Abstract
Based on recent findings indicating that metabolism might be governed by a limit on the rate at which cells can dissipate Gibbs energy, in this Perspective, we propose a new mechanism of how metabolic activity could globally regulate biomolecular processes in a cell. Specifically, we postulate that Gibbs energy released in metabolic reactions is used to perform work, allowing enzymes to self‐propel or to break free from supramolecular structures. This catalysis‐induced enzyme movement will result in increased intracellular motion, which in turn can compromise biomolecular functions. Once the increased intracellular motion has a detrimental effect on regulatory mechanisms, this will establish a feedback mechanism on metabolic activity, and result in the observed thermodynamic limit. While this proposed explanation for the identified upper rate limit on cellular Gibbs energy dissipation rate awaits experimental validation, it offers an intriguing perspective of how metabolic activity can globally affect biomolecular functions and will hopefully spark new research.
Keywords: active matter, enhanced diffusion, Gibbs energy, metabolism, regulation
Subject Categories: Metabolism
Recent findings suggested that metabolism is governed by a limit on the rate at which cells can dissipate Gibbs energy. This Perspective proposes a new mechanism of how metabolic activity and the Gibbs energy released in metabolic reactions could globally regulate biomolecular processes.
A new hypothesis explaining how metabolic activity affects biomolecular functions
Metabolism and other cellular functions are controlled by a plethora of regulatory mechanisms. However, in recent years, it has become clear that metabolism is not only subject to regulation, but metabolic cues themselves can also regulate other cellular functions (Haas et al, 2016; Ryan et al, 2018; Zhu & Thompson, 2019; Orozco et al, 2020). In most of these cases, altered levels of metabolites trigger regulatory action, for instance, by binding to transcription factors (Kochanowski et al, 2017; Lempp et al, 2019), or by allosteric interactions with enzymes or other biomolecules (Hackett et al, 2016; Sander et al, 2019). In this Perspective, we propose that besides these very specific and well‐studied metabolite‐dependent regulation mechanisms, there might be an additional global mechanism of how an active metabolism could affect essentially all biomolecular functions in a cell.
This proposed mechanism stems from our previous work (Niebel et al, 2019), in which we proposed that a thermodynamic limit could govern cellular metabolism, i.e., that a thermodynamic limit could determine the intracellular metabolic flux distribution. In brief, cellular metabolism consists of many different chemical reactions (Fig 1A, left part). Each of these reactions carries a certain flux (v) and exhibits a particular Gibbs energy of reaction (∆ rG) (Fig 1A, middle part, for further explanation of terms cf. Table 1). The product of a reaction’s flux and Gibbs energy defines the rate at which Gibbs energy is dissipated in this reaction. If the Gibbs energy dissipation rates of all the metabolic reactions occurring in a cell are summed up, we obtain the cellular Gibbs energy dissipation rate. In our study (Niebel et al, 2019), we uncovered that a limit might exist on this rate (Fig 1A, right part). Once the glucose uptake rate is high and the limiting rate of Gibbs energy dissipation is reached, Saccharomyces cerevisiae and Escherichia coli start to excrete ethanol and acetate, respectively, in a behavior which is known as aerobic fermentation or overflow metabolism. Remarkably, despite the fact that both organisms have largely different cell volumes, the value of this limit was found to be in the same order of magnitude (when normalized by the cellular dry weight), suggesting that it does not simply scale with cell morphology, but is rather an intrinsic property of the organisms. When we used the identified limit of the Gibbs energy dissipation rate as a constraint in flux balance analysis simulations (Orth et al, 2010) while maximizing biomass production (i.e., growth rate), we obtained excellent predictions of metabolic phenotypes. These predictions included the shift from a respiratory towards a fermentative metabolism at increased glucose uptake rates (as the latter provides a less dissipative way for metabolizing carbon), maximal growth rates on various nutrients (including in complex media), intracellular flux distributions and even predictions of changes in metabolite concentrations (Niebel et al, 2019). While using an additional constraint in flux balance analysis simulations is not a new approach (see Box 1), the excellent agreement of these predictions with experimental data, and their broad scope, suggested that an upper limit on the cellular Gibbs energy dissipation rate could indeed exist and that this limit may govern cellular metabolism and growth.
Figure 1. Cellular Gibbs energy dissipation rate has an upper limit and may be explained by metabolism‐induced molecular motion.
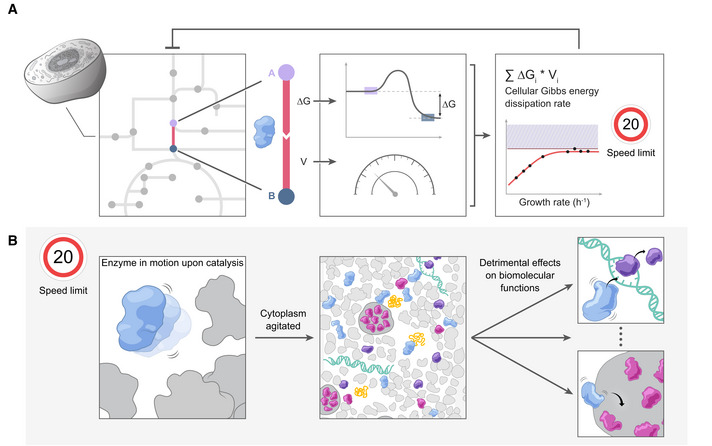
(A) A cell encompasses a large number of interconnected chemical reactions, forming a metabolic network (left panel); each reaction is catalyzed by an enzyme and is characterized by two parameters: its Gibbs energy of reaction, ΔG, and its flux, v (middle panel). The product of the Gibbs energy of reaction and the flux is the Gibbs energy dissipation rate. When this parameter is summed across all the reactions in the metabolic network, the cellular Gibbs energy dissipation rate is obtained. The cellular Gibbs energy dissipation rate has units of J/h, or J/gDW/h if normalized to the cell dry biomass. We previously found that this parameter reaches an upper limit, at moderately high growth rates/substrate uptake rates (Niebel et al, 2019) (right panel). Constrained by this upper limit, cells that have reached the “plateau” may still achieve increased growth rates provided that metabolic fluxes are redirected towards reactions that dissipate less Gibbs energy. The interplay between the upper limit and metabolism is represented by the negative feedback arrow. (B) To explain the existence of this limit, we propose that catalysis leads to enhanced enzyme motion (left panel). In the crowded intracellular environment (middle panel), excessive motion will ultimately cause detrimental effects on biomolecular functions (right panel).
Table 1.
Key terminology.
| Steady‐state | State in which the parameters of the system (metabolite concentration, reaction fluxes, Gibbs energy dissipation rate) remain constant over time. May be achieved even under non‐equilibrium conditions |
| Equilibrium | State in which the thermodynamic driving force (e.g., ∆ rG) is null, hence the net fluxes (e.g., metabolic rate, Gibbs energy dissipation rate) are likewise null. In contrast, non‐equilibrium is characterized by a non‐zero driving force, and may or may not be at steady state |
| Gibbs energy of reaction (∆ rG) | Thermodynamic driving force associated with a chemical reaction. Negative values of this parameter (∆ rG < 0) are associated with thermodynamic feasibility of the reaction to proceed in the forward direction. It is a function of the momentary concentration of substrates and products (ci ), affected by their stoichiometric coefficients (Si ), in the reaction: , with being the Gibbs energy of reaction under standard conditions (a tabulated value) |
| Metabolic flux (v) | Rate at which a metabolic reaction occurs, measuring the amount of substrate(s) consumed or product(s) formed per unit of time |
| Gibbs energy dissipation rate (g) | Defined for a single reaction as the product of its flux, v, and the associated Gibbs energy of reaction, ∆ rG, thus: . It is non‐zero for any reaction that is out of equilibrium |
| Cellular Gibbs energy dissipation rate | Same as above, applied to a whole cell, as represented by a macrochemical equation (“substrates→ biomass + byproducts”). It can be obtained by summing the Gibbs energy dissipation rates of all reactions taking place in the cell |
| Thermal Brownian motion | Random, diffusive motion that a small particle undergoes in a medium due to temperature. The higher the temperature, the faster the particle moves, so that its diffusion coefficient, D, is higher. At thermal equilibrium, the value of the diffusion coefficient can be estimated from the Stokes‐Einstein equation, , where c is a constant, T is the temperature, R is the particle radius and η is the fluid viscosity |
| Newtonian fluid | A fluid with viscosity which is independent of the shear rate (i.e., rate at which the fluid is deformed) |
| Non‐reciprocal conformational changes | Conformational changes (of proteins, for example) which are asymmetric with respect to time. The initial conformation can be reached by a “path” which does not imply the mere reversal of a conformational change |
| Multivalent interactions | Interactions established between two molecules, spread out over multiple regions of each of these molecules |
| Supramolecular structure | A loose aggregate of proteins and other macromolecules, held together by multivalent interactions |
| Phase separation | Phenomenon by which homogeneously mixed molecules separate into distinct, coexisting, liquid phases. While one of the phases is depleted of some of those components, the other is enriched |
Brief explanation of some of the most relevant terms used in the text.
Box 1. Comparison of the upper limit on Gibbs energy dissipation rate with alternative constraints on metabolism.
