Abstract
Gut bacteria modulate the response to immune checkpoint blockade (ICB) treatment in cancer, but the effect of diet and supplements on this interaction is not well studied. We assessed fecal microbiota profiles, dietary habits, and commercially available probiotic supplement use in melanoma patients and performed parallel preclinical studies. Higher dietary fiber was associated with significantly improved progression-free survival in 128 patients on ICB, with the most pronounced benefit observed in patients with sufficient dietary fiber intake and no probiotic use. Findings were recapitulated in preclinical models, which demonstrated impaired treatment response to anti–programmed cell death 1 (anti–PD-1)–based therapy in mice receiving a low-fiber diet or probiotics, with a lower frequency of interferon-γ–positive cytotoxic T cells in the tumor microenvironment. Together, these data have clinical implications for patients receiving ICB for cancer.
Treatment with immune checkpoint blockade (ICB) has revolutionized cancer therapy (1), and the influence of the gut microbiome on therapeutic response has now been demonstrated in numerous human cohorts and in preclinical models (2–7). The human gut microbiome is itself shaped by a wide variety of environmental exposures, including diet (8, 9) and medication use (10–13), with host genetics accounting for <10% of variation (14). However, whether factors such as dietary fiber intake and the use of commercially available probiotics affect immunotherapy responses in cancer patients remains unclear.
To help address this, we profiled the gut (fecal) microbiome and assessed clinicopathologic features and outcomes in a large cohort of melanoma patients (n = 438; Fig. 1A and fig. S1). The majority of these patients were receiving systemic therapy for metastatic melanoma (n = 321), and responses to treatment were assessed with radiographic imaging in those with evaluable treatment responses (n = 293), classifying patients as either responders [(R) complete or partial response or stable disease ≥6 months; n = 193] or non-responders [(NR) stable disease <6 months or progressive disease; n = 100] using Response Evaluation Criteria in Solid Tumors (RECIST 1.1) (15). The majority of patients were treated with ICB (87%), most commonly anti–programmed cell death 1 (anti–PD-1) therapy (Fig. 1A, fig. S1, and table S1). Patients initiating therapy with ICB were asked to co-enroll onto a lifestyle survey protocol, which included baseline assessments of dietary habits and use of probiotic supplements within the past month (n = 158; Fig. 1A, fig. S1, and table S1) (16, 17).
Fig. 1. Profiles of gut microbiota in patients with melanoma and associations with outcomes on therapy.
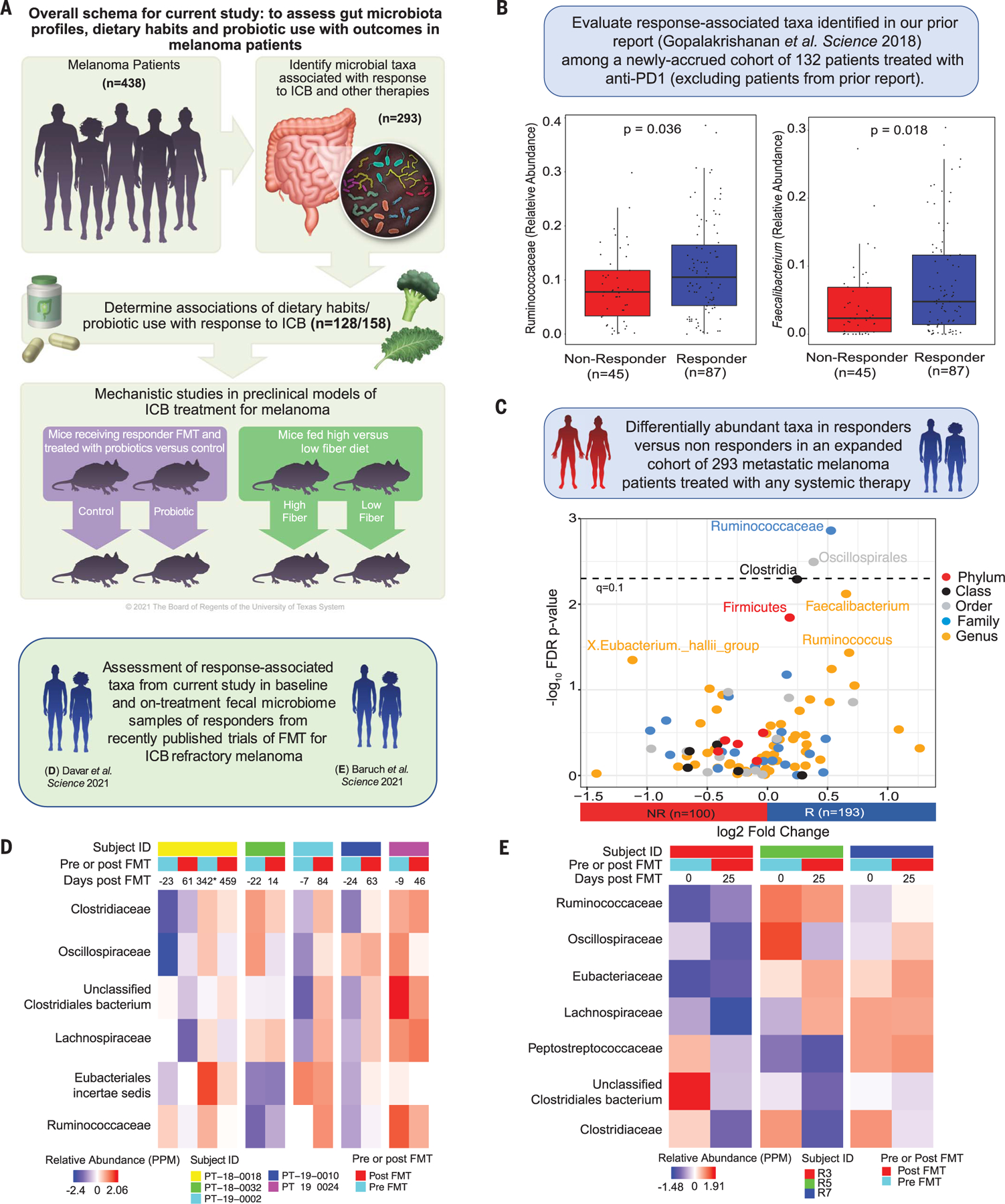
(A) Schema of study design. (B) Box plots comparing the relative abundance of anti–PD-1 response–associated taxa from Gopalakrishnan et al. (4) with a newly recruited cohort (n = 132) of anti–PD-1–treated patients (P = 0.036 and P = 0.018, respectively, for Ruminococcaceae and Faecalibacterium by Wilcoxon rank sum test). Patients included in the prior study were excluded from this analysis. (C) Volcano plot depicting pairwise comparisons of relative abundances of bacterial taxa. The y axis displays the −log10 false discovery rate (FDR)–corrected P value (dashed line, q < 0.1), and the x axis shows the log2 fold change comparing 193 R and 100 NR patients with systemic therapy across the full cohort, including patients from the prior study (by Wilcoxon rank sum test with FDR correction per level). (D) Heatmap of scaled relative abundances [parts per million (PPM)] of bacteria belonging to order Clostridiales and family Ruminococcaceae in pre- and post-FMT samples of anti–PD-1 refractory metastatic melanoma FMT recipients who responded to FMT + anti–PD-1 in Davar et al. (20) [National Center for Biotechnology Information (NCBI) accession no. PRJNA672867]. Number of days from FMT are depicted on the top of each heatmap column, with post-FMT values being the geometric mean days of all post-FMT time points for that patient. The geometric mean of relative abundances of post-FMT samples from each patient were calculated as the single post-FMT mean relative abundance. The exception is patient PT−18−0018, who received two FMTs (denoted by an asterisk). The first post-FMT column for this patient reflects the geometric mean of samples leading up to the second FMT event. (E) Heatmap of scaled relative abundances of bacteria belonging to order Clostridiales and family Ruminococcaceae in pre- and post-FMT samples of anti–PD-1 refractory metastatic melanoma FMT recipients who responded to FMT + anti–PD-1 in Baruch et al. (19) (NCBI accession no. PRJNA678737). Number of days from FMT are depicted on the top of each heatmap column, with post-FMT values being the geometric mean days of all post-FMT time points for that patient. The geometric mean of relative abundances of post-FMT samples from each patient were calculated as the single post-FMT mean relative abundance.
