Abstract
Growing knowledge about crop domestication, combined with increasingly powerful gene-editing toolkits, sets the stage for the continual domestication of crop wild relatives and other lesser-known plant species.
The domestication of wild species led to a wide variety of crops adapted to a range of climatic and edaphic conditions, which allowed expansion of cultivation to larger areas and over longer periods. Subsequent crop breeding led to higher yields and facilitated population growth (Evans, 1998). Until recent times, both domestication and breeding occurred empirically with little understanding of the underlying biological mechanisms. Today, we know that genetic variation is created by mutation and that breeding operates by stacking favorable mutations in a single plant through recombination. Technical advances will eventually allow controlled manipulation of mutation and recombination (Taagen et al., 2020; Nasti and Voytas, 2021), and thus de novo domestication of wild species by targeted modification of their genomes (Gasparini et al., 2021). Such tools will also aid in accelerating the improvement of traditional, semi-domesticated “orphan” crops that perform poorly in modern agricultural systems (Tadele, 2019). The combination of these approaches will help broaden the narrow genetic basis of crops on which humankind currently relies (Milla and Osborne, 2021). Furthermore, recent breakthroughs have shown that targeted control of gene expression is an even faster avenue to produce desirable phenotypes (Pan et al., 2021a), thus bypassing the need for mutation and recombination. However, the deliberate effort to create new crops or improve existing ones requires a thorough understanding of the genetic basis of domestication (Kantar et al., 2017). The synergistic combination of classical archeaobotany and genetics (Denham et al., 2020) with high-throughput genomics (Purugganan and Jackson, 2021) is revealing that a variety of different processes may have operated in the domestication of crops. This knowledge, combined with increasingly powerful gene-editing toolkits, sets the stage for the continual domestication of crop wild relatives and other lesser-known plant species by defining an ideal plant type (“ideotype”) that is more resilient, nutritious, and productive in a given environment (Zsögön et al., 2017). Wild plants that are naturally resistant to biotic (insects and diseases) and tolerant of abiotic (drought and heat) stresses can be selected and manipulated by introducing mutations that mimic the domestication events that led to improved yield and agronomic performance of the major crops of today. Proof-of-concept for the potential of this de novo domestication approach was provided using gene editing to create agronomically important traits in wild relatives of the tomato (Solanum lycopersicum) (Li et al., 2018; Zsögön et al., 2018), the orphan crop Physalis (Lemmon et al., 2018) and, more recently, in a polyploid wild relative of rice (Oryza species) (Yu et al., 2021). Here, we review how recent progress in the understanding of crop domestication and technical breakthroughs in gene-editing technology could be combined to produce better crops for the future.
The evolving understanding of crop domestication
Understanding the dynamics of crop domestication is essential to discover the genomic signature of the phenotypic changes operated in the wild ancestral species. Until recently, it was widely believed that domestication was long and protracted, produced exclusively by human agency, and that convergent traits of the domestication syndrome (the suite of traits common to many crops that are not found in their respective wild ancestors, see Table 1 for examples) would have a similar genetic basis (Purugganan, 2019). Growing evidence is painting a more complex picture. First, although genomic analyses confirmed that in some cases, the domestication process took hundreds or even thousands of years, there are many examples of fairly recent and rapid domestication events: sugar beet (Beta vulgaris) (Dohm et al., 2014), kiwifruit (Actinidia chinensis) (Ferguson, 2013), and African oil palm (Elaeis guineensis) (Zeven, 1972). There are also notable cases of “second cycle domestication”, whereby existing domesticates were quickly adapted to new uses, for instance, changing soybean (Glycine max) from a vegetable to a protein and oil crop (Kofsky et al., 2018) and lupines (Lupinus species) from green manure to a seed crop (Gulisano et al., 2019). Second, operational selection, which occurs mostly because of human operations (hence the name) may have occurred after a first round of genetic changes created by the attraction of animals and plants to human habitations and their adaptation to this human-made environment (Spengler and Mueller, 2019). Pre-domestication of plants by grazing and foraging animals would also have contributed to early selection on and seed and horticultural crops and fruit trees (Zonneveld et al., 2018). Thus, it is possible that at least some traits of the domestication syndrome represent exaptations (“preadaptations”) for farming that were initially fixed by mutualism with animals (Spengler et al., 2021). Finally, studies dissecting the function of “domestication genes” point to a highly diversified genetic basis for convergent domestication traits in crops (Lai et al., 2018; Chen et al., 2021a). For instance, the nonshattering trait selected during domestication was achieved via widely divergent mechanisms in legumes and cereals: pod indehiscence in the former and rachis flexibility in the latter, which are mechanisms controlled by different developmental pathways and genes (Parker et al., 2021). Similarly, other fundamental domestication and breeding traits such as changes in growth habit, altered photoperiodic response for flowering or tuberization, reduced seed or fruit dispersal, and reduced seed dormancy are controlled by a multiplicity of genes from different families (Table 1).
Table 1.
The diversified genetic basis of some relevant crop domestication and breeding traits
| Trait | Species | Gene | Type of variant | References |
|---|---|---|---|---|
| Changes in growth habit (dwarfism, determinate growth and/or side branching) | Wheat (Triticum spp) | Rht genes (gibberellin insensitivity) | C | Peng et al., 1999 |
| Rice (Oryza spp) | sd1 (gibberellin biosynthesis) | C, ID | Sasaki et al., 2002 | |
| Common beans (Phaseolus vulgaris) | TFL1 (CETS) | C, ID, S, T | Kwak et al., 2012 | |
| Pea (Pisum sativum) | TFL1 (CETS) | C, ID, R | Foucher et al., 2003 | |
| Soybean (Glycine max) | Dt1 (CETS) and Dt2 (MADS-box) |
|
Tian et al., 2010; Ping et al., 2014 | |
| Tomato (Solanum lycopersicum) | SP/TFL1 (CETS) | C | ||
| Cotton (Gossypium spp) | GbAF (CETS) | C | Si et al., 2018 | |
| Altered photoperiodic response | Barley (Hordeum vulgare) | pseudo-response regulator PpdH1 | C | Turner et al., 2005 |
| Maize (Zea mays) | CMF transcription factor | R | Yang et al., 2013; Huang et al., 2018 | |
| Sorghum (Sorghum bicolor) | pseudo-response regulator 37 (PRR37) | C | Murphy et al., 2011 | |
| Sunflower (Helianthus annuus) | FT (CETS) | C | Blackman et al., 2010 | |
| Soybean (Glycine max) | pseudo-response regulator 37 (PRR37) | C | Lu et al., 2020a; Wang et al., 2020 | |
| Common beans (Phaseolus vulgaris) | PHYTOCHROME A3 (PHYA3) and CONSTANS-like 2 |
