Abstract
To overcome pathogen infection, plants deploy a highly efficient innate immune system, which often uses hydrogen peroxide (H2O2), a versatile reactive oxygen species, to activate downstream defense responses. H2O2 is a potential substrate of aquaporins (AQPs), the membrane channels that facilitate the transport of small compounds across plasma membranes or organelle membranes. To date, however, the functional relationship between AQPs and H2O2 in plant immunity is largely undissected. Here, we report that the rice (Oryza sativa) AQP OsPIP2;2 transports pathogen-induced apoplastic H2O2 into the cytoplasm to intensify rice resistance against various pathogens. OsPIP2;2-transported H2O2 is required for microbial molecular pattern flg22 to activate the MAPK cascade and to induce the downstream defense responses. In response to flg22, OsPIP2;2 is phosphorylated at the serine residue S125, and therefore gains the ability to transport H2O2. Phosphorylated OsPIP2;2 also triggers the translocation of OsmaMYB, a membrane-anchored MYB transcription factor, into the plant cell nucleus to impart flg22-induced defense responses against pathogen infection. On the contrary, if OsPIP2;2 is not phosphorylated, OsmaMYB remains associated with the plasma membrane, and plant defense responses are no longer induced. These results suggest that OsPIP2;2 positively regulates plant innate immunity by mediating H2O2 transport into the plant cell and mediating the translocation of OsmaMYB from plasma membrane to nucleus.
A rice AQPs acts as a positive immune regulator by transporting H2O2 and modulating a membrane-anchored transcription factor.
Introduction
Reactive oxygen species (ROS) play an important role in plant defenses against pathogen infections, and a major site for ROS production during plant–pathogen interactions is the apoplast (Qi et al., 2017; Zhang et al., 2018; Yang et al., 2020). The apoplastic ROS, especially superoxide (O2−), is mainly produced by the NADPH oxidases (NOXs) located in the plasma membrane (Ryder et al., 2019). Upon perception of microbial molecular patterns such as bacterial flagellin, the plasma membrane-localized pattern-recognition receptor (PRR) complex activates the central immune regulator BOTRYTIS-INDUCED KINASE1 (BIK1). The activated BIK1 is released from the PRR complex and directly phosphorylates NOX to activate apoplastic O2− production (Kimura et al., 2020). The O2− production is followed by its dismutation to hydrogen peroxide (H2O2), which is aided by superoxide dismutase (SOD) present in the apoplast (Baluška and Vivanco, 2009; Sies et al., 2017). H2O2 is a relatively stable component of ROS compared with its precursor. H2O2 can be transported into the cytoplasm by particular members of the aquaporin (AQP) super family (Sies et al., 2017). The cytoplasmic H2O2 acts as a second messenger to mediate multiple defense responses, such as the activation of mitogen-activated protein kinases (MAPKs) and callose deposition (Petrov and Van Breusegem, 2012; Tian et al., 2016; Zhang et al., 2018; Yang et al., 2019).
AQPs were initially reported as water (H2O) channel proteins (Preston and Agre, 1991). More and more studies have revealed that the function of AQPs extends beyond H2O transport. The plasma membrane intrinsic protein (PIP) subfamily members have been shown to transport carbon dioxide (CO2) or H2O2 (Nakhoul et al., 1998; Uehlein et al., 2003; Kaldenhoff and Fischer, 2006; Ludewig and Dynowski, 2009; Bienert and Chaumont, 2014; Zhang et al., 2019a). In mice, AQP3 regulates innate immunity at mucosal surfaces by transporting extracellular H2O2 into the cells (Thiagarajah et al., 2017). Human AQP8 facilitates the diffusional transport of H2O2 across the mitochondrial membrane to regulate cell viability (Marchissio et al., 2012). In plants, AtPIP1;4 contributes to photosynthesis by mediating CO2 transport (Li et al., 2015), and also promotes plant disease resistance by transporting H2O2 from apoplast to cytoplasm (Tian et al., 2016). The cytoplasmic H2O2 transported by AtPIP1;4 activates pattern-triggered immunity (PTI), such as callose deposition and upregulation of defense-related genes (Tian et al., 2016). Interestingly, the enhanced disease resistance by AtPIP1;4 expression in Arabidopsis thaliana (Arabidopsis) is not due to the higher H2O2 accumulation, since NOX and AtPIP1;4 act independently in H2O2 production and transportation (Tian et al., 2016). The role of another AQP AtPIP2;1 is required for cytoplasmic H2O2 accumulation after flg22 or ABA treatment, thus promoting stomatal closure (Rodrigues et al., 2017). In addition to AtPIP1;4 and AtPIP2;1, four other AtPIP2 isoforms (AtPIP2;2, AtPIP2;4, AtPIP2;5, and AtPIP2;7) mediate H2O2 transport in yeast and decrease the viability of yeast (Bienert and Chaumont, 2014). However, it remains unclear if these PIPs also transport H2O2 across plant plasma membranes. It is also largely unknown if PIPs in crops act in a similar way to modulate plant immunity.
The activity of AQPs is modulated by posttranslational modifications (Maurel et al., 2015). Mass spectrometry analyses have revealed that AQPs undergo numerous posttranslational modifications, including phosphorylation, methylation, NH2-terminal acetylation, and ubiquitination (Maurel et al., 2015). Among these modifications, phosphorylation plays a critical role in AQP gating and trafficking. Phosphorylation of serine (S) residues has been identified in the N-terminal region of Arabidopsis PIP2 and maize (Zea mays) PIP1 (Maurel et al., 2009). Phosphorylation of multiple serine and threonine residues has also been identified in the C-terminal tail of Arabidopsis PIP2s (Maurel et al., 2009). An S203A mutation in ZmPIP2;1 reduces its H2O transport activity. The gating activity of spinach (Spinacia oleracea) SoPIP2;1 is activated by the phosphorylation of conserved S197 (Zhang et al., 2019a). Phosphorylation of AtPIP2;1 at S121 promotes H2O2 transport into guard cells to trigger stomatal closure after the treatment of flg22 or ABA (Rodrigues et al., 2017), indicating a role for AQP phosphorylation in response to biotic and abiotic stresses.
In rose (Rosa hybrida), the AQP RhPIP2;1 associates with a membrane-anchored MYB transcriptional factor (maMYB) RhPTM, and the phosphorylation of RhPIP2;1 at S273 promotes the nuclear accumulation of RhPTM C terminus under drought condition. Silencing RhPTM promoted plant growth but decreased the survival rate under drought stress (Zhang et al., 2019b). The role of maMYB in development has also been elucidated (Slabaugh et al., 2011). However, it is still largely unknown if maMYBs play a role during plant–pathogen interactions.
In this study, we identified an AQP OsPIP2;2 from rice, which is strongly induced by pathogen infection and exhibits H2O2 transport activity. We demonstrated that OsPIP2;2 positively regulates plant resistance to bacterial and fungal pathogens partially by transporting apoplastic H2O2 into cytoplasm. In addition, we identified a transcriptional factor OsmaMYB that associates with OsPIP2;2. We demonstrated that OsmaMYB contributes to plant defense and its translocating into the nucleus is regulated by the phosphorylation of OsPIP2;2 at S125. Together, our results indicate that OsPIP2;2 functions as a positive immune regulator by linking H2O2 transport and nuclear accumulation of OsmaMYB, providing a scientific basis for engineering enhanced plant resistance by editing OsPIP2;2 in the future.
Results
OsPIP2;2 acts as a major H2O2-transporting facilitator
We analyzed the expression levels of 11 OsPIP genes in wild-type Nipponbare (NPB) at 8 h after Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99A inoculation or 1 h after 2 mM H2O2 treatment. The expression of most OsPIPs was upregulated by PXO99A or H2O2 treatment, among which the expression levels of OsPIP1;3, OsPIP2;2, OsPIP2;3, and OsPIP2;6 were more obviously induced (Supplemental Figure S1, A and B). Under normal growth conditions, the expression degree of OsPIP2;2 was much higher than that of OsPIP1;3, OsPIP2;3, or OsPIP2;6 (Supplemental Figure S1, C). Given that the total H2O2 content was significantly increased at 8 h postinoculation with PXO99A (Supplemental Figure S1, D), these OsPIPs may link H2O2 signal to plant defense response.
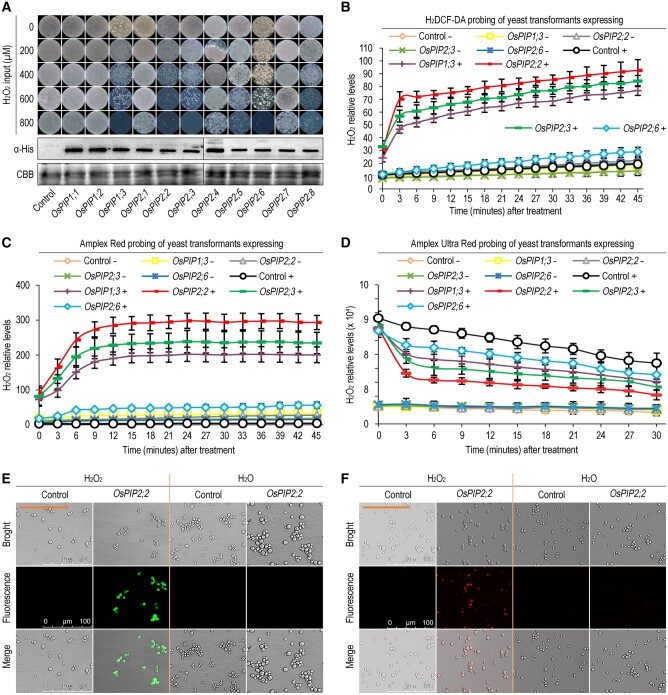
To investigate the H2O2 transport activity of OsPIPs, 11 OsPIPs were cloned and transformed individually into yeast cells. Yeast cells were grown on the SD-Ura medium plate containing different concentrations of H2O2. The results demonstrated that yeast cells transformed with OsPIP1;3, OsPIP2;2, OsPIP2;3, or OsPIP2;6 were more sensitive to H2O2 compared with the empty vector control and other OsPIPs (Figure 1, A), which was caused by toxicity of the transported H2O2 from apoplast to cytoplasm. Western blot assay confirmed that each OsPIP was normally expressed in yeast cells (Figure 1, A). This result indicated that OsPIP1;3, OsPIP2;2, OsPIP2;3, and OsPIP2;6 promote yeast sensitive to H2O2 by mediating H2O2 transport. To further demonstrate the role of these OsPIPs in H2O2 transport, the H2O2 probes 2′,7′-dichlorofluorescin diacetate (H2DCF-DA) and Amplex Red (AR) were employed to evaluate cytoplasmic H2O2, and Amplex Ultra Red (AUR) probe which is impermeable to plasma membrane was used to detect the apoplastic H2O2. The combination of these probes can indicate the transport efficiency of H2O2. The H2O2 in the apoplast and cytoplasm was quantified by a microplate reader. The result indicates that OsPIP1;3, OsPIP2;2, and OsPIP2;3, but not OsPIP2;6, can efficiently transport H2O2 into cytoplasm by using the H2DCF-DA probe, among which OsPIP2;2 exhibited the highest H2O2 transport activity (Figure 1, B and Supplemental Figure S2, A, D, and G). Similar results were obtained by using another two probes AR and AUR (Figure 1, C and D and Supplemental Figure S2, B, C, E, F, H, and I). In addition, OsPIP2;2 exhibits the highest expression level among these three OsPIPs under normal growth conditions (Supplemental Figure S1, C), and its expression was significantly induced by PXO99A inoculation or H2O2 treatment compared to other OsPIPs (Supplemental Figure S1, A and B). Therefore, we focused on OsPIP2;2 for further analyses. To visualize H2O2 transport, laser confocal microscopy was conducted with H2DCF-DA and AR probes. As expected, H2O2 can be efficiently transported into yeast cells by OsPIP2;2 expression as indicated by fluorescent signal (Figure 1, E and F).