We first assessed the relative abundance of gut microbial taxa associated with response to anti–PD-1 immunotherapy in our prior published study (4) within a larger cohort of newly accrued anti–PD-1–treated patients (n = 132 total; n = 87 R and n = 45 NR), excluding patients from the previously published cohort. On the basis of our prior study, we hypothesized that bacteria from the Ruminococcaceae family and Faecalibacterium genus would be associated with response to therapy. We tested this by specifically querying the abundance of these taxa in responders versus nonresponders to anti–PD-1, again observing enrichment of both taxa (Fig. 1B) as well as of Faecalibacterium prausnitzii in the metagenomic subset (n = 111 total; n = 71R and n = 40 NR; fig. S2A) in anti–PD-1 responders. We did not observe significant differences in the alpha and beta diversity of the gut microbiota in responders versus nonresponders (fig. S2, E and F), in contrast to our prior study. This discrepancy may reflect associations driven by a small number of patients in the prior study with improved power and reduced error in the larger cohort (fig. S3 and tables S2 to S4), and it underscores the lack of concordance across numerous studies that implicate gut bacteria in response to cancer immunotherapy (18).
Next, we assessed the composition of the gut microbiome in responders and nonresponders in the full cohort of late-stage melanoma patients with evaluable responses to any systemic therapy (n = 293 total; n = 193 R and n = 100 NR; Fig. 1C), as well as in all patients treated with anti–PD-1 monotherapy (fig. S2), including patients from both the newly accrued and the previously published cohorts. Across the full cohort, we observed a significantly higher abundance of Ruminococcaceae in the gut microbiota of responders versus nonresponders treated with anti–PD-1 or other systemic therapies that remained consistent after adjustment for potential confounders [age, sex, body mass index (BMI), prior treatment, and antibiotic use] (Fig. 1C, fig. S2, and tables S5 to S9). However, we did not observe significant differences in the overall composition of the gut microbiota in responders versus nonresponders in this larger cohort of patients on systemic therapy (fig. S2 and table S9), nor was there strong concordance with response-associated taxa from the prior study—beyond Ruminococcaceae— in the newly accrued cohort (figs. S3 and S4 and tables S3 and S10). We also assessed the abundance of our previously reported response-associated taxa in published datasets from two recently completed clinical trials demonstrating potential efficacy of the use of fecal microbiota transplant (FMT) + anti–PD-1 in immunotherapy-refractory melanoma patients (19, 20), noting that many of our response-associated taxa appeared to be enriched in the post-FMT specimens from patients who responded to this treatment (Fig. 1, D and E, and fig. S5).
Given that cancer patients are increasingly interested in using probiotic supplements to augment gut health, we assessed the use of commercially available probiotics within our cohort and observed that 31% (49 of 158) of late-stage melanoma patients initiating ICB reported that they had taken a probiotic supplement within the past month. Patients who reported taking a probiotic supplement preceding the start of treatment with ICB had a lower BMI, were less likely to take statins, and reported slightly higher intake of vegetables and legumes than patients who did not take probiotic supplements (table S1). The proportion of patients reporting antibiotic use within the past month was markedly similar in those who did (29%) versus did not (28%) report probiotic use. Steroid or proton-pump inhibitor use was also not associated with probiotic use (table S1). We then assessed whether probiotic use was associated with differential outcomes in patients treated with ICB and observed no statistically significant differences in progression-free survival (PFS) (n = 158; Fig. 2A; median PFS 17 versus 23 months; Table 1) or odds of response in patients who reported taking probiotics (59% R) versus those who did not (68% R) (Table 1 and tables S1, S11, and S12). The modest associations of probiotic use and outcomes in this cohort were not surprising to us because limitations existed regarding the overall cohort size, as well as substantial heterogeneity in the specific probiotic supplement(s) reportedly used by patients. Although we did not observe statistically significant differences in outcomes or in microbiota features (fig. S6 and table S9) in patients on ICB by probiotic use, the overall trends observed were intriguing—particularly given the relatively high proportion of patients reporting probiotic supplementation in this cohort. Thus, we sought to examine the effects of probiotic use on response to ICB in preclinical models.
Fig. 2. Effect of probiotic supplement use in patients and in preclinical models of melanoma immunotherapy.
