Highlights
-
•
Intrinsic activation of cGAS-STING signalings potentiate tumorigenesis.
-
•
cGAS-STING signalings promote tumor progression by sustaining cancer stemness.
-
•
STAT3 may act as a downstream effector of cGAS-STING signalings to stimulate cancer stemness.
Keywords: cGAS-STING pathway, Tumorigenesis, Tumor progression, Cancer stemness, STAT3
Abstract
The cytosolic DNA-sensing cGAS-STING pathway has been proved to be involved in tumor progression and influence the effect of cancer immunotherapy. However, little attentions have been paid to the role of cGAS-STING pathway on cancer stemness. Herein, we found that the cGAS-STING pathway was activated in different tumor cells. cGAS- or STING-knockout impaired the capability of tumor formation in vivo and tumorsphere formation in vitro. In addition, loss of cGAS-STING cascade promoted tumor apoptosis, but inhibited tumor growth and metastasis. We further demonstrated that cGAS-STING pathway potentiated tumor formation by sustaining cancer stemness. Moreover, analysis of RNA-seq showed that cGAS-STING pathway maintained cancer stemness probably by activating STAT3. Our findings highlight the role of intrinsic activation of cGAS-STING pathway in tumorigenesis, and reveal a new mechanism of its regulation of tumor progression via sustaining cancer stemness through STAT3 activation.
Introduction
Despite great advances in cancer prevention and treatment in recent years, cancer still ranks as a leading cause of death and imposes a heavy health burden worldwide [1]. Currently, accumulating evidences suggest that a comprehensive and profound understanding of tumor biology offers a golden key [2], [3], [4]. As well-known, genome instability is a typical hallmark of cancer, which can cause spontaneous DNA damage. The development of genome instability renders tumors with selective proliferative advantages and the ability of angiogenesis, thereby expedites tumor growth and invasion [5,6]. However, how does genome instability promote tumor progression?
It has been revealed that the cGMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway, which primarily discovered as an important DNA-sensing machinery in innate immunity, involved in tumor progression and therapeutic responses [7], [8], [9]. Generally, the cGAS-STING pathway exerts tumor-suppressive roles. Cytosolic double-stranded DNAs (dsDNAs) derived from multiple sources of DNA damage, including chemoradiation, reactive oxygen species (ROS), and hyperactivation of oncogene signalings, can be sensed by cGAS and further activate STING to trigger TBK1 (TANK-binding kinase-1)-IRF3 (interferon regulatory factor-3) axis and canonical NF-κB (nuclear factor kappa B) signaling to upregulate the expression of type 1 interferon (IFN), which in turn delay cancer progression [10]. Some agonists of GAS-STING pathway have been developed and used in pre-clinical research [11]. Fu et al. reported that combining STING agonist with PD-1 blockade induced regression of tumor [12]. Radiotherapy or chemotherapy, which elicits cGAS-STING-IFN axis, was also reported to augment the effect of immunotherapy [13].
However, growing evidences proved that cGAS-STING cascade did not always play a positive role. Tumor cells always harbor chromosome instability, which lead to production of micronuclei. The envelopes around micronuclei are prone to rupture during mitosis, which lead to the exposure of DNA fragments [14]. These dsDNAs induce continuous activation of the cGAS-STING pathway to form a chronic inflammatory environment to promote tumor metastasis [15]. Beyond this, in the early stage of DNA damage after radiotherapy, cGAS could competitively bind to PARP1 to prevent the formation of PARP1-Timeless repair complex, aggravating chromosomal instability and tumor progression [16]. Thus, the roles of cGAS-STING pathway in cancer are dichotomous. On the one hand, it leads to production of type 1 IFN to inhibit tumor growth [10]. On the other hand, it suppresses DNA repair and upregulates expression of the ligands of immunosuppressive molecule CCR2, further shaping an immunosuppressive microenvironment, and promoting immune evasion and metastasis [16,17].
Hitherto, researches on cGAS-STING pathway in cancer mainly focused on tumor immunity and immunotherapy, which seemed not enough to elucidate their multiple functions in cancer progression. As we know, cancer stem cells (CSCs), which endow tumors with the capability for self-renew and the potential to seed new tumor, are closely related to tumor progression, metastasis, resistance to therapy, and relapse [18,19]. Patients with high CSCs burden are prone to undergo relapse and distant metastasis after conventional therapy [20,21]. Besides, it has been revealed that CSCs possess more efficient DNA repair, which contributes to their resistant to chemoradiation in advanced tumors [22], [23], [24]. This may mean that DNA damage response (DDR) is somehow involved in cancer stemness. Interestingly, damaged DNA fragments are main triggers of cGAS-STING pathway activation in cancer cells. Hence, there is reason to wonder whether the cGAS-STING pathway is associated with cancer stemness.
Herein, we investigated the role of cGAS-STING pathway in tumorigenesis and found that the cGAS-STING pathway potentiates tumor progression via sustaining cancer stemness through STAT3 activation. Our findings highlight the role of intrinsic activation of cGAS-STING pathway in tumorigenesis, and reveal a new mechanism of its regulation of tumor progression.
Materials and methods
Cancer cell lines and cell culture
Human breast cancer cell line MCF-7 (ATCC® HTB-22), cervical carcinoma cell line HeLa (ATCC® CRM-CCL-2), pancreatic cancer cell line PANC-1 (ATCC® CRL-1469) and human kidney epithelial cell line HEK 293T (ATCC® CRL-3216) were derived from ATCC (American Type Culture Collection). The cells were cultured as follow: MCF-7 was cultured in EMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 0.01 mg/ml insulin (Gibco). HeLa was cultured in RPMI 1640 (Gibco) with 10% FBS. PANC-1 and 293T were cultured in DMEM (High Glucose) (Gibco) supplemented with 10% FBS. Antibiotics (1% penicillin-streptomycin solution, Gibco) was routinely added to the media. All the cell lines were incubated in a 5% CO2 humidified incubator at 37 °C. All the cell lines were identified by STR (short tandem repeat) and free of mycoplasma.
Subcutaneous xenograft mouse model
For tumor formation assay, human cancer cells (5 × 106 cells) were subcutaneously implanted into the left groin of 6-week-old male BALB/c nude mice. Tumor diameter was measured and recorded every 2–4 days, and tumor volume was calculated by the formula [(length × width2)/2]. Five mice were used per group, and tumor growth was continuously monitored for more than a month. Mice were sacrificed when tumor diameter reached 1500 mm.
For tumor initiation assay, cells suspension of HeLa cells was diluted into 3 gradients (5 × 105, 5 × 104 and 5 × 103 cells, respectively). Mice were randomly divided into control and knockout groups, and tumor cell inoculation was carried out as described above. Tumor volume was measured by digital caliper. Animals were executed when the diameter of control tumors arrived at 1500 mm. All animal studies complied with the ARRIVE guidelines and obtained an ethic number (No. 2020AW101) from the Institutional Animal Care and Use Committee (IACUC) of Shanghai General Hospital.
Mouse metastasis model
In brief, cells were digested by trypsin and washed twice using PBS. Next, cells were resuspended in PBS at a concentration of 5 × 107 cells/ml. 5 × 106 cells in 100 μl cell suspensions were injected into mouse through tail vein. Mice were monitored for two to three months and sacrificed at the end of the study. The lungs were collected and weighed. Animal studies complied with ARRIVE guidelines and obtained an ethic number (No. 2020AW101) from the Institutional Animal Care and Use Committee (IACUC) of Shanghai General Hospital.
Soft agar colony formation assay
Soft agar was composed of upper- and lower-layer culture systems. The upper layer was a 1:1 mixture of 0.6% agar and 2 × medium (2 × DMEM/RPMI 1640 with 20% FBS), which contains 5000–8000 cells. The lower layer was made by mixing 1% agar and 2 × medium in equal proportions. Then, the lower layer in a 6-well plate was first spread at volume of 1.5 ml/well. After agar solidification, the upper layer was spread evenly on it. 30 min later, the upper layer was covered with 1 ml of medium. The medium was changed every two days and the number of clones was observed after 14 days.
Tumorsphere formation assay
Tumor cells were diluted into a single cell suspension and seeded in triplicate in an ultra-low attachment 96-well plate at a concentration of less than 1000 cells/well. The cells were cultured in DMEM/1640 medium containing 0.4% BSA, 20 ng/ml human recombinant EGF and 10 ng/ml human recombinant bFGF (Peprotech). After 10–14 days, multiple images of different fields of view were taken. The tumorsphere number and size were compared and drawn as a histogram.
