Significance Statement
Although podocyte detachment is a well-established factor driving the progression of glomerular kidney diseases, the underlying mechanisms initiating podocyte loss remain elusive. In particular, the co-occurrence of podocyte detachment and adaptive reinforcement of the actin cytoskeleton and integrin adhesion complexes presents a conundrum. The authors provide a comprehensive map of the podocyte adhesome and identify an actin-binding adhesome protein, α-parvin (PARVA), as a podocyte-specific mechanical linker. By employing a complementary approach involving both in vivo and in vitro models, they demonstrate that PARVA prevents podocyte detachment via mechano-adaptive remodeling of adhesion complexes. These observations suggest that insufficient linkage of a tensile actin cytoskeleton to integrin adhesion complexes is a causative mechanism in podocyte detachment in glomerular diseases.
Keywords: focal segmental glomerulosclerosis, glomerular epithelial cells, podocyte, proteinuria, integrin adhesion complex, IPP complex, PARVA, glomerular filtration barrier, PARVB, focal adhesion
Visual Abstract
Abstract
Background
The cell-matrix adhesion between podocytes and the glomerular basement membrane is essential for the integrity of the kidney’s filtration barrier. Despite increasing knowledge about the complexity of integrin adhesion complexes, an understanding of the regulation of these protein complexes in glomerular disease remains elusive.
Methods
We mapped the in vivo composition of the podocyte integrin adhesome. In addition, we analyzed conditional knockout mice targeting a gene (Parva) that encodes an actin-binding protein (α-parvin), and murine disease models. To evaluate podocytes in vivo, we used super-resolution microscopy, electron microscopy, multiplex immunofluorescence microscopy, and RNA sequencing. We performed functional analysis of CRISPR/Cas9-generated PARVA single knockout podocytes and PARVA and PARVB double knockout podocytes in three- and two-dimensional cultures using specific extracellular matrix ligands and micropatterns.
Results
We found that PARVA is essential to prevent podocyte foot process effacement, detachment from the glomerular basement membrane, and the development of FSGS. Through the use of in vitro and in vivo models, we identified an inherent PARVB-dependent compensatory module at podocyte integrin adhesion complexes, sustaining efficient mechanical linkage at the filtration barrier. Sequential genetic deletion of PARVA and PARVB induces a switch in structure and composition of integrin adhesion complexes. This redistribution of these complexes translates into a loss of the ventral actin cytoskeleton, decreased adhesion capacity, impaired mechanical resistance, and dysfunctional extracellular matrix assembly.
Conclusions
The findings reveal adaptive mechanisms of podocyte integrin adhesion complexes, providing a conceptual framework for therapeutic strategies to prevent podocyte detachment in glomerular disease.
The glomerular filtration barrier is organized as a three-layered filter, consisting of endothelial cells, the glomerular basement membrane (GBM), and specialized epithelial cells, named podocytes.1,2 Detachment of podocytes from the GBM has been identified as an unifying pathogenic motif in various glomerular pathologies.3,4 These observations and insights from genetic glomerular disease have initiated research to decipher the role of podocyte cell-matrix interactions in health and disease.5,6 Given the evolving complexity of the integrin adhesome,7–9 it is not completely clear how individual components of this multiprotein complex contribute to contextual signal integration and adaptive responses to ensure physiologic homeostasis.
Integrin-based heterodimeric adhesion molecules represent one of the largest classes of cell-matrix adhesion receptors. A multitude of adaptor, kinase, and linker molecules are recruited to the intracytoplasmic integrin portion, commonly termed as the integrin adhesome.10 Interestingly, mutations in either core (ITGA3) or adhesome-associated (CD151, ACTN4) genes have been identified as disease-associated mutations, prominently affecting podocyte function, resulting in severe glomerular pathologies.11–13 Using cell-type–specific proteomics and transcriptome analysis, we and others expanded the knowledge about prototypical adhesion receptor proteins within specialized glomerular cell types.14–18 These studies contributed to a concept of balanced reciprocal crosstalk between the adhesome and GBM composition.17,19 Podocytes are characterized by a highly complex cytomorphological architecture with arborizing cellular protrusions (commonly termed podocyte foot processes; FPs).19,20 Functional and genetic studies highlighted the relevance of titrated cytoskeletal activity and composition to ensure FP morphology and plasticity.21–23 One concept proposes that adaptive responses require a well-balanced interplay between cytoskeletal and adhesome components to ensure efficient mechano-transductive capacity. Previous work already described the pseudo-kinase integrin-linked kinase (ILK) as a component of the podocyte adhesome, and demonstrated the effect of ILK deficiency on GBM structure and podocyte differentiation.24,25 Recent in vivo and in vitro data confirmed that ILK instead exerts more structural scaffolding functions than the previously claimed direct kinase properties.26–28 ILK is recruited to integrin-based adhesion complexes (IACs) in a ternary complex consisting of Pinch and Parvin proteins (IPP complex). Parvin proteins (α, β, γ) interact with other adhesome proteins (including the IPP complex) and contain an actin-binding domain facilitating actin filament recruitment toward IACs.29–31 Global genetic deletion of Parvin-α (Parva) resulted in embryonic lethality.28
In this study, we identify the IPP component α-parvin (PARVA) as a central modulator of podocyte mechano-transductive signaling. We demonstrate that PARVA is essentially required for efficient podocyte cell-matrix adhesion and that inherent compensation within the IPP complex is involved in podocyte extracellular matrix (ECM) contextual responses. These observations provide the framework for a concept of cell-type-specific inherent adhesome modulation for adaptive signal integration in health and disease.
Methods
Animals
Conditional Parva knockout (KO) mice (Parvafl/fl*hNPHS2Cre) were generated by intercrossing of a previously described conditional allele for Parva (Parvafl/fl, generously provided by E. Montanez, University of Barcelona, Spain32) with a podocyte-specific Cre line (generously provided by L. Holzmann, University of Pennsylvania, School of Medicine, Philadelphia, USA33). The Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) reporter strain was purchased from JAX mice and crossed to hNPHS2Cre mice for isolation of primary podocytes. Respective colonies were maintained on a C57BL/6 mixed background. Animal models for nephrotoxic serum nephritis, Epb41l5fl/fl*Nphs1-rtTA-3G*tetOCre, and Nphs2R231Q/A286V were generated and described previously.34–36 All mouse experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the German law governing the welfare of animals. All studies were approved by the Regierungspräsidium Freiburg (G-17/127), Germany.
Cell Culture
Human immortalized podocytes (AB8/13) were generously provided by Moin A. Saleem, Bristol University, United Kingdom.37 Isolation of primary podocytes with genetically proven origin from Parvafl/fl*mT/mG*hNPHS2Cre and Parvawt/wt*mT/mG*hNPHS2Cre reporter mice was performed as described before.21 Podocytes were cultured in RPMI 1640 cell culture medium, supplemented with Glutamine (Thermo Fisher Scientific, 61870036), 10% FCS (Sigma-Aldrich, S0615), 10 µg/ml ITS (Sigma-Aldrich, 11074547001), 5 mM HEPES (Thermo Fisher Scientific, 15630–056), nonessential amino acids (MEM NEAA, Thermo Fisher Scientific, 11140050) 1:1000, 0.1 mM sodium-pyruvate (Thermo Fisher Scientific, 11360–039), and penicillin-streptomycin (Thermo Fisher Scientific, 15140122), according to standard human immortalized podocyte culture procedures at 33°C, 95% air, and 5% CO2.38 Human immortalized podocytes were cultured at 37°C for 8–10 days before seeding into the respective experiments. All functional experiments using podocyte cell lines were performed at 37°C. Primary podocytes were maintained at 37°C. Cells were tested negative for mycoplasma contamination on a regular basis by PCR (Mycoplasma PCR Detection kit, Hiss Diagnostics GmbH, Germany) and by Hoechst 33258 staining.
Antibodies
Antibodies used in this study are described in detail in Supplemental Table 1.
CRISPR/Cas9 Genome Editing
For generation of PARVA and PARVA/PARVB double KO (dKO) cells, the CRISPR/Cas9 genome editing technology was applied on human immortalized podocytes, as described before.38 Single guide RNAs targeting exons of the human PARVA or PARVB gene were designed using the web-based platform CHOPCHOP39: PARVA sgRNA-1: 5′- ATGGTCGAACAAGGTGTCAA(AGG) -3′, PARVA sgRNA-2: 5′- CTGGGAGTCGAATTGGTGCG(CGG) -3′, PARVA sgRNA-3: 5′- GTCTGGGGCATGGTCGAACA(AGG) -3′, PARVA sgRNA-4: 5′- CTTGGATTTGCCGAGACTGG(AGG) -3′, PARVB sgRNA-1: 5′- GCACGTCATTAATCCAGTCG(AGG) -3′, PARVB sgRNA-2: 5′- GATCGAAGCTTTCCGGAGTC(AGG) -3′. gRNAs were cloned in a modified lentiCRISPRv2 plasmid (TLCV2), which was a gift from Adam Karpf (Addgene plasmid 87360; http://n2t.net/addgene:87360; RRID:Addgene_87360).40 Control (wild-type; WT) clones were generated by using TLCV2 plasmids without gRNA sequence or nontargeting (nt) sequences (nt-sgRNA-1: 5′- GCGGGCAGAACGACCCTGAC -3″, nt-sgRNA-2: 5′- GAAGACGTGCTGGCGTCACC -3″).41 Lentiviral particles were produced in HEK293T cells according to standard procedures. Podocytes were transduced with lentiviral particles, followed by puromycin selection and doxycycline induction for 4 days. Single-cell clones were generated, screened for loss of PARVA and PARVB expression, and validated by western blot and immunofluorescence (IF) analysis. Monoclonal podocyte cell lines were generated from the polyclonal parental cell line and evaluated for differentiation capability (growth arrest, morphology), population-doubling time and podocyte cobblestone morphology. This monoclonal background was used for generation of PARVA WT-2 (nt-sgRNA-1), WT-3 (nt-sgRNA-2), KO-2 (PARVA sgRNA-3), KO-3 (PARVA sgRNA-3), and KO-4 (PARVA sgRNA-4) cell lines. In addition, the PARVA WT-1 (lentiCRISPRv2 without gRNA) cell line represents the polyclonal podocyte population and the PARVA KO-1 (PARVA sgRNA-1) clone was independently generated from this polyclonal background. The PARVA KO-2 cell line was transduced with nt-gRNA1&2 or PARVB gRNA1&2 to generate PARVA and PARVB dKO clones (dKO-1/-2/-3) or PARVA single KO (sKO-2) podocytes as control.
Expression Plasmids
The mEmerald-Parvin-C-14 plasmid was a gift from Michael Davidson (Addgene plasmid, 54214; http://n2t.net/addgene:54214; RRID: Addgene 54214). Human PARVB.pcDNA3.1+/C-(K)-DYK plasmids for PARVB transcript variant 1 (PARVB tv1; NM_001003828.2) and transcript variant 3 (PARVB tv-3; NM_001243385.1) were purchased from GeneScript (Hu22058 and Hu12475). PARVA and PARVB cDNAs were subcloned into a Flag-tagged or untagged pWPXLd plasmid, which was a gift from Didier Trono (Addgene plasmid, 12258; http://n2t.net/addgene:12258; RRID: Addgene_12258). The FLAG.pWPXLd plasmid was used as negative control for FLAG-PARVA.pWPXLd and an empty pWPXld plasmid for PARVB-tv1.pWPXLD and PARVB-tv3.pWPXLD. Lentiviral transduction of podocytes was performed according to standard procedures and protein expression was validated by western blot and IF analysis.
Western Blot Analysis
Whole-cell lysates were generated by cell lysis in RIPA buffer at 4°C for 15 minutes. Lysates were cleared by centrifugation at 16,000 g and 4°C for 15 minutes. After clearing, supernatants were mixed with Laemmli buffer (2×) with dithiothreitol, and denaturation was performed at 95°C for 5 minutes. Protein concentration was measured using the Pierce BCA Protein Assay Kit (23225, Thermo Fisher Scientific), following the manufacturer’s instructions. Equalized amounts of protein were loaded and separated by SDS-PAGE, following standard procedures. Western blotting was performed using the Trans-Blot Turbo Transfer System (BioRad) and appropriate polyvinylidene difluoride membranes (1704157, BioRad). Membranes were blocked in 5% BSA in Tris-buffered saline with Tween-20 (TBST) and primary antibodies (Supplemental Table 1) were diluted in TBST and incubated on membranes for 24 hours at 4°C. After washing in TBST, secondary HRP-linked antibodies (Supplemental Table 1) were applied for 1 hour. The HRP/ECL chemiluminescence detection method (32109, Thermo Fisher Scientific) in combination with a chemiluminescence imager was used for digital signal acquisition. Densitometry and quantification of blot bands from independent replicates was performed using Fiji ImageJ v1.52.
