Abstract
The close relationship between the intestine and the skin has been widely stated, seen from gastrointestinal (GI) disorders often accompanied by skin manifestations. Exactly how the gut microbiome is related to skin inflammation and influences the pathophysiology mechanism of skin disorders are still unclear. Many studies have shown a two-way relationship between gut and skin associated with GI health and skin homeostasis and allostasis. This systematic review aimed to explore the associations between the gut microbiome with inflammatory skin disorders, such as acne, psoriasis, atopic dermatitis, and urticaria, and to discover the advanced concept of this relationship. The literature search was limited to any articles published up to December 2020 using PubMed and EBSCOHost. The review followed the PRISMA guidelines for conducting a systematic review. Of the 319 articles screened based on title and abstract, 111 articles underwent full-text screening. Of these, 23 articles met our inclusion criteria, comprising 13 atopic dermatitis (AD), three psoriasis, four acne vulgaris, and four chronic urticaria articles. Acne vulgaris, atopic dermatitis, psoriasis, and chronic urticaria are inflammation skin disorders that were studied recently to ascertain the relationship of these disorders with dysbiosis of the GI microbiome. All acne vulgaris, psoriasis, and chronic urticaria studies stated the association of gut microbiome with skin manifestations. However, the results in atopic dermatitis are still conflicting. Most of the articles agree that Bifidobacterium plays an essential role as anti-inflammation bacteria, and Proteobacteria and Enterobacteria impact inflammation in inflammatory skin disorders.
Key words: Gut microbiome, Microbiota, Acne vulgaris, Atopic dermatitis, Psoriasis, Chronic urticaria, Dysbiosis, Inflammation
Introduction
The human microbiota is a group of symbiotic, commensal, and pathogenic microorganisms that naturally share the human body’s space. They share beneficial interactions essential to human health and ecological or genetic changes that contribute to the outcome of diseases.1 Initially, the term, microbiome, describes the genome within a community of microbes that plays a role in human health by contributing to changes in specific organ functions. It differs from microbiota, which defines a total number of microbes living in a certain environment, denoting taxonomy and abundance of community members.2 Nowadays, the term microbiome has become interchangeable with microbiota to refer to the organisms themselves.3,4
Microbiomes act as an organ, which plays an integral part in regulating the system with other organs through microbiomehost interactions.5 Microbiomes induce local effects in an organ and change the functions in distant organs such as the liver, heart, and central nervous system. The microbiome also interacts with other microbiota between and within body sites. The critical messenger in the crosstalk between microbiota and host cells is the microbial metabolites. The communication is done through the collective set of all genomes of a microbial community called metagenomes.6,7 The microbiota can respond to environmental stimulation through an alternation of DNA methylation and histone modifications, which can have long-term effects on the host’s physiology.8
Grice and Segre9 are the first scientists who introduced the microbiome as our second genome. They reported that the human microbiome had less significant intrapersonal variation despite the broad diversity between each organ site. Thus, each body site has a highly specialized niche characterized by its microbial consortia, community dynamics, and host tissue interactions. 10 The five major areas of the healthy human microbiome (airways, skin, oral cavity, gastrointestinal (GI) tract, and vagina) have been launched by Human Microbiome Projects (HMP) to create the baseline view and make a resource to the broad scientific community. This project makes a tremendous breakthrough to determine the gene from whole communities rather than individual genomes, which is essential as the foundational role of microbial communities in both environmental and human health.11 The HMP data reported that the community microbiome’s stability is least stable in the oral cavity and most durable in the vagina and gut communities.12
The GI tract contains a vast majority of the microbes residing in the human body and harbors probably the most abundant compartment of the immune cells present in the body.13 Lumen epithelial cells play an essential role as the gatekeepers, which translate critical information to the immune cells located in the lamina propria.14 Conversely, there is evidence that many different microbial metabolites such as folate, indoles, secondary bile acids, trimethylamine- N-oxide, and short-chain fatty acid (SDFAs) also influence host metabolism primarily by binding to specific host membranes or nuclear receptors.15 Within this close relationship, we live in perfect harmony with microorganisms. A balanced microbial community in the gut is of tremendous importance in maintaining health and immune functions.16
The recent finding indicates crosstalk between the gut microbiome and the skin, known as the gut-skin axis.17 The concept of gut-brain-skin axis introduced by Arck et al. in 2016 hypothesized that excessive shared signals and cellular protagonists, the complex innervation of both skin and gut, and the prominence of neurogenic inflammation contribute to several gastrointestinal and skin diseases.17 There is a limited number of studies on the relationship of the gut microbiome to skin health. Therefore, we conducted a literature review to describe the gut bacteria communication with the host, the mechanism, and the outcome of gut microbiota imbalance on skin health, which may manifest in atopic dermatitis, acne vulgaris, psoriasis, and urticaria.
Materials and methods
This literature review used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.18 The literature search was limited to any articles published up to December 2020 by using PubMed and EBSCOHost. The search keywords were “gut” “gastrointestinal” “gastrointestinal microbiome” “microbiome” “microbiota” “gut microbiota” “intestinal microbiota” and “acne” “atopic dermatitis” “atopic eczema” “urticaria” “hives” and “psoriasis”. We assessed identified articles to determine whether there was a subject regarding the relationship between the gut microbiome and skin diseases. Inclusion criteria included the relationship between the gut microbiome and skin diseases, the mechanisms, and the comparison of gut microbiota between the subjects with certain skin diseases and healthy controls. Review articles, non-English language, and protocol studies were excluded in the first screening of records. After assessing fulltext articles, the articles that involved animals or in vitro studies and pregnant women as subjects were also excluded.
Results
Of the 319 articles on PubMed and EBSCOHOST databases screened based on title and abstract, 111 articles underwent full-text screening. Of these, 23 articles met our inclusion criteria, with 13 AD articles, three psoriasis, four acne vulgaris, and four urticaria. We excluded 25 articles because they only observed skin microbiome and did not describe the gut microbiota variants. Included articles consisted of cohort, crosssectional, case-control, and meta-analysis studies (Figure 1).