The cellular Gibbs energy dissipation rate is the sum over all fluxes (v) in a metabolic network, where each flux is weighed by the corresponding Gibbs energy of reaction (Δ rG) (Equation 1). Imposing a limit on the cellular Gibbs energy dissipation rate in flux balance analysis models means that an upper constraint, , is imposed on this variable:
| (1) |
From a mathematical point of view, the upper limit on the cellular Gibbs energy dissipation rate is analogous to other constraints that have been used in flux balance analysis to account for “resource allocation”, e.g., in terms of total amount of proteins in the cell, macromolecular crowding, or membrane occupancy (Basan et al, 2015; Vazquez & Oltvai, 2016; Szenk et al, 2017; Elsemman et al, 2022). All such constraints resemble a weighted sum of fluxes. The rationale here is that each reaction comes with a certain “cost” in terms of the given resource (e.g., fraction of the cell proteome, volume, or surface area occupied by the enzymes; represented by w), where the sum of the individual costs must not surpass the limited capacity of the cell, C:
| (2) |
The structural similarity of these constraints (thermodynamic‐ or resource allocation‐based) leads to a similar outcome in the predictions from the models: in either case, overflow metabolism (i.e., simultaneous use of respiratory and fermentative pathways at high substrate uptake rates) is predicted (de Groot et al, 2020). Remarkably, the similarity between the constraints may extend beyond their analogous mathematical formulations. For example, it is possible that the thermodynamic constraint is intrinsically related with the proteome allocation constraint. On the one hand, the biological strategy evolved to cope with reactions that involve a large Δ rG (e.g., in respiration, where the substrates are fully oxidized) is characterized by a splitting of the overall reaction into multiple steps, each associated with an enzyme. As a result, highly dissipating pathways are also pathways that have a larger cost in terms of resources (namely proteins). On the other hand, the causality may also go in the reverse direction: if proteome constraints limit the number of proteins available in a certain pathway, then metabolic flux is decreased, and the overall Gibbs energy dissipation rate in that pathway is similarly reduced (because the dissipation rate is the product of flux and Gibbs energy of reaction).
Future work is needed to further investigate the molecular basis of these constraints and their putative connection. While the “resource allocation”‐based constraints are intuitively apprehended, the constraint on the cellular Gibbs energy dissipation rate is both more challenging to explain mechanistically and more difficult to test experimentally. It is a goal of this perspective to put forward mechanistic ideas and intuition about the thermodynamic constraint, thereby potentially opening a new view on how metabolic activity could globally affect biomolecular functions and ultimately “constrain” metabolism.
While the observation that cellular metabolism might be governed by a thermodynamic limit is intriguing, it also triggers a new question: what could be the molecular mechanism that underlies this upper limit on the cellular Gibbs energy dissipation rate? Or, in other words, why would cells—which, according to Erwin Schrödinger, need to “free [themselves] from all the entropy [they] cannot help producing while alive” (Schrödinger, 1944)—be limited by the rate at which they can do so? Here, dwelling on this question, and pulling together fragmented pieces of evidence from diverse research fields, we developed a bold hypothesis to explain this limit. This hypothesis might explain the physiological behavior of cells from the molecular level of enzymes. Specifically, we hypothesize that during their catalytic action, metabolic enzymes use part of the released Gibbs energy to increase their motion, i.e., to perform work (Fig 1B, left part). This then leads to increased movement of biomolecules in cells (Fig 1B, middle part), which can, in turn, affect regulatory and biomolecular processes, with some being favored and others disfavored, by the increasing rates of collision (Fig 1B, right part). If regulatory processes such as transcription or translation are dependent on molecular motion, a feedback loop can be established, where metabolic activity (and thus Gibbs energy dissipation) is kept under control, resulting in a maximal limit on the cellular Gibbs energy dissipation rate.
In this perspective article, we start by asking what the global effects of the cellular Gibbs energy dissipation could be. Second, we explore whether enzymes can perform work during catalysis. Third, we ask what the consequences of increased molecular motion could be for a cell. Overall, this article offers an intriguing perspective on how metabolic activity could globally affect biomolecular functions.
What could be the global effects of cellular Gibbs energy dissipation?
In a first instance, one could interpret the upper limit on the cellular Gibbs energy dissipation rate as a limit on the heat transfer rate from the cell to its surroundings. Under this assumption, increased metabolic activity, and thus heat production, could lead to increased temperature in the cell or its compartments (e.g., mitochondria), which could potentially damage proteins or other macromolecules, or have other detrimental effects on cellular processes. Supporting this idea, intracellular temperature measurements using a variety of thermosensors (Zohar et al, 1998; Yang et al, 2011; Okabe et al, 2012; Kiyonaka et al, 2013; Takei et al, 2014; Arai et al, 2015; Chrétien et al, 2018; Savchuk et al, 2019) suggested that there are temperature differences between cellular compartments. The mitochondrion, in particular, was suggested to have a temperature higher than that of the cytoplasm (Okabe et al, 2012; Chrétien et al, 2018). Notably, the mitochondrion is a compartment where a high Gibbs energy dissipation rate is expected, with the respiratory chain contributing 50% to the cellular Gibbs energy dissipation rate under certain conditions (Niebel et al, 2019).
However, the conclusions from experiments with thermosensors have been questioned (Baffou et al, 2014). Assuming steady‐state conditions and neglecting entropic effects (so that the change of Gibbs energy equals an enthalpy change, i.e., all of the Gibbs energy dissipated is converted into heat), the temperature in the center of a yeast cell, for example, would only increase by 10−5 K compared to the temperature in the environment. The reported temperature measurements cannot be explained even when accounting for (i) spatially confined heat release (e.g., all heat is released in the mitochondria), (ii) a temporal variation of the release (all heat is released in a temporal burst), or (iii) a finite thermal conductivity of membranes (considering an insulator effect of the cell membrane) (Baffou et al, 2014). This discrepancy has become known as the “105 gap” (Suzuki et al, 2015). While the gap may in part be due to the sensitivity of the thermosensors to molecular events in their surroundings (Lane, 2018), and as recently shown, introducing more accurate estimates of the thermal conductivity in a cell may help reduce this gap (Sotoma et al, 2021)), the difference between predicted and measured temperature values is still far from being negligible (Suzuki & Plakhotnik, 2020). Overall, limited heat transfer, resulting in increased and detrimental intracellular temperature, cannot explain the observed limit on the Gibbs energy dissipation rate.
Alternatively, Gibbs energy released during metabolic activity could be used to perform mechanical work, and cells could be limited by the amount of “work” they can “withstand”. Along these lines, one could hypothesize that the Gibbs energy released during enzyme catalysis is harnessed to increase the motion of the catalyzing enzymes. Such increased motion may potentially be a result of enzymes undergoing self‐propulsion during their catalytic action, by the enzymes breaking free from a supramolecular structure, or a combination of both. If indeed Gibbs energy is harnessed as work, one would expect higher diffusion coefficients in metabolically active cells. It is conceivable that too much intracellular molecular motion is not compatible with proper cellular functioning, thus establishing a limit on the cellular Gibbs energy dissipation rate. This is the idea we want to put forward here.
However, critical readers may wonder how no increase in intracellular temperature and increased molecular movement can be compatible, given that temperature is typically defined as the average kinetic energy of the molecules in a system. As we explain in Box 2 (Fig 2A), these two arguments are not physically inconsistent in the non‐equilibrium situation of a living cell. Thus, increased molecular motion, induced by enzyme catalysis, could indeed occur without significantly increasing intracellular temperature.
Figure 2. Increased molecular motion can occur at hardly increased temperature.
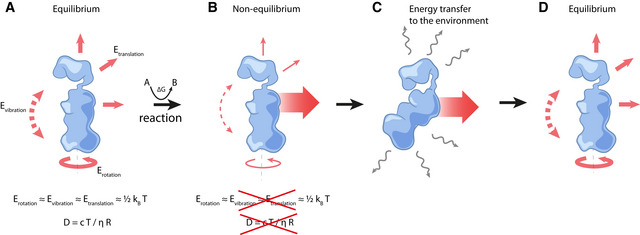
(A) According to the equipartition theorem, the average energy of all degrees of freedom (rotation, vibration, and translation) is the same, and equal to . Under these conditions, the diffusion coefficient can be estimated from the Stokes–Einstein equation, and the enzyme is said to undergo thermal Brownian motion (see definition in Table 1). (B) For an enzyme driven out of equilibrium by the Gibbs energy released during catalysis, the equal distribution of energy among the various degrees of freedom no longer applies, and the diffusion coefficient may not abide by the Stokes–Einstein equation, potentially being larger than the value predicted by this equation. (C) As it moves, the enzyme also dissipates energy to its surroundings as heat, which is swiftly transferred to the environment. (D) The loss of energy brings the enzyme back to its initial equilibrium state with the local surroundings.
Box 2. In a non‐equilibrium system, increased molecular motion can occur at hardly increased temperature.
In statistical physics, the temperature of a system is defined, at equilibrium, as the average kinetic energy of the ensemble of all molecules it contains. Equivalently, and following the equipartition theorem, the energy of each degree of freedom of the system can also be expressed as a function of temperature, T, according to the expression , where kB is the Boltzmann constant. In other words, there is as much kinetic energy associated with the translation of a molecule along any of the three spatial directions as in the rotation or vibration of this molecule (Fig 2A).
Out of equilibrium, which is the condition in which cells operate, however, the equipartition theorem ceases to apply: it is then possible that any one of the degrees of freedom (i.e., translation, rotation, vibration) is associated with more energy than the remaining ones (Casas‐Vázquez & Jou, 2003), and the average energy of each degree of freedom (taken across all molecules of the system) is no longer necessarily given by (Fig 2A and B).
As such, when a biochemical reaction, by releasing Gibbs energy, drives the system out of equilibrium, there can be an enhancement in the mobility of enzymes (see the main text for a discussion on the possible mechanisms), above the levels that would be expected based on thermal Brownian motion. That is, the diffusion coefficient increases above the value that is normally obtained for that enzyme under equilibrium conditions. Yet, in this out of equilibrium state, collisions with surrounding molecules inevitably lead to the dissipation of energy in the form of heat (Fig 2B–D). Nevertheless, the efficient heat transfer across the cell (Baffou et al, 2014), ensures that this is rapidly removed from the system.
There are indications that Gibbs energy is indeed released during enzyme catalysis and leads to increased intracellular molecular motion. Investigations of the in vivo jiggle of chromosomal loci in yeast and bacteria have indicated a correlation between metabolic activity and the diffusion of the tracked loci (Weber et al, 2012): when cells were treated with sodium azide and 2‐deoxyglucose, inhibiting the synthesis of ATP, the apparent diffusion coefficient of the observed chromosomal locus decreased by half compared to untreated cells. Cells only treated with sodium azide, allowing for the synthesis of some ATP through glycolysis, exhibited an intermediate phenotype. If the movement of the chromosomal loci were only due to thermal Brownian motion, a linear relationship between the observed diffusion coefficient and temperature would be expected, according to the Stokes‐Einstein relation. However, the observed diffusion coefficient instead showed an exponential relationship with temperature, in agreement with the Arrhenius equation, which describes the influence of temperature on the rate of chemical reactions. These observations suggest that enzyme catalysis, and thus metabolism, may indeed lead to enhanced translational motion of molecules in the cell.