Scanning Electron Microscopy and Transmission Electron Microscopy Procedures
Preparation of kidney samples for transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were essentially performed as previously described.38 After perfusion and fixation (4% paraformaldehyde (PFA) + 1% glutaraldehyde in PBS for 24 hours at 4°C), samples were stored in 0.1 M sodium cacodylate buffer at 4°C until analysis. TEM samples were postfixed in 0.5% osmium tetroxide in ddH2O for 60 minutes on ice, and washed six times in ddH2O. The tissue was incubated in 1% uranyl acetate in ddH2O at room temperature for 2 hours. Dehydration was performed by 15-minute incubations in increasing concentrations of EtOH and finally acetone. After embedding in Durcupan resin, ultrathin sections were generated using a UC7 Ultramicrotome (Leica), collected on Formvar‐coated copper grids. Imaging was conducted using a Zeiss Leo 912 transmission electron microscope. Embedding, semithin sectioning, and electron microscopy was performed at the EM core facility of the Department of Nephrology, Faculty of Medicine, University of Freiburg. TEM images were analyzed for FP width and GBM thickness (FP to endothelial cell distance) using Fiji ImageJ v1.52, as previously described.35 For SEM, small tissue cubes were prepared and immersion fixed in 4% PFA and 1% glutaraldehyde in PBS for 3 days. Further dehydration was performed in ethanol (50%–100%, each 10% step for 1 hour at room temperature), and subsequently transferred to hexamethyldisilazan (Sigma, Schnelldorf, Germany). Gold sputtering was performed employing a Polaron Cool Sputter Coater E 5100. Samples were imaged using a scanning electron microscope (Leo 1450 VP scanning). SEM was performed at the EM core facility of the Department of Nephrology, Faculty of Medicine, University of Freiburg.
Microscopy
IF stainings were imaged using an inverted Zeiss Axio Observer microscope equipped with an Colibri 7 illumination system, Axiocam 702 mono camera, ApoTome.2 device, scanning stage, 100×, 63×, 40×, 20×, and 10× objectives, and 49 DAPI, 38 GFP, 43 HE dsRed, and 50 Cy5 filter sets (Carl Zeiss AG). Z-stacks were converted to maximum intensity projections using the ZEN software (ZEN 2012 SP1, Carl Zeiss AG). For iterative indirect IF imaging (4i), the scanning stage was used for sample positioning and repetitive imaging. For analysis of immunohistochemistry (IHC) and histology an inverted Zeiss Axio Imager microscope equipped with an Axiocam color and 40×, 63×, 100× objective and a Ventana DP 200 slide scanner (Roche Diagnostics Deutschland GmbH, Mannheim, Germany) equipped with a 20× and a 40× objective was used.
Histology
Preparation and processing of tissue samples, formalin fixation, paraffin embedding, and microtome sectioning were performed using standardized procedures at the Institute of Surgical Pathology, Faculty of Medicine, University of Freiburg. Mouse kidneys were fixed by 4% PFA in PBS after direct perfusion via the renal arteries, followed by further immersion fixation for 24 hours at 4°C.
For IF staining procedures, 2 µm sections of FFPE-tissue were generated, deparaffinized, rehydrated, and underwent established heat-induced antigen retrieval (HIAR; pressure cooker, 5 or 10 minutes, depending on the individual epitope, Supplemental Table 1). Blocking of sections was performed in 5% BSA in PBS for 1 hour, followed by primary antibody incubation in blocking solution for 24 hours. Subsequently, sections were washed in PBS and secondary fluorophore-tagged antibodies (Alexa Fluor Dyes, Thermo Fisher Scientific Inc.) were added for 45 minutes (in blocking solution). Hoechst 33342 staining was used for the visualization of nuclei (Thermo Fisher Scientific). ProLong Gold Antifade solution was used for final embedding (Thermo Fisher Scientific).
Samples for IHC were sectioned and processed as described for IF (except for performing additional peroxidase blocking for 15 minutes using 1% H2O2 after HIAR). HRP-linked secondary antibodies (Dako) were applied for 30 minutes in blocking solution. Visualization and staining were performed using the DAB+ Substrate Chromogen System (Dako) including counterstaining with hematoxylin. After dehydration, slides were mounted using Entellan.
Histology of FFPE-tissue sections was performed from deparaffinized sections. Periodic acid–Schiff reaction was performed by the Department of Pathology, Faculty of Medicine, University of Freiburg, according to standard procedures.
Histologic Assessment (Glomerular Sclerosis and Tubulointerstitial Damage)
Assessment of structural damage has been performed by applying semiquantitative scoring schemes on fully digitized whole slide image data. Level and extent of glomerular sclerosis was scored by employing a five-tiered score (0–4) as previously described.42 A minimum of 50 glomeruli per animal were analyzed and mean glomerular sclerosis score per animal was calculated. Interstitial fibrosis and tubular atrophy were assessed applying a four-tiered scoring system (0–3; 0, <5%; 1, 6%–25%; 2, 26%–50%; 3, >50%) adapted and on the basis of previously described approaches.43,44 Similar grading schemes were used for the assessment of interstitial immune cell infiltrates. At least four animals per genotype and time point were analyzed.
Quantification of Glomerular Podocytes
Estimation of glomerular podocytes was performed according to standard protocols.45 Kidney sections were stained for NPHS1, WT1, and Hoechst 33342, as described above. Glomerular cells with positivity for NPHS1 and WT1 were identified as podocytes. In addition, tuft areas and nuclei diameter were measured using Fiji ImageJ v1.52. Podocyte numbers per tuft area and glomerulus were calculated (≥20 glomeruli per animal were analyzed). Detachment into the urine was additionally visualized by WT1 staining of urinary SDS-PAGE blots. Urine samples were balanced according to respective creatinine concentration, mixed with 2x Laemmli buffer and heat denatured (5 minutes at 95°C). SDS-PAGE was performed as described above.
In vivo Quantification of Podocyte IAC Components
IF staining of individual IAC components was performed as described above. Kidney samples used for IF quantification were processed in parallel (including preparation, fixation, staining, and imaging) to ensure comparability. NPHS1 was used as a podocyte compartment marker for subsequent image analysis and segmentation using Fiji ImageJ v1.52. In brief, threshold levels were applied on NPHS1 IF signals to obtain a segmentation mask of the podocyte compartment for individual glomeruli. Mean fluorescence intensities (MFIs) of IAC components and NPHS1 were measured and acquired within this mask. In addition, line scan profiles of representative capillaries were measured for FERMT2-ILK and ITGB1-ILK costaining. The Duolink in situ Orange Starter Kit (Sigma-Aldrich) was used for proximity ligation assay (PLA), according to the manufacturer’s instructions. For PLA quantification, NPHS1 staining was used for further segmentation and the number of PLA positive dots was automatically counted within this mask of the podocyte compartment. At least 20 glomeruli per animal and marker were analyzed for mean fluorescence intensities of IAC components and 10 glomeruli for line scan analysis.
Super-resolution Microscopy (Podocyte Exact Morphology Measurement Procedure)
The evaluation of the filtration slit density was performed using a recently established super-resolution microscopy-based methodology (structured illumination microscopy, SIM; N-SIM [Nikon] with a 100× objective) termed as podocyte exact morphology measurement procedure (PEMP).21 The three-dimensional (3D) SIM (z-stack) images of slit diaphragms were colorized according to their position on the z axis. Filtration slit density values of 20 glomeruli in three mice per group were quantified.
Multiplex IF Staining and Imaging
The tyramide signal amplification based Opal detection method was used for IF staining of human kidney biopsy samples (Opal Polaris 7-Color Automation IHC Kit; Akoya Biosciences, Marlborough, MA, USA). Optimal Opal-antibody pairs were on the basis of previously optimized protocols in uniplex IF staining. Retrieval, primary antibody application, and detection with Opal fluorophores were performed according to the manufacturer’s instructions. In brief, slides underwent standardized antigen retrieval after prior deparaffinization (Tris-EDTA buffer, pH 9). After further blocking, primary antibody incubations were performed for between 30 minutes and 1 hour, or overnight depending on individual antibodies (on the basis of previous establishment in uniplex staining). Incubation with tyramide signal amplification dyes (10 minutes) was performed after further washing (Opal 7 kit: Opal 480, Opal 520, Opal 570, Opal 620, Opal 690, and Opal 780). Slides were counterstained with DAPI and mounted with ProLong Gold Antifade. Image acquisition was performed on a Vectra Polaris scanner (Akoya Biosciences, Marlborough, MA, USA) using 40× magnification and the implemented MOTiF scan. Further visualization and image analysis was performed on the Phenochart Version 1.1.0 software (Akoya Biosciences, Marlborough, MA, USA). Use of human kidney biopsy and tumor nephrectomy samples was granted by local ethics committee (Ethikkommission, Universitätsklinikum Freiburg) under the license EK512/18.
Iterative Indirect IF Imaging on Formalin-Fixed Paraffin-Embedded Tissue Sections
Iterative indirect IF imaging (4i) is a multiplex imaging technique recently developed by Gut et al. for in vitro samples.39 We have previously adapted this technique for multiplex staining of 2 µm formalin-fixed paraffin-embedded (FFPE) tissue sections, following the published protocol. In brief, FFPE tissue was sectioned, transferred on high precision microscopy glass slides (Ibidi, 10812), and processed for IF staining, as described above. HIAR was performed in Tris-EDTA buffer at pH 9 using a pressure cooker for 10 minutes. After HIAR, a staining/imaging chamber (Ibidi, 80828) was attached to the slide. All of the following buffers were prepared and applied as previously published.46 Elution and staining procedures were repeated for each antibody until required IF plexity for the sample was reached. Image registration was performed using the Fiji ImageJ v1.52 descriptor-based registration plugin and Hoechst 33342 staining for registration. After registration, fluorescence intensities were manually adjusted for proper labeling of positive cells (excluding nonspecific background and autofluorescence signal) and QuPath image analysis software was used for cell segmentation and image analysis.47 For 4i FFPE-tissue imaging, an automated microscope scanning stage and 20× objective was used for sample positioning and repetitive imaging.
In Situ Topological Adhesome Mapping
In situ topological adhesome mapping (ITAM) was utilized to describe the cell-type specific IAC composition in the human kidney. IAC specific antibodies were applied for IF analysis as described above. On the basis of IF signal levels, IAC components were classified as low or not expressed, expressed, or specifically enriched for each analyzed compartment. In addition, recently published murine podocyte RNA sequencing and glomerular proteome datasets were used for external validation of the podocyte ITAM dataset (MES, GEC, and podocyte proteome A, Hatje et al.; podocyte proteome-B, Schell et al.; podocyte proteome C, Rinschen et al.).14,48–50 IAC components were determined as “expressed” with a cutoff expression of >50 transcript per million on mRNA level or with a log2 fold-change cell-type/other glomerular cells between −0.5 and 0.5 on a proteome level. IAC components were determined as cell-type–specific enriched with a statistical significant log2 fold change cell-type/other glomerular cells >0.5 and >50 transcript per million on mRNA level, or a statistical significant log2 fold change podocyte/nonpodocyte >0.5 on proteome level. Proximity interactions of podocyte IAC components were recently published.9
Measurement of Urinary Albumin and Creatinine
Urinary albumin excretion was assessed as a direct indicator of glomerular barrier integrity. Albumin-creatinine ratios were quantified by measuring spot urine from WT and respective KO mice at defined time points. Albumin levels were determined by using a mouse specific albumin ELISA kit (ab108792, Abcam). Urinary creatinine measurements were performed by using an enzymatic creatinine kit (Creatinine PAP LT-SYS LT-CR 0106, Labor & Technik, Eberhard Lehmann, Germany). All assays were used according to manufacturer’s instructions.
Analysis of Murine Blood Serum
Collected serum samples were stored at −80°C until use. For the measurement of creatinine and urea enzymatic kits were used, following the manufacturer’s instructions (Creatinine PAP LT-SYS LT-CR 0106, Urea LT-UR 0010, Labor & Technik, Eberhard Lehmann, Germany).