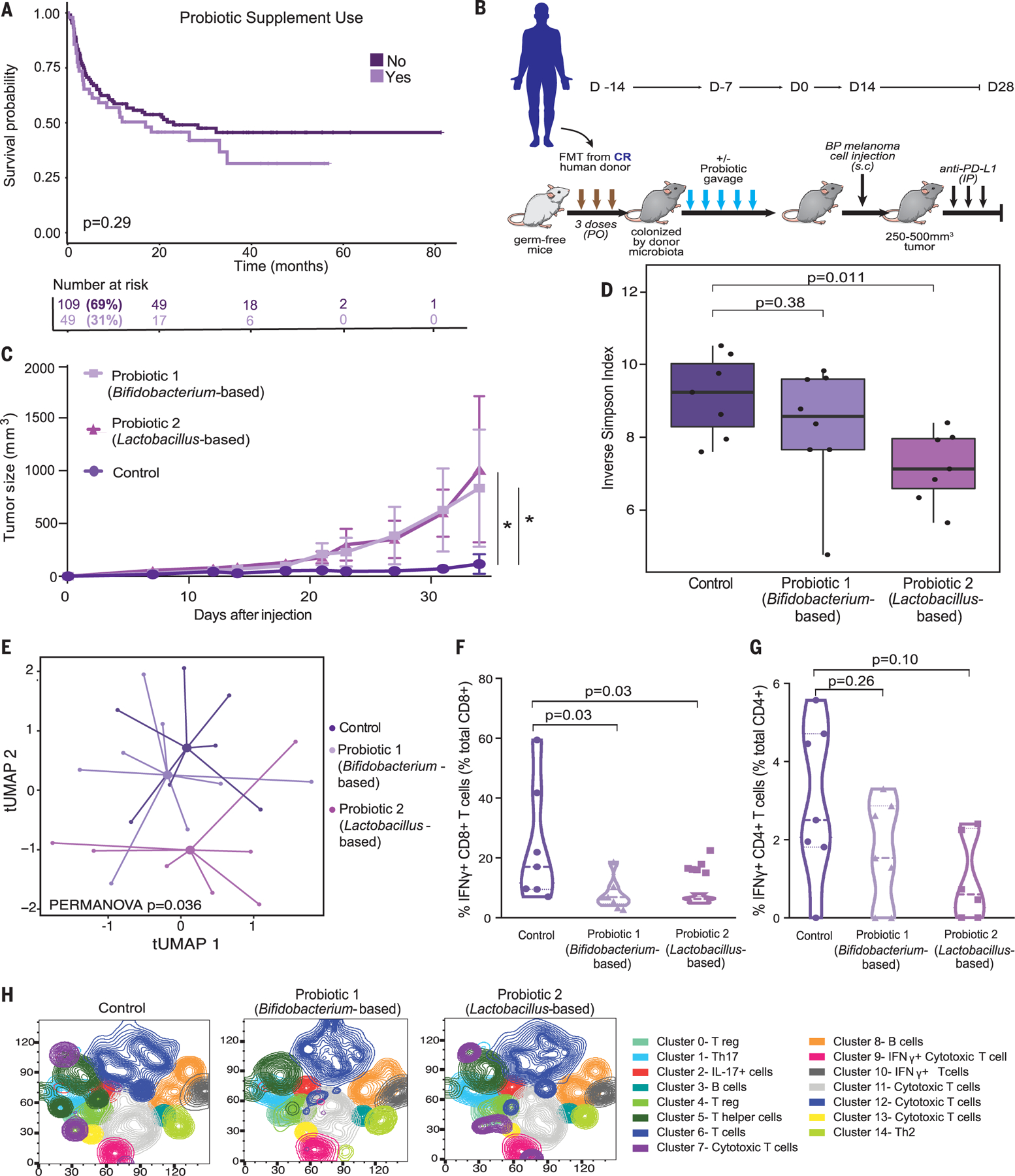
(A) Kaplan-Meier plot comparing progression-free survival intervals by probiotic use among patients who received ICB (n = 158; P = 0.29 by log-rank test). (B) Experimental design of studies in germ-free (GF) mice that received FMT from a complete responder (CR) donor combined with probiotic 1, probiotic 2, or sterile water control before tumor injection [2.5 × 105 to 8 × 105 BRAFV600E/PTEN−/− (BP) tumor cells] and treatment with anti–PD-L1. Time is in days relative to tumor injection [day 0 (D0)]. PO, per orem; s.c, subcutaneous; IP, intraperitoneal. (C) Mouse tumor growth curves comparing volume of tumors in mice who received probiotics or sterile water control (n = 4 to 5 per group); probiotic 1 versus probiotic 2 versus sterile water control. Data are means ± SEM tumor volume. All P values are from a likelihood ratio test in a linear mixed model (P = 0.04 Bifidobacterium longum 35624–based probiotic 1 versus control; P = 0.01 Lactobacillus rhamnosus GG–based probiotic 2 versus control). *P < 0.05. (D) Box plots comparing alpha diversity of the gut microbiome, as measured by the inverse Simpson index in mice treated with control, probiotic 1 (Bifidobacterium longum 35624–based), or probiotic 2 (Lactobacillus rhamnosus GG–based) (pairwise P values compared with control were calculated by Wilcoxon rank sum test). Fecal samples were collected for microbiome analysis (via metagenomic sequencing) from tumor-bearing mice before the anti–PD-L1 therapy (n =7 to 8 per group), mimicking baseline sample collection from patients. (E) Ordination plot by t-distributed uniform manifold approximation and projection (t-UMAP) by Bray-Curtis distance, demonstrating compositional differences of the gut microbiome in mice treated with sterile water control, probiotic 1 (Bifidobacterium longum 35624–based), or probiotic 2 (Lactobacillus rhamnosus GG–based) [permutational multivariate analysis of variance (PERMANOVA) P = 0.036]. (F and G) Pairwise comparisons of sterile water control versus probiotic 1 (Bifidobacterium longum 35624–based) or control versus probiotic 2 (Lactobacillus rhamnosus GG–based) groups (n = 6 per group) via supervised analysis with manual gating for either frequency of IFN-γ+ CD8+ T cells in tumors (percent total tumor CD8+ T cells) (P = 0.03, P = 0.03) (F) or frequency of IFN-γ+ CD4+ T cells in tumors (percent total tumor CD4+ T cells) (P = 0.26, P = 0.10) (G). (H) Unsupervised analysis of flow cytometry data showing density t-distributed stochastic neighbor embedding (t-SNE) plot of tumor-infiltrating immune cells overlaid with color-coded clusters, with an equal number of CD45+ infiltrating leukocytes for each treatment group (control, probiotic 1, and probiotic 2).
