Purpose:
Microbial keratitis is an urgent condition in ophthalmology that requires prompt treatment. This study aimed to apply deep learning algorithms for rapidly discriminating between fungal keratitis (FK) and bacterial keratitis (BK).
Methods:
A total of 2167 anterior segment images retrospectively acquired from 194 patients with 128 patients with BK (1388 images, 64.1%) and 66 patients with FK (779 images, 35.9%) were used to develop the model. The images were split into training, validation, and test sets. Three convolutional neural networks consisting of VGG19, ResNet50, and DenseNet121 were trained to classify images. Performance of each model was evaluated using precision (positive predictive value), sensitivity (recall), F1 score (test's accuracy), and area under the precision–recall curve (AUPRC). Ensemble learning was then applied to improve classification performance.
Results:
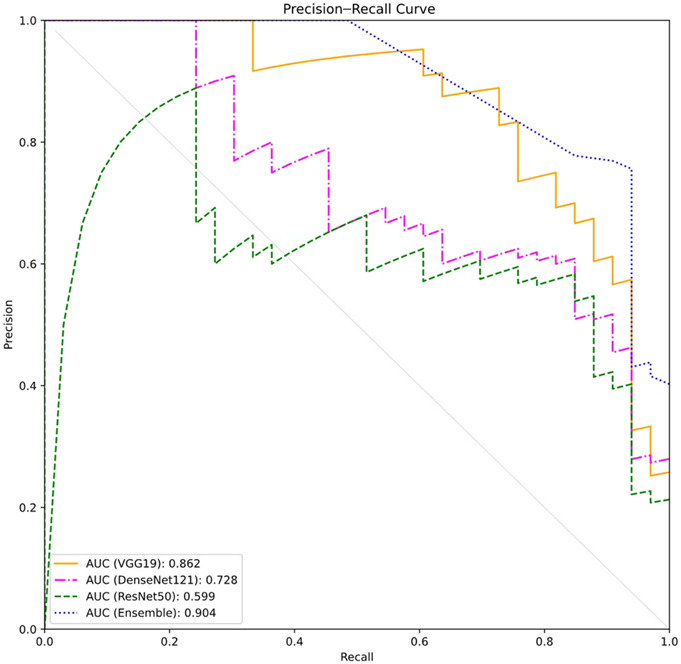
The classification performance in F1 score (95% confident interval) of VGG19, DenseNet121, and RestNet50 was 0.78 (0.72–0.84), 0.71 (0.64–0.78), and 0.68 (0.61–0.75), respectively. VGG19 also demonstrated the highest AUPRC of 0.86 followed by RestNet50 (0.73) and DenseNet (0.60). The ensemble learning could improve performance with the sensitivity and F1 score of 0.77 (0.81–0.83) and 0.83 (0.77–0.89) with an AUPRC of 0.904.
Conclusions:
Convolutional neural network with ensemble learning showed the best performance in discriminating FK from BK compared with single architecture models. Our model can potentially be considered as an adjunctive tool for providing rapid provisional diagnosis in patients with microbial keratitis.
Key Words: deep learning, microbial keratitis, convolutional neural network
Microbial keratitis (MK), a nonviral corneal infection, is an urgent condition in ophthalmology that requires prompt treatment to avoid severe complications of corneal perforation and vision loss.1 The global incidence of MK is estimated to be 1.5 to 2.0 million cases per year,2 ranging from 11 per 100,000 in Minnesota3 to as high as 799 per 100,000 in Nepal.4 The organisms causing MK vary among different countries depending on climate, contact lens use, socioeconomic status, and accessibility to healthcare service.1,5–7 For instance, a large-scale multicountry study in Asia including 6626 eyes from 6563 subjects showed that fungi were the commonly isolated organisms [eg, Fusarium spp. (18.3%) and Aspergillus flavus (8.3%)],5 whereas bacteria were the main causative pathogen in Australia, Europe, and the United States (64.6%–90.6%).6–9 Fungal keratitis (FK) is more severe, requires appropriate antifungal therapy, and has a higher risk of progression to endophthalmitis compared with bacterial keratitis (BK).10,11 Therefore, discriminating between bacterial and fungal infection is a crucial step for proper treatment and preventing further complications of MK.
