Abstract
Invention of DNA origami has transformed the fabrication and application of biological nanomaterials. In this review, we discuss DNA origami nanoassemblies according to their four fundamental mechanical properties in response to external forces: elasticity, pliability, plasticity and stability. While elasticity and pliability refer to reversible changes in structures and associated properties, plasticity shows irreversible variation in topologies. The irreversible property is also inherent in the disintegration of DNA nanoassemblies, which is manifested by its mechanical stability. Disparate DNA origami devices in the past decade have exploited the mechanical regimes of pliability, elasticity, and plasticity, among which plasticity has shown its dominating potential in biomechanical and physiochemical applications. On the other hand, the mechanical stability of the DNA origami has been used to understand the mechanics of the assembly and disassembly of DNA nano-devices. At the end of this review, we discuss the challenges and future development of DNA origami nanoassemblies, again, from these fundamental mechanical perspectives.
1. Introduction
Since DNA origami was invented in 2006,1 its programmable nature and highly precise structure at the nanometer scale have made DNA origami an ideal nanoscale platform adopted in various disciplines across chemistry, physics, and biology fields.2–8 While DNA origami has been recently reviewed9–14 and properties of various biomolecules, DNA in particular, under mechanical force have been reported,15–18 none of the prior work has provided a comprehensive mechanical perspective on DNA origami nanoassemblies. Given that DNA origami is a biomaterial whose mechanical properties are of utmost importance in both applications19,20 and mechanistic studies,21 it is of critical significance and urgency to provide a mechanical perspective to this new nanomaterial.
Like macroscopic materials, there are four basic mechanical regimes in DNA origami structures in response to external forces (Fig. 1).22,23 In the low force regime, a small mechanical force can change the property of Holliday junctions24 or pi–pi base stacking in DNA origami. Because neither the overall frame nor constituting secondary structures are damaged, the bending, stretching, and other deformations of DNA origami are reversible when force is withdrawn. This regime can be understood as the elasticity or pliability of the DNA origami nanoassemblies.
Fig. 1.
Four mechanical regimes of objects under mechanical force.
In the next regime under dozens of picoNewton (pN) mechanical force,24 some Holliday junctions start to get compromised, resulting in an irreversible deformation of the overall DNA origami structure. However, since the majority of the junctions are intact, the DNA origami structure still remains intact. Such an irreversible deformation under force is considered as the plasticity of DNA origami self-assemblies.
In the final regime of even greater mechanical forces, the entire DNA origami structure falls apart.25,26 In this regime, all the Holliday junctions are compromised, resulting in the dissociation of staple strands from the long ssDNA template strand. The dissociated staple strands then release into bulk solution irreversibly. We define the force that disassembles DNA origami as the mechanical stability of DNA nanostructures.19,27,28
In the following sections, we first discuss mechanical properties of basic components in DNA origami structures. We then provide selected examples of DNA origami structures that harness these four force regimes: the reversible pliability and elasticity, as well as the irreversible plasticity and stability. Finally, we discuss the folding mechanism of DNA origamis revealed by subjecting the DNA nanoassemblies under disassembling mechanical forces.
2. Mechanical properties of DNA origamis are determined by duplex DNA and Holliday junctions
Since mechanical force exerted on an object is a localized vector characterized by the force loading rate and the applied direction, the mechanical stability of the object varies with the direction29 and the loading rate30 of external force. Therefore, it is of high importance to specify these two factors when comparing mechanical stabilities of different objects. In this section, we discuss mechanical stabilities of DNA secondary structures that constitute DNA origami nanoassemblies.
2.1. Duplex DNA
There are two basic directions, unzipping and shearing, along which a force can be applied to evaluate the mechanical stability of a duplex DNA (Fig. 2).
Fig. 2.
Schematics of (a) unzipping of dsDNA, (b) shearing of dsDNA, and (c) stacking of two adjacent Watson–Crick base pairs. The arrows in (c) represent stacking interactions and the dashed lines represent hydrogen bonds20 (reprinted with permission from AAAS).
2.1.1. Mechanical stability of dsDNA by unzipping.
Bockelmann et al. applied mechanical force perpendicular to the dsDNA backbone (Fig. 2a).31 They found that such unzipping was able to de-hybridize duplex DNA into two single-stranded DNA (ssDNA) strands. Due to the difference between G/C and A/T Watson–Crick base pairs, the mechanical force changed at this stage, oscillating back and forth around 15 pN.31 The stability of G/C pairs significantly exceeds that of A/T pairs.32–34 For example, Rief et al. showed that mechanically unzipping force of a poly-A/T duplex was 10 pN while that of a poly-G/C duplex was 20 pN.17
2.1.2. Mechanical stability of dsDNA by shearing.
When a force is applied along the long axis of the duplex DNA arranged in an antiparallel fashion (Fig. 2b), shearing of duplex DNA may occur.35 The dsDNA shearing contains two stages: a nonlinear elastic behavior followed by mechanical disintegration.36 The shearing force increases with the length of the dsDNA within a certain limit.35 Strunz group found that the mechanical shearing force of a 30-base pair dsDNA was about 46–50 pN.36 For shorter duplex DNA (10–30 bp), the mechanical shearing force was about 20–50 pN.36
2.1.3. Mechanical stability of pi–pi stacking.
The stacking force exists between adjacent base pairs along two spiral DNA backbone strands (Fig. 2c).20,37 By a dual-beam optical tweezer instrument,20 Dietz group found single base stacking had a mechanical stability on the order of 3 pN. The change in free-energy of base stacking decreases according to the following order: (CG:GC) or (AT:TA) base-pair stacking > (AT:AT) > (CG:AT). The trend is caused by different stacking modes between pyrimidines and purines. They found that larger stacking areas had longer lifetimes and stronger stability with respect to smaller stacking areas. According to different sequence combinations, stacking areas and salt conditions, the expected lifetime of the stacked array can be obtained.38,39
2.2. Holliday junction and other secondary DNA structures in DNA origami
2.2.1. Holliday junctions.
The Holliday junction serves as a basic infrastructure in DNA origami (Fig. 3a). Mechanical stability of individual arms of a Holliday junction by unzipping is equivalent to that of duplex DNA (~15 pN) discussed in Section 2.1. The mechanical force at which two conformation isomers of a Holliday junction (Fig. 3a) reach equilibrium is rather weak, on the order of 0.5 pN.40 On the other hand, a structurally similar DNA structure, cruciform, has a mechanical stability up to 50 pN under torsionally constrained conditions.41 Such a significant difference has been ascribed to the cooperative unfolding of the two cruciform arms in a positively supercoiled template. Mao group systematically investigated the mechanical property of Holliday junctions in different DNA origami structures.24 Overall, they found that the mechanical stability of nanopyramids was weaker than that of nanotubes, both of which (nanopyramids and nanotubes) showed higher mechanical stabilities than nanotiles (Fig. 3b). In addition, compared with longitudinal stretching, higher force was required to disassemble the nanotubes during horizontal stretching. All these suggested that Holliday junctions had anisotropic mechanical properties, which was rationalized by the anisotropic arrangement of Holliday junctions along specific mechanical unfolding directions (Fig. 3c and d).24
Fig. 3.
Mechanical stability of DNA origami structures is determined by Holliday junctions. (a) Schematic of two structural isomers of a Holliday junction13 (reprinted with permission from ref. 13, copyright (2017) American Chemical Society). (b) Comparison of mechanical stabilities of four different DNA origami structures. Tiles: nanotiles, Py: nanopyramid, 8T(L): 8-tube DNA (Longitudinal direction), 6T(L): 6-tube DNA (Longitudinal direction), 6T(H): 6-tube DNA (Horizontal direction) and 8T(H): 8-tube DNA (Horizontal direction)24 (reprinted with permission from Oxford University Press). (c) and (d) show the disintegrations of Holliday junctions under horizontal and longitudinal tensions, respectively24 (reprinted with permission from Oxford University Press).