Table 1. Associations of baseline probiotic supplement use and dietary fiber intake in late-stage melanoma patients treated with ICB and followed for tumor response and progression-free survival.
Dashes indicate not applicable. HR, hazard ratio; CI, confidence interval; OR, odds ratio; N/R, not reached; ref, referent group.
| Comparison | n | Progression-free survival |
Odds of response to ICB |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Events | Median months | HR* | 95% CI | P value† | Responder n (%) | OR* | 95% CI | P value† | ||
| Probiotic supplement use | ||||||||||
| Total | 158 | 85 | – | – | –– | 65% | – | – | – | |
| No | 109 | 56 | 23 | 1.00 | ref | – | 74 (68%) | 1.00 | ref | – |
| Yes | 49 | 29 | 17 | 1.30 | 0.82, 2.07 | 0.27 | 29 (59%) | 0.79 | 0.37, 1.66 | 0.52 |
| Dietary fiber intake | ||||||||||
| Total | 128 | 73 | – | – | – | – | 65% | – | – | – |
| Per 5 g/day increase | – | – | – | 0.71 | 0.52, 0.98 | 0.04 | – | 1.70 | 0.97, 3.00 | 0.06 |
| Insufficient | 91 | 57 | 13 | 1.00 | ref | – | 55 (60%) | 1.00 | ref | – |
| Sufficient | 37 | 16 | N/R | 0.59 | 0.33, 1.04 | 0.07 | 28 (76%) | 2.20 | 0.86, 5.61 | 0.10 |
| Dietary fiber intake + probiotic supplement use | ||||||||||
| Total | 123 | 72 | – | – | – | – | 63% | – | – | – |
| Sufficient fiber + no probiotics | 22 | 8 | N/R | 0.44 | 0.21, 0.92 | 0.03 | 18 (82%) | 2.94 | 0.87, 9.94 | 0.08 |
| Other‡ | 101 | 64 | 13 | 1.00 | ref | – | 60 (59%) | 1.00 | ref | –. |
HR and 95% CI estimated using Cox proportional hazards regression. OR and 95% CI estimated using logistic regression. All models include multivariable adjustment for subtype, stage, lactate dehydrogenase level, and BMI.
P value by Wald test.
Other category includes patients who either reported insufficient fiber intake or probiotic use.
To do this, germ-free mice first received FMT using donor stool from a complete responder (CR) patient to anti–PD-1 blockade. After this, mice were orally gavaged with one of two commercially purchased probiotics (Bifidobacterium longum– or Lactobacillus rhamnosus GG–based) versus sterile water control. Viability and composition of the bacterial strains in the probiotic were confirmed by culture and sequencing (fig. S7). Mice were then challenged with murine melanoma tumors and treated with anti–PD-1 ligand 1 (anti–PD-L1) therapy (because treatment with this antibody is more effective in this particular murine tumor model than anti–PD-1) (Fig. 2B). In these studies, mice receiving probiotics demonstrated impaired antitumor response to treatment with anti–PD-L1 and had significantly larger tumors compared with control mice (Fig. 2C), with findings that were recapitulated in an additional murine tumor model (fig. S8). Notably, similar findings were also observed in non–germ-free and specific pathogen–free (SPF) mice implanted with melanoma tumors (fig. S8) that harbor a microbiota from birth to which they are well coadapted.
We next compared the gut microbiota of mice receiving probiotics versus sterile water control, and we observed differences in gut microbiota diversity in the mice receiving probiotics compared with control (Fig. 2, D and E, and fig. S9). Analysis of tumor-infiltrating immune subsets from anti–PD-L1–treated mice revealed a significantly reduced frequency of interferon-γ (IFN-γ) positive CD8+ T cells in tumors of probiotic-treated mice versus controls (Fig. 2F). A trend toward fewer IFN-g CD4+ T helper 1 (TH1) cells in tumors from mice receiving probiotics versus control was also observed, although this did not reach statistical significance (Fig. 2G). Unsupervised analyses of the flow cytometry data corroborated the findings in immune subsets between probiotic treatment versus control, demonstrating a reduced frequency of cytotoxic T cells in the tumor microenvironment of probiotic-treated mice (Fig. 2H). These data are in line with previously published studies that have demonstrated increased tumorigenesis in murine models of colorectal carcinoma in probiotic-treated mice (21), although other studies have shown a beneficial effect of other probiotic formulations and rationally designed bacterial consortia in preclinical models and patient cohorts (22–24). Together, these studies support the need for more careful investigations of the effects of current commercially available probiotic formulations on immunity and cancer immunotherapy response.
Given that many of the response-associated bacteria identified in our cohort have known roles in starch degradation and fiber fermentation (25–29), we next sought to assess the effect of dietary fiber intake on response to ICB. We asked patients who were initiating treatment with ICB to complete the National Cancer Institute Dietary Screener Questionnaire (NCI-DSQ) (17), and responses were scored to derive dietary fiber intake from 26 queried food items. Dietary fiber intake was assessed per 5-g/day incremental increase and further categorized according to the distribution of reported intake within our cohort with low or insufficient fiber intake corresponding to <20 g/day and sufficiently high fiber intake at or above 20 g/day, a threshold met by ~30% (37 of 128) of ICB patients (Fig. 3A and fig. S10A). As expected, dietary fiber intake was highly correlated with fruit, vegetable, legume, and whole grain intake and, to a lesser extent, with calcium intake (fig. S10B and table S1). Patients with insufficient dietary fiber intake were more likely to be obese— a factor that we and others have previously found to be paradoxically associated with improved response to ICB (30, 31)—and were also more likely to take antihypertensive medications (table S1).
Fig. 3. Effect of dietary fiber intake in patients and in preclinical models of melanoma immunotherapy.
