Abstract
Accurate germplasm characterization is a vital step for accelerating crop genetic improvement, which remains largely infeasible for crops such as bread wheat (Triticum aestivum L.), which has a complex genome that undergoes frequent introgression and contains many structural variations. Here, we propose a genomic strategy called ggComp, which integrates resequencing data with copy number variations and stratified single-nucleotide polymorphism densities to enable unsupervised identification of pairwise germplasm resource-based Identity-By-Descent (gIBD) blocks. The reliability of ggComp was verified in wheat cultivar Nongda5181 by dissecting parental-descent patterns represented by inherited genomic blocks. With gIBD blocks identified among 212 wheat accessions, we constructed a multi-scale genomic-based germplasm network. At the whole-genome level, the network helps to clarify pedigree relationship, demonstrate genetic flow, and identify key founder lines. At the chromosome level, we were able to trace the utilization of 1RS introgression in modern wheat breeding by hitchhiked segments. At the single block scale, the dissected germplasm-based haplotypes nicely matched with previously identified alleles of “Green Revolution” genes and can guide allele mining and dissect the trajectory of beneficial alleles in wheat breeding. Our work presents a model-based framework for precisely evaluating germplasm resources with genomic data. A database, WheatCompDB (http://wheat.cau.edu.cn/WheatCompDB/), is available for researchers to exploit the identified gIBDs with a multi-scale network.
ggComp is a genomic-based approach that enables multi-scale germplasm network construction and promotes germplasm resource utilization in crop breeding.
Introduction
With the advances in plant genomics, assembling crop genomes using genomic resources carrying favorable phenotypes has been proposed and explored in crop breeding programs (Jia et al., 2017). Identifying the inherited pattern within crop species and the genomic segments that have been widely selected with preferred phenotypes from cultivars and landraces during historical breeding is an important way to thoroughly evaluate the existing germplasm resources and lay the foundations of future breeding (Bevan et al., 2017). By combining large-scale high-throughput genomic data and phenotype information with various methods, germplasm resource haplotypes carrying important variation in many crops like rice (Oryza sativa L.; Xie et al., 2010; Zhang et al., 2021), maize (Zea mays L.; Zhang et al., 2018; Coffman et al., 2020; Haberer et al., 2020), soybean (Glycine max [L.] Merr.) (Kim et al., 2014), and wheat (Pont et al., 2019; Hao et al., 2020) has been identified. And genomic-based germplasm networks are gradually attracting attention as a useful tool to solve complex relationships among germplasm resources (Haberer et al., 2020). However, there still exists a confusion about which analytical tools and genotyping approaches to employ, which could limit the amount of information effectively retrieved from complex genomic datasets. Some previous studies based on single-nucleotide polymorphism (SNP) array (Zhang et al., 2018), Genotyping-by-Sequencing (GBS; Romay et al., 2013), or Whole-Genome-Sequencing (Kim et al., 2014; Coffman et al., 2020) data tended to use ad hoc thresholds for pairwise sequence differences to identify haplotypes. Although they could identify haplotypes shared among samples, the performance and accuracy robustness are unclear because of a lack of statistical support. Some studies (Balfourier et al., 2019; Hao et al., 2020) applied Identity-By-Descent detection algorithms which were initially developed in human study, like Beagle/RefinedIBD (Browning et al. 2013) or Plink (Purcell et al. 2007). These algorithms are restricted to pair-wise sample comparisons, which is hard to apply to large sample sets. A recent method, hap-IBD (Zhou et al., 2020), was developed for inferring haplotype-sharing IBD in large sample sets with improved efficiency and accuracy. However, it requires the genetic map as input, which hinders its application in various species. Sequence identity generated by pairwise assembled sequence-alignment has also been used to identify shared haplotypes in crops like wheat (Brinton et al., 2020) and maize (Haberer et al., 2020), but these strategies rely on chromosome-scale genome assemblies and were not suitable for studies at population level.
As a typical self-pollination crop, wheat has a homozygous genome with a high level of linkage disequilibrium (LD; Hao et al., 2020) and frequent genomic structure variations. A multiple-wheat genome comparison study showed that ∼12% genes were the result of presence/absence variations (PAVs) and that 26% genes were the result of tandem duplication variations (Walkowiak et al., 2020). Moreover, the wheat genome is plagued with nonrandomly distributed high-density SNP blocks (Thind et al., 2018), which have been revealed to be the result of frequent interspecies introgression (Cheng et al., 2019; He et al., 2019). The most representative example is the 1RS chromosome from the rye (Secale cereale L.) genome, which has multiple translocation types used in modern wheat breeding (Wang et al., 2017). Taken together, genomic segments with nonrandomly distributed SNP and pervasive structural variations required a more suitable method to unravel underlying haplotypes and find their connections in wheat. Thus a proper statistical model is needed to distinguish variations generated during hundreds years of breeding or thousands years of domestication.
Here, we propose a statistical model-based method, genomic-based germplasm compare (ggComp), by combining the frequent genomic loss and the stratified SNP density to evaluate wheat germplasm resources in a pairwise manner. We showed that ggComp can precisely track the inheritance of genomic regions in accordance with recorded pedigrees and demonstrated low recombination frequencies during modern breeding. To unravel the germplasm utilization network during modern wheat breeding, we constructed a wheat germplasm network that can complement recorded pedigree information, help discover hidden relationships, and identify key founder lines or exotic lines. With inferred shared germplasm genomic blocks on chromosome 1BL, we discriminated the large introgression from rye and showed that all the tested Chinese accessions with the 1RS·1BL translocation can be traced back to Lovrin10 and Aimengniu. We then proposed an Markov Clustering (MCL)-based strategy to identify haplotypes at the germplasm level in a binwise manner, and dissected the haplotypes of the “Green Revolution” genes and investigated the origin and utilization of semi-dwarf alleles. We also found that Ppd-D1a was more preferred among Chinese cultivars (CNCs) than Chinese landraces (CNLs). This work aims to provide an effective framework for characterizing wheat germplasm and directing future breeding design.
Results
Resequencing panel of representative wheat accessions
With 26 instances of new sequencing data and 186 accessions selected from published resequencing data (Cheng et al., 2019; Guo et al., 2020; Walkowiak et al., 2020), we composed a representative panel of 212 worldwide wheat accessions (Supplemental Figure S1; Supplemental Table S1). The average sequencing depth was 6.07×. Reads were mapped to the reference genome (IWGSC RefSeq version 1.0; International Wheat Genome Sequencing Consortium, 2018) and biallelic SNPs were selected by filtering the minor allele frequency (MAF) at 1%. We identified more than 76.96 million SNPs and average value of heterozygosity rate was 2.5%, which is consistent with the self-pollination nature of wheat. Saturation analysis showed a coverage depth of 6× was able to recover 90.8% of homozygous SNPs in the wheat genome (Supplemental Figure S2). Thus, these identified SNPs could satisfy a comprehensive genome-wide investigation of genetic polymorphisms.
Frequent and pervasive genomic deletion blocks in wheat accessions
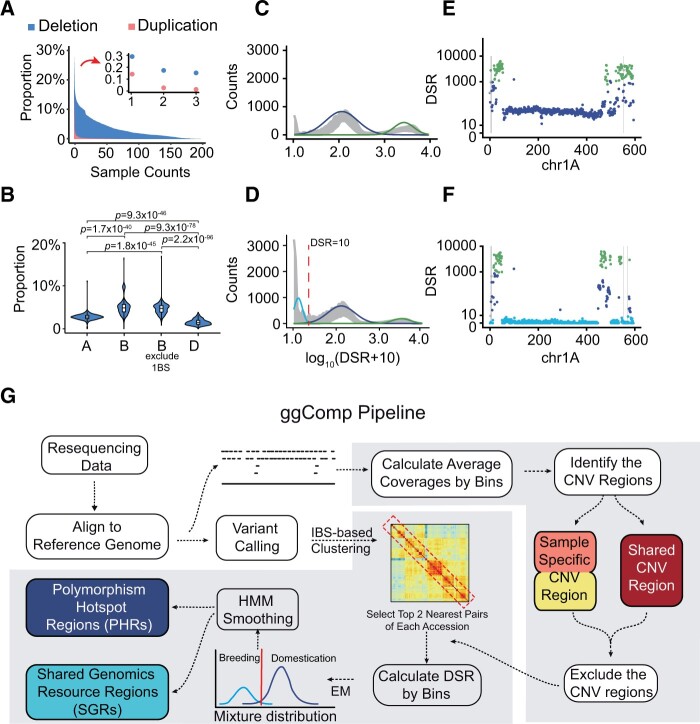
To fully evaluate the genetic diversity in wheat varieties, we identified copy number variation (CNV) blocks and investigated the polymorphisms between pairwise accessions. The normalized average coverage depths in 1-Mbp bins were used to identify CNV blocks (Supplemental Figure S3). A total of 5,566 Mbp of CNV blocks (39.5%), comprising 4,178 Mbp of deletion blocks (29.7%) and 2,167 Mbp of duplication blocks (15.4%), were detected in at least one accession (Figure 1A), which is consistent with previous estimations using a high-density SNP array (Balfourier et al., 2019). For samples harboring the 1RS·1BL translocation, the entire short arm of chromosome 1B could be detected as constituting CNV-deletion blocks due to poorly aligned sequences oriented from rye genomes to the reference genome (Rabanus-Wallace et al., 2021; Supplemental Figure S4). The B subgenome had the highest frequency of CNV-deletion blocks, even after excluding the 1BS chromosome considering the potential impact of the 1RS·1BL translocation, while the D subgenome had the lowest frequency (Figure 1B). A similar trend was observed for CNV-duplication blocks, albeit at much lower frequency (Supplemental Figure S5). The CNV-deletion blocks tended to gather at chromosomal extremities and this broadly distributed pattern of CNV-deletion blocks (Supplemental Figure S6) indicated that frequent loss of genomic segments is an ongoing process for neo-polyploid wheat with a redundant genome. A CNV block-based phylogenetic tree showed that most CNCs were more closely related to foreign accessions than to CNLs (Supplemental Figure S7). A shared CNV-deletion block corresponding to the 1RS·1BL translocation was found in 18 CNCs and 1 foreign accession but not in any CNLs (Supplemental Figure S7), which was consistent with the understanding that the 1RS translocation was introduced from a few European elite accessions (Yang et al., 2004). The high occurrence of CNV blocks across wheat accessions indicated that structural variations should be considered when assessing the genetic differences among wheat accessions.