|
Weller et al., 2019; González et al., 2021 | |
| Beet (Beta vulgaris) | pseudo-response regulator BTC1 | R | Pin et al., 2012 | |
| Tomato (Solanum lycopersicum) | SP5G and SP11b (CETS) |
|
Soyk et al., 2017; Song et al., 2020b | |
| Cassava (Manihot esculenta) | FT (CETS) | R | Adeyemo et al., 2019 | |
| Potato (Solanum tuberosum) | CYCLING DOF FACTOR | ID | Kloosterman et al., 2013 | |
| Cotton (Gossypium spp) | FT (CETS) | R | Prewitt et al., 2018 | |
| Rose (Rosa chinensis) | CONSTANS and CONSTANS-like 4 | R | Lu et al., 2020a | |
| Reduced seed or fruit dispersal | Barley (Hordeum vulgare) | btr1 and btr2 | C | Pourkheirandish et al., 2015 |
| Foxtail millet (Setaria italica) | Les1 (MYB) | T | Mamidi et al., 2020 | |
| Rice (Oryza spp) | qSH1 (homeodomain TF), sh4 (MYB3 binding domain), SH3 (YABBY) |
|
Konishi et al., 2006; Li et al., 2006; Lv et al., 2018 | |
| Sorghum (Sorghum bicolor) | Sh1 (YABBY) | D, S | Lin et al., 2012 | |
| Wheat (Triticum spp) | Q (APETALA2) | C, R | Simons et al., 2006 | |
| Soybean (Glycine max) | Shatt1-5; PDH1 (dirigent-like protein) |
|
Dong et al., 2014; Funatsuki et al., 2014 | |
| Tomato (Solanum lycopersicum) | JOINTLESS-2 (MADS-box) | C, T | Roldan et al., 2017 | |
| Reduced seed dormancy | Barley (Hordeum vulgare) |
|
C | Sato et al., 2016; Nakamura et al., 2016 |
| Rice (Oryza spp) | seed dormancy 4 (Zn-finger transcription factor), G (CAAX amino-terminal protease) | C, D | Sugimoto et al., 2010; Wang et al., 2018 | |
| Wheat (Triticum spp) | Phs1 (MKK3) | C | Torada et al., 2016 | |
| Common beans (Phaseolus vulgaris) | pectin acetylesterase 8 | ID | Soltani et al., 2021 | |
| Soybean (Glycine max) | G (CAAX amino-terminal protease) | C | Wang et al., 2018 | |
| Cotton (Gossypium spp) | GhMFT1 and GhMFT2 (CETS) | R | Yu et al., 2019 |
Type of variant refers to the allele changes found in domesticated versus wild relatives.
C, non-synonymous coding sequence change; ID, insertion–deletion; R, regulatory variant (that alters gene expression); S, splicing variant; T, transposon insertion in coding sequence.
The diversified genetic basis for domestication traits in plants is quite unlike the findings for animals, in which strong loss-of-function mutations in pluripotent embryonic cells of the neural crest lead to multiple traits of the domestication syndrome (Wilkins, 2020). This is not unexpected considering the marked differences in the general system of development between plants and animals (Meyerowitz, 2002). Most animals lack persistent stem cells, whereas plants retain embryonic cells in their meristems (Périlleux et al., 2019). Animals, therefore, have a closed developmental program and respond to environmental cues mainly through movement. Plants, on the other hand, are sessile, and their open and modular developmental pattern is highly flexible (Zsögön and Peres, 2018). The flexibility of the genetic circuitry controlling phenotypic outcomes is a consequence of the enormous environmental variability to which plants are subjected over the course of their lifetime. Thus, the plethora of genes underlying plant domestication traits, compared to the relatively few in animals, may reflect the higher developmental plasticity of plants. Plasticity itself is a quantitative trait strongly shaped by environmental conditions (Stotz et al., 2021), and the initial degree of plasticity could have determined which early domesticants were advanced and which were abandoned (Vilela and González-Paleo, 2015; Piperno et al., 2019). The potential for domestication of a species may thus be directly related to its genomic (Dubcovsky and Dvorak, 2007), physiological (Matesanz and Milla, 2018), and developmental (Chen et al., 2021b) plasticity. Identification of the causal genes for plasticity and functional characterization of their roles in plant fitness will inform future domestication studies and provide new valuable targets for manipulation (Laitinen and Nikoloski, 2019; Liu et al. 2021b).
The molecular mechanisms underlying crop domestication and breeding
Plant breeding is the selection of optimal phenotypes, i.e. the best possible combinations of genotype and environment. The work of breeders began long before it was understood that genotypic variation is created by mutation, whereby new traits arise, and recombination, which allows the stacking of useful traits into a single plant (Evans, 1998). Since both of these biological processes happen by chance, breeders create new varieties by crossing parents with traits of interest and selecting the best individuals among the progenies. The incorporation of powerful genotyping (Bevan et al., 2017), phenotyping (Atefi et al., 2021), and growth acceleration methods (Ghosh et al., 2018) represent valuable contributions to the crop breeding enterprise. However, they still rely on a strong component of chance, as the phenotypes are not predictable and ought to be found among large numbers of variants. The first step to achieve full predictability in the generation of novel phenotypes is by controlling the basic processes that give rise to new genotypes: mutation and recombination.
A mutation is the ultimate source of genetic variation and occurs spontaneously at low frequencies. Soybean (Glycine max), for instance, has a genome of 1.15 Gbp and suffers a spontaneous mutation rate of one bp per ∼5 × 10−8 bp per year. Assuming for simplicity that all mutations will be single-nucleotide polymorphisms (SNPs), one specific SNP of interest will occur in only one out of 100 million plants in one growing season. If the plants are grown at conventional density, this would represent a field area of 400 hectares. One of the major breakthroughs to accelerate the creation of genetic variation was the discovery of artificial mutagenesis by Hermann Müller (1890–1967). Application of physical or chemical treatments increases the mutation rate by many orders of magnitude, for instance, treating soybean seeds with ethyl methanesulfonate can generate one mutation per 74 kb of the genome (Tsuda et al., 2015). Thus, from the early 20th century on, agronomists incorporated mutagenic treatments to increase genetic variation in their breeding programs.
The first crop variety produced via induced mutagenesis was “Chlorina” tobacco (Nicotiana tabacum) in the 1930s (Jankowicz-Cieslak et al., 2017). Today, over 3,200 mutant varieties in 170 species are grown commercially (https://mvd.iaea.org/). However, mutagenesis is an undirected and random process, and in many cases, the identity of the variants and their developmental and physiological bases remained largely unexplored. This lack of knowledge precluded targeted manipulation and extrapolation between different crop species. Furthermore, creating or discovering mutations that produce desirable agronomic traits is only the first step. The next challenge is to combine or stack different beneficial mutations into a single plant (e.g. Chopra et al., 2020). This is achieved through crossing and selection in a segregating population, and the chances of finding the appropriate mutant combinations depend on the frequency of genetic recombination during meiosis (Choi and Henderson, 2015). Unlike the mutation rate, however, the rate of meiotic recombination is highly plastic and notoriously difficult to even determine accurately (Stumpf and McVean, 2003).