IF Staining of Cultured Cells
For IF staining cells were cultured on collagen IV (Sigma-Aldrich, C5533) or (only if indicated) fibronectin (Corning, 354008) coated, eight-well chamber slides (Ibidi, 80827) or on circular RGD micropatterned slides (30 µm and 100 µm diameters) (Ibidi, 83851) at 37°C for 24 hours. Podocytes were washed in PBS with 1 mM CaCl2+ MgCl2+, fixed in 4% PFA (Electron Microscopy Sciences, 15714-S) in PBS (Thermo Fisher Scientific, 10010023) for 20 minutes, washed three times in PBS, and permeabilized applying 0.1% Triton X-100 in PBS for 3 minutes. HIAR in pH 9 Tris-EDTA buffer at 90°C for 40 minutes was performed for some antibodies as indicated in Supplemental Table 1. Cells were washed in PBS and subsequently blocked in 5% BSA in PBS for 1 hour at room temperature. Primary antibodies were diluted in blocking buffer and incubated at 4°C overnight. Samples were washed three times in PBS and fluorophore-tagged secondary antibodies (Alexa Fluor Dyes, Thermo Fisher Scientific) were diluted in blocking buffer and incubated for 45–60 minutes at room temperature. Nuclei were stained using Hoechst 33342 (Thermo Fisher Scientific, H3570) and F-actin using fluorophore-labeled phalloidin (Alexa Fluor, Thermo Fisher Scientific). Cells were washed six times with PBS and imaged in PBS. The 4i staining of fixed cells was essentially performed as described previously.46 Images were acquired by manual positioning of podocytes using a 40× objective.
Cell Spreading and Protrusion Formation Assay
Cell-spreading assays were performed as previously described.48 Briefly, podocytes were detached by trypsinization, three times washed in culture medium, counted, diluted to required concentration, and preincubated in culture medium (floating) at 37°C in a cell culture incubator for 20 minutes. Subsequently, podocytes were spread on collagen IV coated eight-well polymer coverslips (80822, Ibidi) for 30 minutes at 37°C. Coverslips were preincubated in culture medium in parallel to the cells. Podocytes were carefully washed with PBS (1 mM CaCl2+ MgCl2+), fixed in 4% PFA in PBS, permeabilized, and stained by fluorophore-labeled phalloidin (Alexa Fluor, Thermo Fisher Scientific). Fiji ImageJ v1.52 was used for analysis of cell morphology by cell segmentation via thresholding. At least 100 cells per condition and individual replicate were analyzed. The 3D protrusion formation of immortalized podocytes was performed as previously described.48 In brief, WT or pooled dKO (dKO-1,-2, and -3) cells were seeded in Matrigel (Corning), and incubated in 0.2% FCS podocyte culture medium containing 10 µM Y27632 (Selleckchem) for 24 hours at 37°C. Maximum intensity projections of z-stack images were generated and the “main” (longest) protrusion length was measured using Fiji ImageJ v1.52.
Cell-adhesion Assay
Cell adhesion was performed on different extracellular matrix components as previously described.38 Collagen IV (50 μg/ml, Sigma-Aldrich, C5533), collagen I (50 μg/ml, PureCol, Advanced BioMatrix, 5005), fibronectin (50 μg/ml, human fibronectin, Corning, 354008), basement membrane gel (50 μg/ml Matrigel, Corning, 356231), and laminin 521 or laminin 111 (5 μg/ml, Biolaminin LN, BioLamina, LN111, LN521) were used as indicated. Briefly, 24-well cell-culture plates were coated according to the manufacturer’s instructions, overnight at 4°C. Coated wells were washed in PBS and blocked with heat-denatured BSA (1% in PBS) for 1 hour, then washed again in PBS. Cells were trypsinized, counted, diluted in culture medium, and preincubated in a cell-culture incubator at 37°C for 20 minutes (floating). Equal amounts of cells were seeded for 15 minutes on the precoated 24-well cell-culture plates (tissue culture plastic control was uncoated and not blocked with heat-denatured BSA; BSA control was uncoated tissue culture plastic blocked with heat-denatured BSA). For comparison of different ECM components, dKO-1/-2/-3 clones were pooled before seeding. After carefully washing three times in PBS and fixation in 4% PFA, adherent cells were stained with 0.1% crystal violet in ddH2O for 1 hour. The dye was solubilized in 0.5% Triton X-100 in PBS, and absorbance was measured at 570 nm using a microplate reader.
IAC Analysis
Quantification and analysis of IACs was performed as previously described.38 Briefly, podocytes were seeded on 50 µg/ml collagen IV coated eight-well µ-slides (80827, Ibidi) and cultured for 24 hours at 37°C. Imaging and IF staining for the IAC components PXN, vinculin (VCL), PARVB, ILK, zyxin (ZYX), and F-actin (phalloidin) was performed as described above. PXN-ILK, VCL-PARVB, PXN-ZYX-phalloidin were costained on the basis of the host species of antibodies. For analysis of IACs, an image-processing macro for Fiji ImageJ v1.52 was used, as described before.21 In brief, individual IACs were segmented on the basis of PXN or VCL staining and analyzed for morphometric parameters and mean fluorescence intensities. In addition, the corresponding cell size was measured to calculate IAC density and the fraction of IAC covered basal surface area for individual podocytes. At least 30 cells per condition and independent experiment were analyzed. Data from PXN- and VCL-based segmentation were combined for analysis of morphometric IAC parameters.
IAC Mechano-linkage Assay
To assess the effect of extracellular mechanical preload, podocytes were cultured on collagen IV–coated PDMS substrates (CytoSoft, Advanced BioMatrix) with various rigidities (2 kPa, 16 kPa, and 64 kPa) and glass chamber slides (Ibidi). Previous work has shown that nocodazole stabilizes (force dependent) IAC maturation,51,52 whereas the ROCK inhibitor Y-27632 releases intracellular actomyosin tension on IACs.53 Moreover, these mechanisms were demonstrated to control tension-dependent adhesion stability in an ILK-dependent manner.54 Podocytes were precultured at 37°C for 24 hours on substrates with various rigidities, followed by addition of 20 µM nocodazole (S2775, Selleckchem), or 20 µM nocodazole and 10 µM Y-27632 (S1049, Selleckchem), or DMSO as a treatment control for 4 hours. Podocytes were stained by phalloidin and analyzed for cell morphology (cell collapse/rounding). At least 150 podocytes per condition, genotype, and replicate (total over 28,000 cells) were analyzed.
Analysis of Cell-derived Matrices
Synthesis and analysis of cell-derived matrices (CDMs) of podocytes were performed as essentially described before.38 In brief, podocytes were seeded for 7 days on gelatin-coated coverslips. After decellularization, CDMs were fixed in PFA and assayed by indirect IF staining for COL4A2 and fn1 as described above. Semiquantitative scoring of CDMs was performed on the basis of a five-tiered grading scheme, as previously described.38 Microscopy z-stacks were acquired using a 100× objective and processed to maximum intensity projections for image presentation.
RNA Sequencing
Isolation of glomeruli from 8- to 9-week-old male mice (with onset of proteinuria) was performed on the basis of perfusion with magnetic beads (Dynabeads), as previously described in detail.55 RNA was isolated from whole glomeruli using the Qiagen RNAeasy midi Kit (74104, Quiagen) according to the supplier’s instructions. Poly(A) selection, library preparation, and Illumina NovaSeq 6000 2 × 150bp paired-end sequencing was performed by GENEWIZ Germany. The NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs) was used. Demultiplexing and adapter trimming were performed with bcl2fastq v2.19. A total of 15–27 million reads per sample were sequenced. The Galaxy Europe platform was used for data analysis.56 The build in mus musculus (mm10) genome and Cutadapt v1.16.5, HISAT2 v2.1,57 featureCounts v1.6.4,58 and DESeq2 v2.11.40.659 tools were applied for read processing, mapping, and differentially expression analysis. Sequencing data have been deposited in NCBI Gene Expression Omnibus and are accessible via GEO series accession number GSE181690 (adhesome component analysis is enclosed in Supplemental Table 2 and differential expression analysis is enclosed in Supplemental Dataset 1).
Human Microarray Analysis
Human renal biopsy specimens were collected in an international multicenter study, the European Renal cDNA Bank-Kröner-Fresenius biopsy bank (ERCB-KFB60). Biopsies were obtained from patients after receiving informed consent, and with the approval of the local ethics committees. After renal biopsy, the tissue was transferred to an RNase inhibitor, and microdissected into glomeruli and tubulointerstitium. Total RNA was isolated from microdissected glomeruli, reverse transcribed, and linearly amplified according to a protocol previously reported.61 Published datasets of glomerular samples were analyzed for mRNA expression levels (GSE32591, GSE35489, GSE37463, GSE47185, GSE 99340). Analysis included gene expression profiles from patients with diabetic nephropathy (Glom: n=14), hypertensive nephropathy (Glom: n=15), minimal change disease (Glom: n=15), IgA nephropathy (IgA; Glom: n=27), FSGS (Glom: n=23), membranous nephropathy (Glom: n=21), lupus nephritis (SLE; Glom: n=32), ANCA-associated glomerulonephritis (Glom: n=23), and controls (living donors (Glom: n=41). CEL file normalization was performed with the Robust Multichip Average method using RMAExpress (Version 1.20) and the human Entrez‐Gene custom CDF annotation from Brain Array version 25 (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download.asp). To identify differentially expressed genes, the significance analysis of microarrays method62 was applied using the correct function in Multiple Experiment Viewer (TiGR MeV, Version 4.9). A q-value <5% was considered to be statistically significant. Correlation analyses (Supplemental Table 3) were performed using spearman correlations (SPSS 27.0, IBM Corp.). Bootstrapping was applied to obtain 1000 times resampling and derive the corresponding 95% confidence interval for the correlation coefficient. A P value <0.05 was considered statistically significant. In addition, the Nephroseq database (www.nephroseq.org) was used for analysis of ZYX, ACTN4, and TLN1 expression in human glomerular disease (“Ju CKD Glom” dataset).
Quantification and Statistical Analysis
If not stated otherwise, data are expressed as mean±SEM. Scatter dots indicating individual data points were used for statistical analysis. Statistic tests were used on the basis of data distribution and experimental design. Statistical tests and the unit of analysis used for each experiment are collectively shown in Supplemental Table 4. The GraphPad Prism 8 software was used for statistical analysis. Statistical significance was defined as *P<0.05, **P<0.01, ***P<0.001, and ****P< 0.0001, or non-significant (ns). The number of independent experiments and total amount of analyzed cells, mice, or samples are stated in the figures and/or figure legends.
Results
The IPP Complex Is a Central Constituent within the Podocyte Adhesome In Situ
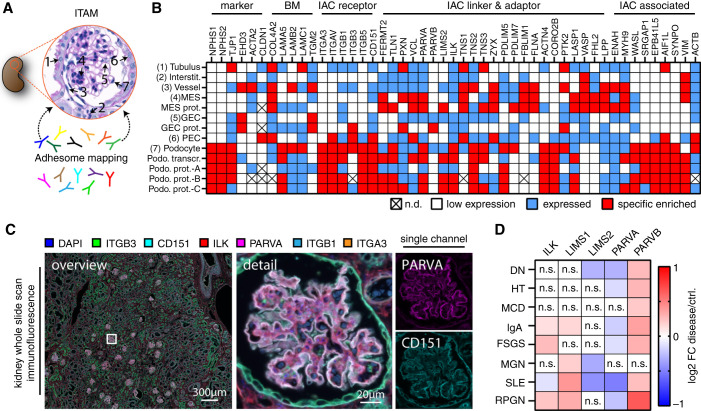
We combined large-scale antibody-based staining approaches with available transcriptomic and proteomic datasets to generate an in situ topological adhesome mapping (ITAM) of the human kidney with a focus on the glomerular compartment (Figure 1, A and B and Supplemental Figure 1). Localization patterns for the majority of selected epitopes correlated with cell-type specific transcriptome and proteome profiles (Figure 1B). These data highlighted a subset of proteins within glomerular podocytes, where the IPP complex appeared as a central motif interconnected with a multitude of other adhesome-related proteins (Supplemental Figure 2). Using multiplex imaging techniques, we validated ITAM profiles by showing a pronounced expression pattern of the IPP component PARVA in human glomeruli (Figure 1C and Supplemental Figure 2). Given the importance of podocyte adhesion, we analyzed the expression of IPP complex genes in a variety of glomerular disease entities. We observed a uniform reduction in PARVA mRNA levels, whereas PARVB appeared with increased abundance (Figure 1D). Decrease of PARVA and increase of PARVB mRNA levels significantly correlated with podocyte damage (as assessed by decrease of podocyte-specific marker genes, Supplemental Table 3).
Figure 1.
