Abstract
Lignin is a complex phenylpropanoid polymer deposited in the secondary cell walls of vascular plants. Unlike most gymnosperm and eudicot lignins that are generated via the polymerization of monolignols, grass lignins additionally incorporate the flavonoid tricin as a natural lignin monomer. The biosynthesis and functions of tricin-integrated lignin (tricin-lignin) in grass cell walls and its effects on the utility of grass biomass remain largely unknown. We herein report a comparative analysis of rice (Oryza sativa) mutants deficient in the early flavonoid biosynthetic genes encoding CHALCONE SYNTHASE (CHS), CHALCONE ISOMERASE (CHI), and CHI-LIKE (CHIL), with an emphasis on the analyses of disrupted tricin-lignin formation and the concurrent changes in lignin profiles and cell wall digestibility. All examined CHS-, CHI-, and CHIL-deficient rice mutants were largely depleted of extractable flavones, including tricin, and nearly devoid of tricin-lignin in the cell walls, supporting the crucial roles of CHS and CHI as committed enzymes and CHIL as a noncatalytic enhancer in the conserved biosynthetic pathway leading to flavone and tricin-lignin formation. In-depth cell wall structural analyses further indicated that lignin content and composition, including the monolignol-derived units, were differentially altered in the mutants. However, regardless of the extent of the lignin alterations, cell wall saccharification efficiencies of all tested rice mutants were similar to that of the wild-type controls. Together with earlier studies on other tricin-depleted grass mutant and transgenic plants, our results reflect the complexity in the metabolic consequences of tricin pathway perturbations and the relationships between lignin profiles and cell wall properties.
Mutating early flavonoid biosynthetic genes differentially affects the structure and content of tricin-incorporated lignin in rice cell walls.
P.Y.L., L.W., A.C.W.L., H.L., Y.T.K., M.X.C., F.Y.Z., and Y.T. performed experiments. P.Y.L., L.W., T.U., J.Z., Y.T., and C.L. conceived research and analyzed data. P.Y.L., Y.T., and C.L. wrote the manuscript with help from all the others.
Introduction
Lignin is a complex phenolic polymer produced in the secondary cell walls of vascular plants. By filling up the spaces between cell wall polysaccharides, that is, cellulose and hemicelluloses, lignin contributes to the essential mechanical and biochemical properties required for the development of land plant vasculature (Boerjan et al., 2003; Bonawitz and Chapple, 2013; Coomey et al., 2020). From an agro-industrial application perspective, lignin has long been considered as the major factor hindering polysaccharide-based lignocellulosic utilization processes, including those used for producing pulp and paper as well as fermentable sugars for downstream biochemicals. More recently, lignin is increasingly being viewed as a viable source of biomass-derived aromatic chemicals (Ragauskas et al., 2014; Rinaldi et al., 2016; Sun et al., 2018; Abu-Omar et al., 2021). Accordingly, there has been considerable interest in the development of bioengineering strategies for controlling lignin quantity and quality for both polysaccharide- and lignin-based lignocellulosic utilization (Rinaldi et al., 2016; Umezawa, 2018; Dixon and Barros, 2019; Dixon and Pasinetti, 2010; Ralph et al., 2019; Umezawa et al., 2020).
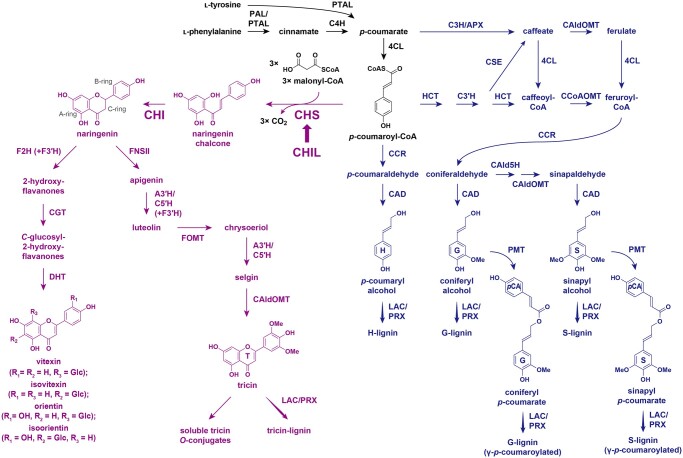
The chemical structures of lignin polymers, which are highly variable among different classes of vascular plants, are primarily defined by the formulation of lignin monomers used for lignin polymerization via oxidative radical coupling in the cell walls (Freudenberg, 1965; Sarkanen and Ludwig, 1971; Ralph et al., 2004, 2019). In general, lignins produced by most gymnosperms, eudicots, and nongrass monocot species are derived from monolignols, namely coniferyl, sinapyl, and p-coumaryl alcohols, which respectively gives rise to the guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H) units in the final lignin polymers (Figure 1). In grasses (i.e. the major monocot family Poaceae), a substantial portion of the monolignols incorporated into lignin polymers are γ-acylated by p-hydroxycinnamates, mainly p-coumarates (pCAs; Ralph et al., 1994; Ralph, 2010; Karlen et al., 2018) as well as ferulates (FAs), albeit at much lower levels (Karlen et al., 2016). Hence, along with the conserved, nonacylated monolignols, the γ-acylated monolignols represent canonical lignin monomers in grasses (Ralph, et al., 2019). The cinnamate/monolignol pathway leading to the formation of these nonacylated and γ-acylated monolignols has been extensively investigated (Figure 1; Umezawa, 2018; Dixon and Barros, 2019; Ralph et al., 2019; Coomey et al., 2020; Umezawa et al., 2020).
Figure 1.
Proposed flavonoid and monolignol biosynthetic pathways in grasses. PTAL, l-phenylalanine/l-tyrosine ammonia-lyase; PAL, l-phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-hydroxycinnamate:CoA ligase; F3ʹH, flavonoid 3ʹ-hydroxylase; FOMT, flavonoid O-methyltransferase; F2H, flavanone 2-hydroxylase; CGT, C-glycosyltransferase; DHT, dehydratase; C3H, p-coumarate 3-hydroxylase; APX, ascorbate peroxidase; HCT, p-hydroxycinnamoyl-CoA:quinate/shikimate hydroxycinnamoyl transferase; C3′H, p-coumaroyl ester 3-hydroxylase; CSE, caffeoyl shikimate esterase; CCoAOMT, caffeoyl-CoA OMT; CAld5H, coniferaldehyde 5-hydroxylase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; PMT, p-coumaroyl-CoA:monolignol transferase; LAC, laccase; PRX, peroxidase. In purple: flavonoid biosynthetic pathway. In blue: monolignol biosynthetic pathway. Thick arrow: enhancer activity.
Another defining feature of grass lignin is the inclusion of tricin, which is a flavone (a class of flavonoid), as an additional natural lignin monomer biosynthesized outside of the cinnamate/monolignol pathway (Figure 1; del Río et al., 2012, 2020; Lan et al., 2015; Lam et al., 2021). During grass cell wall lignification, tricin co-polymerizes with nonacylated and γ-acylated monolignols via oxidative radical coupling and becomes an integrated part of the lignin polymers in the cell walls (Lan et al., 2015, 2016a). Tricin-integrated lignin (tricin-lignin) is ubiquitous among grasses, but it has also been detected in several nongrass monocot species as well as in a dicot alfalfa (Medicago sativa; Lan et al., 2016b). Given the considerable importance of numerous grass crops as lignocellulose feedstocks and the prominent effects of lignin on the utility of lignocellulose, developing bioengineering approaches for modulating tricin-lignin formation in grasses has become an emerging research objective (Halpin, 2019; Coomey et al., 2020; del Río et al., 2020; Umezawa et al., 2020; Lam et al., 2021). Currently, however, our understanding of how tricin-lignin influences the utility of grass biomass remains limited.
The flavonoid biosynthetic pathway that produces various extractable flavonoids, including flavones such as tricin O-conjugates and flavone C-glycosides abundant in grass vegetative tissues (Harborne and Hall, 1964; Brazier-Hicks et al., 2009; Dong et al., 2014), has been characterized by analyses of various grass mutants (Lam et al., 2021); however, the contribution of each pathway enzyme to tricin-lignin formation has not been fully investigated. As are all other flavonoids, tricin can be generated via the conserved entry of the flavonoid biosynthetic pathway in which two committed enzymes, CHALCONE SYNTHASE (CHS) and CHALCONE ISOMERASE (CHI), channel the carbon flux from the phenylpropanoid and polyketide pathways by using p-coumaroyl-CoA and malonyl-CoA, respectively, to generate the flavanone naringenin, a common precursor for all flavonoid classes (Figure 1; Yu et al., 2005; Shih et al., 2008; Eloy et al., 2017; Wang et al., 2020). The downstream flavone-specific pathway leading to tricin involves the construction of the flavone aromatic system via C-ring desaturation catalyzed by FLAVONE SYNTHASE II (FNSII; Lam et al., 2015, 2017) and successive B-ring hydroxylations and O-methylations mediated by APIGENIN 3ʹ-HYDROXYLASE/CHRYSOERIOL 5ʹ-HYDROXYLASE (A3ʹH/C5ʹH; Lam et al., 2019a) and O-methyltransferases (OMTs), including 5-HYDROXYCONIFERALDEHYDE OMT (CAldOMT or CAFFEIC ACID OMT, COMT), a bifunctional OMT involved in both tricin and monolignol biosynthesis in grasses (Figure 1; Lam et al., 2015; Fornalé et al., 2016; Eudes et al., 2017; Lam et al., 2019b). In addition to these committed enzymes, CHI-LIKE (CHIL), which belongs to the class IV CHI family whose members generally lack CHI catalytic activity, has been described to be a component of flavonoid metabolons as a noncatalytic enhancer (Ban et al., 2018; Ni et al., 2020; Waki et al., 2020). Although the CHIL-mediated enhancement of flavonoid production has been reported in many plant species (Morita et al., 2014; Jiang et al., 2015; Clayton et al., 2018; Zhao et al., 2021), its potential role in the biosynthesis of grass flavones, including tricin-lignin, has yet to be examined.
We previously investigated the roles of the downstream tricin biosynthetic enzymes, that is, FNSII, A3ʹH/C5ʹH, and CAldOMT, on tricin-lignin formation in rice (Oryza sativa), a model grass species and a commercially important grain crop. Rice mutant and transgenic plants deficient in OsFNSII (CYP93G1; Lam et al., 2017), OsA3ʹH/C5ʹH (CYP75B4; Lam et al., 2019a), and OsCAldOMT1 (or RICE OMT9; Lam et al., 2019b) produced altered cell walls with lignins largely depleted in tricin units, demonstrating the pivotal roles of these enzymes on tricin-lignin formation. Intriguingly, these tricin-depleted rice plants consistently had decreased total lignin levels, altered S/G lignin unit ratio, and enhanced cell wall digestibility. In stark contrast, Eloy et al. (2017) reported that a maize (Zea mays) Colorless2-inhibitor diffuse (C2-Idf) mutant defective in CHS, displayed a considerable increase in the lignin content and a decrease in cell wall digestibility, along with substantially decreased tricin-lignin levels. Overall, these earlier studies suggest that disrupting the tricin pathway enzymes not only impedes the formation of tricin-lignin but also affects the core lignin polymer units derived from monolignols, although the underlying mechanisms remain unclear.
Following our previous studies focusing on the downstream tricin-specific biosynthetic enzymes (i.e. FNSII, A3ʹH/C5ʹH, and CAldOMT; Lam et al., 2017, 2019a, 2019b), this study was conducted to examine the consequences of disrupting the two committed entry enzymes in the flavonoid biosynthetic pathway, that is, CHS and CHI, as well as the noncatalytic enhancer protein, that is, CHIL, on tricin-lignin formation in rice. Genome-edited rice mutants harboring loss-of-function mutations in the rice CHI (OsCHI) and CHIL (OsCHIL1 and OsCHIL2) genes were generated using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system. These CRISPR mutants along with a T-DNA insertional mutant of rice CHS (OsCHS1; Wang et al., 2020) were analyzed regarding their extractable flavone profiles, cell wall structures with emphasis on the disruptions in tricin-lignin formation, and the concurrent shifts in the profiles of the monolignol-derived lignin units and cell wall saccharification. We demonstrated the crucial roles of CHS, CHI, and CHIL in generating tricin-lignin in rice. Intriguingly, along with the depletion in tricin units, the core lignin polymer units derived from monolignols were differentially altered in the CHS-, CHI-, and CHIL-deficient rice mutants. Together with the data from earlier studies on other tricin-depleted grass mutants (Eloy et al. 2017; Lam et al., 2017, 2019a, 2019b), the complex effects of tricin pathway perturbations on lignin profiles and cell wall properties are discussed.