The gold standard for diagnosis of MK is corneal tissue culture, the results of which take at least 48 hours. Although the test has high specificity, the sensitivity is low and varies among centers, ranging from 34.2% to 65%.9,12 Several adjunctive diagnostic tools have been proposed to help clinicians, including polymerase chain reaction (PCR) and confocal microscopy to make more rapid diagnoses and increase the detection rate while being less invasive for patients. However, these alternative tests have some limitations such as the amount of corneal tissue sample required and high technology/cost, which limit generalizability. Clinical history and corneal findings can be important clues for discrimination between BK and FK. However, previous studies have shown that approximately 30% of corneal infections were misdiagnosed using clinicians' experience based on corneal findings alone.13,14 Incorporating both patient history and clinical findings into a prediction model could improve discrimination performance compared with models with clinical findings alone.15,16 All conventional models still require information on patient history and corneal findings, which, in turn, are based on the clinical experience of the evaluators.
Recently, deep learning (DL) has been applied as an effective choice to solve various problems related to screening, diagnosis, and management of eye disease.17,18 The number of publications in this area is increasing continually.19,20 Many types of DL algorithms can be applied for prediction purposes; among them, transfer learning is one training strategy commonly used when the size of the training samples is relatively small compared with the number of model parameters to be trained. The transfer learning approach leverages parameters from a pretrained model that has previously learned from one data set to initialize the new model on another data set. In addition, several pretrained convolutional neural networks (CNN) for image classifications have been proposed.21–27 Such pretrained models were trained on a very large image data set called ImageNet.28 Although several DL models have been proposed in ophthalmology, few have addressed for infectious keratitis.29,30 Therefore, we conducted this study to apply DL in the classification of FK and BK. Our findings should be helpful in aiding specialists and nonspecialists in diagnosing MK.
METHODS
This study was a retrospective chart review of patients who were diagnosed with either BK or FK in the Department of Ophthalmology, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, from 2012 to 2020. This study had been approved by the Ethics Committee of Ramathibodi Hospital (MURA2019/317).
Patients were eligible if they met the following criteria: 1) diagnosed with either FK or BK by positive culture or PCR or pathological reports or complete response to a definitive therapy (therapeutic diagnosis) and 2) available anterior segment images during the active phase of infection. Patients were excluded for the following reasons: 1) had low-quality images, that is, unfocused, decentered, and/or inadequate light exposure and 2) had mixed infections (bacteria and fungus) or other coinfections (ie, Pythium, Acanthamoeba, and virus).
Data Acquisition
A total of 194 patients met eligibility criteria and were included in our study. We retrieved slit-lamp anterior segment images from the database of the Ophthalmology Department from 2012 to 2020. The images were taken using a slit-lamp biomicroscope (Haag-Streit BX900; Haag-Streit AG, Switzerland) mounted with a digital single-lens reflex camera (Canon EOS 7D) by a single experienced technician. Only images undertook at initial presentation and at the follow-up period with disease progression before receiving definite treatments with good centration, sharp focus, and adequate coverage of corneal lesions judged by an ophthalmologist (P.J.) were included. Multiple images captured by the diffuse illumination technique (with or without slit beam enhancement) from the same patient were also used for developing the model with the following reasons: 1) patient's eye position to the slit lamp and light exposure could be varied accordingly to different shots, 2) different images may contain different aspects of the lesion and also different quality, and 3) using multiple images could be equivalent to realistic data augmentation and thus could add more information to the algorithm.