Figure 1.
OsPIP2;2 mediates H2O2 transport in yeast. A, Yeast colonies grown on SD-Ura medium with different concentrations of H2O2. The expression levels of OsPIPs in yeast cells were evaluated by western blot using an antibody against His tag, and Coomassie brilliant blue (CBB) staining was used to show protein loading. Photographs were taken at 3 d postincubation. In this figure and in yeast-involved figures provided hereafter, “Control” indicates the empty yeast binary vector without any insert of tested genes, in contrast to the recombinant vector carrying each of the OsPIPs. B, C, and D, Chronological changes of H2O2 content in yeast cells after treatment with 0 or 300 μM H2O2 detected by H2DCF-DA, AR, and AUR probes respectively. The symbol “+” indicates supplied with 300 μM H2O2, and “−” indicates supplied with H2O as control. Data are shown as means ± SEM (n = 8). E and F, H2DCF-DA (E) and AR (F) probing of yeast cells 45 min after treatment with 300 μM H2O2.
We generated OsPIP2;2 mutant lines (Ospip2;2) using CRISPR/Cas9 technology and OsPIP2;2 overexpression (OsPIP2;2OE) lines (Supplemental Figure S3) to investigate if OsPIP2;2 is required for H2O2 transport and defense response in rice. Two representative homozygous mutant lines (Ospip2;2#41, Ospip2;2#279) and two independent OsPIP2;2OE lines (OsPIP2;2OE#1, OsPIP2;2OE#2) were obtained. There is a T insertion in the mutant line Ospip2;2#41, causing a frameshift in OsPIP2;2. A point mutation and three continuous nucleotide deletion (or probably three discontinuous nucleotide deletion) were identified in another mutant Ospip2;2#279 (Supplemental Figure S3, B), so that glutamic acid at position 6 (E6) was mutated into glycine (G) and alanine at position 7 (A7) was deleted in OsPIP2;2 (Supplemental Figure S3, B). The mutation of AtPIP2;1 at E6 interferes with its subcellular localization and H2O transported activity (Sorieul et al., 2011). The OsPIP2;2 overexpression lines were firstly confirmed by genomic polymerase chain reaction (PCR) (Supplemental Figure S3, C). Then the increased transcriptional expression of OsPIP2;2 was confirmed by quantitative reverse transcription PCR (qRT-PCR), and OsPIP2;2 protein accumulation was detected by western blot in OsPIP2;2OE lines (Supplemental Figure S3, D and E). Proper H2O2 production is required for cell elongation (Tian et al., 2018). To evaluate if OsPIP2;2 is involved in H2O2 transport in planta, transgenic plants were grown on 1/2 Murashige and Skoog (MS) medium containing 0.5 mM H2O2. The plumule elongation was significantly promoted in NPB upon H2O2 treatment, and the effect was more obvious in OsPIP2;2-transgenic rice (Supplemental Figure S4). However, the Ospip2;2 lines were insensitive to H2O2 treatment (Supplemental Figure S4), suggesting that OsPIP2;2 can efficiently transport H2O2 into cytoplasm to promote cell elongation. The Ospip2;2 and OsPIP2;2OE lines grow similarly to the wild-type in greenhouse (Supplemental Figure S5), suggesting that OsPIP2;2 does not obviously affect plant growth and development.
OsPIP2;2 contributes to rice resistance against bacterial blight, leaf streak, and rice blast
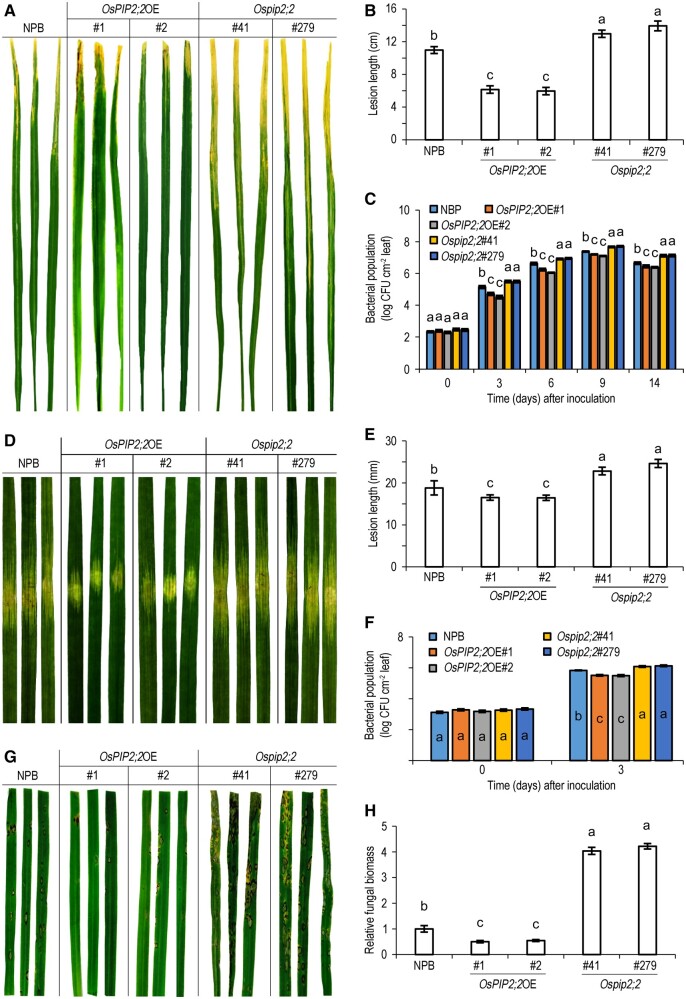
It was widely considered that H2O2 acts as a signal molecule to regulate plant defense (Tian et al., 2016; Yang et al., 2020). Since OsPIP2;2 can transport H2O2, we propose that it modulates plant immunity. To test this hypothesis, we inoculated Ospip2;2, OsPIP2;2OE lines with rice bacterial pathogen Xoo strain PXO99A or X. oryzae pv. oryzicola (Xoc) strain RS105. The OsPIP2;2OE lines were more resistant than the NPB, whereas the Ospip2;2 lines were more susceptible than NPB, as indicated by the lesion length and the bacterial titers after inoculation with PXO99A or RS105 (Figure 2, A–F). To further investigate if OsPIP2;2 enhances rice resistance to fungal pathogens, the transgenic plants were inoculated with Magnaporthe oryzae strain HB3 by spraying a conidia suspension. The OsPIP2;2OE lines showed enhanced resistance to HB3 with scattered lesions and decreased fungal biomass compared to NPB (Figure 2, G and H). In contrast, the Ospip2;2 exhibited enhanced susceptibility to HB3 with increased fungal biomass (Figure 2, H). Together, these results indicate that OsPIP2;2 confers broad-spectrum resistance to multiple pathogens.
Figure 2.
OsPIP2;2 confers resistance to X. oryzae pv. oryzae (Xoo) strain PXO99A, X. oryzae pv. oryzicola (Xoc) strain RS105, and M. oryzae strain HB3. A, Bacterial blight symptoms in leaves 15 d postinoculation with PXO99A using the clipped-end method. In this figure and in plant-involved figures provided hereafter, “NPB” refers to the rice variety NPB, “OsPIP2;2OE” means an OsPIP2;2-overexpressing rice (NPB) line coded with a number during characterization, and “Ospip2;2” indicates an OsPIP2;2-defected NPB line generated by CRISPR/Cas9 and coded with a number. B, Lesion length in (A). Data are shown as means ± SEM (n = 20). C, Titers of PXO99A 3 d postinoculation. Data are shown as means ± SEM (n = 4). D, Bacterial leaf stripe symptoms in leaves 5 d after RS105 infiltration. E, Lesion length in (D). Data are shown as means ± SEMs (n = 20). F, Titers of RS105 3 d postinoculation. Data are shown as means ± SEM (n = 3). G, Rice blast symptoms in leaves 5 d after spray inoculation with HB3 conidia. H, Relative fungal biomass in (G). The fungal biomass was determined using qPCR of the M. oryzae Pot2 gene against the rice OsActin gene. Data are shown as means ± SEM (n = 3). Lowercase letters indicate significant differences at by one-way ANOVA and Duncan’s multiple range tests (P ≤ 0.01).