Results
Rice genes involved in the entry to the flavonoid biosynthetic pathway
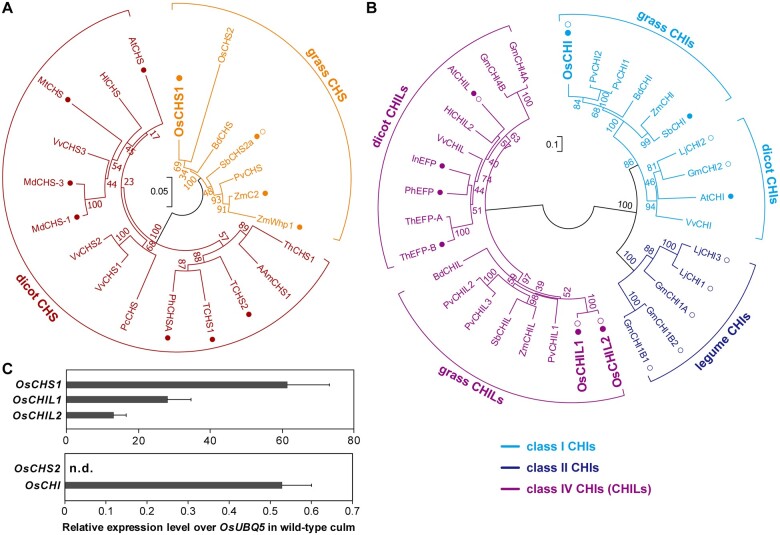
The first committed step of the flavonoid biosynthetic pathway is the conversion of p-coumaroyl-CoA to naringenin chalcone via a reaction catalyzed by CHS (Figure 1). The rice genome encodes two putative CHSs, namely OsCHS1 (LOC_Os11g32650) and OsCHS2 (LOC_Os07g11440). These two CHSs have similar sequences (82% amino acid sequence identity) and clustered with other grass CHSs rather than with the dicot CHSs in the constructed phylogenetic tree (Figure 2A). Previously, OsCHS1 was shown to be able to convert p-coumaroyl-CoA to naringenin chalcone in vitro (Park et al., 2020) and complement the lack of flavonoid accumulation in the CHS-deficient Arabidopsis (Arabidopsis thaliana) transparent testa4 mutant (Shih et al., 2008). Recently, OsCHS1 mutation was reported to prevent the accumulation of various extractable flavonoids in rice anthers (Wang et al., 2020). Here, gene expression analysis by reverse transcription-quantitative PCR (RT-qPCR) indicated that OsCHS1 is highly expressed in wild-type (WT) rice culms (Figure 2C), in which lignification typically occurs, suggesting its potential involvement in the biosynthesis of tricin for lignification. For OsCHS2, although a previous study demonstrated its enzymatic activity in vitro (Park et al., 2020), no OsCHS2 transcripts were detected in WT rice culms (Figure 2C). Only OsCHS1 was therefore analyzed further in this study.
Figure 2.
Major CHS, CHI, and CHIL proteins in rice. A and B, The unrooted phylogenetic trees of CHS (A) and classes I, II, and IV CHI/CHIL (B) proteins in angiosperm were built by neighbor-joining method. Bootstrapping with 1,000 replicates was performed. Scale bars denote 0.05 (A) and 0.1 (B) substitutions per site. Am, A. majus; At, A. thaliana; Bd, B. distachyon; Gm, G. max; Hl, H. Iupulus; In Ipomoea nil; Lj, L. japonicas; Md, M. × domestica; Mt, M. truncatula; Pv, P. virgatum; Pc, P. crispum; Ph, P. hybrida; Sb, S. bicolor; T, tomato, S. lycopersicum; Th, T. hybrid; Vv, V. vinifera; Zm, Z. mays. Filled circle: characterized in planta. Open circle: characterized in vitro. C, Relative gene expression levels over an ubiquitin gene (OsUBQ5) in WT culm tissue. Values refer to means ± standard deviations from biologically independent plants (n = 3). n.d., not detected.
The second step of the flavonoid biosynthetic pathway is the conversion of naringenin chalcone to naringenin, which is catalyzed by CHI (Figure 1). The rice genome encodes only one CHI protein, namely OsCHI or GOLD HULL AND INTERNODE1 (LOC_Os03g60509; Hong et al., 2012), which belongs to the class I CHI family, whose members typically exhibit bona fide CHI catalytic activities (Figure 2B; Shih et al., 2008). Previously, OsCHI was demonstrated to catalyze the conversion of naringenin chalcone to naringenin in vitro (Hong et al., 2012) as well as complement the lack of flavonoid accumulation in the CHI-deficient Arabidopsis tt5 mutant (Shih et al., 2008). Here, gene expression data indicated that OsCHI is expressed in the lignifying WT rice culm tissues (Figure 2C), supporting its potential involvement in tricin-lignin formation.
We further investigated CHIL, which is a class IV CHI protein family member lacking CHI catalytic activity, but able to bind and enhance the catalytic activities of flavonoid biosynthetic enzymes, particularly CHS (Ban et al., 2018; Ni et al., 2020; Waki et al., 2020; Zhao et al., 2021). Two putative CHIL genes, OsCHIL1 (LOC_Os11g02440) and OsCHIL2 (LOC_Os12g02370), are encoded by the rice genome (Figure 2B). Ban et al. (2018) demonstrated that OsCHIL1 and OsCHIL2 similarly enhance OsCHS1 activity in vitro. Considering their high protein sequence identity (96%) and their expression in lignifying rice culms (Figure 2C), we reasoned that both OsCHIL1 and OsCHIL2 contribute to the biosynthesis of tricin for lignification.
Preparation of CHS-, CHI-, and CHIL-deficient mutant rice
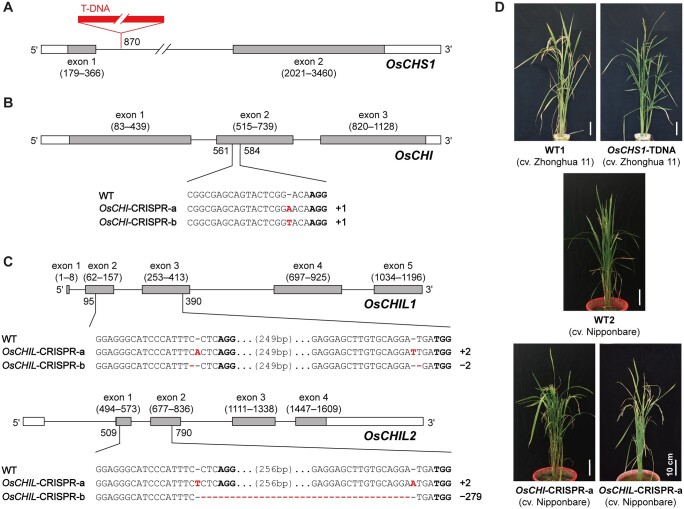
To investigate the in planta functions of the selected rice flavonoid genes, we prepared a set of rice mutant lines harboring knockout mutations in the selected genes. For OsCHS1, we used a rice mutant (O. sativa L. spp. japonica cv. Zhonghua 11) with a T-DNA insertion in the intron of OsCHS1 (Figure 3A). Plants homozygous for the T-DNA insertion (OsCHS1-TDNA) and their near isogenic WT control (WT1) were isolated from the self-progenies of heterozygotes available in Rice Mutant Database (Zhang et al., 2006; Wang et al., 2020).
Figure 3.
CHS-, CHI-, and CHIL-deficient rice mutant lines used in this study. A–C, Gene structures and mutation patterns in OsCHS1-TDNA (A), OsCHI-CRISPR (B) and OsCHIL-CRISPR (C) mutant lines. In red, deletion or insertion. Bold, protospacer adjacent motif site. D, Phenotype of OsCHS1-TDNA, OsCHI-CRISPR-a, and OsCHIL-CRISPR-a mutant lines and their WT control lines, WT1 and WT2. Scale bars denote 10 cm. OsCHS1-TDNA, OsCHS1 knockout line (cv. Zhonghua 11); OsCHI-CRISPR-a and OsCHI-CRISPR-b, OsCHI knockout line (cv. Nipponbare); OsCHIL-CRISPR-a and OsCHIL-CRISPR-b, OsCHIL1 and OsCHIL2 double-knockout line (cv. Nipponbare).
Knockout mutants of OsCHI and OsCHIL (O. sativa L. spp. japonica cv. Nipponbare) were generated by CRISPR/Cas9-mediated mutagenesis. For OsCHI, a CRISPR/Cas9 binary vector (Meng et al., 2017) harboring a single-guide RNA (sgRNA) specific for the second exon of this gene was transformed into rice. A number of T0 transformants containing various insertion and deletion mutations (indels) in the targeted site were identified by direct sequencing (Ma et al., 2015). Consequently, two homozygous OsCHI-CRISPR mutant lines (T2 generation) with different mutations (OsCHI-CRISPR-a and OsCHI-CRISPR-b) were isolated (Figure 3B). We also generated OsCHIL1 and OsCHIL2 double-knockout mutants using a multiplex CRISPR/Cas9 construct (Ma et al., 2015; Ma and Liu, 2016) harboring two sgRNAs that target different conserved sequences in OsCHIL1 and OsCHIL2. As a result, two homozygous mutant lines (T2 generation) with different mutations (OsCHIL-CRISPR-a and OsCHIL-CRISPR-b) were obtained (Figure 3C).
All the OsCHI-CRISPR and OsCHIL-CRISPR mutant plants (T2 and T3 generations) used for further chemotyping and phenotyping studies were fully genotyped and confirmed to harbor consistent mutations as listed in Figure 3. They were further confirmed to have no off-target mutations at least for the top three off-target sites predicted for each sgRNA (Supplemental Table S1). Multiple sequence alignment suggested that the indels in the mutant lines resulted in frameshift mutations, leading to truncated nonfunctional proteins, whereas potential illegitimate translations initiated from in-frame start codons were unlikely to produce functional proteins (Supplemental Figures S1 and S2). Loss-of-function mutations were therefore expected for all OsCHI- and OsCHIL-CRISPR mutants.
Phenotypes of CHS-, CHI-, and CHIL-deficient mutant rice
For phenotypic analyses, fully genotyped homozygous OsCHS1-TDNA (cv. Zhonghua 11), OsCHI-CRISPR-a (cv. Nipponbare), and OsCHIL-CRISPR-a (cv. Nipponbare) plants (T3 generations) were cultivated side-by-side with their corresponding WT controls, namely WT1 (cv. Zhonghua 11) and WT2 (cv. Nipponbare), under controlled conditions (Figure 3D). The OsCHS1-TDNA plants were phenotypically similar to WT1 plants, with the exception of a slightly shorter culm and a lack of seeds (Table 1), which is consistent with a previously reported sterility phenotype (Wang et al., 2020). The growth characteristics of the OsCHI-CRISPR-a and OsCHIL-CRISPR-a plant were similar to those of WT2, although OsCHI-CRISPR-a plants had slightly longer ears, OsCHIL-CRISPR-a plants were slightly shorter, and both mutant lines had a slightly lower fertility rate (Table 1). The culms and hulls of the OsCHI-CRISPR-a plants were yellowish (Supplemental Figure S3), which is consistent with the previously reported phenotype of other OsCHI-deficient mutants (Hong et al., 2012; Hirano et al., 2017). The observed coloration may be the result of an unusual accumulation of naringenin chalcone as further discussed below. In contrast, there were no observable color changes in the OsCHS1-TDNA and OsCHIL-CRISPR-a plant tissues. Overall, our data indicate that disrupting any of the targeted CHS, CHI, and CHIL genes does not drastically impair rice vegetative growth, although CHS-deficiency leads to complete sterility.
Table 1.
