Abstract
Activating transcription factor 4 (ATF4) is a multifunctional transcription regulatory protein in the basic leucine zipper superfamily. ATF4 can be expressed in most if not all mammalian cell types, and it can participate in a variety of cellular responses to specific environmental stresses, intracellular derangements, or growth factors. Because ATF4 is involved in a wide range of biological processes, its roles in human health and disease are not yet fully understood. Much of our current knowledge about ATF4 comes from investigations in cultured cell models, where ATF4 was originally characterized and where further investigations continue to provide new insights. ATF4 is also an increasingly prominent topic of in vivo investigations in fully differentiated mammalian cell types, where our current understanding of ATF4 is less complete. Here, we review some important high-level concepts and questions concerning the basic biology of ATF4. We then discuss current knowledge and emerging questions about the in vivo role of ATF4 in one fully differentiated cell type, mammalian skeletal muscle fibers.
Key words: ATF4, skeletal muscle atrophy, sarcopenia, ursolic acid, tomatidine
Activating Transcription Factor 4 Is an Essential Subunit of Many Different Heterodimeric Basic Leucine Zipper Transcription Factors
Mammals possess >50 basic leucine zipper (bZIP) proteins, including activating transcription factor 4 (ATF4) (1). bZIP proteins form transcription factors by assembling into either homodimeric complexes (2 identical bZIP proteins) or heterodimeric complexes (2 nonidentical bZIP proteins) (2., 3., 4., 5.). bZIP homodimers and heterodimers are capable of binding specific gene regulatory elements, gripping them like a clothespin on a clothesline. Dimerization and DNA binding are mediated by bZIP domains, which are found in all bZIP family members, but are slightly different between bZIP family members.
The bZIP domain typically lies near the COOH terminus of a bZIP protein and usually makes up a minor portion of the overall protein, ∼18% in ATF4. The remaining portion is highly variable between bZIP family members, tends to be intrinsically disordered, and is critical for the regulation and function of the protein. For example, in ATF4, sequence outside of the bZIP domain makes up ∼82% of the protein, contains most of ATF4’s known sites of posttranslational modification, and mediates numerous interactions with non-bZIP proteins that regulate ATF4’s stability, localization, and capacity to activate gene transcription (6., 7., 8., 9., 10., 11., 12., 13., 14., 15., 16., 17.).
A bZIP domain consists of a basic region and a leucine zipper. The basic region mediates DNA binding, and the pair of basic regions in a homo- or heterodimeric bZIP transcription factor dictates which genetic regulatory elements can be targeted by the transcription factor. The leucine zipper is an amphipathic helix that forms a coiled-coil structure with the leucine zipper of an identical and/or nonidentical bZIP protein, thereby mediating homo- and/or heterodimerization. In some bZIP proteins, the leucine zipper is amenable to homodimerization. However, the leucine zipper in ATF4 is structurally unsuited to homodimerization, and thus, ATF4 cannot form stable homodimers (18). On the other hand, the ATF4 leucine zipper is highly amenable to heterodimerization with leucine zippers of other bZIP family members. As a result, ATF4 has a strong propensity to form heterodimeric bZIP transcription factors with many other bZIP family members. This point is well illustrated by in vitro binding studies, which have shown that an individual ATF4 bZIP domain has negligible affinity for another ATF4 bZIP domain but high affinity for the bZIP domains of at least 30 other bZIP family members (19, 20). Consistent with those in vitro findings, the in vivo effects of ATF4 are largely if not entirely mediated by ATF4 heterodimers (21., 22., 23., 24., 25., 26., 27., 28., 29.). Moreover, ATF4 can coexist as multiple distinct heterodimers in a single cell type in vivo (29). Thus, the ATF4 leucine zipper prevents ATF4 from being functional in the absence of heterodimerization partners, but it also imparts versatility and multifunctionality by allowing ATF4 to heterodimerize with many other bZIP family members. The name given to ATF4 (activating transcription factor 4) can be misleading insofar as it suggests that ATF4 is a standalone transcription factor, capable of activating genes by itself. ATF4 is not a functional transcription factor by itself but one-half of many possible heterodimeric transcription factors.
ATF4 Heterodimers Have Unique and Context-Dependent Biological Effects
Heterodimerization of bZIP proteins is a classic biological example of combinatorial control (2., 3., 4., 5.). By combining 2 different bZIP proteins, each ATF4 heterodimer can be subject to a unique array of regulatory mechanisms and can generate a unique set of downstream effects. For example, by having different pairs of basic regions, different ATF4 heterodimers can target different genetic regulatory elements and thereby regulate different genes. By having different pairs of NH2-terminal regions, different ATF4 heterodimers can have different protein–protein interactions, posttranslational regulation, and capacity for gene activation. Thus, the unique structure of each ATF4 heterodimer imparts unique function and regulation.
Although each ATF4 heterodimer has the potential to bind and regulate dozens of specific genes, the actual effects of an ATF4 heterodimer are highly context dependent due to many variables that can directly or indirectly affect the capacity of ATF4 heterodimers to access and regulate each of their potential target genes. Some of the context-dependent factors may be static and intrinsic to the cell type, and others may be dynamic in response to changing intracellular or extracellular conditions. As a result, the effects of an ATF4 heterodimer may be different in different cell types and may be different in the same cell type under different conditions.
Different contexts may also generate different availabilities of bZIP family members that can heterodimerize with ATF4. Because ATF4 can simultaneously participate in multiple distinct heterodimers, the overall set of genes that require ATF4 for maximal expression in a specific context (ATF4-dependent genes) can be a mixture of genes that are regulated by different ATF4 heterodimers, with some ATF4-dependent genes activated by one ATF4 heterodimer and other ATF4-dependent genes activated by other ATF4 heterodimers. Variable combinations and concentrations of ATF4 heterodimers impart a higher level of combinatorial control.
Formation of ATF4 Heterodimers Is Low under Homeostatic Conditions but Can Be Stimulated by Diverse Stress Conditions or Growth Factors
ATF4 heterodimers are formed when ATF4 is expressed in the presence of at least 1 bZIP family member that can heterodimerize with ATF4. The rate-limiting factor in heterodimer formation is typically ATF4 expression, which is always tightly controlled. Although cells can potentially regulate ATF4 expression at several levels (gene transcription, mRNA turnover, mRNA translation, and protein degradation), the regulation of ATF4mRNA translation is thought to be a major site of regulation under most circumstances (17, 30, 31). Under homeostatic conditions, ATF4 mRNA is poorly translated, even when ATF4 mRNA is highly abundant. However, ATF4 translation increases dramatically in response to certain cellular stress conditions and in response to some anabolic hormones and growth factors, leading to the formation of ATF4 heterodimers (Figure 1).
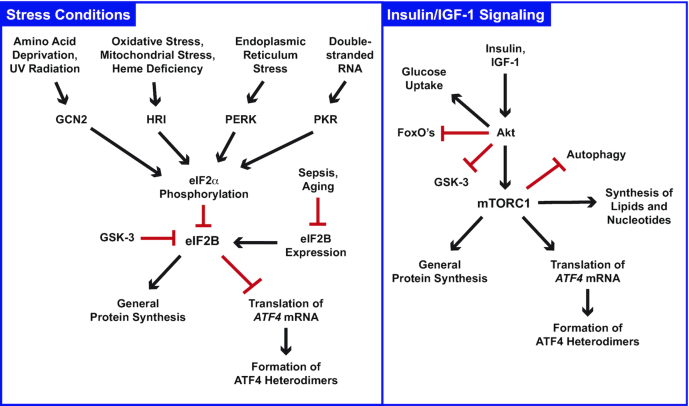
Figure 1.
Schematic illustrating some conditions and signaling pathways that promote translation of ATF4 mRNA. Arrows indicate signaling activation and red bars represent inhibition. Please note that these pathways have been characterized in intricate detail, and many known intermediary molecules and steps are not shown here or discussed in the text. Furthermore, crosstalk between the eIF2α kinase and mTORC1 pathways can occur in some contexts, and ATF4 heterodimers can exert important feedback inhibition on both eIF2α kinase signaling and mTORC1 signaling. For additional information, the reader is referred to several excellent reviews and original research articles on these topics (e.g., 17, 31, 35, 46, 47, 111., 112., 113., 114.). ATF4, activating transcription factor 4; eIF2α, eukaryotic translation initiation factor 2 subunit α; eIF2Β, eukaryotic translation initiation factor 2B; FoxO, forkhead box O; GCN2, general control nonderepressible 2; GSK-3, glycogen synthase kinase 3; HRI, heme-regulated inhibitor kinase; IGF-1, insulin-like growth factor 1; mTORC1, mechanistic target of rapamycin complex 1; PERK, protein kinase RNA-like endoplasmic reticulum kinase; PKR, protein kinase RNA activated.