2.2.2. H-DNA, G-quadruplex (GQ), and i-Motif (iM).
In addition to Holliday junctions, other DNA secondary structures are occasionally employed in DNA origami nanoassemblies. H-DNA is formed between homopurine–homopyrimidine tracts that fold into a triplex (Fig. 4a).42 The structure showed a mechanical stability around 20 pN in physiologically relevant buffers.43,44
Fig. 4.
DNA secondary structures. (a) an H-DNA structure (the oval links indicate Hoogsteen hydrogen bonds), (b) a G-quadruplex structure and (c) an i-Motif structure. The M+ in the chemical structure in (b) refers to a monovalent ion such as Na+.46,47
G-quadruplex is composed of a stack of G-quartets (Fig. 4b),45 which are connected together by Hoogsteen hydrogen bonds. The structure is further stabilized by intercalating monovalent cations such as Na+.46 Mao group and others determined mechanical stabilities of G-quadruplexes were on the order of ~25 pN.47
Another secondary structure, i-Motif,48 is made of a stack of hemiprotonated cytosine–cytosine pairs (Fig. 4c).49 Because of the requirement of hemiprotonated cytosines, i-Motif is mainly formed in a slightly acidic environment around pH 5.5.49 Mao group has determined its mechanical stability around 30 pN.49
Recently, new DNA secondary structures have been used in DNA origami structures. Sen group constructed DNA nanostructures using G-triplex,50 which has presented a mechanical force around 32 pN.51 Li et al. assembled poly(thymine) into antiparallel duplexes in presence of melamine, which connected to two thymines in the same plane via hydrogen bonding.52 The average mechanical stability of a stack of thymine–melamine–thymine was about twice (26.2 pN) as the that of AT pairs, albeit with a much wider unzipping force range.52 These synthetic DNA secondary structures increase the versatility of DNA origami nanoassemblies.
2.3. Significance of the mechanical stability of DNA secondary structures
As a basic component in origami nanoassemblies, DNA is a naturally occurring material in cells. Therefore, it is not surprising that mechanical stabilities of various DNA secondary structures lie within the reach of many biological machineries such as motor proteins53,54 that process DNA templates. This feature renders DNA origami an ideal nano-assembled material to interfere with various biological processes. However, for a nanomaterial to be applied inside cells, it is important that components of DNA origami should withstand the hydrophobic force in cell membranes during the cell entry of these origami nanoassemblies. Here we take transmembrane DNA origami channels as examples to illustrate that the mechanical stabilities of these DNA secondary structures in DNA origami can indeed survive the hydrophobic stress inside phospholipid membranes.55–57
In one approach, Howorka and colleagues used a six-helix bundle (6 HB) to design transmembrane DNA nanopores (Fig. 5).55 The nanopore was modified with tetraphenylporphyrins (TPPs) to be anchored in phospholipid bilayers. Lipid modifications such as cholesterols were also used to anchor nanopores in lipid bilayers.56,57
Fig. 5.
A tetraphenylporphyrins (TPP) labelled DNA origami nanopore was anchored in a phospholipid bilayer55 (reproduced from ref. 55 with permission from John Wiley and Sons, copyright 2013).
These DNA nanopores are rather stable in phospholipid membranes. Given that hydrophobic forces of phospholipid bilayer are on the order of tens of picoNewtons,58 such examples clearly demonstrated that DNA origami can withstand the stress of hydrophobic forces in membranes, which paves the way to carry out biological applications inside cells after DNA origami nanodevices pass through cell membranes.
Once inside cells, DNA secondary structures in DNA origami may be compromised by intracellular proteins including various nucleases.59,60 After studying the interactions between DNA nanostructures and proteins, Castronovo group found that mechanical properties of DNA origamis, such as DNA packing density, local or super structures, and intactness of DNA staples, affected enzymatic activities on DNA nanostructures.59,61 While using non-B DNA structures may inhibit the digestion of various enzymes on DNA origami nanoassemblies, these structures may compromise the efficiency of the loading of non-intercalating drugs.62 Other approaches to maintain structural integrity of DNA origami inside cells include the introduction of sharp shape in the origami structure,61 enzymatic ligations and chemical modifications,63,64 as well as photochemical crosslinking.65,66
3. Applications of DNA origami nanoassemblies by exploiting four mechanical properties
3.1. Reversible elasticity regime
Similar to a macroscopic object, elasticity of DNA origami is originated from the changes in the backbone enthalpy and conformational entropy of a DNA nanoassembly in response to external forces. When the external force is small, such elastic response is fully reversible, facilitating the repetitive usage of DNA origami structures.
To be used as a quantitative force measurement tool, Korber and Dietz designed a force spectrometer in which supporting components and elastic sectors in DNA origami formed a spring clip structure (Fig. 6a).8,67–69 Because of the honeycomb structure in the DNA origami, two origami levers showed strong stiffness.6,19,56 A spring hinge was added to convert these two levers into the force spectrometer. The authors found that the force exerted by the spring can offset the attraction between two nucleosomes.70 The authors also used this force spectrometer to detect salt-induced disassembly of nucleosome core particles, from which they obtained binding constants and the energetic penalties for nucleosome integrations.67
Fig. 6.
DNA origami structures exploiting reversible elasticity properties. (a) A force spectrometer. The red cylinder represents the spring and hinge8,67 (reprinted from ref. 8 of AAAS). (b) Nanoclamps that maintain 0 pN, 6 pN, and 12 pN tension in ssDNA71 (reprinted with permission from AAAS). (c) Adjustable DNA geometry components (Left: relaxed state; Right: tightened state)72 (reprinted with permission from ref. 72, copyright (2014) American Chemical Society). (d) A bistable nanomechanism with three states73 (reprinted with permission from ref. 73, Copyright (2015) American Chemical Society). Circular diagrams to the left of (c) and (d) represent cross sections (helix bundles) of DNA origami backbones.
In other examples, Liedl et al. designed a DNA nanoclamp that can produce 0–50 pN to study the bending of duplex DNA induced by a TATA-binding protein (Fig. 6b).71 Su and Castro developed an adjustable curved DNA device (Fig. 6c),72 which can carry out controllable bending by adjusting the length of a hinged ssDNA.
Elastic levers represent another type of applications. Su and Castro groups prepared a DNA nanodevice with rigid links (blue and green) and a compliant link (red) (Fig. 6d).73 When the nanodevice was deformed by external factors, the mechanical energy could be stored in the compliant link. This energy could be calculated from the bending angle and elastic properties of the origami device.
Elasticity has been exploited to build nanosprings. To observe mechanical movement of myosin VI and associated mechanical force changes, Shih group constructed a DNA nanospring (Fig. 7a)5 with a spring constant 0.012 ± 0.002 pN nm−1. Compared with traditional nanosprings made of ssDNA, this nanospring had a more gradual change in spring constant in the range of several pNs. Recently, an environmentally responsive DNA nanospring was demonstrated in the Mao group. By incorporating i-Motif, this DNA nanospring was responsive to environmental pH variation (Fig. 7b).74 At slightly acidic pH where cancer cells usually experience, the origami was coiled into a nanospring, inhibiting the movement of cancer cells via clustered RGD-integrin interactions. At neutral pH under which healthy cells experience, the nanospring was stretched, which did not affect the motion of cells. Such a device can be used to selectively target metastasis of cancer cells without affecting the property of healthy cells. Measurement of spring constant of this device using force-jump approach revealed that these nanosprings were 50 times stiffer than that obtained in the Shih lab,5 which suggests more mechanobiological applications can be carried out using these nanosprings.75
Fig. 7.
DNA nanosprings. (a) A programmable nanospring is applied to myosin VI heads5 (reprinted with permission from Springer Nature). (b) A pH sensitive DNA nanospring controls cell motions74 (reprinted with permission from ref. 74, copyright (2021) American Chemical Society).