Growth phenotypes of OsCHS-TDNA, OsCHI-CRISPR-a, and OsCHIL-CRISPR-a and their isogenic wild-types
| Growth phenotypes | WT1 (cv. Zhonghua 11) | OsCHS1-TDNA (cv. Zhonghua 11) | WT2 (cv. Nipponbare) | OsCHI-CRISPR-a (cv. Nipponbare) | OsCHIL-CRISPR-a (cv. Nipponbare) |
|---|---|---|---|---|---|
| Plant height (cm)a | 115.1 ± 6.4 | 106.2 ± 8.3 | 89.2 ± 6.2 | 84.6 ± 7.6 | 80.7 ± 4.8* |
| Culm length (cm)b | 88.7 ± 2.6 | 77.3 ± 5.1** | 56.3 ± 7.9 | 55.7 ± 6.1 | 64.0 ± 8.2 |
| Ear length (cm) | 22.7 ± 1.5 | 20.7 ± 1.8 | 18.2 ± 2.0 | 20.9 ± 1.4* | 19.3 ± 1.5 |
| Tiller number | 8.4 ± 3.0 | 10.1 ± 3.1 | 6.2 ± 2.6 | 5.2 ± 0.8 | 6.3 ± 1.0 |
| Ear number | 11.0 ± 1.7 | 14.4 ± 6.0 | 10.7 ± 4.5 | 8.6 ± 1.1 | 10.5 ± 2.5 |
| Fertility rate (%) | 70.9 ± 18.5c | 0.0 ± 0.0**,c | 85.5 ± 1.6 | 63.1 ± 11.3** | 75.2 ± 3.3** |
Values refer to means ± standard deviations from biologically independent plants. Asterisks indicate significant differences between mutant line and wild type (Student’s t test, n = 5, *P < 0.05,
P < 0.01).
Length between the base of aerial part and the tip of top leaf.
Length between the base of aerial part and the base of panicle.
Data retrieved from Wang et al. (2020).
Soluble flavonoid metabolites in CHS-, CHI-, and CHIL-deficient mutant rice
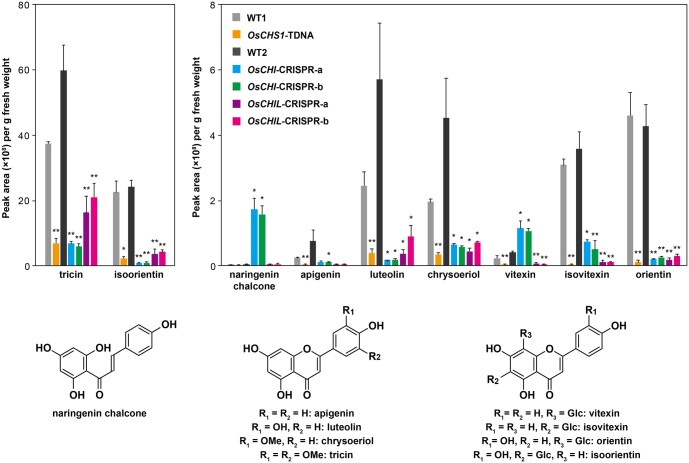
To determine the effects of CHS, CHI, and CHIL genetic mutations on the biosynthesis of soluble flavonoids, metabolites extracted from the leaves of the mutant and WT seedlings were analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). For this analysis, leaves were used because various soluble flavonoids are accumulated at relatively higher concentrations than in other major vegetative tissues of rice seedings (Lam et al., 2019a). As reported earlier (Lam et al., 2019a), various flavones, such as apigenin, luteolin, chrysoeriol, and tricin, and flavone C-glycosides, including vitexin, isovitexin, orientin, and isoorientin, were detected in the WT control plants (Figure 4).
Figure 4.
Soluble flavonoid metabolites in CHS-, CHI-, and CHIL-deficient rice mutant lines. LC–MS/MS analysis of methanol extracts from leaf tissues of rice seedlings was performed for determination of relative accumulation levels of soluble naringenin chalcone, flavones (apigenin, luteolin, chrysoeriol, and tricin), and flavone C-glycosides (vitexin, isovitexin, orientin, and isoorientin). Values refer to means ± standard deviations from biologically independent plants. Asterisks indicate significant differences between mutant line and its isogenic WT (Student’s t test, n = 3, *P < 0.05, **P < 0.01). OsCHS1-TDNA, OsCHS1 knockout line (cv. Zhonghua 11); OsCHI-CRISPR-a and OsCHI-CRISPR-b, OsCHI knockout line (cv. Nipponbare); OsCHIL-CRISPR-a and OsCHIL-CRISPR-b, OsCHIL1 and OsCHIL2 double-knockout line (cv. Nipponbare); WT1 and WT2, WT control lines (WT1, cv. Zhonghua 11; WT2, cv. Nipponbare).
Compared with the corresponding levels in the WT plants, flavone and flavone C-glycoside contents in the OsCHS1-TDNA plants decreased by ˃81%. Likewise, the contents of most flavonoids in OsCHI-CRISPR-a and OsCHI-CRISPR-b plants decreased by ˃79%, relative to the WT control levels. The exception was vitexin, which was, interestingly, more abundant in both mutant lines than in WT2 (Figure 4). Additionally, naringenin chalcone, the CHI substrate (Figure 1), accumulated substantially in the OsCHI-CRISPR-a and OsCHI-CRISPR-b plants, whereas it was barely detectable in the WT control and other mutant lines (Figure 4). Because naringenin chalcone is a yellow pigment (Forkmann and Kuhn, 1979), its elevated accumulation in the OsCHI-CRISPR plants is consistent with the yellowish coloration of the mutant’s tissues (Supplemental Figure S3). Collectively, these results demonstrate that OsCHS1 and OsCHI are, respectively, the predominant CHS and CHI responsible for the biosynthesis of the major soluble flavones in rice.
Similar to the OsCHS1-TDNA and the OsCHI-CRISPR lines, the examined flavone and flavone C-glycoside contents were much lower in the OsCHIL-CRISPR lines than in the WT plants, demonstrating that, together with OsCHS1 and OsCHI, the rice CHILs (OsCHIL1 and/or OsCHIL2) play key roles in the biosynthesis of the soluble flavones in rice. Meanwhile, similar to the OsCHS1-TDNA line but in contrast to the OsCHI-CRISPR lines, the OsCHIL-CRISPR lines did not accumulate naringenin chalcone. These findings are in agreement with the notion that CHIL serves as a noncatalytic enhancer of CHS, but not of CHI (Ban et al., 2018; Ni et al., 2020; Waki et al., 2020; Zhao et al., 2021). This possibility was further corroborated by analyzing the CHI catalytic activities of the crude protein extracts from OsCHIL-CRISPR mutant seedlings, which revealed that the activities were similar to that of the WT control (Supplemental Figure S4). Because CHIL was reported to bind CHS and enhance the production of naringenin chalcone primarily by inhibiting the formation of 4-coumaroyltriacetic acid lactone (CTAL), a derailment product of the CHS-catalyzed reaction (Waki et al., 2020), we also examined whether the OsCHIL-CRISPR lines over-accumulate CTAL. Our LC–MS/MS analysis, however, failed to detect any CTAL in either of the OsCHIL-CRISPR mutant lines or in the WT control. This result indicates low accumulation levels of CTAL even in the absence of CHIL activity, possibly due to its low stability and/or further metabolic conversion in planta.
Cell wall formation in CHS-, CHI-, and CHIL-deficient mutant rice
To investigate the effects of CHS, CHI, and CHIL mutations on cell wall formation, especially the deposition of tricin-lignin, cell wall samples prepared from culm tissues of the mutant (OsCHS1-TDNA, OsCHI-CRISPR-a, and OsCHIL-CRISPR-a) and WT control (WT1 and WT2) plants were subjected to comparative structural analyses based on histochemistry, wet chemistry, and 2D nuclear magnetic resonance (NMR). Culm tissues of mature rice plants were used for these cell wall and lignin analyses as well as the saccharification assay described below as they are rich in lignified secondary cell walls and represent rice biomass used for lignocellulosic utilization.
Histochemistry
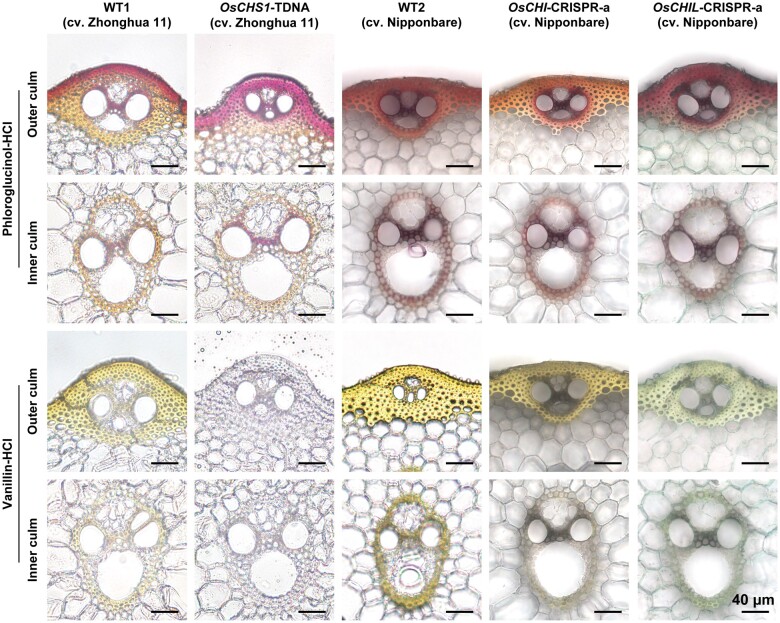
Transverse sections of culm tissues collected at the heading stage were treated with phloroglucinol–HCl and vanillin–HCl, which respectively stain lignin cinnamaldehyde end groups and cell wall-bound flavonoids (Lam et al., 2017), and then subjected to microscopic examinations. There were no obvious morphological changes to the vascular tissues in any of the mutants. The secondary cell walls of the vascular bundles and cortical sclerenchyma fibers in the mutant and WT culms were positively stained with the phloroglucinol–HCl lignin stain (Figure 5). The lignified secondary cell walls of the WT control were also yellowish following the vanillin–HCl flavonoid staining, suggestive of a normal accumulation of cell wall-bound flavonoids, presumably tricin-lignin (Figure 5). In contrast, flavonoid signals were undetectable or relatively weak in the cell walls of the OsCHS1-TDNA and OsCHIL-CRISPR-a plants, reflecting a considerable depletion of cell wall-bound flavonoids, presumably tricin-lignin (Figure 5). Regarding the cell walls of the OsCHI-CRISPR plants, decreased yellow staining was detected in the inner culm vascular bundles, but apparently not in the outer culm cortical fibers (Figure 5). However, in the OsCHI-CRISPR plants, yellowish cortical fiber cell walls were observed even before vanillin–HCl was applied (Supplemental Figure S3). Therefore, histochemical analysis did not enable a conclusive evaluation of the accumulation of cell wall-bound flavonoids in this particular mutant line. As discussed above, the yellowish coloration of the OsCHI-CRISPR tissues may have been due to the abnormal accumulation of chalcone pigments (Figure 4).
Figure 5.
Lignin and flavonoid depositions in the cell walls of CHS-, CHI-, and CHIL-deficient rice mutant lines. Transverse sections of culm tissues were subjected to phloroglucinol–HCl staining and vanillin–HCl staining, which stain monolignol-derived lignins and cell wall-bound flavonoids, respectively. Scale bars denote 40 µm. OsCHS1-TDNA, OsCHS1 knockout line (cv. Zhonghua 11); OsCHI-CRISPR-a, OsCHI knockout line (cv. Nipponbare); OsCHIL-CRISPR-a, OsCHIL1 and OsCHIL2 double-knockout line (cv. Nipponbare).
Wet chemistry
To further investigate the cell wall structures of the rice mutants, extractive-free cell wall residue (CWR) samples prepared from senesced mature culms of the mutant and WT control plants were subjected to a series of wet chemical analyses.
The Klason lignin assay results indicated that the lignin content did not change significantly in the OsCHS1-TDNA cell walls compared with that in the WT control cell walls (Table 2). On the other hand, the lignin contents of the OsCHI-CRISPR-a and OsCHIL-CRISPR-a cell walls decreased by 33.6% and increased by 12.6%, respectively, compared with the WT cell walls. To further examine these differential changes in lignin content, we also determined Klason lignin contents of the additional OsCHI- and OsCHIL-CRISPR mutant lines, that is, OsCHI-CRISPR-b and OsCHIL-CRISPR-b, harboring different mutation types (Figure 3). As a result, although we observed the same tendencies of lignin content changes for both of the mutant lines, that is, 7.0% decrease for OsCHI-CRISPR-b and 4.6% increase for OsCHIL-CRISPR-b on average; we did not detect any statistical significance for either of the two mutant lines (Supplemental Table S2). At this time, we cannot entirely exclude the possibility that the lignin content changes observed for the OsCHI-CRISPR and OsCHIL-CRISPR mutant lines may not be directly attributed to the malfunctions of the OsCHI- and OsCHIL genes.