There is evidence that the domestication process itself may have favored the selection of increased recombination rates (Ross-Ibarra, 2004). In the laboratory model Arabidopsis (Arabidopsis thaliana), crossovers occur in higher frequency at specific regions of the genome (“hotspots”) (Choi and Henderson, 2015), and they can be influenced by environmental parameters, for instance, temperature (Lloyd et al., 2018). Wide crosses to wild crop relatives have been instrumental to regain genetic variation lost during the domestication process (Hajjar and Hodgkin, 2007). However, the introduction of foreign genome segments into crops may also reduce or altogether impair recombination, leading to breeding “blind spots” in the genome, with the additional downside that potentially deleterious alleles may become fixed in these regions (Lenormand and Otto, 2000). A classic example is the introduction of root-knot nematode resistance in tomato from its wild relative S. peruvianum (controlled by the Mi-1 gene) and of resistance to yellow leaf curl virus from S. chilense (via the Ty-1 gene). The loci are located in close proximity on chromosome 6 in a region of suppressed recombination, so only commercial hybrid cultivars harbor both resistance traits (Lin et al., 2014).
The difficulty in combining favorable mutations in a single plant during the domestication process was partially bypassed by selecting genes with pleiotropic effects. For instance, mutations of the Q gene in wheat allows free threshing (separation of the grain from the straw), a key domestication trait in cereals, but also simultaneously confers increased grain weight, roundness and yield (Xie et al., 2018). The Q gene encodes an APETALA2-family transcription factor, and the wild and domesticated alleles diverge by a nonsynonymous SNP that leads to an amino acid substitution and a synonymous SNP in a microRNA target site that leads to higher gene expression (Simons et al., 2006). Classical QTL mapping studies contributed to narrowing down and identifying chromosome regions associated with specific domestication traits in crops (Paterson, 2002). However, both this approach for a causal association of genotype and phenotype and its most modern iteration, genome-wide association studies (GWAS), while very powerful in defining the genes underlying a wide range of agronomic and metabolic traits (Fernie and Gutierrez-Marcos, 2019; Liu and Yan, 2019; Alseekh et al., 2021) have had limited impact in uncovering pleiotropy in domestication traits: most studies unfortunately only addressed a single, or various very similar, phenotypes at a time. The development of multi-phenotype databases such as those developed for Arabidopsis should fill this gap (Togninalli et al., 2020). High-throughput phenomics platforms will allow the creation of similar databases for other crops, with the result that the potential for gene discovery will increase proportionally.
Controlling mutation and recombination to produce predictable phenotypes
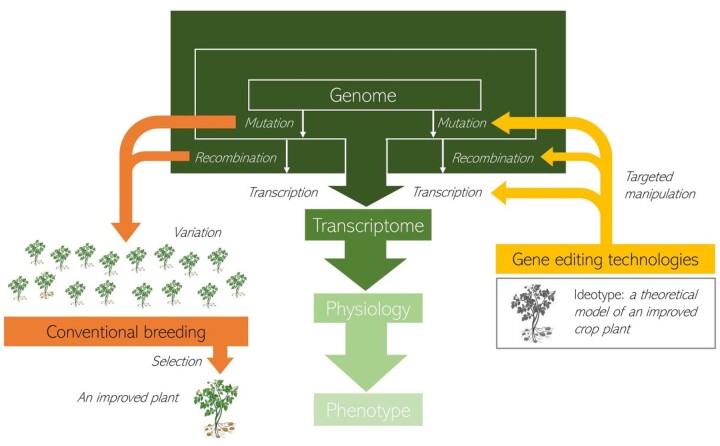
A recurrent theme of the plant biology literature in the late 20th century was the attempts of physiologists and geneticists to uncover the mechanisms underlying agronomic gains produced by breeders in crops (Fischer et al., 2014). Even with the improved technical platforms available today, dissecting the genetic and physiological basis of advantageous phenotypes is a challenging endeavor (Korte and Farlow, 2013). However, the improved understanding of plant function gathered in the last decades constitutes a valuable repertoire that can inform a new, knowledge-based breeding pipeline (Li and Yan, 2020). Traits can now be designed, modeled, engineered, and stacked using a priori knowledge by targeted manipulation of the fundamental processes of mutation and, potentially, very soon also recombination (Taagen et al., 2020). This suggests that the mechanistic understanding of the connection between genotype and phenotype will increasingly be used to inform and direct rather than learn from plant breeding (Figure 1).
Figure 1.
A pipeline that combines recently developed gene-editing technologies and conventional breeding to allow domestication and breeding of orphan crops and crop wild relatives.
Targeted DNA modifications with a level of precision that was not thought possible even a few years ago are now routinely carried out in laboratories around the world (Gao, 2021; Pan et al., 2021a). Tools that modify DNA sequence with specific and predictable changes have undergone rapid improvements over the past decade. Beginning with customized zinc-finger nucleases and closely followed by TAL-effector nucleases, the technology has quickly expanded to multiple CRISPR/Cas platforms (Čermák, 2021). Today, these Cas systems include four variants: nucleases, transposases/recombinases, base editors (BEs), and prime editors (PEs) (Anzalone et al., 2020). Both BE and PE are promising tools for genetic manipulation of crop wild relatives (Molla et al., 2021), as they can perform a variety of tasks including the disruption of specific enhancer or transcription factor motif sequences, microRNA (miRNA) target sites, and precise modification of coding sequences to alter specific amino acid residues of proteins, such as those associated with pathogen resistance. BE works by enzymatically converting DNA nucleotide bases to generate point mutations without generating a double-strand break or requiring a DNA repair template (Koblan et al., 2021). PE is the most recent addition to the gene-editing toolkit and is essentially a Streptococcus pyogenes Cas9 nickase (SpCas9n; a nickase is an enzyme capable of introducing a single-strand cut in the DNA molecule) protein fused to a reverse transcriptase (RT) module (Anzalone et al., 2019). An editing template from a modified guide RNA (gRNA) called “pegRNA” (prime-editing guide RNA) is reverse transcribed by the SpCas9n–RT complex and incorporated into the nicked target locus. Unlike the BE machinery, PE reagents can introduce most types of edits, including short insertions and deletions (Anzalone et al 2019). The reports demonstrating the use of this platform in plants suggest that its efficacy is hitherto limited (Lin et al., 2020), and further optimization is required before it can become a routine tool. Recent encouraging results reported improved efficiency of PEs in maize (Zea mays) from zero to 71.7% by increasing pegRNA expression with a composite 35S:CmYLCV:U6 promoter (Jiang et al., 2020).
The main bottlenecks preventing the widespread implementation of a platform for de novo domestication of wild species is not, however, a lack of gene-editing tools but methods for efficient delivery of the gene-editing reagents into the plant cells and the need for in vitro plant regeneration (Atkins and Voytas, 2020). Highly efficient DNA-free systems to deliver gene-editing reagents using RNA viruses have recently been described and hold promise to avoid the need for transgenes entirely (Ellison et al., 2020; Ma et al., 2020). However, these methods still require plant regeneration from explants using protocols that are time-consuming and labor-intensive and are feasible only for a handful of species (Anjanappa and Gruissem, 2021).