3.2. Reversible pliability regime
Pliability refers to the resistance of a material to reversible deformation under external forces. It can reflect materials’ flexibility, stiffness, and bending resistance. The parameter that can represent the degree of pliability is persistence length.76–79 The greater the persistence length, the greater the stiffness of the material. Persistence lengths of basic origami components such as dsDNA and ssDNA are ~50 nm80 and ~1 nm,81 respectively. In DNA origami nanoassemblies, due to combined helical bundles made of duplex DNA strands, the persistence length is often in the micrometer range.19,20,79 The much-increased persistence length suggests much stiffer DNA origami structures, which has been harnessed for applications that require rigid frameworks.
3.2.1. Rigid levers.
The introduction of optical/magnetic tweezers has provided unprecedented mechanical information on individual macromolecules and assemblies. These targets are often linked to optically and magnetically trapped beads via linkers such as DNA.82 Since DNA is soft, noise in mechanical measurement is significant.
To address this problem, Dietz group used 10–12 HB DNA origami beams as linkers for mechanical force transmission.19,20 A 10 HB beam has persistence length ~3.5 microns.20 These rigid linkers therefore effectively increased the signal-to-noise ratio during mechanical unfolding experiments. Resolutions on the order of 5 nm per 2 ms in the mechanical unfolding of DNA hairpins have been achieved.19
3.2.2. Rigid nanocages.
Mao and collaborators constructed a series of hollow cuboid nanocages (Fig. 8a)83–85 that serve as nanoconfinement to investigate the folding and unfolding of DNA structures such as G-quadruplexes, i-Motifs, and duplex DNA. Their studies revealed that duplex DNA had reduced mechanical stability in nanoconfinement (unzipping force ~9.4 pN vs. ~20.2 pN without confinement) whereas G-quadruplexes and i-Motifs demonstrated nearly 2 times stronger mechanical stabilities (~38 pN, for G-quadruplex) with respect to free structures (~20 pN, for G-quadruplex in solution). Such results suggest a new way to control mechanical properties of DNA origamis structures by using confined environment.
Fig. 8.
Rigid DNA origami nanocages that provide nanoconfinement to the folding and unfolding of biomolecules (a) ref. 83–85 (reproduced from ref. 83 with permission from Springer Nature, copyright 2017), that host proteins to obtain high-resolution structures (b) ref. 6 (reproduced from ref. 6 with permission from PNAS, copyright 2016; the blue bundles indicate the orientation of the nanocage), and that serve as templates to grow nanoparticles (c) ref. 86 (reproduced from ref. 86 with permission from ACS, copyright 2014).
In another example, Dietz and Scheres designed a hexagonal prism-like hollow columnar structure (Fig. 8b),6 in which orientation of transcription factor p53 was constrained. This allowed the decipher of higher resolution structures of p53. Similarly, Seidel group used double-layer structure to manufacture a DNA origami mold (Fig. 8c),86 which served to prepare metal nanoparticles with specific compositions and shapes. Recently, a stiff cage with three-layer DNA origami design was prepared by Bathe and Yin,3 which showed improved nanometer precision for preparations of inorganic nanostructures.
3.2.3. Other rigid structures.
DNA origami has been used as a rigid template to prepare metamaterials and biosensors.87,88 To obtain chiral plasma signals, Wang group first synthesized a 2D DNA origami plate. After modifying the plate with gold nanorods (AuNRs), the plate rolled into a cylinder on the surface of AuNRs (Fig. 9a).89 Then, gold nanoparticles (AuNPs) were attached at different locations of the cylinder, resulting in either left-handed or right-handed AuNP helices. In the sensing application, chiral plasma signals in stiff origami hosting templates have been used to detect adenosine molecules (Fig. 9b). In this sensor, binding of adenosine targets changed the relative position between two origami arms, which varied the chiral plasma of the AuNRs attached to the two arms.90 Similar strategies have been exploited in other applications such as fabrication of AuNR trimers for chirality manipulations and controllable assembly of 3D anisotropic nanomaterials.91–95 In biochemical applications, rigid DNA origami rotor blades with high torsional stiffness have been used to measure rotations caused by nucleic acid processing enzymes.96
Fig. 9.
Rigid DNA structures serving as templates for (a) a chiral plasma device89 (reproduced from ref. 89 with permission from John Wiley and Sons, copyright 2017) and (b) a biosensing device90 (reproduced from ref. 90 with permission from ACS, copyright 2018).
3.2.4. Bending resistance and flexibility.
Bending resistance and flexibility are other examples of pliability. Under appropriate forces, the pliability allows DNA origami to undergo limited deformation while still maintaining the integrity of the overall structure.
Based on the DNA tile-tube assembly,97 Maier et al. prepared artificial flagella by assembly of short ssDNA fragments, which have different mechanical properties in twist diameter, stiffness, bending stiffness, and flexibility (Fig. 10a).98,99 These flagella have shown swimming capabilities, which can serve as biocompatible nanorobots. Inspired by Yin’s work,97 Smith group used structurally tunable DNA nanotubes to form semiflexible polymers with entangled networks.79 Their persistence lengths ranged from 1.2 to 26 μm with other interesting mechanical properties such as adjustable bending stiffness.79
Fig. 10.
Schematics of (a) magnetic beads modified with artificial flagella which move in an external magnetic field (B)98 (reprinted with permission from ref. 98, the direct link is DOI: 10.1021/acs.nanolett.5b03716 and further permissions related to this material excerpted should be directed to the ACS) and (b) temperature-controlled morphological change of nanocages. From left to right, a contracted nanocage (4 °C), a cargo containing nanocage (37 °C), a packaged nanocage (4 °C), and the nanocage with the released cargo (37 °C)100 (reprinted with permission from ref. 100, copyright (2013) American Chemical Society).
Juul et al. designed a DNA nanocage based on temperature responsive DNA structures (Fig. 10b).100 Among six nanocage corners made of 3-nt thymidine linkers, one corner contained four pieces of 32-nt ssDNA.101,102 This corner was tightened by the folding of hairpins in the ssDNA fragments at low temperatures, which would melt at an elevated temperature. In contrast, short thymidine linkers did not show this conformation change, leading to temperature dependent morphology change in the DNA nanocage. This property allowed temperature actuated releasing of cargos contained inside the nanocage.
From these examples, it is clear that bending resistance and flexibility allow ssDNA with ~1 nm persistence length to be assembled into flexible and functional structures, which have greatly enriched the structural complexity and expanded the application scope of DNA origami devices.
3.3. Irreversible plasticity regime
Compared to elastic deformation which is reversible, when an object undergoes irreversible deformation under sufficiently high force, the object demonstrates its plasticity property which is irreversible.103,104 For DNA origami, plasticity can be affected by physical, mechanical and chemical conditions.105,106 The coupling between physicochemical environment and morphology of the DNA origami renders origami nanoassemblies ideal mechanochemical platforms to report changes in the chemical or physical surroundings. Indeed, many applications have exploited these traits in DNA origami devices, which can be categorized by localized plasticity and system plasticity according to the regions affected by external forces.
3.3.1. Localized plasticity.
When physical, chemical, or mechanical stimuli exert on a specific region in an origami device, only the affected region produces irreversible structural and/or functional changes. This response is defined as localized plasticity.
Based on a brick-like nanocage whose cavity surface was modified with photolabile cross-linkers,3,107,108 Kohman et al. packaged a cargo inside a nanocage cavity via these photolabile linkers. The nanocage released the cargo under light by breaking these localized photolabile linkers (Fig. 11a).109 The size of the cavity could be varied to load different cargos ranging from small molecules to proteins.
Fig. 11.