EdU cell proliferation assay
Cells were firstly cultivated in suspension over 48 h. Then, EdU solution (Cell-Light EdU Apollo567 in vitro, RiboBio) was added into culture medium with the ratio of 1:1000. After 2 h culture, cells were fixed using 4% paraformaldehyde for 15 min and neutralize with 2 mg/ml Glycine for 5 min at room temperature. Permeabilizing was performed in 0.5% Triton-X100, followed by EdU staining (10 min, room temperature). Then, cells were washed in PBS and detected by Accuri C6 Flow cytometer (BD Biosciences). For monolayer cultured cells, they needed to be further stained with Hoechst 33258, and imaged by the fluorescence microscope (Leica).
Apoptosis assay
The apoptosis assay was conducted by BD Pharmingen™ PE Annexin V Apoptosis Detection Kit I (BD Biosciences) according to manufacturer's instruction. Briefly, cancer cells were cultivated in suspension for 24 h. Then, they were washed with pre-chilled PBS for 3 times and centrifuged at 300 g for 5 min. Cell pellets were suspended in 100 μl binding buffer, followed by Annexin V and 7-AAD staining in dark at room temperature for 15 min. After that, cells were analyzed by flow cytometer.
Analysis of cancer stem cell markers by flow cytometry
Cells were cultured in media containing EGF and bFGF in ultra-low attachment 6-well plate. After 5 days, suspension cells were collected, washed and counted.
For cell surface markers detection, 1 × 106 cells were taken out and washed with FACS buffer (1 × PBS, 2% FBS, 0.1% NaN3), and resuspended in 100 μl FACS buffer. PE Mouse anti-Human CD24 (BD Biosciences), PE-Cy™7 Mouse anti-Human CD44 (BD Biosciences) were added to the cell suspensions, and incubated for 15 min in dark at room temperature. Cells were washed by FACS buffer again and detected by the Accuri C6 Flow cytometer.
ALDH enzyme activities were also detected by flow cytometry. Following the manufacturer's instructions of ALDEFLUOR™ kit (STEMCELL Technologies), cells were collected and resuspended in ALDEFLUOR assay buffer. Then, cells were evenly divided into two tubes which contained ALDEFLOUR reagent or ALDEFLOUR plus DEAB (ALDH enzyme inhibitor). After incubation in 37 °C water baths for 40 min, cells were analyzed by the Accuri C6 Flow cytometer.
All the results of flow cytometry were further analyzed by FlowJo V10 software.
RNA sequencing
RNA-seq of cancer cells was performed by Majorbio Bio-pharm Technology (Shanghai, China). In short, total RNA was extracted from cancer cells using TRIzol® Reagent according the manufacturer's instructions (Invitrogen), and genomic DNA was removed using DNase I (Takara). Then RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using NanoDrop ND-2000 (Thermo). Only high-quality RNA sample (OD260/280 = 1.8–2.2, OD260/230≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, > 1 μg) was used to construct the sequencing library. Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. Paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq xten/NovaSeq 6000 sequencer (2 × 150 bp read length). DEGs (differential expression genes) between two different samples was analyzed by DESeq2 (version 1.12.4). Moreover, functional-enrichment analysis including GO and GSEA were performed at Bonferroni-corrected P-value ≤ 0.05.
Statistics
Statistical analysis was performed using software GraphPad Prism 8. All data were represented as means ± SEM. Unpaired student's t-test was used for comparisons between control and knockout group in all of the experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 were regarded as significant differences, while n.s means no significant.
Details for other experimental procedures were shown in Supplementary Materials and Methods.
Results
cGAS-STING pathway potentiates tumorigenesis
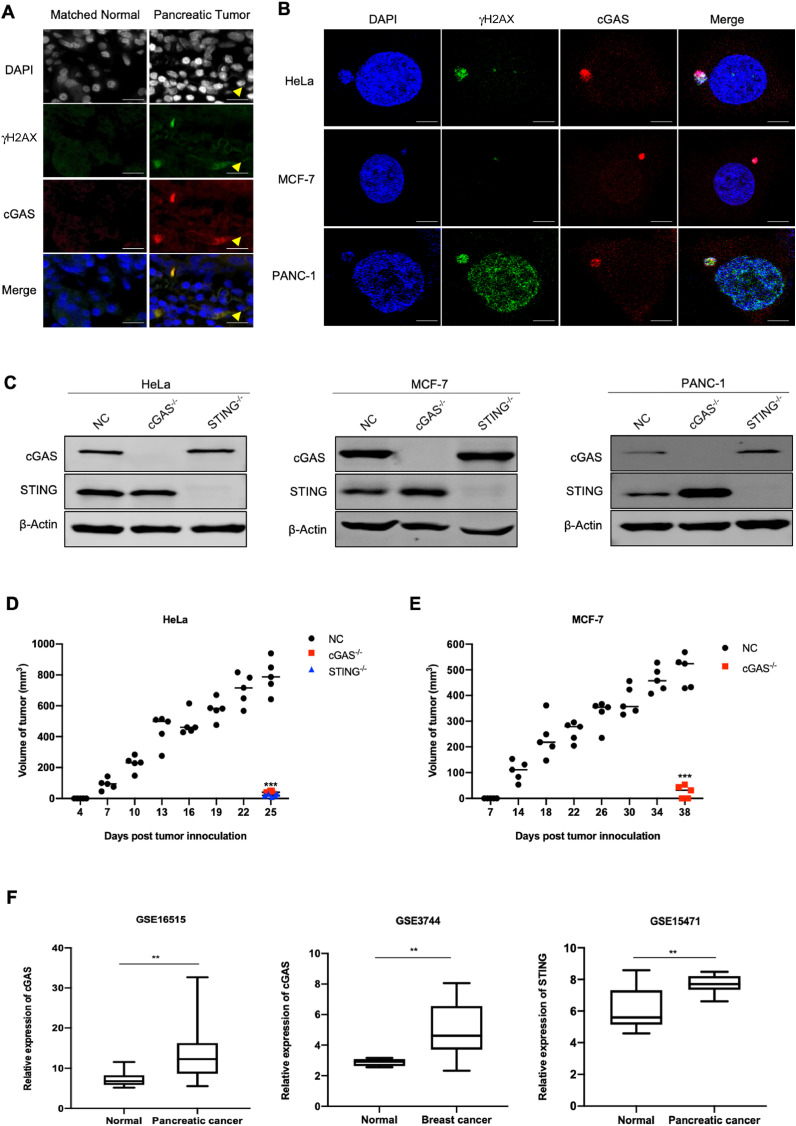
Because of chromosomal instability (CIN), tumors tend to induce the formation of micronuclei. The damaged DNAs in micronuclei are prominent triggers of cGAS [10]. We first investigated whether there was activation of the cGAS-STING pathway in tumor cells by performing immunofluorescence assay on tumor tissues and tumor cell lines. In normal tissues, micronucleus could hardly be observed, but it was obvious in tumor tissues. Meanwhile, the micronucleus was positive with DNA damage marker γH2AX staining, accompanied by the accumulation of cGAS protein (Fig. 1A). Similarly, we also observed the formation of micronuclei in three different cell lines without any stimulation, accompanied by the co-localization of γH2AX and cGAS protein. Although γH2AX positive also presented in the nucleus of PANC-1 cells, the nuclear envelope is generally stable and cannot be penetrated by cGAS. Therefore, there was no significant overlaps between the immunostaining of γH2AX and cGAS in the nucleus (Fig. 1B).
Fig. 1.