Data Preparation
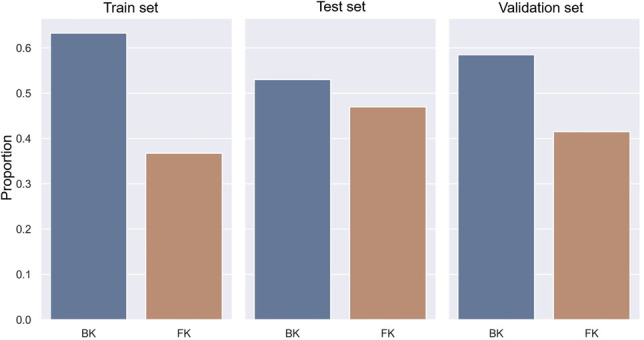
A total of 2167 images of 194 patients were randomly split into training, validation, and test sets with a targeted ratio of 80:10:10 using GroupShuffleSplit in Scikit-learn version 0.23.231 based on the following conditions: 1) distribution of BK and FK was similar among 3 data sets and 2) each patient was assigned in each data set only once. To meet these conditions, the final ratio was 85:5:10 for training, validation, and test sets, respectively. The training set comprised 1159 images of BK and 673 images of FK. The validation comprised 67 images of BK and 45 images of FK. The test set comprised 162 images of BK and 61 images of FK, as shown in Figure 1. Original images had variation in size and resolution (ranging from 4 to 18 megapixels). Taken into account of image quality and model performance, the original images were considered to be resized to 256 × 256 pixels before entering into the DL analysis. Light reflections and other artifacts were handled by performing the mean subtractions following recommendation for each method.21,22,26
FIGURE 1.
Distribution of BK and FK in each data set.
Modeling
We constructed the model called DeepKeratitis on top of 3 pretrained CNNs, which were selected from open-source Keras version 2.3.1, VGG19,21 ResNet50,22 and DenseNet121.26 All of them were trained on the ImageNet data set,28 which is an exceptionally large data set for the image classification task. Each pretrained architecture was extended with 2 more extension layers: GlobalAveragePooling2D and 1 dense layer with 2 outputs. The output returned the probability of BK and FK which were mutually exclusive.
Additional configuration techniques were applied to each model, including early layer freezing, data augmentation with horizontal flip, weight decay, early stopping, and Grad-CAM.32 Early layer freezing aims to use low-level features of pretrained architecture for transfer learning purposes. Because pretrained models had different architecture, the number of early layers to be frozen was different starting from the first to the kth layers. The first 11 layers of 28 layers were frozen in VGG19. The first 141 layers of 433 layers were frozen for DenseNet121. The first 80 layers of 181 layers were frozen for ResNet50. A weight decay (ReduceLROnPlateau) penalty of 10% was applied to the learning rate for every 5 epochs if validation loss did not improve to allow faster model convergence. Early stopping was applied if validation loss did not improve for 20 epochs continuously to prevent overfitting. Grad-CAM was applied to reveal which regions in a given image (represented as heatmap) influenced the model decision for further analysis of errors.32 Our experiment set color map as jet, which consisted of blue–green–red color gradient, indicating a different degree of influence on model prediction in an ascending order. The selected optimizer was Minibatch Gradient Descent33 with hyperparameter tuning by varying batch size (8, 16, and 32) and learning rate (0.001 and 0.0001). The optimum decision thresholds were calibrated to maximize classification performance for VGG19, ResNet50, and DenseNet121. For the ensemble model (random forest), the number of estimators was set at 100. Other hyperparameters were used as the default parameters.
Performance evaluation metrics, including precision (positive predictive value), sensitivity (recall), and F1 score, along with 95% confidence interval (CI), were estimated. The F1 score is the harmonic mean of the precision (P) and recall (R), that is, [2 PR/(P + R)], where precision is the number of true positive results divided by the total number of positive tests. This parameter measures a test's performance; the value closes to 1.0, reflecting high precision and recall. Area under the precision–recall curve (AUPRC) was estimated to evaluate discriminative performance. Curves close to the right upper quadrant indicate a better performance level than those close to the baseline.