Examples of localized plasticity in DNA origami nanoassemblies. (a) Light-sensitive transporting nanocapsules109 (reprinted with permission from American Chemical Society). (b) Scissor-like origami probes110 (reprinted with permission from The Royal Society of Chemistry). (c) A 7-tile DNA origami mechanochemical sensor for PDGF and nucleic acid detections4 (reprinted with permission from John Wiley and Sons). (d) A nanoscale DNA box for drug delivery2 (reprinted with permission from Springer Nature). (e) Nanorobots7 that can be actuated in a logic gate fashion by ligands binding to the aptamers containing locks shown in the left boxes (reprinted with permission from AAAS).
In Yamazaki’s strategy, invasive binding of a peptide nucleic acid (PNA) transformed a stick-like origami structure into an irreversible scissor-like structure (Fig. 11b),110 which can be used to report the binding of nucleotide analogues such as PNA. Mao group designed a 7-tile DNA origami nanoassembly for multiplex mechanochemical sensing of a platelet-derived growth factor (PDGF) and/or a complementary nucleic acid fragment (Fig. 11c).4 When binding to a target, the seven DNA origami tiles would sequentially decouple under 10–25 pN force, generating mechanochemical signals in optical tweezers.7,111
Drug delivery requires efficient transportation of cargos to designated locations where the payload can be released in response to external cues. Andersen et al. constructed a DNA origami box (Fig. 11d),2 which can be opened when oligonucleotides bind and unlock the cover.112 The cavity of these boxes was large enough to contain large biomolecular assemblies such as ribosomes. Likewise, Church et al. synthesized a hollow hexagonal barrel as a nano-transportation robot (Fig. 11e).7,107 The lock for this robot consisted of aptamer-containing DNA duplexes which would be unlocked upon binding of molecular targets to the aptamers arranged according to a specific logic gate pattern (i.e. AND, OR, etc.).
3.3.2. System plasticity.
In contrast to the localized plasticity where the irreversible morphology or functional change occurs only at localized area in response to external stimuli, system plasticity refers to the irreversible topology change of the entire origami device.
The ssDNA probe demonstrated by the Yan group perhaps represents the simplest example of system plasticity (Fig. 12a).105 Made of 20-nt ssDNA, these soft probes did not show distinct signals under AFM scanning. When hybridized with complementary single-stranded RNA, the duplex structure became stiffer, showing a V-shaped rigid structure clearly distinguished by AFM.
Fig. 12.
Examples of system plasticity in DNA origami nanoassemblies. (a) An ssDNA probe105 (reprinted with permission from AAAS). (b) Electrostatically wrapped targets for cell delivery113 (reprinted with permission from ref. 113, Copyright (2014) American Chemical Society). (c) Dynamic morphological change in a DNA domino device115–117 (reprinted with permission from ref. 117, Copyright (2020) American Chemical Society). Arrows in the second panel of (c) depict the locations to bind oligonucleotides that trigger subsequent structural changes in nanodevices.
To encapsulate virus capsid proteins (CPs) more efficiently, Kostiainen et al. let positively charged CPs bind to origami structures (Fig. 12b),113,114 which reduced the repulsion between negatively charged DNA helixes. Since each CP slightly bent the rectangular origami, the origami plate rolled into a column, enhancing the transfer efficiency of the CPs into the cells.
Using concepts of dynamic DNA units known as anti-junctions,115,116 Ke and Song groups prepared reconfigurable DNA origami arrays (Fig. 12c).115 Because anti-junctions could switch between two stable conformations, the structure change in the anti-junction would propagate the change in other anti-junctions, leading to the conversion of entire origami structures just like dominoes. The same idea has led the groups to develop a reconfigurable DNA origami domino array-based dynamic pattern operation (DODADPO) system117 in which structural transformations were achieved with incorporation of more functionalities in nanodevices. This allowed to explore more potential applications such as platforms for chemical syntheses.
3.4. Irreversible disintegration regime
At even higher external forces with respect to those experienced by the DNA origami in the plasticity regime, the DNA nanoassembly may disintegrate irreversibly. This disintegration has been cleverly exploited for sensing applications (Section 3.4.1) as well as to study the mechanism of the assembly and disassembly of DNA origami nanoassemblies (Section 3.4.2).
3.4.1. Applications exploiting disassembly and assembly processes.
Chen et al. used a DNA hairpin to detect traction force of adherent cells (Fig. 13a).118 The hairpin with 5.7–16.5 pN mechanical stability served as a bridge connecting the target cell and the substrate. When a cell moved, its traction force disassembled the hairpin, resulting in longer distance between a fluorophore and a quencher. This decreased FRET (Fluorescence Resonance Energy Transfer) efficiency between the fluorophore and the quencher, causing increased fluorescence signal. Similar strategies have been used to measure tensile forces between adjacent cells.119–122
Fig. 13.
Examples exploiting the mechanical stability of DNA assemblies. (a) The DNA hairpin probe to detect the traction force of cells118 (reprinted with permission from Springer Nature). (b) A DNA origami UV radiometer123 (reprinted with permission from ref. 123, copyright (2020) American Chemical Society).
Another example exploiting the disassembly of DNA origami came from the Liu and Wang groups (Fig. 13b)123 in the construction of an ultraviolet light radiometer. Given that ultraviolet light can damage DNA, the basic component in a DNA origami device, integrities of particular DNA origami nanoassemblies were monitored under AFM to reflect the damaging UV intensity in the environment.
The irreversible disassembly exploited in the radiometer does not allow repetitive usage of DNA origami devices. To address this problem, Scheckenbach et al. exploited the self-repair strategy in DNA origami structures.124 They used strand exchange to facilitate the self-healing of DNA origami structures at damaged locations. However, for origami structures whose damage locations are not known, such a strategy requires a whole set of displacing DNA staples, which is costly and requires special staple designs to facilitate the displacement. In addition, this method does not apply to the case where the damage occurs in the template strand.
3.4.2. Mechanics of assembly and disassembly of DNA origami nanodevices.
To better understand the mechanics of DNA origami structures, it is important to monitor the assembly or disassembly of individual DNA origami nanoassemblies under external forces (Fig. 14). Tracking individual DNA origami structures in the force-based approaches provided much increased temporal and spatial resolution to follow the assembly and disassembly processes. In addition, the use of mechanical unfolding can directly synchronize these processes. Such studies on DNA origami folding mechanisms often start with mechanical unfolding of a particular origami nanoassembly, which disintegrates the device irreversibly due to the loss of DNA staples.
Fig. 14.
Force induced disassembly of a DNA origami structure followed by self-assembly of the DNA structure. Blue staples depict those disassembled first during the mechanical unfolding process.
Using AFM and FRET spectroscopy to follow the assembly process of a DNA device after its mechanical unfolding, Saccà group proposed a dynamic model for the assembly process.125 In this model, the initial folding of DNA origami and the topological stress in the nucleation sites demonstrated their critical roles in DNA origami assembly. While the former process determined the whole energy landscape, the latter profoundly affected the final stability and the topology of the DNA origami.
In another work, Yoon and colleagues first used magnetic tweezers (MT) to mechanically unfold DNA origami nanoassemblies.25 Upon relaxing the mechanical force on the stretched template strand, the lower entropy state of DNA origami then folded into various intermediate structures in the presence of staple strands while avoiding unnecessary secondary structures. Finally, displacement reactions took place to overcome energetic barriers, which helped to remove redundant staple strands. In a mechanical model proposed by Chen et al.,21 these barriers could come from overcoming duplex DNA twisting and accommodating local conformations to desired global structures. The three-step process made DNA origami mechanically stable due to the compliance with entropy.