The IPP complex is a central constituent within the podocyte adhesome in situ. (A) and (B) Schematic depicting antibody-based in situ mapping of adhesome-related proteins within the human kidney (ITAM). On the basis of semiquantitative scoring, expression patterns were correlated to available cell-type–specific transcriptome and proteome data for podocytes (color code indicates semiquantitative scoring as low expressed, expressed, or specifically enriched; n.d., not detected; transcr., transcriptome; prot., proteome; podo., podocyte; MES, mesangial cells; GEC, glomerular endothelial cells; PEC, parietal epithelial cells; see Methods for classification criteria). (C) Multiplex immunostaining for selected adhesome proteins on human kidney tissue validated compartment-enriched expression patterns for PARVA in the glomerulus. (D) mRNA profiles of IPP complex genes in various glomerular disease entities demonstrated almost uniform decreased levels of PARVA mRNA, whereas PARVB appeared with increased levels (n.s., nonsignificant; color-coded heatmap on the basis of log2-fold changes between diseased tissue and healthy controls).
Loss of PARVA Results in a FSGS-like Glomerular Disease with Rapid Progression to ESKD
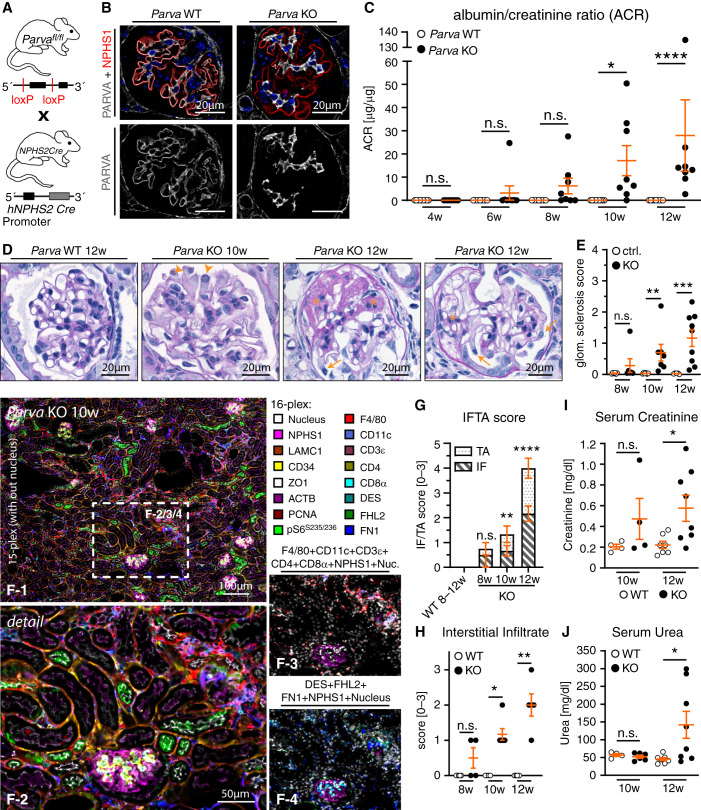
To test the relevance of PARVA in podocytes, we used a recently established conditional allele for Parva.32,63 We crossed the podocyte-specific hNPHS2-Cre line with a floxed allele for Parva and validated KO efficiency in situ (Figure 2, A and B). Interestingly, podocyte-specific KO did not affect glomerular morphogenesis, reflected by normal glomerular filtration barrier function until the age of 8–10 weeks. Evaluation of albumin-creatinine ratios demonstrated a sudden and fast progressive phenotype later in life, as increasing levels of proteinuria were detected until 12 weeks after birth (Figure 2C). Moreover, KO mice showed mesangial hypercellularity and extracellular matrix deposition with the onset of albuminuria. Within only 4 weeks, these histologic lesions progressed to a stage of either global or segmental sclerosis (Figure 2, D and E and Supplemental Figure 3). This severe glomerular damage also affected tubular and interstitial renal parenchyma. Employing multiplex IF imaging demonstrated a predominant presence of interstitial and periglomerular F4/80 positive macrophage populations (Figure 2, F–H and Supplemental Figure 4), whereas increased interstitial fibrosis and tubular atrophy were observed in KO animals (Figure 2, G and H). Podocyte-specific loss of Parva translated into an abrupt onset of FSGS-like glomerular disease and an accelerated progression to ESKD (Figure 2, I and J).
Figure 2.
Loss of PARVA results in FSGS-like glomerular disease with rapid progression to end-stage-kidney disease. (A) Schematic indicating the crossing strategy for the generation of podocyte-specific conditional KO for Parva. (B) IF staining for PARVA and the podocyte-specific marker NPHS1 were used to validate efficient deletion of PARVA. Positive staining remained detectable within the mesangial and parietal cell compartment in respective KO animals. (C) Urinary albumin-creatinine ratio (ACR) measurements indicated significantly increased levels of proteinuria at the age of 10 weeks after birth (each individual dot represents one experimental animal; at least nine WT and eight KO animals were analyzed per time point). (D) and (E) Histologic evaluation of glomerular damage patterns employing Periodic acid–Schiff staining. At 10 weeks of age, only modest expansion of the mesangial compartment and incipient podocyte detachment was detectable (orange arrowheads indicate detaching podocytes). With persistent proteinuria, areas of FSGS were detectable (orange asterisks) at 12 weeks of age, accompanied by prominent podocyte swelling (orange arrows). Assessment of glomerulosclerosis confirmed progressive glomerular damage resembling FSGS-like conditions (each dot represents one individual experimental animal; at least five WT and six KO animals were analyzed per time point). (F) Employing multiplex IF imaging infiltration of individual immune cell populations was evaluated in Parva KO and WT mice at 10 weeks of age. Here a prominent infiltration of F4/80 positive macrophages in interstitial and periglomerular regions was detected. Insert f-3 selectively depicts immune cell infiltrates and insert f-4 mesenchymal structures/markers (see Supplemental Figure 4 for single channels and WT). Color coded index indicates individually assessed markers by multiplex imaging: NPHS1, podocytes; LAMC1, basement membranes; CD34, endothelial cells; ZO1 (TJP1), epithelial cells; ACTB, cellular actin cytoskeleton; PCNA, proliferating cells; pS6S235/236, mTOR pathway activation; F4/80, macrophages; CD11c, conventional dendritic cells; CD3ɛ, T lymphocytes; CD4, regulatory T lymphocytes, CD8α, cytotoxic T lymphocytes; DES, Desmin intermediate filament rich cells (e.g., mesangial cells and fibroblasts); FHL2, mesangial cells and fibroblasts; fn1 (Fibronectin), extracellular matrix. (G) and (H) Semiquantitative assessment of structural damage in terms of tubular atrophy and interstitial fibrosis correlated to the progressive nature of glomerular disease (bar charts present mean values; at least four animals were analyzed per time point; dots present individual animals). (I) and (J) Analysis of serum creatinine and urea levels reflected the observed structural tissue damage in respective Parva KO animals (each individual dot represents one experimental animal; at least four animals were analyzed per time point). *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001, n.s., nonsignificant.
PARVA Prevents Podocyte Detachment from the GBM
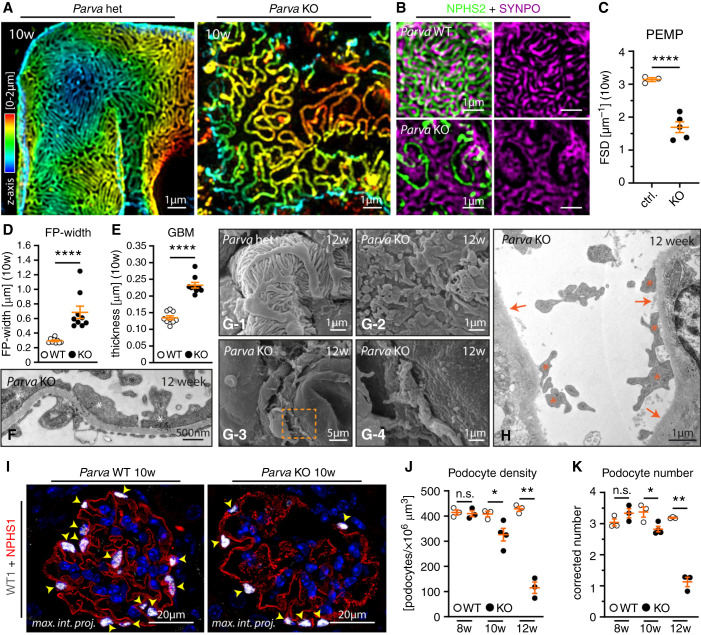
The association of PARVA with IACs implies that podocytes might detach from the GBM translating into the observed phenotype. We used super-resolution microscopy and established ultrastructural analysis via electron microscopy to characterize the effecrt of PARVA on podocyte morphology. Here, we observed that with the onset of proteinuria, respective KO animals presented a more simplified podocyte morphology, including significant retraction of podocyte FPs (Figure 3, A–F and Supplemental Figure 5). Detachment of podocytes from the GBM became increasingly apparent with ongoing disease progression, as demonstrated by SEM and TEM (Figure 3, D–H). At the age of 10–12 weeks, loss of PARVA was characterized by a denudation (Figure 3, G and H and Supplemental Figure 5) and thickening of the GBM (Figure 3E and Supplemental Figure 5). We quantitatively evaluated the glomerular numbers and density of podocytes using IF microscopy. Quantitative analysis showed podocyte numbers decreased at 10 weeks after birth. This trend progressed even further with increasing age (Figure 3, I–K and Supplemental Figure 5). Interestingly, the decrease of podocytes per glomerulus was accompanied by a slight increase in glomerular volume, implying that podocytes exert buttress forces on the underlying glomerular capillary convolute (as also recently described36). Altogether, these data demonstrate that a specific loss of PARVA in glomerular podocytes results in an accelerated loss of podocytes with overt denudation of the GBM.
Figure 3.
PARVA prevents podocyte detachment from the GBM. (A–C) 3D-SIM in respective KO animals identified a reduced filtration slit density (FSD) indicative for aberrant FP architecture (SIM data of 3 control: two WT, one heterozygous, and five KO animals were quantified by PEMP). Z-axis scales of 3D-SIM were color coded as indicated. (D–F) TEM of respective KO and WT animals demonstrated significant retraction and simplification of podocyte FPs in podocytes from KO mice (FP effacement, white asterisk). These changes were accompanied by rarefication of slit diaphragm density (three animals per time point and genotype were analyzed for mean FP width and GBM thickness; dots indicate individual glomeruli; see Supplemental Figure 5 for additional images of WT and KO animals). (G) and (H) SEM and TEM studies at 12 weeks of age showed a pronounced denudation of the GBM due to ongoing podocyte detachment (orange dotted squared box indicates areas of zoom-in; orange asterisks highlight residual fragments of persistently linked podocyte FPs to the GBM; orange arrows indicate areas of completely denuded GBM). (I) IF microscopy employing NPHS1 to highlight the podocyte compartment, and WT1 as a specific nuclear marker for podocytes (yellow arrowheads mark individual podocyte nuclei in WT and respective KO animals). (J) and (K) Quantification of podocyte numbers at 8, 10, and 12 weeks after birth revealed a progressive decrease in parameters such as podocyte density (J) and corrected podocyte numbers (K) (each individual dot represents one experimental animal, at least three animals per time point and genotype were analyzed). *P<0.05, **P<0.01 and ****P<0.0001.