Cellular stress conditions primarily increase ATF4 translation by activating eukaryotic translation initiation factor 2 subunit α (eIF2α) kinases. Mammals have 4 eIF2α kinases: general control nonderepressible 2 (GCN2), heme-regulated inhibitor kinase (HRI), protein kinase RNA-like endoplasmic reticulum kinase (PERK), and protein kinase RNA activated (PKR). Well-characterized physiologic mechanisms that activate eIF2α kinases include but are not limited to amino acid deprivation and ultraviolet radiation (which activate GCN2); oxidative stress, mitochondrial stress, and heme deficiency (which activate HRI); endoplasmic reticulum stress (which activates PERK); and the accumulation of double-stranded RNA in certain viral infections (which activates PKR) (17, 30., 31., 32., 33.). Importantly, eIF2α kinase-mediated stress responses are cell autonomous, meaning that they allow an individual cell to sense and respond to the provocative stress without assistance from other cells. The overall collection of signaling pathways that are mediated by the 4 mammalian eIF2α kinases is known as the integrated stress response (34). Activation of any one of the 4 eIF2α kinases is sufficient to increase ATF4 mRNA translation.
eIF2α kinases (GCN2, HRI, PERK, and PKR) increase translation of ATF4 mRNA by phosphorylating the translation initiation factor eIF2α on a specific serine residue (serine 51 in humans) (17, 30, 31). When eIF2α is phosphorylated, it inhibits the activity of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor whose activity is required for high levels of general (cap-dependent) protein synthesis (30). eIF2B activity can also be inhibited by eIF2α-independent mechanisms that sometimes occur in conditions of cellular stress, such as a reduction of eIF2B expression, or inhibitory phosphorylation of eIF2B by protein kinases such as glycogen synthase kinase 3 (GSK-3) (35).
Inhibition of eIF2B activity (through eIF2α phosphorylation or other mechanisms) increases the translation of ATF4 mRNA, but it also simultaneously decreases translation of almost all other cellular mRNAs (17, 30, 31, 35). As a result, inhibition of eIF2B activity increases formation of ATF4 heterodimers but decreases bulk protein synthesis. The resulting reduction in general protein synthesis typically contributes to growth arrest and conserves energy and nutrient resources. In this context, newly generated ATF4 heterodimers activate stress response genes that encode, among other things, amino acid transporters, aminoacyl-tRNA synthetases, enzymes for the synthesis of amino acids and nucleotides, heat shock proteins and other molecular chaperones, enzymes that alleviate oxidative stress, proteins that inhibit cell growth and division, and additional bZIP family members that can heterodimerize with ATF4 and amplify the stress response. By activating these types of genes, ATF4 heterodimers help the cell try to survive and correct the provocative cellular stress.
In mammals, ATF4 translation can also be stimulated by certain anabolic hormones and growth factors, including insulin, insulin-like growth factor 1 (IGF-1) and transforming growth factor β (TGFβ). These hormones and growth factors can act in a non-cell-autonomous manner, and some, such as insulin, are endocrine hormones that simultaneously act upon several cell types and tissues to coordinate whole-body metabolic responses. Importantly, these hormones and growth factors do not inhibit eIF2B but rather increase ATF4 translation by activating the mechanistic target of rapamycin complex 1 (mTORC1) (36., 37., 38., 39., 40., 41., 42.). Through distinct biochemical mechanisms, mTORC1 activation and eIF2B inhibition allow ribosomes to bypass short upstream open reading frames in the 5' leader of ATF4 mRNA and translate the downstream open reading frame that encodes ATF4 (38, 39, 43, 44). Alongside their effects on ATF4 translation, mTORC1 activation and eIF2B inhibition also modestly increase the concentration of ATF4 mRNA (36, 45).
In contrast to eIF2B inhibition (which increases ATF4 expression and decreases general protein synthesis), mTORC1 activity increases both ATF4 and general protein synthesis (36., 37., 38., 39., 40., 41., 42., 46). In addition, mTORC1 stimulates other anabolic processes, such as the synthesis of lipids and nucleotides, while inhibiting catabolic processes such as autophagy (46). Furthermore, when mTORC1 is activated by anabolic hormones such as insulin and IGF-1, the effects of mTORC1 are accompanied by hormone-induced effects that complement but do not require mTORC1 activity, including inhibition of specific catabolic signaling molecules such as GSK-3 and forkhead box O (FoxO) transcription factors, inhibition of glycogenolysis and gluconeogenesis, and stimulation of glucose uptake and glycogen synthesis (47). In the context of insulin/IGF-1/mTORC1 signaling, ATF4 heterodimers primarily activate genes that promote amino acid uptake and the synthesis of charged tRNAs, amino acids, nucleotides, and glutathione, thereby facilitating a portion of anabolism within a much broader anabolic and anticatabolic response.
How can ATF4 heterodimers facilitate such diverse cellular adaptations, ranging from growth arrest (in the integrated stress response) to anabolism (in insulin/IGF-1 signaling)? This is an important and unresolved question. One possibility is that different ATF4 heterodimers or different combinations of ATF4 heterodimers mediate the different effects of signaling molecules such as eIF2α kinases and mTORC1. Another potential explanation, not mutually exclusive with the first, is that ATF4 heterodimers exist and act in very different contexts during the integrated stress response and insulin/IGF-1 signaling. In the integrated stress response, ATF4 heterodimers exist and act under conditions that typically favor catabolism and prohibit anabolism. In contrast, during insulin/IGF-1 signaling, ATF4 heterodimers exist and act in the setting of nutrient abundance and numerous other hormone-mediated events that promote anabolism and inhibit catabolism.
Although ATF4 heterodimers facilitate changes that are appropriate to the existing conditions, they can have deleterious effects if their expression and activity are sustained for too long. For example, if a cellular stress continues unabated, ATF4 heterodimers can promote degenerative conditions such as skeletal muscle atrophy (29, 48., 49., 50., 51.).
Diverse Stress Conditions Cause Skeletal Muscle Fibers to Undergo Atrophy
Skeletal muscle atrophy is an acquired loss of muscle mass and strength. It typically affects adult skeletal muscles that were previously well developed and healthy. It is also known as “muscle wasting” and can be called “sarcopenia” when it occurs as an effect of aging. Muscle atrophy is also a major feature of whole-body wasting syndromes known as “cachexia.” Skeletal muscle atrophy can affect any person and rarely has a heritable component.
Skeletal muscle atrophy can be localized or generalized (52., 53., 54.). Localized muscle atrophy occurs when a single muscle or group of muscles experiences localized disuse due to conditions such as arthritis, joint injury, joint repair, or local motor neuron trauma or disease. Generalized muscle atrophy has many causes, including whole-body muscle disuse (as in bedrest, generalized neurologic disorders, and spaceflight), nutrient deprivation/malnutrition, advanced age, and almost any severe systemic illness, including critical illness, cancer, diabetes mellitus, heart failure, pulmonary disease, renal failure, cirrhosis, rheumatoid arthritis, male hypogonadism, Cushing syndrome, thyroid disorders, and chronic infections such as tuberculosis and HIV/AIDS. Certain medications also cause generalized muscle atrophy (cancer chemotherapy, high-dose glucocorticoid therapy, antiandrogen prostate cancer therapy, etc.), as do many rare genetic disorders that primarily or secondarily affect skeletal muscle. In many patients, the etiology of skeletal muscle atrophy is multifactorial, caused by various combinations of malnutrition, muscle disuse, systemic illness, and aging.
Histologically, muscle atrophy appears as a reduction in the size of skeletal muscle fibers (52., 53., 54.). Skeletal muscle contains several types of cells, including skeletal muscle fibers, myogenic stem cells (satellite cells), endothelial cells, fibroblasts, adipocytes, and nerve terminals. Skeletal muscle fibers are large, multinucleated, and terminally differentiated cells that comprise most of skeletal muscle mass and are responsible for muscle contraction, force generation, and most of the metabolic functions of skeletal muscle. Because they are postmitotic, skeletal muscle fibers cannot divide or undergo classical senescence. Furthermore, muscle atrophy is not typically associated with the death of skeletal muscle fibers. However, depending on the external conditions, muscle fibers can undergo atrophy (becoming smaller) or hypertrophy (becoming larger). Conditions such as malnutrition, disuse, illness, and aging cause previously well-developed and healthy adult skeletal muscle fibers to undergo atrophy, leading to an anatomic reduction in muscle mass (skeletal muscle atrophy) and a corresponding loss of strength and metabolic capacity.
In humans, muscle fiber atrophy occurs slowly (over years) with aging and rapidly (over days to weeks) with sustained nutrient deficiency, sustained muscle disuse, or severe systemic illness. The precise triggers of muscle fiber atrophy are variable and not always well understood, especially in aging and systemic illness, but can include a lack of sufficient nutrients, a lack of sufficient motor neuron input, ischemia, a relative or absolute deficiency of a hormone with anabolic actions on muscle fibers (e.g., insulin, IGF-1, and testosterone), a relative or absolute excess of a factor with catabolic or antianabolic actions on muscle fibers (e.g., glucocorticoid, myostatin, activin, and certain factors secreted from inflammatory cells, tumor cells, and senescent cells), or any combination thereof (52). These extrinsic triggers act upon muscle fibers to generate numerous cellular changes that promote muscle fiber atrophy, including disruptions in the normal structure and function of mitochondria, myonuclei, and neuromuscular junctions, as well as variable changes in general protein synthesis and/or protein degradation pathways that reduce the quantity and quality of sarcomeric proteins, the major constituents of muscle fibers. These basic characteristics of muscle atrophy are conserved across mammalian species.
