investigating the potential involvement of Hex-deficient (and, by extension, PMP-poor) neutrophils in the pathophysiology of Tay-Sachs and Sandhoff disease are also warranted. Finally, future efforts should be invested to explore the α-mannosidase-mediated truncation processes downstream of the initial Hex trimming reaction in the proposed truncation pathway (producing abundant structures such as M2F), for example, by investigating MAN2B1-deficient patients suffering from alpha-mannosidosis.
Conclusions
Utilizing a set of well-characterized Hex-disrupted HL-60 mutants and primary blood neutrophils and enabled by powerful quantitative glycomics and glycoproteomics approaches, we are here the first to provide experimental evidence for the Hex α and β-mediated generation of PMPs in human neutrophils. Further, we propose that neutrophil glycoprotein substrates that co-traffic with the Hex α and β subunits to the azurophilic granules may be particularly susceptible to undergo Hex-mediated trimming via a glycan processing pathway fundamentally different from the canonical elongation pathway. This putative noncanonical trimming cascade, herein referred to as the truncation pathway, which proposedly exist in and is highly utilized by human neutrophils and possibly by other PMP-rich cell types including monocytes/macrophages (Hinneburg et al. 2020), dendritic cells (Parker et al. 2021) and cancer cells (Chatterjee et al. 2019), mirrors similar well-described Hex-mediated truncation pathways in PMP-rich model organisms including worms, flies and plants (Paschinger and Wilson 2019; Tjondro et al. 2019). Unlike the constitutive and tissue-wide utilization of the truncation pathway in many of these lower organisms, it is likely that humans use the Hex-mediated truncation pathway in a limited set of tissues and (patho)physiological conditions, perhaps explaining why this proposed biosynthetic pathway and PMPs, so far, have received little attention in human glycobiology. Excitingly, however, the roles of neutrophilic PMPs in diverse immune-related processes are rapidly emerging and building evidence points to the functional involvement of PMPs in several key areas of neutrophil-mediated immunity (Loke et al. 2016; Ugonotti et al. 2020). The knowledge of how PMPs are formed in human neutrophils, as communicated in this report, will further enable us to better understand their spatiotemporal expression, function and significance in the human immune system.
Materials and methods
An overview of the experimental workflow is provided in Figure 1B. Methods relating to the quantitative glycomics and glycoproteomics are briefly covered in this section; exhaustive methodological details of those experiments are found in the Extended Methods in the SI.
Hex sequence analysis
FASTA sequences of human Hex α (UniProtKB, P06865), Hex β (P07686) and Hex D (Q8WVB3), C. elegans (worm) hex-1 (Q22492), hex-2 (G5EFL1), hex-3 (G5EDG9), hex-4 (Q8WTK2) and hex-5 (G5EEC8), A. thaliana (plant) HEXO1 (A7WM73), HEXO2 (Q9SYK0) and HEXO3 (Q8L7S6) and D. melanogaster (fruit fly) Hex fdl (Q8WSF3), Hexo1 (Q0E8H9) and Hexo2 (Q9W3C4, all downloaded from UniProtKB, July 2021) were compiled, imported into Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the cluster analysis performed using default settings. For sequence similarity and identity, the FASTA sequences for either human Hex α or β were imported into EMBOSS Needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) in pairs with the other Hex sequences listed above. The default settings were used for the analysis. For the sequence alignment, the FASTA sequences of human Hex α and β, C. elegans hex-1, A. thaliana HEXO1, HEXO2 and HEXO3 and D. melanogaster Hex fdl, Hexo1 and Hexo2 were compiled and imported into T-Coffee (http://tcoffee.crg.cat/apps/tcoffee/do:regular). The default settings were used and the fasta_aln output file was selected and copied into Boxshade (https://embnet.vital-it.ch/software/BOX_form.html) to generate a greyscale representation of the sequence alignment.