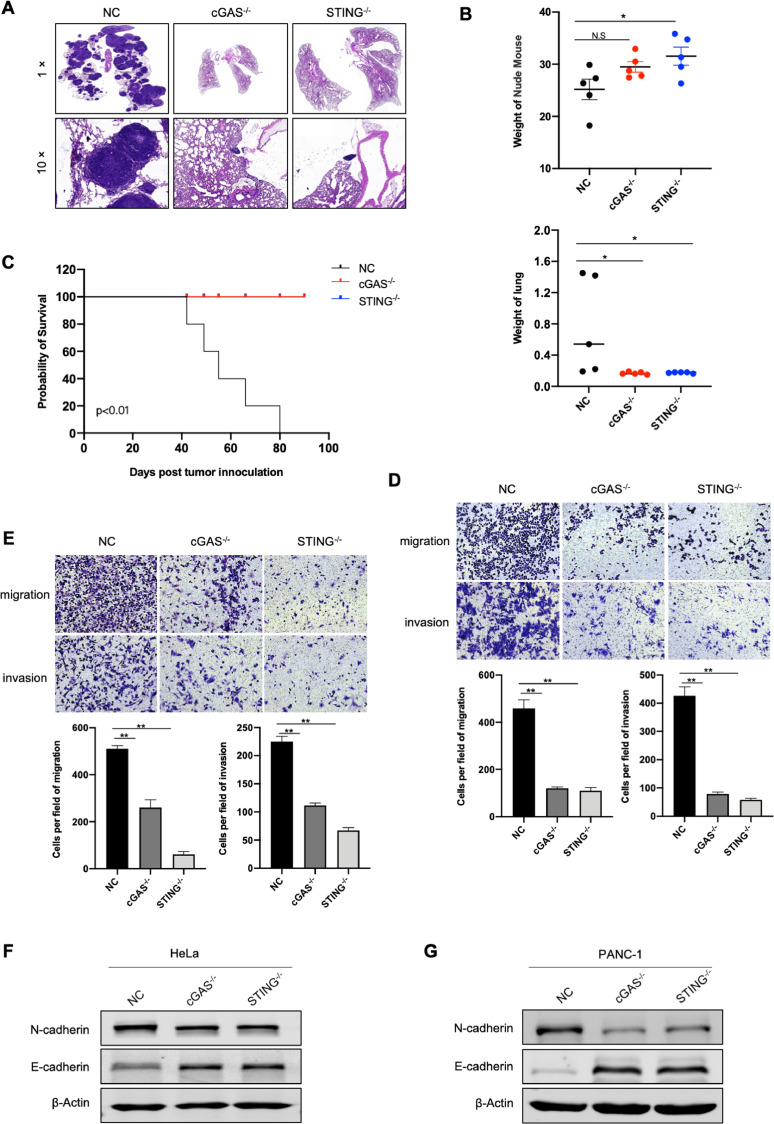
Activation of cGAS-STING pathway potentiates tumorigenesis. (A) Representative images of immunofluorescence staining of γH2AX and cGAS in pancreatic tumor tissues and matched normal tissue. Scale bar: 50 μm. (B) Representative images of immunofluorescence staining of γH2AX and cGAS in different tumor cell lines. Scale bar: 50 μm. (C) Knockout effect of cGAS or STING in HeLa, MCF-7 and PANC-1 cells. (D, E) Tumor growth curve of the tumor-bearing mice injected with negative control (NC), cGAS-/- and STING-/- HeLa or MCF-7 cells. 5 mice were used for each group. (F) The mRNA expression of cGAS and STING in pancreatic tumor tissues (GSE16515, GSE15471), breast tumor tissue (GSE3744) were compared with those of normal tissues. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Then we explored the role of cGAS-STING pathway in tumor in vivo. We first generated cGAS- or STING-knockout cell lines (Figs. 1C and Supplementary S1A), and subsequently inoculated these cancer cells into the groin of nude mice to compare their tumorigenesis capability. We were surprised to find that tumor initiation time was significantly delayed, and the capability of tumorigenesis was dramatically reduced or even lost in cGAS- or STING-depleted group (Figs. 1D, E and Supplementary S1B, C). In addition, we analyzed the mRNA expression of cGAS and STING in different tumor and their adjacent normal tissues [25], [26], [27], [28], [29], [30], [31], [32]. We found that cGAS expression in a variety of tumor tissues (including breast cancer and pancreatic cancer) was higher than that of normal tissues (Figs. 1F, Supplementary S2A–D, and Supplementary Table S2). Similarly, compared to normal tissue, the expression of STING was obviously upregulated in multiple cancers as well (Figs. 1F, Supplementary S2E–G, and Supplementary Table S3). Collectively, these data indicated the essential role of cGAS-STING pathway in tumor formation.
Loss of cGAS-STING cascade impairs cancer cell stemness
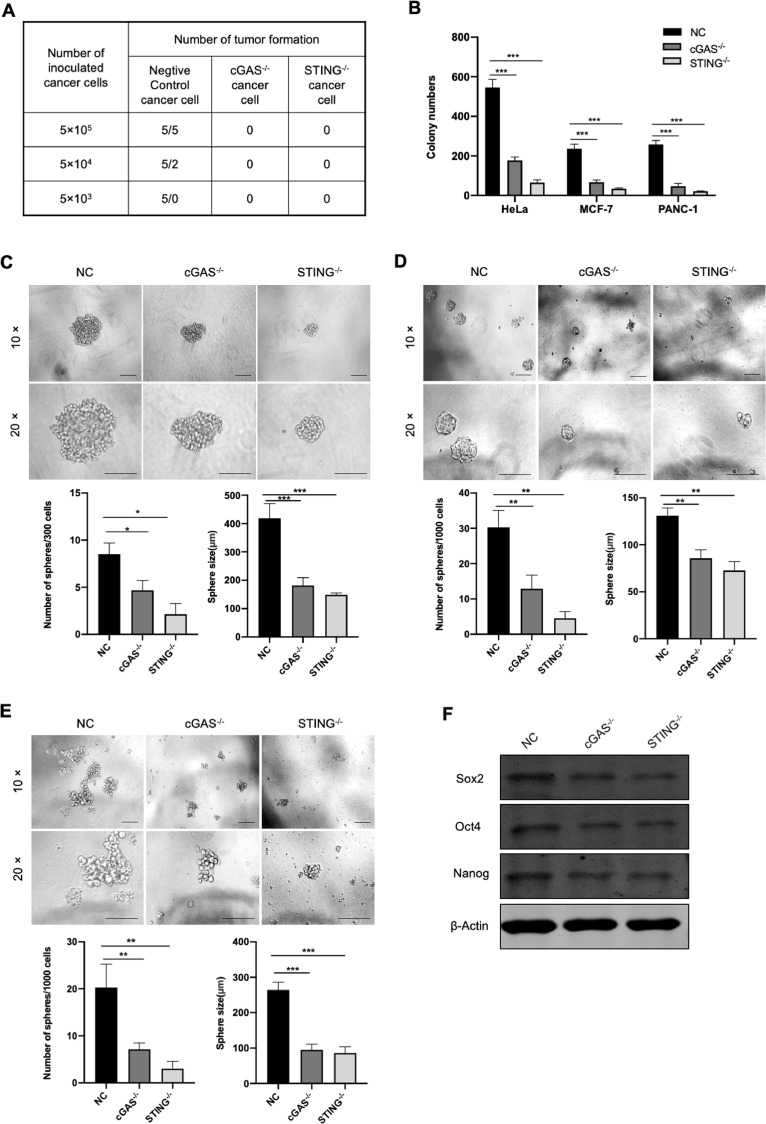
Given that cancer stem cells have a strong potential to promote tumorigenesis. We wondered whether the capability of cGAS-STING pathway in tumorigenesis was associated with cancer stemness or not. So, we conducted a series of experiments to figure out it. We first diluted cancer cells to 5 × 105, 5 × 104, 5 × 103 in 100 μl cell suspensions, respectively, and inoculated these cells into the groin of nude mice. In the regular cancer cells, the rate of tumor formation was 100% when 5 × 105 cells were inoculated, 40% for 5 × 104 cells, and 0 for 5 × 103 cells, whereas the rate was 0 at all these cell concentrations in cGAS- or STING-depleted cells (Fig. 2A). Then, we performed soft-agar colony formation assay and sphere formation assay. We found that loss of cGAS or STING significantly suppressed colony formation ability of different tumor cells in soft-agar (Figs. 2B and Supplementary S1D). Under low adhesion culture conditions, cGAS-STING pathway deficiency dramatically impaired the sphere formation ability of tumor. Compared to control, there was an obviously reduction in the number and size of tumorspheres in cGAS- or STING-depleted cells (Fig. 2C–E). Moreover, the expression of self-renew related transcription factors, such as Nanog, Sox2 and Oct4, were decreased when cGAS or STING was knocked-out (Fig. 2F). Collectively, these results revealed that cGAS-STING pathway possibly sustains the stemness of tumor cells.