Molecular Mechanisms of Skeletal Muscle Fiber Atrophy
The molecular mechanisms of muscle fiber atrophy are numerous, complex, incompletely understood, and still being discovered at a rapid pace (52., 53., 54.). A few general points can be made.
First, these mechanisms involve genes, RNAs, proteins, and lipids that act within muscle fibers and play a causal role in muscle fiber atrophy in at least one context. Some of these mechanisms promote muscle atrophy by stimulating degradative cellular processes within muscle fibers, whereas other mechanisms promote muscle atrophy by inhibiting molecular and cellular processes that are critical for the normal structure and function of muscle fibers.
Second, these mechanisms tend to be relatively dormant in nonstressed adult skeletal muscle fibers, but they become significantly more active at some point during at least one condition that causes muscle atrophy. Many of these mechanisms may also be transiently activated by stress conditions that are too transient to cause muscle atrophy, such as short-term fasting or exercise. However, by exerting a stronger or more sustained stress on muscle fibers, conditions that cause muscle atrophy seem to activate these mechanisms in a stronger or more sustained way, which enables them to contribute to the process of muscle fiber atrophy.
Third, the molecular mechanisms of muscle fiber atrophy are context dependent and can vary based on the nature of stress (i.e., the underlying cause(s) of muscle atrophy) and on the severity and duration of the stress. Although dozens of molecular mechanisms of muscle atrophy have been identified and characterized over the past 2 decades, there are no examples thus far of a molecular mechanism that is universally involved in muscle fiber atrophy under all conditions. In other words, every known molecular mechanism of muscle atrophy has been dissociated from the pathogenesis of muscle atrophy caused by at least 1 atrophy-inducing condition, be it starvation, a type of illness, a type of muscle disuse, or aging. In some cases, mechanisms are dissociated because they do not occur during a particular atrophy-inducing condition. In other cases, mechanisms do occur, but they are nonessential for muscle fiber atrophy in that specific context due to redundancy or due to the acquisition of new roles or functions in that specific context; the best-characterized example in this regard is mTORC1 activity in muscle fibers, which promotes muscle fiber atrophy in some contexts and protects against muscle fiber atrophy in other contexts (e.g., 55., 56., 57., 58., 59., 60., 61., 62., 63., 64., 65., 66., 67., 68., 69., 70.).
Fourth, there are no examples of a single molecular mechanism that is fully responsible for the entire process of muscle fiber atrophy in any context. Rather, every known molecular mechanism of muscle atrophy mediates a portion of muscle atrophy under certain conditions. Thus, in every context, muscle fiber atrophy appears to require multiple molecular mechanisms, some of which operate in an independent and additive manner. This is an important consideration for therapeutic strategies.
A Heterodimer Composed of ATF4 and CCAAT Enhancer Binding Protein β Promotes Skeletal Muscle Fiber Atrophy during Starvation, Immobilization, and Aging
Like other cell types, skeletal muscle fibers do not significantly express ATF4 in the absence of cellular stress. Moreover, ATF4 expression in skeletal muscle fibers is nonessential for the normal development or maintenance of skeletal muscle mass and skeletal muscle function (49., 50., 51.). However, ATF4 expression in skeletal muscle fibers rises in response to many conditions that cause muscle atrophy (48, 49, 71). When ATF4 is expressed in skeletal muscle fibers, it interacts with several different bZIP family members, including CCAAT enhancer binding protein γ (C/EBPγ), CCAAT enhancer binding protein δ (C/EBPδ), MAF bZIP transcription factor (c-MAF), the p30 and p42 isoforms of CCAAT enhancer binding protein α (C/EBPα), and the liver-enriched inhibitory protein (LIP) and liver activator protein (LAP) isoforms of CCAAT enhancer binding protein β (C/EBPβ) (29). As a result of these interactions, ATF4 forms multiple heterodimers that simultaneously coexist within skeletal muscle fibers. The rate-limiting step in formation of these heterodimers is ATF4 expression (Figure 2).
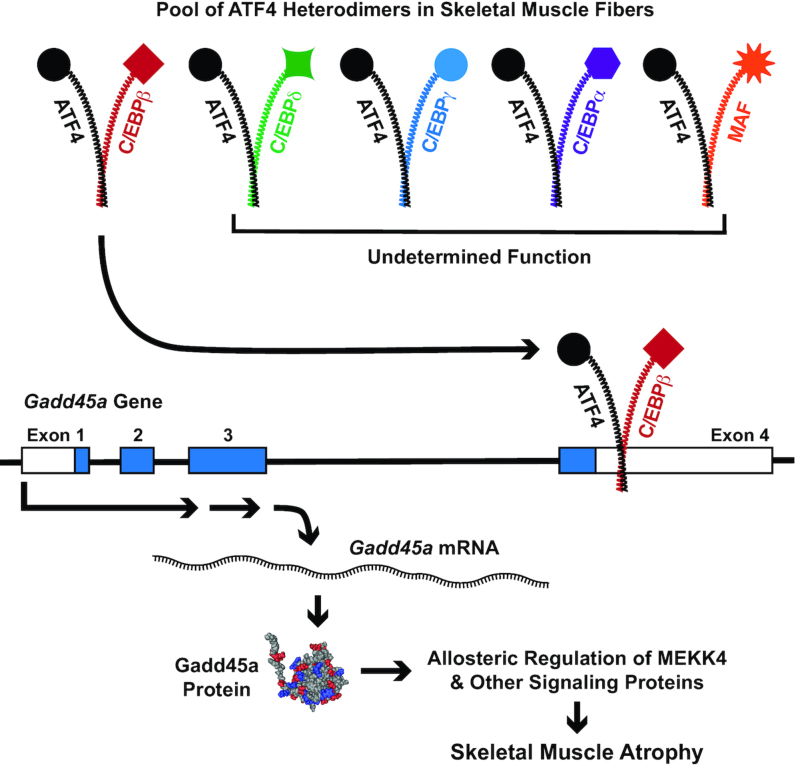
Figure 2.
Schematic illustrating ATF4 heterodimers in skeletal muscle fibers and how the ATF4-C/EBPβ heterodimer activates the Gadd45a gene in skeletal muscle fibers. Reproduced with permission from Ebert et al. (29). ATF4, activating transcription factor 4; C/EBPα, CCAAT enhancer binding protein α; C/EBPβ, CCAAT enhancer binding protein β; C/EBPδ, CCAAT enhancer binding protein δ; C/EBPγ, CCAAT enhancer binding protein γ; Gadd45a, growth arrest and DNA damage inducible α; MEKK4, mitogen-activated protein kinase kinase kinase 4.
One ATF4 heterodimer, composed of ATF4 and the LAP isoform of C/EBPβ, promotes skeletal muscle atrophy. This conclusion is based on several lines of evidence from in vivo mouse models. First, a targeted reduction of ATF4 in skeletal muscle fibers reduces muscle atrophy during starvation, immobilization, and aging (48, 49, 51). Second, forced expression of ATF4 in skeletal muscle fibers induces muscle atrophy in healthy young adult animals (48). Third, a targeted reduction of C/EBPβ in skeletal muscle fibers inhibits ATF4-mediated muscle atrophy (29). In contrast to C/EBPβ, other ATF4 heterodimerization partners are not required for ATF4-mediated muscle atrophy (29). Based on these experimental findings, the ATF4-C/EBPβ heterodimer appears to account for the atrophic effects of ATF4 in skeletal muscle fibers, and expression of the ATF4-C/EBPβ heterodimer in skeletal muscle fibers appears to be sufficient to induce muscle atrophy and partially required for some forms of generalized muscle atrophy (starvation and aging) and localized muscle atrophy (immobilization). The functions of the other ATF4 heterodimers in skeletal muscle fibers are not yet known.