We obtained four articles revealing the relationship between gut microbiome and acne. All of these studies supported the alteration of the gut microbiome contributing to acne. Meanwhile, two studies using high-throughput sequencing demonstrated the decrease of Actinobacteria, an increase of Proteobacteria, and decreased Firmicutes/ Bacteroidetes (F/B) ratio in patients with acne.19,20 In contrast to these findings, Thompson et al. in 2019 reported that the highest proportion of F/B in the gut microbiota was showing in pre-antibiotic acne cases.21 We also included a “letter to the editor” by Reiner et al., which discussed the variable abundance at the phylum level in the F/B ratio in acne patients (Table 1).22
We found sixteen articles that correlated atopic dermatitis (AD) and the gut microbiome. Even though ten reports stated there were non-significant differences in the bacterial families between healthy and AD,23-32 four articles support significant low abundance diversity in AD gut microbiome (Table 2).33-38 The subjects of the all the papers ranged from infants, toddlers, and children. Pregnant women as subjects were excluded from this review.
Nine articles investigated the relationship between gut microbiome and psoriasis. All articles have high agreement on the role of dysbiosis microbiota with psoriasis. Seven articles came up with a relatively large gut microbiome quantity between psoriasis and healthy controls (Table 3). They found a significant difference in psoriatic and healthy controls.39-43 One article compared the abundance of F. prausnitzii and E.coli in psoriatic patients, with inflammatory bowel disease and healthy controls.44
Table 1.
Gut microbiome involvement in acne.
| The relation of the gut-skin microbiome | Increase | Decrease | References |
|---|---|---|---|
| There are significant differences in gut microbial composition and function between patients with acne and health controls. Acne patients have decreased gut microbiota diversity and a decreased ratio in Firmicutes to Bacteroidetes (F/B). | Bacteroidetes | Firmicutes F/B ratio Clostridia Clostridiales Lachnospraceae Ruminococcaceae | Deng et al., 201819 |
| Actinobacteria in acne patients were decreased significantly, while Proteobacteria were increased. At the genus level, there were decrease levels of Bifidobacterium, Butyrocicocus, Coprobacillus, Lactobacillus, and Allobaculum. | Proteobacteria Bifidobacterium Butyrocicocus Coprobacillus Lactobacillus Allobaculum | Actinobacteria | Yan et al., 201819 |
| There were changes of relative abundance variations of phylum level in the Firmicutes to Bacteroidetes (F/B) ratio found in acne patients with moderate to severe acne, which reduced after antibiotics were used. Race and diet show a significant impact on the F/B ratio in acne patients receiving antibiotic therapy. | F/B ratio in baseline acne patient | - | Rainer et al., 202022 |
| After antibiotic acne treatment, there was a reduction of the mean ratio of F/B in the gut microbiota compared to pre-antibiotic acne cases. The Bacteroidetes enriched in the gut microbiota after four weeks of antibiotics treatment compared to baseline acne patients and acne-free controls. | F/B ratio in baseline acne patient Ruminococccus gnavus Bacteroides acidifaciens Firmicutes Bacteroidetes | Lactobacillus inners Lactobacillus zeae Bifidobacterium animalis Rothia mucilaginosa | Thompson et al., 201921 |
Four original articles reported the relationship between chronic urticaria and gut microbiota. The decrease of A. muciniphila, Cl. leptum, F. prausnitzii, and lower genera Enterobacteriaceae, Lactobacillus, and Bifidobacterium have been shown in chronic urticaria (CU) and reported in all articles (Table 4).45-51 Other findings by Lu et al. in 2019 demonstrated Bacteroidetes are reduced in CU patients, in contrast to Actinobacteria and Proteobacteria.48 However, other studies from Rezazadeh et al. and Wang et al. in 2020 reported relative increases in Bacteroidetes compared to controls. 50,51 All studies revealed the same finding that Enterobacteriaceae was increased in CU).48-51 Wang et al. in 2020 also found significant changes in butanoate metabolism from spontaneous urticaria patients.51
Discussion
The four dominant phyla that reside in gut microbiota are Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria.52 These phyla make up 93.5% of isolated human fecal material, and the composition reflects their physiological properties in a specific region. Thus, they have unique characteristics in the skin inflammatory disorder, as discussed below.
Acne vulgaris
Acne vulgaris is a common sebaceous gland disease that mostly occurs on the face. It has a significant impact on the patient’s appearance, self-confidence, and quality of life.53 Acne vulgaris manifests clinically as non-inflammatory comedones, inflammatory papules, pustules, and nodules. 54 The pathogenesis of acne involves hyperseborhea, triggered by internal and external factors; dysbiosis of cutaneous microbiome leading to the disturbed skin barrier and inflammation; as well as activation of innate immune by P. acnes, which induces the expression of protease-activated receptors (PARs), tumor necrosis factor-a (TNF-a), and toll-like receptors (TLRs), and the production of interferon (INF-g), interleukin (IL)-8, IL-12, IL-1 and matrix metalloproteinases (MMPs) by keratinocytes resulting in the follicular hyperkeratinization. 55 Gastrointestinal dysfunction and emotional stress also contribute to acne lesions.53,56,57
Even though the cutaneous microbiome’s role in acne has been elucidated, the gut microbiome’s role in this inflammatory sebaceous unit disease’s pathophysiology has not been primarily studied. Four articles investigated the relationship between gut and acne and found significant differences between healthy controls and acne patients’ microbiota.19-22
Deng in 2018 found patients with acne have a low abundance of Firmicutes and increased Bacteroidetes. Even though the article stated that this is consistent with the Western diet’s enterotype,19 another study opposes that finding. Gupta et al. in 2017 stated that there are low microbial diversities in the Western urban industrialized population, with enriched taxa of Bacteroides, Bifidobacterium, Ruminococcus Blautia, and Dorea (Actinobacteria, Firmicutes, Rickenellaceae).58 These findings demonstrated the complexity of the gut microbiome mechanism in modulating acne. A Western diet includes increased food consumption with a high glycemic index, red meat, dairy, and egg protein. This diet increases sebogenesis, providing an optimal environment for P. acnes and other microorganisms to thrive, resulting in acne formation. 59
Firmicutes are involved in fat production. The increased phylum Firmicutes in obese patients also revealed the evidence that they have a low incidence of acne. This finding is in accordance with Deng et al. in 2018, which reported that Firmicutes were lower in patients with acne than the control group. However, this outcome contrasts with Thompson et al.’s in 2020 findings. They demonstrated that the high Firmicutes were obtained in acne patients before antibiotic treatments.19,21
Figure 1.