OsPIP2;2 transports apoplastic H2O2 into cytoplasm and is required for PTI
To evaluate if OsPIP2;2 promotes total H2O2 accumulation, we measured H2O2 content in rice after PXO99A inoculation. No obvious difference was observed in total H2O2 content among the wild-type NPB, OsPIP2;2OE, and Ospip2;2, as indicated by 3,3-diaminobenzidine (DAB) staining and AR quantification after PXO99A inoculation (Supplemental Figure S6, A and B). The H2O2 production was significantly inhibited by the treatment with diphenylene iodonium (DPI), which reduces the H2O2 levels in plants, further confirming the H2O2 accumulation induced by PXO99A inoculation (Supplemental Figure S6, A and B). The activity of SOD, which catalyzes the dismutation of superoxide anion to H2O2, was similar among the wild-type NPB, OsPIP2;2OE, and Ospip2;2 (Supplemental Figure S6, C). In addition, the expression of H2O2-related genes was also similar among these different lines (Supplemental Figure S6, D). Similar results were obtained when these lines were challenged with RS105 or HB3 (Supplemental Figures S7, S8). Together, the results above suggest that OsPIP2;2 does not enhance plant resistance by promoting total H2O2 accumulation.
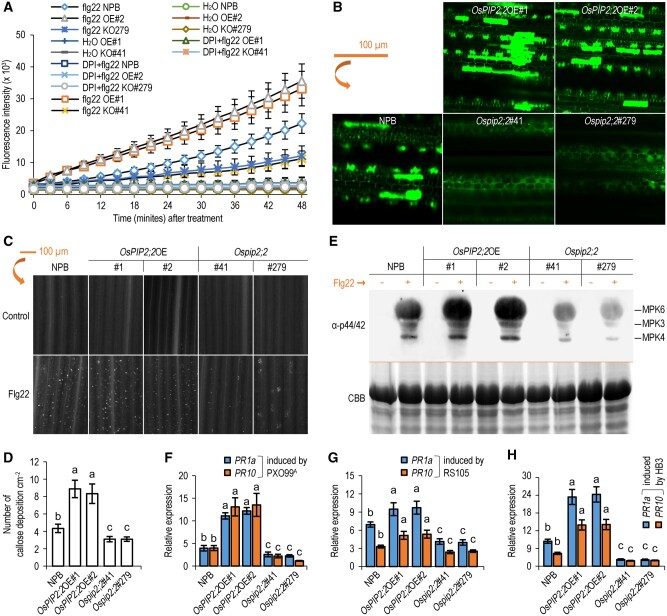
To elucidate if OsPIP2;2 transports H2O2 during plant defense, we analyzed the kinetic changes of AR fluorescent intensity in wild-type NPB, OsPIP2;2OE and Ospip2;2 over 40 min after flg22 treatment. The content of cytoplasmic H2O2 was considerably increased in OsPIP2;2OE compared with the wild-type or Ospip2;2 (Figure 3, A). This result was confirmed by confocal laser scanning microscopy using H2DCF-DA probe (Figure 3, B). These results suggest that OsPIP2;2 transports apoplastic H2O2 into the cytoplasm in planta. Callose deposition is linked to H2O2 signaling, and has been considered as a typical PTI defense response (Tian et al., 2016; Yang et al., 2019). The callose deposition was increased in OsPIP2;2OE but reduced in Ospip2;2 compared with that in the wild-type after flg22 treatment (Figure 3, C and D).
Figure 3.
OsPIP2;2 links PAMP-induced apoplastic H2O2 to the PTI pathway. A, Chronological changes in the H2O2-probing AR fluorescence densities in leaves of 2-week-old rice seedlings after flg22 treatment (means ± SEM, n = 8). OE#1, OsPIP2;2OE#1; OE#2, OsPIP2;2OE#2; KO#41, Ospip2;2#41; KO#279, Ospip2;2#279. B, Cell imaging of the DCF fluorescence by laser confocal microscopy. Leaves were preloaded with H2DCF-DA 45 min before treatment with 300 μM H2O2. C, flg22-induced callose deposition in rice. The leaves of 2-week-old rice seedlings were treated with 10 μM flg22 for 16 h and then stained by aniline blue. B and C, The scale bar representing 100 μm applies to all the images. D, Quantification of callose deposition in leaves in C by ImageJ. Data were shown as means ± SEM (n = 3). E, MAPK activation was detected after flg22 treatment by western blot. The expected identities of the respective bands are marked on the right. The experiment was performed twice with similar results. F–H, Expression levels of pathogenesis-related genes OsPR1a and OsPR10 expression levels in plants with PXO99A, RS105, or HB3 24 h postinoculation. Data are shown as means ± SEM (n = 4). Lowercase letters indicate significant differences by one-way ANOVA and Duncan’s multiple range tests (at P ≤ 0.01).
The MAPK activation, another typical defense response during PTI, was enhanced in OsPIP2;2OE but compromised in Ospip2;2 compared with that in the wild-type (Figure 3, E). Consistently, the expression of defense-related genes was also upregulated in OsPIP2;2OE, but reduced in Ospip2;2 compared with that in the wild-type when plants were challenged with different pathogens (Figure 3, F–H). Collectively, these results suggest that OsPIP2;2 positively regulates PTI by linking H2O2 transport to trigger plant defense response.
The S125 phosphorylation is required for the H2O2 transport activity of OsPIP2;2
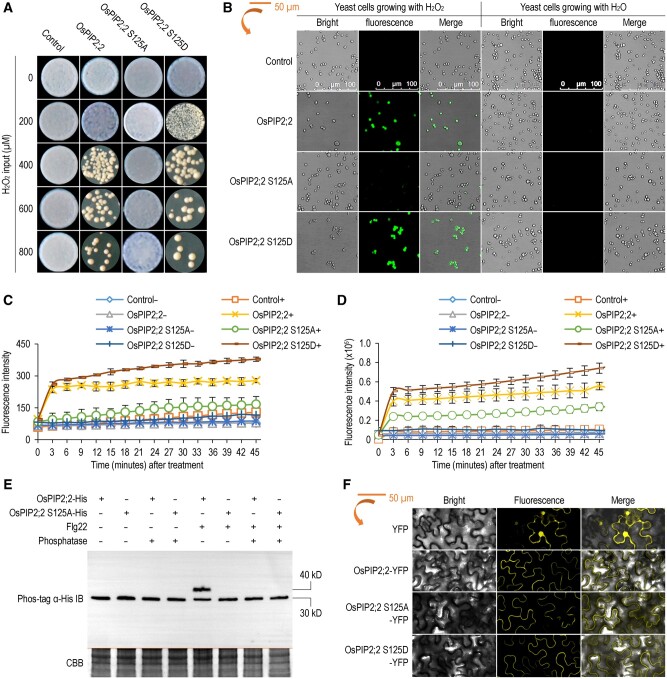
The S121 phosphorylation of AtPIP2;1 is required for flg22- or ABA-induced H2O2 influx in Arabidopsis guard cells (Rodrigues et al., 2017). Multiple protein sequence alignment of AtPIP2;1, OsPIP2;2, ZmPIP2;5, and SoPIP2;1 indicated that the phosphorylation residue is conserved, and the corresponding residue in OsPIP2;2 is S125 (Supplemental Figure S9). To determine the role of S125 phosphorylation in H2O2 transport activity of OsPIP2;2, we generated a phosphorylation deficient mutant (OsPIP2;2 S125A) and a phosphomimetic mutant (OsPIP2;2 S125D). We transformed wild-type OsPIP2;2 and these two phosphorylation mutants into yeast strain NMY51 individually, and evaluated yeast growth on the SD-Ura plate supplemented with H2O2. The yeast transformants expressing OsPIP2;2 S125D were more sensitive to H2O2, whereas the yeast transformants expressing OsPIP2;2 S125A were insensitive to H2O2 compared to the wild-type OsPIP2;2 (Figure 4, A). This result indicates that the S125 phosphorylation of OsPIP2;2 is required for its H2O2 transport activity. In good agreement with this result, we further found that the OsPIP2;2 phosphomimetic mutant can efficiently transport exogenously supplied H2O2 into yeast cells, whereas the phosphorylation deficient mutant cannot (Figure 4, B). Quantification of the cytoplasmic H2O2 further confirmed the role of S125 phosphorylation in OsPIP2;2’s H2O2 transport function (Figure 4, C). To demonstrate the role of S125 phosphorylation in H2O2 transport activity of OsPIP2;2 in planta, we transiently expressed wild-type OsPIP2;2 or its two mutants in tobacco (Nicotiana benthamiana). We confirmed that the OsPIP2;2 phosphomimetic mutant can transport H2O2 efficiently into plant cells, whereas the phosphorylation deficient mutant cannot (Figure 4, D). Similar protein levels were observed among OsPIP2;2, OsPIP2;2 S125A, and OsPIP2;2 S125D as shown by western blot assay (Supplemental Figure S10, A).
Figure 4.
Phosphorylation of OsPIP2;2 at S125 is required for H2O2 transport. A, Yeast transformants grown in SD-Ura medium with H2O2 at 28°C. Photographs were collected at 3 d postincubation. B, Imaging of the DCF fluorescence in yeast cells by laser confocal microscopy using H2DCF-DA dye after treatment with 300 μΜ H2O2. C, Kinetic variations of AR fluorescence densities in yeast transformants after application of 300 μΜ H2O2 over 40 min. Data are shown as means ± SEM (n = 8). D, Kinetic variations of AR fluorescence densities in N. benthamiana expressing OsPIP2;2 or its mutants after H2O2 treatment. Data are shown as means ± SEM (n = 8). E, Immunoblot analysis of OsPIP2;2 phosphorylation at S125 by using His antibody after Phos-tag gel separation. OsPIP2;2 or OsPIP2;2 S125A were expressed in leaves of N. benthamiana, and then the infiltrated leaves were treated with 1 μM flg22 for 30 min before protein extraction. Protein samples treated with (+) or without (–) calf intestinal alkaline phosphatase were separated by a Phos-tag gel. CBB staining was used to show protein loading (bottom panel). F, Subcellular localization of OsPIP2;2 and its mutants in N. benthamiana observed under lasers intensity as in (B).
To clarify if OsPIP2;2 is phosphorylated upon flg22 perception, we expressed OsPIP2;2 and OsPIP2;2 S125A in N. benthamiana leaves. The total protein was separated by a Phos-tag gel, and the phosphorylation was detected by western blot. Increased phosphorylation of OsPIP2;2 was observed upon flg22 treatment in the wild-type OsPIP2;2 but not in the OsPIP2;2 S125A mutant (Figure 4, E), suggesting that the S125 of OsPIP2;2 is phosphorylated after flg22 treatment. Since OsPIP2;2 transports apoplastic H2O2, we evaluated if the S125 phosphorylation of OsPIP2;2 is also induced by H2O2. We transformed OsPIP2;2 and OsPIP2;2 S125A into yeast strain ΔX33, respectively, and detected phosphorylation of these proteins after H2O2 treatment. Similar to flg22 treatment, only the wild-type OsPIP2;2 can be phosphorylated upon H2O2 treatment, but the OsPIP2;2 S125A cannot (Supplemental Figure S10, B). To evaluate if S125 phosphorylation affects OsPIP2;2’s subcellular localization, we expressed yellow-fluorescent protein (YFP)-tagged wild-type OsPIP2;2 and two phosphorylation mutants in N. benthamiana individually, and observed their fluorescent signal using a confocal laser scanning microscope. The wild-type OsPIP2;2 and the phosphorylation mutants exhibited similar subcellular localization, and they were likely to be localized to the plasma membrane (Figure 4, F and Supplemental Figure S10, C).