The ATF4-C/EBPβ Heterodimer Activates Genes That Encode Mediators of Muscle Fiber Atrophy
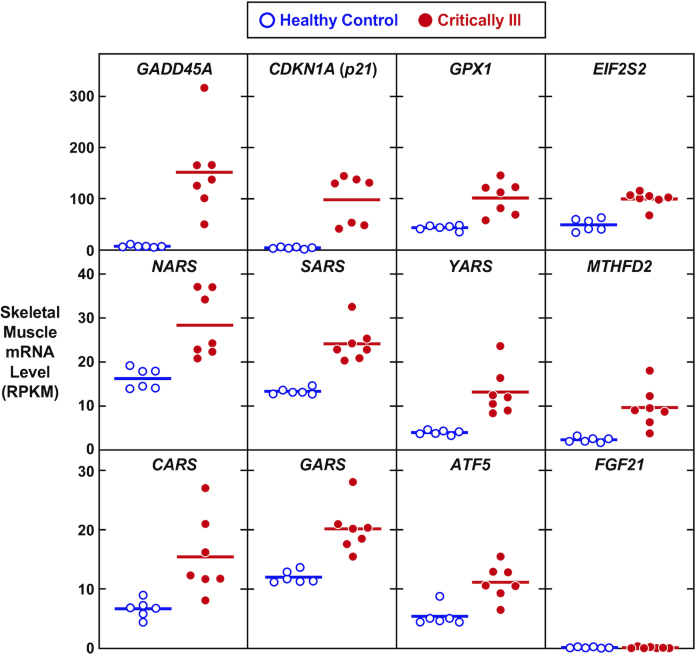
In skeletal muscle fibers, the ATF4-C/EBPβ heterodimer activates at least 2 genes that encode mediators of muscle atrophy, growth arrest and DNA damage inducible α (Gadd45a) and p21. The Gadd45a gene is weakly expressed in nonstressed skeletal muscle fibers, but it is strongly induced during muscle atrophy in humans, mice, and other mammalian species (48., 49., 50., 72., 73., 74., 75., 76., 77., 78., 79., 80.). For example, in muscle biopsy specimens from critically ill patients with severe generalized skeletal muscle atrophy, the concentration of GADD45A mRNA is increased 22-fold relative to muscle biopsy specimens from healthy control human subjects (80) (Figure 3). The ATF4-C/EBPβ heterodimer activates the Gadd45a gene in skeletal muscle fibers by binding an ATF4-C/EBPβ response element in the 3′ untranslated region of Gadd45a exon 4 (29) (Figure 2). This ATF4-C/EBPβ response element in the Gadd45a gene is 100% conserved across all available mammalian genomes, including humans (29).
Figure 3.
Induction of activating transcription factor 4–dependent gene expression in skeletal muscle of human patients with skeletal muscle atrophy due to critical illness. These data were mined from publicly available RNA sequencing data, which compared skeletal muscle biopsy specimens of 7 patients with critical illness myopathy and 6 matched human control subjects (80). Each data point represents 1 subject, and horizontal bars denote the means. P values are <0.01 for all transcripts except Fgf21, where P = 0.77. ATF5, activating transcription factor 5; CARS, cysteinyl-tRNA synthetase 1; CDKN1A, cyclin dependent kinase inhibitor 1A; EIF2S2, eukaryotic translation initiation factor 2 subunit β; FGF21, fibroblast growth factor 21; GADD45A, growth arrest and DNA damage inducible α; GARS, glycyl-tRNA synthetase 1; GPX1, glutathione peroxidase 1; MTHFD2, methylenetetrahydrofolate dehydrogenase (NADP + dependent) 2, methenyltetrahydrofolate cyclohydrolase; NARS, asparaginyl-tRNA synthetase 1; SARS, seryl-tRNA synthetase 1; YARS, tyrosyl-tRNA synthetase 1.
By inducing the Gadd45a gene, the ATF4-C/EBPβ heterodimer increases expression of Gadd45a, an 18-kDa globular protein (29). Gadd45a mediates stress responses by allosterically regulating other signaling molecules in a context-dependent manner (81). In skeletal muscle fibers, Gadd45a directly interacts with a specific group of signaling molecules that includes 11 protein kinases [mitogen-activated protein kinase kinase kinase 4 (MEKK4); Raf-1 proto-oncogene, serine/threonine kinase (Raf-1); A-Raf proto-oncogene, serine/threonine kinase (A-Raf); Janus kinase 1 (JAK1); integrin linked kinase (ILK); ribosomal protein S6 kinase A3 (RSK2); striated muscle-enriched protein kinase (SPEG); ribosomal protein S6 kinase A5 (MSK1); ribosomal protein S6 kinase A4 (MSK2); RIO kinase 3 (RIOK3); and receptor interacting serine/threonine kinase 3 (RIPK3)] and 3 protein tyrosine phosphatases [acid phosphatase 1 (ACP1), protein-tyrosine phosphatase 1B (PTP-1B), and protein tyrosine phosphatase nonreceptor type 6 (SHP-1)] (82).
The interaction between Gadd45a and MEKK4 has been studied in detail (82., 83., 84., 85.). MEKK4 is a member of the mitogen-activated protein (MAP) kinase kinase kinase family, but it lacks protein kinase activity in the absence of Gadd45a. When MEKK4 binds Gadd45a, MEKK4 undergoes a conformational change that activates its kinase domain. Thus, the active MAP kinase kinase kinase is not MEKK4 alone but rather a complex of MEKK4 and Gadd45a. In muscle fibers, the Gadd45a/MEKK4 complex activates 4 downstream MAP kinase kinases, MKK3, MKK4, MKK6, and MKK7 (mitogen-activated protein kinase kinases 3, 4, 6, and 7), which in turn activate p38 MAP kinase. This signal transduction cascade is partially required for Gadd45a-mediated muscle fiber atrophy (82). Through its direct biochemical interactions with signaling molecules such as MEKK4, Gadd45a generates several cellular changes in skeletal muscle fibers that collectively promote muscle atrophy, including a disruption of insulin/IGF-1 signaling, a reduction in general protein synthesis, increased degradation of sarcomeric protein, a loss of mitochondria, and dramatic alterations in myonuclear morphology and gene expression (49).
Another important ATF4-C/EBPβ target in skeletal muscle fibers is the p21 gene, also known as cyclin-dependent kinase inhibitor 1A (Cdkn1a). Like Gadd45a expression, p21 expression is strongly associated with muscle atrophy in humans and other mammalian species, as shown in Figure 3 and other studies (48., 49., 50., 72., 73., 74., 75., 76., 77., 78., 79., 80., 86, 87). In skeletal muscle fibers, the ATF4-C/EBPβ heterodimer induces p21 mRNA expression (29, 48, 50), leading to an increase in the concentration of p21 protein. Similar to Gadd45a expression, increased p21 expression in skeletal muscle fibers is sufficient to induce muscle fiber atrophy and required for ATF4-mediated muscle fiber atrophy (50). The biochemical and cellular mechanisms by which p21 induces muscle fiber atrophy are not yet defined. Although p21 is a well-known cell cycle inhibitor and mediator of replicative senescence (88, 89), its mechanistic role in muscle fibers seems likely to be different, because muscle fibers have exited the cell cycle.
Relation of the ATF4-C/EBPβ Pathway to Other Molecular Mechanisms of Muscle Fiber Atrophy
In skeletal muscle fibers, the ATF4-C/EBPβ heterodimer often operates in parallel to other molecular mechanisms that can promote muscle fiber atrophy, Gadd45a expression, and p21 expression in an ATF4-independent manner (50, 90, 91). As a result of this redundancy, the ATF4-C/EBPβ heterodimer can be partially or wholly dispensable for muscle fiber atrophy, Gadd45a expression, and p21 expression in some conditions of skeletal muscle fiber stress. The existing data indicate that the ATF4-C/EBPβ heterodimer is partially required for muscle fiber atrophy during starvation, immobilization, and aging and not required for muscle atrophy during muscle denervation. It is not yet known whether the ATF4-C/EBPβ heterodimer plays a causal role in muscle fiber atrophy secondary to other conditions, such as the various systemic illnesses that cause generalized skeletal muscle atrophy.
Other Roles of ATF4 in Skeletal Muscle Fibers
ATF4 heterodimers also activate many other genes in skeletal muscle fibers, including genes for amino acid transporters (Slc7a1, solute carrier family 7 member 1; Slc7a5, solute carrier family 7 member 5; Slc38a2, solute carrier family 38 member 2; Slc6a9, solute carrier family 6 member 9); aminoacyl-tRNA synthetases (Aars, alanyl-tRNA synthetase 1; Iars, isoleucyl-tRNA synthetase 1; Gars, glycyl-tRNA synthetase 1; Nars, asparaginyl-tRNA synthetase 1; Sars, seryl-tRNA synthetase 1; Lars, leucyl-tRNA synthetase 1; Mars, methionyl-tRNA synthetase 1; Cars, cysteinyl-tRNA synthetase 1; Yars, tyrosyl-tRNA synthetase 1; Tars, threonyl-tRNA synthetase 1); enzymes involved in amino acid, nucleotide, and glutathione metabolism [Asns, asparagine synthetase; Got1, glutamic-oxaloacetic transaminase 1; Gpt2, glutamic pyruvate transaminase 2; Mthfd2, methylenetetrahydrofolate dehydrogenase (NADP + dependent) 2, methenyltetrahydrofolate cyclohydrolase; Aldh18a1, aldehyde dehydrogenase 18 family member A1; Aldh1l2, aldehyde dehydrogenase 1 family member L2; Gpx1, glutathione peroxidase 1; Mgst1, microsomal glutathione S-transferase 1]; regulators of translation and cell signaling (Eif4ebp1, eukaryotic translation initiation factor 4E binding protein 1; Eif2s2, eukaryotic translation initiation factor 2 subunit β; Eif3c, eukaryotic translation initiation factor 3 subunit C; Grb10, growth factor receptor bound protein 10; Nupr1, nuclear protein 1; Herpud1, homocysteine inducible ER protein with ubiquitin-like domain 1; Igfbp7, insulin-like growth factor binding protein 7; Arhgef2, Rho/Rac guanine nucleotide exchange factor 2; Vegfa, vascular endothelial growth factor A); and other bZIP proteins (Cebpg and Atf5, activating transcription factor 5) (49, 51). Accordingly, these mRNAs are often induced alongside Gadd45a mRNA and p21 mRNA during skeletal muscle atrophy (Figure 3). It is not yet known which ATF4 heterodimers regulate these genes in skeletal muscle fibers or whether any of these genes substantially contribute to muscle atrophy.