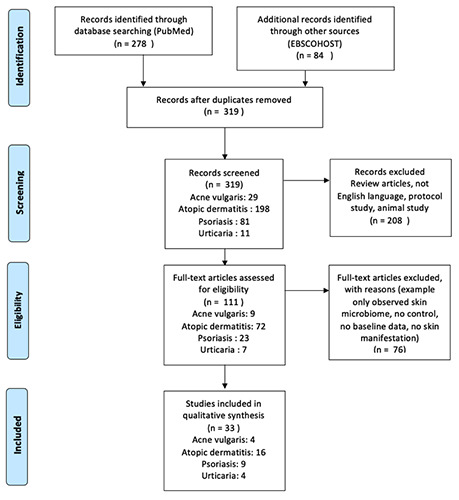
Flow diagram of review using Preferred Reporting Items for Systematic Reviews and Meta-Analyses.18
Table 2.
Diversity of gut microbiota in atopic dermatitis compared to controls.
| Type of study | Increase | Decrease | References |
|---|---|---|---|
| Culture and identification of bacteria in intestinal microbiota from fecal samples of 116 infants. Bacteria were identified through gram staining and biochemical/genetic tests. The authors found a gradual increase in the proportion of gram-negative relative to gram-positive bacteria with age because of the early gram-positive anaerobic colonizers. They did not show the detail of diversity abundance. | Escherichia Coli; Staphylococcus aureus; Bacteroides species; Clostridium species | Klebsiella; Coagulase-negative staphylococci; Enterococcus species; Bifidobacterium; Lactobacillus | Adlerberth, 201737 |
| DNA was extracted from stool samples, and 16S rRNA sequencing was performed to analyze the subjects' gut microbiome profiles. 24 infants with eczema detected have less-abundant genera than 24 healthy controls | Bifidobacterium | Chan, 202033 | |
| These subject at the ages of 9-12 months included 11 healthy controls and seven patients with atopic dermatitis. Fecal samples were analyzed by liquid chromatography-tandem mass spectrometry to conduct taxonomic, functional, and pathway-based protein annotation. The authors found non-significant differences in the bacterial families. | Bifidobacteriaceae; Erysipelotrichaceae; Prevotellaceae | Lactobacillaceae; Streptococcaceae; Bacteroidaceae | Kingkaw, 202023 |
| Analysis of bacteriology of fecal samples from 21 infants using genetic probes. Among the high numbers of Bacteroides and E. coli were associated with the extent of atopic sensitization, they did not show the detail of diversity abundance. | Lactobacilli/Enterococci; Bifidobacterial; Bacteroides | Clostridium histolyticum | Kirjavainen, 200238 |
| The diversity of gut microbiota was increased in 12 infants with AD than 12 healthy infants at six months of age without statistical significance. The 16S ribosomal RNA gene sequencing approach was used to amplify for the V1 to V3 regions. | Clostridia; Escherichia coli; Bacilli | Lee, 201624 | |
| Extracted DNA from 129 six-month-old infants (66 healthy control subjects and 63 infants with AD) was amplified using barcoded primers targeting the V1-V3 region of the 16S rRNA gene. A simple comparison between control and AD samples revealed no differences in members of the gut microbiota. | Clostridium | Bifidobacterium; Akkermansia; Faecalibacterium | Lee, 201825 |
| The study groups comprised 21 toddlers with eczema and 28 age-matched healthy controls. The bacterial counts and identification of the various species were based on growth, color, and the number of colonies on selective media, Gram staining, and cell morphology. A particular count was done using 16SrRNA-specific probes and FISH. This study observed reduced and different patterns of interbacterial relationships in children with eczema compared to healthy controls. | Lactic-acid-producing bacteria; Enterococci | Bifidobacterium; Clostridium | Mah, 200634 |
| The authors analyzed the fecal microbiota composition of a group of young black African children aged 12 to 36 months old with and without AD (29 with AD and nine with control). Early morning feces from the children's diaper were collected and used for microbiome composition assessments using bacterial 16S ribosomal RNA. No significant differences were observed in diversity abundance. | Actinobacteria; Firmicutes; Proteobacteria | Bacteroidetes | Mahdavinia, 201727 |
| Eighty-three children were recruited: 36 with AD and 47 controls without AD. The diversity indices were not different between cases with and without AD. | Faecalibacterium, ClostridiumXI, Ruminococcus, Collinsella, Blautia, unclassified; Lachnospiraceae, Bacteroides | Prevotella; Xylanibacter; Streptococcus | Mahdavinia, 201826 |
| Stools were collected at the age of one-month-old from twelve infants from an at-risk birth cohort in a case-control manner. Clinical follow-up for atopic outcomes was carried out at the ages of 12 and 24 months. Microbial genomic DNA was extracted from stool samples and used for shotgun sequencing. The results revealed no significant difference in diversity and richness between control and eczema. | Escherichia | Bifidobacterium; Enterobacter; Bacteroidetes; Firmicutes; Lactobacillus; Veillonela | Oh, 201728 |
| Fecal samples from 132 infants were analyzed using pyrosequencing, including 84 healthy controls, 22 transient AD, and 26 persistent AD subjects. alpha and beta diversity results did not differ among the three groups | Actinobacteria; Streptococcus; Bifidobacterium; Akkermansia | Clostridium | Park, 202029 |
| The authors conducted a nested case–control study comparing fecal samples of 26 infants who became sensitized and developed eczema within the first year of life with 52 non-sensitized non-eczematous infants. The composition of the fecal samples was examined using PCR combined with denaturing gradient gel electrophoresis. The diversity of the microbiota was similar in cases and controls. | B. Breve (0.15) | B. bifidum; B. infantis/ B. longum; B. adolescentis | Penders, 200630 |
| The authors performed a comparative study of the gut microbiota profile of 19 AD children and 18 healthy individuals. Fecal samples were analyzed by real-time PCR and 16S rRNA targeted metagenomics. AD patients showed a significantly lower level of alpha biodiversity, according to the observed. | Sutterella (Alcaligenaceae); Bacteroides (Bacteroidaceae); Parabacteroides (Porphyromonadace); Oscillospira and Faecalibacterium (Ruminococcaceae, Clostridia Class) | Actinobacteria; Eggerthella (Coriobacteriaceae); Propionibacterium; (Propionibacteriaceae), Enterococcus (Enterococcaceae), Eubacterium (Erysipelotrichaceae), Actinomyces (Actinomycetaceae), Blautia, and Coprococcus (Lachnospiraceae) | Reddel, 201935 |
| The gut microbiome from 132 subjects, including 90 patients with AD, was analyzed using 16S rRNA gene and metagenome sequence analyses. No significant difference was observed in all AD microbiota's microbial diversity than that of all non-AD microbiota. | F. Prausnitzii | Lactobacillus; Bifidobacterium | Song, 201531 |
| Fecal samples from 93 volunteers were analyzed using 16S rRNA sequencing, including 44 patients with AD and 49 healthy control subjects aged 6–22. These results suggest that patients with AD had lower diversity than healthy control subjects significantly. | Porphyromonealaceae; Blautia; Parabacteroides; Bacteroides ovatus; Bacteroides uniformis; Prevotella stercorea | Clostridium | Ye et al., 202036 |
| The authors conducted a case-control study of 50 infants with eczema (cases) and 51 healthy infants (controls). We performed high-throughput sequencing for V3±V4 hypervariable regions of the 16S rRNA genes from the fecal gut material. Neither the bacterial richness nor diversity were significantly different between the two groups. | Enterobacteriaceae; Eschericia Shigella; Faecalibacterium prautzinii | Bifidobacterium bifidum; Bifidobacterium longum; Bifidobacterium fragilis | Zheng et al., 201532 |
Table 3.