Table 2.
Cell wall chemical analyses and enzymatic saccharification assay of OsCHS1-TDNA, OsCHI-CRISPR-a, and OsCHIL-CRISPR-a and their isogenic wild-types
| Cell wall chemotypes | WT1 (cv. Zhonghua 11) | OsCHS1-TDNA (cv. Zhonghua 11) | WT2 (cv. Nipponbare) | OsCHI-CRISPR-a (cv. Nipponbare) | OsCHIL-CRISPR-a (cv. Nipponbare) |
|---|---|---|---|---|---|
| Lignin content | |||||
| Klason lignin (mg/g CWR) | 152.0 ± 5.5 | 137.7 ± 9.3 | 120.4 ± 6.3 | 80.0 ± 10.9** | 135.6 ± 5.5* |
| Lignin composition by thioacidolysis | |||||
| Total monomer yield (µmol/g CWR) | 157.9 ± 9.7 | 161.2 ± 14.3 | 67.2 ± 0.2 | 56.4 ± 3.8* | 81.0 ± 3.5* |
| S/(S + G + H) (mol%) | 35.7 ± 2.7 | 34.7 ± 2.4 | 43.0 ± 1.0 | 42.8 ± 0.4 | 35.2 ± 0.6** |
| G/(S + G + H) (mol%) | 59.9 ± 3.0 | 62.7 ± 2.2 | 52.6 ± 0.7 | 54.3 ± 0.4* | 60.5 ± 0.6** |
| H/(S + G + H) (mol%) | 4.4 ± 0.4 | 2.7 ± 0.3** | 4.4 ± 0.3 | 2.9 ± 0.1** | 4.3 ± 0.0* |
| S/G | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.8 ± 0.0 | 0.8 ± 0.0 | 0.6 ± 0.0** |
| Cell wall-bound hydroxycinnamates | |||||
| pCA (mg/g CWR) | 25.8 ± 0.6 | 25.6 ± 1.6 | 21.5 ± 3.5 | 15.0 ± 2.9 | 24.3 ± 1.2 |
| FA (mg/g CWR) | 3.6 ± 0.2 | 3.3 ± 0.3 | 2.6 ± 0.2 | 2.1 ± 0.1 | 2.7 ± 0.1 |
| Enzymatic saccharification efficiency | |||||
| Glc released after 6 h (mg/g CWR) | 198.0 ± 14.8 | 208.9 ± 3.4 | 226.3 ± 18.7 | 202.1 ± 21.2 | 266.8 ± 47.1 |
| Glc released after 24 h (mg/g CWR) | 211.9 ± 11.5 | 219.0 ± 3.8 | 239.3 ± 16.4 | 210.4 ± 19.3 | 275.8 ± 39.7 |
| Glc released after 6 h (g/g glucan) | 464.8 ± 33.4 | 475.6 ± 9.3 | 552.8 ± 42.0 | 531.5 ± 43.3 | 623.8 ± 116.0 |
| Glc released after 24 h (g/g glucan) | 496.3 ± 24.6 | 498.6 ± 13.1 | 584.7 ± 39.8 | 553.5 ± 36.4 | 644.7 ± 98.7 |
Values refer to means ± standard deviations from biologically independent plants. Asterisks indicate significant differences between mutant line and its isogenic wild type (Student’s t test, n = 3, *P < 0.05,
P < 0.01).
Analytical thioacidolysis, which quantifies lignin-derived monomeric degradation products released via cleavage of the major β-O-4 linkages in lignin polymers (Lapierre et al., 1986), was performed. The total yields of the thioacidolysis-derived S, G, and H monomers were unchanged in OsCHS1-TDNA, but significantly decreased and increased in OsCHI-CRISPR-a and OsCHIL-CRISPR-a, respectively. This result was overall in line with the lignin content changes as determined by the Klason lignin assay (Table 2). The thioacidolysis-derived S/G monomer ratio significantly decreased in the OsCHIL-CRISPR cell walls, but not in the OsCHS1-TDNA and OsCHI-CRISPR cell walls, relative to the WT ratios (Table 2). The neutral sugar analysis suggested there were no drastic changes in the composition of the major cell wall polysaccharides in the OsCHS1-TDNA plants, whereas a slight decrease in crystalline glucan contents and an increase in xylan contents were detected in the OsCHI-CRISPR cell walls, and a slight reduction in mannan was detected in the OsCHIL-CRISPR cell walls (Supplemental Table S3). Furthermore, cell wall-bound pCA and FA were quantified according to the corresponding free acids released by a mild alkaline hydrolysis. Overall, we detected no significant change in the cell wall-bound pCA and FA levels between the mutant and WT plants (Table 2). Taken together, our chemical analysis indicates that OsCHS1, OsCHI, and OsCHIL mutations differentially affect lignin contents and structures as well as the composition of cell wall polysaccharides in rice cell walls, but they minimally affect p-hydroxycinnamates.
2D NMR
For a more in-depth characterization of lignin polymers, we performed 2D 1H-13C correlation heteronuclear single quantum coherence (HSQC) NMR analysis of lignin-enriched CWR samples prepared from culm CWRs by removing a large proportion of cell wall polysaccharides using crude cellulases (Kim and Ralph, 2010; Mansfield et al., 2012).
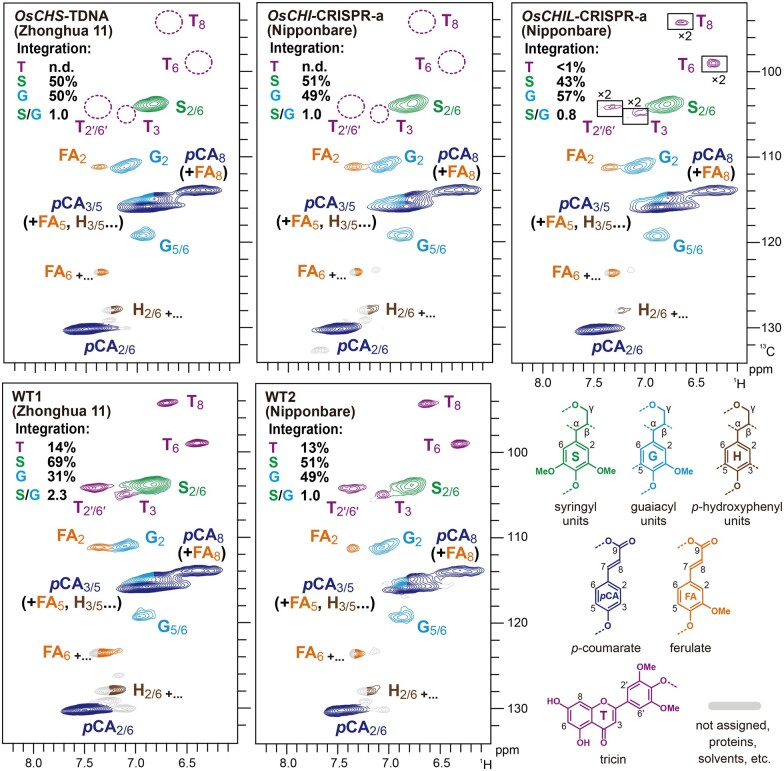
The aromatic sub-regions of the HSQC spectra of all examined rice samples showed intense aromatic signals from typical S (S) and G (G) lignin units (Figure 6). In line with the thioacidolysis-derived S/G monomer ratio data (Table 2), volume integration of the well-resolved C2/6–H2/6 aromatic signals indicated that the S/G lignin signal ratio (½S2/6/G2) decreased in the OsCHIL-CRISPR cell walls, but was unchanged in the OsCHS1-TDNA and OsCHI-CRISPR cell walls, relative to that of the corresponding WT control (Figure 6).
Figure 6.
Aromatic sub-regions of 2D HSQC NMR spectra of lignin-enriched CWRs of OsCHS1-TDNA, OsCHI-CRISPR-a, and OsCHIL-CRISPR-a and their isogenic wild-types. Volume integrals in the spectra refer to percentage of sum of S and G aromatic units. Boxes labeled by ×2 represent regions with scales enlarged by two-fold. Colors of the contours match those of the lignin substructures shown. OsCHS1-TDNA, OsCHS1 knockout line (cv. Zhonghua 11); OsCHI-CRISPR-a, OsCHI knockout line (cv. Nipponbare).
The aromatic sub-regions of the HSQC spectra further resolved a set of flavonoid signals from tricin-lignin present in rice cell walls. In the WT1 and WT2 spectra, diagnostic signals from lignin-integrated tricin units (T3, T6, T8, and T2ʹ/6ʹ; del Río et al., 2012; Lan et al., 2015; Lam et al., 2017) were clearly detected (Figure 6). Volume integrations of tricin C2'/6'–H2'/6' signals (½T2ʹ/6ʹ) accounted for 17% and 13% of the S and G total (G2 + ½S2/6 = 100%) in the spectra of the WT1 and WT2 cell walls, respectively. In contrast, these tricin signals decreased to undetectable levels in the spectra of the OsCHS1-TDNA and OsCHI-CRISPR cell walls. These results established that OsCHS1 and OsCHI are indispensable for the production of tricin serving as lignin monomers in rice cell walls. The tricin signals were also substantially decreased, to <1% (of the G and S total), demonstrating that, along with OsCHS1 and OsCHI, OsCHILs (OsCHIL1 and OsCHIL2) play key roles in the tricin-lignin biosynthesis. Nevertheless, the fact that these tricin signals were still detectable in the spectrum of the OsCHIL-CRISPR cell walls suggested that, unlike OsCHS1 and OsCHI, OsCHILs may not be absolutely essential for the production of tricin monomers for lignification. We also examined the possible incorporation of alternative flavonoids and chalcones into the mutants’ lignins, including naringenin chalcone in the OsCHI-CRISPR lignins, based on NMR data of several flavonoid and chalcone standards (Rencoret et al., 2021a, 2021b). However, our NMR analysis failed to detect the incorporation of any alternative flavonoids or chalcones in our mutants’ lignins.
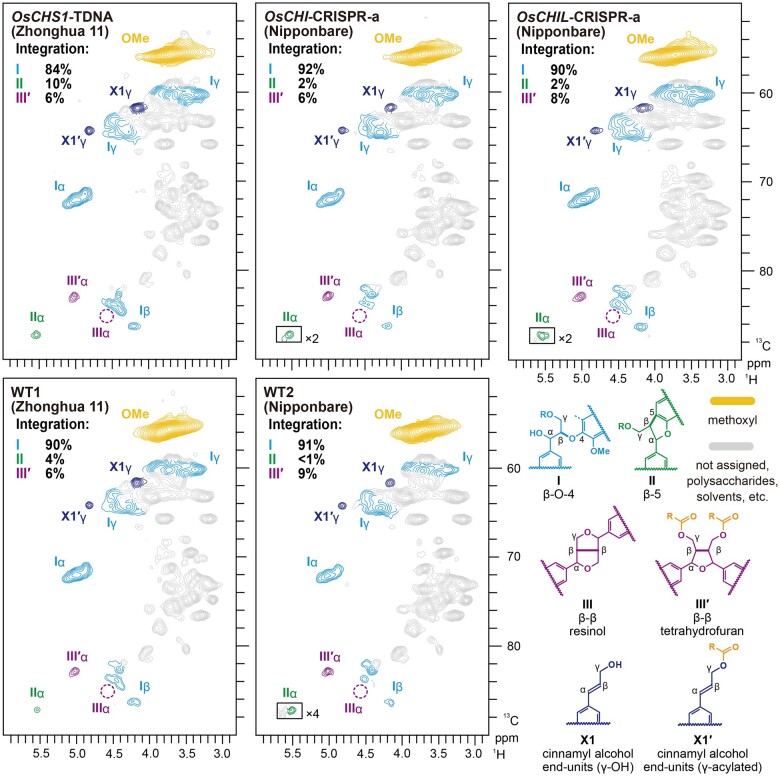
The oxygenated-aliphatic sub-regions of the HSQC spectra provided further insights into the distribution of various inter-monomeric linkage types in lignin polymers. Typical lignin side-chain signals from β-O-4 (I), β-5 (II), and tetrahydrofuran-type β–β (IIIʹ) units (Lu and Ralph, 2002, 2008) were detected in all examined rice cell wall spectra (Figure 7); only trace amounts of resinol-type β–β (III) units derived from nonacylated monolignols were detected, which is in accordance with our previous observations that rice cell wall lignins are highly γ-p-coumaroylated (Takeda et al., 2018). Volume integration using the well-resolved Cα–Hα correlations suggested that β-O-4 (I) is the predominant linkage type, accounting for 84%–92% of the total major lignin linkage signals detected (Iα + IIα + ½IIIʹα = 100%) in all WT and mutant cell wall spectra (Figure 7). Our NMR analysis determined that the major lignin linkage types were similarly distributed among WT2, OsCHI-CRISPR, and OsCHIL-CRISPR, whereas the β-O-4 and β-5 linkages were slightly less and more abundant, respectively, in OsCHS1-TDNA than in the WT1 control. Overall, it is likely that mutations in OsCHS1, OsCHI, and OsCHIL genes do not drastically affect the distribution of the major inter-monomeric linkage types in lignin polymers, although they substantially influence tricin-lignin formation and the S/G aromatic unit ratio, especially the OsCHIL mutations.