OsPIP2;2 associates with a membrane-anchored MYB-like transcription factor OsmaMYB that contributes to plant immunity
We identified potential OsPIP2;2 interactors using a split-ubiquitin membrane yeast two-hybrid (MYTH) system. Among 30 potential OsPIP2;2-interacting protein candidates obtained by screening a cDNA library, an Oryza sativa membrane-anchored MYB-like transcription factor (OsmaMYB) was identified, which is a homolog of the previously reported maMYB in rose (Zhang et al., 2019b; Supplemental Figure S11, B). Full-length cDNA of OsmaMYB was cloned and the association was confirmed in yeast two-hybrid system (Figure 5, A). To verify the OsPIP2;2–OsmaMYB association in planta, we co-expressed OsPIP2;2 and OsmaMYB in N. benthamiana, and confirmed their interaction by co-immunoprecipitation (Co-IP) and luciferase (LUC) complementation assays (Figure 5, B–D).
Figure 5.
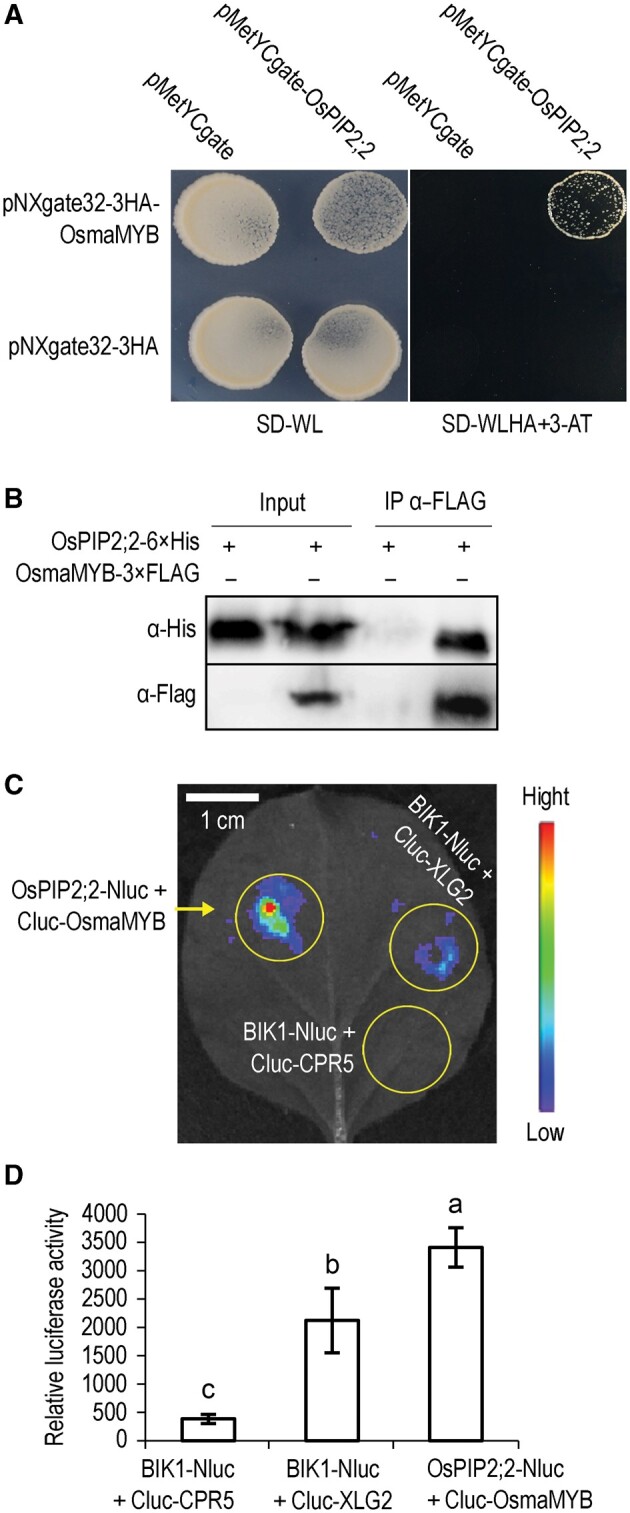
OsPIP2;2 associates with OsmaMYB. A, Associations between OsPIP2;2 and OsmaMYB in the split-ubiquitin yeast two-hybrid system. Yeast cells co-transformed with bait and prey vectors were grown on medium supplemented with 10 mM of 3-amino-1,2,4-triazole. B, Co-IP of OsPIP2;2 and OsmaMYB after co-expression in N. benthamiana. The indicated constructs were co-expressed in N. benthamiana by A. tumefaciens. Total proteins were extracted 2 d postinfiltration, and subjected to anti-FLAG immunoprecipitations. C and D, Association between OsPIP2;2 and OsmaMYB indicated by the split-LUC complementation assay. The indicated constructs were co-expressed in N. benthamiana leaves and then LUC activities were examined in both qualitative (C) and quantitative assays (D). Cluc-CPR5 and BIK1-Nluc were used as the negative control. Cluc-XLG2 and BIK1-Nluc served as the positive control. Data are shown as means ± SEM (n = 8). Lowercase letters indicate significant differences at P ≤ 0.01, Duncan’s multiple range tests and one-way ANOVA.
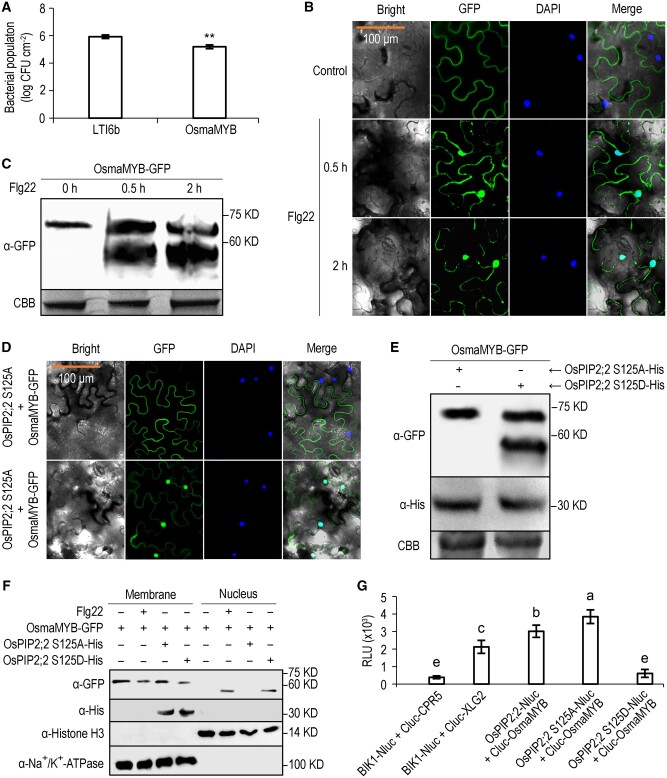
To preliminarily evaluate if OsmaMYB is involved in plant immunity, OsmaMYB was expressed in N. benthamiana and the infiltrated leaves were then inoculated with DC3000 ΔhopQ1-1 48 h postinfiltration. The bacterial titer of DC3000 ΔhopQ1-1 was significantly lower in the leaves expressing OsmaMYB compared with that expressing LTI6b which is a plasma membrane protein as a control (Figure 6, A). This result indicates that OsmaMYB positively regulates plant defense against biotrophic bacterial pathogens.
Figure 6.
OsmaMYB contributes to plant immunity and is activated by phosphorylation of OsPIP2;2 at S125. A, Titers of DC3000 ΔhopQ1-1 3 d post-inoculation. Agrobacterium tumefaciens was used to transiently express OsmaMYB on one-half of an N. benthamiana leaf and LTI6b control on the other. Data are shown as means ± SEM (n = 6). Asterisks indicate significant differences compared to control by Student’s t test (**P ≤ 0.01). The experiment was performed twice with similar results. B, Subcellular localization of OsmaMYB by confocal laser microscopy. OsmaMYB-GFP fusion was transiently expressed in N. benthamiana leaves, and the infiltrated leaves were treated with H2O or flg22. C, Immunoblot analysis of OsmaMYB. The infiltrated leaves were treated with flg22, and samples were collected at three different time points for immunoblot analysis. The OsmaMYB was detected using an anti-GFP antibody. CBB staining is used to show protein loading (bottom panel). D, Subcellular localization of OsmaMYB-GFP after co-expression with OsPIP2;2 S125A or OsPIP2;2 S125D. B and D, orange scale bar representing 100 μm applies to all the images. E, Immunoblot analysis of OsmaMYB-GFP co-expressed with the OsPIP2;2 phosphorylation mutants. F, Subcellular fractionation analysis of the distribution of OsmaMYB. Histone H3 was used as a nuclear marker, Na+/K+-ATPase was used as a membrane marker. G, Quantification of LUC activity. The indicated Nluc and Cluc constructs were co-expressed in N. benthamiana leaves for LUC complementation assay. The relative luminescence unit indicates the strength of protein–protein interaction. Data are shown as mean ± SEM (n = 8). S125A, phosphor-deficient form; S125D, phosphomimetic form. Lowercase letters indicate significant differences at P ≤ 0.01 by Duncan’s multiple range tests and one-way ANOVA. These experiments were performed at least twice with similar results.