Figure 1.
Complex landscape of the wheat genome and the design of the ggComp pipeline. A, Distribution of the CNV blocks shared among accessions. CNV blocks are detected against the CS reference genome with a block length of 1 Mbp. Y-axis, proportion of the wheat genome in which CNV blocks are detected in at least X accessions in our collection. The regions with X ≤ 3 are highlighted in the top-right side. Blue, CNV-deletion blocks. Red, CNV-duplication blocks. B, Distribution of CNV-deletion block ratios in the genome for all accessions among whose subgenomes are compared. The B subgenome, excluding 1BS, shows the highest CNV-deletion ratios. The D subgenome ranks the lowest. The P-values are calculated according to two-tailed t tests. C and D, Distribution of log10(DSR10) in each window (1-Mbp length) for 300 accession pairs randomly selected from all accessions pairs (C). And accession pairs of 212 accessions with their top 2 nearest IBS genetic distances accessions (D). Gray ribbon, ranges of mean ± standard deviation (sd) of density for accession pairs. The curves represent the three subdistributions fitted by the EM algorithm. Light blue curve, low density. Dark blue curve, middle density. Green curve, high density. A DSR = 10 (red dashed line) was selected as the initial threshold to distinguish PHRs and SGRs. E and F, Profiles of the binwise high-density SNP regions between Bima4 and Bima1 (E) and between Bima4 and CS (F) along chromosome 1A. The point colors correspond to the three fitted sub-distributions in (C). The gray shadows indicate CNV blocks. The Y-axis is presented as a log scale. G, Workflow of ggComp algorithm. The resulting data are mapped to the genome, following SNP calling and binwise depth analysis. The CNV blocks are identified based on binwise normalized depth and the IBS-based clustering is used to select accession pairs with closest genetic distance. The DSR between these pairs is calculated after excluding CNV regions and applying EM algorithm to the DSR distribution to identify threshold between PHR and SGR. For a pair of accessions, the CNV blocks are excluded, and PHRs and SGRs were identified with the initial threshold. An HMM smoothing step is applied to generate the final PHRs and SGRs. The colored boxes indicate the output of ggComp.
Genome-wide characterization of genetic diversity reveals stratified genetic distances in genomic blocks
After excluding the identified CNV blocks, we randomly chose 300 accessions pairs from all accession pair to profile genetic diversity by calculating the different SNP ratios (DSRs) in 1-Mbp bins across whole genome (Figure 1C; Supplemental Figure S8A). The log-DSR distribution showed stratified densities of polymorphisms. We further selected accession pairs of 212 accessions with their top 2 nearest accessions by calculating Identical-By-State (IBS) genetic distances to analyze the distribution of DSRs in 1-Mbp bins across genome. Interestingly, compared with random selected accessions pair, the log-DSR distribution of accession pairs with close IBS-distances emerged a new peak (Figure 1D; Supplemental Figure S8B). Accordingly, we applied the expectation–maximization (EM) algorithm for the Gaussian mixture model to dissect the mixture distribution into three components: high-, mid-, and low-density differential SNPs. For example, the majority of the 1A chromosome between the two sibling lines Bima1 and Bima4 was at low-density levels (Figure 1F), while the distance between Bima4 and Chinese Spring (CS) was at a mid- or high-density level (Figure 1E). The distribution differed among chromosomes, while these levels tended to remain unchanged across chromosomes (Supplemental Figures S9 and S10). Considering that differential frequency at low-density level is ∼1 per 100 kbp, which is likely to have accumulated during the recent modern breeding process or to have resulted from sequencing error, we reasoned that these low-density regions shared the same germplasms for accession pairs. The mid- and high-density levels corresponded to the different germplasms. Taken together, our results showed that bin-wise DSR values could be used to discriminate chromosome segments as similar or different germplasm resources for breeding.
Unsupervised method to identify shared genomic resource blocks between wheat accessions
To identify the shared genomic blocks between wheat accessions under a genetically stratified background, we proposed ggComp as an unsupervised method that integrates the EM algorithm and hidden markov model (HMM) algorithm. ggComp combines the identified CNV blocks and binwise DSRs to classify genomic bins into six main categories: Shared Genomic resource Regions (SGRs), polymorphism hotspot regions (PHRs), sample-specific CNV-deletion regions, sample-specific CNV-duplication regions, shared CNV-deletion regions, and shared CNV-duplication regions (Figure 1G). The SGRs and shared CNV regions were considered as germplasm resource-based Identity-By-Descent (gIBD) blocks in this research.
The CNV blocks were first identified for each accession against the reference genome and further distinguished as accession-specific CNV blocks or shared CNV blocks within accession pairs. After excluding CNV blocks, DSR values were calculated for all pairs of accessions in nonoverlapping 1-Mbp windows across chromosomes. The initial threshold used to distinguish PHRs and SGRs were determined based on the density distribution of DSR across all accession pairs with top two closest genetic distance of each accession calculated PLINK (Purcell et al., 2007) and decomposed by the EM algorithm. The regions at low-density level of stratified DSRs represented SGRs and the regions at mid- and high-density levels represented PHRs. The stratified DSR profiles for Bima4 versus CS (Figure 1E; and Supplemental Figure S9) and Bima4 versus Bima1 (Figure 1F; Supplemental Figure S10) indicated that the results were robust enough to unravel genomic relationships between wheat varieties. To reduce potential mistakes introduced by hard thresholds with noise signals, we applied a HMM-based smoothing step and generated final gIBD profiles with soft-corrected PHRs and SGRs (Supplemental Figure S11). Compared with the raw ones, the polished PHRs and SGRs were more consecutive and less likely to be affected by stochastic classification error from independent windows (Supplemental Figure S12). Finally, all 1-Mbp windows were annotated via nine different categories for each accession pair. The ggComp pipeline was developed as an open-source command-line tool that can be accessed at https://zack-young.github.io/ggComp/.