Hex-targeted CRISPR-Cas9-based gene editing of HL-60 cells
Target regions of HEXA and HEXB were selected based on a previously published CRISPR-library (Tzelepis et al. 2016; Zhu et al. 2021). Single guide ribonucleic acid (sgRNA) was synthesized using PCR amplification and in vitro transcription using EnGenR sgRNA Synthesis Kit (New England Biolabs, Ipswich, U.S.) following the manufacturer’s protocol. To this end, briefly, a customized DNA oligo was synthesized with T7 promoter followed by the target sequence and first 17 bases of the sgRNA scaffold. The synthesized DNA was incubated with the sgRNA scaffold oligo and T7 enzyme at 37°C for 2 h to generate the sgRNA. Prior to transfection, wt HL-60 (ATCC, Manassas, U.S.) cells were transferred into fresh complete Iscove’s Modified Dulbecco’s Media (IMDM) (Gibco™ IMDM 1640 Medium, HEPES) with 10% (v/v) fetal bovine serum (FBS), 1% arachidonic acid and 1% glutamax. 1 μg sgRNA was then mixed with 1 μg Cas9 (EnGenR Cas9 NLS, Streptococcus pyogenes, New England Biolabs) at room temperature for 30 min to form the ribonucleoprotein particles. The HL-60 cells (1.5–2.5 × 105/experiment) were then resuspended in 10 μL Resuspension Buffer R (Neon Transfection Kit, Thermo Fisher Scientific), mixed with the ribonucleoprotein particles, and electroporated using the Neon Transfection System (Thermo Fisher Scientific) and the following conditions: 1.6 kV, 10 ms, 3 pulses (Gundry et al. 2016; Zhu et al. 2021). After electroporation, the cells were transferred to a 24-well plate containing 500 μL conditioned IMDM medium per well. The cell population was then single cell sorted into 96 well plates using fluorescence-activated cell sorting as previously described, in order to obtain isogenic clones (Stolfa et al. 2016). A panel of these cells were scaled up, and genomic editing was verified using NGS.
For NGS, genomic DNA was isolated from the sorted single cell clones, and PCR was performed with gene-specific primers to amplify the edited target regions in the Hex genes. PCR purification was performed by gel extraction, amplicons were barcoded as previously described (Zhu et al. 2021) and analyzed using Illumina MiSeq (300 bp, paired end). From these efforts, two HEXA-KO (A1 and A2) and two HEXB-KO (B1 and B2) HL-60 mutant isogenic clones were selected. One hundred percent of the sequence reads of these clones were different from wt unedited control HL-60 cells that were mock transfected with Cas9 (no sgRNA). Unedited HL-60 cells not Cas9 mock transfected were also analyzed in this study, and these appeared highly similar to the mock electroporated HL-60 cells (see SI for data, labeled twt). Multiple attempts were made to generate HEXA/HEXB double KOs using the above workflow, but all the knockouts sequenced contained at least one Hex allele corresponding to wt HL-60.
Culturing, growth profiling and protein extraction of HL-60 cells
The HL-60 cell lines were cultured under stable growth conditions in Roswell Park Memorial Institute (RPMI)-1640 media supplemented with 10% (v/v) FBS in Corning T25 cell culture flasks. The incubator was set to 37°C and 5% (v/v) carbon dioxide levels. To monitor the growth characteristics of the generated HL-60 cell lines, 1 million cells from each line were seeded into 10 mL fresh media. After 24 h and 48 h, the cells were sedimented via centrifugation at 300 × g and live cells were counted using 0.4% (w/v) trypan blue staining (Sigma).