As discussed above, ATF4 heterodimers can be formed in response to insulin/IGF-1/mTORC1 signaling and can contribute to insulin/IGF-1/mTORC1-mediated anabolism by activating genes that promote amino acid uptake and the synthesis of amino acids and charged tRNAs; this has been observed in proliferating cultured cells and in hepatocytes in vivo (36., 37., 38., 39., 40., 41.). In skeletal muscle fibers, mTORC1 signaling is critical for skeletal muscle development and for the maintenance of muscle mass and function into at least middle-aged adulthood (58, 59), and physiologic mTORC1 signaling increases expression of ATF4 and target genes such as SLC7A1, SLC7A5, and SLC38A2 in skeletal muscle of young adult humans (92). However, mice with a targeted, lifelong absence of ATF4 expression in skeletal muscle fibers undergo normal skeletal muscle development and exhibit normal muscle mass and function until they are old, at which point they begin to exhibit protection from age-related muscle atrophy and weakness (49, 51). Thus, the available evidence indicates that ATF4 heterodimers mediate some effects of insulin/IGF-1/mTORC1 signaling in skeletal muscle fibers, but those effects do not seem to play a meaningful role in skeletal muscle anabolism or other cellular processes that are required for skeletal muscle health.
Several genetic myopathies also increase skeletal muscle ATF4 expression (61, 64, 93., 94., 95., 96., 97., 98.). Some of these myopathies are caused by constitutive disruption of either mitochondrial function or autophagy in skeletal muscle fibers, and others are caused by constitutive activation of either mTORC1 or the eIF2α kinase PERK in skeletal muscle fibers. These genetic lesions are toxic to muscle fibers and induce a loss of muscle mass that is associated with increased ATF4 expression. It is not yet known whether these conditions involve the ATF4-C/EBPβ heterodimer or any of the other ATF4 heterodimers that have been identified in nonmyopathic muscle fibers. The relation of these single-gene disorders to skeletal muscle atrophy is also not clear. Although they share some gross features with skeletal muscle atrophy, there are also important differences, and in these disorders, ATF4 activates some genes whose expression is not generally associated with natural causes of skeletal muscle atrophy, such as FGF21 (fibroblast growth factor 21) (Figure 3).
Potential Mechanisms That Activate the ATF4-C/EBPβ Pathway in Skeletal Muscle Fibers
Another important and unresolved question is, what upstream mechanisms activate the ATF4-C/EBPβ pathway in skeletal muscle fibers during conditions such as starvation, immobilization, and aging? As discussed above, ATF4 expression is the rate-limiting step in formation of the ATF4-C/EBPβ heterodimer, and ATF4 expression can be increased by several different upstream mechanisms, including mTORC1 activation, eIF2α kinase signaling, reduced eIF2B expression, and inhibitory phosphorylation of eIF2B. Moreover, studies discussed above have established that both mTORC1 activation and eIF2α kinase activation can increase ATF4 expression in skeletal muscle fibers, as in other cell types. Furthermore, all of the mechanisms that are known to increase ATF4 expression in other cell types (mTORC1 activation, eIF2α kinase signaling, reduced eIF2B expression, and inhibitory phosphorylation of eIF2B) have been observed in skeletal muscle fibers during at least one natural condition that causes skeletal muscle atrophy (e.g., 35, 63, 65). Thus, there are several viable candidate upstream regulators of the ATF4-C/EBPβ pathway in skeletal muscle fibers.
One speculation is that different mechanisms or combinations of mechanisms might be responsible for increasing ATF4 expression in skeletal muscle fibers under different conditions. For example, in advanced aging, mTORC1 activity is increased in skeletal muscle fibers and is thought to contribute to the pathogenesis of age-related skeletal muscle atrophy (e.g., 64, 65, 67, 68, 70). Thus, in the context of advanced aging, mTORC1 may be an important driver of the ATF4-C/EBPβ pathway in skeletal muscle fibers. In contrast, many acute stress conditions repress mTORC1 activity in muscle fibers while inducing a simultaneous reduction in eIF2B activity in muscle fibers via eIF2α kinase signaling, decreased eIF2B expression, and/or inhibitory phosphorylation of eIF2B (35). Thus, in some contexts, such as starvation, eIF2B inhibition may be a major driver of the ATF4-C/EBPβ pathway in skeletal muscle fibers. Consistent with this hypothesis, a general inhibitor of eIF2α kinase signaling (eIF2α-S51A) reduces ATF4 expression and muscle fiber atrophy during starvation (48). Understanding the mechanisms that increase the concentration of ATF4 in skeletal muscle fibers during conditions such as starvation, immobilization, and aging should be a soluble topic for investigation and could provide important new insights into the pathogenesis of skeletal muscle atrophy.
Potential Approaches to Repress the ATF4-C/EBPβ Pathway in Skeletal Muscle Fibers
Skeletal muscle atrophy is a highly prevalent condition that can create serious problems for patients, including weakness, fatigue, impairments in activity and mobility, falls, loss of independent living, impairments in whole-body metabolism, delayed recovery from acute illness and injury, prolonged hospital stays, and increased mortality. In some younger patients, skeletal muscle atrophy can be reversed by correction of its underlying cause. However, in many other patients, the underlying cause cannot be fully corrected, and in older patients, correction of the underlying cause is often not sufficient to restore skeletal muscle mass and function to its baseline level, even with subsequent physical therapy. Thus, there is an important need for pharmacologic and nutritional agents that act directly on skeletal muscle fibers in a way that substantially helps to maintain and/or restore the normal structure and function of skeletal muscle fibers. At present, no such agents are fully developed and approved for human use. However, many agents are under investigation, including some that repress or could potentially repress the ATF4-C/EBPβ pathway in skeletal muscle fibers.
The candidate agents with actual or potential efficacy toward the ATF4-C/EBPβ pathway can be roughly divided into 3 classes based on their sites of action (Figure 4). The first class of agents specifically acts upon the ATF4-C/EBPβ heterodimer or its downstream mediators within skeletal muscle fibers. Proof of concept for this approach comes from studies in mouse models, where targeted inhibition of ATF4, C/EBPβ, Gadd45a, MEKK4, or p21 via muscle fiber–specific gene excision or RNA interference reduces muscle fiber atrophy during conditions where the ATF4-C/EBPβ pathway is active (48., 49., 50., 51., 82, 90, 99). Clinical application of this strategy would require, among other things, development of new methods that permit the targeting of specific genes or mRNA transcripts specifically in skeletal muscle fibers of humans. Such methods are under investigation but not yet available.
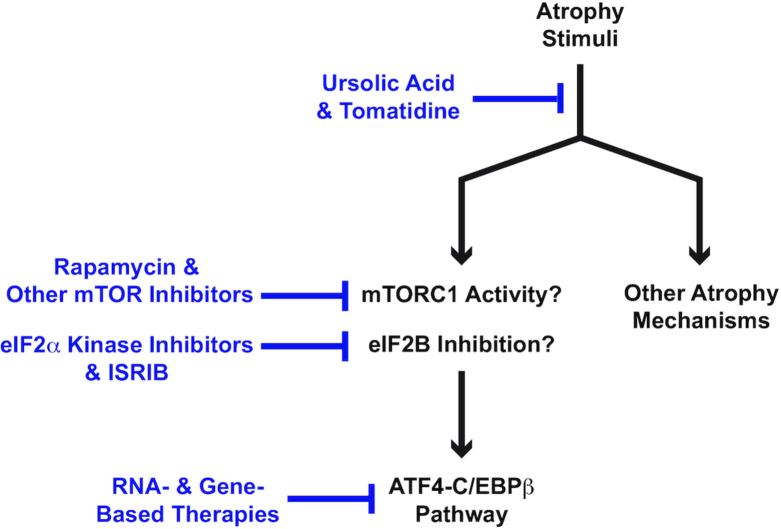
Figure 4.
Schematic illustrating some pharmacologic and nutritional agents that repress or could potentially repress the ATF4-C/EBPβ pathway in skeletal muscle fibers. ATF4, activating transcription factor 4; C/EBPβ, CCAAT enhancer binding protein β; eIF2α, eukaryotic translation initiation factor 2 subunit α; eIF2Β, eukaryotic translation initiation factor 2B; ISRIB, integrated stress response inhibitor; mTOR, mechanistic target of rapamycin; mTORC1, mechanistic target of rapamycin complex 1.