Fig. 2.
cGAS-STING pathway deficiency impedes tumor initiation and tumorsphere formation. (A) Serial dilution of NC and knockout group of HeLa cells was used to test tumor initiation among different groups. n = 5 for each group. (B) Quantitation of soft agar colony numbers of NC and knockout group from different tumors. (C–E) Tumorsphere formation assay of NC and knockout groups from HeLa cells (C), MCF-7 cells (D) and PANC-1 cells (E). The number and size of tumor spheres was determined by microscopy. Scale bar: 50 μm in upper panel, 100 μm in lower panel. (F) The expression of Nanog, Sox2, Oct4 in NC, cGAS-/- and SSTING-/- HeLa cells were detected by Western blot. The quantification of the band intensity of Western blot was provided in the Supplementary Table S5. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
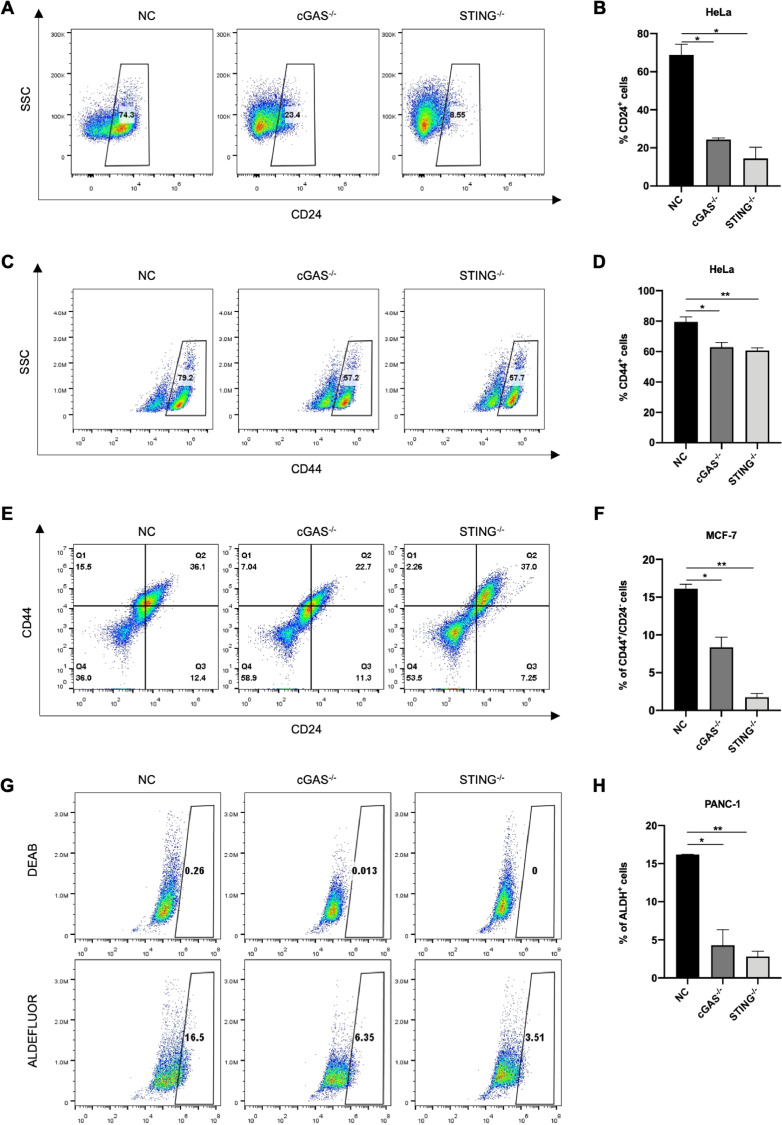
To further validate the role of cGAS-STING pathway in regulating cancer stemness, we analyzed expression of CD24, CD44 and ALDH, which are widely accepted biomarkers of stemness for most solid tumors. After cGAS or STING was depleted, the proportion of CD24+ populations in HeLa cells reduced about 70% (Fig. 3A, B), while the proportion of CD44+ populations reduced about 20% (Fig. 3C, D). In breast cancer, the cell population with high expression of CD44 and low expression of CD24 is generally considered to be cancer stem cells [33, 34]. The flow cytometry assays were performed and revealed that loss of cGAS-STING cascade led to a significant reduction in the proportion of cancer stem cell populations in MCF-7 cells. Compared to that of regular cancer cells, the proportion of CD44+/CD24−/low population dropped by half in cGAS-depleted cells, while the CD44+/CD24−/low cell population nearly disappeared in STING-depleted cells (Fig. 3E, F).
Fig. 3.
cGAS-STING pathway deficiency impairs the expression of cancer stem cell markers. (A–D) The ratio of CD24+ and CD44+ populations in NC, cGAS- or STING-knockout HeLa cells detected by FACS. Scatter spots of CD24+ or CD44+ cells showed in left panels (A, C) while quantification of results showed in right panels (B, D). (E, F) Representative scatter spots of FACS detecting the CD44+/CD24− population in NC, cGAS- or STING-knockout MCF-7 cells (E), and the results were quantified and showed at right (F). (G, H) Representative results and quantification of ALDH+ cells in NC, cGAS- or STING-depleted PANC-1 cells detected by ALDEFLOUR™ assay. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01.
However, we found that there was no obvious alteration in the ratio of CD44+ and CD24+ cells between control and knockout group in pancreatic cancer (Supplementary Fig. S3A, B). These results indicated that the effects of cGAS-STING pathway deficiency on cancer stem cell markers varied from tumor to tumor. Accumulating evidences have shown that pancreatic cancer stem cell is a subpopulation characterized with ALDH+ [35], [36], [37]. Hence, we detected the ratio of ALDH+ cells in pancreatic cancer and found that loss of cGAS-STING pathway impaired ALDH expression. Compared to the control, the proportion of ALDH+ population in cGAS- or STING-depleted pancreatic cancer cells was reduced by more than 80% (Fig. 3G, H). But we did not detect ALDH+ population in HeLa and MCF-7 cells (Supplementary Fig. S3C, D). Together, these results suggested that cGAS-STING cascade was required for the maintenance of cancer stemness.
cGAS-STING pathway deficiency impedes growth and metastasis of cancer
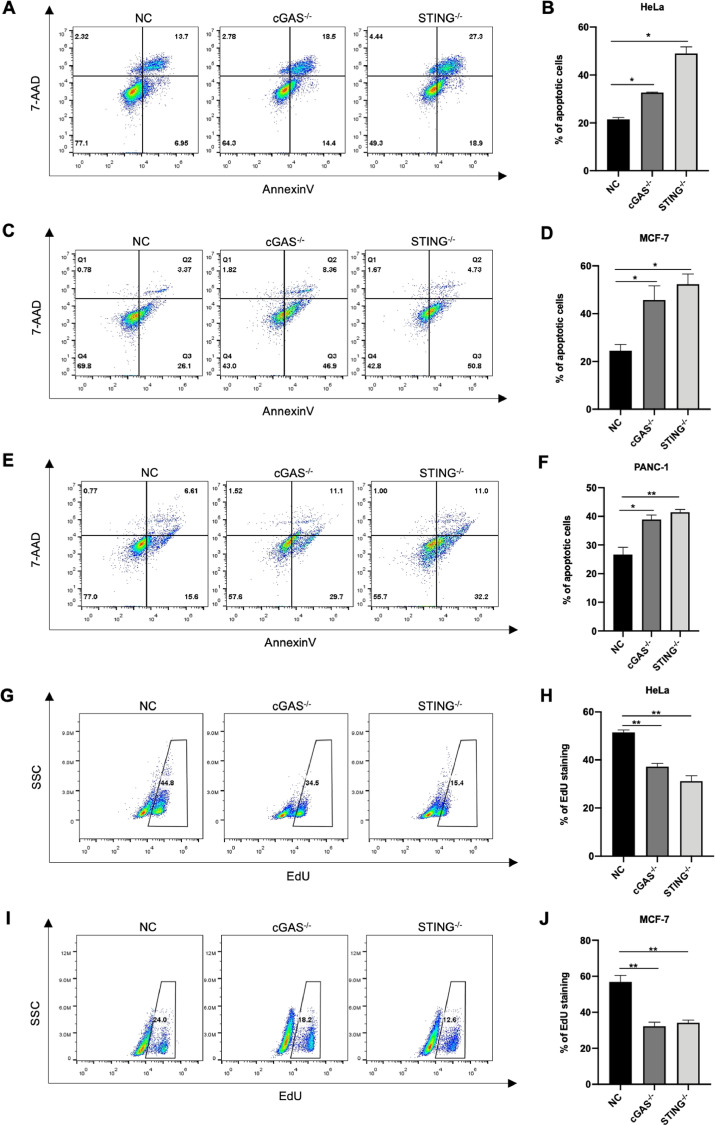
We further explored the effect of cGAS-STING pathway deficiency on tumor survival by Annexin V assays. For HeLa and MCF-7 cells, loss of cGAS-STING cascade nearly doubled the probability of cell apoptosis compared to those with regular cancer cells (Fig. 4A–D). Similar results were also obtained in PANC-1 cells (Fig. 4E, F). These results revealed that cGAS- or STING-depleted cancer cells were prone to apoptosis under suspension condition.