Research that focuses on improved transformation methods, especially for wild and recalcitrant exotic cultivars of certain crops, is paramount for effective gene editing. Two alternative approaches are emerging to overcome this hurdle. The first is the optimization of existing plant transformation and regeneration protocols. For example, using the embryonic axis as explant tissue has seen improved cultivar-independent transformation of legumes including soybean (Glycine max), cowpea (Vigna unguiculata), and common bean (Phaseolus vulgaris) (Paes de Melo et al., 2020; Song et al., 2020a; Che et al., 2021). In addition, regenerated shoots can be produced rapidly in 3–6 weeks and screened for the targeted edits. These protocols are likely to become increasingly more efficient with the introduction of developmental regulator technology that has recently transformed monocot transformation (Lowe et al., 2018). The second aims at avoiding in vitro regeneration steps altogether by delivering gene-editing reagents to somatic cells of whole plants: Fast-Treated Agrobacterium Co-Culture (Fast-TrACC) followed by de novo meristem induction. Fast-TrACC is a protocol whereby an Agrobacterium culture harboring a plasmid with plant developmental regulators (e.g. WUSCHEL, SHOOTMERISTMLESS) that induce meristem formation and a reporter gene (e.g. luciferase, GUS) that may be included to monitor and calibrate the efficiency of reagent delivery (Nasti et al., 2021). The method was demonstrated in Nicotiana benthamiana, a Solanaceae species, and required only 5–15 plants to create gene-edited shoots (Maher et al., 2020). When optimized and extended to other species, this platform will allow gene-edited somatic cells to form meristems that yield seed-producing shoots, increasing throughput and shrinking timescales for creating edited plants. As these approaches are refined and other new ones developed, they will allow crop breeding, metabolic engineering and creation of useful traits in crop wild relatives and orphan crops in a highly predictable and controlled manner (Nasti and Voytas, 2021).
A neat example of the potential of gene editing for crop breeding is provided by the history of canola, the improved varieties of rapeseed (Brassica napus) and turnip rape (B. rapa) that revolutionized Canadian agriculture in the 1960s. Prior to the 1970s, the high level of toxic erucic acid and glucosinolates hampered the use of rapeseed oils and meals for human and animal consumption, respectively (Khachatourians et al., 2001). Low erucic acid varieties of rapeseed and turnip rape were identified in Germany and Poland in 1960 and were used as parental lines to introduce the trait into commercial varieties that were only released in 1968 (B. napus “Oro”) and 1971 (B. rapa “Span”). Today, a thorough understanding of erucic acid biosynthesis in canola and its genetic basis (Chiron et al., 2015) allows creation of varieties with reduced erucic acid content via targeted gene editing (Okuzaki et al., 2018). Furthermore, altered fatty acid profiles in the seeds can be generated by precise mutation at a specific region of a gene in a much shorter timeframe via targeted gene manipulation (Huang et al., 2020). This case in point shows how a deeper understanding of metabolic pathways, in particular, and the genetic basis of agronomic traits in general can be harnessed by state-of-the-art DNA manipulation technologies to yield impressive breeding results. In contrast to the relatively simple goal of inactivating gene function using targeted mutagenesis (e.g. Zsögön et al., 2018), a more ambitious research challenge that will soon become feasible is to modify gene expression patterns in a predictable and controlled way (Pan et al., 2021a).
The next frontier: manipulation of gene expression as a tool to create predictable phenotypes
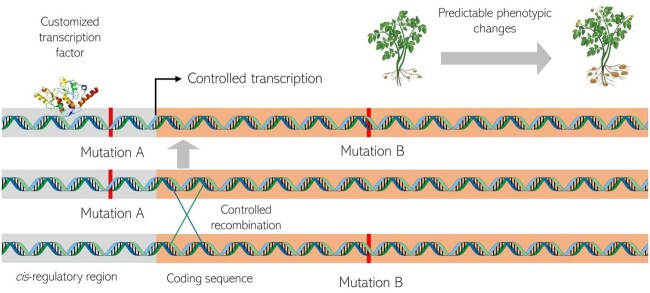
The coordination of gene expression is the foundation of cell identity and tissue function, and thus, ultimately determines the existence of differential traits between species. Engineering of cis-regulatory regions that control gene expression is increasingly gaining traction as an alternative to create desirable agronomic traits that closely resemble those produced by crop domestication and improvement (Rodríguez-Leal et al., 2017). Regulatory variants are the basis of many important domestication traits in crops such as maize (Chen et al., 2021a) and tomato (Alonge et al., 2020). However, two major hurdles exist for this approach. The first is that, in many cases, the causative changes reside in distant regions of functional genes, which poses a challenge to effective genome manipulation. For instance, reduced branching in the maize plant is controlled by a transposon insertion 60-kb upstream of teosinte branched 1 (tb1) that enhances its expression in leaf axils and prevents outgrowth of axillary buds (Studer et al., 2011). The product of tb1 is basic helix–loop–helix-containing transcription factor of the TEOSINTE BRANCHED1, CYCLOIDEA, PCF family. The second is the lack of sufficient knowledge about the relation between promoter sequence and function to allow deliberate manipulation with predictable phenotypic outcomes. Through variation in its expression pattern, the fw2.2 gene controls fruit size in tomato. However, even though the gene was identified more than two decades ago, the causal variant upstream of the coding sequence remains unknown, precluding its manipulation for breeding (Beauchet et al., 2021). Targeted dissection of promoter regions is starting to reveal the complex relationships between cis-regulatory mutations, gene expression patterns and phenotypic outcomes (Hendelman et al., 2021; Liu et al., 2021a; Wang et al., 2021). Application of machine learning on these large datasets (Vo ngoc et al., 2020), coupled with genome-wide analysis through technologies such as ATAC-seq, which allow assaying chromatin accessibility (Buenrostro et al., 2015), make it likely that cis-element editing will soon be better designed to yield more predictable phenotypes (Figure 2).
Figure 2.
Schematic representation showing how manipulating mutation and recombination can create new traits predictably and reliably.
Many relevant crop traits such as yield and drought tolerance are under complex, polygenic control. Thus, to achieve a desirable phenotype during crop improvement it is imperative to regulate the expression of multiple genes. Pinpoint control over endogenous gene expression would be the ultimate tool to design and deploy agronomic traits in plant species. Repurposing of the CRISPR toolkit now allows manipulation of gene expression using synthetic transcriptional activators or repressors. CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) are powerful technologies that allow for the simultaneous fine-tuning expression of multiple genes in plants (Lowder et al., 2015; Zhang et al., 2019; Pan et al., 2021a). For instance, with an improved CRISPRa system, termed CRISPR-Act3.0, multiple genes in the same metabolic pathway or in different biological processes could be upregulated at the same time (Pan et al., 2021b). Another promising application of CRISPRa and CRISPRi is their use in large-scale screening of a pool of genes or the whole genome for identifying a causal relationship between gene expression and a desirable phenotype. Once the gene target is identified, CRISPR-based genome editing tools can then be applied to edit cis-elements of the gene to achieve the expected expression level and create a predictable phenotype. However, exactly how cis-regulatory sequences, epigenetic modifications, and chromatin structure mediate transcription is insufficiently understood (Cramer, 2019). This area of research will benefit from advances in synthetic biology and studies using animal and human cell models (Pandelakis et al., 2020).