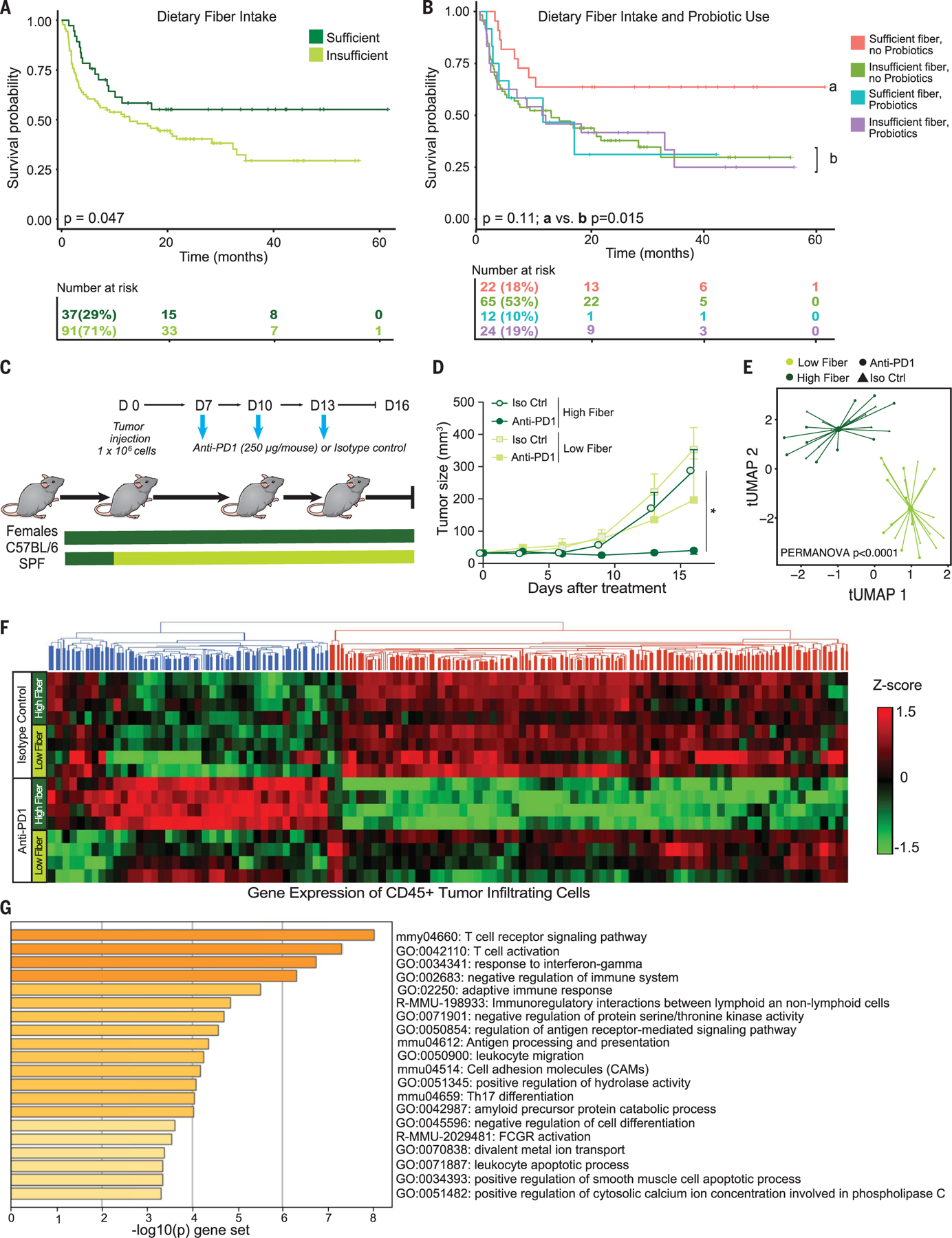
(A) Kaplan-Meier plot comparing progression-free survival intervals by dietary fiber intake among patients who received ICB (n = 128; P = 0.047 by log-rank test). (B) Kaplan-Meier plot comparing progression-free survival intervals by combined dietary fiber and probiotic status among patients who received ICB (n = 123; overall P across four groups = 0.11; P for sufficient dietary fiber intake + no probiotics use versus else = 0.015; both by log-rank test). (C) Experimental design of studies in C57BL/6 SPF mice that received either a high-fiber or low-fiber diet at inoculation of M3 (HCmel1274) melanoma cells (1 × 106 tumor cells) and were then treated with anti–PD-1 or isotype control. Time is in days relative to tumor injection. (D) M3 melanoma growth kinetics of control (high-fiber) diet (circles) or low-fiber (fiber-free) diet (squares) treated four times with intraperitoneal injection of anti–PD-1 antibody (dark green) or of isotype control (Iso Ctrl) (light green). Data are means ± SEM of tumor volume from one representative experiment (n = 5 per group). All P values are from a likelihood ratio test in a linear mixed model (isotype control and high fiber, P = 0.69; anti–PD-1 and high fiber, P = 0.02; anti–PD-1 and low fiber versus isotype control and low fiber, P = 0.08). *P < 0.05. (E) t-UMAP plot comparing the gut microbiome (via shotgun metagenomic sequencing of fecal samples) of mice by treatment and diet group from two experiments (n = 4 to 5 per group) using Bray-Curtis distances (PERMANOVA P < 0.0001) at experimental day 16. (F) Heatmap of gene expression of flow-sorted CD45+ tumor-infiltrating immunocytes in mice fed high-versus low-fiber diets and treated with anti–PD-1 or isotype control. (G) Gene set enrichment analysis depicting pathways enriched in high-fiber diet mice treated with anti–PD-1 versus isotype control which were not differentially expressed by treatment in low-fiber diet mice.
Patients who reported sufficient dietary fiber intake (n = 37 of 128) demonstrated improved PFS over those with insufficient dietary fiber intake (median PFS not reached versus 13 months; Fig. 3A and Table 1). After adjustment for clinical factors, every 5-g increase in daily dietary fiber intake corresponded with a 30% lower risk of progression or death (Table 1). Similar associations were observed when assessing dietary fiber intake in relation to the odds of response to ICB (Table 1). The observed protective effect of dietary fiber intake in relation to PFS and response remained consistent among the subset of patients treated with anti–PD-1 monotherapy, with the exclusion of patients reporting recent antibiotic use given the known impact of these on ICB response (32) (tables S11 and S12). We did not observe substantial differences in the composition of the gut microbiota in those who reported sufficient versus insufficient fiber intake as assessed by 16S and metagenomic sequencing (fig. S11 and table S9); however, this is not unexpected given the known challenges of isolating associations of specific dietary components from other factors known to affect the gut microbiota in observational human cohorts (33, 34).