Articles discussing relationship between gut and skin microbiome in psoriasis.
| Type of study | Increase | Decrease | References |
|---|---|---|---|
| The characterization of gut microbial in active psoriasis was compared with non-psoriatic control and found a significant difference in beta-diversity between two groups | Firmicutes; Actinobacteria; Blautia; Faecalibacterium | Bacteroides; Proteobacteria; Prevotella | Shapiro et al., 201943 |
| Fecal samples were taken from 35 psoriasis patients and 27 healthy controls and sequenced with 16SrRNA. The authors found significant differences between the two groups | Bacteroidetes; Actinobacteria | Firmicutes; Proteobacteria; Bifidobacterium | Huang, et al., 201841 |
| DNA extraction of the fecal sample was taken from 19 psoriasis patients and twenty healthy individuals. Psoriasis patients have low diversity and different relative abundance of bacterial taxa. | Actinobacteria; Firmicutes | Bacteroidetes; Proteobacteria; Faecalibacterium | Hidalgo-Cantabrana et al., 201940 |
| Fecal DNA bacteria from 52 plaque psoriasis patients were analyzed by 16s rRNA and 300 healthy individuals from the human microbiome project. The study found the psoriasis microbiome differs from the healthy population. | Akkermansia; Faecalibacterium; Ruminococcus | Bacteroides | Codoner et al. 201839 |
| Twenty-nine Psoriasis patient's fecal samples were compared with 33 healthy controls by quantitative PCR, and this study reports the lower abundance of bacteria from healthy controls. | E.coli | F. prausnitzii | Eppinga et al., 201644 |
| The 55 psoriasis patients and 27 healthy controls underwent testing with quantitative measures of their DNA fecal by 16s rRNA gene. They demonstrate changes in gut microbiome composition according to their psoriasis status. | Faecalibacterium; Blautia; Firmicutes; Proteobacteria | Bacteroides; Paraprevotella | Dei-Cas, 202045 |
| Fifteen psoriasis patients and 17 healthy control subjects collected their fecal samples of bacterial DNA sequenced using 16SrRNA pyrosequencing. The study found lower diversity in psoriasis patients compared to healthy controls. | - | Coprococcus; Akkermansia; Ruminococcus; Pseudobutyrivibrio; Bacteroidetes | Scher, 201442 |
| Fourteen psoriasis vulgaris patients and 14 health controls underwent testing of their fecal samples and sequenced by 16SrRNA. They report the slightly decreased diversity on psoriasis patients, though not significantly different. | Bacteroides; Enterococcus | Tenericutes; Verruromicobia; Tenericutes; Mollicutes; Akkermansia muciniphila | Tan, 201746 |
| Fecal samples of 20 psoriasis patients and 20 healthy control were sequenced with 16S rRNA and multiplex ELISA cytokine assessment. They reported an alteration in gut Firmicutes, but there is relative diversity in the F/B ratio. | Faecalibacterium | Oscilibacter; Roseburia | Yegorov, 202047 |
Yan et al. in 2017 reported a decrease in Actinobacteria and increased Proteobacteria in patients with moderate and severe acne. They also reported that Bifidobacterium, Butyrocicoocus, Coprobacillus, Lactobacillus, and Allobaculum were decreased at the genus level.20 The Proteobacteria phylum has many pathogenic bacteria such as E. coli, Salmonella, and Vibrio cholerae. Overgrowth of Proteobacteria resulting from the host’s inability to keep commensal Proteobacteria in a minor fraction and reduced resistance to colonization by the microbial community can further facilitate inflammation or invasion by exogenous pathogens.60 They contribute to the overgrowth of pathogen bacteria in the gastrointestinal, which results in proinflammatory cytokine stimulation and gastrointestinal dysfunction. Decreased levels of Actinobacteria and its genera turn into dysbiosis and induce proinflammatory cytokines leading to leaky gut.
Lactobacillus and Bifidobacterium are the most studied species of probiotics. Bifidobacterium and Lactobacillus induce regulatory dendritic cells and CD4+Foxp3+T cells (regulatory T cells), resulting in hyporesponsive B cells and T helper cells without apoptosis along with suppression of cytokines production. Thus, the less abundance of Bifidobacterium and Lactobacillus will result in cytokine production, including tumor necrosis factor- a and interferon-g. The reduction of these bacteria directly affects intestinal epithelial barrier function by decreasing intestinal permeability and enhancing abdominal epithelial resistance.61-63
Atopic dermatitis
Atopic dermatitis is a chronic relapsing eczematous rash, and it is often the first manifestation of the atopic march.64 As a chronic relapsing disorder, current treatment strategies must focus on controlling the symptoms rather than curing them. The clinical appearance of atopic dermatitis may vary depending on several factors, such as the dermatitis stage, the age of patients, skin color, and infection. Atopic dermatitis plaques may appear as blisters (acute), thickened skin with increased skin markings (chronic), or a mixture of both (subacute). The preference for clinical manifestation is varied in every span of age. At infancy, the lesions appear mainly on the face, neck, and extremities’ extensor surface. Contrarily, the flexor surface of the extremity is a common clinical finding in childhood and adulthood. However, the clinical features of AD can be deconstructed into seven general categories, which are inflammation, infection, irritation, itch, ichthyosis (dry skin), immunological influences, and impeding comorbid conditions.64
Even though it is intriguing to focus on the skin when studying AD as the main pathology site, the complexity of immunological changes occurs. It connects with other organs like the gut and lungs. Stimulatory antigens crossing these barriers can give rise to inappropriate immune stimulation. 65 Recent evidence has revealed that dysbiosis microbiota in the human body is associated with AD.66 It has become increasingly evident that gut microbiota play a vital role in regulating innate and adaptive immunity and developing allergic diseases.67
The reduced intensity and diversity of microbes lead to abnormal immune maturation in early childhood. The lower diversity of microbiota could be related to a limited bacterial turnover, resulting in insufficient T helper (Th)1 cell induction and the failure to suppress Th2 responses. The switching of the immune stimulation towards a pronounced Th2-phenotype is a major mechanism to maintain allergy development.30,35 However, there were discrepancies with the findings of our review articles. The disagreement was probably due to inter-ethnic variations, age, and first feeding time.