To primarily explore how OsmaMYB modulates plant immunity, we analyzed the subcellular localization of OsmaMYB upon flg22 treatment. OsmaMYB contains a predicted nuclear localization signal near the C terminus (Supplemental Figure S11, A), suggesting the possibility of its nuclear localization. In nuclear staining with 4,6-diamidino-2-phenylindole (DAPI), no fluorescent signal of OsmaMYB-green-fluorescent protein (GFP) was observed in the nucleus after H2O treatment, but nuclear accumulation of OsmaMYB-GFP was observed after flg22 treatment (Figure 6, B). The result of western blot demonstrated that an additional band of OsmaMYB-GFP appeared after flg22 treatment (Figure 6, C), indicating that OsmaMYB is cleaved after microbial pattern treatment. To further determine if OsmaMYB is released from the plasma membrane to regulate plant defense, we generated an OsmaMYB mutant by deleting its predicted transmembrane domain (TMD), and transiently expressed it in N. benthamiana leaves. As expected, the deletion mutant OsmaMYBΔTMD escaped from the plasma membrane, and was distributed in the cytoplasm and nucleus (Supplemental Figure S12, A). The infiltrated leaves were then inoculated with DC3000 ΔhopQ1-1, and enhanced resistance was observed in leaves expressing OsmaMYBΔTMD compared with the wild-type OsmaMYB or the LTI6b control (Supplemental Figure S12, B).
The transactivation activity of OsmaMYB was evaluated in yeast cells. The deletion mutant OsmaMYBΔTMD was fused to the DNA binding domain (BD) of the yeast transcription factor GAL4, and the recombinant plasmid was introduced into yeast strain AH109. The yeast cells expressing BD-OsmaMYBΔTMD can grow on the selective medium SD-Trp-His, whereas the cells expressing BD empty vector cannot (Supplemental Figure S12C), suggesting that OsmaMYBΔTMD exhibits transactivation activity.
Phosphorylation of OsPIP2;2 at S125 promotes translocation of OsmaMYB into the nucleus
Membrane-anchored transcriptional factors are maintained in an inactive state by associating with membranes, and are released from the plasma membrane and translocated into the nucleus to regulate transcription of target genes in response to certain signals (Zhang et al., 2019b). Upon flg22 treatment, the OsmaMYB C terminus accumulates in the nucleus (Figure 6, B). We also demonstrated that OsPIP2;2 associated with OsmaMYB (Figure 5). To clarify if the translocation of OsmaMYB C terminus to the nucleus is modulated by phosphorylation of OsPIP2;2 at S125, we co-expressed OsmaMYB-GFP with OsPIP2;2 S125A or OsPIP2;2 S125D in N. benthamiana leaves, and observed the subcellular localization of OsmaMYB-GFP by confocal laser scanning microscopy. No fluorescent signal of OsmaMYB-GFP was observed in the nucleus when OsPIP2;2 S125A was co-expressed, whereas nuclear accumulation of OsmaMYB-GFP was found when OsPIP2;2 S125D was present (Figure 6, D). The western blot result confirmed that OsmaMYB was cleaved when OsPIP2;2 S125D, but not OsPIP2;2 S125A, was co-expressed (Figure 6, E). To further confirm that OsmaMYB is translocated into the nucleus upon the perception of flg22 or phosphorylation of OsPIP2;2 at S125, we performed subcellular fractionation and detected OsmaMYB by western blot. OsmaMYB was only detected in the membrane fraction without flg22 treatment or when co-expressed with OsPIP2;2 S125A. However, it was detected in both membrane and nucleus upon flg22 treatment or when co-expressed with OsPIP2;2 S125D. We also noticed that only the truncated OsmaMYB can be detected in the nucleus, further indicating that OsmaMYB is cleaved and translocated into the nucleus upon flg22 perception or phosphorylation of OsPIP2;2 at S125 (Figure 6, F).
Intriguingly, OsPIP2;2 S125A associates stronger with OsmaMYB, whereas OsPIP2;2 S125D has a lower affinity with OsmaMYB compared with wild-type OsPIP2;2 as indicated by LUC complementation assay (Figure 6, G and Supplemental Figure S13, A). This result was confirmed by yeast two-hybrid assay (Supplemental Figure S13, B). These results indicate that OsmaMYB associates with OsPIP2;2 under normal growth conditions and is released from the OsmaMYB–OsPIP2;2 protein complex upon microbial pattern perception to regulate plant defense.
Discussion
AQPs were initially defined as H2O transporting channels and subsequently were demonstrated to transport many other substrates (Ludewig and Dynowski, 2009; Bienert et al., 2014; Bienert and Chaumont, 2014). In addition to transporting H2O, the PIP subfamily members have been shown to transport H2O2 (Tian et al., 2016; Rodrigues et al., 2017). H2O2 transport connects signals between apoplast and cytoplasm, resulting in plant resistance to pathogens (Tian et al., 2016). In this study, we systematically analyzed the expression of OsPIPs upon pathogen infection and their H2O2 transport activity, and demonstrated that the expression of most OsPIPs is upregulated upon pathogen infection, but only a few of OsPIPs are able to transport H2O2 (Figure 1 and Supplemental Figure S1, A). These results strongly suggest a role for OsPIPs in plant defense. Among these OsPIPs, the expression of OsPIP2;2 is strongly induced by pathogen infection, and exhibits a high basal expression level and H2O2 transport activity. Therefore, we focused on OsPIP2;2.
The OsPIP2;2 overexpression lines are more resistant to three different pathogens compared with the wild-type plants (Figure 2), suggesting that OsPIP2;2 is an ideal target for engineering broad-spectrum resistance in rice. No obvious difference was found in total H2O2 production among the wild-type NPB, OsPIP2;2OE and Ospip2;2 lines, suggesting that OsPIP2;2 does not modulate plant defense by promoting total H2O2 accumulation (Supplemental Figures S6–S8). Therefore, we proposed that OsPIP2;2 may regulate plant immunity by linking H2O2 transport. The H2O2 transported from apoplast can act as a signaling molecule to trigger defense responses (Hu et al., 2017; Sies et al., 2017). Indeed, enhanced callose deposition was observed in the OsPIP2;2 overexpression lines. It has been demonstrated in both animals and plants that H2O2 activates MAPK cascades (Zhang et al., 2018). In this study, MAPK cascade was enhanced in the OsPIP2;2 overexpression lines upon flg22 treatment compared to wild-type, but was significantly compromised in Ospip2;2 (Figure 3, E). The MAPK cascade activates the expression of many defense-related genes such as PR1 and PR10 in the salicylic acid signaling pathway (Medina-Puche et al., 2020). As expected, expression of PR1 and PR10 was significantly upregulated in OsPIP2;2OE after pathogen infection (Figure 3, F–H). Our results strongly support that OsPIP2;2 positively modulates plant resistance to bacterial and fungal pathogens by transporting apoplastic H2O2 into the cytoplasm.
It is still unclear how PIPs are regulated during plant–pathogen interactions. The activity of PIPs can be regulated by protein modifications, such as phosphorylation and dephosphorylation (Prak et al., 2008; Qing et al., 2016). The E6 residue was mutated into glycine in the mutant Ospip2;2#279 and the mutant exhibited compromised H2O2 transport and disease resistance, indicating that the E6 residue of OsPIP2;2 is essential for its function in plant defense. It has been shown that mutation of AtPIP2;1 at E6 interferes with its subcellular localization and H2O transported activity (Sorieul et al., 2011). It was also indicated that the E6 residue in AtPIP2;1 can be methylated (Santoni et al., 2006). However, it is still unclear if methylation at E6 modulates AQP protein targeting. It was demonstrated that the AQP proteins retained partial substrate transport activity after the protein kinase inhibitor K252a treatment (Johansson et al., 1998; Azad et al., 2008), indicating that the AQPs exhibit basal activity at a nonphosphorylated state. The H2O transport activity of spinach SoPIP2;1 is activated by phosphorylation at S197 (Johansson et al., 1996, 1998; Zhang et al., 2019a), and is suppressed by dephosphorylation at S115 and S274 under drought stress (Nyblom et al., 2009). Recently, AtPIP2;1 was demonstrated to transport H2O2 and its activity is activated by phosphorylation at S121. Brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1) and open stomata 1/Snf1-related protein kinase 2.6 (SnRK2.6) were both required for flg22-induced AtPIP2;1 phosphorylation at S121 (Grondin et al., 2015; Rodrigues et al., 2017). Interestingly, the S121 is conserved among the homologs of AtPIP2;1. The corresponding residue in OsPIP2;2 is S125. We demonstrated that the OsPIP2;2 phosphorylation deficient mutant failed to transport H2O2, whereas the phosphomimetic mutant transported apoplastic H2O2 into the cytoplasm more efficiently (Figure 4, A–D), highlighting the role of S125 phosphorylation in OsPIP2;2’s activity. In Arabidopsis, both BAK1 and SnRK2.6 are required for AtPIP2;1 phosphorylation at S121 (Rodrigues et al., 2017). The S125 phosphorylation of OsPIP2;2 was detected after flg22 treatment in N. benthamiana (Figure 4, E), indicating that the co-receptor BAK1 is involved in OsPIP2;2 phosphorylation. The SnRK–PIP association has been demonstrated before (Song et al., 2019), so it is not surprising that SnRK family members are required for OsPIP2;2 phosphorylation. The phosphorylation of OsPIP2;2 at S125 is detected in yeast cells after H2O2 treatment, and the SnRK family exists in yeast (Supplemental Figure S10, B). Therefore, we propose that BAK1 and SnRK2.6 homologs in rice are responsible for the S125 phosphorylation of OsPIP2;2.
A membrane-anchored transcription factor OsmaMYB was identified as an interactor of OsPIP2;2 (Figure 5). We also demonstrated that several other OsPIPs with H2O2 transport activity also associated with OsmaMYB (Supplemental Figure S13, A and B), indicating that these OsPIPs utilize a similar mechanism to modulate plant defense. However, OsPIP2;2 associated more strongly with OsmaMYB (Supplemental Figure S13, A and B) and exhibited higher basal expression level (Supplemental Figure S1, C). It was reported that OsPIP2;2 may recruit OsPIP1;3 to the plasma membrane (Liu et al., 2019), indicating that OsPIP2;2 can modulate other OsPIPs. The evidence above together may explain why mutating OsPIP2;2 alone significantly compromised disease resistance and H2O2 transport (Figures 2, 3).