For the quantitative glycomics and glycoproteomics experiments, cells were cultured in FBS-free RPMI-1640 media for 24 h prior to harvesting and protein extraction. Cells were pelleted by centrifugation at 300 × g for 5 min and the media (supernatant) collected. The cell pellet was washed three times in phosphate-buffered saline (PBS) prior to cell lysis via probe sonication (30 A for 10 s, twice) in a radioimmunoprecipitation assay (RIPA) buffer. The cell lysate was then centrifuged at 10,000 × g for 10 min and the supernatant containing the protein extract of the lysate was collected. To obtain the protein extracts from the secretome, the cell media were concentrated and buffer exchanged with RIPA using Amicon® Ultra-15 10 kDa centrifugal filters (Merck Millipore). Protein extracts from both the cell lysate and secretome were subjected to protein quantitation via a bicinchoninic acid (BCA) assay prior to downstream analysis. Proteins from the lysate and secretome of the various HL-60 cell lines were precipitated overnight with 1:4 (v/v) ice cold acetone at −30°C. The proteins were pelleted at 10,000 × g, washed with ice cold acetone and redissolved in 8 M urea. Technical triplicates of each HL-60 variant were used in subsequent sample processing steps.
Protein extracts from primary neutrophils from Sandhoff disease and healthy donors
Unactivated blood neutrophils from an infant (2 year old) clinically diagnosed with Sandhoff disease via Hex genotyping (HEXB−/−, IVS11 + 5G > A) and an age-paired (4 year old) healthy donor were obtained from the EuroBioBank (http://www.eurobiobank.org/). Neutrophils were isolated using Histopaque (density: 1.077 g/mL, Sigma) and 3% (v/v) dextran (Sigma) in 0.15 M sodium chloride. Protein extracts were obtained by neutrophil cell lysis using a lysis buffer comprising 10 mM Tris, 300 mM NaCl, 2 mM EDTA, 0.5% (v/v) Triton X-100, pH 7.4 supplemented with protease inhibitors (Roche). Protein concentrations were determined via a BCA assay. Technical triplicate analyses of the protein extracts from the donor neutrophils were carried out.
Hex activity assays
The Hex activity of the HL-60 cell lysate and secretome was measured using an established protocol with minor modifications (Wendeler and Sandhoff 2009). In brief, 1.5 μg total protein from the cell lysate or secretome (0.1 μg/μL) and 1.5 ng recombinant human Hex A and B (RnD Systems, 0.1 ng/μL, positive controls) were separately incubated with 30 μL prewarmed 3 mM 4-methylumbelliferyl-2-acetamido-2-deoxy-β-D-glucopyranoside (MUG) or 4-methylumbelliferyl-2-acetamido-2-deoxy-β-D-glucopyranoside-6-sulfate (MUGS) substrate (Merck Millipore) in a phosphate–citrate buffer (pH 5.0) at 37°C for 30 min in the dark. The reaction was stopped by the addition of 0.25 M glycine-carbonate stop buffer (pH 10.0). Hex-mediated hydrolysis of MUG and MUGS is known to produce a fluorogenic product (4-methylumbelliferone, 4-MU) allowing fluorometric quantitation of the reaction using a plate reader (FLUOstar Optima, BMG Technologies) with excitation at 360 nm and emission at 450 nm. The absolute quantity of released 4-MU was determined from a premade standard curve created using known concentrations of 4-MU (Merck Millipore).
Hex-centric ELISA
Double-sandwich ELISA kits for both the Hex α and β subunits were purchased (Cloud-Clone Corp., Houston, USA). Each kit includes an ELISA plate precoated with monoclonal antibodies toward human Hex α or β (raised in mice) for capture and biotin-linked human Hex α or β antibodies and avidin-linked horseradish peroxidase for detection. Assays were performed as per the manufacturer’s instructions. In brief, either 10 μg total protein from the cell lysate or 5 μg total protein from the secretome was added in triplicates to the ELISA plate alongside blanks. The plate was sealed and incubated for 1 h at 37°C. The liquid was removed and 100 μL detection reagent A working solution was added to each well. The plate was resealed and incubated for 1 h at 37°C. The liquid was discarded and the wells were washed three times with 300 μL wash solution. Subsequently, 100 μL detection reagent B working solution was added to each well and the plate was reincubated for 30 min at 37°C. The liquid was discarded and the wells rewashed three times with the wash solution. Next, 90 μL substrate solution was added to each well and the plate incubated for 20 min at 37°C. Lastly, 50 μL stop solution was added and absorbance (450 nm) was read in a plate reader (FLUOstar Optima, BMG Technologies). The absolute concentrations of Hex α and β subunits were determined based on a premade standard curve generated using known concentrations of Hex α and β standards provided with the ELISA kit.