Five articles reported the increased abundance of Bacteroides spp., which are common inhabitants of the human gut. However, their increased presence has been associated with food allergies and other atopic manifestations.27,35-38 Indeed, higher levels of Bacteroides in atopy could lead to the continuous production of lipopolysaccharides (LPS), the major component of the gram-negative cell wall, in the gut, which could trigger an inflammatory response. Moreover, Bacteroides species were reported to alter gut permeability.35 Zeng et al. in 2016 reported a decrease of Bacteroides fragilis in AD infants.32
Table 4.
Articles discussing relationship between gut and skin microbiome in urticaria.
| Types of study | CU patient vs. healthy control | References | |
|---|---|---|---|
| Increase | Decrease | ||
| Ten chronic urticaria (CU) patients compared with healthy controls of their fecal microbiota with 16SrRNA sequences. They found a highly different microbial composition between those two. | Proteobacteria Actinobacteria Enterobacteriales, Lactobacilales, Pseudomonadales Veillonella, Stterella, Streptococcus, Clostridium, and Escherichia | Bacteroidetes Faecalibacterium, Prevotella, Lachnobacterium; Faecalibacterium prausnitzii, Prevotella copri, Bacteroides fragilis, Bacteroides plebeius | Lu et al., 201948 |
| Twenty CU patients and 20 healthy individuals collected their fecal samples and samples are sequenced using bacterial PCR. The relative amounts of bacteria were different significantly. | Enterobacteriaceae | Akkermansia muciniphilia, Clostridium leptum, Faecalibacterium prausnitzii | Nabizadel et al., 201749 |
| The fecal sample of twenty CU patients and 20 match individuals were collected and analyzed their bacterial with PCR. They demonstrated the relative amount of Lactobacillus and Bifidobacterium were significantly higher in fecal samples from controls to CU. | Bacteroidetes (p>0.05) | Lactobacillus; Bifidobacterium | Rezazadeh et al. 201850 |
| A hundred chronic spontaneous urticaria and 100 healthy individuals were analyzed their fecal samples and sequence by 16S rRNA. The result shows alterations in gut microbes and metabolites. | Enterobacteriaceae Bacteroidetes (relative increase) Bacteroides | Firmicutes; Bifidobacteriales Faecalibacterium; Bifidobacterium; Ruminococcaceae | Wang et al., 202051 |
Bifidobacterium spp. are assumed to be beneficial for human health due to their effects on vitamin production, immune system stimulation, inhibition of potentially pathogen bacteria, and improvement of food ingredients digestion. Eight articles reported the reduction of Bifidobacterium in AD patients compared to control. This absence of Bifidobacterium in AD children could lead to a lack of anti-inflammatory effect.35
An increased abundance of E. coli was reported in two articles. However, studies on the role of E. coli in the intestine are lacking. One of the plausible hypotheses is that LPS originating from E. coli might play a role in regulating the immune system through the intestinal epithelial cells. Lee et al. in 2018 found a positive association between the relative abundance of E. coli and the percentage of blood eosinophils that play a critical role in developing AD and regulating immune homeostasis microbiota profiles in the intestine.25
Psoriasis
Psoriasis is a chronic inflammatory disease characterized by thickening of the skin, especially in the pressure site.68 Psoriasis is also considered a systemic disease, where the cardiovascular system, insulin homeostasis, psychomotor systems, and lipid metabolism are convoluted.69 This disease affects approximately 2-4% of the world’s population and is influenced by genetics, disorder of the immune system, epigenetic, and environmental factors.43,70
The pathogenesis of psoriasis involves sustained inflammation that leads to uncontrolled keratinocyte proliferation and dysfunctional differentiation.71 T cells are crucial in the pathogenesis of psoriasis, especially the critical role of Th17 cells and, recently, the role of gut and skin microbiome mediator in psoriasis. Naïve T cells are activated by antigen-presenting cells (APC) in the epidermis, which promotes naïve T cells to differentiate into T-helper (Th)1 and Th17, then Th1 and Th17 cell increase while Th2 and regulatory T cells (Treg) decrease. The activated T cells migrate from lymph nodes to the skin, where they are stimulated to produce numerous cytokines. These cytokines interact with the resident epidermal and dermal cells and cause changes, including keratinocyte proliferation and epidermal thickness. Environmental factors appear to induce inflammatory diseases in individuals with latent genetic susceptibility. Diet, microbial infections (virus, bacteria, fungi), chemical irritants or ultraviolet (UV) radiation exposure, and bad habits (drinking and smoking) are environmental factors that could induce inflammation. The interplay between genetic and ecological factors contributes to the onset, development, and present clinical symptoms of psoriasis.72
The gut-skin axis microbiome has been postulated to be involved in the altered immune response in psoriasis.69,73-77 Even though the finding of microbiota associated with psoriasis is conflicting, the studies revealed interesting and important changes in the gut microbiome led to the pathogenesis in psoriasis.
Firmicutes were observed as the most abundant microbiota in psoriasis and psoriasis arthritis and increased as seen in four studies.40,41,43,78 The result of a high level of Firmicutes is the disturbance the ratio of Firmicutes and Bacteroidetes. This imbalance then influences carbohydrate metabolism and alters the production of medium- and short-chain fatty acids (SCFA), resulting in increased acetate and reduced butyrate production, and finally leads to chronic inflammation and compromises the gut epithelial barrier. The lack of any gut epithelial barrier alters immune responses, both locally and systemically.43
Bacteroides produce SCFA such as propionates and butyrates, which are important in regulating colonic T cell regulators (Tregs) directly. Thus, the SCFA regulate intestinal homeostasis and control inflammation by limiting the proliferation of effector CD4+ T cells (Teff).79 There were conflicting data about the role of Bacteroides in psoriasis. Reduction of Bacteroidetes in psoriasis reported in other studies suggest lowering the SCFA.39,40,42,43,45 SCFA are the major metabolic products of anaerobic fermentation of glycans, where Bacteroides bind to complex recalcitrant glycan and stimulate the production of SCFA.45 The articles that demonstrated the high abundance of Bacteroides assumed the role of Bacteroides as a potential pathogen that promotes the pathogenesis of psoriasis.