Figure 7.
Aliphatic sub-regions of 2D HSQC NMR spectra of lignin-enriched CWRs of OsCHS1-TDNA, OsCHI-CRISPR-a, and OsCHIL-CRISPR-a and their isogenic wild types. Volume integrals in the spectra refer to percentage of sum of I, II, and IIIʹ inter-monomeric units. Boxes labeled by ×2 and ×4 represent regions with scales enlarged by two- and four-fold, respectively. Colors of the contours match those of the lignin substructures shown. OsCHS1-TDNA, OsCHS1 knockout line (cv. Zhonghua 11); OsCHI-CRISPR-a, OsCHI knockout line (cv. Nipponbare).
Cell wall digestibility of CHS-, CHI-, and CHIL-deficient mutant rice
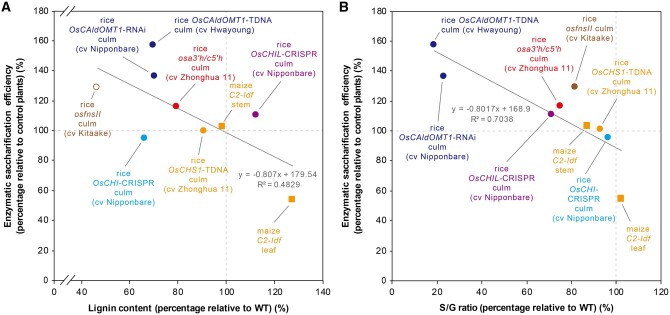
We previously observed considerable increases in the cell wall saccharification efficiency of several tricin-depleted rice mutants (Lam et al., 2017, 2019a, 2019b), which prompted us to investigate the cell wall saccharification of the tricin-depleted OsCHS1-, OsCHI-, and OsCHIL-deficient rice mutants prepared in this study. Destarched culm CWR samples from the mutant and WT plants were digested by a cocktail of cellulolytic enzymes (Hattori et al., 2012), after which the released glucose (Glc) was monitored (Table 2). Consequently, none of the mutants, even the OsCHI-CRISPR lines with significantly decreased lignin contents, exhibited any significant enhancement in the enzymatic saccharification efficiency expressed either as Glc released per mass of CWR or as Glc released per total glucan (Table 2). As further discussed below, these results sharply contrasted with our previous observations regarding other tricin-depleted rice mutant and transgenic plants (Figure 8; Lam et al., 2017, 2019a, 2019b).
Figure 8.
Correlation analyses between changes in enzymatic saccharification efficiency and lignin content/composition of tricin-depleted grass mutant and transgenic plants. A, Correlation analysis between change in enzymatic saccharification efficiency (24-h digestion; without pretreatment) and change in lignin content. Lignin content was determined by Klason lignin assays (filled data points) or thioglycolic acid assay (empty data point). B, Correlation analysis between change in enzymatic saccharification efficiency (24-h digestion; without pretreatment) and change in S/G ratio. S/G ratio was determined by thioacidolysis. Data of rice osfnsII (Lam et al., 2017), osa3ʹh/c5ʹh (Lam et al., 2019a, 2019b), OsCAldOMT1-TDNA and OsCAldOMT1-TDNA (Lam et al., 2019a, 2019b), and maize C2-ldf (Eloy et al., 2017) were retrieved from the literature. The percentages relative to control plants were calculated by dividing the value of the mutant or transgenic plant by that of the corresponding control plant. Equations and R2 values of the best fit lines are shown. Species names, cultivars, plant line names, and tissues used for analysis are indicated. Circles: rice. Squares: maize. Data points of the mutants are labeled with different colors according to the gene mutated.
Discussion
OsCHS1, OsCHI, and OsCHILs play key roles in the biosynthesis of extractable flavones and tricin-lignin in rice
Examination of the OsCHS1-TDNA and OsCHI-CRISPR mutants revealed considerable decreases in the abundance of the major extractable flavones in seedling leaves (Figure 4) and a complete lack of tricin-lignin in the mature culm cell walls (Figure 6); trace amounts of soluble flavonoid metabolites can still be detected in OsCHS1-TDNA mutant (Figure 4), which might be because the T-DNA insertion in the intron could not completely disrupt the transcription of OsCHS1 (Wang, 2008). Additionally, naringenin chalcone, the CHI substrate (Figure 1), over-accumulated in the leaves of the OsCHI-CRISPR seedlings (Figure 4). These observations indicate that OsCHS1 and OsCHI are, respectively, the major source of the CHS and CHI activities for the entry to the conserved flavonoid biosynthetic pathway in rice. As recently reported, the OsCHS1-TDNA plants were completely infertile (Table 1), which may be explained by the loss of specific flavonoids associated with pollen tube development (Wang et al., 2020). In contrast, the OsCHI-CRISPR plants were still fertile, albeit with a slightly decreased fertility rate (Table 1), possibly because these plants were still able to produce sufficient residual flavonoids for pollen tube development via the spontaneous isomerization of naringenin chalcone to naringenin in the absence of CHI activity (Mol et al., 1985).
Furthermore, this study demonstrated that CHIL, a noncatalytic enhancer crucial for flavonoid biosynthesis, is dedicated to the production of extractable flavones as well as tricin-lignin in rice cell walls. Recent biochemical studies suggested that CHIL (class IV CHI) lacks CHI activities because of the absence of the conserved catalytic amino acid residues in class I CHI (Ban et al., 2018; Cheng et al., 2018). Instead, CHIL binds to CHS and rectify its promiscuous catalysis, thereby substantially promoting the CHS activities for naringenin chalcone production (Ban et al., 2018; Ni et al., 2020; Waki et al., 2020; Zhao et al., 2021). The dramatic decrease in the accumulation of extractable flavones (Figure 4) and cell wall tricin-lignin (Figure 6) in the mutants reflected the importance of OsCHILs (OsCHIL1 and OsCHIL2) for flavone biosynthesis in rice. Nevertheless, it is likely that they are not indispensable, at least for supplying tricin monomers for lignification, as detectable amounts of tricin are still present in the mutant cell walls (Figure 6). This was in contrast to the OsCHS1-TDNA and OsCHI-CRISPR mutants, which produced cell walls with no detectable tricin-lignin (Figure 6). The observed lack of changes in the crude CHI activity (Supplemental Figure S4) or abnormal accumulation of naringenin chalcone (Figure 4; Supplemental Figure S3) in the OsCHIL-CRISPR plants are in line with the notion that CHIL promotes flavonoid biosynthesis primarily by affecting the catalytic activity of CHS rather than CHI (Ban et al., 2018; Ni et al., 2020; Waki et al., 2020; Zhao et al., 2021).
Overall, this study further characterized the key components of the flavonoid biosynthetic pathway in rice essential for the production of various flavone-derived metabolites, including tricin-lignin (Figure 1). As CHS, CHI, and CHIL sequences and functions are highly conserved among diverse plant species, including grasses (Shih et al. 2008; Dixon and Pasinetti, 2010; Waki et al., 2020), they represent viable bioengineering targets, along with the downstream pathway enzymes, such as FNSII (Lam et al., 2017), A3ʹH/C5ʹH (Lam et al., 2019a), and CAldOMT (Kim et al., 2006; Lam et al., 2019b), for manipulating the production of tricin-type metabolites, including tricin-lignin, in various grass food and biomass crops.
Differentially altered lignin profiles in CHS-, CHI-, and CHIL-deficient mutant rice cell walls
Mutations in genes encoding monolignol biosynthetic enzymes often result in the incorporation of pathway intermediates or their derivatives into cell wall lignins as noncanonical monomers, indicating the flexibility of lignification regarding which phenolic compounds can serve as lignin monomers in planta (Vanholme et al., 2019). Previously, we demonstrated that deficiencies in the tricin biosynthetic enzymes, that is, FNSII (Lam et al., 2017) and apigenin 3′-hydroxylase/chrysoeriol 5′-hydroxylase (A3′H/C5′H; Lam et al., 2019a), resulted in the incorporation of the pathway intermediates, that is, naringenin and apigenin, respectively (Figure 1), into lignin polymers instead of the canonical tricin monomer in rice. However, it is unlikely that such incorporations of noncanonical flavonoid monomers into lignins occur in the three tricin-depleted rice mutant lines tested in this study. Although the OsCHI-CRISPR plants had highly elevated levels of extractable naringenin chalcone (Figure 4), 2D NMR analysis failed to detect any aromatic signals that support the incorporation of naringenin chalcone or any other flavonoids into the lignin polymers (Figure 6). Likewise, incorporation of noncanonical lignin monomers was not detected in the OsCHS1-TDNA and OsCHIL-CRISPR mutant plants (Figure 6). We recently determined that rice mutant and downregulation line of OsCAldOMT1 do not incorporate selgin, the CAldOMT substrate in the tricin pathway (Figure 1), into cell wall lignins (Lam et al., 2019b), although the extractable selgin content increases considerably (Lam et al., 2015). It is unclear why some of the tricin pathway intermediates, such as naringenin (Lam et al., 2017) and apigenin (Lam et al., 2019a), are readily incorporated into lignin polymers in rice, but other intermediates, including selgin (Lam et al., 2019b) and naringenin chalcone (this study), are not. It is conceivable that these lignification-incompatible flavonoids might fail to meet at least one of the following criteria: sufficient production in lignifying tissues, proper localization in lignifying cell walls, and/or biochemical compatibility for the dehydrogenative polymerization in cell walls (Vanholme et al., 2019). At least for naringenin chalcone, however, a very recent study reported its incorporation into the rind lignin of papyrus (Cyperus papyrus L.), demonstrating its compatibility in lignin polymerization in planta (Rencoret et al., 2021b). Furthermore, a number of alternative flavonoids, such as dihydrotricin, naringenin, apigenin, kaempferol, and/or amentoflavone, were recently implicated as natural lignin monomers participating in lignification of papyrus (Rencoret et al., 2021b) and also several ancestral vascular plants (Rencoret et al., 2021a). These data suggest that not only tricin but also other flavonoids in general, at least those bearing reactive phenolic hydroxyl groups in the B-ring, are compatible in lignin polymerization in planta.
In addition to the depletion of lignin-integrated tricin units, there were notable changes to the content and composition of the monolignol-derived lignin polymer units in the rice mutants. The total lignin content in the culm cell walls showed tendencies to decrease in the OsCHI-CRISPR mutants, but conversely increase in the OsCHIL-CRISPR mutant, whereas no significant change was detected in the OsCHS1-TDNA mutant (Table 2; Supplemental Table S2). In addition, both thioacidolysis and 2D NMR analyses consistently indicated that the S/G lignin unit ratio decreased significantly in the OsCHIL-CRISPR plants, whereas no significant changes were detected in the other two mutant lines (Table 2; Figure 6). Moreover, it was also apparent that the proportion of β-O-4 (I) linkages decreased compared with the other lignin inter-monomeric linkage types particularly in the OsCHS1-TDNA cell walls (Figure 7).