Quantitative N-glycomics
Briefly, N-glycans were released from polyvinylidene difluoride-immobilized protein extracts using peptide:N-glycosidase F (PNGase F, Elizabethkingia miricola, Promega) (Jensen et al. 2012). The detached N-glycans were reduced to alditols using sodium borohydride and profiled using PGC-LC–MS/MS performed in negative ion mode on an LTQ Velos Pro ion trap mass spectrometer (Thermo Fisher Scientific). The glycan structures were manually elucidated based on their molecular mass, PGC-LC retention time and MS/MS fragmentation pattern (Chatterjee et al. 2021). RawMeat v2.1 (Vast Scientific), GlycoMod (Expasy), Xcalibur Qual Browser v2.2 (Thermo Fisher Scientific) and GlycoWorkBench v2.1 aided the characterization process. EIC-based relative quantitation of all observed N-glycans was performed using Skyline v.19.1 (Ashwood et al. 2018; Adams et al. 2020). The relative usage of the canonical elongation and putative noncanonical truncation pathways was established by determining the proportion of observed N-glycans arising only from these two respective processing pathways, i.e. no biosynthetically underprocessed N-glycan intermediates predicted to be upstream of the proposed junction of the two competing pathways including the oligomannosidic and hybrid-type N-glycans were considered for such calculations. See Extended Methods in the SI for details.
Quantitative (N-glyco)proteomics
The label-assisted quantitative (glyco)proteomics experiments were carried out using a recently published method (Kawahara et al. 2021). Briefly, protein extracts from the HL-60 cell lysate and secretome were reduced, alkylated and digested overnight with sequence-grade porcine trypsin (Promega). Peptides were desalted, labeled with TMT (Thermo Fisher Scientific) and desalted again. Glycopeptides were enriched with ion-pairing zwitterionic hydrophilic interaction liquid chromatography solid phase extraction (Mysling et al. 2010). Both the peptide and glycopeptide fractions were subjected to high pH reversed-phase C18 solid phase extraction prefractionation and were desalted prior to LC–MS/MS analysis.
All peptide samples were separated using nanoscale C18 chromatography and detected by a Q-Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) operating in positive ion mode. The LC–MS/MS raw data were searched against the entire human proteome (UniProtKB, all reviewed entries) using MaxQuant v1.6.17 (Tyanova et al. 2016) and Byonic v3.9.4 (Protein Metrics Inc., Cupertino, CA) as a node in Proteome Discoverer v2.4 (Thermo Fisher Scientific). Only glycopeptides with Byonic PEP-2D scores <0.001 were considered. Unmodified peptides and peptides with nonglycan modifications were filtered to peptide-to-spectral matches and protein false discovery rates below 0.01. Glycopeptides were quantified using the Reporter Ion Quantifier node in Proteome Discoverer. Unmodified peptides and peptides with nonglycan modifications were quantified with MaxQuant using the reporter ion intensities. See Extended Methods in the SI for details.
Data representation and statistical analysis
Data arising from multiple technical replicates (n, as indicated in figure legends) have been plotted as the mean. The error bars indicate their standard deviation. For all data comparisons, statistical significance was assessed using unpaired one-tailed Student’s t-tests. Statistical significance has been indicated according to the confidence level (*P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001 whereas ns denotes no significance i.e. P ≥ 0.05).