The role of maMYB in development has been characterized (Slabaugh et al., 2011). Recently, a membrane-anchored MYB protein, RhPTM, was reported to associate with an AQP RhPIP2;1 to regulate the trade-off between growth and stress survival in rose (Zhang et al., 2019b). Although it has been well documented that some MYB transcriptional factors contribute to plant resistance (Chinnusamy and Zhu, 2009), the role of maMYB subfamily in plant–pathogen interactions is still poorly understood. To uncover if OsmaMYB is involved in plant defense, we transiently expressed OsmaMYB in N. benthamiana, and inoculated the infiltrated leaves with bacterial pathogen DC3000 ΔhopQ1-1. The result indicates that OsmaMYB positively modulates plant immunity (Figure 6, A). To preliminarily elucidate how OsmaMYB modulates plant immunity, subcellular localization of OsmaMYB was observed after flg22 treatment. OsmaMYB was localized to the plasma membrane at a resting state (Figure 6, B), but was cleaved and translocated into the nucleus after flg22 treatment (Figure 6, B and C), indicating that it is released from the plasma membrane upon the microbial pattern perception. The truncated OsmaMYBΔTMD conferred more resistance to pathogen infection compared with the full-length OsmaMYB (Supplemental Figure S12, B), further suggesting that OsmaMYB needs to be released from the plasma membrane to regulate plant defense. OsmaMYB was maintained in the plasma membrane when co-expressed with the phosphorylation deficient mutant OsPIP2;2 S125A, but was processed and translocated into the nucleus when co-expressed with the phosphomimic mutant OsPIP2;2 S125D (Figure 6, D–F). This result indicates that OsmaMYB is dynamically modulated by the phosphorylation of OsPIP2;2 at S125. OsmaMYB associated more strongly with OsPIP2;2 S125A than OsPIP2;2 S125D (Figure 6, G and Supplemental Figure S13, A–C), further confirming that OsmaMYB is regulated by the S125 phosphorylation of OsPIP2;2. Under normal growth condition, we did not observe obvious phosphorylation signal by Phos-tag analysis (Figure 4, E and Supplemental Figure S10, B), indicating that OsPIP2;2 is not phosphorylated or phosphorylated at a very low level. We proposed that OsPIP2;2 retains its basal substrate transport activity under normal growth condition, and its activity is enhanced by phosphorylation and results in cleavage of OsmaMYB. Thus, we propose that OsPIP2;2 associates with OsmaMYB without treatment, and is phosphorylated at S125 upon microbial perception, causing the processing and release of OsmaMYB from the plasma membrane (Figure 7). The truncated C terminus of OsmaMYB is then translocated into the nucleus and activates the expression of defense-related genes (Figure 7). Hence, we propose that OsPIP2;2 enhances plant resistance by mediating H2O2 transport into the plant cells and mediating translocation of OsmaMYB from the plasma membrane to the nucleus, providing an effective mechanism to finely modulate plant immunity.
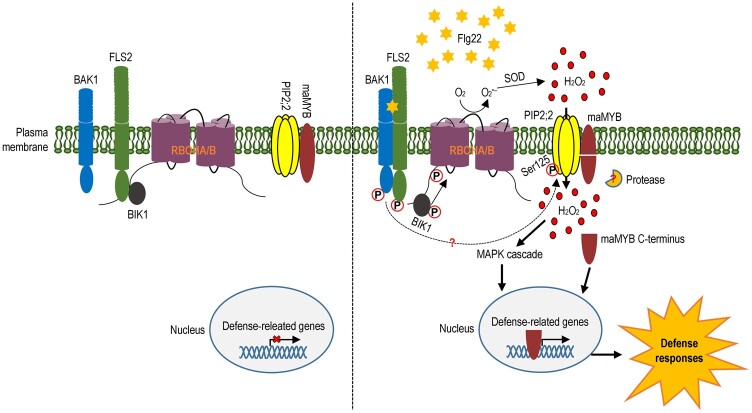
Figure 7.
Model for the modulation of plant immunity by OsPIP2;2 through H2O2 transport and OsmaMYB. At a resting state, OsPIP2;2 associates with the membrane-anchored OsmaMYB, and plant defense cannot be activated. Upon perception of microbial pattern flg22, the membrane-associated enzyme RBOHA/B is activated to catalize H2O2 generation in the apoplast. Upon flg22 perception, moreover, the AQP OsPIP2;2 is phosphorylated at S125 and therefore functions to mediate the H2O2 transport into the cytoplasm. The transported H2O2 triggers a series of defense responses including MAPK cascade, callose deposition, and upregulation of defense-related genes. Meanwhile, OsmaMYB is processed by an unknown protease, and the C terminus is released from the plasma membrane and de-associates with the OsPIP2;2 complex. Thereafter OsmaMYB is translocated into the nucleus and activates the expression of defense-related genes. In this way, OsPIP2;2 enhances plant resistance by linking H2O2 transport and OsmaMYB.
Materials and methods
All the gene constructs were sequenced to make sure they were correct. The primers used in this study are provided in Supplemental Table S1.
Biological materials and growth conditions
Rice NPB and N. benthamiana plants were grown in a growth chamber at 27°C and 70% relative humidity under a 16-h light and 8-h dark photoperiod. Escherichia coli strains were grown in Luria–Bertani (LB) medium containing appropriate antibiotics at 37°C. Agrobacterium tumefaciens strains were cultured in LB medium supplemented with rifampicin at 28°C. Saccharomyces cerevisiae strain NMY51 was grown on yeast extract peptone dextrose agar) medium at 28°C. The Pichia pastoris deletion strain ΔX33 containing a disrupted endogenous AQY1 gene was cultured on yeast extract peptone agar medium at 28°C. Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99A and X. oryzae pv. oryzicola (Xoc) RS105 were grown in nutrient broth (NB) or NB amended with agar at 28°C. The M. oryzae strain HB3 was cultured on potato sucrose agar at 28°C, and conidial formations were induced under light for 2–3 d after removing the surface mycelium.
RNA and DNA isolation
RNA or DNA was extracted from leaves of 2-week-old rice seedlings. Total RNA was isolated using TRIzol according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Genomic DNA was extracted following the cetyltrimethylammonium bromide-based method as described previously (Porebski et al., 1997).
qRT-PCR
One microgram of total RNA was reverse transcribed with an oligo(dT) primer using HiScript Q RT SuperMix for qRT-PCR (Cat#R123-01, Vazyme, Nanjing, China). The OsActin1 gene was used as an internal control. qRT-PCR was performed in a 20-μL reaction using ChamQ SYBR qPCR Master Mix following the manufacturer’s recommendation (Cat#Q341-02; Vazyme). qRT-PCR primers are provided in Supplemental Table S1. Reactions were performed on a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) under the following conditions: 95°C for 30 s, then 40 cycles at 95°C for 10 s and 60°C for 30 s, followed by 95°C for 15 s and 60°C for 1 min, and then 95°C for 15 s to obtain melt curves. The relative expression level of each gene was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Bioinformatics analyses
The amino acid sequence of the OsPIP2;2 protein was aligned with the AtPIP2;1, SoPIP2;1, and ZmPIP2;5 sequences using the NCBI Blast Multiple Alignment tool. The Multiple Alignment tool was also used to align the protein sequences of OsmaMYB and its homologs (ZmmaMYB-like, TdmaMYB-like, RhPTM, SlymaMYB, and AtmaMYB). TMDs in these proteins were predicted with TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and located by Multiple Alignment, along with the known transporter filters and phosphorylation sites in the sequences. A consensus neighbor-joining tree was established between OsmaMYB and its homologs using the MEGA version 6.0 program (https://www.megasoftware.net/).
H2O2 transport assay
To evaluate the H2O2 transport activity of OsPIPs, the OsPIP gene constructs were transformed into S. cerevisiae strain NMY51 individually. The yeast transformants were cultured in SD-Ura medium overnight at 30°C, and yeast cells were collected by centrifugation. The precipitate was washed 3 times with sterile H2O, and resuspended in sterile H2O to a final concentration of OD600 = 0.01. Ten microliter of the suspension was spotted onto SD-Ura solid medium containing different concentrations of H2O2. Three days later, the viability of the yeast cells was scored.
Visualization and quantification of H2O2 in yeast and plant cells were performed as described previously (Tian et al., 2016).
Plant transformation
A full-length OsPIP2;2 coding sequence (CDS) was cloned from NPB, and the fragment fused with His tag was inserted into the binary vector pCAMBIA1301. The recombinant construct was transformed into wild-type NPB to generate OsPIP2;2OE lines. To edit OsPIP2;2 using the CRISPR/Cas9 system, the target sequences were selected within the coding regions of OsPIP2;2 (Supplemental Figure S3A), and sgRNAs were designed with CRISPR MultiTargeter (http://www.multicrispr.net/index.html) and synthesized by Tsingke (Beijing). BLAST program was used to evaluate the target specificity against rice genome sequences. The above constructs were introduced into NPB by the A. tumefaciens-mediated transformation method (Biorun, Wuhan).
Plant inoculations
Virulent Xoo strain PXO99A and Xoc strain RS105 were grown overnight at 28°C, washed twice, and resuspended to OD600 = 0.6 in sterile H2O for inoculation. Leaves of 2-month-old rice plants were cut with scissors and dipped in the PXO99A suspension, and the lesion lengths were measured at 14 d postinoculation (dpi). The leaves of 2-week-old seedlings were infiltrated with RS105 suspension, and the lesion lengths were measured at 5 dpi (Cai et al., 2017; Ma et al., 2021). Bacterial titers were determined by counting colony-forming units (CFUs). To evaluate rice resistance to M. oryzae, 2-week-old seedlings were sprayed inoculated with M. oryzae strain HB3 conidia suspension at a concentration of 1 × 105 conidia/mL in 0.2% v/v Tween 20 (Yang et al., 2019). The fungal biomass was examined by the qPCR assay as described previously (Wang et al., 2020).
Callose deposition assay
Leaves of 2-week-old rice seedlings were sprayed with 10 μΜ flg22 (GeneScript, Nanjing) or H2O containing 0.025% v/v Silwet L-77. Callose deposition was detected 16 h after treatment as described previously (Yang et al., 2019). The leaf samples were observed by using a Nikon microscope under UV light (340–380 nm).
DAB staining
DAB was used for staining H2O2 according to a previously described method (Wang et al., 2016). The leaves of 2-week-old rice seedlings were immersed in 100-mL aqueous DAB solution (50 mg DAB, 0.5 mM NaH2PO4, 20 μL Tween 20) and vacuumed for 20 min. The leaf tissues were then stained in DAB solution for 8 h, and de-stained in a solution containing ethanol: acetic acid (3:1 v/v) for 8 h. The de-stained leaves were observed under a light microscope (Nikon, Tokyo, Japan).