Conclusion
The ability to control mutation and recombination, the two fundamental biological processes underlying crop domestication and breeding, may soon make it possible to engineer novel plant phenotypes with agronomic potential. Technical advances in gene editing promise novel tools that will allow manipulating not only gene coding sequences and their combination into a single plant but also cis-regulatory elements, epigenetic regulation, and quantitative genetic variation. Simultaneously editing multigene families and controlling epistatic interactions between many genes should then be feasible (Soyk et al., 2020). Systems to customize gene expression under different environments will allow specific genetic control of developmental and physiological variation. However, it is important to bear in mind that all of these advances will ultimately rely on fundamental knowledge about the relationship between genotype and phenotype in crops and their wild relatives (see “Outstanding Questions”). Thus, the expanding knowledge about the genetic basis of complex agronomic traits in crops will represent a key asset to allow improvement of orphan crops and domestication of crop wild relatives. A synergistic combination of conventional breeding and gene-editing technology (Figure 1) will contribute to food security in the face of uncertain future conditions.
Advances.
Our understanding of the molecular genetic events contributing toward crop domestication has been greatly enhanced since the advent of next-generation sequencing.
A handful of studies have demonstrated that, in using our knowledge of domestication genes, it is possible to design multi-gene strategies to enhance the agronomic potential of less cultivated species via gene editing or introgression approaches.
More recently, the utility of manipulating gene regulator sequences to modify the expression of suites of genes in tandem has been postulated, and proof-of-concept studies in this research area have been realized.
Outstanding questions.
Is our molecular genetic toolbox already sufficiently diverse to allow the facile development of new crops? More specifically, do we have the tools we need for regeneration/introgression of the genetic diversity which we will require?
Is our understanding of wild or semi-domesticated species sufficient to allow us to predict which changes would be most likely to produce an agronomic impact?
Can increasing knowledge about the molecular basis of transcriptional control at the cis-regulatory, epigenetic, and chromatin structure levels be harnessed to produce predictable phenotypes through the manipulation of gene expression?
Acknowledgments
Mention of any trade names or commercial products in this article is solely to provide specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The United Stated Department of Agriculture (USDA) is an equal opportunity provider and employer, and all agency services are available without discrimination.
Funding
A.Z. is partly funded by a grant from the Foundation for Research Assistance of the Minas Gerais State (FAPEMIG, Brazil) (RED-00053-16) and a CAPES/Alexander von Humboldt Foundation Experienced Researcher Fellowship (88881.472837/2019-01). Y.Q. is supported by National Science Foundation Plant Genome Research Program grants (award no. IOS-1758745 and IOS-2029889), the US Department of Agriculture Biotechnology Risk Assessment Grant Program competitive grants (award no. 2018-33522-28789 and 2020-33522-32274), Emergency Citrus Disease Research and Extension Program (award no. 2020-70029-33161), Agriculture and Food Research Initiative Agricultural Innovations Through Gene Editing Program (award no. 2021-67013-34554), Foundation for Food and Agriculture Research Program (award no. 593603 and 305614-00001), Maryland Innovation Initiative Funding from TEDCO (award no. 1120-012_2), The Maryland Agricultural Experiment Station Competitive Grant Program, and Syngenta. S.J.C. is supported by the US Department of Agriculture, Agricultural Research Service.
Conflict of interest statement. None declared.
Contributor Information
Shaun Curtin, United States Department of Agriculture, Plant Science Research Unit, St. Paul, Minnesota 55108, USA; Center for Plant Precision Genomics, University of Minnesota, St. Paul, Minnesota 55108, USA; Center for Genome Engineering, University of Minnesota, St. Paul, Minnesota 55108, USA; Department of Agronomy and Plant Genetics, University of Minnesota, St. Paul, Minnesota 55108, USA.
Yiping Qi, Department of Plant Science and Landscape Architecture, University of Maryland, College Park, Maryland, USA; Institute for Bioscience and Biotechnology Research, University of Maryland, Rockville, Maryland, USA.
Lázaro E P Peres, Laboratory of Hormonal Control of Plant Development. Departamento de Ciências Biológicas, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, CP 09, 13418-900, Piracicaba, São Paulo, Brazil.
Alisdair R Fernie, Max-Planck-Institute of Molecular Plant Physiology, Am Mühlenberg 1, 14476 Potsdam-Golm, Germany.
Agustin Zsögön, Max-Planck-Institute of Molecular Plant Physiology, Am Mühlenberg 1, 14476 Potsdam-Golm, Germany; Departamento de Biologia Vegetal, Universidade Federal de Viçosa, Viçosa, Minas Gerais, CEP 36570-900, Brazil.
S.C., Y.Q., L.E.P.P., A.R.F., and A.Z. conceived the concept and wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Agustin Zsögön (agustin.zsogon@ufv.br)
References
- Adeyemo OS, Hyde PT, Setter TL (2019) Identification of FT family genes that respond to photoperiod, temperature and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reprod 32: 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. (2020) Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182: 145–161.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S, Kostova D, Bulut M, Fernie AR (2021) Genome-wide association studies: assessing trait characteristics in model and crop plants. Cell Mol Life Sci 78: 5743–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjanappa RB, Gruissem W (2021) Current progress and challenges in crop genetic transformation. J Plant Physiol 261: 153411. [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38: 824–844 [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576: 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atefi A, Ge Y, Pitla S, Schnable J (2021) Robotic technologies for high-throughput plant phenotyping: contemporary reviews and future perspectives. Front Plant Sci 12: 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins PA, Voytas DF (2020) Overcoming bottlenecks in plant gene editing. Curr Opin Plant Biol 54: 79–84 [DOI] [PubMed] [Google Scholar]
- Beauchet A, Gévaudant F, Gonzalez N, Chevalier C (2021) In search of the still unknown function of FW2.2/cell number regulator, a major regulator of fruit size in tomato. J Exp Bot 72: 5300–5311 [DOI] [PubMed] [Google Scholar]
- Bevan MW, Uauy C, Wulff BBH, Zhou J, Krasileva K, Clark MD (2017) Genomic innovation for crop improvement. Nature 543: 346–354 [DOI] [PubMed] [Google Scholar]
- Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH (2010) The role of recently derived FT paralogs in sunflower domestication. Curr Biol 20: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ (2015) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T (2021) Sequence modification on demand: search and replace tools for precise gene editing in plants. Transgenic Res 30: 353–379 [DOI] [PubMed] [Google Scholar]