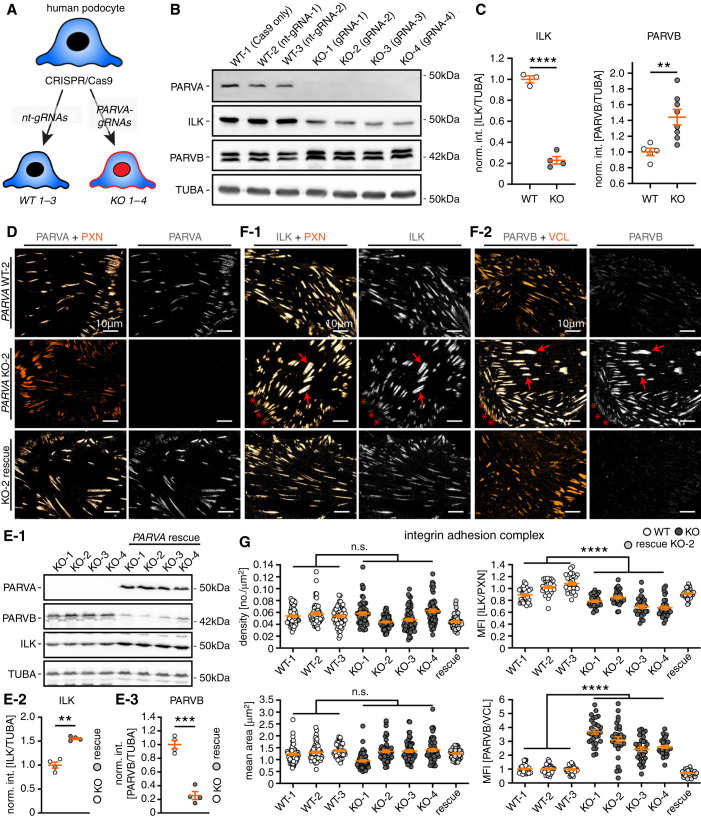
Parvin Proteins Exhibit Compensatory Recruitment toward IACs
To further characterize the mechanistic effect of PARVA on podocytes, we used CRISPR/Cas9 genome engineered podocytes (including nontargeting guide controls, and four individual PARVA gRNAs and KO clones). Initial validation of these cell clones demonstrated the complete loss of PARVA and indicated degradation of ILK protein levels in respective KO clones (Figure 4, A–C). More interestingly, this reduction of ILK protein abundance was accompanied by increased levels of PARVB, indicative of an inherent compensatory mechanism within the IPP complex (Figure 4, B and C). Reintroduction and rescue of PARVA protein reversed this compensatory phenotype for degradation of ILK and upregulation of PARVB (Figure 4, D and E and Supplemental Figure 6). These observations indicated a potential effect on adhesome composition corresponding to respective PARVA protein levels. Therefore, we used a recently established multiplex IF imaging approach to evaluate the in situ distribution and localization patterns of selected IAC components.46 We observed a pronounced signal decrease for ILK at individual IAC sites, other core adhesome components such as ZYX, ACTN4, or paxillin (PXN) appeared unaffected (Supplemental Figure 6).
Figure 4.
Parvin proteins exhibit compensatory recruitment toward IAC. (A) Schematic describing the CRISPR/Cas9-based genome editing strategy to generate specific KOs for PARVA in immortalized human podocytes (nt-gRNA, nontargeting guide RNA; note, numbers 1–3 and 1–4 indicate respective clones for individual gRNAs). (B) and (C) Western blot analysis for PARVA, ILK, and PARVB confirmed complete abolishment of PARVA protein levels in respective individual KO clones from four independent gRNAs. In addition, ILK levels were significantly reduced whereas PARVB protein levels showed a compensatory increase in respective KO cells (dots represent individual cell lines; for PARVB two independent replicates for each cell line were combined for statistical analysis, int., intensity, norm., normalized to WT). (D) The specific localization of PARVA to the subcellular IAC compartment was demonstrated by costaining with PXN in either WT, PARVA deleted, or rescued KO cells (upper and lower panel, respectively). (E) Re-expression of a flag-tagged version of PARVA in PARVA KO 1–4 clones not only restored PARVA protein levels, but also reversed ILK decrease and PARVB compensatory responses (TUBA, tubulin alpha used as a loading control; dots represent individual cell lines; int., intensity; norm., normalized to KO). (F) IF imaging for ILK and PARVB localization patterns revealed only slightly decreased recruitment of ILK toward IACs accompanied by strongly increased recruitment of PARVB (red arrows indicated IACs with strong colocalization patterns for individual IAC proteins; PXN or VCL was used as IAC marker). (G) Morphometric analyses demonstrated that PARVA deficient cells do not exhibit significant alterations in numbers and mean size of IACs (individual dots represent 60 analyzed cells per clone; two independent experiments for morphometric measurements were conducted and 30 cells per clone and experiment were analyzed). Ratiometric imaging for ILK and PARVB mean fluorescence intensity (MFI) levels demonstrated significant differences in localization toward the IAC depending on PARVA (MFIs were normalized to the mean of WT clones; individual dots for ILK and PARVA MFI measurements represent 30 analyzed cells per clone of one representative experiment, respectively). Cells of WT and KO clones were pooled for statistical testing. **P<0.01, ***P<0.001, and ****P<0.0001.
Further evaluation of WT, PARVA KO, and respective rescue clones demonstrated a preferential redistribution of ILK toward centrally localized IACs. In contrast, IACs that are more peripheral showed reduced levels of ILK. More interestingly, the altered subcellular localization of ILK was characterized by a pronounced increase in PARVB signals at internally localized IAC sites (Figure 4F). Ratio- and morphometric imaging studies furthermore demonstrated the sole loss of PARVA had no significant effect on structural parameters of IACs (Figure 4G and Supplemental Figure 7), but instructed the recruitment of IPP complex components such as PARVB, thereby sustaining recruitment of ILK to podocyte IACs (Figure 4G and Supplemental Figure 6).
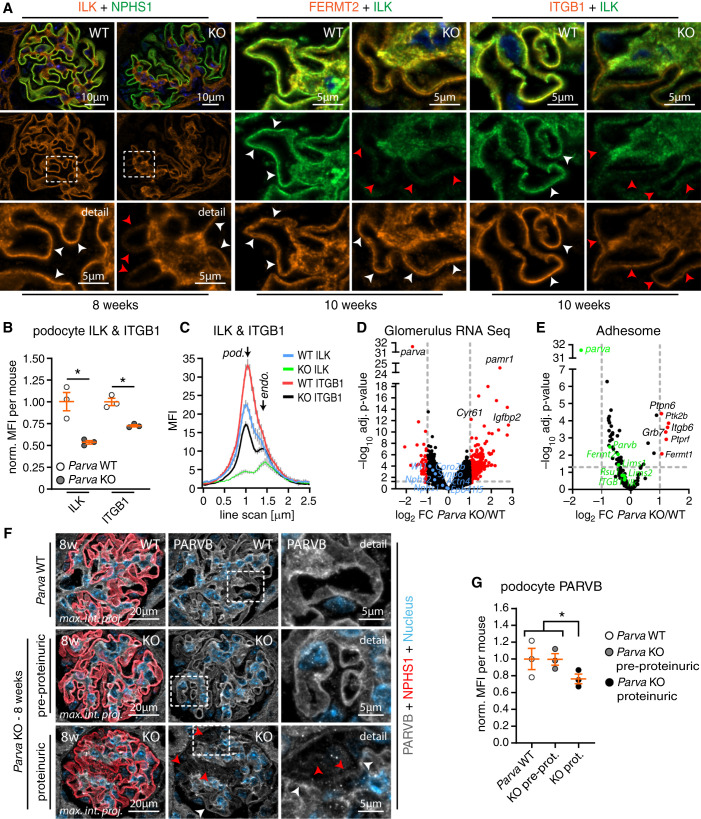
Loss of PARVA Induces Degradation of the IPP Complex In Vivo
The detailed analysis of our in vivo model revealed a sudden disease onset and rapid progression to FSGS due to the pronounced detachment of podocytes (Figures 2 and 3). However, further in vitro modeling highlighted compositional changes at individual IACs (Figure 4). Therefore, we aimed to translate these observations toward the more complex situation in vivo. Employing IF microscopy at different disease time points revealed that decreased signal intensities for ILK only became evident with disease onset, and even preceded changes for NPHS1 (as a highly sensitive marker for podocyte damage, Figure 5A). The heterogeneous pattern of ILK decrease within the capillary convolute might reflect the compensational nature of ongoing IPP dissolution under in vivo conditions. Interestingly, other core adhesome proteins such as FERMT2 were unaffected, overall supporting the specificity of this observed shift in adhesome composition in vivo (Figure 5, A–C and Supplemental Figure 8). To elucidate whether these early effects depend on transcriptional alterations or post-translational events, we isolated glomeruli from WT and Parva KO animals and performed RNA sequencing (Figure 5, D and E, Supplemental Table 2, and Supplemental Dataset 1). These analyses confirmed the very efficient deletion of Parva in respective KO animals (Figure 5D). Interestingly, a subanalysis of adhesome-related genes did not demonstrate pronounced alterations, further indicating that observed changes in ILK, ITGB1, and PARVB are related to post-transcriptional alterations (Figure 5E). This was furthermore supported by PLA showing decreased levels of ITGB1-ILK complexes at IACs within the podocyte compartment in situ (Supplemental Figure 8). Our previous in-vitro modeling approaches implied that PARVB partially compensates for the genetic deletion of PARVA (see Figure 4). Interestingly, PARVB signal levels within the podocyte compartment concomitantly decreased with the onset of albuminuria in respective KO animals, indicating that an intact IPP complex including PARVA/PARVB is required to maintain podocyte function (Figure 5, F and G).
Figure 5.
Loss of PARVA induces degradation of the IPP complex in vivo. (A) IF imaging for ILK, FERMT2, and ITGB1 in WT and Parva KO mice after onset of proteinuria (8 weeks and 10 weeks, respectively). FERMT2 showed preserved signal and localization patterns, whereas ITGB1 and more pronounced ILK were reduced in the podocyte compartment (white dashed boxes indicate zoomed-in areas in the lower panel, white arrowheads indicate preserved signal intensities, red arrowheads mark decreased signals). (B) Quantification of MFIs in the podocyte compartment showed a significant reduction in signal intensities of ITGB1 and ILK (NPHS1 staining was used to segment for the podocyte compartment; dots indicate mean values of individual animals; three WT and three KO animals and ≥20 glomeruli per animal were analyzed; see Supplemental Figure 13 for analysis of ILK, ITGB1, and NPHS1 per glomerulus; norm., normalized to WT). (C) Representative line scans demonstrated reduced signal intensities for ITGB1 and ILK in the podocyte compartment (MFIs of representative line scans of respectively 10 glomeruli from one WT and one KO animal are shown). (D) and (E) Volcano plots of bulk glomerular RNA-sequencing experiments in 8–9-week-old animals (with onset of proteinuria) confirmed the efficient deletion of Parva. Further subfiltering for podocyte adhesome genes did not detect significant downregulations of respective genes at this age (pooled analysis from three WT and three Parva KO animals; blue dots indicate podocyte-specific marker genes, green dots indicate IPP complex proteins and associated IAC components). (F) and (G) IF analysis of PARVB signal intensities in the podocyte compartment of respective 8-week-old Parva KO animals detected decreased PARVB levels in animals at the onset of proteinuria, but not in pre-proteinuric KO animals (dashed boxes highlight areas of zoom-in; red arrowheads indicate glomerular podocytes with reduced signals; white arrowheads highlight podocyte compartments with preserved signal level intensities; NPHS1 staining was used to segment for the podocyte compartment; dots indicate mean values of individual animals; three WT, three preproteinuric, and three proteinuric KO animals and ≥20 glomeruli per animal were analyzed; norm, normalized MFI to WT; nonproteinuric (WT and preproteinuric KO animals were pooled for statistical analysis). *P<0.05.