Taking together, these studies suggested that DNA origami follows a self-assembly path to lower its entropy, which is compensated by enthalpic energy released from the hybridization between the staples and the long scaffold template. Since the initial nucleation determines the topology and mechanical property of the final origami state, it is important to control the preparation conditions for the DNA origamis. For example, to obtain reproducible origami structures each time, thermodynamic equilibrated condition should be maintained for the DNA nanoassembly. On the other hand, kinetic conditions can be explored at the nucleation state to obtain DNA origami structures with desirable mechanical properties.
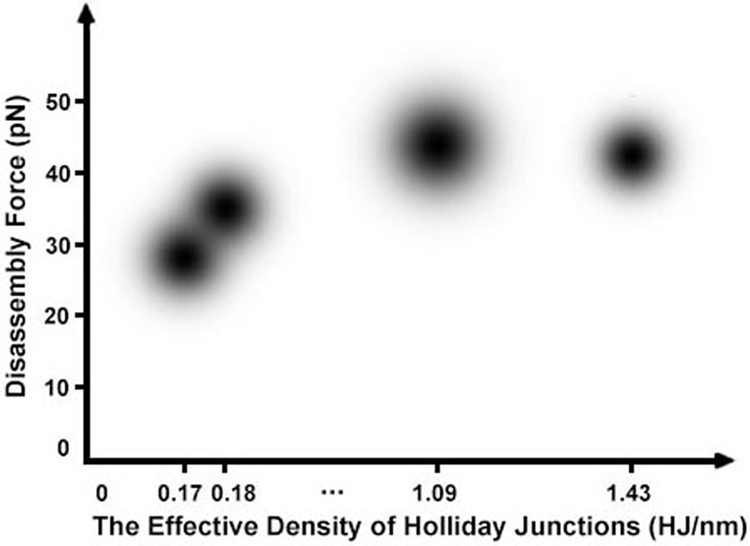
By assembling long DNA template and short strands of DNA staples together, DNA origami reshapes the physical limit of DNA materials. Duplex DNA has a relatively short persistence length of ~50 nm while its mechanical unzipping stability is considered to be low (about 15 pN),31 both of which are compatible with innate constraints of biological environment,53,54,58 For individual Holliday junctions, the mechanical stability and isomerization force are also low.40 However, DNA origami shows at least twice stronger in mechanical stability (>30 pN)4,24 and ~20 times longer in persistence length20,79 compared to duplex DNA. With respect to individual Holliday junctions, the isomerization force of DNA origami is ~60 times higher.24,40 All these indicate that DNA origami as a whole has much stronger and stiffer properties. While persistence length can be explained by the helical bundles made of multiple duplex DNA strands employed in the DNA origami assembly, the mechanical stability can be rationalized by the effective density of Holliday junctions (Fig. 15), which has shown a positive correlation between mechanical stability and the density of Holliday junctions along a particular direction of applied force.24 The critical role of Holliday junctions in the DNA origami assembly has been confirmed by the computer simulation in which removal of some DNA staples leads to easier accessibility of restriction enzymes to compromise the origami structure, likely due to the more flexible origami framework.59
Fig. 15.
Relationship between disassembly force (pN) of DNA origami nanoassemblies and densities of Holliday Junctions (HJ nm−1) along the direction of applied force. This plot uses published data.24
4. Conclusions and prospects
In summary, we have discussed the properties and applications of DNA origami according to the four fundamental mechanical regimes, reversible elasticity and pliability, as well as irreversible plasticity and stability, in response to external stimuli. Almost all applications of DNA origami nanoassemblies can be categorized into these four mechanical regimes. Different properties of these four regimes have been rationalized by the collective assembly of basic components in DNA origami: duplex DNA and Holliday junctions.
Challenges exist for current research and development of DNA origami nanoassemblies. First, for all four mechanical regimes (pliability, elasticity, plasticity and stability), the applicable mechanical force range is limited for DNA origami. It is necessary to expand the force responsive range for DNA origami (either strengthening or weakening) by incorporating other materials such as small molecules,52 polymers,126 nanometallic particles, silica coating,127,128 and carbon nanotubes, among others. In doing so, not only can DNA origami components withstand greater mechanical force, but also their properties can be more precisely regulated at smaller force ranges. Currently, DNA origami has high programmability and spatial precision due to the presence of duplex DNA. Given there are only four bases in DNA, the chemical diversity of DNA origami framework is limited. In addition, it is still expensive to scale up DNA based materials. With the incorporation of other synthetic materials, DNA origami’s mechanical properties will be diversified while cost can be reduced. In particular, in the plasticity regime, incorporation of synthetic functional groups is expected to drastically expand the capability of DNA origami to respond to physical (temperature, light, and force) and chemical stimuli. In another approach, Gerling et al. demonstrated that DNA components assembly by shape-complementarity rather than base pairing can produce sturdy micrometer-scale objects.38 Such shape-complementarity is likely due to the excluded volume effect.129,130 It is interesting to directly measure the mechanical force of this effect, which is yet to be achieved.
Second, it has been successfully demonstrated that a hierarchical network in nanoassembly can drastically improve its mechanical properties.131–133 In a recent report, such hierarchical structures have shown to improve effective Young’s modulus.134 Other studies have shown high tensile elasticity132 and exceptional stiffness135 in hierarchical structures. We propose that similar hierarchical structures can be incorporated in the DNA origami nanoassemblies. In current strategies, Holliday junctions serve as fundamental crosslinks in the DNA origami framework. Since the Holliday junction has weak mechanical properties, mechanical isomerization force in particular,40 we argue other DNA secondary structures can be used. One good example is DNA quadruplexes. These structures are mechanically more stable than duplex DNA.47 The four-stranded topology in G-quadruplex is expected to be compatible with current design in DNA origami. In addition, long range assembly of G-quadruplex junctions is feasible, increasing the level of hierarchical topology. Recent demonstration of using the G-triplex in the building of DNA nanoassemblies has indicted the feasibility of this approach.50
Finally, most mechanical characterization of DNA origami devices uses single-molecule force instruments such as optical tweezers,19,24 magnetic tweezers,25 and AFM.125 All these instruments have low throughput. For scale-up applications of DNA origami nanoassemblies, the molecule-by-molecule characterization and demonstration of DNA nanodevices become a bottleneck to expand the use of this new material. Therefore, new mechanical characterization devices and approaches in a high-throughput manner136 become an imminent call to further the development of DNA origami nanomaterials.
Biography

Jiahao Ji
Jiahao Ji obtained his BS degree from School of Chemistry and Chemical Engineering, Nanjing University. Currently, he is a graduate student in Prof. Hanbin Mao’s research group at the Department of Chemistry and Biochemistry, Kent State University, focusing on single-molecule mechanochemical studies.

Deepak Karna
Deepak is currently a PhD candidate in the Department of Chemistry & Biochemistry, Kent State University, under the supervision of Prof. Hanbin Mao. He obtained his undergraduate degree in Biotechnology from Kathmandu University, Nepal. His major research interest lies in bionanotechnology targeted with DNA origamis.