Fig. 4.
cGAS-STING pathway prevents tumor apoptosis while promotes tumor proliferation. (A, B) Represent results and quantification of apoptotic cells of NC, cGAS- or STING-knockout HeLa cells. (C, D) Representative scatter spots and quantification of apoptotic cells of NC and knockout MCF-7. (E, F) Representative scatter spots determining apoptotic cells in NC, cGAS- or STING-depleted PANC-1 cells (E), and the quantification of results showed at right (F). (G–J) Left panels: Flow cytometry analysis of EdU staining positive cells in NC and knockout groups after cultured in suspension for 48 h (G, I). Right panels: Quantitative result of percentage of EdU staining cells in NC, cGAS- or STING-depleted cancer cells cultured in monolayer for 48 h (H, J). Data were represented as mean ± SEM. *P < 0.05, **P < 0.01.
Moreover, loss of cGAS or STING impaired viability and proliferation of cancer cells. The percentage of cells with EdU incorporation declined in cGAS- or STING-depleted group, no matter they were cultured in suspension (Fig. 4G and I) or monolayer (Figs. 4H and J, Supplementary S4A, B).
We then determined the influences of cGAS- or STING-depletion on tumor metastasis in vivo. Cancer cells were injected into the nude mice through a tail vein. Two to three months later, lung tissues were collected and taken for H&E staining to distinguish tumor tissue from normal one, and the survival curve of nude mice was accordingly drawn. Compared with those of the regular tumor cells, mice injected with cGAS- or STING-depleted cells rarely undergone lung metastasis (Figs. 5A, B and Supplementary S4C), and had a longer survival (Fig. 5C). The same results were also obtained in vitro, that is the tumor migration and invasion ability reduced after cGAS or STING was depleted (Fig. 5D, E).
Fig. 5.
cGAS-STING pathway promotes tumor metastasis. (A) H&E staining of lung tissues from the tumor-bearing mice. The mice were injected with NC, cGAS- and STING-knockout HeLa cells through the tail vein. n = 5 for each group. (B) Weight of lung tissues and mice from NC and knockout group. n = 5 for each group. (C) Survival rate of mice injected with NC, cGAS- or STING-depleted HeLa cells. n = 5 for each group. (D, E) Transwell assay of NC and knockout groups of HeLa cells (D) and PANC-1 cells (E). The bottom panels are quantitative results of migration and invasion. (F, G) Western blot analysis of the expression of N-cadherin and E-cadherin in the indicated HeLa (F) and PANC-1 cells (G). The quantification of the band intensity of Western blot was provided in the Supplementary Table S5. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01. n.s, no significant difference.
Corresponding to above phenomenon, cGAS- or STING-depletion caused upregulation of E-cadherin expression and downregulation of N-cadherin expression, indicating that cGAS-STING pathway might potentiate cancer metastasis via mediating EMT transition (Fig. 5F, G). Besides, cGAS-STING pathway deficiency could increase drug sensitivity of tumor to a certain extent (Supplementary Fig. S4D). Together, these findings suggested that cGAS-STING cascade contributed to sustaining tumor survival, promoting tumor growth and distant metastases.
Recovery of cGAS-STING pathway reinvigorates cancer stemness
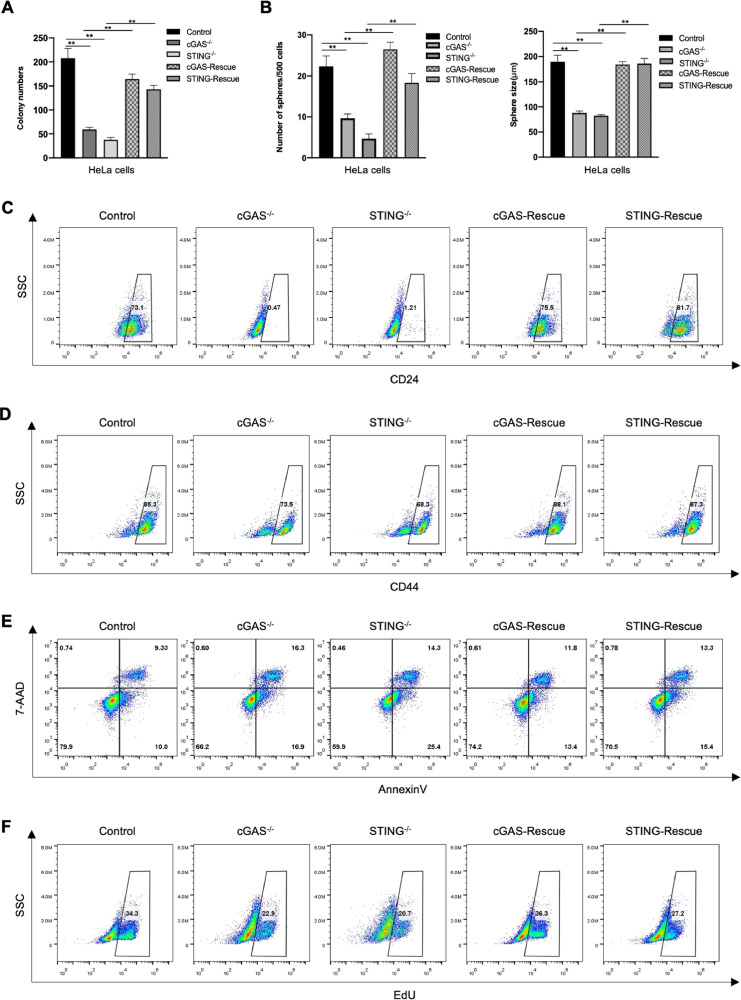
We hereinabove have addressed that cGAS-STING pathway deficiency impaired cancer stemness, thereby delaying tumor progression. So, we wondered whether restoration of cGAS or STING expression could rescue tumor stemness or not. Accordingly, the overexpression plasmids were transferred into the cGAS- or STING-knockout cancer cells, and the recovery effects were confirmed by Western blot (Supplementary Fig. S5A, B). It was found that the cells with recovery expression of cGAS or STING had similar colony-formation capability to that with the control group in soft-agar (Figs. 6A and Supplementary S5C). Similar results were also observed under suspension conditions, which the ability of tumorsphere formation in the recovery group was nearly equal to that of the control group (Figs. 6B and Supplementary S5D).
Fig. 6.
Rescued cGAS or STING expression recovers cancer stemness. (A) Quantification of soft agar colony numbers of Control, cGAS-/- with empty vector, STING-/- with empty vector, cGAS- and STING-rescued HeLa cells. (B) Quantification of sphere numbers and tumorsphere size of the indicated HeLa cells. (C-D) Representative results of FACS detecting CD24+ or CD44+ cells in the indicated HeLa cells. (E) Representative scatter spots of apoptotic cells of the indicated HeLa cells. (F) Representative results of EdU staining positive cells in the indicated HeLa cells after cultured in suspension for 48 h. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01.