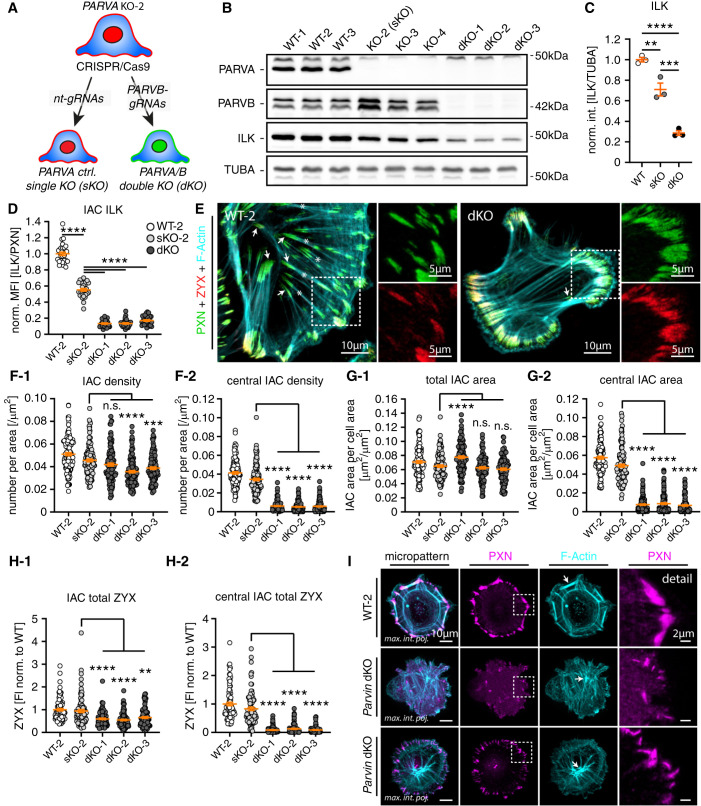
Dissolution of the Podocyte IPP Complex Translates into IAC Class Switch
The complementary analysis of in vitro and in vivo models for genetic deletion of PARVA implied compensational responses at the podocyte IAC. To dissect underlying mechanisms and functional consequences, we further modified our in vitro models by sequential deletion of PARVB in PARVA single KO (sKO) podocytes (Figure 6A and Supplemental Figure 9). We selected three double KO (dKO) clones, which demonstrated a further decrease in ILK protein levels when compared with sKO clones for PARVA or WT cells (Figure 6, B and C). These observations were validated by rescue cell lines for either PARVA or PARVB, and by analysis on the individual IAC ILK levels using IF microscopy, demonstrating an almost complete loss of ILK from IAC sites in dKO cells (Figure 6D and Supplemental Figure 9). Morphometric analysis revealed the overall density and area of IACs not to be consistently altered in all clones on combined deletion of PARVA and PARVB (Figure 6, E–G). However, dKO clones showed a pronounced change of IAC distribution patterns, where central IACs were almost completely absent (Figure 6, E–G and Supplemental Figure 9). These centrally localized IACs are commonly linked to ventral actin cytoskeleton stress fibers (VSFs) and are involved in cellular processes, such as mechano-transduction and force transmission.64,65 To better characterize mechano-linkage in dKO cells, we used ratiometric imaging using IF microscopy for F-actin, ZYX, and PXN. These experiments demonstrated that peripheral IACs in dKO cells have a more confluent morphology accompanied by dysbalanced recruitment of mechano-transductive adhesome components, such as ZYX and F-actin (Figure 6H and Supplemental Figure 9). As the development of VSFs is highly dependent on cellular morphology, we made use of micropatterned surfaces to synchronize cells. Under these harmonized conditions, respective dKO cells formed only punctuate and misaligned IACs at the cellular periphery, accompanied by a weakly interconnected and disordered actin cytoskeleton (Figure 6I and Supplemental Figure 10). Together, these observations demonstrate that PARVA/PARVB critically determine the assembly of IAC subclasses (peripheral versus central) and structure of the related actin cytoskeleton.
Figure 6.
Dissolution of the podocyte IPP complex translates into the IAC class switch. (A) Schematic describing genome engineering to generate control PARVA sKO-2 and dKO podocyte clones for PARVA and PARVB. (B) and (C) Western blot experiments confirmed the efficient deletion for PARVA and PARVB accompanied by progressive decrease in ILK protein abundance (individual dots indicate independent monoclonal clones). (D) Ratiometric analysis for ILK signal levels at individual IACs demonstrated progressive signal reduction, depending on PARVA and PARVB presence (dots represent individual cells; 30 cells per clone of one representative experiment were analyzed; MFIs were normalized to WT-2). (E) IF microscopy for the IAC marker PXN and Zyxin (ZYX) showed a pronounced shift in IAC classes. Control cells exhibited centrally localized IACs coupled to ventral F-actin stress fibers (white asterisks and arrows respectively; F-actin was stained by phalloidin). Respective dKO cells showed only feathered, peripherally localized IACs connected with thin actin fibers (white dashed boxes indicate zoomed-in areas). (F) and (G) Morphometric analysis and clustering of IAC size and distribution demonstrated a nearly complete loss of internal IACs in dKO cells (dots present individual cells derived from four independent experiments; 30 cells per experiment and clone, total of 120 cells per clone, were analyzed; sKO-2 cells were compared with individual dKO clones for statistical testing). (H) Ratiometric imaging indicated overall decreased levels of IAC bound ZYX related to nearly absent binding to central IACs in respective dKO cells (total IAC ZYX FI per cell was analyzed; dots represent individual cells derived from three independent experiments; 30 cells per experiment and clone (total of 90 cells per clone) were analyzed; sKO-2 cells were compared with individual dKO clones for statistical testing). (I) IF microscopy on space restricted micropatterned surfaces revealed diminished IAC formation in dKO cells accompanied by a diffuse and loosely interweaved actin network (white arrows indicate F-actin fibers; white dashed boxes indicate zoomed-in areas). **P<0.01, ***P<0.001, and ****P<0.0001.
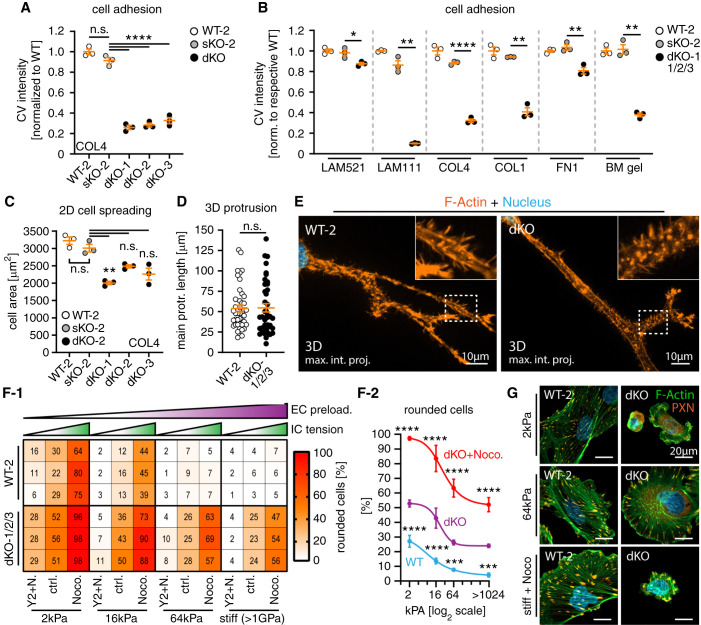
PARVA/B are Critically Required for Establishing Efficient Mechano-linkage
Integrin receptor engagement is one main axis in establishing efficient cell-matrix interactions involving outside-in and inside-out signaling. Evaluating cellular adhesion, dKO cells showed drastically reduced adhesion capabilities when compared with PARVA sKO cells and WT control cells (Figure 7A). Furthermore, we tested different ECM ligands and observed that adhesion defects were influenced in a ligand-dependent manner (Figure 7B and Supplemental Figure 10). However, overall expression levels of integrin receptors and adaptors appeared unaltered in dKO cells, whereas IAC patterns were disrupted for all tested integrin receptors (Supplemental Figures 10 and 11). Therefore, observed adhesion defects might be caused by reduced integrin receptor activation due to IPP complex disruption, than by modulation of specific integrin receptors.66 Next, we used cell-spreading assays to test if impaired adhesion in sKO and dKO podocytes translates into defective protrusion generation. However, loss of only PARVA or PARVA and PARVB exerted mild to moderate effects on cell spreading, most likely explained by delayed adhesion (Figure 7C and Supplemental Figure 12). In line with these observations, no obvious alterations in protrusion generation were detectable in dKO cells under 3D cell culture conditions (Figure 7, D and E). To finally test whether disruption of the IPP complex translates into impaired mechano-linkage in podocytes, we set up a complementary assay evaluating external matrix rigidity levels (external IAC preload) and internal cytoskeletal tension (internal IAC/cytoskeleton tension, Figure 7, F and G and Supplemental Figure 12). Integrative analysis of these two essential parameters allowed to deduce that mechano-adaptive capacities are significantly impaired on deletion of PARVA/B, further supporting the notion that the IPP complex is centrally involved in IAC-cytoskeleton coupling in podocytes (Figure 8, C-D). This model is furthermore substantiated by drastically impaired matrix assembly function in dKO cells (Supplemental Figure 12, a phenotype directly related to IAC-mechano-linkage38).
Figure 7.
PARVA/B are critically required for establishing efficient mechano-linkage. (A) and (B) PARVA/B dKO podocytes showed drastically reduced cell adhesion on specific ECM ligands (each dot presents one individual replicate per genotype, relative numbers of adherent cells were determined by crystal violet [CV] staining). (C) Cell spreading assays demonstrated only slightly impaired dynamic spreading capacity of dKO cells on collagen IV (COL4), whereas sKO-2 showed only mild differences to respective WT-2 controls (each dot presents mean size of one individual replicate and experiment per genotype; ≥100 cells per clone and experiment were analyzed). (D) and (E) Analysis of 3D protrusion generation in basement membrane gels (matrigel) showed no obvious alterations of protrusion formation of dKO podocytes (each dot presents one cell per genotype; dKO-1/2/3 clones were pooled for analysis; ≥40 cells per genotype were analyzed; white dashed boxes indicate zoomed-in areas; cells were stained for F-actin by phalloidin). (F) and (G) Podocyte morphology depending on sufficient mechano-linkage by IACs integrating extracellular (substrate) mechanical preload and intracellular cytoskeleton tension. Disruption of the IPP complex in dKO cells caused impaired adaptation to low substrate preload and high cytoskeleton tension reflected by morphologic collapse (rounding) and detachment of podocytes (values indicate percentage of rounded cells per individual replicate; three independent experimental replicates and ≥205 cells per condition and replicate were analyzed; EC, extracellular; IC, intracellular; Noco. or N., Nocodazole; Y2, Y-27632). Shown statistical significances (f-2) refer to comparison of WT-2 to dKO and dKO-nocodazole to dKO cells for indicated experimental conditions (three independent replicates per experimental condition). Images show representative experimental conditions and cells with collapsed (rounded) or spread morphology stained for F-actin and PXN (see Supplemental Figure 12 for overview images). Error bars indicate SEM (A)–(D) or SD for graph (G); *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
PARVA Controls Adhesome Composition and Mechano-adaptive ZYX Expression In Vivo
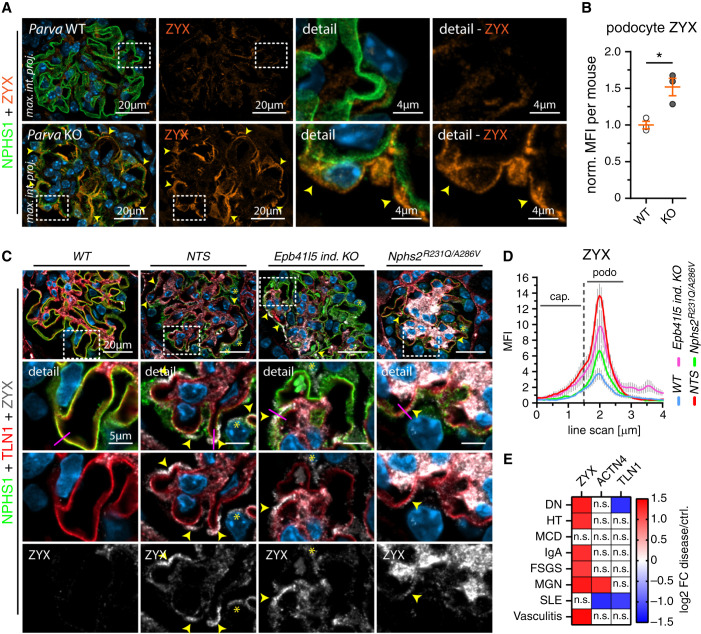
Given the rapid detachment phenotype in our mouse model, we wondered whether an IPP dependent IAC composition switch is also responsible for this significant loss of podocytes in vivo (Figure 3). Notably, we observed increased signal intensities for mechano-transductive IAC components such as ZYX in respective KO animals, whereas other adhesome components were either unaffected or exhibited reduced signal levels, such as integrin receptors (Figure 8, A and B, Figure 5, and Supplemental Figure 13). Moreover, analysis of murine models of glomerular disease and human disease datasets indicated ZYX expression as a common mechano-adaptive response in podocyte disease entities (Figure 8, C - E). Interestingly, ZYX expression was increased in IAC-triggered podocyte disease (Parva KO animals), but only modestly affected in (primarily not detaching) disease entities caused by slit diaphragm dysfunction (Nphs2R231Q/A286V).36 These findings suggest specific alterations of the podocyte IAC composition in vitro and in vivo caused on impaired IPP complex function.
Figure 8.