Earlier studies on other tricin-depleted grass mutant and transgenic plants further emphasize the complexity in the effects of tricin pathway perturbations on monolignol biosynthesis (Lam et al., 2017). The FNSII-deficient (osfnsII; Lam et al., 2017), A3′H/C5′H-deficient (osa3′h/c5′h; Lam et al., 2019a), and CAldOMT-deficient (OsCAldOMT1-TDNA and OsCAldOMT1-RNAi; Lam et al., 2019b) rice plants all have considerably decreased lignin levels (including monolignol-derived units; Figure 8A). All these rice plants also produced lignins with significantly decreased S/G unit ratios (Figure 8B); the decreased S/G ratio in OsCAldOMT1-TDNA and OsCAldOMT1-RNAi could be primarily because CAldOMT is a bifunctional enzyme required for both tricin and S lignin biosynthesis (Lam et al., 2019b). On the other hand, CHS-downregulated maize C2-Idf mutant reportedly has a highly elevated lignin level with no significant change in the S/G unit composition in the leaves, but no notable lignin changes in the stem (Eloy et al., 2017). The elevated lignin content in the C2-Idf leaves is likely at least partially a consequence of the redirection of the carbon flux toward the monolignol pathway following the blockage of the entry of the branching flavonoid pathway (Figure 1; Eloy et al., 2017). However, this scenario is inconsistent with the absence of lignin content changes in the C2-Idf stems (Eloy et al., 2017) as well as in our rice OsCHS1-TDNA mutant culms (Table 2). Overall, it is likely that blockages at different positions of the tricin pathway have diverse effects on the carbon relocation to the parallel monolignol pathways leading to different monolignol types. Such carbon allocation between and within the flavonoid and monolignol pathways can vary between plant species and among tissues and cell types. In addition, cell wall alterations observed in our rice mutant and transgenic lines could be potentially affected by variations in genetic backgrounds. Clearly, the flavonoid- and lignin-associated grass mutants and transgenic plants will need to be more comprehensively investigated to resolve these issues.
Complex relationships between altered lignin profiles and the cell wall digestibility of tricin-depleted grass mutants
The identification of tricin-lignin as a common component of grass cell walls compelled researchers to examine whether tricin-lignin influences the utility of grass biomass (Halpin, 2019; del Río et al., 2020; Lam et al., 2021). In this context, earlier investigations (Eloy et al., 2017; Lam et al., 2017, 2019a), as well as this study (Table 2) assessed the efficiency of cell wall saccharification, which is a key step during the production of fermentable sugars from lignocellulose, in various tricin-depleted grass mutants with different mutated genes associated with the tricin biosynthetic pathway. Notably, the cell wall saccharification in several rice mutant and transgenic lines, including osfnsII (Lam et al., 2017), osa3′h/c5′h (Lam et al., 2019a), OsCAldOMT1-TDNA and OsCAldOMT1-RNAi (Lam et al., 2019b), has been found to be considerably better than that in the corresponding control plants (Figure 8). However, other tricin-depleted mutants, exhibited no significant improvement in their saccharification performance, including the OsCHS1-TDNA, OsCHI-CRISPR, and OsCHIL-CRISPR rice mutants analyzed in this study. In fact, the saccharification performance reportedly decreased in the leaves of the maize C2-Idf mutant (Eloy et al., 2017). Overall, it is likely that the direct effects of tricin-lignin on the enzymatic hydrolysis of cell walls are minimal. Instead, as noted for some of the previously characterized rice (Lam et al., 2017, 2019a, 2019b) and maize (Eloy et al., 2017) mutants, other lignin factors, especially the total lignin content (including tricin-lignin as well as the major monolignol-derived units), may be the major determinants for the notable enhancement (Lam et al., 2017, 2019a, 2019b) or deterioration (Eloy et al., 2017) of the cell wall saccharification in tricin-depleted mutants. Indeed, there are weak negative relationships between cell wall saccharification efficiency and changes in the total lignin content (Figure 8A) as well as the S/G lignin unit ratio (Figure 8B) among the examined tricin-depleted grass mutant and transgenic plants. Similar relationships between the lignin structural profiles and cell wall saccharification efficiency have been reported for other biomass samples (Chen and Dixon, 2007; Li et al., 2016). Nevertheless, the OsCHS1-TDNA, OsCHI-CRISPR, and OsCHIL-CRISPR rice mutants evaluated in this study had no significant changes in their saccharification performances, even with significantly altered lignin content and/or composition (Table 2 and Figure 8). Accordingly, these lignin factors are unlikely to be the only determinants of the cell wall digestibility of the tricin-depleted grass plants. In general, altered lignin profiles affect cell wall digestibility because of the associated changes in the supramolecular organization of lignocellulose (e.g. surface coverage and molecular assembly of cellulose) that eventually affect the accessibility and catalytic activities of cellulolytic enzymes (Ruel et al., 2009; Carmona et al., 2015; Liu et al., 2016; Martin et al., 2019). Therefore, future studies should more precisely elucidate the relationships between the altered lignin profiles and the supramolecular structures of lignocellulose in tricin-depleted grass mutant and transgenic plants and also other lignin-altered plants. Such studies, together with more in-depth investigations on the cell wall functions of the tricin-altered grass mutants and transgenic plants, may enhance our understanding of the still-elusive physiological functions of tricin-lignin and its potential effects on grass biomass utility.
Materials and methods
Phylogenetic analysis
Protein sequences were aligned by Clustal W (Larkin et al., 2007) and corrected manually in MEGAX (Kumar et al., 2018; Lam et al., 2015). Unrooted phylogenetic trees were constructed by neighbor-joining method using MEGAX (Kumar et al., 2018), with 1,000 bootstrap replicates.
Plant materials
The rice (O. sativa) OsCHS1-TDNA mutant (accession: RMD-03Z11BO45; cv. Zhonghua 11) was obtained from the National Centre of Plant Gene Research at Huazhong Agricultural University (Wuhan, China; Zhang et al., 2006). Regarding the CRISPR/Cas9-mediated mutagenesis, sgRNAs were designed using CRISPR-P version 2.0 (Liu et al., 2017) and the Mfold web server (Zuker, 2003). To generate OsCHI-CRISPR mutants, the sgRNA targeting 5′-CGGCGAGCAGTACTCGGACA-3′ (sgRNA-a) was cloned under the control of the OsU6 promoter in the CRISPR/Cas9 binary vector BGK032 (Biogle Technology, Hangzhou, China; Meng et al., 2017). To generate OsCHIL-CRISPR mutants, sgRNAs targeting two conserved regions in OsCHIL1 and OsCHIL2 (i.e. 5′-GGAGGGCATCCCATTTCCTC-3′ [sgRNA-b] and 5′-GAGGAGCTTGTGCAGGATGA-3′ [sgRNA-c]) were cloned under the control of the LacZ-U3 promoter and U6a promoter, respectively, in the CRISPR/Cas9 binary vector pYLCRISPR/Cas9-MH (Ma et al., 2015; Ma and Liu, 2016). Details regarding the primers used are provided in Supplemental Table S4. The constructed CRISPR/Cas9 binary vectors were transformed into Agrobacterium tumefaciens strain EHA105, which were used to transform rice embryonic calli (cv. Nipponbare). All the CRISPR/Cas9 plants used for further analysis in this study were genotyped via direct sequencing (Ma et al., 2015) using the primers listed in Supplemental Table S4. To determine the homozygosity of the mutants, the PCR products used for direct sequencing were ligated to the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA, USA). The resulting recombinant plasmids were transformed into Escherichia coli strain DH5α, after which at least 12 colonies per line were sequenced (Takeda et al., 2019). Potential off-target sites were identified according to the CRISPR-2.0 predictions (Liu et al., 2017). The top three potential off-target sites were analyzed (Supplemental Table S1). Genomic regions containing each potential off-target site were amplified by PCR using specific primers listed in Supplemental Table S4 and then subjected to direct sequencing. Regenerated plants and isolated mutant lines were grown to maturity in a phytotron with a 12-h light (30°C):12-h dark (27°C) cycle. Fully genotyped homozygous CRISPR/Cas9 mutants (T3 generation) were cultivated under the same conditions. Mature plants (45 d after heading) were phenotypically characterized, harvested, and dried at 27°C for 30 d for the subsequent cell wall characterization.
Crude protein extraction and CHI enzyme assay
Leaves were collected from 1-month-old seedlings and immediately frozen in liquid nitrogen before they were ground to fine powders. To extract crude proteins, the ground powders were mixed with 1 mL 0.05 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–NaOH buffer (pH 7.5) and then centrifuged to remove cell residues. The CHI enzyme assay was performed using reaction mixtures comprising 100 µg crude proteins, 0.05-M HEPES–NaOH buffer (pH 7.5), 10 µL ethanol as a co-solvent, and 50-μM naringenin chalcone (Kaltenbach et al., 2018). Reactions were monitored by measuring the absorbance at 390 nm.
Gene expression analysis
Total RNA extracted from culm tissues collected at the heading stage as previously described (Koshiba et al., 2013) was reverse transcribed to cDNA using random hexamer primers (Invitrogen). Gene expression was analyzed using the 7300 Real-time PCR system (Applied Biosystems, Forester City, CA, USA) and gene-specific primers listed in Supplemental Table S4. A ubiquitin gene (OsUBQ5) was used as an internal control.
Soluble flavonoid analysis
Leaf tissue (100 mg) collected from 2-month-old plants was immediately frozen in liquid nitrogen and pulverized using TissueLyser (Qiagen, Hilden, Germany). To extract flavonoid metabolites, 400-µL 80% (v/v) methanol containing 10-µM apigenin-d5 as an internal standard was added. The resulting solution was sonicated in an ice water bath for 30 min. Filtered extracts were separated in a Kinetex C18 100 Å column (100 × 2.1 mm; internal diameter, 2.6 µm; Phenomenex, Torrance, CA, USA) connected to a SCIEX X500R quadrupole time-of-flight mass spectrometer (AB Sciex, Redwood City, CA, USA). A solvent system comprising 0.5% (v/v) formic acid in water (A) and 0.5% (v/v) formic acid in methanol (B) was used. The high performance liquid chromatography analysis was completed using a linear gradient from 10% to 90% B over 20 min. The flow rate was maintained at 0.3 mL min−1. Flavonoids were detected via information-dependent acquisition. Compounds were identified by comparing their exact masses, fragmentation patterns, and retention times with those of authentic standards.
Histochemical analysis
Tissue fixation, sectioning, and histochemical staining using phloroglucinol–HCl and vanillin–HCl were performed as previously described (Lam et al., 2017).
Cell wall chemical analysis
The CWRs for chemical and 2D NMR analyses were prepared from mature rice culm tissues as previously described (Yamamura et al., 2012). Klason lignin assay (Hatfield et al., 1994), analytical thioacidolysis (Yamamura et al., 2011; Yue et al., 2012), cell wall-bound hydroxycinnamate analysis (Yamamura et al., 2011), and cell wall polysaccharide analysis (Lam et al., 2017) were conducted as previously described.
2D NMR spectroscopy
The culm CWR samples were ball-milled in ZrO2 vessels containing ZrO2 ball bearings using the Planetary Micro Mill Pulverisette 7 (Fritsch, Idar-Oberstein, Germany; 600 rpm, 12 cycles of 10 min with 5 min intervals; Kim and Ralph, 2010; Mansfield et al., 2012). Samples were then digested with crude cellulases (Cellulysin; Calbiochem, La Jolla, CA, USA) to produce lignin-enriched CWR samples as previously described (Tobimatsu et al., 2013). Lignin-enriched CWRs (∼20 mg) were dissolved in 500-μL dimethylsulfoxide (DMSO)-d6/pyridine-d5 (4:1, v/v). NMR spectra were acquired using a Bruker Biospin Avance™ III 800US system (Bruker Biospin, Billerica, MA, USA) equipped with a cryogenically cooled 5-mm TCI-gradient probe. Adiabatic HSQC NMR experiments were performed using the standard Bruker implementation (hsqcetgpsp.3). Specific parameters were set as previously described (Kim and Ralph, 2010; Mansfield et al., 2012). Data were processed and analyzed using Bruker TopSpin version 4.0 (Bruker Biospin, Billerica, MA, USA). Central DMSO solvent peaks (δC/δH: 39.5/2.49 ppm) were used as an internal reference. HSQC plots were obtained using the typical matched Gaussian apodization in F2, and squared cosine-bell apodization and one level of linear prediction (16 coefficients) in F1. Volume integrations were completed without using linear prediction and correction factors. For volume integration of lignin and tricin aromatic signals, S2/6, G2, and T2′/6′ signals were used, and the S2/6 and T2′/6′ integrals were logically halved. Regarding the integration of the lignin inter-monomeric linkages, Iα, IIα, and III′α were used, and III′α integrals were logically halved. Values of the relative signal intensities presented in Figures 6 and 7 were calculated as ½S2/6 + G2 = 100 and Iα + IIα + ½III′α = 100 bases, respectively.