Identification of parental descending genomic regions of the wheat cultivar Nongda5181 utilizing gIBD
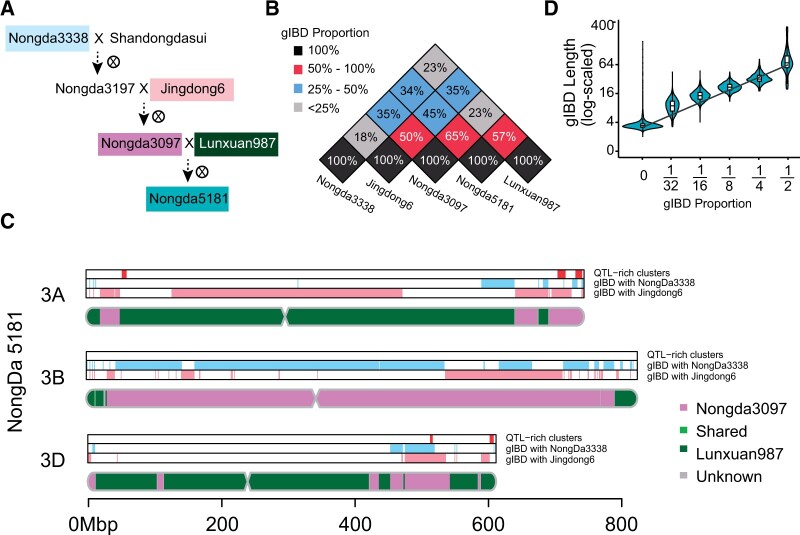
Beyond the traditional pedigree information, there is a need to track the inheritance patterns of haplotypes in breeding pedigrees which could help to visualize the dynamics of chromosomal recombination and identify optimal parents for crosses that contain desired combinations of features (Zhou et al., 2016). The genomic distribution pattern of Nongda5181, a CNC released nationwide in 2017, as derived from its parental lines was inferred by utilizing ggComp. Five lines were included in this study, while the seeds of Nongda3197 and Shandongdasui were not preserved (Figure 2A). We first identified gIBD on Nongda5181 inherited from its parental lines Nongda3097 and Nongda987. The results showed that 65% of genome blocks were gIBD between Nongda5181 and Nongda3097 (Figure 2B; Supplemental Figure S13), including 8,679 Mbp of SGRs and 408 Mbp of CNV regions. 57% of genome blocks were gIBD between Nongda5181 and Lunxuan987, which included 7,453-Mbp SGRs and 619-Mbp CNV regions (Figure 2B; Supplemental Figure S14). A total of 3,390 Mbp (24%) of genomic blocks shared by all three varieties contributed to the same genetic background (Supplemental Figure S15). Excluding the shared background, Nongda3097 and Lunxuan987 contributed 53% and 44% of genomic resources, respectively, to Nongda5181, which is consistent with the expectation that half of genome was derived from each parent. By summarizing the gIBD maps of Nongda5181 with two parental lines, we revealed a clear genomic origin pattern of Nongda5181 (Figure 2C; Supplemental Figure S15). A total of 91 breakpoints were detected between parental lines in Nongda5181 genome, with an estimated average of 4.3 breakpoints on each chromosome, indicating that the inherited parental genomic regions are usually in large chromosome blocks (Supplemental Figure S15; Supplemental Table S2). Moreover, 45% and 34% of Nongda5181 genome were shared with genomes of its grandparental line Jingdong6 and great-grandparental line Nongda3338, respectively (Figure 2B; Supplemental Figures S16 and S17). We used the indirectly related lines Nongda3338, Jingdong6, and Lunxuan987 to estimate that 25% of the background constituted shared genomic resources. Excluding the background, 26.7% and 12.0% of Nongda5181 could be traced to its grandparental line and great-grandparental line, respectively, which are close to the expected ratios of 1/4 and 1/8, respectively. Several QTL-rich clusters within these shared genome blocks were found to be passed through generations (Figure 2C; Supplemental Figure S18). Thus, the gIBD fit well with Nongda5181 and its pedigree, and each generation inherited approximately half of the parental lines. Additionally, we evaluated the gIBD lengths and genome similarity (whole-genome gIBD proportion) for all accession pairs. Interestingly, a positive correlation (Spearman’s rho = 0.69, P < 2.2 × 10−16) was found between genome similarity and gIBD length (Figure 2D), indicating that the inherited genomic blocks broke down exponentially along with increasing generation. Generally, gIBD generated by ggComp gives a theoretical basis for understanding the dynamics of chromosomal recombination and selected genomic regions through the breeding process.
Figure 2.
Dissection of inherited genomic blocks of wheat cultivar along with their pedigree. A, Pedigree of cultivar Nongda5181. The filled boxes indicate that resequencing data are available. The varieties Nongda3197 and Shandongdasui are not preserved. B, Heatmap matrix of pairwise gIBD proportions among Lunxuan987, Nongda3097, Jingdong6, Nongda3338, and Nongda3097. The gIBD proportion is labeled in each cell. C, Dissection of the inherited genomic blocks in the 3A, 3B, and 3D chromosomes of Nongda5181. The bottom chromosome-shaped track shows reconstructed recombination events of Nongda5181 from its parental lines, Lunxuan987 (purple) and Nongda3097 (green). Descended blocks from the great-grandparental line Nongda3338 (blue) and grandparental line Jingdong6 (pink) are shown in independent tracks. QTL-rich clusters annotated by (Cao et al., 2020) are marked (red track). D, Distribution of the gIBD lengths (y-axis) for different accession pair groups (x-axis). Accession pair groups are categorized by gIBD proportions shared by two accessions; for example, the distribution at x = 1/8 indicates a group of accession pairs with gIBD proportions ranging from [1/8 to 1/4). The line is fitted by linear regression on a log-scale axis.
A whole-genome scale gIBD-based network between germplasms provides insights into the wheat breeding process
To investigate the utilization of wheat germplasm resources, gIBD were calculated in all accession pairs. Clustering of gIBD-based distance matrix showed that most cultivars and a few landraces could be assigned to the same clade separated from clade with the majority of landraces (Figure 3A; Supplemental Figure S19), reflecting the genetic diversity bottleneck of modern wheat cultivars derived from limited landraces with respect to the improvement process. Most modern CNCs were mixed with a few European cultivars (EUCs), supporting the previous notion that European germplasm was the primary exotic resource used to broaden the germplasm base for modern Chinese wheat breeding.
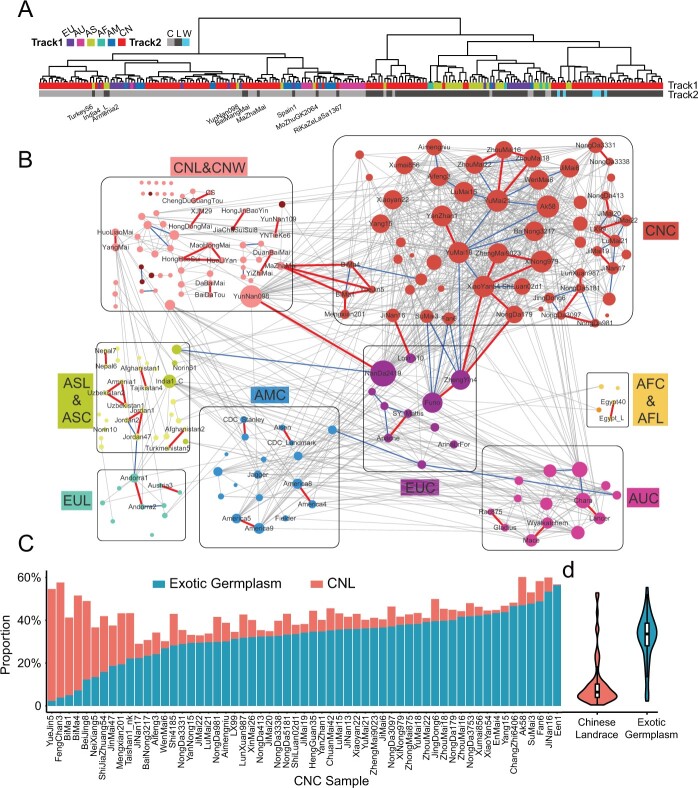
Figure 3.
The whole-genome GGNet reveals relationships among wheat varieties. A, Hierarchical clustering performed with gIBD-based distances. Method, ward. Pairwise distance, -log2(gIBD_ratio). Track1, China (CN), Europe (EU), Australia (AU), Africa (AF), America (AM). Track2, cultivar (C), landrace (L), semi-wild (W). B, The whole-genome scale GGNet. A node represents an accession. The edge colors indicate the ranges of the gIBD ratio (genome similarity) for accession pairs. Only the edges in which the gIBD ratio ≥20% are shown. Gray edges, 40% ≥ gIBD ratio > 20%. Blue edges, 50% ≥ gIBD ratio > 40%. Red edges, gIBD ratio > 50%. The node size corresponds to the node weight, which is the summed weight of all derived edges, that is, the summed gIBD ratios with those of all other accessions in the network. The gray squares indicate variety groups. The three-letter group codes are concatenated by the codes of track1 and track2 in (A). CNW, Chinese semi-wild; EUL, European landrace; AUC, Australian cultivar; AFC, African cultivar; AFL, African landrace; AMC, American cultivar; ASC, Asian cultivar; ASL, Asian landrace. C, Genome composition of CNCs that specifically share material with exotic germplasm (green) and with CNLs (red). D, Violin plot of genetic contributions to CNCs by CNLs (red) and exotic germplasm (blue). The y-axis is consistent with that in (C).
Based on gIBD similarity matrix that records the ratio of gIBD among accessions, a genomic-based germplasm network (GGNet) at whole-genome level was constructed by an unsupervised manner to visualize the potential breeding process around accessions (Figure 3B). Due to the exponential relationship between gIBD proportion and generation, accessions with direct connections (similarity >50%) indicate parental relationships or sibling relationships. For example, the pedigree of Nongda5181 (Figure 2A) can be fully retrieved from GGNet (Figure 3B). Mazhamai is a well-known CNL used as a founder line and has been used to produce many famous derived cultivars, such as Bima1 and Bima4 (Sheng et al., 1983), of whose relationships could also be implied in GGNet. Mazhamai has a direct relationship with Bima1 and Bima4, with similarity ratios of 52% and 60%, and Bima1 and Bima4 are siblings, with 49% similarity (Supplemental Figure S20).
For many wheat accessions, the intermediate generation may not be recorded or is incompletely preserved. GGNet provides a solution to precisely characterize the genomic relationship independently of pedigree records and reveal undocumented or hidden relationships. A CNL in Sichuan Province, Chengduguangtou, is an accession closest to CS (68% similarity) (Supplemental Figure S21), which confirms previous speculations that Chengduguangtou is a potential genome donor for CS (Liu et al., 2018). In contrast, both the Tibetan semi-wild wheat Zang1817 and CS were collected in south-western China and share little similarity in terms of SGRs. The connections for landrace pairs Dabaimai–Baidatou and Huoliaomai–Yangmai also showed similarity percentages higher than 50% but were not documented. Yunnan098 was recorded as a CNL in Yunnan Province, but in the network, Yunnan098 was strongly closely related to exotic accession Nanda2419, with 67% similarity. This indicates that sequenced Yunnan098 accession might not be a pure native landrace but may actually be derived from Nanda2419 (Supplemental Figure S22).