PARVA controls adhesome composition and mechano-adaptive ZYX expression in vivo. (A) and (B) IF analysis revealed altered patterns (yellow arrowheads) and increased intensity levels for ZYX in Parva deficient podocytes (10 weeks of age). Quantitative assessment of MFI in the podocyte compartment confirmed significant increase of ZYX levels (NPHS1 staining was used to segment for the podocyte compartment; dots indicate mean values of individual animals; three WT and three KO animals and ≥20 glomeruli per animal were analyzed; norm., normalized to WT; see Supplemental Figure 13 for analysis of ZYX per glomerulus). (C) and (D) Analysis of ZYX expression in murine disease models indicates upregulation of ZYX as a common pattern in podocyte disease. ZYX accumulation (yellow arrowheads) was detected slightly proximal of the SD and IAC layer as indicated by costaining of NPHS1 and TLN1, respectively (yellow asterisks indicate ZYX expression in the cytoplasm of podocytes; representative line scan analysis [purple lines] of 10 glomeruli per genotype was performed; error bars indicate SEM; NTS, nephrotoxic serum; cap., capillary; podo., podocyte). (E) Analysis of human glomerular disease (Nephroseq database) further substantiated increased expression on mRNA levels of ZYX in varying glomerular disease entities. *P< 0.05.
Discussion
Podocyte detachment is a unifying theme in a variety of glomerular diseases, ranging from genetic to acquired pathologies.67–69 However, it is still not clear whether dynamic compensation occurs at podocyte IACs, or how such modulations might affect efficient mechano-linkage to the GBM and influence the integrity of the glomerular filtration barrier.
In this study, a comprehensive topological in situ staining approach highlighted the central role and enrichment of the IPP complex within the podocyte compartment (Figure 1). Moreover, decreased mRNA levels of PARVA were observed in a variety of glomerular disease entities (Figure 1). Previous work already focused on the role of ILK in podocytes demonstrating that loss of ILK results in podocytopathy and proteinuria starting 2–3 weeks after birth.24,25 Of note, detailed analysis of our conditional KO model for Parva showed a relatively late onset of podocyte disease with albuminuria occurring not before 8–10 weeks of age (Figure 2). The phenotype in Parva-deficient animals rapidly progressed and exhibited classic histologic features resembling human FSGS (Figure 2). This difference in disease onset and progression between Parva- and Ilk-deficient animal models might be attributable to differing molecular function of PARVA in the IPP complex, involved in actin cytoskeleton linkage and IPP complex stabilization. However, also varying genetic backgrounds and/or differential compensation (e.g., via PARVB) at the podocyte IAC site might also influence these phenotypic differences.
One main finding of our attempts to model sequential disruption of the IPP complex is the observation that deletion of PARVA/PARVB resulted in a complete IAC class switch (Figures 4 and 6). Respective dKO cells only assembled peripheral adhesion sites, whereas central IACs linked to the ventral actin cytoskeleton were vastly reduced, compared with single KO cells for PARVA. This switch of IAC classes functionally translated into impaired cellular adhesion, indicating a defect in efficient mechanical linkage between IACs and the cytoskeleton (Figure 7). Although prior studies have observed that the sole deletion of PARVA already resulted in impaired cellular adhesion, altered cytoskeletal architecture, and adhesion site formation,32,63 compensatory upregulation of PARVB in podocytes largely prevented any overt cellular dysfunction. In fact, a counterbalancing regulation of Parvin proteins has been previously demonstrated.70
Integrating the observations from our in vitro studies raises the question of whether this podocyte loss is really caused by dissolution of receptor complexes or, rather, IAC functionality. Conventional ultrastructural studies and very recent super-resolution microscopy approaches described the formation of sarcomeric actin networks in conditions of podocyte foot process effacement.71,72 This adaptive cytoskeletal rearrangement is considered as a proxy for the formation of ventral actin cytoskeleton fibers (VSFs).64 The drastic reduction of VSFs in dKO podocytes due to an IAC class switch might therefore imply insufficient IAC-cytoskeleton coupling as the underlying mechanism for podocyte detachment observed in vivo (Figures 3, 6, and 7). The IAC components ZYX and α-actinin (e.g., ACTN4) have been shown to stabilize actin stress-fiber integrity, and thereby force transmission.73,74 Interestingly, recruitment of α-actinin to VSF occurred in a ZYX-dependent manner and previous studies in human and murine glomerular disease also documented increasing levels of ACTN4,75,76 which potentially reflect increased integrin cytoskeleton coupling to overcome impaired podocyte adhesion.77 In line with that, we discovered similar adaptive responses in IAC components (e.g., ZYX) in our in vitro and in vivo models, further supporting the concept of adaptive VSF and IAC remodeling in podocyte disease (Figures 7 and 8 and Supplemental Figure 13). Despite this significant adaptive response, loss of PARVA led to exaggerated podocyte detachment. These observations indicate insufficient linkage of reinforced tensile VSFs to IACs as a causative mechanism for podocyte detachment in glomerular disease (see also schematic Supplemental Figure 14).
In conclusion, in this investigation we provide combined in vitro and in vivo evidence for balanced compensatory events at podocyte IACs, ensuring the integrity of the glomerular filtration barrier. Future studies on specific compositional signatures in IACs might provide a conceptual framework for subclassification or substratification of glomerular pathologies and lead to the identification of potential therapeutic strategies to prevent podocyte detachment.
Disclosures
E. Montanez reports being supported by the Spanish Ministry of Science, Innovation and Universities (PID2019-108902GB-I00). G. Walz reports being a scientific advisor for or member of the Meona Group. N. Endlich declares that PEMP, which has been used for this manuscript, is registered for a patent; reports being among the founders of the start-up NIPOKA, which commercializes PEMP; and reports having an advisory or leadership role as a Plos One Editor. T. Benzing reports consultancy through advisory activity for Otsuka in the field of cystic kidney disease and hyponatremia; reports receiving research funding for the autosomal dominant polycystic kidney disease registry by Otsuka; reports receiving honoraria and travel support from Amgen, Hexal, Novartis, Otsuka, Roche, and Sanofi-Genzyme; and reports having an advisory or leadership role on the Editorial Board of JASN, Nephrology Dialysis Transplantation, and Science Signaling. T. Huber reports being a consultant for AstraZeneca, Bayer, Boehringer-Ingelheim, DaVita, Deerfield, Fresenius Medical Care, GoldfinchBio, Mantrabio, Novartis, and Retrophin; reports receiving research funding from Amicus Therapeutics, Fresenius Medical Care; reports receiving honoraria from AstraZeneca, Bayer, Boehringer-Ingelheim, DaVita, Deerfield, Fresenius Medical Care, GoldfinchBio, Mantrabio, Novartis, and Retrophin; and reports having an advisory or leadership role with Kidney International (Editorial Board); Nature Review Nephrology (Journal, Advisory Board). All remaining authors have nothing to disclose.
Funding
This study was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, SCHE 2092/1-2, SCHE 2092/3-1, SCHE 2092/4-1 [RP9, CP2, CP3] and CRU329, Project-ID 43198400– SFB1453 (to C. Schell), CRC1192– HU1016/8-2, HU1016/11, HU 1016/12-1 (to T. Huber), CRU329 (to T. Benzing), the Else Kröner-Fresenius-Stiftung (2016_Kolleg.03 to M. Rogg), the Federal Ministry of Education and Research (BMBF grant STOP-FSGS 01GM101C, 01GM1518B (to N. Endlich and T. Huber), Freiburg Institute for Advanced Studies (FRIAS) within the research focus MatrixCode (to C. Schell) and Else Kröner-Fresenius-Stiftung Matriglom (EKFS A_09 to C. Schell).
Supplementary Material
Acknowledgments
We would like to thank Katja Gräwe, Severine Kayser, and Charlotte Meyer for expert technical assistance. We thank Veran Drenic and Nihal Telli (NIPOKA) for excellent work regarding SIM imaging. The ERCB-KFB was supported by the Else Kröner-Fresenius Foundation. We also thank all participating centers of the ERCB-KFB and their patients for their cooperation. For active members at the time of the study see ref.78 In addition, we would like to express our gratitude to all members of our laboratories for helpful discussions and support.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Author Contributions
M. Rogg and C. Schell conceptualized the study; J. I. Maier, M. Lindenmeyer and M. Rogg were responsible for data curation; N. Endlich, J. I. Maier, M. Rogg, and C. Schell were responsible for formal analysis; C. Schell was responsible for the funding acquisition; M. Helmstädter, J. I. Maier, M. Rogg, A. Sammarco, C. Schell, C. Van Wymersch, and P. Zareba were responsible for the investigation; T. Benzing, N. Endlich, M. Lindenmeyer, E. Montanez, and M. Rogg were responsible for the methodology; T. Benzing, T. B. Huber, M. Lindenmeyer, E. Montanez, G. Walz, and M. Werner were responsible for the resources; C. Schell provided supervision; M. Rogg was responsible for the validation and visualization; and M. Rogg and C. Schell wrote the original draft and reviewed and edited the manuscript.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021101319/-/DCSupplemental.
Supplemental Figure 1. Representative stainings for ITAM analysis, corresponding to Figure 1.
Supplemental Figure 2. Illustration of the mapped podocyte adhesome, corresponding to Figure 1.
Supplemental Figure 3. Histological analysis of Parva KO mice, corresponding to Figure 2.
Supplemental Figure 4. 4i multiplex imaging of Parva KO mice, corresponding to Figure 2.
Supplemental Figure 5. 3D-SIM and TEM analysis of Parva KO mice, corresponding to Figure 3.
Supplemental Figure 6. Analysis of human PARVA KO podocytes, corresponding to Figure 4.
Supplemental Figure 7. Analysis of primary murine Parva KO podocytes, corresponding to Figure 4.
Supplemental Figure 8. IPP complex analysis in Parva KO mice, corresponding to Figure 5.
Supplemental Figure 9. Functional analysis of PARVA/B dKO cells, corresponding to Figure 6.
Supplemental Figure 10. Functional analysis of PARVA/B dKO cells, corresponding to Figures 6 and 7.
Supplemental Figure 11. Functional analysis of PARVA/B dKO cells, corresponding to Figure 7.
Supplemental Figure 12. Functional analysis of PARVA/B dKO cells, corresponding to Figure 7.
Supplemental Figure 13. IAC component analysis in Parva KO mice, corresponding to Figure 8.
Supplemental Figure 14. Schematic.
Supplemental Table 1. Antibodies.
Supplemental Table 2. RNAseq: Adhesome analysis of Parva KO mice, corresponding to Figure 5.
Supplemental Table 3. Correlation analysis, corresponding to Figure 1.
Supplemental Table 4. Statistical methods.
Supplemental Dataset 1. RNAseq of Parva KO mice.