After this, we further evaluated whether dietary fiber intake and probiotic use may jointly affect clinical outcomes in patients treated with ICB, given the potential associations between these factors. In this cohort, patients with sufficient dietary fiber intake were somewhat more likely to take probiotics than those reporting insufficient dietary fiber intake (35 versus 27%; table S1). We assessed potential additive effects across a combined variable comparing four groups of patients— including those reporting insufficient dietary fiber intake with no probiotic use (53%), those reporting insufficient dietary fiber intake with probiotic use (19%), those reporting sufficient dietary fiber intake with no probiotic use (18%), and those reporting sufficient dietary fiber intake with probiotic use (10%) (n = 123 patients total; Fig. 3B). Differences in outcomes were noted across the groups (Fig. 3B), with significantly longer PFS observed in patients reporting sufficient dietary fiber intake and no probiotic use compared with all other groups (median PFS not reached versus 13 months; Fig. 3B and Table 1). Similar positive associations were observed for ICB response in patients reporting sufficient dietary fiber intake and no probiotic use compared with all other groups (n = 123; 82 versus 59% responders; Table 1 and tables S11 and S12). Microbial alpha diversity and Ruminococcaceae family and Faecalibacterium genus abundances were also numerically higher in patients with sufficient dietary fiber intake and no probiotic use, although only 18% of patients met these criteria and results did not reach statistical significance (fig. S12).
Intrigued by these findings, we next examined whether dietary fiber modulation could enhance therapeutic response to ICB in preclinical melanoma models. In these studies, conventionally housed C57BL/6 SPF mice were provided with a standard fiber-rich whole grain diet (17.6% fiber) versus a fiber-poor diet (2% fiber) (29), challenged with murine melanoma tumors (35), and treated with anti–PD-1 therapy versus isotype control (Fig. 3C). Mice receiving a fiber-rich diet demonstrated delayed tumor outgrowth compared with mice who received a fiber-poor diet when treated with anti–PD-1 (Fig. 3D). These findings were recapitulated in additional tumor models (figs. S13 and S14). By contrast, there was no effect of fiber-rich versus fiber-poor diet on the response to anti–PD-1 therapy in germ-free mice, which supports the hypothesis that the effect of this dietary intervention on treatment efficacy is microbiota dependent (fig. S13). Profiling of the gut microbiome revealed significant differences in the community structure of mice fed fiber-rich versus fiber-poor diets (Fig. 3E) and taxonomic differences between the groups (fig. S15).
Stool metabolomic profiling also revealed significantly higher levels of the short chain fatty acid (SCFA) propionate in mice receiving a fiber-rich diet, although no significant differences were noted in SCFA levels as a whole (fig. S16). Immune profiling by flow cytometry of tumors in treated mice revealed a significantly higher frequency of CD4+ T cells overall (and those expressing PD-1) in the tumors of mice on high-versus low-fiber diets (fig. S17, A and B). We next conducted RNA sequencing of CD45+ tumor-infiltrating lymphocytes (TILs) and observed significantly higher expression of genes related to T cell activation and interferon response in mice receiving a high-versus low-fiber diet in the setting of treatment with anti–PD-1 (Fig. 3, F and G, and tables S13 and S14). Further, network analysis of murine data suggested that the fiber-fermenting Ruminococcaceae family of bacteria may contribute to the effects of fiber on antitumor immunity by affecting pathways of T cell activation as well as the accumulation of T cells in the tumor, including inducible T cell co-stimulator (ICOS)–expressing CD8+ and CD4+ T cells (fig. S17, C to H).
Together, these data have important implications. We show that dietary fiber and probiotic use, factors known to affect the gut microbiome, are associated with differential outcomes to ICB. Although causality cannot be addressed from the observational human cohort, where unmeasured confounders may exist, our preclinical models support the hypothesis that dietary fiber and probiotics modulate the microbiome and that antitumor immunity is impaired in mice receiving a low-fiber diet and in those receiving probiotics— with suppression of intratumoral IFN-γ T cell responses in both cases.
Numerous challenges exist to decipher how best to leverage the microbiome to optimize patient outcomes, starting with what to target— selected features or community function—and whether this can be safely achieved through supplementation or more comprehensive dietary approaches. Several prior studies have shown that controlled increases in dietary fiber intake can modulate the gut microbiome but also that interindividual variation in the gut microbiome drives differential effects of specific fibers (and prebiotics) on host metabolism (36–40). Ongoing dietary intervention studies in the setting of ICB are critical for establishing whether a targeted and achievable diet change at the initiation of ICB can safely and effectively improve outcomes (NCT04645680). Although our findings suggest that undirected use of commercially available probiotics may be harmful in the setting of ICB, further study of rationally designed and targeted probiotics or bacterial consortia is warranted on the basis of promising early data of this approach (22–24).
Some analyses in the current cohort were not adequately powered to assess the full effect of these factors, and further validation is needed in independent cohorts with more indepth and detailed assessment of dietary intake and the use of specific probiotic supplements, along with further mechanistic studies in preclinical models. Nonetheless, these notable (and perhaps unexpected) findings from studies in this observational patient cohort are corroborated by parallel studies in preclinical models with preliminary mechanistic insights. In light of these collective results, dietary habits and probiotic supplement use should be considered in patients receiving ICB and in efforts to modulate the gut microbiota. These factors should be more thoughtfully evaluated in strategies to improve cancer outcomes.
Supplementary Material
ACKNOWLEDGMENTS
C.N.S. acknowledges the Parker Institute for Cancer Immunotherapy for funding time devoted to continued analysis of this project. She also acknowledges M. D. Swartz, L. B. Piller, and X. Du at UTSPH for their participation on her thesis committee. J.L.M. acknowledges the Transdisciplinary Research in Energetics and Cancer Research Training Workshop R25CA203650. J.L.M. and C.R.D. acknowledge the MDACC Center for Energy Balance in Cancer Prevention and Survivorship. C.R.D. acknowledges the MDACC Bionutrition Core. We thank our collaborators and personnel at the LICI, NCI Microbiome and Genetics Core Facility, and NCI Mouse Gnotobiotic Core Facility, The Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine (including S. J. Javornik Cregeen for her role in microbiome data processing), and Cosmos ID for their high quality and timely microbiome data generation services. We also wish to acknowledge MD Anderson’s Program for Innovative Microbiome and Translational Research (PRIME-TR) for supporting the analysis and interpretation of the microbiome results presented herein (J.A.W. and N.J.A. are the program director and executive scientific director for PRIME-TR, respectively). Most importantly, the study team wishes to thank all patients who contributed their time, samples, and data to this research.