The observation that metabolic activity fluidizes the cytoplasm, which is otherwise in a glass‐like state, provides further evidence that metabolism induces molecular motion inside the cell (Parry et al, 2014; Nishizawa et al, 2017; Åberg & Poolman, 2021). It has been argued that the fluidity of the cytoplasm is dependent on metabolism‐induced physicochemical changes, namely in cytoplasmic pH (Munder et al, 2016), cellular crowding (Joyner et al, 2016; Delarue et al, 2018), or ATP concentration (Patel et al, 2017; Persson et al, 2020). Nevertheless, it can also be envisioned that the catalysis‐induced molecular motion, i.e., the agitation of the cytoplasm by active enzymes, is capable of fluidizing it (Parry et al, 2014), playing a role analogous to that of molecular motors in eukaryotic cells (Lau et al, 2003; Guo et al, 2014). In fact, molecular dynamics analyses where proteins were subjected to changes in volume have shown that even small changes in the diffusion coefficient of proteins can lead to significant changes in the fluidization state of the cytoplasm (Oyama et al, 2019). Further evidence for increased molecular motion in cells comes from statistical thermodynamic analyses of a simplified model of translation initiation in bacteria. Under the assumptions that mRNA‐ribosome complexes are at equilibrium and that the translation rate is proportional to the number of such complexes, relating the abundance of a fluorescent reporter to the calculated Gibbs energy of the ribosome‐mRNA binding step led to the suggestion that mRNA binding to the ribosome requires a system temperature of more than 1,000 K (Salis, 2011), which is clearly outside the range of life‐permitting values. Yet, translation initiation does occur, and therefore, this discrepancy may potentially be explained by non‐equilibrium effects, such as increased molecular motion. One final example of enhanced motion coupled to enzymatic activity is the observation that membranes become softer, showing undulations akin to those arising from an elevated temperature (up to three‐fold enhancement in effective temperature in case of Ca2+‐ATPase), when the membrane‐embedded protein channels are active (Prost & Bruinsma, 1996; Girard et al, 2005).
Taken together, there is evidence that in metabolically active cells, molecules move more than would be expected from thermal Brownian motion alone. This is the case even in bacteria, which lack gliding of macromolecules along the cytoskeletal structures. Changes in physicochemical parameters may also contribute to changes in intracellular diffusion rates and gradients of chemical compounds have been implicated in driving convective flows (Ortiz‐Rivera et al, 2016; Testa et al, 2021). Nevertheless, in the following sections, we argue that the connection between the observed increase in intracellular motion and metabolic activity could be established through enzymes performing work by using the Gibbs energy released during catalysis.
How could enzymes perform work during catalysis?
If, as indicated by several observations, metabolism plays a role in “stirring up” the cytoplasm, then Gibbs energy released in enzymatic reactions must somehow be transduced into mechanical work. Some archetypal examples of mechanical transduction at the molecular scale include motor proteins, which use Gibbs energy to induce movement along cytoskeletal filaments (e.g., kinesin and myosin) and nucleic acids (e.g., polymerases, topoisomerases and gyrases), or to rotate bacterial flagella and F0F1‐ATP synthase (Phillips et al, 2012; Kolomeisky, 2013; Guo et al, 2014). Also in chemistry, it is known that catalytically active asymmetric micro‐/nanoscale objects can self‐propel during catalysis (Ismagilov et al, 2002; Qin et al, 2017; Zhao et al, 2018a; Arqué et al, 2019; Sun et al, 2019; Luo et al, 2020), and that both reactant and solvent molecules can experience increased mobility upon catalysis, even when gas formation and convection are ruled out (Wang et al, 2020).
Analogously, there is evidence that metabolic enzymes show motor‐like behavior. For instance, multiple fluorescence correlation spectroscopy (FCS) experiments, in which the diffusion coefficients of fluorescently labeled enzymes are inferred from fluorescence fluctuations, showed enhanced diffusion of free‐swimming or membrane‐bound enzymes when mixed with their substrates. This was shown for enzymes such as urease (Muddana et al, 2010; Riedel et al, 2015; Jee et al, 2018a, 2018b; Ghosh et al, 2019), catalase (Sengupta et al, 2013; Riedel et al, 2015), alkaline phosphatase (Riedel et al, 2015; Ghosh et al, 2019), fructose bisphosphate aldolase (Illien et al, 2017), acetylcholinesterase (Jee et al, 2018b), ATPase (Ghosh et al, 2019), and hexokinase (Zhao et al, 2018b). Likewise, motility increase in membrane‐bound enzymes was observed with single‐particle tracking techniques (Ghosh et al, 2019; Song et al, 2021). In some cases, the change in diffusion was explained by conformational changes upon substrate binding, where a decrease in hydrodynamic radius would cause the enzyme to diffuse faster (Illien et al, 2017; Agudo‐Canalejo et al, 2018; Kondrat & Popescu, 2019; Agudo‐Canalejo & Golestanian, 2020). In other instances, it was shown that the measured increase in diffusion can be a result of dissociation of oligomeric enzymes into their subunits (Jee et al, 2019). However, neither of these explanations is in line with our hypothesis, since neither implies causality between the rate of Gibbs energy released during catalysis and increased molecular motion.
Yet, as we will illustrate below, there are instances where the Gibbs energy release during catalysis does appear to cause the increase in molecular motion. We do however acknowledge that there is an ongoing discussion about whether increased enzyme diffusion upon catalysis does or does not occur. It has been suggested that experimental artifacts affecting FCS may be responsible for the high diffusion coefficient values reported in the literature (Günther et al, 2018; Feng & Gilson, 2019). Along these lines, alternative techniques such as nuclear magnetic resonance (Günther et al, 2019), dynamic light scattering (Zhang et al, 2018), anti‐Brownian electrokinetic trapping (Chen et al, 2020), and single‐molecule displacement mapping (Choi et al, 2022) have found no diffusion enhancement for some of these enzymes, namely, aldolase and alkaline phosphatase (Riedel et al, 2015; Illien et al, 2017). Nevertheless, other techniques, such as single‐molecule measurements using total internal reflection fluorescence microscopy, have reported an even higher diffusion coefficient for urease than the value obtained with FCS (Xu et al, 2019). These seemingly contradictory results may be partly explained by differences in the experimental conditions: the enzymatic reaction may have not occurred to the same extent in all experiments, e.g., due to the lack of co‐factors essential for the enzymatic reaction; or if the substrate and product concentrations were different, in which case the Gibbs energy of the reaction would have been different when the measurements were made. Along these lines, a recent paper suggested a correlation between enzyme mobility and the rate of Gibbs energy release (Jee et al, 2020).
Despite the uncertainty surrounding the true extent of diffusion enhancement of single enzymes, we still consider it possible that enzymes harness the Gibbs energy released during catalysis to perform work. Here, we illustrate two classes of mechanisms by which this could be accomplished: (i) self‐propulsion of the enzyme or (ii) by the enzyme breaking free from a loose and disordered supramolecular structure.
Self‐propulsion
Enzyme self‐propulsion could be one consequence of work performed during catalysis potentially resulting in increased molecular motion in the cell. Self‐propulsion provides an active translational component to the enzyme’s purely thermal, stochastic motion (i.e., thermal Brownian motion), so that the effective diffusion coefficient increases. Such self‐propulsion can be achieved by phoretic effects (Fig 3A) or conformational changes (Fig 3B). With phoretic effects, enzymes are dragged along a gradient of a relevant thermodynamic parameter such as temperature, electric potential, or species concentration, where the gradient is established by the enzyme’s own catalytic activity. With conformational changes, enzymes alter their shape during catalysis in order to displace the surrounding fluid. In the case of phoretic effects, one would label enzymes as “squirmers”, and those undergoing conformational changes as “swimmers” (Bechinger et al, 2016).
Figure 3. Mechanisms for enzyme self‐propulsion by work performed.
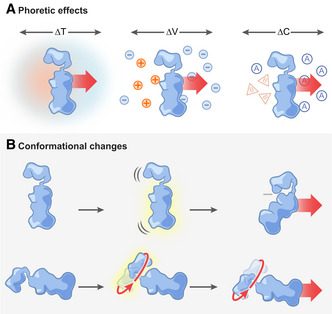
(A) Phoretic effects, in which an enzyme moves without necessarily having to change its conformation. This motion may be accomplished when local gradients around the enzyme are established during catalysis. These gradients can be of different nature: temperature, ΔT (self‐thermophoresis (Golestanian, 2015), left panel); electrostatic potential, ∆V (self‐electrophoresis (Muddana et al, 2010), middle panel), as a result of a differential accumulation of positive (+) and negative (−) charges; or substrate (A) or product (B) concentration, ∆C (self‐diffusiophoresis (Golestanian et al, 2005; Banigan & Marko, 2016), right panel). (B) Conformational changes, leading to active swimming motion, either by asymmetric pressure waves across the enzyme (chemoacoustic effect (Riedel et al, 2015), top panel), or by directional movement of its structural elements around a fixed point or axis (Slochower & Gilson, 2018) (bottom panel).