Supplementary Material
Acknowledgements
J.U. was supported by a Macquarie University Research Excellence Scholarship (MQRES). R.K. was supported by an Early Career Fellowship from the Cancer Institute NSW. H.C.T. was supported by an Australian Cystic Fibrosis postgraduate studentship award and an international MQRES (iMQRES). S.C. was supported by an iMQRES. S.N. was supported by National Institutes of Health grant HL103411. M.T.A. was supported by a Macquarie University Safety Net Grant.
Contributor Information
Julian Ugonotti, Department of Molecular Sciences, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia.
Rebeca Kawahara, Department of Molecular Sciences, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia.
Ian Loke, Cordlife Group Limited, 1 Yishun Industrial Street, Singapore 768160, Singapore.
Yuqi Zhu, Department of Chemical and Biological Engineering, University at Buffalo, State University of New York, 906 Furnas Hall, Buffalo, NY 14260, USA.
Sayantani Chatterjee, Department of Molecular Sciences, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia.
Harry C Tjondro, Department of Molecular Sciences, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia.
Zeynep Sumer-Bayraktar, Department of Molecular Sciences, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia.
Sriram Neelamegham, Department of Chemical and Biological Engineering, University at Buffalo, State University of New York, 906 Furnas Hall, Buffalo, NY 14260, USA.
Morten Thaysen-Andersen, Department of Molecular Sciences, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia; Biomolecular Discovery Research Centre, Macquarie University, Balaclava Road, Macquarie Park, Sydney, NSW 2109, Australia.
Data availability statement
The raw mass spectrometry files generated from the glycomics and glycoproteomics experiments performed for this study have been made publicly available at GlycoPOST (https://glycopost.glycosmos.org/, accession ID GPST000206) (Watanabe et al. 2021) and at the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/, accession ID PXD027664) (Perez-Riverol et al. 2019), respectively.
Conflict of interest statement
None declared.
Abbreviations
- 4-MU
4-methylumbelliferone
- BCA
bicinchoninic acid assay
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- GH
glycoside hydrolase
- Hex
N-acetyl-β-D-hexosaminidase
- HNE
human neutrophil elastase
- IMDM
Iscove’s Modified Dulbecco’s media
- LC
liquid chromatography
- MPO
myeloperoxidase
- MS/MS
tandem mass spectrometry
- MUG
4-methylumbelliferyl-2-acetamido-2-deoxy-β-D-glucopyranoside
- MUGS
4-methylumbelliferyl-2-acetamido-2-deoxy-β-D-glucopyranoside-6-sulfate
- nCG
neutrophil cathepsin G
- NGS
next generation sequencing
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PGC
porous graphitized carbon
- PMG
paucimannosidic glycan
- PMP
paucimannosidic protein
- RIPA
radioimmunoprecipitation assay
- RPMI
Roswell Park Memorial Institute
- sgRNA
single guide ribonucleic acid
- TMT
tandem mass tag
- wt
wildtype
- ZIC-HILIC
zwitterionic hydrophilic interaction liquid chromatography
References
- Adams KJ, Pratt B, Bose N, Dubois LG, St John-Williams L, Perrott KM, Ky K, Kapahi P, Sharma V, MacCoss MJ, et al. 2020. Skyline for small molecules: A unifying software package for quantitative metabolomics. J Proteome Res. 19:1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteen MG, Oehler V, Nemcovicova I, Wilson IB, Vocadlo DJ, Gloster TM. 2016. Mechanism of human nucleocytoplasmic hexosaminidase D. Biochemistry. 55:2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwood C, Lin CH, Thaysen-Andersen M, Packer NH. 2018. Discrimination of isomers of released N- and O-Glycans using diagnostic product ions in negative ion PGC-LC-ESI-MS/MS. J Am Soc Mass Spectrom. 29:1194–1209. [DOI] [PubMed] [Google Scholar]