Faecalibacterium prausnitzii is suggested to be causally related to the induction of psoriasis. Studies showed a reduction of F. prausnitzii linked to systemic inflammation in both skin and intestines due to the ability of F. prausnitzii on secreting anti-inflammatory molecules that modulate the host immune system.40 F. prausnitzii can regulate Th17/ regulatory T cell differentiation and reported consistently as butyrate producers in the intestine.45 However, other studies reported a high abundance of Faecalibacterium in psoriasis. 39,43,45,47 This dynamic role of Faecalibacterium spp. supported the evidence of these bacteria’s pathogenesis in psoriasis. The conflicting data are possibly because of different inclusion and exclusion criteria on recruiting psoriasis patients.
Akkermansia was identified as a mucolytic mucosa-associated bacterium in healthy subjects.42 The abundance of A. muciniphila was significantly reduced in patients with chronic inflammatory diseases, including obesity and Crohn’s disease. The bacterium converts mucin to SCFA acetate and propionate, thus activating host epithelial cells to produce antimicrobial peptides such as defensins and Ctype lectin that play key roles in homeostasis. 42,46
Chronic urticarial
Urticaria is commonly seen in dermatologic practice, clinically characterized by wheals, angioedema, or both. Of the 20% of individuals who experience acute urticaria, 0.1% will develop chronic symptoms, and most of them turn into chronic spontaneous urticaria (CSU).80 Chronic urticaria (CU) occurs when there are recurrent wheals that persist at a loss for six weeks. It divides into chronic spontaneous urticaria and chronic inducible urticaria (chronic physical urticaria).81,82 The pathogenesis of CU includes imbalances in immunity, inflammation, and coagulation. In CU, when the antigen is entering the body its binds to the IgE high affinity (FceRIa) Fc receptor located on the mast cells and stimulates circulating basophils in the skin, which induce degranulation of mast cells. When the same antigen is encountered for the second time, these IgE antibodies that were already presented on the mast cells and basophils immediately bind to the antigen and develop an allergic reaction more quickly.81
CU is also triggered by autoimmunity. The relationship of gut microbiota affects CU’s immune system has been studied in these past years. Some authors hypothesized that these immune changes in CU are also involved in gut microbiome dysbiosis. There was a significant difference in microbial composition in CU or CSU patients compared to healthy controls in all articles. 48-51 Enterobacteria, Bacteroidetes, Firmicutes, and Bifidobacteriales, could be markers for inflammatory diseases. All articles found a higher abundance of Enterobacteria in CU. Studies from Lu et al. in 2019 demonstrated a reduced number of Bacteroidetes and an increase in Proteobacteria. This result was following the research conducted by Polkowska- Pruszynska in 2020 in other inflammatory diseases such as asthma and dermatitis atopic.83 The anti-inflammatory bacteria such as Akkermansia muciniphila, Faecalibacterium prausnitzii, Clostridium leptum, genus Lactobacillus, and Bifidobacterium have shown significant prevention of other allergic and inflammatory diseases.83
The mechanisms of protective effects of those anti-inflammatory bacteria against CU can be due to the induction of regulatory T (Treg) cells by these bacteria. The intestinal microbiota composition could lower the pH level, modulate the integrity of the epithelial cell barrier, and regulate mucus secretion, which altered the function and expression of tight junction protein.49,50 The further studies are needed to reveal the pathway of this observation.
Conclusions
The gut microbiome in inflammatory skin disorders such as atopic dermatitis, acne vulgaris, urticaria, and psoriasis tend to be altered and imbalanced. However, the role of specific microbiota as the marker of skin inflammation still needs further study. The Bifidobacterium tends to decrease in all skin disorder reviewed in these articles and increases in E. coli and Proteobacteria are shown in inflammatory skin disorders. The role of Bacteroidetes and Firmicutes is still conflicting and inconsistent in many articles.
Acknowledgments
We acknowledge Yeniar Fitrianingrum for the excellent assistance with manuscript editing. Thank you to staff of Klinik Bahasa Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada (FKKMK UGM) Yogyakarta. This study is part of the doctoral research dissertation at the Faculty of Medicine, Public Health and Nursing (FKKMK UGM), Universitas Gadjah Mada, Yogyakarta Indonesia, and supported by Program Tugas Belajar, Provincial of Central Java Government.
Funding Statement
Funding: None.