RESULTS
Of 194 included patients with 2167 images, 128 patients (1388 images) were with BK and 66 patients (779 images) were with FK. Most of the included patients (119 patients, 61.3%) were diagnosed based on laboratory/pathological report, whereas 75 patients (38.7%) were classified based on therapeutic criteria.
Classification Performance
VGG19 and ResNet50 were optimized to a batch size of 16, whereas DenseNet121 was optimized to a batch size of 32. All pretrained architectures were optimized to 0.001 of the learning rate except ResNet50, which was optimized to 0.0001. The optimum decision thresholds to classify FK were 0.39, 0.53, and 0.60 for VGG19, DenseNet121, and ResNet50, respectively.
The classification performance of DeepKeratitis (single architecture of the CCN) and ensemble models at the optimal threshold is summarized in Table 1. The model performance metrics were reported at 95% CI in the format of x (y, z), where x is the performance, y is the lower bound, and z is the upper bound. Among pretrained CNN models, VGG19 showed highest overall performance followed by DenseNet121 and RestNet50 with the F1 score, precision, and sensitivity of 0.78 (0.72–084), 0.88 (0.83–0.93), and 0.70 (0.63–0.77), respectively. Although DenseNet121 and RestNet50 yielded lower F1 score and precision, they gained higher sensitivity than VGG19 with the corresponding values of 0.85 (0.80–0.90), 0.85 (0.80–0.90), and 0.70 (0.63–0.77). Applying the ensemble learning model could achieve higher classification performance compared with that from a single architecture with F1 score, precision, and sensitivity of 0.83 (0.77–0.89), 0.91 (0.87–0.95), and 0.77 (0.81–0.83), respectively.
TABLE 1.
Optimum Hyperparameters and Performance of all Models (N = 223 images)
| Model | Optimum Hyperparameters | Classification Performance (95% CI) | ||||
| Batch Size | Learning Rate | Threshold | Precision | Sensitivity | F1 Score | |
| VGG19 | 16 | 0.001 | 0.39 | 0.88 (0.83–0.93) | 0.70 (0.63–0.77) | 0.78 (0.72–0.84) |
| DenseNet121 | 32 | 0.001 | 0.53 | 0.61 (0.54–0.68) | 0.85 (0.80–0.90) | 0.71 (0.64–0.78) |
| RestNet50 | 16 | 0.0001 | 0.60 | 0.57 (0.49–0.65) | 0.85 (0.80–0.90) | 0.68 (0.61–0.75) |
| Ensemble | N/A | N/A | 0.60 | 0.91 (0.87–0.95) | 0.77 (0.81–0.83) | 0.83 (0.77–0.89) |
The AUPRCs were constructed (Fig. 2), indicating that the ensemble model yielded the highest discriminative performance followed by VGG19, DenseNet121, and ResNet50 with the AUPRCs of 0.904, 0.862, 0.728, and 0.599, respectively.
FIGURE 2.
Precision–recall curves in the test data set obtained by 4 different models.