- Babatunde KA, Wang X, Hopke A, Lannes N, Mantel PY, Irimia D. 2021. Chemotaxis and swarming in differentiated HL-60 neutrophil-like cells. Sci Rep. 11:778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiden B, Sandhoff K. 2019. Lysosomal glycosphingolipid storage diseases. Annu Rev Biochem. 88:461–485. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Kawahara R, Tjondro HC, Shaw DR, Nenke MA, Torpy DJ, Thaysen-Andersen M. 2021. Serum N-glycomics stratifies bacteremic patients infected with different pathogens. J Clin Med. 10:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Lee LY, Kawahara R, Abrahams JL, Adamczyk B, Anugraham M, Ashwood C, Sumer-Bayraktar Z, Briggs MT, Chik JHL, et al. 2019. Protein paucimannosylation is an enriched N-glycosylation signature of human cancers. Proteomics. 19:e1900010. [DOI] [PubMed] [Google Scholar]
- Cowland JB, Borregaard N. 2016. Granulopoiesis and granules of human neutrophils. Immunol Rev. 273:11–28. [DOI] [PubMed] [Google Scholar]
- Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, Hsu JI, Hoegenauer KA, Rooney CM, Goodell MA, et al. 2016. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep. 17:1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutternigg M, Kretschmer-Lubich D, Paschinger K, Rendic D, Hader J, Geier P, Ranftl R, Jantsch V, Lochnit G, Wilson IB. 2007. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants, and insects. J Biol Chem. 282:27825–27840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutternigg M, Rendic D, Voglauer R, Iskratsch T, Wilson IB. 2009. Mammalian cells contain a second nucleocytoplasmic hexosaminidase. Biochem J. 419:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. 2007. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 446:1017–1022. [DOI] [PubMed] [Google Scholar]
- Hepbildikler ST, Sandhoff R, Kolzer M, Proia RL, Sandhoff K. 2002. Physiological substrates for human lysosomal beta -hexosaminidase S. J Biol Chem. 277:2562–2572. [DOI] [PubMed] [Google Scholar]
- Hinneburg H, Chatterjee S, Schirmeister F, Nguyen-Khuong T, Packer NH, Rapp E, Thaysen-Andersen M. 2019. Post-column make-up flow (PCMF) enhances the performance of capillary-flow PGC-LC-MS/MS-based Glycomics. Anal Chem. 91:4559–4567. [DOI] [PubMed] [Google Scholar]
- Hinneburg H, Pedersen JL, Bokil NJ, Pralow A, Schirmeister F, Kawahara R, Rapp E, Saunders BM, Thaysen-Andersen M. 2020. High-resolution longitudinal N- and O-glycoprofiling of human monocyte-to-macrophage transition. Glycobiology. 30:679–694. [DOI] [PubMed] [Google Scholar]
- Jensen PH, Karlsson NG, Kolarich D, Packer NH. 2012. Structural analysis of N- and O-glycans released from glycoproteins. Nat Protoc. 7:1299–1310. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Recuero S, Srougi M, Leite KRM, Thaysen-Andersen M, Palmisano G. 2021. The complexity and dynamics of the tissue glycoproteome associated with prostate cancer progression. Mol Cell Proteomics. 20:100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneluk RG, Mahuran DJ, Neote K, Klavins MH, O'Dowd BF, Tropak M, Willard HF, Anderson MJ, Lowden JA, Gravel RA. 1986. Isolation of cDNA clones coding for the alpha-subunit of human beta-hexosaminidase. Extensive homology between the alpha- and beta-subunits and studies on Tay-Sachs disease. J Biol Chem. 261:8407–8413. [PubMed] [Google Scholar]
- Kytzia HJ, Sandhoff K. 1985. Evidence for two different active sites on human beta-hexosaminidase A. Interaction of GM2 activator protein with beta-hexosaminidase A. J Biol Chem. 260:7568–7572. [PubMed] [Google Scholar]