Phoretic effects drive molecule motion by the presence of gradients. Such gradients may be externally imposed. For example, ATP gradients have been suggested to be involved in driving the motion of membrane‐bound molecules by phoretic effects (Ramm et al, 2021). It is however possible that self‐generated catalysis‐induced gradients, present in the vicinity of an enzyme, could set the enzyme in motion. Importantly, only the latter would account for self‐propulsion. Examples of “self‐phoretic” effects include self‐electrophoresis, self‐diffusiophoresis, and self‐thermophoresis (Golestanian, 2015; Feng & Gilson, 2020) (Fig 3A). Self‐electrophoresis was proposed as a mechanism for the enhanced diffusion of urease (Muddana et al, 2010), where the generated ammonium ions would form a local electric field that would generate a piconewton‐scale propulsive force on the enzyme until the ions diffuse away. Yet, this mechanism cannot explain the increased diffusion of enzymes that catalyze reactions involving only neutral species (Feng & Gilson, 2020). Self‐diffusiophoresis of an enzyme would occur by an asymmetric distribution of its reaction products (Golestanian et al, 2005), where the gradient is established by the reaction (Banigan & Marko, 2016). Such phoretic motion, however, is dependent on the rather weak interactions between the enzyme, the substrates, and reaction products, rendering this mechanism insufficient to explain the experimental results of increased enzyme diffusion (Feng & Gilson, 2020). Finally, self‐thermophoresis exploits a local temperature gradient, but its effect was found to be fifteen orders of magnitude too low to account for the experimental observations of enhanced diffusion of catalase (Golestanian, 2015).
Alternatively, enzymes may self‐propel by conformational changes causing them to actively swim across the fluid. According to the chemoacoustic model (Riedel et al, 2015), the energy released during catalysis by an enzyme with asymmetry between its catalytic site and its center‐of‐mass could generate an asymmetric “pressure wave” that moves across the enzyme and deforms it (Fig 3B, top panel). Upon its deformation, the protein exerts a force on the surrounding fluid, which in turn exerts a force back on the protein, propelling it, and thereby enhancing its apparent diffusion (Riedel et al, 2015). The chemoacoustic model has been questioned in later publications, arguing that the heat released by the enzyme‐catalyzed reaction cannot induce a significant acoustic response because of damping by the solvent (Bai & Wolynes, 2015), and that this mechanism would be four orders of magnitude too small to account for the experimental observations (Golestanian, 2015).
Another possibility of how conformational changes during enzyme catalysis could lead to self‐propulsion would be for the enzyme to move some of its domains around fixed points or axes (Fig 3B, bottom panel). Indeed, enzymes can use the released Gibbs energy to drive motion by changing conformational states (Astumian, 2018), with the energy required to drive these conformational changes being comparable to that released during the catalytic reaction (Boehr et al, 2010). Thus, it can be envisioned that the Gibbs energy released during an enzyme’s reaction potentially provides the energy required to let structural elements of the enzyme move in a way that results in its self‐propulsion.
However, micro‐/nanoscopic swimmers, such as enzymes, are faced with restrictions on their motion. In fact, for micro‐/nano‐sized swimmers, inertial forces are negligible compared with the viscous forces imposed by the surrounding fluid. This dominance of the viscous forces leads to a remarkable consequence: objects can only “swim” (achieving a non‐zero net displacement) in Newtonian fluids, such as water, if they change their conformation in a non‐reciprocal way (i.e., with no time‐reversal symmetry) (Purcell, 1977). This is also the case in the cytoplasm, where molecules are surrounded by water: on very short timescales, they move as though they were in a dilute solution (Di Rienzo et al, 2014; Makuch et al, 2020). As enzymes are chiral molecules, they will likely undergo such non‐reciprocal conformational changes (Slochower & Gilson, 2018), and there is thus the possibility for their net‐displacement by “swimming” (Bai & Wolynes, 2015).
While swimming by non‐reciprocal conformational changes resembles a physically sound mechanism, some authors argued that the magnitude of diffusion enhancement to be expected from this mechanism is too low to explain the experimental values (Bai & Wolynes, 2015; Golestanian, 2015). Nevertheless, in the highly crowded intracellular environment, the high concentration of enzymes which might swim in this manner still renders an enhanced overall diffusion possible by means of a collective effect. The small hydrodynamic flow generated by each enzyme’s conformational change can add up to a measurable increase in the mean diffusion coefficient, as suggested by Brownian dynamics simulations (Skóra et al, 2021).
Breaking free from a supramolecular structure
There is a second possible explanation for how enzymes could increase their motion in the cytoplasm upon catalytic action. The cytoplasm is a highly crowded environment (Ellis, 2001; McGuffee & Elcock, 2010), where the short distance between molecules facilitates attractive and repulsive interactions between them (Monteith et al, 2015; Yu et al, 2016). This proximity, and the establishment of transient multivalent interactions, increases the structural complexity of the cytoplasm, effectively creating a loose and disordered supramolecular structure of proteins and other macromolecules (Fig 4A). For example, enzymes, despite not being structural proteins, can be part of supramolecular structures in both prokaryotes and eukaryotes (Noree et al, 2019; Park & Horton, 2019).
Figure 4. Work performed by an enzyme can lead to its breaking free from a supramolecular structure.
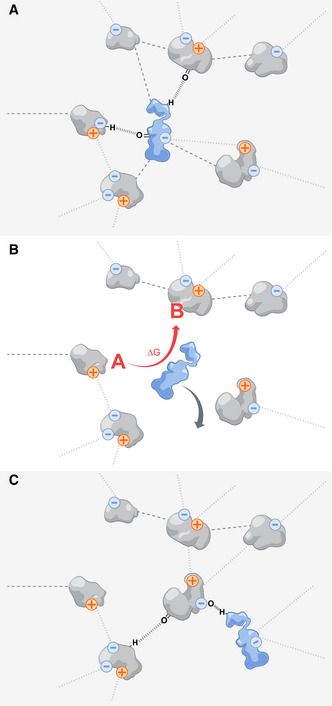
(A) Due to the high concentration of macromolecules in the cell, proteins can establish multivalent interactions with each other and with other molecules. These interactions can arise between charged (electrostatic interactions, dotted lines), or uncharged parts of each molecule (dashed lines); hydrogen bonds may also be established (parallel dashed lines). (B) Such interactions can be counteracted by the release of Gibbs energy in enzymatic reactions, which allow the enzymes to momentarily escape the supramolecular structure and undergo unhindered diffusive motion. (C) When the energy is dissipated to its surroundings, the protein re‐attaches to the supramolecular structure.
With this picture in mind, one can envision that the Gibbs energy released during enzyme catalysis can be used to “break the enzyme free” from such a structure (Fig 4B) until it is once again “integrated” (Fig 4C). This may happen if the Gibbs energy released in the reaction exceeds the energy of the interactions between the enzyme and its neighbors, which is likely since non‐specific protein‐protein interactions are weak (Yu et al, 2016). Upon its release from the influence of surrounding macromolecules, an enzyme can undergo unhindered diffusive motion (in agreement with high time‐resolution raster image correlation spectroscopy measurements of protein diffusion (Di Rienzo et al, 2014)) until it encounters some other molecules, with which it again interacts and to which it binds. The release of the enzyme from such structures, made possible by the performance of work enabled by the Gibbs energy released during catalysis, can constitute another way of increasing the apparent diffusion of enzymes in vivo. This may likewise provide one possible explanation for the observed fluidization of the cytoplasm (Parry et al, 2014; Nishizawa et al, 2017). Multiple enzymes collectively engaging in their catalytic activity would compromise the integrity of this supramolecular structure, which would otherwise contribute to the glass‐like properties of the cytoplasm.
The two mechanisms discussed above, by which Gibbs energy released during an enzymatic reaction could perform work, manifest as increased metabolism‐dependent intracellular diffusion. Even though the role of self‐propulsion as the cause of enhanced enzyme diffusion observed in in vitro experiments has been questioned (Günther et al, 2018; Feng & Gilson, 2019), this might be different in the highly crowded environment of the cytoplasm. Even a small increase in enzyme motion could potentially have a significant impact on overall intracellular motion and could thereby explain the in vivo observations. Indeed, several theoretical and computational studies have shown that the concerted action of many enzymes undergoing conformational changes can lead to enhancement of the diffusion of passive tracer particles (Mikhailov & Kapral, 2015; Kapral & Mikhailov, 2016; Dennison et al, 2017; Hosaka et al, 2020), and at least one study showed that in solutions of active urease and aldolase, tracer microspheres experienced an increase in mobility (Zhao et al, 2017). According to further studies and in line with the idea of a “stirred up” cytoplasm, a generalized increase in diffusion (not just of tracers, but also of the enzymes themselves) is likewise to be expected (Mikhailov & Kapral, 2015; Kapral & Mikhailov, 2016; Koyano et al, 2019; Oyama et al, 2019; Skóra et al, 2021). We do not know whether it is self‐propulsion or breaking free from a supramolecular structure that explains the catalysis‐dependent enhanced enzyme diffusion in cells. One mechanism might apply for some enzymes, and the other for others. It is also possible that both mechanisms occur simultaneously.
What are the consequences of increased molecular motion for the cell?
If, as hypothesized above, Gibbs energy released during an enzymatic reaction is transduced into work and leads to increased molecular motion in cells, this might have consequences for a wide range of cellular processes. In general, molecular motion is important for biomolecular functions, as it allows molecules to interact. However, this motion is strongly reduced when the fluidizing activity of metabolism is inhibited and the cytoplasm transitions into a glass‐like state (Parry et al, 2014; Nishizawa et al, 2017). As such, a catalysis‐induced increase in molecular motion may be important for living cells, for example, for psychrophilic organisms, which inhabit low‐temperature environments and thus need to increase the fluidity of their cytoplasm and membranes (D’Amico et al, 2006). For such organisms, catalytic activity would help keeping the cytoplasm fluidized.
Metabolism‐induced molecular motion may also control phase separation, a phenomenon which has shown to influence regulatory processes (Alberti, 2017; Klosin et al, 2020; Azaldegui et al, 2021). Indeed, studies in colloidal physics showed that actively moving particles tend to cluster (Cates & Tailleur, 2015; Agudo‐Canalejo & Golestanian, 2019; Deblais et al, 2020). It is unclear whether the same can be expected from highly mobile enzymes, not only due to the high complexity of molecular interactions in the cell (Söding et al, 2020) but also due to other phenomena, such as coalescence of phase‐separated droplets and molecule release from the interface of such droplets (Ranganathan & Shakhnovich, 2020). While it remains unclear if catalysis‐induced molecular motion increases or decreases molecular clustering or phase separation, it seems likely that it will affect it in one way or the other.