Analysis of Misclassification Errors
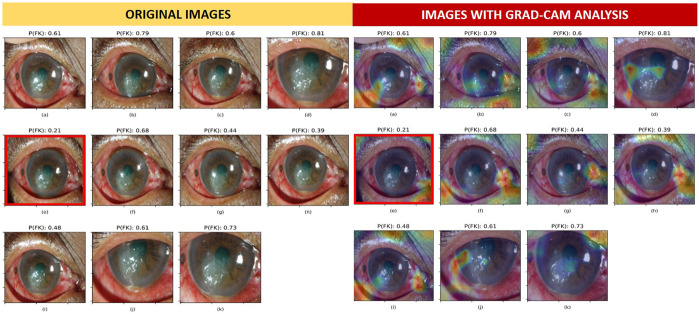
Misclassified images from VGG19, Densenet121, Resnet50, and ensemble were 13, 23, 26, and 12, respectively. Misclassified images from the best single DL architecture (VGG19) were further explored and investigated against the original observed samples using Grad-CAM. Figure 3B demonstrates the dominant areas that contributed to the model classification. All 11 images in Figure 3 were obtained from a patient with FK. The warmer colored gradient indicated the more influencing areas. Image (E) was misclassified as BK because the probability of being FK (0.21) was lower than the decision threshold (0.39). Compared with other correctly classified images, that is, (D), (I), and (K), the heatmap of image (E) highlighted the inferonasal area of the eye without considering the areas on the pathologic cornea. Findings from Grad-CAM analysis implied that image composition was very important for model prediction; however, the factors affecting area selection were unexplained because of the black box nature of DL. However, we also observed that images with similar composition could result in different predictive probabilities. We subsequently investigated the effect of image brightness on the model prediction and found that the probability of FK varied across different adjusted brightness using the misclassified image. Image brightness was defined as the average of pixel value in gray scale, ranging from 0 (darkest) to 225 (brightest). The misclassified image could be correctly classified as FK (probability of FK higher than 0.39) when the image brightness was adjusted to be within the optimal range (optimal brightness category) of 24.70 ± 11.46, as shown in Figure 4.
FIGURE 3.
Misclassification analysis using Grad-CAM in patients with FK. The optimal decision threshold of FK in VGG19 was 0.39. The probability of FK [P(FK)] is presented at the top of each image. The overlay heatmap with Grad-CAM analysis highlighted the areas that influenced model prediction. Image (E) was misclassified as BK with the P(FK) of 0.21; this was lower than the decision threshold.
FIGURE 4.
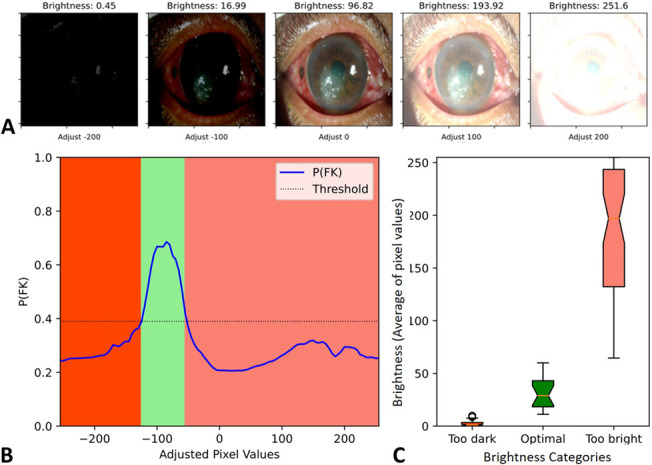
Variation of prediction probability on brightness adjustment in a misclassified image. A, Images with brightness adjustment by adding a constant of −200, −100, 0, 100, and 200 to all pixels and (B) graph demonstrates prediction probability (y axis) at different pixel value adjustments (x axis). C, Box plot illustrates an optimal brightness range of image for achieving correct classification which was 24.70 (range 9.63–45.20). The lower and higher image brightness can lead to image misclassification.
DISCUSSION
We constructed DeepKeratitis on top of pretrained DL architectures to distinguish patients with FK from BK. Our findings indicated that for single DL-based methods, VGG19 showed the highest performance, with the F1 score of 78% followed by DenseNet121 (71%) and ResNet50 (68%). We further adopted the ensemble technique, which used multiple learning algorithms to obtain better predictive performance; the F1 score increased to 83%.