- Che P, Chang S, Simon MK, Zhang Z, Shaharyar A, Ourada J, O’Neill D, Torres-Mendoza M, Guo Y, Marasigan KM, et al. (2021) Developing a rapid and highly efficient cowpea regeneration, transformation and genome editing system using embryonic axis explants. Plant J 106: 817–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Li W, Tan L, Tian F (2021a) Harnessing knowledge from maize and rice domestication for new crop breeding. Mol Plant 14: 9–26 [DOI] [PubMed] [Google Scholar]
- Chen Z, Sun J, Li D, Li P, He K, Ali F, Pan Q, Mi G, Chen F, Yuan L (2021b) Plasticity and domestication of root anatomy in maize-teosinte derived population. J Exp Bot doi: 10.1093/jxb/erab406 (September 6, 2021) [DOI] [PubMed] [Google Scholar]
- Chiron H, Wilmer J, Lucas M-O, Nesi N, Delseny M, Devic M, Roscoe TJ (2015) Regulation of fatty acid elongation 1 expression in embryonic and vascular tissues of Brassica napus. Plant Mol Biol 88: 65–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Henderson IR (2015) Meiotic recombination hotspots—a comparative view. Plant J 83: 52–61 [DOI] [PubMed] [Google Scholar]
- Chopra R, Johnson EB, Emenecker R, Cahoon EB, Lyons J, Kliebenstein DJ, Daniels E, Dorn KM, Esfahanian M, Folstad N, et al. (2020) Identification and stacking of crucial traits required for the domestication of pennycress. Nat Food 1: 84–91 [Google Scholar]
- Cramer P (2019) Organization and regulation of gene transcription. Nature 573: 45–54 [DOI] [PubMed] [Google Scholar]
- Denham T, Barton H, Castillo C, Crowther A, Dotte-Sarout E, Florin SA, Pritchard J, Barron A, Zhang Y, Fuller DQ (2020) The domestication syndrome in vegetatively propagated field crops. Ann Bot 125: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, et al. (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549 [DOI] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang B-H, Liu B-L, Wang Y-Z . (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5: 3352. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison EE, Nagalakshmi U, Gamo ME, Huang P, Dinesh-Kumar S, Voytas DF (2020) Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants 6: 620–624 [DOI] [PubMed] [Google Scholar]
- Evans LT (1998) Feeding the ten billion: plants and population growth. Cambridge University Press, Cambridge, UK [Google Scholar]
- Fernie AR, Gutierrez-Marcos J (2019) From genome to phenome: genome-wide association studies and other approaches that bridge the genotype to phenotype gap. Plant J 97: 5–7 [DOI] [PubMed] [Google Scholar]
- Ferguson AR (2013) Kiwifruit: the wild and the cultivated plants. Adv Food Nutr Res 68: 15–32 [DOI] [PubMed] [Google Scholar]
- Fischer A, Byerlee D, Edmeades G (2014) Crop yields and global food security: will yield increase continue to feed the world? Australian Centre for International Agricultural Research, Canberra, Australia [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C (2003) Determinate and late flowering are two terminal flower1/centroradialis homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15: 2742–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, Komatsu K, Katayama T, Sayama T, Ishimoto M, et al. (2014) Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci U S A 111: 17797–17802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C (2021) Genome engineering for crop improvement and future agriculture. Cell 184: 1621–1635 [DOI] [PubMed] [Google Scholar]
- Gasparini K, Moreira J dos R, Peres LEP, Zsögön A (2021) De novo domestication of wild species to create crops with increased resilience and nutritional value. Curr Opin Plant Biol 60: 102006. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watson A, Gonzalez-Navarro OE, Ramirez-Gonzalez RH, Yanes L, Mendoza-Suárez M, Simmonds J, Wells R, Rayner T, Green P, et al. (2018) Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat Protocols 13: 2944–2963 [DOI] [PubMed] [Google Scholar]
- González AM, Vander Schoor JK, Fang C, Kong F, Wu J, Weller JL, Santalla M (2021) Ancient relaxation of an obligate short-day requirement in common bean through loss of constans-like gene function. Curr Biol doi: 10.1016/j.cub.2021.01.075 [DOI] [PubMed] [Google Scholar]
- Gulisano A, Alves S, Martins JN, Trindade LM (2019) Genetics and breeding of Lupinus mutabilis: an emerging protein crop. Front Plant Sci 10: 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar R, Hodgkin T (2007) The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156: 1–13 [Google Scholar]
- Hendelman A, Zebell S, Rodriguez-Leal D, Dukler N, Robitaille G, Wu X, Kostyun J, Tal L, Wang P, Bartlett ME(2021) Conserved pleiotropy of an ancient plant homeobox gene uncovered by cis-regulatory dissection. Cell 184: 1724–1739.e16 [DOI] [PubMed] [Google Scholar]
- Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, Xu G, Tian J, Wang C, Li D, et al. (2018) ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci U S A 115: E334–E341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Cui T, Zhang L, Yang Q, Yang Y, Xie K, Fan C, Zhou Y (2020) Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor Appl Genet 133: 2401–2411 [DOI] [PubMed] [Google Scholar]
- Jankowicz-Cieslak J, Mba C, Till BJ (2017) Mutagenesis for crop breeding and functional genomics. In Jankowicz-Cieslak J, Tai TH, Kumlehn J, Till BJ, eds, Biotechnologies for Plant Mutation Breeding: Protocols. Springer International Publishing, Cham, pp 3–18 [Google Scholar]
- Jiang Y-Y, Chai Y-P, Lu M-H, Han X-L, Lin Q, Zhang Y, Zhang Q, Zhou Y, Wang X-C, Gao C, et al. (2020) Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol 21: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantar MB, Nashoba AR, Anderson JE, Blackman BK, Rieseberg LH (2017) The genetics and genomics of plant domestication. BioScience 67: 971–982 [Google Scholar]
- Khachatourians GG, Sumner AK, Phillips PWB (2001) The biotechnology revolution in global agriculture: innovation, invention and investment in the canola industry. In PWB Phillips, GG Khachatourians, eds, An introduction to the history of canola and the scientific basis for innovation, CABI, Wallingford, UK, pp 33–47
- Kloosterman B, Abelenda JA, Gomez M del MC, Oortwijn M, Boer JM de, Kowitwanich K, Horvath BM, Eck HJ van, Smaczniak C, Prat S, et al. (2013) Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495: 246–250 [DOI] [PubMed] [Google Scholar]
- Koblan LW, Arbab M, Shen MW, Hussmann JA, Anzalone AV, Doman JL, Newby GA, Yang D, Mok B, Replogle JM, et al. (2021) Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat Biotechnol 39: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofsky J, Zhang H, Song B-H (2018) The untapped genetic reservoir: the past, current, and future applications of the wild soybean (Glycine soja). Front Plant Sci doi: 10.3389/fpls.2018.00949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M, Toro O, Debouck DG, Gepts P (2012) Multiple origins of the determinate growth habit in domesticated common bean (Phaseolus vulgaris). Ann Bot 110: 1573–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Yan L, Lu Y, Schnable JC (2018) Largely unlinked gene sets targeted by selection for domestication syndrome phenotypes in maize and sorghum. Plant J 93: 843–855 [DOI] [PubMed] [Google Scholar]
- Laitinen RAE, Nikoloski Z (2019) Genetic basis of plasticity in plants. J Exp Bot 70: 739–745 [DOI] [PubMed] [Google Scholar]
- Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, Van Eck J, Lippman ZB (2018) Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plants 4: 766–770 [DOI] [PubMed] [Google Scholar]