The germplasm network also revealed that many exotic germplasms have contributed greatly to modern Chinese wheat cultivars, including many well-known founder lines, such as Funo and Lovrin10 (Sheng et al., 1983; Xu et al., 2010). On average, only 10.7% of CNCs genome could be found as the same haplotype block in CNLs, except for a few landraces, such as Mazhamai, which is a direct genome contributor of Bima1, Bima4, and Yuejin5 (Figure 3C). However, compared with these CNLs, exotic germplasm contributes a substantially higher proportion of germplasm to modern Chinese wheat cultivars (Figure 3D).
The node size of network represents the summed weights of connected lines. A relatively heavy weight of accession indicates the potential of being a parental line in the population. For example, Zhoumai16, Zhengyin4 (introduction names: St2422/464), and Nanda2419 (introduced from Italy) were identified as central hubs in the germplasm network, implying their contributions to a number of derived cultivars consistent with records. The germplasm network provides an intuitive framework for exploring the role of founder lines from a broad perspective.
Compared with the inference results of commonly used IBS method, the gIBD-based germplasm network provides a more quantitative way to characterize relationships among wheat accessions. Unlike the kinship matrix generated by the IBS or IBD-based method, GGNet was constructed on a sparse matrix reflecting the breeding process, accounting for stratified genetic distances and frequent CNV blocks. For example, the accession groups with pedigree relations, such as Lunxuan987–Nongda3097–Nongda5181 and Mazhamai–Bima1–Bima4, were separated in the IBS-based clustering results, while they were clustered together by the gIBD-based strategy (Supplemental Figure S23).
Dissecting the origin and descent of 1RS utilization in CNCs with the help of a chromosome scale gIBD-based network
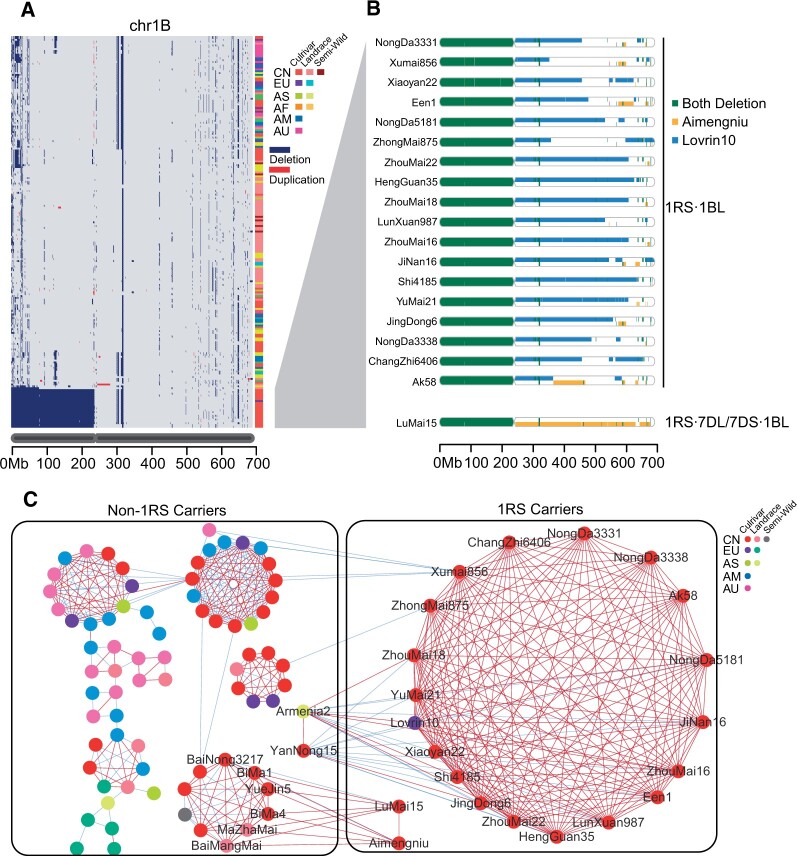
The translocation of 1RS chromosome from rye (S. cereale) into bread wheat chromosome 1B played a vital role during modem wheat breeding (Yang et al., 2004). 1RS·1BL wheat lines were introduced into China in the 1980s and began to be widely used (Sheng et al., 1983). According to pedigree information, Lovrin10 and Aimengniu included in dataset are two major 1RS·1BL contributors in Chinese breeding (Yang et al., 2004). A primitive GGNet of 1B chromosome that did not discriminate the origin of 1RS showed an apparent large cluster contained 20 accessions that all shared >50% similarity with Lovrin10 and also exist sporadic accessions closely related with Aimengniu (Supplemental Figure S24). Based on the CNV heat map of chromosome 1B (Supplemental Figure 4A), besides Lovrin10 and Aimengniu, 18 of preceding 20 accessions and one accession closely related with Aimengniu were found possessing CNV-deletion blocks along the whole 1BS chromosome, nearly all of which are CNCs released after the 1980s and confirmed to carry the 1RS·1BL translocation (Supplemental Table S3). We assessed all these 1RS·1BL carriers by identifying the gIBD with Lovrin10 or Aimengniu on 1BL chromosome. The result showed that most carriers hitchhiked Lovrin10-derived gIBD on 1BL (Figure 4B), supporting the notion that Lovrin10 was the main contributor of 1RS·1BL introgression (Yang et al., 2004). The lengths of Lovrin10 hitchhiking gIBD on 1BL ranged from 106 to 380 Mbp, consistent with this hitchhiking effect previously identified through quantitative trait locus (QTL) mapping (Xu et al., 2010). Lumai15 was the only accession that shared Aimengniu-derived gIBD on 1BL as Aimengniu and Lovrin10 harbored different types of genomic resources on 1BL chromosome (Supplemental Figure S25). This was consistent with a previous study that Aimengniu and Lumai15 lines were identified as having an alternative rearrangement type (1RS·7DL/7DS·1BL) via high-resolution FISH (Huang et al., 2018). By reconstructing the GGNet of 1B chromosome that differentiates the 1RS of Lovrin10 lineage and Aimengniu lineage (Figure 4C), two lineages showed two distinct patterns demonstrated 1BL also inherited from different ancestral. There still exist several non-1RS carriers shared >40% similarity with Lovrin10 or Aimengniu indicated that potential important trait-associated alleles located in 1BL.
Figure 4.
Identification of the 1RS·1BL translocation in wheat accessions and the tracing of its origin by genetic drag. A, CNV block heatmap of the 1B chromosome of all accessions. Each row stands for one accession. Dark blue, CNV-deletion block. Red, CNV-duplication blocks. The 1B chromosome is plotted below. The right annotation bar shows the geographic origin (CN, Chinese accessions; EU, European accessions; AU, Australian accessions; AF, African accessions; AM, American accessions) and historical groups (cultivar, landrace, and semi-wild) of each accession. Accessions harboring the 1RS translocation (showing a fully deleted 1BS arm) are ordered at the bottom. B, SGRs distribution along chromosome 1B for the 18 cultivars detected as having the 1BS deletion compared to Lovrin10 (blue) and Aimengniu (yellow). Compared with Lovrin10, most accessions showed genetic drag for 1BL, indicating that the utilized 1RS·1BL translocations can be traced back to Lovrin10. Lumai15 shared SGRs with Aimengniu on 1BL but otherwise did not differ from Lovrin10, which is consistent with previous findings in which Lumai15 and Aimengniu actually have 1RS·7DL/7DS·1BL-type translocations rather than 1RS·1BL-type translocations. C, The chromosome scale GGNet of 1B chromosome. The edge colors indicate the ranges of the gIBD ratio (genome similarity) for accession pairs. Only the edges in which the gIBD ratio ≥40% are shown. Blue edges, 50% > gIBD ratio ≥ 40%; Red edges, gIBD ratio ≥ 50%. A node represents an accession and only accessions that have direct or indirect connection with Aimengniu or Lovrin10 in the net were shown. The right annotation bar shows the geographic origin (CN, Chinese accessions; EU, European accessions; AU, Australian accessions; AM, American accessions) and historical groups (cultivar, landrace, and semi-wild) of each accession. Two boxes showed accessions were grouped by whether carring 1RS or not.