Increased molecular motion can also promote structural changes in proteins and other biomolecules. As recently shown, albumin adsorbed to a layer of synthetic molecular motors underwent denaturation when the motors were set into motion (Zhou et al, 2020). Similarly, signal transduction and regulatory processes can be perturbed by motion. Molecular movement was suggested to influence signal transduction by mechanically perturbing cytoskeletal elements, which have been speculated to play a role in intracellular signaling (Forgacs et al, 2004). Furthermore, as shown in vitro, the rate of DNA‐loop formation by the lac operon in E. coli doubled when the DNA molecules were forced into oscillations (Chen et al, 2010). One can conceive that increased molecular motion in the cytoplasm may have similar effects on DNA‐loop formation.
Structural changes of biomolecules such as RNA or proteins, potentially induced by altered molecular motion caused by altered metabolic activity, could also affect regulation. Interestingly, changes in the secondary structure of RNAs were found in Bacillus subtilis upon the modification of the metabolic state (Ritchey et al, 2020). As another example, the rpoH transcript of E. coli undergoes temperature‐induced conformational changes that determine whether it is translated or not (Yuzawa et al, 1993). The protein resulting from this translation is a transcription factor, σ32, involved in the heat shock response. Notably, σ32 regulates CreB (Nonaka et al, 2006), and CreB, in turn, activates the enzymes Pta and AckA (Avison et al, 2001). These enzymes are responsible for acetate production and “overflow metabolism”, the type of metabolism associated with the identified limit on cellular Gibbs energy dissipation rate (Niebel et al, 2019). Beyond transcriptional and translational control, the regulation of enzyme clustering is an opportunity for the cell to exert some control over its metabolism (Sweetlove & Fernie, 2018), particularly at branch points of its metabolic pathways (Castellana et al, 2014; Hinzpeter et al, 2019). It is thus tempting to think that, if the stability of these clusters is affected by molecular motion, this would be another way for molecular motion to impact regulation in the cell.
While increased molecular movement can be beneficial for some cellular processes and play a role in regulation, we postulate that excessive intracellular motion can be detrimental for cells. The latter is not self‐evident, and further investigations are required to examine if it holds true. Still, some ideas could point in this direction. As an example, excessive intracellular motion might lead to protein unfolding. If this is the case, a critical limit on intracellular motion can emerge once the negative effects of increased motion (e.g., protein unfolding) cannot be counteracted anymore (e.g., by chaperones). As another example, molecular motion inside a compartment has been suggested to influence the translocation of macromolecules to/from that compartment, on the basis of 2D‐Langevin dynamics simulation results (Tan et al, 2021). Target‐search processes such as transcription factors finding their target DNA‐binding site or pairing of homologous DNA strands constitute an equally interesting example. On the one hand, this search is generally regarded to be sped up by a higher diffusion. On the other hand, dynamic changes in the conformation of the DNA molecules (e.g., unlooping, mentioned above) may lead to an increase in the search time (Felipe et al, 2021). Thus, excessive molecular motion might not be compatible with life. As such, a critical limit on molecular motion, induced by work of enzymes, could explain the identified upper limit on the Gibbs energy dissipation rate (Niebel et al, 2019).
Conclusion and outlook
In summary, we propose that Gibbs energy released during catalysis can be harnessed by enzymes in the form of work, which will ultimately lead to an enhancement of their effective diffusion (Fig 5A). We envision that this increased motion might result from self‐propulsion or from enzymes transiently escaping the influence of neighboring molecules, i.e., breaking free from a supramolecular structure. We also expect that excessively increased molecular motion in the cell will on the whole have a detrimental effect on biomolecular functions (Fig 5B–E), which ultimately imposes an upper limit on the Gibbs energy dissipation rate.
Figure 5. Proposed explanation for the mechanistic basis of the observed limit on the cellular Gibbs energy dissipation rate.
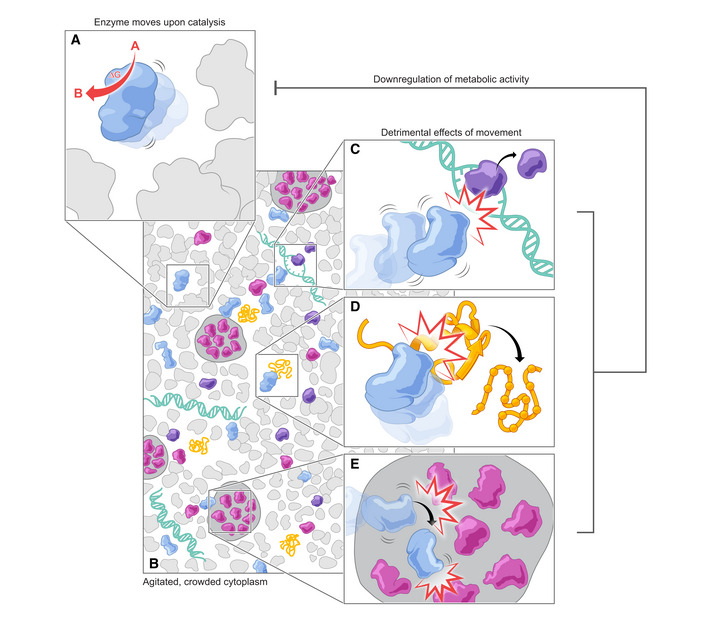
(A) In metabolically active cells, enzyme catalysis leads to the release of Gibbs energy and results in an increase in enzyme motion. (B) Since the intracellular environment (and cytoplasm, in particular) is highly crowded, collisions between enzymes and other molecules will take place. (C) This increase in motion and collisions impacts various biomolecular functions, for example, transcription. (D) Increased motion can be detrimental for protein folding. (E) Phase separation may be affected by increased molecular motion. Together with other effects, this may ultimately regulate the cell’s metabolic activity, as depicted by the gray negative feedback arrow. The regulation of metabolic activity is expected to control the Gibbs energy dissipation rate, gdiss , and maintain it below its upper limit (Niebel et al, 2019).
At the moment, several key questions remain open (summarized in Table 2). Yet, the hypothesis we outline here by combining evidence from different fields, may explain aspects that still remain controversial within a narrower field. For example, the claim that self‐propulsion explains enhanced enzyme diffusion in vitro is still debated (Günther et al, 2018; Feng & Gilson, 2019). Could it be that in some experiments the reaction did not occur as expected? Could it be that the key to enhanced enzyme diffusion is the amount of Gibbs energy released? And could it be that in experiments with different outcome, different substrate or product concentrations were used, leading to different ∆Gs? Our perspective might similarly have the potential to resolve the controversy over intracellular temperature measurements. While studies with molecular thermosensors repeatedly reported increased intracellular “temperature” values (Chrétien et al, 2018), theoretical considerations have dismissed significant temperature increase inside cells as unrealistically high and suggested that the measurements are confounded by experimental artifacts (Baffou et al, 2014; Lane, 2018). Nevertheless, these measurements may be indicative of real phenomena. The reason why molecular thermosensors, which are possibly sensitive to non‐equilibrium molecular motion, report increased “temperature” values may be increased molecular motion, as it has been observed in metabolically active cells (Parry et al, 2014). Such molecular thermosensors might in fact act as motion sensors.
Table 2.
Open questions.
| Question | How to address |
|---|---|
| Can the effect of enhanced enzyme diffusion be confirmed and could conflicting claims be explained by differences in activity and/or thermodynamic potential (i.e., rate of Gibbs energy dissipation) between experiments? | Full control over biochemical aspects of enzyme diffusion measurements (by FCS, DLS, etc.) and complement such measurements by assessing reactant/product concentrations, the reaction rate, and estimates of Gibbs energy of reaction during the analysis |
| Is diffusion enhancement dependent on the rate of Gibbs energy dissipation? | Measure enzyme diffusion as a function of the actual Gibbs energy dissipation rate during the experiment |
| By which mechanisms is increased enzyme motion achieved? | Quantum‐mechanics/molecular dynamics simulations could be used together with experimental techniques (such as high‐precision optical tweezers) to assess dynamic conformational changes of different enzymes undergoing catalysis. High time resolution measurements of enzyme diffusion in cellular environments and novel methods for probing transient protein‐protein interactions |
| Does the degree of molecular motion in cells correlate with the cellular Gibbs energy dissipation rate? | Measure intracellular motion using different probes (of various length scales) under conditions in which the metabolic activity, and thus the cellular Gibbs energy dissipation rate, has been carefully tuned |
| Can we understand the molecular structure of the cytoplasm as an “active bath” and what is the influence of catalytically active enzymes on this structure? | Explore the diffusivity changes in the cell and phase separation phenomena upon sudden inactivation and re‐activation of enzymes (e.g., by optogenetics) that dissipate Gibbs energy at different rates |
| Which regulatory mechanisms controlling metabolism are susceptible to enhanced intracellular motion? | Explore critical regulatory steps in the cell, particularly those that have been previously linked with the response to increased temperature. Assess mRNA and protein conformation under different metabolic conditions, making use of high‐throughput techniques to map in vivo structural changes of macromolecules |
| Is catalysis‐induced increased enzyme motion the cause for the limit on the cellular Gibbs energy dissipation rate, hence the cause for the puzzling metabolic phenotype called “aerobic glycolysis”, “Crabtree effect”, or “overflow metabolism”? | The combined work on the aforementioned aspects will demonstrate whether indeed an active metabolism with enzyme catalysis increasing molecular motion in the cell can explain the inferred limit on the Gibbs energy dissipation rate. Further demonstrating that intracellular motion is dependent on metabolic activity in several organisms and predictive of the onset of changes in metabolic phenotype will show its generality |
Questions that still need to be answered, and some ideas on how this may be achieved.