Because for MEDLINE searching ("corneal ulcer" OR keratitis) AND [artificial intelligence (MeSH Terms), December 2019], we found only 1 study using CNN-based models to diagnose canine ulcerative keratitis and classify lesions into superficial and deep corneal ulcers.29 The authors showed that ResNet and VGGNet achieved accuracies over 90% for classifying normal corneas, corneas with superficial ulcers, and corneas with deep ulcers.29 They suggested that a CNN with multiple image classification models could be used as an effective tool for determining corneal ulcers but did not take this step themselves29 and did not take into account the type of infection. In 2020, Xu et al30 published a study that used 3 classic deep architectures: VGG16, GoogLeNet-v3, and DenseNet on a large data set of 2284 images (867 patients) to classify 4 types of infectious keratitis (ie, BK, FK, herpes simplex keratitis, and others). The authors demonstrated that deep models achieved an accuracy of 50.83% in VGG16, 55.83% in GoogLeNet-v3, and 64.17% in DenseNet.30 Despite using a large image data set, over half of the images were of herpes simplex keratitis and other corneal diseases. Moreover, mixed infection between bacteria and fungus and other infections were not considered. This heterogeneous data set might result in limited model performance when using image-level features. Although the authors applied sequential-ordered sets technique to reform the model, centroid annotation of the lesion and complex partitioning were required. Gu et al34 showed promising results of applying DL for diagnosis corneal disease (ie, infectious keratitis, noninfectious keratitis, corneal dystrophy or degeneration, and corneal neoplasm), although the specific types of infectious keratitis were not taken into account in their algorithm and only 1 CNN architecture (Inception-v3) was studied. Recently, Kuo et al proposed a DL approach based on 288 images using a single architecture (DenseNet algorithm) for diagnosis between FK and non-FK. Their model achieved a diagnostic accuracy at 69.4% with the sensitivity and specificity of 71.1% and 68.4%, respectively.35 Unlike our study, we used a larger data set of MK (2167 images) and compared the performances of 3 different architectures. Although we included cases with both therapeutic and definite diagnosis, we confirmed that all patients with therapeutic diagnosis were resolved by only single treatment approach (either antibacterial or antifungal medication). In addition, the standard regimen for BK in our center is the combination of fortified antibacterial eye drops: cefazolin (50 mg/mL) and gentamicin (14 mg/mL), not fluoroquinolone, which contains antifungal activity.36 As a result, we found that the accuracy for the MK classification was highest in VGG19 (78%), and this value was improved after applying an ensemble model (83%).
Previous estimates of clinician accuracy in classifying infectious keratitis have ranged from 49.29% for the average performance of an ophthalmologist provided with the image only to 57.16% for the average performance of an ophthalmologist provided with the image together with medical history.30 Those with more experience fellows and attending staff performed significantly better than residents, indicating that the human level classification essentially depends on clinical expertise.30 Given a large data set training, a DL-based model can achieve higher performance compared with human ability and conventional predictive models.14–16,30 For images that were misclassified, we performed further analysis and found that the probability of FK varied with image brightness. Therefore, good image composition and optimal brightness were required for achieving the optimal model performance.
For clinical applications, our CNN models, particularly ensemble learning, are potentially helpful for clinicians in discriminating between BK and FK. They can be considered as an adjunctive tool that provides rapid provisional diagnosis apart from laboratory tools, for example, smear, cultures, and PCR. The DL-based models provide several benefits for high-yield, noninvasive, and rapid response (within a few minutes). Moreover, they can be applied simply and in most clinical settings if configured using online interfaces or mobile applications.
There are some limitations to this study. First, our study only focused on BK and FK. Therefore, our model cannot be used for other types of corneal infections, such as viral, protozoal, or mixed corneal infections; excluding these other cases may overestimate the diagnostic properties. Further studies using DL with more complicated classification models are necessary to establish a comprehensive model for diagnosing all kinds of infectious keratitis. Second, we also included images from patients with therapeutic diagnosis (38.7%); thus, misclassification bias could not be ruled out. Third, the DL-based image analysis approach depends greatly on image orientation, focus, light exposure, camera settings, resolution, and overall image quality. The current model was developed based on a single-center data set derived from a high-resolution camera setting. The model accuracy possibly dropped when using low-resolution images or images sent through instant messaging platforms. Furthermore, the model could be sensitive to the brightness of images, in which augmentation for brightness was not applied. Finally, the performance of the model could be reduced if used with eyes that have a different anatomical structure, that is, other populations with different ethnicities, having other ocular pathology (eg, pterygium, pingueculae, and arcus senilis), and patients with previous corneal surgeries (eg, corneal transplantation, limbal stem cell transplantation, and intracorneal ring segment insertion). Clinical expertise and laboratory investigation should be considered.