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007;449:811-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murillo N, Raoult D. Skin microbiota: overview and role in the skin diseases acne vulgaris and rosacea. Future Microbiol 2013;8:209-22. [DOI] [PubMed] [Google Scholar]
- 3.Knight R, Callewaert C, Marotz C, et al. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet 2017;18:65-86. [DOI] [PubMed] [Google Scholar]
- 4.Schlaeppi K, Bulgarelli D. The plant microbiome at work. Mol Plant Microbe Interact 2015;28:212-7. [DOI] [PubMed] [Google Scholar]
- 5.Gill T, Brooks SR, Rosenbaum JT, et al. Novel Inter-omic Analysis Reveals Relationships Between Diverse Gut Microbiota and Host Immune Dysregulation in HLA-B27-Induced Experimental Spondyloarthritis. Arthritis Rheumatol 2019;71:1849-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrosino JF, Highlander S, Luna RA, et al. Metagenomic pyrosequencing and microbial identification. Clin Chem 2009;55:856-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Meulen TA, Harmsen H, Bootsma H, et al. The microbiome-systemic diseases connection. Oral Dis 2016;22:719-34. [DOI] [PubMed] [Google Scholar]
- 8.Qin Y, Wade PA. Crosstalk between the microbiome and epigenome: messages from bugs. J Biochem 2018;163:105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet 2012;13:151-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gevers D, Knight R, Petrosino JF, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol 2012;10:e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cresci GA, Bawden E. Gut Microbiome: What We Do and Don’t Know. Nutr Clin Pract 2015;30:734-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014;14:667-85. [DOI] [PubMed] [Google Scholar]
- 14.Cani PD. Human gut microbiome: hopes, threats and promises. Gut 2018;67:1716-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tralau T, Sowada J, Luch A. Insights on the human microbiome and its xenobiotic metabolism: what is known about its effects on human physiology? Expert Opin Drug Metab Toxicol 2015;11:411-25. [DOI] [PubMed] [Google Scholar]
- 16.Actis GC. The gut microbiome. Inflamm Allergy Drug Targets 2014;13:217-23. [DOI] [PubMed] [Google Scholar]
- 17.Arck P, Handjiski B, Hagen E, et al. Is there a ‘gut-brain-skin axis’? Exp Dermatol 2010;19:401-5. [DOI] [PubMed] [Google Scholar]
- 18.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Y, Wang H, Zhou J, et al. Patients with Acne Vulgaris Have a Distinct Gut Microbiota in Comparison with Healthy Controls. Acta Derm Venereol 2018;98:783-90. [DOI] [PubMed] [Google Scholar]
- 20.Yan HM, Zhao HJ, Guo DY, et al. Gut microbiota alterations in moderate to severe acne vulgaris patients. J Dermatol 2018;45:1166-71. [DOI] [PubMed] [Google Scholar]
- 21.Thompson KG, Rainer BM, Antonescu C, et al. Minocycline and Its Impact on Microbial Dysbiosis in the Skin and Gastrointestinal Tract of Acne Patients. Ann Dermatol 2020;32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainer BM, Thompson KG, Antonescu C, et al. Impact of lifestyle and demographics on the gut microbiota of acne patients and the response to minocycline. J Dermatolog Treat 2020:1-2. [DOI] [PubMed] [Google Scholar]
- 23.Kingkaw A, Nakphaichit M, Suratannon N, et al. Analysis of the infant gut microbiome reveals metabolic functional roles associated with healthy infants and infants with atopic dermatitis using metaproteomics. Peer J 2020;8:e9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee E, Lee SY, Kang MJ, et al. Clostridia in the gut and onset of atopic dermatitis via eosinophilic inflammation. Ann Allergy Asthma Immunol 2016;117:91-2.e1. [DOI] [PubMed] [Google Scholar]
- 25.Lee MJ, Kang MJ, Lee SY, et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol 2018;141:1310-9. [DOI] [PubMed] [Google Scholar]
- 26.Mahdavinia M, Rasmussen HE, Botha M, et al. Effects of diet on the childhood gut microbiome and its implications for atopic dermatitis. J Allergy Clin Immunol 2019;143:1636-7.e5. [DOI] [PubMed] [Google Scholar]
- 27.Mahdavinia M, Rasmussen HE, Engen P, et al. Atopic dermatitis and food sensitization in South African toddlers: Role of fiber and gut microbiota. Ann Allergy Asthma Immunol 2017;118:742-3.e3. [DOI] [PubMed] [Google Scholar]
- 28.Oh S, Yap GC, Hong P-Y, et al. Immune-modulatory genomic properties differentiate gut microbiota of infants with and without eczema. PLoS ONE 2017;12:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park YM, Lee SY, Kang MJ, et al. Imbalance of Gut Streptococcus, Clostridium, and Akkermansia Determines the Natural Course of Atopic Dermatitis in Infant. Allergy Asthma Immunol Res 2020;12:322-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penders J, Stobberingh EE, Thijs C, et al. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy 2006;36:1602-8. [DOI] [PubMed] [Google Scholar]
- 31.Song H, Yoo Y, Hwang J, et al. Faecalibacterium prausnitzii subspecies- level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol 2016;137:852-60. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H, Liang H, Wang Y, et al. Altered Gut Microbiota Composition Associated with Eczema in Infants. PLoS ONE 2016;11:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan CWH, Yuet Wa Chan J, et al. Altered Gut Microbiome and Environmental Factors Associated with Development of Eczema in Hong Kong Infants: A 4-Month Pilot Study. Int J Environ Res Public Health 2020;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mah KW, Björkstén B, Lee BW, et al. Distinct pattern of commensal gut microbiota in toddlers with eczema. Int Arch Allergy Immunol 2006;140:157-63. [DOI] [PubMed] [Google Scholar]
- 35.Reddel S, Del Chierico F, Quagliariello A, et al. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci Rep 2019;9:4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye S, Yan F, Wang H, et al. Diversity analysis of gut microbiota between healthy controls and those with atopic dermatitis in a Chinese population. J Dermatol 2020. [DOI] [PubMed] [Google Scholar]
- 37.Adlerberth I, Strachan DP, Matricardi PM, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol 2007;120:343-50. [DOI] [PubMed] [Google Scholar]
- 38.Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut 2002;51:51-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Codoñer FM, Ramírez-Bosca A, Climent E, et al. Gut microbial composition in patients with psoriasis. Sci Rep 2018;8:3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hidalgo-Cantabrana C, Gómez J, Delgado S, et al. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br J Dermatol 2019;181:1287-95. [DOI] [PubMed] [Google Scholar]
- 41.Huang L, Gao R, Yu N, et al. Dysbiosis of gut microbiota was closely associated with psoriasis. Sci China Life Sci 2019;62:807-15. [DOI] [PubMed] [Google Scholar]
- 42.Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol 2015;67:128-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shapiro J, Cohen NA, Shalev V, et al. Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J Dermatol 2019;46:595-603. [DOI] [PubMed] [Google Scholar]