- Leal AF, Benincore-Florez E, Solano-Galarza D, Garzon Jaramillo RG, Echeverri-Pena OY, Suarez DA, Almeciga-Diaz CJ, Espejo-Mojica AJ. 2020. GM2 Gangliosidoses: Clinical features, pathophysiological aspects, and current therapies. Int J Mol Sci. 21:6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Lin CH, Fanayan S, Packer NH, Thaysen-Andersen M. 2014. Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front Immunol. 5:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN. 2006. Crystallographic structure of human beta-hexosaminidase A: Interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J Mol Biol. 359:913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard R, Rendic D, Rabouille C, Wilson IB, Preat T, Altmann F. 2006. The drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J Biol Chem. 281:4867–4875. [DOI] [PubMed] [Google Scholar]
- Liew PX, Kubes P. 2019. The neutrophil's role during health and disease. Physiol Rev. 99:1223–1248. [DOI] [PubMed] [Google Scholar]
- Liu T, Duan Y, Yang Q. 2018. Revisiting glycoside hydrolase family 20 beta-N-acetyl-d-hexosaminidases: Crystal structures, physiological substrates and specific inhibitors. Biotechnol Adv. 36:1127–1138. [DOI] [PubMed] [Google Scholar]
- Loke I, Kolarich D, Packer NH, Thaysen-Andersen M. 2016. Emerging roles of protein mannosylation in inflammation and infection. Mol Aspects Med. 51:31–55. [DOI] [PubMed] [Google Scholar]
- Loke I, Ostergaard O, Heegaard NHH, Packer NH, Thaysen-Andersen M. 2017. Paucimannose-rich N-glycosylation of spatiotemporally regulated human neutrophil elastase modulates its immune functions. Mol Cell Proteomics. 16:1507–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke I, Packer NH, Thaysen-Andersen M. 2015. Complementary LC-MS/MS-based N-glycan, N-glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil Cathepsin G. Biomolecules. 5:1832–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe DD, Chandrasekaran EV, Lau JT, Matta KL, Neelamegham S. 2008. Systems-level studies of glycosyltransferase gene expression and enzyme activity that are associated with the selectin binding function of human leukocytes. FASEB J. 22:4154–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysling S, Palmisano G, Hojrup P, Thaysen-Andersen M. 2010. Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal Chem. 82:5598–5609. [DOI] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lutteke T, O'Boyle N, Packer NH, Stanley P, Toukach P, et al. 2019. Updates to the symbol nomenclature for Glycans guidelines. Glycobiology. 29:620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Partridge T, Wormald C, Kawahara R, Stalls V, Aggelakopoulou M, Parker J, Powell Doherty R, Ariosa Morejon Y, Lee E, et al. 2021. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 35:109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K, Wilson IBH. 2019. Comparisons of N-glycans across invertebrate phyla. Parasitology. 146:1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinage B, Higgins MA, Abbott DW, Robb C, Dalia AB, Deng L, Weiser JN, Parsons TB, Fairbanks AJ, Vocadlo DJ, et al. 2011. Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis. Structure. 19:1603–1614. [DOI] [PubMed] [Google Scholar]
- Reiding KR, Franc V, Huitema MG, Brouwer E, Heeringa P, Heck AJR. 2019. Neutrophil myeloperoxidase harbors distinct site-specific peculiarities in its glycosylation. J Biol Chem. 294:20233–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorvig S, Ostergaard O, Heegaard NH, Borregaard N. 2013. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: Correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 94:711–721. [DOI] [PubMed] [Google Scholar]
- Rosales C. 2018. Neutrophil: a cell with many roles in inflammation or several cell types? Front Physiol. 9:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sango K, McDonald MP, Crawley JN, Mack ML, Tifft CJ, Skop E, Starr CM, Hoffmann A, Sandhoff K, Suzuki K, et al. 1996. Mice lacking both subunits of lysosomal beta-hexosaminidase display gangliosidosis and mucopolysaccharidosis. Nat Genet. 14:348–352. [DOI] [PubMed] [Google Scholar]
- Schachter H. 2009. Paucimannose N-glycans in Caenorhabditis elegans and Drosophila melanogaster. Carbohydr Res. 344:1391–1396. [DOI] [PubMed] [Google Scholar]