In conclusion, we developed a DL prototype called DeepKeratitis to assist ophthalmologists to rapidly discriminate between FK and BK. Our findings, when developed, should have positive impacts on public health, especially in developing countries where specialists and laboratory facilities are limited. Moreover, this approach could reduce the number of unnecessary cornea specialist consultations and facilitate telehealth consultation. Further studies with large image data sets using various types of infectious keratitis and realistic data augmentation such as brightness randomization would be necessary for making the model more robust before adoption in real-world clinical settings.
ACKNOWLEDGMENTS
This article is an important part of the training of Amit Kumar Ghosh who is a MSc student in Data Science for Healthcare and Clinical Informatics in the Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand.
Footnotes
The authors have no funding or conflicts of interest to disclose.
A. K. Ghosh and R. Thammasudjarit contributed equally as first authors.
Contributor Information
Amit Kumar Ghosh, Email: axizamit@gmail.com.
Ratchainant Thammasudjarit, Email: ratchainant.tha@mahidol.edu.
John Attia, Email: john.attia@newcastle.edu.au.
Ammarin Thakkinstian, Email: ammarin.tha@mahidol.edu.
REFERENCES
- 1.Ung L, Bispo PJM, Shanbhag SS, et al. The persistent dilemma of microbial keratitis: global burden, diagnosis, and antimicrobial resistance. Surv Ophthalmol. 2019;64:255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erie JC, Nevitt MP, Hodge DO, et al. Incidence of ulcerative keratitis in a defined population from 1950 through 1988. Arch Ophthalmol. 1993;111:1665–1671. [DOI] [PubMed] [Google Scholar]
- 3.Vajpayee RB, Dada T, Saxena R, et al. Study of the first contact management profile of cases of infectious keratitis: a hospital-based study. Cornea. 2000;19:52–56. [DOI] [PubMed] [Google Scholar]
- 4.Upadhyay MP, Karmacharya PC, Koirala S, et al. The Bhaktapur eye study: ocular trauma and antibiotic prophylaxis for the prevention of corneal ulceration in Nepal. Br J Ophthalmol. 2001;85:388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khor WB, Prajna VN, Garg P, et al. The Asia cornea society infectious keratitis study: a prospective multicenter study of infectious keratitis in Asia. Am J Ophthalmol. 2018;195:161–170. [DOI] [PubMed] [Google Scholar]
- 6.Truong DT, Bui MT, Dwight Cavanagh H. Epidemiology and outcome of microbial keratitis: private university versus urban public hospital care. Eye Contact Lens. 2016;44:S82–S86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walkden A, Fullwood C, Tan SZ, et al. Association between season, temperature and causative organism in microbial keratitis in the UK. Cornea. 2018;37:1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tena D, Rodríguez N, Toribio L, et al. Infectious keratitis: microbiological review of 297 cases. Jpn J Infect Dis. 2019;72:121–123. [DOI] [PubMed] [Google Scholar]
- 9.Green M, Apel A, Stapleton F. Risk factors and causative organisms in microbial keratitis. Cornea. 2008;27:22–27. [DOI] [PubMed] [Google Scholar]
- 10.Henry CR, Flynn HW, Miller D, et al. Infectious keratitis progressing to endophthalmitis: a 15-year study of microbiology, associated factors, and clinical outcomes. Ophthalmology. 2012;119:2443–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dursun D, Fernandez V, Miller D, et al. Advanced Fusarium keratitis progressing to endophthalmitis. Cornea. 2003;22:300–303. [DOI] [PubMed] [Google Scholar]
- 12.Shalchi Z, Gurbaxani A, Baker M, et al. Antibiotic resistance in microbial keratitis: ten-year experience of corneal scrapes in the United Kingdom. Ophthalmology. 2011;118:2161–2165. [DOI] [PubMed] [Google Scholar]