SOD activity analysis
SOD activity was measured using the Total SOD Assay Kit (Cat#S0103, Beyotime, Shanghai) according to the manufacturer’s instructions. Briefly, 0.2 g of leaf tissues were harvested, ground into fine powder in liquid nitrogen, and resuspended in phosphate buffer. The suspension was centrifuged at 12,000 g for 5 min at 4°C. The supernatant was used to determine total SOD activity.
MAPK assay
Two-week-old rice plants were sprayed with 10 µM flg22 (Genscript, Nanjing, 95% purity) or H2O containing 0.025% v/v Silwet L-77 for 10 min. Leaves were ground into powder in liquid nitrogen and resuspended in extraction buffer (10 mM Tris–HCl, 50 mM NaCl, 5 mM dithhiothreitol [DTT], 0.2% v/v Triton X-100, 1× protease inhibitor cocktail (Cat#CW2200S, CWBIO, Beijing), 1× halt phosphatase inhibitor cocktail [Cat#P1081; Beyotime, Shanghai, China]). The protein concentration was determined using the BCA Protein Assay Kit (Cat#CW0014S; CWBIO, Beijing, China). Equal amounts of total protein were loaded on a 12% w/v SDS-PAGE gel, and the phosphorylation of MPK3, MPK4, and MPK6 was determined by using an anti-phospho-p44/42 monoclonal antibody (Cat#AM071; Beyotime, Shanghai).
Yeast two-hybrid assay
The split-ubiquitin based-MYTH system was used in this study. Full-length CDS of target genes were cloned into the pMetCgate vector, and the CDS of OsmaMYB was inserted into the pNXgate32-3HA. The recombinant constructs were co-transformed into S. cerevisiae strain NMY51. The presence of both plasmids was confirmed by growth on a SD-Trp-Leu plate, and interactions were evaluated by plating on a SD-Trp-Leu-His-Ade (SD-WLHA) plate.
Co-IP
Agrobacteria harboring the corresponding constructs were co-infiltrated into N. benthamiana leaves. The infiltrated leaves were harvested 3 d postinfiltration, ground in liquid nitrogen, and resuspended with IP buffer (25 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% v/v Nonidet P-40, 5% v/v glycerol, 1 mM PMSF, 20 μM MG132, 5 mM DTT, and 1× protease inhibitor cocktail). After centrifugation at 12,000 g for 15 min at 4°C, 20 µL of anti-FLAG agarose beads (Abmart, Shanghai, China) were added and incubated for 3 h at 4°C. The beads were washed 3 times with wash buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.5% v/v Triton X-100, 0.5 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail). Immunoprecipitates were analyzed by western blot.
LUC complementation assay
The LUC complementation assay was performed as described previously (Zhou et al., 2018). Agrobacterium tumefaciens harboring the indicated Nluc and Cluc constructs were co-infiltrated into N. benthamiana leaves. The leaves were infiltrated with 0.5 mM D-luciferin (Cat#L0159; Bivision, Nanjing, China) 48-h postagroinfiltration and kept in the dark for 5 min. The images were captured by a Living Imaging system (IVIS200; Xenogen, Inc., Alameda, CA, USA). To quantify relative LUC activity, leaf discs were taken 2 d after agroinfiltration, incubated with 1 mM luciferin in a 96-well, and relative LUC activity was measured by a microplate reader.
Transactivation assay
To evaluate transactivation activity assay in yeast, the truncated version of OsmaMYB, in which the predicted TMD was deleted, was cloned into pGBKT7 (Clontech) to fuse to the DNA-BD of GAL4. The pGBKT7 empty vector was served as a negative control. The construct was transformed into yeast strain AH109 and screened on the selective medium (lacking either Trp or both Trp and His).
Phosphorylation assay
OsPIP2;2 and its mutant were cloned and inserted into pPICZb and pCAMBIA1300 vector, respectively. To detect OsPIP2;2 phosphorylation in yeast, the recombinant pPICZb plasmids were transformed into yeast strain ΔX33 individually. To detect OsPIP2;2 phosphorylation in N. benthamiana, the pCAMBIA1300 recombinant constructs were separately introduced into Agrobacteria, and the recombinant proteins were transiently expressed in N. benthamiana leaves for 48 h. The infiltrated leaves were treated with 1 μM flg22 30 min before protein extraction.
The above total protein samples were incubated with calf intestinal alkaline phosphatase (Cat#9001-78-9; Solarbio, Beijing, China) at 37°C for 30 min, and then were separated by a 12% w/v SDS-PAGE gel contained biotinylated Mn2+-Phos-tag complex AAL-107 (Kinoshita et al., 2007) purchased from Wako (http://www.Phos-tag.com). The OsPIP2;2 protein was further detected by western blot with His antibody.
Transient expression in tobacco
Agrobacterium tumefaciens strain GV3101 containing different constructs was cultured in liquid LB medium at 28°C in a shaking incubator for 24 h, bacterial cells were collected by centrifugation at 3,500 g for 5 min, and resuspended in infiltration buffer (10 mM MgCl2, 100 mM MES pH 5.7, and 2 mM acetosyringone) to an OD600 = 0.3. The bacterial suspension was infiltrated into leaves of 4-week-old N. benthamiana. After 48 h, the infiltrated areas of the leaves were excised and examined with a laser confocal scanning microscope (LSM880, Zeiss, Jena). The argon laser intensity was set to 6% and 2% for DAPI and YFP, respectively. Wave lengths of excitation filters and emission light were 405 and 410–508 nm for DAPI, and the YFP signal was captured with 514-nm emission and 519–620-nm excitation. Chanel gains were adjusted as 600 and 550 for DAPI and YFP, respectively.
Protein extraction and western blotting
To detect OsPIP2;2 or OsmaMYB protein expression, total protein was extracted from leaves with extraction buffer made of 10 mM Tris–HCl, 50 mM NaCl, 5 mM dithiothreitol (DTT), 0.5% w/v SDS, 1× protease inhibitor cocktail (Cat#CW2200S; CWBIO, Beijing). For subcellular fractionation assay, 0.5 g leaf samples were ground in liquid nitrogen, and resuspended with extraction buffer (10 mM HEPES pH 7.8, 10 mM KCl, 10 mM MgCl2, 5 mM EDTA, 250 mM sucrose, 0.5% v/v Triton X-100, 1 mM DTT, 0.2 mM PMSF), then the extracts were filtered with Miracloth to remove cellular debris. After centrifugation at 3,000 g for 20 min at 4°C, the precipitation was washed with extraction buffer and collected as the nuclear fraction. The supernatant was centrifuged at 16,000 g for 30 min at 4°C, and the precipitation was collected as the membrane faction. Protein extracts were separated on 12% w/v SDS-PAGE gels and then transferred to a polyvinylidene difluoride membrane (Millipore, Burlington, MA, USA). The target proteins were detected by hybridization with the corresponding antibodies used at dilutions: α-His (TransGen Biotech, Beijing, China) or α-GFP (TransGen Biotech) at 1:4,000, α-FLAG (Affinity Biosciences) at 1:5,000, and α-Histone H3 (PhytoAB Inc., San Francisco, CA, USA) or α-Na+/K+-ATPase (PhytoAB Inc.) at 1:2,000. Secondary peroxidase-conjugated anti-mouse or anti-rabbit antibody (TransGen Biotech) was used at 1:5,000 dilution.
Statistical analysis
Quantitative data were analyzed using the NCBI ImageJ program and IBM SPSS version 19.0 (Shi, 2012), respectively. Significance in differences between (among) data from different plants or treatments was estimated by analysis of variance along with Student’s t tests or Duncan’s new multiple range tests.
Accession numbers
Sequence information in this study can be found in GenBank databases under the following accession numbers (proteins): LOC_Os02g44630 (OsPIP1;1), LOC_Os04g47220 (OsPIP1;2), LOC_Os02g57720 (OsPIP1;3), LOC_Os07g26690 (OsPIP2;1), LOC_Os02g41860 (OsPIP2;2), LOC_Os04g44060 (OsPIP2;3), LOC_Os07g26630 (OsPIP2;4), LOC_Os07g26660 (OsPIP2;5), OsPIP2;6 (LOC_Os04g16450), LOC_Os09g36930 (OsPIP2;7), LOC_Os03g64330 (OsPIP2;8), Os05g0438800 (OsActin1), LOC4337055 (OsPPO1), LOC432412 (OsPPO2), LOC4326969 (OsPOD1), LOC4340091 (OsSOD), LOC4342317 (OsPR1a), LOC_Os03g18850 (OsPR10), LOC4344711 (OsmaMYB), and MGG_05850 (MoPot2).
Supplemental data
The following supplemental materials are available.
Supplemental Figure S1. The expression profiles of OsPIPs in response to PXO99A.
Supplemental Figure S2. H2O2 transport mediated by OsPIPs in yeast.
Supplemental Figure S3. Confirmation of the OsPIP2;2 mutation and overexpression lines.
Supplemental Figure S4. Contribution of OsPIP2;2 in H2O2-mediated cell elongation.
Supplemental Figure S5. Growth and development of transgenic lines in the greenhouse.
Supplemental Figure S6. Analysis of the H2O2 content in rice after infection with PXO99A.
Supplemental Figure S7. Analysis of the H2O2 content in rice after infection with RS105.
Supplemental Figure S8. Analysis of the H2O2 content in rice after infection with HB3.
Supplemental Figure S9. Protein alignment of several PIPs proteins showing conserved S125 residue in OsPIP2;2.
Supplemental Figure S10. Phosphorylation of OsPIP2;2 at S125 does not change its expression level.
Supplemental Figure S11. Phylogenetic analysis of maMYBs.
Supplemental Figure S12. Promotion of plant resistance by OsmaMYB C terminus.
Supplemental Figure S13. Association between OsmaMYB and OsPIPs.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Prof. Wenhua Zhang (Nanjing Agricultural University) for suggestions on ROS analysis, Prof. Xinhua Ding (Shandong Agricultural University) for suggestions on the presentation, and Prof. Yuancun Liang for providing microbial cultures. We thank our laboratory colleagues Drs. Lei Chen and Shenshen Zou and Mr. Zeng An Li (Shandong Agricultural University) for rice propagation, and all graduate students studying in the H.D. laboratory for experimental assistance.
Funding
The study was supported by grants from the National Natural Science Foundation of China (31772247, 32072399, and 31672008) and the Open Project Program of State Key Laboratory of Crop Stress Biology for Arid Areas, NWAFU (CSBAA2020008).