The second class of agents could potentially repress the ATF4-C/EBPβ pathway by interfering with upstream mechanisms that promote ATF4 expression. Examples of agents that inhibit ATF4 expression in specific contexts include small-molecule inhibitors of specific eIF2α kinases (e.g., 100., 101., 102., 103.), small molecules that directly promote eIF2B activity such as integrated stress response inhibitor (ISRIB) and its derivatives (e.g., 104, 105), and small molecules that inhibit mTORC1 activity such as rapamycin and other mTOR inhibitors (e.g., 67, 68, 70). Although it remains to be determined if these molecules repress the ATF4-C/EBPβ pathway in skeletal muscle fibers, several agents in this class are available or close to being available for human use, and they may prove to be effective in the treatment of muscle atrophy, perhaps in part by repressing the ATF4-C/EBPβ pathway in muscle fibers. If different upstream mechanisms control ATF4 expression under different conditions, then different agents in this class may be indicated under different conditions.
The third class of agents represses the ATF4-C/EBPβ pathway as part of a broader effect on molecular mechanisms of muscle fiber atrophy. Examples of agents in this class include the natural compounds ursolic acid and tomatidine. As discussed above, muscle fiber atrophy is mediated by a complex network of signaling pathways, including the ATF4-C/EBPβ pathway and others. Collectively, these signaling pathways induce many genes and repress many other genes within muscle fibers, leading to corresponding positive and negative changes in muscle fiber mRNA concentrations. Many of the mRNAs that increase during muscle atrophy promote muscle fiber atrophy, and many of the mRNAs that decrease during muscle atrophy help to protect against muscle fiber atrophy. The entire collection of mRNAs that increase or decrease in muscle fibers as they undergo atrophy is known as an mRNA expression signature of skeletal muscle atrophy. An mRNA expression signature of skeletal muscle atrophy is a molecular signature of mechanistic importance, because it captures many of the molecular changes that are causally related to the pathogenesis of skeletal muscle atrophy (106). Within an mRNA expression signature of muscle atrophy, the ATF4-C/EBPβ heterodimer is responsible for a portion of the positive changes in mRNA concentrations. The remainder of positive changes and all of the negative changes in mRNA concentrations are mediated by other atrophy mechanisms, such as activation of FoxO transcription factors and impairments in insulin/IGF-1 signaling.
Ursolic acid and tomatidine were identified through a systems-based strategy that searched for small molecules whose mRNA expression signatures negatively correlate to the mRNA expression signatures of 2 disparate causes of muscle atrophy (prolonged fasting and spinal cord injury) in 2 species of skeletal muscle (humans and mice) (107, 108). As predicted by the way they were discovered, ursolic acid and tomatidine have very similar effects on gene expression in skeletal muscle fibers, which are opposite to the gene expression changes that occur during muscle fiber atrophy (51, 107, 108). The effects on muscle fiber gene expression are partly mediated through repression of the ATF4-C/EBPβ pathway but also involve inhibition of FoxO-mediated gene expression, increased sensitivity to insulin and IGF-1, increased expression of PPARG coactivator 1α (PGC-1α), and increased mTORC1 signaling in young but not old muscle fibers (51, 107, 108). In preclinical models, these agents reduce muscle fiber atrophy during starvation, muscle disuse, advanced age, spinal cord injury, and renal failure, and they promote muscle fiber hypertrophy, leading to increases in muscle mass, strength, muscle quality, and endurance exercise capacity (51, 107., 108., 109., 110.). Ursolic acid and tomatidine are naturally occurring in foods such as apples and tomatoes, respectively, so they have potential nutritional and pharmacologic uses.
Although pharmacologic and nutritional agents that repress the ATF4-C/EBPβ pathway could be beneficial for many patients with skeletal muscle atrophy, additional approaches will almost certainly be needed. As an analogy, consider patients with osteoporosis, hypertension, type 2 diabetes, or hyperlipidemia. Similar to skeletal muscle atrophy, those other disorders are highly prevalent, especially with advancing age, and their pathophysiology is exceedingly complex and still not fully understood, particularly at the molecular level. Nonetheless, we now have a large and growing repertoire of highly useful therapeutic approaches for patients with osteoporosis, hypertension, type 2 diabetes, and hyperlipidemia, and in each case, we have multiple therapeutic approaches that can be used alone or in combination, depending on the specific circumstances of each patient. Although therapies for muscle atrophy have lagged far behind, important progress is being made, and based on our current knowledge, it already seems clear that a similar multifaceted therapeutic approach will be needed. For patients with muscle atrophy or at high risk for muscle atrophy, the ideal repertoire of therapeutic approaches will likely include multiple classes of pharmaceuticals that can be tailored to the specific molecular pathways responsible for muscle fiber atrophy in a given patient, as well as new types of muscle-focused nutritional agents, personalized physical therapy regimens, and, when possible, specific therapies that target the underlying cause(s) of muscle atrophy. Within that ideal repertoire, methods that repress the ATF4-C/EBPβ pathway within skeletal muscle fibers could play an important role.
Acknowledgments
We thank Dr Kimberlee Potter and her colleagues at the US Department of Veterans Affairs, who have provided invaluable encouragement and contributions to this work since 2008.
The authors’ responsibilities were as follows—All authors were responsible for the design, writing, and final content of the manuscript, and all authors read and approved the final version.
Footnotes
Supported by funding from the NIH (grants R01AR071762 and R01AG060637 to CMA; grants R44AG047684 and R44AR069400 to SME, JJT, and CMA; grant P30AG024832 to BBR and EV; grant R01AR060209 to ARJ; grant R01DK109714 to TGA and RCW; grants R01DK78646, R01DK116231, and R01DK126206 to AV) and the US Department of Veterans Affairs (grant I01BX00976 to CMA and MAY).
Author disclosures: SME, BBR, ARJ, AV, JJT, and CMA are shareholders in Emmyon, Inc., which holds patent rights for uses of ursolic acid and tomatidine in skeletal muscle health. SME, JJT, and CMA serve as officers at Emmyon, Inc., and BBR, ARJ, and AV serve as consultants. All other authors report no conflicts of interest.
References
- 1.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22(18):6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 3.Landschulz WH, Johnson PF, McKnight SL. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 4.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 5.Lamb P, McKnight SL. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- 6.Lassot I, Segeral E, Berlioz-Torrent C, Durand H, Groussin L, Hai T, Benarous R, Margottin-Goguet F. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21(6):2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry syndrome. Cell. 2004;117(3):387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 8.Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J Biol Chem. 2005;280(50):41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 9.Pons J, Evrard-Todeschi N, Bertho G, Gharbi-Benarous J, Benarous R, Girault JP. Phosphorylation-dependent structure of ATF4 peptides derived from a human ATF4 protein, a member of the family of transcription factors. Peptides. 2007;28(12):2253–2267. doi: 10.1016/j.peptides.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ampofo E, Sokolowsky T, Gotz C, Montenarh M. Functional interaction of protein kinase CK2 and activating transcription factor 4 (ATF4), a key player in the cellular stress response. Biochim Biophys Acta. 2013;1833(3):439–451. doi: 10.1016/j.bbamcr.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10(7):634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma K, D'Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8(5):1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 14.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 2014;13(7):1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendriks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21(10):927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagheri-Yarmand R, Sinha KM, Gururaj AE, Ahmed Z, Rizvi YQ, Huang SC, Ladbury JE, Bogler O, Williams MD, Cote GJ, et al. A novel dual kinase function of the RET proto-oncogene negatively regulates activating transcription factor 4-mediated apoptosis. J Biol Chem. 2015;290(18):11749–11761. doi: 10.1074/jbc.M114.619833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podust LM, Krezel AM, Kim Y. Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J Biol Chem. 2001;276(1):505–513. doi: 10.1074/jbc.M005594200. [DOI] [PubMed] [Google Scholar]
- 19.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300(5628):2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 20.Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science. 2013;340(6133):730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339(1):135–141. [PMC free article] [PubMed] [Google Scholar]
- 22.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2002;277(27):24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 23.Lopez AB, Wang C, Huang CC, Yaman I, Li Y, Chakravarty K, Johnson PF, Chiang CM, Snider MD, Wek RC, et al. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem J. 2007;402(1):163–173. doi: 10.1042/BJ20060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gombart AF, Grewal J, Koeffler HP. ATF4 differentially regulates transcriptional activation of myeloid-specific genes by C/EBPepsilon and C/EBPalpha. J Leukoc Biol. 2007;81(6):1535–1547. doi: 10.1189/jlb.0806516. [DOI] [PubMed] [Google Scholar]