- 13.Dalmon C, Porco TC, Lietman TM, et al. The clinical differentiation of bacterial and fungal keratitis: a photographic survey. Invest Ophthalmol Vis Sci. 2012;53:1787–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlgren MA, Lingappan A, Wilhelmus KR. The clinical diagnosis of microbial keratitis. Am J Ophthalmol. 2007;143:940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jongkhajornpong P, Nimworaphan J, Lekhanont K, et al. Predicting factors and prediction model for discriminating between fungal infection and bacterial infection in severe microbial keratitis. PLoS One. 2019;14:e0214076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas PA, Leck AK, Myatt M. Characteristic clinical features as an aid to the diagnosis of suppurative keratitis caused by filamentous fungi. Br J Ophthalmol. 2005;89:1554–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahimy E. Deep learning applications in ophthalmology. Curr Opin Ophthalmol. 2018;29:254–260. [DOI] [PubMed] [Google Scholar]
- 18.Grewal PS, Oloumi F, Rubin U, et al. Deep learning in ophthalmology: a review. Can J Ophthalmol. 2018;53:309–313. [DOI] [PubMed] [Google Scholar]
- 19.Du XL, Li WB, Hu BJ. Application of artificial intelligence in ophthalmology. Int J Ophthalmol. 2018;11:1555–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting DSW, Pasquale LR, Peng L, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations, ICLR 2015–Conference Track Proceedings. 2015; San Diego, CA.
- 22.He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016; Las Vegas, NV.
- 23.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2015; Boston, MA.
- 24.Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings–30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017; Honolulu, HI.
- 25.Howard AG, Zhu M, Chen B, et al. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv Prepr arXiv170404861. 2017.
- 26.Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. In: Proceedings–30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017; Honolulu, HI.
- 27.Zoph B, Vasudevan V, Shlens J, et al. Learning transferable architectures for scalable image recognition. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018; Salt Lake City, UT: 8697–8710 .
- 28.Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. [Google Scholar]
- 29.Kim JY, Lee HE, Choi YH, et al. CNN-based diagnosis models for canine ulcerative keratitis. Sci Rep. 2019;9:14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Kong M, Xie W, et al. Deep sequential feature learning in clinical image classification of infectious keratitis engineering; 2020. Available at: 10.1016/j.eng.2020.04.012. Accessed December 30, 2020. [DOI]
- 31.Scikit-learn 0.23.2. Sklearn.model_selection.GroupShuffleSplit. Available at: https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GroupShuffleSplit.html. Accessed January 15, 2021.
- 32.Selvaraju RR, Das A, Vedantam R, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2019;128:336–359. [Google Scholar]
- 33.Khirirat S, Feyzmahdavian HR, Johansson M. Mini-batch gradient descent: Faster convergence under data sparsity, 2017. In: IEEE 56th Annual Conference on Decision and Control (CDC), 2017; Melbourne, Australia: 2880–2887. doi: 10.1109/CDC.2017.8264077. [DOI]
- 34.Gu H, Guo Y, Gu L, et al. Deep learning for identifying corneal diseases from ocular surface slit-lamp photographs. Sci Rep. 2020;10:17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo MT, Hsu BW, Yin YK, et al. A deep learning approach in diagnosing fungal keratitis based on corneal photographs. Sci Rep. 2020;10:14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matoba AY, Barrett R, Lehmann AE. Cure rate of fungal keratitis with antibacterial therapy. Cornea. 2017;36:578–580. [DOI] [PubMed] [Google Scholar]