Funding:
This study received support from National Institute of Health grant 1R01 CA219896–01A1 (J.A.W.); US-Israel Binational Science Foundation grant 201332 (J.A.W.); the Melanoma Research Alliance grant 4022024 (J.A.W.); American Association for Cancer Research Stand Up to Cancer grant SU2C-AACR-IRG-19–17 (J.A.W.); the Andrew Sabin Family Fellows Program (J.A.W. and C.R.D.); MD Anderson Cancer Center’s Melanoma Moon Shots Program (J.A.W., J.L.M., C.R.D., M.A.D., H.A.T., J.E.G., and E.M.B.); and the Melanoma Research Alliance grant 564449 (L.C., J.A.W., and J.L.M.). The authors additionally received support from Department of Defense grant W81XWH 16 1 0121 (J.A.W.); the MD Anderson Cancer Center Multidisciplinary Research Program grant (J.A.W.); the Parker Institute for Cancer Immunotherapy at MD Anderson Cancer Center (J.A.W., H.A.T., P.S., and J.P.A.); American Society of Clinical Oncology and Conquer Cancer Foundation Career Development award AWD0000627 (J.L.M.); the Elkins Foundation (J.L.M.); the Seerave Foundation (J.L.M.); Rising Tide Foundation grant AWD00004505 (J.L.M.); the Mark Foundation grant AWD00004538 (J.L.M.); the Longenbaugh-Torian Fund (J.L.M.); the MD Anderson Cancer Center SPORE in Melanoma P50CA221703 (M.A.D., H.A.T., I.C.G., J.A.W., S.P.P., J.E.L., J.E.G., A.J.L., and J.L.M.); the MD Anderson Physician Scientist Program (J.L.M.); the Cancer Research Institute Irvington Fellowship Program (M.V.); Cancer Prevention and Research Institute of Texas Research Training Program RP170067 (A.P.C., J.T., and J.Z.); Cancer Prevention and Research Institute of Texas Research Training Program RP210028 (L.M.K.); the US Department of State, Bureau of Educational and Cultural Affairs (A.P.C.); the Fulbright Franco–Américaine Commission (A.P.C.); Cancer Prevention and Research Institute of Texas Research Training Program RP160097 (X.Z.); National Health and Medical Research Council of Australia CJ Martin Early Career Fellowship grant 1148680 (M.C.A.); National Institutes of Health T32 CA 009599 (M.G.W. and B.A.H.); the MD Anderson NCI Cancer Center Support grant P30 CA016672 (M.G.W., B.A.H., C.R.D., K.C.-D., and C.B.P.); National Institute of Health grant 1F32CA260769–01 (G.Mo.); the Charles A. King Trust Postdoctoral Research Fellowship (Y.Y. and C.H.); the John M. Skibber Endowed Professorship (J.E.G.); the Michael and Patricia Melanoma Research Endowment (J.E.G.); National Institute of Health grant R0AI143886 (J.H.); Columbia University Health Sciences NCI Cancer Center Support grant P30 CA013696 (J.H.); National Institute of Health grant R01HL124112 (R.R.J.); Cancer Prevention and Research Institute of Texas grant RR160089 (R.R.J.); National Institute of Health grant R01AI109294 (S.S.W.); National Institute of Health grant R01AI133822 (S.S.W.); the Richard E. Haynes Distinguished Professor in Clinical Cancer Prevention (L.C.); American Cancer Society grant RSG-17–049-01-NEC (C.R.D.); the National Institute of Health Intramural Research Program (E.P.-G., C.-P.D., and G.Me.); FLEX Synergy Award from the National Cancer Institute Center for Cancer Research (E.P.-G., C.-P.D., and G.Me.); the National Cancer Institute Intramural Research Program (J.A.M., M.V., J.H.B., R.R.R., and G.T.); the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (M.A.D.); American Cancer Society/Melanoma Research Alliance grant 134148-MRAT-19–168-01 (M.A.D.); the AIM at Melanoma Foundation (M.A.D.); the National Institute of Health/National Cancer Institute grant 1 P50 CA221703–02 (M.A.D.); the National Institute of Health/National Cancer Institute grant 1U54CA224070–03 (M.A.D.); Cancer Fighters of Houston (M.A.D.); and the Anne and John Mendelsohn Chair for Cancer Research (M.A.D.).
Footnotes
Competing interests: J.A.W., V.G., and M.C.A. are inventors on patent WO2020106983A1 submitted by the Board of Regents, The University of Texas System, and Institut Gustav Roussy that covers methods and compositions for treating cancer and for predicting a subject’s response to combination checkpoint inhibitor therapy. J.A.W. and V.G. are inventors and C.N.S. is a collaborator on a US patent application (PCT/US17/53.717) submitted by the University of Texas MD Anderson Cancer Center that covers methods to enhance ICB responses by modulating the microbiome. J.A.W. and R.R.J. are inventors on patent WO2020150429A1 submitted by the Board of Regents, The University of Texas System, that covers methods and compositions for treating immune checkpoint inhibitor (ICI)–associated colitis in a subject through the administration of fecal matter from a healthy donor to the subject. R.R.J. is an inventor on patent WO2016086161A1 submitted by the Memorial Sloan Kettering Cancer Center that covers compositions and methods for increasing the abundance of commensal bacteria belonging to the order Clostridiales that are associated with reduced lethal graft-versus-host disease and improved overall survival after bone marrow or hematopoietic stem cell transplant. R.R.J. is an inventor on patent WO2017041039A1 submitted by Memorial Sloan Kettering Cancer Center that covers methods and compositions for reducing the risk of cancer relapse in a subject who has received cancer treatment. J.A.W. reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, Physician Education Resource, Med Immune, and Bristol-Myers Squibb and serves as a consultant and/or advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol-Myers Squibb, Merck, Biothera Pharmaceuticals, Microbiome DX, and Micronoma. J.A.W. also receives research support from GlaxoSmithKline, Roche/Genentech, Bristol-Myers Squibb, and Novartis. V.G. reports honoraria from Expert Connect and Kansas Society of Clinical Oncology. J.L.M. reports advisory board participation and honoraria from Bristol-Myers Squibb, Merck, and Roche/Genentech. M.C.A. reports advisory board participation, honoraria, and research funding to their institution from MSD Australia, outside the submitted work, and contract research for BMS Australia, outside the current work. A.P.C. is an employee and equity holder at Immunai. A.P.C. serves as an advisory member and holds equity in Vastbiome. G.Mo. is a coinventor on US patents (PCT/US2019/022194, PCT/US2020/029556, and PCT/US2020/046050) not related to the content of this paper. M.I.R. reports paid consultant position for AMGEN and paid consultant and advisory board membership for MERCK. I.C.G. reports research support from Bristol-Myers Squibb and Merck. I.C.G. serves as a consultant for Bristol-Myers Squibb, Array, and Novartis. R.N.A. receives research funding from Merck, Bristol-Myers Squibb, Genentech, Novartis, and Iovance. J.E.G. is a consultant and/or is on the advisory board for