Tracing gene utilization among accessions by networks of gHaps
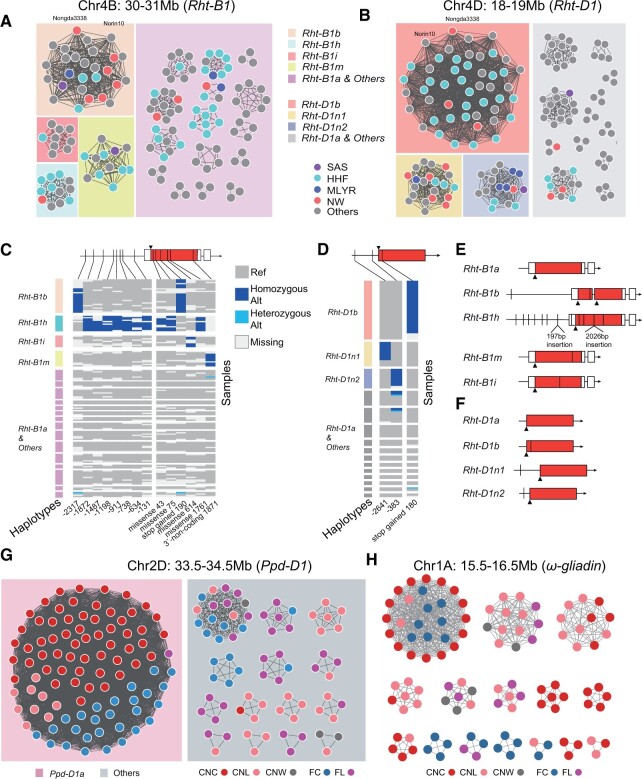
As gIBD between accession pairs across whole genome was identified, we further classified all gIBD of each block into higher-order haplotypes: germplasm resource type-based haplotypes (gHaps) trying to detect resource utilization among our dataset. To validate the reliability of gHap, we compared gHap with the haplotypes detected by genome assembly-based method (Brinton et al., 2020). Haplotypes obtained through two methods are largely concordant that gHap can overlap nearly 90% of haplotypes identified by Brinton et al. (Supplemental Figure S26A). In some specific region, ggComp performed even better, as shown by a detailed analysis in a region of chromosome 6A (Supplemental Figure S26B), where ggComp detected a different haplotype with SY Mattis, which can be confirmed with dot-plot of two assemblies (Supplemental Figure S26C) and distribution of sequence identity in bins shown by pairwise alignment of assemblies (Supplemental Figure S26D). Dwarfing genes Rht-B1b and Rht-D1b were analyzed first as they were predominantly selected to reduce height and improve yields during Green Revolution (Reitz and Salmon, 1968; Gale and Youssefian, 1985; Rebetzke and Richards, 2000; Zhang et al., 2006). The gHap networks of Rht-B1 and Rht-D1 based on gIBD among different accessions were constructed that each cluster corresponds to a specific gHap (Figure 5, A and B). For the bin located at Rht-B1, accessions in the largest cluster harbored Rht-B1b allele (Figure 5A), which perfectly fits the genotype distribution of Rht-B1b (Figure 5C). The gHap also revealed previously identified Rht-B1h, Rht-B1i, and Rht-B1m alleles (Li et al., 2013). For Rht-D1, the largest cluster also corresponded to Rht-D1b (Figure 5D). Two new alleles, Rht-D1n1 and Rht-D1n2, corresponding to the second and third largest clusters, were identified with mutations in upstream regions located 383 bp and 2,640 bp, respectively, from the start codon (Figure 5D). Both of the semi-dwarfing alleles Rht-B1b and Rht-D1b contained stop-gain mutations (Figure 5, E and F), which produced N-terminal truncations through translational reinitiation (Van De Velde et al., 2021). Several accessions identified as harboring Rht-B1h alleles were confirmed by resequencing data to be consistent with known variation forms (Figure 5E). Although the function of new alleles was still unclear, these categories provided useful guidance to determine wild-type alleles and explore potential haplotypes to be further exploited in breeding (Figure 5F). And these distinct gHaps that classified as “Rht-B1a & Others” (Figure 5A) or “Rht-D1a & Others” (Figure 5B) could not be distinguished by the sequence of Rht-B1 and Rht-D1, they could be distinguished by other genes or loci within the blocks.
Figure 5.
Local-scale GGNet helps reveal the selection of gene located blocks during breeding. GGNet of the genomic windows with Rht-B1 (A) and Rht-D1 (B). Each node represents an accession. Node colors, different agro-ecological zones. SAS, Southwestern autumn-sown spring wheat zone. HHF, Huang Huai facultative wheat zone. MLYR, Middle and lower Yangtze River valley autumn-sown spring wheat zone. NW, Northern winter wheat zone. An edge indicates that the two connected nodes share the same gHap. The rectangle background colors indicate major selected clusters and others, with the corresponding alleles labeled in the squared legend. C and D, The heatmap shows sequence variations with groups corresponding to alleles of Rht-B1 (C) and Rht-D1 (D). Dark blue, homozygous variation. Light blue, heterozygous variation. Gray, genotype of the reference genome (CS). White, missing data. The variation positions at the bottom are the relative distances from the start codon. In the left annotation track, the group colors are consistent with the box colors in (A) and (B). The triangles represent start codon positions. Red region in boxes, coding regions. White region in boxes, Untranslated Region (UTR) sequences. E and F, Gene structure features of different alleles of Rht-B1 (E) and Rht-D1 (F). Rht-D1n1 and Rht-D1n2 are two unreported alleles found in this study. GGNets of the genomic windows with Ppd-D1 (G) and ω-gliadin (H). Each node represents an accession. Node colors, CNC, CNL, and CNW and foreign cultivars and landraces. An edge indicates that the two connected nodes share the same gHap. The rectangle background colors in (G) indicate whether accessions carrying Ppd-D1a or not.
The distribution of gHap within populations also provides a useful prospective to dissecting the natural and artificial selection process. The annotation of Rht-B1b and Rht-D1b in the wheat germplasm network revealed asymmetric selection pressure on two homoeologous genes through breeding practices in China (Supplemental Figure S27). The results showed that 2 (out of 57) CNLs and 39 (out of 59) CNCs shared at least one of the Rht-B1b and Rht-D1b alleles (Supplemental Table S4), which is consistent with the fact that Rht-B1b and Rht-D1b alleles are rare in CNLs, but their frequencies are higher in the CNCs. The breeding line Nongda3338 is the only accession that gained both Rht-B1b and Rht-D1b. It should be noted that there were still 20 CNCs in this collection that contained none of the two alleles, indicating the potential of unexplored semi-dwarfing genes used in China (Supplemental Figure S27). We also dissected the gHap of Ppd-D1 and a ω-gliadin gene located blocks chr2D:33.5–34.5 Mb and chr1A:15.5–16.5 Mb, respectively (Figure 5, G and H). Unlike other blocks, an extremely large group of accessions contained 97% CNCs shared one gHap of Ppd-D1 and the allele type of Ppd-D1 is Ppd-D1a. Considering that the landraces are self-contained populations adapted to their geographic origin, the highly enriched photoperiod insensitivity-related Ppd-D1a allele (Figure 5G) in CNCs improved the adaptation to a broader range of environments, which is consistent with previous findings (Yang et al., 2009). This is the critical genomic signature at the initial stage of modern wheat breeding in China, aiming at improving adaptation traits (Hu et al., 2018). Diverse gHap clusters were detected in the ω-gliadin gene-located block even in cultivars (Figure 5H), which is different from the single main cluster detected for Ppd-D1 located block. With regards to the fact that the ω-gliadin gene related to end-use characteristic, it could be selected by breeders in different directions. There were also several gHaps detected only among landraces and had not been used by breeders.
Generally, the gHap consistently fit with known alleles, helping accurately mine beneficial alleles which were under selection. It is worth noting that different accession clusters indicated different germplasm resource types, and this strategy provides an efficient way to trace the utilized gene resources in wheat varieties, independent of pedigree information.
Discussion
In genomics era, utilizing sequencing data to make evaluation of crop germplasms is extraordinarily important to support germplasm management and breeding strategies making. Kinship analysis with limited markers of array- or GBS-based genotyping methods has been widely used for evaluating genetic diversity but is not suited for fully and precisely characterizing relationships at genomic blocks level. Evaluating genomic resources in crops that carry a complex genome (e.g. wheat) is difficult and requires a more appropriate approach. Hexaploid wheat is domesticated from neo-polyploid plants with a high frequency of genomic structural variation and frequent intra- and interspecific introgressions. We systematically characterized the high frequency of CNV blocks across wheat varieties (Figure 1A) and revealed stratified SNP densities via a statistical model (Figure 1D). We developed the ggComp method that can be applied to constructing multi-scale networks by precisely characterizing germplasm relationships from local genomic regions to whole genome among accession pairs (Figure 1G); ggComp integrates statistical models, discovering hard thresholds with the EM algorithm and utilizing the HMM model for soft corrections to reduce noise (Supplemental Figure S11). The comparison process is independent of pedigree information and extra samples. As a reference-based approach, the missing or duplicated regions in the reference assembly could result in large deletion blocks or duplicated blocks in alignment of accessions. Thus, the CNV-block identification was introduced as an intermediate step to eliminate their impact on the downstream of gHap identification. On the other hand, there are some sequences or structure variations that could not be mapped on CS genomes. For such cases, the boundary regions with aligned sequences can be used as indicators for gHap inference by leveraging on the LD. For example, the 1B/1R translocation carriers could be identified by low mapping ability on 1BS chromosome, and the utilization of 1B/1R introgressions inferred by gHap inferred with adjacent regions on 1BL (Figure 4A). Theoretically, ggComp could be applied to other selfing species with long LD decay distances and is suited for dealing with complex genomic landscapes, especially those of polyploid plants with interspecific introgressions.
The gIBD identified by ggComp are purported to be genomic blocks representing the same breeding germplasm resource. As modern breeding requires less time than domestication, a subtler threshold is needed to discriminate different germplasm resources diverged during modern breeding. Therefore, our gIBD based on a model-driven strategy with statistic support is more suitable for breeding purpose compared with traditional IBD-detected under subjective criteria regions. With gIBD, the genomic regions of national cultivar Nongda5181 can be appropriately assigned to parents, demonstrating the reliability of gIBD identified by ggComp (Figure 2). The limited crossover numbers indicated low recombination frequency and selection for large genomic blocks in wheat breeding practices (Supplemental Figure S15), which is consistent with previous study (Hao et al., 2020; Walkowiak et al., 2020).