Cell wall enzymatic saccharification analysis
Enzymatic saccharification was analyzed as previously described (Hattori et al., 2012; Lam et al., 2017). Briefly, CWR samples (∼10 mg) were destarched and hydrolyzed using an enzyme cocktail consisting of Celluclast 1.5 L (Novozymes, Bagsvaerd, Denmark; 1.1 FPU), Novozymes 188 (Novozymes; 2.5 CbU), and Ultraflo L (Novozymes; 65 μg) in 50-mM sodium citrate buffer (pH 4.8). The reaction was completed in a rotary reactor (Heatblock Rotator SN-48BN, Nissin, Saitama, Japan) at 12.5 rpm and 50°C for 6 or 24 h. The liberated Glc was measured using the Glc CII test kit (Wako Pure Chemicals Industries, Osaka, Japan).
Accession numbers
Sequence data can be retrieved under accession number AB00801 (OsCHS1, LOC_Os11g32650), XP_015646206 (OsCHS2, LOC_Os07g11440), AK061390 (OsCHI, LOC_Os03g60509), AK059463 (OsCHIL1, LOC_Os11g02440), AK099443 (OsCHIL2, LOC_Os12g02370), AK061988 (OsUBQ5, LOC_Os01g22490), XM_002450826 (Sorghum bicolor SbCHS2a; Yu et al., 2005), AY728478 (Z. mays ZmC2; Eloy et al., 2017), P24824 (Z. mays ZmWhp1; Mo et al., 1992), XP_003577513 (Brachypodium distachyon BdCHS), Pavir.Ha01439 (Panicum virgatum PvCHS), P13114 (A. thaliana AtCHS; Shirley et al., 1995), X55194 (Solanum lycopersicum TCHS1), X55195 (S. lycopersicum TCHS2; Schijlen et al., 2007), AB015872 (Vitis vinifera VvCHS1), AB066275 (V. vinifera VvCHS2), AB066274 (V. vinifera VvCHS3; Goto-Yamamoto et al., 2002), CAC19808 (Humulus Iupulus HlCHS; Ban et al., 2018), AJ277211 (Medicago truncatula MtCHS1; Wasson et al., 2006), V01538 (Petroselinum crispum PcCHS; Hermann et al., 1988), AB106522 (Torenia hybrid ThCHS1; Fukusaki et al., 2004), X03710 (Antirrhinum majus AmCHS1; Hatayama et al., 2006), X14591 (Petunia hybrida PhCHSA; van der Meer et al., 1992), AAY45748 (Malus × domestica MsCHS-1), EB120544 (M. × domestica MsCHS-2), ACJ54531 (M. × domestica MsCHS-3; Dare et al., 2013), XP_003559241 (B. distachyon BdCHI), XP_00246363 (S. bicolor SbCHI; Liu et al., 2010), Q08704 (Z. mays ZmCHI), P41088 (A. thaliana AtCHI; Shirley et al., 1995), AY595415 (Glycine max GmCHI2; Ralston et al., 2005), Q8H0G1 (Lotus japonicas LjCHI2; Shimada et al., 2003), P51117 (V. vinifera VvCHI), AY595413 (G. max GmCHI1A; Ralston et al., 2005), AY595414 (G. max GmCHI1B1; Ralston et al., 2005), Q53B70 (G. max GmCHI1B2; Ralston et al., 2005), AJ548840 (L. japonicas LjCHI1; Shimada et al., 2003), AB073787 (L. japonicas LjCHI3; Shimada et al., 2003), XP_003559241 (B. distachyon BdCHIL), KXG22932 (S. bicolor SbCHIL), NP_001151452 (Z. mays ZmCHIL), NP_568154 (A. thaliana AtCHIL; Jiang et al., 2015; Ban et al., 2018), NP_001236782 (G. max GmCHI4a; Ralston et al., 2005), NP_001242041 (G. max GmCHI4b; Ralston et al., 2005), MG324005 (Humulus lupulus CHIL2; Ban et al., 2018), BJ578539 (Ipomoea nil InEFP; Morita et al., 2014), BAJ10400 (P. hybrida PhEFP; Morita et al., 2014), BAJ10401 (T. hybrida ThEFP-A; Morita et al., 2014), BAJ10402 (T. hybrida ThEFP-B; Morita et al., 2014), and XP_002280158 (V. vinifera VvCHIL),
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Predicted effects of mutations on OsCHI in OsCHI-CRISPR mutant lines.
Supplemental Figure S2. Predicted effects of mutations on OsCHIL1 and OsCHIL2 in OsCHIL-CRISPR mutant lines.
Supplemental Figure S3. Phenotypes of OsCHI-CRISPR and WT control lines.
Supplemental Figure S4. CHI catalytic activities of crude proteins of OsCHI-CRISPR, OsCHIL-CRISPR, and WT control lines.
Supplemental Table S1. Off-target analysis of T3 generation of OsCHI-CRISPR and OsCHIL-CRISPR mutant lines.
Supplemental Table S2. Klason lignin assay of OsCHI-CRISPR-b, OsCHIL-CRISPR-b, and WT control line.
Supplemental Table S3. Cell wall polysaccharide analysis of OsCHS1-TDNA, OsCHI-CRISPR, OsCHIL-CRISPR, and WT control lines.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr Hironori Kaji, Ms Ayaka Maeno, and Ms Kyoko Yamada for their assistance in NMR analysis. A part of this study was conducted using the facilities at the DASH/FBAS, RISH, Kyoto University, and the NMR spectrometer at JURC, ICR, Kyoto University.
Funding
This work was supported in part by grants from the Research Grants Council of Hong Kong, China (grant no. GRF17126918), HKU seed fund for Basic Research (grant no. 201910159284), the Japan Society for the Promotion of Science (JSPS, grant nos. KAKENHI #JP16H06198 and #JP20H03044), and the Research Institute for Sustainable Humanosphere, Kyoto University (grant no. Mission-linked Research Funding #2016-5-2-1). P.Y.L. and Y.T.K. acknowledge the JSPS fellowship programs (program nos. #17F17103 and #17J0965416, respectively).
Conflict of interest statement. None declared.
Contributor Information
Pui Ying Lam, Research Institute for Sustainable Humanosphere, Kyoto University, Kyoto 611-0011, Japan.
Lanxiang Wang, School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong, China.
Andy C W Lui, School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong, China.
Hongjia Liu, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China.
Yuri Takeda-Kimura, Research Institute for Sustainable Humanosphere, Kyoto University, Kyoto 611-0011, Japan.
Mo-Xian Chen, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China.
Fu-Yuan Zhu, Co-Innovation Center for Sustainable Forestry in Southern China, College of Biology and the Environment, Nanjing Forestry University, Nanjing, 210037 China.
Jianhua Zhang, Department of Biology, Hong Kong Baptist University, Kowloon Tong, Hong Kong, China; State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Sha Tin, Hong Kong, China.
Toshiaki Umezawa, Research Institute for Sustainable Humanosphere, Kyoto University, Kyoto 611-0011, Japan; Research Unit for Realization of Sustainable Society, Kyoto University, Kyoto 611-0011, Japan.
Yuki Tobimatsu, Research Institute for Sustainable Humanosphere, Kyoto University, Kyoto 611-0011, Japan.
Clive Lo, School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong, China.
References
- Abu-Omar MM, Barta K, Beckham GT, Luterbacher JS, Ralph J, Rinaldi R, Román-Leshkov Y, Samec JSM, Sels BF, Wang F (2021) Guidelines for performing lignin-first biorefining. Energy Environ Sci 14: 262–292 [Google Scholar]
- Ban Z, Qin H, Mitchell AJ, Liu B, Zhang F, Weng JK, Dixon RA, Wang G (2018) Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc Natl Acad Sci USA 115: E5223–E5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotech 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona C, Langan P, Smith JC, Petridis L (2015) Why genetic modification of lignin leads to low-recalcitrance biomass. Phys Chem Chem Phys 17: 358–364 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Cheng AX, Zhang X, Han XJ, Zhang YY, Gao S, Liu CJ, Lou HX (2018) Identification of chalcone isomerase in the basal land plants reveals an ancient evolution of enzymatic cyclization activity for synthesis of flavonoids. New Phytol 217: 909–924 [DOI] [PubMed] [Google Scholar]
- Clayton WA, Albert NW, Thrimawithana AH, McGhie TK, Deroles SC, Schwinn KE, Warren BA, McLachlan ARG, Bowman JL, Jordan BR, et al. (2018) UVR8‐mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants. Plant J 96: 503–517 [DOI] [PubMed] [Google Scholar]
- Coomey JH, Sibout R, Hazen SP (2020) Grass secondary cell walls, Brachypodium distachyon as a model for discovery. New Phytol 227: 1649–1667 [DOI] [PubMed] [Google Scholar]
- Dare AP, Tomes S, Jones M, McGhie TK, Stevenson DE, Johnson RA, Greenwood DR, Hellens RP (2013) Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica). Plant J 74: 398–410 [DOI] [PubMed] [Google Scholar]
- del Río JC, Rencoret J, Gutiérrez A, Elder T, Kim H, Ralph J (2020) Lignin monomers from beyond the canonical monolignol biosynthetic pathway: another brick in the wall. ACS Sustain Chem Eng 8: 4997–5012 [Google Scholar]
- del Río JC, Rencoret J, Prinsen P, Martínez AT, Ralph J, Gutiérrez A (2012) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60: 5922–5935 [DOI] [PubMed] [Google Scholar]
- Dixon RA, , Pasinetti GM (2010) Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol 154: 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Barros J (2019) Lignin biosynthesis: old roads revisited and new roads explored. Open Biol 9: 190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Chen W, Wang W, Zhang H, Liu X, Luo J (2014) Comprehensive profiling and natural variation of flavonoids in rice. J Integr Plant Biol 56: 876–886 [DOI] [PubMed] [Google Scholar]
- Eloy NB, Voorend W, Lan W, Saleme MdLS, Cesarino I, Vanholme R, Smith RA, Goeminne G, Pallidis A, Morreel K, et al. (2017) Silencing CHALCONE SYNTHASE impedes the incorporation of tricin into lignin and increases lignin content. Plant Physiol 173: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A, Dutta T, Deng K, Jacquet N, Sinha A, Benites VT, Baidoo EEK, Richel A, Sattler SE, Northen TR, et al. (2017) SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PLoS One 12: e0178160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkmann G, Kuhn B (1979) Genetic control of chalcone isomerase activity in anthers of Petunia hybrida. Planta 144: 189–192 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Rencoret J, García-Calvo L, Encina A, Rigau J, Gutiérrez A, del Río JC, Caparros-Ruiz D (2016) Changes In cell wall polymers and degradability in maize mutants lacking 3'- and 5'-O-methyltransferases involved in lignin biosynthesis. Plant Cell Physiol 58: 240–255 [DOI] [PubMed] [Google Scholar]
- Freudenberg K (1965) Lignin: its constitution and formation from p-hydroxycinnamyl alcohols. Science 148: 595–600 [DOI] [PubMed] [Google Scholar]
- Fukusaki EI, Kawasaki K, Kajiyama S, An CI, Suzuki K, Tanaka Y, Kobayashi A (2004) Flower color modulations of Torenia hybrida by downregulation of chalcone synthase genes with RNA interference. J Biotechnol 111: 229–240 [DOI] [PubMed] [Google Scholar]
- Goto-Yamamoto N, Wan GH, Masaki K, Kobayashi S (2002) Structure and transcription of three chalcone synthase genes of grapevine (Vitis vinifera). Plant Sci 162: 867–872 [Google Scholar]
- Halpin C (2019) Lignin engineering to improve saccharification and digestibility in grasses. Curr Opin Biotechnol 56: 223–229 [DOI] [PubMed] [Google Scholar]
- Harborne JB, Hall E (1964) Plant polyphenols—XII.: The occurrence of tricin and of glycoflavones in grasses. Phytochemistry 3: 421–428 [Google Scholar]
- Hatayama M, Ono E, Yonekura-Sakakibara K, Tanaka Y, Nishino T, Nakayama T (2006) Biochemical characterization and mutational studies of a chalcone synthase from yellow snapdragon (Antirrhinum majus) flowers. Plant biotechnol 23: 373–378 [Google Scholar]
- Hatfield RD, Jung H-JG, Ralph J, Buxton DR, Weimer PJ (1994) A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J Sci Food Agric 65: 51–58 [Google Scholar]
- Hattori T, Murakami S, Mukai M, Yamada T, Hirochika H, Ike M, Tokuyasu K, Suzuki S, Sakamoto M, Umezawa T (2012) Rapid analysis of transgenic rice straw using near-infrared spectroscopy. Plant Biotechnol 29: 359–366 [Google Scholar]
- Hermann A, Schulz W, Hahlbrock K (1988) Two alleles of the single-copy chalcone synthase gene in parsley differ by a transposon-like element. Mol Gen Genet 212: 93–98 [DOI] [PubMed] [Google Scholar]