We then investigated whether the expression of cancer stem cell markers was also reinvigorated after the cGAS-STING pathway was restored. As shown, recovering of cGAS-STING pathway reversed the decrease of ratio of CD44+ and CD24+ cells caused by cGAS- or STING-depletion (Fig. 6C, D). Moreover, the anti-apoptotic ability of tumor cells was also accordingly rescued when cGAS or STING expression was recovered, which was evidenced by the results of Annexin V assay (Fig. 6E). Meanwhile, the proliferation ability of tumor cells in suspension culture was also restored (Fig. 6F). In conclusion, the recovery of the cGAS-STING pathway could rescue the stemness of tumor cells, which further demonstrated the crucial role of cGAS-STING cascade in cancer stemness maintenance.
cGAS-STING pathway sustains cancer stemness through STAT3 activation
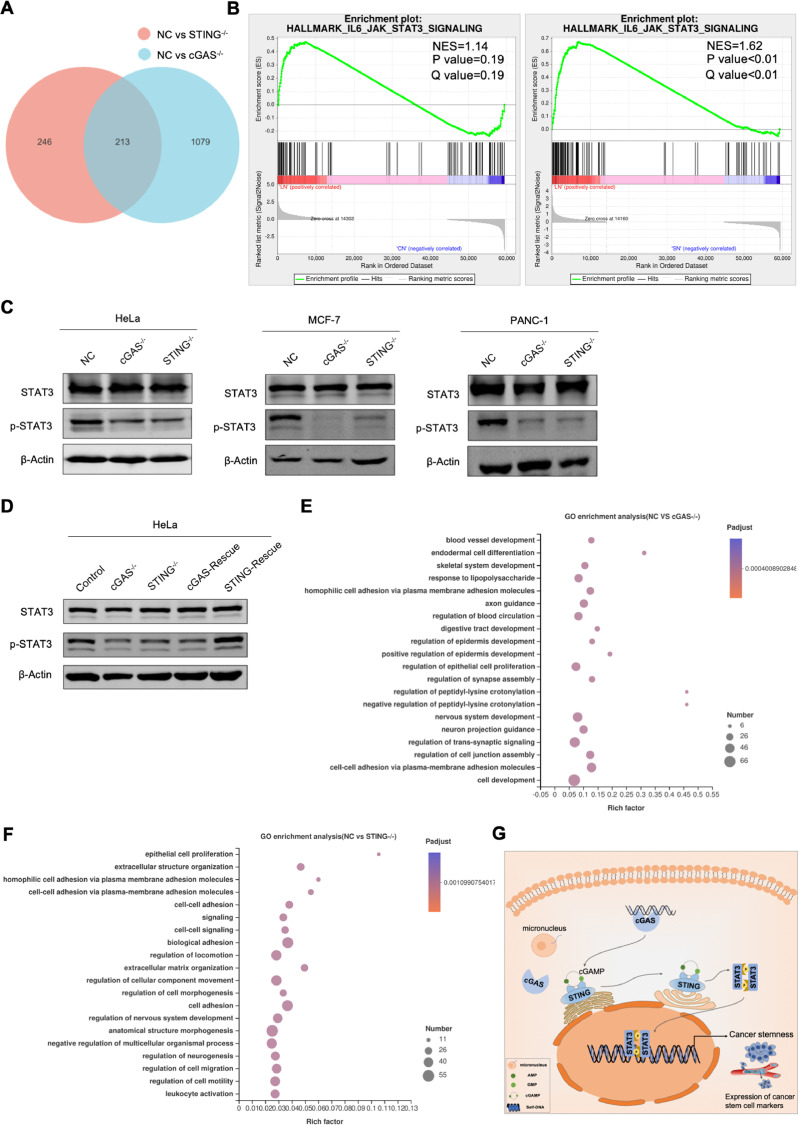
We further explored the molecular mechanisms by which the cGAS-STING pathway sustained cancer stemness. Thereupon, we performed RNA-seq on regular, cGAS- or STING-knockout HeLa cells, and carried out the differential gene expression analysis. DEseq2 analysis showed that, comparing to that of regular cancer cells, around 1300 genes were significantly altered in cGAS-knockout cells, while about 500 genes were significantly altered in STING-knockout cells (p < 0.05) (Figs. 7A and Supplementary S5E).
Fig. 7.
cGAS-STING pathway regulates cancer stemness via improving STAT3 activity. (A) Venn diagram depicts the number of genes whose expression changes significantly in cGAS- or STING-knockout cells compared to the NC (P < 0.05). (B) GSEA analysis of JAK/STAT3 signaling in NC vs cGAS-/- or STING-/- group. (C) Western blot analysis of STAT3 expression and STAT3 phosphorylation in NC, cGAS-/- and STING-/- cells from different cancer cells. (D) Western blot analysis of STAT3 expression and STAT3 phosphorylation in control, cGAS-/- with empty vector, STING-/- with empty vector, cGAS- and STING-rescued HeLa cells. (E, F) GO analysis of significantly changed genes in NC vs cGAS-/- group or NC vs STING-/- group. (G) Schematic diagram of cGAS-STING pathway sustaining cancer stemness by activation of STAT3 activity. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01 from unpaired Student's t-test.
Furthermore, Gene Set Enrichment Analysis (GSEA) showed that genes related to JAK/STAT3 pathway were significantly enriched in NC vs STING-/- group, and their expression was downregulated in STING-knockout cells. This result indicated that cGAS-STING pathway regulated the activity of STAT3 (Fig. 7B and Supplementary Table S4). Many studies showed the importance of the JAK/STAT3 pathway, especially the activation of STAT3, which was closely related to tumorigenesis and cancer progression [38,39]. Thus, we detected phosphorylation of STAT3 by Western blot, and found that loss of cGAS or STING caused the reduction of STAT3 activity (Fig. 7C). Conversely, overexpression of cGAS or STING rescued the inhibition on STAT3 (Fig. 7D).
We further investigated the influence of cGAS- or STING-knockout on other functional pathway via Gene Ontology (GO) enrichment analysis. GO analysis of the differentially expressed genes showed that most of them clustered at pathway influencing cell development, growth and migration, which consisted with the phenomenon observed in vivo and in vitro (Fig. 7E, F). Together, these evidences suggested that the cGAS-STING cascade induced STAT3 activation to sustain the stem-like properties of cancer cells.
Discussion
Hereinabove, we provided solid evidences that activation of cGAS-STING pathway potentiated tumor progression via sustaining cancer stemness. First, cGAS-STING was proved to be activated in different tumor cells. cGAS- or STING-knockout prevented tumorigenesis. Loss of cGAS-STING cascade obstructed tumor metastasis, and extended the survival time of mice. Meanwhile, cGAS-STING pathway deficiency promoted tumor apoptosis, and impeded tumor growth. Mechanistically, we found that cGAS-STING pathway potentiated tumorigenesis via sustaining cancer stemness, and the STAT3 protein might be a bridge between cGAS-STING pathway and cancer stemness (Fig. 7G).
Actually, the cytosolic DNA-sensing cGAS-STING pathway plays an important but dichotomous role in tumor therapy. Damaged DNAs stimulated cGAS-STING cascade, and further mediated the secretion of pro-inflammatory factors and induced cancer cells to enter a state of senescence [40]. Cellular senescence restricted tumor growth [41], yet, chronic SASP-related inflammation enabled immune suppression and metastasis [10]. Autophagy maintained cell homeostasis by removing damaged organelles, misfolded proteins, and oxygen radicals [42,43]. cGAS-STING pathway-derived autophagy potentiated the death of cells undergoing replicative crisis, protecting from neoplastic transformation [44]. However, on the other hand, augment of autophagy supported cancers to survival from stressful threats and fueled metastasis, which in turn promoted tumor progression [45]. While STING agonist has been proved to elevate the sensitivity of cancers to anti-PD1 therapy, it upregulated the expression of PD-L1 and indoleamine 2,3-dioxygenase (IDO) at the same time, which instead contributed to immune escape[12,46]. Thus, cGAS-STING pathway activation might be a double-edged sword. Multiple treatments, such as radiotherapy, chemotherapy and agonists, which triggers cGAS-STING cascade to cure tumors, should be used with more cautions.
Activation of cGAS-STING pathway elicited by external stimulations had attracted much attention, but its basal activation in tumors was neglected. It was known that ongoing chromosome mis-segregation (also known as CIN) was a prominent feature of tumor, and cytosolic DNAs produced by CIN were major triggers of cGAS-STING cascade [47]. The dsDNAs that were not cleared in time stimulated cGAS-STING pathway to continuously produce inflammatory factors to shape an inflammatory microenvironment. In turn, chronic inflammation further aggravated CIN, forming a positive feedback mechanism to potentiate tumor growth and metastasis [48,49]. Our study showed that the activation of cGAS-STING pathway without external stimulations potentiated tumorigenesis and distant metastasis. In line with our observations, a previous study has indicated that STING-knockout attenuated growth of Lewis lung carcinoma [50]. Although STING-knockdown did not exhibit inhibitory effects on tumor growth in some models, we speculated that it may be due to the possibility of shRNA off-target [51,52]. Beyond that, we noticed that DMXAA, a STING agonist, restricted tumor growth, which was contrast to our results [53]. Indeed, DMXAA failed to delay tumor progression in clinical trial, because that DMXAA-STING interaction was mouse-specific [54]. However, the role of cGAS-STING pathway in tumor is still obscure, and the specific mechanism of cGAS-STING pathway regulating tumor development remains to be further explored.