- Stolfa G, Mondal N, Zhu Y, Yu X, Buffone A Jr, Neelamegham S. 2016. Using CRISPR-Cas9 to quantify the contributions of O-glycans, N-glycans and glycosphingolipids to human leukocyte-endothelium adhesion. Sci Rep. 6:30392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker G, Herlant-Peers MC, Fournet B, Montreul J. 1977. Structure of seven oligosaccharides excreted in the urine of a patient with Sandhoff's disease (GM2 gangliosidosis-variant O). Eur J Biochem. 81:165–171. [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M, Kolarich D, Packer NH. 2021. Glycomics & Glycoproteomics: From analytics to function. Mol Omics. 17:8–10. [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M, Packer NH. 2012. Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology. 22:1440–1452. [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M, Venkatakrishnan V, Loke I, Laurini C, Diestel S, Parker BL, Packer NH. 2015. Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J Biol Chem. 290:8789–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjondro HC, Loke I, Chatterjee S, Thaysen-Andersen M. 2019. Human protein paucimannosylation: cues from the eukaryotic kingdoms. Biol Rev Camb Philos Soc. 94:2068–2100. [DOI] [PubMed] [Google Scholar]
- Tjondro HC, Ugonotti J, Kawahara R, Chatterjee S, Loke I, Chen S, Soltermann F, Hinneburg H, Parker BL, Venkatakrishnan V, et al. 2021. Hyper-truncated Asn355- and Asn391-glycans modulate the activity of neutrophil granule myeloperoxidase. J Biol Chem. 296:100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 11:2301–2319. [DOI] [PubMed] [Google Scholar]
- Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, et al. 2016. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid Leukemia. Cell Rep. 17:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugonotti J, Chatterjee S, Thaysen-Andersen M. 2020. Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders. Mol Aspects Med. 79:100882. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan V, Dieckmann R, Loke I, Tjondro HC, Chatterjee S, Bylund J, Thaysen-Andersen M, Karlsson NG, Karlsson-Bengtsson A. 2020. Glycan analysis of human neutrophil granules implicates a maturation-dependent glycosylation machinery. J Biol Chem. 295:12648–12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Aoki-Kinoshita KF, Ishihama Y, Okuda S. 2021. GlycoPOST realizes FAIR principles for glycomics mass spectrometry data. Nucleic Acids Res. 49:D1523–D1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeler M, Sandhoff K. 2009. Hexosaminidase assays. Glycoconj J. 26:945–952. [DOI] [PubMed] [Google Scholar]
- Wendeler M, Werth N, Maier T, Schwarzmann G, Kolter T, Schoeniger M, Hoffmann D, Lemm T, Saenger W, Sandhoff K. 2006. The enzyme-binding region of human GM2-activator protein. FEBS J. 273:982–991. [DOI] [PubMed] [Google Scholar]
- Zhang W, Cao P, Chen S, Spence AM, Zhu S, Staudacher E, Schachter H. 2003. Synthesis of paucimannose N-glycans by Caenorhabditis elegans requires prior actions of UDP-N-acetyl-D-glucosamine:Alpha-3-D-mannoside beta1,2-N-acetylglucosaminyltransferase I, alpha3,6-mannosidase II and a specific membrane-bound beta-N-acetylglucosaminidase. Biochem J. 372:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Groth T, Kelkar A, Zhou Y, Neelamegham S. 2021. A GlycoGene CRISPR-Cas9 lentiviral library to study lectin binding and human glycan biosynthesis pathways. Glycobiology. 31:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry files generated from the glycomics and glycoproteomics experiments performed for this study have been made publicly available at GlycoPOST (https://glycopost.glycosmos.org/, accession ID GPST000206) (Watanabe et al. 2021) and at the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/, accession ID PXD027664) (Perez-Riverol et al. 2019), respectively.