- Lenormand T, Otto SP (2000) The evolution of recombination in a heterogeneous environment. Genetics 156: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311: 1936–1939 [DOI] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C (2018) Domestication of wild tomato is accelerated by genome editing. Nature Biotechnol 36: 1160–1163 [DOI] [PubMed] [Google Scholar]
- Li Q, Yan J (2020) Sustainable agriculture in the era of omics: knowledge-driven crop breeding. Genome Biol 21: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, et al. (2020) Prime genome editing in rice and wheat. Nat Biotechnol 38: 582–585 [DOI] [PubMed] [Google Scholar]
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46: 1220–1226 [DOI] [PubMed] [Google Scholar]
- Lin Z, Li X, Shannon LM, Yeh C-T, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, et al. (2012) Parallel domestication of the Shattering1 genes in cereals. Nat Genet 44: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-J, Yan J (2019) Crop genome-wide association study: a harvest of biological relevance. Plant J 97: 8–18 [DOI] [PubMed] [Google Scholar]
- Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu Q, Bartlett M, Jackson D (2021a) Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat Plants 7: 287–294 [DOI] [PubMed] [Google Scholar]
- Liu N, Du Y, Warburton ML, Xiao Y, Yan J (2021b) Phenotypic plasticity contributes to maize adaptation and heterosis. Mol Biol Evol 38: 1262–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Morgan C, H. Franklin FC, Bomblies K (2018) Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, Voytas DF, Hsieh T-F, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W (2018) Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev Biol-Plant 54: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Sun J, Jiang A, Bai M, Fan C, Liu J, Ning G, Wang C (2020a) Alternate expression of constans-like 4 in short days and constans in long days facilitates day-neutral response in Rosa chinensis. J Exp Bot 71: 4057–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Wu W, Wang M, Meyer RS, Ndjiondjop M-N, Tan L, Zhou H, Zhang J, Fu Y, Cai H, et al. (2018) Genetic control of seed shattering during African rice domestication. Nat Plants 4: 331–337 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat Plants 6: 773–779 [DOI] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF (2020) Plant gene editing through de novo induction of meristems. Nat Biotechnol 38: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi S, Healey A, Huang P, Grimwood J, Jenkins J, Barry K, Sreedasyam A, Shu S, Lovell JT, Feldman M, et al. (2020) A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat Biotechnol 38: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesanz S, Milla R (2018) Differential plasticity to water and nutrients between crops and their wild progenitors. Environ Exp Bot 145: 54–63 [Google Scholar]
- Meyerowitz EM (2002) Plants compared to animals: the broadest comparative study of development. Science 295: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Milla R, Osborne CP (2021) Crop origins explain variation in global agricultural relevance. Nat Plants 7: 598–607 [DOI] [PubMed] [Google Scholar]
- Molla KA, Sretenovic S, Bansal KC, Qi Y (2021) Precise plant genome editing using base editors and prime editors. Nat Plants 7: 1166–1187 [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci U S A 108: 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu J, Ando T, et al. (2016) Mitogen-activated protein kinase kinase 3 regulates seed dormancy in barley. Curr Biol 26: 775–781 [DOI] [PubMed] [Google Scholar]
- Nasti RA,, Voytas DF (2021) Attaining the promise of plant gene editing at scale. Proc Natl Acad Sci U S A 118: e2004846117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasti RA, Zinselmeier MH, Vollbrecht M, Maher MF, Voytas DF (2021) Fast-TrACC: a rapid method for delivering and testing gene editing reagents in somatic plant cells. Front Genome Edit 2: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzaki A, Ogawa T, Koizuka C, Kaneko K, Inaba M, Imamura J, Koizuka N (2018) CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol Biochem 131: 63–69 [DOI] [PubMed] [Google Scholar]
- Paes de Melo B, Lourenço-Tessutti IT, Morgante CV, Santos NC, Pinheiro LB, de Jesus Lins CB, Silva MCM, Macedo LLP, Fontes EPB, Grossi-de-Sa MF (2020) Soybean embryonic axis transformation: combining biolistic and agrobacterium-mediated protocols to overcome typical complications of in vitro plant regeneration. Front Plant Sci 11: 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Sretenovic S, Qi Y (2021a) CRISPR/dCas-mediated transcriptional and epigenetic regulation in plants. Curr Opin Plant Biol 60: 101980. [DOI] [PubMed] [Google Scholar]
- Pan C, Wu X, Markel K, Malzahn AA, Kundagrami N, Sretenovic S, Zhang Y, Cheng Y, Shih PM, Qi Y (2021b) CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat Plants 7: 942–953 [DOI] [PubMed] [Google Scholar]
- Pandelakis M, Delgado E, Ebrahimkhani MR (2020) CRISPR-based synthetic transcription factors in vivo: the future of therapeutic cellular programming. Cell Sys 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker TA, Lo S, Gepts P (2021) Pod shattering in grain legumes: emerging genetic and environment-related patterns. Plant Cell 33: 179–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH (2002) What has QTL mapping taught us about plant domestication? New Phytol 154: 591–608 [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Périlleux C, Bouché F, Randoux M, Orman-Ligeza B (2019) Turning meristems into fortresses. Trends in Plant Sci 24: 431–442 [DOI] [PubMed] [Google Scholar]
- Pin PA, Zhang W, Vogt SH, Dally N, Büttner B, Schulze-Buxloh G, Jelly NS, Chia TYP, Mutasa-Göttgens ES, Dohm JC, et al. (2012) The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Cur Biol 22: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Ping J, Liu Y, Sun L, Zhao M, Li Y, She M, Sui Y, Lin F, Liu X, Tang Z, et al. (2014) Dt2 Is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26: 2831–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno DR, Holst I, Moreno JE, Winter K (2019) Experimenting with domestication: understanding macro- and micro-phenotypes and developmental plasticity in teosinte in its ancestral pleistocene and early holocene environments. J Archaeol Sci 108: 104970 [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The self-pruning gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Pourkheirandish M, Hensel G, Kilian B, Senthil N, Chen G, Sameri M, Azhaguvel P, Sakuma S, Dhanagond S, Sharma R, et al. (2015) Evolution of the grain dispersal system in barley. Cell 162: 527–539 [DOI] [PubMed] [Google Scholar]
- Prewitt SF, Ayre BG, McGarry RC (2018) Cotton centroradialis/terminal flower 1/self-pruning genes functionally diverged to differentially impact plant architecture. J Exp Bot 69: 5403–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD (2019) Evolutionary insights into the nature of plant domestication. Curr Biol 29: R705–R714 [DOI] [PubMed] [Google Scholar]
- Purugganan MD, Jackson SA (2021) Advancing crop genomics from lab to field. Nat Genet 53: 595–601 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480 [DOI] [PubMed] [Google Scholar]