- 25.Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell. 2008;19(12):5373–5386. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20(9):436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen DM, Won KJ, Nguyen N, Lazar MA, Chen CS, Steger DJ. ATF4 licenses C/EBPbeta activity in human mesenchymal stem cells primed for adipogenesis. Elife. 2015;4:e06821. doi: 10.7554/eLife.06821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, Kasoji M, Haines DC, Quinones OA, Johnson PF. C/EBPgamma is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol Cell Biol. 2016;36(5):693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebert SM, Bullard SA, Basisty N, Marcotte GR, Skopec ZP, Dierdorff JM, Al-Zougbi A, Tomcheck KC, DeLau AD, Rathmacher JA, et al. Activating transcription factor 4 (ATF4) promotes skeletal muscle atrophy by forming a heterodimer with the transcriptional regulator C/EBPbeta. J Biol Chem. 2020;295(9):2787–2803. doi: 10.1074/jbc.RA119.012095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wek RC, Jiang HY, Anthony TG. Coping with stress: EIF2 kinases and translational control. Biochem Soc Trans. 2006;34(1):7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 31.Wek RC. Role of eIF2alpha kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol. 2018;10(7):a032870. doi: 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature. 2020;579(7799):433–437. doi: 10.1038/s41586-020-2076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA, Kampmann M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature. 2020;579(7799):427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 35.Gordon BS, Kelleher AR, Kimball SR. Regulation of muscle protein synthesis and the effects of catabolic states. Int J Biochem Cell Biol. 2013;45:2147–2157. doi: 10.1016/j.biocel.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282(23):16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- 37.Malmberg SE, Adams CM. Insulin signaling and the general amino acid control response: two distinct pathways to amino acid synthesis and uptake. J Biol Chem. 2008;283(28):19229–19234. doi: 10.1074/jbc.M801331200. [DOI] [PubMed] [Google Scholar]
- 38.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351(6274):728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 2017;19(6):1083–1090. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, Manning BD. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife. 2021 doi: 10.7554/eLife.63326. 10:e63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byles V, Cormerais Y, Kalafut K, Barrera V, Hughes Hallett JE, Sui SH, Asara JM, Adams CM, Hoxhaj G, Ben-Sahra I, et al. Hepatic mTORC1 signaling activates ATF4 as part of its metabolic response to feeding and insulin. Mol Metab. 2021;53:101309. doi: 10.1016/j.molmet.2021.101309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selvarajah B, Azuelos I, Plate M, Guillotin D, Forty EJ, Contento G, Woodcock HV, Redding M, Taylor A, Brunori G, et al. mTORC1 amplifies the ATF4-dependent de novo serine-glycine pathway to supply glycine during TGF-beta1-induced collagen biosynthesis. Sci Signal. 2019;12(582):eaav3048. doi: 10.1126/scisignal.aav3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 44.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(31):11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285(43):33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 47.Saltiel AR. Insulin signaling in health and disease. J Clin Invest. 2021 doi: 10.1172/JCI142241. 131:e142241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebert SM, Monteys AM, Fox DK, Bongers KS, Shields BE, Malmberg SE, Davidson BL, Suneja M, Adams CM. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol. 2010;24(4):790–799. doi: 10.1210/me.2009-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, Dierdorff JM, Foster ED, Adams CM. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287(33):27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox DK, Ebert SM, Bongers KS, Dyle MC, Bullard SA, Dierdorff JM, Kunkel SD, Adams CM. p53 and ATF4 mediate distinct and additive pathways to skeletal muscle atrophy during limb immobilization. Am J Physiol Endocrinol Metab. 2014;307(3):E245–E261. doi: 10.1152/ajpendo.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ebert SM, Dyle MC, Bullard SA, Dierdorff JM, Murry DJ, Fox DK, Bongers KS, Lira VA, Meyerholz DK, Talley JJ, et al. Identification and small molecule inhibition of an activating transcription factor 4 (ATF4)–dependent pathway to age-related skeletal muscle weakness and atrophy. J Biol Chem. 2015;290(42):25497–25511. doi: 10.1074/jbc.M115.681445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsson L, Degens H, Li M, Salviati L, Lee YI, Thompson W, Kirkland JL, Sandri M. Sarcopenia: aging-related loss of muscle mass and function. Physiol Rev. 2019;99(1):427–511. doi: 10.1152/physrev.00061.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert SM, Al-Zougbi A, Bodine SC, Adams CM. Skeletal muscle atrophy: discovery of mechanisms and potential therapies. Physiology (Bethesda). 2019;34:232–239. doi: 10.1152/physiol.00003.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sartori R, Romanello V, Sandri M. Mechanisms of muscle atrophy and hypertrophy: implications in health and disease. Nat Commun. 2021;12(1):330. doi: 10.1038/s41467-020-20123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin-sensitive pathway. J Nutr. 2000;130(10):2413–2419. doi: 10.1093/jn/130.10.2413. [DOI] [PubMed] [Google Scholar]
- 56.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3(11):1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 57.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 58.Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8(5):411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, Richard-Bulteau H, Vignaud A, Baas D, Defour A, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187(6):859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol. 2009;587(7):1535–1546. doi: 10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Ruegg MA. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013;17(5):731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 62.Bentzinger CF, Lin S, Romanino K, Castets P, Guridi M, Summermatter S, Handschin C, Tintignac LA, Hall MN, Ruegg MA. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skelet Muscle. 2013;3(1):6. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang H, Inoki K, Lee M, Wright E, Khuong A, Khuong A, Sugiarto S, Garner M, Paik J, DePinho RA, et al. mTORC1 promotes denervation-induced muscle atrophy through a mechanism involving the activation of FoxO and E3 ubiquitin ligases. Sci Signal. 2014;7(314):ra18. doi: 10.1126/scisignal.2004809. [DOI] [PubMed] [Google Scholar]
- 64.Guridi M, Tintignac LA, Lin S, Kupr B, Castets P, Ruegg MA. Activation of mTORC1 in skeletal muscle regulates whole-body metabolism through FGF21. Sci Signal. 2015;8(402):ra113. doi: 10.1126/scisignal.aab3715. [DOI] [PubMed] [Google Scholar]
- 65.Markofski MM, Dickinson JM, Drummond MJ, Fry CS, Fujita S, Gundermann DM, Glynn EL, Jennings K, Paddon-Jones D, Reidy PT, et al. Effect of age on basal muscle protein synthesis and mTORC1 signaling in a large cohort of young and older men and women. Exp Gerontol. 2015;65:1–7. doi: 10.1016/j.exger.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon MS. mTOR as a key regulator in maintaining skeletal muscle mass. Front Physiol. 2017;8:788. doi: 10.3389/fphys.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joseph GA, Wang SX, Jacobs CE, Zhou W, Kimble GC, Tse HW, Eash JK, Shavlakadze T, Glass DJ. Partial inhibition of mTORC1 in aged rats counteracts the decline in muscle mass and reverses molecular signaling associated with sarcopenia. Mol Cell Biol. 2019 doi: 10.1128/MCB.00141-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang H, Inoki K, Brooks SV, Okazawa H, Lee M, Wang J, Kim M, Kennedy CL, Macpherson PCD, Ji X, et al. mTORC1 underlies age-related muscle fiber damage and loss by inducing oxidative stress and catabolism. Aging Cell. 2019;18(3):e12943. doi: 10.1111/acel.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graber TG, Fry CS, Brightwell CR, Moro T, Maroto R, Bhattarai N, Porter C, Wakamiya M, Rasmussen BB. Skeletal muscle-specific knockout of DEP domain containing 5 protein increases mTORC1 signaling, muscle cell hypertrophy, and mitochondrial respiration. J Biol Chem. 2019;294(11):4091–4102. doi: 10.1074/jbc.RA118.005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ham DJ, Borsch A, Lin S, Thurkauf M, Weihrauch M, Reinhard JR, Delezie J, Battilana F, Wang X, Kaiser MS, et al. The neuromuscular junction is a focal point of mTORC1 signaling in sarcopenia. Nat Commun. 2020;11(1):4510. doi: 10.1038/s41467-020-18140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21(1):140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 72.Welle S, Brooks AI, Delehanty JM, Needler N, Bhatt K, Shah B, Thornton CA. Skeletal muscle gene expression profiles in 20–29 year old and 65–71 year old women. Exp Gerontol. 2004;39(3):369–377. doi: 10.1016/j.exger.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 73.Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA. Gene expression profile of aging in human muscle. Physiol Genomics. 2003;14(2):149–159. doi: 10.1152/physiolgenomics.00049.2003. [DOI] [PubMed] [Google Scholar]
- 74.Edwards MG, Anderson RM, Yuan M, Kendziorski CM, Weindruch R, Prolla TA. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. 2007;8(1):80. doi: 10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonzalez de Aguilar JL, Niederhauser-Wiederkehr C, Halter B, De Tapia M, Di Scala F, Demougin P, Dupuis L, Primig M, Meininger V, Loeffler JP. Gene profiling of skeletal muscle in an amyotrophic lateral sclerosis mouse model. Physiol Genomics. 2008;32(2):207–218. doi: 10.1152/physiolgenomics.00017.2007. [DOI] [PubMed] [Google Scholar]