While catalysis‐induced motion may be necessary for cells to accomplish a variety of biomolecular functions, we argue that there is a limit on how much of such motion cells can withstand. As a result, molecular motion should be kept under control, likely by means of a feedback loop acting on metabolic activity (Fig 5A–E). Such a feedback loop would have to sense the changes in molecular motion by resorting to some mechanically sensitive structures (Milstein & Meiners, 2011) that directly or indirectly regulate the expression and/or activity of metabolic enzymes. Following this line of thought, an increase in Gibbs energy dissipation rate driven, e.g., by an increase in substrate uptake rate, would increase molecular motion in the cytoplasm. The result of increased motion on transcription factor binding to DNA, mRNAs, and protein folding, etc., would be such that metabolic activity (and thus, the rate of Gibbs energy dissipation) is capped. In addition to mechanisms sensing molecular motion, sensing of metabolic fluxes (Kochanowski et al, 2013) may also play an important role. Through such mechanisms, the cell would reach the upper limit on Gibbs dissipation rate, and cytoplasmic motion be maintained within viable boundaries.
As with any hypothesis, the validity of the ideas we present in this Perspective will have to be tested. A starting point will be to confirm whether intracellular motion correlates with the cellular Gibbs energy dissipation rate. For this to be feasible experimentally, the metabolic conditions must be carefully tuned so that cells operate at a particular cellular Gibbs energy dissipation rate. Then, the motion of differently sized, endogenous or exogenous probes needs to be determined, ideally by various techniques. Another important element is to further demonstrate that enzymes indeed show enhanced motion as a result of their catalytic activity, and to decipher the experimental parameters when it occurs and when not. The mechanisms by which such enhanced motion is achieved should also be clarified: does it occur by self‐propulsion, by breaking free from a supramolecular structure, or by a combination of both? Here, high‐precision optical tweezers may open the possibility to investigate conformational changes that enzymes undergo during catalysis. Experimental evidence is also required to support the second tier of our hypothesis postulating that motion exerts a regulatory effect on metabolism. In that regard, it will be important to assess if the conformation of relevant biomolecules such as mRNA and proteins undergoes changes when exposed to different levels of intracellular motion, potentially harnessing recent technical advances to determine RNA or protein structures in vivo (Cappelletti et al, 2021; Marinus et al, 2021). Importantly, a causal relationship between these conformational changes and changes in metabolic activity and Gibbs energy dissipation needs to be confirmed.
If proven correct, the proposed mechanistic basis for the existing upper limit on Gibbs energy dissipation rate could move our understanding of cellular metabolism and cell functioning to a new level, across different scales. The ideas presented here could build up momentum for “physics of life”, where concepts from physics are used to gain insight into biological and biochemical phenomena. Recent examples include studies on the physicochemistry of cells and its importance for biomolecular processes (Parry et al, 2014; Joyner et al, 2016; Munder et al, 2016; Delarue et al, 2018; Persson et al, 2020; Xiang et al, 2021), phase separation (Alberti & Dormann, 2019; Klosin et al, 2020; Fritsch et al, 2021) or “active matter” (Battle et al, 2016; Gompper et al, 2020), which highlight that we are moving towards an exciting, physics‐based understanding of biology. At the same time, and more importantly, this article also offers a new perspective on how physical principles may lead to low‐level regulatory mechanisms in the cell that act in addition to the well‐established control systems that have been described in recent decades. We hope that the perspective that we put forward here will spark new thoughts and ideas, as well pave the way for new research avenues.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Acknowledgements
This work was in part funded by the “BaSyC—Building a Synthetic Cell” Gravitation grant (No. 024.003.019) of the Netherlands Ministry of Education, Culture, and Science (OCW) and the Dutch Research Council (NWO), the European Commission within the Marie Skłodowska‐Curie Innovative Training Network ALERT (No. 713482), and the Dutch Research Council (NWO) within the NWA‐ORC 2019 call (No. NWA.1292.19.170). We thank L. Janssen, G. Koenderink, B. Mulder, S. Pigolotti, B. Poolman, and P.R. ten Wolde for insightful discussions and helpful comments on the manuscript, as well as S. Knemeyer, V. Yeung, and S. Min Suh for the help with the figures.
Mol Syst Biol. (2022) 18: e10822
References
- Åberg C, Poolman B (2021) Glass‐like characteristics of intracellular motion in human cells. Biophys J 120: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo‐Canalejo J, Golestanian R (2019) Active phase separation in mixtures of chemically interacting particles. Phys Rev Lett 123: 18101 [DOI] [PubMed] [Google Scholar]
- Agudo‐Canalejo J, Golestanian R (2020) Diffusion and steady state distributions of flexible chemotactic enzymes. Eur Phys J Spec Top 229: 2791–2806 [Google Scholar]
- Agudo‐Canalejo J, Illien P, Golestanian R (2018) Phoresis and enhanced diffusion compete in enzyme chemotaxis. Nano Lett 18: 2711–2717 [DOI] [PubMed] [Google Scholar]
- Alberti S (2017) Phase separation in biology. Curr Biol 27: R1097–R1102 [DOI] [PubMed] [Google Scholar]
- Alberti S, Dormann D (2019) Liquid–liquid phase separation in disease. Annu Rev Genet 53: 171–194 [DOI] [PubMed] [Google Scholar]
- Arai S, Suzuki M, Park S‐J, Yoo JS, Wang L, Kang N‐Y, Ha H‐H, Chang Y‐T (2015) Mitochondria‐targeted fluorescent thermometer monitors intracellular temperature gradient. Chem Commun 51: 8044–8047 [DOI] [PubMed] [Google Scholar]
- Arqué X, Romero‐Rivera A, Feixas F, Patiño T, Osuna S, Sánchez S (2019) Intrinsic enzymatic properties modulate the self‐propulsion of micromotors. Nat Commun 10: 2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astumian RD (2018) Trajectory and cycle‐based thermodynamics and kinetics of molecular machines: the importance of microscopic reversibility. Acc Chem Res 51: 2653–2661 [DOI] [PubMed] [Google Scholar]
- Avison MB, Horton RE, Walsh TR, Bennett PM (2001) Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem 276: 26955–26961 [DOI] [PubMed] [Google Scholar]
- Azaldegui CA, Vecchiarelli AG, Biteen JS (2021) The emergence of phase separation as an organizing principle in bacteria. Biophys J 120: 1123–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffou G, Rigneault H, Marguet D, Jullien L (2014) A critique of methods for temperature imaging in single cells. Nat Methods 11: 899–901 [DOI] [PubMed] [Google Scholar]
- Bai X, Wolynes PG (2015) On the hydrodynamics of swimming enzymes. J Chem Phys 143: 165101 [DOI] [PubMed] [Google Scholar]
- Banigan EJ, Marko JF (2016) Self‐propulsion and interactions of catalytic particles in a chemically active medium. Phys Rev E 93: 12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T (2015) Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle C, Broedersz CP, Fakhri N, Geyer VF, Howard J, Schmidt CF, Mackintosh FC (2016) Broken detailed balance at mesoscopic scales in active biological systems. Science 352: 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger C, Di Leonardo R, Löwen H, Reichhardt C, Volpe G, Volpe G (2016) Active particles in complex and crowded environments. Rev Mod Phys 88: 45006 [Google Scholar]
- Boehr DD, McElheny D, Dyson HJ, Wright PE (2010) Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc Natl Acad Sci USA 107: 1373–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti V, Hauser T, Piazza I, Pepelnjak M, Malinovska L, Fuhrer T, Li Y, Dörig C, Boersema P, Gillet L et al (2021) Dynamic 3D proteomes reveal protein functional alterations at high resolution in situ. Cell 184: 545–559.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas‐Vázquez J, Jou D (2003) Temperature in non‐equilibrium states: a review of open problems and current proposals. Reports Prog Phys 66: 1937–2023 [Google Scholar]
- Castellana M, Wilson MZ, Xu Y, Joshi P, Cristea IM, Rabinowitz JD, Gitai Z, Wingreen NS (2014) Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol 32: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates ME, Tailleur J (2015) Motility‐induced phase separation. Annu Rev Condens Matter Phys 6: 219–244 [Google Scholar]
- Chen YF, Milstein JN, Meiners JC (2010) Protein‐mediated DNA loop formation and breakdown in a fluctuating environment. Phys Rev Lett 104: 258103 [DOI] [PubMed] [Google Scholar]
- Chen Z, Shaw A, Wilson H, Woringer M, Darzacq X, Marqusee S, Wang Q, Bustamante C (2020) Single‐molecule diffusometry reveals no catalysis‐induced diffusion enhancement of alkaline phosphatase as proposed by FCS experiments. Proc Natl Acad Sci USA 117: 21328–21335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AA, Park HH, Chen K, Yan R, Li W, Xu K (2022) Displacement statistics of unhindered single molecules show no enhanced diffusion in enzymatic reactions. J Am Chem Soc 144: 4839–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D, Bénit P, Ha H‐H, Keipert S, El‐Khoury R, Chang Y‐T, Jastroch M, Jacobs HT, Rustin P, Rak M (2018) Mitochondria are physiologically maintained at close to 50°C. PLOS Biol 16: e2003992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico S, Collins T, Marx J‐C, Feller G, Gerday C, Gerday C (2006) Psychrophilic microorganisms: challenges for life. EMBO Rep 7: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblais A, Maggs AC, Bonn D, Woutersen S (2020) Phase separation by entanglement of active polymerlike worms. Phys Rev Lett 124: 208006 [DOI] [PubMed] [Google Scholar]
- Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G et al (2018) mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174: 338–349.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison M, Kapral R, Stark H (2017) Diffusion in systems crowded by active force‐dipole molecules. Soft Matter 13: 3741–3749 [DOI] [PubMed] [Google Scholar]
- Ellis RJ (2001) Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 26: 597–604 [DOI] [PubMed] [Google Scholar]
- Elsemman IE, Rodriguez Prado A, Grigaitis P, Garcia Albornoz M, Harman V, Holman SW, van Heerden J, Bruggeman FJ, Bisschops MMM, Sonnenschein N et al (2022) Whole‐cell modeling in yeast predicts compartment‐specific proteome constraints that drive metabolic strategies. Nat Commun 13: 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe C, Shin J, Kolomeisky AB (2021) DNA looping and DNA conformational fluctuations can accelerate protein target search. J Phys Chem B 125: 1727–1734 [DOI] [PubMed] [Google Scholar]
- Feng M, Gilson MK (2019) A thermodynamic limit on the role of self‐propulsion in enhanced enzyme diffusion. Biophys J 116: 1898–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Gilson MK (2020) Enhanced diffusion and chemotaxis of enzymes. Annu Rev Biophys 49: 87–105 [DOI] [PubMed] [Google Scholar]
- Forgacs G, Yook SH, Janmey PA, Jeong H, Burd CG (2004) Role of the cytoskeleton in signaling networks. J Cell Sci 117: 2769–2775 [DOI] [PubMed] [Google Scholar]
- Fritsch AW, Diaz‐Delgadillo AF, Adame‐Arana O, Hoege C, Mittasch M, Kreysing M, Leaver M, Hyman AA, Jülicher F, Weber CA (2021) Local thermodynamics govern formation and dissolution of Caenorhabditis elegans P granule condensates. Proc Natl Acad Sci USA 118: e2102772118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Mohajerani F, Son S, Velegol D, Butler PJ, Sen A (2019) Motility of enzyme‐powered vesicles. Nano Lett 19: 6019–6026 [DOI] [PubMed] [Google Scholar]
- Girard P, Prost J, Bassereau P (2005) Passive or active fluctuations in membranes containing proteins. Phys Rev Lett 94: 88102 [DOI] [PubMed] [Google Scholar]
- Golestanian R (2015) Enhanced diffusion of enzymes that catalyze exothermic reactions. Phys Rev Lett 115: 108102 [DOI] [PubMed] [Google Scholar]
- Golestanian R, Liverpool TB, Ajdari A (2005) Propulsion of a molecular machine by asymmetric distribution of reaction products. Phys Rev Lett 94: 220801 [DOI] [PubMed] [Google Scholar]
- Gompper G, Winkler RG, Speck T, Solon A, Nardini C, Peruani F, Löwen H, Golestanian R, Kaupp UB, Alvarez L et al (2020) The 2020 motile active matter roadmap. J Phys Condens Matter 32: 193001 [DOI] [PubMed] [Google Scholar]
- de Groot DH, Lischke J, Muolo R, Planqué R, Bruggeman FJ, Teusink B (2020) The common message of constraint‐based optimization approaches: overflow metabolism is caused by two growth‐limiting constraints. Cell Mol Life Sci 77: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther JP, Börsch M, Fischer P (2018) Diffusion measurements of swimming enzymes with fluorescence correlation spectroscopy. Acc Chem Res 51: 1911–1920 [DOI] [PubMed] [Google Scholar]
- Günther JP, Majer G, Fischer P (2019) Absolute diffusion measurements of active enzyme solutions by NMR. J Chem Phys 150: 124201 [DOI] [PubMed] [Google Scholar]
- Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott‐Schwartz J, Mackintosh FC, Weitz DA (2014) Probing the stochastic, motor‐driven properties of the cytoplasm using force spectrum microscopy. Cell 158: 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R, Cucchi D, Smith J, Pucino V, Macdougall CE, Mauro C (2016) Intermediates of metabolism: From bystanders to signalling molecules. Trends Biochem Sci 41: 460–471 [DOI] [PubMed] [Google Scholar]
- Hackett SR, Zanotelli VRT, Xu W, Goya J, Park JO, Perlman DH, Gibney PA, Botstein D, Storey JD, Rabinowitz JD (2016) Systems‐level analysis of mechanisms regulating yeast metabolic flux. Science 354: aaf2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinzpeter F, Tostevin F, Gerland U (2019) Regulation of reaction fluxes via enzyme sequestration and co‐clustering. J R Soc Interface 16: 20190444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka Y, Komura S, Mikhailov AS (2020) Mechanochemical enzymes and protein machines as hydrodynamic force dipoles: the active dimer model. Soft Matter 16: 10734–10749 [DOI] [PubMed] [Google Scholar]
- Illien P, Zhao X, Dey KK, Butler PJ, Sen A, Golestanian R (2017) Exothermicity is not a necessary condition for enhanced diffusion of enzymes. Nano Lett 17: 4415–4420 [DOI] [PubMed] [Google Scholar]
- Ismagilov RF, Schwartz A, Bowden N, Whitesides GM (2002) Autonomous movement and self‐assembly. Angew Chemie Int Ed 41: 652–654 [Google Scholar]
- Jee AY, Chen K, Tlusty T, Zhao J, Granick S (2019) Enhanced diffusion and oligomeric enzyme dissociation. J Am Chem Soc 141: 20062–20068 [DOI] [PubMed] [Google Scholar]
- Jee AY, Cho YK, Granick S, Tlusty T (2018a) Catalytic enzymes are active matter. Proc Natl Acad Sci USA 115: E10812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee AY, Dutta S, Cho YK, Tlusty T, Granick S (2018b) Enzyme leaps fuel antichemotaxis. Proc Natl Acad Sci USA 115: 14–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee AY, Tlusty T, Granick S (2020) Master curve of boosted diffusion for 10 catalytic enzymes. Proc Natl Acad Sci USA 117: 29435–29441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner RP, Tang JH, Helenius J, Dultz E, Brune C, Holt LJ, Huet S, Müller DJ, Weis K (2016) A glucose‐starvation response regulates the diffusion of macromolecules. Elife 5: e09376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapral R, Mikhailov AS (2016) Stirring a fluid at low Reynolds numbers: Hydrodynamic collective effects of active proteins in biological cells. Phys D Nonlinear Phenom 318–319: 100–104 [Google Scholar]
- Kiyonaka S, Kajimoto T, Sakaguchi R, Shinmi D, Omatsu‐Kanbe M, Matsuura H, Imamura H, Yoshizaki T, Hamachi I, Morii T et al (2013) Genetically encoded fluorescent thermosensors visualize subcellular thermoregulation in living cells. Nat Methods 10: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Klosin A, Oltsch F, Harmon T, Honigmann A, Jülicher F, Hyman AA, Zechner C (2020) Phase separation provides a mechanism to reduce noise in cells. Science 367: 464–468 [DOI] [PubMed] [Google Scholar]
- Kochanowski K, Gerosa L, Brunner SF, Christodoulou D, Nikolaev YV, Sauer U (2017) Few regulatory metabolites coordinate expression of central metabolic genes in Escherichia coli. Mol Syst Biol 13: 903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanowski K, Volkmer B, Gerosa L, Van Rijsewijk BRH, Schmidt A, Heinemann M (2013) Functioning of a metabolic flux sensor in Escherichia coli . Proc Natl Acad Sci USA 110: 1130–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomeisky AB (2013) Motor proteins and molecular motors: how to operate machines at the nanoscale. J Phys Condens Matter 25: 463101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrat S, Popescu MN (2019) Brownian dynamics assessment of enhanced diffusion exhibited by ‘fluctuating‐dumbbell enzymes’. Phys Chem Chem Phys 21: 18811–18815 [DOI] [PubMed] [Google Scholar]
- Koyano Y, Kitahata H, Mikhailov AS (2019) Diffusion in crowded colloids of particles cyclically changing their shapes. EPL 128: 40003 [Google Scholar]
- Lane N (2018) Hot mitochondria? PLoS Biol 16: e2005113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AWC, Hoffman BD, Davies A, Crocker JC, Lubensky TC (2003) Microrheology, stress fluctuations, and active behavior of living cells. Phys Rev Lett 91: 198101 [DOI] [PubMed] [Google Scholar]
- Lempp M, Farke N, Kuntz M, Freibert SA, Lill R, Link H (2019) Systematic identification of metabolites controlling gene expression in E. coli . Nat Commun 10: 4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Li S, Wan J, Yang C, Chen B, Guan J (2020) Enhanced propulsion of urease‐powered micromotors by multilayered assembly of ureases on Janus magnetic microparticles. Langmuir 36: 7005–7013 [DOI] [PubMed] [Google Scholar]
- Makuch K, Hołyst R, Kalwarczyk T, Garstecki P, Brady JF (2020) Diffusion and flow in complex liquids. Soft Matter 16: 114–124 [DOI] [PubMed] [Google Scholar]
- Marinus T, Fessler AB, Ogle CA, Incarnato D (2021) A novel SHAPE reagent enables the analysis of RNA structure in living cells with unprecedented accuracy. Nucleic Acids Res 49: E34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffee SR, Elcock AH (2010) Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol 6: e1000694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov AS, Kapral R (2015) Hydrodynamic collective effects of active protein machines in solution and lipid bilayers. Proc Natl Acad Sci USA 112: E3639–E3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein JN, Meiners JC (2011) On the role of DNA biomechanics in the regulation of gene expression. J R Soc Interface 8: 1673–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteith WB, Cohen RD, Smith AE, Guzman‐Cisneros E, Pielak GJ (2015) Quinary structure modulates protein stability in cells. Proc Natl Acad Sci USA 112: 1739–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddana HS, Sengupta S, Mallouk TE, Sen A, Butler PJ (2010) Substrate catalysis enhances single‐enzyme diffusion. J Am Chem Soc 132: 2110–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder MC, Midtvedt D, Franzmann T, Nüske E, Otto O, Herbig M, Ulbricht E, Müller P, Taubenberger A, Maharana S et al (2016) A pH‐driven transition of the cytoplasm from a fluid‐ to a solid‐like state promotes entry into dormancy. Elife 5: e09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel B, Leupold S, Heinemann M (2019) An upper limit on Gibbs energy dissipation governs cellular metabolism. Nat Metab 1: 125–132 [DOI] [PubMed] [Google Scholar]