Merck, Regeneron, Syndex, Novartis, and Bristol-Myers Squibb, unrelated to the content of this work. R.R.J. is an advisor and holds equity in Seres Therapeutics and Kaleido Biosciences; serves on the advisory board of MaaT Pharma, LISCure Biosciences, and Prolacta Biosciences; and consults for Davolterra, Merck, Microbiome DX, and Karius. C.H. is on the scientific advisory board for Seres Therapeutics and Empress Therapeutics. M.K.W. is on the advisory boards of Merck, Pfizer, Bristol Myers Squibb, Regeneron, EMD-Serono, ExiCure, Castle Biosciences, and Adagene. H.A.T. is a consultant for BMS, Merck, Novartis, Genentech, Eisai, Iovance, Karyopharm, and Pfizer and reports research funding to institution from BMS, Merck, Novartis, Genentech, GSK, and Dragonfly. A.D. serves as a consultant for Nektar, MultiVir, Idera, Array, and Bristol-Myers Squibb. P.S. reports consulting or stock ownership or advisory board for Achelois, Adaptive Biotechnologies, Affini-T, Apricity, BioAtla, BioNTech, Candel Therapeutics, Catalio, Codiak, Constellation, Dragonfly, Earli, Enable Medicine, Glympse, Hummingbird, ImaginAb, Infinity Pharma, Jounce, JSL Health, Lava Therapeutics, Lytix, Marker, Oncolytics, PBM Capital, Phenomic AI, Polaris Pharma, Sporos, Time Bioventures, Trained Therapeutix, Two Bear Capital, and Venn Biosciences. J.P.A. reports consulting, stock ownership, or advisory board membership for Achelois, Adaptive Biotechnologies, Apricity, BioAtla, BioNTech, Candel Therapeutics, Codiak, Dragonfly, Earli, Enable Medicine, Hummingbird, ImaginAb, Jounce, Lava Therapeutics, Lytix, Marker, PBM Capital, Phenomic AI, Polaris Pharma, Time Bioventures, Trained Therapeutix, Two Bear Capital, and Venn Biosciences. M.A.D. has been a consultant to Roche/Genentech, Array, Pfizer, Novartis, BMS, GSK, Sanofi-Aventis, Vaccinex, Apexigen, Eisai, and ABM Therapeutics, and he has been the principal investigator of research grants to MD Anderson from Roche/Genentech, GSK, Sanofi-Aventis, Merck, Myriad, and Oncothyreon. P.H. is on the scientific advisory board for Dragonfly and Immatics. M.I.R. is on the melanoma advisory board for Merck and is a paid consultant for AMGEN and Merck. S.P.P. reports institutional clinical trial support from NCI, Merck, and Bristol Myers Squibb during the conduct of the study; institutional clinical trial support from Reata Pharmaceuticals, Novartis, Deciphera, Provectus Biopharmaceuticals, Foghorn Therapeutics, TriSalus Life Sciences, and Seattle Genetics; advisory board honoraria from Castle Biosciences and TriSalus Life Sciences; honoraria as Peer Discussion Group Leader for Merck; and honoraria for service as Chair of International Data Monitoring Committee for Immunocore, outside the submitted work.
Data and materials availability: Raw sequencing data and all relevant human data necessary for reproducing results are available in the NCBI Sequence Read Archive under BioProject ID PRJNA770295. All analyzed sequencing data are available in the supplementary materials.
REFERENCES AND NOTES
- 1.Ribas A, Wolchok JD, Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Routy B et al. , Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Matson V et al. , Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gopalakrishnan V et al. , Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel AE et al. , Neoplasia 19, 848–855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters BA et al. , Genome Med 11, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews MC et al. , Nat. Med 27, 1432–1441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu GD et al. , Science 334, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald D et al. , mSystems 3, e00031–18 (2018).29795809 [Google Scholar]
- 10.Maier L et al. , Nature 555, 623–628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suez J et al. , Cell 174, 1406–1423.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Willing BP, Russell SL, Finlay BB, Nat. Rev. Microbiol 9, 233–243 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Asnicar F et al. , Nat. Med 27, 321–332 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothschild D et al. , Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Eisenhauer EA et al. , Eur. J. Cancer 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Thompson FE, Midthune D, Kahle L, Dodd KW, Nutr J 147, 1226–1233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Cancer Institute, Dietary Screener Questionnaire (DSQ) in the NHANES 2009–10: Data processing and scoring procedures (2019); https://epi.grants.cancer.gov/nhanes/dietscreen/scoring.
- 18.Gharaibeh RZ, Jobin C, Gut 68, 385–388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baruch EN et al. , Science 371, 602–609 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Davar D et al. , Science 371, 595–602 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arthur JC et al. , Sci. Rep 3, 2868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita Y et al. , Cancer Immunol. Res 8, 1236–1242 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Sivan A et al. , Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanoue T et al. , Nature 565, 600–605 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E, Gut Microbes 3, 289–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Besten G et al. , J. Lipid Res 54, 2325–2340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poeker SA et al. , Sci. Rep 8, 4318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.So D et al. , Am. J. Clin. Nutr 107, 965–983 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Desai MS et al. , Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McQuade JL et al. , Lancet Oncol 19, 310–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An Y et al. , J. Transl. Med 18, 235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinato DJ et al. , JAMA Oncol 5, 1774–1778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klurfeld DM et al. , Am. J. Physiol. Endocrinol. Metab 315, E1087–E1097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makki K, Deehan EC, Walter J, Bäckhed F, Cell Host Microbe 23, 705–715 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Guijarro E et al. , Nat. Med 26, 781–791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michalak L et al. , Nat. Commun 11, 5773 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver A et al. , mSystems 6, e00115–21 (2021).33727392 [Google Scholar]
- 38.Leshem A, Segal E, Elinav E, mSystems 5, e00665–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murga-Garrido SM et al. , Microbiome 9, 117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel CR, McQuade JL, Trends Cancer 5, 521–524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