- 76.Banduseela VC, Ochala J, Chen YW, Goransson H, Norman H, Radell P, Eriksson LI, Hoffman EP, Larsson L. Gene expression and muscle fiber function in a porcine ICU model. Physiol Genomics. 2009;39(3):141–159. doi: 10.1152/physiolgenomics.00026.2009. [DOI] [PubMed] [Google Scholar]
- 77.Llano-Diez M, Gustafson AM, Olsson C, Goransson H, Larsson L. Muscle wasting and the temporal gene expression pattern in a novel rat intensive care unit model. BMC Genomics. 2011;12(1):602. doi: 10.1186/1471-2164-12-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hulmi JJ, Silvennoinen M, Lehti M, Kivela R, Kainulainen H. Altered REDD1, myostatin, and Akt/mTOR/FoxO/MAPK signaling in streptozotocin-induced diabetic muscle atrophy. Am J Physiol Endocrinol Metab. 2012;302(3):E307–E315. doi: 10.1152/ajpendo.00398.2011. [DOI] [PubMed] [Google Scholar]
- 79.Ibebunjo C, Chick JM, Kendall T, Eash JK, Li C, Zhang Y, Vickers C, Wu Z, Clarke BA, Shi J, et al. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol Cell Biol. 2013;33(2):194–212. doi: 10.1128/MCB.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Llano-Diez M, Fury W, Okamoto H, Bai Y, Gromada J, Larsson L. RNA-sequencing reveals altered skeletal muscle contraction, E3 ligases, autophagy, apoptosis, and chaperone expression in patients with critical illness myopathy. Skelet Muscle. 2019;9(1):9. doi: 10.1186/s13395-019-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salvador JM, Brown-Clay JD, Fornace AJ., Jr Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- 82.Bullard SA, Seo S, Schilling B, Dyle MC, Dierdorff JM, Ebert SM, DeLau AD, Gibson BW, Adams CM. Gadd45a protein promotes skeletal muscle atrophy by forming a complex with the protein kinase MEKK4. J Biol Chem. 2016;291(34):17496–17509. doi: 10.1074/jbc.M116.740308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95(4):521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 84.Mita H, Tsutsui J, Takekawa M, Witten EA, Saito H. Regulation of MTK1/MEKK4 kinase activity by its N-terminal autoinhibitory domain and GADD45 binding. Mol Cell Biol. 2002;22(13):4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyake Z, Takekawa M, Ge Q, Saito H. Activation of MTK1/MEKK4 by GADD45 through induced N-C dissociation and dimerization-mediated trans autophosphorylation of the MTK1 kinase domain. Mol Cell Biol. 2007;27(7):2765–2776. doi: 10.1128/MCB.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stevenson EJ, Giresi PG, Koncarevic A, Kandarian SC. Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. J Physiol. 2003;551(1):33–48. doi: 10.1113/jphysiol.2003.044701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laure L, Suel L, Roudaut C, Bourg N, Ouali A, Bartoli M, Richard I, Daniele N. Cardiac ankyrin repeat protein is a marker of skeletal muscle pathological remodelling. FEBS J. 2009;276(3):669–684. doi: 10.1111/j.1742-4658.2008.06814.x. [DOI] [PubMed] [Google Scholar]
- 88.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14(2):159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 89.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bongers KS, Fox DK, Ebert SM, Kunkel SD, Dyle MC, Bullard SA, Dierdorff JM, Adams CM. Skeletal muscle denervation causes skeletal muscle atrophy through a pathway that involves both Gadd45a and HDAC4. Am J Physiol Endocrinol Metab. 2013;305(7):E907–E915. doi: 10.1152/ajpendo.00380.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milan G, Romanello V, Pescatore F, Armani A, Paik JH, Frasson L, Seydel A, Zhao J, Abraham R, Goldberg AL, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun. 2015;6(1):6670. doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drummond MJ, Fry CS, Glynn EL, Timmerman KL, Dickinson JM, Walker DK, Gundermann DM, Volpi E, Rasmussen BB. Skeletal muscle amino acid transporter expression is increased in young and older adults following resistance exercise. J Appl Physiol. 2011;111(1):135–142. doi: 10.1152/japplphysiol.01408.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 94.Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19(1):83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 95.Keipert S, Ost M, Johann K, Imber F, Jastroch M, van Schothorst EM, Keijer J, Klaus S. Skeletal muscle mitochondrial uncoupling drives endocrine cross-talk through the induction of FGF21 as a myokine. Am J Physiol Endocrinol Metab. 2014;306(5):E469–E482. doi: 10.1152/ajpendo.00330.2013. [DOI] [PubMed] [Google Scholar]
- 96.Miyake M, Nomura A, Ogura A, Takehana K, Kitahara Y, Takahara K, Tsugawa K, Miyamoto C, Miura N, Sato R, et al. Skeletal muscle-specific eukaryotic translation initiation factor 2alpha phosphorylation controls amino acid metabolism and fibroblast growth factor 21-mediated non-cell-autonomous energy metabolism. FASEB J. 2016;30(2):798–812. doi: 10.1096/fj.15-275990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pereira RO, Tadinada SM, Zasadny FM, Oliveira KJ, Pires KMP, Olvera A, Jeffers J, Souvenir R, McGlauflin R, Seei A, et al. OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 2017;36(14):2126–2145. doi: 10.15252/embj.201696179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tezze C, Romanello V, Desbats MA, Fadini GP, Albiero M, Favaro G, Ciciliot S, Soriano ME, Morbidoni V, Cerqua C, et al. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab. 2017;25(6):1374–1389. doi: 10.1016/j.cmet.2017.04.021. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bongers KS, Fox DK, Kunkel SD, Stebounova LV, Murry DJ, Pufall MA, Ebert SM, Dyle MC, Bullard SA, Dierdorff JM, et al. Spermine oxidase maintains basal skeletal muscle gene expression and fiber size and is strongly repressed by conditions that cause skeletal muscle atrophy. Am J Physiol Endocrinol Metab. 2015;308(2):E144–E158. doi: 10.1152/ajpendo.00472.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robert F, Williams C, Yan Y, Donohue E, Cencic R, Burley SK, Pelletier J. Blocking UV-induced eIF2alpha phosphorylation with small molecule inhibitors of GCN2. Chem Biol Drug Design. 2009;74(1):57–67. doi: 10.1111/j.1747-0285.2009.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harding HP, Zyryanova AF, Ron D. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase. J Biol Chem. 2012;287(53):44338–44344. doi: 10.1074/jbc.M112.428987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Axten JM, Romeril SP, Shu A, Ralph J, Medina JR, Feng Y, Li WH, Grant SW, Heerding DA, Minthorn E, et al. Discovery of GSK2656157: an optimized PERK inhibitor selected for preclinical development. ACS Med Chem Lett. 2013;4(10):964–968. doi: 10.1021/ml400228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakamura A, Nambu T, Ebara S, Hasegawa Y, Toyoshima K, Tsuchiya Y, Tomita D, Fujimoto J, Kurasawa O, Takahara C, et al. Inhibition of GCN2 sensitizes ASNS-low cancer cells to asparaginase by disrupting the amino acid response. Proc Natl Acad Sci U S A. 2018;115(33):E7776–E7785. doi: 10.1073/pnas.1805523115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schoof M, Boone M, Wang L, Lawrence R, Frost A, Walter P. eIF2B conformation and assembly state regulate the integrated stress response. Elife. 2021:10:e65703. doi: 10.7554/eLife.65703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adams CM, Ebert SM, Dyle MC. Use of mRNA expression signatures to discover small molecule inhibitors of skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2015;18(3):263–268. doi: 10.1097/MCO.0000000000000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kunkel SD, Suneja M, Ebert SM, Bongers KS, Fox DK, Malmberg SE, Alipour F, Shields RK, Adams CM. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 2011;13(6):627–638. doi: 10.1016/j.cmet.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dyle MC, Ebert SM, Cook DP, Kunkel SD, Fox DK, Bongers KS, Bullard SA, Dierdorff JM, Adams CM. Systems-based discovery of tomatidine as a natural small molecule inhibitor of skeletal muscle atrophy. J Biol Chem. 2014;289(21):14913–14924. doi: 10.1074/jbc.M114.556241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu R, Chen JA, Xu J, Cao J, Wang Y, Thomas SS, Hu Z. Suppression of muscle wasting by the plant-derived compound ursolic acid in a model of chronic kidney disease. J Cachexia Sarcopenia Muscle. 2017;8(2):327–341. doi: 10.1002/jcsm.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bigford GE, Darr AJ, Bracchi-Ricard VC, Gao H, Nash MS, Bethea JR. Effects of ursolic acid on sub-lesional muscle pathology in a contusion model of spinal cord injury. PLoS One. 2018;13(8):e0203042. doi: 10.1371/journal.pone.0203042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ye J, Palm W, Peng M, King B, Lindsten T, Li MO, Koumenis C, Thompson CB. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 2015;29:2331–2336. doi: 10.1101/gad.269324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu D, Dai W, Kutzler L, Lacko HA, Jefferson LS, Dennis MD, Kimball SR. ATF4-mediated upregulation of REDD1 and Sestrin2 suppresses mTORC1 activity during prolonged leucine deprivation. J Nutr. 2020;150(5):1022–1030. doi: 10.1093/jn/nxz309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, Salas Salinas C, Chinga F, Monroy AM, Fu W, et al. ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol. 2019;21(7):889–899. doi: 10.1038/s41556-019-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Misra J, Holmes MJ, Mirek ET, Langevin M, Kim HG, Carlson KR, Watford M, Dong XC, Anthony TG, Wek RC. Discordant regulation of eIF2 kinase GCN2 and mTORC1 during nutrient stress. Nucleic Acids Res. 2021;49(10):5726–5742. doi: 10.1093/nar/gkab362. [DOI] [PMC free article] [PubMed] [Google Scholar]