- Nishizawa K, Fujiwara K, Ikenaga M, Nakajo N, Yanagisawa M, Mizuno D (2017) Universal glass‐forming behavior of in vitro and living cytoplasm. Sci Rep 7: 15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA (2006) Regulon and promoter analysis of the E. coli heat‐shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev 20: 1776–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Begovich K, Samilo D, Broyer R, Monfort E, Wilhelm JE (2019) A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol Biol Cell 30: 2721–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe K, Inada N, Gota C, Harada Y, Funatsu T, Uchiyama S (2012) Intracellular temperature mapping with a fluorescent polymeric thermometer and fluorescence lifetime imaging microscopy. Nat Commun 3: 705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, Lewis CA, Sabatini DM (2020) Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab 2: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Rivera I, Shum H, Agrawal A, Sen A, Balazs AC (2016) Convective flow reversal in self‐powered enzyme micropumps. Proc Natl Acad Sci USA 113: 2585–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama N, Kawasaki T, Mizuno H, Ikeda A (2019) Glassy dynamics of a model of bacterial cytoplasm with metabolic activities. Phys Rev Res 1: 32038 [Google Scholar]
- Park CK, Horton NC (2019) Structures, functions, and mechanisms of filament forming enzymes: a renaissance of enzyme filamentation. Biophys Rev 11: 927–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry BR, Surovtsev IV, Cabeen MT, O’Hern CS, Dufresne ER, Jacobs‐Wagner C (2014) The bacterial cytoplasm has glass‐like properties and is fluidized by metabolic activity. Cell 156: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA (2017) ATP as a biological hydrotrope. Science 356: 753–756 [DOI] [PubMed] [Google Scholar]
- Persson LB, Ambati VS, Brandman O (2020) Cellular control of viscosity counters changes in temperature and energy availability. Cell 183: 1572–1585.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R, Kondev J, Theriot J, Garcia HG (2012) Physical biology of the cell, 2nd edn. New York, NY: Taylor & Francis Inc; [Google Scholar]
- Prost J, Bruinsma R (1996) Shape fluctuations of active membranes. Europhys Lett 33: 321 [Google Scholar]
- Purcell EM (1977) Life at low Reynolds number. Am J Phys 45: 3–11 [Google Scholar]
- Qin W, Peng T, Gao Y, Wang F, Hu X, Wang K, Shi J, Li D, Ren J, Fan C (2017) Catalysis‐driven self‐thermophoresis of Janus plasmonic nanomotors. Angew Chemie Int Ed 56: 515–518 [DOI] [PubMed] [Google Scholar]
- Ramm B, Goychuk A, Khmelinskaia A, Blumhardt P, Eto H, Ganzinger KA, Frey E, Schwille P (2021) A diffusiophoretic mechanism for ATP‐driven transport without motor proteins. Nat Phys 17: 850–858 [Google Scholar]
- Ranganathan S, Shakhnovich EI (2020) Dynamic metastable long‐living droplets formed by sticker‐spacer proteins. Elife 9: e56159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C, Gabizon R, Wilson CAM, Hamadani K, Tsekouras K, Marqusee S, Pressé S, Bustamante C (2015) The heat released during catalytic turnover enhances the diffusion of an enzyme. Nature 517: 227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo C, Piazza V, Gratton E, Beltram F, Cardarelli F (2014) Probing short‐range protein Brownian motion in the cytoplasm of living cells. Nat Commun 5: 5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey LE, Tack DC, Yakhnin H, Jolley EA, Assmann SM, Bevilacqua PC, Babitzke P (2020) Structure‐seq2 probing of RNA structure upon amino acid starvation reveals both known and novel RNA switches in Bacillus subtilis. RNA 26: 1431–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DG, Murphy MP, Frezza C, Prag HA, Chouchani ET, O’Neill LA, Mills EL (2018) Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat Metab 1: 16–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM (2011) The ribosome binding site calculator. Methods in enzymology pp 19–42. Cambridge, MA: Academic Press Inc; [DOI] [PubMed] [Google Scholar]
- Sander T, Farke N, Diehl C, Kuntz M, Glatter T, Link H (2019) Allosteric feedback inhibition enables robust amino acid biosynthesis in E. coli by enforcing enzyme overabundance. Cell Syst 8: 66–75.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchuk OA, Silvestre OF, Adão RMR, Nieder JB (2019) GFP fluorescence peak fraction analysis based nanothermometer for the assessment of exothermal mitochondria activity in live cells. Sci Rep 9: 7535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger E (1944) What is life? The physical aspect of the living cell. Cambridge, UK: Cambridge University Press; [Google Scholar]
- Sengupta S, Dey KK, Muddana HS, Tabouillot T, Ibele ME, Butler PJ, Sen A (2013) Enzyme molecules as nanomotors. J Am Chem Soc 135: 1406–1414 [DOI] [PubMed] [Google Scholar]
- Skóra T, Popescu MN, Kondrat S (2021) Conformation‐changing enzymes and macromolecular crowding. Phys Chem Chem Phys 23: 9065–9069 [DOI] [PubMed] [Google Scholar]
- Slochower DR, Gilson MK (2018) Motor‐like properties of nonmotor enzymes. Biophys J 114: 2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Zwicker D, Sohrabi‐Jahromi S, Boehning M, Kirschbaum J (2020) Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol 30: 4–14 [DOI] [PubMed] [Google Scholar]
- Song S, Mason AF, Post RAJ, De Corato M, Mestre R, Yewdall NA, Cao S, van der Hofstad RW, Sanchez S, Abdelmohsen LKEA et al (2021) Engineering transient dynamics of artificial cells by stochastic distribution of enzymes. Nat Commun 12: 6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotoma S, Zhong C, Kah JCY, Yamashita H, Plakhotnik T, Harada Y, Suzuki M (2021) In situ measurements of intracellular thermal conductivity using heater‐thermometer hybrid diamond nanosensors. Sci Adv 7: eabd7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Mathesh M, Li W, Wilson DA (2019) Enzyme‐powered nanomotors with controlled size for biomedical applications. ACS Nano 13: 10191–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Plakhotnik T (2020) The challenge of intracellular temperature. Biophys Rev 12: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Zeeb V, Arai S, Oyama K, Ishiwata S (2015) The 105 gap issue between calculation and measurement in single‐cell thermometry. Nat Methods 12: 802–803 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Fernie AR (2018) The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat Commun 9: 2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenk M, Dill KA, de Graff AMR (2017) Why do fast‐growing bacteria enter overflow metabolism? Testing the membrane real estate hypothesis. Cell Syst 5: 95–104 [DOI] [PubMed] [Google Scholar]
- Takei Y, Arai S, Murata A, Takabayashi M, Oyama K, Ishiwata S, Takeoka S, Suzuki M (2014) A nanoparticle‐based ratiometric and self‐calibrated fluorescent thermometer for single living cells. ACS Nano 8: 198–206 [DOI] [PubMed] [Google Scholar]
- Tan F, Chen Y, Zhao N (2021) Effects of active crowder size and activity‐crowding coupling on polymer translocation. Soft Matter 17: 1940–1954 [DOI] [PubMed] [Google Scholar]
- Testa A, Dindo M, Rebane AA, Nasouri B, Style RW, Golestanian R, Dufresne ER, Laurino P (2021) Sustained enzymatic activity and flow in crowded protein droplets. Nat Commun 12: 6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Oltvai ZN (2016) Macromolecular crowding explains overflow metabolism in cells. Sci Rep 6: 31007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Park M, Dong R, Kim J, Cho Y‐K, Tlusty T, Granick S (2020) Boosted molecular mobility during common chemical reactions. Science 369: 537–541 [DOI] [PubMed] [Google Scholar]
- Weber SC, Spakowitz AJ, Theriot JA (2012) Nonthermal ATP‐dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci USA 109: 7338–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Surovtsev IV, Chang Y, Govers SK, Parry BR, Liu J, Jacobs‐Wagner C (2021) Interconnecting solvent quality, transcription, and chromosome folding in Escherichia coli. Cell 184: 3626–3642.e14 [DOI] [PubMed] [Google Scholar]
- Xu M, Ross JL, Valdez L, Sen A (2019) Direct single molecule imaging of enhanced enzyme diffusion. Phys Rev Lett 123: 128101 [DOI] [PubMed] [Google Scholar]
- Yang J‐M, Yang H, Lin L (2011) Quantum dot nano thermometers reveal heterogeneous local thermogenesis in living cells. ACS Nano 5: 5067–5071 [DOI] [PubMed] [Google Scholar]
- Yu I, Mori T, Ando T, Harada R, Jung J, Sugita Y, Feig M (2016) Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. Elife 5: e19274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa H, Nagai H, Mori H, Yura T (1993) Heat induction of sigma 32 synthesis mediated by mRNA secondary structure: a primary step of the heat shock response in Escherichia coli . Nucleic Acids Res 21: 5449–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Armstrong MJ, Bassir Kazeruni NM, Hess H (2018) Aldolase does not show enhanced diffusion in dynamic light scattering experiments. Nano Lett 18: 8025–8029 [DOI] [PubMed] [Google Scholar]
- Zhao X, Dey KK, Jeganathan S, Butler PJ, Córdova‐Figueroa UM, Sen A (2017) Enhanced diffusion of passive tracers in active enzyme solutions. Nano Lett 17: 4807–4812 [DOI] [PubMed] [Google Scholar]
- Zhao X, Gentile K, Mohajerani F, Sen A (2018a) Powering motion with enzymes. Acc Chem Res 51: 2373–2381 [DOI] [PubMed] [Google Scholar]
- Zhao X, Palacci H, Yadav V, Spiering MM, Gilson MK, Butler PJ, Hess H, Benkovic SJ, Sen A (2018b) Substrate‐driven chemotactic assembly in an enzyme cascade. Nat Chem 10: 311–317 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chen J, Luan Y, Vainikka PA, Thallmair S, Marrink SJ, Feringa BL, Van Rijn P (2020) Unidirectional rotating molecular motors dynamically interact with adsorbed proteins to direct the fate of mesenchymal stem cells. Sci Adv 6: eaay2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Thompson CB (2019) Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol 20: 436–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar O, Ikeda M, Shinagawa H, Inoue H, Nakamura H, Elbaum D, Alkon DL, Yoshioka T (1998) Thermal imaging of receptor‐activated heat production in single cells. Biophys J 74: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]