Hanbin Mao
Professor Hanbin Mao got his PhD training in Analytical Chemistry at Texas A&M University in 2003. After two years of postdoctoral research at UC Berkeley specializing in single-molecule force spectroscopy, Dr Mao joined the Department of Chemistry and Biochemistry at Kent State University in 2005. His research is focused on biosensing and single molecular biophysics. His research lab in Kent State has developed a new interdisciplinary field, mechano-analytical chemistry, in which mechanical properties of molecules have been exploited for (bio)chemical analyses and mechanochemistry studies.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Rothemund PWK, Nature, 2006, 440, 297–302. [DOI] [PubMed] [Google Scholar]
- 2.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV and Kjems J, Nature, 2009, 459, 73–76. [DOI] [PubMed] [Google Scholar]
- 3.Sun W, Boulais E, Hakobyan Y, Wang WL, Guan A, Bathe M and Yin P, Science, 2014, 346, 1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koirala D, Shrestha P, Emura T, Hidaka K, Mandal S, Endo M, Sugiyama H and Mao HB, Angew. Chem., Int. Ed, 2014, 53, 8137–8141. [DOI] [PubMed] [Google Scholar]
- 5.Iwaki M, Wickham SF, Ikezaki K, Yanagida T and Shih WM, Nat. Commun, 2016, 7, 13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martina TG, Bharata TAM, Joergera AC, Baia X-C, Praetoriusd F, Fershta AR, Dietzd H and Scheres SHW, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, E7456–E7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas SM, Bachelet I and Church GM, Science, 2012, 335, 831–834. [DOI] [PubMed] [Google Scholar]
- 8.Funke JJ, Ketterer P, Lieleg C, Schunter S, Korber P and Dietz H, Sci. Adv, 2016, 2, e1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeman NC, Annu. Rev. Biochem, 2010, 79, 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro AV, Han D, Shih WM and Yan H, Nat. Nanotechnol, 2011, 6, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo M, Yang Y and Sugiyama H, Biomater. Sci, 2013, 1, 347–360. [DOI] [PubMed] [Google Scholar]
- 12.Simmel SS, Nickels PC and Liedl T, Acc. Chem. Res, 2014, 47, 1691–1699. [DOI] [PubMed] [Google Scholar]
- 13.Hong F, Zhang F, Liu Y and Yan H, Chem. Rev, 2017, 117, 12584–12640. [DOI] [PubMed] [Google Scholar]
- 14.Dey S, Fan CH, Gothelf KV, Li J, Lin CX, Liu LF, Liu N, Nijenhuis MAD, Saccà B, Simmel FC, Yan H and Zhan PF, Nat. Rev. Methods Primers, 2021, 1, 13. [Google Scholar]
- 15.Bustamante C, Smith SB, Liphardt J and Smith D, Curr. Opin. Struct. Biol, 2000, 10, 279–285. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S and Li MS, Phys. Rep, 2010, 486, 1–74. [Google Scholar]
- 17.Rief M, Clausen-Schaumann H and Gaub HE, Nat. Struct. Biol, 1999, 6, 346–349. [DOI] [PubMed] [Google Scholar]
- 18.Clausen-Schaumann H, Rief M, Tolksdorf C and Gaub HE, Biophys. J, 2000, 78, 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfitzner E, Wachauf C, Kilchherr F, Pelz B, Shih WM, Rief M and Dietz H, Angew. Chem., Int. Ed, 2013, 52, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilchherr F, Wachauf C, Pelz B, Rief M, Zacharias M and Dietz H, Science, 2016, 353, aaf5508. [DOI] [PubMed] [Google Scholar]
- 21.Chen HR, Weng T-W, Riccitelli MM, Cui Y, Irudayaraj J and Choi JH, J. Am. Chem. Soc., 2014, 136, 6995–7005. [DOI] [PubMed] [Google Scholar]
- 22.Seaton DT, Schnabel S, Landau DP and Bachmann M, Phys. Rev. Lett, 2013, 110, 028103. [DOI] [PubMed] [Google Scholar]
- 23.Jin YL, Urbani P, Zamponi F and Yoshino H, Sci. Adv, 2018, 4, eaat6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha P, Emura T, Koirala D, Cui YX, Hidaka K, Maximuck WJ, Endo M, Sugiyama H and Mao HB, Nucleic Acids Res, 2016, 44, 6574–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae W, Kim K, Min D, Ryu J-K, Hyeon C and Yoon T-Y, Nat. Commun, 2014, 5, 5654. [DOI] [PubMed] [Google Scholar]
- 26.Engel MC, Smith DM, Jobst MA, Sajfutdinow M, Liedl T, Romano F, Rovigatti L, Louis AA and Doye JPK, ACS Nano, 2018, 12, 6734–6747. [DOI] [PubMed] [Google Scholar]
- 27.Liphardt J, Onoa B, Smith SB, Tinoco LJ and Bustamante C, Science, 2001, 292, 733–737. [DOI] [PubMed] [Google Scholar]
- 28.Carrion-Vazquez M, Li HB, Lu H, Marszalek PE, Oberhauser AF and Fernandez JM, Nat. Struct. Mol. Biol, 2003, 10, 738–743. [DOI] [PubMed] [Google Scholar]
- 29.Jonchhe S, Ghimire C, Cui YX, Sasaki S, McCool M, Park S, Iida K, Nagasawa K, Sugiyama H and Mao HB, Angew. Chem., Int. Ed, 2019, 58, 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans E, Annu. Rev. Biophys. Biomol. Struct, 2001, 30, 105–128. [DOI] [PubMed] [Google Scholar]
- 31.Bockelmann U, Thomen P, Essevaz-Roulet B, Viasnoff V and Heslot F, Biophys. J, 2002, 82, 1537–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bockelmann U, Essevaz-Roulet B and Heslot F, Phys. Rev. Lett, 1997, 79, 4489–4492. [Google Scholar]
- 33.Bockelmann U, Essevaz-Roulet B and Heslot F, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top, 1998, 58, 2386–2394. [Google Scholar]
- 34.Essevaz-Roulet B, Bockelmann U and Heslot F, Proc. Natl. Acad. Sci. U. S. A, 1997, 94, 11935–11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash S and Singh Y, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys, 2011, 84, 031905. [DOI] [PubMed] [Google Scholar]
- 36.Strunz T, Oroszlan K, Schäfer R and Güntherodt H-J, Proc. Natl. Acad. Sci. U. S. A, 1999, 96, 11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kool ET, Annu. Rev. Biophys. Biomol. Struct, 2001, 30, 1–22. [DOI] [PubMed] [Google Scholar]
- 38.Gerling T, Wagenbauer KF, Neuner AM and Dietz H, Science, 2015, 347, 1446–1452. [DOI] [PubMed] [Google Scholar]
- 39.Ketterer P, Willner EM and Dietz H, Sci. Adv, 2016, 2, e1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DMJ and Ha T, Science, 2007, 318, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal S, Selvam S, Cui YX, Hoque ME and Mao HB, ChemPhysChem, 2018, 19, 2627–2634. [DOI] [PubMed] [Google Scholar]
- 42.Tiner WJS, Potaman VN, Sinden RR and Lyubchenko YL, J. Mol. Biol, 2001, 314, 353–357. [DOI] [PubMed] [Google Scholar]
- 43.Mao HB, unpublished data.