- 44.Eppinga H, Sperna Weiland CJ, Thio HB, et al. Similar Depletion of Protective Faecalibacterium prausnitzii in Psoriasis and Inflammatory Bowel Disease, but not in Hidradenitis Suppurativa. J Crohns Colitis 2016;10:1067-75. [DOI] [PubMed] [Google Scholar]
- 45.Dei-Cas I, Giliberto F, Luce L, et al. Metagenomic analysis of gut microbiota in non-treated plaque psoriasis patients stratified by disease severity: development of a new Psoriasis- Microbiome Index. Sci Rep 2020;10:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan L, Zhao S, Zhu W, et al. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp Dermatol 2018;27:144-9. [DOI] [PubMed] [Google Scholar]
- 47.Yegorov S, Babenko D, Kozhakhmetov S, et al. Psoriasis Is Associated With Elevated Gut IL-1α and Intestinal Microbiome Alterations. Front Immunol 2020;11:571319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu T, Chen Y, Guo Y, et al. Altered Gut Microbiota Diversity and Composition in Chronic Urticaria. Dis Markers 2019;2019:6417471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nabizadeh E, Jazani NH, Bagheri M, Shahabi S. Association of altered gut microbiota composition with chronic urticaria. Ann Allergy Asthma Immunol 2017;119:48-53. [DOI] [PubMed] [Google Scholar]
- 50.Rezazadeh A, Shahabi S, Bagheri M, et al. The protective effect of Lactobacillus and Bifidobacterium as the gut microbiota members against chronic urticaria. Int Immunopharmacol 2018;59:168-73. [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Guo S, He H, et al. Gut Microbiome and Serum Metabolome Analyses Identify Unsaturated Fatty Acids and Butanoate Metabolism Induced by Gut Microbiota in Patients With Chronic Spontaneous Urticaria. Front Cell Infect Microbiol 2020;10:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J 2017;474:1823-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Liao W, Chao W, et al. Risk factors for sebaceous gland diseases and their relationship to gastrointestinal dysfunction in Han adolescents. J Dermatol 2008;35:555-61. [DOI] [PubMed] [Google Scholar]
- 54.Eichenfield LF, Del Rosso JQ, Mancini AJ, et al. Evolving perspectives on the etiology and pathogenesis of acne vulgaris. J Drugs Dermatol 2015;14:263-72. [PubMed] [Google Scholar]
- 55.Dreno B. What is new in the pathophysiology of acne, an overview. J Eur Acad Dermatol Venereol 2017;31:8-12. [DOI] [PubMed] [Google Scholar]
- 56.Bowe W, Patel NB, Logan AC. Acne vulgaris, probiotics and the gut-brainskin axis: from anecdote to translational medicine. Benef Microbes 2014;5:185-99. [DOI] [PubMed] [Google Scholar]
- 57.Bowe WP, Logan AC. Acne vulgaris, probiotics and the gut-brain-skin axis - back to the future? Gut Pathog 2011;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta VK, Paul S, Dutta C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front Microbiol 2017;8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maarouf M, Platto JF, Shi VY. The role of nutrition in inflammatory pilosebaceous disorders: Implication of the skingut axis. Australas J Dermatol 2019;60:e90-e8. [DOI] [PubMed] [Google Scholar]
- 60.Shin N-R, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 2015;33:496-503. [DOI] [PubMed] [Google Scholar]
- 61.Kwon HK, Lee CG, So JS, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA 2010;107:2159-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madsen K, Cornish A, Soper P, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 2001;121:580-91. [DOI] [PubMed] [Google Scholar]
- 63.Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNF-alphaand IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology 2006;130:731-46. [DOI] [PubMed] [Google Scholar]
- 64.Dinulos JG, Trickett A, Crudele C. New science and treatment paradigms for atopic dermatitis. Curr Opin Pediatr 2018;30:161-8. [DOI] [PubMed] [Google Scholar]
- 65.Thomas CL, Fernández-Peñas P. The microbiome and atopic eczema: More than skin deep. Australas J Dermatol 2017;58:18-24. [DOI] [PubMed] [Google Scholar]
- 66.Brüssow H. Turning the inside out: the microbiology of atopic dermatitis. Environ Microbiol 2016;18:2089-102. [DOI] [PubMed] [Google Scholar]
- 67.Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy 2009;39:518-26. [DOI] [PubMed] [Google Scholar]
- 68.Zákostelská Z, Málková J, Klimešová K, et al. Intestinal Microbiota Promotes Psoriasis-Like Skin Inflammation by Enhancing Th17 Response. PLoS One 2016;11:e0159539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komine M. Recent Advances in Psoriasis Research; the Clue to Mysterious Relation to Gut Microbiome. Int J Mol Sci 2020;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jinrong Zeng SL, Yumeng Huang, Qianjin Lu. Critical role of environmental factors in the pathogenesis of psoriasis. J Dermatol 2017:10. [DOI] [PubMed] [Google Scholar]
- 71.Rendon A, Schakel K. Psoriasis Pathogenesis and Treatment. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng J, Luo S, Huang Y, Lu Q. Critical role of environmental factors in the pathogenesis of psoriasis. J Dermatol 2017;44:863-72. [DOI] [PubMed] [Google Scholar]
- 73.Algazina T, Yermekbayeva B, Batpenova G, Kushugulova A. Features of microbiota in psoriatic disease: from skin and gut perspectives (review) Georgian Med News 2019:98-104. [PubMed] [Google Scholar]
- 74.Alesa DI, Alshamrani HM, Alzahrani YA, et al. The role of gut microbiome in the pathogenesis of psoriasis and the therapeutic effects of probiotics. J Family Med Prim Care 2019;8:3496-503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Langan EA, Griffiths CEM, Solbach W, et al. The role of the microbiome in psoriasis: moving from disease description to treatment selection? Br J Dermatol 2018;178:1020-7. [DOI] [PubMed] [Google Scholar]
- 76.Chimenti MS, Perricone C, Novelli L, et al. Interaction between microbiome and host genetics in psoriatic arthritis. Autoimmun Rev 2018;17:276-83. [DOI] [PubMed] [Google Scholar]
- 77.Eppinga H, Konstantinov SR, Peppelenbosch MP, Thio HB. The microbiome and psoriatic arthritis. Curr Rheumatol Rep 2014;16:407. [DOI] [PubMed] [Google Scholar]
- 78.Chen YJ, Ho HJ, Tseng CH, et al. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp Dermatol 2018;27:1336-43. [DOI] [PubMed] [Google Scholar]
- 79.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, shortchain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fine LM, Bernstein JA. Guideline of Chronic Urticaria Beyond. Allergy Asthma Immunol Res 2016;8:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kayiran MA, Akdeniz N. Diagnosis and treatment of urticaria in primary care. North Clin Istanb. 2019;6:93-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zuberbier T, Aberer W, Asero R, et al. The EAACI/GA(2) LEN/EDF/WAO Guideline for the definition, classification, diagnosis, and management of urticaria: the 2013 revision and update. Allergy 2014;69:868-87. [DOI] [PubMed] [Google Scholar]
- 83.Polkowska-Pruszyńska B, Gerkowicz A, Krasowska D. The gut microbiome alterations in allergic and inflammatory skin diseases - an update. J Eur Acad Dermatol Venereol 2020;34:455-64. [DOI] [PubMed] [Google Scholar]