Herein, we proposed that cGAS-STING pathway potentiated tumorigenesis by sustaining cancer stemness. Stemness is often used as a literature term to refer to the molecular program that maintains the state of stem cells [55]. The inherent activation of cGAS-STING cascade has been proved to preserve the stemness of CD8+ T cells, thereby promoting T cell therapy [56]. We found that cGAS- or STING-knockout impaired tumor initiation and tumorsphere formation, while recovery of cGAS or STING expression in the knockout cells rescued this inhibitory effect. Loss of cGAS-STING pathway decreased CSC markers expression. Conversely, recovering cGAS-STING signaling retrieved expression of these marker. Collectively, our findings identified that activation of cGAS-STING signalings preserved cancer stemness.
Moreover, we examined gene expression in cGAS- and STING-knockout cells by next-generation sequencing, and found that the expression of JAK-STAT3 pathway-associated genes were down-regulated in cGAS- or STING-depleted cells, indicating the inhibitor role of cGAS-STING pathway deficiency on STAT3 activity. The alterations in STAT3 phosphorylation confirmed this result. It has been reported that STAT3 mediated self-renew of pluripotent stem cells and promoted tumorigenesis, and targeting STAT3 restrained tumor survival and invasion [57,58]. Thus, we proposed that STAT3 might be an effector protein of cGAS-STING pathway to maintain cancer stemness. Of course, there is no doubt that certain limitations remain in our study, and more research is highly needed.
In short, our study demonstrated that cell-intrinsic activation of cGAS-STING pathway potentiated tumor progression via preserving cancer stemness. The regulation of cGAS-STING pathway on cancer stemness might be achieved by increasing STAT3 activity. Previous studies have shown that activation of cGAS-STING pathway induced type I IFNs and inflammatory factors, and then promoted immune infiltration, and delayed tumor progression [8]. Moreover, prophylactic use of STING agonists increases tumor sensitivity to anti-PD-1 therapy [12]. However, others reported that the activation of cGAS-STING pathway provided opportunities for tumor immune evasion and distant metastasis [10], impairing long time benefits for patients. Therefore, the strategy of activating cGAS-STING pathway to treat tumors needs to be carefully selected, and the potential risks need to be strictly evaluated in clinical research.
Conclusion
In conclusion, we have found that intrinsic activation of cGAS-STING pathway potentiates tumor progression by preserving cancer stemness. Our results provide a new insight for the study of cGAS-STING pathway in cancer.
CRediT authorship contribution statement
Fu-rao Liu: Conceptualization, Methodology, Formal analysis, Visualization, Writing – original draft, Investigation. Ming-jie Jiang: Software, Methodology, Formal analysis. Zhu Mei: Methodology, Resources. Chen-jing Lin: Formal analysis, Software. Ling Tian: Conceptualization, Supervision, Writing – original draft, Writing – review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgment
This work is supported by the National Natural Science Foundation of China (Grant No. 82073203). We thank Mrs Sun Pan, and Mrs Zhang Na from Department of Central Laboratory of Shanghai Chest Hospital for their technical help.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101404.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Bejarano L., Jordao M.J.C., Joyce J.A. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11:933–959. doi: 10.1158/2159-8290.CD-20-1808. [DOI] [PubMed] [Google Scholar]
- 3.Fridman W.H., Zitvogel L., Sautes-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 4.Jiang M.J., Gu D.N., Dai J.J., Huang Q., Tian L. Dark side of cytotoxic therapy: chemoradiation-induced cell death and tumor repopulation. Trends Cancer. 2020;6:419–431. doi: 10.1016/j.trecan.2020.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Fouad Y.A., Aanei C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017;7:1016–1036. [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A., Alegre M.L., Gajewski T.F. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motwani M., Pesiridis S., Fitzgerald K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 10.Kwon J., Bakhoum S.F. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 2020;10:26–39. doi: 10.1158/2159-8290.CD-19-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang M., Chen P., Wang L., Li W., Chen B., Liu Y., Wang H., Zhao S., Ye L., He Y., Zhou C. cGAS-STING, an important pathway in cancer immunotherapy. J. Hematol. Oncol. 2020;13:81. doi: 10.1186/s13045-020-00916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu J., Kanne D.B., Leong M., Glickman L.H., McWhirter S.M., Lemmens E., Mechette K., Leong J.J., Lauer P., Liu W., Sivick K.E., Zeng Q., Soares K.C., Zheng L., Portnoy D.A., Woodward J.J., Pardoll D.M., Dubensky T.W., Kim Y. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 2015;7:283ra52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevtsov M., Sato H., Multhoff G., Shibata A. Novel approaches to improve the efficacy of immuno-radiotherapy. Front. Oncol. 2019;9:156. doi: 10.3389/fonc.2019.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatch E.M., Fischer A.H., Deerinck T.J., Hetzer M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakhoum S.F., Cantley L.C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L., Jiang Y., Fei Y., Zhu C., Tan R., Jungblut P., Pei G., Dorhoi A., Yan Q., Zhang F., Zheng R., Liu S., Liang H., Liu Z., Yang H., Chen J., Wang P., Tang T., Peng W., Hu Z., Xu Z., Huang X., Wang J., Li H., Zhou Y., Liu F., Yan D., Kaufmann S.H.E., Chen C., Mao Z., Ge B. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 17.Liang H., Deng L., Hou Y., Meng X., Huang X., Rao E., Zheng W., Mauceri H., Mack M., Xu M., Fu Y.X., Weichselbaum R.R. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 2017;8:1736. doi: 10.1038/s41467-017-01566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pattabiraman D.R., Weinberg R.A. Tackling the cancer stem cells - what challenges do they pose? Nat. Rev. Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lytle N.K., Barber A.G., Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer. 2018;18:669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merlos-Suarez A., Barriga F.M., Jung P., Iglesias M., Cespedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Munoz P., Clevers H., Sancho E., Mangues R., Batlle E. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Skvortsova I., Debbage P., Kumar V., Skvortsov S. Radiation resistance: cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Semin. Cancer Biol. 2015;35:39–44. doi: 10.1016/j.semcancer.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Abad E., Graifer D., Lyakhovich A. DNA damage response and resistance of cancer stem cells. Cancer Lett. 2020;474:106–117. doi: 10.1016/j.canlet.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Vitale I., Manic G., De Maria R., Kroemer G., Galluzzi L. DNA damage in stem cells. Mol. Cell. 2017;66:306–319. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Blanpain C., Mohrin M., Sotiropoulou P.A., Passegue E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8:16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Badea L., Herlea V., Dima S.O., Dumitrascu T., Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. Dataset. [PubMed] [Google Scholar]
- 26.Pei H., Li L., Fridley B.L., Jenkins G.D., Kalari K.R., Lingle W., Petersen G., Lou Z., Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. Dataset. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson A.L., Wang Z.C., De Nicolo A., Lu X., Brown M., Miron A., Liao X., Iglehart J.D., Livingston D.M., Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. Dataset. [DOI] [PubMed] [Google Scholar]