The whole-genome scale GGNet (Figure 3B) was constructed by pairwise comparisons and the main advantage of this network over pedigree- or marker-based kinship-derived networks is that it successfully connects wheat germplasm accessions with genomic resource blocks. GGNet serves as a framework for presenting the trends of genetic flow during breeding, demonstrating closely related accessions, assisting the inference of pedigree relationships, and evaluating the contributions of founder lines in the context of networks. Additionally, it would be helpful to discover unintentionally duplicated collections. GGNet thus provides a valuable framework for studying wheat genetic diversity and can serve as an evolving platform to manage wheat germplasm and guide the design of breeding processes. Taken together, the results indicate that ggComp is effective at evaluating wheat germplasms at the genomic level.
For the slow LD decay property of the wheat genome, the gIBD-based method has been shown to be an effective way to dissect gene haplotypes or allelic groups. Haplotype inference based on genetic diversity in the genomic region is more resilient to sequencing error and random mutation noise than traditional methods relying on limited SNPs. The investigation of four genes Rht-B1, Rht-D1, Ppd-D1, and ω-gliadin showed that the allelic identification results perfectly fit the understanding of the data in a previous study (Figure 5) and expanded the knowledge of allele type diversity. Our results indicate that breeders are actually selecting conserved linkage blocks with target gene alleles in wheat breeding, and gHaps can help trace the origin of utilized alleles. Especially for poorly assembled genes or repeat-enriched genomic loci, the gHap-based strategy utilizes the boundary context for haplotype inference, providing an effective solution for mining beneficial alleles without knowing exact sequences. This method also constitutes an intuitive way to discover beneficial alleles and assist gene mining in QTL studies. Germplasm resource types serve as an effective measure to estimate trait-associated effects for alleles and provide guidance for marker selection when designing breeding strategies.
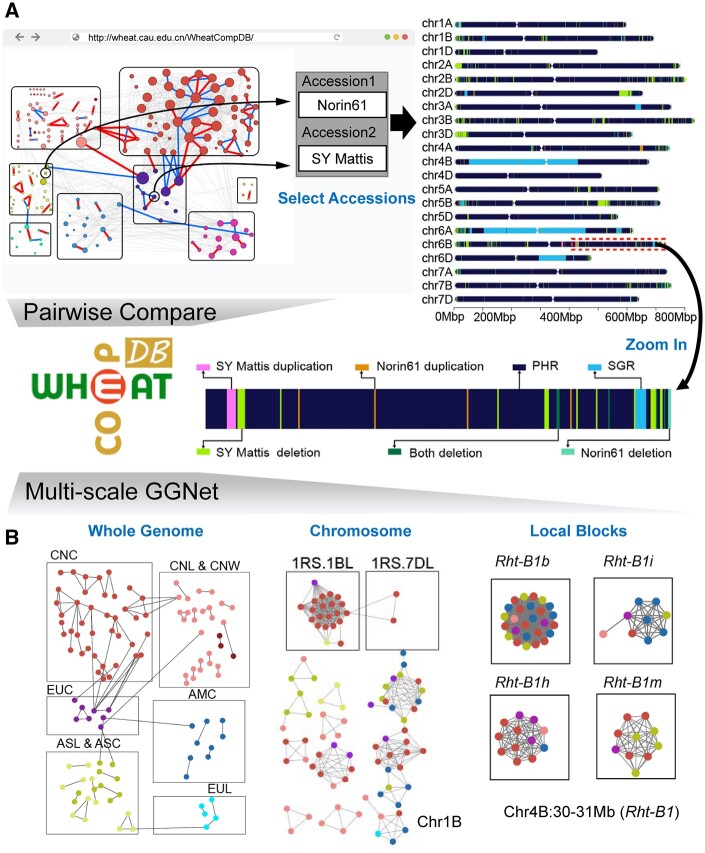
All pairwise accession comparison results and GGNet in multi-scale (whole-genome scale, chromosome scale, and single-block scale) can be accessed in WheatCompDB (http://wheat.cau.edu.cn/WheatCompDB/; Figure 6).
Figure 6.
WheatComp database with pairwise comparison and multi-scale GGNet functions assists germplasm evaluation and breeding. A, The pairwise compare function supports genomic comparison and visualization between any two accessions in the database. B, The multi-scale GGNet function supports the construction of germplasm network at whole-genome, chromosome and local blocks scales.
Conclusion
We developed a uniform method to perform unsupervised characterization of the stratified genomic diversity among wheat varieties and provided a preliminary framework to construct a comprehensive modern wheat germplasm network. This work provides a valuable resource for facilitating efficient germplasm utilization and directing wheat breeding programs in the future.
Materials and methods
Sample collection and whole-genome resequencing
Whole-genome sequencing data of 212 accessions were collected with worldwide distribution (Supplemental Table S1). Raw data of 186 previous published accessions were reanalyzed from raw data (Cheng et al., 2019; Hao et al., 2020; Walkowiak et al., 2020) or BAM files (Guo et al., 2020). The raw reads of previously published re-sequenced accessions are available under NCBI Sequence Read Archive accession PRJNA476679, PRJNA596843, PRJNA597250, and PRJNA544491. For the 26 new sequenced accessions, the genomic DNA from young roots was extracted by the standard cetyltrimethylammonium bromide-based protocol (Murray and Thompson, 1980). Pair-end sequencing was performed using the Illumina Novaseq 6000 platform, with read length of 150 bp and insertion size around 500-bp.
Genomic alignment, variation calling, and annotation
Trimmomatic (Bolger et al., 2014) were used to trim raw reads and BWA-MEM (Li and Durbin, 2009) was used to map retained high-quality clean reads to the CS reference genome (IWGSC RefSeq version 1.0) (International Wheat Genome Sequencing Consortium, 2018). Read pairs with abnormal insert sizes (more than 10,000 or less than −10,000 or =0) or low mapping qualities (<1) were filtered by bamtools (version 2.4.168) (Barnett et al., 2011). Potential PCR duplicates reads were removed by samtools (version 1.3.169) (Li et al., 2009). SNPs and INDELs were identified by the HaplotypeCaller module of GATK (version 3.870) (McKenna et al., 2010) in GVCF model. All GVCF files were performed joint call by GATK GenotypeGVCFs module. SNPs in VCF was filtered using GATK VariantFiltration function with the settings “–filterExpressionQD < 2.0 ‖‖ FS > 60.0 ‖‖ MQRankSum<−12.5 ‖‖ Read-PosRankSum<−8.0 ‖‖ SOR > 3.0 ‖‖ MQ < 40.0 ‖‖ DP > 30 ‖‖ DP < 3.” The filtering parameters for INDELs were “QD < 2.0, FS > 200.0,” and “ReadPosRankSum<−20.0 ‖‖ DP > 30 ‖‖ DP < 3.” SNPs were further filtered by following criteria: (1) MAF ≥ 1% and (2) bi-allelic sites. SnpEff (version 4.371) (Cingolani et al., 2012) was used to annoated SNPs and INDELs.
Identification of the threshold between PHRs and SGRs
Genetic distance between each pair of accessions was calculated first by PLINK (version 1.9) (Purcell et al., 2007) with parameter “–distance square 1-ibs.” Each accession and its top two closest samples were chosen to perform DSR calculations in every nonoverlapping 1-Mbp window across the whole genome after excluding all CNV blocks. The DSR is calculated as follows,
where denotes the count of differential homozygous SNPs, denotes the count of missing sites in either accession, and denotes the window size. The heterozygous sites were ignored in DSR calculation by default, considering that wheat is a self-pollinated crop and heterozygous SNPs detected in wheat likely to be resulted from sequencing or mapping error. in denominator was used to eliminate potential effect of missing sites. The density distribution of could be considered following a composited gaussian distribution. EM algorithm was used to fit the means and variances of three sub-distributions by R package mixtools (Benaglia et al., 2009). A hard threshold between PHRs and SGRs was selected according to Minimum-Error-Rate classification of Bayesian decision theory that
where is the posterior probability of the state of nature being SGR given the threshold x and is the posterior probability of the state of nature being PHR given the threshold x.
The pipeline of ggComp
All of the analysis steps described below were developed and wrapped as an open-source command-line tool that can be accessed at https://zack-young.github.io/ggComp/.
(1) Detection of the CNV blocks. CNV blocks were identified via the sliding window method with 1-Mbp windows. The average read depths (ARDs) were calculated by the ARD in each window via “coverage” function of bedtools (Quinlan and Hall, 2010). The ARDs were then normalized by dividing the median value of total ARDs in each accession. Then windows with normalized ARD ≤ 0.5 or normalized ARD ≥ 1.5 were defined as CNV-deletion block or CNV-duplication block. The threshold selection was based on the distribution of whole genome binwise normalized ARDs (Supplemental Figure S3).