References
- 1.Grahammer F, Schell C, Huber TB: The podocyte slit diaphragm--from a thin grey line to a complex signalling hub. Nat Rev Nephrol 9: 587–598, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Benzing T, Salant D: Insights into glomerular filtration and albuminuria. N Engl J Med 384: 1437–1446, 2021 [DOI] [PubMed] [Google Scholar]
- 3.Trimarchi H: Mechanisms of podocyte detachment, podocyturia, and risk of progression of glomerulopathies. Kidney Dis 6: 324–329, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriz W, Lemley KV: A potential role for mechanical forces in the detachment of podocytes and the progression of CKD. J Am Soc Nephrol 26: 258–269, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sever S, Schiffer M: Actin dynamics at focal adhesions: A common endpoint and putative therapeutic target for proteinuric kidney diseases. Kidney Int 93: 1298–1307, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lennon R, Randles MJ, Humphries MJ: The importance of podocyte adhesion for a healthy glomerulus. Front Endocrinol (Lausanne) 5: 160, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries JD, Chastney MR, Askari JA, Humphries MJ: Signal transduction via integrin adhesion complexes. Curr Opin Cell Biol 56: 14–21, 2019 [DOI] [PubMed] [Google Scholar]
- 8.Horton ER, Byron A, Askari JA, Ng DHJ, Millon-Frémillon A, Robertson J, et al. : Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 17: 1577–1587, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chastney MR, Lawless C, Humphries JD, Warwood S, Jones MC, Knight D, et al. : Topological features of integrin adhesion complexes revealed by multiplexed proximity biotinylation. J Cell Biol 219: 202003038, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michael M, Parsons M: New perspectives on integrin-dependent adhesions. Curr Opin Cell Biol 63: 31–37, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Has C, Spartà G, Kiritsi D, Weibel L, Moeller A, Vega-Warner V, et al. : Integrin α3 mutations with kidney, lung, and skin disease. N Engl J Med 366: 1508–1514, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karamatic Crew V, Burton N, Kagan A, Green CA, Levene C, Flinter F, et al. : CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood 104: 2217–2223, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, et al. : Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 24: 251–256, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Schell C, Rogg M, Suhm M, Helmstädter M, Sellung D, Yasuda-Yamahara M, et al. : The FERM protein EPB41L5 regulates actomyosin contractility and focal adhesion formation to maintain the kidney filtration barrier. Proc Natl Acad Sci U S A 114: E4621–E4630, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogg M, Yasuda-Yamahara M, Abed A, Dinse P, Helmstädter M, Conzelmann AC, et al. : The WD40-domain containing protein CORO2B is specifically enriched in glomerular podocytes and regulates the ventral actin cytoskeleton. Sci Rep 7: 15910, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda-Yamahara M, Rogg M, Yamahara K, Maier JI, Huber TB, Schell C: AIF1L regulates actomyosin contractility and filopodial extensions in human podocytes. PLoS One 13: e0200487, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasuda-Yamahara M, Rogg M, Frimmel J, Trachte P, Helmstaedter M, Schroder P, et al. : FERMT2 links cortical actin structures, plasma membrane tension and focal adhesion function to stabilize podocyte morphology. Matrix Biol 68-69: 263–279, 2018 [DOI] [PubMed] [Google Scholar]
- 18.Randles MJ, Lausecker F, Humphries JD, Byron A, Clark SJ, Miner JH, et al. : Basement membrane ligands initiate distinct signalling networks to direct cell shape. Matrix Biol 90: 61–78, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell C, Huber TB: The evolving complexity of the podocyte cytoskeleton. J Am Soc Nephrol 28: 3166–3174, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriz W, Shirato I, Nagata M, LeHir M, Lemley KV: The podocyte’s response to stress: The enigma of foot process effacement. Am J Physiol Renal Physiol 304: F333–F347, 2013 [DOI] [PubMed] [Google Scholar]
- 21.Schell C, Sabass B, Helmstaedter M, Geist F, Abed A, Yasuda-Yamahara M, et al. : ARP3 controls the podocyte architecture at the kidney filtration barrier. Dev Cell 47: 741–757.e8, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda J, Asano-Matsuda K, Kitzler TM, Takano T: Rho GTPase regulatory proteins in podocytes. Kidney Int 99: 336–345, 2021 [DOI] [PubMed] [Google Scholar]
- 23.Schiffer M, Teng B, Gu C, Shchedrina VA, Kasaikina M, Pham VA, et al. : Pharmacological targeting of actin-dependent dynamin oligomerization ameliorates chronic kidney disease in diverse animal models. Nat Med 21: 601–609, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai C, Stolz DB, Bastacky SI, St-Arnaud R, Wu C, Dedhar S, et al. : Essential role of integrin-linked kinase in podocyte biology: Bridging the integrin and slit diaphragm signaling. J Am Soc Nephrol 17: 2164–2175, 2006 [DOI] [PubMed] [Google Scholar]
- 25.El-Aouni C, Herbach N, Blattner SM, Henger A, Rastaldi MP, Jarad G, et al. : Podocyte-specific deletion of integrin-linked kinase results in severe glomerular basement membrane alterations and progressive glomerulosclerosis. J Am Soc Nephrol 17: 1334–1344, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Vaynberg J, Fukuda K, Lu F, Bialkowska K, Chen Y, Plow EF, et al. : Non-catalytic signaling by pseudokinase ILK for regulating cell adhesion. Nat Commun 9: 4465, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghatak S, Morgner J, Wickström SA: ILK: A pseudokinase with a unique function in the integrin-actin linkage. Biochem Soc Trans 41: 995–1001, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Lange A, Wickström SA, Jakobson M, Zent R, Sainio K, Fässler R: Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature 461: 1002–1006, 2009 [DOI] [PubMed] [Google Scholar]
- 29.Mishra YG, Manavathi B: Focal adhesion dynamics in cellular function and disease. Cell Signal 85: 110046, 2021 [DOI] [PubMed] [Google Scholar]
- 30.Revach OY, Grosheva I, Geiger B: Biomechanical regulation of focal adhesion and invadopodia formation. J Cell Sci 133: jcs244848, 2020 [DOI] [PubMed] [Google Scholar]
- 31.Olski TM, Noegel AA, Korenbaum E: Parvin, a 42 kDa focal adhesion protein, related to the alpha-actinin superfamily. J Cell Sci 114: 525–538, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Fraccaroli A, Pitter B, Taha AA, Seebach J, Huveneers S, Kirsch J, et al. : Endothelial alpha-parvin controls integrity of developing vasculature and is required for maintenance of cell-cell junctions. Circ Res 117: 29–40, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeller MJ, Sanden SK, Soofi A, Wiggins RC, Holzman LB: Two gene fragments that direct podocyte-specific expression in transgenic mice. J Am Soc Nephrol 13: 1561–1567, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Schell C, Kretz O, Bregenzer A, Rogg M, Helmstädter M, Lisewski U, et al. : Podocyte-specific deletion of murine CXADR does not impair podocyte development, function or stress response. PLoS One 10: e0129424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier JI, Rogg M, Helmstädter M, Sammarco A, Walz G, Werner M, et al. : A novel model for nephrotic syndrome reveals associated dysbiosis of the gut microbiome and extramedullary hematopoiesis. Cells 10: 1509, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butt L, Unnersjö-Jess D, Höhne M, Edwards A, Binz-Lotter J, Reilly D, et al. : A molecular mechanism explaining albuminuria in kidney disease. Nat Metab 2: 461–474, 2020 [DOI] [PubMed] [Google Scholar]
- 37.Saleem MA, O’Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, et al. : A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 13: 630–638, 2002 [DOI] [PubMed] [Google Scholar]
- 38.Maier JI, Rogg M, Helmstädter M, Sammarco A, Schilling O, Sabass B, et al. : EPB41L5 controls podocyte extracellular matrix assembly by adhesome-dependent force transmission. Cell Rep 34: 108883, 2021 [DOI] [PubMed] [Google Scholar]
- 39.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E: CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47[W1]: W171–W174, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barger CJ, Branick C, Chee L, Karpf AR: Pan-cancer analyses reveal genomic features of FOXM1 overexpression in cancer. Cancers (Basel) 11: 251, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. : Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184–191, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.el Nahas AM, Bassett AH, Cope GH, Le Carpentier JE: Role of growth hormone in the development of experimental renal scarring. Kidney Int 40: 29–34, 1991 [DOI] [PubMed] [Google Scholar]
- 43.Bienaimé F, Muorah M, Yammine L, Burtin M, Nguyen C, Baron W, et al. : Stat3 controls tubulointerstitial communication during CKD. J Am Soc Nephrol 27: 3690–3705, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariani LH, Martini S, Barisoni L, Canetta PA, Troost JP, Hodgin JB, et al. : Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant 33: 310–318, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkatareddy M, Wang S, Yang Y, Patel S, Wickman L, Nishizono R, et al. : Estimating podocyte number and density using a single histologic section. J Am Soc Nephrol 25: 1118–1129, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gut G, Herrmann MD, Pelkmans L: Multiplexed protein maps link subcellular organization to cellular states. Science 361: eear7042, 2018 [DOI] [PubMed] [Google Scholar]
- 47.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. : QuPath: Open source software for digital pathology image analysis. Sci Rep 7: 16878, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogg M, Maier JI, Dotzauer R, Artelt N, Kretz O, Helmstädter M, et al. : SRGAP1 controls small Rho GTPases to regulate podocyte foot process maintenance. J Am Soc Nephrol 32: 563–579, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinschen MM, Gödel M, Grahammer F, Zschiedrich S, Helmstädter M, Kretz O, et al. : A multi-layered quantitative in vivo expression atlas of the podocyte unravels kidney disease candidate genes. Cell Rep 23: 2495–2508, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatje FA, Wedekind U, Sachs W, Loreth D, Reichelt J, Demir F, et al. : Tripartite separation of glomerular cell types and proteomes from reporter-free mice. J Am Soc Nephrol 32: 2175–2193, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaverina I, Krylyshkina O, Small JV: Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J Cell Biol 146: 1033–1044, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ezratty EJ, Partridge MA, Gundersen GG: Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol 7: 581–590, 2005 [DOI] [PubMed] [Google Scholar]
- 53.Lavelin I, Wolfenson H, Patla I, Henis YI, Medalia O, Volberg T, et al. : Differential effect of actomyosin relaxation on the dynamic properties of focal adhesion proteins. PLoS One 8: e73549, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanchi F, Grashoff C, Nguemeni Yonga CF, Grall D, Fässler R, Van Obberghen-Schilling E: Molecular dissection of the ILK-PINCH-parvin triad reveals a fundamental role for the ILK kinase domain in the late stages of focal-adhesion maturation. J Cell Sci 122: 1800–1811, 2009 [DOI] [PubMed] [Google Scholar]
- 55.Boerries M, Grahammer F, Eiselein S, Buck M, Meyer C, Goedel M, et al. : Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int 83: 1052–1064, 2013 [DOI] [PubMed] [Google Scholar]
- 56.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. : The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44[W1]: W3–W10, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D, Langmead B, Salzberg SL: HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y, Smyth GK, Shi W: featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930, 2014 [DOI] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen CD, Frach K, Schlöndorff D, Kretzler M: Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int 61: 133–140, 2002 [DOI] [PubMed] [Google Scholar]
- 61.Cohen CD, Klingenhoff A, Boucherot A, Nitsche A, Henger A, Brunner B, et al. : Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc Natl Acad Sci U S A 103: 5682–5687, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altstätter J, Hess MW, Costell M, Montanez E: α-parvin is required for epidermal morphogenesis, hair follicle development and basal keratinocyte polarity. PLoS One 15: e0230380, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lehtimäki JI, Rajakylä EK, Tojkander S, Lappalainen P: Generation of stress fibers through myosin-driven reorganization of the actin cortex. eLife 10: e60710, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burridge K, Guilluy C: Focal adhesions, stress fibers and mechanical tension. Exp Cell Res 343: 14–20, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huet-Calderwood C, Brahme NN, Kumar N, Stiegler AL, Raghavan S, Boggon TJ, et al. : Differences in binding to the ILK complex determines kindlin isoform adhesion localization and integrin activation. J Cell Sci 127: 4308–4321, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naik AS, Le D, Aqeel J, Wang SQ, Chowdhury M, Walters LM, et al. : Podocyte stress and detachment measured in urine are related to mean arterial pressure in healthy humans. Kidney Int 98: 699–707, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng D, Notbohm J, Benjamin A, He S, Wang M, Ang LH, et al. : Disease-causing mutation in α-actinin-4 promotes podocyte detachment through maladaptation to periodic stretch. Proc Natl Acad Sci U S A 115: 1517–1522, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng D, DuMontier C, Pollak MR: Mechanical challenges and cytoskeletal impairments in focal segmental glomerulosclerosis. Am J Physiol Renal Physiol 314: F921–F925, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Chen K, Tu Y, Wu C: Distinct roles of two structurally closely related focal adhesion proteins, alpha-parvins and beta-parvins, in regulation of cell morphology and survival. J Biol Chem 279: 41695–41705, 2004 [DOI] [PubMed] [Google Scholar]
- 71.Shirato I, Sakai T, Kimura K, Tomino Y, Kriz W: Cytoskeletal changes in podocytes associated with foot process effacement in Masugi nephritis. Am J Pathol 148: 1283–1296, 1996 [PMC free article] [PubMed] [Google Scholar]
- 72.Suleiman HY, Roth R, Jain S, Heuser JE, Shaw AS, Miner JH: Injury-induced actin cytoskeleton reorganization in podocytes revealed by super-resolution microscopy. JCI Insight 2: e94137, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith MA, Blankman E, Gardel ML, Luettjohann L, Waterman CM, Beckerle MC: A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev Cell 19: 365–376, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC: Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol Biol Cell 23: 1846–1859, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goode NP, Shires M, Khan TN, Mooney AF: Expression of alpha-actinin-4 in acquired human nephrotic syndrome: A quantitative immunoelectron microscopy study. Nephrol Dial Transplant 19: 844–851, 2004 [DOI] [PubMed] [Google Scholar]
- 76.Smoyer WE, Mundel P, Gupta A, Welsh MJ: Podocyte alpha-actinin induction precedes foot process effacement in experimental nephrotic syndrome. Am J Physiol 273: F150–F157, 1997 [DOI] [PubMed] [Google Scholar]
- 77.Dandapani SV, Sugimoto H, Matthews BD, Kolb RJ, Sinha S, Gerszten RE, et al. : Alpha-actinin-4 is required for normal podocyte adhesion. J Biol Chem 282: 467–477, 2007 [DOI] [PubMed] [Google Scholar]
- 78.Shved N, Warsow G, Eichinger F, Hoogewijs D, Brandt S, Wild P, et al. : Transcriptome-based network analysis reveals renal cell type-specific dysregulation of hypoxia-associated transcripts. Sci Rep 7: 8576, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.