- 28.Bowen N.J., Walker L.D., Matyunina L.V., Logani S., Totten K.A., Benigno B.B., McDonald J.F. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med. Genom. 2009;2:71. doi: 10.1186/1755-8794-2-71. Dataset. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pita J.M., Figueiredo I.F., Moura M.M., Leite V., Cavaco B.M. Cell cycle deregulation and TP53 and RAS mutations are major events in poorly differentiated and undifferentiated thyroid carcinomas. J. Clin. Endocrinol. Metab. 2014;99:E497–E507. doi: 10.1210/jc.2013-1512. Dataset. [DOI] [PubMed] [Google Scholar]
- 30.Reyes I., Reyes N., Suriano R., Iacob C., Suslina N., Policastro A.M.A., Schantz S., Tiwari R.K., Geliebter J. Gene expression profiling identifies potential molecular markers of papillary thyroid carcinoma. Cancer Biomark. 2019;24:71–83. doi: 10.3233/CBM-181758. Dataset. [DOI] [PubMed] [Google Scholar]
- 31.Khamas A., Ishikawa T., Shimokawa K., Mogushi K., Iida S., Ishiguro M., Mizushima H., Tanaka H., Uetake H., Sugihara K. Screening for epigenetically masked genes in colorectal cancer using 5-Aza-2’-deoxycytidine, microarray and gene expression profile. Cancer Genom. Proteom. 2012;9:67–76. Dataset. [PubMed] [Google Scholar]
- 32.Hong Y., Ho K.S., Eu K.W., Cheah P.Y. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin. Cancer Res. 2007;13:1107–1114. doi: 10.1158/1078-0432.CCR-06-1633. Dataset. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim S.A., Gadalla R., El-Ghonaimy E.A., Samir O., Mohamed H.T., Hassan H., Greve B., El-Shinawi M., Mohamed M.M., Gotte M. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, notch and EGFR signaling pathways. Mol. Cancer. 2017;16:57. doi: 10.1186/s12943-017-0621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhat-Nakshatri P., Kumar B., Simpson E., Ludwig K.K., Cox M.L., Gao H., Liu Y., Nakshatri H. Breast cancer cell detection and characterization from breast milk-derived cells. Cancer Res. 2020;80:4828–4839. doi: 10.1158/0008-5472.CAN-20-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lytle N.K., Ferguson L.P., Rajbhandari N., Gilroy K., Fox R.G., Deshpande A., Schurch C.M., Hamilton M., Robertson N., Lin W., Noel P., Wartenberg M., Zlobec I., Eichmann M., Galvan J.A., Karamitopoulou E., Gilderman T., Esparza L.A., Shima Y., Spahn P., French R., Lewis N.E., Fisch K.M., Sasik R., Rosenthal S.B., Kritzik M., Von Hoff D., Han H., Ideker T., Deshpande A.J., Lowy A.M., Adams P.D., Reya T. A multiscale map of the stem cell state in pancreatic adenocarcinoma. Cell. 2019;177:572–586. doi: 10.1016/j.cell.2019.03.010. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimmakayala R.K., Leon F., Rachagani S., Rauth S., Nallasamy P., Marimuthu S., Shailendra G.K., Chhonker Y.S., Chugh S., Chirravuri R., Gupta R., Mallya K., Prajapati D.R., Lele S.M., Caffrey T.C., Grem J.L., Grandgenett P.M., Hollingsworth M.A., Murry D.J., Batra S.K., Ponnusamy M.P. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene. 2021;40:215–231. doi: 10.1038/s41388-020-01518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang M.J., Chen Y.Y., Dai J.J., Gu D.N., Mei Z., Liu F.R., Huang Q., Tian L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer. 2020;19:68. doi: 10.1186/s12943-020-01178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson D.E., O'Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He W., Wu J., Shi J., Huo Y.M., Dai W., Geng J., Lu P., Yang M.W., Fang Y., Wang W., Zhang Z.G., Habtezion A., Sun Y.W., Xue J. IL22RA1/STAT3 signaling promotes stemness and tumorigenicity in pancreatic cancer. Cancer Res. 2018;78:3293–3305. doi: 10.1158/0008-5472.CAN-17-3131. [DOI] [PubMed] [Google Scholar]
- 40.Yang H., Wang H., Ren J., Chen Q., Chen Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J., Simithy J., Lan Y., Lin Y., Zhou Z., Capell B.C., Xu C., Xu M., Kieckhaefer J.E., Jiang T., Shoshkes-Carmel M., Tanim K., Barber G.N., Seykora J.T., Millar S.E., Kaestner K.H., Garcia B.A., Adams P.D., Berger S.L. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550:402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N., Levine B. Autophagy in human diseases. N. Engl. J. Med. 2020;383:1564–1576. doi: 10.1056/NEJMra2022774. [DOI] [PubMed] [Google Scholar]
- 43.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nassour J., Radford R., Correia A., Fuste J.M., Schoell B., Jauch A., Shaw R.J., Karlseder J. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vera-Ramirez L., Vodnala S.K., Nini R., Hunter K.W., Green J.E. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 2018;9:1944. doi: 10.1038/s41467-018-04070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng A.N., Cheng L.C., Kuo C.L., Lo Y.K., Chou H.Y., Chen C.H., Wang Y.H., Chuang T.H., Cheng S.J., Lee A.Y. Mitochondrial lon-induced mtDNA leakage contributes to PD-L1-mediated immunoescape via STING-IFN signaling and extracellular vesicles. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., Osborn R.T., Wheeler A.P., Nowotny M., Gilbert N., Chandra T., Reijns M.A.M., Jackson A.P. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comaills V., Kabeche L., Morris R., Buisson R., Yu M., Madden M.W., LiCausi J.A., Boukhali M., Tajima K., Pan S., Aceto N., Sil S., Zheng Y., Sundaresan T., Yae T., Jordan N.V., Miyamoto D.T., Ting D.T., Ramaswamy S., Haas W., Zou L., Haber D.A., Maheswaran S. Genomic instability is induced by persistent proliferation of cells undergoing epithelial-to-mesenchymal transition. Cell Rep. 2016;17:2632–2647. doi: 10.1016/j.celrep.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.A., Murphy C.J., Ly P., Shah P., Sriram R.K., Watkins T.B.K., Taunk N.K., Duran M., Pauli C., Shaw C., Chadalavada K., Rajasekhar V.K., Genovese G., Venkatesan S., Birkbak N.J., McGranahan N., Lundquist M., LaPlant Q., Healey J.H., Elemento O., Chung C.H., Lee N.Y., Imielenski M., Nanjangud G., Pe'er D., Cleveland D.W., Powell S.N., Lammerding J., Swanton C., Cantley L.C. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemos H., Mohamed E., Huang L., Ou R., Pacholczyk G., Arbab A.S., Munn D., Mellor A.L. STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Res. 2016;76:2076–2081. doi: 10.1158/0008-5472.CAN-15-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranoa D.R.E., Widau R.C., Mallon S., Parekh A.D., Nicolae C.M., Huang X., Bolt M.J., Arina A., Parry R., Kron S.J., Moldovan G.L., Khodarev N.N., Weichselbaum R.R. STING promotes homeostasis via regulation of cell proliferation and chromosomal stability. Cancer Res. 2019;79:1465–1479. doi: 10.1158/0008-5472.CAN-18-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayman T.J., Baro M., MacNeil T., Phoomak C., Aung T.N., Cui W., Leach K., Iyer R., Challa S., Sandoval-Schaefer T., Burtness B.A., Rimm D.L., Contessa J.N. STING enhances cell death through regulation of reactive oxygen species and DNA damage. Nat. Commun. 2021;12:2327. doi: 10.1038/s41467-021-22572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts Z.J., Ching L.M., Vogel S.N. IFN-beta-dependent inhibition of tumor growth by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) J. Interferon Cytokine Res. 2008;28:133–139. doi: 10.1089/jir.2007.0992. [DOI] [PubMed] [Google Scholar]
- 54.Conlon J., Burdette D.L., Sharma S., Bhat N., Thompson M., Jiang Z., Rathinam V.A., Monks B., Jin T., Xiao T.S., Vogel S.N., Vance R.E., Fitzgerald K.A. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreso A., Dick J.E. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Li W.W., Lu L., Lu J.J., Wang X.R., Yang C., Jin J.S., Wu L.L., Hong X.C., Li F.L., Cao D.Q., Yang Y.Q., Wu M., Su B., Cheng J.K., Yang X.M., Di W., Deng L. cGAS-STING–mediated DNA sensing maintains CD8 T cell stemness and promotes antitumor T cell therapy. Sci. Transl. Med. 2020;12:1–14. doi: 10.1126/scitranslmed.aay9013. [DOI] [PubMed] [Google Scholar]
- 57.Jin W. Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells. 2020;9:217. doi: 10.3390/cells9010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang T., Fahrmann J.F., Lee H., Li Y.J., Tripathi S.C., Yue C., Zhang C., Lifshitz V., Song J., Yuan Y., Somlo G., Jandial R., Ann D., Hanash S., Jove R., Yu H. JAK/STAT3-regulated fatty acid beta-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 2018;27:136–150. doi: 10.1016/j.cmet.2017.11.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.