(2) Identification and HMM smoothing between PHRs and SGRs. After excluding CNV blocks, SGRs and PHRs were distinguished by a hard threshold, which is determined by performing EM strategy on the DSR density distribution (DSR = 10 by default). We then applied an HMM-based strategy to correct noise signal (Supplemental Figure S11). The raw types (that are SGR, PHR, and CNV) of accession genomes were used as the observations, and the underlying states (sgr, phr, and cnv) inferred by HMM are assumed to be the real types. To build the initial model, a set of “transition frequency” calculated from 20% of all pairwise comparison results (Supplemental Table S5) was used as both of the initial state transition probability matrix and the emission probability matrix. Then the model was trained using the Baum–Welch iterative re-estimation procedure provided by python library hmmlearn (https://pypi.org/project/hmmlearn/) on raw sequences with parameters “n_iter = 100” and “tol = 0.001.” The emission probabilities from state sgr and phr to observation CNV were set to 0 as to avoid the emission from cnv to SGR and PHR or otherwise in the model. Then emission probabilities were normalized accordingly (Supplemental Table S6). After training, hidden state sequences (i.e. the denoized sequences) could be estimated from observation sequences using function “MultinomialHMM.decode()” from hmmlearn, with parameter “algorithm=viterbi.”
(3) Visualizing the distribution of CNV blocks, SGRs, and PHRs between pairwise accessions. To get a better sense of genomic relationship between accessions, we developed a visualization function to present the distribution of SGRs, PHRs, sample-specific CNV-deletion regions, sample-specific CNV-duplication regions, shared CNV-deletion regions, and shared CNV-duplication regions along 21 chromosomes of wheat. The length of chromosomes and position of centromeres were acquired from International Wheat Genome Sequencing Consortium (2018).
Saturation analysis of SNP calling
We performed the saturation analysis based on resequencing data of Aimengniu, which had a total genome coverage of around 14×, to dissect the relationship between wheat re-sequencing coverage and number of identified SNP, and result indicated our current dataset was qualified to perform a genomic resource dissection through pairwise comparison strategy. Samtools (version 1.3.169) (Li et al., 2009) with the parameter “view -s” was used to randomly extract alignments from BAM files of Aimengniu to produce multiple datasets at different coverage levels. SNP calling was conducted by using GATK (version 3.870) (McKenna et al., 2010) pipeline.
Haplotype calls based on genome assemblies
For the comparison between the genome assemblies of SY Mattis and Jagger, we applied samtools-1.9 faidx to extract individual chromosomes from the assemblies and then generated the dot-plot. Pairwise alignments were performed for each chromosome, following the method in Brinton et al. (2020). The NUCmer program of MUMmer-3.2332 (Kurtz et al. 2004) was used for pairwise alignment with –mum option. The raw delta files were filtered using the delta-filter command with the options −l 20,000, −r, and –q. The comparison results were processed through the Rscript provided by Brinton et al. (2020) in https://github.com/Uauy-Lab/pangenome-haplotypes.
Nongda5181 parental-descend genome region identification
To obtain more consecutive gIBD between Nongda5181 and its parents, we further determined the source of genomic block shared between Nongda3097 and Lunxuan987 in Nongda5181 by the donor of regions in the vicinity of them. We assumed that parental shared block could be redirected to one parent if the vicinities of this block belong to the same parent.
Phylogenetic analysis
SNP sites with <10% missing rate were used to for phylogenetic analysis. And the IBS-based hierarchical phylogenetic tree was obtained by calculating the pairwise genetic distances using PLINK (version 1.971) (Purcell et al., 2007) with parameter “--distance square 1-ibs.” The IBS-based and gIBD-based tree were constructed using the hclust method and ggtree (Yu et al., 2017) in the R package.
Genomic-based germplasm network construction
The proportion of gIBD between accessions on the whole genome was used to build GGNet. gIBD percentage was sorted into three levels (1) 20–40%; (2) 40–50%, and (3) >50%. gIBD percentage ˂20% was not shown in the network. After converting gIBD percentage information into a distance matrix, the matrix was imported into Cytoscape (Shannon et al., 2003) to generate a germplasm utilization network. The clustered network was plotted by artificial adjustment in Cytoscape and the node size was adjusted by the degree of each node produced by the network analysis function in Cytoscape.
Germplasm resource gHap identification
The gHaps were calculated based on gIBDs in each block. We firstly separated gIBD into SGR, “both deletion” and “both duplication.” The SGR information was transformed into adjacency list and processed by MCL software (Enright et al., 2002) with parameter (-abc -I 2.0). The accessions that sorted into a single cluster were considered as sharing the same gHap.
Statistical analyses
Spearman’s rank correlation coefficient test statistic was performed using the “cor.test” function in R (parameters, method = “spearman”; exact = TRUE). Two-tailed Student’s t tests were executed using “t.test” in R (parameters, alternative = “two.sided”; paired = FALSE).
Data access
The ggComp pipeline and corresponding manual are available as open-source code under the MIT License at https://zack-young.github.io/ggComp/. And the database WheatCompDB is available at http://wheat.cau.edu.cn/WheatCompDB/.
Accession numbers
The raw sequence data reported in this paper have been have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA722149 and to the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number CRA004026. Please refer to the attached table (Supplemental Table S1) for details.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Geographic distribution of wheat accessions by country or region of origin.
Supplemental Figure S2. The effect of mapping depth on SNP recall rate.
Supplemental Figure S3. Distribution of normalized read depth per bin along the whole genome of all accessions.
Supplemental Figure S4. CNV block distribution of Aikang58 and Lovrin10.
Supplemental Figure S5. Distribution of CNV-duplication blocks ratios in A, B, and D subgenomes.
Supplemental Figure S6. Frequency of CNV-deletion blocks (blue) and CNV-duplication blocks (red) in each window of all accessions along the whole genome compared with the CS reference genome.
Supplemental Figure S7. Profile of CNV segment distribution in wheat accessions.
Supplemental Figure S8. Distribution of log10(DSR10) in each window (1-Mbp length).
Supplemental Figure S9. Distribution of binwise SNP density between Bima4 and Bima1 along the whole genome.
Supplemental Figure S10. Distribution of binwise SNP density between Bima4 and CS along the whole genome.
Supplemental Figure S11. Layout of the HMM.
Supplemental Figure S12. Comparison of the distributions of PHR, SGR, and CNV blocks between Lancer and Norin61 before and after applying HMM strategy.
Supplemental Figure S13. Distribution of PHR, SGR, and CNV blocks between Nongda5181 and Nongda3097.
Supplemental Figure S14. Distribution of PHR, SGR, and CNV blocks between Nongda5181 and Lunxuan987.
Supplemental Figure S15. Dissecting the inherited genomic blocks of Nongda5181 from parents.
Supplemental Figure S16. Distribution of PHR, SGR, and CNV blocks between Nongda3097 and Jingdong6.
Supplemental Figure S17. Distribution of PHR, SGR, and CNV blocks between Nongda3097 and Nongda3338.
Supplemental Figure S18. Dissecting the inherited genomic blocks of Nongda5181 from parents and grandparents.
Supplemental Figure S19. Genome similarity hierarchical clustering based on ward’s hierarchical clustering method on a scale of log2(gIBD proportion).
Supplemental Figure S20. Distribution of PHR, SGR, and CNV blocks between Bima1 and Bima4.
Supplemental Figure S21. Distribution of PHR, SGR, and CNV blocks between Chengduguangtou and CS.
Supplemental Figure S22. Distribution of PHR, SGR, and CNV blocks between Yunnan098 and Nanda2419.
Supplemental Figure S23. Comparison between gIBD-based (left) and IBS-based (right) hierarchical clustering results.
Supplemental Figure S24. The chromosome scale GGNet of 1B chromosome.
Supplemental Figure S25. Distribution of PHR, SGR, and CNV blocks between Aimengniu and Lovrin10.
Supplemental Figure S26. The comparison between gHap and haplotypes identified by Brinton et al.
Supplemental Figure S27. The trajectories of semi-dwarf alleles Rht-B1b and Rht-D1b utilization in CNCs were presented in the context of GGNet.
Supplemental Table S1. Detailed information of the whole-genome resequencing data of wheat used in this study.
Supplemental Table S2. Chromosomal crossover counts in Nongda5181 between its parents.
Supplemental Table S3. List of accessions that carried the 1RS chromosome and their released time.
Supplemental Table S4. The identified gHap types of Rht-B1 and Rht-D1 in wheat accessions.
Supplemental Table S5. Initial transition frequency matrix between observations for the HMM model.
Supplemental Table S6. Trained state transition probability matrix and emission probability matrix for the HMM model.
Supplementary Material
Acknowledgments
We thank Zengjun Qi (Nanjing Agricultural University) and Dr Zhaoyan Chen (Chinese Agricultural University) for helpful discussion in this work. We thank Lv Sun and Yongfa Wang for sample preparation, thank Yongming Chen and Zhen Qin for providing technological support in developing database.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 91935303) to H.P. and supported by the Major Program of the National Natural Science Foundation of China (grant no. 31991210) to Q.S. This work was also supported by the Chinese Agricultural University Fund for Joint Research Project with Partner University (2019TC153 and 2020SF002) to W.G., and by The 2115 Talent Development Program of China Agricultural University.
Conflict of interest statement. China Agricultural University has filed a patent application on the method of characterizing germplasm relationship in genomic blocks in wheat.
Contributor Information
Zhengzhao Yang, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Zihao Wang, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Wenxi Wang, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Xiaoming Xie, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Lingling Chai, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Xiaobo Wang, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Xibo Feng, Tibet Key Experiments of Crop Cultivation and Farming/College of Plant Science, Tibet Agriculture and Animal Husbandry University, Linzhi 860000, China.