- Hirano K, Masuda R, Takase W, Morinaka Y, Kawamura M, Takeuchi Y, Takagi H, Yaegashi H, Natsume S, Terauchi R, et al. (2017) Screening of rice mutants with improved saccharification efficiency results in the identification of CONSTITUTIVE PHOTOMORPHOGENIC 1 and GOLD HULL AND INTERNODE 1. Planta 246: 61–74 [DOI] [PubMed] [Google Scholar]
- Hong L, Qian Q, Tang D, Wang K, Li M, Cheng Z (2012) A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1 phenotype. Planta 236: 141–151 [DOI] [PubMed] [Google Scholar]
- Jiang W, Yin Q, Wu R, Zheng G, Liu J, Dixon RA, Pang Y (2015) Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J Exp Bot 66: 7165–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach M, Burke JR, Dindo M, Pabis A, Munsberg FS, Rabin A, Kamerlin SCL, Noel JP, Tawfik DS (2018) Evolution of chalcone isomerase from a noncatalytic ancestor. Nat Chem Biol 14: 548–555 [DOI] [PubMed] [Google Scholar]
- Karlen SD, Free HCA, Padmakshan D, Smith BG, Ralph J, Harris PJ (2018) Commelinid monocotyledon lignins are acylated by p-coumarate. Plant Physiol 177: 513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen SD, Zhang C, Peck ML, Smith RA, Padmakshan D, Helmich KE, Free HCA, Lee S, Smith BG, Lu F, et al. (2016) Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci Adv 2: e1600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67: 387–394 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Hirose N, Mukai M, Yamamura M, Hattori T, Suzuki S, Sakamoto M, Umezawa T (2013) Characterization of 5-hydroxyconiferaldehyde O-methyltransferase in Oryza sativa. Plant Biotechnol 30: 157–167 [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY,, Liu H,, Lo C (2015) Completion of Tricin Biosynthesis Pathway in Rice: Cytochrome P450 75B4 Is a Unique Chrysoeriol 5'-Hydroxylase. Plant Physiol 168: 1527–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Lui ACW, Wang L, Liu H, Umezawa T, Tobimatsu Y, Lo C (2021) Tricin biosynthesis and bioengineering. Front Plant Sci 12: 733198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Lui ACW, Yamamura M, Wang L, Takeda Y, Suzuki S, Liu H, Zhu FY, Chen MX, Zhang J, et al. (2019a) Recruitment of specific flavonoid B-ring hydroxylases for two independent biosynthesis pathways of flavone-derived metabolites in grasses. New Phytol 223: 204–219 [DOI] [PubMed] [Google Scholar]
- Lam PY, Tobimatsu Y, Matsumoto N, Suzuki S, Lan W, Takeda Y, Yamamura M, Sakamoto M, Ralph J, Lo C, et al. (2019b) OsCAldOMT1 is a bifunctional O-methyltransferase involved in the biosynthesis of tricin-lignins in rice cell walls. Sci Rep 9: 11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Tobimatsu Y, Takeda Y, Suzuki S, Yamamura M, Umezawa T, Lo C (2017) Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol 174: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Morreel K, Lu F, Rencoret J, del Río JC, Voorend W, Vermerris W, Boerjan W, Ralph J (2016a) Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol 172: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Rencoret J, Lu F, Karlen SD, Smith BG, Harris PJ, del Río JC, Ralph J (2016b) Tricin-lignins: occurrence and quantitation of tricin in relation to phylogeny. Plant J 88: 1046–1057 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C (1986) Preparative thioacidolysis of spruce lignin: isolation and identification of main monomeric products. Holzforschung 40: 47–50 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li M, Pu Y, Ragauskas AJ (2016) Current understanding of the correlation of lignin structure with biomass recalcitrance. Front Chem 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10: 530–532 [DOI] [PubMed] [Google Scholar]
- Liu H, Du Y, Chu H, Shih CH, Wong YW, Wang M, Chu IK, Tao Y, Lo C (2010) Molecular dissection of the pathogen-inducible 3-deoxyanthocyanidin biosynthesis pathway in sorghum. Plant Cell Physiol 51: 1173–1185 [DOI] [PubMed] [Google Scholar]
- Liu J, Im Kim J, Cusumano JC, Chapple C, Venugopalan N, Fischetti RF, Makowski L (2016) The impact of alterations in lignin deposition on cellulose organization of the plant cell wall. Biotechnol Biofuels 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Ralph J (2002) Preliminary evidence for sinapyl acetate as a lignin monomer in kenaf. Chem Commun 90–91 [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J (2008) Novel tetrahydrofuran structures derived from β–β-coupling reactions involving sinapyl acetate in Kenaf lignins. Org Biomol Chem 6: 3681–3694 [DOI] [PubMed] [Google Scholar]
- Ma X, Liu Y-G (2016) CRISPR/Cas9‐based multiplex genome editing in monocot and dicot plants. Curr Protoc Mol Biol 115: 31.36.1–31.36.21 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Martin AF, Tobimatsu Y, Kusumi R, Matsumoto N, Miyamoto T, Lam PY, Yamamura M, Koshiba T, Sakamoto M, Umezawa T (2019) Altered lignocellulose chemical structure and molecular assembly in CINNAMYL ALCOHOL DEHYDROGENASE-deficient rice. Sci Rep 9: 17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Yu H, Zhang Y, Zhuang F, Song X, Gao S, Gao C, Li J (2017) Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol Plant 10: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol JNM, Robbinst MP, Dixon RA, Veltkamp E (1985) Spontaneous and enzymic rearrangement of naringenin chalcone to flavanone. Phytochemistry 24: 2267–2269 [Google Scholar]
- Morita Y, Takagi K, Fukuchi‐Mizutani M, Ishiguro K, Tanaka Y, Nitasaka E, Nakayama M, Saito N, Kagami T, Hoshino A, et al. (2014) A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J 78: 294–304 [DOI] [PubMed] [Google Scholar]
- Ni R, Zhu TT, Zhang XS, Wang PY, Sun CJ, Qiao YN, Lou HX, Cheng AX (2020) Identification and evolutionary analysis of chalcone isomerase fold proteins in ferns. J Exp Bot 71: 290–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HL, Yoo Y, Bhoo SH, Lee TH, Lee SW, Cho MH (2020) Two chalcone synthase isozymes participate redundantly in UV-induced sakuranetin synthesis in rice. Int J Mol Sci 21: 3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, et al. (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344: 1246843. [DOI] [PubMed] [Google Scholar]
- Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung HJG (1994) Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc 116: 9448–9456 [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochemistry Rev 3: 29–60 [Google Scholar]
- Ralph J (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Lapierre C, Boerjan W (2019) Lignin structure and its engineering. Curr Opin Biotechnol 56: 240–249 [DOI] [PubMed] [Google Scholar]
- Ralston L, Subramanian S, Matsuno M, Yu O (2005) Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol 137: 1375–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencoret J, Gutiérrez A, Marques G, del Rio JC, Tobimatsu Y, Lam PY, Pérez-Boada M, Ruiz-Dueñas FJ, Barrasa JM, Martinez AT (2021a) New insights on structures forming the lignin-like fractions of ancestral plants. Front Plant Sci 12: 740923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencoret J, Rosado MJ, Kim H, Timokhin VI, Gutiérrez A, Bausch F, Rosenau T, Potthast A, Ralph J, del Río JC (2022b) Flavonoids naringenin chalcone, naringenin, dihydrotricin, and tricin are lignin monomers in papyrus. Plant Physiol 188: 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi R, Jastrzebski R, Clough MT, Ralph J, Kennema M, Bruijnincx PCA, Weckhuysen BM (2016) Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew Chem Int Ed 55: 8164–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K, Berrio‐Sierra J, Derikvand MM, Pollet B, Thévenin J, Lapierre C, Jouanin L, Joseleau JP (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Sarkanen KV, Ludwig CH (1971) Lignins. Occurrence, Formation, Structure, and Reactions. Wiley-Interscience, New York, NY [Google Scholar]
- Schijlen EGWM, de Vos CHR, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144: 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Wang M, Lo C (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe SI (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol 131: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Sun Z, Fridrich B, de Santi A, Elangovan S, Barta K (2018) Bright side of lignin depolymerization: toward new platform chemicals. Chem Rev 118: 614–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Suzuki S, Tobimatsu Y, Osakabe K, Osakabe Y, Ragamustari SK, Sakamoto M, Umezawa T (2019) Lignin characterization of rice CONIFERALDEHYDE 5-HYDROXYLASE loss-of-function mutants generated with the CRISPR/Cas9 system. Plant J 97: 543–554 [DOI] [PubMed] [Google Scholar]
- Takeda Y, Tobimatsu Y, Karlen SD, Koshiba T, Suzuki S, Yamamura M, Murakami S, Mukai M, Hattori T, Osakabe K, et al. (2018) Downregulation of p-COUMAROYL ESTER 3-HYDROXYLASE in rice leads to altered cell wall structures and improves biomass saccharification. Plant J 95: 796–811 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Chen F, Nakashima J, Escamilla-Treviño LL, Jackson L, Dixon RA, Ralph J (2013) Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T (2018) Lignin modification in planta for valorization. Phytochem Rev 17: 1305–1327 [Google Scholar]
- Umezawa T, Tobimatsu Y, Yamamura M, Miyamoto T, Takeda Y, Koshiba T, Takada R, Lam PY, Suzuki S, Sakamoto M (2020) Lignin metabolic engineering in grasses for primary lignin valorization. Lignin 1: 30–41 [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JN, Stuitje AR (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, De Meester B, Ralph J, Boerjan W (2019) Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol 56: 230–239 [DOI] [PubMed] [Google Scholar]
- Waki T, Mameda R, Nakano T, Yamada S, Terashita M, Ito K, Tenma N, Li Y, Fujino N, Uno K, et al. (2020) A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat Commun 11: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? J Biochem Technol 1: 11–20 [Google Scholar]
- Wang L, Lam PY, Lui ACW, Zhu FY, Chen MX, Liu H, Zhang J, Lo C (2020) Flavonoids are indispensable for complete male fertility in rice. J Exp Bot 71: 4715–4728 [DOI] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura M, Hattori T, Suzuki S, Shibata D, Umezawa T (2012) Microscale thioacidolysis method for the rapid analysis of β-O-4 substructures in lignin. Plant Biotechnol 29: 419–423 [Google Scholar]
- Yamamura M, Wada S, Sakakibara N, Nakatsubo T, Suzuki S, Hattori T, Takeda M, Sakurai N, Suzuki H, Shibata D, et al. (2011) Occurrence of guaiacyl/p-hydroxyphenyl lignin in Arabidopsis thaliana T87 cells. Plant biotechnol 28: 1–8 [Google Scholar]
- Yu CKY, Springob K, Schmidt J, Nicholson RL, Chu IK, Yip WK, Lo C (2005) A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in sorghum. Plant Physiol 138: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Lu F, Sun R-C, Ralph J (2012) Syntheses of lignin-derived thioacidolysis monomers and their uses as quantitation standards. J Agr Food Chem 60: 922–928 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S (2006) RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34: D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu X, Gong Q, Cao J, Shen W, Yin X, Grierson D, Zhang B, Xu C, Li X, Chen K, Sun C (2021) Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol J 19: 671–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.