Conflict of interest statement. The authors declare no conflict interest.
Contributor Information
Mou Zhang, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Haotian Shi, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Ningning Li, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Nana Wei, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Yan Tian, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Jinfeng Peng, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Xiaochen Chen, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Liyuan Zhang, National Key Laboratory of Crop Biology, Shandong Agricultural University, Taian, China; Department of Plant Pathology, Shandong Agricultural University, Taian, China.
Meixiang Zhang, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China.
Hansong Dong, Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China; National Key Laboratory of Crop Biology, Shandong Agricultural University, Taian, China; Department of Plant Pathology, Shandong Agricultural University, Taian, China.
M.Z. and H.D. conceived and supervised this project. M.Z. performed most of the experiments, analyzed the data, and wrote the manuscript. H.S., N.L., N.W., Y.T., and L.Z. helped identify the mutant. J.P. and X.C. helped manage rice. M.Z. and H.D. analyzed the data and wrote the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) are: Meixiang Zhang (meixiangzhang@njau.edu.cn) and Hansong Dong (hsdong@njau.edu.cn).
References
- Azad AK, Katsuhara M, Sawa Y, Ishikawa T, Shibata H (2008) Characterization of four plasma membrane aquaporins in tulip petals: a putative homolog is regulated by phosphorylation. Plant Cell Physiol 49: 1196–1208 [DOI] [PubMed] [Google Scholar]
- Baluška F, Vivanco J (2009) Reactive Oxygen Species in Plant Signaling, Springer, Berlin, Germany [Google Scholar]
- Bienert GP, Chaumont F (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta 1840: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Bienert GP, Heinen RB, Berny MC, Chaumont F (2014) Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta 1838: 216–222 [DOI] [PubMed] [Google Scholar]
- Cai L, Cao Y, Xu Z, Ma W, Zakria M, Zou L, Cheng Z, Chen G (2017) A transcription activator-like effector Tal7 of Xanthomonas oryzae pv. oryzicola activates rice gene Os09g29100 to suppress rice immunity. Sci Rep 7: 5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015) Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Cao J, Zhang J, Xia F, Ke Y, Zhang H, Xie W, Liu H, Cui Y, Cao Y, et al. (2017) Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat Plants 3: 1–9 [DOI] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Shukla VK, Chrispeels MJ, Larsson C, Kjellbom P (1998) Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell 10: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Larsson C, Kjellbom BEP (1996) The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell 8: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldenhoff R, Fischer M (2006) Functional aquaporin diversity in plants. Biochim Biophys Acta 1758: 1134–1141 [DOI] [PubMed] [Google Scholar]
- Kimura S, Hunter K, Vaahtera L, Tran HC, Citterico M, Vaattovaara A, Rokka A, Stolze SC, Harzen A, Meissner L, et al. (2020) CRK2 and C-terminal phosphorylation of NADPH oxidase RBOHD regulate reactive oxygen species production in Arabidopsis. Plant Cell 32: 1063–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T (2007) Specific recognition and detection of phosphorylated proteins using characteristics of metal ion. Yakugaku Zasshi 127: 1897–1913 [DOI] [PubMed] [Google Scholar]
- Li L, Wang H, Gago J, Cui H, Qian Z, Kodama N, Ji H, Tian S, Shen D, Chen Y, et al. (2015) Harpin Hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Sci Rep 5: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Fukumoto T, Gena P, Feng P, Sun Q, Li Q, Matsumoto T, Kaneko T, Zhang H, Zhang Y, et al. (2019) Ectopic expression of a rice plasma membrane intrinsic protein (OsPIP1;3) promotes plant growth and water uptake. Plant J 102: 779–796 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ludewig U, Dynowski M (2009) Plant aquaporin selectivity: where transport assays, computer simulations and physiology meet. Cell Mol Life Sci 66: 3161–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Li J, Ma L, Wang P, Xue Y, Yin P, Xiao J, Wang S (2021) Pathogen-inducible OsMPKK10.2-OsMPK6 cascade phosphorylates the Raf-like kinase OsEDR1 and inhibits its scaffold function to promote rice disease resistance. Mol Plant 14: 620–632 [DOI] [PubMed] [Google Scholar]
- Marchissio MJ, Frances DE, Carnovale CE, Marinelli RA (2012) Mitochondrial aquaporin-8 knockdown in human hepatoma HepG2 cells causes ROS-induced mitochondrial depolarization and loss of viability. Toxicol Appl Pharmacol 264: 246–254 [DOI] [PubMed] [Google Scholar]
- Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucq L (2015) Aquaporins in plants. Physiol Rev 95: 1321–1358 [DOI] [PubMed] [Google Scholar]
- Maurel C, Santoni V, Luu DT, Wudick MM, Verdoucq L (2009) The cellular dynamics of plant aquaporin expression and functions. Curr Opin Plant Biol 12: 690–698 [DOI] [PubMed] [Google Scholar]
- Medina-Puche L, Tan H, Dogra V, Wu M, Rosas-Diaz T, Wang L, Ding X, Zhang D, Fu X, Kim C, et al. (2020) A defense pathway linking plasma membrane and chloroplasts and co-opted by pathogens. Cell 182: 1109–1124 [DOI] [PubMed] [Google Scholar]
- Nakhoul NL, Davis BA, Romero MF, Boron WF (1998) Effect of expressing the water channel aquaporin-1 on the CO2 permeability of Xenopus oocytes. Am J Physiol Cell Physiol 274: C543–C548 [DOI] [PubMed] [Google Scholar]
- Nyblom M, Frick A, Wang Y, Ekvall M, Hallgren K, Hedfalk K, Neutze R, Tajkhorshid E, Tornroth-Horsefield S (2009) Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating. J Mol Biol 387: 653–668 [DOI] [PubMed] [Google Scholar]
- Petrov VD, Van Breusegem F (2012) Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants 2012: pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high colysaccharide and polyphenol components. Plant Mol Biol Rep 15: 8–15 [Google Scholar]
- Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V (2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Preston GM, Agre P (1991) Isolation of the cDNA for erythrocyte integral membrane protein of 28 kilodaltons: member of an ancient channel family. Proc Natl Acad Sci USA 88: 11110–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Gong Z, Zhou JM (2017) Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol 38: 92–100 [DOI] [PubMed] [Google Scholar]
- Qing D, Yang Z, Li M, Wong WS, Guo G, Liu S, Guo H, Li N (2016) Quantitative and functional phosphoproteomic analysis reveals that ethylene regulates water transport via the C-terminal phosphorylation of aquaporin PIP2;1 in Arabidopsis. Mol Plant 9: 158–174 [DOI] [PubMed] [Google Scholar]
- Rodrigues O, Reshetnyak G, Grondin A, Saijo Y, Leonhardt N, Maurel C, Verdoucq L (2017) Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc Natl Acad Sci USA 114: 9200–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder LS, Dagdas YF, Kershaw MJ, Venkataraman C, Madzvamuse A, Yan X, Cruz-Mireles N, Soanes DM, Oses-Ruiz M, Styles V, et al. (2019) A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature 574: 423–427 [DOI] [PubMed] [Google Scholar]
- Santoni V, Verdoucq L, Sommerer N, Vinh J, Pflieger D, Maurel C (2006) Methylation of aquaporins in plant plasma membrane. Biochem J 400: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W (2012) SPSS19.0 Statistical Analysis from Accidence to Conversance (in Chinese). Tsinghua University Press, Beijing, China [Google Scholar]
- Slabaugh E, Held M, Brandizzi F (2011) Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J 67: 395–405 [DOI] [PubMed] [Google Scholar]
- Sies H, Berndt C, Jones DP (2017) Oxidative stress. Annu Rev Biochem 86: 715–748 [DOI] [PubMed] [Google Scholar]
- Song Y, Zhang H, You H, Liu Y, Chen C, Feng X, Yu X, Wu S, Wang L, Zhong S, et al. (2019) Identification of novel interactors and potential phosphorylation substrates of GsSnRK1 from wild soybean (Glycine soja). Plant Cell Environ 42: 145–157 [DOI] [PubMed] [Google Scholar]
- Sorieul M, Santoni V, Maurel C, Luu DT (2011) Mechanisms and effects of retention of over-expressed aquaporin AtPIP2;1 in the endoplasmic reticulum. Traffic 12: 473–482 [DOI] [PubMed] [Google Scholar]
- Thiagarajah JR, Chang J, Goettel JA, Verkman AS, Lencer WI (2017) Aquaporin-3 mediates hydrogen peroxide-dependent responses to environmental stress in colonic epithelia. Proc Natl Acad Sci USA 114: 568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Wang X, Li P, Wang H, Ji H, Xie J, Qiu Q, Shen D, Dong H (2016) Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol 171: 1635–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian YC, Fan M, Qin ZX, Lv HJ, Wang MM, Zhang Z, Zhou WY, Zhao N, Li XH, Han C, et al. (2018) Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat Commun 9: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425: 734–737 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang R, Fang H, Zhang C, Zhang F, Hao Z, You X, Shi X, Park CH, Hua K, et al. (2020) Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice. Mol Plant 14: 253–266 [DOI] [PubMed] [Google Scholar]
- Wang R, Ning Y, Shi X, He F, Zhang C, Fan J, Jiang N, Zhang Y, Zhang T, Hu Y, et al. (2016) Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr Biol 26: 2399–2411 [DOI] [PubMed] [Google Scholar]
- Yang C, Yu Y, Huang J, Meng F, Pang J, Zhao Q, Islam A, Xu N, Tian Y, Liu J (2019) Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 31: 172–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Huai B, Lu Y, Cai K, Guo J, Zhu X, Kang Z, Guo J (2020) A stripe rust effector Pst18363 targets and stabilises TaNUDX23 that promotes stripe rust disease. New Phytol 225: 880–895 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen L, Dong H (2019a) Plant aquaporins in infection by and immunity against pathogens–a critical review. Front Plant Sci 10: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chiang YH, Toruno TY, Lee D, Ma M, Liang X, Lal NK, Lemos M, Lu YJ, Ma S, et al. (2018) The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Feng M, Chen W, Zhou X, Lu J, Wang Y, Li Y, Jiang CZ, Gan SS, Ma N, et al. (2019b) In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat Plants 5: 290–299 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Bi G, Zhou JM (2018) Luciferase complementation assay for protein-protein interactions in plants. Curr Protoc Plant Biol 3: 42–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.