Jinghui Li, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China; Wheat Center, Henan Institute of Science and Technology/Henan Provincial Key Laboratory of Hybrid Wheat, Xinxiang 453003, China.
Huiru Peng, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Zhenqi Su, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Mingshan You, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Yingyin Yao, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Mingming Xin, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Zhaorong Hu, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Jie Liu, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Rongqi Liang, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Zhongfu Ni, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Qixin Sun, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Weilong Guo, State Key Laboratory for Agrobiotechnology/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement/College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
W.G., H.P., Z.N., and Q.S. conceived this work. Z.Y., Z.W., W.W., and W.G. designed and implement the algorithm. W.W. and Z.Y. built up the database. X.X., L.C., X.W., X.F., and R.L. prepared the data. Z.Y. and Z.W. performed analysis. L.C., J.L., H.P., Z.S., M.Y., Y.Y., M.X., Z.N., Z.H., J.L., Q.S., and W.G. interpreted the data. W.G., Z.Y., Z.W., and W.W. wrote the manuscript and revised it. All authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Weilong Guo (guoweilong@cau.edu.cn).
References
- Balfourier F, Bouchet S, Robert S, De Oliveira R, Rimbert H, Kitt J, Choulet F, Paux E (2019) Worldwide phylogeography and history of wheat genetic diversity. Sci Adv 5: eaav0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT (2011) BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27: 1691–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaglia T, Chauveau D, Hunter DR, Young DS (2009) mixtools: an R package for analyzing mixture models. J Stat Softw 32: 1–29 [Google Scholar]
- Bevan MW, Uauy C, Wulff BB, Zhou J, Krasileva K, Clark MD (2017) Genomic innovation for crop improvement. Nature 543: 346–354 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton J, Ramirez-Gonzalez RH, Simmonds J, Wingen L, Orford S, Griffiths S, Wheat Genome P, Haberer G, Spannagl M, Walkowiak S, et al. (2020) A haplotype-led approach to increase the precision of wheat breeding. Commun Biol 3: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR (2013) Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Xu D, Hanif M, Xia X, He Z (2020) Genetic architecture underpinning yield component traits in wheat. Theor Appl Genet 133: 1811–1823 [DOI] [PubMed] [Google Scholar]
- Cheng H, Liu J, Wen J, Nie X, Xu L, Chen N, Li Z, Wang Q, Zheng Z, Li M. , et al. (2019) Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol 20: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman SM, Hufford MB, Andorf CM, Lubberstedt T (2020) Haplotype structure in commercial maize breeding programs in relation to key founder lines. Theor Appl Genet 133: 547–561 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Youssefian S (1985) Chapter 1 - Dwarfing genes in wheat. In Russell GE, ed, Progress in Plant Breeding–1, Butterworth-Heinemann, Oxford, pp 1–35 [Google Scholar]
- Guo W, Xin M, Wang Z, Yao Y, Hu Z, Song W, Yu K, Chen Y, Wang X, Guan P. , et al. (2020) Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat Commun 11: 5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, Spannagl M, Marcon C, Ruban A, Urbany C, et al. (2020) European maize genomes highlight intraspecies variation in repeat and gene content. Nat Genet 52: 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C, Jiao C, Hou J, Li T, Liu H, Wang Y, Zheng J, Liu H, Bi Z, Xu F. , et al. (2020) Resequencing of 145 landmark cultivars reveals asymmetric sub-genome selection and strong founder genotype effects on wheat breeding in China. Mol Plant 13: 1733–1751 [DOI] [PubMed] [Google Scholar]
- He F, Pasam R, Shi F, Kant S, Keeble-Gagnere G, Kay P, Forrest K, Fritz A, Hucl P, Wiebe K. , et al. (2019) Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet 51: 896–904 [DOI] [PubMed] [Google Scholar]
- Hu HZ, Sheng ZQ, He CS, Wen YZ, Dong ZZ, Xu L (2018) Wheat production and technology improvement in China. J Agric 8: 99–106 [Google Scholar]
- Huang X, Zhu M, Zhuang L, Zhang S, Wang J, Chen X, Wang D, Chen J, Bao Y, Guo J. , et al. (2018) Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor Appl Genet 131: 1967–1986 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Jia J, Li H, Zhang X, Li Z, Qiu L (2017) Genomics-based plant germplasm research (GPGR). Crop J 5: 166–174 [Google Scholar]
- Kim YH, Park HM, Hwang TY, Lee SK, Choi MS, Jho S, Hwang S, Kim HM, Lee D, Kim BC. , et al. (2014) Variation block-based genomics method for crop plants. BMC Genomics 15: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Yang W, Lou X, Liu D, Sun J, Guo X, Wang J, Li Y, Zhan K, Ling HQ. , et al. (2013) Novel natural allelic variations at the Rht-1 loci in wheat. J Integr Plant Biol 55: 1026–1037 [DOI] [PubMed] [Google Scholar]
- Liu D, Zhang L, Hao M, Ning S, Yuan Z, Dai S, Huang L, Wu B, Yan Z, Lan X. , et al. (2018) Wheat breeding in the hometown of Chinese Spring. Crop J 6: 82–90 [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. , et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goue N, Balfourier F. , et al. (2019) Tracing the ancestry of modern bread wheats. Nat Genet 51: 905–911 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. , et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanus-Wallace MT, Hackauf B, Mascher M, Lux T, Wicker T, Gundlach H, Baez M, Houben A, Mayer KFX, Guo L. , et al. (2021) Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat Genet 53: 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebetzke GJ, Richards RA (2000) Gibberellic acid-sensitive dwarfing genes reduce plant height to increase kernel number and grain yield of wheat. Austral J Agric Res 51: 235–246 [Google Scholar]
- Reitz LP, Salmon SC (1968) Origin, history, and use of norin 10 wheat. Crop Sci 8: 686–689 cropsci1968.0011183X000800060014x. [Google Scholar]
- Romay MC, Millard MJ, Glaubitz JC, Peiffer JA, Swarts KL, Casstevens TM, Elshire RJ, Acharya CB, Mitchell SE, Flint-Garcia SA. , et al. (2013) Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol 14: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng ZQ, Su WZ, Jia BY, Bao JS (1983) Chinese Wheat Varieties and Their Pedigree(in Chinese), China Agricultural Publishing House, Beijing, China [Google Scholar]
- Thind AK, Wicker T, Muller T, Ackermann PM, Steuernagel B, Wulff BBH, Spannagl M, Twardziok SO, Felder M, Lux T. , et al. (2018) Chromosome-scale comparative sequence analysis unravels molecular mechanisms of genome dynamics between two wheat cultivars. Genome Biol 19: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Velde K, Thomas SG, Heyse F, Kaspar R, Van Der Straeten D, Rohde A (2021) N-terminal truncated RHT-1 proteins generated by translational reinitiation cause semi-dwarfing of wheat Green Revolution alleles. Mol Plant 14: 679–687 [DOI] [PubMed] [Google Scholar]
- Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D. , et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588: 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu Y, Su H, Guo X, Han F (2017) Centromere structure and function analysis in wheat-rye translocation lines. Plant J 91: 199–207 [DOI] [PubMed] [Google Scholar]
- Xie W, Feng Q, Yu H, Huang X, Zhao Q, Xing Y, Yu S, Han B, Zhang Q (2010) Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci USA 107: 10578–10583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Li X J, Li X Q, Yang X M, Liu W H, Gao A N, Li L H (2010) Inheritance of 1BL/1RS of founder parent Lovrin 10 in its progeny. J Triticeae Crops 30: 221–226 [Google Scholar]
- Yang Z, Hu HZ, Sheng ZG, Qin XL, Min CX, Chao GY, Bin JZ, Jun YG (2004) Utilization of 1BL/1RS translocation in wheat breeding in China. Acta Agron Sin 30: 531–535 [Google Scholar]
- Yang FP, Zhang XK, Xia XC, Laurie DA, Yang WX, He ZH (2009) Distribution of the photoperiod insensitive Ppd-D1a allele in Chinese wheat cultivars. Euphytica 165: 445–452 [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY (2017) ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Method Ecol Evol 8: 28–36 [Google Scholar]
- Zhang F, Wang C, Li M, Cui Y, Shi Y, Wu Z, Hu Z, Wang W, Xu J, Li Z (2021) The landscape of gene-CDS-haplotype diversity in rice: properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol Plant 14: 787–804 [DOI] [PubMed] [Google Scholar]
- Zhang R, Xu G, Li J, Yan J, Li H, Yang X (2018) Patterns of genomic variation in Chinese maize inbred lines and implications for genetic improvement. Theor Appl Genet 131: 1207–1221 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang S, Zhou Y, He Z, Xia X (2006) Distribution of the Rht-B1b, Rht-D1b and Rht8 reduced height genes in autumn-sown Chinese wheats detected by molecular markers. Euphytica 152: 109–116 [Google Scholar]
- Zhou Y, Browning SR, Browning BL (2020) A fast and simple method for detecting identity-by-descent segments in large-scale data. Am J Hum Genet 106: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Chen W, Lin Z, Chen H, Wang C, Li H, Yu R, Zhang F, Zhen G, Yi J. , et al. (2016) Pedigree-based analysis of derivation of genome segments of an elite rice reveals key regions during its breeding. Plant Biotechnol J 14: 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.