- 44.Li N, Wang JL, Ma KK, Liang L, Mi LP, Huang W, Ma XF, Wang ZY, Zheng W, Xu LY, Chen J-H and Yu ZB, Nucleic Acids Res, 2019, 47, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neidle S and Parkinson GN, Curr. Opin. Struct. Biol, 2003, 13, 275–283. [DOI] [PubMed] [Google Scholar]
- 46.Williamson JR, Raghuraman MK and Cech TR, Cell, 1989, 59, 871–880. [DOI] [PubMed] [Google Scholar]
- 47.Koirala D, Ghimire C, Bohrer C, Sannohe Y, Sugiyama H and Mao HB, J. Am. Chem. Soc, 2013, 135, 2235–2241. [DOI] [PubMed] [Google Scholar]
- 48.Gehring K, Leroy J-L and Guéron M, Nature, 1993, 363, 561–565. [DOI] [PubMed] [Google Scholar]
- 49.Selvam S, Mandal S and Mao HB, Biochemistry, 2017, 56, 4616–4625. [DOI] [PubMed] [Google Scholar]
- 50.Lat PK, Schultz CW, Yu H-Z and Sen D, Angew. Chem., Int. Ed, 2021, 60, 8722–8727. [DOI] [PubMed] [Google Scholar]
- 51.Koirala D, Mashimo T, Sannohe Y, Yu ZB, Mao HB and Sugiyama H, Chem. Commun, 2012, 48, 2006–2008. [DOI] [PubMed] [Google Scholar]
- 52.Li Q, Zhao JM, Liu LF, Jonchhe S, Rizzuto FJ, Mandal S, He HW, Wei SS, Sleiman HF, Mao HB and Mao CD, Nat. Mater, 2020, 19, 1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M and Bustamante C, Nature, 2007, 446, 820–823. [DOI] [PubMed] [Google Scholar]
- 54.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J and Block SM, Science, 1998, 282, 902–907. [DOI] [PubMed] [Google Scholar]
- 55.Burns JR, Gopfrich K, Wood JW, Thacker VV, Stulz E, Keyser UF and Howorka S, Angew. Chem., Int. Ed, 2013, 52, 12069–12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H and Simmel FC, Science, 2012, 338, 932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Göpfrich K, Li CY, Ricci M, Bhamidimarri SP, Yoo J, Gyenes B, Ohmann A, Winterhalter M, Aksimentiev A and Keyser UF, ACS Nano, 2016, 10, 8207–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stetter FWS, Cwiklik L, Jungwirth P and Hugel T, Biophys. J, 2014, 107, 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suma A, Stopar A, Nicholson AW, Castronovo M and Carnevale V, Nucleic Acids Res, 2020, 48, 4672–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramakrishnan S, Shen BX, Kostiainen MA, Grundmeier G, Keller A and Linko V, ChemBioChem, 2019, 20, 2818–2823. [DOI] [PubMed] [Google Scholar]
- 61.Stopar A, Coral L, Giacomo SD, Adedeji AF and Castronovo M, Nucleic Acids Res, 2018, 46, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kollmann F, Ramakrishnan S, Shen BX, Grundmeier G, Kostiainen MA, Linko V and Keller A, ACS Omega, 2018, 3, 9441–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramakrishnan S, Schärfen L, Hunold K, Fricke S, Grundmeier G, Schlierf M, Keller A and Krainer G, Nanoscale, 2019, 11, 16270–16276. [DOI] [PubMed] [Google Scholar]
- 64.Kalinowski M, Haug R, Said H, Piasecka S, Kramer M and Richert C, ChemBioChem, 2016, 17, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 65.Gerling T, Kube M, Kick B and Dietz H, Sci. Adv, 2018, 4, eaau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajendran A, Endo M, Katsuda Y, Hidaka K and Sugiyama H, J. Am. Chem. Soc, 2011, 133, 14488–14491. [DOI] [PubMed] [Google Scholar]
- 67.Funke JJ, Ketterer P, Lieleg C, Korber P and Dietz H, Nano Lett, 2016, 16, 7891–7898. [DOI] [PubMed] [Google Scholar]
- 68.Funke JJ and Dietz H, Nat. Nanotechnol, 2016, 11, 47–52. [DOI] [PubMed] [Google Scholar]
- 69.Castro CE, Kilchherr F, Kim D-N, Shiao EL, Wauer T, Wortmann P, Bathe M and Dietz H, Nat. Methods, 2011, 8, 221–229. [DOI] [PubMed] [Google Scholar]
- 70.Bertin A, Mangenot S, Renouard M, Durand D and Livolant F, Biophys. J, 2007, 93, 3652–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nickels PC, Wünsch B, Holzmeister P, Bae W, Kneer LM, Grohmann D, Tinnefeld P and Liedl T, Science, 2016, 354, 305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou LF, Marras AE, Su H-J and Castro CE, ACS Nano, 2014, 8, 27–34. [DOI] [PubMed] [Google Scholar]
- 73.Zhou LF, Marras AE, Su H-J and Castro CE, Nano Lett, 2015, 15, 1815–1821. [DOI] [PubMed] [Google Scholar]
- 74.Karna D, Stilgenbauer M, Jonchhe S, Ankai K, Kawamata I, Cui YX, Zheng Y-R, Suzuki Y and Mao HB, Bioconjugate Chem, 2021, 32, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karna D, Pan W, Pandey S, Suzuki Y and Mao HB, Nanoscale, 2021, 13, 8425–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hyeon CB, Dima RI and Thirumalai D, J. Chem. Phys, 2006, 125, 194905. [DOI] [PubMed] [Google Scholar]
- 77.Liedl T, Högberg B, Tytell J, Ingber DE and Shih WM, Nat. Nanotechnol, 2010, 5, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kauert DJ, Kurth T, Liedl T and Seidel R, Nano Lett, 2011, 11, 5558–5563. [DOI] [PubMed] [Google Scholar]
- 79.Schuldt C, Schnauß J, Händler T, Glaser M, Lorenz J, Golde T, Käs JA and Smith DM, Phys. Rev. Lett, 2016, 117, 197801. [DOI] [PubMed] [Google Scholar]
- 80.Bustamante C, Marko JF, Siggia ED and Smith S, Science, 1994, 265, 1599–1600. [DOI] [PubMed] [Google Scholar]
- 81.Tinland B, Pluen A, Sturm J and Weill G, Macromolecules, 1997, 30, 5763–5765. [Google Scholar]
- 82.Cecconi C, Shank EA, Marqusee S and Bustamante C, Methods Mol. Biol, 2011, 749, 255–271. [DOI] [PubMed] [Google Scholar]
- 83.Shrestha P, Jonchhe S, Emura T, Hidaka K, Endo M, Sugiyama H and Mao HB, Nat. Nanotechnol, 2017, 12, 582–588. [DOI] [PubMed] [Google Scholar]
- 84.Jonchhe S, Pandey S, Emura T, Hidaka K, Hossain MA, Shrestha P, Sugiyama H, Endo M and Mao HB, Proc. Natl. Acad. Sci. U. S. A, 2018, 115, 9539–9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jonchhe S, Pandey S, Karna D, Pokhrel P, Cui YX, Mishra S, Sugiyama H, Endo M and Mao HB, J. Am. Chem. Soc, 2020, 142, 10042–10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Helmi S, Ziegler C, Kauert DJ and Seidel R, Nano Lett, 2014, 14, 6693–6698. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y, Xu J, Wang YW and Chen HY, Chem. Soc. Rev, 2013, 42, 2930–2962. [DOI] [PubMed] [Google Scholar]
- 88.Valev VK, Baumberg JJ, Sibilia C and Verbiest T, Adv. Mater, 2013, 25, 2517–2534. [DOI] [PubMed] [Google Scholar]
- 89.Shen CQ, Lan X, Zhu CG, Zhang W, Wang LY and Wang QB, Adv. Mater, 2017, 29, 1606533. [DOI] [PubMed] [Google Scholar]
- 90.Huang YK, Nguyen M-K, Natarajan AK, Nguyen VH and Kuzyk A, ACS Appl. Mater. Interfaces, 2018, 10, 44221–44225. [DOI] [PubMed] [Google Scholar]
- 91.Zhu CG, Wang M, Dong JY, Zhou C and Wang QB, Langmuir, 2018, 34, 14963–14968. [DOI] [PubMed] [Google Scholar]
- 92.Chen Z, Choi CK and Wang QB, ACS Appl. Mater. Interfaces, 2018, 10, 26835–26840. [DOI] [PubMed] [Google Scholar]