- Roldan MVG, Périlleux C, Morin H, Huerga-Fernandez S, Latrasse D, Benhamed M, Bendahmane A (2017) Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci Rep 7: 4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J (2004) The evolution of recombination under domestication: a test of two hypotheses. Am Nat 163: 105–112 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Sato K, Yamane M, Yamaji N, Kanamori H, Tagiri A, Schwerdt JG, Fincher GB, Matsumoto T, Takeda K, Komatsuda T (2016) Alanine aminotransferase controls seed dormancy in barley. Nat Comm 7: 11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Z, Liu H, Zhu J, Chen J, Wang Q, Fang L, Gao F, Tian Y, Chen Y, Chang L, et al. (2018) Mutation of SELF-PRUNING homologs in cotton promotes short-branching plant architecture. J Exp Bot 69: 2543–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva WB, Vicente MH, Robledo JM, Reartes DS, Ferrari RC, Bianchetti RE, Araújo WL, Freschi L, Peres LEP, Zsögön A (2018) Self-pruning acts synergistically with diageotropica to guide auxin responses and proper growth form. Plant Physiol 176: 2904–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai Y-S, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani A, Walter KA, Wiersma AT, Santiago JP, Quiqley M, Chitwood D, Porch TG, Miklas P, McClean PE, Osorno JM, et al. (2021) The genetics and physiology of seed dormancy, a crucial trait in common bean domestication. BMC Plant Biol 21: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Han X, Wiersma AT, Zong X, Awale HE, Kelly JD (2020a) Induction of competent cells for Agrobacterium tumefaciens-mediated stable transformation of common bean (Phaseolus vulgaris L.). PLOS One 15: e0229909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang S, Wang X, Sun S, Liu Z, Wang K, Wan H, Zhou G, Li R, Yu H, et al. (2020b) Variations in both FTL1 and SP5G, two tomato FT paralogs, control day-neutral flowering. Mol Plant 13: 939–942 [DOI] [PubMed] [Google Scholar]
- Soyk S, Benoit M, Lippman ZB (2020) New Horizons for Dissecting Epistasis in Crop Quantitative Trait Variation. Ann Rev Genetics 54: 287–307 [DOI] [PubMed] [Google Scholar]
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB (2017) Variation in the flowering gene self pruning 5G promotes day-neutrality and early yield in tomato. Nat Genet 49: 162–168 [DOI] [PubMed] [Google Scholar]
- Spengler RN, Mueller NG (2019) Grazing animals drove domestication of grain crops. Nat Plants 5: 656–662 [DOI] [PubMed] [Google Scholar]
- Spengler RN, Petraglia M, Roberts P, Ashastina K, Kistler L, Mueller NG, Boivin N (2021) Exaptation traits for megafaunal mutualisms as a factor in plant domestication. Front Plant Sci 12: 649394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz GC, Salgado-Luarte C, Escobedo VM, Valladares F, Gianoli E (2021) Global trends in phenotypic plasticity of plants. Ecol Lett 24: 2267–2281 [DOI] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genetics 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf MPH, McVean GAT (2003) Estimating recombination rates from population-genetic data. Nat Rev Genet 4: 959–968 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, et al. (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci U S A 107: 5792–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taagen E, Bogdanove AJ, Sorrells ME (2020) Counting on crossovers: controlled recombination for plant breeding. Trends Plant Sci 25: 455–465 [DOI] [PubMed] [Google Scholar]
- Tadele Z (2019) Orphan crops: their importance and the urgency of improvement. Planta 250: 677–694 [DOI] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci U S A 107: 8563–8568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togninalli M, Seren Ü, Freudenthal JA, Monroe JG, Meng D, Nordborg M, Weigel D, Borgwardt K, Korte A, Grimm DG (2020) AraPheno and the AraGWAS Catalog 2020: a major database update including RNA-Seq and knockout mutation data for Arabidopsis thaliana. Nucl Acids Res 48: D1063–D1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torada A, Koike M, Ogawa T, Takenouchi Y, Tadamura K, Wu J, Matsumoto T, Kawaura K, Ogihara Y (2016) A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr Biol 26: 782–787 [DOI] [PubMed] [Google Scholar]
- Tsuda M, Kaga A, Anai T, Shimizu T, Sayama T, Takagi K, Machita K, Watanabe S, Nishimura M, Yamada N, et al. (2015) Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genomics 16: 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Vilela AE, González-Paleo L (2015) Changes in resource-use strategy and phenotypic plasticity associated with selection for yield in wild species native to arid environments. J Arid Environ 113: 51–58 [Google Scholar]
- Vo ngoc L, Huang CY, Cassidy CJ, Medrano C, Kadonaga JT (2020) Identification of the human DPR core promoter element using machine learning. Nature 585: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sun S, Wu T, Liu L, Sun X, Cai Y, Li J, Jia H, Yuan S, Chen L, et al. (2020) Natural variation and CRISPR/Cas9‐mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol J 18: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li W, Fang C, Xu F, Liu Y, Wang Z, Yang R, Zhang M, Liu S, Lu S, et al. (2018) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 50: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Wang X, Aguirre L, Rodríguez-Leal D, Hendelman A, Benoit M, Lippman ZB (2021) Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat Plants 7: 419–427 [DOI] [PubMed] [Google Scholar]
- Weller JL, Vander Schoor JK, Perez-Wright EC, Hecht V, González AM, Capel C, Yuste-Lisbona FJ, Lozano R, Santalla M (2019) Parallel origins of photoperiod adaptation following dual domestications of common bean. J Exp Bot 70: 1209–1219 [DOI] [PubMed] [Google Scholar]
- Wilkins AS (2020) A striking example of developmental bias in an evolutionary process: the “domestication syndrome.” Evol Dev 22: 143–153 [DOI] [PubMed] [Google Scholar]
- Xie Q, Li N, Yang Y, Lv Y, Yao H, Wei R, Sparkes DL, Ma Z (2018) Pleiotropic effects of the wheat domestication gene Q on yield and grain morphology. Planta 247: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, Li K, Yang N, Li Y, Zhong T, et al. (2013) CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci U S A 110: 16969–16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lin T, Meng X, Du H, Zhang J, Liu G, Chen M, Jing Y, Kou L, Li X, et al. (2021) A route to de novo domestication of wild allotetraploid rice. Cell 184: 1156–1170.e14 [DOI] [PubMed] [Google Scholar]
- Yu X, Liu H, Sang N, Li Y, Zhang T, Sun J, Huang X (2019) Identification of cotton mother of FT and TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. PLOS One 14: e0215771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeven AC (1972) The partial and complete domestication of the oil palm (Elaeis guineensis). Econom Bot 26: 274–279 [Google Scholar]
- Zhang Y, Malzahn AA, Sretenovic S, Qi Y (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants 5: 778–794 [DOI] [PubMed] [Google Scholar]
- Zonneveld M van, Larranaga N, Blonder B, Coradin L, Hormaza JI, Hunter D (2018) Human diets drive range expansion of megafauna-dispersed fruit species. Proc Natl Acad Sci U S A 115: 3326–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Zsögön A, Peres LEP (2018) Molecular control of plant shoot architecture. Plant Cell 30: tpc.118.tt1218–tpc.118.tt1218 [Google Scholar]
- Zsögön A, Cermak T, Voytas D, Peres LEP (2017) Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: case study in tomato. Plant Sci 256: 120–130 [DOI] [PubMed] [Google Scholar]