- 93.Nguyen M-K and Kuzyk A, ACS Nano, 2019, 13, 13615–13619. [DOI] [PubMed] [Google Scholar]
- 94.Lan X, Liu TJ, Wang ZM, Govorov AO, Yan H and Liu Y, J. Am. Chem. Soc, 2018, 140, 11763–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lan X, Lu XX, Shen CQ, Ke YG, Ni WH and Wang QB, J. Am. Chem. Soc, 2015, 137, 457–462. [DOI] [PubMed] [Google Scholar]
- 96.Kosuri P, Altheimer BD, Dai MJ, Yin P and Zhuang XW, Nature, 2019, 572, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin P, Hariadi RF, Sahu S, Choi HM, Park SH, Labean TH and Reif JH, Science, 2008, 321, 824–826. [DOI] [PubMed] [Google Scholar]
- 98.Maier AM, Weig C, Oswalds P, Frey E, Fischer P and Liedl T, Nano Lett, 2016, 16, 906–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dietz H, Douglas SM and Shih WM, Science, 2009, 325, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Juul S, Iacovelli F, Falconi M, Kragh SL, Christensen B, Frøhlich R, Franch O, Kristoffersen EL, Stougaard M, Leong KW, Ho Y-P, Sørensen ES, Birkedal V, Desideri A and Knudsen BR, ACS Nano, 2013, 7, 9724–9734. [DOI] [PubMed] [Google Scholar]
- 101.Andersen FF, Knudsen B, Oliveira CL, Frøhlich RF, Krüger D, Bungert J, Agbandje-McKenna M, McKenna R, Juul S, Veigaard C, Koch J, Rubinstein JL, Guldbrandtsen B, Hede MS, Karlsson G, Andersen AH, Pedersen JS and Knudsen BR, Nucleic Acids Res, 2008, 36, 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oliveira CL, Juul S, Jorgensen HL, Knudsen B, Tordrup D, Oteri F, Falconi M, Koch J, Desideri A, Pedersen JS, Andersen FF and Knudsen BR, ACS Nano, 2010, 4, 1367–1376. [DOI] [PubMed] [Google Scholar]
- 103.Tangyunyong P, Thomas RC, Houston JE, Michalske TA, Crooks RM and Howard AJ, Phys. Rev. Lett, 1993, 71, 3319. [DOI] [PubMed] [Google Scholar]
- 104.Ebrahem F, Bamer F and Markert B, Phys. Rev. E, 2020, 102, 033006. [DOI] [PubMed] [Google Scholar]
- 105.Ke YG, Lindsay S, Chang Y, Liu Y and Yan H, Science, 2008, 319, 180–183. [DOI] [PubMed] [Google Scholar]
- 106.Ijäs H, Hakaste I, Shen BX, Kostiainen MA and Linko V, ACS Nano, 2019, 13, 5959–5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM and Shih WM, Nucleic Acids Res, 2009, 37, 5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao YX, Shaw A, Zeng XH, Benson E, Nyström AM and Högberg B, ACS Nano, 2012, 6, 8684–8691. [DOI] [PubMed] [Google Scholar]
- 109.Kohman RE, Cha SS, Man HY and Han X, Nano Lett, 2016, 16, 2781–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamazaki T, Aiba Y, Yasuda K, Sakai Y, Yamanaka Y, Kuzuya A, Ohya Y and Komiyama M, Chem. Commun, 2012, 48, 11361–11363. [DOI] [PubMed] [Google Scholar]
- 111.Green LS, Jellinek D, Jenison R, Östman A, Heldin C-H and Janjic N, Biochemistry, 1996, 35, 14413–14424. [DOI] [PubMed] [Google Scholar]
- 112.Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN and Turberfield AJ, Nat. Nanotechnol, 2008, 3, 93–96. [DOI] [PubMed] [Google Scholar]
- 113.Mikkilä J, Eskelinen AP, Niemelä EH, Linko V, Frilander MJ, Törmä P and Kostiainen MA, Nano Lett, 2014, 14, 2196–2200. [DOI] [PubMed] [Google Scholar]
- 114.Douglas T and Young M, Nature, 1998, 393, 152–155. [Google Scholar]
- 115.Fan SS, Wang DF, Cheng J, Liu Y, Luo T, Cui DX, Ke YG and Song J, Angew. Chem., Int. Ed, 2020, 59, 12991–12997. [DOI] [PubMed] [Google Scholar]
- 116.Du SM, Zhang Sand Seeman NC, Biochemistry, 1992, 31, 10955–10963. [DOI] [PubMed] [Google Scholar]
- 117.Fan SS, Cheng J, Liu Y, Wang DF, Luo T, Dai B, Zhang C, Cui DX, Ke YG and Song J, J. Am. Chem. Soc, 2020, 142, 14566–14573. [DOI] [PubMed] [Google Scholar]
- 118.Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR and Chen CS, Nat. Methods, 2014, 11, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Glazier R, Shinde P, Ogasawara H and Salaita K, ACS Appl. Mater. Interfaces, 2021, 13, 2145–2164. [DOI] [PubMed] [Google Scholar]
- 120.Zhao B, Li NW, Xie TF, Bagheri Y, Liang CW, Keshri P, Sun YB and You MX, Chem. Sci, 2020, 11, 8558–8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brockman JM, Su HQ, Blanchard AT, Duan YX, Meyer T, Quach ME, Glazier R, Bazrafshan A, Bender RL, Kellner AV, Ogasawara H, Ma R, Schueder F, Petrich BG, Jungmann R, Li RH, Mattheyses AL, Ke YG and Salaita K, Nat. Methods, 2020, 17, 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao B, O’Brien C, Mudiyanselage APKKK, Li NW, Bagheri Y, Wu R, Sun YB and You MX, J. Am. Chem. Soc, 2017, 139, 18182–18185. [DOI] [PubMed] [Google Scholar]
- 123.Fang WN, Xie M, Hou XL, Liu XG, Zuo XL, Chao J, Wang LH, Fan CH, Liu HJ and Wang LH, J. Am. Chem. Soc, 2020, 142, 8782–8789. [DOI] [PubMed] [Google Scholar]
- 124.Scheckenbach M, Schubert T, Forthmann C, Glembockyte V and Tinnefeld P, Angew. Chem., Int. Ed, 2021, 60, 4931–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kosinski R, Mukhortava A, Pfeifer W, Candelli A, Rauch P and Saccà B, Nat. Commun, 2019, 10, 1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hannewald N, Winterwerber P, Zechel S, Ng DYW, Hager MD, Weil T and Schubert US, Angew. Chem., Int. Ed, 2021, 60, 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nguyen L, Döblinger M, Liedl T and Heuer-Jungemann A, Angew. Chem., Int. Ed, 2019, 58, 912–916. [DOI] [PubMed] [Google Scholar]
- 128.Nguyen M-K, Nguyen VH, Natarajan AK, Huang YK, Ryssy J, Shen BX and Kuzyk A, Chem. Mater, 2020, 32, 6657–6665. [Google Scholar]
- 129.Wills PR, Georgalis Y, Dijk J and Winzor DJ, Biophys. Chem, 1995, 57, 37–46. [DOI] [PubMed] [Google Scholar]
- 130.Minton AP, Methods Enzymol, 1998, 295, 127–149. [DOI] [PubMed] [Google Scholar]
- 131.Bauer J, Meza LR, Schaedler TA, Schwaiger R, Zheng XY and Valdevit L, Adv. Mater, 2017, 29, 1701850. [DOI] [PubMed] [Google Scholar]
- 132.Zheng XY, Smith W, Jackson J, Moran B, Cui HC, Chen D, Ye JC, Fang N, Rodriguez N, Weisgraber T and Spadaccini CM, Nat. Mater, 2016, 15, 1100–1106. [DOI] [PubMed] [Google Scholar]
- 133.Meza LR, Zelhofer AJ, Clarke N, Mateos AJ, Kochmann DM and Greer JR, Proc. Natl. Acad. Sci. U. S. A, 2015, 112, 11502–11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi S, Li Y, Ngo-Dinh B-N, Markmann J and Weissmüller J, Science, 2021, 371, 1026–1033. [DOI] [PubMed] [Google Scholar]
- 135.Muth JT, Dixon PG, Woish L, Gibson LJ and Lewis JA, Proc. Natl. Acad. Sci. U. S. A, 2017, 114, 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu CP, Jonchhe S, Pokhrel P, Karna D and Mao HB, Chem. Sci, 2021, 12, 10159–10164. [DOI] [PMC free article] [PubMed] [Google Scholar]